Abstract
Group II introns are novel genetic elements that have properties of both catalytic RNAs and retroelements. Initially identified in organellar genomes of plants and lower eukaryotes, group II introns are now being discovered in increasing numbers in bacterial genomes. Few of the newly sequenced bacterial introns are correctly identified or annotated by those who sequenced them. Here we have compiled and thoroughly analyzed group II introns and their fragments in bacterial DNA sequences reported to GenBank. Intron distribution in bacterial genomes differs markedly from the distribution in organellar genomes. Bacterial introns are not inserted into conserved genes, are often inserted outside of genes altogether and are frequently fragmented, suggesting a high rate of intron gain and loss. Some introns have multiple natural homing sites while others insert after transcriptional terminators. All bacterial group II introns identified to date encode reverse transcriptase open reading frames and are either active retroelements or derivatives of retroelements. Together, these observations suggest that group II introns in bacteria behave primarily as retroelements rather than as introns, and that the strategy for group II intron survival in bacteria is fundamentally different from intron survival in organelles.
INTRODUCTION
Overview
Group II introns were initially discovered and studied in organelles of plants, fungi and other lower eukaryotes where they are relatively abundant. Studies of these introns, particularly yeast mitochondrial introns, uncovered their fundamental properties. The introns were found to be catalytic RNAs that self-splice in vitro with a mechanism analogous to nuclear pre-mRNA splicing (1,2). Some group II introns were observed to encode reverse transcriptase open reading frames (RT ORFs) (3), and were shown to be active retroelements (4,5) that utilize a mobility mechanism similar to nuclear non-long teminal repeat (non-LTR) retroelements (6,7).
In 1993, group II introns were discovered in bacteria (8). Because the bacterial hosts (Azotobacter and Calothrix) were related to the ancestors of mitochondria and chloroplasts, the finding prompted the idea that mobile group II introns originated in bacteria and subsequently spread to the organelles (8,9). By now, as a consequence of the genome sequencing projects, it is clear that group II introns are very widely distributed among bacteria, and are not confined to relatives of mitochondria and chloroplasts. Over 20 full-length group II introns have been reported in diverse bacteria (10,11) and at current rates of reporting, bacterial group II introns may eventually exceed organellar group II introns both in number and structural types. Because of the growing number of reported bacterial group II introns and many incorrect annotations in the public databases, it is useful to compile accurate information about their presence in bacteria.
Mobility properties of group II introns
Group II introns exist in ORF-less and ORF-containing forms. ORF-less introns consist of only the six domains of the catalytic RNA structure, and have a size of ∼600 nt, although some introns have sizeable extraneous insertions. ORF-containing introns encode an RT-related reading frame that is always inserted into domain 4 of the RNA structure, giving a total intron size of 2–3 kb. The ORF has three domains: an RT domain, domain X and the Zn (nuclease) domain, which is optional in some introns (11–13). Domain X is associated with maturase, or splicing activity. During splicing, the intron-encoded protein binds to unspliced intron and the maturase activity stimulates the self-splicing reaction. All three domains (RT, X and Zn) participate in the mobility reaction.
The main mobility event of group II introns is homing, which is the site-specific insertion of introns into intron-less alleles. Homing occurs in yeast mitochondria during genetic crosses when only one strain contains the intron, or in bacteria when homing site DNA is introduced into a bacterial cell that contains an intron. Homing is highly site specific and occurs at an ~30 bp target site, spanning roughly from –20 to +10 relative to the insertion site (14–16). Because the target site contains both upstream and downstream flanking sequences, the intron can only insert into homing sites that are not already filled by intron.
The mechanism of homing, known as target-primed reverse transcription (TPRT), is carried out by the RNP particle that is the end product of splicing (RT bound to intron lariat). First the intron reverse splices into the DNA target site, a reaction inherently RNA catalyzed but assisted by the intron-encoded protein. The Zn domain nicks the bottom strand 9 or 10 nt downstream of the insertion site, and the RT reverse transcribes the intron. Recombination functions are thought to complete the insertion reaction. Much more thorough detail on the transposition mechanism can be found in other reviews (12,13).
Group II introns also insert into non-cognate sites at low frequencies (retrotransposition), which can lead to colonization of new genomic locations. Retrotransposition has been reported for yeast cox1I1 (17,18), Podospora anserina cox1I1 (19), L.l.I1 (20) and S.me.I1 (21), and is thought to occur by the same basic mechanism as TPRT but into cryptic homing sites (18). (L.l.I1 and S.me.I1 are named Ll.ltrB and RmInt1 elsewhere, but are abbreviated here for consistency according to Table 1.)
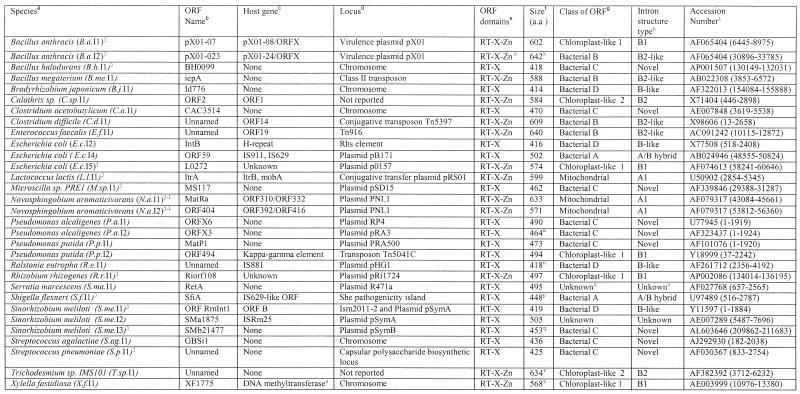
Table 1. Full-length group II introns.
aColumn headings are as for Table 1 except where noted.
bThe presence of intron RNA structural domains 1–6. Parentheses indicate a partial domain.
cThe presence of intron-encoded ORF domains: RT domains 0–7, domain X and Zn domain. Parentheses indicate a partial domain.
dSize of the intron fragment in either amino acids or base pairs.
eClosest full-length intron relative based on BLASTN or BLASTP matches.
fRedundant GenBank entries or equivalent fragments in closely related strains are: E.c.F2, D83536 (80931–82728), AE000133 (7985–9748); M.le.F2, Z96801 (~8394–~9153); M.lo.F1, NC_002678 (4897521–4898300); M.t.F1, AL021428 (~16487–17122); S.f.F2, NC_002698 (29835–31623); AL391753 (32145–33934); S.f.F3, NC_002698 (151605–153082), AL391753 (153916–155393); S.p.F1, AE008411 (8266–8453); S.p.F4, AE008431 (3560–4014); Y.p.F1, NC_001883 (82626–83752), AF053947 (82626–83752), NC_001976 (54417–55543), AL117211 (76455–77581).
gThe fragment is repeated additional times in the same genome in the following entries: M.le.F1, AL583924 (6211–7354, 131992–133135), AL583920 (106679–107822), AL583917 (205253–206396), AL023514 (33633–34776), AL008609 (36594–37737), AL583923 (210031–211174), Z98756 (33658–34801), L78817 (27897–29039); M.le.F2, AL583923 (~5217–~5976), AL023635 (~11576–~12335), L78825 (~19705–~20465).
So far, mobility mechanisms of bacterial group II introns are essentially the same as for mitochondrial introns (22,23). The main difference is that yeast mitochondrial introns rely heavily on host repair activities and usually co-convert flanking markers (24,25), whereas the bacterial introns L.l.I1 and S.me.I1 do not require recA function and do not co-convert flanking markers (26,27). The distinction has been explained by the complete reverse transcription of reverse spliced intron for bacterial introns, but incomplete reverse transcription for yeast introns followed by host-encoded double-strand break repair processes, which co-convert one or both exons (24,25). Surprisingly, yeast introns are mobile at levels of ∼40% when RT activity is eliminated by mutations (5,25), indicating that the most critical activity for mobility in yeast mitochondria is double-stranded DNA cleavage (reverse splicing plus antisense strand cleavage). In contrast, all bacterial introns contain the YADD motif but few encode Zn domains that cut the antisense strand (10,11), suggesting that RT is the most important activity for mobility in bacteria. These differences are most easily rationalized by differences in host activities rather than inherent differences in intron mobility mechanisms.
Interestingly, the highly studied bacterial L.l.I1 intron is closely related to mitochondrial introns and belongs to the same phylogenetic class, while it is more distantly related to most other bacterial introns (10,11). It is not surprising, then, that biochemical activities of L.l.I1 are very similar to those of mitochondrial introns. The only non-mitochondrial-type intron characterized for mobility is S.me.I1. Like most bacterial group II introns, S.me.I1 lacks a Zn domain; however, it is still efficiently mobile in vivo (10–30%). This has been rationalized by functional substitution of cellular nucleases for the Zn domain’s activity (27). Other splicing and mobility properties of S.me.I1 are mainly consistent with activities of L.l.I1 (21,27,28). Previously we speculated that some phylogenetic classes of introns may have mobility properties diverging from those of mitochondrial-type introns (11). While experimental data so far do not demonstrate this, observations discussed below support the possibility that there may be variations in mobility properties for different types of bacterial introns, or a continuum of features.
A model for the evolution and spread of group II introns
In efforts to illuminate how group II introns have spread among bacteria and organelles, phylogenetic characterization of the intron-encoded RTs categorized seven classes of ORFs (10,11). In Zimmerly et al. (11) and here, these classes are denoted the mitochondrial class, chloroplast-like classes 1 and 2, and bacterial classes A, B, C and D. The compositions of the phylogenetic classes are mixed, suggesting horizontal transfers among organelles and bacteria, particularly within the chloroplast-like classes (11). Bacterial introns are in fact found in all seven phylogenetic classes, indicating great diversity of group II introns in bacteria. While the phylogenetic data are consistent with an origin of mobile group II introns in bacteria followed by spread to organelles, the poor statistical support for a branching order prevents clear definition of a history (11). Subsequent analysis of the intron RNA secondary structures showed that the RNA structure co-evolved with the RT ORF (29), with each phylogenetic class of ORF being associated with a distinct RNA structure. For simplicity in this manuscript, we use the phylogenetic class names to refer to both ORF and intron RNA structural classes, although strictly, the names refer to ORF phylogenetic classes.
COMPILATION OF GROUP II INTRONS IN BACTERIAL GENOMES
Group II introns and their fragments were identified in GenBank using standard BLAST searches available at the NCBI web site (http://www.ncbi.nlm.nih.gov/BLAST). The database was first searched using TBLASTN (protein query versus translated nucleotide database) using diverse group II intron ORFs as queries, which identified ORFs and ORF fragments. Next, the database was searched using BLASTN (nucleotide query versus nucleotide database) using intron RNA sequences as queries (excluding the ORF sequences), thereby identifying fragments of intron RNAs or possibly ORF-less introns. Because RNA structural sequence is poorly conserved, the RNA sequences of each full-length and fragmented intron were separately used as queries (about 40 queries in all). All identified ORF fragments were scanned for frame shifts that might extend the reading frames due to mutations or sequencing errors (noted in Table 1 footnote). Intron boundaries were located by folding the RNA structures, mainly according to previously determined foldings of group II introns (29). Without exception, related ORFs were associated with related intron RNAs, which easily allowed identification of RNA boundaries for these introns, or points of fragmentation. RNA domains 5 and 6 were located readily because they are highly conserved in sequence and structure, while domains 1–4 were more difficult to assess. Sequences that could not be folded into group II intron structures were considered fragments (alternative RNA foldings were considered, but are very difficult to evaluate). In most cases, an intron categorized as a fragment was supported by either ORF fragmentation or an identifiable break in alignment to a closely related RNA sequence. Exceptions are A.g.F1, P.ae.F1 and S.ma.F1 (all identical), and B.j.F1, which all lack identifiable RNA domains 1–4 and could not be folded into group II intron structures. Still, it cannot be excluded that these ‘fragments’ may be full-length introns having atypical or unrecognized RNA structures. Similarly, S.ma.I1 is full length but has mispairing in domain 5 and cannot be folded into a convincing structures, suggesting that S.ma.I1 is either a degenerate intron structure or an unrecognized subclass of RNA structure.
Full-length introns
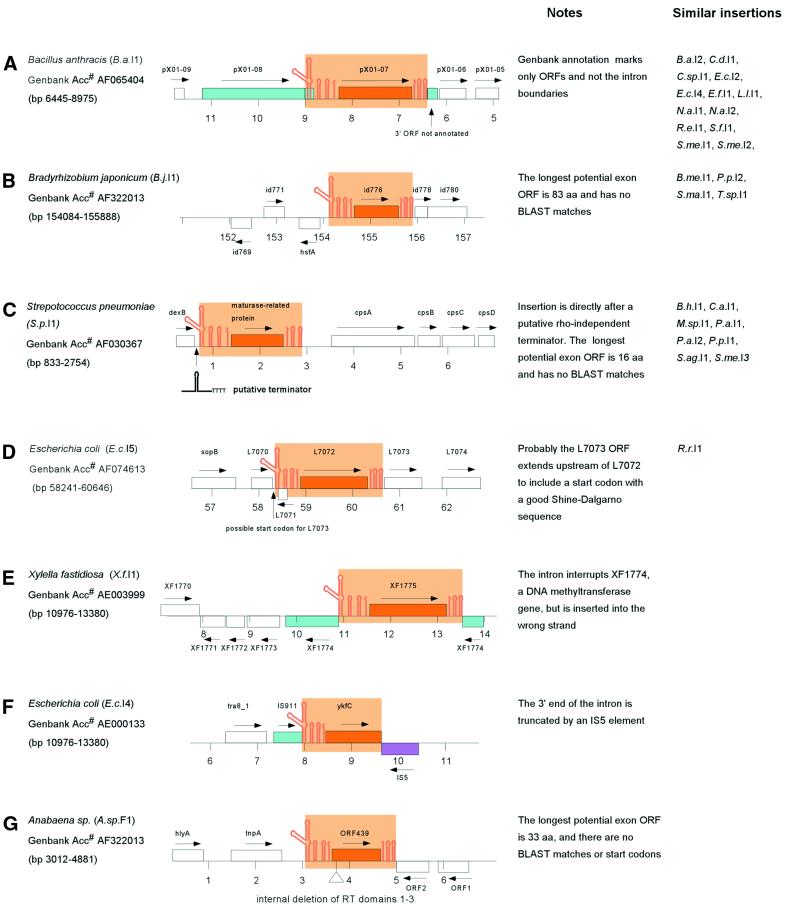
Thirty-one full-length group II introns were identified having complete intron RNA structures (domains 1–6) and full RT ORFs [domains 0–7, X; domain Zn is considered optional; see Zimmerly et al. (11) for domain definitions; Table 1]. Of these, only 12 are correctly identified as group II introns in GenBank with accurate annotation of ORF and intron/exon boundaries. As noted previously (10,11,30), bacterial group II introns are mostly located in plasmids or other mobile DNAs. At least 23 of the 31 introns are located in mobile DNAs, including pathogenicity islands (S.f.I1) and virulence plasmids (B.a.I1, B.a.I2, E.c.I5). Interestingly, none of the 31 introns is inserted into a predicted essential or housekeeping gene, except X.f.I1 (discussed below). Insertion sites fall into five categories, depicted in Figure 1A–E. In the first category, the introns are clearly inserted into ORF-encoding exons. Unexpectedly, this only accounts for 14 of the 31 introns. Correct identification of intron boundaries for these introns often redefines the annotated protein products of the host gene. The B.a.I1 intron, for example, is reported simply as an RT ORF (pX01-07), but identification of the intron boundaries links together its upstream exon (pX01-08) with a short unannotated downstream reading frame (Fig. 1A). In the second class, introns are apparently inserted between genes (e.g. B.j.I1; Fig. 1B). The criterion for this class is that the largest potential ORF surrounding the intron encodes a protein of less than 100 amino acids with no significant BLAST matches. The third class constitutes all members of bacterial class C, whose introns insert directly after rho-independent terminator sequences (discussed below, and shown in detail in Fig. 2C). The S.p.I1 intron, for example, is positioned directly after a putative terminator structure (Figs 1C and 2C), and the longest potential ORF surrounding it is 18 amino acids with no BLAST matches. For introns of the fourth class, it is not clear whether or not the intron is located within a gene. The E.c.I5 intron is inserted outside of a gene according to GenBank annotation, but the downstream reading frame could be extended upstream of the intron to utilize a start codon with a credible Shine–Dalgarno sequence (Fig. 1D). Similarly, the R.r.I1 intron is located in a hypothetical ORF of 189 amino acids, but the ORF has no significant BLAST matches. The final type of insertion is exemplified by the X.f.I1 intron, which is inserted into a housekeeping gene, but in the wrong orientation (Fig. 1E). The intron cannot be spliced out from the host gene’s mRNA and intron insertion has effectively ‘knocked out’ the host DNA methyltransferase gene. Other full-length introns not illustrated in Figure 1 are available as Supplementary Material.
Figure 1.
Examples of group II intron insertions in bacterial genomes. Orange color denotes group II intron RNA domains (stem–loops) and RT ORF (outlined box). Blue boxes represent ORFs that are interrupted by the intron; the purple box indicates an IS element interrupting an intron. Arrows show gene orientations. GenBank accession numbers with correct intron boundaries are on the left, and numbering in the diagram is in kilobases according to the GenBank entries. Introns with similar insertions are shown on the right. (A) The intron interrupts an ORF. (B) The intron has apparently inserted between ORFs. (C) The intron has inserted after a putative teminator structure. (D) It is unclear whether or not the intron is inserted into an ORF. (E) The intron has inserted into an ORF but in the wrong orientation. (F) The intron is truncated by an IS element. (G) The intron is internally deleted.
Figure 2.
Multiple natural insertion sites for group II introns. For each intron, the flanking sequence is shown for 50 bp upstream and 50 bp downstream, with the intron sequence abbreviated gtgcg---ac (or a variation). GenBank accession numbers are on the right, with redundant entries in parentheses. When the same intron is inserted into multiple sites, a consensus sequence is shown beneath the set. Color shadings denote introns (orange), the experimentally determined boundaries of the L.l.I1 homing site (purple), 5S sequence (green) and the terminator stem–loops (blue). T residues following the terminator stem–loop sequences are in upper case. Abbreviations for introns and intron fragments are according to Tables 1–3. (A) Alignment of the flanking sequences of the L.l.I1 intron. The L.l.I1 intron copies are ≥99% identical. (B) Alignment of the flanking sequences of S.f.I1, E.c.I4 and Y.p.F1. Intron sequences are >93% identical. (C) Alignment of flanking sequences of introns of bacterial class C. All introns are inserted after potential terminator sequences (blue shading and upper-case T residues). (D) Example RNA secondary structure of the terminator for the B.h.I1 intron.
Fragments of introns
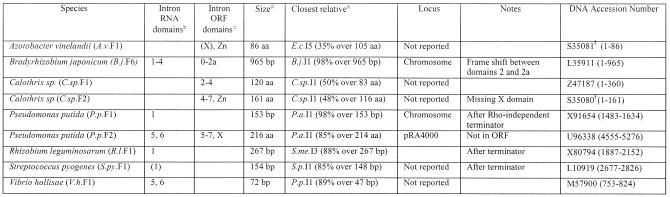
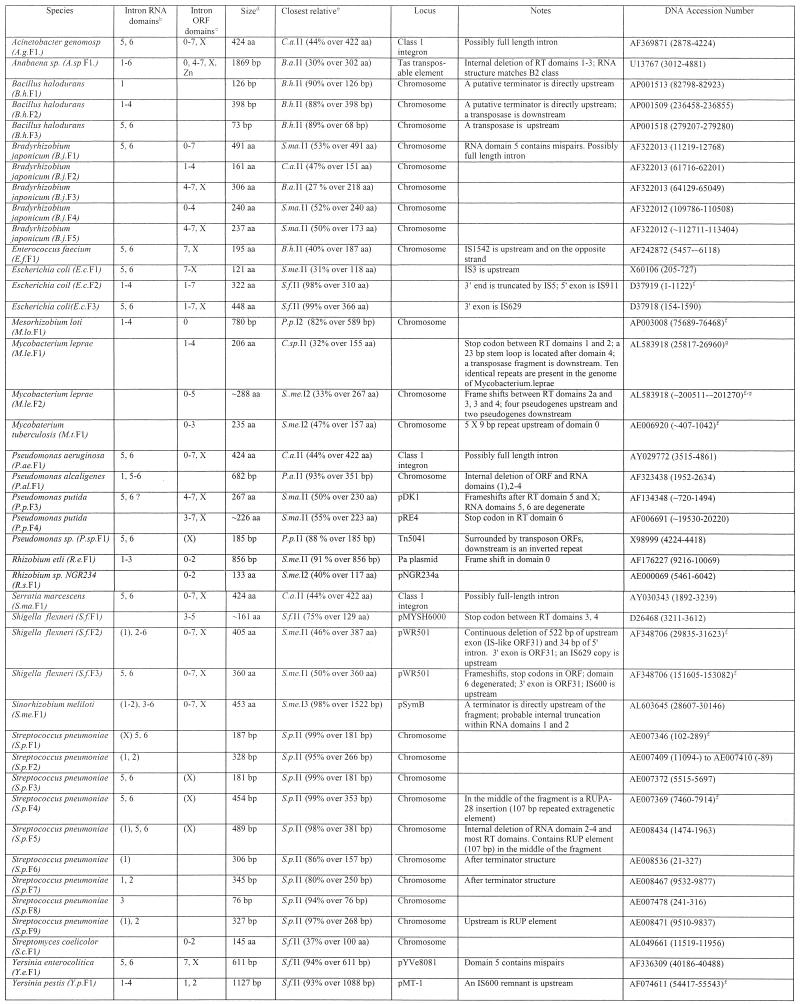
Nine partial group II intron sequences were found which are incompletely sequenced introns or intron fragments (Table 2). Other than these, 42 true fragments were found for which flanking sequences are available (Table 3). Classification of introns as full length and fragments is somewhat oversimplistic because there is a continuum of degeneration. A number of ‘full length’ introns contain structural deviations that suggest impaired function. Mismatches in intron domain 5, the ribozyme’s presumed active site, are found in S.ma.I1, B.me.I1, B.j.I1, E.f.I1, R.e.I1, Sme.I2 and C.a.I1. Frame shifts or stop codons within full-length introns are found in R.e.I1, S.me.I3, S.f.I1, B.a.I2, P.a.I2, T.sp.I1 and X.f.I1. While some ‘mutations’ might be due to sequencing errors, it is likely that most of these ‘full-length’ introns are compromised in function. Similarly, the ‘fragment’ class spans a continuum from nearly full-length introns truncated by only 34 bp at the 5′ end (S.f.F2), to short, internal deletions (A.sp.F1, deleted for RT domains 1–3) to very short remnants (<80 bp; B.h.F3, S.p.F5).
Table 2. Incompletely sequenced intronsa.
aColumn headings are as for Table 1 except where noted.
bThe presence of intron RNA structural domains 1–6. Numbers in parentheses indicate the presence of a partial domain.
cThe presence of intron-encoded ORF domains: RT domains 1–7, domain X and Zn domain. Numbers in parentheses indicate the presence of a partial domain.
dSize of the intron fragment in either amino acids or base pairs.
eClosest full-length intron relative based on BLASTN or BLASTP matches.
fGenBank protein accession number.
Table 3. Intron fragmentsa.
aColumn headings are as for Table 1 except where noted.
bThe presence of intron RNA structural domains 1–6. Numbers in parentheses indicate the presence of a partial domain.
cThe presence of intron-encoded ORF domains: RT domains 0–7, domain X and Zn domain. Numbers in parentheses indicate the presence of a partial domain.
dSize of the intron fragment in either amino acids or base pairs.
eClosest full-length intron relative based on BLASTN or BLASTP matches.
fRedundant GenBank entries or equivalent fragments in closely related strains are: E.c.F2, D83536 (80931–82728), AE000133 (7985–9748); M.le.F2, Z96801 (~8394–~9153); M.lo.F1, NC_002678 (4897521–4898300); M.t.F1, AL021428 (~16487–17122); S.f.F2, NC_002698 (29835–31623); AL391753 (32145–33934); S.f.F3, NC_002698 (151605–153082), AL391753 (153916–155393); S.p.F1, AE008411 (8266–8453); S.p.F4, AE008431 (3560–4014); Y.p.F1, NC_001883 (82626–83752), AF053947 (82626–83752), NC_001976 (54417–55543), AL117211 (76455–77581).
gThe fragment is repeated additional times in the same genome in the following entries: M.le.F1, AL583924 (6211–7354, 131992–133135), AL583920 (106679–107822), AL583917 (205253–206396), AL023514 (33633–34776), AL008609 (36594–37737), AL583923 (210031–211174), Z98756 (33658–34801), L78817 (27897–29039); M.le.F2, AL583923 (~5217–~5976), AL023635 (~11576–~12335), L78825 (~19705–~20465).
Like full-length introns, the fragments are often located either within plasmids or mobile DNAs (at least 15 of 42), or are flanked by mobile DNAs (7 of 42). Host elements again include virulence plasmids and pathogenicity islands (S.f.F1, S.f.F2, S.f.F3, Y.e.F1, Y.p.F1). In most cases the mechanism of fragmentation is not apparent. Of the 42 fragments, 16 are 5′ truncated, 10 are 3′ truncated, 12 are truncated at both ends and four are internally deleted. Perhaps the most obvious candidate mechanism for fragmentation is incomplete reverse transcription of the intron during TPRT, which would cause 5′ truncations and would be analogous to truncations of almost all copies of non-LTR retroelements in eukaryotic genomes. Only 38% of bacterial introns are 5′ truncated, and so incomplete reverse transcription may account for a portion of truncations, but is not the primary source. For some introns, specific fragmentation events are evident. The E.c.I4 intron is reported in full-length form in GenBank accession nos AB024946 and U97489, but in AE000133 is truncated at its 3′ end by the insertion of a full-length IS5 element in the opposite orientation (Fig. 1F). S.f.F2 contains a continuous deletion of 522 bp of upstream exon and 34 bp of intron, which might be due to a single deletion event after intron insertion, or possibly inaccurate 5′ resolution during intron insertion. Other fragments are positioned next to repeat sequences that might be remnants of the truncation event. Directly upstream of the M.t.F1 fragmentation boundary is a 5 × 9 bp microsatellite repeat. Downstream of the deletion point in RT domain 4 of M.le.F1 is a 53 bp inverted repeat that could form a perfect 23 bp stem loop. M.le.F1 has apparently propagated within the genome as part of a larger, uncharacterized repetitive DNA because the fragment is present 10 times in the Mycobacterium leprae genome in a 2382 bp repeat, each of which contains RT domains 1–4 with a stop codon between domains 1 and 2 and a transposase fragment downstream of the inverted repeat.
PREVALENCE OF GROUP II INTRONS IN BACTERIAL GENOMES
To gain a more accurate picture of the prevalence of group II introns in bacterial genomes, we examined the set of fully sequenced genomes. Of 63 sequenced eubacterial genomes (NCBI: http://www.ncbi.nlm.nih.gov/PMGifs/Genomes/micr.html; October 31, 2001), 32% (20 of 63) contain either a group II intron or fragment; 14% (9 of 63) contain a full-length intron while 24% (15 of 63) contain at least one fragment. The most frequent hosts are gamma proteobacteria (29 introns and fragments) and the Bacillus/Clostridium group of Gram-positive bacteria (24 introns and fragments). However, these two groups have been the most heavily sequenced, and so there is no clear bias of group II introns to particular classes of bacteria. Similarly, there is no apparent segregation of intron classes to particular bacterial groups, consistent with substantial horizontal transfers of the introns among eubacteria (11). So far, no group II introns have been reported in archaebacteria out of 11 archaeal genomes sequenced. If group II introns are present in archaebacteria, they are probably less common than in eubacteria, or too divergent to recognize easily.
It is important to note that the absence of group II introns in a sequenced genome does not mean introns are absent from all strains of the species. For example, five group II introns have been reported so far in various Escherichia coli strains (here referred to as E.c.I1, E.c.I2, E.c.I3, E.c.I4 and E.c.I5). [E.c.1 and E.c.3 are not included in Table 1 because their sequences are not available in GenBank; partial sequences are available in Ferat et al. (30)]. However, of the 72 E.coli strains in the ECOR collection, only 18 strains contain an intron, and at most there are two of the five introns in a strain (30). On the other hand, the S.me.I1 intron is present in 83 of 85 isolates of Sinorhizobium meliloti, with 1–11 copies per strain (28). The C.d.I1 intron is present in just one copy in each of five natural isolates examined (31). So, intron content among strains is very spotty for some bacterial species, but more uniform for others.
MULTIPLE HOMING SITES
Since group II introns are highly site-specific mobile elements with target sites of ~30 bp, it is surprising to find that some bacterial introns are inserted into multiple natural targets. Multiple target sites for bacterial introns may be due to either a larger genome size that presents more potential target sites, or to relaxed target site specificity. The L.l.I1 intron is found in two genes, a relaxase of plasmid pRS01 and mobilization protein A of plasmid pAH82 (Fig. 2A). While the introns are 99% identical in sequence, the insertion sites are only 69% identical (24 of 35 bp) in the experimentally defined homing site (–26 to +9) (16). However, DNA sequence is conserved beyond the homing site, suggesting that the introns have inserted into homologous locations of related mobilization genes. In this case, the most critical positions for target recognition are conserved, and the two targets are consistent with rules of recognition defined experimentally for this intron (32,33). Similarly, the E.c.I4 intron has three target sites (Fig. 2B). GenBank reports two examples each of E.c.I4 introns inserted into IS629 and IS911 sites in E.coli or closely related Shigella; a third example comes from Y.p.F1, which is 93% identical to E.c.I4 but has a different upstream sequence. The three insertion sites are only 57% identical (17 of 30 bp) upstream of the intron (Y.p.F1 is missing the 3′ exon). Here again, the sequence common to all three suggests a consensus sequence for the homing site.
More unusual are the multiple insertion sites of bacterial class C introns. All are inserted directly after potential rho-independent terminators, but otherwise the target sequences vary in sequence. The genome of Bacillus halodurans contains five full-length and three fragmented copies of the B.h.I1 intron, all >99% identical. Four of the full-length introns are located at analogous locations at the end of rRNA clusters, while one copy is located in an unrelated position between two hypothetical genes. Of the four rRNA insertion sites, each is completely identical upstream of the intron, and non-identical downstream (Fig. 2C), a pattern inconsistent with conventional group II intron homing sites that span from approximately –20 to +10. The pattern is explained by the observation that the introns are positioned directly after a putative terminator [Fig. 2D; first observed in Granlund et al. (34)]. Two fragments of B.h.I1 are also located downstream of a putative terminator, and the fifth B.h.I1 copy is located after an imperfect terminator structure. In fact, all introns of bacterial class C are inserted behind potential rho-independent terminators (Fig. 2C), but apart from the potential stem–loop structure, there is little sequence conservation for a given intron. The lack of target sequence conservation raises questions about insertion site specificity for this class of introns, and suggests that different principles may be at work compared with other group II introns characterized to date.
It is interesting to consider the consequences of intron insertion behind a transcriptional terminator. Retroelements have evolved ingenious strategies to prevent host damage. Group II introns, for example, generally avoid host damage because their splicing property removes them at the RNA level, which allows them to insert nearly anywhere in a genome without greatly affecting host gene expression. Insertion after a terminator is another way to prevent damage, because the introns will insert outside of genes, and simultaneously will be transcribed at a low level. The perplexing question is this: If introns of bacterial class C never insert into genes, why do they need to splice? The answer would appear to be that the only purpose of splicing is for transposition, since splicing and reverse splicing are expected to be required for insertion into a new site.
SPREAD OF INTRON FAMILIES
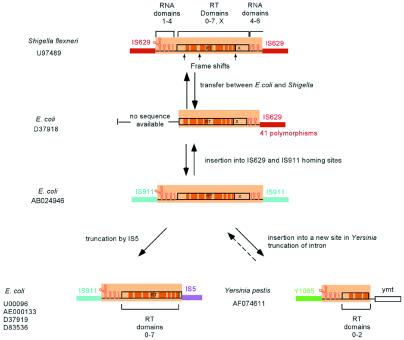
A number of closely related introns have been independently reported in different strains, genomic locations and bacterial species, or in full-length or fragmented form, and suggest specific examples of intron spread. The best example is the E.c.I4 family, whose members are >93% identical in E.coli, Shigella and Yersinia. Figure 3 shows a model for intron spread among the species and homing sites. At some point the intron transferred between E.coli and Shigella, in either direction. Because the IS629 sequences in E.coli and Shigella differ by 41 polymorphisms out of 436 bp, while the introns have only 14 polymorphisms over 2 kb, it appears that the intron independently inserted into the two IS629 sites rather than transferring between the two species via the host IS629 element. In Shigella, the intron ORF accumulated three frame shifts, which presumably inactivated its mobility function. In E.coli, the intron inserted into both IS911 and IS629 sites, with one intron in IS629 becoming truncated by the insertion of an IS5 element. At some point the intron was passed between E.coli and Yersinia, again in either direction. In Yersinia, one copy became truncated at its 3′ end by an unknown mechanism. Another copy in Yersinia enterocolitica became truncated for all but the 3′ portion (Table 3) and is not shown in the figure.
Figure 3.
Model for the spread of the E.c.I4 intron among three species and three homing sites. All introns in the family are >93% identical. See text for description.
Other families include introns of bacterial class C: B.h.I1 (five copies)/B.h.F1–F3 (>90% identity); P.p.I1/P.a.I1/P.a.I2/P.sp.F1/P.p.F1/P.p.F2 (>88% identity); S.p.I1/S.ag.I1./S.p.F1–F5 (>90% identity); and S.me.I3 (two copies)/S.me.F1 (>98% identity). All introns in these families are found in closely related species and are inserted after non-identical terminators (Fig. 2C). Although there are many intron fragments in Bradyrhizobium japonicum (Table 3), they are not closely related and do not form a coherent family.
COMPARISON OF BACTERIAL AND ORGANELLAR GROUP II INTRONS
The insertion patterns described here differ markedly from intron distribution in organelles, where the introns are inserted in conserved genes and are very rarely fragmented. These differences could be due to biochemical properties of the introns themselves, or may be due to effects of the host cells such as available host cofactors or competing biochemical processes. At this point we will consider differences between bacterial and organellar group II introns.
Location of introns
While almost all organellar group II introns are located in conserved genes that are critical to cellular function (e.g. subunits of cytochrome oxidase, NAD dehydrogenase, rubisco), nearly all bacterial introns are associated with mobile DNAs and, strikingly, no bacterial group II introns have yet been found in highly conserved or housekeeping genes, with the exception of X.f.I1 which has inactivated its host gene. Nearly half of bacterial group II introns are located outside of genes, and even with the exclusion of bacterial class C, >20% of bacterial introns are putatively outside of genes. Given that the gene density of bacterial genomes is ∼90% (35,36) it appears that there is moderate selection against insertion into genes, and strong selection for insertion into mobile DNAs. Why the non-random distribution? One possibility is that insertion into mobile DNAs gives a replicative advantage. While this may be true, it seems unlikely to account entirely for the distribution. IS elements do not transpose frequently, and even introns that have inserted outside of mobile DNAs are not inserted into conserved genes. A more likely explanation is that the introns interfere with host gene expression on some level. Group I introns in bacterial genomes have been observed to be similarly excluded from ORFs and instead are inserted into tRNA and rRNA genes (37). This has been rationalized as interference with splicing due to simultaneous transcription and translation, with ribosomes progressing through the intron before it can be spliced out (37). Bacterial group II introns also might splice poorly for the same reason, or for other reasons such as inefficient catalytic RNAs, inefficient maturase activities or absence of host splicing cofactors. Previously, we noted that at least some bacterial introns have poorly conserved maturase (X) domains (11), which might indicate less developed splicing function. There may be other less obvious ways that an intron might compromise function of its host gene, such as a greater metabolic cost of selfish DNAs in a conserved region than in a poorly transcribed region.
Fragmentation
Another difference of bacterial group II introns is that they are very frequently fragmented. While intron fragmentation in organelles is rare [1 in 51 based on compilation in Zimmerly et al. (11)], >50% (42 of 73) of bacterial group II introns are fragmented. The high frequency of intron fragments in bacterial genomes suggests a much higher rate of intron gain and loss compared with organellar genomes. Bacterial introns may move to new locations to balance out intron loss due to degenerations, while organellar introns occupy a genomic position for a longer time, and cannot afford to become fragmented because the host genes are highly conserved.
Why are there no reported ORF-less introns in bacteria?
While the majority of organellar group II introns are ORF-less, so far no ORF-less introns have been identified in bacteria. The closest example is P.al.F1 (38), which superficially resembles an ORF-less intron, but instead appears to be a derivative of P.a.I1 deleted for the ORF and also intron RNA domains 2–4 and part of domain 1. Why have no ORF-less introns been identified in bacteria? One reason may be that they would be very easily overlooked if they are not inserted into conserved genes. We have thoroughly searched without success for ORF-less introns related to ORF-containing introns, but have only identified fragments (Table 3). Additional BLAST searches based on the highly conserved domain 5 identified only ORF-containing introns in bacteria and no ORF-less introns (N.Toor and S.Zimmerly, unpublished results). It is probably safe to say there are no ORF-less introns in bacteria closely related to currently known ORF-containing introns, and that if ORF-less group II introns exist in bacteria, they are rare. Of course, it is impossible to discount the existence of bacterial ORF-less introns because they cannot be found with a given search strategy.
Are there differences in mobility and splicing activities?
Previously we speculated that different phylogenetic classes of introns may have different mobility properties compared with characterized mitochondrial introns (11). The introns most likely to support this prediction are introns of bacterial class C, which insert exclusively after terminator structures. These introns are unique structurally because their of abbreviated and unusual RNA secondary structures with a shortened domain 5 (10,29,34). One bacterial class C intron was shown to self-splice in vitro (S.ag.I1), although through hydrolysis rather than a lariat (34). Therefore, at least one intron of this class is competent for self-splicing even though no intron in the class is located in a gene.
Target site specificity for bacterial class C appears to be relaxed compared with other group II introns. Some degree of site specificity is suggested by B.h.I1 because four introns have inserted directly after identical 5S rRNA terminator sequences; however, the downstream sequences are non-identical, which goes against principles of conventional homing sites. Other than these four B.h.I1 copies, all other bacterial class C introns insert into different DNA sequences, but always after a terminator. The terminator sequences might influence DNA recognition through its inverted repeats, analogous to the recognition of inverted repeats by other proteins (34). It is also possible that the terminator structure is recognized at the RNA level, since almost all of the putative stem–loops contain G-U base pairs (Fig. 2). An intriguing possibility is that intron mobility for this class may occur at the RNA level with a mechanism distinct from TPRT, possibly by reverse splicing into an RNA transcript at its terminator, reverse transcription of intron and upstream exon, and integration of the cDNA into the genome.
Other than bacterial class C, there is little indication for differences in mobility mechanisms or properties, although there may well be variation in site specificities, efficiencies of homing or splicing efficiencies. Variation in homing efficiency is probably expected due to host environment. The L.l.I1 intron is efficiently mobile in Lactococcus (10–30%) (39), but when the intron is introduced into E.coli, mobility drops at least 100-fold and is detectable only using a selectable marker within the intron (26). This difference in mobility has been attributed to endogenous nucleases in E.coli, which nick the intron RNA and prevent complete reverse transcription of intron during mobility (22,33). Such differences in host environment may explain why introns have essentially saturated some genomes (S.meliloti; 28) and not others (E.coli; 30).
Another possible difference in mobility is suggested by the observation that the S.me.I1 intron is highly mobile after the initial introduction of homing site DNA to a cell, but mobility is shut off after several cell divisions (27). The mechanism for this burst followed by immobility is not fully explained. It is not known how general an effect this will be across bacterial species, but it could contribute to unexpected dynamics of intron spread in different organisms.
DO GROUP II INTRONS IN BACTERIA BEHAVE MORE LIKE RETROELEMENTS OR INTRONS?
The insertion patterns of bacterial group II introns raise fundamental issues about their role in bacteria. Comparison of bacterial and organellar introns suggests that the strategies for intron survival are intrinsically different. We predict that introns in bacteria have adapted to function mainly as retroelements, while introns in organelles have adapted to function mainly as introns.
Organellar group II introns can be considered to function primarily as introns because most organellar introns lack ORFs and are not mobile. Splicing of organellar introns is required to be highly efficient because the introns are located in housekeeping genes (e.g. cytochrome oxidase) with up to a dozen introns per gene. Organelles have recruited many proteins to aid splicing of group I and group II introns. Such protein factors function either directly or indirectly, and with varying degrees of intron specificity (13,40–43). Even introns that encode active maturase activities (e.g. Saccharomyces cerevisiae cox1I1 and cox1I2) require additional accessory proteins for efficient splicing in vivo (43). Thus, group II introns in organelles are required to splice efficiently, and evolution has recruited multiple host proteins to optimize the splicing reaction.
In contrast, bacterial introns are not required to splice efficiently because they are not present in conserved genes and are often located outside of genes. It is not clear that splicing cofactors will be provided by the bacterial host cells, particularly if the introns are transferred among distantly related bacteria. No introns have yet been identified in bacteria that are expected to function only in splicing (i.e. ORF-less introns or introns with degenerate ORFs); however, there are many examples of introns that resemble retroelements due to insertion into the wrong strand of a gene, insertion outside of genes or insertions after a terminator structure. Group II introns in bacteria can be considered ‘guilty by association’ in that they are often inserted within IS elements, truncated by IS elements or flanked by IS elements, suggesting that bacterial group II introns are subject to similar selective pressures as IS elements. Finally, the high rate of fragmentation suggests that bacterial introns are gained and lost frequently. In contrast to organellar introns, bacterial group II introns have many characteristics of retroelements, and their intronic character is less prominent.
These differences suggest fundamentally different strategies for intron survival in bacteria versus organelles. In organelles, intron mobility is highly efficient, highly site specific and targeted to conserved genes, a strategy suited to a small, tightly packed genome. A possible life cycle for an organellar intron might be gain of intron by homing, degeneration of ORF or intron but maintenance of splicing function, possibly ORF loss, and finally precise intron deletion. In bacteria, on the other hand, the frequent fragmentation of introns suggests a life cycle of constant insertions into new locations followed by fragmentation and loss. Survival of group II introns in bacteria appears to rely on constant movement, whereas intron survival in organelles appears to rely on a comparatively stable position in conserved genes with concomitantly efficient splicing function.
EVOLUTIONARY SPECULATIONS
If bacterial introns are indeed the most primitive of mobile group II introns, then their retroelement features would suggest an ancestral state. Previously, we proposed a model for the evolution of group II introns, the retroelement ancestor hypothesis, which predicts that all currently known group II introns were derived from mobile bacterial group II introns (29). The catalytic RNA structures were proposed to have differentiated in bacteria as components of retroelements, followed by ORF loss in mitochondria and chloroplasts to form the numerous organellar ORF-less introns. The predominant retroelement features of bacterial group II introns discussed here are consistent with this model, although other explanations can be invoked such as migration of introns from organelles to bacteria with subsequent enhancement of retroelement character as an adaptation to the new environment. Assuming that bacterial group II introns are the most primitive, then the absence of ORF-less group II introns in bacteria would also be consistent with a previous speculation that the catalytic RNA structure itself could have been derived from a retroelement as a way of preventing host damage (29,44). Finally, the retroelement character of bacterial introns casts an intriguing light on spliceosomal introns. If group II introns began as retroelements and subsequently evolved into spliceosomal introns, then splicesomal introns might be viewed as derivative retrotransposons that very successfully invaded and colonized higher eukaryotic genomes.
NOTE ADDED IN REVISION
Since this manuscript was submitted, the following introns have been reported to GenBank: Streptococcus agalactiae, accession no. AF380672 (3850–5706), bacterial class C, same intron as S.ag.I1 but inserted after a different terminator; Nostoc sp., accession nos AP003604 (45422–47907), AP003599 (30737–33044), AP003600 (259212–261419), all chloroplast-like class 2; Nostoc sp., accession no. AP003600 (333632–335504), chloroplast-like class 2, fragmented intron identical to A.sp.F1.
NOTE ADDED IN PROOF
The fragments A.g.F1, P.ae.F1 and S.ma.F1 have been determined to be full-length introns of bacterial class C with the following boundaries: A.g.F1, GenBank accession nos AF369871 (2878–4803); P.ae.F1 AY029772 (3515–5441); S.ma.F1 AY030343 (1893–3818). Additional introns reported to GenBank since revision are: Serratia marcescens, accession nos AF453998 (534–2504), bacterial class C; and Pseudomonas putida accession nos AY065966 (2474–4399), bacterial class C, identical to A.g.F1, P.ae.F1 and S.ma.F1.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank Alan Lambowitz and Michael Hynes for helpful comments on the manuscript, and Nav Toor, Rob Olson and Josh Christianson for help in folding intron structures. This work was supported by the National Science and Engineering Research Council (Canada) and the Alberta Heritage Foundation for Medical Research.
REFERENCES
- 1.van der Veen R., Arnbert,A.C., van der Horst,G., Bonen,L., Tabak,H.F. and Grivell,L.A. (1986) Excised group II introns in yeast mitochondria are lariats and can be formed by self-splicing in vitro. Cell, 44, 225–234. [DOI] [PubMed] [Google Scholar]
- 2.Peebles C.L., Perlman,P.S., Mecklenburg,K.L., Petrillo,M.L., Tabor,J.H., Jarrell,K.A. and Cheng,H.-L. (1986) A self-splicing RNA excises an intron lariat. Cell, 44, 213–223. [DOI] [PubMed] [Google Scholar]
- 3.Michel F. and Lang,B.F. (1985) Mitochondrial class II introns encode proteins related to the reverse transcriptases of retroviruses. Nature, 316, 641–643. [DOI] [PubMed] [Google Scholar]
- 4.Lazowska J., Meunier,B. and Macadre,C. (1994) Homing of a group II intron in yeast mitochondrial DNA is accompanied by unidirectional co-conversion of upstream-located markers. EMBO J., 13, 4963–4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moran J.V., Zimmerly,S., Eskes,R., Kennell,J.C., Lambowitz,A.M., Butow,R. and Perlman,P.S. (1995) Mobile group II introns of yeast mitochondrial DNA are novel site-specific retroelements. Mol. Cell. Biol., 15, 2828–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zimmerly S., Guo,H., Perlman,P.S. and Lambowitz,A.M. (1995) Group II intron mobility occurs by target DNA-primed reverse transcription. Cell, 82, 545–554. [DOI] [PubMed] [Google Scholar]
- 7.Zimmerly S., Guo,H., Eskes,R., Yang,J., Perlman,P.S. and Lambowitz,A.M. (1995) A group II intron RNA is a catalytic component of a DNA endonuclease involved in intron mobility. Cell, 83, 529–538. [DOI] [PubMed] [Google Scholar]
- 8.Ferat J.-L. and Michel,F. (1993) Group II self-splicing introns in bacteria. Nature, 364, 358–361. [DOI] [PubMed] [Google Scholar]
- 9.Michel F. and Ferat,J.-L. (1995) Structure and activities of group II introns. Annu. Rev. Biochem., 64, 435–461. [DOI] [PubMed] [Google Scholar]
- 10.Martinez-Abarca F. and Toro,N. (2000) Group II introns in the bacterial world. Mol. Microbiol., 38, 917–926. [DOI] [PubMed] [Google Scholar]
- 11.Zimmerly S., Hausner,G. and Wu,X. (2001) Phylogenetic relationships among group II intron ORFs. Nucleic Acids Res., 29, 1238–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonen L. and Vogel,J. (2001) The ins and outs of group II introns. Trends Genet., 17, 322–331. [DOI] [PubMed] [Google Scholar]
- 13.Lambowitz A.M., Caprara,M., Zimmerly,S. and Perlman,P.S. (1999) Group I and group II ribozymes as RNPs: clues to the past and guides to the future. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 451–485.
- 14.Guo H., Zimmerly,S., Perlman,P.S. and Lambowitz,A.M. (1997) Group II intron endonucleases use both RNA and protein subunits for recognition of specific sequences in double-stranded DNA. EMBO J., 16, 6835–6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang J., Mohr,G., Perlman,P.S. and Lambowitz,A.M. (1998) Group II intron mobility in yeast mitochondria: target DNA-primed reverse transcription activity of aI1 and reverse splicing into DNA transposition sites in vitro. J. Mol. Biol., 282, 505–523. [DOI] [PubMed] [Google Scholar]
- 16.Singh N.N. and Lambowitz,A.M. (2001) Interaction of a group II intron ribonucleoprotein endonuclease with its DNA target site investigated by DNA footprinting and modification interference. J. Mol. Biol., 309, 361–386. [DOI] [PubMed] [Google Scholar]
- 17.Mueller M.W., Allmaier,M., Eskes,R. and Schweyen,R.J. (1993) Transposition of group II intron aI1 in yeast and invasion of mitochondrial genes at new locations. Nature, 366, 174–176. [DOI] [PubMed] [Google Scholar]
- 18.Dickson L., Huang,H.-R., Liu,L., Matsuura,M., Lambowitz,A.M. and Perlman,P.S. (2001) Retrotransposition of a yeast group II intron occurs by reverse splicing directly into ectopic DNA sites. Proc. Natl Acad. Sci. USA, 98, 13207–13212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sellem C.H., Lecellier,G. and Belcour,L. (1993) Transposition of a group II intron. Nature, 366, 176–178. [DOI] [PubMed] [Google Scholar]
- 20.Cousineau B., Lawrence,S., Smith,D. and Belfort,M. (2000) Retrotransposition of a bacterial group II intron. Nature, 404, 1018–1021. [DOI] [PubMed] [Google Scholar]
- 21.Martinez-Abarca F. and Toro,N. (2000) RecA-independent ectopic transposition in vivo of a bacterial group II intron. Nucleic Acids Res., 28, 4397–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuura M., Saldanha,R., Ma,H., Wank,H., Yang,J., Mohr,G., Cavanagh,S., Dunny,G.M., Belfort,M. and Lambowitz,A.M. (1997) A bacterial group II intron encoding reverse transcriptase, maturase, and DNA endonuclease activities: biochemical demonstration of maturase activity and insertion of new genetic information within the intron. Genes Dev., 11, 2910–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saldanha R., Chen,B., Wank,H., Matsuura,M., Edwards,J. and Lambowitz,A.M. (1999) RNA and protein catalysis in group II intron splicing and mobility reactions using purified components. Biochemistry, 38, 9069–9083. [DOI] [PubMed] [Google Scholar]
- 24.Eskes R., Yang,J., Lambowitz,A.M. and Perlman,P.S. (1997) Mobility of yeast mitochondrial group II introns: engineering a new site specificity and retrohoming via full reverse splicing. Cell, 88, 865–874. [DOI] [PubMed] [Google Scholar]
- 25.Eskes R., Liu,L., Ma,H., Chao,M.Y., Dickson,L., Lambowitz,A.M. and Perlman,P.S. (2000) Multiple homing pathways used by yeast mitochondrial group II introns. Mol. Cell Biol., 20, 8432–8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cousineau B., Smith,D., Lawrence,S., Mueller,J.E., Yang,J., Mills,D., Manias,D., Dunny,G., Lambowitz,A.M. and Belfort,M. (1998) Retrohoming of a bacterial group II intron: mobility via complete reverse splicing, independent of homologous DNA recombination. Cell, 94, 451–462. [DOI] [PubMed] [Google Scholar]
- 27.Martinez-Abarca F. and Toro,N. (2000) Homing of a bacterial group II intron with an intron-encoded protein lacking a recognizable endonuclease domain. Mol. Microbiol., 35, 1405–1412. [DOI] [PubMed] [Google Scholar]
- 28.Martinez-Abarca F., Zekri,S. and Toro,N. (1998) Characterization and splicing in vivo of a Sinorhizobium meliloti group II intron associated with particular insertion sequences of the IS630-Tc1/IS3 retroposon superfamily. Mol. Microbiol., 28, 1295–1306. [DOI] [PubMed] [Google Scholar]
- 29.Toor N., Hausner,G. and Zimmerly,S. (2001) Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA, 7, 1142–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferat J.-L., Le Gouar,M. and Michel,F. (1994) Multiple group II self-splicing introns in mobile DNA from Escherichia coli. C.R. Acad. Sci. Paris, 317, 141–148. [PubMed] [Google Scholar]
- 31.Mullany P., Pallen,M., Wilks,M., Stephen,J.R. and Tabaqchali,S. (1996) A group II intron in a conjugative transposon from the gram-positive bacterium Clostridium difficile. Gene, 174, 145–150. [DOI] [PubMed] [Google Scholar]
- 32.Mohr G., Smith,D., Belfort,M. and Lambowitz,A.M. (2000) Rules for DNA target-site recognition by a lactococcal group II intron enable retargeting of the intron to specific DNA sequences. Genes Dev., 14, 559–573. [PMC free article] [PubMed] [Google Scholar]
- 33.Guo H., Karberg,M., Long,M., Jones,J.P.,III, Sullenger,B. and Lambowitz,A.M. (2000) Group II introns designed to insert into therapeutically relevant DNA target sites in human cells. Science, 289, 452–457. [DOI] [PubMed] [Google Scholar]
- 34.Granlund M., Michel,F. and Norgren,M. (2001) Mutually exclusive distribution of IS1548 and GBSi1, an active group II intron identified in human isolates of group B streptococci. J. Bacteriol., 183, 2560–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blattner F.R. et al. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- 36.Kunst F. et al. (1997) The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature, 390, 249–256. [DOI] [PubMed] [Google Scholar]
- 37.Edgell D.R., Belfort,M. and Shub,D.A. (2000) Barriers to intron promiscuity in bacteria. J. Bacteriol., 182, 5281–5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yeo C.C., Yiin,S., Tan,B.H. and Poh,C.L. (2001) Isolation and characterization of group II introns from Pseudomonas alcaligenes and Pseudomonas putida. Plasmid, 45, 233–239. [DOI] [PubMed] [Google Scholar]
- 39.Mills D.A., Manias,D.A., McKay,L.L. and Dunny,G.M. (1997) Homing of a group II intron from Lactococcus lactis subsp. lactis ML3. J. Bacteriol., 179, 6107–6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jenkins B.D. and Barkan,A. (2001) Recruitment of a peptidyl-tRNA hydrolase as a facilitator of group II intron splicing in chloroplasts. EMBO J., 20, 872–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waldherr M., Ragnini,A., Jank,B., Teply,R., Wiesenberger,G. and Schweyen,R.J. (1993) A multitude of suppressors of group II intron-splicing defects in yeast. Curr. Genet., 24, 301–306. [DOI] [PubMed] [Google Scholar]
- 42.Weisenberger G., Waldherr,M. and Schweyen,R.J. (1992) The nuclear gene MRS2 is essential for the excision of group II introns from yeast mitochondrial transcripts in vivo. J. Biol. Chem., 267, 6963–6969. [PubMed] [Google Scholar]
- 43.Seraphin B., Simon,M., Boulet,A. and Faye,G. (1989) Mitochondrial splicing requires a protein from a novel helicase family. Nature, 337, 84–87. [DOI] [PubMed] [Google Scholar]
- 44.Curcio M.J. and Belfort,M. (1996) Retrohoming: cDNA-mediated mobility of group II introns requires a catalytic RNA. Cell, 84, 9–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.