Abstract
Background
Accurately determining arrhythmia mechanism from a 12-lead electrocardiogram (ECG) of supraventricular tachycardia can be challenging. We hypothesized a convolutional neural network (CNN) can be trained to classify atrioventricular re-entrant tachycardia (AVRT) vs atrioventricular nodal re-entrant tachycardia (AVNRT) from the 12-lead ECG, when using findings from the invasive electrophysiology (EP) study as the gold standard.
Methods
We trained a CNN on data from 124 patients undergoing EP studies with a final diagnosis of AVRT or AVNRT. A total of 4962 5-second 12-lead ECG segments were used for training. Each case was labeled AVRT or AVNRT based on the findings of the EP study. The model performance was evaluated against a hold-out test set of 31 patients and compared to an existing manual algorithm.
Results
The model had an accuracy of 77.4% in distinguishing between AVRT and AVNRT. The area under the receiver operating characteristic curve was 0.80. In comparison, the existing manual algorithm achieved an accuracy of 67.7% on the same test set. Saliency mapping demonstrated the network used the expected sections of the ECGs for diagnoses; these were the QRS complexes that may contain retrograde P waves.
Conclusion
We describe the first neural network trained to differentiate AVRT from AVNRT. Accurate diagnosis of arrhythmia mechanism from a 12-lead ECG could aid preprocedural counseling, consent, and procedure planning. The current accuracy from our neural network is modest but may be improved with a larger training dataset.
Keywords: Artificial intelligence, Machine learning, Electrocardiogram, Atrioventricular re-entrant tachycardia, Atrioventricular nodal re-entrant tachycardia, Electrophysiology study, Ablation
Key Findings.
-
•
A convolutional neural network can be trained to differentiate atrioventricular re-entrant tachycardia from atrioventricular nodal re-entrant tachycardia using the 12-lead electrocardiogram (ECG).
-
•
Accurate diagnosis of arrhythmia mechanism from a 12-lead ECG could aid preprocedural counseling, consent, and procedure planning.
-
•
Saliency mapping demonstrated the neural network used the primarily the QRS complexes to make classification decisions.
Introduction
Ablation for supraventricular tachycardia (SVT) is a highly effective therapy, considered first line in patients with recurrent SVTs or who do not wish to take long-term medical therapy.1 SVT in general refers to all tachycardias other than ventricular tachycardia and atrial fibrillation, and includes atrial tachycardia (AT), atrioventricular re-entrant tachycardias (AVRT), and atrioventricular nodal re-entrant tachycardias (AVNRT). AT can usually be diagnosed where there are more P waves than QRS complexes on the 12-lead electrocardiogram (ECG) and where adenosine administration reveals underlying atrial wave activity with transient high-grade atrioventricular-ventricular block.1 AVRT and AVNRT, however, may be more difficult to differentiate, as the RP interval may be suggestive but ultimately not definitive for distinguishing the tachycardia mechanism.1 Several additional ECG criteria have been described but are vulnerable to interobserver and intraobserver variability.2, 3, 4, 5, 6, 7, 8, 9
Although ablation for AVNRT and AVRT is highly effective,1 a definitive diagnosis is generally required at the time of an electrophysiology (EP) study. Prior knowledge of the tachycardia mechanism could aid preprocedural counseling, consent, and procedure planning. Occasionally the SVT has been previously documented, but cannot be induced in the EP laboratory.10 In these cases, empirical ablation may be performed—eg, ablation of an accessory pathway if one is found, or slow pathway modification if dual atrioventricular nodal physiology (DAVNP) is identified.11, 12, 13
Deep learning, particularly using convolutional neural networks (CNN), has been used extensively for classification tasks using the 12-lead ECG, with great accuracy.14, 15, 16 Several deep learning models have been built to identify SVT from the 12-lead ECG,17 and 1 to predict the presence of SVT from a sinus rhythm ECG.18 We hypothesized a CNN could be trained for the specific task of differentiating AVNRT from AVRT, allowing for automated preprocedural rhythm determination.
Methods
Patient selection
Patients were selected from a clinical database of all invasive EP procedures at Hammersmith Hospital, Imperial College Healthcare NHS Trust, United Kingdom. A search was performed for any slow pathway modification or accessory pathway ablations for the years 2012–2021. The patients all had procedures using an EP recording system, LABSYSTEM™ PRO (Boston Scientific). The ECG and the intracardiac electrogram recordings for each case (preablation) were visually examined for lengths that contained sustained tachycardias without diagnostic pacing maneuvers or excessive noise. The intracardiac electrograms were inspected to ensure the selected segments contained AVRT or AVNRT and not another tachycardia (eg, sinus tachycardia or AT). Using the EP recording system, digital 12-lead ECG recordings were exported (Figure 1).
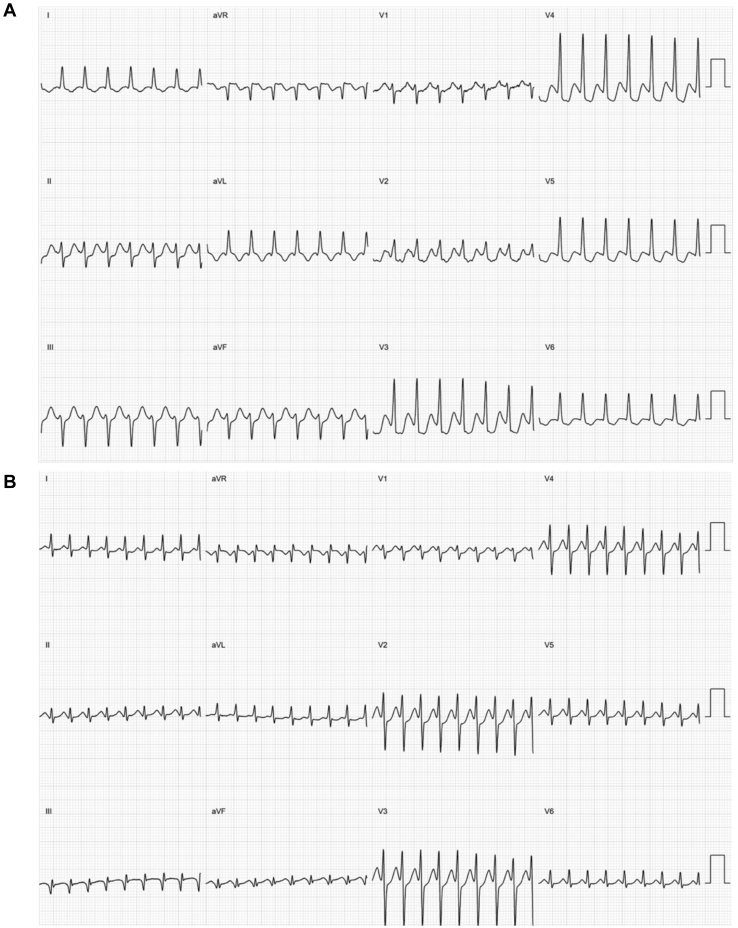
Figure 1.
Electrocardiogram (ECG) class examples. Example 12-lead ECGs of the classes used to train the neural network. A: An ECG showing atrioventricular re-entrant tachycardia. B: An ECG showing atrioventricular nodal re-entrant tachycardia.
Data labeling
For each case, the procedure report was reviewed to determine the diagnosis. Diagnoses were made by the clinical team carrying out the ablation procedure and were based on maneuvers performed during tachycardia. The final diagnoses could have included all types of AVNRT (slow-fast, fast-slow, slow-slow) or orthodromic AVRT. QRS complexes in tachycardia could have been narrow or broad (eg, owing to rate-related aberrancy). Antidromic AVRT was not included. Atrial fibrillation, AT, and atrial flutter were not included. Cases with little or no sustained arrhythmia, or without a clear diagnosis, were not included.
12-lead ECG extraction
Digital 12-lead surface ECGs were extracted at a sampling rate of 1000 Hz. For patients with multiple procedures, within 2012–2021, only 1 procedure was included. Patients were randomly assigned to training and testing datasets. In the training set, each patient had up to 5 minutes of data exported in up to 1-minute segments. Each 1-minute segment was further split into 5-second segments. Segmentation into 5-second segments was performed in an automated fashion, with no visual inspection steps. Given the likely significant similarities between consecutive 5-second recordings, the test set used only one 5-second segment for each patient. The starting 5-second segment was used. Importantly, no data from the test dataset were used in training dataset and the patients in the test set were distinct from those in the training set. Data flow charts are shown in Supplemental Figures S1 and S2.
Data preprocessing
Multichannel exports from the EP recording system were converted into time series data.19 Each ECG recording was downsampled to 200 Hz. This allowed a reduction in the dataset size, thereby reducing model training time while maintaining accuracy.
CNN structure, training, and testing
CNN are a type of deep learning based on biological neural networks.20 The convolutional layers detect features on the input data that can be used in subsequent layers of the neural network to predict a classification for the input data.
In this work, a CNN was trained on 12-lead ECGs for arrhythmia classification. Our approach to model development has been previously described21 and is detailed in the Supplemental Methods.
Our final network was selected based on performance in the validation set; this was a modified version of that used by Attia and colleagues22 (Supplemental Figure S3). The remaining tested networks, including those using residual blocks, were inferior to this network when evaluated using the validation dataset. The complete architecture of the neural network is detailed in Supplemental Figure S3.
The model input was 12-lead ECGs of 5-second duration. The ECG data were zero padded to 1024 × 12. The learning rate was 0.01. The binary cross-entropy was minimized using the Adam optimizer. The neural network was trained for a maximum of 40 epochs with early termination set to when there was no further improvement in validation set loss for 7 consecutive epochs. The training batch size was 16. During hyperparameter tuning, the optimal value for dropout was found to be 0; therefore the dropout layer was removed from the final model architecture. The filter numbers and sizes are detailed in Supplemental Figure S3. To create the final model the whole training dataset (including the 20% validation set used for hyperparameter tuning) was split into 10 folds. Each patient’s data was assigned to only 1 fold to avoid training and validation against the same patient’s data. In order to use the whole training dataset for model training, 10-fold cross-validation was performed. In this process a model was trained on 9 folds of data and validated on the 10th fold. This is repeated 10 times so that each fold is used as a validation fold once, to create 10 models. The 10 models were then averaged to create an ensemble model for use on the unseen test set.
To explore how the neural network arrived at each classification, we used the method of saliency mapping. We used the vanilla gradient algorithm in tf-keras-vis (version 0.5.5) package in Python (version 3.9),23 where each data point of the ECG is mathematically tested to quantitatively determine how much it contributes to the output produced. A map can then be created where the highlighted sections are the most significant, and salient, areas. This is important, as it can allow us to understand the processes by which the network chooses the class in the testing phase to ensure unexpected factors are not influencing the network.24
Further information concerning the ECG preprocessing and architecture selection process is detailed in the Supplemental Methods.
Comparison to existing algorithm
We compared the performance of our CNN to an existing algorithm, described by Jaeggi and colleagues5 and evaluated in adults by Arya and colleagues2 (Supplemental Figure S4). Given the differing inclusion criteria in their studies and our own, the algorithm did not apply to some ECGs in our dataset, owing to the presence of rate-related aberrancy in 2 patients in our study test set. For these cases a diagnosis of AVNRT was assigned, based on the higher prevalence of AVNRT. The algorithm was applied to the test set ECGs by a clinician blinded to the CNN and actual diagnosis.
Statistical analysis
The performance of the neural network was evaluated using the overall accuracy, F1 score, and area under curve (AUC) metrics. Binomial proportion confidence intervals are reported. Statistical analysis was carried out in Python (version 3.9). The McNemar test was used to compare model performance with the ECG algorithm.
Results
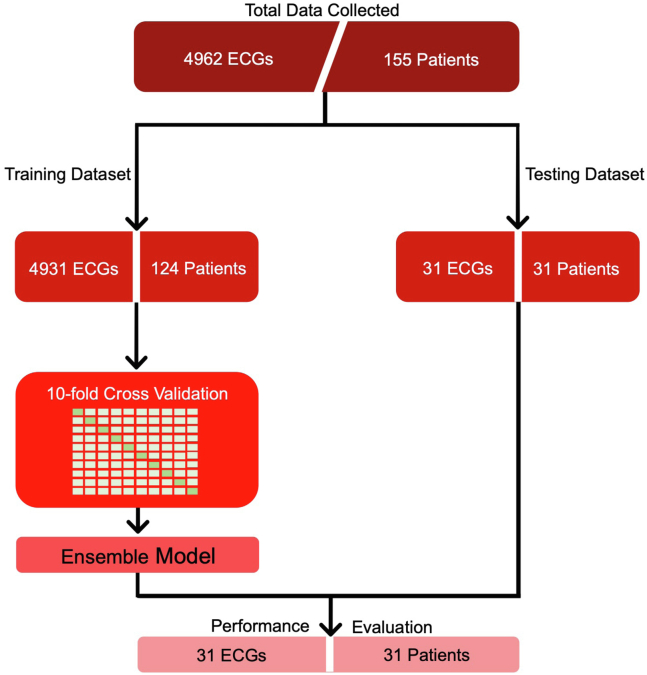
There were 155 patients’ ECGs analyzed; 45% of patients were male and the average age was 54 ± 18.2 years. There were 106 AVNRT cases and 49 AVRT cases. Patients were randomly assigned to the training and test datasets (124 in the training dataset and 31 in the testing dataset; Figure 2). There were 10 patients with atypical AVNRT, 9 fast-slow and 1 slow-slow. In the AVRT cases there were 12 right-sided accessory pathways and 37 left-sided. In the test dataset there were 21 AVNRT cases and 10 AVRT.
Figure 2.
Study flowchart. Data from 155 patients were collected, 106 atrioventricular nodal re-entrant tachycardia cases and 49 atrioventricular re-entrant tachycardia cases. Twelve-lead electrocardiograms (ECGs) were segmented into 5-second recordings. For the training dataset, multiple recordings were used per patient; the test set used only 1 recording per patient. An ensemble model was created from the 10 models through cross-validation. Model performance was evaluated on the unseen test set.
Neural network performance
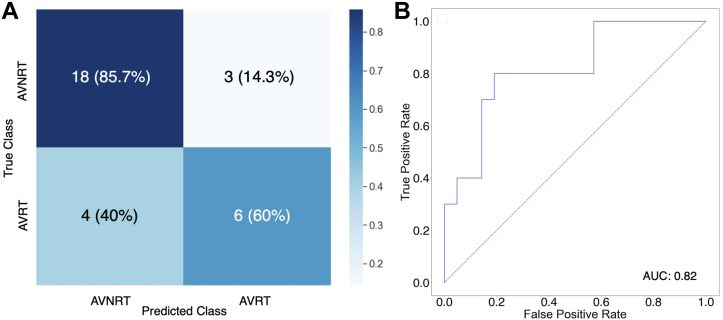
For the testing dataset, the accuracy was 77.4% (95% confidence interval 0.63–0.92). The F1 score was 0.63 and Cohen kappa 0.47. Figure 3A displays the confusion matrix. The model correctly identified AVNRT 85.7% of the time and AVRT 60% of the time. Figure 3B shows the receiver operating characteristic curve; the AUC was 0.82 for distinguishing the classes. At the default threshold of 0.5, the sensitivity and specificity for classifying AVNRT and AVRT were 85.7% and 60%, respectively. On the test set the processing time for the trained model was a total of 5.37 seconds for the total 31 ECGs in the test set; this equates to an average of 173 ms per 5-second ECG.
Figure 3.
Model performance. A: Confusion matrix demonstrating the accuracy of the model in classifying between the 2 groups. Atrioventricular nodal re-entrant tachycardia (AVNRT) and atrioventricular re-entrant tachycardia (AVRT) cases were correctly classified at 85.7% and 60%, respectively. B: Receiver operating characteristic (ROC) curves for model performance; diagnosis of AVRT was considered a positive case. The area under the ROC (AUC) was 0.82.
Comparison to existing algorithm
The accuracy of the algorithm by Jaeggi and colleagues5 was 67.7%. Two of the cases in the test set had rate-related aberrancy and therefore Jaeggi and colleagues’ algorithm did not apply. A diagnosis of AVNRT was therefore applied based on the higher prevalence of AVNRT. The true diagnosis in these 2 cases was in fact AVNRT. Excluding the 2 cases with rate-related aberrancy yielded an accuracy of 65.5%; this was not statistically significantly different from CNN accuracy (P = .29). The accuracy of Jaeggi and colleagues’ algorithm was 73.7% for AVNRT and 50% for AVRT in our test set.
Saliency mapping
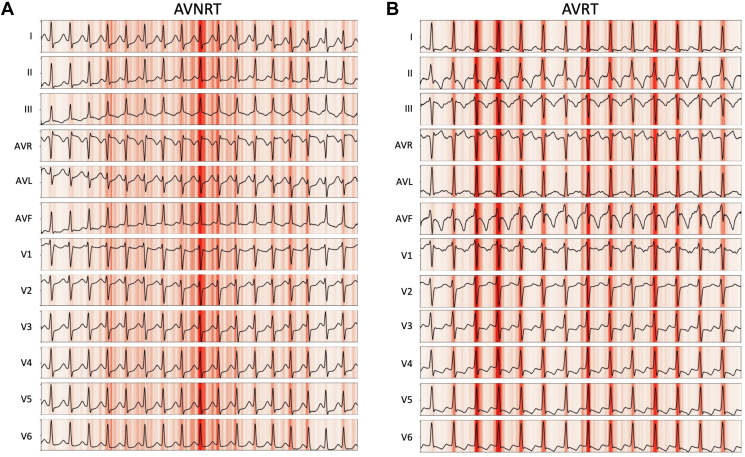
A saliency map can be used to help understand why a CNN predicted a particular outcome. This is achieved by mapping the outcome back to key areas of the input that most influenced the network in producing the classification result. Figure 4 presents the saliency mappings of an example 12-lead ECG for each class, AVNRT and AVRT. The network used the expected sections of the ECGs for diagnoses; these were the QRS complexes that may contain retrograde P-wave activity.
Figure 4.
Saliency maps of each electrocardiogram (ECG) class. Saliency maps showing the most prominent sections of ECG recordings in influencing the output of the neural network for the ECG classes of A: atrioventricular nodal re-entrant tachycardia (AVNRT) and B: atrioventricular re-entrant tachycardia (AVRT). Darker red indicates higher saliency. The QRS in each case appears to be salient; this may relate to the presence and timing of retrograde P waves in the QRS complex and is biologically plausible.
Discussion
We report the first deep learning model trained to distinguish AVRT from AVNRT using only the surface 12-lead ECG, using the ground truth labels derived from the gold standard of an invasive EP study. Although the accuracies we report are modest, this proof of concept may allow future development of a more accurate classification model. Incorporation of artificial intelligence (AI)–enabled ECG may help guide treatment in patients with AVNRT or AVRT.
Traditional ECG criteria for distinguishing AVRT from AVNRT
There are several manual ECG criteria that may be used to attempt to elucidate SVT mechanism from the 12-lead ECG.9 Initiation and termination of tachycardia is particularly helpful but may not always be captured.1 ECGs of sustained tachycardia can be examined to determine the P/QRS relationship and RP interval, and response to adenosine may also be considered.1 Pseudo r deflection in lead V1 and pseudo S in the inferior leads is particularly specific for typical AVNRT (91%–100%) but lacks sensitivity (15%–58%).9 Tai and colleagues8 also described a manual stepwise approach. They use manual inspection for retrograde P-wave morphology to differentiate AVRT and AVNRT and also predict the location of concealed accessory pathways. They report an accuracy of 97.8% in differentiating AVRT and AVNRT; however, a major limitation was that clearly visible P waves were required for inclusion in their study. Others have reported modified criteria and algorithms,2, 3, 4, 5, 6, 7 with varying accuracies. Importantly, these studies have varying exclusion criteria, making comparison challenging; some exclude atypical forms of AVNRT or cases where the RP interval is greater than the PR interval. Jaeggi and colleagues5 originally described an algorithm in children. Arya and colleagues2 subsequently evaluated the algorithm’s performance in adults, reporting an accuracy of 81.4%, while Di Toro and colleagues3 report AUC of 0.72 for their aVL notch criteria. Additionally, a manual approach is likely to depend on the reading clinician’s experience and lead to interobserver and intraobserver variability. Depending on the algorithm, interobserver agreement can be as low as 70% and even intraobserver agreement may be as low as 75%.7
In our study, the CNN had a numerically higher accuracy than the algorithm approach, although this did not reach statistical significance in this small dataset. Interobserver variability and the differences in inclusion criteria in the 2 studies may explain the difference in performance of Jaeggi and colleagues’5 algorithm in our study compared to when evaluated by Arya and colleagues.2
Automated analysis of the ECG to distinguish AVRT from AVNRT
Perlman and colleagues25 have reported a tree scheme for the classification of SVT diagnoses from the 12-lead ECG. Through signal processing techniques, they extract features of atrial electrical activity from the ECG and then apply a clinically based tree algorithm. Additionally, they employ a 5-nearest-neighbors classifier to specifically distinguish AVNRT and AVRT. The advantage of this approach is explainability regarding the features used to make each classification decision. The automated approach also removes the challenge of interobserver and intraobserver variability present with human interpretation. They also report very high accuracies of 93% for correct classification of AVRT/AVNRT. However, their report included only 11 patients with AVRT or AVNRT for training and 19 in validation. Therefore, the performance of their model is likely to be highly dependent on the patterns found in the 11 patients used for algorithm training, with the potential for very poor generalizability to other cohorts.
Potential clinical applications
Rapid and automated ECG interpretation with AI has the potential to transform the care of patients with arrhythmia. AI-ECG platforms are ready for prospective clinical evaluation, with additional deep learning algorithms being added regularly.26 In particular, for patients undergoing EP procedures, AI-enabled ECG could be incorporated into clinical workflows, complementing electrophysiologist ECG interpretation.21
Catheter ablation for AVNRT is a highly effective therapy that has been shown to be superior to antiarrhythmic drugs for reducing hospitalizations while also avoiding antiarrhythmic drugs’ side effects.27 The treatment of choice for recurrent AVRT is also catheter ablation.1 Orthodromic AVRT is the cause of >90% of AVRTs and 20%–30% of all sustained SVTs.1
Patients undergoing EP studies often have documented SVT on an ECG. There may be some clues as to the rhythm; however, particularly for AVNRT vs AVRT, the diagnosis is often unclear. Accessory pathways may be concealed, which adds to the diagnostic difficulty. Currently, it is only during the EP study, if SVT is inducible, that the diagnosis can be made.
Accurate knowledge of tachycardia mechanism prior to an EP study may aid preprocedural counseling and consent. In particular, where the likely diagnosis is AVNRT, a more detailed discussion regarding the small but notable risk of AV node damage could take place. Conversely, where AVRT is the likely diagnosis, discussion regarding likely need for trans-septal puncture (given that the majority of concealed accessory pathways are left sided28) and the related risks of bleeding and thromboembolism may be discussed in more detail. Lastly, some operators may prefer to map accessory pathways with the aid of 3D mapping.29, 30, 31 Preprocedural knowledge of tachycardia mechanism could allow placement of 3D mapping patches in the appropriate cases without wasting these on cases where AVNRT is likely.
In some cases, no tachycardia can be induced.10 Empirical slow pathway modification has been considered a possible approach where DAVNP can be demonstrated.11, 12, 13 However, this involves subjecting the patient to the risks of ablation, in particular the risk of complete AV block, with uncertainty regarding potential benefit.
We propose that a deep learning model could be applied to the preprocedure SVT ECG and used to guide empirical ablation when SVT cannot be induced on the day of the EP procedure. This could take the form of slow pathway modification when DAVNP is demonstrated in a patient (who may or may not have a bystander accessory pathway). An alternative scenario would be the presence of a concealed accessory pathway with no high-risk features and DAVNP; identification of the rhythm on a preprocedure SVT ECG could then allow the appropriate ablation to be performed.
Limitations
Our model performance was modest, with specificity for AVRT in particular being relatively low. This is likely to be due to the small dataset in our study. However, we hope this proof of concept will allow studies with larger datasets to improve on model performance. Our data were additionally from a single center and single recording system. Ideally, external validation is required to ensure our model generalizes to other populations and ECG recording systems. The input to our model requires electrophysiologist involvement, as only AVRT or AVNRT cases were included. This could alternatively take the form of other, pre-existing models (such as that proposed by Perlman and colleagues25) that could be applied first to exclude other diagnoses. Lastly, the input requires a digitized 12-lead ECG. In many healthcare settings ECGs are in paper or PDF format; however, with the growing applications of AI-enabled ECGs we hope ECG machines capable of recording the raw digital signals will become more prevalent.
Conclusion
We describe the first neural network trained to differentiate AVRT from AVNRT. Accurate diagnosis of arrhythmia mechanism from a 12-lead ECG could aid preprocedural counseling, consent, and procedure planning. Additionally, where AVNRT is diagnosed on the ECG, empirical slow pathway ablation could be performed where sustained arrhythmia cannot be induced during the EP study. The current accuracy from our neural network is modest but may be improved with a larger training dataset.
Acknowledgments
Funding Sources
A.S. is funded by a British Heart Foundation (BHF) clinical research training fellowship (FS/CRTF/21/24183) and was supported by an NIHR Academic Clinical Fellowship (ACF-2019-21-001). F.S.N. and N.S.P. are supported by the BHF (RG/16/3/32175, RG/F/22/110078 and PG/16/17/32069). F.S.N. is also supported by the National Institute for Health Research Imperial Biomedical Research Centre.
Disclosures
The authors have no conflicts to disclose.
Authorship
All authors attest they meet the current ICMJE criteria for authorship.
Patient Consent
For this retrospective study using anonymized data, the regional ethics board determined individual patient consent was not required.
Data Availability Statement
The datasets generated or analyzed, or both, during this study are not publicly available owing to ethical restrictions.
Ethics Statement
This retrospective study was approved by the regional ethics board (IRAS ID: 293374). The authors designed the study and gathered and analyzed the data according to the Helsinki Declaration guidelines on human research.
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.cvdhj.2023.01.004
Appendix. Supplementary data
References
- 1.Brugada J., Katritsis D.G., Arbelo E., et al. 2019 ESC Guidelines for the management of patients with supraventricular tachycardia. The Task Force for the management of patients with supraventricular tachycardia of the European Society of Cardiology (ESC) Eur Heart J. 2020;41:655–720. doi: 10.1093/eurheartj/ehz467. [DOI] [PubMed] [Google Scholar]
- 2.Arya A., Kottkamp H., Piorkowski C., et al. Differentiating atrioventricular nodal reentrant tachycardia from tachycardia via concealed accessory pathway. Am J Cardiol. 2005;95:875–878. doi: 10.1016/j.amjcard.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 3.Di Toro D., Hadid C., Lopez C., Fuselli J., Luis V., Labadet C. Utility of the aVL lead in the electrocardiographic diagnosis of atrioventricular node re-entrant tachycardia. Europace. 2009;11:944–948. doi: 10.1093/europace/eup130. [DOI] [PubMed] [Google Scholar]
- 4.Filgueiras Medeiros J., Nardo-Botelho F.M., Felix-Bernardes L.C., et al. Diagnostic accuracy of several electrocardiographic criteria for the prediction of atrioventricular nodal reentrant tachycardia. Arch Med Res. 2016;47:394–400. doi: 10.1016/j.arcmed.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Jaeggi E.T., Gilljam T., Bauersfeld U., Chiu C., Gow R. Electrocardiographic differentiation of typical atrioventricular node reentrant tachycardia from atrioventricular reciprocating tachycardia mediated by concealed accessory pathway in children. Am J Cardiol. 2003;91:1084–1089. doi: 10.1016/s0002-9149(03)00153-x. [DOI] [PubMed] [Google Scholar]
- 6.Maury P. Distinction between atrioventricular reciprocating tachycardia and atrioventricular node re-entrant tachycardia in the adult population based on P wave location Should we reconsider the value of some ECG criteria according to gender and age? Europace. 2003;5:57–64. doi: 10.1053/eupc.2002.0280. [DOI] [PubMed] [Google Scholar]
- 7.Zhong Y.M., Guo J.H., Hou A.J., Chen S.J., Wang Y., Zhang H.C. A modified electrocardiographic algorithm for differentiating typical atrioventricular node re-entrant tachycardia from atrioventricular reciprocating tachycardia mediated by concealed accessory pathway. Int J Clin Pract. 2006;60:1371–1377. doi: 10.1111/j.1368-5031.2006.00839.x. [DOI] [PubMed] [Google Scholar]
- 8.Tai C.-T., Chen S.-A., Chiang C.-E., et al. A new electrocardiographic algorithm using retrograde P waves for differentiating atrioventricular node reentrant tachycardia from atrioventricular reciprocating tachycardia mediated by concealed accessory pathway. J Am Coll Cardiol. 1997;29:394–402. doi: 10.1016/s0735-1097(96)00490-1. [DOI] [PubMed] [Google Scholar]
- 9.Kalbfleisch S.J., El-Atassi R., Calkins H., Langberg J.J., Morady F. Differentiation of paroxysmal narrow QRS complex tachycardias using the 12-lead electrocardiogram. J Am Coll Cardiol. 1993;21:85–89. doi: 10.1016/0735-1097(93)90720-l. [DOI] [PubMed] [Google Scholar]
- 10.Huycke E.C., Lai W.-T., Nguyen N.X., Keung E.C., Sung R.J. Role of intravenous isoproterenol in the electrophysiologic induction of atrioventricular node reentrant tachycardia in patients with dual atrioventricular node pathways. Am J Cardiol. 1989;64:1131–1137. doi: 10.1016/0002-9149(89)90865-5. [DOI] [PubMed] [Google Scholar]
- 11.Blomström-Lundqvist C., Scheinman M.M., Aliot E.M., et al. ACC/AHA/ESC guidelines for the management of patients with supraventricular arrhythmias—executive summary: a report of the American college of cardiology/American heart association task force on practice guidelines and the European society of cardiology committee for practice guidelines (writing committee to develop guidelines for the management of patients with supraventricular arrhythmias) developed in collaboration with NASPE-Heart Rhythm Society. J Am Coll Cardiol. 2003;42:1493–1531. doi: 10.1016/j.jacc.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 12.Laish-Farkash A., Shurrab M., Singh S., et al. Approaches to empiric ablation of slow pathway: results from the Canadian EP web survey. J Interv Card Electrophysiol. 2012;35:183–187. doi: 10.1007/s10840-012-9696-z. [DOI] [PubMed] [Google Scholar]
- 13.Shurrab M., Szili-Torok T., Akca F., et al. Empiric slow pathway ablation in non-inducible supraventricular tachycardia. Int J Cardiol. 2015;179:417–420. doi: 10.1016/j.ijcard.2014.10.043. [DOI] [PubMed] [Google Scholar]
- 14.Ribeiro A.H., Ribeiro M.H., Paixao G.M.M., et al. Automatic diagnosis of the 12-lead ECG using a deep neural network. Nat Commun. 2020;11:1760. doi: 10.1038/s41467-020-15432-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hannun A.Y., Rajpurkar P., Haghpanahi M., et al. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat Med. 2019;25:65–69. doi: 10.1038/s41591-018-0268-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu H., Cheng C., Yin H., et al. Automatic multilabel electrocardiogram diagnosis of heart rhythm or conduction abnormalities with deep learning: a cohort study. Lancet Digit Health. 2020;2:e348–e357. doi: 10.1016/S2589-7500(20)30107-2. [DOI] [PubMed] [Google Scholar]
- 17.Somani S., Russak A.J., Richter F., et al. Deep learning and the electrocardiogram: review of the current state-of-the-art. Europace. 2021;23:1179–1191. doi: 10.1093/europace/euaa377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jo Y.-Y., Kwon J.-M., Jeon K.-H., et al. Artificial intelligence to diagnose paroxysmal supraventricular tachycardia using electrocardiography during normal sinus rhythm. Eur Heart J Digit Health. 2021;2:290–298. doi: 10.1093/ehjdh/ztab025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howard JP. Custom Digital ECG Segmentation Software Github2020. https://github.com/jphdotam/BARD-labeller
- 20.Howard J.P., Francis D.P. Machine learning with convolutional neural networks for clinical cardiologists. Heart. 2022;108:973–981. doi: 10.1136/heartjnl-2020-318686. [DOI] [PubMed] [Google Scholar]
- 21.Sau A., Ibrahim S., Ahmed A., et al. Artificial intelligence-enabled electrocardiogram to distinguish cavotricuspid isthmus dependence from other atrial tachycardia mechanisms. Eur Heart J Digit Health. 2022;3:405–414. doi: 10.1093/ehjdh/ztac042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Attia Z.I., Kapa S., Lopez-Jimenez F., et al. Screening for cardiac contractile dysfunction using an artificial intelligence-enabled electrocardiogram. Nat Med. 2019;25:70–74. doi: 10.1038/s41591-018-0240-2. [DOI] [PubMed] [Google Scholar]
- 23.Kubota Y. tf-keras-vis 2020. https://github.com/keisen/tf-keras-vis
- 24.Ribeiro M.T., Singh S., Guestrin C. KDD ’16: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. ACM; 2016. “Why should I trust you?”: explaining the predictions of any classifier. [Google Scholar]
- 25.Perlman O., Katz A., Amit G., Zigel Y. Supraventricular tachycardia classification in the 12-lead ECG using atrial waves detection and a clinically based tree scheme. IEEE J Biomed Health Inform. 2016;20:1513–1520. doi: 10.1109/JBHI.2015.2478076. [DOI] [PubMed] [Google Scholar]
- 26.Siontis K.C., Noseworthy P.A., Attia Z.I., Friedman P.A. Artificial intelligence-enhanced electrocardiography in cardiovascular disease management. Nat Rev Cardiol. 2021;18:465–478. doi: 10.1038/s41569-020-00503-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katritsis D.G., Zografos T., Katritsis G.D., et al. Catheter ablation vs. antiarrhythmic drug therapy in patients with symptomatic atrioventricular nodal re-entrant tachycardia: a randomized, controlled trial. Europace. 2017;19:602–606. doi: 10.1093/europace/euw064. [DOI] [PubMed] [Google Scholar]
- 28.Kuck K.H., Friday K.J., Kunze K.P., Schlüter M., Lazzara R., Jackman W.M. Sites of conduction block in accessory atrioventricular pathways. Basis for concealed accessory pathways. Circulation. 1990;82:407–417. doi: 10.1161/01.cir.82.2.407. [DOI] [PubMed] [Google Scholar]
- 29.Dengke Z., Lan L., Xiangli S., Shubin J. Treatment of left accessory cardiac pathway conduction disorders using radiofrequency catheter ablation under the guidance of the Ensite NavX 3D mapping system: a retrospective study. Int J Cardiovasc Imaging. 2019;35:387–392. doi: 10.1007/s10554-018-1449-3. [DOI] [PubMed] [Google Scholar]
- 30.Chen G., Wang Y., Proietti R., et al. Zero-fluoroscopy approach for ablation of supraventricular tachycardia using the Ensite NavX system: a multicenter experience. BMC Cardiovasc Disord. 2020;20:48. doi: 10.1186/s12872-020-01344-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma Y., Qiu J., Yang Y., Tang A. Catheter ablation of right-sided accessory pathways in adults using the three-dimensional mapping system: a randomized comparison to the conventional approach. PLoS One. 2015;10 doi: 10.1371/journal.pone.0128760. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated or analyzed, or both, during this study are not publicly available owing to ethical restrictions.