Abstract
We report the identification and characterization of the gene encoding the eighth and final human ribonuclease (RNase) of the highly diversified RNase A superfamily. The RNase 8 gene is linked to seven other RNase A superfamily genes on chromosome 14. It is expressed prominently in the placenta, but is not detected in any other tissues examined. Phylogenetic analysis suggests that RNase 7 is the closest relative of RNase 8 and that the pair likely resulted from a recent gene duplication event in primates. Further analysis reveals that the RNase 8 gene has incorporated non-silent mutations at an elevated rate (1.3 × 10–9 substitutions/site/year) and that orthologous RNase 8 genes from 6 of 10 primate species examined have been deactivated by frameshifting deletions or point mutations at crucial structural or catalytic residues. The ribonucleolytic activity of recombinant human RNase 8 is among the lowest of members of this superfamily and it exhibits neither antiviral nor antibacterial activities characteristic of some other RNase A ribonucleases. The rapid evolution, species-limited deactivation and tissue-specific expression of RNase 8 suggest a unique physiological function and reiterates the evolutionary plasticity of the RNase A superfamily.
INTRODUCTION
The initial analysis of the draft human genome sequence reveals several gene families that are unique to vertebrate species (1). Among them, the ribonuclease (RNase) A gene superfamily (2) is the only one that encodes proteins with enzymatic activity (1). The evolution of this vertebrate-specific enzyme family has received increasing attention because it serves as an excellent model for understanding the origin of new genes and novel gene function (3–9). To date, seven members of the RNase A superfamily have been identified in humans: RNase 1 (pancreatic RNase), RNase 2 (eosinophil-derived neurotoxin, EDN), RNase 3 (eosinophil cationic protein, ECP), RNase 4, RNase 5 (angiogenin), RNase 6 (k6) and RNase 7 (GenBank accession no. XM_033539; its function is under investigation). RNase A superfamily RNases exhibit diverse expression patterns and have varying catalytic activities against specific RNA substrates. They also exhibit a variety of physiological functions (2,10–13), including digestion of RNA released from foregut bacteria of herbivorous mammals (RNase 1), angiogenesis (RNase 5) and inhibition of viral infection (RNases 2 and 3).
Evolutionary analysis of the RNase A gene superfamily has provided significant insight into several of these diverse physiological functions. This is particularly so in the case of the primate eosinophil RNases (RNases 2 and 3) and their counterparts, the eosinophil-associated RNases (EARs) in rodents. In previous work we demonstrated that RNases 2 and 3 are among the most rapidly evolving coding sequences of primates (3), with positive Darwinian selection promoting the rapid amino acid substitutions observed in RNase 3 (6). The rodent EARs have evolved via a process of rapid gene duplication and deactivation, resulting in markedly different gene inventories in closely related species, similar to what has been observed in the immunoglobulin, major histocompatibility complex and T cell receptor gene families (7). Despite the rapid incorporation of mutations, nearly all have retained the capacity for RNase activity, findings which, taken together, have provided the impetus for our studies on the role of eosinophils and their RNases in host defense against single-stranded RNA virus pathogens (reviewed in 14). Similarly, our study on a duplication of the pancreatic RNase gene in the leaf-eating douc langur has provided us with insights into the molecular basis of digestive physiology of colobus monkeys, which have evolved a unique ruminant-type alimentary tract and use bacterial fermentation to digest leaves (15).
With the recent discovery of RNase 7, and now RNase 8, we anticipate that evolutionary analysis will continue to elucidate novel activities and unique molecular adaptations in this highly diversified gene superfamily.
MATERIALS AND METHODS
Identification and evolutionary analysis of RNase 8
The draft human genome sequence (1) was searched for new RNase genes using the seven known RNase genes as queries. Our initial search had low power for the detection of homologous genes probably because RNase genes are highly divergent in sequence and are relatively small. Since the human genes for RNases 1–7 are all linked on chromosome 14, we focused our search and examined all open reading frames (ORFs) of ∼450 nt in this region for sequence features of RNases. The identified sequence (RNase 8) was aligned with known RNases using CLUSTAL V (16). The phylogenetic tree of the encoded amino acid sequences was reconstructed by the neighbor joining method (17) with protein p distances, which are good for highly divergent sequences (18). One thousand bootstrap replications (19) were conducted to assess the reliability of the reconstructed tree. The synonymous (silent) and non-synonymous (non-silent) nucleotide substitution rates of gene sequences were estimated by the modified Nei–Gojobori method described by Zhang et al. (6). The MEGA2 program (20) was used for the evolutionary analysis.
The coding region of RNase 8 was amplified from genomic DNAs from human (Homo sapiens), chimpanzee (Pan troglodytes), gorilla (Gorilla gorilla), rhesus monkey (Macaca mulatta), pig-tailed macaque (Macaca nemestrina), baboon (Papio hamadryas), African green monkey (Cercopithecus aethiops), talapoin monkey (Miopithecus talapoin), tamarin (Saguinus oedipus) and owl monkey (Aotus trivirgatus) by PCR with primers 337 (5′-CTCCTAAGAGAGATGGCACCGGCC) and 338 (5′-CAAAGAGCAAGCCAGTCTGGAAACCTA). The PCR products were cloned into the pCR II TA cloning vector (Invitrogen, San Diego, CA) and sequenced in both directions by the dideoxy chain termination method with a Perkin-Elmer 377 automatic sequencer.
Northern blotting
A human multiple tissue northern membrane was obtained from Clontech (Palo Alto, CA). The membrane was prehybridized and hybridized following the manufacturer’s instructions. The hybridization was performed with a radiolabeled human RNase 8-specific oligonucleotide probe of sequence 5′-cacaggaacaagtgggtaccctgggtcaccctgttgtggagggtcacagg-3′, which corresponds to nt 395–444 of the coding region. The membrane was then washed following the manufacturer’s instructions; autoradiograms were developed after 5 day exposures at –80°C. As a control, hybridization of the human actin gene was performed with a probe provided by Clontech.
Recombinant RNase 8 and its enzymatic activity
The signal peptide region of RNase 8 was determined according to the known cleavage sites in other RNases. The mature peptide region of the RNase 8 gene was subcloned into the bacterial expression vector pFLAG CTS (Kodak, New Haven, CT) and was verified by sequencing. The vector adds the octapeptide DYKDDDDK (FLAG) to the C-terminus of the recombinant protein, which facilitates its purification and detection with M2 anti-FLAG monoclonal antibody (Sigma) without altering ribonucleolytic activity (4). Recombinant proteins were isolated, purified and quantified as described previously (4). The RNase activity of the recombinant proteins against a standard yeast tRNA substrate was measured in 40 mM sodium phosphate buffer pH 7.4, at 25°C. Purified RNase (∼1.0 pmol) was added to 0.8 ml of the aforementioned buffer with 1.42 nmol tRNA. The reaction was stopped by addition of 0.5 ml of 20 mM lanthanum nitrate with 3% perchloric acid, and insoluble tRNA was removed by centrifugation. The amount of solubilized tRNA was determined by UV absorbance at 260 nm. The catalytic activity of the RNase was determined as nmol RNA digested/s/nmol RNase (4).
Bacterial growth assay
Following Rosenberg (21), we examined whether induction of the production of recombinant proteins in Escherichia coli inhibits growth of the host bacteria. Overnight cultures of bacteria transformed with a plasmid harboring the RNase 8 gene or the RNase 3 gene (positive control) or the pFLAG vector alone (negative control) were diluted 1:40 in LB broth with 100 µg/ml ampicillin. Optical densities (600 nm) were recorded at t = 0 and hourly thereafter. When exponential phase growth was achieved, 0.1 mM IPTG was added to half of the culture to induce the production of recombinant protein. Optical densities were recorded hourly thereafter. Each experiment was repeated three times.
Antiviral assays
The activity of recombinant RNase 8 and other RNases in reducing the infectivity of respiratory syncytial virus (RSV) on Hep-2 human epithelial cells was examined by the quantitative shell vial amplification technique following the published procedure (22). Briefly, recombinant RNases were added to Hep-2 monolayers growing on coverslips (50 000 cells/coverslip) followed by ∼2000 plaque-forming units (= infectious units) of RSV-B (American Type Culture Collection, Manassas, VA). The vials containing virus and target cells were centrifuged for 60 min at 500 g at room temperature (a technique known as spin amplification, which enhances infectivity in vitro; 22) and then incubated at 37°C overnight, after which the coverslips were washed, acetone fixed and stained with FITC–anti-RSV with methylene blue counterstain (Chemicon, Temecula, CA). Infected cells were identified via fluorescence microscopy.
RESULTS
Human RNase 8 and its relationships with other RNase A superfamily RNases
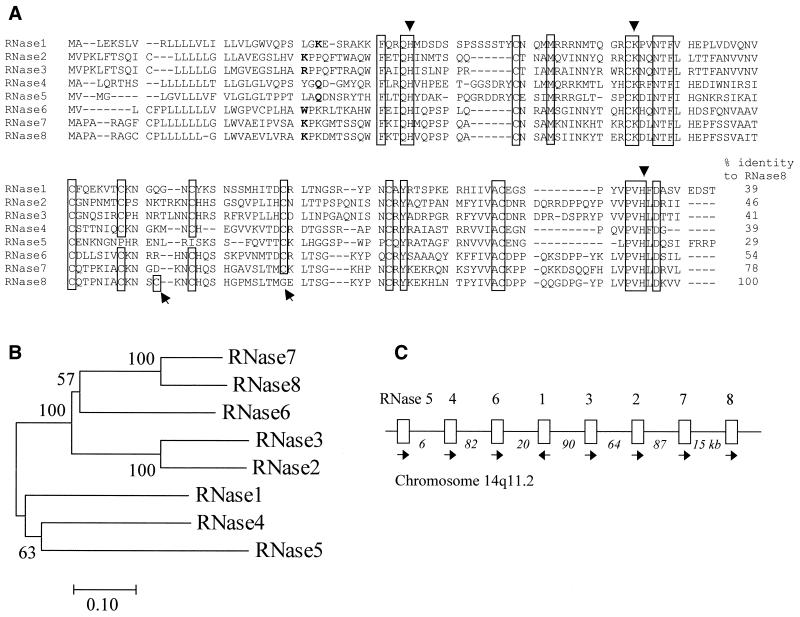
We identified an ORF of 462 nt encoding RNase 8 in the draft human genome sequence in the region of human chromosome 14 that encodes all other known RNase A superfamily members (the contig sequence that contains the RNase 8 gene has GenBank accession no. AL161668). Figure 1A depicts the encoded protein sequence alignment of all eight human RNases. RNase 8 is most similar to RNase 7 (78% sequence identity) and least similar to RNase 5 (29%). It has 154 amino acids, including a signal peptide of 27 amino acids. The isoelectric point of the mature peptide is 8.6. RNase 8 has eight cysteines, one catalytic lysine and two catalytic histidines that are all among the characteristic features of all mammalian RNase A ribonucleases (except for RNase 5, with only six cysteines). The RNase 8 gene also encodes the RNase A superfamily invariant ‘signature’ motif, CKXXNTF, which includes the catalytic lysine residue. One unusual feature of RNase 8 is that a cysteine residue (at position 81 of the human RNase 8 mature protein) that is conserved among all other RNases has changed to glycine, while a new cysteine residue has appeared at position 66 (Fig. 1A).
Figure 1.
Sequence, evolutionary origin and chromosomal location of RNase 8 in relation to other RNase A ribonuclease genes. (A) Amino acid sequence alignment of human members of the RNase A superfamily. Gaps are indicated by –. The conserved amino acids are boxed, with the three catalytic residues indicated by triangles. A positional change of a cysteine in RNase 8 is indicated by arrows beneath the alignment. The starting residue of the mature RNase 8 protein is inferred by its homology to other RNases. The alignment was generated by CLUSTAL V without correction and it may be improved with additional biological and structural information. (B) Phylogenetic tree of the eight RNase A proteins. The neighbor joining method with protein p distance was used. Bootstrap percentages are shown on interior branches. (C) The chromosomal locations of the RNase A genes in humans. Arrows show the transcriptional direction. The distances between genes are indicated (kb). RNase 8 is closest to the telomere and RNase 5 is closest to the centromere. The entire RNase A gene cluster is ∼368 kb.
To clarify the evolutionary relationships among these RNases, we constructed a phylogenetic tree that includes all eight human RNase A ribonucleases (Fig. 1B). The phylogenetic analysis strongly suggests a close relationship between RNases 7 and 8 (bootstrap = 100%), similar to that observed between the two eosinophil RNases (RNases 2 and 3). The tree also indicates that RNases 2, 3, 6, 7 and 8 are more closely related to one another, forming a separate subgroup from RNases 1, 4 and 5. The linear arrangement of the RNase A superfamily genes along human chromosome 14 is shown in Figure 1C.
RNase 8 genes of primates
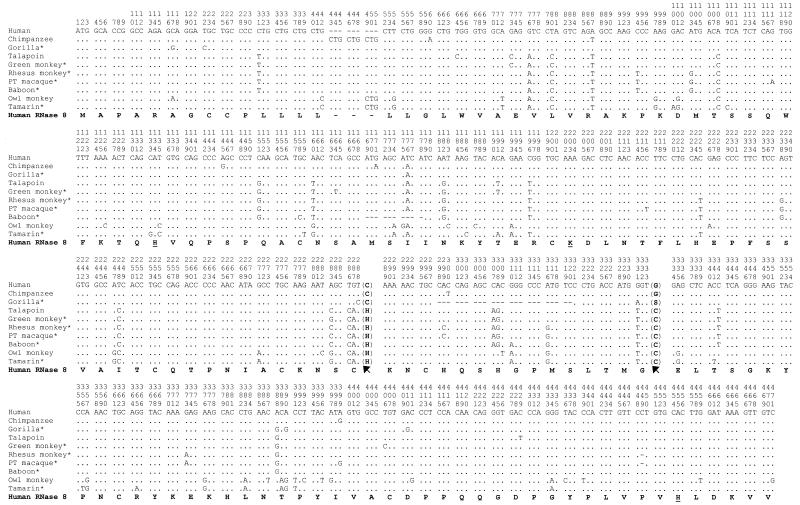
The pronounced sequence similarity between RNases 7 and 8 suggests that they have originated from a relatively recent gene duplication, similar to that reported previously for RNases 2 and 3 (3). To examine its species distribution, we amplified and sequenced RNase 8 genes from 10 higher primate species, including three hominoids, five Old World monkeys and two New World monkeys (Fig. 2). Our phylogenetic analysis of the 10 gene sequences strongly suggests that they are orthologous (data not shown). The human sequence we obtained was identical to that in the draft human genome sequence. However, to our surprise, the RNase 8 genes of four primates (gorilla, baboon, rhesus monkey and pig-tailed macaque) have deletions that either interrupt the ORF or drastically alter the encoded amino acid sequence distal to the deletion. For instance, the gorilla RNase 8 gene has a deletion of 23 nt, which results in a premature stop codon. The RNase 8 genes of the rhesus monkey and pig-tailed macaque each have a single nucleotide deletion close to the end of the ORF, which alters the stop codon and the final eight amino acids of the protein, including a catalytic histidine. Furthermore, the first catalytic histidine has been changed to aspartic acid in the tamarin RNase 8 and a structural cysteine changed to arginine in the RNase 8 from African green monkey (Fig. 2). These deletions and replacements necessarily abolish the RNase activity of the proteins encoded. In other words, 6 of the 10 primates examined have deactivated RNase 8 genes.
Figure 2.
Nucleotide sequences of primate RNase 8 genes. The identical nucleotide sequence to human RNase 8 is as indicated by dots, whereas gaps are indicated by –. The translated amino acid sequence of human RNase 8 is shown in bold, with the three catalytic residues underlined. Potential pseudogenes bear an astersik after the organism name. The evolutionary change of a cysteine position is indicated by arrows, with the corresponding amino acids shown next to the nucleotides. Human, chimpanzee and gorilla belong to the hominoids; talapoin, green monkey, rhesus monkey, pig-tailed macaque and baboon belong to the Old World monkeys; owl monkey and tamarin belong to the New World monkeys.
As noted earlier, the position typically assumed by a cysteine residue is taken by a glycine in human RNase 8, with a replacement cysteine located more proximally. An evaluation of nine additional primate RNase 8 sequences indicates that this pattern is observed among hominoids (human, chimpanzee and gorilla) only and is shared by all three hominoids examined. We were not, however, able to determine whether gain of the new cysteine residue predated loss of the old cysteine in evolution, as none of the sequences examined showed the occurrence of only one of the two events.
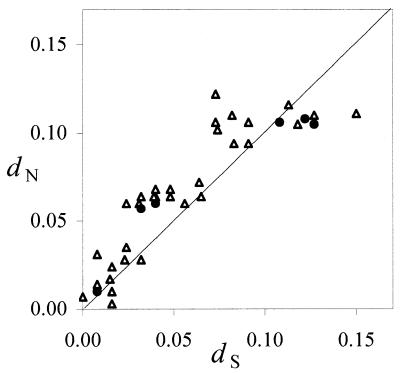
Comparison of rates of synonymous, or silent (dS), and non-synonymous, or non-silent (dN), nucleotide substitutions between orthologous RNase 8 sequences is shown in Figure 3. Interestingly, of the six pairwise comparisons among the four complete coding sequences (human, chimpanzee, talapoin and owl monkey), three showed dN > dS and three showed dN < dS. However, in none of the comparisons is the difference between dN and dS statistically significant. Even when all six comparisons are considered together, the average dN (0.081 ± 0.019) and dS (0.075 ± 0.018) values are not significantly different. When all 10 species are considered, the average dN (0.066 ± 0.010) and dS (0.056 ± 0.012) values are likewise not significantly different from one another. In these latter analyses, the codons involving deletions were removed. Functional genes are usually under purifying selection that inhibits non-synonymous substitution (dN < dS). In rare cases, one can identify a gene under positive selection, a condition under which non-synonymous substitution is promoted (dN > dS), such as that observed in primate RNase 3 (6). In the present case, dN and dS are virtually identical, thus it is possible that the RNase 8 gene is evolving under relaxed purifying selection. It is also possible that a mixture of purifying selection and positive selection resulted in a dN value that is close to dS.
Figure 3.
Pairwise synonymous (dS) and non-synonymous (dN) nucleotide distances of the 10 primate RNase 8 gene sequences. Closed circles indicate comparisons among the four functional genes (human, chimpanzee, talapoin and owl monkey), whereas open triangles indicate all other comparisons. The diagonal line indicates dN = dS.
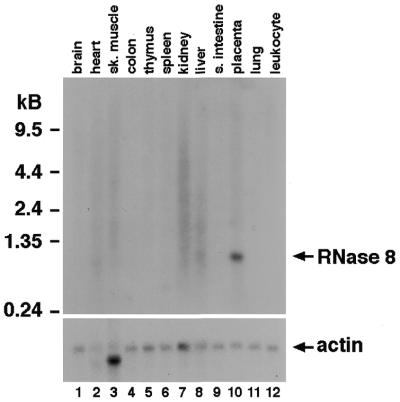
Unique expression pattern
The expression pattern of human RNase 8 was examined via northern analysis of mRNAs from 12 tissues (Fig. 4). RNase 8 mRNA (∼1 kb) was detected in placenta, but not in any of the other 11 human tissues examined. We further confirmed the expression of RNase 8 by amplification and sequencing of cDNAs prepared from human placental mRNAs (Clontech, Palo Alto, CA) using primers designed for specific amplification of RNase 8. The sequenced cDNA, however, has one nucleotide difference from the genomic sequence obtained. This may either be a polymorphism or an error generated in cDNA synthesis.
Figure 4.
Northern analysis of RNase 8. Total RNAs from the normal human tissues indicated were probed with a 32P-radiolabeled oligonucleotide of the human RNase 8 gene. The same blot was also probed with a human actin gene fragment to control for relative loading. Sk. muscle, skeletal muscle; s. intestine, small intestine.
Enzymatic activity and function of RNase 8
We prepared recombinant human RNase 8 from bacteria and examined its activity against yeast tRNA in a standard assay. We found that RNase 8 was ribonucleolytically active, but the activity is lower than that observed for RNases 1–3, 6 and 7 (Table 1). The RNase activity of RNase 5 is known to be extremely low (23), but the available data were obtained in a somewhat different experimental setting (24), thus prohibiting a direct comparison. Similarly, the ribonucleolytic activity of RNase 4 was reported to be similar to that of RNase 1 (25).
Table 1. RNase activity against yeast tRNA.
| Enzyme | Activitya | References |
|---|---|---|
| 0.65 | 4 | |
| RNase 3 | 0.048 | 12 |
| RNase 4 | ∼RNase 1 | 25 |
| RNase 5 | Extremely low | 23, 24 |
| RNase 6 | 0.034 | 32 |
| RNase 7 | 1.14 | J. Zhang and H.F. Rosenberg, unpublished |
| RNase 8 | 0.012 | This paper |
aActivity was measured as per nmol substrate digested/nmol enzyme/s. The activities reported in the literature were transformed to reflect the expected values under the present experimental conditions. However, these estimates are approximate and should be treated with caution.
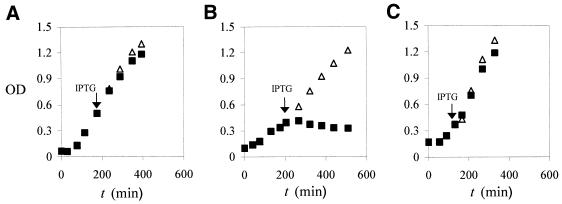
The closest relatives of RNase 8 whose functions have been studied are the eosinophil RNases, RNases 2 and 3. Interestingly, RNases 2 and 3 also arose as a gene pair relatively recently in primate evolution (3,6) and thus may have some functional features in common with RNases 7 and 8. RNase 2 has antiviral activity against RSV and HIV studied in vitro (13,26) and RNase 3 has a similar, though lower, activity against RSV (12). We thus examined whether RNase 8 has activity against RSV. As shown in Table 2, RNase 2 reduced the infectivity of RSV significantly when compared to the control, whereas RNase 8 had no effect on RSV infection even at a higher concentration. RNase 3 is known to be toxic to bacteria (27,28) and we examined whether RNase 8 has the same activity. As shown in Figure 5, induction of expression of the RNase 3 gene encoded by the pFLAG plasmid of E.coli inhibits growth of the host bacteria, whereas no inhibitory effect is seen when the plasmid-encoded RNase 8 gene is expressed.
Table 2. Antiviral activity of RNases.
| RNase (concentration) | Infectious units/ml (± SEM) |
|---|---|
| BSA control (20 ng/µl) | 2409 ± 83 |
| RNase 2 (25 ng/µl) | 1122 ± 73a |
| RNase 8 (66 ng/µl) | 2463 ± 57 |
The antiviral activities of RNase 2 and RNase 8 were studied using the quantitative shell vial assay described in Domachowske and Bonville (22), which has been used to study the extent to which RNase A RNases reduce the infectivity of RSV for target HEp-2 human epithelial cells in vitro. RNase 2 mediates the dose-dependent, RNase activity-dependent reduction in the number of RSV-infected target cells, reported as a reduction in apparent infectious units/ml (13).
aP < 0.001 (compared with the BSA control).
Figure 5.
Growth of bacterial transformatant cultures. (A) Vector only. (B) RNase 3, which is known to have antibacterial activity in this assay (21). (C) RNase 8. Optical densities (600 nm) were recorded at t = 0 and hourly thereafter. When exponential phase growth was achieved, 0.1 mM IPTG was added to half of the culture to induce the production of recombinant protein. Optical densities were recorded hourly thereafter. The experiments were repeated three times and the mean values are reported. Solid squares, no IPTG; open triangles, after IPTG induction.
DISCUSSION
In this work, we report the isolation and characterization of the gene encoding human RNase 8, the eighth and likely final human member of the RNase A gene superfamily. RNase 8 was the only novel sequence of the superfamily identified after an extensive search of candidate ORFs in the draft human genome sequence of chromosome 14 and as such it is unlikely that any additional RNase A superfamily genes will be found in this region. Given that all eight reported RNase A genes map to this region, the chance that new superfamily members will be found on other human chromosomes seems small. Thus, we tentatively conclude that all human RNase A superfamily members have now been identified.
Much of our work on the RNase A superfamily has focused on determining the biological implications of specific evolutionary patterns and paradigms (14). Here, our phylogenetic analysis suggests that the RNase 7–RNase 8 gene pair emerged as a result of a relatively recent gene duplication event, one quite similar to that reported for the eosinophil RNases, RNases 2 and 3 (3,6). Assuming that hominoids and New World monkeys diverged 40 million years ago (29) and that the evolutionary rates of RNase 7 and RNase 8 were identical and constant after the duplication, we estimate that the duplication event occurred ∼44 million years ago, shortly before the separation of New World monkeys from Old World monkeys. Also similar to what has been observed for the eosinophil RNases, RNase 8 has been incorporating non-silent mutations at a very rapid rate (1.3 × 10–9 substitutions/site/year), among the highest rates in any coding sequences studied in primates (3,30). However, in the case of RNases 2 and 3, intact coding sequences were identified in all primate species, with all elements necessary for at least minimal RNase activity maintained throughout; the findings relating to sequence diversity and RNase activity prompted us to consider a role for these proteins in host defense against single-stranded RNA virus pathogens (14). In contrast, the gene encoding RNase 8 is deactivated by frameshift or functionally deleterious mutations in 6 of 10 primate species examined (discussed further below). Thus, although the human ortholog of RNase 8 is intact, the fact that many of the primate orthologs are not suggests that RNase 8 is likely to have a more specific, species-limited function that may not yet be completely realized in evolutionary terms.
The tissue-specific expression of human RNase 8 may provide some clues with respect to function. We show here that RNase 8 is expressed uniquely in the placenta, while remaining undetectable in 11 other human tissues examined. In contrast, most RNase A genes are expressed in a wide range of tissues, save for RNase 3, which is only found in eosinophils. A few RNases are also found in the placenta, although none exclusively (31). For instance, RNase 6 is expressed in the lung, heart and kidney in addition to placenta (32). When one considers the overall physiology of placental function, it is interesting to consider the possibility that RNase 8 may be developing into a tissue-specific angiogenic factor, reminiscent of the more generalized role in angiogenesis described for RNase 5 (11,23), although there is little precise sequence similarity between the two. As a final note, in ruminants, a duplicate of the RNase 1 gene, known as the seminal RNase, is exclusively expressed in seminal vesicles (33). It has a variety of activities, including digestion of double-stranded RNA and cytotoxicity (reviewed in 34). Interestingly, the seminal RNase gene has also been deactivated in most ruminant species, but it has remained functional in cattle (35,36). Equally interesting is the fact that, analogous to seminal RNase, which has additional cysteines that form physiological interchain disulfide bonds (34), RNase 8 likewise has an unusual cysteine 66 that may not participate in prototypical disulfide bond formation. While the true structure of RNase 8 awaits crystallographic analysis, it is possible that cysteine 66 can form interchain disulfide bonds, perhaps by pairing with the otherwise uncoupled cysteine 23.
The most unexpected feature of RNase 8 is its frequent deactivation in multiple species, with disrupted or dysfunctional coding sequences identified in gorilla, African green monkey, rhesus monkey, pig-tailed macaque, baboon and tamarin. If the progenitor of the RNase 8 gene was functional, at least three deactivation events must have occurred, one in hominoids, one in Old World monkeys and one in New World monkeys. However, given the nature of the mutations that occurred in the four Old World monkey species examined (Fig. 2), it is more likely that three independent deactivations occurred in Old World monkeys alone, bringing the total to five. It is also possible, though very unlikely, that the gene was deactivated in the common ancestor of Old World monkeys, but was reactivated later in the talapoin monkey. All the gene deactivation events appear relatively recent, as most of the coding sequences remain completely, or at least nearly, intact. The cause of this massive deactivation of RNase 8 genes is unclear.
In summary, the exclusive expression in placenta, rapid evolution and the unusual pattern of gene deactivation in non-human primates suggests a unique physiological function of RNase 8 that awaits further exploration.
Acknowledgments
ACKNOWLEDGEMENTS
This work was partially supported by a start-up fund and a Rackham grant of the University of Michigan to J.Z.
DDBJ/EMBL/GenBank accession nos AF473854–AF473863
REFERENCES
- 1.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 2.D’Alessio G. and Riordan,J.F. (1997) Ribonucleases, Structures and Functions. Academic Press, San Diego, CA.
- 3.Rosenberg H.F., Dyer,K.D., Tiffany,H.L. and Gonzalez,M. (1995) Rapid evolution of a unique family of primate ribonuclease genes. Nature Genet., 10, 219–223. [DOI] [PubMed] [Google Scholar]
- 4.Rosenberg H.F. and Dyer,K.D. (1995) Eosinophil cationic protein and eosinophil-derived neurotoxin. Evolution of novel function in a primate ribonuclease gene family. J. Biol. Chem., 270, 21539–21544. [DOI] [PubMed] [Google Scholar]
- 5.Beintema J.J. and Kleineidam,R.G. (1998) The ribonuclease A superfamily: general discussion. Cell. Mol. Life Sci., 54, 825–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J., Rosenberg,H.F. and Nei,M. (1998) Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc. Natl Acad. Sci. USA, 95, 3708–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J., Dyer,K.D. and Rosenberg,H.F. (2000) Evolution of the rodent eosinophil-associated RNase gene family by rapid gene sorting and positive selection. Proc. Natl Acad. Sci. USA, 97, 4701–4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J. and Rosenberg,H.F. (2002) Diversifying selection of the tumor-growth promoter angiogenin of higher primates. Mol. Biol. Evol., in press. [DOI] [PubMed] [Google Scholar]
- 9.Jermann T.M., Opitz,J.G., Stackhouse,J. and Benner,S.A. (1995) Reconstructing the evolutionary history of the artiodactyl ribonuclease superfamily. Nature, 374, 57–59. [DOI] [PubMed] [Google Scholar]
- 10.Barnard E.A. (1969) Biological function of pancreatic ribonuclease. Nature, 221, 340–344. [DOI] [PubMed] [Google Scholar]
- 11.Fett J.W., Strydom,D.J., Lobb,R.R., Alderman,E.M., Bethune,J.L., Riordan,J.F. and Vallee,B.L. (1985) Isolation and characterization of angiogenin, an angiogenic protein from human carcinoma cells. Biochemistry, 24, 5480–5486. [DOI] [PubMed] [Google Scholar]
- 12.Domachowske J.B., Dyer,K.D., Adams,A.G., Leto,T.L. and Rosenberg,H.F. (1998) Eosinophil cationic protein/RNase 3 is another RNase A-family ribonuclease with direct antiviral activity. Nucleic Acids Res., 26, 3358–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Domachowske J.B., Dyer,K.D., Bonville,C.A. and Rosenberg,H.F. (1998) Recombinant human eosinophil-derived neurotoxin/RNase 2 functions as an effective antiviral agent against respiratory syncytial virus. J. Infect. Dis., 177, 1458–1464. [DOI] [PubMed] [Google Scholar]
- 14.Rosenberg H.F. and Domachowske,J.B. (2001) Eosinophils, eosinophil ribonucleases and their role in antiviral host defense. J. Leukoc. Biol., 70, 691–698. [PubMed] [Google Scholar]
- 15.Zhang J., Zhang,Y.P. and Rosenberg,H.F. (2002) Adoptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nature Genet., in press. [DOI] [PubMed] [Google Scholar]
- 16.Higgins D.G., Bleasby,A.J. and Fuchs,R. (1992) CLUSTAL V: improved software for multiple sequence alignment. Comput. Appl. Biosci., 8, 189–191. [DOI] [PubMed] [Google Scholar]
- 17.Saitou N. and Nei,M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol., 4, 406–425. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J. and Nei,M. (1996) Evolution of Antennapedia-class homeobox genes. Genetics, 142, 295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felsenstein J. (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution, 39, 783–791. [DOI] [PubMed] [Google Scholar]
- 20.Kumar S., Tamura,K. Jakobsen,I. and Nei,M. (2000) MEGA2,Molecular Evolutionary Genetics Analysis. Arizona State University, Tempe, AZ.
- 21.Rosenberg H.F. (1995) Recombinant human eosinophil cationic protein. Ribonuclease activity is not essential for cytotoxicity. J. Biol. Chem., 270, 7876–7881. [DOI] [PubMed] [Google Scholar]
- 22.Domachowske J.B. and Bonville,C.A. (1998) Overnight titration of human respiratory syncytial virus using quantitative shell vial amplification. Biotechniques, 25, 644–647. [DOI] [PubMed] [Google Scholar]
- 23.Strydom D.J. (1998) The angiogenins. Cell. Mol. Life Sci., 54, 811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harper J.W. and Vallee,B.L. (1988) Mutagenesis of aspartic acid-116 enhances the ribonucleolytic activity and angiogenic potency of angiogenin. Proc. Natl Acad. Sci. USA, 85, 7139–7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sorrentino S. and Libonati,M. (1997) Structure-function relationships in human ribonucleases: main distinctive features of the major RNase types. FEBS Lett., 404, 1–5. [DOI] [PubMed] [Google Scholar]
- 26.Lee-Huang S., Huang,P.L., Sun,Y., Huang,P.L., Kung,H.F., Blithe,D.L. and Chen,H.I. (1999) Lysozyme and RNases as anti-HIV components in beta-core preparations of human chorionic gonadotropin. Proc. Natl Acad. Sci. USA, 96, 2678–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young J.D., Peterson,C.G., Venge,P. and Cohn,Z.A. (1986) Mechanism of membrane damage mediated by human eosinophil cationic protein. Nature, 321, 613–616. [DOI] [PubMed] [Google Scholar]
- 28.Lehrer R.I., Szklarek,D., Barton,A., Ganz,T., Hamann,K.J. and Gleich,G.J. (1989) Antibacterial properties of eosinophil major basic protein and eosinophil cationic protein. J. Immunol., 142, 4428–4434. [PubMed] [Google Scholar]
- 29.Kumar S. and Hedges,S.B. (1998) A molecular timescale for vertebrate evolution. Nature, 392, 917–920. [DOI] [PubMed] [Google Scholar]
- 30.Li W.-H. (1997) Molecular Evolution. Sinauer Associates, Sunderland, MA.
- 31.Futami J., Tsushima,Y., Murato,Y., Tada,H., Sasaki,J., Seno,M. and Yamada,H. (1997) Tissue-specific expression of pancreatic-type RNases and RNase inhibitor in humans. DNA Cell Biol., 16, 413–419. [DOI] [PubMed] [Google Scholar]
- 32.Rosenberg H.F. and Dyer,K.D. (1996) Molecular cloning and characterization of a novel human ribonuclease (RNase k6): increasing diversity in the enlarging ribonuclease gene family. Nucleic Acids Res., 24, 3507–3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sasso M.P., Lombardi,M., Confalone,E., Carsana,A., Palmieri,M. and Furia,A. (1999) The differential pattern of tissue-specific expression of ruminant pancreatic type ribonucleases may help to understand the evolutionary history of their genes. Gene, 227, 205–212. [DOI] [PubMed] [Google Scholar]
- 34.D’Alessio G., Di Donato,A., Mazzarella,L. and Piccoli,R. (1997) Seminal ribonuclease: the importance of diversity. In D’Alessio,G. and Riordan,J.F. (eds), Ribonucleases, Structures and Functions. Academic Press, San Diego, CA, pp. 383–423.
- 35.Trabesinger-Ruef N., Jermann,T., Zankel,T., Durrant,B., Frank,G. and Benner,S.A. (1996) Pseudogenes in ribonuclease evolution: a source of new biomacromolecular function? FEBS Lett., 382, 319–322. [DOI] [PubMed] [Google Scholar]
- 36.Kleineidam R.G., Jekel,P.A., Beintema,J.J. and Situmorang,P. (1999) Seminal-type ribonuclease genes in ruminants, sequence conservation without protein expression? Gene, 231, 147–153. [DOI] [PubMed] [Google Scholar]