Abstract
Background
Extensive studies have revealed the function and mechanism of lncRNAs in development and differentiation, but the majority have focused on those lncRNAs adjacent to protein-coding genes. In contrast, lncRNAs located in gene deserts are rarely explored. Here, we utilize multiple differentiation systems to dissect the role of a desert lncRNA, HIDEN (human IMP1-associated "desert" definitive endoderm lncRNA), in definitive endoderm differentiation from human pluripotent stem cells.
Results
We show that desert lncRNAs are highly expressed with cell-stage-specific patterns and conserved subcellular localization during stem cell differentiation. We then focus on the desert lncRNA HIDEN which is upregulated and plays a vital role during human endoderm differentiation. We find depletion of HIDEN by either shRNA or promoter deletion significantly impairs human endoderm differentiation. HIDEN functionally interacts with RNA-binding protein IMP1 (IGF2BP1), which is also required for endoderm differentiation. Loss of HIDEN or IMP1 results in reduced WNT activity, and WNT agonist rescues endoderm differentiation deficiency caused by the depletion of HIDEN or IMP1. Moreover, HIDEN depletion reduces the interaction between IMP1 protein and FZD5 mRNA and causes the destabilization of FZD5 mRNA, which is a WNT receptor and necessary for definitive endoderm differentiation.
Conclusions
These data suggest that desert lncRNA HIDEN facilitates the interaction between IMP1 and FZD5 mRNA, stabilizing FZD5 mRNA which activates WNT signaling and promotes human definitive endoderm differentiation.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13059-023-02925-w.
Keyword: Desert lncRNA, Human pluripotent stem cell, Endoderm differentiation, IMP1, FZD5
Background
Long noncoding RNAs (lncRNAs) are transcripts longer than 200 nucleotides without evident protein coding capacity. LncRNAs can cross-talk with DNA, RNA, and proteins, and exert biological function through chromatin remodeling, transcriptional or post-transcriptional gene regulation. The inspection of the genomic positions of lncRNA loci actively transcribed in human embryonic stem cells (ESCs) has revealed that 89% are associated with the promoters, enhancers, or bodies of protein-coding genes (PCGs) [1]. These lncRNAs could affect the expression of adjacent PCGs in a manner of base pair complementarity or acting as scaffolds to mediate local protein-DNA interaction [2]. However, the functional annotation of distal lncRNAs, especially those located in gene deserts (named as “desert lncRNAs”), is far less studied, mainly due to the difficulty of dissecting the biological function and revealing the downstream target.
Growing evidence has shown that lncRNAs are vital regulators in development and differentiation. LncRNAs participate in regulating lineage commitment and differentiation potential of pluripotent stem cells (PSCs), including neural differentiation, myogenesis, cardiogenesis, and endodermal lineage differentiation [3–5]. Definitive endoderm (DE), which arises at gastrulation when three germ layers emerge, is the innermost layer and later develops into the respiratory tract, the digestive tract, and their derivatives. During embryonic patterning and germ layer formation, high Activin or Nodal and modest WNT signaling together activate the DE program via the endoderm transcription factors SOX17, FOXA2, EOMES and GATA4/6 [6–10]. Besides signaling pathways and transcription factors, epigenetic regulators and noncoding RNAs are also important contributors to precise and exquisite endoderm differentiation.
Most studies of endoderm lncRNAs focused on those physically located nearby lineage transcription factors, which illustrated the importance of the coordinated expression of lncRNA/mRNA gene pairs during development [1]. For instance, antisense lncRNA Evx1as, which can regulate EVX1 transcription through Mediator binding and chromatin looping, orchestrated mesendoderm differentiation of mouse ESCs [11]. DEANR1 and GATA6-AS1 facilitated SMAD2/3 binding on the promoter of its nearby PCG, FOXA2 and GATA6 respectively, to promote endoderm differentiation from human PSCs [12, 13]. Another lncRNA DIGIT, divergently transcribed from mesendoderm transcription factor GSC, regulated GSC expression to increase endoderm commitment [14]. A follow-up study further showed that DIGIT interacted with BRD3 and promoted phase-separated condensates of BRD3, which mediated BRD3 binding at H3K18ac-enriched promoter regions of endoderm transcription factors [15]. Of note, these reported endoderm-functional lncRNAs are all adjacent to PCGs and regulate the transcription of neighboring endoderm genes, thus contributing to endoderm differentiation. Nevertheless, whether and how desert lncRNAs located far away from PCGs regulate early germ layer differentiation is largely unknown. Besides, the biological roles of the evolution-extended desert lncRNAs in gene-poor regions need further detection but are often neglected due to the lack of large-scale screening methods and difficulty in mechanism study.
Our RNA-seq profiling of human ESCs and the derived purified DE cells revealed many DE-specific lncRNAs with unknown functions [12], including dozens of desert lncRNAs (i.e., without PCGs nearby in the genomic range of 50 kb). Here, we studied the function of a cytoplasm-localized desert lncRNA HIDEN (human IMP1-associated "desert" definitive endoderm lncRNA), which was highly expressed during human endoderm differentiation. Loss-of-function assay of HIDEN through shRNA or promoter deletion demonstrated that HIDEN was necessary for DE differentiation. We then identified the essential role of HIDEN-interacting protein IMP1 for DE differentiation. Further studies showed HIDEN physically interacted with IMP1 protein and together facilitated the mRNA stability of a WNT receptor Frizzled 5 (FZD5). Disruption of HIDEN resulted in lower FZD5 mRNA expression and impaired WNT activation, which caused abnormal DE differentiation.
Results
Desert lncRNAs are highly expressed during human endoderm differentiation
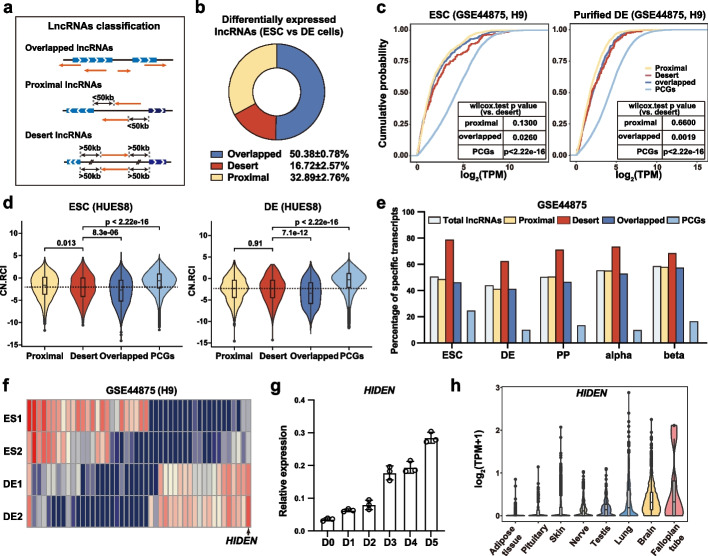
To achieve a systematic understanding of the characteristics and functional relevance of lncRNAs in early embryo development, we first divided lncRNAs into three categories based on the genomic distance between lncRNAs and nearby PCGs from GENCODE V29 annotation [16]: the overlapped lncRNAs sharing at least one nucleotide with PCGs in the genome, the proximal lncRNAs locating close to PCGs within 50 kb but with no overlap with PCGs, and the desert lncRNAs being far away from PCGs more than 50 kb in the genome (Fig. 1a). We found that about half of lncRNAs were overlapped lncRNAs, while the percentage of proximal and desert lncRNAs was 27.92% and 23.72% separately (Additional file 1: Fig. S1a). All these lncRNAs showed minimal coding potential compared with PCGs (Additional file 1: Fig. S1b). To further explore the features of these lncRNAs, we systematically investigated the lncRNA expression level, subcellular localization and cell expression specificity by re-analyzing multiple RNA-seq data. We compared differentially expressed lncRNAs between ESCs and differentiated DE cells in three datasets (Additional file 2: Table S1), and about 16.72% were desert lncRNAs (Fig. 1b).
Fig. 1.
Desert lncRNAs are highly expressed during human endoderm differentiation. a The classification of overlapped lncRNAs, proximal lncRNAs and desert lncRNAs. b Pie chart showing the average proportion of differentially expressed overlapped, proximal, and desert lncRNAs identified between human PSCs and DE cells (data from three PSC lines). c The expression level of lncRNAs and PCGs in H9 ESCs and DE cells. The curves were colored by the category of lncRNAs. The p value between desert lncRNAs and other subsets were listed in the chart. d The subcellular localization of lncRNAs and PCGs, calculated by “relative concentration index” (RCI) in HUES8 ESCs and DE cells. CN.RCI = log2(CE/NE) + log2(CE/NM) (CE: cytoplasmic elution component, NE: nuclear elution component, NM: nuclear insoluble component). e The cell expression specificity of lncRNAs and PCGs in ESCs, DE, pancreatic endocrine (PP), pancreatic alpha and beta cells, calculated by specificity score. f Heatmap of differentially expressed desert lncRNAs between ESCs and DE cells. Red indicates higher expression while blue indicates lower expression. The lncRNAs list was shown in Additional file 2: Table S1. g Time course expression of HIDEN during endoderm differentiation from human HUES8 ESCs was detected by RT-qPCR (n = 3). h The expression of HIDEN in 30 human tissues from GTEx database. The top eight tissues with high expression were shown
Compared to PCGs, all these lncRNAs showed lower RNA expression level in both PSCs and DE cells, but the desert lncRNAs were apparently higher than the other two (Fig. 1c, Additional file 1: Fig. S1c). Given that the function of lncRNAs is always linked to their subcellular localization [17, 18], we further dissected the subcellular localization of these lncRNAs based on the relative concentration index (RCI) [19]. The results showed that PCGs were mostly located in cytoplasm while lncRNAs showed significant nuclear localization tendency, especially for the overlapped lncRNAs (Fig. 1d, Additional file 1: Fig. S1d), which may be connected to its cis-regulatory model [1]. This nuclear localization tendency was also conserved in human lung cancer cell line A594 (Additional file 1: Fig. S1d). Previous reports showed that lncRNAs were expressed in a more cell-type-specific manner than PCGs [20]. Using a stage-specificity score [12], we found that lncRNAs exhibited more specific expression patterns not only in the continuous pancreatic lineage differentiation processes but also in different cell types, while desert lncRNAs showed the highest stage-specificity score among these lncRNAs (Fig. 1e). To further confirm the cell expression specificity of lncRNAs, we quantified the Normalized Difference (ND) of expression in the continuous pancreatic differentiation data [21]. The result was consistent with the stage-specificity score (Additional file 1: Fig. S1e). The cell-stage-specific expression feature of desert lncRNAs was also observed in another dataset of different cell types (Additional file 1: Fig. S1f-g). Altogether, these results indicated that lncRNAs were transcripts with low coding potential, nuclei-localized tendency, and significant cell-type-specific expression pattern.
Within all lncRNAs, the desert lncRNAs show higher expression level and significant cell-type-specific expression pattern, but the function and regulatory mechanism are largely uncharted in early embryo development. To explore the role of desert lncRNAs during human early differentiation, we identified the differentially expressed desert lncRNAs by transcriptome analysis of H9 ESC and its purified differentiated DE cells. Among the 46 differentially expressed desert lncRNAs (Additional file 2: Table S1), HIDEN (ENSG00000253507, AC104257.1) was highly expressed in DE cells (Fig. 1f). A time-course analysis demonstrated that HIDEN was gradually upregulated during DE differentiation (Fig. 1g), and with low coding potential predicted by two online software CPAT and CPC2 [22, 23] (Additional file 1: Fig. S1h). Furthermore, we analyzed the expression of HIDEN in 30 human tissues from the GTEx database and found HIDEN was highly expressed in germ lineages and brain as well as endoderm-derived lung (Fig. 1h).
HIDEN is a desert lncRNA required for endoderm differentiation
To better annotate the transcript information of HIDEN, rapid amplification of cDNA ends (RACE) was performed to obtain the full-length and map the genomic location of HIDEN in HUES8-derived endoderm cells. Based on the 5’ and 3’ RACE results (Additional file 1: Fig. S2a), we cloned the full-length of HIDEN transcript from cDNA of HUES8 DE cells and HIDEN was annotated as 1045 nucleotides with four exons (sequences were shown in Additional file 3). Interestingly, there was another isoform in PGP1 cells (Additional file 1: Fig. S2b): 1169 nucleotides with one additional small exon (Additional file 3). The genome sequence of HIDEN exhibited low species conservation (Additional file 1: Fig. S2b).
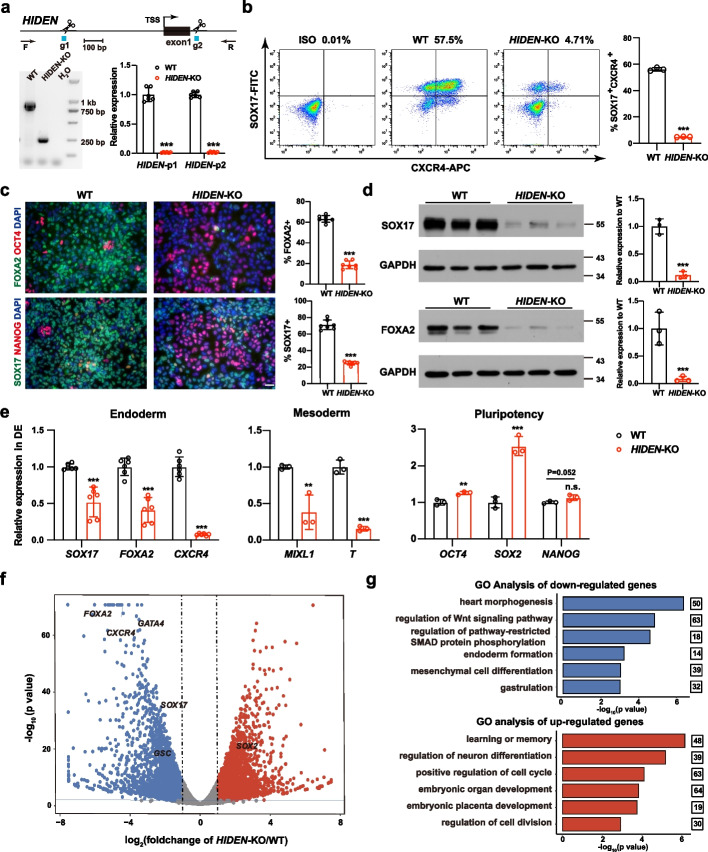
To investigate the biological function of HIDEN during DE differentiation from human PSCs, we established a HIDEN-knockout cell line in PSC line PGP1 with dual-sgRNA guided CRISPR/Cas9 system by deleting the promoter and the first exon existing in both isoforms (Fig. 2a). PX459 plasmid was constructed to allow tandem expression of two sgRNAs whose target sites were indicated in Fig. 2a [24], and the co-expression of two sgRNAs enhanced chances of the fragment deletion between two sgRNA target sites. Genomic PCR and RT-qPCR results validated successful knockout and complete deletion of HIDEN (Fig. 2a). HIDEN-knockout PSCs maintained normal expression of pluripotency markers, such as OCT4 and SSEA4, as shown in immunofluorescent staining results (Additional file 1: Fig. S2c). RT-qPCR results further confirmed that the expression of pluripotency genes, such as SOX2, OCT4 and NANOG, were not disrupted in HIDEN-knockout PSCs (Additional file 1: Fig. S2d). We also examined the cell proliferation based on cell counting kit (CCK) and the results showed HIDEN-knockout PSCs exhibited a comparable proliferation rate as wildtype PSCs (Additional file 1: Fig. S2e). Together, we demonstrated that the HIDEN deletion had no impact on pluripotent genes expression or self-renewal, indicating HIDEN was not required for pluripotency maintenance which was consistent with the low expression of HIDEN in undifferentiated state (Fig. 1g).
Fig. 2.
HIDEN is a desert lncRNA required for endoderm differentiation. a Generation of HIDEN knockout (KO) PSCs by CRISPR/Cas9. Top: sgRNAs used to delete the promoter of HIDEN and genomic PCR primers used to detect the promoter deletion. Bottom: The HIDEN RNA level was quantified using two sets of primers in HIDEN-KO DE cells compared to wildtype (n = 6). b Flow cytometric analysis of SOX17+CXCR4+ cells in wildtype and HIDEN-KO DE cells. The statistical results were shown on the right (n = 3). c Immunofluorescent staining of DE markers (SOX17, FOXA2) and pluripotency markers (OCT4, NANOG) in wildtype and HIDEN-KO DE cells. Quantitative results were shown on the right (n = 7). Scale bar, 50 m. d The protein levels of DE markers (SOX17 and FOXA2) were determined in wildtype and HIDEN-KO DE cells (n = 3). GAPDH was used as internal control. e The RNA expression levels of representative endoderm genes (n = 6), mesoderm genes (n = 3) and pluripotency genes (n = 3) in wildtype and HIDEN-KO DE cells. f Scatterplot showing differentially expressed genes identified by RNA-seq of wildtype and HIDEN-KO DE cells (n = 3). Upregulated and downregulated genes upon HIDEN-KO were shown in red and blue, respectively. g GO analysis of the downregulated or upregulated genes in HIDEN-KO DE cells compared to wildtype
To determine whether HIDEN was essential for DE differentiation, we subjected the wildtype and HIDEN-knockout PSCs to DE differentiation. We then examined the endoderm differentiation efficiency by CXCR4 (CD184)- or SOX17-based flow cytometric analysis [25] and key endoderm gene expression (SOX17, FOXA2, CXCR4, GATA4/6) at day 3 [6–10]. In vitro endoderm differentiation was induced by activin signaling [25, 26], which usually produced cultures consisting of up to 60–80% definitive endoderm cells after 3 days’ differentiation in our system [13]. Flow cytometric analysis showed an obvious decrease of SOX17- and CXCR4- double positive endoderm cells in the HIDEN-knockout group: 4.71% in the knockout group compared to 57.5% in wildtype cells (Fig. 2b). Similarly, immunofluorescent staining results showed fewer FOXA2- and SOX17-positive and more OCT4- and NANOG-positive cells in HIDEN-deleted differentiated cells compared to wildtype (Fig. 2c). Western blot also indicated that the protein levels of SOX17 and FOXA2 were significantly decreased in HIDEN knockout differentiated cells (Fig. 2d). Furthermore, RT-qPCR assay showed the endoderm genes (SOX17, FOXA2, CXCR4) were much less activated and the RNA levels of pluripotency genes were not that rapidly downregulated in HIDEN-deleted differentiated cells compared with wildtype (Fig. 2e). Taken together, the deletion of HIDEN caused an obvious defect in endoderm differentiation of human PSCs.
We then investigated the transcriptome in HIDEN-deficient DE cells (Additional file 4: Table S2). Again, the transcript of HIDEN was completely gone based on RNA-seq data (Additional file 1: Fig. S2b). RNA-seq results identified 2844 downregulated genes and 2700 upregulated genes. The volcano plot displayed a remarkable decrease in expression of endoderm genes, such as SOX17, FOXA2, CXCR4 and GATA4 (Fig. 2f). In addition, gene ontology (GO) analysis of downregulated genes in HIDEN-knockout cells exhibited a significant enrichment in terms of heart morphogenesis, regulation of WNT signaling pathway, regulation of pathway-restricted SMAD protein phosphorylation, endoderm formation, mesenchymal cell differentiation and gastrulation (Fig. 2g, Additional file 5: Table S3). On the other hand, GO terms of upregulated genes in HIDEN-knockout cells included learning or memory, regulation of neuron differentiation, positive regulation of cell cycle, embryonic organ development, embryonic placenta development, and regulation of cell division (Fig. 2g, Additional file 5: Table S3), supporting that the deletion of HIDEN affected key signaling pathways and endoderm development process. These results further demonstrated that HIDEN was an important regulator of the endoderm-specific transcriptome.
To confirm the importance of HIDEN for endoderm differentiation, we conducted spontaneous embryonic bodies (EB) differentiation and RT-qPCR was used to quantify the expression of representative markers of three germ layers. Compared to wildtype cells, endoderm genes (SOX17, FOXA2, CXCR4) were decreased in HIDEN-knockout cells, while pluripotency genes (OCT4, NANOG), mesoderm genes (MIXL1, T) and ectoderm genes (PAX6, SOX1) were increased, which was consistent with the necessary role of HIDEN in DE differentiation (Additional file 1: Fig. S2f). In addition, we performed endodermal pancreatic differentiation in wildtype and HIDEN-deleted PGP1 cells with the previously reported protocol [27], including ESC, DE, pancreatic progenitor 1 (PP1), and pancreatic progenitor 2 (PP2) stages, to further confirm that HIDEN indeed was crucial for endoderm specification. The positive staining of key pancreatic marker PDX1 in both PP1 and PP2 were significantly reduced in HIDEN-deleted differentiated cells compared to wildtype cells (Additional file 1: Fig. S2g). Consistently, the RNA expression of pancreas-specific transcription factors was also decreased in HIDEN-deleted differentiated cells, such as PDX1, FOXA2, GATA6 in PP1 stage and PDX1, NKX6-1, PTF1A, NKX2-2, NGN3 in PP2 stage (Additional file 1: Fig. S2h). These results illustrated the pivotal role of HIDEN in human definitive endoderm differentiation.
To further confirm the necessary role of HIDEN in DE differentiation, we generated two stable cell lines with HIDEN knockdown in HUES8 using short hairpin RNAs (shRNAs), achieving at least 63% efficiency (Additional file 1: Fig. S3a). Pluripotent markers, such as SOX2, OCT4, NANOG and SSEA4, showed no significant difference between control and HIDEN-KD ESCs demonstrated by either RT-qPCR or immunofluorescence results (Additional file 1: Fig. S3b-c). Consistently, we observed that HIDEN knockdown severely impaired human ESC differentiation toward DE (Additional file 1: Fig. S3d-g).
HIDEN physically interacts with IMP1
Next, we questioned how HIDEN contributed to endoderm differentiation from human PSCs, starting with determination of HIDEN expression in different cellular fractions in DE cells. While nuclear-localized MALAT1 and NEAT1 and cytoplasm-localized GAPDH, ACTB, and SOX17 mRNAs showed expected subcellular localization in our assay, we found HIDEN was mainly localized in the cytoplasmic fraction of differentiated endoderm cells (Fig. 3a), implying that HIDEN probably regulated gene expression at the post-transcriptional level by interacting with RNA-binding proteins.
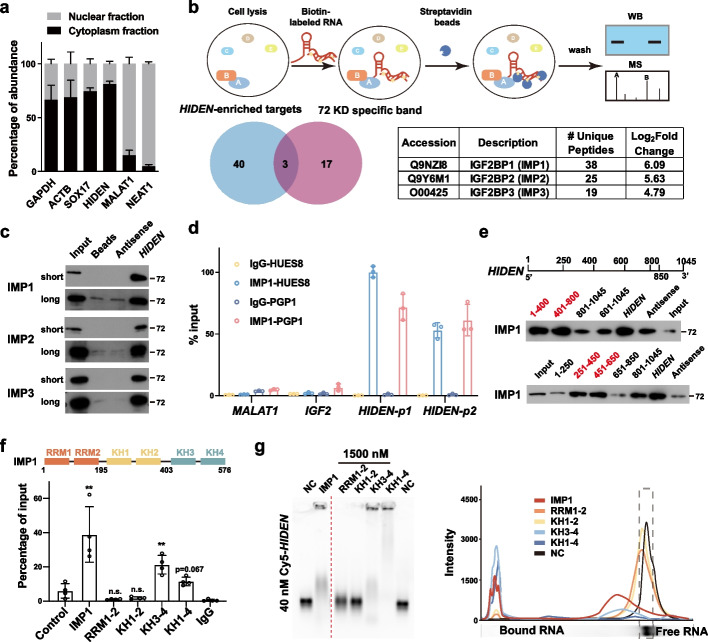
Fig. 3.
HIDEN physically interacted with IMP1. a Subcellular localization of HIDEN in DE cells determined by RT-qPCR following nucleo-cytoplasmic separation (n = 6). b The schematic diagram of RNA pulldown was shown on the top. Venn diagram indicates the overlapped hints identified by mass spectrometry of the specific band around 72 kDa (unique peptides ≥ 4, red circle) and whole extracts (unique peptides ≥ 10 in HIDEN-pulldown group and log2(fold-change (HIDEN/Antisense)) > 2.32, blue circle). The unique peptides and log2(fold-change) of IMP1/2/3 compared to antisense in mass spectrometry data of HIDEN-pulldown group were shown in the table. c Immunoblot for IMP1, IMP2, and IMP3 after RNA pulldown in DE cells. Beads and antisense were used as negative controls. Pictures captured for short and long exposure time were shown. d IMP1 RIP followed by RT-qPCR in DE cells of two PSC lines, PGP1 and HUES8 (n = 3). RNA levels were normalized to input. e Mapping the IMP1-binding region in HIDEN in 293 T cells. Top, diagrams of full-length HIDEN and the deletion fragments used in RNA pulldown. Bottom, immunoblot for IMP1 in protein samples pulled down by different HIDEN fragments. f Mapping the HIDEN-binding domain in IMP1 protein. Domain structure of IMP1 protein (top). FLAG-tagged IMP1 or IMP1 mutants and HIDEN were co-overexpressed in 293 T cells and FLAG RIP was performed to examine the enrichment of HIDEN (bottom). g Electrophoretic mobility shift assay (EMSA) results of in vitro binding assays. Cy5-labeled HIDEN RNA (40 nM) were transcribed in vitro and His-tagged IMP1 proteins (1500 nM) were purified from E. coli
To identify the interacting proteins of HIDEN, we conducted RNA pull-down assay using biotin-labeled HIDEN or its antisense RNA or other unrelated RNAs including luciferase and GATA6-AS1 as controls in DE cells (Fig. 3b). Silver staining showed an enriched band around 72 kDa captured specifically by HIDEN (Additional file 1: Fig. S4a), so this specific band was cut for mass spectrometry analysis (Additional file 6: Table S4). Meanwhile, whole immunoprecipitated extracts of HIDEN and its antisense were sent for quantitative mass spectrometry analysis and fold-change of HIDEN/antisense was calculated (Additional file 6: Table S4). The overlapped hints of mass spectrometry data based on the specific 72 kDa band and whole IP extracts were IGF2BP1 (IMP1), IGF2BP3 (IMP3), and IGF2BP2 (IMP2) (Fig. 3b). Western blot following RNA pulldown also confirmed the interaction of HIDEN with IMP1, IMP2, and IMP3 in DE cells (Fig. 3c). All these three proteins belong to the IMP family and are highly conserved RNA-binding proteins. Given that IMP1 was the most enriched protein in mass spectrometry analysis (Fig. 3b), we decided to take IMP1 for further study. RIP-qPCR analysis showed IMP1 was significantly bound with HIDEN, rather than mRNA IGF2 or lncRNA MALAT1 in DE cells from both PSC lines (Fig. 3d). These results indicated that HIDEN physically interacted with IMP1 protein.
To explore the direct binding region of HIDEN for IMP1, we used RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) and Mfold (http://www.unafold.org) to model the secondary structure of HIDEN. We obtained a similar secondary structure model of HIDEN from both tools (Additional file 1: Fig. S4b), and based on the predicted structure, we roughly divided HIDEN into three fragments: 1–400 nt, 401–800 nt and 801–1045 nt. We accordingly generated different biotin-labeled truncated HIDEN fragments for RNA pulldown assay in HEK293T cells. The result indicated that 1-400 nt and 401-800 nt fragments of HIDEN were responsible for the interaction between HIDEN and IMP1 (Fig. 3e). The second round of deletion-mapping analysis using smaller fragments of HIDEN revealed that two regions (251-450nt, 451-650nt) were mainly responsible for the interaction of HIDEN with IMP1 (Fig. 3e). IMP1 protein is composed of six canonical RNA-binding domains, including two RNA recognition motif (RRM) domains and four K homology (KH) domains (Fig. 3f). To clarify which domain of IMP1 interacted with HIDEN, we performed the co-expression experiment of truncated FLAG-tagged IMP1 and HIDEN in HEK293T cells. The RIP-qPCR results showed the highest enrichment of HIDEN in truncated KH3-4 domain compared to other mutants (Fig. 3f). We also purified various truncated IMP1 proteins (Additional file 1: Fig. S4c) and incubated with full-length Cy5-labeled HIDEN transcript for electrophoretic mobility shift assay (EMSA). Consistently, EMSA results showed that the KH3-4 domains of IMP1 were necessary for IMP1 binding to HIDEN at different concentrations (Fig. 3g and Additional file 1: Fig. S4d). Taken together, the 251–650 nt fragment of HIDEN and the KH3-4 domains of IMP1 were identified to be responsible for the interaction between HIDEN and IMP1 protein separately.
IMP1 deficiency inhibits endoderm differentiation
As the role of the HIDEN-interacting protein IMP1 in endoderm differentiation was unknown, we next examined whether IMP1 functioned in endoderm differentiation as well. We performed a loss-of-function assay by CRISPR/Cas9 in H9 ESCs and finally generated three IMP1 knockout clones with different genotypes (Additional file 1: Fig. S5a), which displayed normal colony morphology (Additional file 1: Fig. S5b). We confirmed the complete loss of IMP1 protein in these IMP1-knockout cell lines by immunoblot (Additional file 1: Fig. S5c). The expression of pluripotency genes (SOX2, NANOG, and OCT4) was almost unchanged in IMP1-knockout ESCs compared to wildtype (Additional file 1: Fig. S5d).
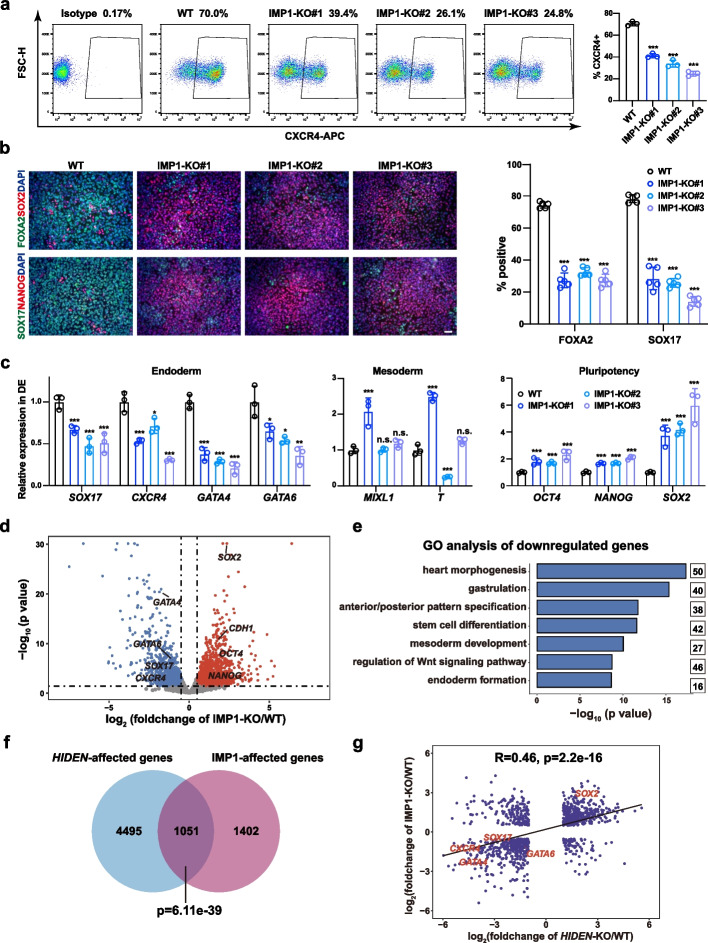
Then, we subjected these IMP1-knockout ESCs to endoderm differentiation. Flow cytometric results showed a significant decrease of CXCR4-positive cells in IMP1-knockout cells (Fig. 4a). Consistently, immunostaining results revealed that IMP1 knockout resulted in reduced expression of endoderm markers (FOXA2, SOX17) and remaining expression of pluripotency markers (SOX2, NANOG) (Fig. 4b). Moreover, RT-qPCR results showed a consistent decreased expression of endoderm genes, such as SOX17, CXCR4, GATA4 and GATA6, along with higher pluripotency genes expression (OCT4, NANOG, SOX2) and disrupted mesoderm genes expression (MIXL1, T), in IMP1-knockout DE cells (Fig. 4c). In addition, we performed RNA-seq analysis of wildtype and IMP1-knockout DE cells (Additional file 4: Table S2). From the volcano plot, we observed the downregulated endoderm genes (SOX17, CXCR4, GATA4, GATA6) and upregulated pluripotency genes (SOX2, OCT4, NANOG) (Fig. 4d). The top GO terms of downregulated genes in IMP1-deficient differentiated cells were heart morphogenesis, gastrulation, anterior/posterior pattern specification, stem cell differentiation, regulation of WNT signaling pathway, endoderm formation (Fig. 4e, Additional file 5: Table S3), similar to the results in HIDEN-knockout endoderm cells (Fig. 2g). We next compared the differentially expressed genes upon HIDEN- or IMP1-knockout identified by transcriptome analysis. We found 1051 genes co-regulated by HIDEN/IMP1, including endoderm genes such as SOX17, GATA6, GATA4 and CXCR4 (Fig. 4f). Moreover, the linear correlation result of co-regulated genes by HIDEN/IMP1 indicated HIDEN and IMP1 acted in the same gene regulatory loop during DE differentiation (Fig. 4g).
Fig. 4.
IMP1 deficiency inhibits endoderm differentiation. a Flow cytometric analysis of CXCR4-positive cells in wildtype (WT) and IMP1-KO DE cells. The statistical results were shown on the right (n = 3). b Immunofluorescent staining of DE markers (SOX17, FOXA2) and pluripotency markers (NANOG, SOX2) in WT and IMP1-KO DE cells. Quantitative results were shown on the right (n = 5). Scale bar = 50 μm. c RNA levels of representative endoderm genes, mesoderm genes and pluripotency genes in WT and IMP1-KO DE cells determined by RT-qPCR (n = 3). d Scatterplot showing differentially expressed genes in RNA-seq of WT and IMP1-KO DE cells. Upregulated and downregulated genes upon IMP1 knockout were shown in red and blue, respectively. e GO analysis of the downregulated genes in DE cells upon IMP1 knockout. f The overlap of differentially expressed genes upon HIDEN-KO and IMP1-KO. g The correlation of HIDEN-KO and IMP1-KO affected genes
To further exclude the off-target effect of CRISPR/Cas9 and confirm the important role of IMP1 in endoderm differentiation, we generated three stable IMP1-knockdown ESC lines using shRNAs (Additional file 1: Fig. S5e). These IMP1-knockdown ESCs exhibited obvious endoderm differentiation defects in RNA expression of endoderm markers (SOX17, FOXA2, CXCR4) (Additional file 1: Fig. S5f) and the percentage of CXCR4 positive cells (Additional file 1: Fig. S5g). Overall, these results indicated that the depletion of IMP1 resulted in impaired endoderm differentiation from human PSCs.
WNT signaling pathway acts as downstream of HIDEN/IMP1
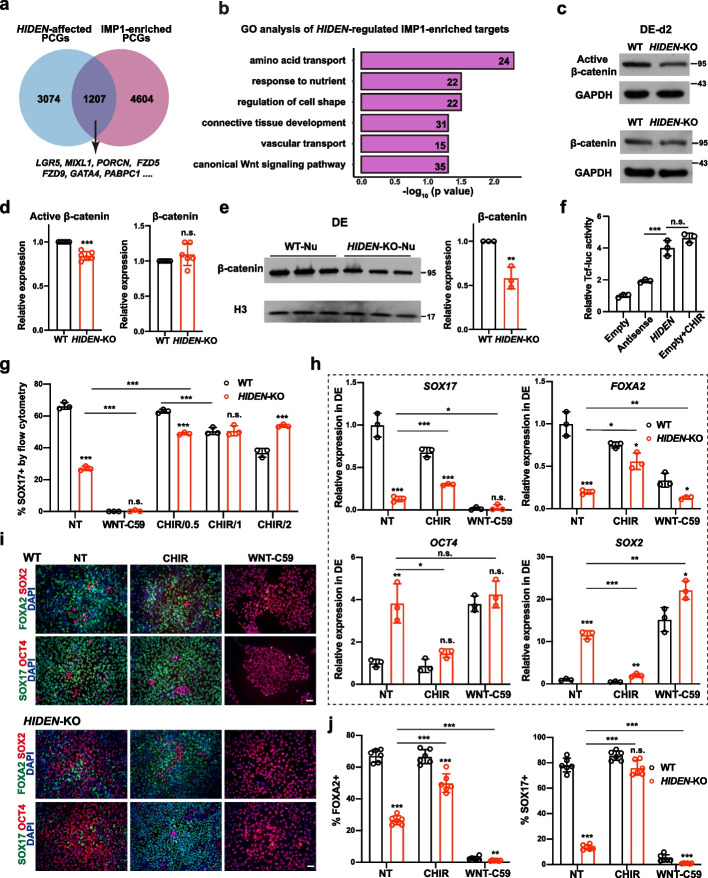
To understand how HIDEN and IMP1 affected human endoderm differentiation, we first examined the expression of IMP1 upon HIDEN depletion. The protein level of IMP1 remained unchanged in HIDEN-knockout or -knockdown DE cells compared to control cells (Additional file 1: Fig. S6a-b). Since IMP1 is a conserved RNA-binding protein and regulates mRNA stability, translation efficiency or cellular localization [28, 29], we examined the RNA-binding capacity of IMP1 in HIDEN knockout DE cells by RIP-seq. We integrated RNA-seq and RIP-seq data and identified the candidate target genes of HIDEN/IMP1 by overlapping the IMP1-bound RNAs and the differentially expressed protein-coding genes in DE cells upon HIDEN deletion (Fig. 5a). GO analysis of HIDEN/IMP1 target genes enriched in terms of nutrient response and transport, regulation of cell shape, connective tissue development, and canonical WNT signaling pathway (Fig. 5b, Additional file 5: Table S3). More importantly, by comparing IMP1-bound between wildtype and HIDEN-knockout DE cells, we identified 4880 genes with reduced IMP1 binding upon depleting HIDEN (Additional file 1: Fig. S6c, Additional file 7: Table S5). The GO analysis of these genes enriched terms of regulation of mRNA metabolic process, RNA splicing, WNT signaling pathway, canonical WNT signaling pathway, stem cell population maintenance, and endoderm development (Additional file 1: Fig. S6d, Additional file 5: Table S3). Considering the importance of the WNT signaling pathway in endoderm differentiation [7, 30, 31] and the fact that the enriched GO term of regulation of WNT signaling pathway upon HIDEN or IMP1 knockout, we hypothesized that WNT pathway acted as the downstream of HIDEN/IMP1. Therefore, we collected wildtype or HIDEN-deleted DE cells during endoderm differentiation to evaluate the protein level of active (unphosphorylated, nuclear-located) β-catenin, the key effector of WNT signal pathway. Compared to wildtype cells, we observed a significant decrease of active β-catenin and unaltered expression of total β-catenin at both differentiation day 2 (Fig. 5c, d) and day 4 (Additional file 1: Fig. S6e) in HIDEN-knockout cells. Consistently, β-catenin was significantly reduced in the nuclear fraction (Fig. 5e), which further proved the reduced WNT signaling activity in HIDEN-depleted DE cells. We also observed the similarly decreased active β-catenin in differentiated HIDEN-knockdown cells (Additional file 1: Fig. S6f). In addition, we performed the β-catenin/TCF-responsive luciferase reporter assay in HEK293T cells, showing that the overexpression of HIDEN led to elevated TCF luciferase activity (Fig. 5f). These results suggested that HIDEN depletion indeed resulted in impaired WNT activity during endoderm differentiation of PSCs.
Fig. 5.
WNT signaling pathway acts as downstream of HIDEN/IMP1. a Venn diagram indicates the overlapped genes of differentially expressed PCGs upon HIDEN knockout in DE cells and IMP1-enriched targets identified by IMP1 RIP-seq in DE cells. b GO analysis of the overlapped genes from a. c, d HIDEN knockout led to reduced active β-catenin and unaltered total β-catenin level compared to wildtype after two days’ DE differentiation, as shown by Western blot (c) and statistical results (d) (n = 6). e The protein level of β-catenin in nuclear fraction of WT or HIDEN-KO DE cells (n = 3). f The TCF-luciferase activity in 293 T when transfected with HIDEN, antisense control or empty vector (n = 3). Cells treated with 1 μM CHIR-99021 were used as positive controls. g-j Flow cytometric analysis of SOX17-positive cells (g), the presentative endoderm genes expression revealed by RT-qPCR (h) and immunostaining (i-j) in wildtype or HIDEN-KO cells after manipulating WNT signaling through small molecular inhibitors during DE differentiation (n = 3). WNT signaling activator CHIR-99021, with different concentrations (0.5 μM, 1 μM and 2 μM) at (g) and 1 μM at (h-j), and WNT signaling inhibitor WNT-C59 (1 μM) were used. NT indicated for non-treated group. i Scale bar = 50 μm. j Quantitative results of SOX17- and FOXA2-positive cells were shown (n = 6)
We next explored whether the reduced WNT signaling activity accounted for the decreased endoderm differentiation caused by HIDEN deletion. WNT signal activation is necessary for highly efficient DE differentiation, especially for the early phase of mesendoderm differentiation [30, 32, 33]. Therefore, we applied WNT activator CHIR-99021 and inhibitor Wnt-C59 respectively to rescue or phenocopy the defected DE differentiation upon HIDEN deletion. Indeed, we observed CHIR treatment could elevate the SOX17-positive cells specifically in HIDEN-knockout cells (Fig. 5g), while Wnt-C59 treatment significantly impeded endoderm differentiation in both groups (Fig. 5g). RT-qPCR results also showed increased endoderm gene expression (SOX17, FOXA2) in CHIR-treated HIDEN-deleted DE cells compared to the non-treated group (Fig. 5h), along with decreased pluripotency gene expression (OCT4, SOX2) (Fig. 5h) and disrupted mesoderm gene expression (MIXL1, T) (Additional file 1: Fig. S6g). Wnt-C59 treatment led to severe blocked DE differentiation in both wildtype and HIDEN-deleted DE cells, indicated by low expression of endoderm genes, high expression of pluripotency genes (Fig. 5h). Immunostaining results showed similar results (Fig. 5i-j). These results together indicated that manipulating WNT signaling pathway could partially rescue the endoderm differentiation deficiency caused by HIDEN disruption, indicating WNT signaling pathway acted as the functional downstream of HIDEN to regulate endoderm differentiation.
We also tested the functional link between IMP1 and WNT signaling pathway in endoderm differentiation. Similar to HIDEN-knockout cells, the WNT activator CHIR or inhibitor Wnt-C59 was able to rescue or phenocopy the failed DE differentiation due to IMP1 knockout, respectively. Flow cytometry analysis exhibited increased CXCR4-positive DE cells in CHIR-treated IMP1-deleted DE cells compared to the non-treated group (Additional file 1: Fig. S6h). Consistent with this functional assay, overexpression of IMP1 could activate the transcriptional activity of β-catenin/TCF-responsive luciferase reporter (Additional file 1: Fig. S6i). Taken together, these results supported that WNT signaling pathway largely contributed to HIDEN/IMP1-mediated endoderm regulation.
HIDEN/IMP1 promotes WNT signal pathway through FZD5
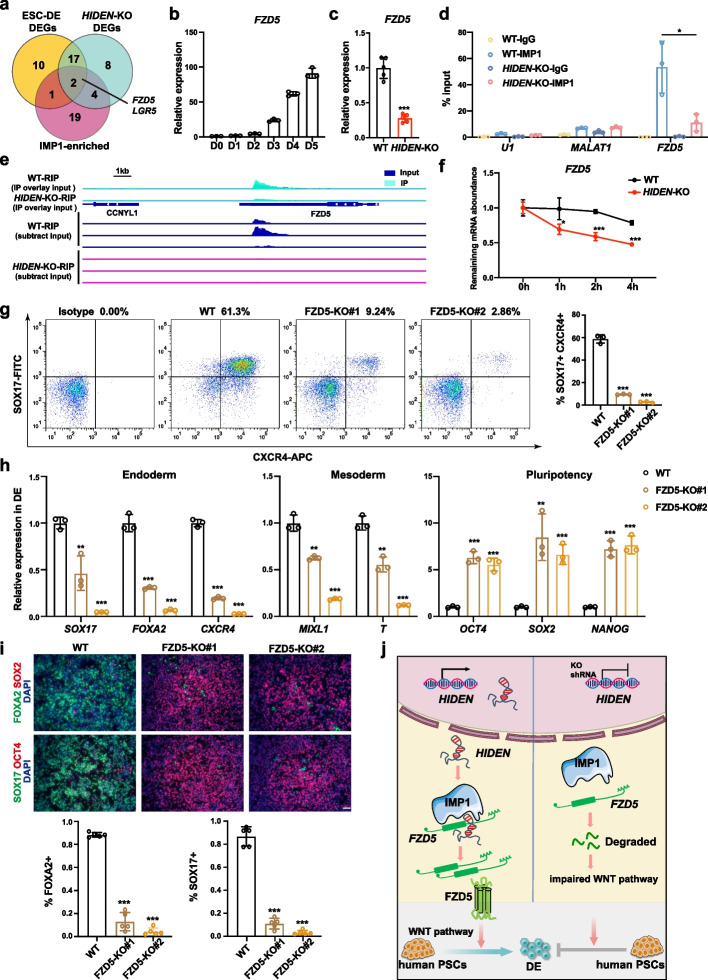
To find out the specific HIDEN/IMP1 direct targets to exert WNT-promoting effects, we studied the WNT-associated genes (listed in Additional file 8: Table S6), by the overlap among differentially expressed genes during DE differentiation (Additional file 1: Fig. S7a), differentially expressed genes upon HIDEN deletion in DE cells (Additional file 1: Fig. S7b), and IMP1-bound genes defined by IMP1 RIP-seq in DE cells (Fig. 6a). The overlapped genes were FZD5 and LGR5 (Fig. 6a). The RNA-seq analysis indicated FZD5 and LGR5 exhibited increased expression during endoderm differentiation and were downregulated upon HIDEN knockout (Additional file 1: Fig. S7c-d). Since the expression level of LGR5 was relatively lower than FZD5 during DE differentiation (Additional file 1: Fig. S7c-d), we mainly focused on FZD5. The upregulation of FZD5 expression during DE differentiation from human PSCs and the downregulation of FZD5 expression upon HIDEN deletion in endoderm cells were confirmed by RT-qPCR results (Fig. 6b, c). More importantly, IMP1 could highly enrich FZD5 mRNA in DE cells, and this interaction between IMP1 protein and FZD5 mRNA largely depended on HIDEN (Fig. 6d). Compared to differentiated wildtype cells, the binding of IMP1 to FZD5 mRNA was significantly decreased and the expression of FZD5 was much lower upon HIDEN knockout (Fig. 6e). The relative higher enrichment of FZD5 mRNA pulled down by HDIEN compared to other controls, such as lncRNA MALAT1, mRNA GAPDH and IMP1 (Additional file 1: Fig. S7e), further proved the association of HIDEN and FZD5 mRNA. By transcription inhibition assay using Actinomycin D treatment, we found FZD5 mRNA was more destabilized in HIDEN-knockout DE cells compared to wildtype cells (Fig. 6f). In addition, upon IMP1 knockout, we observed the lower expression of FZD5 mRNA in differentiated cells (Additional file 1: Fig. S7f), indicating the role of HIDEN in regulating the stability of FZD5 mRNAs via IMP1.
Fig. 6.
HIDEN-IMP1 stabilized FZD5 mRNA to contribute to DE differentiation. a Venn diagram indicated the overlapping of WNT-associated differentially expressed genes during DE differentiation (yellow circle), WNT-associated HIDEN-regulated genes (green circle), and IMP1-enriched WNT-associated genes (pink circle). The WNT-associated genes were listed in Additional file 8: Table S6 (from online WNT website: http://web.stanford.edu/group/nusselab/cgi-bin/wnt/). b The time course expression of FZD5 during DE differentiation, as shown by RT-qPCR (n = 3). c The relative expression of FZD5 in wildtype or HIDEN-KO DE cells was determined by RT-qPCR (n = 3). d RT-qPCR following IMP1 RIP was performed for determination of the indicated transcripts enrichment by IMP1 in wildtype or HIDEN-KO DE cells (n = 3). RNA levels were normalized to input. U1 and MALAT1 were considered as negative controls. e The visualization of IMP1 binding on FZD5 mRNA identified by IMP1 RIP-seq in wildtype or HIDEN-knockout DE cells. A detailed description was in Methods. f RNA stability assay of FZD5 in wildtype or HIDEN-KO DE cells treated with Actinomycin D (n = 3). g Flow cytometric analysis of SOX17+CXCR4+ cells in wildtype or FZD5-KO DE cells (n = 3). h RNA levels of representative endoderm genes, mesoderm genes and pluripotency genes in wildtype or FZD5-KO DE cells, determined by RT-qPCR (n = 3). i Immunofluorescent staining of DE markers (SOX17, FOXA2) and pluripotency markers (OCT4, SOX2) in wildtype or FZD5-KO DE cells. Quantitative results of SOX17- and FOXA2-positive cells were shown on the bottom (n = 5). Scale bar = 50 μm. j The functional model of HIDEN in human DE differentiation
To determine whether the HIDEN/IMP1-regulated FZD5 was the functional target in DE differentiation, we generated two FZD5 knockout human ESC lines in HUES8 (Additional file 1: Fig. S7g) which showed remarkably decreased expression of FZD5 (Additional file 1: Fig. S7h). Importantly, when subjected to endoderm differentiation conditions, FZD5 knockout led to blocked endoderm differentiation, evidenced by the reduced endoderm markers in flow cytometry analysis (Fig. 6g), RT-qPCR (Fig. 6h), and immunostaining results (Fig. 6i). In addition, the knockdown ESCs by shRNAs targeting FZD5, albeit maintained the normal expression of pluripotency genes demonstrated by immunofluorescence results (Additional file 1: Fig. S7i-j), indeed resulted in impaired endoderm differentiation, which was indicated by reduced endoderm gene expression in immunostaining, flow cytometry analysis, and RT-qPCR results (Additional file 1: Fig. S7k-m). These results suggested that the decreased FZD5 expression caused by HIDEN/IMP1 depletion inhibited endoderm differentiation from human PSCs.
Discussion
In this study we have characterized a subclass of lncRNAs (“desert” lncRNAs) and investigated the biological role and underlying mechanism of a specific desert lncRNA HIDEN in human endoderm differentiation (Fig. 6j). HIDEN is highly expressed during human endoderm differentiation. The depletion of HIDEN, both shRNA-mediated knockdown and knockout by deleting the promoter region, severely delays DE differentiation. HIDEN is mainly localized in cell cytoplasm and physically interacts with IMP1 protein, which is an important regulator of endoderm differentiation as well. Moreover, the observations that the disruption of HIDEN leads to reduced WNT signaling activity, and the impaired DE differentiation due to HIDEN or IMP1 deletion could be restored by WNT signaling activator, synergistically suggest that the WNT signaling pathway acts as the main downstream of HIDEN/IMP1 to contribute to endoderm differentiation. In detail, HIDEN/IMP1 promotes the mRNA stability of WNT receptor gene FZD5 through enhancing the interaction of IMP1 and FZD5 mRNA, and loss of FZD5 significantly blocks endoderm differentiation, together indicating that FZD5 is a functional target of HIDEN/IMP1 in human endoderm differentiation.
LncRNAs have been extensively studied in the last decade, and actively participate in many biological contexts, including pluripotency maintenance and lineage differentiation. However, most current studies of lncRNAs focus on those overlapped lncRNAs and proximal lncRNAs, including divergent lncRNAs, antisense lncRNAs, and enhancer-associated lncRNAs, but desert lncRNAs are often neglected. Here using transcriptome sequencing, RNA-seq following nuclear and cytoplasmic separation during human PSC differentiation, we pay attention to desert lncRNAs and characterize the expression level, subcellular localization, and cell-type specificity. Compared to overlapped lncRNAs and proximal lncRNAs, which are genomically located close to PCGs, desert lncRNAs tend to have higher expression, more cytoplasm-localized, and higher cell-type expression specificity, indicating important biological roles. Thereafter, we have identified a novel desert lncRNA HIDEN and revealed the function and mechanism in human endoderm differentiation. As a cytoplasm-localized desert lncRNA, HIDEN interacts with IMP1 and together regulate FZD5 mRNA stability. This illustrated the vital role of lncRNA in a multi-level gene regulation network composed of lncRNA, RNA-binding protein, and signaling pathway, which contributes to the fine-tuned spatial and temporal gene expression programs during lineage specification. It is also noteworthy that HIDEN is the first reported functional desert lncRNA in human endoderm differentiation, providing more insights on understanding and efficient acquisition of endoderm-derived functional cells or tissue.
IMP1 is a classical RNA-binding protein and highly associated with tumorigenesis and cancer metastasis [29, 34], but the function of IMP1 in early embryonic development is largely unknown. IMP1 is mainly expressed in the embryonic stage and with negotiable levels in adult tissues [29, 35]. The high expression of IMP1 during embryogenesis indicates a potential role of IMP1 in development and lineage specification. Indeed, IMP1-deficient mice have a smaller size (about 40%) and significant perinatal mortality than normal littermates mainly due to intestinal development defects [36], indicating an important role of IMP1 in endoderm-derived lineage development. A previous report showed that human ESCs with IMP1 knockdown exhibited a reduction in cell adhesion and increase in cell death through its effects on stabilizing ITGB5 mRNAs and maintaining BCL2 expression [37]. In our study, we did not observe obvious cell death in either IMP1 knockdown or knockout PSCs; however, we found that the depletion of IMP1 led to an impaired DE differentiation (Fig. 4). Moreover, we revealed that IMP1 interacted with HIDEN and together positively regulated WNT signaling pathway by stabilizing FZD5 mRNA in endoderm differentiation (Figs. 5 and 6). As WNT receptors, frizzled family members such as FZD5, FZD7, and FZD3 could transduce WNT/β-catenin signal to affect human ESC self-renewal and induce differentiation [38]. For example, the disruption of FZD7 impaired the pluripotent state of human ESCs, while selective activation of FZD7 signaling is sufficient to promote mesendodermal differentiation [39]. Here we provided the data that deletion of FZD5 could severely block endoderm differentiation (Fig. 6), together demonstrating the important role of frizzled family in early germ layer differentiation. In addition, these results about loss-of-function of IMP1 (Fig. 4) are consistent with the observation of endodermal intestine defect in IMP1-knockout mice [36], providing a likely mechanism as WNT was critical for endoderm and intestine development [40, 41].
IMP1 binds to massive mRNAs and regulates the turnover during cancer research and embryonic development, but the underlying molecular mechanism, particularly how the target specificity is achieved, is not fully revealed yet [29, 42]. IMP1 contains two RNA-recognition motifs (RRM1, RRM2) in the N-terminal region and four KH domains (KH1-4) in the C-terminal region [29]. However, the in vitro assay indicates the KH domains, rather than RRM domains, of IMP1 are directly responsible for RNA binding, especially KH3 and KH4, which form an intramolecular pseudodimer and create RNA-binding surfaces. More in detail, the highly conserved GXXG loop located in the KH domain is important for RNA binding [43]. The long half-life of IMP-mRNA complexes in vitro supports the notion that the interaction between IMP1 and mRNAs enhances mRNA stability [44, 45]. In our study, HIDEN transcript interacts with the KH3-4 domain of IMP1 and enhances FZD5 mRNA stability through facilitating the interaction of IMP1 protein and FZD5 mRNA (Fig. 6), which is in accordance with the importance of KH3-4 domains in IMP1 and further indicates the lncRNA-mediated target selectivity of IMP1. Growing evidence indicates that IMP1 binds to mRNAs and together form mRNP granules in cell cytoplasm [45–48], regulating mRNA homeostasis by incorporating its target transcripts into mRNP granules, protecting target mRNAs from miRNA-mediated silencing or releasing them to initiate translation at an appropriate time [49]. In addition, recent studies identified IMP1 as an N6-methyladenosine (m6A) reader recognizing the consensus GG(m6A)C sequence of target RNAs [44]. More interestingly, the recognition of m6A is mediated by the KH3-4 domains of IMP1 [44]. Whether the involvement of HIDEN in IMP1-mRNA regulation is related to m6A is an interesting question for the following study, and more studies should clarify the structural basis of how lncRNA participates in IMP1-regulated mRNAs stability.
Conclusions
In summary, we have characterized the biological function of a desert lncRNA HIDEN in human endoderm differentiation. HIDEN interacts with IMP1 protein and together regulates the stability of FZD5 mRNA to activate the WNT signal pathway. We further provide functional evidence that IMP1 and FZD5 are required for human DE differentiation. Our findings thus have not only characterized a subset of lncRNAs (i.e., desert lncRNAs), but also revealed the HIDEN/IMP1-FZD5 axis and its important role in human DE differentiation, providing insights in understanding cell fate determination.
Methods
Cell culture and differentiation
Two human ESC lines, HUES8 and H9, and one iPSC line PGP1 were used in this study. They were cultured in mTeSR™ medium (STEMCELL Technologies, #05850) on Matrigel-coated plates. Human embryonic kidney 293T (HEK293T) cells were cultured with DMEM medium containing 10% fetal bovine serum (FBS; Gibco, #10270–106) and 1% penicillin–streptomycin (Gibco, #15140163). The endoderm differentiation protocol of human PSCs was based on the previous reports [13, 30] with small adjustments. For HUES8 endoderm differentiation, IMDM (Gibco, #C12440500BT) and F12 (Gibco, #C11765500BT) were mixed at the ratio of 1:1 (IMDM/F12), supplemented with 0.2% BSA (YEASEN, #36106ES76), 1% B27 (without Vitamin A, Shanghai BasalMedia Technologies, S441J7) and 1% penicillin–streptomycin. Activin A (100 ng/ml, PeproTech, #120-14P) was added for 3 or 4 days. For H9 endoderm differentiation, DMEM was used as basal medium instead of IMDM/F12 and the rest ingredients were the same as above. For PGP1 endoderm differentiation, we removed B27 from the medium and treated cells with 1 M JNK-IN-8 (Selleck, S4901) [50] and 100 ng/ml Activin A for day 1, followed by 100 ng/ml Activin A treatment for the next 2 or 3 days. The following pancreatic lineage differentiation was based on the previous report with minor adjustments [27, 51]. Generally, human PSCs were induced into definitive endoderm for four days according to the above method. Then the differentiated cells were cultured in MCDB131 (Gibco, #10372019) supplemented with 0.5% BSA, 1.5% NaHCO3, 1% ITS-X (BasalMedia, # S452J7), 1% GlutaMAX (Gibco, #35050061), 1% penicillin–streptomycin, 10 mM Glucose (Invitrogen, #A2494001), 0.25 mM ascorbic acid (Sigma, #A5960), 50 ng/mL KGF (PeproTech, #100–19), 2 M IWR-1 (Selleck, #S7086) for 2 days. Next, cells were treated with MCDB131 supplemented with 2% BSA, 2.5% NaHCO3, 1% ITS-X, 1% GlutaMAX, 1% penicillin–streptomycin, 10 mM Glucose, 0.25 mM Vitamin C, 50 ng/ mL KGF, 2 M IWR-1, 0.25 M SANT1 (Selleck, #S7092), 200 nM LDN193189 (Selleck, #S7507), 100 nM TTNPB (Selleck, #S4627) and 500 nM PDBU (Sigma, #P1269) for 4 days (pancreatic progenitors 1, PP1 stage). During pancreatic progenitors 2 stage (PP2), 400 nM LDN193189, 10 nM TTNPB and 250 nM PDBU were used, and the rest gradients were the same as PP1 stage for 4 days. For EB differentiation, human PSCs were digested into single cells by Accutase and then counted and resuspended at the density of 200 cells/L in mTeSR1 with 10 μM Y-27632. Then the single cell drops (10 μL) were hanging on the lid of Petri dishes. The aggregated EBs were collected into 6-well low attachment plates after one day culture. EB medium (DMEM, 10% FBS, 1% penicillin–streptomycin) was changed every two days. After 9 days’ spontaneous differentiation, the differentiated EBs were harvested for qPCR assay. Other chemicals used in this study included CHIR-99021 (also CHIR, Selleck, #S2924), Wnt-C59 (Selleck, #S7037).
Plasmid constructs of shRNA knockdown
The shRNAs specifically against HIDEN, IMP1, FZD5 and scramble control were cloned into lentiviral vector pTY plasmid. HEK293T were transfected with lentiviral plasmid expressing shRNA or scramble control shRNA (shC) and lentiviral helping vectors (pNHP, pCEP-TAT, pHEF-VSVG) for lentivirus packaging. The stable PSC lines were established by selection with 2 g/mL puromycin for two weeks. The targeting sequences of the effective shRNAs were provided in Additional file 9: Table S7.
CRISPR-Cas9 mediated knockout
Genome editing was performed by electroporating human PSC cells with pX459 plasmid expressing target sgRNAs. Cells were selected with 2 g/mL puromycin for 2 days, followed by single cell sorting (BD FACS Aria) and genotyping. The sequences of all sgRNAs and the primers for genomic sequencing were listed in Additional file 9: Table S7.
RACE and cDNA cloning
After 4 days of endoderm differentiations, HUES8 cells were harvested and total RNA was isolated using Hipure Total RNA Mini Kit (Magen, #R4111-03). The 5’ and 3’ fragments of HIDEN were amplified using SMARTer RACE 5'/3' Kit (TAKARA, #634858) according to the manufacturer's instructions, then PCR products were cloned for Sanger sequencing. The gene-specific primers and primers used to clone full length of HIDEN were listed in Additional file 9: Table S7. The full length of HIDEN transcript was shown in Additional file 3.
Cell proliferation assay
Human PSCs were treated with Accutase, then counted and seeded in 96-well plates coated with Matrigel at a density of 3000 cells/well followed by culturing for 48, 72, or 96 h. Before detection, the plates were replenished with fresh medium containing 10 μl CCK for each well and incubated for 4 h at 37 °C according to the manufacturer's instructions. After shaking on an orbital shaker, the optical density (OD) was measured at 450 nm with MD SpectraMax i3x.
Quantitative RT-qPCR
Total RNA was isolated from cultured cells using Hipure Total RNA Mini Kit (Magen) or TriPure isolation reagent (Roche, #11667165001) according to the manufacturer's instructions. First strand cDNA was synthesized by reverse transcription of 1 g RNA using ABScript II RT Master MIX (ABclonal, #RK20402). 2 SYBR Green qPCR Master Mix (Bimake, #B21203) was used for quantification of gene expression on a CFX384 qPCR machine (Bio-Rad). GAPDH served as the internal control for normalization. One-tail unpaired t-test was performed to obtain p-values for RT-qPCR experiments. The primers used in all qPCR assays were listed in Additional file 9: Table S7.
Flow cytometric analysis
Differentiated PSCs were digested into single cells by TrypLE (Gibco, #12604021) and washed twice with DPBS containing 2% FBS. Cells were then incubated with CD184-APC (BD, #555976) for 30 min. As for the following intracellular flow cytometry, cells were fixed according to the manufacturer's instructions of Transcription Factor Buffer Set (BD, #562574), and then incubated with SOX17-Alexa488 (BD, #562205) antibody. Corresponding isotype was used as control. The SOX17+ or CXCR4+ cells were detected by FACSCelesta flow cytometer (BD) or CytoFLEX flow cytometer (Beckman Coulter) and analyzed by FlowJo software.
Western blot
Cells were lysed in RIPA buffer (Beyotime, #P0013C) with cocktail (Roche, #4693132001). The lysates were separated by 10% SDS-PAGE and immunoblotted with indicated antibodies. The primary antibodies used in this study included: SOX17 (R&D, #AF1924, 1:1000), FOXA2 (R&D, #AF2400, 1:1000), IGF2BP1 (IMP1, ABclonal, #A1517, 1:1000), IGF2BP2 (IMP2, ABclonal, #A2749, 1:1000), IGF2BP3 (IMP3, ABclonal, #A4444, 1:1000), GAPDH (Proteintech, #10494-1-AP, 1:5000), H3 (Proteintech, #17,168-1-AP, 1:3000), active β-catenin (CST, #8814, 1:1000), β-catenin (CST, #8480, 1:1000). After the overnight incubation of primary antibodies at 4 °C, the membrane was washed and incubated with secondary antibodies at room temperature for 2 h. The proteins were visualized by the ECL detection reagents (Millipore, #WBUSLS0100).
Immunofluorescence staining
Cells were fixed in 4% paraformaldehyde after PBS washing, then blocked and permeabilizated by blocking buffer (PBS with 10% donkey serum and 0.3% Triton X-100). Cells were incubated overnight at 4 °C by blocking buffer with the primary antibodies at proper concentration. The used primary antibodies in the experiment were: SOX17 (R&D, #AF1924, 1:200), FOXA2 (R&D, #AF2400, 1:200), OCT4 (CST, #2750, 1:200), SOX2 (BD, #561,469, 1:200), NANOG (CST, #4903, 1:200), SSEA4 (Millipore, #MAB4304, 1:400), TRA-1-60 (Millipore, #MAB4360, 1:400), PDX1 (R&D, #AF2419, 1:200). After washing with PBS for three times, cells were incubated with blocking buffer with the corresponding secondary fluorescent antibodies. The results of immunofluorescence were visualized and imaged under an Olympus IX53 microscope.
Cytosolic/nuclear fractionation
Differentiated endoderm cells were grown on 6-well plates and harvested at day 4. Cells were lysed in CE buffer (10 mM Hepes, 60 mM KCl, 1 mM EDTA, 0.34 M sucrose, 0.3% NP-40, 1 mM DTT) with cocktails for 5 min on ice, then centrifugated at 3000 rpm for 15 min to separate nuclear and cytoplasmic compartments. TriPure isolation reagent (Roche, #11667165001) was used to extract nuclear or cytoplasmic RNA, followed by RT-qPCR analysis. The GAPDH, ACTB and SOX17 mRNAs were used as cytoplasmic controls, while lncRNA MALAT1 and NEAT1 were used as nuclear controls.
RNA immunoprecipitation (RIP)
RIP was performed with the Magna RNA-binding protein immunoprecipitation kit (Millipore, #17–700) following the manufacturer's instructions. Cells lysis from 1 107 cells and 6 μg IMP1 or IgG antibody were used. RNA was isolated with TRIzol (Invitrogen, #10296010) and phenol/chloroform/isoamyl alcohol and then analyzed by qRT-PCR.
RNA pull-down
For RNA pull-down assay, full-length HIDEN and its antisense and other control RNAs were transcribed in vitro with HiScribe T7 High Yield RNA Synthesis Kit (NEB, #E2040S) and biotin-16-UTP (Roche, #11388908910) according to the manufacturer's instructions. 10 μg biotin-labeled HIDEN or control RNAs was incubated with total cell lysate of differentiated endoderm cells from PSCs or HEK293T cells. Then 40 μL Dynabeads MyOne Streptavidin C1 (Invitrogen, #65001) were added to isolate the RNA-protein complex, followed by silver staining / mass spectra or Western blot. RNA pulled down by RNA were conducted as above, then digested by proteinase K at 55 °C incubator and extracted by phenol chloroform.
RNA sequencing and RIP-sequencing data analysis
For RNA-sequencing, total RNA was isolated with HiPure Total RNA Mini Kit. RNA libraries were sequenced on an Illumina Hiseq X Ten platform with paired-end reads at Geekgene Technology. RNA-seq raw data that contained adapters were removed and trimmed by Trim Galore (v0.6.6). The clean data were aligned to human GRCh38 genome reference with the HISAT2 [52]. The GENCODE V29 gene transfer format (GTF) was used to count reads by the FeatureCounts (v2.0.1). All counts were further normalized with TPM (Transcripts Per Million) in R software. Differential expression analysis was performed for binary comparisons using the R package DESeq2 [53]. For cutoff threshold, we set P value < 0.05, abs (log2(fold-change)) > 1.0 in HIDEN-KO dataset and abs (log2 (fold-change)) > 0.5 in IMP1-KO dataset separately. Gene Ontology analysis was executed by R package clusterProfile [54].
To investigate the expression level of different kinds of lncRNAs during endoderm differentiation, we reanalyzed three RNAseq datasets in different PSC lines and the DE derivates, including H9 dataset from GSE44875 [12], HUES8 dataset from GSE137208 [55] and GSE143499 [13], PGP1 dataset from GSE173690 [56] and GSE188501 [57]. The subcellular localization of lncRNAs were calculated using the “relative concentration index” (RCI) with slight modification [19]. The expression level of different components was normalized as FPKM and the RCI was calculated as below: CN.RCI = log2(CE/NE) + log2(CE/NM) (CE: cytoplasmic elution component, NE: nuclear elution component, NM: nuclear insoluble component). In addition, we reanalyzed the H1 and A549 datasets from GSE30567 [58] to verify the results. To quantify the stage- or cell-specificity of PCGs and lncRNAs expression, we calculated the specificity score in two different manners according to the references [12, 21]. The dataset from GSE44875 [12] and GSE134743 [59] was reanalyzed by the same pipeline. The coding potential of gencode.v29. transcripts were predicted using the software GeneID (v1.4.5) (https://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_human/release_29/gencode.v29.transcripts.fa.gz) with the parameters in file human.070123.param, and additional -s -3 parameters [21, 60]. To investigate the expression level of HIDEN in human tissues, we downloaded and reanalyzed the 22,952 tissues expression data including 30 human tissues (https://www.gtexportal.org/). The gene expression data (TPMs) (https://storage.googleapis.com/gtex_analysis_v8/rna_seq_data/GTEx_Analysis_2017-06-05_v8_RNASeQCv1.1.9_gene_tpm.gct.gz) and the sample annotation file (https://storage.googleapis.com/gtex_analysis_v8/annotations/GTEx_Analysis_v8_Annotations_SampleAttributesDS.txt) were integrated and further analyzed in R software.
For IMP1-RIP-seq, the raw data that contained adapters were removed and trimmed by Trim Galore (v0.6.6) and then aligned to human GRCh38 genome reference with the HISAT2. To identify differential peaks between Input and IP group, MACS2 (v2.1.1) was used with default parameters in IMP1-RIPseq, which uses a dynamic Poisson distribution to effectively capture local biases in the genome [61–63], allowing for more robust predictions. To find IMP1 reduced enrichment genes in HIDEN-KO, we compared IMP1-RIPseq peaks in wild type and HIDEN-KO group using macs2 bdgdiff (a subcommand of MACS2). Then the peaks were annotated to genes by R package ChIPseeker [64]. The peak visualization was performed in IGV (v2.6.2). In order to make the data comparison more obvious, we overlaid the IP track to input track in IGV and subtracted input signal from IP signal by bigwigCompare command of deepTools [65] respectively (Fig. 6e).
Protein expression and purification
IMP1 and its mutants were inserted into pET28a plasmid containing His tag. The fusion proteins were expressed in E. coli strain Rosetta (Shanghai Weidi Biotechnology, #EC1010) under 0.5 mM IPTG, 25 °C for 4-6 h induction after grown to OD600 0.6-0.8. After IPTG induction, E. coli cell pellets were collected, then resuspended with His Lysis buffer (50 mM Tris-HCl pH8.0, 500 mM NaCl, 10% glycerol, 1.0% Triton X-100, 10 mM Imidazol, 0.2 mM PMSF), and further crushed using high pressure homogenizer. Protein supernatant was collected by centrifugation at 12,500 rpm for 15 min at 4 °C. About 150-200 Ni-NTA Agarose beads (Abclonal, #AS045) were washed twice using His Lysis buffer and then incubated with protein supernatant at 4 °C for 3-4 h. The beads were collected and washed twice for 10 min using 5 mL His Washing buffer (50 mM Tris-HCl pH 8.0, 500 mM NaCl, 10% glycerol, 20 mM Imidazol). Finally, the protein was eluted from the beads using 500 His Elution buffer (20 mM Tris-HCl pH 8.0, 500 mM NaCl, 10% glycerol, 300 mM Imidazol). The eluted protein was concentrated using Amicon Ultracentrifuge filters (Merck, #UFC201024). Protein purity and concentration were determined with SDS-PAGE followed by Coomassie blue staining. BSA was used as protein standard sample.
EMSA (electrophoretic mobility shift assay)
EMSA was performed as reference [15] with slight modifications. Briefly, RNA was labelled with Cy5-UTP (APExBIO, #B8333) using HiScribe T7 High Yield RNA Synthesis Kit (NEB, #E2040S). The 40 nM Cy5-labelled RNA probes were denatured by heating at 95 °C for 1 min and cooling on ice, and then adding an equal volume of 2 EMSA binding buffer (40 mM Tris-HCl pH 7.9, 20% glycerol, 300 mM KCl, 5 mM MgCl2, 2 mM DTT, 0.2 mg/ml BSA and 10 U of RNase inhibitor) and incubated at room temperature for 30 min. Purified IMP1 or mutant proteins was added and then incubated with RNA at room temperature for 30 min. After incubation, the RNA-protein complexes were separated and analyzed using 0.5% native agarose gel. The images were captured using Biorad ChemiDoc MP.
Statistical analysis
Statistical analysis was conducted using PRISM 8.0 software. The results were shown as means SD from at least three independent experiments. Single comparison between two groups was analyzed by two-tailed unpaired t-test. Comparisons between multiple groups were determined using One-Way ANOVA analysis. Pearson's correlation analysis was used to evaluate the correlation between two variables. P value < 0.05 is considered statistically significant (* means p < 0.05, ** means p < 0.01, and *** means p < 0.001), while “n.s.” stands for not statistically significant.
Supplementary Information
Fig. S1. Desert lncRNAs are highly expressed during human endoderm differentiation. Fig. S2. HIDEN knockout is not essential for PSC pluripotency but required for endoderm lineage differentiation. Fig. S3. HIDEN knockdown exhibits decreased endoderm differentiation. Fig. S4. HIDEN physically interacts with IMP1. Fig. S5. IMP1 is required for DE differentiation. Fig. S6. WNT pathway is regulated by HIDEN/IMP1. Fig. S7. HIDEN promotes WNT pathway through FZD5.
Table S1. ESC and DE differentially expressed lncRNAs in three datasets. ESC and DE differentially expressed deserted lncRNAs in H9 cells described in Figure 1f.
HIDEN sequence.
Table S2. The transcript per millionvalue of gene expression in wildtype or HIDEN-KO DE cells. The transcript per millionvalue of gene expression in wildtype or IMP1-KO DE cells.
Table S3. HIDEN-KO upregulated or downregulated GO terms. IMP1-KO downregulated GO terms. GO terms of the overlap between HIDEN-KO DEGs and IMP1 RIP-seq enriched genes. GO analysis of HIDEN-KO reduced genes.
Table S4. The mass spectrometry results of specific 72 KD band and whole IP extracts.
Table S5. IMP1-bound genes identified by IMP1 RIP-seq. The IMP1-enriched genes whose binding with IMP1 was reduced by HIDEN-KO.
Table S6. WNT associated gene list.
Table S7. Oligos used in this study. shRNA target sequences, CRISPR sgRNA target sequence, primers used in genomic genotyping, gene-specific primers used in RACE assays and full-length cloning, and qPCR primers were shown.
Table S8. GEO information used in this study.
Uncropped images for western blots in this manuscript.
Review history.
Acknowledgements
We sincerely thank the core facility of the Medical Research Institute at Wuhan University for the technical support. We would like to thank Dr. Linfeng Huang at Duke Kunshan University for insightful discussion, Dr. Yu Zhou at Wuhan University for help in generating the cytoplasm-nuclei RNA-seq data, Dr. Cheguo Cai at Wuhan University for providing plasmid, Dr. Donghui Zhang at Hubei University for providing PGP1 cells. We would like to thank Lai Jiang, Yinglei Li and other laboratory members for technical help and discussion.
Peer review information
Andrew Cosgrove was the primary editor of this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Review history
The review history is available as Additional file 12.
Authors’ contributions
Jiang conceived and supervised the project, and designed the experiment together with Lu and Yang. Yang and Zhang analyzed the NGS data with the help from Yan. Lu performed the loss-of-function assays and identified the interaction of HIDEN-IMP1. Yang and Lu performed the biochemistry experiment of HIDEN with help from Wen. Li performed the phenotype assay of FZD5-KO, and conducted the 7 TCF-luciferase assay. Liu provided support in cell culture. Lu drafted the manuscript, and Jiang, Yang, Xiao and Wang revised and finalized the manuscript. All authors contributed to and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 32270857, 31970608, 91740102), Seed Fund Program for Sino-Foreign Joint Scientific Research Platform of Wuhan University (WHUDKUZZJJ202205), the Science and Technology Department of Hubei Province (2021CFA049), and the Fundamental Research Funds for the Central Universities in China (2042022dx0003).
Availability of data and materials
Three RNA-seq datasets in different human PSC lines and the DE derivates, including H9 dataset from GSE44875 [12], HUES8 dataset from GSE137208 [55] and GSE143499 [13], PGP1 dataset from GSE173690 [56] and GSE188501 [57], H1 and A549 datasets from GSE30567 [58], other cell-types (ARPE-19–1, H1, H9, HepG2 and Jurkat) dataset from GSE134743 [59] were used in this study, the detailed accession numbers for each sample are listed in Additional file 10: Table S8. The RNA-seq, RIP-seq data generated in this study are available under accession number of GSE188501 [57].
Declarations
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Pei Lu, Jie Yang and Mao Li contributed equally to this work.
References
- 1.Sigova AA, et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci U S A. 2013;110:2876–2881. doi: 10.1073/pnas.1221904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engreitz JM, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539:452–455. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu P, Li M, Zhang D, Jiang W. Lnc-ing pluripotency maintenance and early differentiation in human pluripotent stem cells. FASEB J. 2021;35:e21438. doi: 10.1096/fj.202002278R. [DOI] [PubMed] [Google Scholar]
- 4.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 5.Flynn RA, Chang HY. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14:752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sui L, Bouwens L, Mfopou JK. Signaling pathways during maintenance and definitive endoderm differentiation of embryonic stem cells. Int J Dev Biol. 2013;57:1–12. doi: 10.1387/ijdb.120115ls. [DOI] [PubMed] [Google Scholar]
- 7.Loh KM, et al. Efficient endoderm induction from human pluripotent stem cells by logically directing signals controlling lineage bifurcations. Cell Stem Cell. 2014;14:237–252. doi: 10.1016/j.stem.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sumi T, Tsuneyoshi N, Nakatsuji N, Suemori H. Defining early lineage specification of human embryonic stem cells by the orchestrated balance of canonical Wnt/beta-catenin, Activin/Nodal and BMP signaling. Development. 2008;135:2969–2979. doi: 10.1242/dev.021121. [DOI] [PubMed] [Google Scholar]
- 9.Rankin SA, et al. Timing is everything: Reiterative Wnt, BMP and RA signaling regulate developmental competence during endoderm organogenesis. Dev Biol. 2018;434:121–132. doi: 10.1016/j.ydbio.2017.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villegas VE, Zaphiropoulos PG. Neighboring gene regulation by antisense long non-coding RNAs. Int J Mol Sci. 2015;16:3251–3266. doi: 10.3390/ijms16023251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo S, et al. Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell. 2016;18:637–652. doi: 10.1016/j.stem.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 12.Jiang W, Liu Y, Liu R, Zhang K, Zhang Y. The lncRNA DEANR1 facilitates human endoderm differentiation by activating FOXA2 expression. Cell Rep. 2015;11:137–148. doi: 10.1016/j.celrep.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J, et al. GATA6-AS1 Regulates GATA6 Expression to Modulate Human Endoderm Differentiation. Stem Cell Reports. 2020;15:694–705. doi: 10.1016/j.stemcr.2020.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daneshvar K, et al. DIGIT Is a conserved long noncoding RNA that regulates GSC expression to control definitive endoderm differentiation of embryonic stem cells. Cell Rep. 2016;17:353–365. doi: 10.1016/j.celrep.2016.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daneshvar K, et al. lncRNA DIGIT and BRD3 protein form phase-separated condensates to regulate endoderm differentiation. Nat Cell Biol. 2020;22:1211–1222. doi: 10.1038/s41556-020-0572-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frankish A, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47:D766–D773. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen LL. Linking long noncoding RNA localization and function. Trends Biochem Sci. 2016;41:761–772. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 19.Mas-Ponte D, et al. LncATLAS database for subcellular localization of long noncoding RNAs. RNA. 2017;23:1080–1087. doi: 10.1261/rna.060814.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol. 2018;19:143–157. doi: 10.1038/nrm.2017.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexanian M, et al. A transcribed enhancer dictates mesendoderm specification in pluripotency. Nat Commun. 2017;8:1806. doi: 10.1038/s41467-017-01804-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, et al. CPAT: coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41:e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang YJ, et al. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017;45:W12–W16. doi: 10.1093/nar/gkx428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D'Amour KA, et al. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 26.Guo S, Mao X, He F, Liu H, Ming L. Activin A supplement in the hESCs culture enhances the endoderm differentiation efficiency. Cell Biol Int. 2014;38:849–856. doi: 10.1002/cbin.10274. [DOI] [PubMed] [Google Scholar]
- 27.Tan M, Jiang L, Li Y, Jiang W. Dual inhibition of BMP and WNT signals promotes pancreatic differentiation from human pluripotent stem cells. Stem Cells Int. 2019;2019:5026793. doi: 10.1155/2019/5026793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du QY, Zhu ZM, Pei DS. The biological function of IGF2BPs and their role in tumorigenesis. Invest New Drug. 2021 doi: 10.1007/s10637-021-01148-9. [DOI] [Google Scholar]
- 29.Bell JL, et al. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cell Mol Life Sci. 2013;70:2657–2675. doi: 10.1007/s00018-012-1186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang W, Wang J, Zhang Y. Histone H3K27me3 demethylases KDM6A and KDM6B modulate definitive endoderm differentiation from human ESCs by regulating WNT signaling pathway. Cell Res. 2013;23:122–130. doi: 10.1038/cr.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dziedzicka D, et al. Endogenous suppression of WNT signalling in human embryonic stem cells leads to low differentiation propensity towards definitive endoderm. Sci Rep. 2021;11:6137. doi: 10.1038/s41598-021-85447-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naujok O, Diekmann U, Lenzen S. The generation of definitive endoderm from human embryonic stem cells is initially independent from activin A but requires canonical Wnt-Signaling. Stem Cell Rev Rep. 2014;10:480–493. doi: 10.1007/s12015-014-9509-0. [DOI] [PubMed] [Google Scholar]
- 33.Diekmann U, Lenzen S, Naujok O. A reliable and efficient protocol for human pluripotent stem cell differentiation into the definitive endoderm based on dispersed single cells. Stem Cells Dev. 2015;24:190–204. doi: 10.1089/scd.2014.0143. [DOI] [PubMed] [Google Scholar]
- 34.Huang X, et al. Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) in cancer. J Hematol Oncol. 2018;11:88. doi: 10.1186/s13045-018-0628-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nielsen J, et al. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol. 1999;19:1262–1270. doi: 10.1128/MCB.19.2.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen TV, et al. Dwarfism and impaired gut development in insulin-like growth factor II mRNA-binding protein 1-deficient mice. Mol Cell Biol. 2004;24:4448–4464. doi: 10.1128/mcb.24.10.4448-4464.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conway AE, et al. Enhanced CLIP uncovers IMP Protein-RNA targets in human pluripotent stem cells important for cell adhesion and survival. Cell Rep. 2016;15:666–679. doi: 10.1016/j.celrep.2016.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernandez A, et al. The WNT receptor FZD7 is required for maintenance of the pluripotent state in human embryonic stem cells. P Natl Acad Sci USA. 2014;111:1409–1414. doi: 10.1073/pnas.1323697111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gumber D. et al. Selective activation of FZD7 promotes mesendodermal differentiation of human pluripotent stem cells. Elife. 2020;9. 10.7554/eLife.63060. [DOI] [PMC free article] [PubMed]
- 40.Gregorieff A, Clevers H. Wnt signaling in the intestinal epithelium: from endoderm to cancer. Genes Dev. 2005;19:877–890. doi: 10.1101/gad.1295405. [DOI] [PubMed] [Google Scholar]
- 41.Mah AT, Yan KS, Kuo CJ. Wnt pathway regulation of intestinal stem cells. J Physiol. 2016;594:4837–4847. doi: 10.1113/JP271754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biswas J, et al. The structural basis for RNA selectivity by the IMP family of RNA-binding proteins. Nat Commun. 2019;10:4440. doi: 10.1038/s41467-019-12193-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chao JA, et al. ZBP1 recognition of beta-actin zipcode induces RNA looping. Genes Dev. 2010;24:148–158. doi: 10.1101/gad.1862910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang H, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nielsen J, Kristensen MA, Willemoes M, Nielsen FC, Christiansen J. Sequential dimerization of human zipcode-binding protein IMP1 on RNA: a cooperative mechanism providing RNP stability. Nucleic Acids Res. 2004;32:4368–4376. doi: 10.1093/nar/gkh754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jonson L, et al. Molecular composition of IMP1 ribonucleoprotein granules. Mol Cell Proteomics. 2007;6:798–811. doi: 10.1074/mcp.M600346-MCP200. [DOI] [PubMed] [Google Scholar]
- 47.Zeng WJ, et al. Initiation of stress granule assembly by rapid clustering of IGF2BP proteins upon osmotic shock. Biochim Biophys Acta Mol Cell Res. 2020;1867:118795. doi: 10.1016/j.bbamcr.2020.118795. [DOI] [PubMed] [Google Scholar]
- 48.Mateu-Regue A, et al. Single mRNP analysis reveals that small cytoplasmic mRNP granules represent mRNA singletons. Cell Rep. 2019;29:736. doi: 10.1016/j.celrep.2019.09.018. [DOI] [PubMed] [Google Scholar]
- 49.Degrauwe N, Suva ML, Janiszewska M, Riggi N, Stamenkovic I. IMPs: an RNA-binding protein family that provides a link between stem cell maintenance in normal development and cancer. Genes Dev. 2016;30:2459–2474. doi: 10.1101/gad.287540.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li QV, et al. Genome-scale screens identify JNK-JUN signaling as a barrier for pluripotency exit and endoderm differentiation. Nat Genet. 2019;51:999. doi: 10.1038/s41588-019-0408-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pagliuca FW, et al. Generation of functional human pancreatic beta cells in vitro. Cell. 2014;159:428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu T, et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (N Y) 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng R, et al. Derivation of feeder-free human extended pluripotent stem cells. Stem Cell Rep. 2021;16:1686–1696. doi: 10.1016/j.stemcr.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lan X, et al. PCGF6 controls neuroectoderm specification of human pluripotent stem cells by activating SOX2 expression. Nat Commun. 2022;13:4601. doi: 10.1038/s41467-022-32295-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu P, Yang J, Li M, Wen S, Zhang T, Yan C, Liu R, Xiao Y, Wang X, Jiang W. A desert lncRNA HIDEN regulates human endoderm differentiation via interacting with IMP1 and stabilizing FZD5 mRNA. Datasets.Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE188501 (2023). [DOI] [PMC free article] [PubMed]
- 58.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grubert F, et al. Landscape of cohesin-mediated chromatin loops in the human genome. Nature. 2020;583:737–743. doi: 10.1038/s41586-020-2151-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alioto T, Blanco E, Parra G, Guigo R. Using geneid to Identify Genes. Curr Protoc Bioinformatics. 2018;64:e56. doi: 10.1002/cpbi.56. [DOI] [PubMed] [Google Scholar]
- 61.Bertero A, et al. The SMAD2/3 interactome reveals that TGFbeta controls m(6)A mRNA methylation in pluripotency. Nature. 2018;555:256–259. doi: 10.1038/nature25784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zambelli F, Pavesi G. RIP-Seq data analysis to determine RNA-protein associations. Methods Mol Biol. 2015;1269:293–303. doi: 10.1007/978-1-4939-2291-8_18. [DOI] [PubMed] [Google Scholar]
- 64.Yu G, Wang LG, He QY. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- 65.Ramirez F, Dundar F, Diehl S, Gruning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Desert lncRNAs are highly expressed during human endoderm differentiation. Fig. S2. HIDEN knockout is not essential for PSC pluripotency but required for endoderm lineage differentiation. Fig. S3. HIDEN knockdown exhibits decreased endoderm differentiation. Fig. S4. HIDEN physically interacts with IMP1. Fig. S5. IMP1 is required for DE differentiation. Fig. S6. WNT pathway is regulated by HIDEN/IMP1. Fig. S7. HIDEN promotes WNT pathway through FZD5.
Table S1. ESC and DE differentially expressed lncRNAs in three datasets. ESC and DE differentially expressed deserted lncRNAs in H9 cells described in Figure 1f.
HIDEN sequence.
Table S2. The transcript per millionvalue of gene expression in wildtype or HIDEN-KO DE cells. The transcript per millionvalue of gene expression in wildtype or IMP1-KO DE cells.
Table S3. HIDEN-KO upregulated or downregulated GO terms. IMP1-KO downregulated GO terms. GO terms of the overlap between HIDEN-KO DEGs and IMP1 RIP-seq enriched genes. GO analysis of HIDEN-KO reduced genes.
Table S4. The mass spectrometry results of specific 72 KD band and whole IP extracts.
Table S5. IMP1-bound genes identified by IMP1 RIP-seq. The IMP1-enriched genes whose binding with IMP1 was reduced by HIDEN-KO.
Table S6. WNT associated gene list.
Table S7. Oligos used in this study. shRNA target sequences, CRISPR sgRNA target sequence, primers used in genomic genotyping, gene-specific primers used in RACE assays and full-length cloning, and qPCR primers were shown.
Table S8. GEO information used in this study.
Uncropped images for western blots in this manuscript.
Review history.
Data Availability Statement
Three RNA-seq datasets in different human PSC lines and the DE derivates, including H9 dataset from GSE44875 [12], HUES8 dataset from GSE137208 [55] and GSE143499 [13], PGP1 dataset from GSE173690 [56] and GSE188501 [57], H1 and A549 datasets from GSE30567 [58], other cell-types (ARPE-19–1, H1, H9, HepG2 and Jurkat) dataset from GSE134743 [59] were used in this study, the detailed accession numbers for each sample are listed in Additional file 10: Table S8. The RNA-seq, RIP-seq data generated in this study are available under accession number of GSE188501 [57].