Abstract
CjCas9 is one of the smallest CRISPR-associated (Cas9) nucleases for mammalian genome editing. However, it requires a long N4RYAC (R = A or G; Y = C or T) protospacer-adjacent motif (PAM), limiting its DNA targeting scope. In this study, we investigated the PAMs of three CjCas9 orthologs, including Hsp1Cas9, Hsp2Cas9, and CcuCas9, by performing a GFP-activation assay. Interestingly, Hsp1Cas9 and CcuCas9 recognized unique N4RAA and N4CNA PAMs, respectively. We further generated an Hsp1Cas9-Hsp2Cas9 chimeric Cas9 (Hsp1-Hsp2Cas9), which recognized a simple N4CY PAM. Genome-wide off-target analysis revealed that Hsp1-Hsp2Cas9 has very few off-targets compared to SpCas9. By analyzing the crystal structure of CjCas9, we identified eight mutations that can improve the specificity and generate a high-fidelity Hsp1-Hsp2Cas9-Y. Hsp1-Hsp2Cas9-Y enables the knockout of B4GALNT2 and CMAH in porcine fetal fibroblasts (PFFs). Moreover, we developed a high-fidelity Hsp1-Hsp2Cas9-KY which displayed undetectable off-targets revealed by GUIDE-seq at four tested loci. These natural and engineered Cas9 nucleases enabled efficient genome editing in multiple mammalian cells, expanding the DNA targeting scope.
Keywords: CRISPR/Cas9, CjCas9, chimeric Cas9, high-fidelity, compact Cas9, Hsp1-Hsp2Cas9, Compact Cas9
Graphical abstract
CjCas9 is one of the smallest Cas9 nucleases for mammalian genome editing, but it requires a long PAM, limiting DNA targeting scope. Gao et al. discovery and engineer CjCas9 orthologs for a large targeting scope and high-precision genome editing.
Introduction
Clustered regularly interspaced short palindromic repeats (CRISPR) and the accompanying Cas proteins comprise adaptive immune systems against invading viruses, plasmids, and other mobile genetic elements in many bacteria and archaea.1,2,3,4 The type II CRISPR system has been studied extensively. In this system, a CRISPR RNA (crRNA):transactivating crRNA (tracrRNA) hybrid is combined with a Cas9 nuclease to cleave invading DNA targets that contain (1) a complementary sequence with the crRNA guide (protospacer) and (2) a protospacer-adjacent motif (PAM) immediately downstream of the protospacer.5,6,7,8 The PAM allows these prokaryotic immune systems to distinguish between the invading DNA target (non-self) and the same DNA sequence encoded within CRISPR arrays (self) that produce the RNA guides.9
The CRISPR-Cas9 system has been repurposed as a powerful genome editing tool for various cell types and organisms.9,10,11,12 The PAM requirement increases CRISPR-Cas9 targeting specificity but also reduces the number of targetable sites. To broaden the targeting scope, in one strategy, the existing Cas9 tool is engineered to recognize novel PAMs. For example, the most extensively applied Streptococcus pyogenes Cas9 (SpCas9) recognizing an NGG PAM has been engineered to recognize an NG PAM13 or near any PAM14; the smaller Staphylococcus aureus Cas9 (SaCas9) recognizing an NNGRRT PAM15 has been engineered to recognize an NNNRRT PAM.16 In another strategy, multiple natural Cas9 nucleases are used for genome editing, with each nuclease recognizing a defined PAM. For example, we and others developed SaCas9 (NNGRRT PAM),15 SauriCas9 (NNGG PAM),17 SlugCas9 (NNGG PAM),18 and SchCas9 (NNGR PAM) PAMs.19 Recently, more compact RNA-guided nucleases, including Cas9d, HEARO effectors, and IscB have shown promising for genome editing and may recognize different PAMs.20,21
Type II-C Cas9 nucleases account for nearly half of the total type II Cas9s,22 but only a few of them, including NmeCas9 (N4GAYW/N4GYTT/N4GTCT PAMs),10,23 GeoCas9 (N4CNAA PAM),24 CjCas9 (N4RYAC PAM),25 Nme2Cas9 (N4CC PAM),26 BlatCas9 (N4CNAA PAM),27 PpCas9 (N4RTT PAM),28 and Nsp2Cas9,29 have been developed for genome editing. Recently, Gasiunas et al. biochemically identified diverse PAMs among type II-C Cas9 orthologs,30 suggesting that this type of Cas9s is not fully developed. CjCas9 is one of the smallest Cas9 orthologs (984 amino acids) characterized to date and exhibits higher targeting specificity than SpCas9 and SaCas9.25 However, it recognizes a long N4RYAC PAM (R = A or G; Y = C or T), limiting its targeting scope. In this study, we employed a GFP-activation assay to investigate the PAMs of three CjCas9 orthologs. Interestingly, these orthologs recognize distinct PAMs. We generated a chimeric Cas9 that recognized a simple N4CY PAM. We further engineered it to improve the specificity. These newly identified and engineered Cas9 nucleases expand the DNA targeting scope.
Results
Investigation of PAMs for three CjCas9 orthologs
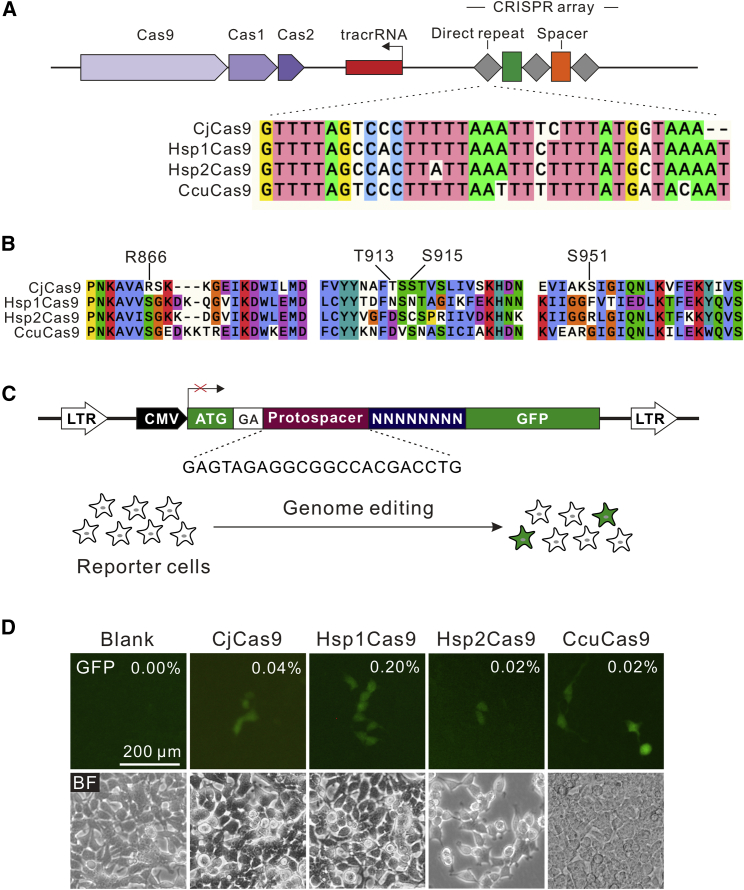
To identify new Cas9 nucleases for genome editing, we used CjCas9 as a reference and searched in UniProt for related orthologs with ∼50% amino acid identity shared with CjCas9.31 We selected three Cas9s from Campylobacter cuniculorum DSM 23162 (CcuCas9), Helicobacter sp. MIT 11-5569 (Hsp1Cas9), and Helicobacter sp. MIT 14-3879 (Hsp2Cas9) for characterization (Table 1). These orthologs have the same gene organization and moderately conserved repeat sequences (Figure 1A). Notably, these Cas9 orthologs differ in three or four residues corresponding to residue Arg866, Thr913, Ser915, or Ser951, which are crucial for PAM recognition by the CjCas9 PAM-interacting (PI) domain (Figure 1B),32 implying that these CjCas9 orthologs may recognize distinct PAMs.
Table 1.
Three CjCas9 orthologs selected from an UniProt search
| Nuclease name | Host strain | Length (aa) | Identity to CjCas9 (%) | UniProt ID |
|---|---|---|---|---|
| Hsp1Cas9 | Helicobacter sp. MIT 11-5569 | 1,057 | 530 | A0A4U8SFT5 |
| Hsp2Cas9 | Helicobacter sp. MIT 14-3879 | 1,067 | 48.9 | A0A3D8IIS5 |
| CcuCas9 | Campylobacter cuniculorum DSM 23162 | 1,032 | 53.8 | A0A1W6BVC1 |
Figure 1.
A GFP-activation assay for the test of Cas9 activity
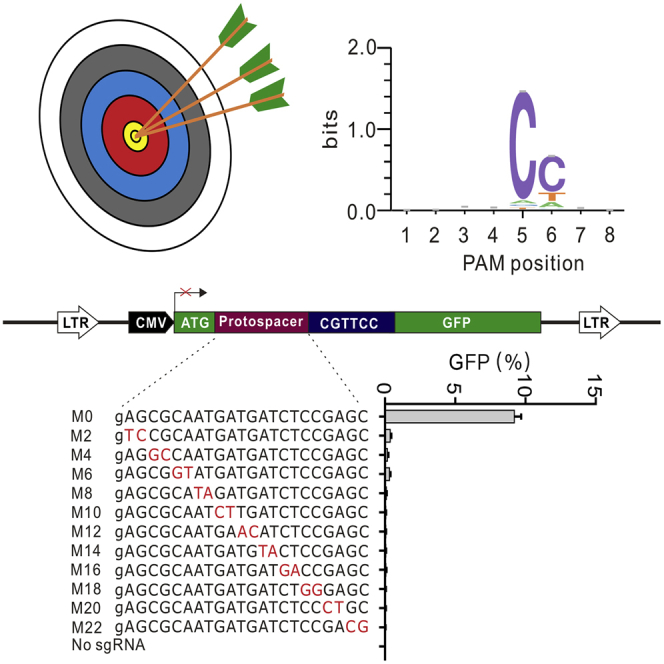
(A) CjCas9 ortholog gene organization. The repeat sequences are shown below. (B) PI domain sequence alignment of CjCas9 orthologs. Amino acids crucial for PAM recognition are shown above. (C) Schematic diagram of the GFP-activation assay. A lentiviral vector contains a CMV-driven GFP, which is disrupted by a protospacer followed by an 8 bp random sequence between the ATG start codon and the GFP coding sequence. The reporter library is stably integrated into HEK293T cells. Genome editing induces in-frame mutations in a portion of cells, resulting in GFP expression. (D) Transfection of CjCas9 orthologs and sgRNAs (CjCas9-sgRNA scaffold) resulted in GFP expression. The proportion of GFP-positive cells is shown. Blank, the reporter cells without Cas9 transfection. BF, bright field; GFP, green fluorescent protein.
We next used a previously developed GFP-activation assay17 to test these ortholog activities. In this approach, a protospacer with an 8 bp random downstream sequence is inserted between the ATG start codon and GFP coding sequence, which interrupts GFP expression. The reporter gene is integrated into the HEK293T cells. If a Cas9 enables genome editing, it will generate insertions or deletions (indels) and induce GFP expression in a portion of cells (Figure 1C). We synthesized each human codon-optimized CjCas9 ortholog and cloned it into a mammalian expression plasmid construct. The CjCas9 single guide RNA (sgRNA) scaffold with a 22 bp guide sequence that is optimal for CjCas9 was expressed in a separate plasmid.25 CjCas9 was used as a positive control. Three days after transfection of each Cas9 ortholog with an sgRNA, GFP-positive cells could be observed for all three orthologs (Figure 1D), demonstrating that these orthologs were active in mammalian cells.
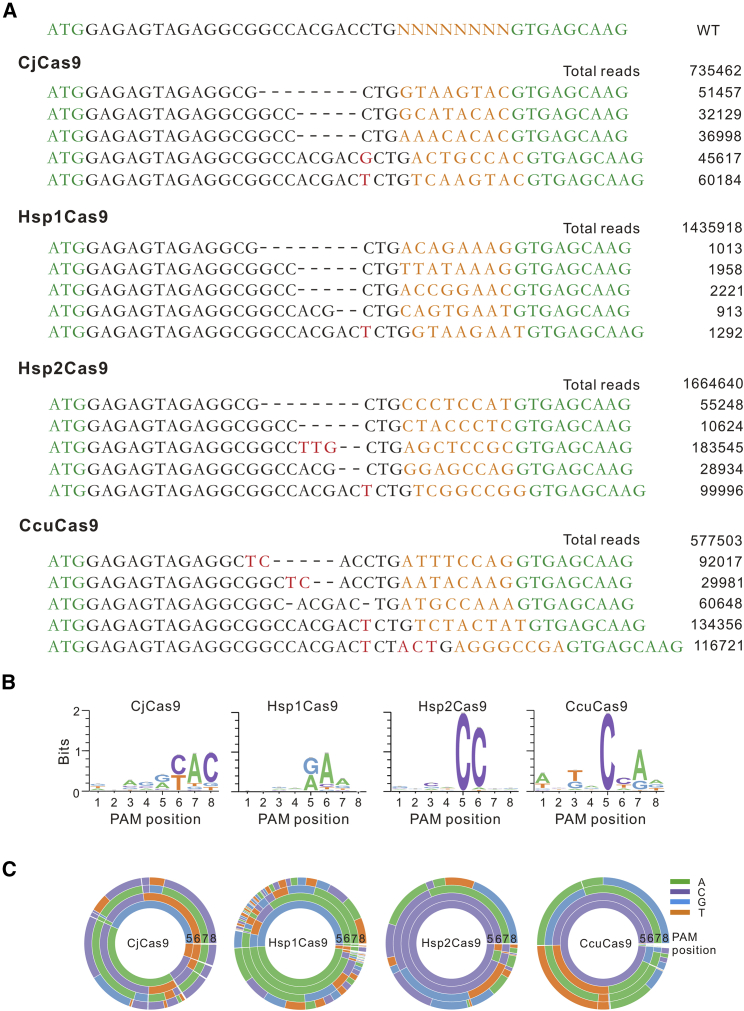
Next, the GFP-positive cells were sorted by flow cytometry, and the target sequence was PCR amplified and used for deep sequencing. The sequencing analysis revealed that these orthologs had indeed generated indels at the target sites (Figure 2A). The WebLogo and a PAM wheel showed that CjCas9 recognized an N4RYAC PAM, consistent with a previous study (Figures 2B and 2C).25 Hsp2Cas9 recognized an N4CC PAM. Interestingly, Hsp1Cas9 and CcuCas9 recognized N4RAA and N4CNA PAMs that are distinct from previous Cas9 nucleases for mammalian genome editing (Figures 2B and 2C).
Figure 2.
PAM sequence analysis for CjCas9 orthologs
(A) Representative sequences of targets after genome editing analyzed by deep sequencing. The GFP-coding sequence is shown in green; 8 bp random sequences are shown in orange; deleted bases are shown in black dashes; and insertion mutations are shown in red. The number of reads for each type of indel is listed on the right. (B) WebLogo analysis revealed consensus PAMs for four CjCas9 orthologs based on deep sequencing data. (C) PAM wheels of four CjCas9 orthologs. The numbers represent the PAM positions.
CjCas9 orthologs enable genome editing at endogenous loci
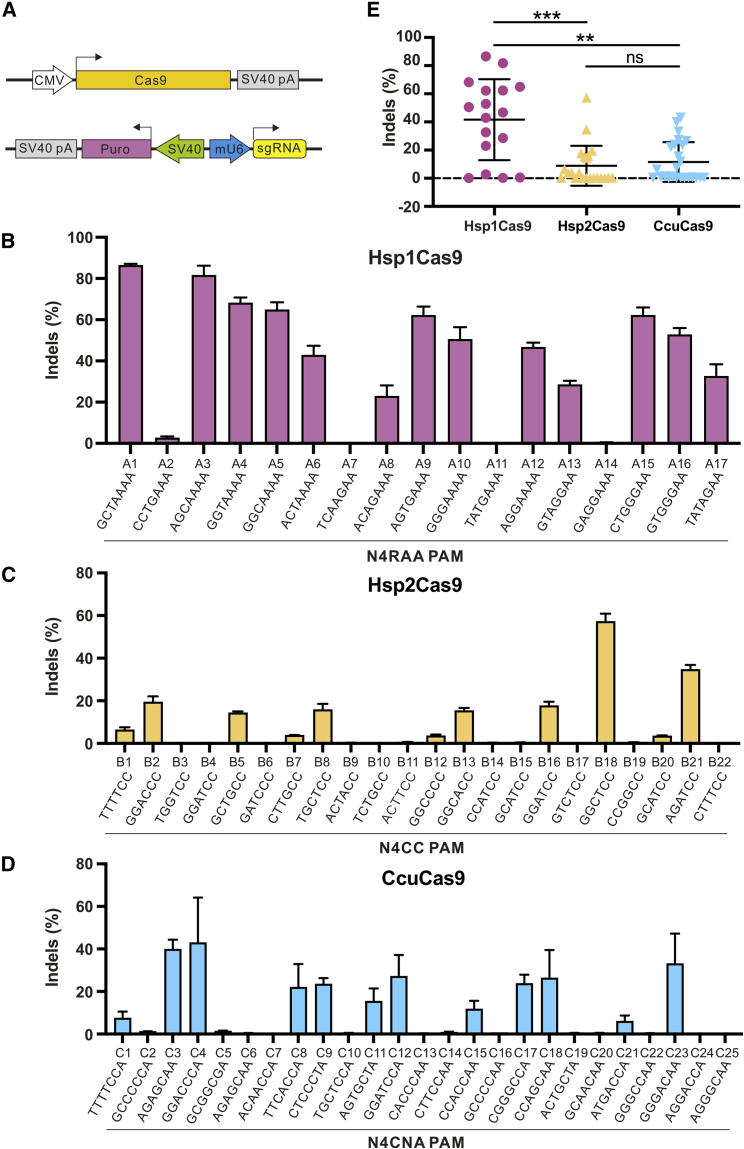
We next tested the editing ability of these orthologs at endogenous loci. We selected a panel of endogenous target sites with PAMs corresponding to each ortholog. Cas9 and sgRNA (CjCas9-sgRNA scaffold) expression plasmids were cotransfected into HEK293T cells, followed by puromycin selection (Figure 3A). Seven days after transfection, the cells were harvested, and genomic DNA was extracted for targeted deep sequencing. All three Cas9 nucleases induced indels at the respective sites with varying efficiencies (Figures 3B–3D). Hsp1Cas9 displayed higher efficiency than Hsp2Cas9 and CcuCas9 (Figure 3E).
Figure 3.
Genome editing capability of CjCas9 orthologs
(A) Schematic of Cas9 and sgRNA (CjCas9-sgRNA scaffold) expression plasmid constructs. pA, polyA; Puro, puromycin resistant gene; mU6, mouse U6 promoter. (B–D) CjCas9 orthologs enable genome editing at endogenous loci in HEK293T cells. Cells were treated with puromycin. Indel frequencies were quantified by targeted deep sequencing. The PAM sequences are shown below. R = A or G. The data represent the mean ± SD; n = 3. (E) Quantification of indel efficiencies. One-way ANOVA. ns, not significant; ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
To assess whether these Cas9 nucleases enable genome editing guided by their sgRNA scaffold, we identified direct repeats and tracrRNAs and designed an sgRNA scaffold for each ortholog by fusing the 3′ end of a direct repeat with the 5′ end of the respective tracrRNA via a 4-nt linker (Figure S1A). The phylogenetic tree revealed that the CjCas9 sgRNA scaffold sequence is closer to Hsp1Cas9 sgRNA scaffold sequence, followed by the CcuCas9 and Hsp2Cas9 sgRNA scaffold sequences (Figure S1B). Each ortholog was transfected with its own sgRNA scaffold into HEK293T cells. The CjCas9 sgRNA scaffold was used as a control. The Hsp1Cas9 sgRNA scaffold displayed comparable activity to the CjCas9 sgRNA scaffold (Figure S1C). The Hsp2Cas9 sgRNA scaffold did not work, probably due to a 3-nt deletion occurring (Figure S1D). CcuCas9 sgRNA scaffold displayed higher activity than the CjCas9 sgRNA scaffold, demonstrating that the CjCas9 sgRNA scaffold was not the optimized scaffold for CcuCas9 (Figure S1E). These data revealed that CjCas9 orthologs enabled mammalian genome editing.
Analysis of CjCas9 ortholog specificity
Next, we evaluated the specificity of these three orthologs by employing the GFP-activation approach.17 Notably, a fixed PAM was used in this assay. We generated a panel of sgRNAs with dinucleotide mutations to detect the specificity of each ortholog, and an on-target sgRNA was used as a control (Figures S2A–S2C). The activity of each sgRNA was analyzed by the percentage of GFP-positive cells. All three Cas9 nucleases showed robust off-target activity with mismatches at PAM-distal regions. Hsp1Cas9 tolerated dinucleotide mismatches at positions 1–18, with the PAM covering positions 23–29. Hsp2Cas9 tolerated dinucleotide mismatches at positions 1–16, with the PAM covering positions 23–28. CcuCas9 tolerated dinucleotide mismatches at positions 1–16, with the PAM covering positions 23–29. These data demonstrated that these three CjCas9 orthologs displayed low specificity.
Generation of chimeric Cas9 nucleases useful for genome editing
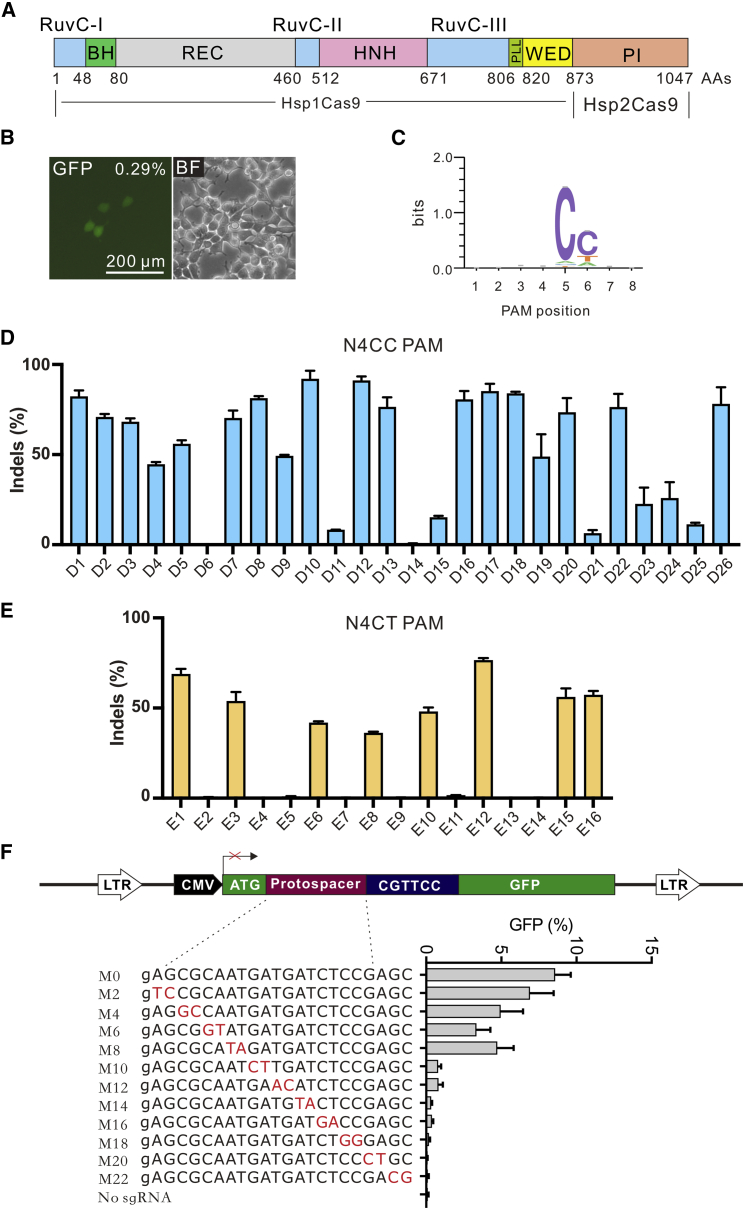
The PI domain of a Cas9 nuclease is critical for PAM recognition.33 Swapping the PI domain between closely related Cas9 nucleases may generate a chimeric nuclease that possesses both positive Cas9 characteristics.17,26,33 Since Hsp1Cas9 displayed high activity, we replaced the Hsp1Cas9 PI domain with the Hsp2Cas9 PI domain, generating Hsp1-Hsp2Cas9 (Figure 4A). The GFP-activation assay revealed that the Hsp1-Hsp2Cas9 was active in mammalian cells (Figure 4B). Deep sequencing revealed that Hsp1-Hsp2Cas9 strongly preferred an N4CC PAM, followed by an N4CT PAM (Figure 4C). We selected a panel of 26 endogenous loci with N4CC PAM across AAVS1, EMX1, VEGFA, and GRIN2B loci often tested for genome editing. Hsp1-Hsp2Cas9 could efficiently generate indels at most sites (Figure 4D). We further selected a panel of 16 endogenous loci with N4CT PAMs. Hsp1-Hsp2Cas9 could generate indels at eight sites (Figure 4E). We used the GFP-activation approach to evaluate the specificity of Hsp1-Hsp2Cas9 by using a panel of mismatched sgRNAs (CjCas9-sgRNA scaffold; Figure 4F). The results showed that Hsp1-Hsp2Cas9 generated substantial off-target cleavage at the PAM-distal region. We also replaced the Hsp1Cas9 PI domain with the CcuCas9 PI domain, generating Hsp1-CcuCas9 (Figure S3A). Hsp1-CcuCas9 induced very few GFP-positive cells in the GFP-activation assay and required an N4CNA PAM (Figures S3B and S3C). In summary, Hsp1-Hsp2Cas9 enabled mammalian genome editing and required an N4CY PAM.
Figure 4.
Characterization of chimeric Hsp1-Hsp2Cas9
(A) Schematic diagram of chimeric Hsp1-Hsp2Cas9 nuclease. The Hsp1Cas9 PI domain was replaced with the Hsp2Cas9 PI domain. (B) GFP-activation assay revealed that Hsp1-Hsp2Cas9 induced GFP expression. CjCas9-sgRNA scaffold was used. The proportion of GFP-positive cells is shown. BF, bright field; GFP, green fluorescent protein. (C) WebLogo for Hsp1-Hsp2Cas9 is generated based on deep sequencing data. (D) Hsp1-Hsp2Cas9 enables genome editing at endogenous loci with the N4CC PAM. CjCas9-sgRNA scaffold was used. Cells were treated with puromycin. Indel efficiencies were determined by targeted deep sequencing. (E) Hsp1-Hsp2Cas9 enables genome editing at endogenous loci with the N4CT PAM. CjCas9-sgRNA scaffold was used. Cells were treated with puromycin. Indel efficiencies were determined by targeted deep sequencing. (F) Hsp1-Hsp2Cas9 specificity was evaluated by the GFP-activation assay. A panel of sgRNAs with dinucleotide mutations (red bases) is shown below. CjCas9-sgRNA scaffold is used. An additional G at the 5′ terminal is added for U6 promoter transcription. The data represent the mean ± SD; n = 3.
Engineering Hsp1-Hsp2Cas9 for high specificity
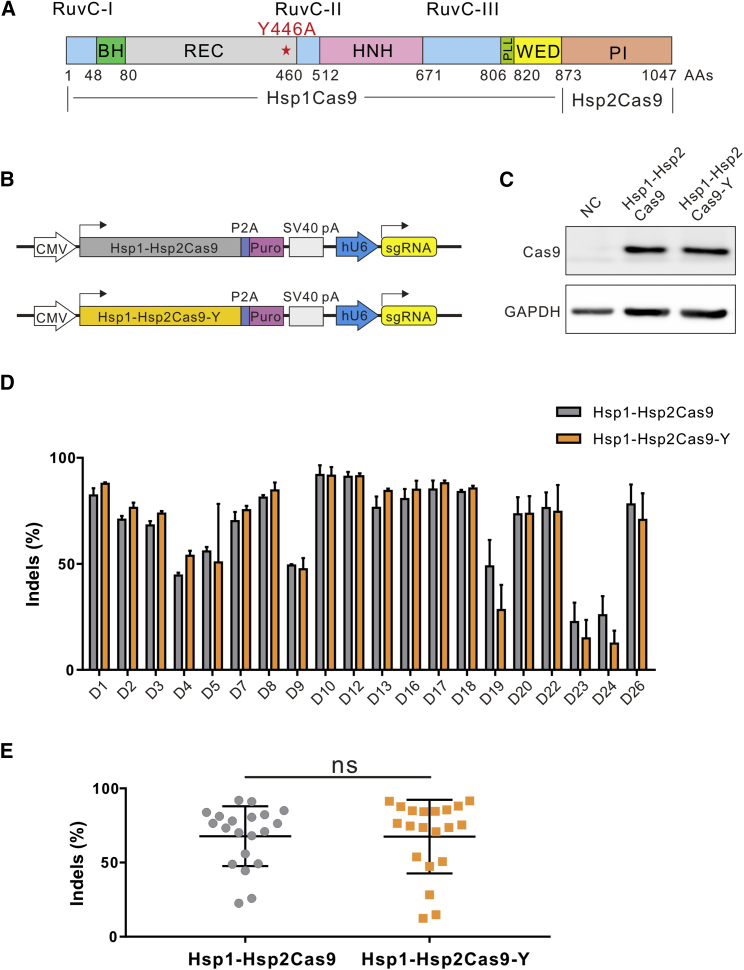
Previous studies have shown that Cas9 specificity can be improved through the modification of amino acid residues that form hydrogen bonds at the target DNA-sgRNA interface.34,35,36 To enhance the specificity of Hsp1-Hsp2Cas9, we first analyzed the crystal structure of CjCas9 and identified nine amino acid residues that potentially form hydrogen bonds at the target DNA-sgRNA interface32 (Figures S4A and S4B). We then aligned Hsp1-Hsp2Cas9 to CjCas9 and identified the corresponding amino acid residues (R269, N292, N295, K390, Q419, T420, R445, Y446, and Q679) that potentially form hydrogen bonds at the target DNA-sgRNA interface (Figure S4C). We replaced individual residues with alanine and tested the specificity of the nuclease. The GFP-activation assay revealed that all mutations except Q419A could improve nuclease specificity, and Y446A improved the most (Figures S5A and S5B). We focused on the Y446A variant, which we named Hsp1-Hsp2Cas9-Y (Figure 5A) in the following study.
Figure 5.
Genome editing capability of Hsp1-Hsp2Cas9 and Hsp1-Hsp2Cas9-Y
(A) Schematic diagram of chimeric Hsp1-Hsp2Cas9-Y nuclease. It contains a Y446A mutation. (B) Schematic of the expressing plasmid constructs of Cas9 nuclease and sgRNA with CjCas9-sgRNA scaffold. pA, polyA; Puro, puromycin resistant gene; hU6, human U6 promoter. (C) Protein expression levels of Hsp1-Hsp2Cas9 and Hsp1-Hsp2Cas9-Y were determined by western blot analysis. NC, negative control, HEK293T cells without Cas9 transfection; Hsp1-Hsp2, Hsp1-Hsp2Cas9; Y446A, Hsp1-Hsp2Cas9-Y. (D) Comparison of Hsp1-Hsp2Cas9 and Hsp1-Hsp2Cas9-Y genome editing activity at 20 endogenous sites with the N4CC PAM in HEK293T cells. Cells were treated with puromycin. The data represent the mean ± SD; n = 3. (E) Quantification of the editing efficiencies of Hsp1-Hsp2Cas9 and Hsp1-Hsp2Cas9-Y. ns, not significant. Indel efficiencies were determined by targeted deep sequencing. The data represent the mean ± SD.
Besides, we tested whether different sgRNA scaffolds could influence Cas9 specificity. We transfected the Hsp1-Hsp2Cas9-Y plasmid and mismatched sgRNA plasmids with the CjCas9, Hsp1Cas9, or CcuCas9 sgRNA scaffold into the GFP-activation reporter cells. The results showed that scaffolds did not influence Cas9 specificity (Figures S6A–S6D).
To further evaluate Cas9 specificity, we performed the genome-wide unbiased identification of double-stranded breaks enabled by sequencing (GUIDE-seq)37 to evaluate Cas9 specificity in HEK293T cells. We selected four sgRNAs targeting the AAVS1 loci. These target sites contained NGGNCC PAM, which can be recognized by SpCas9, Hsp1-Hsp2Cas9, and Hsp1-Hsp2Cas9-Y. GUIDE-seq analysis showed that robust on-target cleavage occurred for all Cas9 nucleases (Figure S7). We detected 28 off targets for SpCas9, three off targets for Hsp1-Hsp2Cas9, and one off target for Hsp1-Hsp2Cas9-Y. These data indicated that the Y446A variation improved Hsp1-Hsp2Cas9 specificity.
Hsp1-Hsp2Cas9-Y enables genome editing at endogenous loci
Next, we employed Hsp1-Hsp2Cas9 and Hsp1-Hsp2Cas9-Y for genome editing at endogenous loci in HEK293T cells. The Hsp1-Hsp2Cas9 and Hsp1-Hsp2Cas9-Y were cloned into identical plasmid constructs for a side-by-side comparison (Figure 5B). Western blot analysis revealed that their protein expression levels were comparable (Figure 5C). We selected 20 endogenous target sites with the N4CC PAM. Targeted deep sequencing analysis revealed that Hsp1-Hsp2Cas9 and Hsp1-Hsp2Cas9-Y exhibited similar editing efficiencies (Figures 5D and 5E).
To better understand the activity of Hsp1-Hsp2Cas9-Y, we compared the activity of Hsp1-Hsp2Cas9-Y to the near-PAMless SpRY.38 We cloned SpRY to the Hsp1-Hsp2Cas9-Y expression construct (Figure S8A), and similar gene expression levels were detected by qRT-PCR (Figure S8B). We tested the activities of these nucleases at 10 endogenous loci with N4CY PAMs. Hsp1-Hsp2Cas9-Y could generate indels (>5%) at five loci with activity higher than SpRY at three loci (Figure S8C). In contrast, SpRY could generate indels at eight loci. Overall, Hsp1-Hsp2Cas9-Y displayed lower activity than SpRY (Figure S8D). Taken together, Hsp1-Hsp2Cas9-Y offered a site-specific supplement to existing Cas9 nucleases for mammalian genome editing.
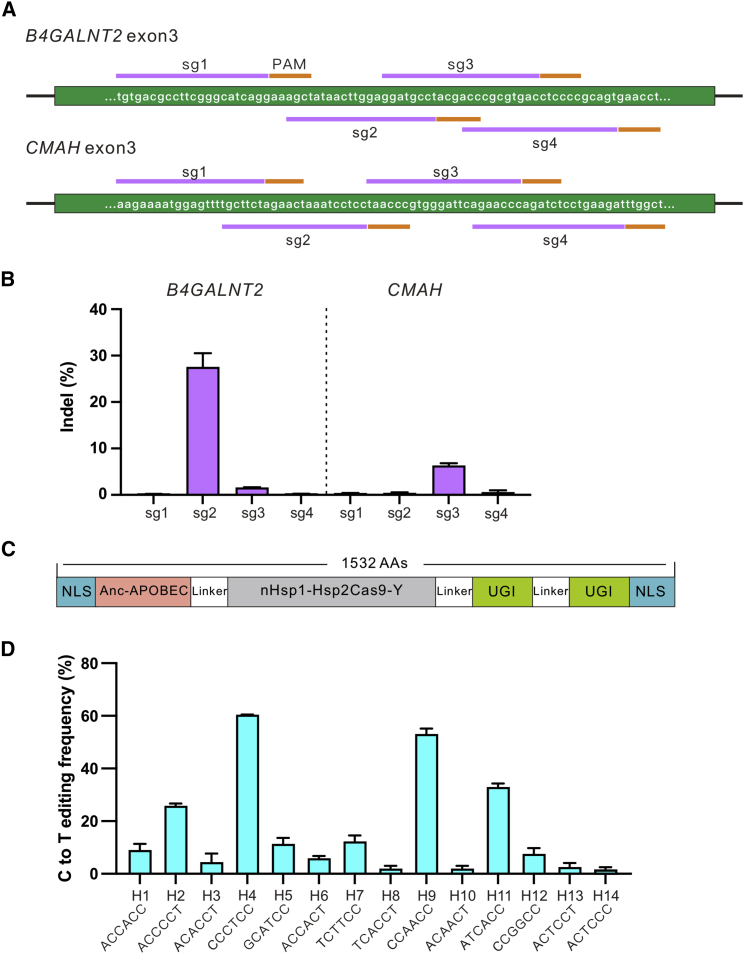
We also assessed the genome editing capability of Hsp1-Hsp2Cas9-Y in other cell types, including HeLa, SH-SY5Y, C33A, and N2a (mouse neuroblastoma cell line) cells. Indels were detected in all these cell types with varying efficiencies (Figures S9A–S9D). Next, we used Hsp1-Hsp2Cas9-Y to knock out B4GalNT2 and CMAH genes in porcine fetal fibroblasts (PFFs). These two genes have been reported to induce hyperacute rejection of the transplanted organ due to natural antibodies.39 We designed four sgRNAs for each gene (Figure 6A). One sgRNA achieved 27.57% indel efficiency for B4GalNT2, and one sgRNA achieved 6.32% indel efficiency for CMAH (Figure 6B).
Figure 6.
Practical applications of Hsp1-Hsp2Cas9-Y
(A) sgRNA design for the knockout of B4GALNT2 exon3 and CMAH exon3 in PFFs. (B) Hsp1-Hsp2Cas9-Y enables the knockout of B4GALNT2 exon3 and CMAH exon3. CjCas9-sgRNA scaffold was used. Cells were treated with puromycin. Indel efficiencies were determined by targeted deep sequencing. The data represent the mean ± SD; n = 3. (C) Schematic of Hsp1-Hsp2Cas9-YCBE. (D) Hsp1-Hsp2Cas9-YCBE enables the C-to-T conversion in HEK293T. CjCas9-sgRNA scaffold was used. Cells were treated with puromycin. C-to-T editing efficiencies were determined by targeted deep sequencing. The data represent the mean ± SD; n = 3.
Next, we used Hsp1-Hsp2Cas9-Y to generate a cytidine base editor (Hsp1-Hsp2Cas9-Y-CBE) and tested its activity at 14 endogenous sites (Figure 6C). C-to-T conversions (>5%) were observed at nine sites with both N4CC PAM and N4CY PAM (Figure 6D). Therefore, Hsp1-Hsp2Cas9-Y-CBE offered an alternative platform for base editing.
Hsp1-Hsp2Cas9-KY enables highly specific genome editing at endogenous loci
To test whether double mutations have a combined effect in improving specificity, we introduced the K390A mutation into Hsp1-Hsp2Cas9-Y, resulting in a Cas9 nuclease that we named Hsp1-Hsp2Cas9-KY (Figure S10A). The GFP-activation assay revealed that the specificity of Hsp1-Hsp2Cas9-KY was further improved (Figure S10B). Genome-wide specificity was analyzed by GUIDE-seq, but no off targets were detected at four loci (Figure S10C). To test the genome editing capability of Hsp1-Hsp2Cas9-KY, we selected 20 endogenous target sites with the N4CC PAM. Similar gene expression levels were detected by qRT-PCR (Figure S10D). Hsp1-Hsp2Cas9-KY generated indels at these loci with similar or lower efficiencies to Hsp1-Hsp2Cas9 (Figure S10E).
Next, we tested the activity of Hsp1-Hsp2Cas9-Y and Hsp1-Hsp2Cas9-KY with different sgRNA scaffolds (CjCas9 scaffold, Hsp1Cas9 scaffold, and CcuCas9 scaffold) at three endogenous loci. Targeted deep sequencing results revealed that these three sgRNA scaffolds displayed comparable activity for both nucleases (Figures S11A and S11B).
To further test whether other mutation combinations could result in high activity and specificity, we introduced K390A and R269A mutations into Hsp1-Hsp2Cas9, resulting in a Cas9 nuclease that we named Hsp1-Hsp2Cas9-KR (Figure S12A). We compared the activity of Hsp1-Hsp2Cas9-KR to Hsp1-Hsp2Cas9. Similar gene expression levels were detected by qRT-PCR (Figures S12B and S12C). We selected a panel of 20 endogenous loci in HEK293T cells. Targeted deep sequencing results revealed that Hsp1-Hsp2Cas9-KR displayed much lower activity than Hsp1-Hsp2Cas9 (Figures S12D and S12E). Therefore, we did not further test Hsp1-Hsp2Cas9-KR specificity.
Discussion
Researchers have put much effort into expanding the Cas9 targeting scope. One strategy is to mine natural Cas9 orthologs for altered PAM recognition. For example, we and others have characterized several type II-A Cas9 orthologs, such as SpCas9,7 SaCas9,15 St1Cas9,40 SauriCas9,17 and SlugCas9,18 for use in mammalian genome editing. However, type II-A Cas9 orthologs strongly prefer purine-rich PAMs. Alternatively, researchers developed type II-C orthologs, including NmeCas9,41 Nme2Cas9,26 CjCas9,25 and BlatCas9,27 for mammalian genome editing. In this study, we developed three type II-C CjCas9 orthologs for mammalian genome editing. Two of them recognized unique N4RAA and N4CNA PAMs. We further generated a chimeric Cas9 nuclease recognizing an N4CY PAM. These natural and engineered orthologs further expand the DNA targeting scope.
Off-target effects are major concerns when CRISPR-Cas9 is used for biomedical and clinical applications. Off-target mutations may induce genomic instability and disrupt the functionality of otherwise normal genes.42 Rational design, where structure-guided protein engineering is used to modify amino acid residues in close contact with the target DNA strand or the non-target DNA strand, is a rapid strategy to improve specificity. Others and we have previously used rational design to improve the specificity of several type II-A Cas9, such as SpCas9,9 SaCas9,15 and SlugCas9.18 In this study, we demonstrated that rational design is also valuable for improving type II-C Cas9 specificity. Genome-wide unbiased analysis revealed that Hsp1-Hsp2Cas9-Y and Hsp1-Hsp2Cas9-KY displayed minimal or undetectable off targets. Current rational design often cannot keep the high specificity and activity of a Cas9 nuclease. Molecular-directed evolution allows simultaneous evaluation of on- and off-target activity,43 offering a strategy for the future engineering of Cas9 nucleases. The Cas9 nucleases developed in this study together with previous Cas9 nucleases form a large Cas9 toolbox. We summarized these Cas9 properties so that users can select a suitable nuclease for their specific applications (Table S1).
Materials and methods
Cell culture
HEK293T, HeLa, SH-SY5Y, C33A, and N2a cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Gibco) and 1% penicillin-streptomycin (Gibco). All cell lines were cultured in a 37°C incubator with 5% CO2.
Plasmid DNA constructs
The DNA sequences of plasmids used in this study are listed in Table S2. The amino acid substitutions were generated by standard PCR. The human codon-optimized Cas9 genes are listed in Table S3. The target sequences are listed in Table S4. The primer sequences are listed in Table S5.
Construction of GFP-activation system
We synthesized a lentiviral plasmid library in which a random 8 bp sequence was inserted between the translation initiation codon (ATG) followed by a target DNA (GAGTAGAGGCGGCCACGACCTG) and GFP coding sequence to prevent GFP expression. The plasmid library was packed into the lentivirus, and the titration of the lentivirus library was detected with qPCR. HEK293T was infected at a multiplicity of infection (MOI) ≤1. The cells were selected with puromycin. Then, the GFP-positive cells induced by mutations were removed by MoFlo XDP machine. The GFP-activation cell library was cultured in 10 cm dishes to keep the integrity of the library.
PAM identification using GFP-activation assay
For PAM sequence screening, the PAM library cells were transfected with Cas9 plasmid (10 μg) and sgRNA plasmid (5 μg) using Lipofectamine 2000 (Invitrogen). Five days after transfection, the GFP-positive cells were sorted with a MoFlo XDP flow cytometer (Beckman Coulter). Genomic DNA was isolated using a TIANamp genomic DNA kit (TIANGEN), and target sites were amplified by two rounds of nested PCR to add the Illumina adaptor sequence. The PCR products were purified with a QIAquick gel extraction kit (QIAGEN) and used for deep sequencing.
Twenty base-pair sequences (AAGCCTTGTTTGCCACCATG/GTGAGCAAGGGCGAGGAGCT) flanking the target sequence (GAGAGTAGAGGCGGCCACGACCTGNNNNNNNN) were used to fix the target sequence. CTG and GTGAGCAAGGGCGAGGAGCT were used to fix 8 bp random sequences. Only target sequences with in-frame mutations were used in the PAM analysis. The random sequence was extracted and visualized by WebLogo44 and PAM wheel.45
Genome editing at endogenous sites
HEK293T and HeLa cells used in endogenous site-editing detection were seeded into 48-well plates (Thermo Fisher Scientific) and transfected with Cas9 plasmid (300 ng) and sgRNA plasmid (200 ng) using Lipofectamine 2000. Puromycin (InvivoGen) was added at a final concentration of 1 μg/mL 1 day after transfection. SH-SY5Y or C33A cells were seeded into 24-well plates (Thermo Fisher Scientific) and transfected with a single plasmid encoding both Cas9 and sgRNA (800 ng) using Lipofectamine 3000 (Invitrogen). Puromycin was added at a final concentration of 0.5 μg/mL 1 day after transfection. N2a cells were seeded into 24-well plates and transfected with Cas9 plasmid (300 ng) and sgRNA plasmid (200 ng) using Lipofectamine 2000. Puromycin was added at a final concentration of 2 μg/mL 1 day after transfection. All cells were harvested 7 days after transfection, and genomic DNA was extracted using QuickExtract DNA extraction solution (Epicentre). The target sites were amplified by two rounds of nested PCR to add the Illumina adaptor sequence. The PCR products were purified by a QIAquick gel extraction kit and used for deep sequencing.
Test of Cas9 specificity
To test the specificities of Hsp1Cas9, Hsp2Cas9, and CcuCas9, we generated GFP reporter cell lines with GCTAAAA PAM, GGATCC PAM, and AGAGCAA PAM. To test the specificity of Hsp1-Hsp2Cas9 and its variants, we generated a GFP reporter cell line with CGTTCC PAM. GFP reporter cell lines were seeded into 48-well plates and transfected with Cas9 plasmid (300 ng) and sgRNA plasmid (200 ng) using Lipofectamine 2000. The cells were trypsinized 5 days after transfection and centrifuged at 900 RPM for 4 min. The supernatant was removed, and the cells were resuspended in phosphate-buffered saline (PBS) (WISENT). Flow cytometry was performed with a FACSCalibur flow cytometer (Becton Dickinson). The data analysis was performed using FlowJo software.
GUIDE-seq
GUIDE-seq experiments were performed as described previously,37 with minor modifications. Briefly, 2 × 105 HEK293T cells were transfected with a single plasmid encoding both Cas9 and sgRNA (1 μg), along with 100 pmol annealed GUIDE-seq double-stranded oligodeoxynucleotides (dsODNs) by electroporation and then seeded into 6-well plates (Thermo Fisher Scientific). The electroporation voltage, width, and number of pulses were 1,150 V, 20 ms, and two pulses, respectively. Genomic DNA was extracted with the DNeasy Blood and Tissue kit (QIAGEN) 5–7 days after transfection according to the manufacturer’s protocol. Restriction fragment-length polymorphism (RFLP) assays were used to assess oligo tag integration rates as previously described.14 The genome library was prepared and subjected to deep sequencing.
Western blot analysis
HEK293T cells were seeded into 24-well plates and transfected with Cas9-expressing plasmid (800 ng) using Lipofectamine 2000 (Invitrogen). Three days after transfection, the cells were harvested and lysed by NP-40 buffer (Beyotime) in the presence of 1 mM phenylmethanesulfonyl fluoride (PMSF) (Beyotime). The cell samples were centrifuged at 12,000 RPM for 10 min at 4°C, and the supernatant was mixed with loading buffer and then boiled at 95°C for 10 min. Cas9 was detected with an anti-HA antibody (1: 1,000; Abcam), and GAPDH was detected with an anti-GAPDH antibody (1: 1,000; Cell Signaling Technology). A secondary goat anti-rabbit antibody (1: 3,000; Cell Signaling Techology) was used for signal visualization of chemiluminescence. A Tanon 5200 chemiluminescent imaging system (Tanon, Shanghai, CHina) was used for the immunoblot analysis.
qRT-PCR
Total RNAs were extracted using TRIzol reagent (Invitrogen), and reverse transcription was performed using RT SuperMix for qPCR (APExBIO). qPCR was performed to measure the expression of Cas proteins relative to GAPDH expression using 2× SYBR Green qPCR Master Mix (APExBIO).
Statistical analysis
All data are presented as the mean ± SD. Statistical analysis was conducted using GraphPad Prism 7. Student’s t test or one-way analysis of variance (ANOVA) was used to determine statistical significance between two or more groups, respectively. A value of p <0.05 was considered to be statistically significant (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
Acknowledgments
This work was supported by grants from the National Key Research and Development Program of China (2021YFC2701103 and 2021YFA0910602); the National Natural Science Foundation of China (82070258, 31700571, and 82270313); Open Research Fund of State Key Laboratory of Genetic Engineering of Fudan University (no. SKLGE-2104); Science and Technology Research Program of Shanghai (19DZ2282100); the Natural Science Fund of Shanghai Science and Technology Commission (19ZR1406300); and Shanghai 2022 “Science and Technology Innovation Action Plan” Medical Innovation Research Project (22Y11909300).
Author contributions
S.G., Y.W., T.Q., J.W., Z.H., and J.L. performed experiments; Y.W. and T.Q. analyzed the data; S.S., H.L., and Y.W. provided experimental guidance; Y.W. designed experiments and wrote the manuscript.
Declaration of interests
Fudan University has filed a patent application based on this work.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2023.01.029.
Contributor Information
Shuna Sun, Email: sun_shuna@fudan.edu.cn.
Huihui Liu, Email: liuhuihui800@caf.ac.cn.
Yongming Wang, Email: ymw@fudan.edu.cn.
Supplemental information
Data availability
All data needed to evaluate the conclusions in the paper are presented in Table S6. The information on raw sequencing data is listed in Table S7. The raw sequencing data have been submitted to the NCBI Sequence Read Archive (SRA PRJNA: 917081 (https://www.ncbi.nlm.nih.gov/bioproject/917081)).
References
- 1.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2.Marraffini L.A., Sontheimer E.J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sorek R., Lawrence C.M., Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- 4.Barrangou R., Marraffini L.A. CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol. Cell. 2014;54:234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garneau J.E., Dupuis M.È., Villion M., Romero D.A., Barrangou R., Boyaval P., Fremaux C., Horvath P., Magadán A.H., Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 6.Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A., Eckert M.R., Vogel J., Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mojica F.J.M., Díez-Villaseñor C., García-Martínez J., Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology (Reading) 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 9.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie Y., Wang D., Lan F., Wei G., Ni T., Chai R., Liu D., Hu S., Li M., Li D., et al. An episomal vector-based CRISPR/Cas9 system for highly efficient gene knockout in human pluripotent stem cells. Sci. Rep. 2017;7:2320. doi: 10.1038/s41598-017-02456-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang B., Wang Z., Wang D., Zhang B., Ong S.G., Li M., Yu W., Wang Y. krCRISPR: an easy and efficient strategy for generating conditional knockout of essential genes in cells. J. Biol. Eng. 2019;13:35. doi: 10.1186/s13036-019-0150-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishimasu H., Shi X., Ishiguro S., Gao L., Hirano S., Okazaki S., Noda T., Abudayyeh O.O., Gootenberg J.S., Mori H., et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science. 2018;361:1259–1262. doi: 10.1126/science.aas9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z., Gonzales A.P.W., Li Z., Peterson R.T., Yeh J.R.J., et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S., et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleinstiver B.P., Prew M.S., Tsai S.Q., Nguyen N.T., Topkar V.V., Zheng Z., Joung J.K. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat. Biotechnol. 2015;33:1293–1298. doi: 10.1038/nbt.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu Z., Wang S., Zhang C., Gao N., Li M., Wang D., Wang D., Liu D., Liu H., Ong S.G., et al. A compact Cas9 ortholog from Staphylococcus Auricularis (SauriCas9) expands the DNA targeting scope. Plos Biol. 2020;18:e3000686. doi: 10.1371/journal.pbio.3000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Z., Zhang C., Wang S., Gao S., Wei J., Li M., Hou L., Mao H., Wei Y., Qi T., et al. Discovery and engineering of small SlugCas9 with broad targeting range and high specificity and activity. Nucleic Acids Res. 2021;49:4008–4019. doi: 10.1093/nar/gkab148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang S., Mao H., Hou L., Hu Z., Wang Y., Qi T., Tao C., Yang Y., Zhang C., Li M., et al. Compact SchCas9 recognizes the simple NNGR PAM. Adv. Sci. 2022;9:e2104789. doi: 10.1002/advs.202104789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altae-Tran H., Kannan S., Demircioglu F.E., Oshiro R., Nety S.P., McKay L.J., Dlakić M., Inskeep W.P., Makarova K.S., Macrae R.K., et al. The widespread IS200/IS605 transposon family encodes diverse programmable RNA-guided endonucleases. Science. 2021;374:57–65. doi: 10.1126/science.abj6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aliaga Goltsman D.S., Alexander L.M., Lin J.L., Fregoso Ocampo R., Freeman B., Lamothe R.C., Perez Rivas A., Temoche-Diaz M.M., Chadha S., Nordenfelt N., et al. Compact Cas9d and HEARO enzymes for genome editing discovered from uncultivated microbes. Nat. Commun. 2022;13:7602. doi: 10.1038/s41467-022-35257-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shmakov S., Smargon A., Scott D., Cox D., Pyzocha N., Yan W., Abudayyeh O.O., Gootenberg J.S., Makarova K.S., Wolf Y.I., et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol. 2017;15:169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hou Z., Zhang Y., Propson N.E., Howden S.E., Chu L.F., Sontheimer E.J., Thomson J.A. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc. Natl. Acad. Sci. USA. 2013;110:15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrington L.B., Paez-Espino D., Staahl B.T., Chen J.S., Ma E., Kyrpides N.C., Doudna J.A. A thermostable Cas9 with increased lifetime in human plasma. Nat. Commun. 2017;8:1424. doi: 10.1038/s41467-017-01408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim E., Koo T., Park S.W., Kim D., Kim K., Cho H.Y., Song D.W., Lee K.J., Jung M.H., Kim S., et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017;8:14500. doi: 10.1038/ncomms14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edraki A., Mir A., Ibraheim R., Gainetdinov I., Yoon Y., Song C.Q., Cao Y., Gallant J., Xue W., Rivera-Pérez J.A., Sontheimer E.J. A compact, high-accuracy Cas9 with a dinucleotide PAM for in vivo genome editing. Mol. Cell. 2019;73:714–726.e4. doi: 10.1016/j.molcel.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao N., Zhang C., Hu Z., Li M., Wei J., Wang Y., Liu H. Characterization of brevibacillus laterosporus Cas9 (BlatCas9) for mammalian genome editing. Front. Cell Dev. Biol. 2020;8:583164. doi: 10.3389/fcell.2020.583164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fedorova I., Vasileva A., Selkova P., Abramova M., Arseniev A., Pobegalov G., Kazalov M., Musharova O., Goryanin I., Artamonova D., et al. PpCas9 from Pasteurella pneumotropica - a compact Type II-C Cas9 ortholog active in human cells. Nucleic Acids Res. 2020;48:12297–12309. doi: 10.1093/nar/gkaa998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei J., Hou L., Liu J., Wang Z., Gao S., Qi T., Gao S., Sun S., Wang Y. Closely related type II-C Cas9 orthologs recognize diverse PAMs. Elife. 2022;11:e77825. doi: 10.7554/eLife.77825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gasiunas G., Young J.K., Karvelis T., Kazlauskas D., Urbaitis T., Jasnauskaite M., Grusyte M.M., Paulraj S., Wang P.H., Hou Z., et al. A catalogue of biochemically diverse CRISPR-Cas9 orthologs. Nat. Commun. 2020;11:5512. doi: 10.1038/s41467-020-19344-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamada M., Watanabe Y., Gootenberg J.S., Hirano H., Ran F.A., Nakane T., Ishitani R., Zhang F., Nishimasu H., Nureki O. Crystal structure of the minimal Cas9 from Campylobacter jejuni reveals the molecular diversity in the CRISPR-cas9 systems. Mol. Cell. 2017;65:1109–1121.e3. doi: 10.1016/j.molcel.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 33.Nishimasu H., Ran F.A., Hsu P.D., Konermann S., Shehata S.I., Dohmae N., Ishitani R., Zhang F., Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen J.S., Dagdas Y.S., Kleinstiver B.P., Welch M.M., Sousa A.A., Harrington L.B., Sternberg S.H., Joung J.K., Yildiz A., Doudna J.A. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550:407–410. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan Y., Chu A.H.Y., Bao S., Hoang D.A., Kebede F.T., Xiong W., Ji M., Shi J., Zheng Z. Rationally engineered Staphylococcus aureus Cas9 nucleases with high genome-wide specificity. Proc. Natl. Acad. Sci. USA. 2019;116:20969–20976. doi: 10.1073/pnas.1906843116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai S.Q., Zheng Z., Nguyen N.T., Liebers M., Topkar V.V., Thapar V., Wyvekens N., Khayter C., Iafrate A.J., Le L.P., et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walton R.T., Christie K.A., Whittaker M.N., Kleinstiver B.P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science. 2020;368:290–296. doi: 10.1126/science.aba8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martens G.R., Reyes L.M., Li P., Butler J.R., Ladowski J.M., Estrada J.L., Sidner R.A., Eckhoff D.E., Tector M., Tector A.J. Humoral reactivity of renal transplant-waitlisted patients to cells from GGTA1/CMAH/B4GalNT2, and SLA class I knockout pigs. Transplantation. 2017;101:e86–e92. doi: 10.1097/TP.0000000000001646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agudelo D., Carter S., Velimirovic M., Duringer A., Rivest J.F., Levesque S., Loehr J., Mouchiroud M., Cyr D., Waters P.J., et al. Versatile and robust genome editing with Streptococcus thermophilus CRISPR1-Cas9. Genome Res. 2020;30:107–117. doi: 10.1101/gr.255414.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo Y., Wei X., Das J., Grimson A., Lipkin S.M., Clark A.G., Yu H. Dissecting disease inheritance modes in a three-dimensional protein network challenges the "Guilt-by-Association" principle. Am. J. Hum. Genet. 2013;93:78–89. doi: 10.1016/j.ajhg.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Höijer I., Emmanouilidou A., Östlund R., van Schendel R., Bozorgpana S., Tijsterman M., Feuk L., Gyllensten U., den Hoed M., Ameur A. CRISPR-Cas9 induces large structural variants at on-target and off-target sites in vivo that segregate across generations. Nat. Commun. 2022;13:627. doi: 10.1038/s41467-022-28244-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Casini A., Olivieri M., Petris G., Montagna C., Reginato G., Maule G., Lorenzin F., Prandi D., Romanel A., Demichelis F., et al. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat. Biotechnol. 2018;36:265–271. doi: 10.1038/nbt.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leenay R.T., Maksimchuk K.R., Slotkowski R.A., Agrawal R.N., Gomaa A.A., Briner A.E., Barrangou R., Beisel C.L. Identifying and visualizing functional PAM diversity across CRISPR-cas systems. Mol. Cell. 2016;62:137–147. doi: 10.1016/j.molcel.2016.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are presented in Table S6. The information on raw sequencing data is listed in Table S7. The raw sequencing data have been submitted to the NCBI Sequence Read Archive (SRA PRJNA: 917081 (https://www.ncbi.nlm.nih.gov/bioproject/917081)).