Abstract
We have determined the kinetics of up-regulation of the homologous recombination gene RAD51, one of the genes induced following DNA damage in isogenic haploid DNA repair-deficient mutants of Saccharomyces cerevisiae, using treatment with the DNA crosslinking agent 8-methoxypsoralen. We show that RAD51 is up-regulated concomitantly, although independently, with a shift from the G1 cell cycle phase to G2/M arrest. This up-regulation is absent in homologous recombination repair-deficient mutants and increased in mutants deficient in nucleotide excision repair and polζ-dependent translesion synthesis. We demonstrate that the Rad53-dependent DNA damage signal transduction cascade is active in RAD51 non-inducing mutants. However, when independently eliminated, it too abolishes RAD51 up-regulation. We present a model in which RAD51 up-regulation requires two signals: one depending on the Rad53-dependent DNA damage signal transduction cascade and the other on homologous recombination repair.
INTRODUCTION
Cellular responses to DNA damage in eukaryotic cells include the arrest of DNA synthesis and cell cycle progression, the expression of DNA damage-inducible genes and the repair of DNA damage (1,2). These responses are coordinated by the Rad53-dependent DNA damage signal transduction cascade. Numerous components of this signal transduction cascade have been identified, including 3-phosphoinositol kinases (e.g. Mec1 and Tel1) and their targets (e.g. Dun1, Chk1, Tof1 and Rad53) (3,4), as well as factors such as Rad9, Rad17, Mec3 and Rad24, (5–7).
The DNA damage-inducible gene RAD51, a homolog of RecA in Escherichia coli, plays an essential role in homologous recombination and double-strand break (DSB) repair by promoting strand exchange (8,9). RAD51 is up-regulated by UV and by ionizing radiation, alkylating agents and psoralens (8,10–12). Little is known about the sensors of damage or about how these DNA damage-responsive genes, such as RAD51, are induced. Sequence analyses of some inducible genes, including RAD51, have identified several families of related upstream regulatory sequences (12–15), and factors (activators and suppressors) that are associated with some of these elements have been found (16,17).
The up-regulation of DNA repair genes that share damage-responsive elements (DREs) with RAD51 has been characterized in two ways: (i) by changing the upstream regulatory sequences or (ii) by changing or deleting a component of the Rad53-dependent DNA damage signal transduction cascade, such as Rad9 (5) or Rad53 (18). Changes in RAD51 up-regulation have also been noted when transcription proteins, such as Fcp1 (RNA pol II TAF) (19), Upf (mRNA decay) (20) and MAP kinases (21), were disrupted.
We used the crosslinking agent 8-methoxypsoralen (8-MOP) as the DNA-damaging agent since it has been shown to be a very powerful inducer of RAD51 (11). In the presence of UVA, 8-MOP forms highly stable DNA monoadducts and interstrand crosslinks at pyrimidine sites (22,23). These present obstacles to both the transcription and replication machineries and must be repaired or bypassed for the cell to survive 8-MOP damage (reviewed in 24).
It has been suggested that only members of this signal transduction cascade and its putative transcription factor targets can influence regulation of a gene such as RAD51. We asked whether the process of DNA repair influences transcriptional regulation of a DNA damage-inducible gene such as RAD51. We show here that this is indeed the case. We demonstrate that in the absence of homologous recombination repair, up-regulation of RAD51 is eliminated. Furthermore, nucleotide excision repair (NER) and polζ-dependent translesion synthesis (TLS)-deficient mutants overexpress RAD51. We also show that up-regulation of RAD51 corresponds to, but is not dependent on, a shift from the G1 to the G2/M cell phase via a delayed S phase. Our results are consistent with a model in which the processing of 8-MOP photolesions via homologous recombination repair acts as a novel signal for RAD51 up-regulation that appears to be equally important as the Rad53-dependent cascade.
MATERIALS AND METHODS
Yeast strains
We used the haploid wild-type and deletion mutant strains listed in Table 1. Strain CA-Δrad14 and the rad14rad50 and rad14rev3 double mutants carrying the rad14Δ::URA2 cassette (see Table 1) were constructed by transforming either the wild-type haploid strain BY4741 or the respective single mutant with a rad14Δ::URA2 disruption cassette. The rad18 mutant was in the FY1679-11C background (we checked that the BY4741 and FY1679-11C wild-type strains exhibit identical sensitivity to 8-MOP plus UVA). All strains were obtained from EUROSCARF (Heidelberg).
Table 1. Saccharomyces cerevisiae strains used.
Cell culture and α factor synchronization
Haploid cells were pre-cultured three times in minimal medium (Difco) corresponding to the requirements of the strain (25). Cells were grown overnight to exponential phase (0.7–1.0 × 107 cells/ml) in YEPD (Difco). To obtain cells synchronized in G1 phase, cells were incubated with α factor (Sigma) at 5 µg/ml for 2 h at 30°C.
8-MOP plus UVA treatment, cell survival and canavanine assays
Cells were treated as described previously (7). Briefly, cells were washed in water and suspended at 5 × 106 cells/ml. 8-MOP (Sigma) was added to a final concentration of 5 µM and the cells were incubated in the dark at room temperature for 30 min. The cells were irradiated at 365 nm using a HPW125 UVA lamp at 15 J/m2 (Philips) fitted with a water filter. At given times, cells were then spun at 4000 r.p.m. for 10 min in an Eppendorf 5810R centrifuge and resuspended at their original concentration in fresh YEPD, incubated at 30°C and shaken at 200 r.p.m. Aliquots were taken for flow cytometry analysis, survival studies and canavanine mutation assays.
Survival was determined as previously described (26). The canavanine mutation assay was carried out as follows. Cultures were grown to stationary phase for 48 h in YEPD. Samples at 2.5 × 107 cells/ml were irradiated and plated at 107 cells/canavanine plate and 2.5 × 102 cells/YEPD plate. Canavanine plates contained minimal medium (Difco) without arginine and with canavanine (Merck) at a final concentration of 60 µg/ml (27).
Flow-assisted cell sorting (FACS)
Between 0.35 and 0.5 × 107 cells were harvested and treated as described previously (28). Cells were sonicated to avoid clumping, as checked by light microscopy. Analysis was carried out using a FACSCalibur apparatus (Becton-Dickinson). Samples were analyzed using the CellQuest software. Each histogram corresponds to 20 000 cells.
Microscopy
To verify cell cycle changes, cell aliquots were taken for staining with 4,6-diamino-2-phenylindole (DAPI) (29). Five microliters of DAPI staining solution were added to 5 µl of cells on a glass slide. Samples were examined using a Zeiss microscope (Axiophot) fitted with a MicroMax CCD camera (Princeton Technologies) and analyzed using IP-Lab software (Scanalytics Inc.).
RNA extraction and northern blotting
Total RNA was extracted from 0.5–1.0 × 108 cells using the FastRNA-Red kit (Bio101) according to the manufacturer’s instructions. Gel loading buffer [48% deionized formamide, 17% formaldehyde (37% stock solution), 1× 3(N-morpholonol)propanesulfonic acid (MOPS) (10× MOPS = 0.4 M MOPS, 0.1 M sodium acetate, 10 mM EDTA, pH 7.2), 12% water, 6% glycerol, 5% bromophenol blue] was added to RNA samples at 20% final volume and samples were incubated at 65°C for 15 min and then on ice for 5 min. Samples were loaded on a 1% agarose gel in 18% formaldehyde (37% stock solution) and run at 5 V/cm in 1× MOPS buffer. All reagents were from Sigma-Aldrich. Gels were washed for 3 × 20 min with water, followed by a brief soak in 20× SSC (3 M NaCl, 0.3 M sodium citrate, pH 7.2) and the RNA was transferred to a GeneScreen Plus nylon membrane (NEN) using a VacuGene vacuum transfer apparatus (Amersham Pharmacia Biotech) according to the manufacturer’s instructions. RNA was crosslinked to the membrane using a UV Stratalinker (Stratagene).
The RAD51 DNA probe (335 bp) was generated by performing a ClaI + BstXI double digestion of pTZ51 (F.Fabre, CEA). The ACT1 probe (587 bp) was generated by HindIII + XbaI digestion of pSK-ACT (M.Vedel, Institut Curie).
Twenty-five nanograms of DNA probe were labeled using the ReadyPrime system (Amersham Pharmacia Biotech) with 5 µCi [α-32P]dCTP as per instructions and unincorporated nucleotides were removed using a NICK column system (Amersham Pharmacia Biotech). Labeled probes were added to blots already pre-hybridized at 42°C with UltraHyb buffer (Ambion) and incubated at 42°C for 4–16 h. Blots were washed twice with 2× SSC, 0.1% SDS for 5 min at 42°C and three times with 0.1× SSC, 0.1% SDS at 42°C. Blots were scanned using a PhosphorImager system (Molecular Dynamics) and analyzed using Image Quant v.5.1 software (Molecular Dynamics).
We measured up-regulation of RAD51 as follows. Values corresponding to RAD51 transcript levels were divided by those corresponding to actin levels (ACT1) for each time point. The ratio calculated for the treated sample was compared to the ratio for the untreated sample for each time point. Levels at time 0 were taken to be equal to 1.
Detection of Rad53 phosphorylation
Total cell extracts were prepared as described above and run on SDS–PAGE (stacking, 4% at acrylamide:bisacrylamide 29:1; separating, 10% at 30:0.4) and then semi-dry blotted onto nitrocellulose. An anti-Rad53 goat antibody (yC-19; Santa Cruz Biotechnology) was incubated at 1:1000 dilution with the membrane in 0.1% Tween, 5% (w/v) milk in phosphate-buffered saline, followed by rinsing and incubation with anti-goat horseradish peroxidase-conjugated antibody (Santa Cruz Biotechnology) at 1:5000 dilution. Proteins were detected by chemiluminescence (Renaissance western blotting reagent; NEN).
RESULTS
Determination of the sensitivity to 8-MOP plus UVA of isogenic repair-deficient mutants
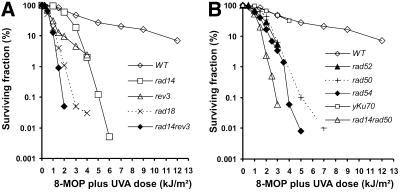
In order to characterize the role of DNA repair in up-regulation of RAD51 we first had to establish which repair pathways were important for repair of 8-MOP photolesions. Survival studies were carried out on haploid, isogenic deletion mutants deficient in DNA repair. The 8-MOP-sensitive mutants included NER-deficient rad14, homologous recombination repair-deficient rad50, rad52 and rad54, post-replication repair-deficient rad18 and polζ-dependent translesion synthesis-deficient rev3 (see Table 1 and Fig. 1A and B). We also included non-homologous end joining (NHEJ)-deficient (30) yku70, as a non-sensitive mutant. The rad14rad50 and rad14rev3 double mutants were more sensitive compared with their corresponding single mutants. The deletion mutants rad30 (an ‘error-free’, polη-dependent translesion synthesis-deficient mutant), ogg1 (base excision repair-deficient), top1 (topoisomerase 1) and sgs1 and srs2 (helicases) were found not to be sensitive to 8-MOP plus UVA and were not tested in further assays.
Figure 1.
Survival data for isogenic DNA repair-deficient mutants following 8-MOP plus UVA treatment. Cells were grown and treated as described in Materials and Methods.
RAD51 up-regulation is not dependent on the damage itself or on the cell cycle checkpoints but is triggered by repair
We characterized the up-regulation of RAD51 following 8-MOP treatment in repair-deficient mutants. We also studied changes in the cell cycle to understand the relationship between these two phenomena.
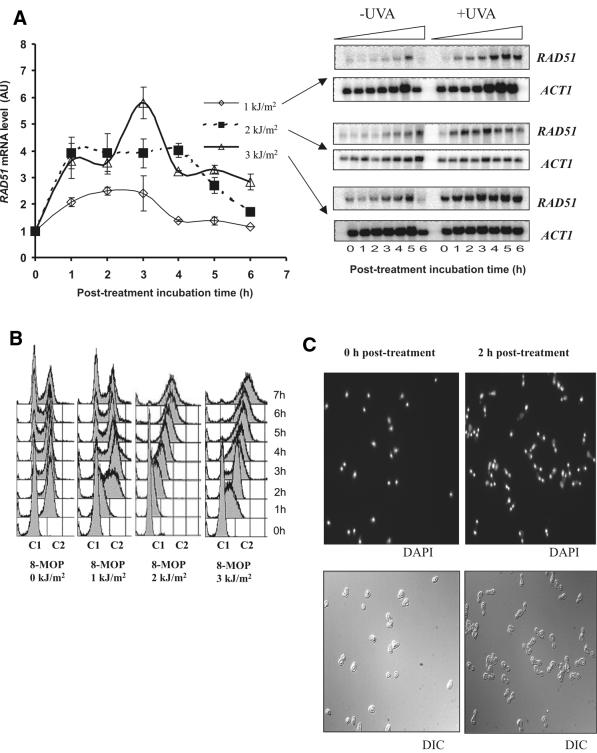
RAD51 up-regulation in the wild-type. Figure 2 shows that in G1 synchronized haploid wild-type cells, RAD51 mRNA transcript levels increased immediately after treatment in a dose-dependent manner, peaking between 2 and 3 h post-treatment and at levels of between 2.5- and 6-fold depending on the UVA dose used, then gradually dropping to basal levels (Fig. 2A). FACS analysis showed that in the untreated control population, S phase was completed at 1 h post α-factor release and a new G1 phase commenced at 2 h post-release (Fig. 2B). In contrast, treated cells shifted to the G2/M cell phase at a slower rate and cells were still predominantly in S phase at 1 h post-treatment, suggesting activation of the S phase checkpoint. The delay in exit from the subsequent G2/M phase was proportional to the dose used (compare 5 h post-treatment). High levels of RAD51 mRNA following treatment corresponded to exit from G1 via an elongated S phase and, in the case of the highest dose, entry into G2/M. Return to basal levels of RAD51 coincided with resumption of a normal cell cycle, i.e. exit from G2/M phase with the appearance of a new G1 peak (∼4 h for the 1 kJ/m2 dose and >7 h for the 3 kJ/m2 dose). Fluorescence microscopy using DAPI staining showed that at 2 h post-treatment the majority of cells partitioned their nuclei and increased dramatically in size, thus suggesting a G2/M checkpoint arrest (Fig. 2C).
Figure 2.
Responses to 8-MOP damage in G1 synchronized wild-type cells. (A) Northern blot analysis of RAD51 mRNA levels following 8-MOP plus three different UVA doses (1, 2 and 3 kJ/m2). Induction was normalized using actin (ACT1) transcript levels and is given in arbitrary units (see Materials and Methods). (B) Cell cycle changes following damage as monitored by FACS analysis. C1 denotes single content (equivalent to haploid G1) and C2 denotes double content (equivalent to haploid G2). (C) Interferential (DIC) and fluorescence (DAPI staining) light microscopy of treated cells at times 0 and 2 h post-treatment (1 kJ/m2).
In control experiments we verified that UVA exposure alone at 5 kJ/m2 and 8-MOP treatment alone did not contribute to the up-regulation of RAD51 (11).
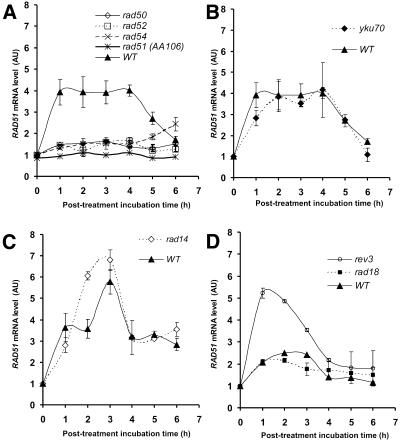
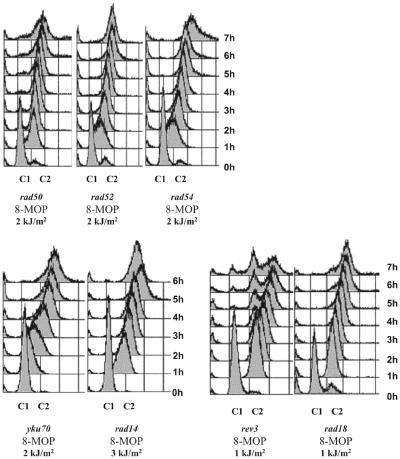
RAD51 up-regulation in homologous recombination repair-deficient mutants. In contrast to the wild-type, all three homologous recombination deficient-mutants, rad50, rad52 and rad54, exhibited no significant up-regulation of RAD51 (Fig. 3A). Area integration analysis showed that up-regulation in the rad54 mutant amounted to less than one-third of the wild-type (30%). The figure for the rad50 and rad52 mutants was similar (24%). Despite the lack of RAD51 up-regulation, all three mutants showed a strong G2/M arrest (Fig. 4) and the rad52 and rad54 mutants exhibited a prolonged S phase (Fig. 4, at 1 h). These changes in the cell cycle were insufficient to up-regulate RAD51 to wild-type levels. The possible role of Rad51 protein in its own up-regulation was tested. The recombination-deficient RAD51 point mutant rad51-E221K lacking ATP-binding activity (31) showed only 13% of the wild-type up-regulation (Fig. 3A). We conclude that RAD51 up-regulation is prevented in the homologous recombination repair-deficient mutants and that the shift from G1 phase to the eventual G2/M phase does not trigger up-regulation of RAD51 but is only a concomitant event.
Figure 3.
Up-regulation of RAD51 in G1 synchronized DNA repair-deficient mutants following 8-MOP plus UVA treatment. (A) Homologous recombination-deficient rad50, rad52, rad54 and rad51-E221K (strain AA106) mutants (2 kJ/m2). (B) NHEJ-deficient yku70 mutant (2 kJ/m2). (C) NER-deficient rad14 mutant (3 kJ/m2). (D) Polζ-dependent TLS-deficient rev3 and post-replication repair-deficient rad18 mutants (1 kJ/m2).
Figure 4.
Cell cycle changes following 8-MOP damage. FACS profiles of G1 synchronized DNA repair-deficient mutants following treatment with 8-MOP plus UVA at different UVA doses (indicated). In all cases, the FACS profile for the untreated sample was identical to the wild-type (see Fig. 2B). C1 denotes single content (equivalent to haploid G1) and C2 denotes double content (equivalent to haploid G2).
RAD51 is up-regulated in the NHEJ-deficient mutant as in the wild-type. The repair of 8-MOP photolesions involves the repair of DSBs (32–34). DSBs can be repaired by either homologous recombination or NHEJ. As shown, the yku70 mutant is as resistant as the wild-type (Fig.1). Nevertheless, we checked whether it played a role in RAD51 up-regulation and found that in the absence of NHEJ RAD51 up-regulation after 8-MOP was identical to that of the wild-type (Fig. 3B). Cells delayed entering G2/M phase following treatment (Fig. 4), indicating activation of the S phase checkpoint. Interestingly, there was a longer arrest within G2/M phase than seen in wild-type cells for the same dose. These delays in the cell cycle may be due to the role of yKu70 in telomere upkeep (35).
Post-replication repair-deficient mutant rad18 exhibits wild-type RAD51 up-regulation. Of all the single mutants we tested, the rad18 post-replication repair-deficient mutant was the most sensitive to 8-MOP plus UVA (Fig. 1), demonstrating the vital (even if as yet unclear) role of post-replication repair in the processing of 8-MOP photolesions. We therefore expected rad18 to have a considerable influence on RAD51 up-regulation. Surprisingly, in spite of its high sensitivity, rad18 expressed RAD51 mRNA at wild-type levels (Fig. 3D). Curiously, the FACS analysis also showed no delay in entry into a strong G2/M arrest (Fig. 4), indicating absence or shortening of the S phase checkpoint.
The NER-deficient mutant rad14 over-induces RAD51. Since NER plays an important role in crosslink repair (Fig. 1), we characterized up-regulation of RAD51 in the NER-deficient rad14 mutant. At 2 and 3 h post-treatment with 8-MOP plus UVA at 3 kJ/m2, RAD51 was over-induced in rad14 relative to the wild-type (Fig. 3C). In particular, the level of RAD51 mRNA at 2 h post-treatment was 1.7-fold higher than the corresponding wild-type level. This up-regulation was accompanied by delayed transition into the G2/M phase followed by a long G2/M arrest (Fig. 4).
The polζ-TLS-deficient mutant rev3 over-induces RAD51. Analysis of RAD51 up-regulation in the highly sensitive polζ-dependent TLS mutant rev3 showed that RAD51 mRNA was greatly overexpressed compared with the wild-type for the same dose of 1 kJ/m2 UVA plus 8-MOP (Fig. 3D). Point-per-point, at 2 and 3 h post-treatment, overexpression was 2.5- and 1.9-fold over that seen in the wild-type, respectively. Integrating the area of the curve yielded an overexpression of 2.1-fold over that of the wild-type. Curiously, the rev3 cells consistently followed an irregular post-treatment cell cycle (Fig. 4). The majority of these cells were not viable since survival of rev3 cells at the dose used is of the order of 7%. Such cell cycle behavior was not observed by McHugh et al. (36) or Grossmann et al. (37), who investigated the cellular responses of the rev3 mutant following treatment with the crosslinking agent cisplatin. It remains to be seen if this discrepancy is due to the genotoxic agent used, the conditions of treatment or perhaps the mutant strain used.
In summary, interference with NER and polζ-dependent TLS increases RAD51 up-regulation while interference with NHEJ and post-replication repair does not.
Cumulative and dominant negative effects on RAD51 up-regulation in double mutants
These results led us to theorize that RAD51 up-regulation increased when alternative repair pathways to homologous recombination (such as NER and polζ-dependent TLS) were eliminated, thus channeling more lesions into this pathway (see Discussion).
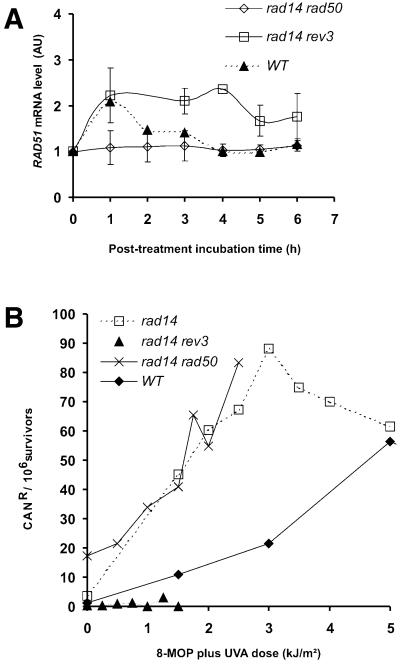
To test this hypothesis, we examined whether the effects of these pathway deletions were cumulative by investigating double mutants deficient in two different repair pathways. As compared with their respective single mutants, rad14rev3 and rad14rad50 exhibited increased sensitivity to 8-MOP plus UVA (Fig. 1). Due to their slow growth, the experiments in these mutants were carried out under asynchronous growth conditions and compared with the asynchronous wild-type (Fig. 5A). We obtained a 2.1-fold total increase in RAD51 up-regulation in rad14rev3. In contrast, the rad14rad50 mutant did not up-regulate RAD51 (nor did a rad52rev3 mutant; data not shown). FACS profiles showed a complete G2/M arrest for both strains (data not shown). These results demonstrate that the requirement for homologous recombination activity for RAD51 up-regulation is dominant over the overexpression seen in the absence of NER or polζ-dependent TLS. Furthermore, in the absence of both NER and polζ-dependent TLS (rad14rev3), up-regulation of RAD51 is further increased, compared with the respective single mutants, demonstrating the cumulative effect predicted by our hypothesis.
Figure 5.
(A) RAD51 up-regulation in rad14rev3 and rad14rad50 mutants following treatment with 8-MOP plus 1 kJ/m2 UVA. The assay was carried out on asynchronous populations. (B) Induction of CANR mutations after treatment with 8-MOP plus UVA (see Materials and Methods).
Interplay between NER, homologous recombination and TLS in repairing 8-MOP plus UVA damage is confirmed by the canavanine mutation assay
Our results indicated to us that RAD51 up-regulation depends on the passage of lesions through the homologous recombination repair pathway. Furthermore, up-regulation is positively affected under conditions that predict an increase in the number of lesions processed by this pathway and vice versa. To prove this interplay between NER, homologous recombination and polζ-dependent TLS, we determined their interaction by using the mutagenic activity of polζ-dependent TLS repair as a marker. For this purpose, we used a mutation assay that measures the increase in canavanine resistance following DNA damage (27). Our results show that both rad14 and rad14rad50 were hyper-mutagenic following treatment, in contrast to rad14rev3 (Fig. 5B). The spontaneous mutation frequency was also higher in the rad14rad50 than in any other strain tested (likely due to the accumulation of endogenous damage). These results show that in the absence of NER and homologous recombination repair, there is an increase in repair via polζ-dependent TLS, whereas in the absence of NER and polζ-dependent TLS, the cell relies on the non-mutagenic homologous recombination pathway to resolve the 8-MOP photolesions. Therefore, these results are in accordance with our hypothesis, demonstrating that lesions are channeled from one repair pathway to another depending on the availability of these pathways.
The Rad53-dependent DNA damage signal transduction cascade is necessary but not sufficient to up-regulate RAD51
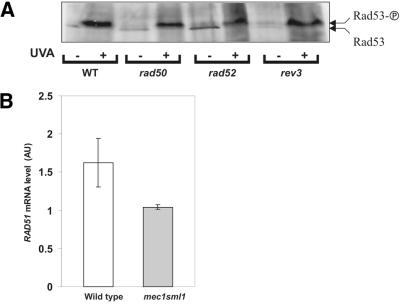
A central player in the DNA damage signal transduction cascade is Rad53 (see Introduction and Fig. 7, left), which is activated by phosphorylation and is responsible for up-regulation of DNA damage-inducible genes as well as cell cycle arrest (2). Several authors have demonstrated that this cascade is also responsible for up-regulation of RAD51 following different types of damage (5,38). To test whether the lack of RAD51 up-regulation in homologous recombination-deficient mutants was due to inactivation of the Rad53-dependent DNA damage signal transduction cascade, we examined the phosphorylated state of Rad53 by western blotting (Fig. 6A). The results clearly show that Rad53 is activated following damage even in the RAD51 non-inducing mutants rad50 and rad52. The rev3 mutant was used as an additional control and was found to be comparable to the wild-type. This result demonstrated that the absence of up-regulation in the homologous recombination-deficient mutants took place despite activation of the Rad53-dependent DNA damage signal transduction cascade.
Figure 7.
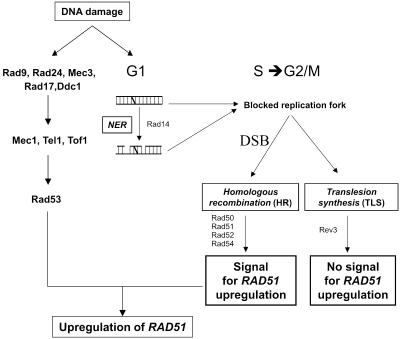
A model for up-regulation of RAD51 in 8-MOP-treated cells. The Rad53-dependent DNA damage signal transduction cascade (left) and repair events (right) leading to up-regulation of RAD51. The 8-MOP photolesion (represented as a diagonal bar between the DNA strands) is either not processed or is incompletely processed by NER, leading to a blocked replication fork resulting in a DSB followed by channeling into homologous recombination or TLS. Passage through the homologous recombination pathway generates intermediates that act as a signal for up-regulation of RAD51. The absence of homologous recombination eliminates RAD51 up-regulation but has no effect on Rad53 phosphorylation or cell cycle changes (see Discussion). Lesions processed by TLS do not generate a signal for RAD51 up-regulation. Proteins lacking in relevant mutants are marked in accordance with their respective pathways. At the same time, a signal is transmitted through the Rad53-dependent DNA damage signal transduction cascade, resulting in cell cycle arrest. This signal is necessary, but insufficient, for RAD51 up-regulation. Neither signal can compensate for the other, demonstrating that both are equally needed.
Figure 6.
(A) Phosphorylation of Rad53 following 8-MOP plus UVA damage in wild-type, rad50, rad52 and rev3 repair-deficient mutants treated with 2 kJ/m2 UVA plus 8-MOP. For the conditions of western blotting see Materials and Methods. (B) RAD51 up-regulation in the mec-sml1 mutant following DNA damage. Northern blot analysis was carried out on asynchronous cells treated with 3 kJ/m2 UVA plus 8-MOP.
We also tested whether the Rad53-dependent signal transduction cascade was necessary for RAD51 up-regulation by examining the mec1-sml1 mutant, which has an inactive Mec1 and, thus, a defective DNA damage signal transduction cascade (39). We found that RAD51 up-regulation was eliminated in this mutant. We therefore conclude that activation of the Rad53-mediated, Mec1-dependent signal transduction cascade is indeed essential for up-regulation of RAD51 but that it is not sufficient, requiring the presence of the homologous recombination pathway. This indicates an equally important, Rad53-independent but homologous recombination-dependent signal which is needed to up-regulate RAD51 following 8-MOP damage.
DISCUSSION
DNA damage leads to the induction of many genes in Saccharomyces cerevisiae, a process regulated by means of a DNA damage signal transduction cascade that is also responsible for cell cycle arrest and DNA repair (1). In recent years, numerous studies have been carried out on genomic induction following DNA damage using microarray technology (18,40,41). We focused rather on a single DNA damage-inducible gene in order to establish its precise mechanism of regulation. We chose to characterize up-regulation of the homologous recombination gene RAD51, the yeast homolog of the E.coli recombinase RecA, which has already been shown to be regulated by means of the Rad53-dependent DNA damage signal transduction cascade (5). We wished to test whether DNA repair played any role in up-regulation of RAD51, a notion that was previously not included in our understanding of DNA damage gene induction. Treatment with the DNA crosslinking agent 8-MOP plus UVA was used since it has previously been observed to be a strong up-regulator of RAD51 (11). In addition, DNA crosslinks are of interest to us since their repair involves cooperation between different repair pathways (42). Survival studies on repair-deficient isogenic mutants established that nucleotide excision repair, homologous recombination, polζ-dependent translesion synthesis (‘error prone’) and post-replication repair all play a role in the processing of 8-MOP photolesions (Fig. 1). In contrast, NHEJ, polη-dependent translesion synthesis (‘error free’) and base excision repair (BER) had no effect on survival. Similarly, elements of the replication machinery such as the topoisomerases Top1 and Top2 and the helicases Sgs1 and Srs2 exhibited survival similar to the wild-type.
Homologous recombination is essential for up-regulation of RAD51 following 8-MOP plus UVA
In G1 synchronized repair-competent wild-type cells, we observed dose-dependent up-regulation of the RAD51 gene after 8-MOP plus UVA damage (Fig. 2A). Monitoring cell cycle progression demonstrated that up-regulation of RAD51 coincided with a delayed shift from G1 to G2/M (Fig. 2B). This suggested a possible connection between the two phenomena, as has been proposed by Aboussekha et al. in the case of ionizing and UV radiation (5). However, this was not always the case in DNA repair-deficient mutants. While all the repair-deficient mutants exhibited a strong G2/M arrest (Fig. 4), the homologous recombination-deficient mutants rad50, rad52 and rad54 and the point mutant rad51-E221K, failed to up-regulate RAD51 to wild-type levels (Fig. 3A). The same was true for the homologous recombination-deficient mutants rad55 and rad57 (data not shown).
This clearly shows that passage through the cell cycle checkpoints does not act as a signal per se for up-regulation of RAD51. Our results also demonstrate that the initial damage caused by 8-MOP plus UVA (monoadducts and crosslinks) cannot be sufficient to trigger up-regulation of RAD51 since it is present in all the mutants assayed. We therefore conclude that a specific event that follows the initial damage acts as the signal for RAD51 up-regulation and that this event depends on the presence of homologous recombination.
What is the nature of the up-regulation signal for RAD51? In the current model, DNA damage is ‘sensed’ by unknown means and this signal is transmitted through the Rad53/Mec1 DNA damage signal transduction cascade (Fig. 7). We first established that this cascade was indeed activated by showing that Rad53 was present in its active, poly-phosphorylated form, even in those mutants that did not up-regulate RAD51 (Fig. 6A). We also demonstrated that in the absence of a functional signal cascade, as is the case in the mec1-sml1 mutant, up-regulation of RAD51 was eliminated (Fig. 6B). We therefore conclude that the DNA damage signal transduction cascade is essential but not sufficient for RAD51 up-regulation following 8-MOP damage. The complementary presence of the homologous recombination pathway is indispensable.
The role of recombination intermediates in providing a signal for RAD51 up-regulation
How might the cell ‘sense’ its need for Rad51? None of the repair proteins absent in the mutants assayed is believed to act as a transcription factor. It is therefore unlikely that these repair proteins act at the level of the RAD51 promoter. We propose instead that homologous recombination repair intermediates act as the signal for up-regulation of RAD51. It has previously been shown that DSBs arise when the replication machinery encounters NER intermediates (as both bacterial and mammalian models suggest) (42) or because of replication stalling (43). Treatment with 8-MOP has been shown to give rise to DSBs (32,33). These are not the result of direct damage but of lesion processing.
The repair of a DSB by homologous recombination is believed to take place in a sequential manner (24). First, the DSB is end-digested by the Rad50 complex (Rad50, Mre11 and Xrs2) with RPA binding to the exposed ssDNA. Next, RPA is replaced by Rad52, which is positioned on the extremities of the DSB (44,45). A polymer of Rad51 forms a filament around a homologous single-stranded (ss)DNA and invades the target duplex, allowing strand exchange to take place with the assistance of Rad54, Rad55 and Rad57. We have shown that interference with any of these steps, including the predicted formation of the Rad51 filament, eliminates RAD51 up-regulation. It is noteworthy that in E.coli the RecA pre-synaptic filament has been proposed to play an important role in up-regulation of the DNA damage SOS response (46).
A model describing up-regulation of RAD51 following 8-MOP damage
Our understanding of the mechanism of crosslink removal in yeast is incomplete (reviewed in 42). From the available data, one can envisage that unresolved crosslinks or monoadducts can form effective blocks to replication forks. Several mechanisms are available to ‘unblock’ a replication fork. Specialized polymerases can be recruited by the replication machinery to allow synthesis past the block, with potential mutagenic consequences (for a review see 47) or homologous recombination can be used (if a donor is available) or a combination of both. Blocked replication also gives rise to DSBs (particularly if they encounter ssDNA nicks), which can be repaired via homologous recombination or break-induced replication in a non-mutagenic manner (43,48,49).
We envisage a model (Fig. 7, right) in which the repair of 8-MOP photolesions (monoadducts and crosslinks) includes the interaction of all three pathways. Since the cells were G1 synchronized at the time of damage, lesions would be dealt with first by NER (to varying degrees of success) in a narrow time window as the cells prepare to replicate. Unrepaired or partially repaired lesions would lead to blocked replication forks and DSBs that will be processed either by homologous recombination or by translesion synthesis (or both). By using specific repair-deficient mutants, we change this repair equilibrium. Thus, more lesions lead to blocked replication forks in the NER-deficient rad14. Similarly, in the translesion synthesis deficient rev3, lesions could only be dealt with by NER and homologous recombination, whereas the opposite applies to the homologous recombination-deficient mutants (such as rad50, rad52, rad54 and rad51-E221K). If up-regulation of RAD51 is dependent on successful passage through the homologous recombination repair pathway, then we can predict an increase in up-regulation of RAD51 when more lesions are ‘channeled’ into this pathway, as seen in the rad14 and rev3 mutants as well as in the rad14rev3 mutant. It also follows that RAD51 up-regulation would be absent in the rad50, rad52 and rad54 as well as rad51-E221K mutant strains since they are unable to successfully perform homologous recombination. In line with this, up-regulation of RAD51 is also absent in rad14rad50 (Fig. 5A) and rad52rev3 (data not shown). The wild-type level of RAD51 mRNA obtained in the yku70 mutant (Fig. 3) is also in agreement with this model, since NHEJ is not involved in the repair of 8-MOP photolesions and its absence should not interfere with the kinetics of lesion processing by the three repair pathways.
The rad18 mutant is curious since it is very sensitive to 8-MOP plus UVA but shows wild-type up-regulation of RAD51. In addition, there is an absence of S phase elongation but a strong G2/M arrest. At present, the role of Rad18 (and its partner Rad6) is unclear, although it is assigned to the post-replication repair pathway. It is generally accepted that it is a ubiquitinating complex, but its targets (possibly chromatin or even polymerases) remain unknown. A possible explanation is that Rad18 might act at a later stage, i.e. after the choice between homologous recombination and translesion synthesis has been carried out, or perhaps it is needed equally by both pathways. Thus, its absence, while greatly affecting overall repair and survival, would not affect up-regulation of RAD51.
In conclusion, we have shown that up-regulation of RAD51 is not dependent on the primary damage or on the cell cycle response. Rather, it is equally dependent on the presence of a functional homologous recombination repair pathway and the Rad53/Mec1 DNA damage signal transduction cascade. It remains to be seen which specific recombination intermediates act as the trigger for RAD51 up-regulation and how the signal is transmitted to the promoter of the target gene. The possibility that RAD51 is not the only inducible DNA damage repair gene to function under such a model is intriguing.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank L. Prakash, F. Fabre, M. Brendel, W. Siede, L. Prakash, M. Vedel and A. Yasui for their kind donations of various plasmids, C. Mann and C. Leroy for donation of anti-Rad53 antibodies and S. Urbach for assistance with the protein work. Warmest thanks are due to Mrs S. Averbeck for her invaluable assistance and advice throughout the course of this work. We are very grateful to Kathleen Smith and Raymond Devoret for proofreading this manuscript. Financial support by the Institut Curie, Section de Recherche (Paris) and by the CNRS and the LNCC (Comité de l’Oise, France) is gratefully acknowledged. Y.C. is supported by a bursary from the French Ministry of Foreign Affairs (BGF).
REFERENCES
- 1.Weinert T. (1998) DNA damage checkpoints update: getting molecular. Curr. Opin. Genet. Dev., 8, 185–193. [DOI] [PubMed] [Google Scholar]
- 2.Elledge S.J. (1996) Cell cycle checkpoints: preventing an identity crisis. Science, 274, 1664–1672. [DOI] [PubMed] [Google Scholar]
- 3.Foss E.J. (2001) Tof1p regulates DNA damage responses during S phase in Saccharomyces cerevisiae. Genetics, 157, 567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanchez Y., Desany,B.A., Jones,W.J., Liu,Q., Wang,B. and Elledge,S.J. (1996) Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science, 271, 357–360. [DOI] [PubMed] [Google Scholar]
- 5.Aboussekhra A., Vialard,J.E., Morrison,D.E., de la Torre-Ruiz,M.A., Cernakova,L., Fabre,F. and Lowndes,N.F. (1996) A novel role for the budding yeast RAD9 checkpoint gene in DNA damage-dependent transcription. EMBO J., 15, 3912–3922. [PMC free article] [PubMed] [Google Scholar]
- 6.Foiani M., Pellicioli,A., Lopes,M., Lucca,C., Ferrari,M., Liberi,G., Muzi Falconi,M. and Plevani,P. (2000) DNA damage checkpoints and DNA replication controls in Saccharomyces cerevisiae. Mutat. Res., 451, 187–196. [DOI] [PubMed] [Google Scholar]
- 7.Melo J.A., Cohen,J. and Toczyski,D.P. (2001) Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev., 15, 2809–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shinohara A., Ogawa,H. and Ogawa,T. (1992) Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell, 69, 457–470. [DOI] [PubMed] [Google Scholar]
- 9.Haber J.E. (2000) Recombination: a frank view of exchanges and vice versa. Curr. Opin. Cell Biol., 12, 286–292. [DOI] [PubMed] [Google Scholar]
- 10.Aboussekhra A., Chanet,R., Adjiri,A. and Fabre,F. (1992) Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol. Cell. Biol., 12, 3224–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Averbeck D. and Averbeck,S. (1998) DNA photodamage, repair, gene induction and genotoxicity following exposures to 254 nm UV and 8-methoxypsoralen plus UVA in a eukaryotic cell system. Photochem. Photobiol., 68, 289–295. [PubMed]
- 12.Basile G., Aker,M. and Mortimer,R.K. (1992) Nucleotide sequence and transcriptional regulation of the yeast recombinational repair gene RAD51. Mol. Cell. Biol., 12, 3235–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao W., Singh,K.K., Chen,B. and Samson,L. (1993) A common element involved in transcriptional regulation of two DNA alkylation repair genes (MAG and MGT1) of Saccharomyces cerevisiae. Mol. Cell. Biol., 13, 7213–7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cole G.M., Schild,D., Lovett,S.T. and Mortimer,R.K. (1987) Regulation of RAD54- and RAD52-lacZ gene fusions in Saccharomyces cerevisiae in response to DNA damage. Mol. Cell. Biol., 7, 1078–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hurd H.K. and Roberts,J.W. (1989) Upstream regulatory sequences of the yeast RNR2 gene include a repression sequence and an activation site that binds the RAP1 protein. Mol. Cell. Biol., 9, 5359–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elledge S.J. and Davis,R.W. (1989) Identification of the DNA damage-responsive element of RNR2 and evidence that four distinct cellular factors bind it. Mol. Cell. Biol., 9, 5373–5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shim Y.S., Jang,Y.K., Lim,M.S., Lee,J.S., Seong,R.H., Hong,S.H. and Park,S.D. (2000) Rdp1, a novel zinc finger protein, regulates the DNA damage response of rhp51(+) from Schizosaccharomyces pombe. Mol. Cell. Biol., 20, 8958–8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Sanctis V., Bertozzi,C., Costanzo,G., Di Mauro,E. and Negri,R. (2001) Cell cycle arrest determines the intensity of the global transcriptional response of Saccharomyces cerevisiae to ionizing radiation. Radiat. Res., 156, 379–387. [DOI] [PubMed] [Google Scholar]
- 19.Kobor M.S., Archambault,J., Lester,W., Holstege,F.C., Gileadi,O., Jansma,D.B., Jennings,E.G., Kouyoumdjian,F., Davidson,A.R., Young,R.A. and Greenblatt,J. (1999) An unusual eukaryotic protein phosphatase required for transcription by RNA polymerase II and CTD dephosphorylation in S. cerevisiae. Mol. Cell, 4, 55–62. [DOI] [PubMed] [Google Scholar]
- 20.Lelivelt M.J. and Culbertson,M.R. (1999) Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol. Cell. Biol., 19, 6710–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts C.J., Nelson,B., Marton,M.J., Stoughton,R., Meyer,M.R., Bennett,H.A., He,Y.D., Dai,H., Walker,W.L., Hughes,T.R., Tyers,M., Boone,C. and Friend,S.H. (2000) Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science, 287, 873–880. [DOI] [PubMed] [Google Scholar]
- 22.Cimino G.D., Gamper,H.B., Isaacs,S.T. and Hearst,J.E. (1985) Psoralens as photoactive probes of nucleic acid structure and function: organic chemistry, photochemistry and biochemistry. Annu. Rev. Biochem., 54, 1151–1193. [DOI] [PubMed] [Google Scholar]
- 23.Bankmann M. and Brendel,M. (1989) Molecular dosimetry of 8-MOP + UVA-induced DNA photoadducts in Saccharomyces cerevisiae: correlation of lesions number with genotoxic potential. J. Photochem. Photobiol. B, 4, 57–74. [DOI] [PubMed] [Google Scholar]
- 24.Kupiec M. (2000) Damage-induced recombination in the yeast Saccharomyces cerevisiae. Mutat. Res., 451, 91–105. [DOI] [PubMed] [Google Scholar]
- 25.Sherman F., Fink,G.R. and Ficks,E.B. (1983) Methods in Yeast Genetics: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 26.Averbeck D. and Averbeck,S. (1994) Induction of the genes RAD54 and RNR2 by various DNA damaging agents in Saccharomyces cerevisiae. Mutat. Res., 315, 123–138. [DOI] [PubMed] [Google Scholar]
- 27.Lemontt J.F. (1977) Pathways of ultraviolet mutability in Saccharomyces cerevisiae. IV. The relation between canavanine toxicity and ultraviolet mutability to canavanine resistance. Mutat. Res., 43, 339–355. [DOI] [PubMed] [Google Scholar]
- 28.Pringle J.R., Adams,A.E., Drubin,D.G. and Haarer,B.K. (1991) Immunofluorescence methods for yeast. Methods Enzymol., 194, 565–602. [DOI] [PubMed] [Google Scholar]
- 29.Richmond K.M. and Williamson,D.H. (1983) Residual cell division measurements are unreliable as indicators of the timing of events in the Saccharomyces cerevisiae cell cycle. J. Cell Sci., 64, 307–322. [DOI] [PubMed] [Google Scholar]
- 30.Friedl A.A., Kiechle,M., Fellerhoff,B. and Eckardt-Schupp,F. (1998) Radiation-induced chromosome aberrations in Saccharomyces cerevisiae: influence of DNA repair pathways. Genetics, 148, 975–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chanet R., Heude,M., Adjiri,A., Maloisel,L. and Fabre,F. (1996) Semidominant mutations in the yeast Rad51 protein and their relationships with the Srs2 helicase. Mol. Cell. Biol., 16, 4782–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dardalhon M. and Averbeck,D. (1995) Pulsed-field gel electrophoresis analysis of the repair of psoralen plus UVA induced DNA photoadducts in Saccharomyces cerevisiae. Mutat. Res., 336, 49–60. [DOI] [PubMed] [Google Scholar]
- 33.Dardalhon M., de Massy,B., Nicolas,A. and Averbeck,D. (1998) Mitotic recombination and localized DNA double-strand breaks are induced after 8-methoxypsoralen and UVA irradiation in Saccharomyces cerevisiae. Curr. Genet., 34, 30–42. [DOI] [PubMed] [Google Scholar]
- 34.Magana-Schwencke N., Henriques,J.A., Chanet,R. and Moustacchi,E. (1982) The fate of 8-methoxypsoralen photoinduced crosslinks in nuclear and mitochondrial yeast DNA: comparison of wild-type and repair-deficient strains. Proc. Natl Acad. Sci. USA, 79, 1722–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bailey S.M., Meyne,J., Chen,D.J., Kurimasa,A., Li,G.C., Lehnert,B.E. and Goodwin,E.H. (1999) DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc. Natl Acad. Sci. USA, 96, 14899–14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McHugh P.J., Sones,W.R. and Hartley,J.A. (2000) Repair of intermediate structures produced at DNA interstrand cross-links in Saccharomyces cerevisiae. Mol. Cell. Biol., 20, 3425–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grossmann K.F., Ward,A.M. and Moses,R.E. (2000) Saccharomyces cerevisiae lacking Snm1, Rev3 or Rad51 have a normal S-phase but arrest permanently in G2 after cisplatin treatment. Mutat. Res., 461, 1–13. [DOI] [PubMed] [Google Scholar]
- 38.de la Torre Ruiz M.A. and Lowndes,N.F. (2000) DUN1 defines one branch downstream of RAD53 for transcription and DNA damage repair in Saccharomyces cerevisiae. FEBS Lett., 485, 205–206. [DOI] [PubMed] [Google Scholar]
- 39.Weinert T.A., Kiser,G.L. and Hartwell,L.H. (1994) Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev., 8, 652–665. [DOI] [PubMed] [Google Scholar]
- 40.Jelinsky S.A., Estep,P., Church,G.M. and Samson,L.D. (2000) Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol. Cell. Biol., 20, 8157–8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tusher V.G., Tibshirani,R. and Chu,G. (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA, 98, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dronkert M.L. and Kanaar,R. (2001) Repair of DNA interstrand cross-links. Mutat. Res., 486, 217–247. [DOI] [PubMed] [Google Scholar]
- 43.Michel B., Ehrlich,S.D. and Uzest,M. (1997) DNA double-strand breaks caused by replication arrest. EMBO J., 16, 430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beernink H.T. and Morrical,S.W. (1999) RMPs: recombination/replication mediator proteins. Trends Biochem. Sci., 24, 385–389. [DOI] [PubMed] [Google Scholar]
- 45.Bressan D.A., Baxter,B.K. and Petrini,J.H. (1999) The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 7681–7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sassanfar M. and Roberts,J.W. (1990) Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J. Mol. Biol., 212, 79–96. [DOI] [PubMed] [Google Scholar]
- 47.Baynton K. and Fuchs,R.P. (2000) Lesions in DNA: hurdles for polymerases. Trends Biochem. Sci., 25, 74–79. [DOI] [PubMed] [Google Scholar]
- 48.Mu D., Bessho,T., Nechev,L.V., Chen,D.J., Harris,T.M., Hearst,J.E. and Sancar,A. (2000) DNA interstrand cross-links induce futile repair synthesis in mammalian cell extracts. Mol. Cell. Biol., 20, 2446–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bierne H. and Michel,B. (1994) When replication forks stop. Mol. Microbiol., 13, 17–23. [DOI] [PubMed] [Google Scholar]