Abstract
Histone acetylation is important for regulating chromatin structure and gene expression. Three classes of mammalian histone deacetylases have been identified. Among class II, there are five known members, namely HDAC4, HDAC5, HDAC6, HDAC7 and HDAC9. Here we describe the identification and characterization of a novel class II member termed HDAC10. It is a 669 residue polypeptide with a bipartite modular structure consisting of an N-terminal Hda1p-related putative deacetylase domain and a C-terminal leucine-rich domain. HDAC10 is widely expressed in adult human tissues and cultured mammalian cells. It is enriched in the cytoplasm and this enrichment is not sensitive to leptomycin B, a specific inhibitor known to block the nuclear export of other class II members. The leucine-rich domain of HDAC10 is responsible for its cytoplasmic enrichment. Recombinant HDAC10 protein possesses histone deacetylase activity, which is sensitive to trichostatin A, a specific inhibitor for known class I and class II histone deacetylases. When tethered to a promoter, HDAC10 is able to repress transcription. Furthermore, HDAC10 interacts with HDAC3 but not with HDAC4 or HDAC6. These results indicate that HDAC10 is a novel class II histone deacetylase possessing a unique leucine-rich domain.
INTRODUCTION
In eukaryotic cells, nuclear DNA is packaged with histones and other chromosomal proteins into chromatin. A fundamental question is how chromatin structure is regulated when and where it is necessary. Various recent studies have established that such regulation is achieved through ATP-dependent chromatin remodeling and covalent chromatin modification (1). The latter includes DNA methylation and post-translational modifications of histones and other chromosomal proteins (2–5). Histones can be modified by acetylation, methylation, phosphorylation, ubiquitination and ADP-ribosylation (6). These modifications occur in an interdependent manner (7,8) and are regulated by cellular signaling pathways (9). Histone acetylation, transfer of the acetyl moiety from acetylcoenzyme A to the ɛ-amino groups of specific lysine residues located at the flexible N-terminal tails of core histones, plays important roles in modulating chromatin activity in transcription, replication, recombination and repair. Mechanistically, histone acetylation affects nucleosome stability, internucleosomal interaction and/or specific protein–protein interactions. Besides histones, non-histone proteins such as transcription factors are also subject to regulation by acetylation. Histone acetylation is catalyzed by histone acetyltransferases (HATs). Various HATs have been identified (10) and several known HATs also acetylate non-histone proteins and regulate their functions (10,11).
Histone deacetylases (HDACs) are the enzymes responsible for the removal of acetyl groups from acetylated histone and non-histone proteins (12). Since the identification of HDAC1 (13), over 10 proteins have been found to possess intrinsic deacetylase activity. Known mammalian HDACs are grouped into three classes (14–16). Class I consists of HDAC1 (13), HDAC2 (17), HDAC3 (18–20) and HDAC8 (21–23). These HDACs show significant sequence similarity to yeast Rpd3p (24). Class II members include HDAC4 (25–28), HDAC5 (26,29), HDAC6 (26,29), HDAC7 (30) and HDAC9 (31). They possess catalytic domains with significant sequence homology to the N-terminal half of yeast Hda1p (32). Class III comprises proteins with catalytic domains similar to that of the yeast NAD+-dependent deacetylase Sir2p (33). While class I deacetylases function as transcriptional co-repressors, Sir2p-related proteins appear to be involved in gene silencing.
Among class II members, HDAC4, HDAC5, HDAC7 and HDAC9 share sequence similarity in their N-terminal domains in addition to the C-terminal Hda1p-related domains (34–36). HDAC6 is unique in that it possesses tandem Hda1p-related domains and a C-terminal zinc finger motif that is able to bind ubiquitin (37). It has been clearly shown that HDAC4, HDAC5, HDAC7 and HDAC9 play important roles in transcriptional repression. When artificially tethered to promoters, they function as repressors (27,28,30,38). They interact with the MEF2 transcription factors to repress MEF2-dependent transcription (27,28,31,36,38–41), and the interaction appears to be regulated during muscle differentiation (40,42,43). Furthermore, emerging evidence suggests that some of these HDACs also interact with other transcriptional regulators, including SMRT/N-CoR (30,44), BcoR (BCL-6 interacting co-repressor) (45), CtBP (E1A C-terminal-binding protein) (46) and Evi-1 (ecotropic viral integration site 1) (47). Class II HDACs are regulated by nucleocytoplasmic trafficking (27,41,48–56).
For Rpd3p and Sir2p there are mammalian homologs with significant similarity in their entire sequences (14,33). It is very puzzling that none of the known class II HDACs contain the C-terminal half of Hda1p, raising the intriguing question whether there are real Hda1p homologs in mammals. A related question is whether there are additional class II HDACs to be identified. Here we describe the identification and characterization of HDAC10, a novel class II deacetylase that has a bipartite structure consisting of an N-terminal Hda1p-related catalytic domain and a C-terminal leucine-rich domain. HDAC10 is enriched in the cytoplasm and widely expressed in human tissues and cultured mammalian cells. Its enzymatic activity is sensitive to trichostatin A (TSA), a specific deacetylase inhibitor with known antitumor activity. When tethered to a promoter, it is able to repress transcription. It interacts with HDAC3 but not with HDAC4 or HDAC6. These results indicate that HDAC10 is a new class II deacetylase that contains an N-terminal catalytic domain and C-terminal leucine-rich domain.
MATERIALS AND METHODS
Molecular cloning
Plasmid construction and DNA sequencing were performed following standard procedures. For characterization of HDAC10 cDNA clones, PCR primers were first designed based on the genomic clone dJ402G11.7 (GenBank accession nos AL022328.7 and CAB63049). Four PCR primers were used: DAC67 (5′-AAA TTT GAA TTC CCC GAG TGC GAG ATC GAG CG-3′), DAC68 (5′-TTT AAA AAG CTT AGG TCA CAT TTG CTG CTG GAG-3′), DAC69 (5′-AAA TTT GAA TTC GCT GTG CCG ATG AGC CCC AG-3′) and DAC70 (5′-AAA TTT AAG CTT AGG AGG TGC CTC TCC ATT-3′). With DAC67/DAC68 and DAC69/DAC70, PCR fragments of expected length were amplified from cDNA mixtures isolated from λ phages propagated from human fetal bone marrow, brain and liver cDNA libraries (Clontech). PCR fragments were cloned into the EcoRI and HindIII sites of pAW48, a Flag-tagged mammalian expression vector derived from pcDNA3.1(–) (Invitrogen) by insertion of the coding sequence for the Flag epitope (Sigma) (28). DNA sequencing confirmed the identity of cloned PCR fragments. The PCR fragment amplified with primers DAC67 and DAC68 was used as a probe for screening a human fetal bone marrow cDNA library (Clontech). Sequence analyses of the resulting cDNA clones revealed that most clones possessed a 69 bp intronic insert after the first nucleotide of codon 21 of the putative HDAC10 sequence (GenBank accession no. CAB63049). This insert was not found in PCR fragments amplified from a human fetal brain cDNA library (Clontech) with primers DAC67 and DAC68, so it was deleted from a full-length cDNA clone to generate expression vectors for HDAC10. Mammalian protein expression plasmids were derived from pAW48. GFP (enhanced green fluorescent protein) constructs were based on pEGFP-C2 (Clontech). The luciferase reporter Gal4–tk–Luc contains five copies of the Gal4-binding site upstream of the minimal thymidine kinase promoter and the luciferase (Luc) coding sequence, and Gal4–SV40–Luc was constructed from pGL2-Control (Promega) by insertion of the Gal4-binding sites from Gal4–tk–Luc (28,57). Expression plasmids for the fusion proteins Gal4–HDAC10 and Gal4–nls–HDAC10 were derived from the expression vectors pM1 (58) and pM1-nls, respectively. For construction of pM1-nls, an oligonucleotide duplex, consisting of DAC71 (5′-AAT TGC CCA AGA AAA AGA GAA AGG TGG AAT TCC CGG G-3′) and DAC72 (5′-GAT CCC CGG GAA TTC CAC CTT TCT CTT TTT CTT GGG C-3′), was inserted downstream of the coding sequence for the Gal4 DNA-binding domain (residues 1–147) in pM1. The inserted sequence encodes a strong nuclear localization signal (NLS, AKKKRKVE) from the SV40 large T antigen.
Northern analysis
Multiple Tissue Northern blots (Clontech) were hybridized with an HDAC10 fragment amplified with primers DAC67 and DAC68 in ExpressHyb hybridization solution (Clontech) according to the manufacturer’s instructions. The probe was labeled with [α-32P]dCTP using the Ready-To-Go DNA labeling kit (–dCTP) (Amersham Pharmacia Biotech).
RT–PCR
Total RNA was isolated from confluent 293 and HeLa cells using TRIzol reagent (Gibco BRL). RNA quality was assessed by agarose gel electrophoresis and subsequent ethidium bromide staining. A One-step RT–PCR kit (Qiagen) was used to obtain HDAC10 cDNA fragments with primers DAC70 and DAC84 (5′-CAG GTG AAC AGT GGT ATA GC-3′). As a negative control, only primer DAC70 was used. An aliquot of 1 µg total RNA was used in a total volume of 10 µl per RT–PCR reaction. After 30–35 cycles of amplification in a GeneAmp PCR system 9600 (Perkin Elmer), amplified products were analyzed by agarose gel electrophoresis.
Protein expression and antibody production
For bacterial expression of the N- and C-terminal domains of HDAC10 as maltose-binding protein (MBP) fusion proteins, PCR fragments amplified with primer pairs DAC67/DAC68 and DAC69/DAC70 were cloned into pMAL-c2 (New England Biolabs). DH5α or BL21 cells harboring the constructs were grown and induced for expression by addition of IPTG (0.5 mM) according to the manufacturer’s instructions. Induced cells were harvested by centrifugation, washed twice with PBS and suspended in buffer B (20 mM Tris–HCl pH 8.0, 10% glycerol, 5 mM MgCl2, 0.1% NP-40 and protease inhibitors) containing 0.5 M KCl. The cells were lysed by sonication. Soluble extracts were used for purification of MBP fusion proteins on amylose agarose (New England Biolabs). After extensive washing, bound proteins were eluted in buffer B containing 0.15 M KCl supplemented with 10 mM maltose. The eluate was subsequently dialyzed extensively against PBS and used to immunize rabbits for polyclonal antiserum production.
For expression of Flag-tagged HDAC10 proteins in Sf9 cells, bacmids were generated using the Bac-to-Bac baculovirus expression system (Gibco BRL) and transfected into Sf9 cells to obtain recombinant baculoviruses. These viruses were then amplified and used to infect Sf9 cells to express Flag-tagged HDAC10 proteins. Infected cells were washed twice with cold phosphate-buffered saline (PBS) and lysed in buffer B containing 0.5 M KCl via one freeze–thaw cycle and subsequent rotation at 4°C for 30 min. Soluble extracts were used to purify the expressed protein on M2 agarose (Sigma). After extensive washing with buffer B containing 0.5 M KCl, bound proteins were eluted in buffer B containing 0.15 M KCl supplemented with 0.1 mg/ml Flag peptide.
For expression of Flag-HDAC10 protein in mammalian cells, its expression plasmids (10 µg) were transfected into 293 or 293T cells (1.5 × 106 cells in a 10 cm dish) using 24 µl of SuperFect transfection reagent (Qiagen). After 48 h, cells were washed twice with cold PBS and collected in buffer B containing 0.5 M KCl. Soluble extracts were used to purify the expressed protein on M2 agarose. The same buffer was used for washing M2 agarose beads, and buffer B containing 0.15 M KCl supplemented with 0.1 mg/ml Flag peptide was used for elution of bound Flag-HDAC10.
Deacetylase assay
Deacetylase activity was determined by measuring the release of [3H]acetate from [3H]acetylhistones as previously described (28,59). Assays were carried out in 0.2 ml of buffer H (50 mM Tris–HCl pH 7.5, 100 mM NaCl, 0.1 mM EDTA and 0.1 mM PMSF) containing [3H]acetylhistones (25 000 d.p.m.). The reaction was allowed to proceed at 37°C for 2 h and stopped by addition of 0.1 ml of 0.1 M HCl, 0.16 M acetic acid. Released [3H]acetate was extracted with 0.9 ml ethyl acetate. After centrifugation, 0.6 ml of the upper organic phase was quantified by liquid scintillation counting.
Green fluorescence microscopy
Expression plasmids for GFP fusion proteins were transfected, with SuperFect transfection reagent, into NIH 3T3 or 293 cells cultured in DMEM (Gibco BRL) supplemented with 10% fetal bovine serum and antibiotics. Sixteen hours post-transfection, transfected cells were analyzed by live green fluorescence microscopy as described (53), using a Nikon Eclipse TE300 microscope equipped with a temperature-adjustable platform and linked to a CCD camera (Hamamatsu) controlled by a Dell computer running Isee imaging software (Inovision Corp.). Images were taken and and exported to a PowerMac computer for further processing with Adobe Photoshop.
Reporter gene assays
SuperFect transfection reagent was used to transiently transfect a luciferase reporter plasmid (50–200 ng) and/or mammalian expression plasmids (50–200 ng) into 293 cells as described (28,57). pBluescript KSII(+) was used to normalize the total amount of plasmids used in each transfection and CMV-β-Gal (50 ng) was co-transfected for normalization of transfection efficiency. After 24 h, cells were lysed in situ and luciferase reporter activity was determined using D-(–)-luciferin (Boehringer Mannheim) as the substrate. Galactosidase activity was measured using Galacto-Light Plus (Tropix) reagent as the substrate. Chemiluminescence from activated luciferin or Galacto-Light Plus reagent was measured on a Luminometer Plate Reader (Dynex).
Immunoprecipitation
For analysis of the interaction between HDAC10 and other HDACs in vivo, HDAC10 (Flag-tagged) and/or other HDAC (HA-tagged) expression plasmids were co-transfected into 293 cells. After 48 h, the extracts were used for immunoprecipitation with anti-Flag M2 agarose beads (Sigma). Beads with bound immunocomplexes were washed four times with buffer B containing 0.15 M KCl and bound proteins were eluted with Flag peptide or 0.1 M glycine–HCl pH 2.5. After separation by SDS–PAGE, proteins were transferred to nitrocellulose membranes for western blot analysis with anti-Flag or anti-HA antibody. Blots were developed with Supersignal chemiluminescent substrate (Pierce).
RESULTS
Identification and molecular cloning of HDAC10
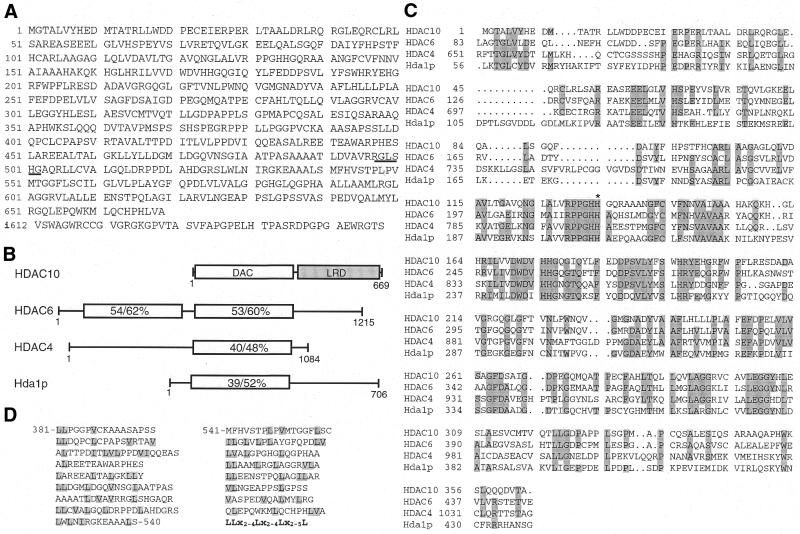
To explore whether there are additional class II mammalian histone deacetylases, the amino acid sequences of HDAC4 and HDAC6 were utilized as baits for BLAST searches against the GenBank database. These searches revealed three hypothetical polypeptides (accession nos CAB63048, CAB63049 and XP_001264.1). These putative polypeptides were predicted from the sequence of one locus at 22q13.31–13.33 (accession nos HS402G11 and BG678925) and they all contain an identical domain that displays significant sequence similarity to the deacetylase domains of HDAC4 and HDAC6. Interestingly, in entry XP_001264 the deacetylase-like domain was considered to be part of a protein kinase. Moreover, 22q13 is frequently deleted in ovarian cancer cell lines (60). These observations prompted us to investigate whether the corresponding transcripts are really produced in vivo. For this, two pairs of PCR primers were designed based on GenBank entry CAB63049 to amplify cDNA fragments from human cDNA libraries. PCR fragments were obtained as expected (data not shown), indicating that at least one transcript is truly produced. To fully elucidate the identity of the transcript(s), a human fetal bone marrow cDNA library was screened with a PCR fragment corresponding to the putative deacetylase domain as probe. From a majority of clones isolated through cDNA library screening, HDAC10 was predicted to be a 669 residue polypeptide consisting of an N-terminal putative deacetylase domain and a C-terminal leucine-rich domain (Fig. 1A and B). Except for a deletion of four residues after Q337, this sequence is the same as the hypothetical polypeptide CAB63048. During the screening, a 658 residue isoform with a distinct C-terminal end (Fig. 1A) was also isolated. Except for the aforementioned four residue deletion, this isoform is the same as the hypothetical polypetide CAB63049. None of the cDNA clones isolated was found to encode the hypothetical polypeptide XP_001264.1, suggesting that it may not be produced in vivo.
Figure 1.
Amino acid sequence and domain organization of HDAC10. (A) Predicted amino acid sequence of HDAC10 and a 658 residue isoform. The first 611 residues of HDAC10 and the isoform are identical, but they differ after residue 611. The distinct C-terminal sequence of the isoform is shown in the bottom line. A putative 14-3-3 binding motif is underlined. The GenBank accession no. for HDAC10 is AF426160. (B) Domain organization of HDAC10, HDAC6, HDAC4 and Hda1p. The putative deacetylase domain (DAC) of HDAC10 is depicted as a plain rectangle and the leucine-rich domain (LRD) is illustrated as a shaded rectangle. Also indicated are sequence identity and similarity (%) of the putative deacetylase domain compared to the corresponding regions of HDAC6, HDAC4 and Hda1p. Except for the deacetylase domains, HDAC6, HDAC4 and Hda1p share no sequence similarity. (C) Sequence comparison of the putative catalytic domain of HDAC10 with the catalytic domains of HDAC6, HDAC4 and Hda1p. Identical residues (three or four of four sequences) are shaded. H135, a residue that may be important for the deacetylase activity of HDAC10, is marked with an asterisk. (D) Repeat-like structure of the leucine-rich domain of HDAC10. This domain consists of 17 repeats loosely conforming to the consensus sequence shown in bold. x, any amino acid residue.
The putative deacetylase domain of HDAC10 is highly similar to those of HDAC6, HDAC4 and Hda1p (amino acid sequence identity 39–54%, similarity 52–62%; Fig. 1B and C), so HDAC10 is a putative class II deacetylase. The C-terminal domain of HDAC10 is leucine rich (leucine content 18%). Moreover, this domain consists of 17 repeats loosely conforming to the consensus sequence LLx2–4Lx2–4Lx2–5L, where x is any amino acid residue (Fig. 1D). This domain is different from well-known leucine-rich repeats (LRR) found in many proteins (61). As indicated in Figure 1A, the RxxSxG motif of HDAC10 constitutes a putative 14-3-3 binding site (62). HDAC10 does not have any putative NLSs, although it possesses several putative leucine-rich nuclear export signals.
Expression of HDAC10 in human tissues and mammalian cell lines
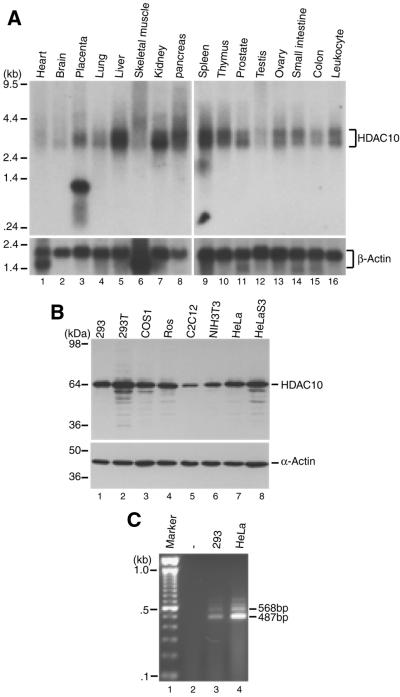
To determine the expression of HDAC10 in human tissues, poly(A) RNA blots were hybridized with a probe corresponding to its N-terminal domain. As shown in Figure 2A, a 3 kb transcript was detected. This transcript is most highly expressed in liver, kidney, pancreas and spleen. Its expression is low in heart, brain and testis. A 3.5 kb transcript was also detected in various tissues. Moreover, an abundant 1.3 kb transcript was found in placenta (lane 3). This transcript is too short for full-length HDAC10, so it may encode a placenta-specific HDAC10 variant whose identity is presently unclear. Therefore, northern analyses indicate that HDAC10 is widely expressed in adult human tissues.
Figure 2.
Expression of HDAC10. (A) Expression of HDAC10 in adult human tissues. Poly(A) RNA blots (Clontech; 2 µg/lane) were probed with a HDAC10 cDNA fragment corresponding to its putative deacetylase domain (top). As a loading control, the same blots were re-probed with a β-actin cDNA probe (bottom). Note the presence of 3 and 3.5 kb transcripts in most tissues and of a 1.3 kb species in placenta (lane 3, top). Molecular size markers are shown at left. (B) Expression of HDAC10 in cultured mammalian cells. Total cell extracts (10 µg/lane) were separated on SDS–polyacrylamide gels, electro-transferred to nitrocellulose and probed with anti-HDAC10 antibody raised against the C-terminal domain (top). As a loading control, the same blots were analyzed with anti-α-actin antibody (Roche) (bottom). (C) RT–PCR analysis of total RNA from 293 and HeLa cells. Primers DAC70 and DAC84 amplify the coding sequences for the C-terminal regions of HDAC10 and the 658 residue isoform as 487 and 568 bp fragments, respectively. Lane 1, 100 bp ladder (Gibco BRL); lane 2, amplification reaction of total RNA from 293 cells with only one primer.
Expression of HDAC10 in cultured cells was also examined. For this, a rabbit anti-HDAC10 antibody raised against its C-terminal domain was utilized. As shown in Figure 2B, this antibody detected an expected 64 kDa band in 293, 293T, COS1, ROS17/2.8, C2C12, NIH 3T3, HeLa and HeLa S3 cell extracts, suggesting that HDAC10 is also widely expressed in cultured mammalian cells.
In the cDNA library screening, two distinct forms of HDAC10 were found, so we investigated whether both forms are expressed in cultured cells. For this, RT–PCR analyses were performed with total RNA isolated from 293 and HeLa cells. As shown in Figure 2C, fragments of different length were amplified (Fig. 1A). The 487 and 568 bp bands correspond to cDNA fragments amplified from the 669 and 658 residue forms of HDAC10, respectively. Therefore, the RT–PCR results are consistent with the conclusion drawn from the cDNA library screening that different HDAC10 transcripts are produced and the predominant one encodes 669 residues.
Histone deacetylase activity of HDAC10
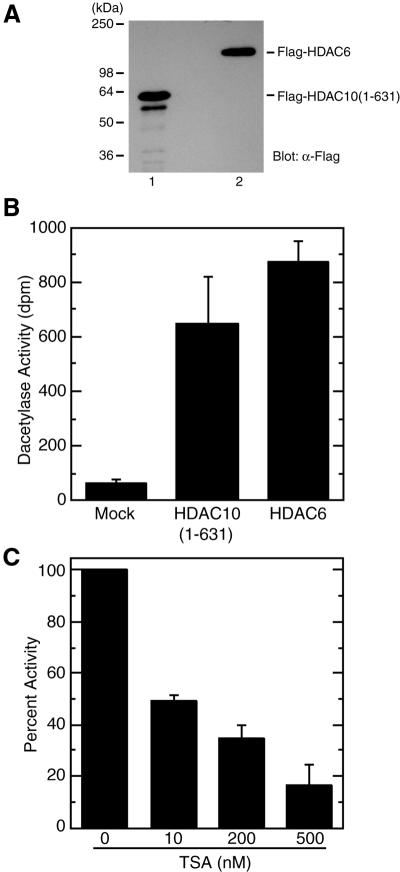
Since HDAC10 possesses a putative deacetylase domain with significant sequence similarity to those of Hda1p, HDAC4 and HDAC6 (Fig. 1), it is important to demonstrate experimentally that HDAC10 really possesses histone deacetylase activity. For this, Flag-HDAC10 was expressed in 293 and 293T cells. Perhaps due to low protein expression levels, not enough Flag-HDAC10 protein could be obtained for reliable determination of deacetylase activity. We then tried to express Flag-HDAC10 in Sf9 cells via a baculovirus expression system. For unknown reasons, full-length HDAC10 was difficult to express in these cells. However, we succeeded in highly expressing Flag-HDAC10(1–631) as a Flag-tagged fusion protein in Sf9 insect cells using a baculovirus expression system. As a positive control, HDAC6 was also expressed as a Flag-tagged fusion protein. Both Flag-HDAC10(1–631) and Flag-HDAC6 were affinity purified on anti-Flag M2 agarose and eluted with Flag peptide. As shown in Figure 3A, the affinity-purified fusion proteins were correctly expressed. Subsequently, deacetylase activities of the fusion proteins were determined. As shown in Figure 3B, Flag-HDAC10(1–631) possessed deacetylase activity that is comparable to that of Flag-HDAC6, indicating that HDAC10 has histone deacetylase activity.
Figure 3.
Deacetylase activity of HDAC10. (A) Expression of HDAC10 from Sf9 cells. HDAC10(1–631) was expressed in Sf9 cells as a Flag-tagged fusion protein. This fusion protein was then immunoprecipitated on anti-Flag M2 agarose, washed extensively with buffer B containing 0.5 M KCl and eluted with buffer B containing 0.15 KCl supplemented with Flag peptide. For comparison, Flag-HDAC6 was similarly expressed and affinity purified. Eluted proteins were analyzed by western blotting with anti-Flag antibody. (B) Deacetylase activity of Flag-HDAC10 and Flag-HDAC6. Equivalent amounts of Flag-HDAC10(1–631) and Flag-HDAC6 were used for deacetylating [3H]acetylhistones and the release of [3H]acetate (d.p.m.) from [3H]acetylhistones was quantified as deacetylase activity. As a control, extracts from uninfected Sf9 cells were subjected to similar purification. Flag-HDAC6 proteins expressed in and affinity purified from Sf9 and 293 cells were found to possess similar deacetylase activity (N.R.Bertos and X.-J.Yang, unpublished data). Average values from three independent experiments are shown with standard deviations. (C) Effect of TSA on the deacetylase activity of HDAC10(1–631). Deacetylase assays were carried out in the presence of the indicated TSA concentrations. The release of [3H]acetate in the absence of TSA was set to 100%. Average values from three independent experiments are shown with standard deviations.
A sequence comparison indicated that HDAC10 is a class II deacetylase (Fig. 1). It is well established that other class II members, such as HDAC4, HDAC5 and HDAC7, are sensitive to TSA, a known antitumor agent (63). So, it was necessary to determine whether the deacetylase activity of HDAC10 is also inhibited by TSA. As shown in Figure 3C, TSA inhibited the deacetylase activity of Flag-HDAC10(1–631) in a dose-dependent manner. TSA at 10 nM reduced the deacetylase activity by 50%. This is similar to what was reported for HDAC4 (28). These results indicate that HDAC10 is a new deacetylase whose activity is inhibited by TSA.
Subcellular localization of HDAC10
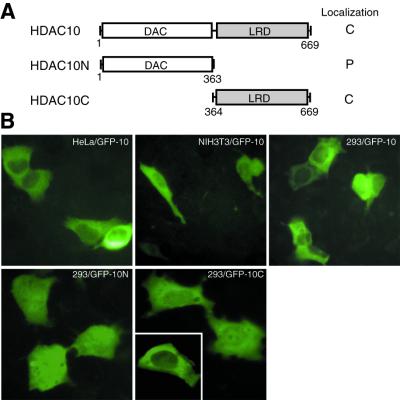
One unique feature of known class II members is that they are also found in the cytoplasm (27,41,48–52,54–56). For example, HDAC4, HDAC5 and HDAC7 are regulated by nucleocytoplasmic shuttling dependent on association with 14-3-3 proteins. Therefore, we asked whether HDAC10 also exists in the cytoplasm and, if so, whether its subcellular localization is regulated. To address this, HDAC10 was expressed as a GFP fusion protein (Fig. 4A). The subcellular distribution of GFP–HDAC10 was analyzed by green fluorescence microscopy. As shown in Figure 4B, GFP–HDAC10 was mainly cytoplasmic in HeLa, NIH 3T3 and 293 cells. HDAC10 possesses putative leucine-rich signals for CRM1-dependent nuclear export (Fig. 1A), suggesting that its cytoplasmic enrichment may be dependent on CRM1. To test this possibility, 293 cells expressing GFP–HDAC10 were treated with leptomycin B, a specific CRM1 inhibitor known to block the nuclear export of HDAC4, HDAC5, HDAC6 and HDAC7 (27,48,50,52,54–56). Under the same conditions, leptomycin B was able to block the nuclear export of GFP–HDAC4, but had no apparent effects on the cytoplasmic enrichment of GFP–HDAC10 (data not shown).
Figure 4.
Subcellular localization of HDAC10. (A) Schematic illustration of HDAC10 and deletion mutants HDAC10N and HDAC10C. Their localization is summarized at right. C, enriched in the cytoplasm; P, pancellular. (B) Representative green fluorescence images of GFP–HDAC10 and its mutants. Expression plasmids for indicated GFP fusion proteins were transfected into indicated cell lines. Green fluorescence of living cells was examined 16 h after transfection.
To examine which part of HDAC10 is responsible for its cytoplasmic localization, the N- and C-terminal domains were expressed separately as GFP fusion proteins (Fig. 4A), and their subcellular localization was examined. As shown in Figure 4B, GFP–HDAC10N is pancellular and GFP–HDAC10C is enriched in the cytoplasm, suggesting that the leucine-rich domain of HDAC10 is responsible for its cytoplasmic enrichment.
Transcriptional repression potential of HDAC10
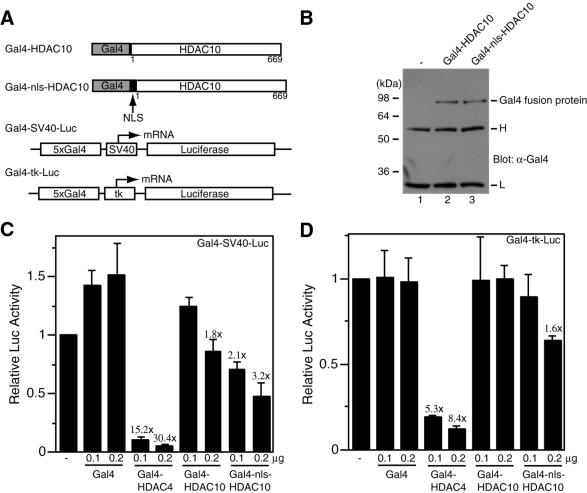
Class I HDACs are well-known transcriptional co-repressors (14). The known class II members, HDAC4, HDAC5, HDAC7 and HDAC9, have been implicated in transcriptional repression (16,34), so we decided to determine whether HDAC10 is able to repress transcription. For this, HDAC10 was expressed as a protein fused to the DNA-binding domain of the yeast transcription factor Gal4 (Fig. 5A). As shown in Figure 5B (compare lanes 1 and 2), this fusion protein was expressed as expected. Its ability to regulate transcription was assessed using the luciferase reporter Gal4–SV40–Luc, in which the luciferase gene is under the control of the SV40 minimal promoter and five Gal4-responsive elements (Fig. 5A). As previously reported (28), Gal4–HDAC4 reduced luciferase reporter activity in a dose-dependent manner (Fig. 5C). Under the same conditions, Gal4–HDAC10 was able to weakly inhibit luciferase reporter activity (Fig. 5C). Since HDAC10 was found to be enriched in the cytoplasm (Fig. 4), a strong NLS from the SV40 large T antigen was inserted into Gal4–HDAC10 to generate Gal4–nls–HDAC10 (Fig 5A). As shown in Figure 5B, this fusion protein was expressed as expected. It was found to be more potent in repressing the activity of Gal4–SV40–Luc (Fig. 5C). With the luciferase reporter Gal4–tk–Luc (Fig. 5A), Gal4–HDAC10 and Gal4–nls–HDAC10 had smaller effects (Fig. 5D), suggesting that the repression is promoter dependent. Taken together, these results indicate that HDAC10 is able to repress transcription.
Figure 5.
Transcriptional repression by tethered HDAC10. (A) Schematic representation of Gal4 fusion proteins (Gal4–HDAC10 and Gal4–nls–HDAC10) and luciferase reporters (Gal4–SV40–Luc and Gal4–tk–Luc). NLS, nuclear localization signal; 5×Gal4, five copies of the Gal4-binding site; tk, thymidine kinase core promoter. (B) Expression of Gal4–HDAC4 and Gal4–nls–HDAC10. Extracts (10 µg/lane), prepared from 293 cells transfected with expression plasmids for the indicated fusion proteins, were subjected to immunoprecipitation with anti-Gal4 antibody (RK5C1; Santa Cruz Biotechnology) and western blotting analysis with the same antibody. Lane 1, similar immunoprecipitation with extracts from non-transfected cells. Molecular size markers are shown at left. H, IgG heavy chain; L, IgG light chain. (C and D) Transcriptional repression by Gal4–HDAC10 and Gal4–nls–HDAC10. Mammalian constructs expressing HDAC4 and HDAC10 fused to the C-terminus of Gal4(1–147) were transfected into 293 cells along with the reporter Gal4–SV40–Luc (C) or Gal4–tk–Luc (D). Note that expression of Gal4 fusion proteins is under the control of an SV40 promoter. Luciferase activities were normalized to the internal β-galactosidase control; the normalized luciferase activity from the transfection without any effector plasmid was arbitrarily set to 1.0. Average values from three independent experiments are shown with standard deviations. The different repression abilities of Gal4–HDAC4 and Gal4–HDAC10 fusion proteins may be due to their distinct expression levels, since Gal4–HDAC4 was expressed to a much higher level (data not shown).
Interaction of HDAC10 with other proteins
HDAC10 has a putative 14-3-3 binding motif (Fig. 1A), so whether HDAC10 interacts with 14-3-3 proteins was first tested. Immunoprecipitation experiments indicated that HDAC10 does not interact with 14-3-3 proteins (data not shown). Interaction among different HDACs has been observed. HDAC1 and HDAC2 form the catalytic core of the Sin3 and NuRD complexes (64–69). HDAC4, HDAC5 and HDAC7 have been found to interact with HDAC1 or HDAC3 (26,70). Moreover, HDAC4 and HDAC5 associate with each other (46). These findings raise the interesting possibility that HDAC10 may bind to class I and known class II members. To address this issue, immunoprecipitation and in vitro binding assays were performed. These assays revealed that HDAC10 does not interact with HDAC4 or HDAC6 (data not shown), suggesting that HDAC10 does not associate with other class II members.
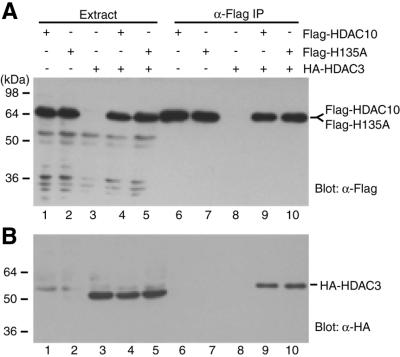
To test whether HDAC10 interacts with HDAC3, Flag-HDAC10 and HA-HDAC3 were co-expressed in 293 cells. Cell extracts were subjected to affinity purification on anti-Flag M2 agarose. Eluted proteins were then analyzed by western blotting with anti-Flag or anti-HA antibody. As shown in Figure 6A, Flag-HDAC10 was expressed (lanes 1 and 4) and could be affinity purified (lanes 6 and 9). Importantly, HA-HDAC3 was co-immunoprecipitated (Fig. 6B, compare lanes 6 and 9), indicating that HDAC3 is able to interact with HDAC10. It has been reported that the catalytic center of HDAC4 is important for association with some of its interacting partners (30,44), so whether the mutation H135A of HDAC10 affects its binding to HDAC3 was tested. An equivalent histidine residue has been found to be critical for the deacetylase activity of Rpd3p, HDAC1 and HDAC4 (27,28,71,72). As shown in Figure 6 (lanes 7 and 10), the point mutant was found to bind HDAC3 as strongly as wild-type HDAC10. Deletion analyses indicated that both its N- and C-terminal domains can bind HDAC3 independently (data not shown). Taken together, these results indicate that HDAC10 is able to interact with HDAC3.
Figure 6.
Interaction of HDAC10 with HDAC3. Flag-tagged HDAC10 (lanes 1, 4, 6 and 9) or the mutant H135A (lanes 2, 5, 7 and 10) were expressed with (lanes 4 and 5) or without (lanes 1–2) HA-HDAC3 in 293 cells and immunoprecipitated with anti-Flag M2 agarose. Extracts (lanes 1–5) and immunoprecipitated proteins (lanes 6–10), which were eluted with Flag peptide, were subjected to western blotting analysis with anti-Flag (A) or anti-HA (B) antibody.
DISCUSSION
The results presented herein demonstrate that HDAC10 is a novel deacetylase with its N-terminal half showing significant sequence similarity to the deacetylase domains of known class II HDACs. cDNA library screening and RT–PCR results indicate that there are two alternatively spliced transcripts, the predominant form of which encodes 669 residues (Fig. 1A). The other transcript encodes a 658 residue polypeptide with a distinct C-terminal end (Fig. 1A). The deacetylase domain of HDAC10 is more similar to those of other class II HDACs than to those of class I HDACs (Fig. 1) and its deacetylase activity is comparable to that of HDAC6 (Fig. 3B). Reminiscent of other class II HDACs, HDAC10 possesses TSA-sensitive deacetylase activity (Fig. 3), is enriched in the cytoplasm (Fig. 4) and has transcriptional repression potential (Fig. 5). Moreover, like HDAC4, HDAC5 and HDAC7 (70), HDAC10 interacts with HDAC3 (Fig. 6). These findings indicate that besides sequence similarity, HDAC10 shares functional properties with other class II HDACs.
As a new class II member, HDAC10 also possesses unique properties. Its leucine-rich domain does not show sequence similarity to any known proteins (Fig. 1) and its tissue distribution is wide and different from those of other class II HDACs. For example, human HDAC10 is highly expressed in liver, kidney, pancreas and spleen (Fig. 2A), whereas human HDAC4 and mouse HDAC6 are most abundant in skeletal muscle and testis, respectively (25–29), suggesting that HDAC10 may have distinct biological functions. HDAC4 and HDAC6 are predominantly cytoplasmic, whereas HDAC5 and HDAC7 are mainly nuclear (27,30,48–50,52,54–56). Distinct from these class II members, HDAC10 is enriched in the cytoplasm (Fig. 4). It has been established that the cytoplasmic localization of known class HDACs is sensitive to leptomycin B (27,48,50,52,54–56). In contrast, the cytoplasmic enrichment of HDAC10 is not sensitive to this compound (data not shown). Analysis of HDAC10 deletion mutants revealed that the leucine-rich domain is important for its cytoplasmic enrichment (Fig. 4). These findings suggest that HDAC10 may function outside the nucleus. HDAC10 does not have a putative NLS, but it may enter the nucleus through passive diffusion or association with nuclear proteins. HDAC10 can deacetylate histones (Fig. 3), repress transcription (Fig. 5) and interact with HDAC3 (Fig. 6), so it may also play a role in the nucleus. Further investigation is needed to clarify these interesting issues.
Like HDAC10, HDAC6 has a unique domain organization. Different from other HDACs, HDAC6 contains two deacetylase domains and a C-terminal zinc finger motif (34,37). Human HDAC6 also possesses an SE14 repeat domain (34). Interestingly, the zinc finger motif of HDAC6 binds to ubiquitin, suggesting that HDAC6 may be involved in regulating ubiquitin-dependent pathways (37). Therefore, HDAC6 may play a role in the cytoplasm. HDAC4, HDAC5, HDAC7 and HDAC9 are similar to each other. It has been proposed that these class II HDACs constitute a subclass (IIa) (34). Compared to HDAC6 and HDAC10, much more is known about the function and regulation of class IIa members. While there is evidence suggesting that HDAC7 is involved in regulating endothelin signaling in the cytoplasm (73), several independent reports indicate that class IIa members function as transcriptional co-repressors whose functions are negatively regulated by binding of 14-3-3 proteins (48,49,51,56). HDAC10 possesses a putative 14-3-3 binding site (Fig. 1A), but is unable to interact with 14-3-3 proteins (data not shown). It is presently unclear whether the subcellular localization of HDAC10 is regulated. The identification and initial characterization of HDAC10 reiterate the notion that compared to class I HDACs, class II members are more diverse in their sequences and biological functions.
Three other groups have recently independently cloned and characterized HDAC10 (74–76). While we have shown that HDAC10 is enriched in the cytoplasm (Fig. 4), others found it in the nucleus and/or cytoplasm (74–76), suggesting that its subcellular localization may be dependent on cellular context and assay conditions. This is similar to what was reported for HDAC4 (48; and references therein). We found that HDAC10 is able to weakly repress transcription (Fig. 5), whereas others described HDAC10 as having potent repression ability (74,75). The different transcriptional potency observed could be due to the distinct assay conditions employed, e.g. different promoters used for expression of Gal4–HDAC10. Clearly, all studies support the proposal that HDAC10 is able to repress transcription. Related to this, HDAC4, HDAC5, HDAC7 and HDAC9 are known to function as transcriptional co-repressors (16,34,35). Consistent with our finding that HDAC10 interacts with HDAC3 (Fig. 6), others reported that HDAC10 binds to SMRT, a subunit of the HDAC3 complex (76).
Like class I and other class II members, HDAC10 is sensitive to TSA (Fig. 3C). TSA and related deacetylase inhibitors have emerged as new antitumor agents that are being tested in clinical trials (77). Remarkably, these inhibitors have recently been found to reduce polyglutamine-dependent neurodegeneration (78), raising the hope that they can be used as a therapy for Huntington’s disease and other polyglutamine diseases. Therefore, identification of HDAC10 sets the stage for its further functional characterization, which should shed light on how to improve the efficacy of these inhibitors in the treatment of cancers and neurodegenerative disorders.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr E. Seto for human HDAC3 cDNA. This work was supported by grants from the National Cancer Institute of Canada and the Canada Foundation for Innovation and by a scholarship from the Canadian Institutes of Health Research (to X.-J.Y.).
REFERENCES
- 1.Workman J.L. and Kingston,R.E. (1998) Alteration of nucleosome structure as a mechanism of transcriptonal regulation. Annu. Rev. Biochem., 67, 545–579. [DOI] [PubMed] [Google Scholar]
- 2.Grunstein M. (1997) Histone acetylation in chromatin structure and transcription. Nature, 389, 349–352. [DOI] [PubMed] [Google Scholar]
- 3.Struhl K. (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev., 12, 599–606. [DOI] [PubMed] [Google Scholar]
- 4.Turner B.M. (2000) Histone acetylation and an epigenetic code. Bioessays, 22, 836–845. [DOI] [PubMed] [Google Scholar]
- 5.Jenuwein T. and Allis,C.D. (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- 6.Wolffe A. (1998) Chromatin: Structure and Function, 3rd Edn. Academic Press, San Diego, CA.
- 7.Zhang Y. and Reinberg,D. (2001) Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev., 15, 2343–2360. [DOI] [PubMed] [Google Scholar]
- 8.Berger S.L. (2001) An embarrassment of niches: the many covalent modifications of histones in transcriptional regulation. Oncogene, 20, 3007–3013. [DOI] [PubMed] [Google Scholar]
- 9.Davie J.R. and Spencer,V.A. (2000) Signal transduction pathways and the modification of chromatin structure. Prog. Nucleic Acid Res. Mol. Biol., 65, 299–341. [DOI] [PubMed] [Google Scholar]
- 10.Sterner D.E. and Berger,S.L. (2000) Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev., 64, 435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kouzarides T. (2000) Acetylation: a regulatory modification to rival phosphorylation? EMBO J., 19, 1176–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hassig C.A. and Schreiber,S.L. (1997) Nuclear histone acetylases and deacetylases and transcriptional regulation: HATs off to HDACs. Curr. Opin. Chem. Biol., 1, 300–308. [DOI] [PubMed] [Google Scholar]
- 13.Taunton J., Hassig,C.A. and Schreiber,S.L. (1996) A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science, 272, 408–411. [DOI] [PubMed] [Google Scholar]
- 14.Cress W.D. and Seto,E. (2000) Histone deacetylases, transcriptional control and cancer. J. Cell Physiol., 184, 1–16. [DOI] [PubMed] [Google Scholar]
- 15.Gray S.G. and Ekstrom,T.J. (2001) The human histone deacetylase family. Exp. Cell Res., 262, 75–83. [DOI] [PubMed] [Google Scholar]
- 16.Khochbin S., Verdel,A., Lemercier,C. and Seigneurin-Berny,D. (2001) Functional significance of histone deacetylase diversity. Curr. Opin. Genet. Dev., 11, 162–166. [DOI] [PubMed] [Google Scholar]
- 17.Yang W.M., Inouye,C., Zeng,Y., Bearss,D. and Seto,E. (1996) Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc. Natl Acad. Sci. USA, 93, 12845–12850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang W.M., Yao,Y.L., Sun,J.M., Davie,J.R. and Seto,E. (1997) Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J. Biol. Chem., 272, 28001–28007. [DOI] [PubMed] [Google Scholar]
- 19.Emiliani S., Fischle,W., Van Lint,C., Al-Abed,Y. and Verdin,E. (1998) Characterization of a human RPD3 ortholog, HDAC3. Proc. Natl Acad. Sci. USA, 95, 2795–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dangond F., Hafler,D.A., Tong,J.K., Randall,J., Kojima,R., Utku,N. and Gullans,S.R. (1998) Differential display cloning of a novel human histone deacetylase (HDAC3) cDNA from PHA-activated immune cells. Biochem. Biophys. Res. Commun., 242, 648–652. [DOI] [PubMed] [Google Scholar]
- 21.Hu E., Chen,Z., Fredrickson,T., Zhu,Y., Kirkpatrick,R., Zhang,G.F., Johanson,K., Sung,C.M., Liu,R. and Winkler,J. (2000) Cloning and characterization of a novel human class I histone deacetylase that functions as a transcription repressor. J. Biol. Chem., 275, 15254–15264. [DOI] [PubMed] [Google Scholar]
- 22.Buggy J.J., Sideris,M.L., Mark,P., Lorimer,D.D., Mcintosh,B. and Clark,J.M. (2000) Cloning and characterization of a novel human histone deacetylase, HDAC8. Biochem. J., 350, 199–205. [PMC free article] [PubMed] [Google Scholar]
- 23.Van de Wyngaert I., de Vries,W., Kremer,A., Neefs,J., Verhasselt,P., Luyten,W.H. and Kass,S.U. (2000) Cloning and characterization of human histone deacetylase 8. FEBS Lett., 478, 77–83. [DOI] [PubMed] [Google Scholar]
- 24.Vidal M. and Gaber,R.F. (1991) RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol. Cell. Biol., 11, 6317–6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischle W., Emiliani,S., Hendzel,M.J., Nagase,T., Nomura,N., Voelter,W. and Verdin,E. (1999) A new family of human histone deacetylases related to Saccharomyces cerevisiae HDA1p. J. Biol. Chem., 274, 11713–11720. [DOI] [PubMed] [Google Scholar]
- 26.Grozinger C.M., Hassig,C.A. and Schreiber,S.L. (1999) Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc. Natl Acad. Sci. USA, 96, 4868–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miska E.A., Karlsson,C., Langley,E., Nielsen,S.J., Pines,J. and Kouzarides,T. (1999) HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J., 18, 5099–5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang A.H., Bertos,N.R., Vezmar,M., Pelletier,N., Crosato,M., Heng,H.H., Th’ng,J., Han,J. and Yang,X.J. (1999) HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol. Cell. Biol., 19, 7816–7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verdel A. and Khochbin,S. (1999) Identification of a new family of higher eukaryotic histone deacetylases. Coordinate expression of differentiation-dependent chromatin modifiers. J. Biol. Chem., 274, 2440–2445. [DOI] [PubMed] [Google Scholar]
- 30.Kao H.Y., Downes,M., Ordentlich,P. and Evans,R.M. (2000) Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev., 14, 55–66. [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou X., Marks,P.A., Rifkind,R.A. and Richon,V.M. (2001) Cloning and characterization of a novel histone deacetylase, HDAC9. Proc. Natl Acad. Sci. USA, 98, 10572–10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rundlett S.E., Carmen,A.A., Kobayashi,R., Bavykin,S., Turner,B.M. and Grunstein,M. (1996) HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl Acad. Sci. USA, 93, 14503–14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guarente L. (2000) Sir2 links chromatin silencing, metabolism and aging. Genes Dev., 14, 1021–1026. [PubMed] [Google Scholar]
- 34.Bertos N.R., Wang,A.H. and Yang,X.J. (2001) Class II histone deacetylases: structure, function and regulation. Biochem. Cell Biol., 79, 243–252. [PubMed] [Google Scholar]
- 35.Fischle W., Kiermer,V., Dequiedt,F. and Verdin,E. (2001) The emerging role of class II histone deacetylases. Biochem. Cell Biol., 79, 337–348. [PubMed] [Google Scholar]
- 36.McKinsey T.A., Zhang,C.L. and Olson,E.N. (2001) Control of muscle development by dueling HATs and HDACs. Curr. Opin. Genet. Dev., 11, 497–504. [DOI] [PubMed] [Google Scholar]
- 37.Seigneurin-Berny D., Verdel,A., Curtet,S., Lemercier,C., Garin,J., Rousseaux,S. and Khochbin,S. (2001) Identification of components of the murine histone deacetylase 6 complex: link between acetylation and ubiquitination signaling pathways. Mol. Cell. Biol., 21, 8035–8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lemercier C., Verdel,A., Galloo,B., Curtet,S., Brocard,M.P. and Khochbin,S. (2000) mHDA1/HDAC5 histone deacetylase interacts with and represses MEF2A transcriptional activity. J. Biol. Chem., 275, 15594–15599. [DOI] [PubMed] [Google Scholar]
- 39.Sparrow D.B., Miska,E.A., Langley,E., Reynaud-Deonauth,S., Kotecha,S., Towers,N., Spohr,G., Kouzarides,T. and Mohun,T.J. (1999) MEF-2 function is modified by a novel co-repressor, MITR. EMBO J., 18, 5085–5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu J., McKinsey,T.A., Nicol,R.L. and Olson,E.N. (2000) Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc. Natl Acad. Sci. USA, 97, 4070–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dressel U., Bailey,P.J., Wang,S.C., Downes,M., Evans,R.M. and Muscat,G.E. (2001) A dynamic role for HDAC7 in MEF2-mediated muscle differentiation. J. Biol. Chem., 276, 17007–17013. [DOI] [PubMed] [Google Scholar]
- 42.Youn H.D., Grozinger,C.M. and Liu,J.O. (2000) Calcium regulates transcriptional repression of myocyte enhancer factor 2 by histone deacetylase 4. J. Biol. Chem., 275, 22563–22567. [DOI] [PubMed] [Google Scholar]
- 43.Lu J., McKinsey,T.A., Zhang,C.L. and Olson,E.N. (2000) Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol. Cell, 6, 233–244. [DOI] [PubMed] [Google Scholar]
- 44.Huang E.Y., Zhang,J., Miska,E.A., Guenther,M.G., Kouzarides,T. and Lazar,M.A. (2000) Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev., 14, 45–54. [PMC free article] [PubMed] [Google Scholar]
- 45.Huynh K.D., Fischle,W., Verdin,E. and Bardwell,V.J. (2000) BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev., 14, 1810–1823. [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang C.L., McKinsey,T.A., Lu,J. and Olson,E.N. (2001) Association of COOH-terminal-binding protein (CtBP) and MEF2-interacting transcription repressor (MITR) contributes to transcriptional repression of the MEF2 transcription factor. J. Biol. Chem., 276, 35–39. [DOI] [PubMed] [Google Scholar]
- 47.Chakraborty S., Senyuk,V., Sitailo,S., Chi,Y. and Nucifora,G. (2001) Interaction of EVI1 with cAMP-responsive element-binding protein-binding protein (CBP) and p300/CBP-associated factor (P/CAF) results in reversible acetylation of EVI1 and in co-localization in nuclear speckles. J. Biol. Chem., 276, 44936–44943. [DOI] [PubMed] [Google Scholar]
- 48.Wang A.H., Kruhlak,M.J., Wu,J., Bertos,N.R., Vezmar,M., Posner,B.I., Bazett-Jones,D.P. and Yang,X.J. (2000) Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Mol. Cell. Biol., 20, 6904–6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grozinger C.M. and Schreiber,S.L. (2000) Regulation of histone deacetylase 4 and 5 transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl Acad. Sci. USA, 97, 7835–7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McKinsey T.A., Zhang,C.L., Lu,J. and Olson,E.N. (2000) Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature, 408, 106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McKinsey T.A., Zhang,C.L. and Olson,E.N. (2000) Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc. Natl Acad. Sci. USA, 97, 14400–14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verdel A., Curtet,S., Brocard,M.P., Rousseaux,S., Lemercier,C., Yoshida,M. and Khochbin,S. (2000) Active maintenance of mHDA2/mHDAC6 histone-deacetylase in the cytoplasm. Curr. Biol., 10, 747–749. [DOI] [PubMed] [Google Scholar]
- 53.Wang A.H. and Yang,X.J. (2001) Histone deacetylase 4 possesses intrinsic nuclear import and export signals. Mol. Cell. Biol., 21, 5992–6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao X., Ito,A., Kane,C.D., Liao,T.S., Bolger,T.A., Lemrow,S.M., Means,A.R. and Yao,T.P. (2001) The modular nature of histone deacetylase HDAC4 confers phosphorylation-dependent intracellular trafficking. J. Biol. Chem., 276, 35042–35048. [DOI] [PubMed] [Google Scholar]
- 55.Miska E.A., Langley,E., Wolf,D., Karlsson,C., Pines,J. and Kouzarides,T. (2001) Differential localization of HDAC4 orchestrates muscle differentiation. Nucleic Acids Res., 29, 3439–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kao H.Y., Verdel,A., Tsai,C.C., Simon,C., Juguilon,H. and Khochbin,S. (2001) Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J. Biol. Chem., 276, 47496–47507. [DOI] [PubMed] [Google Scholar]
- 57.Champagne N., Bertos,N.R., Pelletier,N., Wang,A.H., Vezmar,M., Yang,Y., Heng,H.H. and Yang,X.J. (1999) Identification of a human histone acetyltransferase related to monocytic leukemia zinc finger protein. J. Biol. Chem., 274, 28528–28536. [DOI] [PubMed] [Google Scholar]
- 58.Sadowski I., Bell,B., Broad,P. and Hollis,M. (1992) GAL4 fusion vectors for expression in yeast or mammalian cells. Gene, 118, 137–141. [DOI] [PubMed] [Google Scholar]
- 59.Hendzel M.J., Delcuve,G.P. and Davie,J.R. (1991) Histone deacetylase is a component of the internal nuclear matrix. J. Biol. Chem., 266, 21936–21942. [PubMed] [Google Scholar]
- 60.Lin H., Pizer,E.S. and Morin,P.J. (2000) A frequent deletion polymorphism on chromosome 22q13 identified by representational difference analysis of ovarian cancer. Genomics, 69, 391–394. [DOI] [PubMed] [Google Scholar]
- 61.Kobe B. and Deisenhofer,J. (1994) The leucine-rich repeat: a versatile binding motif. Trends Biochem. Sci., 19, 415–421. [DOI] [PubMed] [Google Scholar]
- 62.Rittinger K., Budman,J., Xu,J., Volinia,S., Cantley,L.C., Smerdon,S.J., Gamblin,S.J. and Yaffe,M.B. (1999) Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol. Cell, 4, 153–166. [DOI] [PubMed] [Google Scholar]
- 63.Yoshida M., Horinouchi,S. and Beppu,T. (1995) Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays, 17, 423–430. [DOI] [PubMed] [Google Scholar]
- 64.Hassig C.A., Fleischer,T.C., Billin,A.N., Schreiber,S.L. and Ayer,D.E. (1997) Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell, 89, 341–347. [DOI] [PubMed] [Google Scholar]
- 65.Zhang Y., Sun,Z.W., Iratni,R., Erdjument-Bromage,H., Tempst,P., Hampsey,M. and Reinberg,D. (1998) SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol. Cell, 1, 1021–1031. [DOI] [PubMed] [Google Scholar]
- 66.Wade P.A., Jones,P.L., Vermaak,D. and Wolffe,A.P. (1998) A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated SNF2 superfamily ATPase. Curr. Biol., 8, 843–846. [DOI] [PubMed] [Google Scholar]
- 67.Zhang Y., LeRoy,G., Seelig,H.P., Lane,W.S. and Reinberg,D. (1998) The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell, 95, 279–289. [DOI] [PubMed] [Google Scholar]
- 68.Tong J.K., Hassig,C.A., Schnitzler,G.R., Kingston,R.E. and Schreiber,S.L. (1998) Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature, 395, 917–921. [DOI] [PubMed] [Google Scholar]
- 69.Xue Y., Wong,J., Moreno,G.T., Young,M.K., Cote,J. and Wang,W. (1998) NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell, 2, 851–861. [DOI] [PubMed] [Google Scholar]
- 70.Fischle W., Dequiedt,F., Fillion,M., Hendzel,M.J., Voelter,W. and Verdin,E. (2001) Human HDAC7 histone deacetylase activity is associated with hDAC3 in vivo. J. Biol. Chem., 276, 35826–35835. [DOI] [PubMed] [Google Scholar]
- 71.Hassig C.A., Tong,J.K., Fleischer,T.C., Owa,T., Grable,P.G., Ayer,D.E. and Schreiber,L. (1998) A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc. Natl Acad. Sci. USA, 95, 3519–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kadosh D. and Struhl,K. (1998) Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev., 12, 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee H.J., Chun,M. and Kandror,K.V. (2001) Tip60 and HDAC7 interact with the endothelin receptor a and may be involved in downstream signaling. J. Biol. Chem., 276, 16597–16600. [DOI] [PubMed] [Google Scholar]
- 74.Kao H.Y., Lee,C.H., Komarov,A., Han,C.C. and Evans,R.M. (2002) Isolation and characterization of mammalian HDAC10, a novel histone deacetylase. J. Biol. Chem., 277, 187–193 [DOI] [PubMed] [Google Scholar]
- 75.Guardiola A.R. and Yao,T.P. (2002) Molecular cloning and characterization of a novel histone deacetylase HDAC10. J. Biol. Chem., 277, 3350–3356 [DOI] [PubMed] [Google Scholar]
- 76.Fischer D.D., Cai,R., Bhatia,U., Asselbergs,F.A., Song,C., Terry,R., Trogani,N., Widmer,R., Atadja,P. and Cohen,D. (2002) Isolation and characterization of a novel class II histone deacetylase, HDAC10. J. Biol. Chem., in press. [DOI] [PubMed] [Google Scholar]
- 77.Marks P.A., Richon,V.M., Breslow,R. and Rifkind,R.A. (2001) Histone deacetylase inhibitors as new cancer drugs. Curr. Opin. Oncol., 13, 477–483. [DOI] [PubMed] [Google Scholar]
- 78.Steffan J.S., Bodai,L., Pallos,J., Poelman,M., McCampbell,A., Apostol,B.L., Kazantsev,A., Schmidt,E., Zhu,Y.Z., Greenwald,M., Kurokawa,R., Housman,D.E., Jackson,G.R., Marsh,J.L. and Thompson,L.M. (2001) Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature, 413, 739–743. [DOI] [PubMed] [Google Scholar]