Abstract
Aedes aegypti is the primary vector of the arboviruses dengue (DENV) and chikungunya (CHIKV). These viruses exhibit key differences in their vector interactions, the latter moving more quicky through the mosquito and triggering fewer standard antiviral pathways. As the global footprint of CHIKV continues to expand, we seek to better understand the mosquito’s natural response to CHIKV—both to compare it to DENV:vector coevolutionary history and to identify potential targets in the mosquito for genetic modification. We used a modified full-sibling design to estimate the contribution of mosquito genetic variation to viral loads of both DENV and CHIKV. Heritabilities were significant, but higher for DENV (40%) than CHIKV (18%). Interestingly, there was no genetic correlation between DENV and CHIKV loads between siblings. These data suggest Ae. aegypti mosquitoes respond to the two viruses using distinct genetic mechanisms. We also examined genome-wide patterns of gene expression between High and Low CHIKV families representing the phenotypic extremes of viral load. Using RNAseq, we identified only two loci that consistently differentiated High and Low families: a long non-coding RNA that has been identified in mosquito screens post-infection and a distant member of a family of Salivary Gland Specific (SGS) genes. Interestingly, the latter gene is also associated with horizontal gene transfer between mosquitoes and the endosymbiotic bacterium Wolbachia. This work is the first to link the SGS gene to a mosquito phenotype. Understanding the molecular details of how this gene contributes to viral control in mosquitoes may, therefore, also shed light on its role in Wolbachia.
Author summary
The virus chikungunya (CHIKV) that causes long term arthritis symptoms in humans is transmitted to through the bite of the Aedes aegypti mosquito. CHIKV, for which there is no vaccine, is becoming increasingly common across the globe. We therefore need to understand the mosquito’s own ability to control CHIKV, as we may use that knowledge to create resistant mosquitoes through genetic modification. We show that the mosquito has very little ability to respond genetically to CHIKV, indicating low potential for the mosquito to evolve resistance. We also found that the genetic basis of CHIKV viral loads appears distinct from dengue, another common virus. As such, any strategy for engineering virus-resistant mosquitoes may need to be virus-specific or focus on the few overlapping genes in the mosquito response. Last, when we examined the mosquito genes whose expression differed between high and low-load virus lineages, we discovered a gene that was highly expressed in low-load families and therefore, potentially acting as a virus controller. Interestingly, a homolog of this gene has been discovered in the genome of the Wolbachia endosymbiont, itself known to limit virus replication inside its insect hosts. The functional importance of this homolog in virus control should therefore be explored in both mosquitoes and Wolbachia.
Introduction
Arthropod-borne pathogens account for more than 17% of all infectious diseases globally [1]. Among the most prevalent are dengue (DENV) and chikungunya (CHIKV) viruses, with over half of the world’s population at risk. There are no effective vaccines or treatments for either pathogen [2] and morbidity can be severe—with some cases leading to dengue shock syndrome [3], or neurological conditions [4], and debilitating, chronic arthralgia [5]. Both viruses inflict significant socioeconomic harm as well.
Both DENV and CHIKV are single-stranded positive-sense RNA viruses belonging to the Flaviviridae and Togaviridae viral families, respectively, and both are transmitted primarily through the bites of Aedes aegypti mosquitoes [6,7]. However, the two pathogens have distinct histories. DENV has a long association with Ae. aegypti: virus and vector together becoming ubiquitous across the tropics [8,9] with descriptions of DENV-like illness (aka ‘break bone fever’) appearing as early as 1635 in the Americas [10]. In contrast, CHIKV has remained a relatively rare pathogen, causing only small outbreaks on some islands of the Indian Ocean until recently [11]. The two viruses have thus had unequal opportunities to coevolve with Ae. aegypti, their shared primary mosquito host. However, CHIKV cases have recently exploded, with extensive range expansion in some areas of the world [12]. The virus was first reported in the Americas in 2013, and by 2018 there had been more than 3 million cases in 45 countries [13,14]. Despite this sharp rise in CHIKV infections around the globe, our understanding of how this pathogen infects and interacts with Ae. aegypti remains incomplete.
Arboviral infection in the mosquito is a complex and dynamic process shaped by the genetic variation of both the mosquito and the virus [15]. Natural Ae. aegypti populations can vary in their susceptibility to arboviruses, including DENV and CHIKV [16–20], yet diverse populations from across the tropics are highly susceptible to CHIKV infection [20–24]. For example, Ae. aegypti populations from the Americas, where CHIKV appeared less than a decade ago, transmitted CHIKV in their saliva at rates as high as 83% [24]. Moreover, CHIKV infectious particles have been detected in Ae. aegypti salivary glands as early as 2 days post-infection (DPI), indicating that the dissemination of the virus within the mosquito is rapid [25]. In contrast, DENV particles are not detected in salivary glands until later [26,27]. Such differences may stem from variation in viral replication inside the mosquito, where both mosquito genetic factors and specific pathogen infection mechanisms shape transmission patterns.
After ingesting a virus-laden blood meal, molecular interactions between mosquito cells and the pathogen begin. First, viral particles attach to cellular receptors, initiating endocytosis or activating specific pathways to access cells [26]. How DENV and CHIKV enter mosquito cells is not completely understood, though the two arboviruses are thought to differ. For DENV, the specific identity of mosquito cellular receptors remains elusive, although several putative receptors have been suggested [27,28]. One of these, Prohibitin, is a ubiquitously expressed, conserved protein shown to interact with DENV-2 [29], and there is mixed evidence for a role for clathrin [30,31]. CHIKV cellular internalization is thought to be mediated through clathrin-mediated endocytosis (CME), a key process in vesicular trafficking [32–34]. Both viruses are icosahedral in shape, however, CHIKV presents a T = 4 geometry while DENV has a pseudo T = 3. CHIKV genome is organized with 4 non-structural proteins at the 5’ followed by 4 structural proteins, while DENV genome organization is inverted, with the structural proteins at the 5’ end followed by non-structural proteins. Upon entry, both viruses are uncoated through intracellular specific conditions, such as acidic pH, and replicate in the cytoplasm [35]. Viral RNA is then translated into a polyprotein (or polyproteins in the case of CHIKV [36]) which is processed during maturation by host and viral enzymes. Glycoproteins are then inserted in the endoplasmic reticulum, where virion assembly occurs. For DENV, immature virions are further processed through the Golgi membrane system by carbohydrate addition and modification and then follow the exocytosis pathway and are released into the extracellular space by fusion of virion-containing vesicles with the plasma membrane. CHIKV is post transcriptionally modified and glycosylated in the endoplasmic reticulum and translocated to the Golgi apparatus to be packed in vesicles and delivered to the cell membrane. Here, further maturation occurs by acquisition of membrane envelope, then virions are released via exocytosis. In sum, there are both similarities and differences in how the two viruses gain entry to cells and use cellular pathways to replicate [37,38].
Cellular tropism for both DENV and CHIKV appears widespread to multiple mosquito organs, including midgut, salivary glands, fat body tissue, nervous, tracheal, and reproductive systems [39–45]. Interestingly, variation in the replication rates between of the two viruses inside Ae. aegypti have been reported. Specifically, a study measuring viral RNA levels found CHIKV had higher copy number than DENV in midguts, and CHIKV was also detected far earlier than DENV in salivary glands [46]. Moreover, co-infection with DENV and CHIKV increases overall mosquito infection rates up to 100% for both viruses, while also enhancing CHIKV and DENV viral replication in the midgut and salivary glands, compared to each virus alone [46]. These findings suggest the cellular and molecular mechanisms by which Ae. aegypti responds to DENV and CHIKV infection may interact or overlap, though enhancement during co-infection may also arise from energetic constraints.
Viral infections trigger cellular and humoral immune responses inside the mosquito. The core pathways of the insect immune response are the Janus Kinase-Signal Transducer Activator of transcription (JAK/STAT), Toll, Immune Deficiency (IMD) and RNA interference (RNAi) [26]. Viral components also interact with pattern recognition receptors, triggering processes such as melanization and the production of anti-microbial peptides (AMP), including attacin, defensin and cecropin, among others [47–49]. Interestingly, some of the traditional innate immune pathways (Toll, JAK/STAT, IMD) that have been shown to contribute to the reduction of DENV replication both in vitro and in vivo play little role in limiting CHIKV infection. Specifically, exogenous activation of the JAK-STAT pathway has been shown to modulate DENV infection but did not enhance resistance to CHIKV [50]. Moreover, CHIKV infection significantly represses the Toll pathway, limiting its efficacy, and the IMD pathway also does not mediate CHIKV infections [51]. Of the major insect immune pathways known to control infection of arboviruses such as DENV, only the RNAi pathway has been shown to play a vital role in limiting CHIKV replication. Specifically, knockdown of Argonaute2 (AGO2), an RNAi effector molecule, resulted in significant increase of viral RNA replication and titers [51]. Consistent with this evidence that mosquito responses to the two viruses are distinct, transcriptomic profiles of DENV and CHIKV infected mosquitoes revealed little overlap between responsive genes [52].
One path to resolving these heterogenous findings is to exploit natural genetic variation in infection response in Ae. aegypti. By examining population level genetic variation in antiviral response, several recent studies have discovered novel, non-canonical antiviral genes in Drosophila [53,54] and in Ae. aegypti [55]—demonstrating the promise of this approach. Here, we used a modified full-sib breeding design to assess the family-level variation in CHIKV and DENV loads, using a Mexican population of Ae. aegypti. By examining viral loads in a common set of mosquito families, our design can uncover the contribution of genetic variation to viral load variance as well as assess the extent to which mosquito responses to these viruses are due to shared or independent genetic mechanisms. We also examined transcriptional differences in mosquito families representing the phenotypic extremes for CHIKV load (lowest and highest mean total body loads), allowing us to identify individual genes that may underlie this CHIKV variation. Our work thus provides insights into the basis of the vector’s own natural immune response to CHIKV and DENV, and points to specific candidate antiviral genes to target with emerging insect genetic tools, such as CRISPR-Cas9 [56,57].
Results
Family-level variation in the heritability of DENV and CHIKV viral loads
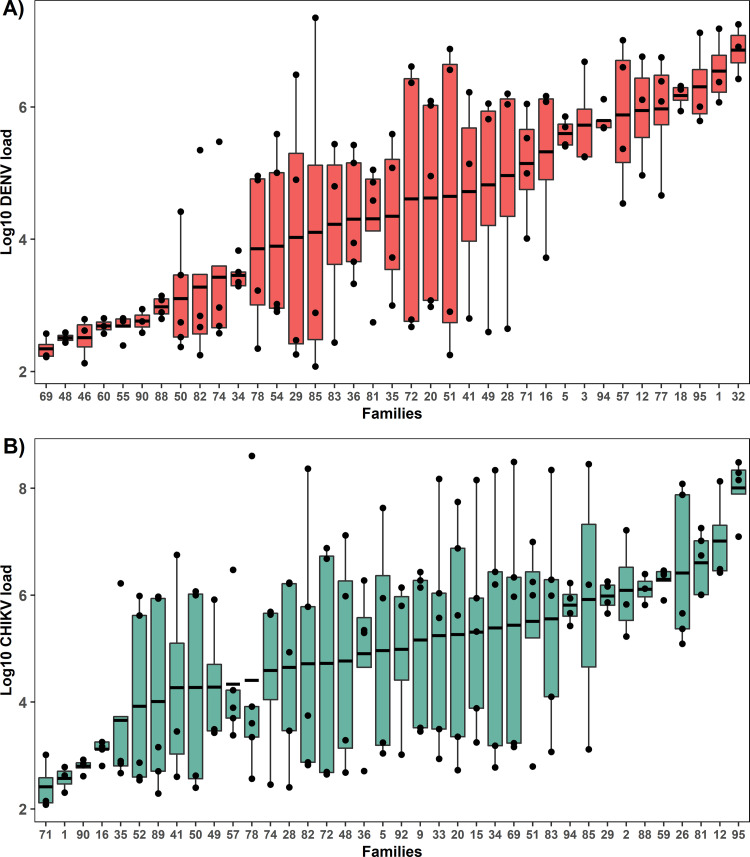
To estimate the heritability of DENV and CHIKV loads, we examined viral loads in whole bodies of female mosquitoes previously fed either virus (Fig 1, experiment 1). The data came from mosquitoes representing 37 mosquito families bred within a modified full-sib breeding design. After starting with over 600 individual mating pair crosses, we obtained data for 37 because most families either did not produce enough eggs or blood feed sufficiently. The siblings in each family were split in half, and each set was fed one virus, allowing us to test for genetic correlation. Mosquitoes were assayed for viral loads 7 days post-infection by extracting RNA and performing DENV or CHIKV-specific RT-qPCR on 3–5 individuals per family. Viral loads were extremely variable across families, spanning from hundreds to millions of copies; 102−107 per body for DENV and 102−108 per body for CHIKV. Both H2 values were significantly greater than zero, indicating a genetic basis underlies variation in viral loads for both DENV and CHIKV. The broad-sense heritability (H2) of DENV load was estimated to be 0.40 (Fig 2A. LRT: X2 = 24.8, P<0.001). For CHIKV load, H2 was 0.18 (Fig 2B. LRT: X2 = 5.9, P = 0.015). Both viral load distributions exhibit some bimodality, a common feature of infection in mosquitoes we have explored previously [15].
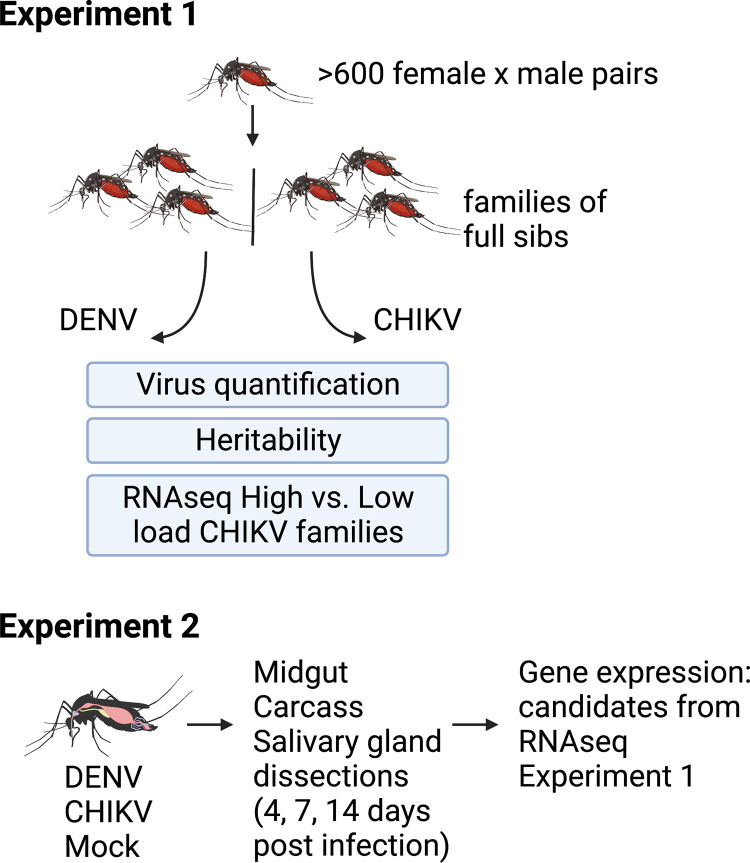
Fig 1. Overview of experimental design.
In experiment 1, we studied CHIKV and DENV loads in siblings from single female x male pair crosses and calculated the heritabilities and genetic correlations of the loads. We then carried out RNAseq comparing two groups; families with the four highest and four lowest mean CHIKV loads. In experiment 2, we reared a separate set of mosquitoes from the same population and exposed them to CHIKV, DENV, or mock (uninfected cell culture control) infection through blood-feeding treatments and then dissected key tissues (midgut, carcass, salivary glands) at three timepoints post-infection (4, 7, 14). We then examined patterns of gene expression of the top candidate gene from the RNAseq analysis in experiment 1 across treatments and tissues x time. Created with BioRender.com.
Fig 2. Ranked families by viral load.
We ultimately obtained data from 37 families from >600 initial paired crosses. A) DENV load-ranked families and B) CHIKV load-ranked families. Each point represents a single infected mosquito. Box plots show mean viral load ± SEM of each distinct Ae. aegypti family. Whole mosquito viral load was log10 transformed. N = 3–5 individuals per family.
Genetic correlation between DENV and CHIKV across mosquito families
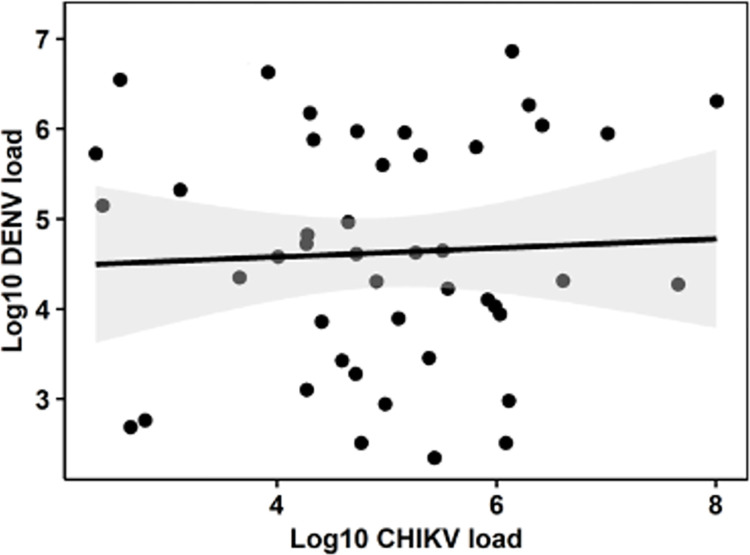
We estimated the genetic correlation, rDENV,CHIKV, to assess the extent to which genetic variation underlying the control of DENV and CHIKV loads was shared. A value indistinguishable from 1 would indicate they are effectively the same genetic ‘trait’ whereas values closer to zero indicate that the traits are genetically independent. We estimated rDENV,CHIKV = 0.1 and this value was significantly less than 1.0 (LRT: χ2 = 5.7, df = 1, P = 0.017) but could not be distinguished from zero (LRT: χ2 = 0.1, df = 1, P = 0.752). We found no evidence of genetic correlation between DENV and CHIKV loads between siblings of the same family based on our analysis of 37 families (Fig 3. Pearson’s r = 0.0018, P = 0.99). Furthermore, the correlation was not significantly greater than zero (LRT: X2 = >0.001, P = 1.00). This is underscored by the lack of any overlap between the five families representing the extremes of the distributions for both viruses (Fig 2). Specifically, high-load families for DENV were families 77, 18, 95, 1, and 32, while the same for CHIKV were families 59, 26, 81, 12, and 95. The family numbers were also different for the five low-load families. The independence between viral load traits for siblings indicates a lack of shared control mechanisms for the two viruses.
Fig 3. Scatter plot showing the relationship between mean DENV and CHIKV loads from individual families.
Gray area represents the 95% confidence interval of the regression line. N = 37 mosquito families.
Gene expression of families representing the phenotypic extremes of viral loads for both DENV and CHIKV
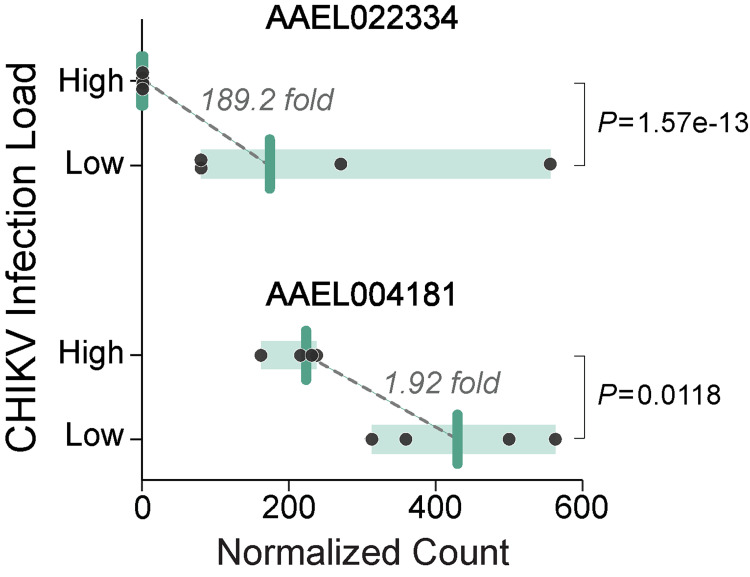
We then tested whether the mean viral loads differed between each virus’s four lowest and four highest families. For both DENV (w = 256, df = 1, P<0.0001) and CHIKV (w = 393, df = 1, P<0.0001), families at the phenotypic extremes differed. Because there has already been substantial profiling of gene expression for Ae. aegypti with differences in susceptibility and viral loads for DENV (reviewed in [58]), we focused on understanding if there were clear differences in gene expression between high and low families for the less well-studied CHIKV. We obtained RNA-Seq expression data for 10,014 genes in the families, each from the high and low family categories (L: 1, 16, 71, 90, H:12, 26, 59, 81). Following multiple test corrections, we identified only two loci that differed consistently (adjusted P<0.05) between the families representing the extremes (Fig 4). The first, LOC110676965, corresponds to AAEL026917, encoding a Zinc finger-containing protein that was identified previously as upregulated in an RNAseq dataset for dengue-infected Ae. aegypti [59]. Similarly, this same gene’s expression responds to Zika infection [60]. The second locus, LOC5564245, encodes AAEL004181-PA, an uncharacterized protein. AAEL004181-PA has no previously reported phenotypes in mosquitoes but is a distant relative in a family of SGS or salivary gland-specific proteins [61]. Interestingly, this locus has been identified as a horizontally transferred gene with the insect endosymbiont, Wolbachia [61–63]. This explains why the next six hits resulting from a protein blast search [64] identify genes in several strains of Wolbachia from Culex species, Drosophila melanogaster, and Drosophila yakuba. Similarities range from 77 to 99%. While there is some controversy over the direction of the horizontal transfer, the preponderance of the evidence is that AAEL004181 is fundamentally a mosquito gene that was transferred into Wolbachia multiple times [61,63]. In Wolbachia, there are two genes, positioned side by side in the genome (known as loci WD0512 and WD0513 in the wMel genome [62], that together are homologous to the mosquito gene. Both WD0512 and WD0513 are actively transcribed [65], suggesting they are functional in Wolbachia.
Fig 4. RNAseq counts for the two loci with significant expression differences between high and low CHIKV-load families.
Points depict normalized read counts of individual families, bars show group medians, and shaded area demarcates the range for each group. Significance was evaluated using Wald tests while correcting for multiple comparisons.
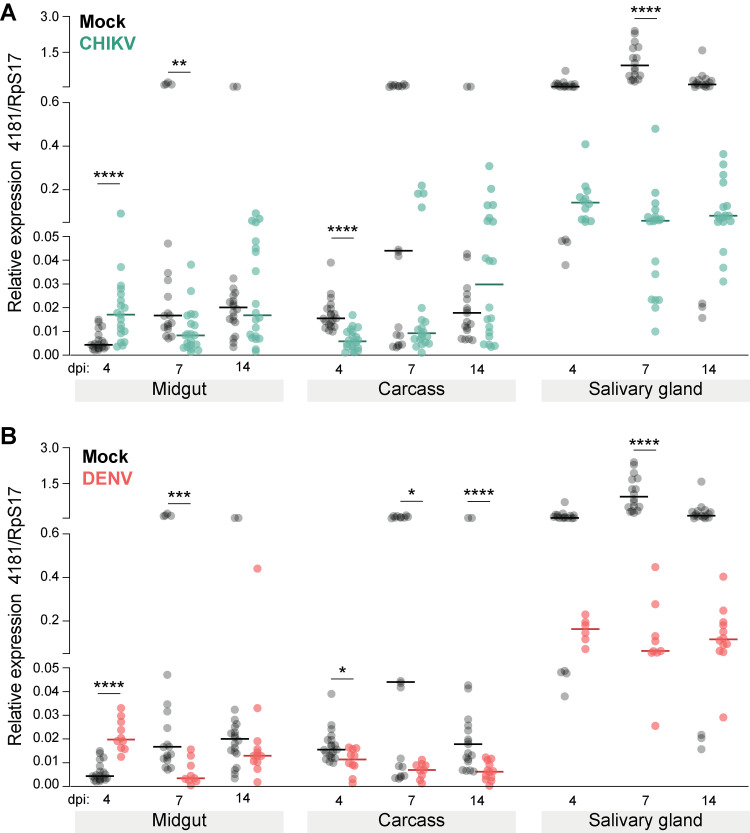
Despite multiple attempts to design primers for the LOC110676965, we were unable to find a set that amplified the locus. This could have resulted from a poor understanding of the underlying intronic structure or, more likely, simply very low-level expression. In our high CHIKV families, for example, the RNAseq read counts were very low (median 0.92 reads/sample) compared to the low families (median 80.7). We were able to design primers to amplify AAEL004181 that detected expression in our samples. We then carried out a separate and more detailed map of expression (Fig 1, experiment 2) over time and across several mosquito tissues (without family structure) to understand its responsiveness to infection. During CHIKV infection, expression of AAEL004181 rises early in the midgut compared to virus-free blood-fed controls (4 dpi, Mann-Whitney U = 55, P<0.0001), but then declines with time in the midgut (7 dpi, Mann-Whitney U = 78, P = 0.0012) and other tissues (carcass, salivary glands, see Fig 5A for full statistical results). A similar pattern is seen for DENV-infected mosquitoes (Fig 5B).
Fig 5.
Tissue-specific expression of AAEL004181 across time in (A) CHIKV- and (B) DENV-infected mosquitoes compared to expression in mock controls. Points depict relative expression level of AAEL004181 in individual samples, while horizontal lines mark group medians. Significance was evaluated by Mann-Whitney U tests between mock and control. * P<0.05, ** P<0.01, *** P<0.001, **** P<0.0001. Note that a day 7 salivary gland outlier in the mock treatment group has been omitted from both panels (value = 7.618). dpi, days post-infection. n = 20 per treatment.
Discussion
We performed a modified full-sib breeding design to study the contribution of the mosquito’s genetic variation while limiting environmental variation, to loads for each virus. By exploring loads for the viruses in siblings of the same family, we were also able to measure the genetic correlation between the individual loads. We used the distribution of loads to select families representing the phenotypic extremes for CHIKV and tested whether there were genes whose expression differed. Our results are consistent with a polygenic basis for both DENV and CHIKV viral loads. Additionally, the lack of genetic correlation between DENV and CHIKV viral loads across siblings demonstrated the complete independence of the genetic response to these two viruses. Finally, our RNAseq comparisons identified only two genes whose expression differed consistently between high and low CHIKV load families.
The heritability values for viral susceptibility for both DENV and CHIKV were significant but fell within different ranges. For DENV, the H2 was 40%. This is in accordance with previous studies examining susceptibility in Ae. aegypti using multiple tissues as proxy. Both Bosio et al. [66] and Ye et al. [67] reported heritabilities of ~40% in the midgut, head, and saliva of DENV-infected mosquitoes. Taken together, this suggests that for DENV infection load, mosquito genetic variation contributes to vector competence almost as much as all other effects combined, including environmental effects. In our experiment, the heritability of CHIKV load was 18%, with almost half of that estimated for siblings in the same family. The lower heritability for CHIKV suggests that environmental or stochastic factors, as well as viral genotype, have a greater role than the mosquito’s own genetic factors in determining variation in load, at least in the tested population [68–70]. CHIKV-infected mosquitoes also exhibited a wider range of viral loads and reached higher absolute viral loads than DENV-infected mosquitoes. The wider range of viral loads in CHIKV was a key contributor to the overall low heritability for this trait. The genetic correlation between siblings for the DENV and CHIKV load trait was not significantly different from zero. This shows independence between the genes that control load for the two viruses. In general, the primary source of genetic correlation is pleiotropy, though linkage disequilibrium can be a contributing factor [71,72]. Our results, therefore, suggest that the genes controlling CHIKV and DENV viral loads are distinct, unlinked and that they lack pleiotropic effects [73].
Phylogenetic studies on Ae. aegypti and DENV have revealed a coevolutionary process between the virus and the co-indigenous mosquito populations [26,47,74] which has in turn shaped susceptibility to the virus. Since CHIKV is an emerging disease, the lack of coevolutionary history with naïve Ae. aegypti populations (i.e., in the Americas) could be a source of the differences between the mechanisms that control infection inside the vector [26]. The global spread of CHIKV and DENV was preceded by the global expansion of Ae. aegypti and Ae. albopictus. While the expansion of Ae. aegypti started over 400 years ago [8], the geographic expansion of Ae. albopictus, a secondary vector of DENV and CHIKV, started in the 1960’s and was previously limited to Southeast Asia [75]. Among the evolutionary forces that drive the evolution of arboviruses, vector interactions may play an important role in selecting viral genotypes [76]. CHIKV emergence in the Indian Ocean was linked to a single mutation that enhanced transmission by Ae. albopictus over Ae. aegypti [77], leading to the hypothesis that the emergence of arboviruses is partially caused by pathogen adaptations to native vector populations. However, invasive viral genotypes that outcompete local genotypes tend to have enhanced viral replication in vectors [78]. These studies highlight how both mosquito and pathogen genetic variation—and their coevolution—can interact to shape the course of pathogen emergence. Such effects are expected to differ between DENV and CHIKV due to their distinct histories with Ae. aegypti.
For a virus such as CHIKV or DENV to act as a selective agent on mosquitoes, it must induce a fitness cost—either directly (i.e., mortality), or indirectly (e.g., energetically, due to immune or stress responses). Fitness changes associated with viral infection in Ae. aegypti have been previously reported. Specifically, CHIKV infection in Aedes mosquitoes decreased the survival of adults and eggs [79] and disrupted the transcription of genes related to the gonotrophic cycle [80]. Infection with another alphavirus, Mayaro virus (MAYV), reduced fecundity without altering longevity in Ae. aegypti [81]. DENV infection in Ae. aegypti has been shown to influence feeding behavior and to reduce longevity, fecundity, and oviposition success [82,83]. These and other fitness costs imposed by infection can act as evolutionary forces and may have shaped vector-virus interactions, though they are potentially limited by low infection rates in wild populations: ~1% for DENV [84–86] and ~5–6% for CHIKV [87,88]. Over time, fitness costs related to infection may have contributed to the observed differences in DENV and CHIKV viral loads in Ae. aegypti, as the two viruses have very different histories of association with this vector.
A more constant evolutionary force that mosquitoes face and that might have potentially shaped the evolution of insect immunity and susceptibility against medically important arboviruses are insect-specific viruses (ISVs), particularly mosquito-specific viruses (MSVs), which cause no obvious fitness cost in mosquitoes [89], though they may have in the past. The majority of Ae. aegypti MSVs are grouped in the Flaviviridae viral family and infect both Ae. aegypti and Ae. albopictus [90]. Other MSV viral families include Bunyaviridae, Mesonviridae and Togaviridae, the last of which has the fewest known MSVs [91]. Interestingly, many of these viruses cluster closely with medically important arboviruses [92]. There is evidence that they interact with mosquito immune pathways such as the RNAi pathway [93], and it is possible MSVs prime the mosquito’s immunity to viruses of public health concern, such as DENV and CHIKV, that are not commonly encountered by mosquitoes in the field. The differences in the heritability estimates and the lack of genetic correlation between DENV and CHIKV viral loads observed in our mosquito populations suggest that this could be the case. Different MSVs could trigger different immune responses. Since the mosquito has likely encountered more MSVs from the Flaviviridae (like DENV) than the Togaviridae (like CHIKV) across evolutionary time, their immune response could be more ‘tailored’ to Flaviviruses like DENV, and this could possibly contribute to the overall lower viral loads measured for DENV relative to CHIKV in our experiment.
Interestingly, our RNAseq screen returned a locus that has been heavily studied in Wolbachia. The Wolbachia genes homologous to AAEL004181, WD0512 and WD0513, both sit within an eight-gene operon called Octomom that underlies striking differences in how strains wMel and wMelPop affect their arthropod hosts. wMelPop is associated with much higher host mortality but also greater antiviral protection in infected Drosophila—effects both explicitly linked to greater Octomom copy number and correlated with higher expression levels of Octomom genes [94–96]. This raises the possibility that this homolog inherently produces an antiviral effect, whether it is expressed from a Wolbachia genome or a mosquito one, though the mechanism may be more complex [95]. However, molecular details of how AAEL004181/WD0512-WD0513 act in cells—and ultimately upon viruses—are currently lacking and should be investigated in future work. Octomom-associated sequences have been found in several Wolbachia strains, but not all, and a few other endosymbiotic bacteria, and mosquitoes [97]. Interestingly, the WD0512-WD0513 genes were lost out of the wMelPop-CLA (cell line adapted) strain adapted through long-term serial passage in Aedes albopictus cells [98]. The mosquito homolog belongs to a gene family that encodes receptors mediating the invasion of malaria sporozoites into the mosquito salivary gland [63]. Tools for genetic manipulation are sophisticated in mosquitoes [99] but are not available in Wolbachia, limiting research progress. However, our results suggest the study of this gene in mosquitoes could potentially also shed light on its role in Wolbachia.
Our experimental design has some caveats that may limit the scope of its overall interpretation. Immune effector expression can vary depending on the time post-infection, and it is possible that the expression levels of specific genes might peak, e.g., immediately after blood feeding, and therefore would be missed by our sampling regime [47,100]. However, given the scale of the family-based design, we could only sample a single time point. The expression results shown here could therefore exclude some genes with time- or tissue-constrained differences and would also miss effects stemming from variation in coding sequence as we focused on expression level. Additionally, these are whole body viral loads. More focused work may wish to focus on disseminated virus as a closer proxy for transmissibility [101].
Quantitative genetic estimates—such as we report here—have real-world importance in vector biology as they can help us predict the strength and impact of new outbreaks as well as infection dynamics between vectors and pathogens. Specifically, the higher heritability of load and more homogeneous viral loads of DENV (compared to CHIKV) indicate that selection for higher or lower viral loads in a mosquito population could occur faster for DENV than for CHIKV [68]. The limited genetic variation for modulating CHIKV infections in Ae. aegypti may also shed light on the explosive re-emergence of this virus in naïve populations on the American continent, though it remains unclear if the low heritability of CHIKV load is a cause and/or consequence of this recent explosion. Nevertheless, we find the mechanisms that control viral load in mosquito populations are markedly different between DENV and CHIKV. From an epidemiological perspective, if the genes that control viral load for multiple viruses are different and unlinked, this introduces a challenge to vector control methods aiming to target and modify universal loci that could be used against multiple arboviruses. In this case, more virus-specific gene editing approaches would be necessary [102]. Last, the appearance of a Wolbachia-associated horizontally transferred homolog in our candidates that associate with CHIKV load is interesting, particularly given the capacity for Wolbachia to limit viral replication via a trait known as pathogen blocking. Unfortunately, without the ability to genetically modify Wolbachia, it is hard to test the functional role of this homolog in the symbiont. However, future studies could take advantage of genetic tools in mosquitoes to better characterize their role in viral infections.
Methods
Mosquito line and stock-rearing practices
An F3 Ae. aegypti mosquito line based on thousands of eggs collected over multiple months from traps placed over the urban environment of Monterrey, Mexico, was reared in the laboratory for three additional generations (i.e., to F6) to expand in preparation for the experiments. Mosquitoes were reared under standard conditions: 26°C, 65% relative humidity, and a 12 h light/dark cycle. Larvae were maintained on fish food (Tetramin, Tetra). Adults were fed with 10% sucrose solution ad libitum. Mosquitoes were fed human blood (BioIVT, Hicksville, NY, USA) for egg collection using a Hemotek system (Hemotek).
Breeding design
A modified full–sib breeding design [57,67,68] was carried out on the mosquitoes in combination with DENV and CHIKV infection. After expanding the mosquito line over three generations, F6 eggs were hatched in synchrony and reared at low density (~150 larvae per 3 L of RO water). After pupation, males and females were separated and transferred to 30 × 30 × 30 cm cages at a density of ~250 individuals per cage. Six to seven- day old virgin adult females were blood-fed and then 250 virgin males were then added for mating. To achieve data from ~40 families ultimately, which is sufficient for such a design [67], we had to set up a > 600 blood-fed and mated females given the proportions of females that either do not lay or that would lay insufficient eggs or that blood fed poorly. Such females were placed in small individual housings containing moist filter paper. Egg papers were collected and dried 3–4 days later for short-term storage. Only female lineages that laid > 60 eggs could be used for the downstream design, given that half of the eggs would be male, conservative estimates of 80% blood feeding participation, and the need to have individuals in both DENV and CHIKV treatments. The eggs of each family were hatched separately, and after pupation, females were separated and split into two cups with a minimum of 8 individuals per cup. These females were maintained on sucrose until vector competence experiments.
Virus culture
All experiments were carried out using the CHIKV strain 20235-St Martin 2013 (NR49901, BEI Resources), that is a member of the Asian genotype [103], and the DENV-2 strain 429557-Mex 2005 (NR12216, BEI Resources), both from Latin American outbreaks. Virus was cultured in C6/36 cells, as previously reported [15,57,67,104]. Briefly, C6/36 Ae. albopictus cells were grown in RPMI 1640 media (Life Technologies, Carlsband, CA, USA) and supplemented with 10% heat-inactivated fetal bovine serum (FBS, Life Technologies) containing glutamine and 20 mM HEPES (Sigma-Aldrich, St. Louis, MO, USA). Cells were grown to a confluency of 80% and then independently infected with DENV-2 or CHIKV. Infected culture flasks were incubated at 27°C. For DENV-2, after 7 days post-infection, supernatant was harvested at a titer of 1.0 x 105 focus forming units per ml (FFU/ml). For CHIKV, supernatant was harvested at 2 days post-infection at a titer of 8.7 x 105 and diluted to a final concentration of 1.0 x 105 FFU/ml. This starting infectious dose was intentionally chosen to be low enough to capture variation in mosquito viral loads at 5 days post-infection, particularly for CHIKV that replicates quickly [25]. Both viruses were stored at -80°C in 1 ml single-use aliquots for vector competence experiments.
Mosquito infections
Methods for oral mosquito infections have been fully described previously [15,57,105]. Prior to infections, mosquitoes were starved for 24 h. Half of the 6–7-day old females of each family were challenged with DENV-2 (1st cup) and the other half (2nd cup) with CHIKV at equal viral titers using a 1:1 mix of the frozen titrated aliquots and human blood. Mosquitoes were allowed to feed for 30 minutes. Shortly after, they were anesthetized with CO2 and unfed individuals were removed and discarded. Feeding rates were very high, 80–90% for both DENV and CHIKV-laden blood. All CHIKV work was carried out at the Penn State Eva J. Pell ABSL-3 laboratory and DENV work in the McGraw BSL-2 lab.
Viral quantification
Seven days post-infection, whole mosquitoes were collected, homogenized, and stored in TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) at -80°C. RNA was extracted according to the manufacturer’s protocol. RNA was eluted in 25 μl of DNA/RNA free water and treated with DNAse (Life Technologies, Carlsbad, CA, USA). From this RNA, DENV and CHIKV loads were quantified using 4× TaqMan Fast Virus 1-Step Master Mix (Applied Biosystems, Foster City, CA, USA) in individual 10 μl reactions containing virus-specific primers and probes (Table 1). RT-qPCR reactions were run in a LightCycler 480 instrument (Roche Applied Science, Switzerland). The thermal cycling conditions were 50°C for 5 min for reverse transcription, 95°C for 10s for RT inactivation/denaturation followed by 50 amplification cycles of 95°C for 3s, 60°C for 30s, and 72°C for 1s. Standard curves for both DENV and CHIKV were generated as described elsewhere [67,106] and contained a ~100 bp fragment of the 3’UTR of the DENV genome and a ~140 bp fragment of the CHIKV genome, respectively. The standard curve spanned from 10 to 107 copies/reaction with a limit of detection of 100 copies for both viruses. The viral load in each sample was extrapolated from the standard curve as copies per mosquito.
Table 1. Primers and probes for virus quantification.
| GenBank ID | Virus | Direction | Sequence (5’- 3’) |
|---|---|---|---|
| NC_001474.2 | DENV-2 | Forward | AAGGACTAGAGGTTAGAGGAGACCC |
| Reverse | CGTTCTGTGCCTGGAATGATG | ||
| Probe | FAM-AACAGCATATTGACGCTGGGAGAGACCAGA-BHQ1 | ||
| MT228632 | CHIKV | Forward | CACCCGAAGTAGCCCTGAATG |
| Reverse | TCCGAACATCTTTCCTCCCG | ||
| Probe | 5CY5-GAGAATAGCCCGCTGTCTAGATCCAC-3BHQ2 |
Estimation of viral load heritability
We tested for genetic variation and broad-sense heritability (H2) for DENV and CHIKV loads across our family design. We estimated the parameters using a modified full-sib breeding design and the random effect linear model previously described [57,67]:
| (1) |
Zij is the trait value for the jth female from the ith family, fi is the random effect of the ith family, and Eij the unexplained error. To test if the genetic variation was significant, the family term was fitted as a random effect and the model (1) was compared to a reduced model without the family term. Loglikelihoods of both models were compared and twice the difference was tested against a chi-squared distribution with a single degree of freedom. Broad sense heritability was estimated as twice the family variance component (σ family) divided by the total phenotypic variance (σ family + σ error), [107]. All models were constructed in SAS Studio version 3.8 (SAS Institute, Cary, NC, USA). Genetic correlations between DENV and CHIKV infected siblings were estimated using a bivariate version of model (1) and fitting unrestrictive covariance correlations at the family level (TYPE = UNR option) using the SAS Proc MIXED command. The significance of the genetic correlation was tested by loglikelihood ratio tests.
RNA library preparation and sequencing
We selected four extreme families (Fig 1) from the high (families 59, 26, 81, and 12) and low (families 71, 1, 90, 16) ends of the CHIKV viral load distribution and pooled equal amounts of total body RNA from 4–5 mosquitoes per family. RNA concentrations were quantified using a Qubit 2.0 Fluorometer (ThermoFisher Scientific, Waltham, MA, USA), and RNA integrity was checked with a 4200 TapeStation (Agilent Technologies, Palo Alto, CA, USA) following manufacturer protocols. rRNA was depleted using a QIAGEN FastSelect rRNA HMR Kit (Qiagen, Hilden, Germany). We then used an NEBNext Ultra II RNA Library Preparation Kit for Illumina, following the manufacturer’s recommendations (NEB, Ipswich, MA, USA), to prepare RNA sequencing libraries. Briefly, enriched RNAs were fragmented for 15 m at 94°C. First-strand and second-strand cDNAs were subsequently synthesized, end-repaired, adenylated at 3’ends, and universal adapters were ligated to cDNA fragments followed by index addition and library enrichment with limited-cycle PCR. Sequencing libraries were validated using the Agilent Tapestation 4200 (Agilent Technologies, Palo Alto, CA, USA), and quantified using a Qubit 2.0 Fluorometer (ThermoFisher Scientific, Waltham, MA, USA) as well as by quantitative PCR (KAPA Biosystems, Wilmington, MA, USA). The sequencing libraries were multiplexed and clustered on one flowcell. After clustering, the flowcell was loaded on the Illumina HiSeq instrument according to the manufacturer’s instructions. The samples were sequenced using a 2x150 Pair-End (PE) configuration. Raw sequence data (.bcl files) generated from Illumina HiSeq were converted into fastq files and de-multiplexed using Illumina bcl2fastq program version 2.20. One mismatch was allowed for index sequence identification.
Data analysis
After demultiplexing, sequence data were checked for overall quality and yield. Sequence reads were then trimmed to remove adapter sequences and nucleotides with poor quality using Trimmomatic v.0.36 [108,109]. The trimmed reads were mapped to the reference genomes using the STAR aligner v.2.5.2b [109]. The STAR aligner is a splice-aware aligner that detects and incorporates splice junctions to help align the entire read sequences. BAM files were generated as a result of this step. Unique gene hit counts were calculated by using featureCounts from the Subread package v.1.5.2 [110]. Only unique reads within exon regions were counted. After extraction of gene hit counts, the gene hit counts table was used for downstream differential expression (DE) analysis. Using DESeq2 [111], we compared gene expression between groups. We used Wald tests to evaluate the significance and calculated log2 fold changes. Genes with adjusted P values < 0.05 and absolute log2 fold changes >1 were called as differentially expressed genes for each comparison.
Gene expression
In a subsequent experiment, so that different tissues and time points could be explored, mosquitoes were fed CHIKV, DENV, or mock (virus-free culture media) under the same conditions as for the heritability measures (as above, virus load 105/ml), and then tissues (midgut, carcass, salivary glands) were dissected at 4-, 7-, and 14-days post-infection (dpi). Rather than demonstrating expression patterns across family groups, we sought to characterize the baseline and induced expression of the candidate genes. Tissues were dissected on ice in PBS and then homogenized in Trizol with a glass bead as above and as previously described [15]. RNA extraction was then carried out as above. Retrotranscription of RNA to cDNA and gene expression analysis was carried out on a LightCycler 480 instrument (Roche) using the Script One-step SYBR Green qRT-PCR (Quantabio, Beverly, MA, USA) according to the manufacturer’s protocol. All CT values for AAEL004181 (Forward 5’- GCCATCGCCGCAACTTCAGC -3’, Reverse 5’- CACCCATGGCTCCCGATCCG -3’) were normalized to the housekeeping Ae. aegypti gene RpS17 or AAEL004175 (Forward 5’- TCCGTGGTATCTCCATCAAGCT-3’, Reverse 5’-CACTTCCGGCACGTAGTTGTC-3’) as per previous [112]. Primers for AAEL004181 were designed using Primer3 v 0.4.0 Gene expression ratios were obtained using the ΔΔCt method [113]. PCR amplification cycled 45 times at 95°C for 3 sec and 60°C for 30 sec, and the final cycle was followed by a melting curve analysis. Expression of RpS17 did not vary between virus-infected vs mock-infected control mosquitoes for any of the tissue x time point treatments examined (S1 Fig).
Supporting information
Three tissues, midgut (MG), carcass (Car), and salivary glands (SG), were dissected on three different days post-infection (dpi).
(PDF)
Acknowledgments
The authors would like to thank Michael Cannon for his assistance with mosquito rearing.
Data Availability
All viral load data, RNAseq read count data, and BAM files are available through figshare DOI 10.6084/m9.figshare.20483691.
Funding Statement
The work was supported by an NIH R01 grant to SFC and EAM (AI143758). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kading RC, Brault AC, Beckham JD. Global perspectives on arbovirus outbreaks: a 2020 snapshot. Trop Med Infect Dis. 2020;5: 142. doi: 10.3390/tropicalmed5030142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilder-Smith A, Gubler DJ, Weaver SC, Monath TP, Heymann DL, Scott TW. Epidemic arboviral diseases: priorities for research and public health. Lancet Infect Dis. 2017;17: e101–e106. doi: 10.1016/S1473-3099(16)30518-7 [DOI] [PubMed] [Google Scholar]
- 3.Rajapakse S. Dengue shock. J Emerg Trauma Shock. 2011;4: 120. doi: 10.4103/0974-2700.76835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mehta R, Gerardin P, de Brito CAA, Soares CN, Ferreira MLB, Solomon T. The neurological complications of chikungunya virus: A systematic review. Rev Med Virol. 2018;28: e1978. doi: 10.1002/rmv.1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suhrbier A. Rheumatic manifestations of chikungunya: emerging concepts and interventions. Nat Rev Rheumatol. 2019;15: 597–611. doi: 10.1038/s41584-019-0276-9 [DOI] [PubMed] [Google Scholar]
- 6.Huang Y-J, Higgs S, Horne K, Vanlandingham D. Flavivirus-mosquito interactions. viruses. 2014;6: 4703–4730. doi: 10.3390/v6114703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim E, Lee W, Madzokere E, Herrero L. Mosquitoes as suitable vectors for alphaviruses. Viruses. 2018;10: 84. doi: 10.3390/v10020084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Powell JR, Gloria-Soria A, Kotsakiozi P. Recent history of Aedes aegypti: Vector genomics and epidemiology records. Bioscience. 2018;68: 854–860. doi: 10.1093/biosci/biy119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, et al. The global distribution and burden of dengue. Nature. 2013;496: 504–507. doi: 10.1038/nature12060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brathwaite Dick O, San Martín JL, Montoya RH, del Diego J, Zambrano B, Dayan GH. The history of dengue outbreaks in the Americas. Am J Trop Med Hyg. 2012;87: 584–593. doi: 10.4269/ajtmh.2012.11-0770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rougeron V, Sam I-C, Caron M, Nkoghe D, Leroy E, Roques P. Chikungunya, a paradigm of neglected tropical disease that emerged to be a new health global risk. J Clin Virol. 2015;64: 144–152. doi: 10.1016/j.jcv.2014.08.032 [DOI] [PubMed] [Google Scholar]
- 12.Wimalasiri-Yapa BMCR, Stassen L, Huang X, Hafner LM, Hu W, Devine GJ, et al. Chikungunya virus in Asia–Pacific: a systematic review. Emerg Microbes Infect. 2019;8: 70–79. doi: 10.1080/22221751.2018.1559708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeller H, Van Bortel W, Sudre B. Chikungunya: Its history in Africa and Asia and its spread to new regions in 2013–2014. J Infect Dis. 2016;214: S436–S440. doi: 10.1093/infdis/jiw391 [DOI] [PubMed] [Google Scholar]
- 14.Yactayo S, Staples JE, Millot V, Cibrelus L, Ramon-Pardo P. Epidemiology of chikungunya in the Americas. J Infect Dis. 2016;214: S441–S445. doi: 10.1093/infdis/jiw390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Novelo M, Hall MD, Pak D, Young PR, Holmes EC, McGraw EA. Intra-host growth kinetics of dengue virus in the mosquito Aedes aegypti. PLOS Pathog. 2019;15: e1008218. doi: 10.1371/journal.ppat.1008218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gubler DJ, Nalim S, Saroso JS, Saipan H, Tan R. Variation in susceptibility to oral infection with dengue viruses among geographic strains of Aedes aegypti. Am J Trop Med Hyg. 1979;28: 1045–1052. doi: 10.4269/ajtmh.1979.28.1045 [DOI] [PubMed] [Google Scholar]
- 17.Ye YH, Ng TS, Frentiu FD, Walker T, van den Hurk AF, O’Neill SL, et al. Comparative susceptibility of mosquito populations in North Queensland, Australia to oral infection with dengue virus. Am J Trop Med Hyg. 2014;90: 422–430. doi: 10.4269/ajtmh.13-0186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaye A, Faye O, Diagne CT, Faye O, Diallo D, Weaver SC, et al. Oral susceptibility of Aedes aegypti (Diptera: Culicidae) from Senegal for dengue serotypes 1 and 3 viruses. Trop Med Intl Health. 2014;19: 1355–1359. doi: 10.1111/tmi.12373 [DOI] [PubMed] [Google Scholar]
- 19.Reiskind MH, Pesko K, Westbrook CJ, Mores CN. Susceptibility of Florida mosquitoes to infection with chikungunya virus. Am J Trop Med Hyg. 2008;78: 422–5. [PMC free article] [PubMed] [Google Scholar]
- 20.Gokhale MD, Paingankar MS, Sudeep AB, Parashar D. Chikungunya virus susceptibility & variation in populations of Aedes aegypti (Diptera: Culicidae) mosquito from India. Indian J Med Res. 2015;142: 33. doi: 10.4103/0971-5916.176614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen T-H, Jian S-W, Wang C-Y, Lin C, Wang P-F, Su C-L, et al. Susceptibility of Aedes albopictus and Aedes aegypti to three imported chikungunya virus strains, including the E1/226V variant in Taiwan. J Formosan Med Assoc. 2015;114: 546–552. doi: 10.1016/j.jfma.2014.12.002 [DOI] [PubMed] [Google Scholar]
- 22.Alto BW, Wiggins K, Eastmond B, Velez D, Lounibos LP, Lord CC. Transmission risk of two chikungunya lineages by invasive mosquito vectors from Florida and the Dominican Republic. PLOS Negl Trop Dis. 2017;11: e0005724. doi: 10.1371/journal.pntd.0005724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tesh RB, Rosen L, Gubler DJ. Variation among geographic strains of Aedes albopictus in susceptibility to infection with chikungunya virus. Am J Trop Med Hyg. 1976;25: 326–335. doi: 10.4269/ajtmh.1976.25.326 [DOI] [PubMed] [Google Scholar]
- 24.Vega-Rúa A, Zouache K, Girod R, Failloux A-B, Lourenço-de-Oliveira R. High level of vector competence of Aedes aegypti and Aedes albopictus from ten American countries as a crucial factor in the spread of chikungunya virus. J Virol. 2014;88: 6294–6306. doi: 10.1128/JVI.00370-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dubrulle M, Mousson L, Moutailler S, Vazeille M, Failloux A-B. Chikungunya virus and Aedes mosquitoes: saliva is infectious as soon as two days after oral infection. PLOS One. 2009;4: e5895. doi: 10.1371/journal.pone.0005895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alonso-Palomares LA, Moreno-García M, Lanz-Mendoza H, Salazar MI. Molecular basis for arbovirus transmission by Aedes aegypti mosquitoes. Intervirol. 2018;61: 255–264. doi: 10.1159/000499128 [DOI] [PubMed] [Google Scholar]
- 27.Smith DR. An update on mosquito cell expressed dengue virus receptor proteins. Insect Mol Biol. 2012;21: 1–7. doi: 10.1111/j.1365-2583.2011.01098.x [DOI] [PubMed] [Google Scholar]
- 28.Cruz-Oliveira C, Freire JM, Conceição TM, Higa LM, Castanho MARB, Da Poian AT. Receptors and routes of dengue virus entry into the host cells. FEMS Microbiol Rev. 2015;39: 155–170. doi: 10.1093/femsre/fuu004 [DOI] [PubMed] [Google Scholar]
- 29.Kuadkitkan A, Wikan N, Fongsaran C, Smith DR. Identification and characterization of prohibitin as a receptor protein mediating DENV-2 entry into insect cells. Virol. 2010;406: 149–161. doi: 10.1016/j.virol.2010.07.015 [DOI] [PubMed] [Google Scholar]
- 30.Mosso C, Galván-Mendoza IJ, Ludert JE, del Angel RM. Endocytic pathway followed by dengue virus to infect the mosquito cell line C6/36 HT. Virol. 2008;378: 193–199. doi: 10.1016/j.virol.2008.05.012 [DOI] [PubMed] [Google Scholar]
- 31.Piccini LE, Castilla V, Damonte EB. Dengue-3 virus entry into Vero cells: Role of clathrin-mediated endocytosis in the outcome of infection. PLOS One. 2015;10. doi: 10.1371/journal.pone.0140824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong S, Balaraman V, Kantor AM, Lin J, Grant DG, Held NL, et al. Chikungunya virus dissemination from the midgut of Aedes aegypti is associated with temporal basal lamina degradation during bloodmeal digestion. PLOS Negl Trop Dis. 2017;11: e0005976. doi: 10.1371/journal.pntd.0005976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoornweg TE, van Duijl-Richter MKS, Ayala Nuñez N V., Albulescu IC, van Hemert MJ, Smit JM. Dynamics of chikungunya virus cell entry unraveled by single-virus tracking in living cells. J Virol. 2016;90: 4745–4756. doi: 10.1128/JVI.03184-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee RCH, Hapuarachchi HC, Chen KC, Hussain KM, Chen H, Low SL, et al. Mosquito cellular factors and functions in mediating the infectious entry of chikungunya virus. PLOS Negl Trop Dis. 2013;7: e2050. doi: 10.1371/journal.pntd.0002050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez AK, Muñoz AL, Segura NA, Rangel HR, Bello F. Molecular characteristics and replication mechanism of dengue, zika and chikungunya arboviruses, and their treatments with natural extracts from plants: An updated review. EXCLI J. 2019;18: 988–1006. doi: 10.17179/excli2019-1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Metz SW, Geertsema C, Martina BE, Andrade P, Heldens JG, Van Oers MM, et al. Functional processing and secretion of Chikungunya virus E1 and E2 glycoproteins in insect cells. Virol J. 2011;8: 1–12. doi: 10.1186/1743-422X-8-353/FIGURES/7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Halstead SB, editor. Dengue. London: Imperial College Press; 2008. [Google Scholar]
- 38.Burt FJ, Chen W, Miner JJ, Lenschow DJ, Merits A, Schnettler E, et al. Chikungunya virus: an update on the biology and pathogenesis of this emerging pathogen. Lancet Infect Dis. 2017;17: e107–e117. doi: 10.1016/S1473-3099(16)30385-1 [DOI] [PubMed] [Google Scholar]
- 39.Wong HV, Chan YF, Sam I-C, Sulaiman WYW, Vythilingam I. Chikungunya virus infection of Aedes mosquitoes. 2016. pp. 119–128. doi: 10.1007/978-1-4939-3618-2_11 [DOI] [PubMed] [Google Scholar]
- 40.Vega-Rúa A, Schmitt C, Bonne I, Krijnse Locker J, Failloux A-B. Chikungunya virus replication in salivary glands of the mosquito Aedes albopictus. Viruses. 2015;7: 5902–5907. doi: 10.3390/v7112917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kantor A, Grant D, Balaraman V, White T, Franz A. Ultrastructural analysis of chikungunya virus dissemination from the midgut of the yellow fever mosquito, Aedes aegypti. Viruses. 2018;10: 571. doi: 10.3390/v10100571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ziegler SA, Nuckols J, McGee CE, Huang Y-JS, Vanlandingham DL, Tesh RB, et al. In vivo imaging of Chikungunya virus in mice and Aedes mosquitoes using a renilla luciferase clone. Vector-Borne Zoonot Dis. 2011;11: 1471–1477. doi: 10.1089/vbz.2011.0648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salazar MI, Richardson JH, Sánchez-Vargas I, Olson KE, Beaty BJ. Dengue virus type 2: replication and tropisms in orally infected Aedes aegypti mosquitoes. BMC Microbiol. 2007;7: 9. doi: 10.1186/1471-2180-7-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhan X, He A, Gan M, Zhang M, Liu J, Li Z, et al. Quantitative analysis of replication and tropisms of dengue virus type 2 in Aedes albopictus. Am J Trop Med Hyg. 2010;83: 700–707. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matusali G, Colavita F, Bordi L, Lalle E, Ippolito G, Capobianchi M, et al. Tropism of the chikungunya virus. Viruses. 2019;11: 175. doi: 10.3390/v11020175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Le Coupanec A, Tchankouo-Nguetcheu S, Roux P, Khun H, Huerre M, Morales-Vargas R, et al. Co-Infection of mosquitoes with chikungunya and dengue viruses reveals modulation of the replication of both viruses in midguts and salivary glands of Aedes aegypti mosquitoes. Int J Mol Sci. 2017;18: 1708. doi: 10.3390/ijms18081708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sim S, Dimopoulos G. Dengue Virus inhibits immune responses in Aedes aegypti cells. PLOS One. 2010;5: e10678. doi: 10.1371/journal.pone.0010678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Agaisse H, Perrimon N. The roles of JAK/STAT signaling in Drosophila immune responses. Immunol Rev. 2004;198: 72–82. doi: 10.1111/j.0105-2896.2004.0133.x [DOI] [PubMed] [Google Scholar]
- 49.Antonova Y, Alvarez KS, Kim YJ, Kokoza V, Raikhel AS. The role of NF-κB factor REL2 in the Aedes aegypti immune response. Insect Biochem Mol Biol. 2009;39: 303–314. doi: 10.1016/j.ibmb.2009.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jupatanakul N, Sim S, Angleró-Rodríguez YI, Souza-Neto J, Das S, Poti KE, et al. Engineered Aedes aegypti JAK/STAT pathway-mediated immunity to dengue virus. PLOS Negl Trop Dis. 2017;11: e0005187. doi: 10.1371/journal.pntd.0005187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McFarlane M, Arias-Goeta C, Martin E, O’Hara Z, Lulla A, Mousson L, et al. Characterization of Aedes aegypti innate-immune pathways that limit chikungunya virus replication. PLOS Negl Trop Dis. 2014;8: e2994. doi: 10.1371/journal.pntd.0002994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shrinet J, Srivastava P, Sunil S. Transcriptome analysis of Aedes aegypti in response to mono-infections and co-infections of dengue virus-2 and chikungunya virus. Biochem Biophys Res Commun. 2017;492: 617–623. doi: 10.1016/j.bbrc.2017.01.162 [DOI] [PubMed] [Google Scholar]
- 53.Magwire MM, Fabian DK, Schweyen H, Cao C, Longdon B, Bayer F, et al. Genome-wide association studies reveal a simple genetic basis of resistance to naturally coevolving viruses in Drosophila melanogaster. PLOS Genet. 2012;8: e1003057. doi: 10.1371/journal.pgen.1003057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Piontkivska H, Matos LF, Paul S, Scharfenberg B, Farmerie WG, Miyamoto MM, et al. Role of host-driven mutagenesis in determining genome evolution of sigma virus (DMelSV; Rhabdoviridae) in Drosophila melanogaster. Genome Biol Evol. 2016;8: 2952–2963. doi: 10.1093/gbe/evw212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ford SA, Allen SL, Ohm JR, Sigle LT, Sebastian A, Albert I, et al. Selection on Aedes aegypti alters Wolbachia-mediated dengue virus blocking and fitness. Nat Microbiol. 2019;4: 1832–1839. doi: 10.1038/s41564-019-0533-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Terradas G, McGraw EA. Using genetic variation in Aedes aegypti to identify candidate anti-dengue virus genes. BMC Infect Dis. 2019;19: 580. doi: 10.1186/s12879-019-4212-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Terradas G, Allen SL, Chenoweth SF, McGraw EA. Family level variation in Wolbachia-mediated dengue virus blocking in Aedes aegypti. Parasit Vectors. 2017;10: 622. doi: 10.1186/s13071-017-2589-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sigle LT, McGraw EA. Expanding the canon: Non-classical mosquito genes at the interface of arboviral infection. Insect Biochem Mol Biol. 2019;109: 72–80. doi: 10.1016/j.ibmb.2019.04.004 [DOI] [PubMed] [Google Scholar]
- 59.Bartholomay LC, Cho W-L, Rocheleau TA, Boyle JP, Beck ET, Fuchs JF, et al. Description of the transcriptomes of immune response-activated hemocytes from the mosquito vectors Aedes aegypti and Armigeres subalbatus. Infect Immun. 2004;72: 4114–4126. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bishop-Lilly KA, Turell MJ, Willner KM, Butani A, Nolan NME, Lentz SM, et al. Arbovirus detection in insect vectors by rapid, high-throughput pyrosequencing. PLOS Negl Trop Dis. 2010;4: e878. doi: 10.1371/journal.pntd.0000878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Woolfit M, Iturbe-Ormaetxe I, McGraw EA, O’Neill SL. An ancient horizontal gene transfer between mosquito and the endosymbiotic bacterium Wolbachia pipientis. Mol Biol Evol. 2009;26: 367–374. doi: 10.1093/molbev/msn253 [DOI] [PubMed] [Google Scholar]
- 62.Klasson L, Kambris Z, Cook PE, Walker T, Sinkins SP. Horizontal gene transfer between Wolbachia and the mosquito Aedes aegypti. BMC Genomics. 2009;10: 33. doi: 10.1186/1471-2164-10-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Korochkina S, Barreau C, Pradel G, Jeffery E, Li J, Natarajan R, et al. A mosquito-specific protein family includes candidate receptors for malaria sporozoite invasion of salivary glands. Cell Microbiol. 2006;8: 163–175. doi: 10.1111/j.1462-5822.2005.00611.x [DOI] [PubMed] [Google Scholar]
- 64.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215: 403–410. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 65.Iturbe-Ormaetxe I, Burke GR, Riegler M, O’Neill SL. Distribution, expression, and motif variability of ankyrin domain genes in Wolbachia pipientis. J Bacteriol. 2005;187: 5136–5145. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bosio CF, Beaty BJ, Black WC 4th. Quantitative genetics of vector competence for dengue-2 virus in Aedes aegypti. Am J Trop Med Hyg. 1998;59: 965–970. doi: 10.4269/ajtmh.1998.59.965 [DOI] [PubMed] [Google Scholar]
- 67.Ye YH, Chenoweth SF, Carrasco AM, Allen SL, Frentiu FD, van den Hurk AF, et al. Evolutionary potential of the extrinsic incubation period of dengue virus in Aedes aegypti. Evolution (N Y). 2016;70: 2459–2469. doi: 10.1111/evo.13039 [DOI] [PubMed] [Google Scholar]
- 68.Falconer DS, MacKay TFC. Introduction to Quantitative Genetics. 4th ed. Essex, UK: Longmans Green; 1996. [Google Scholar]
- 69.Charmantier A, Garant D. Environmental quality and evolutionary potential: lessons from wild populations. Proc R Soc B. 2005;272: 1415–1425. doi: 10.1098/rspb.2005.3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hill WG, Goddard ME, Visscher PM. Data and theory point to mainly additive genetic variance for complex traits. PLOS Genet. 2008;4: e1000008. doi: 10.1371/journal.pgen.1000008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sgrò CM, Hoffmann AA. Genetic correlations, tradeoffs and environmental variation. Heredity (Edinb). 2004;93: 241–248. doi: 10.1038/sj.hdy.6800532 [DOI] [PubMed] [Google Scholar]
- 72.Slatkin M. Linkage disequilibrium—understanding the evolutionary past and mapping the medical future. Nat Rev Genet. 2008;9: 477–485. doi: 10.1038/nrg2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hill WG. Genetic Correlation. Second. In: Maloy S, Hughes K, editors. Brenner’s Encyclopedia of Genetics. Second ed. San Diego: Academic Press; 2013. pp. 237–239. [Google Scholar]
- 74.Barón OL, Ursic-Bedoya RJ, Lowenberger CA, Ocampo CB. Differential gene expression from midguts of refractory and susceptible lines of the mosquito, Aedes aegypti, infected with dengue-2 virus. J Insect Sci. 2010;10: 1–23. doi: 10.1673/031.010.4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lambrechts L, Scott TW, Gubler DJ. Consequences of the expanding global distribution of Aedes albopictus for dengue virus transmission. PLOS Negl Trop Dis. 2010;4: e646. doi: 10.1371/journal.pntd.0000646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lambrechts L, Chevillon C, Albright RG, Thaisomboonsuk B, Richardson JH, Jarman RG, et al. Genetic specificity and potential for local adaptation between dengue viruses and mosquito vectors. BMC Evol Biol. 2009;9: 160. doi: 10.1186/1471-2148-9-160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S. A Single Mutation in chikungunya virus affects vector specificity and epidemic potential. PLOS Pathog. 2007;3: e201. doi: 10.1371/journal.ppat.0030201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chevillon C, Failloux A-B. Questions on viral population biology to complete dengue puzzle. Trends Microbiol. 2003;11: 415–421. doi: 10.1016/s0966-842x(03)00206-3 [DOI] [PubMed] [Google Scholar]
- 79.Resck MEB, Padilha KP, Cupolillo AP, Talyuli OAC, Ferreira-de-Brito A, Lourenço-de-Oliveira R, et al. Unlike Zika, Chikungunya virus interferes in the viability of Aedes aegypti eggs, regardless of females’ age. Sci Rep. 2020;10: 13642. doi: 10.1038/s41598-020-70367-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sirisena PDNN, Kumar A, Sunil S. Evaluation of Aedes aegypti (Diptera: Culicidae) life table attributes upon chikungunya virus replication reveals impact on egg-laying pathways. J Med Entomol. 2018;55: 1580–1587. doi: 10.1093/jme/tjy097 [DOI] [PubMed] [Google Scholar]
- 81.Alto BW, Civana A, Wiggins K, Eastmond B, Shin D. Effect of oral infection of Mayaro virus on fitness correlates and expression of immune related genes in Aedes aegypti. Viruses. 2020;12: 719. doi: 10.3390/v12070719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sylvestre G, Gandini M, Maciel-de-Freitas R. Age-dependent effects of oral infection with dengue virus on Aedes aegypti (Diptera: Culicidae) feeding behavior, survival, oviposition success and fecundity. PLOS One. 2013;8: e59933. doi: 10.1371/journal.pone.0059933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Platt KB, Lerdthusnee K, Linthicum KJ, Myint KSA, Vaughn DW, Innis BL. Impact of dengue virus infection on feeding behavior of Aedes aegypti. Am J Trop Med Hyg. 1997;57: 119–125. doi: 10.4269/ajtmh.1997.57.119 [DOI] [PubMed] [Google Scholar]
- 84.Rahayu A, Saraswati U, Supriyati E, Kumalawati DA, Hermantara R, Rovik A, et al. Prevalence and distribution of dengue virus in Aedes aegypti in yogyakarta city before deployment of Wolbachia infected Aedes aegypti. Int J Environ Res Public Health. 2019;16: 1742. doi: 10.3390/ijerph16101742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Anders KL, Nga LH, Thuy NT Van, Van Ngoc T, Tam CT, Tai LTH, et al. Households as foci for dengue transmission in highly urban Vietnam. PLOS Negl Trop Dis. 2015;9: e0003528. doi: 10.1371/journal.pntd.0003528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Loroño-Pino MA, García-Rejón JE, Machain-Williams C, Gomez-Carro S, Nuñez-Ayala G, Nájera-Vázquez M del R, et al. Towards a Casa Segura: a consumer product study of the effect of insecticide-treated curtains on Aedes aegypti and dengue virus infections in the home. Am J Trop Med Hyg. 2013;89: 385–97. doi: 10.4269/ajtmh.12-0772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thavara U, Tawatsin A, Pengsakul T, Bhakdeenuan P, Chanama S, Anantapreecha S, et al. Outbreak of chikungunya fever in Thailand and virus detection in field population of vector mosquitoes, Aedes aegypti (L.) and Aedes albopictus Skuse (Diptera: Culicidae). Southeast Asian J Trop Med Public Health. 2009;40: 951–962. [PubMed] [Google Scholar]
- 88.Kirstein OD, Ayora-Talavera G, Koyoc-Cardeña E, Chan Espinoza D, Che-Mendoza A, Cohuo-Rodriguez A, et al. Natural arbovirus infection rate and detectability of indoor female Aedes aegypti from Mérida, Yucatán, Mexico. PLOS Negl Trop Dis. 2021;15: e0008972. doi: 10.1371/journal.pntd.0008972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Öhlund P, Lundén H, Blomström A-L. Insect-specific virus evolution and potential effects on vector competence. Virus Genes. 2019;55: 127–137. doi: 10.1007/s11262-018-01629-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chiuya T, Masiga DK, Falzon LC, Bastos ADS, Fèvre EM, Villinger J. A survey of mosquito-borne and insect-specific viruses in hospitals and livestock markets in western Kenya. PLOS One. 2021;16: e0252369. doi: 10.1371/journal.pone.0252369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Agboli E, Leggewie M, Altinli M, Schnettler E. Mosquito-specific viruses—transmission and interaction. Viruses. 2019;11: 873. doi: 10.3390/v11090873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Atoni E, Zhao L, Karungu S, Obanda V, Agwanda B, Xia H, et al. The discovery and global distribution of novel mosquito-associated viruses in the last decade (2007–2017). Rev Med Virol. 2019;29. doi: 10.1002/rmv.2079 [DOI] [PubMed] [Google Scholar]
- 93.Leggewie M, Schnettler E. RNAi-mediated antiviral immunity in insects and their possible application. Curr Opin Virol. 2018;32: 108–114. doi: 10.1016/j.coviro.2018.10.004 [DOI] [PubMed] [Google Scholar]
- 94.Chrostek E, Teixeira L. Mutualism breakdown by amplification of Wolbachia genes. PLOS Biol. 2015;13: e1002065. doi: 10.1371/journal.pbio.1002065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Duarte EH, Carvalho A, López-Madrigal S, Costa J, Teixeira L. Forward genetics in Wolbachia: Regulation of Wolbachia proliferation by the amplification and deletion of an addictive genomic island. PLOS Genet. 2021;17: e1009612. doi: 10.1371/journal.pgen.1009612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chrostek E, Marialva MSP, Esteves SS, Weinert LA, Martinez J, Jiggins FM, et al. Wolbachia variants induce differential protection to viruses in Drosophila melanogaster: a phenotypic and phylogenomic analysis. PLOS Genet. 2013;9: e1003896. doi: 10.1371/journal.pgen.1003896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Baião GC, Janice J, Galinou M, Klasson L. Comparative genomics reveals factors associated with phenotypic expression of Wolbachia. Genome Biol Evol. 2021;13. doi: 10.1093/gbe/evab111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Woolfit M, Iturbe-Ormaetxe I, Brownlie JC, Walker T, Riegler M, Seleznev A, et al. Genomic evolution of the pathogenic Wolbachia Strain, wMelPop. Genome Biol Evol. 2013;5: 2189. doi: 10.1093/GBE/EVT169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Matthews BJ, Vosshall LB. How to turn an organism into a model organism in 10 ‘easy’ steps. J Exp Biol. 2020;223. doi: 10.1242/jeb.218198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Erler S, Popp M, Lattorff HMG. Dynamics of immune system gene expression upon bacterial challenge and wounding in a social insect (Bombus terrestris). PLOS One. 2011;6: e18126. doi: 10.1371/journal.pone.0018126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gloria-Soria A, Brackney DE, Armstrong PM. Saliva collection via capillary method may underestimate arboviral transmission by mosquitoes. Parasit Vectors. 2022;15: 1–9. doi: 10.1186/S13071-022-05198-7/FIGURES/5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Scott DA, Zhang F. Implications of human genetic variation in CRISPR-based therapeutic genome editing. Nat Med. 2017;23: 1095–1101. doi: 10.1038/nm.4377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nakajima 6, Tsuji M, Oda K, Zamoto-Niikura A, Wei Q, -Kurata K. Transcontinental movement of asian genotype chikungunya virus. Emerg Infect Dis. 2014;20: 1400–1402. doi: 10.3201/eid2008.140268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Frentiu FD, Robinson J, Young PR, McGraw EA, O’Neill SL. Wolbachia-mediated resistance to dengue virus infection and death at the cellular level. PLOS One. 2010;5: e13398. doi: 10.1371/journal.pone.0013398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Amuzu HE, Simmons CP, McGraw EA. Effect of repeat human blood feeding on Wolbachia density and dengue virus infection in Aedes aegypti. Parasit Vectors. 2015;8: 246. doi: 10.1186/s13071-015-0853-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rückert C, Weger-Lucarelli J, Garcia-Luna SM, Young MC, Byas AD, Murrieta RA, et al. Impact of simultaneous exposure to arboviruses on infection and transmission by Aedes aegypti mosquitoes. Nat Commun. 2017;8: 15412. doi: 10.1038/ncomms15412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Saxton AM. Genetic analysis of complex traits using SAS. SAS Publishing; 2004. [Google Scholar]
- 108.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30: 2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29: 15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. NAR. 2013;41: e108–e108. doi: 10.1093/nar/gkt214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15: 550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Terradas G, Joubert DA, McGraw EA. The RNAi pathway plays a small part in Wolbachia-mediated blocking of dengue virus in mosquito cells. Sci Rep. 2017/03/07. 2017;7: 43847. doi: 10.1038/srep43847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25: 402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]