Abstract
Activation of the small GTPase Rab7 by its cognate guanine nucleotide exchange factor Mon1-Ccz1 (MC1) is a key step in the maturation of endosomes and autophagosomes. This process is tightly regulated and subject to precise spatiotemporal control of MC1 localization, but the mechanisms that underly MC1 localization have not been fully elucidated. We here identify and characterize an amphipathic helix in Ccz1, which is required for the function of Mon-Ccz1 in autophagy, but not endosomal maturation. Furthermore, our data show that the interaction of the Ccz1 amphipathic helix with lipid packing defects, binding of Mon1 basic patches to positively charged lipids, and association of MC1 with recruiter proteins collectively govern membrane recruitment of the complex in a synergistic and redundant manner. Membrane binding enhances MC1 activity predominantly by increasing enzyme and substrate concentration on the membrane, but interaction with recruiter proteins can further stimulate the guanine nucleotide exchange factor. Our data demonstrate that specific protein and lipid cues convey the differential targeting of MC1 to endosomes and autophagosomes. In conclusion, we reveal the molecular basis for how MC1 is adapted to recognize distinct target compartments by exploiting the unique biophysical properties of organelle membranes and thus provide a model for how the complex is regulated and activated independently in different functional contexts.
Keywords: GTPase, guanine nucleotide exchange factor, endosome, autophagy, membrane recruitment
Organization of intracellular processes in eukaryotes relies on the compartmentalization of biochemical processes to organelles and suborganellar domains (1). Consequently, the targeting of proteins to the appropriate microcompartments underlies strict spatiotemporal control. Molecular on/off switches, namely reversible phosphorylation by kinases and phosphatases (2) or cycling of GTPases between GTP- and GDP-bound form (3), can regulate the activity and interactions of biomolecules. However, rather than control by single all-or-nothing signals, many systems utilize multiple cues and coincidence mechanisms for adaptive regulation (4, 5). The same functional modules can thus fulfill different functions in different settings. Unraveling the molecular basis that governs these processes is required for a mechanistic understanding of cellular functions.
The Mon1-Ccz1 (MC1) complex is a guanine nucleotide exchange factor (GEF) that activates the endosomal GTPase Rab7 (Ypt7 in yeast) (6, 7, 8, 9). MC1-dependent activation of Rab7 is required for fusion of multivesicular bodies/late endosomes and of autophagosomes to lysosomes (vacuole in yeast) (6, 10, 11, 12, 13). Recently, MC1 was also implicated to support mitophagy (14). In yeast, the interactions of MC1 with Vps21 (a Rab5-like GTPase) (10, 15), Atg8 (the homolog of mammalian LC3) (11, 16), and phosphatidylinositol phosphate (PIP) lipids (13, 17, 18) were shown to be required for complex function. Homologous interactions were also described in mammalian cells. Likely, the differential association of MC1 to these localization cues contributes to the spatiotemporal control of its activity.
The structural characterization of MC1 provides a framework for a mechanistic analysis of its function. The complex belongs to the family of Tri Longin domain RabGEFs, which are characterized by a heterodimeric core complex (19, 20) (Fig. 1A). The core subunits, Mon1 and Ccz1 in the case of MC1, are each composed of three conserved longin-type domains (LDs) that are arranged in a triangular fashion. Together, they form a complex with pseudo two-fold symmetry (21) resulting in a two-layered architecture. The bottom layer, formed by the LD2s and LD3s, is required for proper subcellular localization of the complex. A large positively charged patch on the bottom layer mediates membrane binding, defining the orientation of MC1 on the membrane. The top layer, comprised of the LD1s, constitutes the catalytic center that promotes nucleotide exchange of Rab7/Ypt7 (22).
Figure 1.
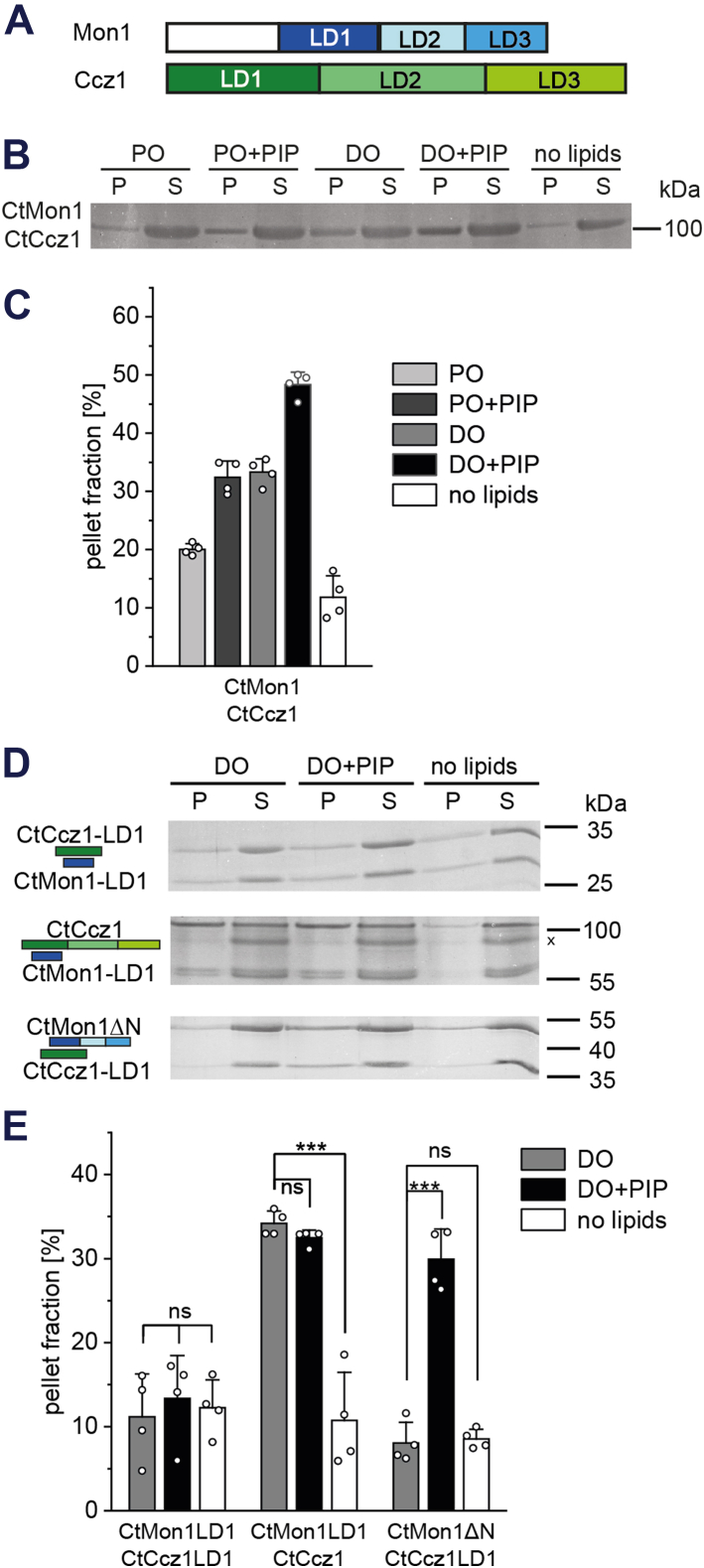
Lipid interactions of the MC1 complex.A, domain architecture of Mon1 and Ccz1. B, sedimentation assays of full-length CtMC1 with liposomes (400 nm) from a PO (palmitoyl-oleoyl) and DO (di-oleoyl) lipid mixes with or without PIP. C, quantification of B (n = 4). D, sedimentation assays of different CtMC1 subcomplexes with liposomes from a DO lipid mix with or without PIP. A Ccz1 degradation band is marked (x). E, quantification of D (n = 4). Quantification data are presented as mean ± SD, and the significance was calculated using Student’s t test (∗∗∗p < 0.001, n.s. not significant). CtMC1, MC1 complex from Chaetomium thermophilum; MC1, Mon1-Ccz1; PIP, phosphatidylinositol phosphate.
Our previous work addressed the GEF activity of MC1, revealing a unique lysine-insertion mechanism (22) and activation of the complex by its recruiter GTPases on model membranes (15). In this study, we focused on how MC1 membrane interactions and targeting to distinct cellular compartments are realized on a molecular level. We performed a detailed analysis of MC1 recruitment to membranes using in vitro reconstitutions and functional analysis in vivo. We identify binding to lipid packing defects as an additional targeting mechanism of MC1 that is essential for its function in autophagy. Interestingly, different recruiter proteins and lipid binding cues act on MC1 in a synergistic, but also redundant manner, providing a mechanism how fine-tuning of activity and differential targeting to distinct organelles can be achieved.
Results
MC1 binding to negative charges and packing defects in membranes
In previous work with the MC1 complex from Chaetomium thermophilum (CtMC1), we demonstrated that CtMon1 LD2/3 are required for the recruitment of MC1 to PIP-containing liposomes and subsequent co-sedimentation (21). We noticed that CtMC1 also bound weakly, but reproducibly to neutral liposomes containing palmitoyl (16:0)—oleoyl (18:1) (PO) phospholipids (Fig. 1, B and C). This effect was more pronounced with liposomes that were composed of di-oleoyl (DO) phospholipids (Fig. 1, B and C). DO liposomes contain more unsaturated acyl chains and have more lipid packing defects than PO liposomes. Thus, CtMC1 likely interacts with packing defects in neutral liposomes, in addition to its binding to charged phospholipids as observed before (21) (Fig. 1, B and C).
We used different truncation constructs of MC1 in liposome sedimentations to map the lipid binding sites. The minimal catalytic core complex of CtMon1LD1/CtCcz1LD1 did not show any lipid interaction (Fig. 1, D and E), suggesting that membrane binding is mediated by the LD2s and LD3s of Mon1 and Ccz1. Indeed, a complex of CtMon1LD1 and CtCcz1fl, which additionally contains LD2/3 of Ccz1, interacted with liposomes with packing defects (Fig. 1, D and E), suggesting that CtCcz1LD2/3 contain a lipid packing sensing motif. In contrast, a minimal complex of CtMon1ΔN together with CtCcz1LD1, which now contained LD2/3 of Mon1, specifically bound to liposomes containing 2% phosphatidylinositol-3-phosphate/1% phosphatidylinositol-3,5-bisphosphate (PIP) (Fig. 1, D and E). This confirmed the mapping of the PIP interaction site to CtMon1LD2/3 (21) and showed that the interaction of the complex with membrane charges occurs independent of its recognition of packing defects. In summary, CtMon1 and CtCcz1 can both interact with membranes, recognize negative charges and lipid packing defects on membranes, and jointly mediate membrane binding of the complex and its recruitment onto lipid bilayers.
Synergistic recruitment of MC1 by lipids and proteins
Next, we asked if the lipid binding properties of MC1 are conserved and therefore investigated the interaction of the MC1 complex from Saccharomyces cerevisiae (ScMC1) with liposomes (Fig. 2, A and B). The yeast complex also showed robust binding to DO liposomes (high packing defects), but only weak interaction with PO liposomes (low packing defects). Furthermore, ScMC1 was recruited to PO+PIP liposomes (negative charges), recapitulating the findings obtained with CtMC1.
Figure 2.
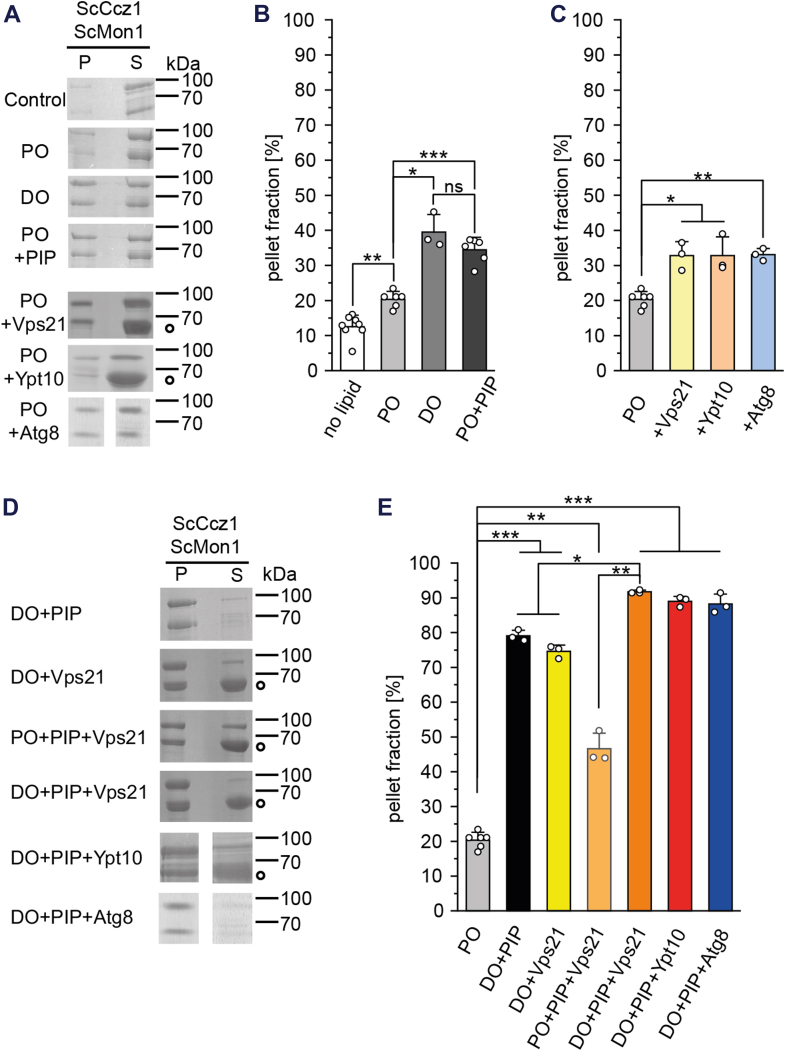
Triple targeting mechanism of the MC1 complex.A, binding of ScMC1 to liposomes from a PO, DO, and PO-PIP mix and to PO liposomes with Vps21, Ypt10, or Atg8. The Mrs6 Rab escort protein from the prenylation reaction is marked by a circle. Sample without liposomes serves as control. B, quantification of ScMC1 binding to PO, DO, and PO-PIP liposomes (n = 3–8). C, quantification of ScMC1 binding to PO liposomes with Vps21, Ypt10, or Atg8 (n=3–6). D, binding of ScMC1 to PO or DO liposomes containing PIPs and recruiter proteins as indicated. E, quantification of D (n=3–6). Quantification data are presented as mean ± SD, and the significance was calculated using Student’s t test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, n.s. not significant). DO, di-oleoyl; MC1, Mon1-Ccz1; PIP, phosphatidylinositol phosphate; PO, palmitoyl (16:0)—oleoyl (18:1); ScMC1, MC1 complex from Saccharomyces cerevisiae.
ScMC1 requires the interaction with recruiter proteins for its function, namely the autophagosomal marker Atg8 and the endosomal Rab5-like GTPases Vps21 and Ypt10 (11, 15). To test their influence on membrane targeting of ScMC1, we incorporated the naturally lipidated forms (prenylated Vps21 or Ypt10 and Atg8 coupled onto phosphatidylethanolamine) of the recruiter proteins into PO liposomes and measured binding in liposome sedimentation assays. Each recruiter promoted membrane binding to a similar extend, but overall relatively weakly (Fig. 2, A and C).
Neither recruitment cue alone—negative charges, lipid packing defects, or a recruiter protein—was able to efficiently target ScMC1 to liposomes. We thus tested different combinations of liposomes with packing defects (DO lipids), negative charges (PIPs) and the recruiter GTPase Vps21. Two recruitment cues remarkably enhanced ScMC1 membrane binding, in particular when liposomes contained packing defects (Figs. 2, D and E and S1A). In particular, the binding observed with DO+PIP liposomes (high packing defects, negative charges) was much stronger, indicative of two independent lipid interaction sites that act synergistically. Membrane binding was further increased when all three recruitment cues were present on the liposomes (Fig. 2, D and E). The triple targeting set-up also led to almost complete ScMC1 membrane binding when Ypt10 or Atg8 were used as recruiter proteins.
Membrane recruitment supports MC1 activity
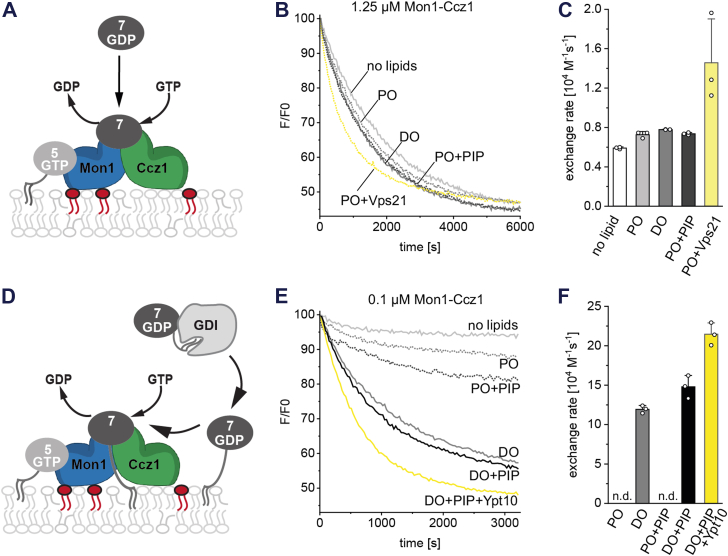
It has been described that MC1 and other RabGEFs have increased catalytic activity toward their substrates in the presence of membranes (15, 17, 23, 24). We thus asked how different recruitment factors influence MC1-stimulated nucleotide exchange of Ypt7. To test if membrane interactions directly activate MC1 by allosteric mechanisms, we performed GEF assays with MC1 and soluble Ypt7 in the presence of liposomes (Fig. 3A). Liposome composition were chosen to mediate MC1 membrane binding to the same extend, either by packing defects (DO liposomes), charges (PO+PIP liposomes), or recruiter GTPase (PO+Vps21 liposomes) and compared with PO liposomes that only weakly bind MC1 (Fig. 2, B and C). Lipid interactions via packing defects or charges did not affect MC1 activity but binding to Vps21 led to a ∼2-fold stimulation of nucleotide exchange (Fig. 3, B and C). This is consistent with earlier observations that Rab5 GTPases not only act as recruiters but also as activators of MC1 (15). We then performed GEF assays with prenylated, GDI-bound pYpt7 (Fig. 3D). Although the GDI keeps pYpt7 predominantly in solution, a small fraction of pYpt7 partitions onto the liposomes where it encounters MC1 and is trapped after nucleotide exchange (15). We observed that the initial GDP-pYpt7 membrane binding strongly depends on packing defects (Fig. S2B). Because pYpt7 does not efficiently partition into PO liposomes, we observe very low MC1 GEF activity with PO and PO+PIP lipid mixes and could not determine exchange rates (Fig. 3, E and F). However, we observe a stimulation of GEF activity of ∼15-fold with DO liposomes with prenylated Ypt7 compared to soluble Ypt7 (Fig. 3, C and F). Nucleotide exchange is slightly increased when PIPs and the Rab5-like Ypt10 are added to the DO liposomes, with Ypt10 showing the strongest effect (Fig. 3, E and F). These data show that MC1 activity depends primarily on the concurrent recruitment with Ypt7 to the confined 2D environment of a membrane and on a minor contribution by stimulation through recruiter GTPase binding.
Figure 3.
Influence of membrane composition on MC1 GEF activity.A, schematic of a GEF assay with MC1 in the presence of liposomes and soluble Ypt7. B, GEF assays with MC1 (1.25 μM) and MANT-GDP Ypt7 (2 μM) in the presence of 200 μM GTP and liposomes of varying compositions (final lipid concentration 500 μM). C, nucleotide exchange rates calculated from B. D, schematic of a recruitment GEF assay with MC1 in the presence of liposomes and GDI-bound prenylated Ypt7. E, GEF assays with MC1 (0.1 μM) and MANT-GDP pYpt7 in complex with GDI (0.5 μM) in the presence of 200 μM GTP and liposomes of varying compositions (final lipid concentration 500 μM). F, nucleotide exchange rates calculated from E. n.d.: not determined because exponential fit of reaction curves failed. Data from three replicates are presented as mean ± SD. GEF, guanine nucleotide exchange factor; MC1, Mon1-Ccz1.
Role of PIP binding in vivo
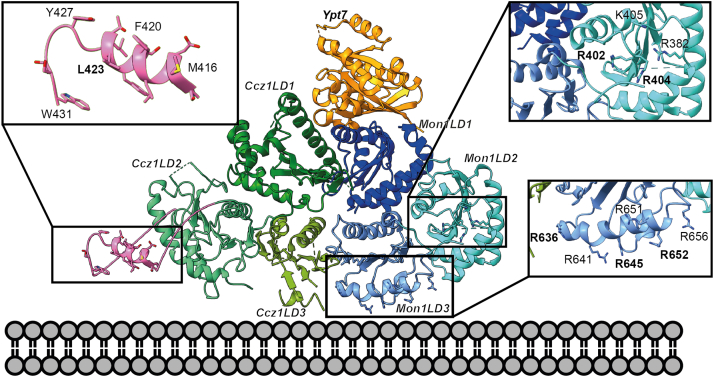
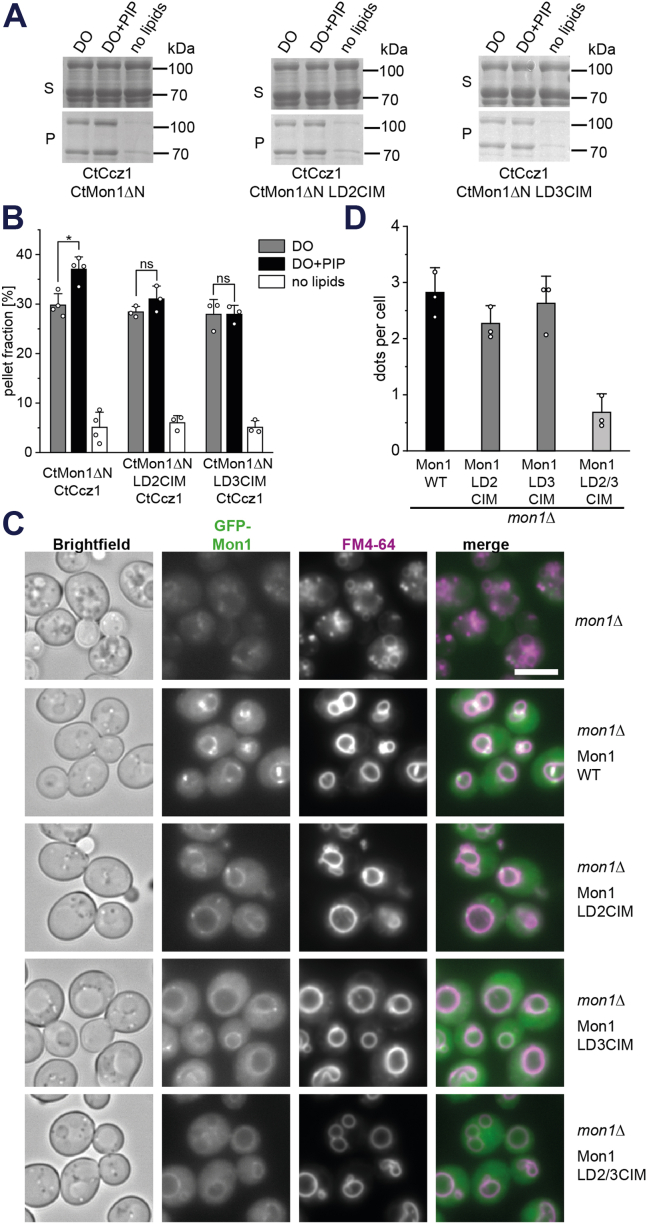
To gain insight into the relevance of PIP binding in physiology, we sought to identify mutations that specifically abolish the interaction of MC1 with negatively charged phospholipid head groups. The structure of CtMC1 revealed a large positive patch on the CtMon1 ‘localization layer’ that likely represents the interaction site (Fig. S2A) (21). We generated two charge inversion mutants (CIMs) of CtMon1, which had three basic residues exchanged to glutamate in either LD2 (CtMon1LD2CIM: R402E, R404E, R551E) or LD3 (CtMon1LD3CIM: R636E, R645E, R652E) (Figs. 4 and S2A). In sedimentation assays, both mutants showed loss of specific binding to PIP-containing liposomes (Fig. 5, A and B), confirming the model that the basic surface of Mon1 is responsible for the interaction with charged membranes.
Figure 4.
Structural model of the CtMon1 (blue)-CtCcz1 (green) complex bound to Ypt7 (orange) shown in its putative orientation on a membrane. Inserts show the residues contributing to positive charged patches on LD2 and LD3 of Mon1 and the putative amphipathic helix (pink) in the α2-β3 loop of CtCcz1 LD2 as modeled by AlphaFold2 (26), respectively. Mutated residues in CtMon1 and CtCcz1 variants are labeled bold. LD, longin-type domain.
Figure 5.
Role of the positively charged patch on Mon1 for membrane binding in vivo.A, sedimentation assays of full-length CtMC1 with CtMon1WT or CtMon1CIM variants. Liposomes were generated from a DO lipid mix with or without PIP. B, quantification of A (n = 3). Data are presented as mean ± SD, and the significance was calculated using Student’s t test (∗p < 0.05, n.s. not significant). C, fluorescence microscopy images of mon1Δ yeast complemented with GFP-ScMon1WT or different GFP-ScMon1CIM variants. Vacuoles are stained with FM4-64. Scale bar: 5 μm. D, quantification of the number of perivacuolar Mon1 dots per cell from three independent experiments. Data are presented as mean ± SD. CtMC1, MC1 complex from Chaetomium thermophilum; DO, di-oleoyl; PIP, phosphatidylinositol phosphate.
In analogy, we created mutants of ScMon1 that had charge inversions in LD2 (ScMon1LD2CIM: R374E, R376E), LD3 (ScMon1LD3CIM: K620E, K624E, K631E) and a combination of both (ScMon1LD2/3CIM: R374E, R376E, K620E, R624E, K631E). We introduced these variants as GFP fusion proteins into a mon1Δ yeast strain. In the absence of Mon1, yeast cells have fragmented vacuoles (Fig. 5C). This phenotype was rescued by the re-introduction of ScMon1WT, which localized to perivacuolar puncta that likely represent late endosomes and the vacuolar membrane. Expression of the CIMs was able to restore vacuolar morphology. However, localization of ScMon1 was impaired (Fig. 5, C and D). Perivacuolar puncta were strongly reduced for ScMon1LD2CIM and ScMon1LD3CIM and completely absent for ScMon1LD2/3CIM, while some vacuolar membrane staining was still observed. Thus, the basic patch on Mon1 is required for proper localization of the protein to endosomal structures.
To investigate the functionality of ScMon1CIM mutants in autophagy, we introduced the respective plasmids into a mon1Δ yeast strain expressing mCherry-Atg8. During starvation, mCherry-Atg8 was properly delivered to the vacuole (Fig. S2B), showing that autophagy was not impaired in these cells.
Identification of a conserved amphipathic helix in Ccz1
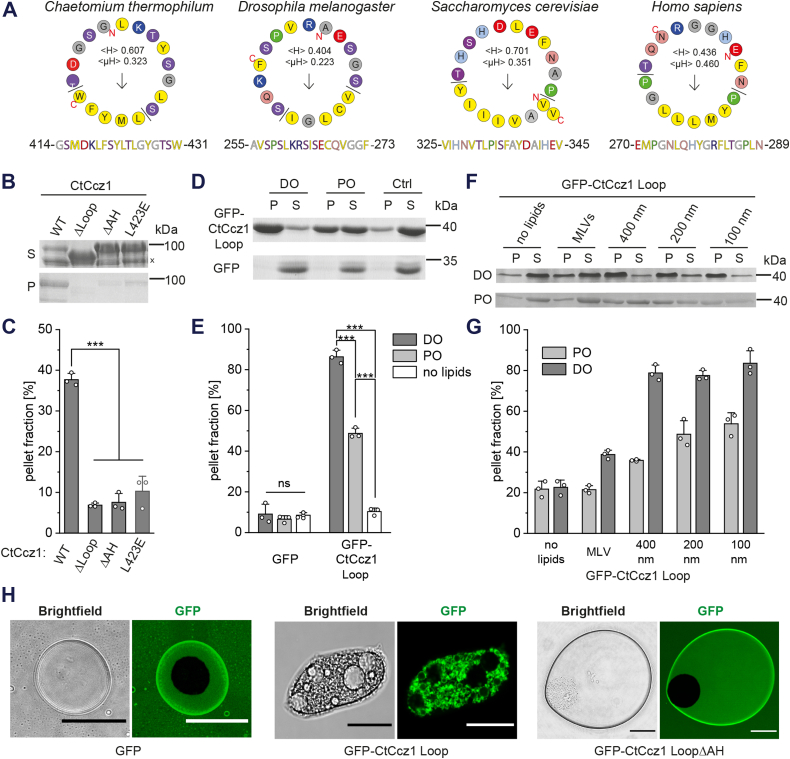
We next asked for the possible molecular basis of the interaction of Ccz1 with lipid packing defects. Interestingly, the truncated CtMC1Δ complex we had used for cryo-EM studies was unable to recognize packing defects (Fig. S3, A and B). Compared to the complexes we used in the mapping experiments (Fig. 1), a predicted loop in the LD2 of CtCcz1 (between helix α2 and strand β3, residues 360–460) was removed. Secondary structure analysis and structure predictions of CtCcz1 suggest that this loop contains an α helix with amphipathic character (residues 416–427) (25, 26, 27) (Fig. 4). Ccz1 homologs from other species also contain a large insertion with a predicted amphipathic helix between α-helix 2 and β-strand 3 of LD2, indicating a functional significance of this motif (Fig. 6A). To test if the putative amphipathic helix of Ccz1 can serve as a lipid packing sensor, we generated several CtCcz1 mutants. While CtCcz1WT robustly bound to DO liposomes with packing defects, deletion constructs without the α2-β3 loop (CtCcz1ΔLoop, lacking residues 361–460) and without the putative amphipathic helix (CtCcz1ΔAH, lacking residues 408–440) lost this ability (Fig. 6, B and C). Also, introduction of a negative charge in the hydrophobic face of the helix (CtCcz1L423E) led to loss of interaction with DO liposomes (Fig. 6, B and C), as to be expected for a lipid packing sensor function of this helix. A similar effect was also observed with additional point mutants F420E and W431E (Fig. S3C).
Figure 6.
Interaction of the MC1 amphipathic helix with lipids.A, helical wheel projection of the putative amphipathic helix from different model organisms (27). B, sedimentation assays of full-length CtCcz1WT or CtCcz1 variants. Liposomes were generated from a DO lipid mix. A Ccz1 degradation band is marked (x). C, quantification of B (n = 3). D, sedimentation assays of GFP-CtCcz1Loop and GFP with liposomes generated from a PO and DO lipid mix. E, quantification of D (n = 3). F, sedimentation assays of GFP-CtCcz1Loop with multilamellar vesicles (MLVs) or liposomes with a defined diameter generated from a PO and DO lipid mix. G, quantification of F (n = 4). H, oil droplets were incubated with GFP, GFP-CtCcz1Loop, or GFP-CtCcz1LoopΔAH variants and visualized by confocal fluorescence microcopy. Scale bar: 20 μm. Quantification data are presented as mean ± SD, and the significance was calculated using Student’s t test (∗∗∗p < 0.001, n.s. not significant). DO, di-oleoyl; MC1, Mon1-Ccz1; PO, palmitoyl (16:0)—oleoyl (18:1).
A fusion of the α2-β3 loop to GFP (GFP-CtCcz1Loop, residues 360–460) strongly interacted with DO liposomes and to lesser extend with PO liposomes that have fewer packing defects (Fig. 6, D and E). GFP alone did not interact with liposomes. Thus, the α2-β3 loop is not only required but also sufficient for binding to lipid packing defects. For both PO and DO lipid mixes, curvature-induced lipid packing defects also increased liposome binding of GFP-CtCcz1Loop (Fig. 6, F and G). Finally, we also tested the effect of lipid packing defects created by different phospholipid head group geometries by including 20% phosphatidylethanolamine (induces negative curvature) or 20% phosphatidylinositol (induces positive curvature) in phosphatidylcholine (no curvature induction) liposomes. In this case, binding of GFP-CtCcz1Loop to liposomes was not changed (Fig. S3, D and E).
To confirm the amphipathic helix properties of CtCcz1Loop, we incubated the GFP fusion protein and GFP alone with an oil suspension (Fig. 6H). GFP-CtCcz1Loop, but not GFP, acted as an emulsifier of oil droplets, causing the consumption of oil drops into small droplets. The resulting droplets were covered with GFP-CtCcz1Loop, showing that the protein has surfactant properties and interacts at the oil–water interface with the hydrophobic surface of the droplets. This behavior is typically observed for amphipathic helices (28). Deletion of the putative amphipathic helix in the Ccz1 loop abolished its ability to emulsify oil droplets. Taken together, these data demonstrate that an amphipathic helix in the α2-β3 loop of Ccz1 LD2 acts as a lipid packing sensor.
Ccz1 amphipathic helix is required for MC1 function in autophagy
The sedimentation assays with simplified lipid mixes clearly demonstrated the ability of Ccz1 to bind to lipid packing defects, but we wondered if this mechanism would be relevant with more complex lipid mixes that reflect the composition of the target organelles of MC1. Based on lipidomics data of the phospholipid distribution in endosomes and autophagosomes (29, 30, 31), we created liposomes that mimic the endosomal and autophagosomal phospholipid composition. These mixtures recapitulate the reported distribution of acyl chains with different degrees of saturation as well as different head group (Fig. S3F and Table S1). ScMC1 bound to both lipid mixes, suggesting that the phospholipid compositions of endosomes and autophagosomes in principle generate enough lipid packing defects to support MC1 membrane recruitment (Fig. 7, A and B). Interestingly, ScMC1 interacted moderately but significantly stronger with the autophagosomal lipid mix, which contains acyl chains with more double bonds and thus has more packing defects. Although these differences are subtle, they apparently suffice to modulate membrane binding. When PIPs were included in the liposomes, ScMC1 recruitment could be further enhanced (Fig. 7, A and B). The difference between the endosomal and autophagosomal lipid mix, however, was diminished under these conditions.
Figure 7.
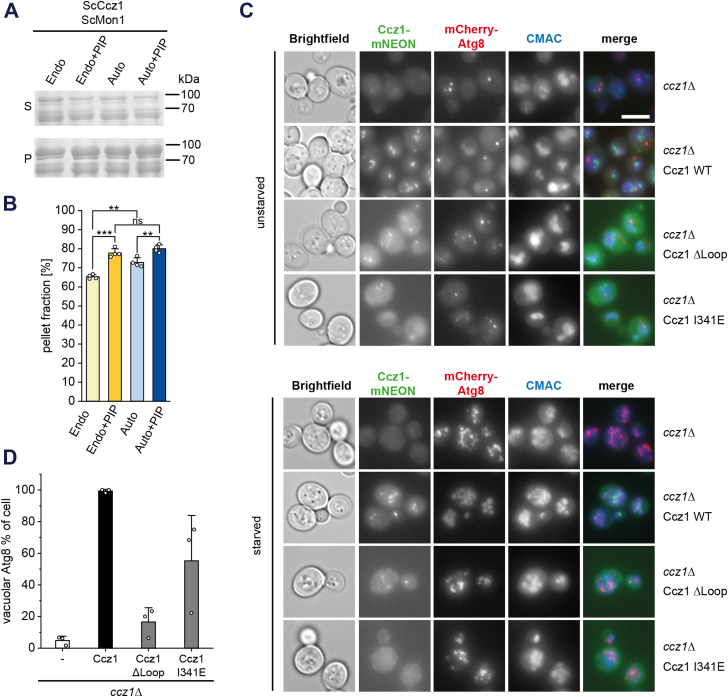
The MC1 complex amphipathic helix is required for autophagy.A, sedimentation assays of ScMC1 with liposomes generated from an endosomal (endo) or autophagosomal (auto) lipid mix with or without PIP. B, quantification of A (n = 4). Data are presented as mean ±SD, and the significance was calculated using Student’s t test (∗∗p < 0.01, ∗∗∗p < 0.001, n.s. not significant). C, immunofluorescence images of unstarved and starved ccz1Δ yeast expressing mCherry-Atg8 and complemented with GFP-ScCcz1WT or different GFP-ScCcz1 variants. Vacuoles are stained with CMAC. Scale bar: 5 μm. D, quantification of the percentage of cells with vacuolar Atg8 under starvation conditions from three independent experiments. Data are presented as mean ± SD. MC1, Mon1-Ccz1; ScMC1, MC1 complex from Saccharomyces cerevisiae.
For functional testing, we generated in analogy to CtCcz1 a loop deletion mutant ScCcz1ΔLoop (lacking residues 270–403) and a variant ScCcz1I341E, which has a charged residue introduced in the hydrophobic face of the amphipathic helix. Loss of Ccz1 in yeast caused vacuolar fragmentation, which was rescued by reintroduction of ScCcz1WT. The same effect was observed with ScCcz1ΔLoop and ScCcz1I341E, which localized like ScCcz1WT to punctate structures (Fig. 6C). Thus, the amphipathic helix is not required for endosomal recruitment of ScMC1 or endosomal maturation. However, when we investigated the functionality of ScCcz1 mutants in starved cells, an autophagy defect was observed. Like in ccz1Δ cells, Atg8 did not reach the vacuole after complementation with ScCcz1ΔLoop and ScCcz1I341E, whereas ScCcz1WT was able to restore autophagic flux (Fig. 6, C and D). The ScCcz1ΔLoop and ScCcz1I341E strains also showed a defect in Ape1 processing, an assay monitoring the uptake of oligomeric Ape1 peptidase via the autophagy-related cytosol-to-vacuole targeting pathway (32) (Fig. S3G). The data therefore show that the amphipathic helix is necessary for the function of ScMC1 in autophagy.
Discussion
Here, we identify an amphipathic helix of the MC1 complex that mediates binding to membranes by recognizing lipid packing defects. This describes a novel targeting mechanism by which the complex is recruited to its proper localization in the cell. This activity is conserved and could be observed for both CtMC1 and ScMC1. A putative amphipathic helix motif is also found in other Ccz1 homologs at the same position in the α2-β3 loop of LD2 (Fig. 6A). Thus, the lipid packing sensing function may also play a role in other species.
Lipid packing defects act synergistically with negative charges of phospholipid head groups and recruiter proteins in recruitment of MC1 to model membranes. While each interaction individually is relatively weak, two targeting cues together strongly promote membrane binding. A close to stoichiometric recruitment, however, was only achieved if all three cues were present. This behavior is consistent with independent binding events where affinities are multiplicative rather than additive. Alternatively, the observed effects could potentially also arise, in parts, from cooperativity between binding events.
The observed synergistic binding could be the basis for an ultrasensitive behavior in complex recruitment that encodes spatiotemporal regulation (33, 34). In particular, the levels of PIPs and recruiter proteins accumulate during the maturation of both endosomes and autophagosomes (35, 36). It is thus tempting to speculate that coincidence detection employing multiple targeting mechanisms with individually weak affinities allows for tightly controlled, yet rapid recruitment and activation of the MC1 GEF and the downstream pathways when thresholds are reached.
The importance of membrane recruitment for the function of MC1 is nicely demonstrated in nucleotide exchange activity assays in the presence of liposomes. Binding to a recruiter GTPase led to a moderate MC1 activation, but interaction with packing defects or charged lipids by itself did not directly activate the GEF complex. This supports the concept that MC1 activity is modulated by Rab5 binding (15), possibly through allosteric effects, but argues against a similar lipid-induced mechanism. However, co-recruitment of GEF and prenylated Ypt7 onto liposomes was the strongest stimulant for GEF activity, suggesting that the identified lipid interactions will be essential to bring enzyme and substrate together. Accordingly, loss of lipid interactions led to defects of MC1 localization and function in yeast cells. We suspect that subcellular targeting requires affinity for lipids and specificity encoded by recruiter proteins, which together ensure proper function.
Functionality of MC1—regarding both membrane binding and activity in a recruiter GEF assay—was strikingly different whether we use low packing defect PO liposomes or high packing defect DO liposomes. This shows that it is important to not only consider lipid head group but also acyl chain composition of liposomes used for in vitro studies, as they can also dramatically impact functionality. To mimic the different properties of distinct organelles in yeast, we used a PO/DO ratio of ∼ 60%/40% for a complex endosomal lipid mix and ∼ 40%/60% for an autophagosomal mix. These lipid mixes are based on recent lipidomics data (30) and probably better suited for in vitro studies than pure PO or DO mixes. Strikingly, these relatively small variations caused measurable effects on protein binding in our liposome sedimentation assays. It is thus worthwhile to experimentally test the influence of bilayer packing defects on membrane-associated processes in reconstitution studies. However, the lipid mixes we used for our in vitro system still do not fully recapitulate the complex properties of biological membranes with membrane proteins, sterols, or sphingolipids.
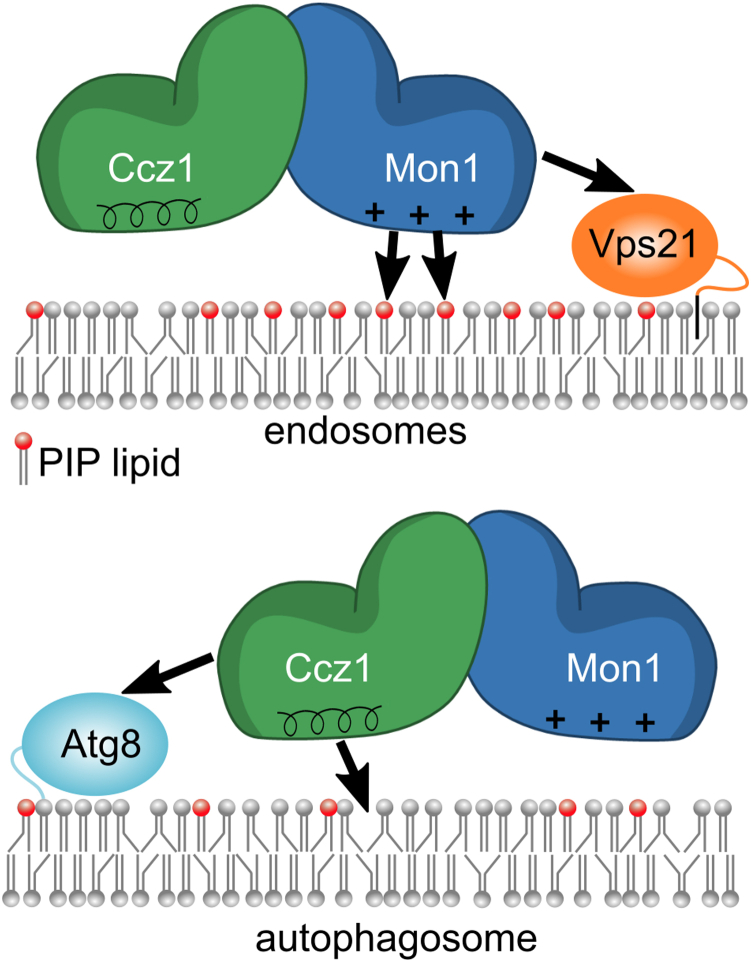
The different targeting cues may not all be required (to the same extend) for all functions of MC1. In fact, our functional analyses indicate that binding to charged lipids is required for endosomal localization of ScMC1 and interaction with packing defects for its function in autophagy but not vice versa. This is consistent with the concept that protein targeting to endosomal organelles is dominated by charges, while targeting to autophagosomes is dominated by packing defect (37). By exploiting distinct membrane properties for differential recruitment, MC1 targeting is different from the mechanism that drives localization of the Rab1 GEF TRAPPIII to the Golgi and autophagosomes. TRAPPIII also requires binding to packing defects and charges for proper function, but both cues are needed at both organelles (38). It is interesting to note that the endosomal Rab5 GTPases bind Mon1 (15, 18) and the autophagosome marker Atg8 binds Ccz1 (11). This suggests a model were Mon1 and Ccz1 together are required for catalytic activity of the complex (22), but each subunit is responsible to mediate targeting to a distinct localization through the recognition of unique protein and lipid cues (Fig. 8).
Figure 8.
Model for the differential targeting of MC1 to endosomes and autophagosomes. Mon1 interacts with Rab5 family GTPases (Vps21) and PIPs to recruit the complex to endosomes. For binding to autophagosomes, Ccz1 instead interacts with Atg8 and lipid packing defects. MC1, Mon1-Ccz1; PIP, phosphatidylinositol phosphate.
Thus, the complex has adapted to the different requirements for targeting in distinct pathways: MC1 functions on endosomes, which have high PI3P levels that are produced during endosomal maturation. This membrane property is exploited by Mon1 through charge interactions. In contrast, MC1 localizes early during autophagosome formation when PIP levels are low and the requirement for recognition of an alternative lipid cue arises. Similar to other autophagosomal proteins that employ amphipathic helices for recruitment (39), Ccz1 utilizes a membrane packing/curvature sensing motif for association with the phagophore. Our findings support the concept of a mechanistic role for the high degree of unsaturation in autophagosomal phospholipid acyl chains observed before (30).
The targeting of MC1 can likely also be fine-tuned by posttranslational modifications, e.g., phosphorylation that was reported for Mon1 (15), which may promote or inhibit targeting. Importantly, distinct protein and lipid binding mechanisms to endosomes and autophagosomes allow differential and independent regulation of both MC1 functions. We expect that this concept may also be realized by other regulatory trafficking complexes that serve multiple functions in the cell.
Experimental procedures
Protein expression and purification
CtMon1 constructs were cloned into pCDF6P (N-terminal GST-tag, PreScission protease cleavage site) and CtCcz1 constructs into pET28HS (N-terminal 6xHis-SUMO tag) (Tables S2 and S3). E. coli BL21 (DE3) cells were co-transformed with different construct combinations, or CtCcz1 alone, and expression was induced after cold shock with 0.25 mM isopropyl-β-D-thiogalactoside for 16 h shaking at 16 °C. Cells were lysed in buffer A (50 mM NaH2PO4, 500 mM NaCl, 1 mM MgCl2, 5% (v/v) glycerol, pH 7.5) with protease inhibitor mix HP (Serva). Cleared supernatants were incubated with glutathione agarose, beads were washed, and proteins were eluted by sequential proteolytic cleavage of the tags with SUMO protease (2 h) and PreScission protease (overnight), or by addition of with 20 mM glutathione and 12 mM DTT. Eluates were further purified by size-exclusion chromatography (SEC650, Bio-Rad) pre-equilibrated with buffer B (25 mM Hepes, 250 mM NaCl, 1 mM MgCl2, 0.5 mM TCEP, pH 7.3). CtCcz1 single expressions were purified via Ni-NTA affinity chromatography, elution with 250 mM imidazole and size-exclusion chromatography as described above.
ScMC1 was expressed under the GAL1 promotor in yeast (CUY2470: BY4732; CCZ1::TRP1-GAL1pr MON1::HIS3MX6-GAL1pr CCZ1::TAP-URA3) (6). Culture were grown in YP+galactose media at 30 °C to A600 of five and harvested by centrifugation. Cells were lysed in buffer Y (50 mM Hepes-NaOH, pH 7.4, 150 mM NaCl, 1.5 mM MgCl2, FY protease inhibitor mix (Serva), 0.5 mM PMSF, 1 mM DTT). Cleared lysates were incubated with IgG Sepharose (GE Healthcare), washed with buffer Y, and eluted by overnight TEV cleavage.
GST-TEV-Vps21 and GST-TEV-Ypt10 were purified with glutathione affinity chromatography, on-column TEV-protease cleavage overnight and dialyzed against assay buffer (50 mM Hepes, NaOH pH 7.4, 150 mM NaCl, 1 mM MgCl2) (15). His-TEV-Atg8 was purified via Ni-NTA affinity chromatography, followed by TEV cleavage in solution, cation-exchange chromatography and gel filtration (running buffer 50 mM Hepes pH 7.5, 150 mM NaCl, 1 mM DTT) (40).
Liposome preparation
Lipids (Avanti Polar Lipids, except PIPs purchased from Echelon Biosciences) were mixed in chloroform and dried in a SpeedVac. The compositions of the different lipid mixes used are listed in Table S1. The lipid film was dissolved in 1 ml of buffer L (25 mM Hepes, 250 mM NaCl, 1 mM MgCl2, 5% sucrose, pH 7.3) to a final lipid concentration of 2 mM. Multilamellar lipid vesicles were generated by five cycles of freezing in liquid nitrogen and thawing at 56 °C and stored at −80 °C. Prior to use, multilamellar vesicles were extruded 21 times through a polycarbonate membrane to generate liposomes of 400 nm diameter unless stated otherwise.
Prenylation of Vps21 and Ypt10
Prior to prenylation, GTPases (40 μM) were loaded with GDP (80 μM) by incubation in the presence of 20 mM EDTA for 30 min at 30 °C, followed by the addition of 25 mM MgCl2. The prenylation was performed according to a protocol modified from (24). Each reaction contained 3 μM GTPase (preloaded with GDP), 3 μM Mrs6 (Rab escort protein), 1 μM Bet2/Bet4 (Rab-GGtase), 9 μM geranylgeranylpyrophosphate in buffer P (20 mM Hepes, 150 mM NaCl, 1 mM DTT, pH 7.3) was incubated for 1 h at 37 °C, and samples were stored at −80°C. To bind prenylated GTPases onto liposomes, prenylated GTPases were incubated with liposomes for 20 min at room temperature in the presence of 1 mM GTP.
Lipidation of Atg8
Multilamellar vesicles were extruded to generate 400 nm liposomes. Lipidation of Atg8 was preformed essentially as described before (40). 1 mM liposomes were mixed with 1 μM Atg7, 1.25 μM Atg3, 2.5 μM Atg8, 0.5 μM Atg5-12, 0.5 μM Atg16, 2 mM ATP, 1 mM MgCl2, and 1 mM DTT (all final concentrations) and incubated for 30 min at 30 °C. Samples were flash-frozen and stored at −80 °C.
Liposome sedimentation assays
Proteins and liposomes were mixed in buffer B to final concentrations of 1 μM and 0.5 mM, respectively, in a final volume of 200 μl. Reactions were incubated for 20 min at room temperature, and liposomes were pelleted at 20,000g for 20 min at 4 °C. The soluble supernatant fraction was precipitated with acetone on ice, and supernatant and pellet fractions were analyzed by SDS-PAGE and Coomassie staining. For quantifications, gel band intensities were measured with ImageJ to calculate the liposome bound fraction of protein. In samples with Mrs6, which migrates at the same molecular weight as Mon1 in SDS-PAGE, only Ccz1 bands were quantified. A two-tailed heteroscedastic t test was used for significance analyses.
Nucleotide exchange assays
Guanine nucleotide exchange assays with soluble Ypt7 were performed by loading purified Ypt7 with 1.5 M excess MANT-GDP in the presence of 20 mM EDTA for 30 min at 30 °C. After quenching the loading reaction with 25 mM MgCl2, the Ypt7-MANT-GDP complex was separated from excess MANT-GDP by size-exclusion chromatography in Buffer B. Liposomes of varying composition, with or without recruiter GTPase, were mixed with MC1 and the nucleotide exchange reaction was started by adding 0.2 mM GTP. The reaction was monitored in microplate reader (M1000 Pro, Tecan) by following the decrease of fluorescence at λem 448 nm (λex 354 nm) in intervals of 30 s at 25 °C. Data were fitted with a first-order exponential decay function y=y0 + A ∙ e−x/t to calculate kobs = τ−1 (s−1). The exchange rate was determined by dividing kobs by the concentration of MC1.
Guanine nucleotide exchange assays with prenylated Ypt7 were performed essentially as described (15). Liposomes of varying composition were preincubated with 0.2 mM GTP, with or without recruiter GTPase, mixed with MANT-GDP loaded prenylated Ypt7:GDI complex, and the volume was adjusted to 150 μl and transferred to microplate reader. 0.1 μM MC1 were added to start the nucleotide exchange reaction of prenylated Ypt7. Data were measured and analyzed as described above.
Oil emulsion assay
Purified GFP, GFP-CtCcz1Loop GFP-CtCcz1LoopΔAH (500 μl of at a concentration of 2 mg ml−1) were mixed with 20 μl olive oil (REWE Beste Wahl, extra virgin) and vortexed for 2 min. After incubation at room temperature for 20 min, phase separation was observed with GFP and GFP-CtCcz1LoopΔAH. For GFP-CtCcz1Loop, formation of an emulsion phase was observed. A 10 μl sample of the emulsion or separation phase, respectively, was transferred to a glass slide and covered with a cover slip. Samples were visualized using a DMI-8-CS l microscope (Leica) with HC PL APO UVIS CS2 63 × 1.20 water or HC PC FLUOTAR 10 × 0.30 dry objective lens and the PMT trans detector. Images were acquired using the Leica software and prepared with ImageJ.
Yeast strains
The yeast strains used in this study carry a deletion of either Mon1 or Ccz1 and express mCherry-Atg8 under the control of the ADH promotor (CUY10469 mCherry-Atg8 ccz1Δ (MATalpha leu2-3112 ura3-52 his3-Δ200 trp-Δ901 lys2-801 suc2-Δ9 GAL ATG8::ADHpr-mCherry-natNT2 ccz1Δ::hphNT1) and CUY13522 mCherry Atg8 mon1Δ (MATalpha leu2-3112 ura3-52 his3-Δ200 trp-Δ901 lys2-801 suc2-Δ9 GAL MON1::hphNT1 ATG8::ADHpr-mCherry-natNT2)). ScMon1 variants were cloned into a pRS406 plasmid with an N-terminal GFP-tag under the control of the NOP1 promotor. ScCcz1 and variants were cloned into a pRS406 plasmid with a C-terminal mNEON-tag under the control of the CCZ1 promotor. The plasmids (Table S2) were linearized and transformed into the respective mon1Δ or ccz1Δ strains by integration into the URA3 locus, respectively.
Microscopy of starved cells
Cells were grown in yeast extract peptone medium containing glucose (YPD) overnight, diluted to A600 of 0.2 in the morning and grown for 2.5 h. Cells were washed either in minimal synthetic medium lacking nitrogen or in YPD and then grown for another 2 h in the respective medium before preparation for microscopy. Cells were collected by centrifugation (5000g, 3 min, 20 °C) and washed in synthetic media once. For staining of the vacuole by CMAC, cells were incubated in synthetic media with or without nitrogen source containing 0.1 μM CMAC for 15 min at 30 °C, washed twice in fresh media without dye. Images were acquired directly afterward at room temperature using a Delta Vision Elite (GE HealthCare) equipped with an inverted microscope (model IX-71; Olympus), an UAPON X 100 (1.49 numerical aperture) oil immersion, an InsightSSI light source (Applied Precision), and an sCMOS camera (PCO). Data were processed using ImageJ 2.1.0. Shown pictures are maximum intensity projections of medial planes of yeast cells. For quantification, vacuolar Atg8 localization was assessed in 48 to 112 cells per data point.
Microscopy of Mon1CIM mutants
Cells were grown in YPD overnight, diluted to A600 of 0.25 in the morning and grown until an A600 of around 1. Cells were collected by centrifugation (5000g, 3 min, 20 °C) and washed in synthetic media once. For staining of the vacuole by FM4-64, cells were incubated in synthetic media containing 30 μM FM4-64 for 30 min at 30 °C, washed twice in fresh media, and incubated another 45 min in media without dye. Images were acquired directly afterward using a Zeiss Axioscope five FL (Zeiss) equipped with Plan-Apochromat 100× (1.4 numerical aperture) oil immersion objective and an Axiocam 702 mono camera. Data were obtained using the ZEN 3.1 pro software and processed using ImageJ 2.1.0. Shown pictures are single medial planes of yeast cells. For quantification, Mon1 dots in 60 to 90 cells were counted per data point.
Ape1 processing assay
Cells were grown overnight in YPD medium, diluted in the morning to an A600 of 0.3 and grown until an A600 of around 1. They were washed either in minimal synthetic medium lacking nitrogen or in YPD media and then incubated for another 90 min. Cells corresponding to two A600 were harvested, resuspended in 500 μl ice-cold H2O, and lysed by immediate addition of 75 μl 1,85 M NaOH containing 1M beta-mercaptoethanol and 1 mM PMSF. After incubation for 10 min at 4 °C, a final concentration of 13% (v/v) trichloroacetic acid was added and incubated again on ice for 15 min. Samples were centrifuged for 15 min at 20,000g, and pellet was washed in 1 ml ice-cold acetone followed by centrifugation for 15 min at 20,000g. Pellet was dried and applied to SDS-PAGE and Western blot for further analysis by a polyclonal antibody directed against Ape1.
Data availability
All data supporting the findings of this study are available from the corresponding author upon request.
Supporting information
This article contains supporting information.
Conflict of interests
The authors declare that they have no conflict of interest with the contents of this article.
Acknowledgments
We are grateful to Carmen Gelze and Jesse Tönjes for excellent technical assistance and to Abdou Rachid Thiam and Katharina Höffgen for helpful discussions. We thank Sascha Martens (University of Vienna, Austria) for providing us with the lipidation machinery for Atg8.
Author contributions
E. H., L. L., K. A. investigation; E. H., L. L., K. A. methodology; D. K. conceptualization; D. K. writing–original draft; E. H., L. L., C. U. writing—review & editing; C. U. and D. K. funding acquisition.
Funding and additional information
This work was supported by grants from the DFG (Deutsche Forschungsgemeinschaft, Germany) to C. U. (SFB 944-P11 & UN111/13–1) and D. K. (SFB 944-P17).
Edited by Phyllis Hanson
Supporting information
References
- 1.Holthuis J.C.M., Ungermann C. Cellular microcompartments constitute general suborganellar functional units in cells. Biol. Chem. 2013;394:151–161. doi: 10.1515/hsz-2012-0265. [DOI] [PubMed] [Google Scholar]
- 2.Ardito F., Giuliani M., Perrone D., Troiano G., Muzio L. Lo. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy (Review) Int. J. Mol. Med. 2017;40:271–280. doi: 10.3892/ijmm.2017.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bos J.L., Rehmann H., Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 4.Carlton J.G., Cullen P.J. Coincidence detection in phosphoinositide signaling. Trends Cell Biol. 2005;15:540–547. doi: 10.1016/j.tcb.2005.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Paolo G., De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 6.Nordmann M., Cabrera M., Perz A., Bröcker C., Ostrowicz C., Engelbrecht-Vandré S., et al. The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr. Biol. 2010;20:1654–1659. doi: 10.1016/j.cub.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Gerondopoulos A., Langemeyer L., Liang J.-R., Linford A., Barr F.A. BLOC-3 mutated in Hermansky-Pudlak syndrome is a Rab32/38 guanine nucleotide exchange factor. Curr. Biol. 2012;22:2135–2139. doi: 10.1016/j.cub.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kucharczyk R., Kierzek A.M., Slonimski P.P., Rytka J. The Ccz1 protein interacts with Ypt7 GTPase during fusion of multiple transport intermediates with the vacuole in S. cerevisiae. J. Cell Sci. 2001;114:3137–3145. doi: 10.1242/jcs.114.17.3137. [DOI] [PubMed] [Google Scholar]
- 9.Kucharczyk R., Dupre S., Avaro S., Haguenauer-Tsapis R., Slonimski P.P., Rytka J. The novel protein Ccz1p required for vacuolar assembly in Saccharomyces cerevisiae functions in the same transport pathway as Ypt7p. J. Cell Sci. 2000;113:4301–4311. doi: 10.1242/jcs.113.23.4301. [DOI] [PubMed] [Google Scholar]
- 10.Poteryaev D., Datta S., Ackema K., Zerial M., Spang A. Identification of the switch in early-to-late endosome transition. Cell. 2010;141:497–508. doi: 10.1016/j.cell.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 11.Gao J., Langemeyer L., Kümmel D., Reggiori F., Ungermann C. Molecular mechanism to target the endosomal Mon1-Ccz1 GEF complex to the pre-autophagosomal structure. Elife. 2018;7 doi: 10.7554/eLife.31145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van den Boomen D.J.H., Sienkiewicz A., Berlin I., Jongsma M.L.M., van Elsland D.M., Luzio J.P., et al. A trimeric Rab7 GEF controls NPC1-dependent lysosomal cholesterol export. Nat. Commun. 2020;11:5559. doi: 10.1038/s41467-020-19032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hegedűs K., Takáts S., Boda A., Jipa A., Nagy P., Varga K., et al. The Ccz1-Mon1-Rab7 module and Rab5 control distinct steps of autophagy. Mol. Biol. Cell. 2016;27:3132–3142. doi: 10.1091/mbc.E16-03-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan B.-R., Li T., Coyaud E., Laurent E.M.N., St-Germain J., Zhou Y., et al. C5orf51 is a component of the Mon1-CCZ1 complex and controls RAB7A localization and stability during mitophagy. Autophagy. 2021;18(4):829–840. doi: 10.1080/15548627.2021.1960116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langemeyer L., Borchers A.-C., Herrmann E., Füllbrunn N., Han Y., Perz A., et al. A conserved and regulated mechanism drives endosomal Rab transition. Elife. 2020;9 doi: 10.7554/eLife.56090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaites L.P., Paulo J.A., Huttlin E.L., Harper J.W. Systematic analysis of human cells lacking ATG8 proteins uncovers roles for GABARAPs and the CCZ1/Mon1 regulator C18orf8/RMC1 in macroautophagic and selective autophagic flux. Mol. Cell Biol. 2017;38 doi: 10.1128/MCB.00392-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cabrera M., Nordmann M., Perz A., Schmedt D., Gerondopoulos A., Barr F., et al. The Mon1-Ccz1 GEF activates the Rab7 GTPase Ypt7 via a longin-fold-Rab interface and association with PI3P-positive membranes. J. Cell Sci. 2014;127:1043–1051. doi: 10.1242/jcs.140921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrence G., Brown C.C., Flood B.A., Karunakaran S., Cabrera M., Nordmann M., et al. Dynamic association of the PI3P-interacting Mon1-Ccz1 GEF with vacuoles is controlled through its phosphorylation by the type 1 casein kinase Yck3. Mol. Biol. Cell. 2014;25:1608–1619. doi: 10.1091/mbc.E13-08-0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerondopoulos A., Strutt H., Stevenson N.L., Sobajima T., Levine T.P., Stephens D.J., et al. Planar cell polarity effector proteins inturned and fuzzy form a Rab23 GEF complex. Curr. Biol. 2019;29:3323–3330. doi: 10.1016/j.cub.2019.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanchez-Pulido L., Ponting C.P. Hexa-longin domain scaffolds for inter-Rab signalling. Bioinformatics. 2019;36:990–993. doi: 10.1093/bioinformatics/btz739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klink B.U., Herrmann E., Antoni C., Langemeyer L., Kiontke S., Gatsogiannis C., et al. Structure of the Mon1-Ccz1 complex reveals molecular basis of membrane binding for Rab7 activation. Proc. Natl. Acad. Sci. U. S. A. 2022;119 doi: 10.1073/pnas.2121494119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiontke S., Langemeyer L., Kuhlee A., Schuback S., Raunser S., Ungermann C., et al. Architecture and mechanism of the late endosomal Rab7-like Ypt7 guanine nucleotide exchange factor complex Mon1-Ccz1. Nat. Commun. 2017;8 doi: 10.1038/ncomms14034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langemeyer L., Perz A., Kümmel D., Ungermann C. A guanine nucleotide exchange factor (GEF) limits Rab GTPase–driven membrane fusion. J. Biol. Chem. 2018;293:731–739. doi: 10.1074/jbc.M117.812941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas L.L., Fromme J.C. GTPase cross talk regulates TRAPPII activation of Rab11 homologues during vesicle biogenesis. J. Cell Biol. 2016;215:499–513. doi: 10.1083/jcb.201608123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zimmermann L., Stephens A., Nam S.Z., Rau D., Kübler J., Lozajic M., et al. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 2018;430:2237–2243. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 26.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gautier R., Douguet D., Antonny B., Drin G. HELIQUEST: a web server to screen sequences with specific -helical properties. Bioinformatics. 2008;24:2101–2102. doi: 10.1093/bioinformatics/btn392. [DOI] [PubMed] [Google Scholar]
- 28.Chorlay A., Thiam A.R. Neutral lipids regulate amphipathic helix affinity for model lipid droplets. J. Cell Biol. 2020;219 doi: 10.1083/jcb.201907099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zinser E., Daum G. Isolation and biochemical characterization of organelles from the yeast,Saccharomyces cerevisiae. Yeast. 1995;11:493–536. doi: 10.1002/yea.320110602. [DOI] [PubMed] [Google Scholar]
- 30.Schütter M., Giavalisco P., Brodesser S., Graef M. Local fatty acid channeling into phospholipid synthesis drives phagophore expansion during autophagy. Cell. 2020;180:135–149.e14. doi: 10.1016/j.cell.2019.12.005. [DOI] [PubMed] [Google Scholar]
- 31.Li L., Tong M., Fu Y., Chen F., Zhang S., Chen H., et al. Lipids and membrane-associated proteins in autophagy. Protein Cell. 2021;12:520–544. doi: 10.1007/s13238-020-00793-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klionsky D.J., Abdel-Aziz A.K., Abdelfatah S., Abdellatif M., Abdoli A., Abel S., et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition) Autophagy. 2021;17:1. doi: 10.1080/15548627.2020.1797280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barr F.A. Review series: rab GTPases and membrane identity: causal or inconsequential? J. Cell Biol. 2013;202:191–199. doi: 10.1083/jcb.201306010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bezeljak U., Loya H., Kaczmarek B., Saunders T.E., Loose M. Stochastic activation and bistability in a Rab GTPase regulatory network. Proc. Natl. Acad. Sci. U. S. A. 2020;117:6540–6549. doi: 10.1073/pnas.1921027117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huotari J., Helenius A. Endosome maturation. EMBO J. 2011;30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yorimitsu T., Klionsky D.J. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12(Suppl 2):1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bigay J., Antonny B. Curvature, lipid packing, and electrostatics of membrane organelles: defining cellular territories in determining specificity. Dev. Cell. 2012;23:886–895. doi: 10.1016/j.devcel.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 38.Joiner A.M., Phillips B.P., Yugandhar K., Sanford E.J., Smolka M.B., Yu H., et al. Structural basis of TRAPPIII-mediated Rab1 activation. EMBO J. 2021;40 doi: 10.15252/embj.2020107607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen N., Shteyn V., Melia T.J. Sensing membrane curvature in macroautophagy. J. Mol. Biol. 2017;429:457–472. doi: 10.1016/j.jmb.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zens B., Sawa-Makarska J., Martens S. In vitro systems for Atg8 lipidation. Methods. 2015;75:37–43. doi: 10.1016/j.ymeth.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available from the corresponding author upon request.