Abstract
Thymine DNA glycosylase (TDG) is a multifaceted enzyme involved in several critical biological pathways, including transcriptional activation, DNA demethylation, and DNA repair. Recent studies have established regulatory relationships between TDG and RNA, but the molecular interactions underlying these relationships are poorly understood. Herein, we now demonstrate that TDG binds directly to RNA with nanomolar affinity. Using synthetic oligonucleotides of defined length and sequence, we show that TDG has a strong preference for binding G-rich sequences in single-stranded RNA but binds weakly to single-stranded DNA and duplex RNA. TDG also binds tightly to endogenous RNA sequences. Studies with truncated proteins indicate that TDG binds RNA primarily through its structured catalytic domain and that its disordered C-terminal domain plays a key role in regulating TDG’s affinity and selectivity for RNA. Finally, we show that RNA competes with DNA for binding to TDG, resulting in the inhibition of TDG-mediated excision in the presence of RNA. Together, this work provides support for and insights into a mechanism wherein TDG-mediated processes (e.g., DNA demethylation) are regulated through the direct interactions of TDG with RNA.
Keywords: thymine DNA glycosylase, RNA binding protein, base excision repair, G-rich RNA, DNA demethylation
DNA glycosylases initiate the base excision repair pathway by recognizing specific nucleobase lesions in DNA and catalyzing cleavage of the corresponding glycosidic bond (1, 2, 3). Thymine DNA glycosylase (TDG), a member of the uracil DNA glycosylase superfamily, has been shown to recognize mismatched pyrimidine bases from G•T and G•U pairs and initiate BER at these sites (4, 5). Like all enzymes in this family, TDG uses a base-flipping mechanism to place the damaged base into its active site to catalyze the cleavage of the glycosidic bond. Regardless of the identity of the substrate, the resulting abasic site (AP) is processed by the canonical BER pathway.
TDG is the only glycosylase known to be capable of removing 5-formylcytosine (5fC) and 5-carboxycytosine (5caC) from DNA in mammals (6, 7) and thus plays a central role in the active DNA demethylation pathway responsible for reversing 5-methylcytosine (5mC) (8, 9). Deletion of TDG from embryonic stem cells results in a five- to tenfold increase in the levels of 5fC and 5caC(10), which preferentially accumulate at genomic features associated with important gene-regulatory functions, such as promoters and enhancers (10, 11, 12, 13, 14, 15). During mammalian development, TDG’s catalytic activity is required for maintaining epigenetic stability of many developmental and tissue-specific genes, and deletion of TDG causes embryonic lethality due, in part, to hypermethylation of promoters within these genes (16, 17). Aside from its catalytic role, TDG has also been shown to potentiate transcription through physical interactions with various transcription factors and activating histone modifiers (18, 19). Notably, TDG is essential for recruiting the histone acetyltransferases (HATs) CBP/p300 to a subset of hormone-responsive genes to facilitate histone acetylation and subsequent transcriptional activation (16, 17, 20). Together, these findings support the notion that TDG plays an important role in regulating and maintaining appropriate genetic and epigenetic states. However, the mechanisms surrounding these important regulatory functions of TDG remain poorly understood.
RNAs, such as long noncoding RNAs (lncRNA) and enhancer RNAs (eRNAs), are known to play key roles in transcriptional regulation and DNA methylation control. Therefore, it is not surprising that emerging evidence indicates potential regulatory relationships between TDG and RNA. TDG was shown to be essential in mediating the transcription of eRNAs at the enhancers of a subset of E2-responsive genes (21), which have also been shown to assemble into RNA-dependent ribonucleoprotein complexes (22). Furthermore, the lncRNA TARID was shown to target a TDG/GADD45A (growth arrest and DNA-damage-inducible, alpha) complex to the TCF21 gene promoter for subsequent DNA demethylation, demonstrating the ability of lncRNAs to guide TDG-mediated activities (23). A very recent study provided evidence that the lncRNA TETILA physically interacts with TDG to target a DNA demethylation complex to specific genes (24). Although TDG lacks a canonical RNA-binding domain, it does have several positively-charged surfaces that could facilitate electrostatic interactions with RNA (Fig. S1). Moreover, TDG’s disordered N- and C-terminal domains are enriched in cationic and/or polar residues, sequence features that are known to promote RNA binding and the self-assembly of ribonucleoprotein complexes (Fig. S1) (25, 26, 27, 28). Despite these observations and the link between RNA and TDG-mediated processes, direct evidence that TDG binds to RNA is lacking.
Herein, we carry out quantitative in vitro binding studies with synthetic RNAs of defined length and sequence and demonstrate, for the first time, that TDG is a bona fide RNA binding protein. We show that TDG binds preferably and tightly to G-rich sequences in single-stranded RNAs but has a weak affinity for ssDNA and duplex RNA. Studies with truncated proteins indicate that TDG binds RNA primarily through its structured catalytic domain and that its disordered C-terminal domain plays a key role in regulating TDG’s affinity and selectivity for RNA. Finally, we demonstrate that RNA competes with DNA for binding to TDG, providing a novel mechanism through which RNA can regulate TDG-mediated excision.
Results
TDG binds RNA with a preference for G-rich sequences in vitro
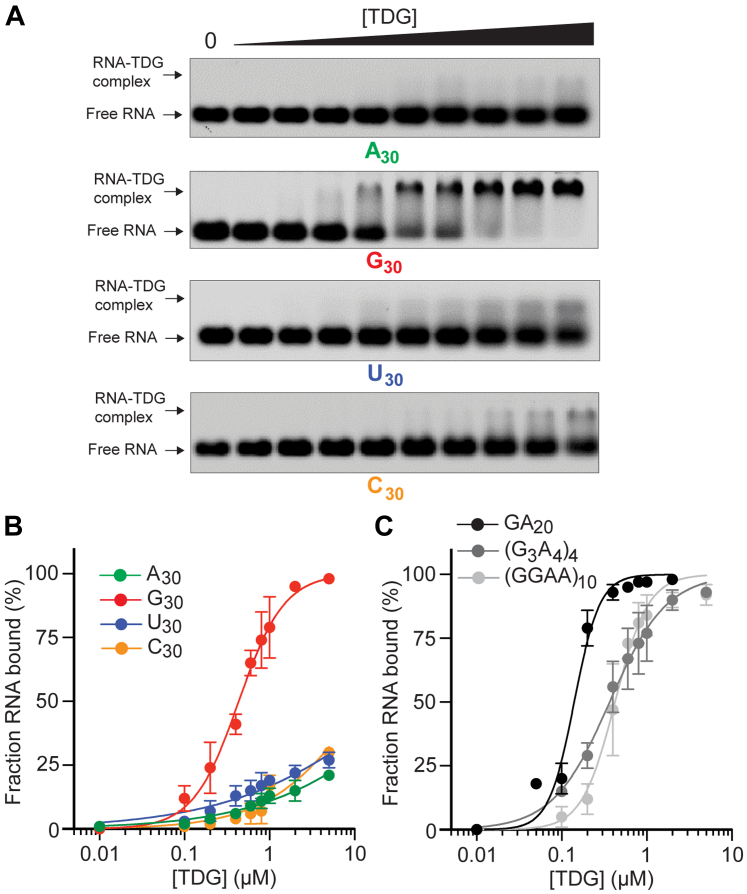
To begin our investigation, we synthesized a series homopolymer 30-mer RNA oligonucleotides of different base compositions and measured TDG binding by an agarose gel-based electrophoretic mobility shift assay (EMSA) (Fig. 1). Because the affinity of RBPs for RNA can be dependent on RNA length, especially for promiscuous RBPs, RNA of the same length was compared. Furthermore, all synthetic RNAs used in this study were cyanine 3 (Cy3)-labeled at their 5′ ends in an identical manner (Table S1). Interestingly, within this series, only poly[G] (G30) had a measurable affinity for TDG, with an apparent Kd of 434 nM (95% confidence interval (95% CI), 396–472 nM) (Fig. 1 and Table 1). While we were able to detect a small TDG-RNA shift in the EMSAs for A30, C30, and U30, the Kd for these interactions greatly exceeded 5 μM, which was the highest protein concentration tested (Fig. 1A). While these data suggested that guanines promoted TDG binding, it was also possible that other bases (e.g., adenines) prevented binding. To test this, we asked whether the insertion of Gs into A30 could rescue binding by TDG. We prepared a series of RNAs containing different arrangements of Gs and As, (GA)20, (GGAA)10, and (G3A4)4 and found that they all bound TDG at least as well as G30 (Fig. 1C and Table 1). Their binding affinities were measured as GA20 > (G3A4)4 > (GGAA)10. The narrow range of dissociation constants among these sequences (∼100–400 nM) relative to TDG’s affinity for A30 (>5 μM) indicated that the specific arrangement of Gs is not a critical factor for binding. Another sequence containing only G and U, (GU)15, also bound tightly to TDG (Fig. S2 and Table 1). Thus, we concluded that TDG binds preferentially to G-rich RNAs.
Figure 1.
TDG binds preferentially to G-rich RNA in vitro.A, Representative EMSA data for homopolymeric RNA sequences binding to TDG (0–5 μM). Uncropped gel images are presented in Fig. S11. B, Saturation plots for binding of TDG to homopolymeric RNA. Data are mean ± SD (n = 3). C, saturation plots for binding of TDG to RNAs containing different arrangements of Gs and As. Data are mean ± SD (n = 3). Representative EMSA data for panel (C) are shown in Fig. S2A. EMSA, electrophoretic mobility shift assay; TDG, Thymine DNA glycosylase
Table 1.
Equilibrium dissociation constants and Hill coefficients (h) for TDG binding to various RNA and DNA strands.
| Sequence | Kd (nM) | 95% CI | Hill (h) | 95% CI |
|---|---|---|---|---|
| G30 | 434 | 396–473 | 1.6 | 1.4–1.9 |
| C30 | >5000 | --- | --- | --- |
| A30 | >5000 | --- | --- | --- |
| U30 | >5000 | --- | --- | --- |
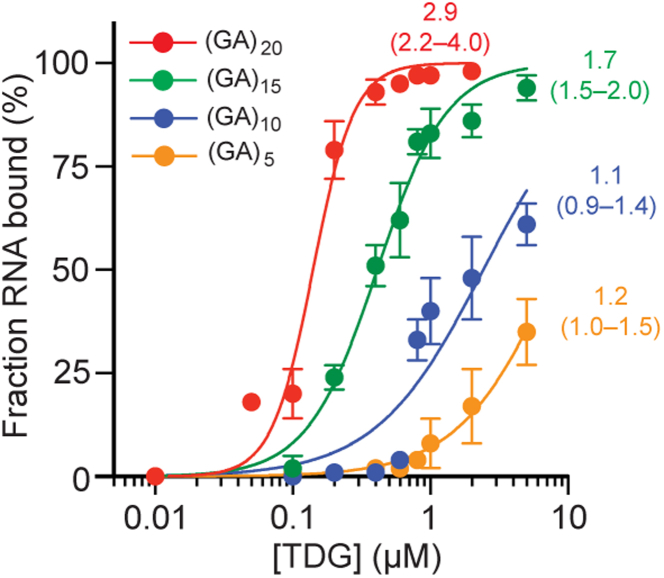
| (GA)20 | 140 | 118–165 | 2.9 | 2.2–4.0 |
| (GA)15 | 410 | 375–447 | 1.7 | 1.5–2.0 |
| (GA)10 | 2420 | 1890–3278 | 1.1 | 0.9–1.4 |
| (GA)5 | >8097 | 6704–10,506 | 1.2 | 1.0–1.5 |
| d(GA)20 | 1448 | 1289–1656 | 0.9 | 0.9–1.0 |
| (GU)20 | 319 | 301–338 | 2.2 | 1.9–2.5 |
| (G3A4)4 | 366 | 338–394 | 1.3 | 1.2–1.4 |
| (GGAA)10 | 423 | 376–472 | 2.2 | 1.7–2.9 |
| d(GGAA)10 | 2287 | 2106–2494 | 2.3 | 2.0–2.7 |
| L-(GGAA)10 | 532 | 505–559 | 3.4 | 2.9–4.0 |
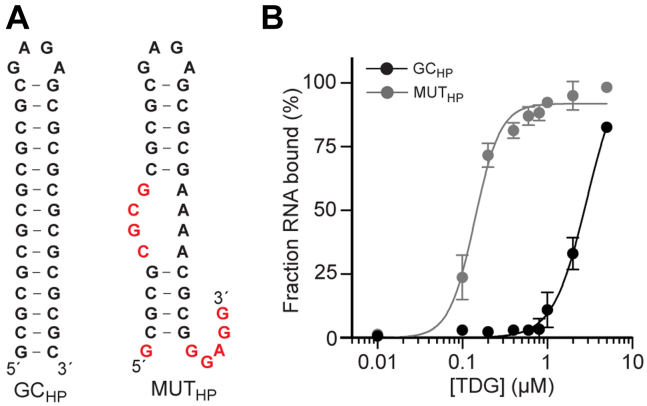
| GCHP | 2896 | 3242–4842 | 2.2 | 1.7–2.9 |
| MUTHP | 139 | 127–153 | 2.9 | 2.0–3.8 |
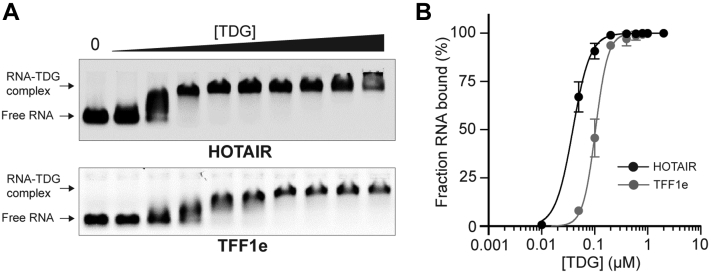
| HOTAIR | 39 | 36–41 | 2.7 | 2.3–3.2 |
| TFF1e | 104 | 100–108 | 3.7 | 3.2–4.4 |
95% CI, 95% confidence interval; TDG, Thymine DNA glycosylase; TFF1e, TFF1 enhancer RNA transcript; HOTAIR, HOX antisense intergenic RNA.
TDG has a low affinity for double-stranded RNA
In addition to its sequence, the secondary structure of RNA is an important factor to consider when studying protein–RNA interactions. RBPs often exhibit structure-specific RNA binding that is important for function (29). In particular, a number of RBPs have been shown to bind preferentially to G-quadruplex (G4)-forming RNA sequences (30, 31, 32). G-rich RNAs having at least four closely spaced G-tracks can form intramolecular G4 structures comprised of planar stacks of Hoogsteen-bonded G-quartets. Previous studies (30), including our own work (33), have shown that (GGAA)10 and (G3A4)4, but not (GA)20, form G4 structures under buffer conditions similar to those used in our binding assays. Indeed, circular dichroism (CD) spectroscopy confirmed that (GGAA)10 and (G3A4)4 form strong G4 structures under our EMSA buffer conditions, whereas (GA)20 does not (Fig. S3A). The fact that TDG bound tightly to these sequences with a narrow range of affinities (Fig. 1C and Table 1) suggested that the presence of a G4 had little influence on TDG’s ability to bind G-rich RNAs.
In addition to G4s, we also examined whether TDG could bind perfect double-stranded RNA (dsRNA) (34). We prepared a G-rich RNA hairpin, GCHP, designed to form a 14 bp stem connected by a tetraloop (Fig. 2A). GCHP had exclusively G/C base pairs within its stem and an overall base composition of 50% G. Hairpin formation was confirmed by CD spectroscopy (Fig. S3B). The affinity of GCHP for TDG (Kd ∼ 3 μM) was drastically reduced relative to the other G-rich sequences tested, which have similar G content (Fig. 2B and Table 1). This result indicated that TDG has low affinity for dsRNA, even if it is G-rich. Consistently, insertion of a few helix defects into GCHP, which resulted in an internal loop and 3′ tail giving ∼40% single-stranded nucleotides (MUTHP), was sufficient to greatly increase its affinity for TDG (Kd = 139 nM, 95% CI 127–153 nM) (Fig. 2 and Table 1).
Figure 2.
TDG avoids binding to double-stranded RNA.A, sequence and secondary structures of CGHP and MUTHP. Differences in MUTHP relative to CGHP are emphasized in red text. B, saturation plots for binding of TDG to CGHP and MUTHP. Data are mean ± SD (n = 3). Representative EMSA data are shown in Fig. S4. EMSA, electrophoretic mobility shift assay; TDG, Thymine DNA glycosylase.
RNA length dependence on TDG binding
We next examined the relationship between RNA length and TDG binding. We prepared a series of incrementally shorter versions of (GA)n, where n = 20, 15, 10, or 5. (Table 1). As shown in Figure 3, as the length of the RNA decreased so too did its affinity for TDG. The Kd increased >10-fold when the length of the RNA was halved from 40-nt to 20-nt and the 10-mer (GA)5 had a Kd > 5 μM (Table 1). Thus, in addition to its structure (above), the length of the RNA is an important factor for binding. This direct relationship between dissociation constant and RNA length suggests that in the context of a larger RNA, such as a lncRNA or eRNA, the length of a G-rich motif (and potentially the density of G within it) will play an important role in determining where and how tightly TDG binds. Interestingly, the Hill coefficient, h, used to describe the cooperativity of binding in systems with multiple simultaneous binding events, also decreased as the RNA was shortened. This overall behavior is consistent with a protein binding non-sequence-specifically and cooperatively to multiple sites on the RNA (35, 36). Indeed, similar behavior has been observed for other promiscuous RNA-binding proteins, including those that bind preferentially to G-rich sequences (37, 38). Crystal structures show that TDG binds DNA with a footprint of ∼10 bps, with the majority of interactions occurring at the periphery (39, 40). One intriguing possibility is that TDG binds RNA using the same surface as for DNA, resulting in an RNA footprint of similar length. This could explain the very weak affinity of TDG for short RNAs (≤10 nt), as well as its higher affinity and cooperativity for longer RNAs (≥30 nt), which can more easily accommodate multiple TDG molecules. Future studies are needed to test this hypothesis.
Figure 3.
TDG binds RNA in a length-dependent manner. Saturation plots for binding of TDG to (GA)20 and its truncations. Binding curves were fit to three independent replicates. The Hill coefficient (h) is given, with 95% CIs in parentheses. Representative EMSA data are shown in Fig. S5. EMSA, electrophoretic mobility shift assay; TDG, Thymine DNA glycosylase.
TDG binds endogenous RNA sequences
Up to this point, our experiments had primarily employed short, homopolymeric sequences. Therefore, we sought to demonstrate TDG’s ability to bind long, native RNA sequences that have mixed base composition and are capable of folding into intricate secondary structures in vivo. For these experiments, we chose the lncRNA HOX antisense intergenic RNA (HOTAIR) and the TFF1 enhancer RNA transcript (TFF1e) (Table S3). HOTAIR has been shown to function as a molecular scaffold for several RNA-binding proteins (41), including the G-rich RNA-binding polycomb repressive complex 2 (PRC2) (30, 37), making it a good model for studying such interactions. TFF1e RNA is transcribed from the TFF1 gene enhancer that is also bound by TDG, suggesting the possibility of an interaction in vivo. We transcribed a 419-nt fragment of HOTAIR and a 316-nt fragment of TFF1e that each contained ∼60% G content (Table S3). As shown in Figure 4 and Table 1, TDG bound tightly to both RNAs, but its Kd for HOTAIR (39 nM, 95% CI 36–41 nM) was ∼threefold lower than for TFF1e (104 nM, 95% CI 100–108 nM). Based on our observations above, this likely can be attributed to differences in sequence and folded secondary structure between these RNAs. These results confirm that TDG has the capacity to bind endogenous RNA sequences and that our prior results are not artifacts resulting from the use of short, homopolymeric sequences.
Figure 4.
TDG binds endogenous RNA sequences.A, representative EMSA data for HOTAIR and TFF1e RNA sequences binding to TDG (0–2 μM). Uncropped gel images are presented in Fig. S12. B, saturation plots for binding of TDG to HOTAIR and TFF1e RNAs. Data are mean ± S.D. (n = 3). EMSA, electrophoretic mobility shift assay; HOTAIR, HOX antisense intergenic RNA; TDG, Thymine DNA glycosylase; TFF1e, TFF1 enhancer RNA transcript.
TDG discriminates between single-stranded RNA and single-stranded DNA
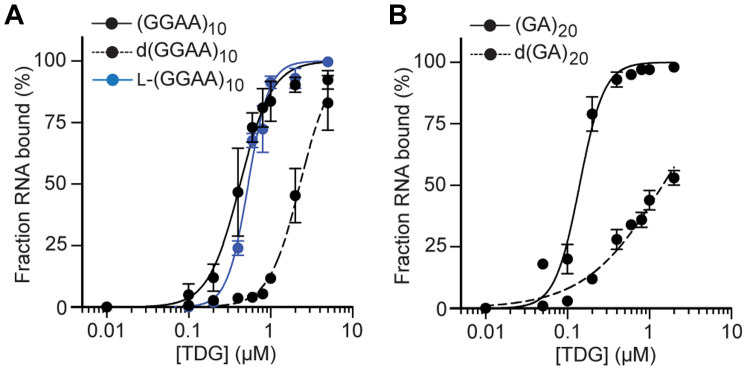
Having shown TDG’s preference for single-stranded RNA (ssRNA), we considered whether TDG could bind single-stranded DNA (ssDNA). Although ssDNA occurs rarely in cells, we expected that any potential differences in binding between these polymers could provide further insights into the nature of TDG-RNA interactions. Therefore, we compared the RNAs (GGAA)10 and (GA)20 to their DNA counterparts, d(GGAA)10 and d(GA)20, respectively, for binding to TDG. We found that TDG had much less affinity for the ssDNAs than their corresponding RNAs (Fig. 5 and Table 1), indicating that TDG has a preference for ssRNA over ssDNA.
Figure 5.
TDG prefers binding RNA over ssDNA. Saturation plots for binding of TDG to (GGAA)10 (A) and (GA)20 (B) RNAs and their variants. Data are mean ± SD (n = 3). Representative EMSA data are shown in Fig. S6. TDG, Thymine DNA glycosylase; ssDNA, single-stranded DNA; EMSA, electrophoretic mobility shift assay. EMSA, electrophoretic mobility shift assay; HOTAIR, HOX antisense intergenic RNA; TDG, Thymine DNA glycosylase; TFF1e, TFF1 enhancer RNA transcript.
Although poorly understood, the ability of RBPs to discriminate ssRNA from ssDNA of the same sequence are most often attributed to specific interactions with the ribose sugar (e.g.; hydrogen bonding with the 2′-hydroxyl group) and/or differences in conformational flexibility between the two polymers, with ssRNA being less flexible than ssDNA (42, 43, 44, 45). In order to probe the involvement of specific interactions with the ribose sugar, we examined whether TDG could bind the enantiomer of (GGAA)10, L-(GGAA)10, composed entirely of L-ribose sugar units. If contacts with the sugar play an important role in binding, then we expected that its complete stereochemical inversion would alter the formation of these interactions and potentially TDG’s ability to bind RNA. Instead, we found that TDG bound L-(GGAA)10 about as well as native (GGAA)10 (Fig. 5A). Assuming TDG binds both enantiomers through a similar set of residues, these data suggest that TDG’s ability to discriminate between ssDNA and ssRNA is not due to structure-specific interactions with the ribose sugar.
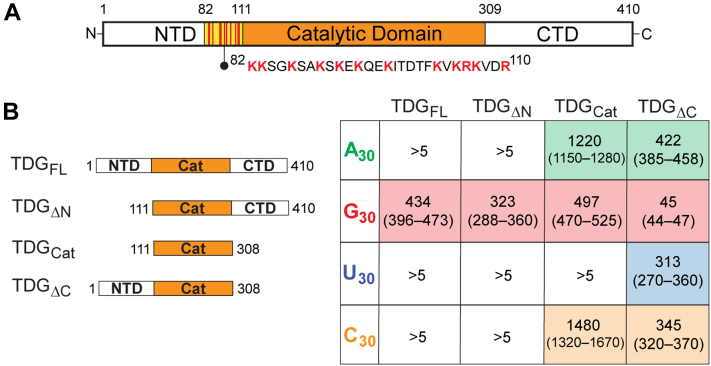
TDG’s N- and C-terminal domains modulate RNA affinity and selectivity
Work from our laboratory (46) and others (39, 47, 48) have demonstrated the importance of TDG’s disordered N-terminal domain (NTD; residues 1–111) and C-terminal domain (CTD: residues 309–410) for DNA binding and catalysis (Fig. 6A). To determine whether the NTD and/or CTD contribute to RNA binding, and to gain insight into the overall RNA-binding mechanism, we examined TDG variants lacking one or both of these disordered domains. As shown in Figure 6B, deletion of TDG’s NTD alone (TDGΔN) had little effect on its affinity and selectivity for RNA. Thus, despite the cation nature of the NTD, especially residues 82 to 110, which are known to promote nonspecific DNA binding (39, 47, 48), the NTD does not appear to contribute to RNA binding in the context of the CTD. Similarly, deletion of both the NTD and CTD from TDG (i.e., the catalytic domain; TDGCat) had little effect on poly[G] binding, although this truncation had increased affinity for A30 and C30 relative to TDGFL and TDGΔN (Fig. 6B). This result also indicates that the primary interactions between TDG and G-rich RNA occur through the structured catalytic domain. The most dramatic effects on RNA binding were observed upon deletion of the CTD alone (TDGΔC). The affinity of TDGΔC for G30 was increased nearly tenfold (Kd = 45 nM, 95% CI 44–47 nM) compared to the other variants tested (Fig. 6B). Moreover, TDGΔC bound tightly (Kd < 500 nM) and indiscriminately to the other, non-G homopolymers A30, C30, and U30. Together, these data suggest that, while TDG-RNA interactions occur primarily through the catalytic domain in the context of the full-length protein, deletion of the CTD results in an additional, nonspecific RNA binding capacity through the cationic NTD. This model is consistent with the known ability of the CTD to destabilize interactions between cationic residues within the NTD and negatively charged DNA (46, 47), resulting in impaired DNA binding. Our results now suggest that the CTD functions similarly in the context of RNA binding by preventing the cationic NTD from engaging RNA. The increased affinity of TDGCat for A30 and C30 relative to TDGFL and TDGΔN also suggests that the CTD influences RNA binding by the structured catalytic domain. Thus, the CTD plays an important role in modulating TDG’s affinity and selectivity for RNA through several mechanisms.
Figure 6.
The influence of TDG’s NTD and CTD on RNA binding.A, TDG domains discussed in this work. The basic patch within the NTD (residues 82–110) is highlighted. B, TDG truncations tested (left) and their affinity for homopolymeric RNAs (right). Shown are Kd values (nM) with 95% CIs in parentheses (n = 3). Representative EMSA data and saturation binding plots are shown in Figs. S7–S9. EMSA, electrophoretic mobility shift assay; TDG, Thymine DNA glycosylase; NTD, N-terminal domain.
RNA competes with DNA for binding to TDG
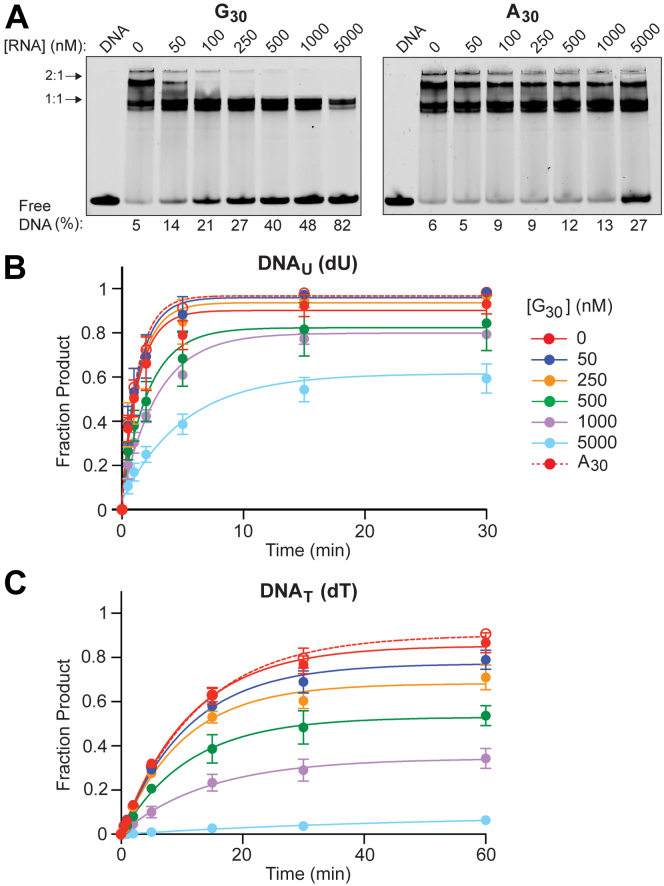
The observation that TDG’s catalytic domain alone (TDGCat) binds tightly to RNA raised the possibility that RNA and DNA compete for the same binding site on TDG. Therefore, we carried out a competition experiment by titrating unlabeled RNA with pre-formed TDG-DNA complexes (Fig. 7A). For these experiments, we employed a DNA duplex (DNAFU) containing the non-cleavable substrate analogue 2′-deoxy-2′-fluoroarabinouridine (FU) to monitor TDG binding in the absence of base excision (Table S1) (49). Combining DNAFU (100 nM) with twofold excess TDG (200 nM) resulted in the formation of both 1:1 and 2:1 TDG/DNAFU complexes as observed by EMSA (Fig. 7A). Previous studies have shown that the 1:1 complex corresponds to a single TDG subunit bound tightly to the FU site (substrate complex), whereas the 2:1 complex corresponds to a second TDG subunit bound weakly to an adjacent undamaged site (nonspecific complex) (40, 50). Importantly, both the substrate and nonspecific complexes are formed through the same binding site on their respective TDGs. As shown in Figure 7A, the addition of G30 RNA led to a concentration-dependent decrease in both the 1:1 and 2:1 TDG-DNAFU complexes. In agreement with affinity measurements (50), disruption of the tighter 1:1 complex between TDG and FU required much higher RNA concentrations when compared to the weaker, nonspecific 2:1 complex. In contrast to G30, A30, which binds very weakly to TDG, had a very modest effect on TDG-DNAFU complexes, even at the highest RNA concentration tested. Together, these data support the conclusion that DNA and RNA share the same or mutually exclusive binding sites on TDG.
Figure 7.
RNA and DNA compete for binding to TDG. A, preformed complexes between DNAFU (100 nM) and TDG (200 nM) are disrupted by G30 RNA (left) but not A30 RNA (right). The “DNA” marker indicates DNAFU in the absence of TDG and RNA. The percent free (unbound) DNAFU is indicated underneath each lane. B and C, TDG-mediated excision of DNAU (B) and DNAT (C) is inhibited by RNA. For each reaction, the DNA substrate (100 nM) was mixed with the indicated concentration of G30 RNA or 5 μM A30 RNA (A30 in the legend) followed by the addition of TDG (200 nM). Data are mean ± S.D. (n = 3). TDG, Thymine DNA glycosylase.
The competition between DNA and RNA for binding to TDG suggested that RNA can inhibit base excision by competing against DNA for the enzyme. Therefore, we monitored TDG-mediated excision of DNA containing G•T and G•U mismatches (DNAT and DNAU, respectively) in the presence of RNA (Fig. 7B and C and Table S1). For both lesions, we found that the extent of excised DNA was reduced by the presence of G30 RNA in a concentration-dependent manner. The effect of RNA on TDG-mediated excision was greater for G•T mismatches (DNAT) compared to G•U mismatches (DNAU), with the presence of 5 μM G30 leading to nearly complete inhibition of G•T processing. Again, this observation is consistent with the much higher affinity of TDG for G•U relative to G•T mismatches (Kd = 0.6 nM and 18 nM, respectively) and further supports a competitive binding model (50). Excision of both mismatches by TDG was also inhibited by native TFF1e RNA (Fig. S10), demonstrating that these effects are not an artifact of G30 RNA. As expected, A30, which binds very weakly to TDG and competes weakly for DNA binding, had little effect on TDG-mediated excision of both mismatches at the highest RNA concentration tested (5 μM) (Fig. 7, B and C).
Discussion
In this study, we provided the first quantitative measurements of the affinity of TDG to RNA, clearly demonstrating that RNA binding is a fundamental property of TDG. We showed that TDG’s interactions with RNA are dependent on its sequence and secondary structure, with TDG binding preferentially to G-rich RNA sequences. Moreover, we demonstrated that RNA competes with DNA for binding to TDG, providing a novel mechanism through which RNA can regulate TDG-mediated excision. While a link between RNA and TDG-mediated processes (e.g., DNA demethylation) has been established in cells (21, 23, 24, 51), the molecular interactions underlying these relationships have remained poorly understood. The results presented herein provide support for and new insights into a mechanism wherein TDG-mediated processes are regulated through the direct interactions of TDG with RNA.
The average human cell contains around 10 pg total RNA (52). Assuming a cellular volume of ∼3000 fl (for HeLa cells) (53, 54, 55), we estimate the cellular concentration of a 40-mer unit of RNA to be ∼250 μM. Thus, the affinity of TDG to G-rich RNA (Kd ∼ 200–500 nM) is likely to be biologically meaningful, even if only a small fraction of this RNA is available for binding. Moreover, our data show that TDG binds G-rich RNA as well as non-specific double-stranded DNA in vitro (Kd ∼ 300 nM). Considering that the vast majority of cellular DNA is wrapped into nucleosomes, which inhibit DNA binding by TDG, it is reasonable to predict that the competition between DNA and RNA binding could be widespread in cells and may play an important role in localizing TDG to specific sites throughout the genome (discussed below). A competitive binding model also has important implications for the regulation of TDG activity. As we showed previously, RNA had a much greater influence on G•T processing compared to G•U processing, suggesting a role for RNA in directing TDG’s substrate selectivity. Regardless of the nucleobase substrate, however, TDG binds very tightly (Kd ∼ 1 nM) to its AP site product (50, 56, 57). Thus, even in the presence of very high RNA concentrations, TDG is likely to remain bound to the AP site following excision to facilitate the handover of this reactive intermediate to the downstream-acting enzymes of BER. Of course, it is entirely likely that we have not yet identified the ideal RNA substrate for TDG and that sequences with much higher affinity exist in cells. Our results also do not consider the influence of other proteins or ribonucleoproteins. Therefore, particular biological situations may exist wherein TDG’s affinity for RNA is substantially greater than that reported herein.
While further biochemical studies are underway to uncover the molecular mechanisms of RNA recognition by TDG, our results provide initial insight into how TDG binds RNA. In particular, TDG’s strong preference for binding to G-rich RNA sequences and its inability to differentiate between a D- and L-ribose sugar backbone strongly indicate that nucleobases play a more dominant role than the sugar/phosphate backbone in TDG-RNA interactions. Our observation that TDG binds weakly to double stranded RNA is also consistent with a binding mode that is reliant on interactions with the nucleobases, which are disfavored with duplex RNA (58, 59). This also implies that the interactions between TDG and RNA are not primarily electrostatic. Instead, TDG may rely more heavily on hydrophobic and/or π-interactions to bind RNA. RNA–protein π interactions, which are predominantly formed between RNA nucleobases and π-containing amino acids, have been shown to play particularly central roles in RNA–protein complexes and contribute considerable stability and selectivity to protein–RNA binding (42, 44, 60). Analyses of π interactions occurring in protein-RNA crystal structures consistently find that most contacts occur with phenylalanine (F) and tyrosine (Y), which prefer a stacked orientation (i.e., π-stacking) relative to the RNA nucleobases (44, 61, 62). Interestingly, the catalytic core of TDG, which appears to be the primary binding site for RNA, has an abundance of these residues (F = 11, Y = 8; 10% of the total amino acids). The catalytic domain also contains eight arginine (R) residues, including R275 within the “insertion loop”, which contacts with both strands of the DNA substrate (40). Coincidentally, the insertion loop appears to confer CpG sequence specificity through its interactions with guanine. Like F and Y, R has the propensity to form π–π interactions with nucleobases (44), as well as cation–π interactions, which have been shown to favor G (63). Arginine is also among the most frequent amino acid to form base-specific hydrogen bonds with RNA, providing a potential strategy for G selectivity (58, 59, 64, 65). These observations suggest a potential starting point for identifying key TDG-RNA contacts.
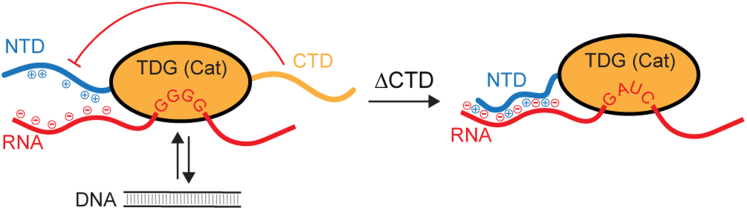
Our results also point to an important role for the CTD in modulating TDG’s affinity and selectivity for RNA. We found that deletion of the CTD resulted in an overall increase in RNA affinity, especially when the cationic NTD was present (Fig. 6). The CTD has been shown to destabilize interactions between cationic residues within the NTD and negatively charged DNA, possibly through direct interactions, resulting in impaired DNA binding (47). Our results now suggest that the CTD functions similarly in the context of RNA binding by preventing the cationic NTD from engaging RNA. The proposed model for RNA binding depicted in Figure 8 summarizes our findings. This model raises the intriguing possibility that posttranslational modification of the CTD (e.g., SUMOylation) and/or its interactions with other proteins could enable TDG-RNA interactions to be tuned to fit specific biological contexts (48). Additional studies are needed to explore these ideas further.
Figure 8.
Proposed model for TDG binding to RNA. TDG binds preferentially to G-rich RNA sequences through its structure catalytic domain, resulting in a competition between RNA and DNA for binding to TDG. The CTD prevents nonspecific electrostatic interactions between RNA and the cationic NTD, possibly through direct interactions between the two domains, promoting G-selectivity at the cost of RNA affinity. Upon deletion of the CTD, electrostatic interactions between the NTD and RNA are thus increased, leading to tighter overall binding but reduced selectivity for G. CTD, C-terminal domain; NTD, N-terminal domain; TDG, Thymine DNA glycosylase.
Finally, our findings have important implications for the role of RNA in the active DNA demethylation pathway, in which TDG plays an essential role (8, 9). Active DNA demethylation occurs at specific promoter and enhancer sequences in response to developmental or environmental signals and is often restricted to a few CpG dinucleotides (16, 17, 21). Yet, it remains unclear how this precision is achieved. While interactions between sequence-specific transcription factors and proteins involved in DNA demethylation, including TDG, are known to play a role in targeting DNA demethylation to specific genes, emerging evidence also supports the involvement of RNA (18, 19, 66). Several studies have shown that lncRNAs, which can bind to complementary DNA sequences, can serve as guides to recruit components of the DNA demethylation machinery to specific locations on the genome (23, 24, 67). The lncRNA TETILA, for example, was shown to function as a scaffold to recruit TET2 and TDG to the matrix metalloproteinase-9 promoter (MMP-9), leading to promoter-specific demethylation and MMP-9 expression (24). Our results provide further support for this model and, specifically, a role for RNA in directing TDG activity. TDG’s selectivity toward single-stranded G-rich sequences also supports this view, as any model involving RNA-mediated targeting implies some level of specificity. In addition to recruitment, our results suggest a role for RNA in modulating TDG-mediated removal of 5fC/5caC from DNA through a competitive binding mechanism. Indeed, TDG binds 5fC/5caC-containing CpG dinucleotides with a Kd similar to G•T mismatches (50), indicating that RNA will also compete with these substrates for binding to TDG. Thus, for genes that require TDG-mediated demethylation for activation, the local concentration of RNA may prove to be important not only for TDG occupancy but also for coordinating the timing of 5fC/5caC removal.
Experimental Procedures
Reagents
Oligonucleotides were either purchased from Integrated DNA Technologies (IDT) or prepared in-house by solid-phase synthesis on an Expedite 8909 DNA/RNA synthesizer using protocols recommended by the manufacturer. Nucleoside phosphoramidites and other solid-phase synthesis reagents were purchased from Glen Research. Sulfo-Cyanine3 (Cy3) and Cyanine5 (Cy5) hydroxysuccinimide (NHS) ester dyes (cat. nos. 21,320, 23,320) were purchased from Lumiprobe Life Science Solutions, In vitro transcribed RNA was purified using the Monarch RNA Cleanup Kit (cat. no. T2030L) purchased from New England BioLabs.
Expression and purification of TDG
Full-length human TDG and truncated TDG variants used herein were expressed and purified as described previously (68, 69). Purified proteins were stored at −80 °C in HP50 buffer (50 mM HEPES, pH 8, 50 mM NaCl, 10% glycerol, 10 mM BME, 1 mM PMSF) until use.
RNA synthesis and purification
All synthetic RNA oligonucleotides (Table S1) were prepared in house using protocols recommended by the manufacturer. Prior to use, oligonucleotides were purified by 20% denaturing PAGE (19:1 acrylamide:bisacrylamide). Excised bands containing the purified oligonucleotides were eluted overnight at 23 °C in a buffer containing 200 mM NaCl, 10 mM EDTA, and 10 mM Tris (pH 7.6). The gel fragments were removed from the solution by centrifugation, and the eluted oligonucleotides were concentrated using a 3 kDa pore-size Amicon Ultra Centrifugal Filter (MilliporeSigma). The concentrated samples were then desalted by ethanol precipitation, and the final concentration was determined using the absorbance at 260 nm on a NanoDrop 2000c (ThermoFisher). The identity of all novel oligonucleotides was confirmed by MS (Figs. S20–S27).
Oligonucleotides were fluorescently labeled using Sulfo-Cy3 and -Cy5 NHS ester dyes via conjugation to the 3ʹ end of oligonucleotides using a 3′-amino modification (3′-amino-modifier C6 CPG) incorporated during synthesis. The conjugation was completed by combining the amino-modified oligonucleotide at 100 μM with a 5 mM final concentration of dye NHS ester in 0.1 M sodium bicarbonate buffer (pH 8.5). The conjugation reaction was shaken overnight at 23 °C. To remove excess dye, samples were ethanol precipitated and purified once more via 20% denaturing PAGE, as described previously. Stock solutions were prepared by dilution of the oligonucleotide to 100 nM in folding buffer (25 mM NaCl, 10 mM HEPES (pH 7.8), 0.1 mM PMSF, and 50 mM KCl).
HOTAIR and TFF1e RNAs were prepared by in vitro transcription from the corresponding DNA templates (Tables S2 and S3). The DNA template used to transcribe TFF1e RNA, referred to as TFF1e-DNA, was generated by PCR amplification of 150 ng of human genomic DNA using primers TFF1eFWD and TFF1eREV (Table S2) employing Phusion High-Fidelity DNA Polymerase according to the manufacturer’s instructions. Similarly, the DNA template used to transcribe HOTAIR RNA, referred to as HOTAIR-DNA, was generated by PCR amplification of 150 ng of human genomic DNA using primers HOTAIRFWD and HOTAIRREV (Table S2). Following PCR amplification, TFF1e-DNA and HOTAIR-DNA were purified and desalted using the GenCatch Advanced PCR Extraction kit and used in an in vitro transcription reaction without further purification. Transcription reactions were carried out using 200 pmol of DNA template per 100 μl total reaction volume. The reaction mixture consisted of 10 U/μl T7 RNA polymerase, 0.001 U/μl Inorganic pyrophosphatase (IPP), 25 mM MgCl2, 2 mM spermidine, 10 mM DTT, 40 mM Tris (pH 7.9), 5 mM of each of the four NTPs, and 0.5 mM 5-aminoallyl-UTP. The reaction mixture was incubated at 37 °C for 2 h followed by the addition of 2 U of Turbo DNAses (Life Tech, Carlsbad, CA). After 30 min at 37 °C, the transcribed RNA was purified using the Monarch RNA Cleanup Kit. Labelling was carried out as described earlier using Sulfo-Cy5 NHS ester dye. After the labeling was complete, RNAs were ethanol precipitated and purified via 20% denaturing PAGE as described above.
Electrophoretic mobility shift assays
The dissociation constant (Kd) of TDG and its truncated variants for the various RNA ligands were determined by EMSA as described previously (30). Briefly, 5 to 50 nM Cy5-labeled RNA was mixed with various concentration of TDG in a reaction mixture containing 37.5 mM NaCl, 12.5 mM KCl, 10 mM HEPES (pH 7.8), 2.5 mM BME, and 5% glycerol. After incubating for 30 min at 23 °C, an aliquot was removed and loaded on a nondenaturing 1% agarose gel buffered with 1×TBE at 4 °C. Electrophoresis was carried out for 45 min at 6 to 8 V/cm (0.75 mm thick gel) and the gel visualized using a Typhoon FLA-9500 Molecular Imager (General Electric Co). Images were quantified using ImageQuant TL software imager (GE Healthcare Lifesciences). The area around the unbound species was tightly boxed, whereas the area for the bound species included both the discretely shifted band(s) and the area between the bands. Thus, any intermediate species were included in the bound fraction. Equations for specific binding with Hill slope were fit using GraphPad Prism 9 Version 9.4.1. Uncropped gel images are presented in the Supporting Information (Figs. S11–S19).
Circular dichroism spectroscopy
Oligonucleotides (5 μM) were folded in a buffer containing 37.5 mM NaCl, 12.5 mM KCl, 10 mM HEPES (pH 7.8), 2.5 mM BME, and 5% glycerol. Circular dichroism spectra were obtained from a 350 μl sample in a quartz cuvette using an Applied Photophysics Chirascan spectrophotometer (Leatherhead) at 1 nm intervals from 220 to 370 nm at room temperature.
DNA competition experiments
The double-stranded DNA substrate DNAFU was prepared by annealing 6 μM each FU-FWD and FU-REV (Table S1) in a buffer consisting of 50 mM NaCl and 20 mM HEPES (pH 7.8). The reaction mixture was heated at 95 °C for 1 min and slowly cooled down to room temperature. DNA competition experiments were analyzed using electrophoretic mobility shift assays (EMSAs). To generate preformed DNAFU/TDG complexes, DNAFU (100 nM) was mixed with TDGFL (200 nM) in a buffer consisting of 25 mM NaCl, 10 mM Tris (pH 7.6), and 5% glycerol. The reaction mixture was incubated at 30°C for 30 min before the indicated concentrations (50–5000 nM) of RNA (G30 or A30) were added. After incubating at 30 °C for another 30 min, an aliquot was loaded onto a 10% native PAGE (19:1 acrylamide:bisacrylamide) containing 0.5× TBE and 5% glycerol. Electrophoresis was carried out for 45 min at 6 to 8 V/cm (0.75 mm thick gel) at 4 °C, and the gel was visualized using a Typhoon FLA-9500 Molecular Imager as described above.
Glycosylase assays
The double-stranded DNA substrates DNAU and DNAT were prepared as described above by annealing 6 μM each dU-FWD with dU-REV and dT-FWD with dT-REV, respectively (Table S1). Glycosylase reactions were prepared by mixing 100 nM DNA substrate with the indicated concentration of RNA in a buffer consisting of 25 mM NaCl, 2.5 mM MgCl2, and 10 mM HEPES (pH 7.8). Experiments were initiated by adding TDG (200 nM) to the buffered substrates and were allowed to incubate at 30 °C. Aliquots (2 μl) were removed at the indicated times and quenched by the addition of a solution (2 μl) of 1% SDS in water. The AP site product was then cleaved by the addition of an equivalent volume of 0.2 M NaOH, followed by the addition of 8 μl of denaturing loading buffer (90% formamide, 10 mM EDTA, pH 8). Products were resolved by 20% denaturing PAGE (19:1 acrylamide:bisacrylamide), and the gel was visualized using a Typhoon FLA-9500 Molecular Imager as described previously.
Data Availability
The data generated during all experiments is available from the author upon reasonable request.
Supporting information
This article contains supporting information. Figures S1–S27 and Tables S1–S3 (33, 70).
Conflict of interests
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
L. A. M. and B. Z.: Methodology, Investigation, Formal analysis, Writing – Review & Editing; A. G.: Investigation, Resources; J. T. S.: Conceptualization, Supervision, Funding acquisition, Writing – Original draft, Methodology.
Funding and additional infromation
This material is based upon work supported by the National Science Foundation [NSF2126416]. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
Reviewed by members of the JBC Editorial Board. Edited by Patrick Sung
Supporting information
References
- 1.Schermerhorn K.M., Delaney S. A chemical and kinetic perspective on base excision repair of DNA. Acc. Chem. Res. 2014;47:1238–1246. doi: 10.1021/ar400275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brooks S.C., Adhikary S., Rubinson E.H., Eichman B.F. Recent advances in the structural mechanisms of DNA glycosylases. Biochim. Biophys. Acta. 2013;1834:247–271. doi: 10.1016/j.bbapap.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drohat A.C., Coey C.T. Role of base excision “repair” enzymes in erasing epigenetic marks from DNA. Chem. Rev. 2016;116:12711–12729. doi: 10.1021/acs.chemrev.6b00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiebauer K., Jiricny J. Mismatch-specific thymine DNA glycosylase and DNA polymerase beta mediate the correction of G.T mispairs in nuclear extracts from human cells. Proc. Natl. Acad. Sci. U. S. A. 1990;87:5842–5845. doi: 10.1073/pnas.87.15.5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neddermann P., Jiricny J. Efficient removal of uracil from G.U mispairs by the mismatch-specific thymine DNA glycosylase from HeLa cells. Proc. Natl. Acad. Sci. U. S. A. 1994;91:1642–1646. doi: 10.1073/pnas.91.5.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maiti A., Drohat A.C. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J. Biol. Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He Y.-F., Li B.-Z., Li Z., Liu P., Wang Y., Tang Q., et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuermann D., Weber A.R., Schär P. Active DNA demethylation by DNA repair: facts and uncertainties. DNA Repair. 2016;44:92–102. doi: 10.1016/j.dnarep.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 9.Wu X., Zhang Y. TET-Mediated active DNA demethylation: mechanism, function and beyond. Nat. Rev. Genet. 2017;18:517–534. doi: 10.1038/nrg.2017.33. [DOI] [PubMed] [Google Scholar]
- 10.Shen L., Wu H., Diep D., Yamaguchi S., D’Alessio Ana C., Fung H.-L., et al. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153:692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neri F., Incarnato D., Krepelova A., Rapelli S., Anselmi F., Parlato C., et al. Single-base resolution analysis of 5-Formyl and 5-carboxyl cytosine reveals promoter DNA methylation dynamics. Cell Rep. 2015;10:674–683. doi: 10.1016/j.celrep.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 12.Wu H., Wu X., Shen L., Zhang Y. Single-base resolution analysis of active DNA demethylation using methylase-assisted bisulfite sequencing. Nat. Biotechnol. 2014;32:1231–1240. doi: 10.1038/nbt.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song C.-X., Szulwach K.E., Dai Q., Fu Y., Mao S.-Q., Lin L., et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 2013;153:678–691. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia B., Han D., Lu X., Sun Z., Zhou A., Yin Q., et al. Bisulfite-free, base-resolution analysis of 5-formylcytosine at the genome scale. Nat. Met. 2015;12:1047–1050. doi: 10.1038/nmeth.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun Z., Dai N., Borgaro J.G., Quimby A., Sun D., Corrêa I.R., et al. A sensitive approach to map genome-wide 5-Hydroxymethylcytosine and 5-formylcytosine at single-base resolution. Mol. Cell. 2015;57:750–761. doi: 10.1016/j.molcel.2014.12.035. [DOI] [PubMed] [Google Scholar]
- 16.Cortázar D., Kunz C., Selfridge J., Lettieri T., Saito Y., MacDougall E., et al. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature. 2011;470:419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- 17.Cortellino S., Xu J., Sannai M., Moore R., Caretti E., Cigliano A., et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cortázar D., Kunz C., Saito Y., Steinacher R., Schär P. The enigmatic thymine DNA glycosylase. DNA Repair. 2007;6:489–504. doi: 10.1016/j.dnarep.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 19.Sjolund A.B., Senejani A.G., Sweasy J.B. MBD4 and TDG: multifaceted DNA glycosylases with ever expanding biological roles. Mutat. Res. 2013;743-744:12–25. doi: 10.1016/j.mrfmmm.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tini M., Benecke A., Um S.-J., Torchia J., Evans R.M., Chambon P. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol. Cell. 2002;9:265–277. doi: 10.1016/s1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
- 21.Kolendowski B., Hassan H., Krstic M., Isovic M., Thillainadesan G., Chambers A.F., et al. Genome-wide analysis reveals a role for TDG in estrogen receptor-mediated enhancer RNA transcription and 3-dimensional reorganization. Epigenetics Chromatin. 2018;11:5. doi: 10.1186/s13072-018-0176-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nair S.J., Yang L., Meluzzi D., Oh S., Yang F., Friedman M.J., et al. Phase separation of ligand-activated enhancers licenses cooperative chromosomal enhancer assembly. Nat. Struct. Mol. Biol. 2019;26:193–203. doi: 10.1038/s41594-019-0190-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arab K., Park Y.J., Lindroth A.M., Schäfer A., Oakes C., Weichenhan D., et al. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol. Cell. 2014;55:604–614. doi: 10.1016/j.molcel.2014.06.031. [DOI] [PubMed] [Google Scholar]
- 24.Zhou L., Ren M., Zeng T., Wang W., Wang X., Hu M., et al. TET2-interacting long noncoding RNA promotes active DNA demethylation of the MMP-9 promoter in diabetic wound healing. Cell Death Dis. 2019;10:813. doi: 10.1038/s41419-019-2047-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiedner H.J., Giudice J. It’s not just a phase: function and characteristics of RNA-binding proteins in phase separation. Nat. Struct. Mol. Biol. 2021;28:465–473. doi: 10.1038/s41594-021-00601-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B.M., Strein C., et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 27.Wang J., Choi J.-M., Holehouse A.S., Lee H.O., Zhang X., Jahnel M., et al. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell. 2018;174:688–699. doi: 10.1016/j.cell.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alshareedah I., Kaur T., Ngo J., Seppala H., Kounatse L.-A.D., Wang W., et al. Interplay between short-range attraction and long-range repulsion controls reentrant liquid condensation of ribonucleoprotein–RNA complexes. J. Am. Chem. Soc. 2019;141:14593–14602. doi: 10.1021/jacs.9b03689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seemann S.E., Mirza A.H., Hansen C., Bang-Berthelsen C.H., Garde C., Christensen-Dalsgaard M., et al. The identification and functional annotation of RNA structures conserved in vertebrates. Genome Res. 2017;27:1371–1383. doi: 10.1101/gr.208652.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X., Goodrich K.J., Gooding A.R., Naeem H., Archer S., Paucek R.D., et al. Targeting of polycomb repressive complex 2 to RNA by short repeats of consecutive guanines. Mol. Cell. 2017;65:1056–1067. doi: 10.1016/j.molcel.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 31.Huang Z.-L., Dai J., Luo W.-H., Wang X.-G., Tan J.-H., Chen S.-B., et al. Identification of G-quadruplex-binding protein from the exploration of RGG motif/G-quadruplex interactions. J. Am. Chem. Soc. 2018;140:17945–17955. doi: 10.1021/jacs.8b09329. [DOI] [PubMed] [Google Scholar]
- 32.Dumas L., Herviou P., Dassi E., Cammas A., Millevoi S. G-quadruplexes in RNA biology: recent advances and Future directions. Trends Biochem. Sci. 2021;46:270–283. doi: 10.1016/j.tibs.2020.11.001. [DOI] [PubMed] [Google Scholar]
- 33.Deckard C.E., Sczepanski J.T. Polycomb repressive complex 2 binds RNA irrespective of stereochemistry. Chem. Commun. 2018;54:12061–12064. doi: 10.1039/c8cc07433j. [DOI] [PubMed] [Google Scholar]
- 34.Sanchez de Groot N., Armaos A., Graña-Montes R., Alriquet M., Calloni G., Vabulas R.M., et al. RNA structure drives interaction with proteins. Nat. Commun. 2019;10:3246. doi: 10.1038/s41467-019-10923-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kowalczykowski S.C., Paul L.S., Lonberg N., Newport J.W., McSwiggen J.A., Von Hippel P.H. Cooperative and noncooperative binding of protein ligands to nucleic acid lattices: experimental approaches to the determination of thermodynamic parameters. Biochemistry. 1986;25:1226–1240. doi: 10.1021/bi00354a006. [DOI] [PubMed] [Google Scholar]
- 36.Epstein I.R. Kinetics of nucleic acid–large ligand interactions: exact Monte Carlo treatment and limitingl cases of reversible binding. Biopolymers. 1979;18:2037–2050. doi: 10.1002/bip.1979.360180815. [DOI] [PubMed] [Google Scholar]
- 37.Davidovich C., Zheng L., Goodrich K.J., Cech T.R. Promiscuous RNA binding by Polycomb repressive complex 2. Nat. Struct. Mol. Biol. 2013;20:1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X., Schwartz J.C., Cech T.R. Nucleic acid-binding specificity of human FUS protein. Nucl. Acids Res. 2015;43:7535–7543. doi: 10.1093/nar/gkv679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coey C.T., Malik S.S., Pidugu L.S., Varney K.M., Pozharski E., Drohat A.C. Structural basis of damage recognition by thymine DNA glycosylase: key roles for N-terminal residues. Nucl. Acids Res. 2016;44:10248–10258. doi: 10.1093/nar/gkw768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maiti A., Morgan M.T., Pozharski E., Drohat A.C. Crystal structure of human thymine DNA glycosylase bound to DNA elucidates sequence-specific mismatch recognition. Proc. Natl. Acad. Sci. U. S. A. 2008;105:8890–8895. doi: 10.1073/pnas.0711061105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhan A., Mandal S.S. LncRNA HOTAIR: a master regulator of chromatin dynamics and cancer. Biochim. Biophys. Acta - Rev. Cancer. 2015;1856:151–164. doi: 10.1016/j.bbcan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pal A., Levy Y. Structure, stability and specificity of the binding of ssDNA and ssRNA with proteins. PLoS Comput. Biol. 2019;15 doi: 10.1371/journal.pcbi.1006768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dickey T.H., Altschuler S.E., Wuttke D.S. Single-stranded DNA-binding proteins: multiple domains for multiple functions. Structure. 2013;21:1074–1084. doi: 10.1016/j.str.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson K.A., Holland D.J., Wetmore S.D. Topology of RNA-protein nucleobase-amino acid π-π interactions and comparison to analogous DNA-protein π-π contacts. RNA. 2016;22:696–708. doi: 10.1261/rna.054924.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen H., Meisburger Steve P., Pabit Suzette A., Sutton Julie L., Webb Watt W., Pollack L. Ionic strength-dependent persistence lengths of single-stranded RNA and DNA. Proc. Natl. Acad. Sci. U. S. A. 2012;109:799–804. doi: 10.1073/pnas.1119057109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deckard C.E., III, Sczepanski J.T. Reversible chromatin condensation by the DNA repair and demethylation factor thymine DNA glycosylase. Nucl. Acids Res. 2021;49:2450–2459. doi: 10.1093/nar/gkab040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smet-Nocca C., Wieruszeski J.-M., Chaar V., Leroy A., Benecke A. The Thymine−DNA glycosylase regulatory domain: residual structure and DNA binding. Biochemistry. 2008;47:6519–6530. doi: 10.1021/bi7022283. [DOI] [PubMed] [Google Scholar]
- 48.Steinacher R., Schär P. Functionality of human thymine DNA glycosylase requires SUMO-regulated changes in protein conformation. Curr. Biol. 2005;15:616–623. doi: 10.1016/j.cub.2005.02.054. [DOI] [PubMed] [Google Scholar]
- 49.Maiti A., Morgan M.T., Drohat A.C. Role of two strictly conserved residues in nucleotide flipping and N-glycosylic bond cleavage by human thymine DNA glycosylase. J. Biol. Chem. 2009;284:36680–36688. doi: 10.1074/jbc.M109.062356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morgan M.T., Maiti A., Fitzgerald M.E., Drohat A.C. Stoichiometry and affinity for thymine DNA glycosylase binding to specific and nonspecific DNA. Nucl. Acids Res. 2011;39:2319–2329. doi: 10.1093/nar/gkq1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boland M.J., Christman J.K. Characterization of dnmt3b:thymine-DNA glycosylase interaction and stimulation of thymine glycosylase-mediated repair by DNA methyltransferase(s) and RNA. J. Mol. Biol. 2008;379:492–504. doi: 10.1016/j.jmb.2008.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Finka A., Sood V., Quadroni M., Rios P.D.L., Goloubinoff P. Quantitative proteomics of heat-treated human cells show an across-the-board mild depletion of housekeeping proteins to massively accumulate few HSPs. Cell Stress Chaperones. 2015;20:605–620. doi: 10.1007/s12192-015-0583-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao L., Kroenke C.D., Song J., Piwnica-Worms D., Ackerman J.J.H., Neil J.J. Intracellular water-specific MR of microbead-adherent cells: the HeLa cell intracellular water exchange lifetime. NMR Biomed. 2008;21:159–164. doi: 10.1002/nbm.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cohen L.S., Studzinski G.P. Correlation between cell enlargement and nucleic acid and protein content of hela cells in unbalanced growth produced by inhibitors of DNA synthesis. J. Cell Physiol. 1967;69:331–339. doi: 10.1002/jcp.1040690309. [DOI] [PubMed] [Google Scholar]
- 55.Park K., Jang J., Irimia D., Sturgis J., Lee J., Robinson J.P., et al. ‘Living cantilever arrays’ for characterization of mass of single live cells in fluids. Lab Chip. 2008;8:1034–1041. doi: 10.1039/b803601b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waters T.R., Gallinari P., Jiricny J., Swann P.F. Human thymine DNA glycosylase binds to apurinic sites in DNA but is displaced by human apurinic endonuclease 1. J. Biol. Chem. 1999;274:67–74. doi: 10.1074/jbc.274.1.67. [DOI] [PubMed] [Google Scholar]
- 57.Fitzgerald M.E., Drohat A.C. Coordinating the initial steps of base excision repair: Apurinic/apyrimidinic endonuclease 1 actively stimulates thymine dna glycosylase by disrupting the product complex. J. Biol. Chem. 2008;283:32680–32690. doi: 10.1074/jbc.M805504200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gupta A., Gribskov M. The role of RNA sequence and structure in RNA–protein interactions. J. Mol. Biol. 2011;409:574–587. doi: 10.1016/j.jmb.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 59.Allers J., Shamoo Y. Structure-based analysis of protein-RNA interactions using the program ENTANGLE. J. Mol. Biol. 2001;311:75–86. doi: 10.1006/jmbi.2001.4857. [DOI] [PubMed] [Google Scholar]
- 60.Corley M., Burns M.C., Yeo G.W. How RNA-binding proteins interact with RNA: molecules and mechanisms. Mol. Cell. 2020;78:9–29. doi: 10.1016/j.molcel.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morozova N., Allers J., Myers J., Shamoo Y. Protein–RNA interactions: Exploring binding patterns with a three-dimensional superposition analysis of high resolution structures. Bioinform. 2006;22:2746–2752. doi: 10.1093/bioinformatics/btl470. [DOI] [PubMed] [Google Scholar]
- 62.Jones S., Daley D.T.A., Luscombe N.M., Berman H.M., Thornton J.M. Protein–RNA interactions: a structural analysis. Nucl. Acids Res. 2001;29:943–954. doi: 10.1093/nar/29.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang H., Li C., Yang F., Su J., Tan J., Zhang X., et al. Cation-pi interactions at non-redundant protein-RNA interfaces. Biochemistry (Moscow) 2014;79:643–652. doi: 10.1134/S0006297914070062. [DOI] [PubMed] [Google Scholar]
- 64.Luscombe N.M., Laskowski R.A., Thornton J.M. Amino acid–base interactions: a three-dimensional analysis of protein–DNA interactions at an atomic level. Nucl. Acids Res. 2001;29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Han K., Nepal C. PRI-Modeler: extracting RNA structural elements from PDB files of protein–RNA complexes. FEBS Lett. 2007;581:1881–1890. doi: 10.1016/j.febslet.2007.03.085. [DOI] [PubMed] [Google Scholar]
- 66.Hassan H.M., Kolendowski B., Isovic M., Bose K., Dranse H.J., Sampaio A.V., et al. Regulation of active DNA demethylation through RAR-mediated recruitment of a TET/TDG complex. Cell Rep. 2017;19:1685–1697. doi: 10.1016/j.celrep.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 67.Arab K., Karaulanov E., Musheev M., Trnka P., Schäfer A., Grummt I., et al. GADD45A binds R-loops and recruits TET1 to CpG island promoters. Nat. Genet. 2019;51:217–223. doi: 10.1038/s41588-018-0306-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deckard C.E., Banerjee D.R., Sczepanski J.T. Chromatin structure and the pioneering transcription factor FOXA1 regulate TDG-mediated removal of 5-formylcytosine from DNA. J. Am. Chem. Soc. 2019;141:14110–14114. doi: 10.1021/jacs.9b07576. [DOI] [PubMed] [Google Scholar]
- 69.Neddermann P., Jiricny J. The purification of a mismatch-specific thymine-DNA glycosylase from HeLa cells. J. Biol. Chem. 1993;268:21218–21224. [PubMed] [Google Scholar]
- 70.Maiti A., Noon M.S., MacKerell A.D., Jr, Pozharski E., Drohat A.C. Lesion processing by a repair enzyme is severely curtailed by residues needed to prevent aberrant activity on undamaged DNA. Proc. Natl. Acad. Sci. USA. 2012;109:8091–8096. doi: 10.1073/pnas.1201010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated during all experiments is available from the author upon reasonable request.