Abstract
Background
Thymic squamous cell carcinoma and type B3 thymoma are primary neoplasms of the anterior mediastinum that are sometimes difficult to differentiate from one another histologically. However, only a few immunohistochemical markers are available for the differential diagnosis. The purpose of this study was to discover a novel marker for differentiating between thymic squamous cell carcinoma and type B3 thymoma.
Methods
We used histological samples of thymic carcinomas (n = 26) and type B3 thymomas (n = 38) which were resected between 1986 and 2017. To search for candidates of differential markers, gene expression levels were evaluated in samples using promoter analysis by cap analysis of gene expression (CAGE) sequencing.
Results
Promoter level expression of CALML5 genes was significantly higher in thymic carcinomas than in type B3 thymomas. We further validated the results of the CAGE analysis in all 26 thymic carcinomas and 38 type B3 thymomas by immunohistochemistry (IHC). CALML5 was strongly expressed in the cytoplasm in 19 of 26 cases with thymic carcinoma, whereas positivity at the protein level was shown in two of 38 type B3 thymomas. Thus, the sensitivity (73.1%) and specificity (94.7%) of CALML5 as markers for immunohistochemical diagnosis of thymic carcinoma were extremely high.
Conclusion
We identified CALML5 as a potential marker for differentiating thymic squamous cell carcinoma from type B3 thymoma. It is assumed that future clinical use of CALML5 may improve the diagnostic accuracy of differentiating between these two diseases.
Keywords: CALML5, immunohistochemistry, thymic carcinoma, thymoma
Type B3 thymoma, the most malignant type of thymoma, is associated with cytological atypia, making its differentiation from thymic carcinoma difficult. CD5, c‐kit, and GLUT‐1 have been used as markers for differentiating thymic carcinoma from thymoma, although their sensitivity and specificity are not sufficient. We performed cap analysis of gene expression (CAGE) sequencing and immunohistochemistry (IHC) using histological samples of thymic carcinomas (n = 26) and type B3 thymomas (n = 38) which were surgically resected. CALML5 was identified as a potential marker for differentiating thymic squamous cell carcinoma from type B3 thymoma.
INTRODUCTION
Thymic epithelial tumors, including thymoma and thymic carcinoma, are rare tumors, with a prevalence of 0.15 cases per 100 000 people every year. 1 The World Health Organization (WHO) classification pathologically classifies thymoma into type A, AB, B1, B2, and B3 based on the morphology of the tumor cells and the relative quantity of immature T lymphocytes. 2 Moreover, these classifications reflect the invasive nature and prognosis of thymoma. 3 Thymic carcinoma is known to have a remarkably worse prognosis than thymoma. 4 Type B3 thymoma, the most malignant type of thymoma, is associated with cytological atypia, making its differentiation from thymic carcinoma difficult; however, this differentiation is essential because the treatments of these two diseases are distinct. Patients with advanced stage or recurrent thymic epithelial tumor are treated with chemotherapy. 5 The National Comprehensive Cancer Network Guidelines version 2.2022 recommends regimens comprising anthracycline anticancer drugs, such as cisplatin, doxorubicin, and cyclophosphamide (CAP) for thymoma and regimens containing carboplatin/paclitaxel for thymic carcinoma. Recent reports have indicated the efficacy of treatment using anti‐PD‐1 antibodies for patients with thymic carcinoma. 6 , 7 A phase II clinical trial using the anti‐PD‐1 antibody pembrolizumab reported more frequent grade 3 or higher immune‐related adverse events in patients with thymoma than in those with thymic carcinoma 7 ; hence, it is presumed that the clinical application of anti‐PD‐1 antibodies should be recommended for thymic carcinoma only.
CD5, c‐kit, and GLUT‐1 have been used as markers for differentiating thymic carcinoma from thymoma. The sensitivity of CD5 as marker for thymic carcinoma identification ranges from 30% to 70%, whereas that of c‐kit is 70%–80%. However, c‐kit immunohistochemistry (IHC) is positive in 5%–15% of thymoma cases. The sensitivity of GLUT‐1 is 70%–100%, and the specificity is 50%–100%, hence better markers are required to improve diagnostic accuracy. 8 , 9 , 10 , 11 , 12 , 13
The diverse functions of cells are determined via different combinations of numerous RNA strands transcribed from genomic DNA. RNA is translated into various proteins, which are responsible for various cellular functions. Therefore, understanding the amount and type of RNA is essential for inferring cell functions in different diseases. The cap analysis of gene expression (CAGE) is a genome‐wide profiling protocol of gene expressions at promoter level by high‐throughput sequencing of capped 5′‐ends of mRNAs and long noncoding RNAs. 14 , 15 It was used in the FANTOM5 project to analyze gene expression in more than 1800 human samples, including human primary cells and tissue and cancer cell lines. 16 , 17 Moreover, CAGE has also been used to characterize various cancer cells, including identification of estrogen‐regulated genes in breast cancer cells and to resolve androgen receptor signaling in prostate cancer cells. 18 , 19 Furthermore, it has been used to find biomarkers that differentiate lung adenocarcinoma from squamous cell lung carcinoma. 20 Pathologically, thymoma and thymic carcinoma have been regarded as part of a continuum of diseases; however, Radovich et al. indicated that these diseases are distinct biological entities, with completely different gene expression patterns, suggesting that it may be possible to base the search for biomarkers on differences in the gene expression. 21
To identify genes as candidate differential markers for diagnosing thymic carcinoma and type B3 thymoma, we initially performed CAGE on RNA extracts obtained from a limited number of clinical samples of thymic carcinoma and type B3 thymoma, and subsequently performed IHC on an extended cohort as its validation. We further examined its function by using a thymic carcinoma cell line to understand the contribution of the candidate marker to cell proliferation and anti‐cancer drug sensitivity.
METHODS
Case selection of CAGE and IHC
CAGE was performed on available frozen samples from thymic carcinoma (n = 4) and type B3 thymoma (n = 3), collected at Juntendo University Hospital between March 2010 and October 2012 (Table 1). These seven tumor tissue specimens were collected following a protocol approved by the Institutional Review Board of Juntendo University. The tissue donors provided written informed consent. In the operating room, 3–5 mm3 cubes of fresh tumor tissue were dissected and immediately placed in 1.0 ml of RNA stabilization reagent (Qiagen GmbH) for 24–48 h at 4°C. Thereafter, the specimens were stored at −80°C until RNA extraction. Total RNA was extracted from frozen tissue sections according to the standard protocol. IHC was performed on specimens from 64 cases (thymic carcinoma, n = 26; type B3 thymoma, n = 38), resected at Juntendo University Hospital between May 1986 and November 2017 including specimens subjected to CAGE sequencing. The 26 thymic carcinoma cases were classified histologically as squamous cell carcinoma. IHC was also performed on specimens from 22 cases of lung squamous cell carcinoma, resected at Juntendo University Hospital between January 2010 and January 2011. The histological diagnosis in the present study was made in accordance with the fifth edition of the WHO classification of thymic epithelial tumors. 2 Two pathologists (TH and KH), blinded to the clinical data, reviewed all stained sections independently. When discrepancies arose, the slides were reviewed using a multiheaded microscope to reach a consensus.
TABLE 1.
Characteristics of patients whose samples were used for cap analysis of gene expression (CAGE).
| Characteristics | Thymic carcinoma (%) | Type B3 thymoma (%) |
|---|---|---|
| No. of patients (n = 7) | 4 (57.1) | 3 (42.9) |
| Age (years) | ||
| Median (range) | 62 (52–78) | 54 (45–62) |
| Sex | ||
| Male | 1 (25.0) | 1 (33.3) |
| Female | 3 (75.0) | 2 (66.7) |
| Masaoka‐Koga staging | ||
| Stage I | 0 (0.0) | 1 (33.3) |
| Stage II | 2 (50.0) | 1 (33.3) |
| Stage III | 0 (0.0) | 0 (0.0) |
| Stage IV | 2 (50.0) | 1 (33.3) |
| WHO TNM staging | ||
| Stage I | 2 (50.0) | 2 (66.7) |
| Stage II | 0 (0.0) | 0 (0.0) |
| Stage III | 0 (0.0) | 0 (0.0) |
| Stage IV | 2 (50.0) | 1 (33.3) |
Abbreviation: WHO, World Health Organization.
CAGE assay
CAGE libraries were prepared following the previously described protocol. 15 In brief, the total RNA extracts were subjected to a reverse transcription reaction with SuperScript III (Life Technologies). After purification using RNAclean XP (Beckman Coulter), double stranded‐RNA/cDNA hybrids were oxidized with sodium periodate to generate aldehydes from the diols of the ribose at the cap structure and 3′‐end, and these were biotinylated with biotin hydrazide (Vector Laboratories). The remaining single‐stranded RNA was digested with RNase I (Promega) before capturing the biotinylated cap structure with magnetic streptavidin beads (Dynal Streptavidin M‐270; Life Technologies). Single‐stranded cDNA was recovered by heat denaturation and was ligated with the 3′‐end and 5′‐end adaptors specific to the samples, subsequently. Double‐stranded cDNAs were prepared by using a primer and DeepVent (exo−) DNA polymerase (New England Biolabs) and were mixed so that sequencing with one lane could produce data from eight samples. Three nanograms of the mixed samples were used to prepare 120 μl of loading sample, 15 which was loaded on c‐Bot, and sequenced by an Illumina HiSeq2500 sequencer (Illumina).
Computational analysis of CAGE data to identify candidate markers
The original samples from which individual reads were obtained were identified with the ligated adaptor sequences. After discarding reads including a base “N” or that hit a ribosomal RNA sequence (U13369.1) with rRNAdust, 22 the reads were aligned to the reference genome (hg19) using BWA (version 0.7.10) 23 and poorly aligned reads (mapping quality <20) were discarded using SAMtools (version 0.1.19). 24 Only libraries with more than 2 million mapped reads were used for further analyses. The robust peak set 15 was used as a reference set for transcription starting site (TSS) regions, and mapped reads starting from these regions were used as raw signals for the promoter activities. Differential analyses were conducted using the Deseq2 25 in the Galaxy software ecosystem (https://usegalaxy.org).
Immunohistochemistry (IHC)
IHC was performed on representative formalin‐fixed paraffin‐embedded (FFPE) tissues. The sections (thickness: 4 μm) were deparaffinized and hydrated. Immunohistochemical examinations were performed using antibodies against CALML5 (A‐3; Santa Cruz Biotechnology, 1:50 dilution); CD5 (4C7; Leica Biosystems, 1:100 dilution); c‐kit (Polyclonal; Dako Cytomation, 1:100 dilution);GLUT‐1 (18 901; Immuno‐Biological Laboratories Co., Ltd, 1:300 dilution); terminal deoxynucleotidyl transferase (TdT) (EP266; Agilent Technologies, prediluted), following the manufacturer's recommendations. Immunohistochemical staining was assessed independently by two independent pathologists (K.H. and T.H.) without prior knowledge of the clinicopathological data. A case was recorded as positive when more than 90% of the tumor cells stained positive for CALML5, more than 10% of the membrane of tumor cells stained positive for CD5, c‐kit, and GLUT‐1 and more than 10% of the lymphocyte nuclei stained positive for TdT.
Cell culture
The human thymic carcinoma cell line ThyL‐6 was established at the University of Fukui (Fukui, Japan), as previously described 26 and maintained under 5% CO2 at 37°C in RPMI‐1640 medium (Wako) supplemented with 10% fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 μg/ml). A431 cells, MDA‐MB‐468 cells, and Lenti‐X 293T cells were obtained from the American Type Culture Collection (Manassas)and maintained under 5% CO2 at 37°C in Dulbecco's Modified Eagle's medium (DMEM; Sigma‐Aldrich) supplemented with 10% fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 μg/ml).
Transfection and construction of the recombinant lentiviral vector
The vector of CALML5 was purchased from the DNASU plasmid repository (Plasmid ID HsCD00506164). The vector of CALML5 and a green fluorescent protein (GFP) control construct were subcloned into the pLX307 lentiviral expression vector (Addgene) under the control of an EF‐1α promoter. The recombinant gene was transfected into the Lenti‐X 293T cell line with PSPAX2 and PMD.2G to produce a virus supernatant. The virus supernatant was harvested at 48 h and concentrated by Lenti‐X concentrator (Takara Bio Inc.). Viral fluid and polybrene were added to ThyL‐6 cells. The viral fluid and polybrene were replaced by medium after 24 h. Puromycin (2 μg/ml) was added after 24 h. The medium and puromycin were replaced after 72–96 h.
RNA sequencing
Total RNA was extracted from the cells using the Rneasy Mini Kit (Qiagen). Total RNA of each sample was quantified and quality checked by an Agilent 2100 Bioanalyzer (Agilent Technologies), NanoDrop (Thermo Fisher Scientific Inc.), and 1% agarose gel. About 1 μg of total RNA with RNA integrity number (RIN) above 7 was used for the following library preparations. Next generation sequencing library preparations were constructed according to the manufacturer's protocol (NEBNext Ultra RNA Library Prep Kit for Illumina, New England Biolabs). The poly(A) mRNA isolation was performed using the NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs) or Ribo‐Zero rRNA removal Kit (Illumina). The mRNA fragmentation and priming were performed using NEBNext First Strand Synthesis Reaction Buffer and NEBNext Random Primers (New England Biolabs). First strand cDNA was synthesized using ProtoScript II Reverse Transcriptase (New England Biolabs) and the second‐strand cDNA was synthesized using Second Strand Synthesis Enzyme Mix (New England Biolabs). The double‐stranded cDNA purified by AxyPrep Mag PCR Clean‐up (Axygen) was then treated with End Prep Enzyme Mix (New England Biolabs) to repair both ends and add a dA‐tail in one reaction, and finally T‐A ligated to add adaptors to both ends. Size selection of the adaptor‐ligated DNA was then performed using AxyPrep Mag PCR Clean‐up (Axygen), and fragments of ~360 bp (with the approximate insert size of 300 bp) were recovered. Each sample was then amplified by PCR for 11 cycles using P5 and P7 primers, with both primers carrying sequences which can anneal with the flow cell primer to perform bridge PCR and a P7 primer carrying a six‐base index allowing for multiplexing. The PCR products were cleaned up using AxyPrep Mag PCR Clean‐up (Axygen), validated using an Agilent 2100 Bioanalyzer (Agilent Technologies), and quantified by using a Qubit 2.0 Fluorometer (Invitrogen).
Then libraries with different indices were multiplexed and loaded on an Illumina NovaSeq instrument according to the manufacturer's instructions (Illumina). Sequencing was carried out using a 2 × 150 paired‐end (PE) configuration; image analysis and base calling were conducted by the HiSeq Control Software (HCS) + OLB + GAPipeline‐1.6 (Illumina) on the NovaSeq instrument. The sequences were processed and analyzed by GENEWIZ.
We analyzed the obtained FASTQ files using the Galaxy software ecosystem (https://usegalaxy.org). Quality check was conducted using FastQ Quality Control. Trimmomatic was used to remove the low‐quality reads and adapter sequences. Mapping was conducted and count data were acquired with htseq‐count. Multidimensional scaling and differential analyses were conducted using the Deseq2 package.
Quantitative real‐time PCR
Total RNA was extracted from the cells using the Rneasy Mini Kit (Qiagen). cDNA was generated from 1 μg of RNA using the Revertra cDNA synthesis kit (Toyobo Life Science), according to the manufacturer's protocol. Quantitative real‐time PCR (qPCR) was performed using the Fast SYBR Green Master Mix (Applied Biosystems) under the following thermal cycling conditions: denaturation at 95°C for 20 s and 40 amplification cycles (denaturation at 95°C for 3 s, annealing and extension at 60°C for 30 s), concurrently with melt‐curve analysis. Actin was used as an internal control.
The sequences of primers used for the analyses were as follows:
CALML5
Forward: 5′‐GGTTGACACGGATGGAAACG‐3′
Reverse: 5′‐ACTCCTGGAAGCTGATTTCGC‐3′
Actin
Forward: 5′‐CTCTTCCAGCCTTCCTTCCT‐3′
Reverse: 5′‐AGCACTGTGTTGGCGTACAG‐3′
Western blot analysis
Cells were washed with iced‐cold PBS and lysed with 2% SDS buffer (50 mM Tris–HCl, pH 6.8%, 2% SDS, and 10% glycerol) supplemented with protease and phosphatase inhibitors (Roche). The protein concentration was measured using the DC protein assay (Bio‐Rad). Equal amounts of whole cell lysates (10–20 μg) were loaded onto 4%–20% Mini‐PROTEAN TGX Precast Gels (Bio‐Rad). After blocking with polyvinylidene difluoride (PVDF) blocking reagent for Can Get Signal (Toyobo Life Science), the blots were incubated overnight with the indicated primary antibodies: anti‐CALML5 antibody (ab154631) used at 1:1000 dilution and anti‐actin antibody (A5316) used at 1:5000 dilution. The membranes were incubated with the appropriate horseradish peroxidase‐conjugated secondary antibody (diluted 1:3000) (GE Healthcare), and this was followed by detection with enhanced chemiluminescence (ECL; GE Healthcare). All dilutions were made in Can Get Signal Immunoreaction Enhancer Solution (Toyobo Life Science). Images of the western blot signals were acquired by Chemidoc and Chemidoc MP imaging systems with Image Lab Touch Software (Bio‐Rad).
Statistical analysis
Statistical analyses were performed using GraphPad Prism 7.0 (GraphPad Software). The two‐tailed Student's t‐test and ANOVA were used to compare the values. Statistically significant differences between the means were considered at p < 0.05.
RESULTS
Identification of genome‐wide differential biomarkers for thymic carcinoma and type B3 thymoma using CAGE
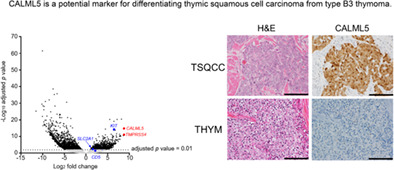
We assessed promoter activity levels in thymic carcinoma (n = 4) and type B3 thymoma (n = 3) tissues using the CAGE protocol with a next‐generation sequencer (HiSeq2500). Radovich et al. reported that thymic carcinoma and type B3 thymoma with the same origin have different expression patterns. 21 It suggests that focusing on the differences in gene expression would lead to the discovery of an appropriate biomarker. The results of our CAGE data analysis confirmed the existence of appropriate biomarkers. Figure 1 shows a volcano plot of our CAGE data showing the difference in gene expression patterns between thymic squamous cell carcinoma and type B3 thymoma. In thymic squamous cell carcinoma compared with type B3 thymoma, the TMPRSS4, CALML5, HEPACAM2, and POU2F3 genes were the top four differentially expressed genes (log2 fold change >8.17) in our samples (Table 2) and the CD5, KIT, and SLC2A1 genes were the already reported differentially expressed genes (log2 fold change >1.47) (Table 3). The log2 fold change and adjusted p‐value for CD5, KIT, and SLC2A1 are shown in Table 3. Only KIT had an adjusted p‐value of less than 0.01 and the highest log2 fold change. The promoter activities of TMPRSS4, CALML5, CD5, KIT, and SLC2A1 are shown in scatter plots of CAGE data (Figure 2a). Expression of the novel candidates (TMPRSS4 and CALML5) and the already reported markers (KIT and SLC2A1) was significantly greater in thymic squamous cell carcinoma than in type B3 thymoma. However, no significant difference was observed in CD5 expression. The mRNA expression levels of the novel (TMPRSS4 and CALML5) and known (CD5, KIT, and SLC2A1) markers obtained by analyzing Cancer Genome Atlas (TCGA) data are represented in bar graphs (as mean with standard error of the mean) (Figure 2b). Thymic squamous cell carcinoma was diagnosed in three cases and type B3 thymoma in 11 cases. There was a significant difference in CALML5, CD5, and KIT mRNA expression, but not in TMPRSS4 and SLC2A1 mRNA expression.
FIGURE 1.
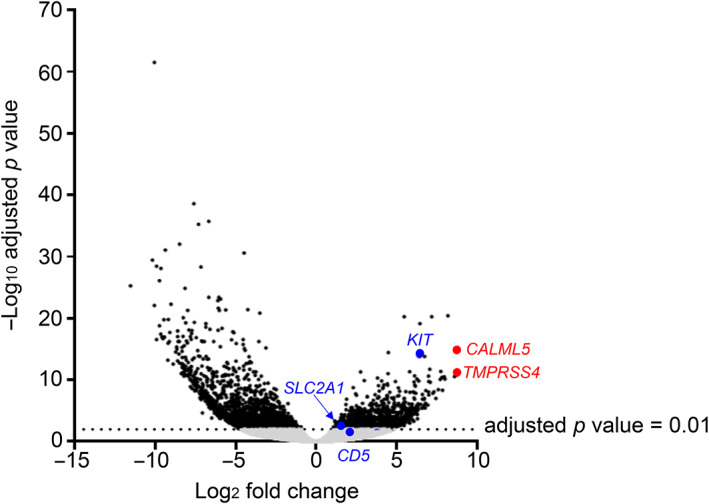
Promoter‐level expression analyses between thymic squamous cell carcinoma and type B3 thymoma. The log2‐fold change in the average expression levels in count per million (CPM) (x‐axis) is plotted against the −log10 adjusted p‐value. Individual dots represent the activities of individual promoters, and the gray dots indicate an adjusted p‐value of 0.01 or higher, and black dots indicate an adjusted p‐value of less than 0.01, and blue dots indicates known markers, and red dot indicates a novel candidate.
TABLE 2.
TOP four genes that were differentially expressed in thymic squamous cell carcinoma compared with type B3 thymoma according to log2 fold change.
| Gene name | Log2 fold change | Adjusted p‐value |
|---|---|---|
| TMPRSS4 | 8.74 | 8.66E−12 |
| CALML5 | 8.71 | 1.91E−15 |
| HEPACAM2 | 8.68 | 5.15E−12 |
| POU2F3 | 8.17 | 3.61E−21 |
TABLE 3.
Known markers for thymic carcinoma were differentially expressed in thymic squamous cell carcinoma compared with type B3 thymoma.
| Gene name | Log2 fold change | Adjusted p‐value |
|---|---|---|
| CD5 | 2.12 | 0.08 |
| KIT | 6.42 | 8.44E−15 |
| SLC2A1 | 1.47 | 0.01 |
FIGURE 2.
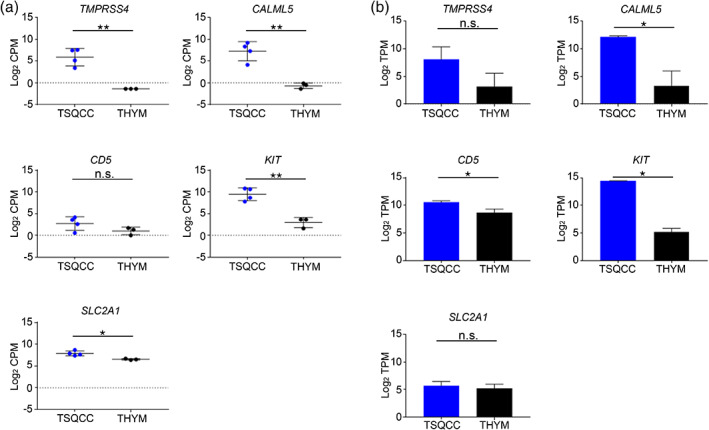
Promoter activity and mRNA expression levels of novel candidates and known markers. (a) The promoter activities of two novel candidates and three known markers for thymic squamous cell carcinoma in scatter plots with cap analysis of gene expression (CAGE) data. (b) The mRNA expression levels of novel candidates and known markers for thymic squamous cell carcinoma with the Cancer Genome Atlas (TCGA) data are shown as bar graphs (mean with standard error of the mean). CAGE, cap analysis of gene expression; CPM, count per million; THYM, type B3 thymoma; TPM, transcripts per million; TSQCC, thymic squamous cell carcinoma. *p < 0.05, **p < 0.005.
CALML5, in contrast to existing markers, effectively differentiates between thymic carcinoma and type B3 thymoma
We identified CALML5 as a diagnostic marker that is able to distinguish thymic carcinoma from type B3 thymoma. CALML5 mRNA was significantly more expressed in thymic squamous cell carcinoma than in type B3 thymoma according to both our CAGE and TCGA data analysis. CALML5, CD5, c‐kit, and GLUT‐1 were examined at the protein level with IHC to confirm their clinical utility for differentiating thymic carcinoma from type B3 thymoma (Figure 3). The patient characteristics are shown in Table 4. The proportion of patients with stage 4 tended to be higher in the thymic carcinoma group than in the type B3 thymoma group, and it is consistent with the difference of patient characteristics between them in daily clinical practice.
FIGURE 3.
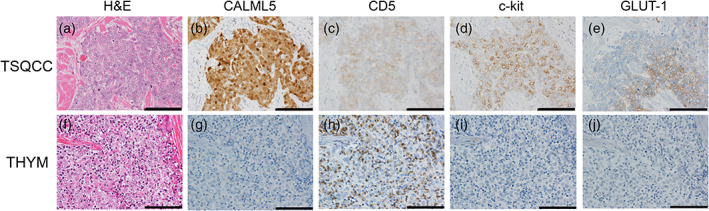
Immunohistochemistry (IHC) for thymic carcinoma and type B3 thymoma with CALML5, CD5, c‐kit, and GLUT‐1. (a–e) A case of thymic carcinoma. (a) Hematoxylin and eosin (H&E). (b) CALML5 is expressed in the cytoplasm and the nuclei. (c) CD5 is expressed in the membrane. (d) C‐kit is expressed in the membrane. (e) GLUT‐1 is expressed in the membrane. (f–j) A case of type B3 thymoma. (f) H&E. The tumor cells are (g) negative for CALML5, (h) negative for CD5, (i) negative for c‐kit, and (j) negative for GLUT‐1. Scale bar is 100 μm. IHC, immunohistochemistry; TdT, terminal deoxynucleotidyl transferase; THYM, type B3 thymoma; TSQCC, thymic squamous cell carcinoma.
TABLE 4.
Characteristics of the patients whose samples were used for immunohistochemistry (IHC).
| Characteristics | Thymic carcinoma (%) | Type B3 thymoma (%) |
|---|---|---|
| No. of patients (n = 64) | 26 (40.6) | 38 (59.4) |
| Age (years) | ||
| Median (range) | 57 (36–79) | 56 (18–76) |
| Sex | ||
| Male | 14 (53.8) | 21 (55.3) |
| Female | 12 (46.2) | 17 (44.7) |
| Masaoka‐Koga staging | ||
| Stage I | 0 (0.0) | 10 (26.3) |
| Stage II | 11 (42.3) | 19 (50.0) |
| Stage III | 3 (11.5) | 7 (18.4) |
| Stage IV | 12 (46.2) | 2 (5.3) |
| WHO TNM staging | ||
| Stage I | 11 (42.3) | 29 (76.3) |
| Stage II | 0 (0.0) | 2 (5.3) |
| Stage III | 3 (11.5) | 5 (13.2) |
| Stage IV | 12 (46.2) | 2 (5.3) |
Abbreviation: WHO, World Health Organization.
While CALML5 was expressed in the cytoplasm and the nuclei of thymic carcinoma, it was not expressed in type B3 thymoma. The samples of thymic carcinoma used for CAGE was positive for CALML5 in three of four cases. Only thymic carcinoma with the lowest CPM of the four cases was negative for CALML5. All three cases of type B3 thymoma were also negative. CALML5 as a diagnostic marker had a sensitivity of 73.1% (19/26 cases) and specificity of 94.7% (36/38 cases) for thymic carcinoma (Table 5). The sensitivity of CALML5 was higher than that of CD5, which was 69.2% (18/26 cases), and the specificity of CALML5 was higher than that of GLUT‐1, which was 60.5% (23/38 cases), and the same as the specificity of c‐kit, which was 94.7% (36/38 cases). Previous studies have reported that CD5 and c‐kit had sensitivities of 30%–70% and 70%–80% for thymic carcinoma, respectively, whereas c‐kit showed a specificity of 85%–95%. Therefore, the results indicated that CALML5 has a higher sensitivity than CD5 and a specificity equal to or higher than that of c‐kit. 8 , 9 , 10 , 11 Furthermore, the tumor cells stained diffusely, which makes it easy to confirm the presence of CALML5 expression even with a smaller number of tumor cells. There was also a single case of CD5− c‐kit− CALML5+ thymic carcinoma (Figure S1). When used in combination, CALML5, CD5, c‐kit, and GLUT‐1 had a sensitivity of 100% (26/26 cases) and specificity of 100% (38/38 cases). IHC for CALML5 was performed in four specimens (one thymic adenocarcinoma, and three thymic carcinoid). All four cases were negative for CALML5.
TABLE 5.
Expression of CALML5, CD5, c‐kit, and GLUT‐1 in thymic squamous cell carcinoma (TSQCC) and type B3 thymoma (THYM).
| No. of positive cases (%) | |||||
|---|---|---|---|---|---|
| Tumor type | No. of patients | CALML5 | CD5 | c‐kit | GLUT‐1 |
| TSQCC | 26 | 19 (73.1) | 18 (69.2) | 24 (92.3) | 26 (100.0) |
| THYM | 38 | 2 (5.3) | 1 (2.6) | 2 (5.3) | 15 (39.5) |
Thymic squamous cell carcinoma can invade the lungs, and lung squamous cell carcinoma can invade the mediastinum, making it difficult to distinguish between the two tumors. IHC was also performed in 22 cases of lung squamous cell carcinoma using with antibodies against CALML5 (Figure S2). The sensitivity was 4.5% (1/22 cases). CALML5 may be useful in differentiating between thymic squamous cell carcinoma and lung squamous cell carcinoma.
CALML5 is involved in cell proliferation and increases cisplatin sensitivity
To study the functional relevance of CALML5 to thymic carcinoma progression, we established CALML5 overexpressing thymic carcinoma cell lines (Figure S3A,B). RNA sequencing was performed using CALML5 overexpressing ThyL‐6 cells and the enhanced green fluorescent protein (EGFP) expressing ThyL‐6 cells (as control). When comparing them in gene set enrichment analysis (GSEA), the most upregulated of the Hallmark gene sets in CALML5 overexpressing ThyL‐6 cells was the E2F gene set (Figure S3C). Since the results of the RNA sequencing suggested that CALML5 may be involved in the cell cycle, we compared cell proliferation between CALML5 overexpressing ThyL‐6 cells and EGFP‐expressing ThyL‐6 cells. Cell proliferation was significantly faster (Figure S3D) and the sensitivity to cisplatin was significantly higher (Figure S3E) in CALML5 overexpressing ThyL‐6 cells.
DISCUSSION
We analyzed RNA expression data using CAGE, and selected candidate biomarkers for differentiating type B3 thymoma from thymic carcinoma. Thereafter, we identified the protein with IHC, and showed that higher CALML5 expression is consistent with the CAGE data and useful for differentiating thymic carcinoma from type B3 thymoma. Our results demonstrated that CALML5 was a more sensitive biomarker than CD5, and our results and comparisons with previous reports on CD5, which are already used to differentiate between thymoma and thymic carcinoma, showed that CALML5 was as specific or more specific than c‐kit. 8 , 9 , 10 , 11 Moreover, CD5, c‐kit and GLUT‐1 are expressed only on the cell membrane; thus, the staining area is small, lowering the sensitivity of detection in small biopsy samples, which are frequently used for diagnosing thymic carcinoma. However, the diffuse distribution of CALML5 in the cytoplasm enlarges the staining area, making evaluation easier than with CD5, c‐kit and GLUT‐1. No single marker has 100% sensitivity and 100% specificity for differentiating thymic carcinoma from thymoma. However, when used in combination, CALML5, CD5, c‐kit and GLUT‐1 increased the sensitivity to 100% (26/26 cases) and the specificity to 100% (38/38 cases). CALML5 is also presumed to improve diagnostic accuracy when combined with CD5, c‐kit, and/or GLUT‐1 IHC.
Because thymic squamous cell carcinoma can invade the lungs, and lung squamous cell carcinoma can invade the mediastinum, distinguishing between the two tumors is difficult. The sensitivity of CALML5 expression as a biomarker of lung squamous cell carcinoma was 4.5%. Therefore, CALML5 may help distinguish between thymic squamous cell carcinoma and lung squamous cell carcinoma.
According to the deduced amino acid sequence, CALML5 has 52% homology with calmodulin, the major calcium‐binding protein; because of this, it is also called calmodulin‐like skin protein (CLSP), as it is expressed in the epidermis. 27 CALML5 is a ZNF750‐ and TINCR‐induced protein that binds stratifin to regulate epidermal differentiation. 28 We assumed that CALML5 is not only involved in epidermal differentiation, but also in the differentiation of thymic epithelial cells, and CALML5 IHC may stain thymic carcinoma. IHC results showed protein expression of CALML5 in thymic squamous cell carcinoma, but almost no protein expression of CALML5 in lung squamous cell carcinoma, thymic adenocarcinoma, thymic carcinoid. Therefore, CALML5 is useful for differentiating between thymoma and thymic carcinoma, rather than between low‐grade tumor and carcinoma based on the results of IHC.
CALML5 is a poor prognostic factor for HPV‐associated oropharyngeal cancer and lung adenocarcinoma, and K63‐linked ubiquitination of CALML5 is found in breast cancer tissue, but not in the surrounding healthy tissue. 29 , 30 , 31 We created a CALML5 overexpressing cell line, ThyL‐6, to investigate the role of CALML5 in thymic carcinoma. RNA sequencing was performed, and it was found that CALML5 may be involved in cell proliferation. Our results suggest that CALML5 may be involved in the proliferation of thymic carcinoma cells and enhance cisplatin sensitivity and that it may be a therapeutic target for thymic carcinoma. Further investigation is warranted in the future.
The present study had certain limitations. Due to the rarity of thymic carcinoma and thymoma, the amount of CAGE data was small because of the small number of cases, indicating that other useful biomarkers for differentiation may have been overlooked. As we could not find thymic squamous cell carcinoma cell lines, we used ThyL‐6, a thymic undifferentiated carcinoma cell line. Also, we could not find CALML5‐overexpressing thymic carcinoma cell lines, and therefore we could not confirm that suppressed CALML5 expression in thymic carcinoma cell lines with high CALML5 expression reduces cell proliferation and cisplatin sensitivity.
In conclusion, in the present study, we discovered that CALML5 is a potential biomarker for differentiating type B3 thymoma from thymic carcinoma, using CAGE and IHC. Further studies are warranted to validate our results, and we expect that clinical use of CALML5 will improve the accuracy of diagnosis in the future.
AUTHOR CONTRIBUTION
Koichiro Kanamori and Kentaro Suina provided formal analysis, investigation, methodology, and writing—original draft preparation; Takehito Shukuya performed study conceptualization, data curation, methodology, project administration, writing—original draft preparation; Takuo Hayashi, Yoichiro Mitsuishi, Shoko Sonobe Shimamura, Wira Winardi, Masayoshi Itoh, and Hideya Kawaji provided data curation, methodology, investigation and writing—review & editing; Ken Tajima, Ryo Ko, Tetsuhiko Asao, Fumiyuki Takahashi, Kazuya Takamochi and Kenji Suzuki provided resources, supervision, and writing—review and editing. Kazuya Takamochi provided funding acquisition, supervision, and writing—review and editing. All authors read and approved the final manuscript.
CONFLICT OF INTEREST STATEMENT
The authors have no conflicts of interest directly relevant to the content of this article.
Supporting information
FIGURE S1. Thymic squamous cell carcinoma shown with H&E and IHC for CALML5, CD5, c‐kit, and GLUT‐1. The tumor cells are diffusely positive for CALML5 and negative for CD5, c‐kit. GLUT‐1 is expressed in the membrane. Scale bar is 100 μm. TSQCC, thymic squamous cell carcinoma; H&E, hematoxylin and eosin.
FIGURE S2. Lung squamous cell carcinoma shown with H&E and IHC for CALML5. The tumor cells are negative for CALML5. Scale bar is 100 μm. LSQCC, lung squamous cell carcinoma; H&E, hematoxylin and eosin; IHC, immunohistochemistry.
FIGURE S3. CALML5 overexpression experiments with ThyL‐6. (A) mRNA expression levels of CALML5 in ThyL‐6 cells. The expression levels of CALML5 are lower in ThyL‐6 than in A431 and MDA‐MB‐468. (B) Western blot analysis of CALML5 overexpression in ThyL‐6. (C) Using the hallmark gene set in gene set enrichment analysis (GSEA), target genes of E2F are positively correlated with the highest CALML5 overexpression in ThyL‐6 cells. (D) Effects of CALML5 overexpression on ThyL‐6 cell proliferation. The cell number on day 5 relative to day 0 is shown. CALML5 overexpression significantly promotes cell proliferation. **p < 0.005. (E) The effect of CALML5 overexpression on the chemosensitivity of ThyL‐6 cells to cisplatin (CDDP). Results are presented as the average of three independent experiments ± standard deviation. NES, normalized enrichment score; EGFP, enhanced green fluorescent protein.
ACKNOWLEDGMENT
This work was supported by JSPS KAKENHI grant no. 18K08185 (to Kazuhisa Takahashi).
Kanamori K, Suina K, Shukuya T, Takahashi F, Hayashi T, Hara K, et al. CALML5 is a novel diagnostic marker for differentiating thymic squamous cell carcinoma from type B3 thymoma. Thorac Cancer. 2023;14(12):1089–1097. 10.1111/1759-7714.14853
REFERENCES
- 1. Engels EA, Pfeiffer RM. Malignant thymoma in the United States: demographic patterns in incidence and associations with subsequent malignancies. Int J Cancer. 2003;105(4):546–51. [DOI] [PubMed] [Google Scholar]
- 2. WHO Classification of Tumors Editorial Board . Thoracic tumours. WHO classification of tumours. 5th ed. Lyon: International Agency for Research on Cancer; 2021. [Google Scholar]
- 3. Okumura M, Ohta M, Tateyama H, Nakagawa K, Matsumura A, Maeda H, et al. The World Health Organization histologic classification system reflects the oncologic behavior of thymoma: a clinical study of 273 patients. Cancer. 2002;94(3):624–32. [DOI] [PubMed] [Google Scholar]
- 4. Kondo K, Monden Y. Therapy for thymic epithelial tumors: a clinical study of 1,320 patients from Japan. Ann Thorac Surg. 2003;76(3):884–75. [DOI] [PubMed] [Google Scholar]
- 5. Berghmans T, Durieux V, Holbrechts S, Jungels C, Lafitte JJ, Meert AP, et al. Systemic treatments for thymoma and thymic carcinoma: a systematic review. Lung Cancer. 2018;126:25–31. [DOI] [PubMed] [Google Scholar]
- 6. Giaccone G, Kim C, Thompson J, McGuire C, Kallakury B, Chahine JJ, et al. Pembrolizumab in patients with thymic carcinoma: a single‐arm, single‐centre, phase 2 study. Lancet Oncol. 2018;19(3):347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cho J, Kim HS, Ku BM, Choi YL, Cristescu R, Han J, et al. Pembrolizumab for patients with refractory or relapsed thymic epithelial tumor: an open‐label phase II trial. J Clin Oncol. 2019;37(24):2162–70. [DOI] [PubMed] [Google Scholar]
- 8. Nakagawa K, Matsuno Y, Kunitoh H, Maeshima A, Asamura H, Tsuchiya R. Immunohistochemical KIT (CD117) expression in thymic epithelial tumors. Chest. 2005;128(1):140–4. [DOI] [PubMed] [Google Scholar]
- 9. Kojika M, Ishii G, Yoshida J, Nishimura M, Hishida T, Ota SJ, et al. Immunohistochemical differential diagnosis between thymic carcinoma and type B3 thymoma: diagnostic utility of hypoxic marker, GLUT‐1, in thymic epithelial neoplasms. Mod Pathol. 2009;22(10):1341–50. [DOI] [PubMed] [Google Scholar]
- 10. Yamada Y, Tomaru U, Ishizu A, Kiuchi T, Marukawa K, Matsuno Y, et al. Expression of proteasome subunit β5t in thymic epithelial tumors. Am J Surg Pathol. 2011;35(9):1296–304. [DOI] [PubMed] [Google Scholar]
- 11. Khoury T, Chandrasekhar R, Wilding G, Tan D, Cheney RT. Tumour eosinophilia combined with an immunohistochemistry panel is useful in the differentiation of type B3 thymoma from thymic carcinoma. Int J Exp Pathol. 2011;92(2):87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Du MJ, Shen Q, Yin H, Rao Q, Zhou MX. Diagnostic roles of MUC1 and GLUT1 in differentiating thymic carcinoma from type B3 thymoma. Pathol Res Pract. 2016;212(11):1048–51. [DOI] [PubMed] [Google Scholar]
- 13. Su XY, Wang WY, Li JN, Liao DY, Wu WL, Li GD. Immunohistochemical differentiation between type B3 thymomas and thymic squamous cell carcinomas. Int J Clin Exp Pathol. 2015;8(5):5354–62. [PMC free article] [PubMed] [Google Scholar]
- 14. Shiraki T, Kondo S, Katayama S, Waki K, Kasukawa T, Kawaji H, et al. Cap analysis gene expression for high‐throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci U S A. 2003;100(26):15776–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Murata M, Nishiyori‐Sueki H, Kojima‐Ishiyama M, Carninci P, Hayashizaki Y, Itoh M. Detecting expressed genes using CAGE. Methods Mol Biol. 2014;1164:67–85. [DOI] [PubMed] [Google Scholar]
- 16. FANTOM Consortium and the RIKEN PMI and CLST (DGT) , Forrest ARR, Kawaji H, Rehli M, Baillie JK, de Hoon MJL, et al. A promoter‐level mammalian expression atlas. Nature. 2014;507(7493):462–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lizio M, Abugessaisa I, Noguchi S, Kondo A, Hasegawa A, Hon CC, et al. Update of the FANTOM web resource: expansion to provide additional transcriptome atlases. Nucleic Acids Res. 2019;47:D752–d758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yamaga R, Ikeda K, Boele J, Horie‐Inoue K, Takayama KI, Urano T, et al. Systemic identification of estrogen‐regulated genes in breast cancer cells through cap analysis of gene expression mapping. Biochem Biophys Res Commun. 2014;447(3):531–6. [DOI] [PubMed] [Google Scholar]
- 19. Takayama K, Tsutsumi S, Katayama S, Okayama T, Horie‐Inoue K, Ikeda K, et al. Integration of cap analysis of gene expression and chromatin immunoprecipitation analysis on array reveals genome‐wide androgen receptor signaling in prostate cancer cells. Oncogene. 2011;30(5):619–30. [DOI] [PubMed] [Google Scholar]
- 20. Takamochi K, Ohmiya H, Itoh M, Mogushi K, Saito T, Hara K, et al. Novel biomarkers that assist in accurate discrimination of squamous cell carcinoma from adenocarcinoma of the lung. BMC Cancer. 2016;16(1):760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Radovich M, Pickering CR, Felau I, Ha G, Zhang H, Jo H, et al. The integrated genomic landscape of thymic epithelial tumors. Cancer Cell. 2018;33(2):244–258.e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hasegawa A, Daub C, Carninci P, Hayashizaki Y, Lassmann T. MOIRAI: a compact workflow system for CAGE analysis. BMC Bioinformatics. 2014;15:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H, Durbin R. Fast and accurate long‐read alignment with Burrows‐Wheeler transform. Bioinformatics. 2010;26(5):589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Inai K, Takagi K, Takimoto N, Okada H, Imamura Y, Ueda T, et al. Multiple inflammatory cytokine‐productive ThyL‐6 cell line established from a patient with thymic carcinoma. Cancer Sci. 2008;99(9):1778–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Méhul B, Bernard D, Simonetti L, Bernard MA, Schmidt R. Identification and cloning of a new calmodulin‐like protein from human epidermis. J Biol Chem. 2000;275(17):12841–7. [DOI] [PubMed] [Google Scholar]
- 28. Sun BK, Boxer LD, Ransohoff JD, Siprashvili Z, Qu K, Lopez‐Pajares V, et al. CALML5 is a ZNF750‐ and TINCR‐induced protein that binds stratifin to regulate epidermal differentiation. Genes Dev. 2015;29(21):2225–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Misawa K, Imai A, Matsui H, Kanai A, Misawa Y, Mochizuki D, et al. Identification of novel methylation markers in HPV‐associated oropharyngeal cancer: genome‐wide discovery, tissue verification and validation testing in ctDNA. Oncogene. 2020;39(24):4741–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ke H, Wu Y, Wang R, Wu X. Creation of a prognostic risk prediction model for lung adenocarcinoma based on gene expression, methylation, and clinical characteristics. Med Sci Monit. 2020;26:e925833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Debald M, Schildberg FA, Linke A, Walgenbach K, Kuhn W, Hartmann G. Specific expression of k63‐linked ubiquitination of calmodulin‐like protein 5 in breast cancer of premenopausal patients. J Cancer Res Clin Oncol. 2013;139(12):2125–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1. Thymic squamous cell carcinoma shown with H&E and IHC for CALML5, CD5, c‐kit, and GLUT‐1. The tumor cells are diffusely positive for CALML5 and negative for CD5, c‐kit. GLUT‐1 is expressed in the membrane. Scale bar is 100 μm. TSQCC, thymic squamous cell carcinoma; H&E, hematoxylin and eosin.
FIGURE S2. Lung squamous cell carcinoma shown with H&E and IHC for CALML5. The tumor cells are negative for CALML5. Scale bar is 100 μm. LSQCC, lung squamous cell carcinoma; H&E, hematoxylin and eosin; IHC, immunohistochemistry.
FIGURE S3. CALML5 overexpression experiments with ThyL‐6. (A) mRNA expression levels of CALML5 in ThyL‐6 cells. The expression levels of CALML5 are lower in ThyL‐6 than in A431 and MDA‐MB‐468. (B) Western blot analysis of CALML5 overexpression in ThyL‐6. (C) Using the hallmark gene set in gene set enrichment analysis (GSEA), target genes of E2F are positively correlated with the highest CALML5 overexpression in ThyL‐6 cells. (D) Effects of CALML5 overexpression on ThyL‐6 cell proliferation. The cell number on day 5 relative to day 0 is shown. CALML5 overexpression significantly promotes cell proliferation. **p < 0.005. (E) The effect of CALML5 overexpression on the chemosensitivity of ThyL‐6 cells to cisplatin (CDDP). Results are presented as the average of three independent experiments ± standard deviation. NES, normalized enrichment score; EGFP, enhanced green fluorescent protein.


