Summary
Background
The Asian lineage Zika virus (ZIKV) emerged as a public health emergency in 2016 causing severe neurological pathologies with no apparent historical correlate to the mild, disease-causing innocuous member of the mosquito-borne flavivirus genus that was discovered in Africa in 1947. Replication error rate of RNA viruses combined with viral protein/RNA structural plasticity can lead to evolution of virus-induced pathogenicity that is critical to identify and validate.
Methods
Infection studies in cells and A129 interferon alpha/beta receptor deficient mice with ZIKV French Polynesian H/PF/2013 clinical isolate, plaque-purified isogenic clone derivatives as well as infectious cDNA clone derived wild-type and site-specific mutant viruses, were employed together with Next-Generation Sequencing (NGS) to pin-point the contributions of specific viral variants in neurovirulence recapitulated in our ZIKV mouse model.
Findings
NGS analysis of the low-passage inoculum virus as well as mouse serum, brain and testis derived virus, revealed specific enrichment in the mouse brain that were not found in the other tissues. Specifically, non-structural (NS) protein 2A variant at position 117 along with changes in NS1 and NS4B were uniquely associated with the mouse brain isolate. Mutational analysis of these variants in cDNA infectious clones identified the NS2A A117V as the lethal pathogenic determinant with potential epistatic contribution of NS1 and NS4B variants in ZIKV brain penetrance.
Interpretation
Our findings confirm that viral subpopulations drive ZIKV neuropathogenicity and identify specific sequence variants that expand in the mouse brain that associates with this phenotype which can serve as predictors of severe epidemics.
Funding
Duke-NUS Khoo Post-doctoral Fellowship Award 2020 (KWKC) and National Medical Research Council of Singapore grants MOH-000524 (OFIRG) (SW) and MOH-OFIRG20nov-0002 (SGV).
Keywords: Zika virus, Non-consensus viral subpopulations, cDNA infectious clone, Non-structural protein 2A, Zika neuropathogenesis, NextGen sequencing
Research in context.
Evidence before this study
The determinants of ZIKV severe congenital pathogenesis were largely identified through comparative analysis of consensus genomes pre- and post-epidemic. Recent study has indicated a natural polymorphism in NS2A of ZIKV isolates that is responsible for increased viral virulence in vivo. However the contributions of viral subpopulations within ZIKV isolates have not been systematically explored.
Added value of this study
This study showed the spatiotemporal landscape of ZIKV variants following infection in mice by Next-Generation Sequencing which revealed a tissue-specific expansion of non-consensus viral subpopulations only in the brain but not in other tissues (serum or testis) that correlated with the disease morbidity. The previously reported ZIKV NS2A:V117 virulent determinant, originally present at low frequency in the infecting inoculum became a consensus variant in the brain of infected mice, demonstrating its association with neuropathogenesis. The enrichment of the other low allele frequency variants in the brain compared to the serum of infected mice also revealed the possible cooperative effect of viral proteins in mediating ZIKV penetrance in the brain.
Implications of all the available evidence
This study has highlighted the importance of virus evolution and sequence dynamics that could become selected through host environmental conditions that favour the dominance of specific variants in disease severity. Hence, detailed examination of the non-consensus variants within the clinical isolates should be considered when exploring the enhanced disease pathogenesis of epidemic outbreak strains. This can lead to the identification of virulence markers as predictors of severe epidemics as well as attenuating determinants that can be employed in vaccine development.
Introduction
Zika virus (ZIKV), a mosquito-borne flavivirus has emerged as a public health concern causing large epidemics in the Pacific Islands and the Americas.1 Following the rapid spread into the Americas, increasing cases of severe neurological abnormalities associated with ZIKV infection; congenital Zika syndrome (CZS) that results in brain malformation (microcephaly) in neonates born to mothers infected during early pregnancy and Guillian Barré syndrome in adults2,3 became the hallmark of a virus infection that was previously thought to be self-limiting and mild. Phylogenetic analysis indicated two major lineages of ZIKV, the African and Asian lineages, where the latter was found to be associated with the neurological syndromes linked to ZIKV infection reported in the outbreaks in the Americas.4 The emergence of the increased disease severity has raised global attention to understand the driving force of ZIKV virulence and pathogenesis.
Comparative genome analysis of the contemporary and ancestral ZIKV strains revealed acquisition of mutations that appear to correlate with the epidemic potential and disease severity through enhanced virus transmission and neurovirulence.5,6 An evolutionary variant mutation A188V in NS1 was shown to increase mosquito infectivity that could facilitate better transmissibility of the virus in the endemic population.7 It was further demonstrated that the same NS1 mutation improved viral fitness through suppression of the Type I IFN response.8 Another field-identified variant mutation S139N in prM has been shown to strongly correlate with fetal microcephaly through enhanced infectivity of neuronal progenitor cells.9 Additionally a M protein polymorphism that arose from mammalian cell infection was found to impair mouse fetal development through dysregulation of metabolism.10 More recently a NS2A polymorphism was shown to contribute to increased ZIKV virulence in vivo.11 It should be noted that most of the studies on elucidating the critical determinants of viral virulence have been based on comparing consensus sequence/proteome of strains (i.e. publicly deposited GenBank sequences). However viral genome diversity have been implicated in clinical fitness of viruses that could impact on disease manifestation.12,13 Despite the fact that several genetic polymorphisms have been described in natural occurring ZIKV population,13,14 the contribution of these variants in ZIKV disease pathogenesis still remains understudied.
In this work we sought to investigate the role of genetic polymorphisms in driving ZIKV pathogenesis using the French Polynesian strain H/PF/2013, a lineage which showed significantly higher genetic diversity than the Asian lineage ZIKV Paraiba01/2015 strain. Next-Generation Sequencing (NGS) of virus isolated from mice that resulted in the observed lethality of H/PF/2013 revealed much greater genetic polymorphisms compared to the starting virus inoculum that had been passaged in C6/36 mosquito cell line. Importantly there was a selective expansion of variants specifically in the brain relative to the serum that appear to correlate with the neuropathogenesis caused by ZIKV infection. Through in vitro and in vivo analysis of independent plaque-purified virus populations and reverse genetics of isogenic recombinant cDNA clone, we narrowed down to a polymorphism in NS2A that appears to be clearly associated with the lethal phenotype displaying high brain infectivity and neuroinflammation in our A129 mouse infection model.
Methods
Cell lines and virus
BHK-21 cells (baby hamster kidney fibroblast cells, ATCC: CCL-10) were cultured in RPMI1640 medium (Gibco) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (P/S) at 37 °C in 5% CO2. LN-229 cells (human glioblastoma cells, ATCC: CRL-2611) were cultured in DMEM medium (Gibco) with 5% FBS, 4.5 g/L glucose and 1% P/S at 37 °C in 5% CO2. C6/36, an Aedes albopictus cell line (ATCC: CRL-1660), was maintained in RPMI1640 medium (Gibco) with 10% FBS, 25 mM HEPES and 1% P/S at 28 °C in the absence of CO2. These cells lines were tested negative for mycoplasma by PCR.
The parental ZIKV French Polynesia clinical isolate H/PF/2013 (GenBank Accession: KJ776791) was obtained from the European Virus Archive (EVAg). ZIKV Brazilian clinical isolate Paraiba01/2015 (GenBank Accession: KX280026) was a gift from the European Chagas Institute. All viruses were grown in C6/36 cells, and the supernatants were stored in −80 °C after filtration through 0.2 μm membrane. Virus titer was determined by standard BHK-21 plaque assay.
Ethics statement
Institutional approval has been granted by Duke-NUS Medical School to perform experiments with ZIKV strain. All animal experiments (protocol 2016/SHS/1197 and 2021/SHS/1651) were approved by the Institutional Animal Care and Use Committee at Singapore Health Services and conformed to the National Institutes of Health (NIH) guidelines and public law.
Cloning and generation of ZIKV recombinant viruses
Viral RNA extracted from the C6/36 expanded H/PF/2013 virus stock was used to synthesize cDNA using Improm II reverse transcription system (Promega). Conventional cloning techniques of assembling the ZIKV PCR fragments into a single full-length cDNA infectious clone was not successful due to the presence of cryptic E. coli promoters (CEPs) in the structural/NS1 genes in the viral genome that induced plasmid instability. In this respect, the cDNA was used as a template to generate fragments 2, 3 and 4 by PCR using primers described in Table S1. A HDVR sequence was engineered at the end of the viral 3′UTR in fragment 4 by overlapping PCR for generation of authentic 3′ end of the RNA transcript. A modified plasmid pACYC177 was used to clone the individual fragments 2, 3, 4 and 2 + 3 + 4. Fragment 1 containing synonymous nucleotide changes to silence the putative CEPs (Table S2) was synthesized by GenScript with a T7 promoter sequence at the 5′UTR for in vitro transcription, and was cloned into the plasmid pACYC177 containing fragments 2 + 3 + 4 to obtain the full-length H/PF/2013 wild-type (WT) cDNA clone. The full-length ZIKV cDNA plasmid was further propagated in E. coli cells for five passages and the genetic stability was assessed by Sanger sequencing. Additionally, several variants in NS1, NS2A and NS4B were reverse-engineered into the WT cDNA infectious clone by site-directed mutagenesis using the following primers:
NS1:M264V_F: 5′-GAGGGCTACAGGACCCAAGTGAAAGGGCCATGGCACAG-3′
NS1:M264V_R: 5′- CTGTGCCATGGCCCTTTCACTTGGGTCCTGTAGCCCTC-3′
NS2A:A117V_F: 5′-CGTGTCTTTTGCAAACTGTGATCTCCGCCTTGGAAGG-3′
NS2A:A117V_R: 5′-CCTTCCAAGGCGGAGATCACAGTTTGCAAAAGACACG-3′
NS4B:I26M_F: 5′-GAGAGGAGGGGGCAACCATGGGATTCTCAATGGACATTG-3′
NS4B:I26M_R: 5′-CAATGTCCATTGAGAATCCCATGGTTGCCCCCTCCTCTC-3′
The underlined nucleotide corresponds to the indicated mutations.
RNA was in vitro transcribed from the full-length cDNA clone using T7 mMESSAGE mMACHINE kit (Ambion) and transfected into C6/36 cells as previously described.15,16 Supernatants from the transfected C6/36 cells were collected on day 7 post-transfection (P0 virus) and passaged once in C6/36 to obtain the P1 virus stock (rZIKV-FP WT and mutant viruses) for in vitro and/or in vivo infection studies.
Virus Next-Generation Sequencing (NGS)
Viral genomes extracted from culture supernatants or infected mouse sera were directly used to construct NGS libraries using the NEBNext Ultra II directional RNA Library Prep kit for Illumina (New England Biolabs). Additional PCR amplification of the viral genome extracted from infected mouse brains or testis was performed prior to library construction for viral enrichment. The primers used for amplification of the viral fragments are described previously.17 The five different amplified fragments (∼2kb/fragment) spanning the whole viral genome were pooled together in equal amounts and subjected to Covaris Shearing to obtain fragment size of ∼300bp for library construction. The quality of the libraries were assessed by DNA1000 Bioanalyzer chip (Agilent Technologies) prior to paired-end sequencing on the Illumina MiSeq (2 × 250bp) sequencing system in Duke-NUS Genome Biology Facility. Reads were de-multiplexed and full viral genomes were assembled as previously described.18 The typical MiSeq coverage for each nucleotide position ranges from 6000 to 8000 reads/nucleotide and samples with an overall read depths less than 3000 were excluded from variant call analysis. The sample replicates were pooled and aligned to the same H/PF/2013 reference sequence (GenBank accession: KJ776791) for variant calling rather than averaging the individual replicates as previously described.12 Variant allele frequency was analyzed by lofreq software as previously described.19 The Simpson's diversity index or Shannon entropy are computed for all single nucleotide variants (SNVs) with no threshold cutoff on the allele frequencies.
Virus plaque purification
Confluent monolayer of BHK-21 cells was infected with 10-fold serially diluted H/PF/2013 clinical isolate virus inoculums for 1 h. Infected cells were then overlaid with 2% low melting agarose in growth medium and incubated at 37 °C for 4 days. Single plaques were isolated and grown in C6/36 cells for two rounds. Virus clones obtained were subjected to Sanger sequencing to segregate the clones based on prM and NS2A SNVs for in vitro and in vivo infection studies.
In vitro virus replication assay
1 × 105 LN-229 cells were infected with H/PF/2013 clinical isolate, rZIKV-FP or plaque purified virus clones at MOI 1. After 1 h incubation at 37 °C, virus inoculums were removed and replaced with growth medium. Supernatants were collected at the indicated timepoints for plaque and extracellular viral RNA quantification, and cells were washed once with PBS prior to lysing with TRizol (Invitrogen) for intracellular viral RNA quantification.
Immunofluorescence assay
Cells on coverslips were fixed with ice-cold methanol at −20 °C for 15 min. Antibody against dsRNA (J2, Scicons: 10010200) was used for detection of infected cells. Digitized images were captured using Zeiss LSM710 upright confocal microscopy (Carl Zeiss, Germany) at 64× magnification. Image processing was done with ImageJ software.20
A129 mouse infection studies
Sv/129 mice deficient of Type I IFN receptor (A129) purchased from B&K Universal (UK), were housed in BSL-2 animal facility in Duke-NUS, Singapore and all animal experiments were carried out as outlined in an approved protocol (see Ethics Statement). Male mice were used in this study to examine the ZIKV-induced pathology on testis. The mice were mixed and randomly distributed into five mice per experimental group for survival studies as previously described in16 or otherwise stated. Eight to eleven-week old male mice were inoculated intravenously (i.v.) with the indicated infection dose of H/PF/2013 clinical isolate, rZIKV-FP (WT or mutants) or representative plaque-purified virus clones and mice survival was monitored over a course of 15 days or otherwise stated. Blood samples were collected on days 1–4, day 6 and day 8 post-infection by submandidular bleeding and serum samples were subjected to viremia measurement by real-time RT-PCR.
To examine the pathology induced by different plaque purified H/PF/2013 virus clones, three mice per group (uninfected, parental H/PF/2013 isolate, #2, #12, #23, #11 and #20) were sacrificed on day 6 post-infection by CO2 inhalation and perfused with 10 ml PBS after blood collection (∼500–700 μl) from the postcaval vein. Serum samples were subjected to measurements of viral load by real-time RT-PCR and cytokines using Ready-SET-Go! ELISA kits (IL6 and TNFα; eBioscience). Tissues were collected and snap-frozen in liquid nitrogen after removal of luminal content from the intestine. These tissues were homogenized in PBS and the homogenates were subjected to viral load analysis by real-time RT-PCR. The brain homogenates were subjected to transcriptional analysis of inflammatory genes expression by NanoString®, pro-inflammatory cytokines IL6 and TNFα ELISA and Next-Generation Sequencing for variants analysis of the viral proteome.
Inflammatory gene profiling using NanoString technologies
Total RNA was isolated from the brain homogenates using RNeasy Micro Kit (Qiagen) following manufacturer's instructions. The RNA concentration was determined by Quant-iT RiboGreen RNA assay kit (Life Technologies). 100 ng of total RNA was subjected to inflammatory genes profiling using a customized nCounter® Mouse Inflammation v2 codeset (NanoString Technologies) according to manufacturer's instructions. The add-on genes in the nCounter® Mouse Inflammation v2 codeset can be found in Table S3. The gene expression profile obtained from nCounter® was analyzed using nSolver 3.0 and presented as heatmap.
RNA extraction and real-time RT-PCR
Total RNA was isolated from cell lysates or tissue homogenates by TRizol (Invitrogen) extraction method. Viral RNA from mouse serum was extracted using QIAamp viral RNA Mini kit (Qiagen) according to manufacturer's instructions. Real-time RT-PCR was carried out in Bio-Rad Real-time thermal cycler CFX96.
For detection of viral RNA from cellular infections, cDNA was first generated using Improm II reverse transcription system (Promega) according to manufacturer's instructions prior to real-time RT-PCR using SYBR green supermix (Bio-Rad). PCR products of ZIKV E gene and housekeeping human actin were used to generate the respective standard curves for absolute quantification of genome copies using the Ct values obtained. The values of intracellular viral genome are normalized to the absolute copies of actin gene and reported as actin-normalized viral RNA genome copies per microgram total RNA as previously described.15
For quantification of viral load in infected mouse serum or tissues, iTaq Universal SYBR green one-step kit (Bio-Rad) was used. The viral load in the serum was reported as absolute genome copies per ml of serum computed based on the standard curve generated from the in vitro transcribed RNA product of ZIKV E gene. The tissue viral load was quantitated using the comparative Ct method as described elsewhere21 and reported as relative viral RNA expression per microgram total RNA. Real-time RT-PCR primers for detection of viral genome were described elsewhere.22
Statistical analysis
Statistical analysis was performed using GraphPad Prism v5.0 software. Comparison of mouse survival rates was performed using the log-rank Mantel–Cox test. Comparison of in vitro infectious virus production from LN-229 cells infection between parental and recombinant viruses was performed using non-parametric Mann–Whitney's test. Significant difference in the viremia kinetics between the parental H/PF/2013 and rZIKV-FP infection groups were compared by 2-way ANOVA with Bonferroni's correction. Significance in the infectious virus production from LN-229 cells infection, expression of immune genes measured by real-time PCR and ELISA measurements between the different plaque purified virus clones groups were determined by a non-parametric Kruskal–Wallis test. P < 0.05 was considered significant. All data were obtained from duplicates of two independent experiments or otherwise stated.
Role of the funders
This study was supported by Duke-NUS Khoo Post-doctoral Fellowship Award 2020 (KWKC) and National Medical Research Council of Singapore grants MOH-000524 (OFIRG) (SW) and MOH-OFIRG20nov-0002 (SGV). The funding authorities did not have any role in study design, data collection, data analysis, interpretation or the preparation of this manuscript.
Results
High genome diversity of ZIKV H/PF/2013 correlates with its lethal phenotype in mice
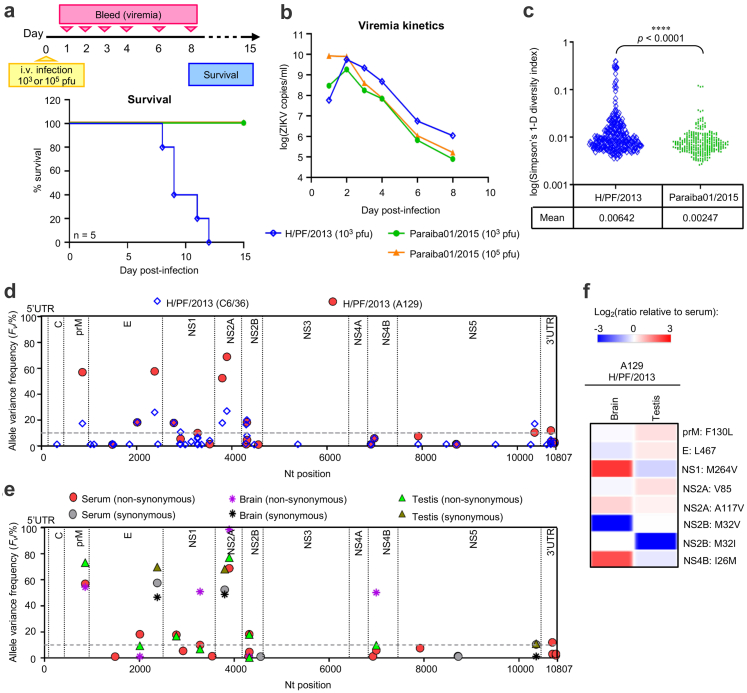
We first examined the disease phenotype of two Asian lineage ZIKV strains- French Polynesian H/PF/2013 (GenBank accession: KJ77679123;) and Brazilian Paraiba01/2015 (GenBank accession: KX280026;14) in A129 mice by scoring for survival over 15 days and systemic viremia (schematics of Fig. 1a). An inoculation of 103 pfu (plaque forming units) (∼7 × 107 genome equivalent (GE)) of the viruses resulted in 100% mortality of the H/PF/2013 infected mice starting from day 8 post-infection while all Paraiba01/2015 infected mice survived (Fig. 1a). Interestingly, even a 100-fold increase in the inoculation dose of Paraiba01/2015 to 105 pfu did not cause lethality in mice despite a comparable day 2 peak viremia to that of the 103 pfu H/PF/2013-infected mice (Fig. 1a and b). Whole genome and proteome sequence alignment of the representative ZIKV strains from the African (MR766; GenBank accession: LC002520) and Asian (pre-epidemic FSS13025; GenBank accession: KU955593, epidemic strains H/PF/2013 and Paraiba01/2015) lineages showed high sequence identity amongst the strains (>89% and >95% identity in genome and proteome respectively), with the African MR766 that is distantly separated from the three Asian strains (Figure S1a). Notably the two Asian strains H/PF/2013 and Paraiba01/2015 which exhibited different in vivo disease phenotype in our mouse model (Fig. 1a) had >99% identity in both its consensus genome and proteome sequences (Figure S1a). In perspective, increased genetic diversity of ZIKV strains has been proposed to be implicated in viral fitness and virulence.24,25 To this end, we next examined the genome diversity of the two Asian ZIKV strains amplified from C6/36 using Next-Generation Sequencing paired with single nucleotide variant (SNV) analysis. A total of 46 SNVs with variant frequency (Fv) > 1% were found in H/PF/2013 (Table S4) compared to 11 SNVs in Paraiba01/2015 (Table S5) (Figure S1b), which results in a corresponding 3-fold higher Simpson's diversity index for the former (Fig. 1c). Together these data suggest that a higher genetic diversity within a viral population in H/PF/2013 may be associated with the lethal disease phenotype.
Fig. 1.
Comparison of in vivo A129 mouse infection phenotype and viral diversity of H/PF/2013 and Paraiba01/2015 ZIKV clinical strains. (a and b) A129 mice (n = 5/group) were i.v. inoculated with either 1000 pfu (∼7 × 107 GE) or 105 pfu of H/PF/2013 and/or Paraiba01/2015 ZIKV clinical strains for survival and viremia kinetics analyses over a period of 15 days. The experimental timeline is shown in (a). (a) Mouse survival (log rank Mantel–Cox test) of A129 infected with H/PF/2013 or Paraiba01/2015 over 15 days at the indicated inoculation dose. (b) Average viremia kinetics measured by real-time RT PCR on the pooled serum collected from the infected mice. (c) Viral RNA extracted from the low passaged C6/36 amplified virus stocks of H/PF/2013 (passage 6, P6) and Paraiba01/2015 (passage 6, P6) were subjected to NGS paired to SNVs analysis. Diversity of all SNVs found in the infecting inoculum of the virus stocks as measured by Simpson's diversity index. The mean diversity of the two viruses is indicated and the difference was compared by Student's t-test ∗∗∗∗P < 0.0001. (d) Viral RNA was extracted from virus-infected C6/36 supernatants (two different batches of H/PF/2013 clinical isolate) or day 2H/PF/2013 infected mice sera from (b) (n = 5) and subjected to paired-end 2 × 250bp MiSeq (Illumina). SNVs analysis was performed by lofreq software to determine the variant frequency (Fv). Shown is a scatter plot of all SNVs with Fv > 1% in H/PF/2013 virus from C6/36 expansion (blue-open diamond symbol) or infected A129 mice sera (red-filled circle symbol) plotted along the viral genome. The grey dotted line represents the threshold of Fv > 10%. (e and f) Brain or testis homogenates (n = 3) of the H/PF/2013 clinical isolated infected mice from (b) were subjected to paired-end 2 × 250bp MiSeq (Illumina). The nucleotide variants and its associated allele frequencies were analyzed using the lofreq software. (e) SNVs of the H/PF/2013 clinical isolate from the infected mice brains (purple and black asterisk symbols) or testis (green and brown-filled triangle symbols) plotted along the viral genome. Shown were the infected brain- or testis-associated SNVs that is also found in the H/PF/2013 clinical isolate infected sera as in (d) (red- and grey-filled circle symbols; included in the same scatter plot for reference). (f) Heatmap showing the fold change of specific variants in the brains or testis of H/PF/2013 infected mice relative to the infected mice sera. Data in (d) or (e and f) are presented as mean value obtained from five independent mice sera or three independent mice brain/testis samples respectively.
Variant frequency of H/PF/2013 isolate in the serum, brain and testis of mice shows tissue-specific enrichment of critical virulent determinants
To explore the basis of the virulence observed for the clinical isolate H/PF/2013 in mice, we performed Next-Generation Sequencing on the viral genome extracted from the infected mouse sera on day 2 post-infection (day of peak viremia) and compared the SNV profile with the C6/36 amplified virus infection inoculum. Of the 46 SNVs (Fv > 1%) found in the H/PF/2013 virus used in infection, there were ten SNVs with Fv > 10% (three synonymous and seven non-synonymous changes; Fig. 1d and inset table of Figure S1b). Interestingly a ∼3-fold enrichment was observed in four of the ten SNVs present in the initial infection inoculum; particularly two were non-synonymous changes in the viral genes prM (F130L) and NS2A (A117V) (Table 1) that have been implicated in ZIKV virulence and neuropathogenesis.10,26
Table 1.
Summary of variant frequencies (Fv) of the various SNVs found in H/PF/2013 infected mice sera, brain and testis.
| Viral protein | aa variant | Variant frequency (Fv/%) |
Testis (A129)c | ||
|---|---|---|---|---|---|
| Virus stock (C6/36)a | Serum (A129)b | Brain (A129)c | |||
| prM | F130L | 16.7 (1.0) | 56.0 (2.5) | 54.4 (19.2) | 73.2 (4.5) |
| E | L467d | 25.5 (0.6) | 56.8 (3.7) | 46.6 (22.1) | 69.8 (2.9) |
| NS1 | M264V | 6.0 (0.2) | 12.1 (3.0) | 50.8 (12.0) | 6.9 (3.4) |
| NS2A | V85d | 17.5 (0.5) | 52.5 (3.1) | 48.8 (19.3) | 68.2 (2.1) |
| A117V | 27.0 (0.1) | 70.2 (3.9) | 98.6 (1.0) | 77.1 (1.9) | |
| NS2B | M32V | 19.6 (0.6) | 18.2 (1.3) | 1.0 (−) | 18.1 (0.9) |
| M32I | 8.1 (0.2) | 4.9 (1.5) | – | 3.2 (−) | |
| NS4B | I26M | 5.6 (0.3) | 14.5 (1.9) | 50.2 (10.4) | 9.8 (4.2) |
SNVs presented in this table is tabulated from Fig. 1e. The Fv of these SNVs in the original C6/36 amplified virus stock is included for reference.
Abbreviations: aa; amino acid.
The indicated Fv in each sample condition is tabulated from variant call analysis performed on the merged replicates alignment files instead of averaging the individual frequencies. Values in parentheses are the standard deviations computed from individual frequencies.
n = 2 independent virus stocks.
n = 5 independent mice sera.
n = 3 independent mouse brains or testis.
Synonymous change.
Brain and testis are the key target organs implicated in ZIKV pathogenesis,27 we therefore interrogated the SNV profiles of the viral RNA extracted from the infected mouse brains and testes with the infected sera profile with focus on the prM and NS2A variants. Of the viral variants found in the H/PF/2013 infected sera (n = 24, Fig. 1d), nine SNVs (three synonymous and six non-synonymous changes) were present in the infected brains (Fig. 1e, Table 1). Notably the NS2A V117 variant that was expanded in the serum was found to be further enriched by 1.4-fold (P = 0.033) in the brains of the infected mice, with a Fv > 98% (Fig. 1e and f) while the prM:F130L variant was not further enhanced in the brains compared to the infected sera (Fig. 1d) (sera: Fv = 57% vs brain: Fv = 54%) (Fig. 1f, Table 1). In contrast, there were 11 common SNVs between the infected testes and sera with comparable Fv, and the prM:F130L (1.3-fold) and NS2A:A117V (1.1-fold) SNVs were not significantly expanded in the testis compared to the serum (Fig. 1f, Table 1), suggesting a selective enrichment of the NS2A variant in the brain.
Apart from the NS2A:V117 variant, we also found two other additional variants NS1:M264V and NS4B:I26M (the Fv of these two variants were 5–6% in the C6/36 amplified H/PF/2013 virus stock (Table 1)) that were significantly expanded in the brain (5.1-fold, P = 0.046 for NS1 variant and 4.2-fold, P = 0.042 for NS4B variant; Fig. 1f, Table 1) compared to its ∼10% prevalence level in the infected serum. Interestingly the enrichment appears to be brain-specific since deep-sequencing of the virus from the infected testis showed that these changes were negatively selected relative to the serum (Fig. 1f). Notably, the Fv for these two variants were of similar levels to each other in the infecting inoculum or the various tissues examined (Table 1), suggesting that the NS1 and NS4B variants may be found together in the virus sequence. Hence these findings can be interpretated that the NS1 and NS4B variants may act in concert to mediate ZIKV brain penetrance/neurotropism. Together our results indicate a tissue specific enrichment of variants that potentially drives ZIKV pathogenesis.
Plaque-purification of virus clones from H/PF/2013 clinical isolate
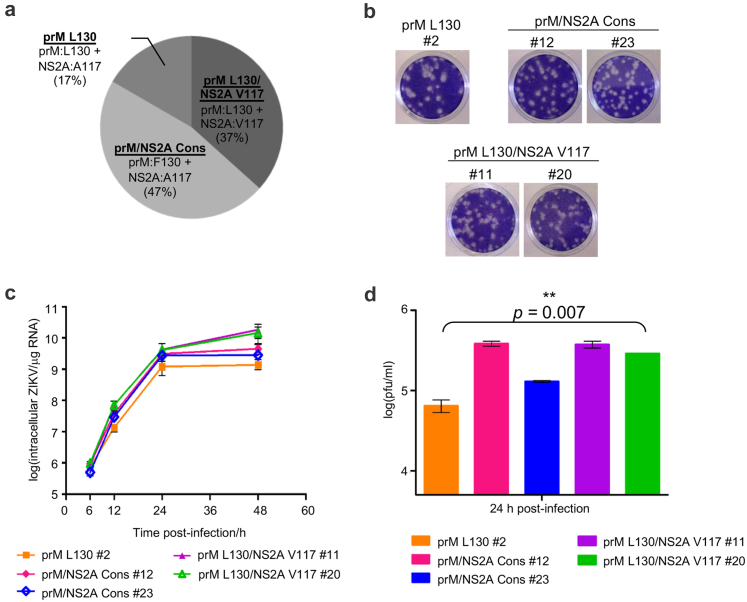
Since deep-sequencing of the virus isolated from the H/PF/2013-infected mice revealed tissue-specific enrichment of viral variants (Fig. 1f), we sought to pinpoint specific and/or epistatic SNV(s) that contribute to ZIKV virulence in our mouse model by isolating independent virus populations from the H/PF/2013 clinical isolate. A total of 30 plaques were isolated, expanded in C6/36 and Sanger-sequenced at the specific positions of the enriched SNVs (prM, NS1, NS2A and NS4B) found in the infected mice. No virus clones harbouring the NS1:M264V and/or NS4B:I26M variants were isolated, likely due to its low variant frequency (5–6%) in the virus stock. On the other hand all isolated clones contain SNVs in prM and/or NS2A genes, and these were segregated into three groups based on the identity of the amino acid residues at positions 130 in prM and 117 in NS2A: prM L130 group (5 clones) carrying the prM variant with NS2A:A117; prM/NS2A Consensus (Cons) group (14 clones) carrying prM:F130 and NS2A:A117; prM L130/NS2A V117 group (11 clones) carrying both variants in prM and NS2A (Fig. 2a). We selected five representative clones - clone #2 from prM L130 group, clones #12 and #23 from the prM/NS2A Consensus group, clones #11 and #20 from prM L130/NS2A V117 group– to examine the in vitro replication characteristics. There were no observable differences in plaque size and morphology by standard plaque assay in BHK-21 cells between clones from the three groups (Fig. 2b). However both the prM/NS2A Consensus and prM L130/NS2A V117 groups exhibited a >½ log10 higher intracellular viral RNA replication (Fig. 2c), with a corresponding 2–6 fold higher production of infectious virus (Fig. 2d) compared to the prM L130 clone #2 in LN-229 cells.
Fig. 2.
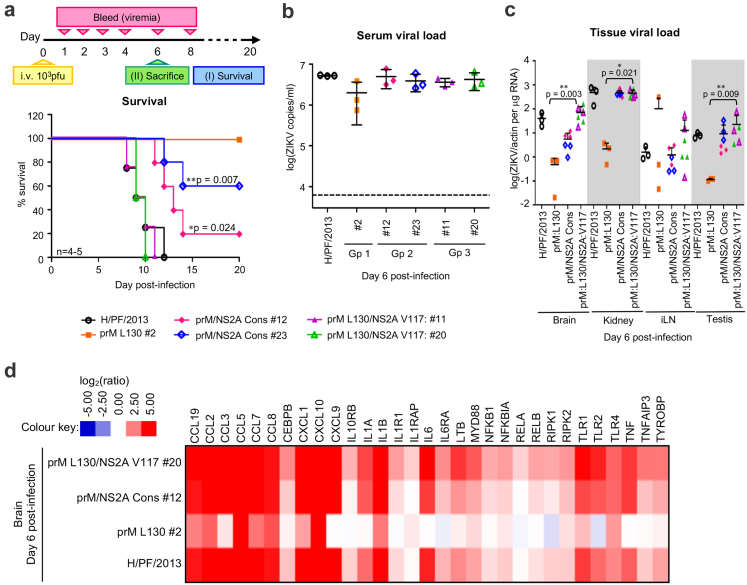
In vitro characterization of the plaque-purified H/PF/2013 clinical isolate virus clones in LN-229 cells. H/PF/2013 clinical isolate was subjected to plaque purification on BHK-21 cells and virus plaques were recovered in C6/36 cells. 30 virus plaques were isolated and grouped based on the amino acid residues 130 in prM and 117 in NS2A as determined by Sanger sequencing. (a) Chart distribution of the 30 plaque-purified H/PF/2013 virus clones according to the specific prM and NS2A amino acid residues (prM L130 group-prM:L130 + NS2A:A117; prM/NS2A Consensus (Cons) group-prM:F130 + NS2A:A117; prM L130/NS2A V117 group-prM:L130 + NS2A:V117). (b) Plaque morphologies of the representative clones from the three virus groups in (a). One representative clone from prM L130 (#2) and two representative clones from prM/NS2A Consensus group (#12 and #23) and prM L130/NS2A V117 group (#11 and #20). (c and d) LN-229 cells were infected with the representative virus clones from (b) at MOI 1 for replication kinetics analyses. (c) Real-time PCR quantification of the representative virus clones genome replication. The viral genome expression is reported as actin-normalized absolute viral RNA genome copies per microgram total RNA. (d) Infectious virus titers of the infected supernatants measured by standard plaque assay. Data are presented as mean ± SD from two independent experiments and the difference in the mean infectious virus production of all the five representative plaque-purified virus clones were compared by Kruskal–Wallis test, ∗∗P < 0.01.
Gene polymorphisms in NS2A and NS2B increased in vivo virulence of H/PF/2013 by inducing high inflammatory response in the brain of mice
We next examined the disease pathogenesis of the independent virus populations with the selected representative plaque-purified clones from Fig. 2b in A129 mice infections by monitoring their survival over 20 days and serum viral load (viremia) kinetics (schematic in Fig. 3a). An inoculation dose of 103 pfu of prM L130/NS2A V117 virus clones resulted in 100% lethality between days 8–11 post-infection and high serum viremia kinetics (clones #11 and #20: median survival of 9.5 days), mirroring the parental H/PF/2013 clinical isolate infection group (Fig. 3a and Figure S2a). On the contrary mice infected with prM L130 clone #2 virus resulted in 100% survival with >1 log10 lower peak viremia on day 2 post-infection compared to prM L130/NS2A V117 virus clones or parental H/PF/2013 clinical isolate (Fig. 3a and Figure S2a). These survival data can be interpreted to suggest that NS2A:V117 but not prM:L130 variant may be responsible for the lethal phenotype of H/PF/2013 in our mouse model since the amino acid L at position 130 of prM protein is present in both plaque-purified groups. The lethality associated with NS2A:V117 is also supported by deep-sequencing data which showed a specific clonal expansion of NS2A:V117 allele with variant frequency close to 70% in A129 mice serum to almost consensus (>98%) in the brain, compared to its 27% occurrence level in the C6/36 derived inoculum (Fig. 1d and e). We further examined the viral load in various tissues (brain, liver, spleen, kidney, small intestine (S. intestine), iliac lymph nodes (iLN), mesentery lymph nodes (mLN) and testis) on day 6 post-infection (schematic in Fig. 3a). Consistent with the higher serum viral load of the prM L130/NS2A V117 virus clones compared to the non-lethal prM L130 clone #2 (Fig. 3b), mice from the former exhibited 1–2 log10 higher tissue viral loads in the brain (P = 0.003), testis (P = 0.007) and kidney (P = 0.021) (Fig. 3c).27 No significant difference was noted in the viral loads in the iLN, liver, spleen and S. intestine between the prM L130 and prM L130/NS2A V117 virus groups (Fig. 3c and Figure S2b). It is noteworthy that the higher viral load in specific tissues of the prM L130/NS2A V117 virus infected mice were similar to that observed for H/PF/2013 clinical isolate infected mice (Fig. 3c).
Fig. 3.
In vivo A129 mice infection with plaque-purified H/PF/2013 clinical isolate virus clones. A129 mouse infection with the representative plaque-purified H/PF/2013 virus clones (from Fig. 2b) using 103 pfu i.v. inoculation (n = 4–5 mice/group). Mouse survival was monitored for 20 days and blood was sampled at indicated timepoints for serum viral load analysis. Additionally, three mice were independently infected and sacrificed on day 6 post-infection for tissue viral load and pathological markers analysis. This experiment timeline is shown in (a). (a) Survival plots (log rank Mantel–Cox test) of A129 mice infected with H/PF/2013 clinical isolate or the representative virus clones. (b) Serum viral load of individual mouse from each infection group on day 6 post-infection measured by real-time PCR. (c) Viral load in mouse tissues (brain, kidney, iliac lymph nodes (iLN) and testis) measured by real-time PCR. The viral RNA expression is presented as actin-normalized relative expression per microgram total RNA. The overall limit of detection of the uninfected mouse tissues is 10−4 per microgram total RNA. The overall difference in the mean viral load of the respective tissues amongst the three plaque-purified groups were compared by Kruskal–Wallis test, ∗P < 0.05; ∗∗P < 0.01. (d) Heatmap of NFκB-mediated signaling pathway genes expression in the infected brain homogenates (average from n = 3 infected brain homogenates per virus groups) from (c) analyzed using nCounter® Mouse Inflammation v2 panel (NanoString Technologies). Gene counts are normalized to the panel's internal housekeeping genes and expressed as log2 (fold change) with respect to uninfected brain samples. Gene clusters of the heatmap is based on KEGG/GO Biological Processes analysis. Data points shown as scatter plots in (b) and (c) are presented as mean ± SD from three independent mice.
Since ZIKV-induced neuro-inflammation is correlated with severe neurological complications of Zika disease,28 we profiled the expression of inflammatory genes in the brain homogenates of the infected mice using the NanoString gene counting platform.29 Infection with H/PF/2013 clinical isolate resulted in an upregulation (log2 > 2.5) of NFκB-associated genes (chemokines (CCLs, CxCLs)/cytokines (IL1α, IL1β, IL6, TNF)) in the brain compared to mock infected (Fig. 3d). Notably infected brains from the representative prM L130/NS2A V117 clone #20 exhibited similarly enhanced expression profile for these NFκB-associated genes to that of the parental H/PF/2013, while the non-lethal prM L130 clone #2 showed lower level of expression of these genes (Fig. 3d). The upregulation of these genes in the prM L130/NS2A V117 virus group infected mice resulted in higher systemic- and neuro-inflammation as determined by the secretion levels of the pro-inflammatory cytokines TNFα and IL6, than prM L130 virus group infected mice (Figure S2c–f). Taken together these results suggest that clonal expansion of NS2A:V117 mediates higher infectivity and pronounced inflammation in the brain compared to NS2A:A117 virus, that correlated with the observed in vivo ZIKV virulence.
Additionally we found that mice infected with the prM/NS2A Consensus group virus clones carrying the consensus amino acids (prM:F130 + NS2A:A117) showed delayed mortality (clone #12: median survival of 13 days, 80% lethality; clone #23: 40% lethality) compared to the parental H/PF/2013 (median survival of 9.5 days, 100% lethality, P = 0.042 compared to #12, P = 0.007 compared to #23) (Fig. 3a). Similar to the prM L130/NS2A V117 viruses, these prM/NS2A Consensus group infected mice exhibited higher peak viremia (Figure S2a), brain and testis viral load (Fig. 3c) than the non-lethal prM L130 virus clone, coupled with enhanced levels of proinflammatory response (Fig. 3d, Figure S2c–f). To identify potential variants accounting for the virulent phenotype of prM/NS2A Consensus group viruses, we deep-sequenced both the non-lethal virus clone prM L130 clone #2 and representative prM/NS2A Consensus clone #12 for SNVs comparative analysis. Interestingly a non-synonymous variant in NS2B (G4319→A, M32I) appeared only in clone #12 (Table 2). It is noteworthy that the NS2B position 32 of the parental H/PF/2013 isolate contains several genetic variations (8.1% G4319→A, M32I; 19.6% A4317→G, M32V; Table S4). Interestingly the prM/NS2A Consensus group clone #23 examined in the infection phenotype studies contains the other NS2B variant M32V that is different from #12. Hence the differences observed in the infectious virus production (Fig. 2d) and the mice survivability (Fig. 3a) from infection of the two representative virus clones (#12 and #23) within the prM/NS2A Consensus group could be attributed to the genetic variation at position 32 of NS2B. Collectively these findings suggest that NS2B polymorphism may be another virulent determinant within the virus population.
Table 2.
Consensus variantsa changes present in the plaque-purified Group 1 clone #2 and Group 2 clone #12.
| Gene | Consensus nt | Variant nt | Amino acid | Variant frequency (%) |
|
|---|---|---|---|---|---|
| Group 1 #2 | Group 2 #12 | ||||
| prM | T861 | C | F130L | 99.4 | – |
| E | C2378 | T | L467 | 99.2 | – |
| NS1 | T2923 | C | L145P | 99.8 | 99.4 |
| NS2B | G4319 | A | M32I | – | 99.6 |
| NS5 | A9107 | G | L480 | – | 99.4 |
Consensus variants are defined as nucleotide changes with frequencies >90%.
NS2A:A117V mutation is critical in inducing in vivo virulence of the non-lethal recombinant isogenic ZIKV H/PF/2013 virus in mice
So far our results infer clonal expansion of specific viral variants in the viral population (Fig. 1) and natural variants in NS2A or NS2B isolated from the H/PF/2013 clinical virus (Fig. 3) could contribute to its in vivo virulence. To explore this further, a full-length ZIKV H/PF/2013 cDNA infectious clone (rZIKV-FP) was constructed by mutational silencing of 15 predicted CEPs within the prM-E-NS1 gene region of the viral genome without introducing any amino acids changes in order to overcome stability issues of cDNA containing plasmids in E. coli (Fig. 4a and Table S2).30, 31, 32 The stability of the resultant rZIKV-FP virus obtained from transfection of the in vitro transcribed RNA from the cDNA clone into C6/36 cells followed by expansion was evaluated by viral serial passaging coupled to NGS. Deep-sequencing of the passage 1 (P1) rZIKV-FP virus showed low genetic diversity with SNVs having Fv < 5% except for the 15 synonymous changes introduced for silencing CEPs during cDNA clone construction (Fig. 4b and c, see Table S6 for full account of SNVs found in the rZIKV-FP). Serial passaging of the P1 rZIKV-FP thrice in C6/36 cells (passage 4, P4) did not further increase the viral diversity as revealed by the similar Fv (Table S6) and Simpson's diversity index (Fig. 4c) of all SNVs in the P1 and P4 viruses, implying genetic stability of rZIKV-FP. Despite of the silent mutations introduced for genetic stability of the cDNA clone, the isogenic rZIKV-FP virus replicates similarly as the parental H/PF/2013 clinical isolate in LN-229 human glioblastoma cells as demonstrated by similar levels of intracellular viral RNA replication (Fig. 4d) and 40–50% cellular infectivity at 24 h post-infection (Fig. 4e). Strikingly mice infected with the isogenic rZIKV-FP virus survived through the infection course that is in contrast to the highly lethal parental clinical isolate (Fig. 4f). Notably the difference in survival between H/PF/2013 and isogenic rZIKV-FP infected mice was coupled with a 10–60 fold higher peak serum viral load for the former compared to the isogenic infectious clone derived virus (P < 0.0001; Fig. 4g). This survivability of the isogenic rZIKV-FP infected mice is in agreement with the correlation of low viral diversity with viral virulence (Figs. 1a–c and 4c).
Fig. 4.
Genome diversity and phenotypic characterization of the ZIKV-FP infectious clone (rZIKV-FP) virus. (a) Schematic of constructing a stable ZIKV-FP (GenBank accession: KJ776791) infectious clone. The genetic stability of the cDNA infectious clone plasmids during prokaryotic amplification is indicated. (i) The original ZIKV-FP genome is cloned as four fragments flank with a T7 promoter and a hepatitis D virus ribozyme (HDVR) sequences at the 5′ and 3′ ends of the viral genome respectively into a modified low copy plasmid vector pACYC177 to obtain the full-length ZIKV cDNA infectious clone. (ii) Stable full-length ZIKV-FP cDNA infectious clone with the putative CEPs in the structural and NS1 genes silenced in fragment 1. (b) Allele frequency of SNVs >1% identified in passage 1 of rZIKV-FP expanded in C6/36 cells plotted along the viral genome. Red triangles represent the nucleotide variants introduced into the rZIKV-FP viral genome to silence the putative CEPs shown in Table S4 (n = 15). Orange triangles represent the inherent SNVs in rZIKV-FP expanded in C6/36 cells (n = 28). SNVs with Fv > 1% in H/PF/2013 clinical isolate from C6/36 expansion (blue-open diamond symbols) as in Fig. 2a were included for comparison. Grey dotted line represent the threshold of Fv > 10%. (c) Diversity of SNVs excluding the introduced variants in passage 1 and 4 of rZIKV-FP as measured by Simpson's diversity index. The mean diversity for the respective passages is indicated. (d–g) rZIKV-FP virus was generated by transfecting the in vitro transcribed (IVT) infectious clone RNA into C6/36 cells for in vitro and in vivo infection studies. (d) Real-time PCR quantification of H/PF/2013 clinical isolate and rZIKV-FP virus genome replication at MOI 1 in infected LN-229 cells. The viral genome expression is reported as actin-normalized absolute viral RNA genome copies per microgram total RNA. Grey dotted line represents the detection level of uninfected LN-229 cells. (e) Confocal images of infected LN-229 cells as in (d) showing dsRNA staining taken at 24 h post-infection using 63× oil immersion lens. The percentage infectivity of the viruses observed at 10× magnification is indicated. (f) A129 mice (n = 5–6/group) were i.v. inoculated with either 50 pfu (∼3.5 × 105 GE) or 1000 pfu (∼7 × 106 GE) of H/PF/2013 clinical isolate or rZIKV-FP virus. Survival plots (log rank Mantel–Cox test) of A129 mice infected with H/PF/2013 clinical isolate or rZIKV-FP. (g) Average viremia kinetics measured by real-time RT-PCR in pooled serum collected from the infected mice as in (f) and the differences between the parental H/PF/2013 and rZIKV-FP infection groups were compared by 2-way ANOVA with Bonferroni's correction. Data are presented as mean ± SD from two independent experiments and significance is indicated (∗P < 0.05; ∗∗∗∗P < 0.0001).
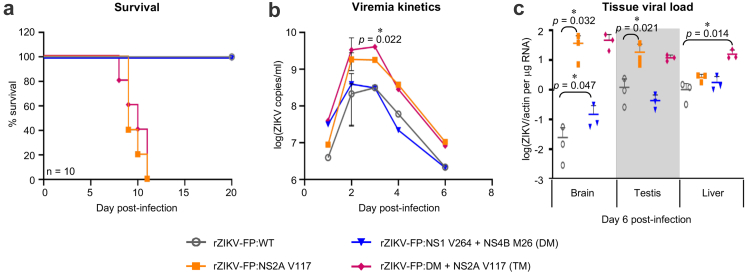
We next reverse-engineered the non-synonymous SNVs that were enriched in the infected mice brain (Fig. 1f and Table 1) into the non-lethal rZIKV-FP cDNA clone to examine the impact of these mutations on ZIKV neuropathogenesis. The NS2B variant at position 32 identified from plaque-purified virus clones (Consensus group virus clones, Fig. 3) was not introduced into the rZIKV-FP cDNA since it was found to be negatively selected (M32V: sera Fv = 18% vs brain Fv = 1%; M32I: sera Fv = 4.6% vs brain Fv = 0%, Table 1) during in vivo infection (Fig. 1f). Analogous to the in vivo disease phenotype of the plaque-purified virus clones (Fig. 3), mice infected with rZIKV-FP mutant virus containing the single NS2A:V117 mutation (SM) demonstrated 100% lethality by day 11 post-infection (Fig. 5a), that is accompanied by significantly higher viremia (1 log10; P = 0.022) and viral load in the brain (3 log10; P = 0.032) and testis (1 log10; P = 0.021) than wild-type (WT) non-lethal rZIKV-FP carrying NS2A:A117 (Fig. 5b and c). No difference is observed in the viral load of the liver between WT and NS2A:A117V mutant virus (Fig. 5c). Interestingly rZIKV-FP double mutant virus (DM) with both NS1:V264 and NS4B:M26 did not cause mortality to mice (Fig. 5a). It is noteworthy that the DM virus induced a ∼6-fold higher viral load in the brain than the WT virus (P = 0.047; Fig. 5c), suggesting a potential epistatic role of these NS1 and NS4B variants in ZIKV brain penetrance that is indeed supported by the brain-specific clonal expansion of these two variants in the parental H/PF/2013 clinical virus infection (Fig. 1f). Nevertheless the lethality of the DM virus was restored upon introduction of the NS2A:V117 mutation, forming a triple mutant (TM) virus that induced comparably high serum and brain viral load as the single NS2A mutant virus (Fig. 5). Surprisingly the TM virus also appears to induce a significantly increased viral load in the liver that is not observed in DM virus (>5-fold increase, P = 0.014; Fig. 5c), suggesting that NS2A:V117 may act epistatically with NS1 and NS4B mutations to drive a different pathological profile apart from neurovirulence. Nonetheless with the NGS data of the infected mouse brains from Fig. 1 that showed the NS2A:V117 variant becoming the consensus residue (Fv > 98%) of the viral proteome, our data can be interpreted that the NS2A SNV is the key determinant for the observed severe neuropathology of the parental H/PF/2013 clinical isolate in our mouse model.
Fig. 5.
In vivo disease phenotype of A129 mice infected with WT and mutants of rZIKV-FP viruses. A129 mice were i.v. inoculated with 1000 pfu of the WT and respective mutants (single NS2A:A117V mutant; double mutant containing NS1:M264V and NS4B:I26M, DM; and triple mutant containing DM and NS2A:A117V, TM) of the rZIKV-FP viruses (n = 10 mice/group). Mouse survival was monitored for 20 days and blood was sampled at indicated timepoints for serum viral load analysis. Three additional mice were independently infected and sacrificed on day 6 post-infection for tissue sampling of viral load. (a) Survival plots (log rank Mantel–Cox test) of the different mouse groups infected with WT or different mutants of rZIKV-FP viruses. (b) Average viremia kinetics measured from pooled serum of the infected mice by real-time PCR. The difference between the viremia kinetics of the various infection groups were determined by 2-way ANOVA with Bonferroni's correction and significance is indicated (∗∗∗∗P < 0.001). (c) Viral load of mouse tissues (brain, testis and liver) measured by real-time PCR. The viral RNA expression is presented as actin-normalized relative expression per microgram total RNA. The overall limit of detection of the uninfected mouse tissues is 10−4 per microgram total RNA. Data points shown as scatter plots are presented as mean ± SD from three independent mice and the difference between the two indicated groups were compared by non-parametric Mann–Whitney's test ∗P < 0.05; ∗∗P < 0.01.
Discussion
Quasispecies of viruses are thought to be critical in ensuring the genetic robustness of the viral population to establish efficient infection and transmission.33,34 In this respect, viral variants have been reported in several naturally circulating ZIKV strains and studies have demonstrated the role of genetic diversity in maintaining the replicative fitness of the virus in different cells13,14 that could in turn impact on the disease pathogenesis. However as the contribution of genetic diversity in ZIKV pathogenesis and disease severity remains poorly defined, we adopted a systematic approach of combining Next-Generation Sequencing, virus plaque purification and reverse genetics to unravel and validate potential virulence markers within the virus population pool of the ZIKV H/PF/2013 virus isolate in comparison to the isogenic rZIKV-FP derived from infectious cDNA clone. The synonymous nucleotide mutations introduced in the cDNA clone to address instability issues in this study did not alter the translated sequence of rZIKV-FP, which was identical to consensus viral proteome of H/PF/2013 (KJ776791)23 deposited in the public database. From this perspective, our results demonstrate that: (1) Positive correlation of tissue-specific expansion of minor variants in H/PF/2013 clinical isolate with disease pathogenesis that drives its lethality in mice; (2) The in vivo virulent phenotype of H/PF/2013 correlated with the clonal expansion of NS2A:V117 variant in mice that induced a higher inflammation in the brain coupled with better replication fitness which is reinforced by our data showing the virulence of rZIKV-FP carrying the NS2A V117 mutation; (3) The variant in NS2B residue 32 resulted in an intermediate virulent phenotype in mice that is characterized by delayed mortality; (4) Critical virulence determinant NS2A:V117 variant in potential epistasis with NS1 and NS4B in the isolates from the brain may drive the severe neuropathogenesis.
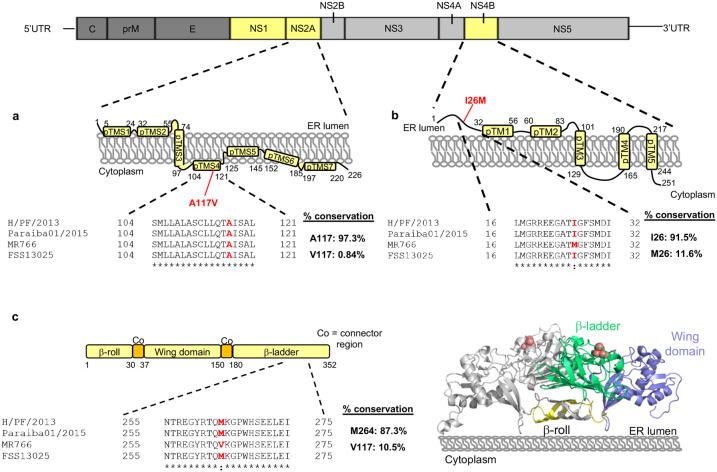
Flavivirus NS2A is a multifunctional membrane protein that is involved in viral RNA synthesis,35,36 virion assembly36, 37, 38 and modulation of host immune response.39,40 Recently specific residues of NS2A involved in viral RNA synthesis and virion assembly have been identified.36, 37, 38 Notably the NS2A:A117V variant that was identified to be the key determinant in causing the lethal in vivo disease phenotype similar to the H/PF/2013 clinical isolate showed higher replication fitness in LN-229 cells compared to prM L130 virus clone #2 (Fig. 2c and d), which appears to be in agreement with the critical function of NS2A in vRNA synthesis and virion assembly. Alanine at position 117 (within transmembrane domain 4) is highly conserved (97.3% of 1187 publicly available ZIKV NS2A sequences in the Virus Pathogen Database and Analysis Resource [ViPR database41]) and is located near the cytoplasmic loop formed by residues 97–104 that have been implicated in NS2A-viral RNA interaction for virion assembly (Figure S3a).38 Additionally ZIKV NS2A has been shown to interact with adherens junctions that promote its lysosomal degradation in the neural cells.26 Taken together it is conceivable that the substitution to valine could alter the interaction with viral RNA38 and host proteins.42 It is beyond the scope of this study to directly verify this mechanism but is implied by our observation of the increased replication fitness of virus clones harbouring NS2A:V117 (prM L130/NS2A V117 clones #11 and #20) compared with prM L130 virus clone #2 with NS2A:A117 in LN-229 cells (Fig. 2d). Notably the replicative advantage of NS2A V117 variant could explain the positive expansion observed in the brain (Fig. 1f) and consequently the NFκB mediated inflammation and increased mortality of H/PF/2013 clinical isolate and prM L130/NS2A V117 virus isolates (Fig. 3).
Apart from the NS2A:V117 variant that demonstrated clear association with the in vivo ZIKV virulence, the combined approach of examining independent virus populations from plaque purification of H/PF/2013 clinical isolate and NGS analyses has revealed other variants in the virus swarm that may contribute to ZIKV pathogenesis. Specifically, our findings of the prM/NS2A Consensus plaque purified clones (#12 and #23, Figs. 2 and 3) identified variants in NS2B residue that could also increase viral virulence due to higher genome replication that enhanced systemic infection compared to the non-lethal prM L130 virus clone (Fig. 3). Interestingly the NS1 and NS4B variants (M264V and I26M respectively) that were present in low frequencies in the infecting inoculum and infected mouse sera were highly enriched in the infected brains (Fig. 1), indicating a possible epistasis leading to ZIKV brain penetrance/neurotropism. Indeed a recent study demonstrated the specific contribution of ZIKV NS1 in triggering brain microvascular endothelial dysfunction that is reflective of its neurotropism.43 Furthermore NS1 has been shown to interact with NS4A-2K-4B precursor that is required for viral RNA replication.44 Interestingly the variants NS1 V264 and NS4B M26 are present in 10–12% of the publicly available ZIKV NS1 (n = 1195) and NS4B (n = 1182) sequence database respectively41 (Figure S3b and c). Of note these two variants were also found in the ancestral African MR766 strain that is historically thought to cause mild disease. However recent studies showed that African-lineage ZIKV also caused severe congenital Zika syndrome in experimental models.45, 46, 47 Notably the position 264 of NS1 is located in the “spaghetti loop” region of the β-ladder domain that is held by numerous H-bonds (Figure S3c),48,49 which is believed to make the region an attractive site for protein interactions. Hence the NS1:M264V and NS4B:I26M variants identified in H/PF/2013 virus isolate infected mouse brain may have an impact on the viral protein–protein interaction and/or viral-host protein interactions leading to higher neuro-infectivity. Further investigations are warranted to fully understand the links between NS2A, NS1 and NS4B that drives neuropathogenesis following infection in the mouse model.
It is noteworthy that our study has independently demonstrated that the non-consensus NS2A:V117 variant previously reported as a virulent determinant of ZIKV Paraiba01/2015 strain,11 is also a major contributor of the disease morbidity of the H/PF/2013 clinical isolate. This is evident in the specific expansion of the NS2A:V117 variant from its low frequency (∼27%) in the initial inoculum to a consensus level (>98%) in the brain, that correlated with the lethal neurological phenotype of H/PF/2013 infection in mice (Figs. 1 and 5). In contrast to Ávila-Pérez et al.,11 it is noted that our ZIKV Paraiba01/2015 does not carry this pathogenic NS2A variant (Table S5) which corroborated with its non-lethal phenotype in mice (Fig. 1a). It should also be noted that the viral diversity of the ZIKV isolates used in this study was lower compared to reported values.24,25 This is likely due to the differences in the NGS and variant calling pipeline used, where lofreq package is more sensitive to low frequency alleles12,19 and no threshold cutoff on the allele frequency was applied in the computation of diversity index. One interpretative limitation of our study is that only male mice are used in the in vivo infection studies. Furthermore the interpretation of the variant(s) enrichment in the tissues from infected mice could potentially be confounded by the level of serum virus from the blood in the organs despite of PBS perfusion prior to tissue collection.
In summary, our comparative variant enrichment analysis between the various tissues with the initial virus inoculum have indicated that non-consensus viral subpopulations play a critical role in driving the pathogenesis of ZIKV. Of note, the virulence of ZIKV appears to correlate with the evolution of variants in NS2A and NS2B viral membrane proteins that enabled better replication fitness. Notably our study highlights the importance of looking beyond the viral consensus sequence and paying attention to low lying variants within the virus swarm that could contribute to disease pathogenesis. Indeed this is evident in the correlation of ZIKV pathogenicity with the tissue-specific expansion of low allele frequency variants (NS1 and NS4B SNVs) in the brain that could serve as a predictive marker of future potential ZIKV outbreaks and severity. Lastly we posit that detailed study of isolates present in clinical isolates could lead to developing vaccine candidates that are less pathogenic and genotypically stable in our in vivo models.
Contributors
K.W.K.C, O.E.E and S.G.V conceived the study. K.W.K.C performed and analyzed the in vitro infection studies in cell-lines. K.W.K.C and S.W performed and analyzed the in vivo mouse studies. K.W.K.C, M.M.C and A.B prepared the libraries for Next-Generation Sequencing and variant call analysis were performed by A.B and M.M.C and O.E.E. K.W.K.C and S.G.V accessed and verified the underlying data and were responsible for the decision to submit the manuscript. K.W.K.C, O.E.E and S.G.V wrote the manuscript. All authors read and commented on the final manuscript.
Data sharing statement
Data in this study is available upon request from the corresponding author at subhash.vasudevan@duke-nus.edu.sg.
Declaration of interests
The authors declared no conflicts of interest.
Acknowledgments
We acknowledged the Advanced Bioimaging Core of Duke-NUS Singapore Health Services for the scientific and technical assistance on CLSM. We also thank Professor Salvatore Albani, Mr. Camillus J. H. Chua and Ms. Sharifah N. Hazirah for assistance and access to the NanoString Technology platform. This work is supported by Duke-NUS Khoo Post-doctoral Fellowship Award 2020 to K.W.K.C and National Medical Research Council of Singapore grants (MOH-000524 (OFIRG) to S.W.; MOH-OFIRG20nov-0002 to S.G.V.).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104570.
Contributor Information
Eng Eong Ooi, Email: engeong.ooi@duke-nus.edu.sg.
Subhash G. Vasudevan, Email: subhash.vasudevan@duke-nus.edu.sg.
Appendix A. Supplementary data
Figure S1.
Figure S2.
Figure S3.
References
- 1.Musso D., Gubler D.J. Zika virus. Clin Microbiol Rev. 2016;29(3):487–524. doi: 10.1128/CMR.00072-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campos G.S., Bandeira A.C., Sardi S.I. Zika virus outbreak, Bahia, Brazil. Emerg Infect Dis. 2015;21(10):1885–1886. doi: 10.3201/eid2110.150847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mlakar J., Korva M., Tul N., et al. Zika virus associated with microcephaly. N Engl J Med. 2016;374(10):951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- 4.Weaver S.C., Costa F., Garcia-Blanco M.A., et al. Zika virus: history, emergence, biology, and prospects for control. Antiviral Res. 2016;130:69–80. doi: 10.1016/j.antiviral.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pettersson J.H., Eldholm V., Seligman S.J., et al. How did Zika virus emerge in the Pacific Islands and Latin America? mBio. 2016;7(5) doi: 10.1128/mBio.01239-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J., Liu Y., Shan C., et al. Role of mutational reversions and fitness restoration in Zika virus spread to the Americas. Nat Commun. 2021;12(1):595. doi: 10.1038/s41467-020-20747-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y., Liu J., Du S., et al. Evolutionary enhancement of Zika virus infectivity in Aedes aegypti mosquitoes. Nature. 2017;545(7655):482–486. doi: 10.1038/nature22365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia H., Luo H., Shan C., et al. An evolutionary NS1 mutation enhances Zika virus evasion of host interferon induction. Nat Commun. 2018;9(1):414. doi: 10.1038/s41467-017-02816-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan L., Huang X.Y., Liu Z.Y., et al. A single mutation in the prM protein of Zika virus contributes to fetal microcephaly. Science. 2017;358(6365):933–936. doi: 10.1126/science.aam7120. [DOI] [PubMed] [Google Scholar]
- 10.Yau C., Low J.Z.H., Gan E.S., et al. Dysregulated metabolism underpins Zika-virus-infection-associated impairment in fetal development. Cell Rep. 2021;37(11) doi: 10.1016/j.celrep.2021.110118. [DOI] [PubMed] [Google Scholar]
- 11.Avila-Perez G., Nogales A., Park J.G., et al. A natural polymorphism in Zika virus NS2A protein responsible of virulence in mice. Sci Rep. 2019;9(1) doi: 10.1038/s41598-019-56291-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bifani A.M., Choy M.M., Tan H.C., Ooi E.E. Attenuated dengue viruses are genetically more diverse than their respective wild-type parents. NPJ Vaccines. 2021;6(1):76. doi: 10.1038/s41541-021-00340-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Boheemen S., Tas A., Anvar S.Y., et al. Quasispecies composition and evolution of a typical Zika virus clinical isolate from Suriname. Sci Rep. 2017;7(1):2368. doi: 10.1038/s41598-017-02652-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsetsarkin K.A., Kenney H., Chen R., et al. A full-length infectious cDNA clone of Zika virus from the 2015 epidemic in Brazil as a genetic platform for studies of virus-host interactions and vaccine development. mBio. 2016;7(4) doi: 10.1128/mBio.01114-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tay M.Y., Saw W.G., Zhao Y., et al. The C-terminal 50 amino acid residues of dengue NS3 protein are important for NS3-NS5 interaction and viral replication. J Biol Chem. 2015;290(4):2379–2394. doi: 10.1074/jbc.M114.607341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan K.W.K., Watanabe S., Jin J.Y., et al. A T164S mutation in the dengue virus NS1 protein is associated with greater disease severity in mice. Sci Transl Med. 2019;11(498) doi: 10.1126/scitranslmed.aat7726. [DOI] [PubMed] [Google Scholar]
- 17.Kwek S.S., Watanabe S., Chan K.R., et al. A systematic approach to the development of a safe live attenuated Zika vaccine. Nat Commun. 2018;9(1):1031. doi: 10.1038/s41467-018-03337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pompon J., Morales-Vargas R., Manuel M., et al. A Zika virus from America is more efficiently transmitted than an Asian virus by Aedes aegypti mosquitoes from Asia. Sci Rep. 2017;7(1):1215. doi: 10.1038/s41598-017-01282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilm A., Aw P.P., Bertrand D., et al. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012;40(22):11189–11201. doi: 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins T.J. ImageJ for microscopy. Biotechniques. 2007;43(1 Suppl):25–30. doi: 10.2144/000112517. [DOI] [PubMed] [Google Scholar]
- 21.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Lanciotti R.S., Kosoy O.L., Laven J.J., et al. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis. 2008;14(8):1232–1239. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baronti C., Piorkowski G., Charrel R.N., Boubis L., Leparc-Goffart I., de Lamballerie X. Complete coding sequence of Zika virus from a French Polynesia outbreak in 2013. Genome Announc. 2014;2(3) doi: 10.1128/genomeA.00500-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collins N.D., Widen S.G., Li L., et al. Inter- and intra-lineage genetic diversity of wild-type Zika viruses reveals both common and distinctive nucleotide variants and clusters of genomic diversity. Emerg Microb Infect. 2019;8(1):1126–1138. doi: 10.1080/22221751.2019.1645572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson K.E.E., Noval M.G., Rangel M.V., et al. Mapping the evolutionary landscape of Zika virus infection in immunocompromised mice. Virus Evol. 2020;6(2):veaa092. doi: 10.1093/ve/veaa092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoon K.J., Song G., Qian X., et al. Zika-virus-encoded NS2A disrupts mammalian cortical neurogenesis by degrading adherens junction proteins. Cell Stem Cell. 2017;21(3):349–358.e6. doi: 10.1016/j.stem.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miner J.J., Diamond M.S. Zika virus pathogenesis and tissue tropism. Cell Host Microbe. 2017;21(2):134–142. doi: 10.1016/j.chom.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lum F.M., Low D.K., Fan Y., et al. Zika virus infects human fetal brain microglia and induces inflammation. Clin Infect Dis. 2017;64(7):914–920. doi: 10.1093/cid/ciw878. [DOI] [PubMed] [Google Scholar]
- 29.Xu W., Solis N.V., Filler S.G., Mitchell A.P. Pathogen gene expression profiling during infection using a nanostring nCounter platform. Methods Mol Biol. 2016;1361:57–65. doi: 10.1007/978-1-4939-3079-1_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pu S.Y., Wu R.H., Yang C.C., et al. Successful propagation of flavivirus infectious cDNAs by a novel method to reduce the cryptic bacterial promoter activity of virus genomes. J Virol. 2011;85(6):2927–2941. doi: 10.1128/JVI.01986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi P.Y., Tilgner M., Lo M.K., Kent K.A., Bernard K.A. Infectious cDNA clone of the epidemic west Nile virus from New York City. J Virol. 2002;76(12):5847–5856. doi: 10.1128/JVI.76.12.5847-5856.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munster M., Plaszczyca A., Cortese M., et al. A reverse genetics system for Zika virus based on a simple molecular cloning strategy. Viruses. 2018;10(7) doi: 10.3390/v10070368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elena S.F., Carrasco P., Daros J.A., Sanjuan R. Mechanisms of genetic robustness in RNA viruses. EMBO Rep. 2006;7(2):168–173. doi: 10.1038/sj.embor.7400636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Visser J.A., Hermisson J., Wagner G.P., et al. Perspective: evolution and detection of genetic robustness. Evolution. 2003;57(9):1959–1972. doi: 10.1111/j.0014-3820.2003.tb00377.x. [DOI] [PubMed] [Google Scholar]
- 35.Mackenzie J.M., Khromykh A.A., Jones M.K., Westaway E.G. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology. 1998;245(2):203–215. doi: 10.1006/viro.1998.9156. [DOI] [PubMed] [Google Scholar]
- 36.Xie X., Zou J., Puttikhunt C., Yuan Z., Shi P.Y. Two distinct sets of NS2A molecules are responsible for dengue virus RNA synthesis and virion assembly. J Virol. 2015;89(2):1298–1313. doi: 10.1128/JVI.02882-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie X., Zou J., Zhang X., et al. Dengue NS2A protein orchestrates virus assembly. Cell Host Microbe. 2019;26(5):606–622.e8. doi: 10.1016/j.chom.2019.09.015. [DOI] [PubMed] [Google Scholar]
- 38.Zhang X., Xie X., Xia H., et al. Zika virus NS2A-mediated virion assembly. mBio. 2019;10(5) doi: 10.1128/mBio.02375-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munoz-Jordan J.L., Sanchez-Burgos G.G., Laurent-Rolle M., Garcia-Sastre A. Inhibition of interferon signaling by dengue virus. Proc Natl Acad Sci U S A. 2003;100(24):14333–14338. doi: 10.1073/pnas.2335168100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu W.J., Wang X.J., Clark D.C., Lobigs M., Hall R.A., Khromykh A.A. A single amino acid substitution in the West Nile virus nonstructural protein NS2A disables its ability to inhibit alpha/beta interferon induction and attenuates virus virulence in mice. J Virol. 2006;80(5):2396–2404. doi: 10.1128/JVI.80.5.2396-2404.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pickett B.E., Sadat E.L., Zhang Y., et al. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012;40(Database issue):D593–D598. doi: 10.1093/nar/gkr859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Norbury A.J., Jolly L.A., Kris L.P., Carr J.M. Vav proteins in development of the brain: a potential relationship to the pathogenesis of congenital Zika syndrome? Viruses. 2022;14(2):386. doi: 10.3390/v14020386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Puerta-Guardo H., Glasner D.R., Espinosa D.A., et al. Flavivirus NS1 triggers tissue-specific vascular endothelial dysfunction reflecting disease tropism. Cell Rep. 2019;26(6):1598–15613.e8. doi: 10.1016/j.celrep.2019.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plaszczyca A., Scaturro P., Neufeldt C.J., et al. A novel interaction between dengue virus nonstructural protein 1 and the NS4A-2K-4B precursor is required for viral RNA replication but not for formation of the membranous replication organelle. PLoS Pathog. 2019;15(5) doi: 10.1371/journal.ppat.1007736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shao Q., Herrlinger S., Zhu Y.N., et al. The African Zika virus MR-766 is more virulent and causes more severe brain damage than current Asian lineage and dengue virus. Development. 2017;144(22):4114–4124. doi: 10.1242/dev.156752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crooks C.M., Weiler A.M., Rybarczyk S.L., et al. African-lineage Zika virus replication dynamics and maternal-fetal interface infection in pregnant rhesus macaques. J Virol. 2021;95(16) doi: 10.1128/JVI.02220-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aubry F., Jacobs S., Darmuzey M., et al. Recent African strains of Zika virus display higher transmissibility and fetal pathogenicity than Asian strains. Nat Commun. 2021;12(1):916. doi: 10.1038/s41467-021-21199-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akey D.L., Brown W.C., Dutta S., et al. Flavivirus NS1 structures reveal surfaces for associations with membranes and the immune system. Science. 2014;343(6173):881–885. doi: 10.1126/science.1247749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown W.C., Akey D.L., Konwerski J.R., et al. Extended surface for membrane association in Zika virus NS1 structure. Nat Struct Mol Biol. 2016;23(9):865–867. doi: 10.1038/nsmb.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.