Summary
Mayer-Rokitansky-Küster-Hauser (MRKH) syndrome is characterized by aplasia of the female reproductive tract; the syndrome can include renal anomalies, absence or dysgenesis, and skeletal anomalies. While functional models have elucidated several candidate genes, only WNT4 (MIM: 603490) variants have been definitively associated with a subtype of MRKH with hyperandrogenism (MIM: 158330). DNA from 148 clinically diagnosed MRKH probands across 144 unrelated families and available family members from North America, Europe, and South America were exome sequenced (ES) and by family-based genomics analyzed for rare likely deleterious variants. A replication cohort consisting of 442 Han Chinese individuals with MRKH was used to further reproduce GREB1L findings in diverse genetic backgrounds. Proband and OMIM phenotypes annotated using the Human Phenotype Ontology were analyzed to quantitatively delineate the phenotypic spectrum associated with GREB1L variant alleles found in our MRKH cohort and those previously published. This study reports 18 novel GREB1L variant alleles, 16 within a multiethnic MRKH cohort and two within a congenital scoliosis cohort. Cohort-wide analyses for a burden of rare variants within a single gene identified likely damaging variants in GREB1L (MIM: 617782), a known disease gene for renal hypoplasia and uterine abnormalities (MIM: 617805), in 16 of 590 MRKH probands. GREB1L variant alleles, including a CNV null allele, were found in 8 MRKH type 1 probands and 8 MRKH type II probands. This study used quantitative phenotypic analyses in a worldwide multiethnic cohort to identify and strengthen the association of GREB1L to isolated uterine agenesis (MRKH type I) and syndromic MRKH type II.
Keywords: rare variant gene enrichment, exonic deletion CNV, Alu-Alu mediated rearrangement, developmental genomics, genitourinary development, infertility, sex-limited trait, transcriptional regulation, human phenotype ontology
Rare variant enrichment analysis was performed on a multiethnic cohort of 148 MRKH individuals, identifying GREB1L as the only gene approaching exome-wide significance. Screening for rare, predicted deleterious variants across this cohort and a cohort of 442 Han Chinese MRKH probands identified 16 of 590 (2.7%) cases with GREB1L variants.
Introduction
Mayer-Rokitansky-Küster-Hauser (MRKH) syndrome (MIM: 277000) represents a developmental disorder of the female reproductive tract characterized by a spectrum of hypoplasia or agenesis of the uterus, cervix, and upper two-thirds of the vagina that causes infertility. Thus, it may appear as a sex-limited trait. Without uterine transplantation or surrogacy, affected women cannot conceive.1
There are two types of MRKH with an overall prevalence of 1 in 5,000.2,3 MRKH type I (MIM: 277000) represents individuals with abnormal development processes limited to the female reproductive tract; i.e., isolated uterine anomalies observed. In addition to uterine and vaginal maldevelopment, MRKH type II (MIM: 610076) variably includes maldevelopment of the kidneys (30%–40%) and vertebrae (30%–40%), and more rarely includes auditory defects (10%–25%) and cardiac malformations (<3%).4 Müllerian Renal Cervical Somite, or MURCS Association (an acronym used for MRKH type II), encompasses genitourinary malformation and cervico-thoracic vertebral defects as well. Ovarian development and secondary sex characteristics are often unaffected; however, primary amenorrhea and infertility are characteristically due to maldevelopment of the uterus and vagina.3,5,6,7
The etiology of MRKH remains enigmatic, and both environmental and genetic causes have been proposed. While environmental factors have been implicated,8 Griffin et al. reported 4 of 14 (29%) of cases had a positive family history. 2A study of 1,397 Müllerian anomalies cases, approximately 4.7% of which were Müllerian aplasia cases based on a randomly sampled subset of the 1,397, that underwent Kinship Analysis revealed that ∼10% had a familial association.9 This familial association was interpreted as shared genetic, environmental, or combined genetic and environmental factors within those families.9 Based on studies recording family history for uterine malformations in general, a recurrence risk of 1%–5% for a first-degree relative is estimated.10 Clustering of MRKH within families suggests an autosomal dominant pattern, albeit with incomplete penetrance, variable expressivity, and the observation of a potential sex-limited trait.11,12,13,14
The process of genitourinary development is complex and involves interactions between the developing epithelium and mesenchyme, as well as among the ureteric buds, metanephric duct, and paramesonephric duct.15 The molecular genetics of genitourinary development have been studied in animal models, and as examples both WNT signaling and HOX genes have been firmly associated with homeostatic, normotypical development of the female reproductive tract.16,17 Despite these research efforts, contributing deleterious variant alleles in most MRKH candidate genes have been identified in only a small percentage of clinically diagnosed cases and families.10
In addition to single gene variant allele associations with MRKH, copy number variants (CNVs) are also implicated as an underlying genetic mutational etiology. For example, recurrent, de novo, disease-associated copy number variation at 1q21.1, 16p11.2, 17q12, and 22q11 has been reported in MRKH probands. Genes mapping within critical regions of these CNVs were determined to contribute to the MRKH phenotype as candidate “driver genes” based on several factors. These include evidence of a gene dosage abnormality (e.g., the gene mapped within defined CNV intervals) and the previous association of a gene with urogenital development biology (i.e., TBX6 in 16p11.2, LHX1 and HNF1B in 17q12) in model organism or genotype-phenotype association studies.18,19,20 Family-based, exome-sequencing (ES) studies have also identified candidate genes for MRKH, such as GREB1L, which was first described as a renal hypodysplasia/aplasia (MIM: 617805) gene, but was more recently implicated in families with predominantly MRKH type II.21,22 However, only WNT4 has been definitively associated with a phenotype of AD MRKH with hyperandrogenism (MIM: 158330).
Here, we used ES on a cohort (N = 148) of MRKH subjects across 144 unrelated families from North/South America and Europe (Table 1 and materials and methods). In addition to previously published candidate gene approaches,23 the large cohort size was leveraged to perform statistical rare variant enrichment analyses that revealed evidence for damaging alleles of GREB1L as a candidate causative gene for MRKH type I and type II. The mutational burden analyses were further validated by using established ultra-rare variant parsing cutoffs to elucidate additional cases from a replication cohort of 442 Chinese Han MRKH proband exomes (Table 1 and materials and methods).
Table 1.
Patient phenotypic features
| Classification | North/South American and European MRKH cohort (n = 148) | Han Chinese MRKH cohort (n = 442) |
|---|---|---|
| MRKH type I | 85 (57.4%) | 330 (74.7%) |
| MRKH type II | 50 (33.8%) | 109 (25.3%) |
| MURCS | 13 (8.8%) | 3 (0.7%) |
| Extragenital Malformations | North/South American and European MRKH cohort (n = 136)a | Han Chinese MRKH cohort (n = 442) |
| Skeletal anomaly | 28 (20.6%) | 90 (20.1%) |
| Kidney anomaly | 41 (30.1%) | 46 (10.4%) |
| Ocular/Hearing | 3 (2.2%) | 2 (0.5%) |
| Cardiac | 2 (1.4%) | 1 (0.2%) |
Abbreviations: MRKH, Mayer–Rokitansky–Küster–Hauser syndrome; MURCS, Müllerian Renal Cervical Somite.
Detailed phenotypic information on extragenital malformations not present for 12 MRKH Type II probands which were excluded from extragenital malformation counts and percentages.
Materials and methods
Patient recruitment
Patients were separately recruited by obstetricians/gynecologists and clinical geneticists at Tübingen University Hospital (Germany), Texas Children’s Hospital (USA), Aretaieion Hospital (Greece), Sophia Children’s Hospital (Greece), and Peking Union Medical College Hospital (PUMCH; China); ES data were later harmonized for cohort analysis. All patients volunteered written, informed consent for research studies. At Tübingen University Hospital, patients undergoing vaginoplasty were considered for inclusion. Uterine anatomy was assessed preoperatively by pelvic magnetic resonance imaging (MRI), and uterine rudiments were excised if >1 cm endometrium was visualized. At Texas Children’s Hospital, Aretaeieion Hospital, Sophia Children’s Hospital, and PUMCH, patients were recruited at time of examination based on the presence of a confirmed Müllerian anomaly by imaging (pelvic ultrasound or MRI) and the absence of biochemical, hormonal, and physical exam features revealing confounding diagnostic features, including pregnancy, cloacal malformations, or disorders of sexual differentiation. As such, all patients with an abnormal female karyotype (i.e., not 46,XX) were excluded from this study. At PUMCH, phenotyping of the urogenital system was independently performed by two obstetricians/gynecologists and all patients underwent renal ultrasound. This study was overseen by the Institutional Review Board of Baylor College of Medicine under an approved research study protocol (IRB approval number: H-1843).
ES and variant calling
ES was performed at multiple centers. Of the 148 MRKH probands used in rare variant enrichment analyses, DNA extracted from blood for 76 probands and their available family members were sequenced by the Baylor College of Medicine Human Genome Sequencing Center using a custom-designed capture reagent, VCRome2.1 (Nimblegen). The Genome Quebec Innovation Center sequenced DNA extracted from blood for 25 probands and available family members by using the NimbleGen SeqCap EZ Human Exome Library v3.0. DNA extracted from uterine tissue for 20 probands and from blood for 31 probands and their available family members were separately sequenced by using the SureSelect Human All Exon V5 kit. Finally, Han Chinese samples were sequenced at PUMCH by sequential capture using SureSelect Human All Exon V6+UTR r2 core design capture kits. All samples were sequenced on an Illumina HiSeq 2000 platform. Raw data processing of FASTQ files from all sequencing centers except PUMCH was performed using the Mercury pipeline, available via DNANexus to achieve a harmonized variant call set for aggregate analyses (http://blog.dnanexus.com/2013-10-22-run-mercury-variant-calling-pipleine/).28 Briefly, an FASTQ file is generated from raw read data (bcl file) before reads are mapped to reference using the Burrows-Wheeler-Alignment tool and merged into a sample-level BAM file.29 For PUMCH samples, processing of sequencing data up to an FASTQ file was performed using the Peking Union Medical College Hospital Pipeline.30 The BAM file is further refined using SAMtools, Picard, and GATK.31,32,33 ATLAS2 was used for variant calling and a custom Cassandra pipeline that uses ANNOVAR was applied for variant annotation.34,35 CNVs were called from ES data using XHMM and HMZDelFinder tools.36,37
Combined cohort analysis for novel candidate genes
Post-sequence processed data, generated as described above, was batched for all MRKH probands as the case cohort (N = 148). The control cohort was generated using post-sequence processed data for parents of trios, or probands only if trio data were unavailable, for individuals within the Baylor-Hopkins Center for Mendelian Genomics (BHCMG) without phenotypes overlapping those of MRKH (e.g., cohorts without uterine, renal, skeletal, and auricular abnormalities; N = 7,956). Rare variants were parsed using the following criteria: gnomAD exome minor allele frequency (MAF) = 0, ExAC MAF = 0, internal CMG MAF ≤0.0002, Combined Annotation Dependent Depletion (CADDv1.3) Phred Score ≥20 for non-in/del variants, mutation type as nonsynonymous, stop-gain, frameshift, non-frameshift, or splicing (± 5 base pairs [bp] from intron/exon boundary), and variant read count ≥10. A Fisher’s exact test was used to determine whether there was an enrichment of rare variants within a particular gene in the case cohort. Holm-Bonferroni adjusted p values were used to control for potential errors introduced by multiple testing.
Probands were screened for CNVs encompassing top candidate genes elucidated by enrichment analysis. Variants meeting the above parsing criteria and within genes with model organism or additional human variant data supporting a role in MRKH were orthogonally confirmed by dideoxy Sanger sequencing (SNVs) or EvaGreen dye based ddPCR (CNVs) according to standard protocols. Screening of scoliosis probands in the BHCMG cohort was completed on a data freeze set of variants from November 2021.
Quantitative deep phenotyping analyses
For 56 MRKH type II probands for which detailed phenotype information was available, a symmetric Lin score was used to compare patient phenotypes after conversion to Human Phenotype Ontology (HPO) terms.38 The score was calculated by leveraging the hierarchical HPO term structure as previously described, except that instead of comparing each patient’s set of phenotype terms to a set of disease-associated phenotype terms, each patient’s set of phenotype terms was compared with the set of disease-associated phenotype terms of each other patient.39,40,41 This generated a quantitative phenotypic similarity score, which was used to construct a heatmap that allowed visualization of clusters of patients with overlapping patterns of phenotypes. The same approach was used to compare all previously reported GREB1L variant-associated phenotypes, as well as GREB1L variant-associated phenotypes reported here quantitatively using HPO. This set of all previously reported GREB1L cases and probands reported here was queried against the publicly available OMIM/Orphanet HPO annotated disease database (12,314 diseases; http://purl.obolibrary.org/obo/hp/hpoa/phenotype.hpoa). p values were calculated for phenotypic similarity between probands and HPO annotated diseases by comparing pairwise similarity between a proband and a particular disease against the phenotypic similarity of 100,000 random pairs of HPO annotated probands and disease. A list of HPO terms annotated to each variant is included (Table S1).
Array comparative genomic hybridization
An Agilent-designed high-resolution 1 million probe whole genome oligonucleotide microarray (AMADID: 021529) was performed on sample BH9101-1 to orthogonally validate the deletion initially detected by XHMM and HMZDelFinder, rule out other potential CNV complexities in this GREB1L region, and refine the genomic interval of the deletion event. Array experiments were performed according to the manufacturers’ protocol for probe labeling and hybridization with minor modifications.42
Short-read Illlumina whole genome sequencing
Short-read Illumina whole genome sequencing (WGS) was performed at the Baylor College of Medicine Human Genome Sequencing Center (HGSC) through the Genomics Research to Elucidate the Genetics of Rare Disease (GREGoR) consortium. Briefly, KAPA Hyper reagents were used to prepare sequencing libraries prior to pooling for multiplex sequencing. The Illumina HiSeqX platform was used to sequence pooled samples generating 150-bp paired-end reads. Demultiplexed data (on average 130 Gb per sample) then underwent post-sequencing processing via the HGSC pipeline.28,43 Average coverage was 35x. Fluidigm SNPtrace and Error-Rate-in-Sequencing (ERIS) platforms were then used for post-sequencing quality control analysis.44,45
VizCNV visualization of short-read WGS data
The short-read WGS read depth was calculated and normalized using previously described methods.46 In short, read depth was calculated using mosdepth (v 0.3.4)47 with the “-- by 1,000” option to produce a bed file storing read depth coverage in 1kb bins. The output of the mosdepth was then visualized with the in-house developed visualization tool VizCNV (https://github.com/BCM-Lupskilab/VizCNV). VizCNV uses the median read depth of each chromosome to normalize the read depth of that chromosome and calculates the log2 ratio for each 1-kb bin. The log2 ratio profiles were segmented using the “SLM” option build to allow for manual inspection of CNVs with sizes larger than 3 kb.
Breakpoint junction Sanger sequencing
For the BH9101-1 GREB1L deletion CNV case, approximate genomic intervals identified by aCGH were further determined and validated at single bp resolution via breakpoint junction analysis using Sanger dideoxynucleotide sequencing. The junction primer pair was designed to produce an amplicon for the deleted allele present only in the proband. “CACACAACCTGGGAAATGGG” and “TGACACCCTGCACATCTGAA” were designed as the forward and the reverse primers, respectively.
Results
Rare variant enrichment testing to identify potentially pathogenic variants in MRKH
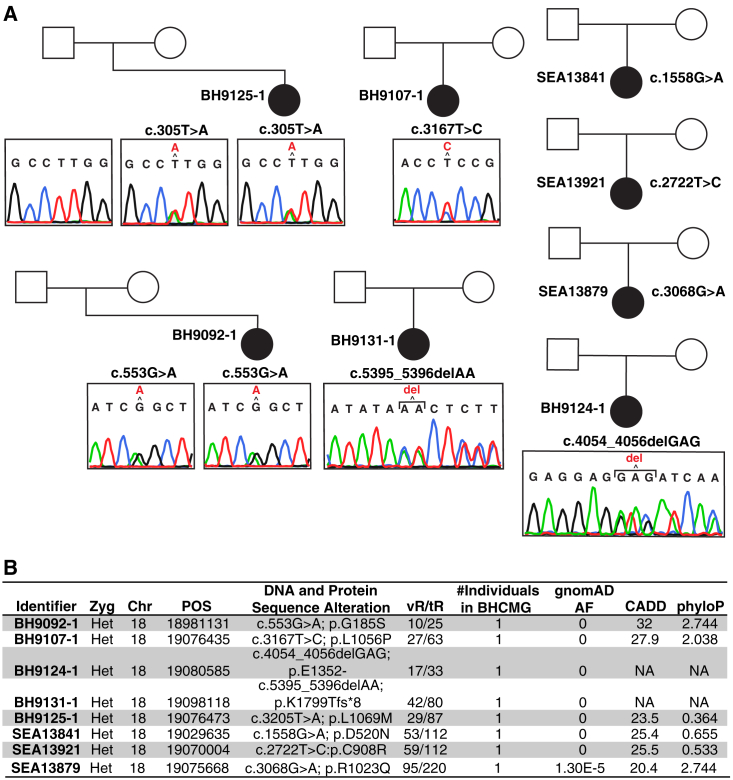
Unbiased gene-based rare variant enrichment analysis in the MRKH cohort studied identified GREB1L as the only gene nearing exome-wide significance (7 of 148, 4.7% cases; 31 of 7,956, 0.4% controls; p value = 4.6 × 10−6, adj. p value = 0.014, odds ratio = 12.7, 95% confidence interval = 4.6–30.0) (Figure 1). The MRKH cohort was then screened for probands with rare predicted deleterious variation in GREB1L that did not meet the stringent gnomAD MAF cutoff of 0 used in rare variant enrichment analysis. Eight heterozygous variant alleles in GREB1L were observed in seven MRKH probands by rare variant enrichment analysis (c.553G>A:p.G185S, c.1558G>A:p.D520N, c.2722T>C:p.C908R, c.3167T>C:p.L1056P, c.3205T>A:p.L1069M, c.4054_4056delGAG: p.1352_1352del, c.5395_5396delAA:p.1799_1799del) and one additional proband (c.3068G>A:p.R1023Q) by screening (Figure 2 and Table 2). These eight GREB1L variants include a 2-bp and a 3-bp indel, and six missense alleles (Figure 2). Parental DNA was available for two probands with a variant in GREB1L, BH9125-1 (paternal and maternal), and BH9092-1 (maternal only). Segregation analysis showed a maternal inheritance pattern of the c.3205T>A:p.L1069M (BH9125-1) and c.553G>A:p.G185S (BH9092-1) variants, consistent with previously reported pathogenic variants in GREB1L, which were largely maternally inherited from asymptomatic mothers.26 However, it is possible that mothers had subclinical phenotypes that were not recognized.
Figure 1.
Genes with significant rare variant mutational enrichment in the Americas and European MRKH cohort
The y axis represents the -log of the adjusted p value and x axis the ratio of the frequency of rare variants passing parsing steps in the MRKH cohort to the control cohort. p value testing was done using a Fisher’s exact test and p values were adjusted using the Holm-Bonferroni method. A total of 3,003 genes, which were identified to have rare variants present in the MRKH cohort using the parsing criteria described (materials and methods), were tested. Of these, 3,001 genes had an adjusted p value of 1.
Figure 2.
Pedigrees representing families with predicted deleterious GREB1L variant alleles observed in the MRKH cohort
Identifiers for probands are located to the left of each respective proband in the pedigree.
(A) Sanger dideoxy traces for each variant allele from individuals with DNA available for testing are represented below the respective pedigree symbol.
(B) Variant data are represented in a table at the bottom of the figure. All variants refer to the GRCh37/Hg19 genome assembly build and to RefSeq transcript ID NM_001142966.3. AF, allele frequency; BHCMG, Baylor-Hopkins Center for Mendelian Genomics Database; CADD, combined annotation dependent depletion score; CHR, chromosome; POS, position; tR, total reads; vR, variant reads; Zyg, zygosity.
Table 2.
GREB1L variant alleles patient ascertainment
| Location | C Change | P Change | Exon | Gnomad (het/hom) | Renal phenotype | Uterine phenotype | Auricular phenotype | Skeletal phenotype | Inheritance | CADD | GERP-RS | Study (ID if this study) | Reported Phenotype |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 18:18963516 | c.37C>T | p.R13∗ | 3 | 0 | + | – | – | – | NK | 37 | 4.5 | Sanna-Cherchi et al.24 | CAKUT |
| 18:18964302 | c.293C>G | p.S98∗ | 4 | 0 | + | – | – | – | de novo | 36 | 5.35 | Boissel et al. 201848 | CAKUT |
| 18:18964356 | c.347C>T | p.T116I | 4 | 0 | – | – | + | – | maternal | 29.6 | 5.25 | Schrauwen et al. 202049 | Auricular |
| 18:18975373 | c.383G>A | p.R128H | 5 | 0 | + | + | – | – | NK | 34 | 4.99 | Sanna-Cherchi et al.24 | CAKUT |
| 18:18981131 | c.553G>A | p.G185S | 6 | 0 | + | + | – | – | maternal | 32 | 5.5 | This Study (BH9092-1) | MRKH 2 |
| 18:18981153 | c.575G>A | p.R192Q | 6 | 0 | – | + | – | – | maternal | 27.1 | 5.8 | This Study (MRK17039400102) | MRKH 1 |
| 18:18981153 | c.575G>T | p.R192L | 6 | 0 | + | – | – | – | maternal | 26.1 | 5.8 | De Tomasi et al.26 | CAKUT |
| 18:18981283 | c.705G>T | p.W235C | 6 | 0 | + | + | – | – | paternal/maternal | 30 | 5.8 | Herlin et al.21 | MRKH 2 |
| 18:18983941 | c.818G>T | p.G273V | 7 | 0 | + | – | – | – | NK | 24.8 | 5.07 | Sanna-Cherchi et al.24 | CAKUT |
| 18:19019497 | c.848A>G | p.N283S | 8 | 0 | – | – | + | – | paternal (affected) | 9.921 | 3.44 | Schrauwen et al. 202049 | Auricular |
| 18:19020262 | c.982C>T | p.R328∗ | 9 | 0 | – | – | + | – | de novo | 38 | 4.52 | Schrauwen et al.25 | Auricular |
| 18:19020263 | c.983G>A | p.R328Q | 9 | 0 | + | – | – | – | maternal | 34 | 5.48 | De Tomasi et al.26 | CAKUT |
| 18:19029567 | c.1490C>G | p.A497G | 12 | 0 | + | – | – | – | NK | 14.35 | 4.4 | Sanna-Cherchi et al.24 | CAKUT |
| 18:19029635 | c.1558G>A | p.D520N | 12 | 0 | + | + | – | – | NK | 25 | 5.57 | This Study (SEA13841) | MRKH 2 |
| 18:19029659 | c.1582delC | p.Q528Rfs∗12 | 12 | 0 | + | + | – | + | maternal | – | – | De Tomasi et al.26 | CAKUT |
| 18:19031043 | c.1780G>T | p.E594∗ | 13 | 0 | + | + | – | – | maternal | 40 | 6.04 | De Tomasi et al.26 | CAKUT |
| 18:19031076 | c.1813A>C | p.S605R | 13 | 0 | + | + | – | – | paternal | 24.8 | 5.88 | De Tomasi et al.26 | CAKUT |
| 18:19032046 | c.1852G>A | p.D618N | 14 | 2.1E-5 (4/0) | + | + | – | + | – | 27.3 | 5.68 | Jacquinet et al.22 | MRKH 2 |
| 18:19032130 | c.1936T>C | p.C646R | 14 | 0 | – | + | – | – | – | 11.14 | 4.67 | Jacquinet et al.22 | MRKH 1 |
| 18:19034490 | c.2148 G>T | p.L716F | 15 | 0 | + | – | – | – | NK | 24.5 | 4.43 | De Tomasi et al.26 | CAKUT |
| 18:19053036 | c.2227delC | p.Q743Rfs∗10 | 16 | 0 | + | + | – | + | maternal | – | – | Jacquinet et al.22 | MRKH 2 |
| 18:19053060 | c.2251C>T | p.R751C | 16 | 0 | + | + | – | – | maternal | 34 | 5.21 | De Tomasi et al.26 | CAKUT |
| 18:19053061 | c.2252 G>A | p.R751H | 16 | 2.6E-5 (4/0) | + | – | – | – | maternal | 34 | 6.08 | De Tomasi et al.26 | CAKUT |
| 18:19053090 | c.2281G>C | p.E761Q | 16 | 0 | + | – | – | – | paternal | 23.8 | 5.2 | Sanna-Cherchi et al.24 | CAKUT |
| 18:19053090 | c.2281G>C | p.E761Q | 16 | 0 | + | – | – | – | paternal | 23.8 | 5.2 | Sanna-Cherchi et al.24 | CAKUT |
| 18:19053121 | c.2312C>T | p.P771L | 16 | 3.2E-5 (1/0) | + | + | – | + | – | 34 | 6.08 | Jacquinet et al.22 | MRKH 2 |
| 18:19053130 | c.2321T>A | p.L774Q | 16 | 0 | – | – | – | + | NA | 29.8 | 6.08 | This Study (WGC098981D) | Scoliosis |
| 18:19070004 | c.2722T>C | p.C908R | 19 | 0 | – | + | – | – | NK | 25.5 | 5.71 | This Study (SEA13921) | MRKH 1 |
| 18:19070069 – 19070070 |
c.2787_2788 delCG |
p.D930Pfs∗12 | 19 | 0 | + | + | – | – | both | – | – | Jacquinet et al.22 | MRKH 2 |
| 18:19070185 | c.2903C>T | p.A968V | 19 | 0 | + | – | – | – | de novo | 34 | 5.98 | Bossei et al. 2018 | CAKUT |
| 18:19070208 | c.2926C>T | p.Q976∗ | 19 | 0 | + | – | – | – | maternal | 40 | 5.98 | De Tomasi et al.26 | CAKUT |
| 18:19070229 | c.2947G>A | p.G983S | 19 | 0 | – | – | – | + | NA | 29.4 | 5.98 | This Study (BH12128-1) | Scoliosis |
| 18:19075668 | c.3068G>A | p.R1023Q | 20 | 1.3E-5 (2/0) | – | + | – | – | NK | 24.1 | 4.55 | This Study (SEA13879) | MRKH 1 |
| 18:19075685 | c.3085G>A | p.D1029N | 20 | 6.4E-6 (1/0) | – | + | – | – | NK | 23.6 | 5.42 | This Study (MRK17026300102) | MRKH 1 |
| 18:19075747 | c.3146 + 1G>A | splicing | 20 | 0 | – | + | – | – | NK | – | – | This Study (MRK17161200102) | MRKH 1 |
| 18:19076435 | c.3167T>C | p.L1056P | 21 | 0 | + | + | – | – | NK | 27.9 | 5.4 | This Study (BH9107-1) | MRKH 2 |
| 18:19076465 | c.3197G>C | p.R1066P | 21 | 1.3E-5 (2/0) | + | – | – | – | NK | 34 | 5.4 | Sanna-Cherchi et al.24 | CAKUT |
| 18:19076473 | c.3205T>A | p.L1069M | 21 | 0 | + | + | – | – | maternal | 23.5 | 1.8 | This Study (BH9125-1) | MRKH 2 |
| 18:19076563 | c.3295C>T | p.Q1099∗ | 21 | 0 | + | – | – | – | maternal | 37 | 3.78 | Sanna-Cherchi et al.24 | CAKUT |
| 18:19076621 | c.3353G>A | p.R1118Q | 21 | 0 | – | + | – | – | NK | 28.6 | 5.59 | This Study (MRK17167400102) | MRKH 1 |
| 18:19080481 | c.3970-20A>G | p.V1324Lfsa34 | 23 | 0 | + | + | – | + | paternal | – | – | Jacquinet et al.22 | MRKH 2 |
| 18:19080514 | c.3983G>A | p.G1328D | 23 | 0 | + | + | – | – | – | 29.1 | 4.68 | Jacquinet et al.22 | MRKH 2 |
| 18:19080529 | c.3998insC | p.L1334Pfsa18 | 23 | 0 | + | – | – | – | maternal | – | – | Sanna-Cherchi et al.24 | CAKUT |
| 18:19080585 – 19080587 |
c.4054_4056 delGAG |
p.E1352- | 23 | 0 | + | + | – | – | NK | – | – | This Study (BH9124-1) | MRKH 2 |
| 18:19085910 | c.4368G>T | Splicing | 25 | 0 | – | – | + | – | de novo | – | – | Schrauwen et al.25 | Auricular |
| 18:19088078 | c.4369-1G>C | Splicing | 26 | 0 | + | + | – | + | maternal | – | – | De Tomasi et al.26 | CAKUT |
| 18:19088215 | c.4505T>C | p.M1502T | 26 | 0 | + | – | – | + | maternal | 24.8 | 4.39 | De Tomasi et al.26 | CAKUT |
| 18:19088236 | c.4526A>T | p.D1509V | 26 | 6.4E-6 (1/0) | + | – | – | – | NK | 26.3 | 5.55 | De Tomasi et al.26 | CAKUT |
| 18:19088393 | c.4576C>T | p.R1526a | 27 | 0 | – | + | – | – | paternal | 48 | 5.83 | This Study (MRK18134900102) | MRKH 1 |
| 18:19088424 | c.4607A>G | p.H1536R | 27 | 0 | + | + | – | + | maternal | 23.3 | 5.83 | De Tomasi et al.26 | CAKUT |
| 18:19088463 | c.4646T>C | p.V1549A | 27 | 0 | + | + | – | – | NK | 26.1 | 5.83 | Sanna-Cherchi et al.24 | CAKUT |
| 18:19088489 | c.4672C>A | p.R1558S | 27 | 5.4E-6 (2/0) | + | – | – | – | maternal | 34 | 5.83 | De Tomasi et al.26 | CAKUT |
| 18:19088497 | c.4680C>A | p.Y1560a | 27 | 0 | + | + | – | – | maternal | 45 | 5.83 | Sanna-Cherchi et al.24 | CAKUT |
| 18:19088517 | c.4700T>C | p.L1567P | 27 | 0 | + | – | – | – | de novo | 28.2 | 5.83 | Sanna-Cherchi et al.24 | CAKUT |
| 18:19088544 | c.4727C>T | p.A1576V | 27 | 0 | + | – | + | – | NK | 33 | 5.83 | De Tomasi et al.26 | CAKUT |
| 18:19093889 | c.4843G>A | p.V1615I | 28 | 2.3E-5 (2/0) | + | – | – | – | maternal | 24.5 | 5.88 | Sanna-Cherchi et al.24 | CAKUT |
| 18:19095440 | c.4964T>C | p.I1655T | 29 | 0 | + | – | + | + | paternal | 23.8 | 6.06 | Sanna-Cherchi et al.24 | CAKUT |
| 18:19095467 | c.4991A>C | p.Y1664S | 29 | 0 | + | – | – | – | NK | 29 | 6.06 | Sanna-Cherchi et al.24 | CAKUT |
| 18:19095544 | c.5068G>A | p.V1690M | 29 | 0 | + | – | – | – | paternal | 29.6 | 6.06 | Sanna-Cherchi et al.24 | CAKUT |
| 18:19096642 | c.5198A>G | p.N1733S | 30 | 0 | + | + | – | – | both | 26.5 | 5.89 | Jacquinet et al.22 | MRKH 2 |
| 18:19098046 | c.5323 G>A | p.D1775N | 31 | 0 | + | – | + | – | NK | 34 | 5.75 | De Tomasi et al.26 | CAKUT |
| 18:19098101 | c.5378T>G | p.L1793R | 31 | 0 | + | – | – | – |
de novo/mat ernal |
31 | 5.97 | Brophy et al.27 | CAKUT |
| 18:19098118 – 19098119 |
c.5395_5396 delAA |
p.K1799Tfsa8 | 31 | 0 | + | + | – | – | NK | – | – | This Study (BH9131-1) | MRKH 2 |
| 18:19098164 | c.5441C>A | p.A1814D | 31 | 0 | + | + | – | – | NK | 34 | 5.97 | This Study (MRK18134100102) | MRKH 2 |
| 18:19102618 | c.5608+1delG | p.G1780Vfsa21 | 33 | 0 | + | – | – | – |
de novo/mat ernal |
– | – | Brophy et al.27 | CAKUT |
| 18:19102660 | c.5650C>T | p.R1884C | 33 | 3.2E-5 (4/0) | – | + | – | – | NK | 34 | 5.99 | This Study (MRK17147200102) | MRKH 1 |
| 18:19102661 | c.5651G>A | p.R1884H | 33 | 3.2E-5 (4/0) | + | – | – | – | NK | 32 | 5.99 | Sanna-Cherchi et al.24 | CAKUT |
| 18:18963479–19154805 | del | CNV | 3–33 | 0 | + | + | – | – | NK | – | – | This Study (BH9101-1) | MRKH 2 |
Abbreviations: +, present; −, absent or not confirmed; CADD, Combined Annotation Dependent Depletion; CNV, Copy-Number-Variant; GERP-RS, Genomic Evolutionary Rate Profiling Rejected Substitution; NK, not known. All variants refer to the GRCh37/Hg19 genome assembly build and to RefSeq transcript ID NM_001142966.3.
To date, only one GREB1L variant has been reported in MRKH type I; a c.1963T>C:p.C646R missense variant allele that was noted to have inconsistent variant effect functional assessment scores.22 In our Americas and European cohort, six of eight (75%) probands with predicted deleterious variation in GREB1L were noted to have MRKH type II (Table S2). The only two probands noted to have MRKH type I were SEA13921 in whom a c.2722T>C:p.C908R (CADD = 25.5) variant was identified and SEA13853 in whom a c.3068G>A:p.R1023Q (CADD = 21.3) variant was found. In contradistinction to the previously reported MRKH type I associated variant (p.C646R), most variant functional assessment scores predicted the p.C908R variant to be deleterious (Table S2). The p.R1023Q variant, like the C646R variant, had inconsistent variant functional assessment scores (Table S2). The GREB1L p.C646R variant allele was not found in our MRKH cohort or the BHCMG controls.
Quantitative phenotyping of MRKH probands to visualize phenotypic overlap
Detailed clinical phenotyping information was available for 71 of the 148 MRKH probands in whom the ES data were used for rare variant enrichment study. To visualize phenotypic overlap quantitatively, clinical phenotype descriptors for MRKH type II probands (33 MRKH type II probands and 38 MRKH type I probands) from these 71 were converted to HPO terms and used in a symmetric computational analysis, in which phenotypic similarity is calculated between the query and target HPO term sets and vice versa as described.50,51,52 Using the Lin method for symmetric phenotypic similarity calculation, each proband’s term set was compared with each other proband’s phenotypic term set (Figure 3 and materials and methods).50,51,52 Of the five GREB1L probands included in this HPO analysis, four clustered into a group (Figure 3, top left) representing patients with an additional phenotype of renal agenesis while one was present in a group (Figure 3, bottom right) of patients with an additional phenotype of renal ectopia.
Figure 3.
GREB1L probands cluster within a phenotypic group including uterine and renal malformations
Heatmap represents symmetric Lin score for proband HPO terms queried against each other. Clusters are demarcated by black outline. Top left cluster: Includes probands (cluster #1) that share unilateral renal agenesis as a feature. Bottom right cluster: Includes probands that share scoliosis (top left group of bottom right cluster; i.e., cluster #2) or ectopic kidney (bottom right group of bottom right cluster; cluster #3). GREB1L probands are represented by red font.
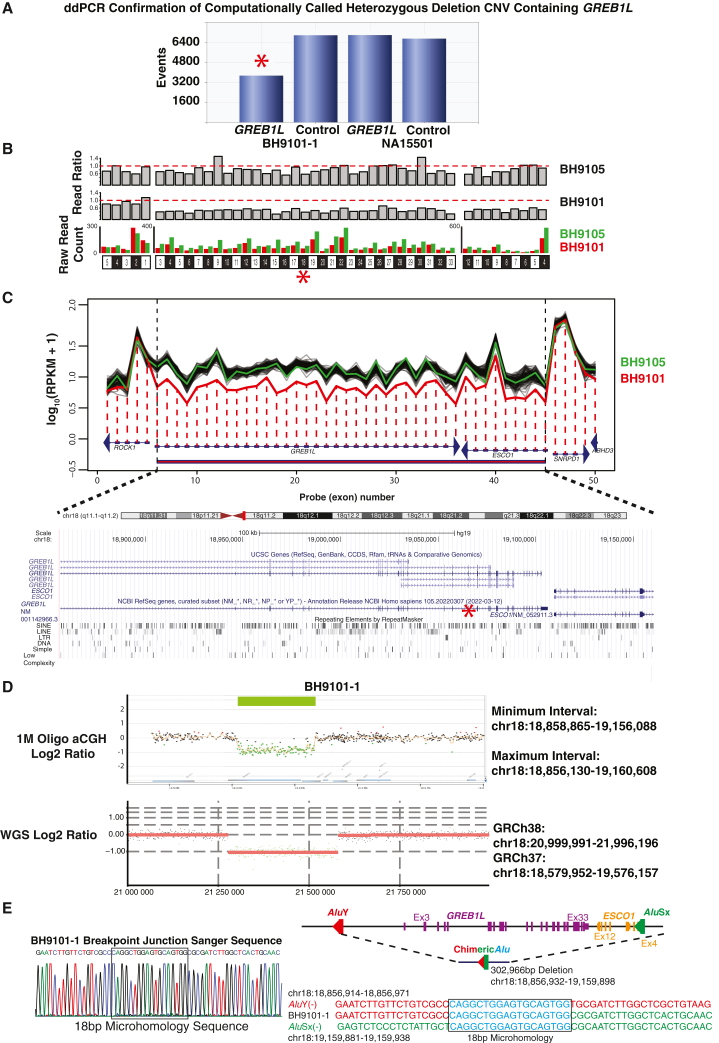
A GREB1L deletion copy number variant allele in a proband with MRKH type II
Additional variant screening studies of the MRKH cohort, consisting of review of CNV called computationally from ES study data using XHMM, revealed a heterozygous GREB1L deletion. This deletion CNV was supported computationally by a second algorithm, HMZDelFinder (Figure 4), in subject BH9101-1 in whom uterine aplasia and unilateral renal agenesis were observed. Heterozygous deletion CNV of GREB1L was orthogonally confirmed by ddPCR of exon 18; the size, extent, and GREB1L exonic content further delineated by 1M genome-wide Agilent array comparative genomic hybridization (aCGH) and Illumina short-read WGS (Figures 4 and S1). Breakpoint junction sequencing was performed using inward facing primers flanking the apparent breakpoint junctions determined by aCGH. Sanger sequencing revealed 18-bp microhomology and enabled base-pair resolution of deletion CNV genomic coordinates to chr18:18,856,932-19,159,898 (Figure 4E). This 302,966-bp genomic deletion encompasses the entirety of GREB1L coding exons, i.e., represents a null allele, and also includes all coding exons of a neighboring gene (ESCO1). Furthermore, both proximal and distal breakpoints mapped within Alu elements in the same orientation, AluY and AluSx respectively, implicating Alu-Alu mediated rearrangement (AAMR) as the mechanism for CNV formation (Figure 4E).53 Thus, for the North/South American plus European studies, seven of nine GREB1L variant cases (six SNV and one CNV) had an MRKH type II phenotype and two of nine probands, both with missense GREB1L alleles, had MRKH type I.23,54
Figure 4.
GREB1L deletion CNV in proband BH9101-1
(A) Droplet digital PCR results for BH9101-1 and NA15501 (an unaffected control sample) for exon 18 of GREB1L and a control biallelic gene, RPP30, are shown. Event number is displayed on the y axis and BH9101-1 and NA15501 results for GREB1L and RPP30 are displayed on the x axis. Exon 18, denoted by a red star throughout Figure 4, ddPCR of BH9101-1 resulted in significantly fewer positive droplets (events) than controls.
(B) Visualization of the read ratio of BH9101-1 and BH9105-1 (a control from the MRKH cohort without a deletion called) raw reads to the average raw reads across 100 samples with the most similar read profile across the exome to BH9101-1 or BH9105-1, and raw read count of BH9101-1 and BH9105-1 across GREB1L and ESCO1 exons called by XHMM to be deleted and the gene closest to the 3′ end of GREB1l, ROCK1. The exon structure of ROCK1, GREB1L, and ESCO1 are shown at the bottom. Raw read count for each exon is shown in red for BH9101-1 and green for BH9105-1.
(C) HMZDelFinder visualization of RPKM across the coding exons of GREB1L and flanking genes is shown in red for BH9101-1, green for BH9105-1, and black for the 100 samples with the most similar read profile to BH9101-1. RPKM across exons 3–33 of GREB1L and 4–12 of ESCO1—representing all coding exons of both genes—is significantly reduced in BH9101-1. Log10(RPKM +1) is shown on the y axis and exon structure and number of exons shown across the x axis. Arrowheads show the direction of transcription. The region with a called CNV is represented by a red bar along the x axis. Below the HMZDelFinder visualization, the UCSC Genome Browser of the region corresponding to deletion is shown. UCSC and RefSeq gene transcripts are shown above and repetitive elements are depicted at bottom. The scale at top represents 100kb.
(D) Top: Genome-wide 1M Probe aCGH results for BH9101-1 are shown for the region of chromosome 18 with deletion. Minimum and maximum deletion intervals are shown based on the last adjacent probes with a deletion call and normal copy number state on 3′ and 5′ ends of the deletion. Log2 ratio is shown on the y axis and chromosome position on the x axis. Genes within the region are shown at bottom and the deletion region highlighted by a green bar at top. Bottom: Visualization of Illumina short-read whole genome sequencing read depth data in BH9101-1 was performed using VizCNV (https://github.com/BCM-Lupskilab/VizCNV). A red line depicts the average log2 ratio of read depth at a locus to the median read depth of chromosome 18 (y axis) within the genomic region chr18:20,999,991-21,996,196 (GRCh38; x axis). Black dots represent the log2 ratio at each locus with a diploid copy number state. Green dots represent the log2 ratio at each locus within the deletion CNV.
(E) Breakpoint junction Sanger sequencing chromatogram for BH9101-1 is shown at left. The 18-bp microhomology sequence is denoted by a black rectangle. On the right, a simplified genomic view of the deletion CNV encompassing GREB1L coding exons (3–33, shown in purple) and ESCO1 coding exons (4–12, shown in orange) is depicted. The Alu elements AluY and AluSx are shown in red and green, respectively. A representation of the AAMR generated deletion and recombinant chimeric Alu (red/green hybrid) is shown below the simplified genomic view, with the individual Alu involved in the recombination shown in red and green, respectively. The deletion spans 302,966 bp (chr18:18,856,932-19,159,898). At bottom right, the sequences of the proximal (red) and distal breakpoints (green) are shown above and below the AAMR product sequence (red and green). Microhomology sequence is shown in blue and denoted by a black rectangle. Genomic coordinates for the proximal breakpoint are shown above the proximal breakpoint sequence, and those for the distal breakpoint are shown below the distal breakpoint sequence.
GREB1L variant alleles in an MRKH replication cohort of Han Chinese probands
To further explore the possibility of damaging GREB1L variant alleles as a cause of congenital uterine malformations, a large cohort of 442 Chinese MRKH probands recruited at PUMCH under the Deciphering disorders Involving Scoliosis and COmorbidities (DISCO study, http://www.discostudy.org/) were investigated. The ES data available were queried for ultra-rare variants in GREB1L (gnomAD MAF ≤0.0001).55 Of the 15 variants found, predicted damaging, loss-of-function (LoF) variant or protein-truncating variant (PTV), and missense variant alleles with CADD score ≥15 were retained (n = 7, c.575G>A:p.R192Q, c.3085G>A:p.D1029N, c.3146 + 1G>A:splicing, c.3353G>A:p.R1118Q, c.4576C>T:p.R1526∗, c.5441C>A:p.A1814D, c.5650C>T:p.R1884C) as putatively pathogenic variants contributing to the MRKH phenotype (Table S2). Notably, six of seven (85.7%) of these variants were found in probands with isolated uterine anomalies or an MRKH I phenotype. We also observed that in the two Chinese cases (MRK18134900102 and MRK17039600102) with parental DNA available, segregation analysis revealed paternal inheritance in one proband (MRK18134900102; c.4576C>T:p.R1526∗) and maternal inheritance of the c.575G>A:p.R192Q variant (MRK17039600102) in the other.
The finding of eight probands with isolated uterine anomalies, two from Europe and six from China (Table S2), further expands the phenotypic spectrum observed in GREB1L to include isolated uterine anomalies or MRKH type I. The implication of predicted damaging heterozygous variants in GREB1L as a cause for isolated ear, renal, and uterine phenotypes, as well as combinations of these phenotypes, resembles the phenotypic spectrum described for MRKH. However, skeletal phenotypes that are common in MRKH type II, have not previously been screened for GREB1L variants to our knowledge. Therefore, the BHCMG was searched for rare, predicted deleterious variants in GREB1L (gnomAD MAF = 0, CADD score ≥20) associated with scoliosis phenotypes. Two probands of the 721 scoliosis probands screened (0.3%), BH12128-1 and WGC098981D (Table S2) from a scoliosis cohort,30 were found to have heterozygous missense variant alleles of GREB1L that could possibly contribute to somitogenesis and their scoliosis phenotype (Table S2). While not enriched in the scoliosis cohort (0.3%) compared with the control cohort (0.4%), the finding of nine previously reported GREB1L cases with skeletal phenotypes, including one with scoliosis (Figure 5A) lends support to a possible contributing pathogenic role for GREB1L variation in these scoliosis probands.
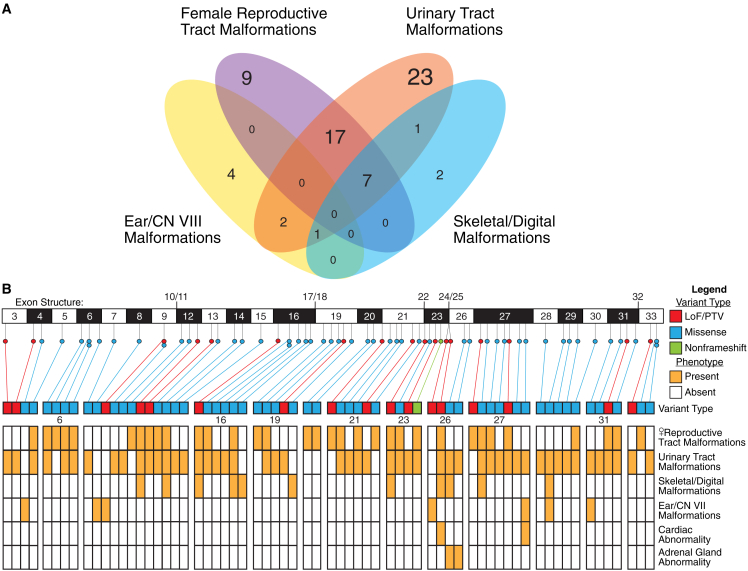
Figure 5.
Rare variants of GREB1L are associated with female reproductive tract, urinary tract, skeletal/digital, and ear/CN VII malformations
(A) Venn diagram depicting the number and overlap of variants associated with each phenotypic trait. Purple denotes female reproductive tract malformations, orange urinary tract malformations, blue skeletal/digital malformations, and yellow ear/CN VIII malformations.
(B) A representation of disease-associated variant type, with LoF/PTV variants colored red, missense blue, and nonframeshift green, within coding exons 3–33 of GREB1L. Phenotypes associated with variants are depicted in a heatmap below variant type diagram, and orange shading within a row marks association of a variant with a phenotype trait. Exons 6, 16, 19, 21, 23, 26, 27, and 31 are separated from adjacent exons in the heatmap to better delineate phenotype overlap as these exons encompass four or more disease-associated variants. CNV deletion GREB1L null allele not shown.
Breakdown of phenotypes associated with GREB1L variation by organ system and exon
While GREB1L was initially described as a renal agenesis gene,24,26,27 a summary of previously reported data (N = 50 variant alleles), and newly reported candidate disease-associated variant allele data herein (N = 18 variant alleles), implicate an association of GREB1L with both isolated and combined renal, uterine, ear, and skeletal malformation phenotypes (Figure S2). The most frequently reported associated phenotypes are female reproductive tract malformations and urinary tract malformations, while ear malformations (including cochlear agenesis and sensorineural hearing impairment) and skeletal malformation associations are less frequently reported. (Figure 5A, Tables 2 and S2).
A robust genotype/phenotype association of particular GREB1L variant alleles (e.g.. LoF, PTV, or missense mutations) or a particular exon with a certain phenotypic feature is not readily apparent. However, there are eight of 30 coding exons having four or more reported pathogenic variants (Figure 5B), i.e., potential MRKH hotspots. Rare, predicted damaging variants in exon 6, for example, are associated with female reproductive tract malformations (four of four) and include a high percentage of urinary tract malformation (three of four; 75%). Exon 16 in addition to having 50% of variants (three of six) associated with uterine malformation and 83.3% of variants (five of six) associated with renal malformations, has 50% of variants (three of six) that are found in probands with skeletal malformations. Last, cardiac and adrenal gland abnormalities have only been reported in association with variants in exons 26 and 27.
Further exploration of the phenotype-genotype associations across GREB1L alleles
A dissection of the phenotypic spectrum by quantitative clustering of variant-associated phenotypes in this study and across literature reports was performed to rigorously investigate genotype-phenotype associations. Variant-associated phenotypes in reported families and those studied herein were annotated with HPO terms and then compared with each other and publicly available OMIM/Orphanet HPO annotated disease database (12,314 diseases; http://purl.obolibrary.org/obo/hp/hpoa/phenotype.hpoa). Clustering analysis included OMIM and Orphanet disease phenotypes found to have a significant (p < 0.0002) match to variant phenotype sets after querying for phenotypic similarity between variant phenotypes against OMIM/Orphanet HPO annotated diseases (materials and methods).
Analysis using the gap statistic was performed for cluster numbers 1 to 18 and cluster number 2 was used due to the high drop off in slope after this point (Figure S3). Heatmap analysis showed that all probands containing a phenotype that did not solely affect the auricular system clustered together (red dendrogram branch), while auricular only probands formed a separate cluster (green dendrogram branch; Figure S4). Heatmap findings also highlight the GREB1L phenotypic spectrum and its variability, as even within the large cluster containing GREB1L variants that affect renal, uterine, and skeletal system development, there are distinct subclusters of variant-associated phenotypes. These data show that no GREB1L variant allele is associated with all phenotype systems implicated in MRKH (female reproductive, renal, skeletal, and auricular), but variably affect one or a few of these organ systems. A higher proportion of LoF/PTV variation (9 of 19, 47% vs. 20 of 49, 41% for missense and nonframeshift variants) causing multiple systems to be affected can be observed, including the LoF allele due to deletion CNV encompassing GREB1L in BH9101-1 that has a phenotype of MRKH type II. However, there are LoF/PTV variants that cause a phenotype in only one of the associated organ systems, the extent to which cases were phenotyped for all organ systems described here is not known, and the number of cases ascertained is limited.
A caveat to these analyses is the depth of phenotypic information available on GREB1L probands. Across previous studies, these probands were phenotyped and ascertained for the organ system of interest, for example some studies focused on renal phenotypes26,27,24 and others on auricular phenotypes.25 The extent of evaluation for extrarenal or extra-auricular phenotypes in those studies, especially for non-obvious phenotypes such as subtle hearing loss, may be limited. Reevaluation of probands with GREB1L variation for additional phenotypes would be required for a more precise evaluation of the phenotypic spectrum and consequences of deleterious variants in GREB1L.
Grid visualization of the phenotypic spectrum associated with GREB1L alleles
A data visualization grid of variants and their HPO annotated phenotype clinical synopsis sets was compiled, along with a frequency for each given HPO phenotype term, to better visualize the spectrum of GREB1L-associated phenotypes (Figure 6). While originally described as a renal hypodysplasia/aplasia gene, and where this observation remains true when considered as a group (e.g., when renal aplasia, multicystic dysplasia, and others are aggregated into one association group of kidney and urinary tract), renal disorders are most common (Figure 6, right), but the most frequent variant-associated phenotypes when each term is considered separately are uterine abnormalities and uterine aplasia (Figure 6, left). These data further support a possible role for GREB1L in isolated uterine abnormalities/aplasia in addition to a role for GREB1L in syndromic presentations of MRKH.
Figure 6.
Phenotypic grid of GREB1L variant-associated HPO terms
Left: GREB1L pathogenic variant alleles are labeled at left, and HPO phenotype terms labeled at the bottom of the phenotype grid. Red denotes the presence of a term and gray absence. Terms are ordered by frequency, which is displayed by black bars on a 0–1 scale at the top of the heatmap. Asterisk (right of grid) represents the specific row for the GREB1L deletion CNV null allele. Right: The phenotypic grid for GREB1L probands has been collapsed into broader phenotypic categories labeled at the bottom of the heatmap. Frequency is displayed at top by black bars on a 0–1 scale. Asterisk (right of grid) represents the specific row for the GREB1L deletion CNV null allele.
Discussion
The role of GREB1L in urogenital development
GREB1L, a 33-exon gene with 31 coding exons that maps to 18q11.1q11.2, was first associated with human disease in 2017, when three separate studies found rare variants in GREB1L to be associated with congenital malformations of the kidney and urinary tract (CAKUT) in an incompletely penetrant manner using an ES-based approach.24,26,27 In addition to this GREB1L CAKUT association,24,26,27 homozygous knockout of GREB1L in mice is lethal with embryonic phenotyping demonstrating Müllerian and Wolffian duct agenesis.26 And, a cell-based tubulomorphogenesis using mlMCD-3 cells suggested that GREB1L was required for the formation of tubular structures.26 Morpholino-based knockdown of GREB1L induced pronephric defects in zebrafish.24 Furthermore, co-injection of human wild-type GREB1L mRNA was able to rescue these defects, whereas co-injection of human GREB1L mRNA containing patients’ variants did not.24 These studies established GREB1L as a known disease gene for CAKUT, renal organogenesis, and renal hypodysplasia/aplasia 3 (MIM: 617805)24,26,27
Despite the description of affected individuals found to have Müllerian anomalies within these original CAKUT cohorts, only more recently was an association between GREB1L and MRKH suggested by analysis of a three-generation MRKH family in which the putative molecular diagnosis is a heterozygous c.705G>T missense variant allele of GREB1L.21 ES study of a cohort of nine families and 68 probands with MRKH reported eight candidate GREB1L disease variants, seven associated with MRKH type II and one with MRKH type I, suggesting an association of GREB1L with MRKH type II.22 The association of GREB1L with an MRKH type II phenotype was supported by previous studies in cells and model organisms. GREB1L has been identified as a paralog of GREB1, which is involved in growth regulation by estrogen in breast cancer.27,56,57,58 GREB1L had also been shown to have a 32-fold induction of expression in response to 24-h treatment with all-trans retinoic acid in F9 cells,59 suggesting it may be an important downstream effector in retinoic acid signaling. Retinoic acid signaling had been implicated in renal development,60 and uterine development including the determination of a utero-vaginal border.61 The studies identifying GREB1L as a renal agenesis gene in humans have also shown that it is expressed in the intermediate mesoderm, a precursor to renal and uterine structures, and that GREB1L plays an important role in urogenital development in both zebrafish and mouse models.26,27
Rare variants in GREB1L must be interpreted with caution as the gene is large (∼280 kb with 30 coding exons) and therefore there is a higher chance of having multiple probands with similar phenotypes carrying a rare variant by chance. The results of unbiased gene-based rare variant enrichment analysis presented here suggests rare GREB1L variants are not present in multiple MRKH probands just by chance due to gene size. Finding 16 GREB1L cases within a multiethnic cohort from around the world supports the two recent reports of GREB1L as a novel candidate gene for MRKH Type II21,22 and contentions outlined herein.
GREB1L-associated MRKH has an atypical inheritance pattern
One important difference between the described GREB1L disease-associated alleles and previous candidate genes, such as PAX8, is the prevalence of GREB1L missense, rather than LoF/PTV alleles. While data such as the finding of a CNV null allele reported here support a LoF in GREB1L as a driver of phenotypic manifestation, the description of incomplete penetrance and variable expressivity for GREB1L disease-associated alleles suggest that a haploinsufficiency model with AD inheritance does not fit observed inheritance patterns. Both background variation, such as a haplotype with more/less susceptibility to damaging variation in GREB1L, and sex-influenced factors, such as differences in the developmental molecular milieu between males and females, could contribute to the incomplete penetrance and variable expressivity observed.
For example, background variation may contain modifiers or “second hits” that contribute to this observation. This phenomenon was observed for TBX6 with 16p11.2 deletion CNV and a common T-C-A haplotype more commonly observed in Han Chinese, that only when present in trans, leads to a congenital scoliosis (CS) phenotype.62 These TACS, TBX6-associated congenital scoliosis, observations led to what is now commonly referred to as the compound inheritance gene dosage (CIGD) model for malformation syndromes.63,64,65 However, analysis for such a modifying variant in MRKH has, thus far, not been successful.22 Genomic sequencing of the MRKH patients with GREB1L genomic deletion CNV null alleles could be an approach to test the CIGD hypothesis in MRKH and potentially aid in elucidating contributory haplotypes, as it is possible that a second genomic variant, particularly one in trans that modifies expression or function of the non-deleted GREB1L allele modifies the phenotype.
Given that part of the proposed GREB1L-associated phenotype is sex-limited, it is potentially surprising to find that for non-de novo GREB1L disease-associated variant alleles previously reported or studied here and described in two generation pedigrees where parental DNA was tested,24,26,27 20 of 27 (74%) are maternally inherited (Table S1). Nevertheless, it is unclear as to whether that maternally inherited variant allele represents a de novo germline mutation or potentially represents a postzygotic new mutation during early development.
The expanded phenotypic spectrum of GREB1L-associated disease
MRKH represents a disease diagnosis with several phenotypic features, and these features overlap with other disease diagnoses. For example, a phenotype of isolated renal anomalies may have the same molecular etiology as MRKH; however, a proband with isolated renal anomalies would not be diagnosed clinically as an MRKH patient. MRKH can been associated with infertility (MIM: 277000). It is interesting to note that MRKH has also played a speculated historical role in both the human populations and geopolitical outcomes of the European continent and the royalty of Greece.66
By building a phenotypic series, we can better ascertain cases in which genetic testing may help improve the targeting of clinical care to a group of patients with the same molecular etiology—a molecularly refined classification that cannot be made by clinical evaluation alone.67 A review of previously reported variants and those we now describe implicate that GREB1L is associated with a spectrum of phenotypic traits. This spectrum includes not only CAKUT and renal agenesis, but also uterine agenesis, and more rarely skeletal and ear malformations, as is consistent with the described clinical features of MRKH also known as GRES (genital, renal, ear, and skeletal) syndrome.12,21,22 A small percentage of patients are also observed to have a cardiac phenotype as historically reported for MRKH; however, the phenotypes (hypertrophic cardiomyopathy and left aortic arch with retroesophageal right subclavian artery) have a high prevalence so their association with GREB1L variants is uncertain.7
The cohort reported here is unique from those previously reported, as there are eight probands with isolated uterine anomalies from the MRKH cohort and two probands with apparently isolated skeletal anomalies from the BHCMG scoliosis cohort with deleterious variation in GREB1L, demonstrating the phenotypic spectrum that can be associated with variant alleles of this gene. In addition to the previously mentioned studies suggesting GREB1L variants as causative for uterine and renal agenesis, these data support GREB1L as a strong candidate gene for not only MRKH type II, but also MRKH type I.21,22,24,26,27,68
It is important to note, however, that six of eight (75%) MRKH type I probands with variants in GREB1L were identified in the Han Chinese cohort. There are several possible explanations for this finding. One is that the Han Chinese cohort analyzed had a much higher number (330 vs. 85) and proportion (74.7% vs. 54.7%) of MRKH type I probands analyzed. Another factor that may play a role is the development of subclinical renal phenotypes. All 442 Han Chinese probands underwent renal ultrasonography to evaluate for renal hypoplasia, agenesis, or malformation; however, it is possible that subtle anomalies of the kidney and urinary tract may not be recognizable via ultrasound study. Yet another possibility is that modifiers common to ethnicity play a role in phenotypic variability. The example described above, of a common T-C-A haplotype affecting expression of TBX6 in the Chinese Han population and comprising the “second hit,” which explains penetrance of the 16p11.2 deletion CNV in CS,62 could also apply to protective modifiers that would contribute to non-penetrance of certain phenotypic features (e.g., renal abnormalities). A prevalent protective modifier in the Han Chinese population could also contribute to the lower proportion of renal abnormalities, which were less prevalent than skeletal abnormalities, observed in Han Chinese MRKH probands.69
Taken together, SNV and indel variants affecting GREB1L have been described predominantly leading to isolated uterine anomalies (9 of 67, 13.4%), isolated renal anomalies (23 of 67, 34.3%), and combined uterine/renal anomalies (17 of 67, 25.4%) (Figure 4A). However, there are also 4 of 67 (6%) SNV variants that have been previously associated with isolated sensorineural hearing loss and ear malformations and we propose 2 of 67 (3%) SNV variants to be associated with isolated skeletal features. While this supports GREB1L-associated disease to include both isolated and syndromic manifestations of MRKH, it is important to note that all cases described here and previously may have certain phenotypic features that were not assessed clinically or evaluated for by clinical testing potentially resulting in an ascertainment bias. We hypothesize that careful phenotyping of additional cases and further amalgamation of GREB1L probands into an allelic series will assist, in a data-driven way, the elucidation of the specific phenotypic spectrum associated with this gene and the biology of the disease process. Moreover, whole genome studies in the remaining “unsolved” MRKH probands may elucidate other genes and noncoding variants that may contribute to MRKH. Such studies may also further elucidate developmental genomics and inheritance models, such as the CIGD and Allele specific Gene Dosage (AsGD) models,70,71 that underlie birth defects and enable more informative genetic counseling for families with children having rare birth defects.
Conclusion
Here we report 18 GREB1L variants in 16 unrelated individuals from a cohort of 590 MRKH probands and two individuals from a CS cohort. The aggregate data from previous studies suggest that (1) GREB1L is a retinoic acid responsive gene,59 (2) is expressed in the intermediate mesoderm that gives rise to the uterus and kidney,27 and (3) is a cause of uterine and/or renal agenesis in zebrafish, mice, and humans.26,27 Here GREB1L is highlighted in association with a spectrum of human disease including isolated uterine agenesis in a large multiethnic cohort from around the world. Together, previously reported studies of GREB1L and the findings reported here strongly support the contention that GREB1L pathogenic variation causes MRKH Types I and II.
Data and code availability
The code supporting the current study has not been deposited in a public repository because it is being updated locally to accommodate further analysis, but it is available from the corresponding author on request.The dbGAP accession number for all exome sequences reported in this paper and with consent for data sharing in controlled-access databases is dbGAP: phs000711v5.p1. The ClinVar accession numbers for variants reported are ClinVar: SCV003842228, ClinVar: SCV003842229, ClinVar: SCV003842230, ClinVar: SCV003842231, ClinVar: SCV003842232, ClinVar: SCV003842233, ClinVar: SCV003842234, ClinVar: SCV003842235, ClinVar: SCV003842236, ClinVar: SCV003842237, ClinVar: SCV003842238, ClinVar: SCV003842239, ClinVar: SCV003842240, ClinVar: SCV003842241, ClinVar: SCV003842242, ClinVar: SCV003842243, ClinVar: SCV003842244, and ClinVar: SCV003842215.
Acknowledgments
This research was funded by the U.S. National Institutes of Health, National Institute of Neurological Disorders and Stroke (NINDS R35 NS105078 to J.R.L), National Human Genome Research Institute/National Heart, Lung, and Blood Institute (NHGRI/NHLBI UM1 HG006542 to J.R.L), the National Human Genome Research Institute (NHGRI K08 HG008986 to J.E.P) and the Genomic Research Elucidates the Genetics of Rare disease (GREGoR) program (UM1 HG011758 to J.E.P., J.R.L., and R.A.G.). A.J. was partially supported by the Howard Hughes Medical Institute and by the BCM Medical Scientist Training Program. This study was additionally supported by funds from the Beijing Natural Science Foundation (JQ20032 to N.W.), CAMS Innovation Fund for Medical Sciences (CIFMS, 2021-I2M-1-051 to N.W.), and the Non-profit Central Research Institute Fund of the Chinese Academy of Medical Sciences (No. 2019PT320025 to N.W.), the Foundation Santé Biomedical Research Grant, Latsis Foundation Research Grant, Stavros Niarchos Foundation, the Deutsche Forschungsgemeinschaft (DFG 351381475), European Research Council (ERC 2199678), European Research Council (ERC 249968) to S.E.A., and an institutional grant from the Faculty of Medicine at the University of Geneva Medical School to S.E.A. Eunice Kennedy Shriver National Institute of Child & Health Development of the National Institutes of Health (P50HD103555) also funded this study. The Foundation Sante Biomedical Research Grant, Latsis Foundation Research Grant, and Stavros Niarchos Foundation grant were awarded to A.S.D.
Declaration of interests
J.R.L. has stock ownership in 23andMe and is a co-inventor on multiple U.S. and European patents related to molecular diagnostics for inherited neuropathies, genomic disorders, eye diseases, and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the chromosomal microarray analysis and clinical genomic sequencing (both ES and WGS) offered in the Baylor Genetics Laboratory (http://bmgl.com). J.R.L. and I.B.V. serve on the Scientific Advisory Board of BG. S.E.A. is the co-founder and CEO of Medigenome, The Swiss Institute of Genomic Medicine.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2023.100188.
Web resources
The URLs for data presented herein are as follows:
ARIC database, http://www2.cscc.unc.edu/aric/
BCM-HGSC Mercury analysis pipeline, https://www.hgsc.bcm.edu/software/mercury.
BCM-HGSC ERIS pipeline, https://www.hgsc.bcm.edu/software/eris.
GATK HaplotypeCaller, https://gatk.broadinstitute.org/hc/en-us/articles/360037225632-HaplotypeCaller.
gnomAD, https://gnomad.broadinstitute.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
SIFT, https://sift.bii.a-star.edu.sg/
CADD, https://cadd.gs.washington.edu/
HPO resource page, https://hpo.jax.org/app/download/annotation.
dbGaP, http://www.ncbi.nlm.nih.gov/gap.
NHLBI ESP Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
Supplemental information
References
- 1.Herlin M.K., Petersen M.B., Brännström M. Mayer-Rokitansky-Küster-Hauser (MRKH) syndrome: a comprehensive update. Orphanet J. Rare Dis. 2020;15:214. doi: 10.1186/s13023-020-01491-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griffin, J.E., Edwards, C., Madden, J.D., Harrod, M.J., and Wilson, J.D. (1976). Congenital absence of the vagina. The Mayer-Rokitansky-Kuster-Hauser syndrome. Ann. Intern. Med. 85, 224–236. 10.7326/0003-4819-85-2-224. [DOI] [PubMed]
- 3.Herlin M., Bjørn A.M.B., Rasmussen M., Trolle B., Petersen M.B. Prevalence and patient characteristics of Mayer-Rokitansky-Küster-Hauser syndrome: a nationwide registry-based study. Hum. Reprod. 2016;31:2384–2390. doi: 10.1093/humrep/dew220. [DOI] [PubMed] [Google Scholar]
- 4.Morcel K., Camborieux L., Programme de Recherches sur les Aplasies Müllériennes. Guerrier D. Recherches sur les Aplasies Mulleriennes. Orphanet J. Rare Dis. 2007;2:13. doi: 10.1186/1750-1172-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oppelt P.G., Lermann J., Strick R., Dittrich R., Strissel P., Rettig I., Schulze C., Renner S.P., Beckmann M.W., Brucker S., et al. Malformations in a cohort of 284 women with Mayer-Rokitansky-Küster-Hauser syndrome (MRKH) Reprod. Biol. Endocrinol. 2012;10:57. doi: 10.1186/1477-7827-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan H.x., Luo G.n. Phenotypic and clinical aspects of Mayer-Rokitansky-Küster-Hauser syndrome in a Chinese population: an analysis of 594 patients. Fertil. Steril. 2016;106:1190–1194. doi: 10.1016/j.fertnstert.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Fontana L., Gentilin B., Fedele L., Gervasini C., Miozzo M. Genetics of Mayer–Rokitansky–Küster–Hauser (MRKH) syndrome. Clin. Genet. 2017;91:233–246. doi: 10.1111/cge.12883. [DOI] [PubMed] [Google Scholar]
- 8.Petrozza J.C., Gray M.R., Davis A.J., Reindollar R.H. Congenital absence of the uterus and vagina is not commonly transmitted as a dominant genetic trait: outcomes of surrogate pregnancies. Fertil. Steril. 1997;67:387–389. doi: 10.1016/S0015-0282(97)81927-9. [DOI] [PubMed] [Google Scholar]
- 9.Hammoud A.O., Gibson M., Peterson C.M., Kerber R.A., Mineau G.P., Hatasaka H. Quantification of the familial contribution to müllerian anomalies. Obstet. Gynecol. 2008;111:378–384. doi: 10.1097/01.AOG.0000267219.10869.9f. [DOI] [PubMed] [Google Scholar]
- 10.Jacquinet A., Millar D., Lehman A. Etiologies of uterine malformations. Am. J. Med. Genet. A. 2016;170:2141–2172. doi: 10.1002/ajmg.a.37775. [DOI] [PubMed] [Google Scholar]
- 11.Voorhoof L., Vandekerckhove D. Congenital absence of the vagina. Tijdschr. Geneeskd. 1979;35:95–106. [Google Scholar]
- 12.Guerrier D., Mouchel T., Pasquier L., Pellerin I. The Mayer-Rokitansky-Küster-Hauser syndrome (congenital absence of uterus and vagina) - phenotypic manifestations and genetic approaches. J. Negat. Results Biomed. 2006;5:1. doi: 10.1186/1477-5751-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herlin M., Højland A.T., Petersen M.B. Familial occurrence of Mayer-Rokitansky-Küster-Hauser syndrome: a case report and review of the literature. Am. J. Med. Genet. A. 2014;164A:2276–2286. doi: 10.1002/ajmg.a.36652. [DOI] [PubMed] [Google Scholar]
- 14.Ma X., Yao B., Pan Q., Xu W., Xu K., Ma F. Familial occurrence of Mayer–Rokitansky–Küster–Hauser syndrome. J. Obstet. Gynaecol. 2016;36:817–818. doi: 10.3109/01443615.2016.1157149. [DOI] [PubMed] [Google Scholar]
- 15.Chiga M., Ohmori T., Ohba T., Katabuchi H., Nishinakamura R. Preformed wolffian duct regulates Müllerian duct elongation independently of canonical Wnt signaling or Lhx1 expression. Int. J. Dev. Biol. 2014;58:663–668. doi: 10.1387/ijdb.140261rn. [DOI] [PubMed] [Google Scholar]
- 16.Du H., Taylor H.S. The role of Hox genes in female reproductive tract development, adult function, and fertility. Cold Spring Harb. Perspect. Med. 2015;6:a023002. doi: 10.1101/cshperspect.a023002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heikkil M., Peltoketo H., Vainio S. Wnts and the female reproductive system. J. Exp. Zool. 2001;290:616–623. doi: 10.1002/jez.1112. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi A., Shawlot W., Kania A., Behringer R.R. Requirement of Lim1 for female reproductive tract development. Development. 2004;131:539–549. doi: 10.1242/dev.00951. [DOI] [PubMed] [Google Scholar]
- 19.Horikawa Y., Iwasaki N., Hara M., Furuta H., Hinokio Y., Cockburn B.N., Lindner T., Yamagata K., Ogata M., Tomonaga O., et al. Mutation in hepatocyte nuclear factor-1ß gene (TCF2) associated with MODY. Nat. Genet. 1997;17:384–385. doi: 10.1038/ng1297-384. [DOI] [PubMed] [Google Scholar]
- 20.White P.H., Farkas D.R., McFadden E.E., Chapman D.L. Defective somite patterning in mouse embryos with reduced levels of Tbx6. Development. 2003;130:1681–1690. doi: 10.1242/dev.00367. [DOI] [PubMed] [Google Scholar]
- 21.Herlin M.K., Le V.Q., Højland A.T., Ernst A., Okkels H., Petersen A.C., Petersen M.B., Pedersen I.S. Whole-exome sequencing identifies a GREB1L variant in a three-generation family with Müllerian and renal agenesis: a novel candidate gene in Mayer-Rokitansky-Küster-Hauser (MRKH) syndrome. A case report. Hum. Reprod. 2019;34:1838–1846. doi: 10.1093/humrep/dez126. [DOI] [PubMed] [Google Scholar]
- 22.Jacquinet A., Boujemla B., Fasquelle C., Thiry J., Josse C., Lumaka A., Brischoux-Boucher E., Dubourg C., David V., Pasquier L., et al. GREB1L variants in familial and sporadic hereditary urogenital adysplasia and Mayer-Rokitansky-Kuster-Hauser syndrome. Clin. Genet. 2020;98:126–137. doi: 10.1111/cge.13769. [DOI] [PubMed] [Google Scholar]
- 23.Chen N., Zhao S., Jolly A., Wang L., Pan H., Yuan J., Chen S., Koch A., Ma C., Tian W., et al. Perturbations of genes essential for Müllerian duct and Wölffian duct development in Mayer-Rokitansky-Küster-Hauser syndrome. Am. J. Hum. Genet. 2021;108:337–345. doi: 10.1016/j.ajhg.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanna-Cherchi S., Khan K., Westland R., Krithivasan P., Fievet L., Rasouly H.M., Ionita-Laza I., Capone V.P., Fasel D.A., Kiryluk K., et al. Exome-wide association study identifies GREB1L mutations in congenital kidney malformations. Am. J. Hum. Genet. 2017;101:789–802. doi: 10.1016/j.ajhg.2017.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schrauwen I., Kari E., Mattox J., Llaci L., Smeeton J., Naymik M., Raible D.W., Knowles J.A., Crump J.G., Huentelman M.J., et al. De novo variants in GREB1L are associated with non-syndromic inner ear malformations and deafness. Hum. Genet. 2018;137:459–470. doi: 10.1007/s00439-018-1898-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Tomasi L., David P., Humbert C., Silbermann F., Arrondel C., Tores F., Fouquet S., Desgrange A., Niel O., Bole-Feysot C., et al. Mutations in GREB1L cause bilateral kidney agenesis in humans and mice. Am. J. Hum. Genet. 2017;101:803–814. doi: 10.1016/j.ajhg.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brophy P.D., Rasmussen M., Parida M., Bonde G., Darbro B.W., Hong X., Clarke J.C., Peterson K.A., Denegre J., Schneider M., et al. A gene implicated in activation of retinoic acid receptor targets is a novel renal agenesis gene in humans. Genetics. 2017;207:215–228. doi: 10.1534/genetics.117.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reid J.G., Carroll A., Veeraraghavan N., Dahdouli M., Sundquist A., English A., Bainbridge M., White S., Salerno W., Buhay C., et al. Launching genomics into the cloud: deployment of Mercury, a next generation sequence analysis pipeline. BMC Bioinformatics. 2014;15:30. doi: 10.1186/1471-2105-15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao S., Zhang Y., Chen W., Li W., Wang S., Wang L., Zhao Y., Lin M., Ye Y., Lin J., et al. Diagnostic yield and clinical impact of exome sequencing in early-onset scoliosis (EOS) J. Med. Genet. 2021;58:41–47. doi: 10.1136/jmedgenet-2019-106823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Broad Institute . Broad Institute; 2009. Picard Tools. [Google Scholar]
- 32.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji H.P. Improving bioinformatic pipelines for exome variant calling. Genome Med. 2012;4:7. doi: 10.1186/gm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fromer M., Purcell S.M. Using XHMM software to detect copy number variation in whole-exome sequencing data. Curr. Protoc. Hum. Genet. 2014;81:7.23.1–7.23.21. doi: 10.1002/0471142905.hg0723s81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gambin T., Akdemir Z.C., Yuan B., Gu S., Chiang T., Carvalho C.M.B., Shaw C., Jhangiani S., Boone P.M., Eldomery M.K., et al. Homozygous and hemizygous CNV detection from exome sequencing data in a Mendelian disease cohort. Nucleic Acids Res. 2017;45:1633–1648. doi: 10.1093/nar/gkw1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson P.N., Köhler S., Bauer S., Seelow D., Horn D., Mundlos S. The human phenotype Ontology: a tool for annotating and analyzing human hereditary disease. Am. J. Hum. Genet. 2008;83:610–615. doi: 10.1016/j.ajhg.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.James R.A., Campbell I.M., Chen E.S., Boone P.M., Rao M.A., Bainbridge M.N., Lupski J.R., Yang Y., Eng C.M., Posey J.E., et al. A visual and curatorial approach to clinical variant prioritization and disease gene discovery in genome-wide diagnostics. Genome Med. 2016;8:13. doi: 10.1186/s13073-016-0261-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Köhler S., Schulz M.H., Krawitz P., Bauer S., Dölken S., Ott C.E., Mundlos C., Horn D., Mundlos S., Robinson P.N. Clinical diagnostics in human genetics with semantic similarity searches in ontologies. Am. J. Hum. Genet. 2009;85:457–464. doi: 10.1016/j.ajhg.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu P., Meng L., Normand E.A., Xia F., Song X., Ghazi A., Rosenfeld J., Magoulas P.L., Braxton A., Ward P., et al. Reanalysis of clinical exome sequencing data. N. Engl. J. Med. 2019;380:2478–2480. doi: 10.1056/NEJMc1812033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carvalho C.M.B., Zhang F., Liu P., Patel A., Sahoo T., Bacino C.A., Shaw C., Peacock S., Pursley A., Tavyev Y.J., et al. Complex rearrangements in patients with duplications of MECP2 can occur by fork stalling and template switching. Hum. Mol. Genet. 2009;18:2188–2203. doi: 10.1093/hmg/ddp151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Regier A.A., Farjoun Y., Larson D.E., Krasheninina O., Kang H.M., Howrigan D.P., Chen B.J., Kher M., Banks E., Ames D.C., et al. Functional equivalence of genome sequencing analysis pipelines enables harmonized variant calling across human genetics projects. Nat. Commun. 2018;9:4038. doi: 10.1038/s41467-018-06159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang-Chu M.M.Y., Yu M., Haverty P.M., Koeman J., Ziegle J., Lee M., Bourgon R., Neve R.M. Human biosample authentication using the high-throughput, cost-effective SNPtraceTM system. PLoS One. 2015;10:e0116218. doi: 10.1371/journal.pone.0116218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jun G., Flickinger M., Hetrick K.N., Romm J.M., Doheny K.F., Abecasis G.R., Boehnke M., Kang H.M. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am. J. Hum. Genet. 2012;91:839–848. doi: 10.1016/j.ajhg.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du H., Jolly A., Grochowski C.M., Yuan B., Dawood M., Jhangiani S.N., Li H., Muzny D., Fatih J.M., Coban-Akdemir Z., et al. The multiple de novo copy number variant (MdnCNV) phenomenon presents with peri-zygotic DNA mutational signatures and multilocus pathogenic variation. Genome Med. 2022;14:122. doi: 10.1186/s13073-022-01123-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pedersen B.S., Quinlan A.R. Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics. 2018;34:867–868. doi: 10.1093/bioinformatics/btx699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boissel S., Fallet-Bianco C., Chitayat D., Kremer V., Nassif C., Rypens F., Delrue M.A., Dal Soglio D., Oligny L.L., Patey N., et al. Genomic study of severe fetal anomalies and discovery of GREB1L mutations in renal agenesis. Genet. Med. 2018;20:745–753. doi: 10.1038/gim.2017.173. [DOI] [PubMed] [Google Scholar]
- 49.Schrauwen I., Liaqat K., Schatteman I., Bharadwaj T., Nasir A., Acharya A., Ahmad W., Van Camp G., Leal S.M. Autosomal dominantly inherited GREB1L variants in individuals with profound sensorineural hearing impairment. Genes (Basel) 2020;11:1–18. doi: 10.3390/genes11060687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herman I., Jolly A., Du H., Dawood M., Abdel-Salam G.M.H., Marafi D., Mitani T., Calame D.G., Coban-Akdemir Z., Fatih J.M., et al. Quantitative dissection of multilocus pathogenic variation in an Egyptian infant with severe neurodevelopmental disorder resulting from multiple molecular diagnoses. Am. J. Med. Genet. A. 2022;188:735–750. doi: 10.1002/ajmg.a.62565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang C., Jolly A., Shayota B.J., Mazzeu J.F., Du H., Dawood M., Soper P.C., Ramalho de Lima A., Ferreira B.M., Coban-Akdemir Z., et al. Novel pathogenic variants and quantitative phenotypic analyses of Robinow syndrome: WNT signaling perturbation and phenotypic variability. HGG Adv. 2022;3:100074. doi: 10.1016/j.xhgg.2021.100074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lima A.R., Ferreira B.M., Zhang C., Jolly A., Du H., White J.J., Dawood M., Lins T.C., Chiabai M.A., van Beusekom E., et al. Phenotypic and mutational spectrum of ROR2-related Robinow syndrome. Hum. Mutat. 2022;43:900–918. doi: 10.1002/humu.24375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song X., Beck C.R., Du R., Campbell I.M., Coban-Akdemir Z., Gu S., Breman A.M., Stankiewicz P., Ira G., Shaw C.A., et al. Predicting human genes susceptible to genomic instability associated with Alu/Alu-mediated rearrangements. Genome Res. 2018;28:1228–1242. doi: 10.1101/gr.229401.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma C., Chen N., Jolly A., Zhao S., Coban-Akdemir Z., Tian W., Kang J., Ye Y., Wang Y., Koch A., et al. Functional characteristics of a broad spectrum of TBX6 variants in Mayer-Rokitansky-Küster-Hauser syndrome. Genet. Med. 2022;24:2262–2273. doi: 10.1016/j.gim.2022.08.012. [DOI] [PubMed] [Google Scholar]
- 55.Hansen A.W., Murugan M., Li H., Khayat M.M., Wang L., Rosenfeld J., Andrews B.K., Jhangiani S.N., Coban Akdemir Z.H., Sedlazeck F.J., et al. A genocentric approach to discovery of Mendelian disorders. Am. J. Hum. Genet. 2019;105:974–986. doi: 10.1016/j.ajhg.2019.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ghosh M.G., Thompson D.A., Weigel R.J. PDZK1 and GREB1 are estrogen-regulated genes expressed in hormone-responsive breast cancer. Cancer Res. 2000;60:6367–6375. [PubMed] [Google Scholar]
- 57.Rae J.M., Johnson M.D., Scheys J.O., Cordero K.E., Larios J.M., Lippman M.E. GREB1 is a critical regulator of hormone dependent breast cancer growth. Breast Cancer Res. Treat. 2005;92:141–149. doi: 10.1007/s10549-005-1483-4. [DOI] [PubMed] [Google Scholar]
- 58.Mohammed H., D’Santos C., Serandour A.A., Ali H.R., Brown G.D., Atkins A., Rueda O.M., Holmes K.A., Theodorou V., Robinson J.L.L., et al. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 2013;3:342–349. doi: 10.1016/j.celrep.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ochsner S.A., Watkins C.M., McOwiti A., Xu X., Darlington Y.F., Dehart M.D., Cooney A.J., Steffen D.L., Becnel L.B., McKenna N.J. Transcriptomine, a web resource for nuclear receptor signaling transcriptomes. Physiol. Genomics. 2012;44:853–863. doi: 10.1152/physiolgenomics.00033.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rosselot C., Spraggon L., Chia I., Batourina E., Riccio P., Lu B., Niederreither K., Dolle P., Duester G., Chambon P., et al. Non-cell-autonomous retinoid signaling is crucial for renal development. Development. 2010;137:283–292. doi: 10.1242/dev.040287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nakajima T., Iguchi T., Sato T. Retinoic acid signaling determines the fate of uterine stroma in the mouse Müllerian duct. Proc. Natl. Acad. Sci. USA. 2016;113:14354–14359. doi: 10.1073/pnas.1608808113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu N., Ming X., Xiao J., Wu Z., Chen X., Shinawi M., Shen Y., Yu G., Liu J., Xie H., et al. TBX6 null variants and a common hypomorphic allele in congenital scoliosis. N. Engl. J. Med. 2015;372:341–350. doi: 10.1056/NEJMoa1406829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lupski J.R. Clan genomics: from OMIM phenotypic traits to genes and biology. Am. J. Med. Genet. A. 2021;185:3294–3313. doi: 10.1002/ajmg.a.62434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang N., Wu N., Zhang L., Zhao Y., Liu J., Liang X., Ren X., Li W., Chen W., Dong S., et al. TBX6 compound inheritance leads to congenital vertebral malformations in humans and mice. Hum. Mol. Genet. 2019;28:539–547. doi: 10.1093/hmg/ddy358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu J., Wu N., Deciphering Disorders Involving Scoliosis and COmorbidities DISCO study. Yang N., Takeda K., Chen W., Li W., Du R., Liu S., Zhou Y., Zhang L., et al. TBX6-associated congenital scoliosis (TACS) as a clinically distinguishable subtype of congenital scoliosis: further evidence supporting the compound inheritance and TBX6 gene dosage model. Genet. Med. 2019;21:1548–1558. doi: 10.1038/s41436-018-0377-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Poulakou-Rebelakou E., Tsiamis C., Tompros N., Creatsas G. The lack of a child, the loss of a throne: the infertility of the first royal couple of Greece (1833-62) J. R. Coll. Physicians Edinb. 2011;41:73–77. doi: 10.4997/JRCPE.2011.115. [DOI] [PubMed] [Google Scholar]
- 67.White J., Beck C.R., Harel T., Posey J.E., Jhangiani S.N., Tang S., Farwell K.D., Powis Z., Mendelsohn N.J., Baker J.A., et al. POGZ truncating alleles cause syndromic intellectual disability. Genome Med. 2016;8:3. doi: 10.1186/s13073-015-0253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kyei Barffour I., Kyei Baah Kwarkoh R. GREB1L as a candidate gene of Mayer-Rokitansky-Küster-Hauser Syndrome. Eur. J. Med. Genet. 2021;64:104158. doi: 10.1016/j.ejmg.2021.104158. [DOI] [PubMed] [Google Scholar]
- 69.Chen N., Pan H., Luo G., Wang P., Xie Z., Hua K., Luo X., Huang X., Liu Q., Sun L., et al. Clinical characteristics of 1,055 Chinese patients with Mayer-Rokitansky-Küster-Hauser syndrome: a nationwide multicentric study. Fertil. Steril. 2021;116:558–565. doi: 10.1016/j.fertnstert.2021.02.033. [DOI] [PubMed] [Google Scholar]
- 70.Lupski J.R. Biology in balance: human diploid genome integrity, gene dosage, and genomic medicine. Trends Genet. 2022;38:554–571. doi: 10.1016/j.tig.2022.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Duan R., Hijazi H., Gulec E.Y., Eker H.K., Costa S.R., Sahin Y., Ocak Z., Isikay S., Ozalp O., Bozdogan S., et al. Developmental genomics of limb malformations: allelic series in association with gene dosage effects contribute to the clinical variability. HGG Adv. 2022;3:100132. doi: 10.1016/j.xhgg.2022.100132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code supporting the current study has not been deposited in a public repository because it is being updated locally to accommodate further analysis, but it is available from the corresponding author on request.The dbGAP accession number for all exome sequences reported in this paper and with consent for data sharing in controlled-access databases is dbGAP: phs000711v5.p1. The ClinVar accession numbers for variants reported are ClinVar: SCV003842228, ClinVar: SCV003842229, ClinVar: SCV003842230, ClinVar: SCV003842231, ClinVar: SCV003842232, ClinVar: SCV003842233, ClinVar: SCV003842234, ClinVar: SCV003842235, ClinVar: SCV003842236, ClinVar: SCV003842237, ClinVar: SCV003842238, ClinVar: SCV003842239, ClinVar: SCV003842240, ClinVar: SCV003842241, ClinVar: SCV003842242, ClinVar: SCV003842243, ClinVar: SCV003842244, and ClinVar: SCV003842215.