Summary
The ability to detect cancer at an early stage in patients who would benefit from effective therapy is a key factor in increasing survivability. This work proposes an evolutionary supervised learning method called CancerSig to identify cancer stage-specific microRNA (miRNA) signatures for early cancer predictions. CancerSig established a compact panel of miRNA signatures as potential markers from 4,667 patients with 15 different types of cancers for the cancer stage prediction, and achieved a mean performance: 10-fold cross-validation accuracy, sensitivity, specificity, and area under the receiver operating characteristic curve of 84.27% ± 6.31%, 0.81 ± 0.12, 0.80 ± 0.10, and 0.80 ± 0.06, respectively. The pan-cancer analysis of miRNA signatures suggested that three miRNAs, hsa-let-7i-3p, hsa-miR-362-3p, and hsa-miR-3651, contributed significantly toward stage prediction across 8 cancers, and each of the 67 miRNAs of the panel was a biomarker of stage prediction in more than one cancer. CancerSig may serve as the basis for cancer screening and therapeutic selection..
Keywords: Artificial Intelligence, Machine learning, Cancer diagnosis prediction, pan-cancer analysis, cancer early stage detection
Introduction
Cancer is one of the major health problems in the world and causes millions of deaths every year. According to the Cancer Statistics, in 2019, there were 1,806,590 new cancer cases and 606,520 cancer deaths that were estimated to occur in the United States alone.1 The cure for this complex disease remains elusive. Detecting cancer at an early stage in patients who would benefit from effective therapy is a key factor to increase survival. As regulators of gene expression in health and disease, microRNAs (miRNAs) may have potential roles as predictive biomarkers in cancer.2
MicroRNAs are a class of noncoding RNAs involved in the regulation of gene expression and control a number of diverse biological processes, including but not limited to differentiation, development, and growth, and are expressed in wide variety of organisms. These RNAs are transcribed from DNA sequences and consist of an average length of 22 nucleotides. Currently, there are 48,860 known mature miRNA sequences from 271 organisms, including 2,654 mature miRNA sequences from humans listed in miRBase,3 although the functions of many of these miRNAs have yet to be discovered.
Next-generation sequencing has made it possible to examine the expression levels of numerous miRNAs in various cancers and investigate their association with cancer development and progression. Over the last two decades, crucial evidence has demonstrated that miRNAs and miRNA biogenesis mechanisms are involved in the development of various cancers.4,5,6 There is considerable evidence that the expression of miRNAs has been linked to a number of human cancers.4,5,6 Depending on their target genes and under certain conditions, miRNAs either have oncogenic or tumor suppressor properties. MicroRNA expression profiles can also define cancer subtypes and are associated with varying treatment responses7 and overall survival.8 Identifying cancer-specific miRNA signatures and corresponding changes in gene expression over time is important for understanding the molecular basis of cancer and detecting early-stage cancers.
Accumulating evidence suggests that miRNA biomarkers can be effective in predicting early stages of cancer. For instance, miR-205, miR-210, and miR-708 have been used for early-stage detection of squamous cell lung cancer,9 and 34 miRNAs as a signature for early-stage detection in breast cancer.10,11 However, numerous miRNAs are expressed in cancers, and variability among different patients makes it challenging to determine true association between cancer and miRNA from spurious associations. Artificial Intelligence/machine learning methods may surmount these challenges by integrating and analyzing large datasets from numerous sources. Previously, we developed various machine learning-based cancer prediction models that contributed to survival prediction8,12,13,14 and early-stage detection10 in different cancer types.
Here, in pursuit of identifying the miRNA signatures that could aid in early-stage detection and may serve as general biomarkers for multiple cancer types, we present an evolutionary learning method called CancerSig. CancerSig is a machine learning method using an inheritable bi-objective combinatorial genetic algorithm (IBCGA)15 to identify cancer stage-specific miRNA signatures from 15 cancer types. Consequently, the miRNAs in each signatures were ranked based on their contribution to the prediction of early and advanced stages, and the top-ranked miRNAs analyzed using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and Gene Ontology (GO) annotations to evaluate the biological significance of these connections. Pan-cancer analysis revealed similarities and differences in miRNA signatures across the 15 cancer types. CancerSig established a compact panel of 242 significant miRNAs across the 15 cancer types with a mean cross-validation accuracy of 84.27%. Analysis of the identified panel of miRNAs may serve as a predictive measure for early-stage diagnosis of cancer and have important implications in biomarker-based cancer therapeutics.
Material and methods
MicroRNA expression profiles
The dataset was retrieved from TCGA. We initially considered all 33 cancer types available in the TCGA database; however, the amount of miRNA data and clinical information for certain cancer types was limited and therefore excluded. Cancer types with a minimum number of 80 patients with miRNA expression profiles were included for analysis and limited the number of cancer types to 15 for further analysis. With this criterion, our dataset contained 6,578 clinical samples with miRNA expression profiles across 15 cancer types. After removing the samples without cancer staging information and miRNA sequence data, each cancer type contained an average of 311 clinical samples with 311 corresponding miRNA expression profiles where each miRNA profile contained an average of 474.46 miRNAs. Overall, there were 4,667 clinical samples with miRNA expression profiles, which included 7,117 miRNAs (with duplication) for 15 cancer types. Normalized mature miRNA sequence data for analysis was obtained using the Illumina HiSeq 2000 platform. For prediction purposes, the dataset was divided into early (stages I and II) and advanced stages (stages III and IV) based on stage information. The number of samples and miRNAs for each cancer type are shown in Table 1.
Table 1.
TCGA datasets considered and used for miRNA and cancer stage association analyses after filtration
| Dataset | Abbreviation | Original samples | Final samples | miRNAs |
|---|---|---|---|---|
| Bladder urothelial carcinoma | BLCA | 412 | 407 | 477 |
| Breast invasive carcinoma | BRCA | 1097 | 386 | 503 |
| Colon adenocarcinoma | COAD | 458 | 221 | 444 |
| Esophageal carcinoma | ESCA | 185 | 162 | 459 |
| Head and neck squamous cell carcinoma | HNSC | 528 | 420 | 498 |
| Kidney renal clear cell carcinoma | KIRC | 537 | 256 | 420 |
| Kidney renal papillary cell carcinoma | KIRP | 291 | 261 | 438 |
| Liver hepatocellular carcinoma | LIHCC | 377 | 348 | 540 |
| Lung adenocarcinoma | LUAD | 522 | 452 | 477 |
| Lung squamous cell carcinoma | LUSC | 504 | 339 | 494 |
| Skin cutaneous melanoma | SKCM | 470 | 389 | 483 |
| Stomach adenocarcinoma | STAD | 443 | 381 | 459 |
| Thyroid carcinoma | THCA | 503 | 500 | 474 |
| Rectum adenocarcinoma | READ | 171 | 66 | 465 |
| Uveal melanoma | UVM | 80 | 79 | 486 |
Artificial intelligence-based prediction method CancerSig
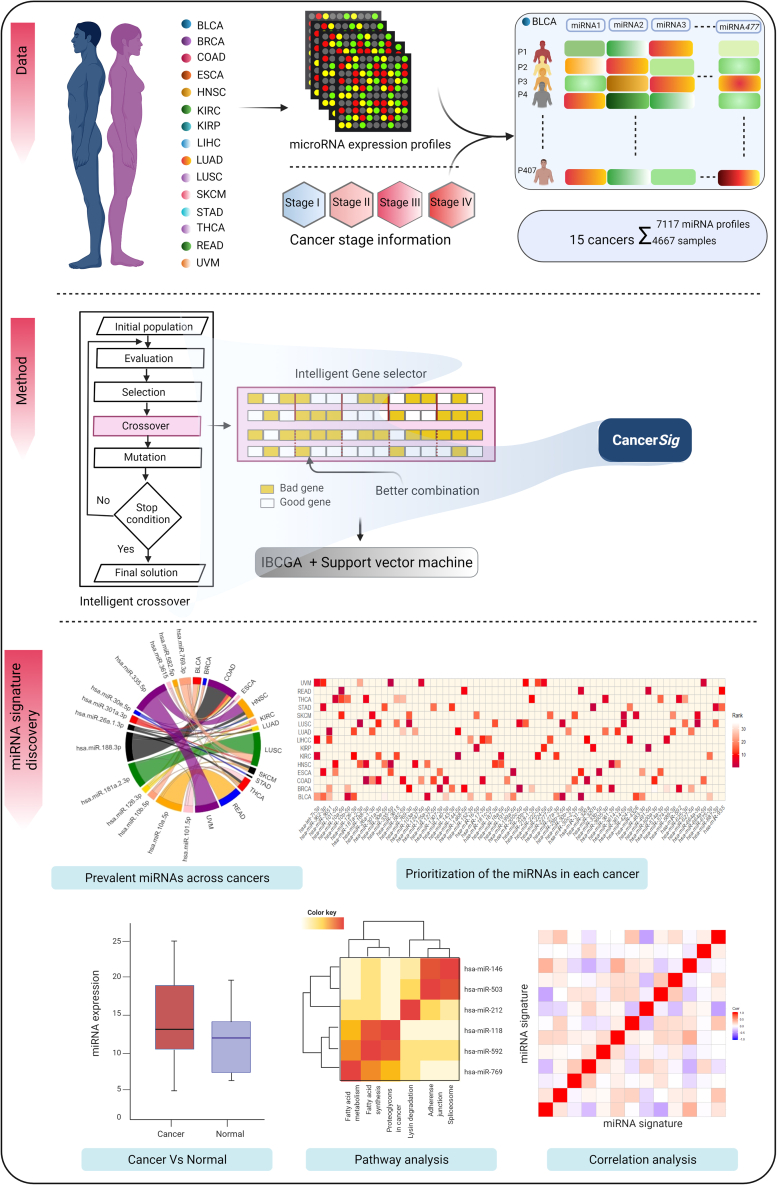
The novel cancer stage prediction method CancerSig identifies miRNA signatures to distinguish the early stage from advanced stage of various cancer types using their miRNA expression profiles with stage labels as input data. The output of CancerSig is the cancer-specific miRNA signatures and the panel of miRNA biomarkers for predicting cancer stages of multiple cancers. High performance of CancerSig arises mainly from an optimal feature selection algorithm IBCGA15 incorporated with a support vector machine (SVM) classifier,16,17 and pan-analysis of the miRNA signature. A brief summary of the methods is presented in the following paragraphs. The schematic diagram of CancerSig method is shown in Figure 1.
Figure 1.
Schematic diagram of the CancerSig method and analysis of the panel of miRNAs
MicroRNA expression profiles of 15 cancer types along with cancer stage information are input in the workflow of the CancerSig method to identify miRNA signatures.
Identifying a minimal set of m miRNAs, i.e., a miRNA signature from a large set of n candidate miRNAs while maximizing the prediction performance, is a bi-objective combinatorial optimization problem C(n, m) where the best value of m is not known in advance. The feature selection algorithm IBCGA uses an intelligent evolutionary algorithm18 to solve a large combinatorial optimization problem and an inheritance mechanism to efficiently identify a robust set of m features in a single run. IBCGA uses an orthogonal array crossover operation with a systematic reasoning ability to reproduce better offspring instead of random recombination used for traditional crossover operations. Accumulated evidence has indicated that IBCGA is good at identifying informative signatures in various cancers8,10,12,13,19 and other bioinformatics problems.20,21
SVM is a well-known powerful classifier, which has been applied to a wide variety of biological applications.17 SVM uses nonlinear transformation to map data from an input space to a higher-dimensional space to establish an accurate prediction model, especially when the training sample size is relatively small. We utilized the LibSVM22 package with the radial basis function kernel to implement CancerSig. The scoring function of the RBF kernel is computed in the feature space between the two data points, xi and xj, defined as follows:
| (Equation 1) |
To identify the miRNA signature and design an optimal classifier, the feature selection of IBCGA and parameter setting (cost C and kernel γ) of SVM play a vital role in modeling. IBCGA solves the problem C(n, m) for each cancer type independently. Taking bladder urothelial carcinoma (BLCA) with 477-miRNA expression profiles as an example, n = 477, and the value of m and the corresponding m miRNAs are determined by IBCGA. The parameter setting of IBCGA was rstart = 10 and rend = 50 meaning that the search for the m value is from 10 to 50. The fitness function is to maximize prediction accuracy of 10-fold cross-validation (10-CV). The tuning parameters of the CancerSig were independent for the each cancer type. The detailed description of parameters and IBCGA algorithm can refer to the studies.10,12 The main steps of IBCGA for identifying a signature of m miRNAs and the SVM classifier are as follows.
-
Step 1
Randomly generate a population of Npop individuals. In this work, Npop = 50, Gmax = 60, rstart = 10, rend = 50, r = rstart.
-
Step 2
Evaluate the fitness value of all individuals using the fitness function.
-
Step 3
Use a tournament selection method that selects the winner from two randomly selected individuals to generate a mating pool.
-
Step 4
Select two parents from the mating pool to perform an orthogonal array crossover operation.
-
Step 5
Apply a conventional mutation operator to the randomly selected individuals in the new population. To prevent the highest fitness value from deteriorating, mutation is not applied to the best individuals.
-
Step 6
If the stopping condition of Gmax generation is satisfied, the best individual is the solution Sr. Otherwise, go to step 2.
-
Step 7
If r < rend, randomly change one bit in the binary genes for each individual from 0 to 1; increase the number r by one, and go to step 2. Otherwise, output the solution Sm with m miRNAs as a signature where Sm is the most accurate solution among the Sr solutions and stop the algorithm.
Robust signature
The selection of a robust signature is necessary when using the nondeterministic algorithm IBCGA in which the solutions of multiple runs are not always the same. For each cancer type, the robust signature among R = 30 solutions Sm had the largest appearance score using the following procedure.
-
Step 1
Perform R independent runs of IBCGA to obtain R signatures. There are features (or miRNAs) in the t-th signatures, t = 1, …, R.
-
Step 2
The appearance score of a signature is calculated as follows:
-
a)
Calculate the feature frequency score f(p) for each miRNA p that ever appears in the R signatures.
-
b)
Calculate the score Ft, t = 1, …, R where Sti is the i-th miRNA in the t-th signature:
| (Equation 3) |
-
Step 3
Output the t-th signature with the largest appearance score Ft as the robust signature.
KEGG pathway and GO annotation analysis
We utilized the DIANA-miRPath web-based server to analyze the miRNA signatures for downstream biological pathway analyses using KEGG and GO.23 Plausible miRNA targets identified from the DIANA-TarBase/microT-CDs algorithm analyzed via hypergeometric distribution method using Fisher’s exact test for enrichment analysis. A p value of <0.05 was used as the threshold to describe statistical significance. To estimate the specificity of the results, we performed another pathway analysis for all identified miRNA signatures across the 15 cancer types. The GO annotations of the miRNA signatures were analyzed to identify miRNAs belonging to the specific GO categories of biological process, cellular components, and molecular function.
Results
Identification of miRNA signatures across cancers
To identify the miRNA signatures associated with early and advanced stages across cancers, we obtained miRNA expression profiles from the clinical samples of 6,578 patients with cancers from TCGA. The TCGA network contains clinical and molecular information on 33 cancer types from tumor samples collected from 68 primary sites. After preprocessing the data, which included removal of duplicate samples, samples without stage information, and miRNAs that were not expressed in more than 80% of samples, the final dataset consisted of 4,667 clinical samples with cancer stage information. The clinical samples, miRNAs, and cancer types used in this study are summarized in Table 1.
To distinguish between early-stage and advanced-stage cancers via miRNA expression profiles, we proposed an evolutionary learning method, CancerSig, based on the feature selection algorithm IBCGA and SVM. Identifying a minimum number of features from a large number of candidate features while maximizing the prediction performance is a bi-objective combinatorial optimization problem that is effectively solved by IBCGA.20,24 Because IBCGA is a nondeterministic method, we performed 30 independent runs and measured the appearance frequency of selected features to select one robust feature set of miRNAs as an miRNA signature for each of 15 cancers.10 The schematic diagram of the CancerSig method is depicted in Figure 1.
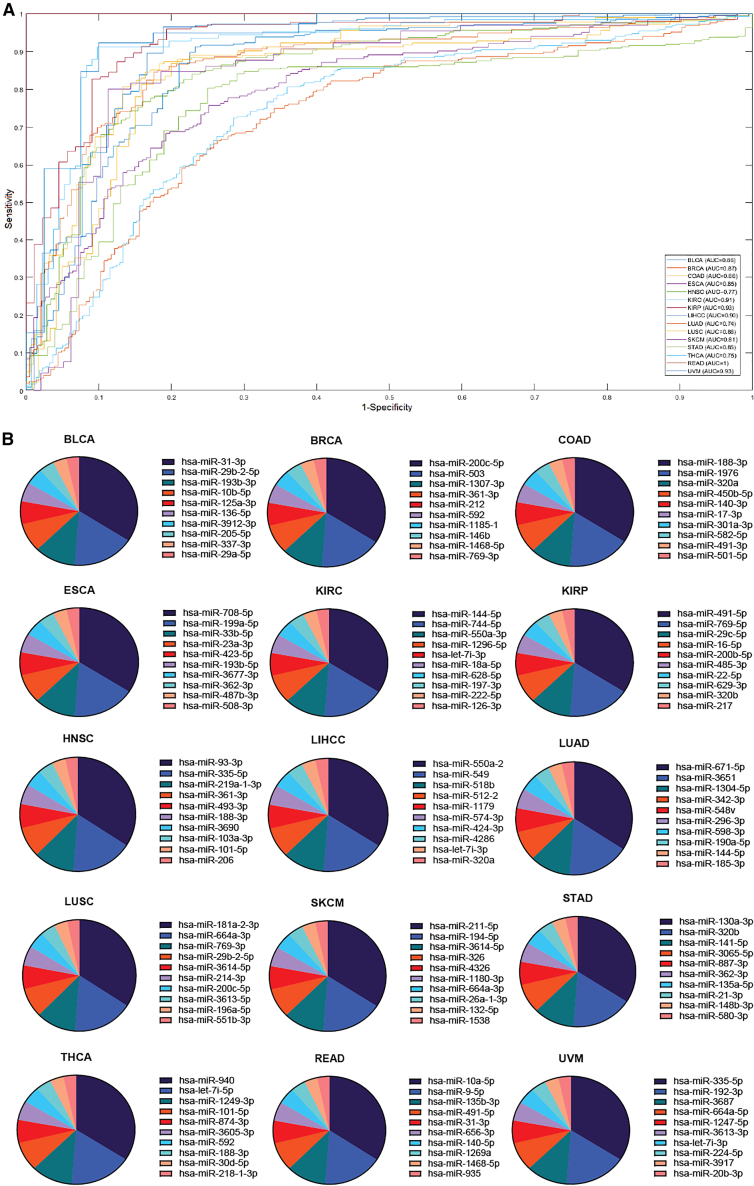
The prediction performance of CancerSig across 15 cancer types is shown in Table 2. CancerSig identified 15 signatures with an average size of 21.93 miRNAs from the profiles with an average of 474.5 miRNAs. CancerSig achieved a mean performance: 10-CV accuracy, sensitivity, specificity, Matthews correlation coefficient (MCC), and area under the receiver operating characteristic (ROC) area under the curve (AUC) of 84.27% ± 6.31%, 0.81 ± 0.12, 0.80 ± 0.10, 0.67 ± 0.11, and 0.80 ± 0.06, respectively. The prediction performance was evaluated using ROC curves and AUC in the range of 0.70–0.95 for 15 cancers, as shown in Figure 2A.
Table 2.
Prediction performance of CancerSig across 15 cancers
| Dataset | miRNA signature | 10-CV accuracy | Sensitivity | Specificity | MCC | AUC | |
|---|---|---|---|---|---|---|---|
| 1 | BLCA | 35 | 84.40 ± 1.27 | 0.64 ± 0.04 | 0.93 ± 0.01 | 0.65 ± 0.02 | 0.82 ± 0.01 |
| 2 | BRCA | 34 | 80.38 ± 1.55 | 0.79 ± 2.7 | 0.81 ± 2.26 | 0.60 ± 0.03 | 0.81 ± 0.02 |
| 3 | COAD | 21 | 86.67 ± 2.40 | 0.89 ± 0.02 | 0.82 ± 0.03 | 0.73 ± 0.04 | 0.81 ± 0.02 |
| 4 | ESCA | 22 | 87.97 ± 2.35 | 0.93 ± 0.02 | 0.78 ± 0.06 | 0.75 ± 0.04 | 0.78 ± 0.03 |
| 5 | HNSC | 20 | 85.20 ± 1.4 | 0.52 ± 0.06 | 0.95 ± 0.01 | 0.57 ± 0.05 | 0.71 ± 0.03 |
| 6 | KIRC | 18 | 87.14 ± 1.76 | 0.85 ± 0.02 | 0.87 ± 0.02 | 0.75 ± 0.03 | 0.87 ± 0.01 |
| 7 | KIRP | 12 | 89.43 ± 2.01 | 0.96 ± 0.01 | 0.73 ± 0.06 | 0.76 ± 0.04 | 0.87 ± 0.03 |
| 8 | LIHCC | 23 | 89.56 ± 1.27 | 0.94 ± 0.01 | 0.73 ± 0.03 | 0.71 ± 0.03 | 0.86 ± 0.02 |
| 9 | LUAD | 29 | 74.29 ± 1.33 | 0.80 ± 0.02 | 0.65 ± 0.03 | 0.53 ± 0.02 | 0.7 ± 0.01 |
| 10 | LUSC | 18 | 79.11 ± 2.28 | 0.74 ± 0.06 | 0.81 ± 0.02 | 0.61 ± 0.03 | 0.80 ± 0.02 |
| 11 | SKCM | 27 | 78.71 ± 2.47 | 0.79 ± 0.03 | 0.76 ± 0.04 | 0.60 ± 0.03 | 0.75 ± 0.02 |
| 12 | STAD | 20 | 77.03 ± 1.9 | 0.70 ± 0.03 | 0.79 ± 0.13 | 0.58 ± 0.03 | 0.78 ± 0.03 |
| 13 | THCA | 26 | 76.37 ± 1.20 | 0.86 ± 0.02 | 0.61 ± 0.04 | 0.55 ± 0.02 | 0.71 ± 0.01 |
| 14 | READ | 11 | 94.57 ± 2.8 | 0.90 ± 0.05 | 0.97 ± 0.02 | 0.88 ± 0.05 | 0.95 ± 0.03 |
| 15 | UVM | 13 | 93.33 ± 3.5 | 0.93 ± 0.04 | 0.90 ± 0.13 | 0.86 ± 0.06 | 0.85 ± 0.03 |
| Mean ± SD | 21.93 ± 7.30 | 84.27 ± 6.31 | 0.81 ± 0.12 | 0.80 ± 0.10 | 0.67 ± 0.11 | 0.80 ± 0.06 |
10-CV, 10-fold cross-validations; MCC, Matthews correlation coefficient; AUC, area under the ROC curve; SD, standard deviation.
Figure 2.
CancerSig prediction performance across cancers
(A) Evaluating the prediction performance of CancerSig using receiver operating characteristic (ROC) across 15 cancers. CancerSig obtained a mean area under the curve (AUC) of 0.80 across all cancers.
(B) Ranking of the relative miRNAs within the signature using MED analysis.
We compared the prediction performance of CancerSig with various machine learning algorithms using the same number of features based on feature importance. LightGBM25 achieved a mean performance: 10-CV accuracy, sensitivity, specificity, and AUC of 72.22 ± 0.04, 0.83 ± 0.06, 0.51 ± 0.14, and 0.72 ± 0.06, respectively, while distinguishing early and advanced stages of BLCA. XGBoost26 achieved a mean 10-CV accuracy, sensitivity, specificity, and AUC of 72.70 ± 0.06, 0.88 ± 0.06, 0.41 ± 0.16, and 0.73 ± 0.05, respectively. Random Forest27 achieved a mean 10-CV accuracy, sensitivity, specificity, and AUC of 70.98 ± 0.05, 0.77 ± 0.07, 0.59 ± 0.16, and 0.74 ± 0.05, respectively. CatBoost28 achieved a mean 10-CV accuracy, sensitivity, specificity, and AUC of 73.93 ± 0.06, 0.91 ± 0.07, 0.39 ± 0.12, and 0.74 ± 0.05, respectively. Extra Trees achieved a mean 10-CV accuracy, sensitivity, specificity, and AUC of 72.72 ± 0.04, 0.78 ± 0.07, 0.63 ± 0.11, and 0.74 ± 0.06, respectively. CancerSig achieved a mean 10-CV accuracy, sensitivity, specificity, and AUC of 84.40 ± 1.27, 0.64 ± 0.04, 0.93 ± 0.01, and 0.82 ± 0.01, respectively, shown in Table S1. The prediction performance of CancerSig is better than other machine learning methods in predicting cancer stage of BLCA. In addition, the prediction performance of the optimizing technique in distinguishing breast invasive carcinoma (BRCA) and liver hepatocellular carcinoma (LIHCC) was compared with other machine learning methods in our previous studies.10,29
A panel of miRNAs across 15 cancers
A panel is designed as a compact set of informative miRNAs obtained from the 15 miRNA signatures, which has 329 miRNAs in total. After removing duplication, the union has 242 informative miRNAs in the designed cancer-stage panel. The panel can predict the cancer stage across 15 cancer types. In predicting the stage of a specific cancer, the same prediction model with the corresponding signature in the panel can be used. Therefore, the prediction performance using the panel of miRNAs for predicting the 15 cancers achieved 10-CV accuracy, sensitivity, specificity, MCC, and AUCs of 84.27% ± 6.31%, 0.81 ± 0.12, 0.80 ± 0.10, 0.67 ± 0.11, and 0.80 ± 0.06, respectively, as shown in Table 2.
Prioritization of the miRNA signatures
Each cancer type had an average of about 22 miRNAs in a signature. To rank the miRNAs of a signature according to the degree of contribution to prediction performance, a main effect difference (MED) analysis8,30 was performed. A higher MED score represents a greater contribution of the specific miRNA to the stage prediction. The miRNAs with the greatest predictive ability can be ranked highest in the signature. The identified miRNA signatures, ranks of their miRNAs, and corresponding MED scores are listed in Tables S2.1–S2.15. The top 10 ranked miRNAs across 15 cancers are shown in Figure 2B.
Prevalent miRNAs across cancers
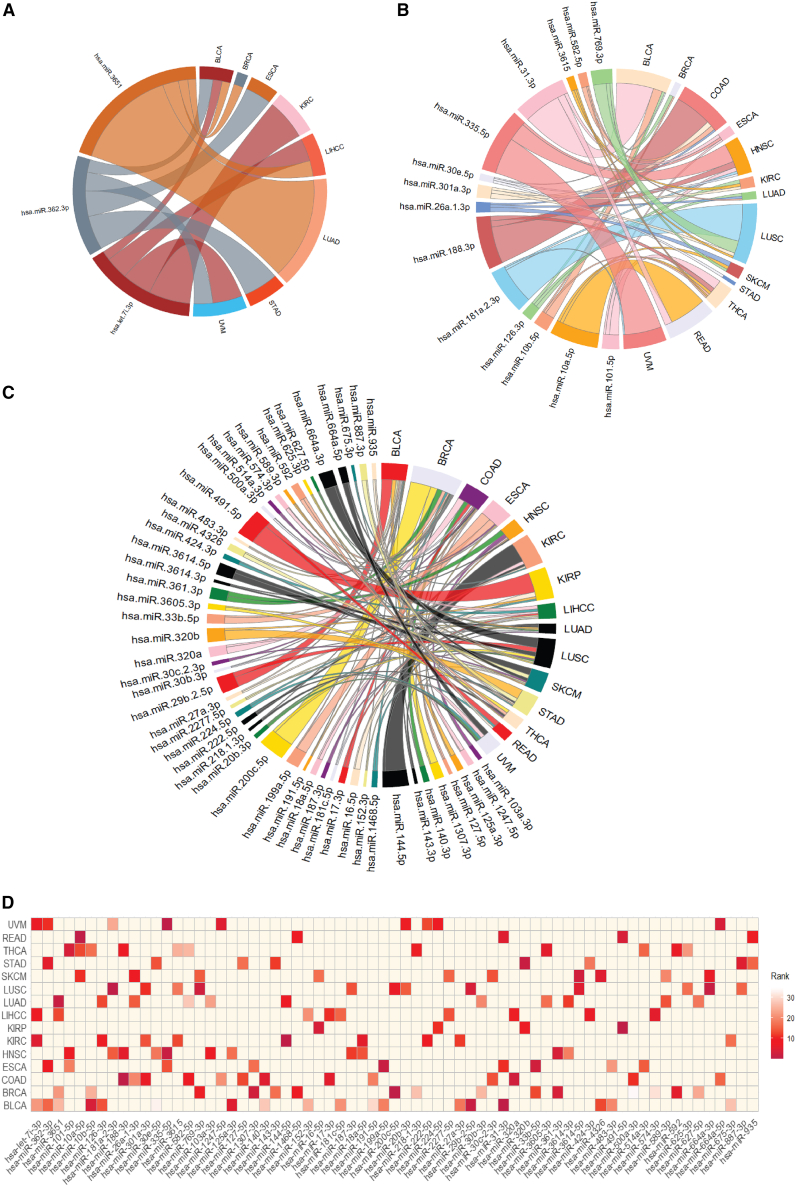
Next, we determined if there are any similarities among the miRNAs of each signature in addition to distinguishing stages across the 15 cancers. In this context, we identified some common miRNAs that were found to be expressed in more than one cancer. Three miRNAs, hsa-let-7i-3p, hsa-miR-362-3p, and hsa-miR-3651, contributed more toward stage prediction across eight cancer types (BLCA, BRCA, esophageal carcinoma [ESCA], kidney renal clear cell carcinoma [KIRC], LIHCC, lung adenocarcinoma [LUAD], stomach adenocarcinoma [STAD], and uveal melanoma [UVM]) than any other miRNAs. According to the MED analysis, hsa-let-7i-3p ranked 5th, 7th, 9th, and 22nd in predicting cancer stage in KIRC, UVM, LIHCC, and BLCA, respectively; hsa-miR-362-3p ranked 6th, 8th, 11th, and 16th in STAD, ESCA, UVM, and BLCA, respectively; and hsa-miR-3651 ranked 2nd, 13th, 22nd, and 26th in LUAD, LIHCC, BRCA, and BLCA, respectively. These miRNAs were annotated with miRBase accession numbers and used in subsequent pathway analyses.
The three target miRNAs were first analyzed via prediction interaction networks supported by Cytoscape v.3.731 to explore the proposed target gene interactions. From the three miRNA target prediction databases, miRTarBase, MicroCosm, and TargetScan, 1,839 predicted miRNA-target interactions were identified. The predicted miRNA-gene target network is shown in Figure S1. In addition to the three primary miRNAs, 64 additional miRNAs were identified in the signatures where each was involved in more than one cancer. Among the 64 miRNAs, each of 14 miRNAs was involved in 3 cancers while each of the remaining 50 miRNAs was involved in 2 cancers. These prevalent miRNAs and their contribution in predicting the stage across cancers are depicted in Figures 3A–3C.
Figure 3.
The predictive ability of miRNAs as a biomarker for cancer stage across cancer types
(A) Three signature miRNAs and their contributions to stage prediction across eight cancer types. Each miRNA contributed to at least four cancers. The size of the line is proportional to the percent contribution toward the stage prediction.
(B) Fourteen miRNAs contributed across cancers, and each miRNA contributed to at least three cancers.
(C) Fifty miRNAs contributed to at least two cancers.
(D) Heatmap showing 67 miRNAs and their ranks based on their predictive ability across 15 cancer types.
To confirm that the expression levels of the identified target miRNAs differ significantly between tumor and non-tumor samples for each cancer type, we compared the relative expression difference of the three miRNAs, hsa-let-7i-3p, hsa-miR-362-3p, and hsa-miR-3651. Significant differences in expression levels between tumor samples and normal samples for all miRNAs analyzed were reported for BRCA, lung squamous cell carcinoma (LUSC), and STAD. Hsa-let-7i-3p and hsa-miR-362-3p were significantly expressed in BLCA, KIRC, LIHCC, and LUAD, while hsa-miR-362-3p and hsa-miR-3651 had significant differences in head and neck squamous cell carcinoma. A statistically significant difference in expression level for kidney renal papillary cell carcinoma (KIRP) was only detected for hsa-miR-362-3p; similarly, significant differences in expression for hsa-miR-3651 were only noted for ESCA and thyroid carcinoma. The relative expression levels of these miRNAs and corresponding comparisons are shown in Table S3.
As the MED analysis, the top-ranked miRNAs are potential predictors of cancer stage. However, some miRNAs had low ranks in some cancers yet high ranks in other cancer types. For instance, hsa-let-7i-3p ranked 22nd in BLCA but ranked 5th in KIRC meaning that its contribution to cancer stage was higher for KIRC. This analysis revealed a panel of 242 miRNAs that are associated with the cancer stage in more than 1 cancer. The heatmap of the most prevalent miRNA rankings across the 15 cancer types is depicted in Figure 3D.
Co-expression analysis of the miRNA signatures
Though IBCGA identified critical miRNA signatures for cancer stage prediction, the algorithm might exclude some informative miRNAs from the signatures to select a small set of candidate miRNAs to enhance prediction performance. To ensure a robust set of miRNAs selected by IBCGA, co-expression analysis was performed via series of correlational analyses. First, correlation coefficient (R) between the miRNAs in the signatures was measured and then the coefficients between all the miRNAs (an average 474.46) and individual miRNAs in the signature for each cancer type were measured. The miRNAs pairs with R ≥ 0.80 were considered for further analysis. There were 154 miRNA pairs that co-expressed with the miRNAs of 15 cancer stage-specific miRNA signatures. Of the signatures analyzed by cancer type, the miRNA signature for LUSC did not have co-expressed miRNAs with R ≥ 0.8. The correlation heatmap of miRNAs in each of the 15 signatures is shown in Figure S2, and the correlation coefficients of the154 miRNA pairs for the 15 miRNA signatures are listed in Table S4.
Significance of the identified miRNA signatures in cancers
We evaluated the significance of the identified miRNAs for each cancer type based on experimentally validated literature and, of these miRNAs, only the top 10 ranked miRNAs of each signature were considered, as shown in Tables S5–S19. Among the identified 15 cancer stage-specific miRNA signatures, most of have experimentally validated evidence to support their dysregulation and potential role in various cancers. However, of the 15 miRNA signatures, the role of 34 miRNAs was not reported in the earlier literature leading us to believe that these 34 miRNAs are novel biomarkers for predicting the cancer stage (listed in Table S20). The roles of these miRNAs in cancer stage detection need to be validated to further determine their significance.
To confirm that the selected miRNAs had differing expression levels by stage within each cancer, we measured the expression differences of the identified top 10 ranked miRNAs between the early- and advanced-stage groups across 15 cancers, as shown in Figure S3. All of the identified miRNA signatures are significantly different between early- and late-stage cancers, which supports the use of these miRNAs in predicting early-stage cancers.
Biological relevance of miRNA signatures across cancers
Employing KEGG analysis, we found that each miRNA signature was involved in several cancers and signaling pathways, including transforming growth factor β, hippo, and the thyroid hormone signaling pathways as well as signaling pathways related to axon guidance (see Table S21). Of the cancer types analyzed, the proteoglycans in cancer pathway was observed in all 15 cancer types, which is consistent with the role of proteoglycans and extracellular matrix components in cancer development and progression.32,33,34,35,36,37,38,39,40,41 In addition, the second KEGG pathway commonly found in 14 cancers was “Signaling pathways regulating the pluripotency of stem cells,” which has a wide range of applications in regenerative medicine and significance in cancer.42 A summary of the miRNA signatures that appeared in more than five cancer types and were identified as having potential involvement in biological pathways by KEGG pathway analysis is provided in Table 3. The identified miRNA signatures enriched in KEGG pathways across the 15 cancer types are shown in Figures S4.1–S4.15.
Table 3.
KEGG pathways commonly appearing in more than five cancers
| KEGG pathways | No. of cancers |
|---|---|
| Proteoglycans in cancer | 15 |
| Signaling pathways regulating pluripotency of stem cells | 14 |
| Beta signaling pathway | 13 |
| Axon guidance | 11 |
| Glioma | 10 |
| Hippo signaling pathway | 10 |
| Renal cell carcinoma | 10 |
| ErbB signaling pathway | 9 |
| FoxO signaling pathway | 9 |
| Pathways in cancer | 9 |
| Prion diseases | 9 |
| Rap1 signaling pathway | 9 |
| Long-term depression | 8 |
| Ras signaling pathway | 8 |
| Circadian rhythm | 7 |
| Focal adhesion | 7 |
| Thyroid hormone signaling pathway | 7 |
| Adherens junction | 6 |
| Colorectal cancer | 6 |
| ECM-receptor interaction | 6 |
| Estrogen signaling pathway | 6 |
| Melanoma | 6 |
| Morphine addiction | 6 |
| Adrenergic signaling in cardio myocytes | 5 |
| Choline metabolism in cancer | 5 |
| Fatty acid biosynthesis | 5 |
| Mucin type O-glycan biosynthesis | 5 |
| Oxytocin signaling pathway | 5 |
| Phosphatidylinositol signaling system | 5 |
| Prolactin signaling pathway | 5 |
| Prostate cancer | 5 |
| Wnt signaling pathway | 5 |
To support the biological validity of our findings, we performed GO enrichment analysis and identified GO terms that were enriched by the identified miRNA signatures. Among the 15 cancer types analyzed, 9 GO terms consistently appeared. The most frequent and significant GO terms that appeared were biosynthetic process, cellular component assembly, cellular nitrogen compound metabolic process, cellular protein modification process, epidermal growth factor receptor signaling pathway, Fc-epsilon receptor signaling pathway, gene expression, neurotrophin TRK receptor signaling pathway, transcription, and DNA template synthesis, listed in Tables 4 and S22. The identified miRNA signatures enriched in the various GO categories across 15 cancers are shown in Figures S5.1–S5.15.
Table 4.
GO category frequency in more than five cancers
| GO category | No. of cancers |
|---|---|
| Biosynthetic process | 15 |
| Cellular component assembly | 15 |
| Cellular nitrogen compound metabolic process | 15 |
| Cellular protein modification process | 15 |
| Epidermal growth factor receptor signaling pathway | 15 |
| Fc-epsilon receptor signaling pathway | 15 |
| Gene expression | 15 |
| Neurotrophin TRK receptor signaling pathway | 15 |
| Transcription, DNA template | 15 |
| Catabolic process | 14 |
| Cytosol | 14 |
| Enzyme binding | 14 |
| Ion binding | 14 |
| Nucleic acid binding transcription factor activity | 14 |
| Nucleoplasm | 14 |
| Organelle | 14 |
| Protein binding transcription factor activity | 14 |
| Protein complex | 14 |
| Small-molecule metabolic process | 14 |
| Blood coagulation | 13 |
| Cytoskeletal protein binding | 13 |
| Symbiosis, encompassing mutualism through parasitism | 13 |
| Viral process | 13 |
| Macromolecular complex assembly | 12 |
| Fibroblast growth factor receptor signaling pathway | 11 |
| Response to stress | 11 |
| Cell death | 10 |
| Phosphatidylinositol-mediated signaling | 10 |
| Nucleobase-containing compound catabolic process | 9 |
| Synaptic transmission | 9 |
| Mitotic cell cycle | 7 |
| Protein complex assembly | 7 |
| Enzyme regulator activity | 6 |
| Transcription initiation from RNA polymerase II promoter | 6 |
Discussion
Due to the different types of cancers and the frequent emergence of cancer symptoms at advanced stages, it is challenging to detect cancer at an early stage. Identifying broad and reliable biomarkers for cancer detection and prediction in the early stage may provide timely treatment for patients.
In this study, we describe the development and preliminary predictive ability of cancer stage prediction method CancerSig using SVM combined with an optimal feature selection algorithm IBCGA to identify miRNA signatures associated with staging across various cancers. CancerSig identified 15 cancer stage-specific miRNA signatures for 15 different cancer types that are associated with the stage of patients with cancers. There were 242 miRNAs that showed promising predictive ability as a panel for stage detection across the 15 cancer types. Rankings of miRNAs via MED analysis highlighted the contribution of each miRNA to stage prediction. Of these, three miRNAs, hsa-let-7i-3p, hsa-miR-362-3p, and hsa-miR-3651, consistently had statistically significantly different expression levels between tumor and non-tumor samples and by cancer stage within the 15 cancer types analyzed. The biological plausibility of these miRNAs as reliable predictors of cancer development and progression are supported by the following evidences in the literature. Hu et al., Cai et al., and Zhao et al. reported that the hsa-let-7 family of miRNAs are dysregulated in several cancer types, such as, breast,43 ovarian,44 and non-small cell lung cancer.45 Hsa-miR-362-3p and hsa-miR-3651 are also found to be regulated in different cancer types.46,47
Analysis of prevalent miRNAs within the total set of 242 miRNAs highlighted 67 miRNAs that contributed to 2 or more cancers. Out of the 67 miRNAs, 3 miRNAs each contributed to four cancers, 14 miRNAs each contributed to 3 cancers, and 50 miRNAs each contributed to 2 cancers. The 15 miRNA signatures were all significantly involved in various signaling pathways, extracellular matrix-associated signaling, and stem cell pluripotency according to KEGG pathway analysis and GO term assignment post-enrichment analysis. One hundred and fifty-four miRNAs co-expressed with the miRNA signatures across cancers were associated with cancer progression and early-stage detection. Of the miRNAs detected within the signature and associated with cancer stage, 34 miRNAs that we describe here have not been reported before. Further research of these miRNAs may provide new avenues for therapeutic and diagnostic test development.
Across all cancer types, miRNA signatures were enriched for proteins associated with proteoglycans. Proteoglycans are macromolecules and the major component of the extracellular matrix. They act as co-receptors for enhancing proliferative signaling and tumor growth.34 Notably, the altered expression of proteoglycans correlates with prognosis in various malignant neoplasms.33,35 In addition, proteoglycan-dependent pathways are involved in promoting metastasis and cell motility in breast cancer.36,37 The miRNAs regulate enzymes that are directly linked to proteoglycan function and are involved in tumor progression.38,39 The aberrant expression of miRNAs affects the expression patterns of laminins, proteoglycans, and proteases in the tumor microenvironment40; consequently, cell adhesion, migration, and apoptosis and cancer stem cell properties are affected.41 In addition, multiple roles of miRNAs in pluripotency have been investigated, including but not limited to cell fate during embryogenesis48 and the regulation of stem cells.49 The upregulation of miR-495 was observed in breast cancer stem cells,50 and miR-34a is downregulated and regulates cancer stem cells in prostate cancer.51 The finding suggests that a specific miRNA signature regulates proteoglycans in the tumor microenvironment and stem cell pluripotency, which may have a profound impact on early-stage cancer detection.
High performance of CancerSig arises mainly from an optimal feature selection algorithm IBCGA incorporated with an SVM classifier. IBCGA is effective at solving bi-objective combinatorial optimization problems and has been proven to be efficient at identifying suitable biomarkers in various cancers.8,10,12,13,52 CancerSig achieved a promising accuracy while predicting the cancer stage across 15 different cancer types; and obtained a mean performance of 10-CV accuracy, sensitivity, specificity, MCC, AUC of 84.27 ± 6.31, 0.81 ± 0.12, 0.80 ± 0.10, 0.67 ± 0.11, and 0.80 ± 0.06, respectively. The limitation of the current method is using the TCGA data alone for the experiments due to the availability of similar extraction methods for miRNA expression profiling and clinical information. However, CancerSig showed better performance on all cancer types irrespective of the data size. This method can be customized based on the availability of miRNA expression data and clinical samples.
In conclusion, identification of the novel miRNA signatures via CancerSig may serve as the basis for predicting the development and stage of various types of cancer. Use of this method may aid in early identification of cancer and cancer stage, which would facilitate clinician decision making for treatment plans and provide patients with timely treatment for cancer. The designed novel panel of miRNA signatures across cancers would guide the development of stage detection chips and miRNA-based target therapies to treat cancer.
Acknowledgments
This work was supported in part by the Marshfield Clinic Research Institute, Marshfield, WI, United States. The Ministry of Science and Technology ROC under contract numbers MOST 111-2740-B-400-002- and 110-2221-E-A49-099–MY3, and was financially supported by the “Center For Intelligent Drug Systems and Smart Bio-devices (IDS2B)” from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors also acknowledge Emily Andreae, PhD, from the Marshfield Clinic Research Institute for manuscript editing assistance.
Author contributions
S.Y.S. and S.-Y.H. designed the system and carried out the detailed study. S.-Y.S. participated in the design of the system and implemented programs. M.-J.T., S.K.S., S.-Y.H., and S.-Y.S. participated in analysis and discussed the results. All authors participated in the manuscript preparation and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2023.100190.
Contributor Information
Srinivasulu Yerukala Sathipati, Email: sathipathi.srinivasulu@marshfieldclinic.org.
Shinn-Ying Ho, Email: syho@nctu.edu.tw.
Web resources
CancerSig, https://github.com/mingjutsai/CancerSig
Supplemental information
Data and code availability
All the data used in this analysis can be found in the TCGA data portal: https://portal.gdc.cancer.gov/ CancerSig is available at Github: https://github.com/mingjutsai/CancerSig.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA. Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.van Schooneveld E., Wildiers H., Vergote I., Vermeulen P.B., Dirix L.Y., Van Laere S.J. Dysregulation of microRNAs in breast cancer and their potential role as prognostic and predictive biomarkers in patient management. Breast Cancer Res. 2015;17:21. doi: 10.1186/s13058-015-0526-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kozomara A., Birgaoanu M., Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peng Y., Croce C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016;1 doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Croce C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 7.Kim T.M., Huang W., Park R., Park P.J., Johnson M.D. A developmental taxonomy of glioblastoma defined and maintained by MicroRNAs. Cancer Res. 2011;71:3387–3399. doi: 10.1158/0008-5472.CAN-10-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yerukala Sathipati S., Ho S.-Y. Identifying the miRNA signature associated with survival time in patients with lung adenocarcinoma using miRNA expression profiles. Sci. Rep. 2017;7:7507. doi: 10.1038/s41598-017-07739-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xing L., Todd N.W., Yu L., Fang H., Jiang F. Early detection of squamous cell lung cancer in sputum by a panel of microRNA markers. Mod. Pathol. 2010;23:1157–1164. doi: 10.1038/modpathol.2010.111. [DOI] [PubMed] [Google Scholar]
- 10.Yerukala Sathipati S., Ho S.-Y. Identifying a miRNA signature for predicting the stage of breast cancer. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-34604-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimomura A., Shiino S., Kawauchi J., Takizawa S., Sakamoto H., Matsuzaki J., Ono M., Takeshita F., Niida S., Shimizu C., et al. Novel combination of serum microRNA for detecting breast cancer in the early stage. Cancer Sci. 2016;107:326–334. doi: 10.1111/cas.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yerukala Sathipati S., Sahu D., Huang H.-C., Lin Y., Ho S.-Y. Identification and characterization of the lncRNA signature associated with overall survival in patients with neuroblastoma. Sci. Rep. 2019;9:5125. doi: 10.1038/s41598-019-41553-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yerukala Sathipati S., Huang H.L., Ho S.Y. Estimating survival time of patients with glioblastoma multiforme and characterization of the identified microRNA signatures. BMC Genom. 2016;17:1022. doi: 10.1186/s12864-016-3321-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sathipati S.Y., Ho S.Y. Identification of the miRNA signature associated with survival in patients with ovarian cancer. Aging (Albany NY) 2021;13:12660–12690. doi: 10.18632/aging.202940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho S.-Y., Chen J.-H., Huang M.-H. Inheritable genetic algorithm for biobjective 0/1 combinatorial optimization problems and its applications. IEEE Trans. Syst. Man Cybern. B Cybern. 2004;34:609–620. doi: 10.1109/tsmcb.2003.817090. [DOI] [PubMed] [Google Scholar]
- 16.Vapnik V.N. An overview of statistical learning theory. IEEE Trans. Neural Netw. 1999;10:988–999. doi: 10.1109/72.788640. [DOI] [PubMed] [Google Scholar]
- 17.Noble W.S. What is a support vector machine? Nat. Biotechnol. 2006;24:1565–1567. doi: 10.1038/nbt1206-1565. [DOI] [PubMed] [Google Scholar]
- 18.Ho S.-Y., Shu L.-S., Chen J.-H. Intelligent evolutionary algorithms for large parameter optimization problems. IEEE Trans. Evol. Comput. 2004;8:522–541. [Google Scholar]
- 19.Yerukala Sathipati S., Tsai M.J., Shukla S.K., Ho S.Y., Liu Y., Beheshti A. MicroRNA signature for estimating the survival time in patients with bladder urothelial carcinoma. Sci. Rep. 2022;12:4141. doi: 10.1038/s41598-022-08082-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J.-R., Huang W.-L., Tsai M.-J., Hsu K.-T., Huang H.-L., Ho S.-Y. ESA-UbiSite: accurate prediction of human ubiquitination sites by identifying a set of effective negatives. Bioinformatics. 2017;33:661–668. doi: 10.1093/bioinformatics/btw701. [DOI] [PubMed] [Google Scholar]
- 21.Tsai M.-J., Wang J.-R., Ho S.-J., Shu L.-S., Huang W.-L., Ho S.-Y. GREMA: modelling of emulated gene regulatory networks with confidence levels based on evolutionary intelligence to cope with the underdetermined problem. Bioinformatics. 2020;36:3833–3840. doi: 10.1093/bioinformatics/btaa267. [DOI] [PubMed] [Google Scholar]
- 22.Chang C.-C., Lin C.-J. LIBSVM: a library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011;2:1–27. [Google Scholar]
- 23.Vlachos I.S., Zagganas K., Paraskevopoulou M.D., Georgakilas G., Karagkouni D., Vergoulis T., Dalamagas T., Hatzigeorgiou A.G. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43:W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tung C.-W., Ho S.-Y. POPI: predicting immunogenicity of MHC class I binding peptides by mining informative physicochemical properties. Bioinformatics. 2007;23:942–949. doi: 10.1093/bioinformatics/btm061. [DOI] [PubMed] [Google Scholar]
- 25.Ke G., Meng Q., Finley T., Wang T., Chen W., Ma W., Ye Q., Liu T.-Y. NIPS. 2017. LightGBM: a highly efficient gradient boosting decision tree. [Google Scholar]
- 26.Chen T., Guestrin C. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. Association for Computing Machinery); 2016. XGBoost: a scalable tree boosting system; pp. 785–794. [Google Scholar]
- 27.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 28.Dorogush A.V., Ershov V., Gulin A. CatBoost: gradient boosting with categorical features support. arXiv. 2018 doi: 10.48550/arXiv.1810.11363. Preprint at. [DOI] [Google Scholar]
- 29.Yerukala Sathipati S., Ho S.-Y. Novel miRNA signature for predicting the stage of hepatocellular carcinoma. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-71324-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tung C.W., Ho S.Y. Computational identification of ubiquitylation sites from protein sequences. BMC Bioinf. 2008;9:310. doi: 10.1186/1471-2105-9-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 33.Theocharis A.D., Skandalis S.S., Tzanakakis G.N., Karamanos N.K. Proteoglycans in health and disease: novel roles for proteoglycans in malignancy and their pharmacological targeting. FEBS J. 2010;277:3904–3923. doi: 10.1111/j.1742-4658.2010.07800.x. [DOI] [PubMed] [Google Scholar]
- 34.Barash U., Cohen-Kaplan V., Dowek I., Sanderson R.D., Ilan N., Vlodavsky I. Proteoglycans in health and disease: new concepts for heparanase function in tumor progression and metastasis. FEBS J. 2010;277:3890–3903. doi: 10.1111/j.1742-4658.2010.07799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sofeu Feugaing D.D., Götte M., Viola M. More than matrix: the multifaceted role of decorin in cancer. Eur. J. Cell Biol. 2013;92:1–11. doi: 10.1016/j.ejcb.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 36.Hassan H., Greve B., Pavao M.S.G., Kiesel L., Ibrahim S.A., Götte M. Syndecan-1 modulates beta-integrin-dependent and interleukin-6-dependent functions in breast cancer cell adhesion, migration, and resistance to irradiation. FEBS J. 2013;280:2216–2227. doi: 10.1111/febs.12111. [DOI] [PubMed] [Google Scholar]
- 37.Ibrahim S.A., Yip G.W., Stock C., Pan J.W., Neubauer C., Poeter M., Pupjalis D., Koo C.Y., Kelsch R., Schüle R., et al. Targeting of syndecan-1 by microRNA miR-10b promotes breast cancer cell motility and invasiveness via a Rho-GTPase- and E-cadherin-dependent mechanism. Int. J. Cancer. 2012;131:E884–E896. doi: 10.1002/ijc.27629. [DOI] [PubMed] [Google Scholar]
- 38.Götte M., Yip G.W. Heparanase, hyaluronan, and CD44 in cancers: a breast carcinoma perspective. Cancer Res. 2006;66:10233–10237. doi: 10.1158/0008-5472.CAN-06-1464. [DOI] [PubMed] [Google Scholar]
- 39.Ibrahim S.A., Hassan H., Götte M. MicroRNA regulation of proteoglycan function in cancer. FEBS J. 2014;281:5009–5022. doi: 10.1111/febs.13026. [DOI] [PubMed] [Google Scholar]
- 40.Götte M., Kovalszky I. Extracellular matrix functions in lung cancer. Matrix Biol. 2018;73:105–121. doi: 10.1016/j.matbio.2018.02.018. [DOI] [PubMed] [Google Scholar]
- 41.Asuthkar S., Velpula K.K., Nalla A.K., Gogineni V.R., Gondi C.S., Rao J.S. Irradiation-induced angiogenesis is associated with an MMP-9-miR-494-syndecan-1 regulatory loop in medulloblastoma cells. Oncogene. 2014;33:1922–1933. doi: 10.1038/onc.2013.151. [DOI] [PubMed] [Google Scholar]
- 42.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 43.Hu X., Guo J., Zheng L., Li C., Zheng T.M., Tanyi J.L., Liang S., Benedetto C., Mitidieri M., Katsaros D., et al. The heterochronic microRNA let-7 inhibits cell motility by regulating the genes in the actin cytoskeleton pathway in breast cancer. Mol. Cancer Res. 2013;11:240–250. doi: 10.1158/1541-7786.MCR-12-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cai J., Yang C., Yang Q., Ding H., Jia J., Guo J., Wang J., Wang Z. Deregulation of let-7e in epithelial ovarian cancer promotes the development of resistance to cisplatin. Oncogenesis. 2013;2:e75. doi: 10.1038/oncsis.2013.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao B., Han H., Chen J., Zhang Z., Li S., Fang F., Zheng Q., Ma Y., Zhang J., Wu N., Yang Y. MicroRNA let-7c inhibits migration and invasion of human non-small cell lung cancer by targeting ITGB3 and MAP4K3. Cancer Lett. 2014;342:43–51. doi: 10.1016/j.canlet.2013.08.030. [DOI] [PubMed] [Google Scholar]
- 46.Shen H., Li W., Tian Y., Xu P., Wang H., Zhang J., Li Y. Upregulation of miR-362-3p modulates proliferation and anchorage-independent growth by directly targeting Tob2 in hepatocellular carcinoma. J. Cell. Biochem. 2015;116:1563–1573. doi: 10.1002/jcb.25110. [DOI] [PubMed] [Google Scholar]
- 47.Pellatt D.F., Stevens J.R., Wolff R.K., Mullany L.E., Herrick J.S., Samowitz W., Slattery M.L. Expression profiles of miRNA subsets distinguish human colorectal carcinoma and normal colonic mucosa. Clin. Transl. Gastroenterol. 2016;7:e152. doi: 10.1038/ctg.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Houbaviy H.B., Murray M.F., Sharp P.A. Embryonic stem cell-specific MicroRNAs. Dev. Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 49.Li N., Long B., Han W., Yuan S., Wang K. microRNAs: important regulators of stem cells. Stem Cell Res. Ther. 2017;8:110. doi: 10.1186/s13287-017-0551-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hwang-Verslues W.W., Chang P.H., Wei P.C., Yang C.Y., Huang C.K., Kuo W.H., Shew J.Y., Chang K.J., Lee E.Y., Lee W.H. miR-495 is upregulated by E12/E47 in breast cancer stem cells, and promotes oncogenesis and hypoxia resistance via downregulation of E-cadherin and REDD1. Oncogene. 2011;30:2463–2474. doi: 10.1038/onc.2010.618. [DOI] [PubMed] [Google Scholar]
- 51.Kashat M., Azzouz L., Sarkar S.H., Kong D., Li Y., Sarkar F.H. Inactivation of AR and Notch-1 signaling by miR-34a attenuates prostate cancer aggressiveness. Am. J. Transl. Res. 2012;4:432–442. [PMC free article] [PubMed] [Google Scholar]
- 52.Yerukala Sathipati S., Tsai M.-J., Carter T., Allaire P., Shukla S.K., Beheshti A., Ho S.-Y. Survival estimation in patients with stomach and esophageal carcinoma using miRNA expression profiles. Comput. Struct. Biotechnol. J. 2022;20:4490–4500. doi: 10.1016/j.csbj.2022.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data used in this analysis can be found in the TCGA data portal: https://portal.gdc.cancer.gov/ CancerSig is available at Github: https://github.com/mingjutsai/CancerSig.