Summary
Background
CD8+ T cells equipped with a full arsenal of antiviral effector functions are critical for effective immune control of HIV-1. It has nonetheless remained unclear how best to elicit such potent cellular immune responses in the context of immunotherapy or vaccination. HIV-2 has been associated with milder disease manifestations and more commonly elicits functionally replete virus-specific CD8+ T cell responses compared with HIV-1. We aimed to learn from this immunological dichotomy and to develop informed strategies that could enhance the induction of robust CD8+ T cell responses against HIV-1.
Methods
We developed an unbiased in vitro system to compare the de novo induction of antigen-specific CD8+ T cell responses after exposure to HIV-1 or HIV-2. The functional properties of primed CD8+ T cells were assessed using flow cytometry and molecular analyses of gene transcription.
Findings
HIV-2 primed functionally optimal antigen-specific CD8+ T cells with enhanced survival properties more effectively than HIV-1. This superior induction process was dependent on type I interferons (IFNs) and could be mimicked via the adjuvant delivery of cyclic GMP-AMP (cGAMP), a known agonist of the stimulator of interferon genes (STING). CD8+ T cells elicited in the presence of cGAMP were polyfunctional and highly sensitive to antigen stimulation, even after priming from people living with HIV-1.
Interpretation
HIV-2 primes CD8+ T cells with potent antiviral functionality by activating the cyclic GMP-AMP synthase (cGAS)/STING pathway, which results in the production of type I IFNs. This process may be amenable to therapeutic development via the use of cGAMP or other STING agonists to bolster CD8+ T cell-mediated immunity against HIV-1.
Funding
This work was funded by INSERM, the Institut Curie, and the University of Bordeaux (Senior IdEx Chair) and by grants from Sidaction (17-1-AAE-11097, 17-1-FJC-11199, VIH2016126002, 20-2-AEQ-12822-2, and 22-2-AEQ-13411), the Agence Nationale de la Recherche sur le SIDA (ECTZ36691, ECTZ25472, ECTZ71745, and ECTZ118797), and the Fondation pour la Recherche Médicale (EQ U202103012774). D.A.P. was supported by a Wellcome Trust Senior Investigator Award (100326/Z/12/Z).
Keywords: CD8+ T cells, HIV-1, HIV-2, STING, Type I IFN
Research in context.
Evidence before this study
Natural control of viral replication is rare in people infected with HIV-1 but common in people infected with HIV-2. Many host and viral factors have been proposed to account for this dichotomy, including the generation and maintenance of qualitatively superior virus-specific CD8+ T cell responses in people infected with HIV-2. The mechanistic basis of this latter observation has nonetheless remained unclear, although it is known that HIV-2 activates the cyclic GMP-AMP synthase-stimulator of interferon genes (cGAS-STING) pathway in dendritic cells (DCs), resulting in the production of type I interferons (IFNs).
Added value of this study
To address this knowledge gap, we developed an unbiased in vitro system that allowed us to compare the de novo induction of antigen-specific CD8+ T cells after exposure to HIV-1 or HIV-2. We found that HIV-2 primed functionally potent antigen-specific CD8+ T cells with enhanced survival properties and greater memory potential more effectively than HIV-1. Our data also linked this phenomenon with activation of the cGAS-STING pathway and the production of type I IFNs in DCs. In line with these results, the enhanced priming efficacy associated with HIV-2-derived viral factors in vitro was largely recapitulated in the presence of the STING agonist cyclic GMP-AMP (cGAMP), even among unfractionated peripheral blood mononuclear cells obtained from people living with HIV-1.
Implications of all the available evidence
HIV-2 primes qualitatively and quantitatively superior antiviral CD8+ T cells as a consequence of direct activation of the cGAS-STING pathway and the induction of type I IFNs. This process can be mimicked via the adjuvant delivery of cGAMP, providing a rational framework for the development of therapeutic strategies that could suppress or even functionally cure HIV-1.
Introduction
CD8+ T cells are critical for effective immune control of human immunodeficiency virus type 1 (HIV-1). As a consequence, extensive studies have been performed to identify the key features of these cells that associate with antiviral efficacy, with the overall aim of informing vaccine design and the development of potentially curative interventions in people living with HIV-1 (PLWH). Early work demonstrated that HIV-1-specific CD8+ T cells were functionally suboptimal and poorly cytolytic in most PLWH.1,2 Subsequently, it was recognized that immune control of viral replication could be linked with the qualitative properties of HIV-1-specific CD8+ T cells,3,4 specifically the deployment of multiple effector functions in response to antigen engagement,5,6 expression of the transcription factor T-bet7 and various cytolytic proteins, such as granzyme B and perforin,8 and enhanced in vitro suppression of HIV-1.9 Many of these attributes have further been shown to depend on the sensitivity of antigen recognition, also referred to as functional avidity.10,11 However, it has remained unclear how best to elicit such optimally equipped virus-specific CD8+ T cells reproducibly, and this knowledge gap has limited the development of effective prophylactic and therapeutic approaches to combat HIV-1. It is notable in this context that effector functionality appears to be set early during infection. In line with this notion, the decline in plasma viremia after primary infection has been shown to coincide with the emergence of potent virus-specific CD8+ T cells, which help limit the viral reservoir in vivo.12,13 Antigen-driven priming events therefore likely shape the functional efficacy of virus-specific CD8+ T cells soon after exposure to HIV-1.
Natural control of HIV-1 is rare and typically confined to individuals with certain human leukocyte antigen (HLA) genotypes.14, 15, 16 Unlike HIV-1, which has spread globally and caused more than 36 million deaths since the start of the pandemic in the 1980s, HIV-2 has been associated with a relatively mild disease course and remains largely confined to West Africa.17 People infected with HIV-2 exhibit slower rates of CD4+ T cell loss18 and progression to AIDS19 and control viral replication more readily than people infected with HIV-1.20,21 A detailed understanding of the mechanisms that underlie this dichotomy could therefore inform the development of new therapeutic strategies to suppress or even functionally cure HIV-1.22 Previous studies have shown that infection with HIV-2 typically elicits virus-specific CD8+ T cells with potent effector functions and the capacity to suppress viral replication ex vivo.23, 24, 25 Moreover, viral factors likely contribute to this pattern of induction, given that natural control of HIV-2 is common and occurs in an HLA-independent manner.22
HIV-2 triggers potent innate antiviral responses in conventional dendritic cells (DCs)26 and also encodes the accessory protein Vpx, which degrades the dNTP phosphohydrolase and restriction factor SAM domain and HD domain-containing protein 1 (SAMHD1), facilitating the production of viral DNA in infected DCs.27,28 Cytosolic viral DNA subsequently activates cyclic GMP-AMP synthase (cGAS), resulting in the formation of cyclic GMP-AMP (cGAMP), a ligand for the adaptor protein stimulator of interferon genes (STING), which activates nuclear factor-κB (NF-κB) and drives the induction of type I interferons (IFNs).29 Moreover, the HIV-2 capsid binds strongly to non-POU domain-containing octamer-binding protein (NONO), a sensor found in DCs, enabling the recognition of viral cDNA by cGAS.30 Conversely, silent replication in myeloid cells can be facilitated by various capsid modifications found among pandemic strains of HIV-1, which act mechanistically to limit innate recognition by cGAS and tripartite motif-containing protein 5 (TRIM5).31
On the basis of these collective observations, we postulated that innate sensing of viral replication could directly influence the functionality of de novo CD8+ T cell responses, potentially explaining the differential immune control of HIV-1 versus HIV-2. Formal demonstration of this possibility was nonetheless complicated by several factors. In particular, there is only limited nucleotide sequence identity (∼50%) between the two viruses,32 and there are no known epitopes that are conserved between HIV-1 and HIV-2. Additional biases may arise in vivo. For example, antigen-presenting cell functions are likely heterogeneous among individuals, and repertoire differences can impact naive precursor frequencies, which almost certainly diversify further with amino acid substitutions in the corresponding epitopes derived from HIV-1 or HIV-2. It is also difficult to identify people undergoing primary infection with HIV-2, which is relatively rare and usually far less symptomatic than primary infection with HIV-1. To circumvent these drawbacks, we designed an unbiased in vitro approach, enabling direct comparisons of priming efficacy upon exposure to HIV-1 or HIV-2. We found that HIV-2 promoted the induction of antigen-specific CD8+ T cells more effectively in terms of quality and quantity than HIV-1. This observation prompted us to test the idea that HIV-1-specific effector memory CD8+ T cells with optimal antiviral functions could be primed by mimicking the HIV-2-associated innate immune response, characterized by direct activation of the NONO-cGAS-STING pathway in DCs. In line with this hypothesis, we found that adjuvant delivery of the STING agonist cGAMP facilitated the induction of potent CD8+ T cells in the context of infection with HIV-1, demonstrating a key therapeutic role for type I IFNs.
Methods
Cells
Peripheral blood mononuclear cells (PBMCs) for use in priming assays were obtained from healthy donors (age range, 24–59 years) via the Établissement Français du Sang (Paris, France) or from HIV-1+ donors receiving antiretroviral therapy (ART) (Table S1). Age/sex-matched donors were selected for experimental comparisons of healthy uninfected individuals versus PLWH (median age, 48 years; sex, all male). Cells were screened via flow cytometry for reactivity with anti-HLA-A2–FITC (clone BB7.2, BD Biosciences, RRID:AB_394130). PBMCs were isolated using Ficoll-Paque PLUS (Cytiva, CAT#17144002) and then cryopreserved in Roswell Park Memorial Institute (RPMI) 1640 medium (Thermo Fisher Scientific, CAT#61870010) containing 10% dimethyl sulfoxide (DMSO, Sigma–Aldrich, CAT#D2650) and 20% fetal bovine serum (FBS, Thermo Fisher Scientific, CAT#15377636). GHOST X4R5 (NIH AIDS Reagent Program, CAT#3942, RRID:CVCL_1E10) and 293FT cells (Thermo Fisher Scientific, CAT#R70007, RRID:CVCL_6911) were cultured in Dulbecco's Modified Eagle Medium (DMEM, Thermo Fisher Scientific, CAT#61965026) supplemented with 10% FBS (Thermo Fisher Scientific, CAT#15377636) and 1% penicillin/streptomycin (Thermo Fisher Scientific, CAT#15140122). CD14+ cells were enriched from PBMCs via positive selection using CD14 MicroBeads (Miltenyi Biotec, CAT#130-042-401). Purity determined via flow cytometry was consistently >99%. CD14+ cells were cultured in RPMI 1640 medium (Thermo Fisher Scientific, CAT#61870010) supplemented with 10% FBS (Eurobio Scientific, CAT#CVFSVF0001), 1% penicillin/streptomycin (Thermo Fisher Scientific, CAT#15140122), 50 μg/mL gentamicin (Thermo Fisher Scientific, CAT#10131027), 10 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES, Thermo Fisher Scientific, CAT#15630080), 10 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF, Miltenyi Biotec, CAT#130-093-867), and 50 ng/mL interleukin-4 (IL-4, Miltenyi Biotec, CAT#130-093-922) to generate monocyte-derived dendritic cells (MDDCs). Experiments were performed on day 4.
Constructs
pNLAD8 (HIV-1) and pJK7312As (HIV-2) were generated as described previously.33,34 pNLAD8 ELA20, pNLAD8 ELA20 Δenv, pJK7312As ELA20, and pJK7312As ELA20 Δenv were generated via overlapping PCR mutagenesis (Table S2). CMV-VSVG and pSIV3+ were generated as described previously.35 All constructs were sequence-verified prior to experimentation. Plasmid DNA was purified using a PureLink HiPure Plasmid Midiprep Kit (Thermo Fisher Scientific, CAT#K210005).
Virus production
Viral particles were produced by transfection of 293FT cells with 0.4 μg of CMV-VSVG DNA, 2.6 μg of pSIV3+, pNLAD8, or pJK7312As DNA, and 8 μL of TransIT-293 (Mirus Bio, CAT# MIR2700) per well in 6-well culture plates (Dutscher, CAT#9206). Medium was replaced after 24 h. Viral supernatants were harvested the next day, filtered at 0.45 μm, and either used fresh or concentrated via ultracentrifugation and then frozen at −80 °C. Viral titers were measured using GHOST X4R5 cells as described previously35 or via HIV-1 p24 or SIV p27 ELISAs (XpressBio, CAT#XB-1010 or CAT#SK845, respectively) using a CLARIOstar Microplate Reader (BMG Labtech) with MARS Data Analysis Software (BMG Labtech). The pNLAD8 ELA20 Δenv and pJK7312As ELA20 Δenv viruses were purified and concentrated on a 20% sucrose cushion via ultracentrifugation at 4 °C. Pellets were resuspended in AIM V medium (Thermo Fisher Scientific, CAT#10297582) and then frozen at −80 °C.
Infection of dendritic cells
MDDCs were harvested on day 4 and resuspended in fresh medium with the addition of 5 μg/mL protamine (Sigma–Aldrich, CAT#P4020) in the absence or presence of 25 μM azidothymidine (AZT, Sigma–Aldrich, CAT#A2169) or 10 μM nevirapine (NVP, Sigma–Aldrich, CAT#SML0097) at a concentration of 1 × 106 cells/mL. Infections were performed using 1 × 106 cells per well in 6-well culture plates (Thermo Fisher Scientific, CAT#140685). Medium or SIVmac VLPs were added to the relevant wells, followed by medium or freshly harvested viral supernatants, normalized for input titer using HIV-1 p24 or SIV p27 ELISAs (XpressBio, CAT#XB-1010 or CAT#SK845, respectively). The final volume per well for all infections was 6 mL. After 24 h, approximately 1 × 105 cells per condition were removed and transferred to 96-well round-bottom culture plates (Thermo Fisher Scientific, CAT#262162), leaving approximately 9 × 105 cells per condition for real-time PCR and intracellular quantification of SAMHD1. After a further 24 h, cells were analyzed via flow cytometry, and supernatants were ultraviolet-irradiated and used to quantify IFN-λ1.
Western blotting
Lysates were generated from 0.5–2 × 106 cells per condition. Cells were lysed in 100 μL of RIPA buffer (50 mM Tris HCl, 150 mM NaCl, 0.1% SDS, 0.5% DOC, 1% NP-40) containing EDTA-free cOmplete Protease Inhibitor Cocktail (Roche, CAT#11873580001). After incubation for 30 min on ice, lysates were cleared via centrifugation at 8000×g for 8 min at 4 °C, supplemented with 20 μL of Laemmli buffer 6× (12% SDS, 30% glycerol, 0.375 M Tris-HCl pH 6.8, 30% 2-mercaptoethanol, 1% bromophenol blue), and boiled for 15 min at 95 °C. Viral supernatants were filtered at 0.45 μm and then centrifuged at 16,000×g for 90 min at 4 °C. Pellets were lysed in 20 μL of RIPA buffer as above, supplemented with 4 μL of Laemmli buffer 6×, and boiled for 15 min at 95 °C. Protein lysates were resolved on 4–20% Mini-PROTEAN TGX Precast Protein Gels (Bio-Rad, CAT#4561096) and then transferred onto membranes using a Trans-Blot Turbo Mini 0.2 μm PVDF Transfer Pack (Bio-Rad, CAT#1704156). Membranes were saturated in phosphate-buffered saline (PBS) containing 5% non-fat dry milk and 0.1% Tween prior to blotting with the following monoclonal antibodies: unconjugated anti-Gag (clone 183-H12-5C, NIH AIDS Reagent Program, RRID:AB_2819250), unconjugated anti-tubulin (clone DM1A, Thermo Fisher Scientific, RRID:AB_1210456), and unconjugated anti-CypA (clone 817815, R&D Systems, RRID: N/A).
Real-time PCR
Real-time PCR analysis was performed as described previously.26 Each sample was measured in triplicate. Cycling conditions were 1 × 5 min at 95 °C and 40 × 10 s at 95 °C, 20 s at 65 °C, and 30 s at 72 °C. Relative concentrations of late RT were calculated with reference to β-globin using the ΔCt method. Primer sequences are listed in Table S3.
In vitro priming of antigen-specific CD8+ T cells
CD8+ T cells were primed in vitro using a modified version of a previously described protocol36, 37, 38, 39, 40, 41, 42 that allowed antigen presentation in the context of HIV-1 or HIV-2. Briefly, thawed PBMCs from HIV-uninfected HLA-A2+ donors were resuspended at 5 × 106 cells/well in 48-well culture plates (Thermo Fisher Scientific, CAT#150787) containing AIM V medium (Thermo Fisher Scientific, CAT#12055-091) supplemented with 50 ng/mL FMS-like tyrosine kinase 3 ligand (FLT3L, R&D Systems, CAT#308-FK-005). After 24 h (day 1), cultures were spinoculated with 1 or 10 ng of ELA20 HIV-1 (based on p24) or ELA20 HIV-2 (based on p27) at 2200×g for 100 min at room temperature in the presence of 5 ng/mL protamine (Sigma–Aldrich, CAT#P4020). In some assays, cells were stimulated with 1 μM ELA20 (Melan-A21–40/A27L, YTAAEELAGIGILTVILGVL), synthesized at >95% purity (Bio-Synthesis Inc.), and 10 μg/mL 3′3′-cGAMP (InvivoGen, CAT#tlrl-nacga), 0.1 μg/mL IFN-α 2a (ImmunoTools, CAT#11343506), or a cocktail of proinflammatory cytokines comprising IL-1β (10 ng/mL, R&D Systems, CAT#201-LB-005), tumor necrosis factor (TNF, 1000 U/mL, R&D Systems, CAT#210-TA-005), and prostaglandin E2 (PGE2, 1 μM, Merck, CAT#538904), together with IL-7 (0.5 ng/mL, R&D Systems, CAT#207-IL005). A combination of anti-IFN-α (20,000 NU/mL, Tebu-Bio, CAT#31101-1) and anti-IFN-β (5000 NU/mL, Tebu-Bio, CAT#31410-1) was used on day 1 as indicated to block the activity of type I IFNs. On day 2, FBS was added at a final v/v ratio of 10%. Medium was replaced every 3 days thereafter with fresh RPMI 1640 medium (Thermo Fisher Scientific, CAT#31870025) supplemented with 10% FBS (Thermo Fisher Scientific, CAT#10270-106) and 1% penicillin/streptomycin (Thermo Fisher Scientific, CAT#15140122). Supernatants were collected on day 3 and then frozen for subsequent chemokine/cytokine measurements. Antigen-specific CD8+ T cells were analyzed on day 11.
Flow cytometry
MDDCs were stained for surface markers of activation 48 h after infection using anti-CD86–PE (clone IT2.2, eBioscience, RRID:AB_10732345) or anti-Siglec-1–APC (clone 7-239, Miltenyi Biotec, RRID:AB_2655546). SAMHD1 was quantified intracellularly 24 h after infection. Briefly, 3 × 105 cells were fixed in 1% paraformaldehyde (PFA)/PBS for 10 min at 37 °C, centrifuged at 400×g for 5 min at 4 °C, resuspended in 100 μL of glycine/PBS (7.5 mg/mL) for 10 min at room temperature, and centrifuged again at 400×g for 5 min at 4 °C. Cells were then resuspended in 100 μL of 80% methanol/H2O (v/v) for 30 min on ice, washed in 1 mL of 5% bovine serum albumin (BSA)/PBS for 5 min at 4 °C, and resuspended in 50 μL of 10% BSA/PBS. After 10 min at room temperature, cells were labeled with unconjugated rabbit anti-SAMHD1 antibodies (1/100, Atlas Antibodies, RRID:AB_2679921), added in a further 50 μL of 10% BSA/PBS supplemented with 10% goat serum, for 1 h at room temperature. Cells were then washed again in 1 mL of 5% BSA/PBS for 5 min at 4 °C, resuspended in 50 μL of goat anti-rabbit IgG–Alexa Fluor 647 (Thermo Fisher Scientific, RRID:AB_2535813) diluted 1/500 in 5% BSA/PBS, incubated for 30 min at room temperature, washed twice in 1 mL of 5% BSA/PBS, centrifuged at 1000×g for 5 min at 4 °C, resuspended in 100 μL of PBS, and analyzed via flow cytometry using a FACSVerse (BD Biosciences) with FlowJo software version 10.6.2 (FlowJo LLC).
PBMCs were harvested from priming assays on day 11, washed in PBS, labeled with LIVE/DEAD Fixable Aqua (Thermo Fisher Scientific, CAT#L34957) for 10 min at room temperature, washed again in PBS, and stained with PE-labeled tetrameric complexes of HLA-A∗02:01/ELA (Melan-A26–35/A27L, ELAGIGILTV)43 for 15 min at 37 °C. Cells were then washed again in PBS and further stained with anti-CCR7–PE-Cy7 (clone 3D12, BD Biosciences, RRID:AB_396765), which was added 3 min before the other surface antibodies, anti-CD8–APC-Cy7 (clone SK1, BD Biosciences, RRID:AB_396892), anti-CD27–Alexa Fluor 700 (clone O323, BioLegend, RRID:AB_493757), and anti-CD45RA–PerCP-Cy5.5 (clone HI100, eBioscience, RRID:AB_10718536). After 15 min at room temperature, cells were fixed/permeabilized using a Transcription Factor Buffer Set (BD Biosciences, CAT#562574), stained intracellularly with anti-granzyme B–V450 (clone GB11, BD Biosciences, RRID:AB_10565977), anti-perforin–FITC (clone B-D48, BioLegend, RRID:AB_2571967), and anti-T-bet–eFluor 660 (clone 4B10, eBioscience, RRID:AB_10596655) for 40 min on ice, washed in Perm/Wash Buffer (BD Biosciences, CAT#562574), and fixed with 2% PFA. Data were analyzed via flow cytometry using an LSR Fortessa (BD Biosciences) with FlowJo software version 10.6.2 (FlowJo LLC).
Cell sorting
Viable tetramer+ CD8+ T cells were sorted directly into 96-well PCR plates (Eppendorf, CAT#0030129504) containing VILO Reaction Mix (Thermo Fisher Scientific, CAT#11754250), SUPERase In RNase Inhibitor (Thermo Fisher Scientific, CAT#AM2696), and 10% NP40 (Thermo Fisher Scientific, CAT#28324) using a FACSAria (BD Biosciences). Plates were snap-frozen and then stored at −80 °C.
Gene expression
The expression levels of 96 selected genes (Table S4) were quantified as described previously.44 Briefly, plates containing sorted cells were thawed on ice, and RNA was denatured for 90 s at 65 °C, snap-chilled on ice, and reverse transcribed using Superscript Enzyme Mix (Thermo Fisher Scientific, CAT#11754250) and T4 Gene 32 Protein (Merck, CAT#10972983001). Cycling conditions were 1 × 5 min at 25 °C, 30 min at 50 °C, 25 min at 55 °C, 5 min at 60 °C, and 10 min at 70 °C. Targets were amplified using specific primers with TaqMan PreAmp Master Mix (Thermo Fisher Scientific, CAT#4488593). Cycling conditions were 1 × 10 min at 96 °C and 20 × 5 s at 96 °C and 4 min at 60 °C. Samples were then treated with Exonuclease I (New England BioLabs, CAT#M0293L) for 30 min at 37 °C and 15 min at 80 °C, diluted 5-fold in DNA Suspension Buffer (Teknova, CAT#T0221), mixed with EvaGreen Supermix with Low ROX (Bio-Rad, CAT#1725211) and DNA Binding Dye (Fluidigm, CAT#100-7609) or with Assay Loading Reagent (Fluidigm, CAT#100-7611) and DNA Suspension Buffer (Teknova, CAT#T0221), and transferred to a primed 96.96 Dynamic Array IFC (Fluidigm, CAT#BMK-M-96.96). Data were acquired using a Biomark HD (Fluidigm) with linear derivative mode baseline correction. Gene expression levels were plotted as log 2−Δct versus the GAPDH gene for normalization. Samples lacking a GAPDH signal were excluded from the analysis.
Measurement of soluble factors
Chemokines and cytokines in culture supernatants were measured using a ProcartaPlex 10-Plex Kit (Thermo Fisher Scientific, CAT#PPX-10) in conjunction with a Bio-Plex 200 System (Bio-Rad). All concentrations were determined as the mean of two replicates. IFN-λ1 concentrations in supernatants from infected MDDCs were quantified using a LEGENDplex Human Type 1/2/3 Interferon Panel (BioLegend, CAT#BLE740396B). Data were analyzed via flow cytometry using a FACSVerse (BD Biosciences) with LEGENDplex Software (BioLegend).
Polyfunctionality assay
ELA20-primed PBMCs containing approximately 2 × 104 HLA-A∗02:01/ELA tetramer+ CD8+ T cells were incubated with ELA (ELAGIGILTV) peptide-pulsed HLA-A2+ B-lymphoblastoid cell line (B-LCL) target cells at an effector-to-target ratio of >1:10 in the presence of anti-CD107a–PE-Vio770 (clone REA792, Miltenyi Biotec, RRID:AB_2654484) for 6 h at 37 °C. Parallel assays were conducted with unpulsed HLA-A2+ B-LCL target cells to evaluate background activation. Protein transport was blocked for the final 5 h with GolgiStop (1:1500, BD Biosciences, CAT#554724). Cells were then fixed/permeabilized using a BD Fixation/Permeabilization Kit (BD Biosciences, CAT#554714). Intracellular staining was performed using anti-IFN-γ–VioBlue (clone REA600, Miltenyi Biotec, RRID:AB_2751736), anti-TNF–Alexa Fluor 700 (clone MAb 11, BioLegend, RRID:AB_2561315), anti-MIP-1β–APC (clone REA511, Miltenyi Biotec, RRID:AB_2857772), and anti-IL-2–Vio770 (clone REA689, Miltenyi Biotec, RRID:AB_2652425) as described previously.11 Data were analyzed via flow cytometry using an LSR Fortessa (BD Biosciences) with FlowJo software version 10.6.2 (FlowJo LLC). Pie charts were constructed using SPICE software version 6.1 (NIAID, NIH).
Reagent validation
All antibodies used in this work were commercial ones, used according to the manufacturer recommendations. All cell lines were regularly tested for being mycoplasma free.
Ethics
The use of blood products from anonymized healthy donors was approved by the Comité de Protection des Personnes of the Pitié Salpétrière Hospital, Paris, France. Written informed consent was obtained from all HIV-1-infected donors under ethical approval from the McGill University Health Centre and the Centre Hospitalier de l’Université de Montréal Institutional Review Boards, Montreal, Canada (REF#2017-6516, MP-CUSM-15-303).
Statistics
Statistical analyses were performed using Prism software version 8.4.3 (GraphPad). Distribution normality was assessed using the Shapiro–Wilk test. Simple group comparisons were performed using the Wilcoxon signed rank test or the Mann–Whitney U test. Parametric alternatives were used only if the data were distributed normally. Multiple group comparisons were performed using paired or non-paired one-way repeated measures ANOVAs with the Šídák test or the Holm-Šídák test, respectively. Gene expression analyses were performed using a mixed-effect model that included HIV exposure (HIV-1 vs HIV-2) or priming conditions (cytokine vs IFN vs cGAMP), and their interaction as fixed effects. Participant and technical replicates were also considered as random effects. These analyses were performed in R software version 3.6.3. Significance was assigned as follows: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Role of funders
The funders did not participate in study design, data acquisition, data analysis, manuscript preparation, or the decision to publish.
Results
A model of antigen-specific CD8+ T cell priming upon exposure to HIV
We previously established an in vitro model based on an accelerated DC culture protocol to assess the efficacy of CD8+ T cell priming among unfractionated human PBMCs and test the added value of adjuvant delivery.36, 37, 38, 39, 40, 41, 42 A key element of this assay is the minimization of stochastic effects via the use of the heteroclitic Melan-A26–35/A27L epitope ELAGIGILTV (ELA) as a target antigen,45 which is consistently recognized by extraordinarily high frequencies of naive precursors in HLA-A2+ individuals.46,47 In the present study, we adapted this approach to develop an unbiased method enabling us to compare the attributes of antigen-specific CD8+ T cells primed by HIV-1 or HIV-2.
For this purpose, we engineered ELA as a common target antigen, contained within the longer sequence YTAAEELAGIGILTVILGVL (ELA20), between the matrix and capsid genes of HIV-1 (pNLAD8) or HIV-2 (pJK7312As) (Fig. 1A and Figure S1A). To focus on innate sensing events and eliminate bias arising from differential viral replication, these constructs were pseudotyped with vesicular stomatitis virus glycoprotein (VSV-G) to allow entry into DCs. Replication-incompetent viral particles with the expected maturation profile were produced by pNLAD8 ELA20 Δenv (ELA20 HIV-1) and pJK7312As ELA20 Δenv (ELA20 HIV-2) (Figure S1B). Sensitive p24 and p27 matrix ELISAs were used to normalize the input titers of ELA20 HIV-1 and ELA20 HIV-2, respectively (Figure S1C), ensuring the delivery of equivalent amounts of ELA. These modified viruses were less infectious than the parent constructs (Figure S1C and D), likely reflecting some form of interference arising from the insertion of ELA, but nonetheless recapitulated the key differences between HIV-1 and HIV-2. Infection of human MDDCs confirmed that ELA20 HIV-2 downregulated SAMHD1 (p < 0.0001, repeated measures ANOVA), unlike ELA20 HIV-1 (Fig. 1B). Higher levels of AZT/NVP-sensitive late reverse transcriptase (RT) products were also observed after infection with ELA HIV-2 versus ELA HIV-1 (p = 0.0458, repeated measures ANOVA), consistent with the presence of Vpx (Figure S1E). In addition, HIV-2 induced the maturation of MDDCs, evidenced by the upregulation of CD86, albeit most obviously after infection with the parental strain, and Siglec-1 and the secretion of IFN-λ1, whereas Vpx was required in trans to elicit similar effects after infection with HIV-1 (Fig. 1C).
Fig. 1.
Virus production and experimental validation. (A) Graphical representation of Δenv HIV-1 and Δenv HIV-2 expressing ELA. (B) SAMHD1 expression in MDDCs 24 h after infection with the indicated pNLAD8 (HIV-1, n = 3) or pJK7312As (HIV-2, n = 4) viruses. NI, no infection. Left: representative flow cytometry plots. Right: data summary. Each dot represents one donor. Bars indicate mean values. ∗∗∗∗p < 0.0001 (unpaired repeated measures ANOVA). (C) Expression of CD86 and Siglec 1 on MDDCs and production of IFN-λ1 by MDDCs 48 h after infection with the indicated pNLAD8 (HIV-1, n = 4) or pJK7312As (HIV-2, n = 4) viruses. SIVmac VLPs were added to provide Vpx. NI, no infection. Each dot represents one donor. ∗p < 0.05, ∗∗p < 0.01, ns = not significant (paired repeated measures ANOVA). (D) Schematic representation of the in vitro priming approach using ELA20 Δenv HIV-1 and ELA20 Δenv HIV-2. (E) Left: representative flow cytometry plots showing ELA-specific CD8+ T cells expanded in the presence of 10 ng of ELA20 Δenv HIV-1 or ELA20 Δenv HIV-2. Right: HLA-A∗02:01/ELA tetramer+ CD8+ T cell frequencies after priming with 1 or 10 ng of ELA20 Δenv HIV-1 or ELA20 Δenv HIV-2. Each connected dot represents one donor. ∗p < 0.05 (Wilcoxon signed rank test).
The modified in vitro priming approach is depicted in Fig. 1D. Briefly, DCs present among HIV-uninfected HLA-A2+ donor PBMCs were mobilized using FLT3L and then exposed to ELA20 HIV-1 or ELA20 HIV-2, and primed antigen-specific CD8+ T cells were enumerated and characterized on day 11. The dynamic range of this assay was established using 1 or 10 ng of each virus to elicit CD8+ T cells specific for ELA (Fig. 1E), and the latter dose was used in subsequent assays to compare the priming efficacy of ELA20 HIV-1 versus ELA20 HIV-2.
HIV-2 primes antigen-specific naive CD8+ T cells more effectively than HIV-1
In our controlled experimental setting with equivalent doses of antigen, HIV-2 induced higher frequencies of ELA-specific CD8+ T cells than HIV-1 (p = 0.007, Wilcoxon signed rank test) (Fig. 2A). All primed ELA-specific CD8+ T cells exhibited a memory phenotype, lacking dual expression of CCR7 and CD45RA (Figure S2). To extend this comparison, we analyzed the transcriptomes of flow-sorted ELA-specific CD8+ T cells elicited by exposure to HIV-1 versus HIV-2, focusing on the expression of 96 genes with important roles in cell function, differentiation, metabolism, and survival, as described previously.44 Principal component analysis (PCA) revealed that ELA-specific CD8+ T cells primed by HIV-1 clustered distinctly from ELA-specific CD8+ T cells primed by HIV-2 (Fig. 2B). Segregation was driven primarily by 24 differentially expressed genes, 22 of which were upregulated in ELA-specific CD8+ T cells primed by HIV-2 relative to ELA-specific CD8+ T cells primed by HIV-1 (Fig. 2C).
Fig. 2.
Qualitative and quantitative analyses of CD8+T cells primed by HIV-1 or HIV-2. (A) Frequencies of ELA-specific CD8+ T cells after priming with 10 ng of ELA20 Δenv HIV-1 or ELA20 Δenv HIV-2. Each connected dot represents one donor. ∗∗p < 0.01 (Wilcoxon signed rank test). (B) PCA map showing the spatial distribution of ELA-specific CD8+ T cells primed with ELA20 Δenv HIV-1 (red) or ELA20 Δenv HIV-2 (blue), grouped according to gene expression. Each symbol represents one donor. (C) Heatmap showing genes that were differentially expressed between ELA-specific CD8+ T cells primed with ELA20 Δenv HIV-1 (n = 5) or ELA20 Δenv HIV-2 (n = 5). (D) ELA-specific CD8+ T cell frequencies and intracellular expression of granzyme B, perforin, and T-bet among tetramer+ CD8+ T cells after priming with ELA20 Δenv HIV-1 or ELA20 Δenv HIV-2. Each connected dot represents one donor. ∗p < 0.05 (Wilcoxon signed rank test). Representative flow cytometry plots are shown in the top left panel. Numbers indicate percentages in the drawn gates. (E) Viability of ELA-specific CD8+ T cells after priming with ELA20 Δenv HIV-1 or ELA20 Δenv HIV-2 (n = 5). Top: representative flow cytometry histograms. Numbers indicate percentages in the drawn gates. Bottom: data summary. Data are shown as median ± IQR. ∗p < 0.05 (Wilcoxon signed rank test).
Among the genes upregulated in ELA-specific CD8+ T cells primed by HIV-2, many were associated with activation and/or functional maturity, including CASP8, CDKN1B, CDKN2A, CTLA4, CCL3L1, BATF, TNFSF10, and MIF (Figure S3). For example, CCL3L1 encodes macrophage inflammatory protein (MIP)-1α, which has been linked with CD8+ T cell-mediated suppression of HIV-1,48 and more generally, BATF is known to enhance the quality and quantity of virus-specific CD8+ T cells in murine models.49 In addition, ELA-specific CD8+ T cells primed by HIV-2 expressed higher levels of IFNA1 and IFNB1 and various genes associated with metabolism and the cell cycle, including VHL and CDC42, than ELA-specific CD8+ T cells primed by HIV-1.
Our model precluded direct measurements of antiviral efficacy due to insufficient numbers of primed cells necessary for this assay. However, ELA-specific CD8+ T cells primed by HIV-2 expressed higher levels of granzyme B (p = 0.04, Wilcoxon signed rank test), perforin (p = 0.03, Wilcoxon signed rank test) and T-bet (p = 0.04, Wilcoxon signed rank test) than ELA-specific CD8+ T cells primed by HIV-1 (Fig. 2D), consistent with a greater capacity to suppress viral replication.50 In addition, ELA-specific CD8+ T cells primed by HIV-2 expressed relatively high levels of genes associated with cell survival, including IL7R, BCL2, and ID2, and the generation of functional memory, including LEF1 and FOXO1,51,52 as well as BAG-6, which is known to repress T cell immunoglobulin mucin-3 (TIM-3), an inhibitory receptor associated with exhaustion.53 ELA-specific CD8+ T cells primed by HIV-2 also displayed enhanced viability compared with ELA-specific CD8+ T cells primed by HIV-1 (p = 0.03, Wilcoxon signed rank test) (Fig. 2E).
Collectively, these results indicated that HIV-2 primed CD8+ T cells with strong activation and effector profiles and enhanced survival properties, all of which have been linked with natural control of HIV-1.
Optimal priming of antigen-specific CD8+ T cells is mediated by type I IFNs
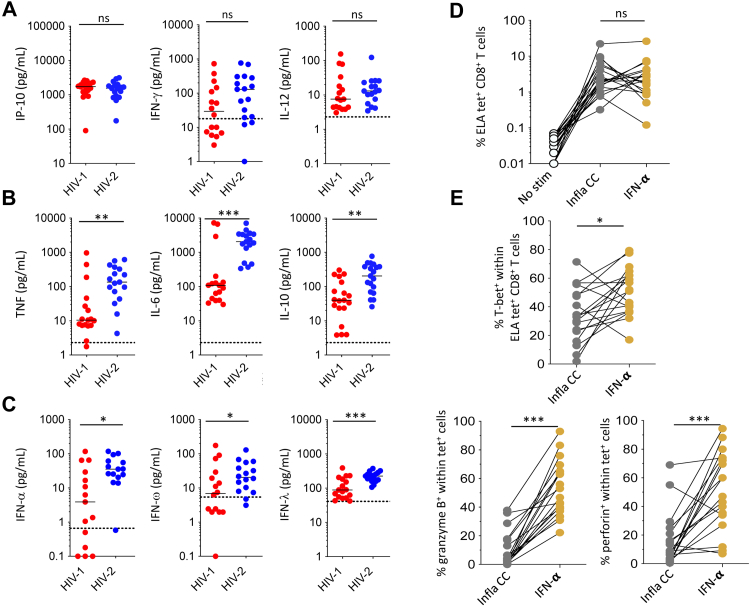
To gain insights into the factors that underlie the effective induction of antigen-specific CD8+ T cells with optimal functionality, we measured the secretion of various chemokines/cytokines in supernatants harvested from priming assays incorporating HIV-1 or HIV-2. No differences were observed with respect to the concentrations of IFN-γ, IL-12, or IP-10 (Fig. 3A). Higher levels of the inflammatory cytokines IL-6 (p = 0.0003, Mann–Whitney U test), IL-10 (p = 0.006, Mann–Whitney U test), and TNF (p = 0.0002, Mann–Whitney U test) (Fig. 3B), as well as IFN-α (type I IFN, p = 0.01, Mann–Whitney U test), IFN-ω (type I IFN, p = 0.04, Mann–Whitney U test), and IFN-λ (type III IFN, p = 0.0006, Mann–Whitney U test) (Fig. 3C), were nonetheless detected after exposure to HIV-2 versus HIV-1, consistent with the preferential activation of NF-κB and the NONO-cGAS-STING pathway in DCs.30
Fig. 3.
CD8+T cell priming efficacy as a function of type I IFNs. (A–C) Concentrations of various chemokines/cytokines in culture supernatants 3 days after exposure of PBMCs to ELA20 Δenv HIV-1 or ELA20 Δenv HIV-2. Each dot represents one donor. Bars indicate median values. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns = not significant (Mann–Whitney U test). (D) ELA-specific CD8+ T cell frequencies after priming with ELA20 in the presence of proinflammatory cytokines (IL-1β, TNF, and PGE2) or IFN-α. Each connected dot represents one donor. Bars indicate median values. ns = not significant (Wilcoxon signed rank test). (E) Intracellular expression of granzyme B, perforin, and T-bet among tetramer+ CD8+ T cells after priming with ELA20 in the presence of proinflammatory cytokines (IL-1β, TNF, and PGE2) or IFN-α. Each connected dot represents one donor. Bars indicate median values. ∗p < 0.05, ∗∗∗p < 0.001 (Wilcoxon signed rank test).
To determine which of these soluble factors most influenced priming efficacy, we compared the properties of CD8+ T cells induced by exposure to ELA20 in the presence of IFN-α or a cocktail of proinflammatory cytokines comprising IL-1β, TNF, and PGE2, which are known to activate NF-κB (Figure S4A). Each of these conditions promoted the priming of ELA-specific CD8+ T cells equivalently (Fig. 3D). However, ELA-specific CD8+ T cells primed in the presence of IFN-α more frequently expressed granzyme B (p = 0.0003, Wilcoxon signed rank test), perforin (p = 0.0006, Wilcoxon signed rank test), and T-bet (p = 0.02, Wilcoxon signed rank test) than ELA-specific CD8+ T cells primed in the presence of IL-1β, TNF, and PGE2 (Fig. 3E). The transcriptomes of ELA-specific CD8+ T cells primed in the presence of IFN-α versus IL-1β, TNF, and PGE2 were also distinct (Fig. 4A) and clustered separately in the corresponding PCA (Fig. 4B). Segregation was driven primarily by 28 genes that were differentially upregulated in the context of IFN-α. These differentially upregulated genes included TBX21, which encodes T-bet, and PRF1, which encodes perforin, the interferon-stimulated gene (ISG) MX1, and SLC2A1 and SLC2A3, which encode the glucose transporter GLUT3 (Fig. 4A and Figure S5). Of note, many of the genes that were upregulated in the context IFN-α were also upregulated after priming with HIV-2 (Fig. 2C), indicating exposure to type I IFNs, consistent with direct activation of the NONO-cGAS-STING pathway in DCs. In line with this observation, ELA-specific CD8+ T cells primed by HIV-2 expressed higher levels of two notable ISGs, namely IFITM1, which encodes interferon-induced transmembrane protein 1, and IRF1, which encodes interferon regulatory factor 1, than ELA-specific CD8+ T cells primed by HIV-1 (Fig. 2C and Figure S3).
Fig. 4.
Comparative transcriptomic analyses of CD8+T cells primed under different conditions. (A) Heatmap showing genes that were differentially expressed among ELA-specific CD8+ T cells primed in the presence of proinflammatory cytokines (IL-1β, TNF, and PGE2, n = 7), IFN-α (n = 7), or cGAMP (n = 7). (B) PCA map showing the spatial distribution of ELA-specific CD8+ T cells primed with ELA20 in the presence of proinflammatory cytokines (IL-1β, TNF, and PGE2, grey), IFN-α (orange), or cGAMP (green), grouped according to gene expression. Each symbol represents one donor. (C) PCA map showing the spatial distribution of ELA-specific CD8+ T cells primed with ELA20 Δenv HIV-1 (red), ELA20 Δenv HIV-2 (blue), or ELA20 in the presence of cGAMP (green), grouped according to gene expression. Each symbol represents one donor. (D) ELA-specific CD8+ T cell frequencies and intracellular expression of granzyme B and perforin after priming with ELA20 Δenv HIV-1, ELA20 Δenv HIV-2, or ELA20 in the presence of cGAMP ± anti-IFN-α and anti-IFN-β. Each connected dot represents one donor. ∗p < 0.05, ∗∗p < 0.01 (Wilcoxon signed rank test).
To extend these findings, we performed similar experiments with cGAMP, which activates STING. The transcriptomes of ELA-specific CD8+ T cells primed in the presence of IFN-α versus cGAMP were very similar (Fig. 4A) and clustered together in the corresponding PCA (Fig. 4B). ELA-specific CD8+ T cells primed in the presence of cGAMP also overlapped transcriptionally with ELA-specific CD8+ T cells primed by HIV-2 but not by HIV-1 (Fig. 4C). Moreover, type I IFN blocking antibodies impaired the expansion and functional maturation of ELA-specific CD8+ T cells primed either in the presence of cGAMP (p = 0.007, Wilcoxon signed rank test) or by HIV-2 (p = 0.01, Wilcoxon signed rank test) but not by HIV-1 (p = 0.97, Wilcoxon signed rank test) (Fig. 4D).
Collectively, these data indicated that the enhanced priming efficacy associated with HIV-2-derived viral factors could be reproduced in the presence of cGAMP, highlighting a key role in this process for type I IFNs.
Functionally replete antigen-specific CD8+ T cells can be primed from people living with HIV-1 by mimicking viral factors associated with HIV-2
The data presented above suggested that antigen-specific CD8+ T cells endowed with functional properties associated with enhanced immune control of HIV-1 could be primed by activating STING. In line with this notion, we found that cGAMP promoted the expression of granzyme B, perforin, and T-bet among ELA-specific CD8+ T cells primed in the presence of IL-1β, TNF, and PGE2 (Figure S6A), consistent with our earlier work.39 Similar effects were observed after priming directly with ELA20 HIV-1 (Figure S6B).
However, immune alterations in PLWH, including the chronic production of type I IFNs54 as well as naive T cell55 and DC dysfunction,56 could feasibly impact priming events and impair the adjuvant efficacy of cGAMP. We therefore extended our in vitro approach to HLA-A2+ PLWH on ART (Table S1). Initially, we measured the secretion of various chemokines/cytokines in supernatants harvested from priming assays incorporating cGAMP or IL-1β, TNF, and PGE2, comparing HIV-uninfected donors (HUDs) with PLWH. Stimulation with cGAMP yielded higher levels of IFN-α and IFN-ω and lower levels of proinflammatory cytokines, such as IL-6, IL-10, and TNF, than stimulation with IL-1β, TNF, and PGE2, and no significant differences were detected between HUDs and PLWH (Fig. 5A and B, and Figure S7). This suggests that the engaged stimulatory pathways are functioning similarly well in HUD and treated PLWH. We then assessed priming efficacy in the presence of cGAMP or IL-1β, TNF, and PGE2 (Figure S4B). Higher frequencies of ELA-specific CD8+ T cells were primed in HUDs versus PLWH in the presence of IL-1β, TNF, and PGE2 (Fig. 5C), likely reflecting baseline immune dysfunction in PLWH. This difference was less apparent after stimulation with cGAMP, which increased the frequencies of ELA-specific CD8+ T cells primed from PLWH (p = 0.002, Wilcoxon signed rank test) (Fig. 5C). Gene expression analyses further showed that the transcriptomes of ELA-specific CD8+ T cells primed in vitro clustered by experimental condition rather than donor source in a PCA comparing HUDs versus PLWH (Fig. 5D). ELA-specific CD8+ T cells primed with cGAMP also more frequently expressed granzyme B (p = 0.01, Wilcoxon signed rank test), perforin (p = 0.004, Wilcoxon signed rank test), and T-bet (p = 0.02, Wilcoxon signed rank test) after priming in the presence of cGAMP versus IL-1β, TNF, and PGE2, with no significant differences between HUDs and PLWH (Fig. 5E).
Fig. 5.
Qualitative and quantitative analyses of antigen-specific CD8+T cells primed from people living with HIV-1. (A, B) Radar plots showing the mean extracellular concentrations (pg/mL) of various chemokines/cytokines in culture supernatants 3 days after stimulation of PBMCs from HUDs (n = 9) (A) or PLWH on ART (n = 10) (B) with proinflammatory cytokines (IL-1β, TNF, and PGE2) or cGAMP. ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 (Mann–Whitney U test). (C) ELA-specific CD8+ T cell frequencies primed from HUDs or PLWH on ART with ELA20 in the presence of proinflammatory cytokines (IL-1β, TNF, and PGE2) or cGAMP. Each connected dot represents one donor. ∗∗p < 0.01 (Wilcoxon signed rank test). (D) PCA map showing the spatial distribution of ELA-specific CD8+ T cells primed from HUDs or PLWH on ART with ELA20 in the presence of proinflammatory cytokines (IL-1β, TNF, and PGE2) or cGAMP, grouped according to gene expression. Each symbol represents one donor. (E) Intracellular expression of granzyme B, perforin, and T-bet among tetramer+ CD8+ T cells primed from HUDs or PLWH with ELA20 in the presence of proinflammatory cytokines (IL-1β, TNF, and PGE2) or cGAMP. Each connected dot represents one donor. ∗p < 0.05, ∗∗p < 0.01 (Wilcoxon signed rank test).
In a final series of experiments, we assessed the antigen sensitivity and functionality of ELA-specific CD8+ T cells primed in the presence of cGAMP or IL-1β, TNF, and PGE2. These assays were performed using flow cytometry to quantify the surface mobilization of CD107a, a marker of cytolytic degranulation, and the intracellular production of IFN-γ, MIP-1β, TNF, and IL-2 (Fig. 6A). Higher frequencies of polyfunctional ELA-specific CD8+ T cells were observed after priming in the presence of cGAMP versus IL-1β, TNF, and PGE2, with no obvious differences between HUDs and PLWH (Fig. 6B). Antigen sensitivity was evaluated using cognate peptide titrations to elicit the secretion of IFN-γ and MIP-1β. ELA-specific CD8+ T cells primed in the presence of cGAMP were approximately one order of magnitude more sensitive to antigen stimulation than ELA-specific CD8+ T cells primed in the presence of IL-1β, TNF, and PGE2, again with no obvious differences between HUDs and PLWH (Fig. 6C). A similar pattern was observed across the full spectrum of effector functions, enabling ELA-specific CD8+ T cells primed in the presence of cGAMP to respond at peptide concentrations as low as 10−8 M (Fig. 6D), equivalent to antigen densities achieved via physiological presentation.11
Fig. 6.
Antigen sensitivity and functionality of CD8+T cells primed from people living with HIV-1. (A) Representative flow cytometry plots showing the mobilization of CD107a and the production of IFNγ, MIP-1β, TNF, and IL-2 in response to antigen stimulation. ELA-specific CD8+ T cells primed with ELA20 in the presence of proinflammatory cytokines (IL-1β, TNF, and PGE2) or cGAMP were incubated with medium alone (top) or HLA-A2+ B-LCL target cells pulsed with ELA at concentration of 10−7 M (middle and bottom). Function plots were gated on viable tetramer+ CD3+ CD8+ lymphocytes (center and right). Numbers indicate percentages in the drawn gates. (B) Combinatorial analysis of effector functions among ELA-specific CD8+ T cells primed from HUDs (n = 2) or PLWH (n = 2). Data are shown as mean ± SEM. Details as in (A). (C) Normalized IFN-γ and MIP-1β production curves for ELA-specific CD8+ T cells primed from HUDs or PLWH as in (A). HLA-A2+ B-LCL target cells were pulsed with the indicated concentrations of ELA. Data are representative of two independent experiments. (D) Functional profiles of ELA-specific CD8+ T cells primed from HUDs or PLWH as in (A). HLA-A2+ B-LCL target cells were pulsed with the indicated concentrations of ELA. Pie chart segments and colors represent the proportions of ELA-specific CD8+ T cells expressing the indicated number of functions, respectively. Data are representative of two independent experiments.
Collectively, these data indicated that highly sensitive and polyfunctional antigen-specific CD8+ T cells could be primed in PLWH via adjuvant delivery of the STING agonist cGAMP, which essentially recapitulated the effects of HIV-2-derived viral factors.
Discussion
In the present study, we found that HIV-2 primed functionally replete antigen-specific CD8+ T cells with enhanced survival properties more effectively than HIV-1, potentially facilitating effective antiviral immunity and the generation of long-term immunological memory. Moreover, this phenomenon could be recapitulated to a substantial extent via the adjuvant delivery of cGAMP, which activates STING. HIV-2 is known to trigger the cGAS-STING pathway via the activity of Vpx, which counteracts SAMHD1,27,28 and via direct interactions between the capsid protein and NONO, which enables the recognition of viral DNA by cGAS.30,31 These processes result in the induction of potent innate immune responses in DCs, including the production of type I IFNs. Our data showed that type I IFNs were largely responsible for the enhanced priming efficacy of HIV-2 and further explained the surrogate effects of exogenously administered cGAMP. Several host and viral factors have been proposed to account for the relatively innocuous disease course of infection with HIV-2.57, 58, 59, 60, 61, 62 The results presented here suggested another contributory mechanism, namely the prompt induction of potent antiviral CD8+ T cell immunity, which could enable the rapid and sustained control of viral replication and limit the early and profound damage to the immune system that typically occurs in the context of acute infection with HIV-1.
The functional properties gained by CD8+ T cells upon priming are greatly influenced by the different signals these cells receive, including from soluble factors, which is dependent on their direct cytokine environment. Exposure to HIV-2 or cGAMP triggers the secretion of a variety of cytokines, among which type I IFNs. Our work highlights a central role of type I IFNs, in particular IFN-α, in promoting the induction of functionally optimal CD8+ T cells during priming, although it is likely that type I IFNs may not act alone, but in combination with other cytokines in this process. Type I IFNs activate the JAK/STAT pathway and promote the upregulation of ISGs.63,64 It is important to mention that this type I interferon must remain transient and localized at the level of T cell priming, as chronic and systemic type I IFNs may in contrast negatively impact the immune response. Although generally beneficial in terms of antiviral immunity, type I IFNs can indeed have adverse systemic effects in the context of infection with HIV/SIV,65,66 potentially accelerating disease progression.67,68 These complexities likely reflect different spatiotemporal patterns in the overall innate immune response.69 Anatomically localized delivery would therefore seem imperative for any translational efforts focused on the use of type I IFNs, such as IFN-α, for the purposes of enhancing immune control of HIV-1. In line with our data, type I IFNs are known to link the innate and adaptive immune systems and promote the generation of effector memory CD8+ T cells.70,71 Moreover, several genes that were upregulated in antigen-specific CD8+ T cells primed in vitro either with ELA20 in the presence of cGAMP or with HIV-2 were also found to be upregulated in HIV-1-specific CD8+ T cells isolated and profiled directly ex vivo from elite controllers, including BCL2, CDC42, CCL3L1, CTLA4, ID2, IFNA1, IFNB1, TNFSF10, and VHL.44 Although this observation needs to be interpreted with caution, especially given the inevitable caveats that apply to in vitro versus in vivo comparisons and the relatively limited number of genes analyzed in our study (n = 96), the noted transcriptional similarities potentially identify a common signature of protective CD8+ T cells that could be explained by type I IFN-mediated effects operating at the level of DCs. In line with this notion, DCs from elite controllers can harbor relatively low levels of SAMHD1, enabling sustained cGAS-STING-mediated production of type I IFNs, associated with the potent stimulation of antigen-specific CD8+ T cells.72 This may confer HIV controllers a superior capacity to mount effective HIV-1-specific CD8+ T cell responses from the very early days of infection, thus helping to establish long-term control of the virus.
The balance between the activity of restriction factors and the induction of a potent type I IFN response appears to be key in the fight against pathogens, as epitomized by SAMHD1, which limits innate and adaptive immune responses against HIV-1.35,73 On the one hand, innate mechanisms that intrinsically suppress viral replication may control initial spread and limit the size of the tissue reservoir more effectively than type I IFNs. On the other hand, type I IFNs are likely more important for the induction and maintenance of adaptive immune responses that effectively control persistent viruses, such as HIV-2. In this context, it is tempting to speculate that HIV-1 has evolved to escape immune responses driven by type I IFNs, potentially at the cost of partial susceptibility to restriction by SAMHD1. A similar scenario has been described recently for herpes simplex virus type 1 in the context of the restriction factor MORC3.74
Our study has several limitations, primarily related to the use of an in vitro experimental system. To focus on viral sensing events and eliminate bias arising from differences in viral replication, we generated pseudotyped HIV-1 and HIV-2, which precluded an assessment of issues related to viral tropism and the potentially immunopathogenic properties of Env. Moreover, our in vitro approach did not recapitulate the complexity of the cellular interactions and the unique tissue architecture that characterize secondary lymphoid organs, where priming events occur in vivo. Future studies using human lymphoid organoids may prove better suited for this purpose.75 In addition, our model was poorly equipped to predict the long-term maintenance of immunological memory in vivo, which relies on numerous factors beyond the functional, phenotypic, and transcriptional properties of antigen-specific CD8+ T cells primed in vitro. Non-human primate studies may therefore be necessary for a more reliable assessment of the benefits of harnessing the cGAS-STING pathway for the induction of functionally replete memory CD8+ T cells to enhance immune control of HIV/SIV.
Type I IFNs hold great therapeutic potential to enhance adaptive immunity in the context of infection with HIV-1.76 Indirect approaches that mirror natural innate responses in the relevant immune compartments may nonetheless be more effective, because the systemic use of type I IFNs, such as IFN-α, can be complicated by side effects and a lack of target cell specificity. These concerns are especially pertinent for any such therapies designed to treat PLWH, in whom chronic infection is typically characterized by elevated levels of type I IFNs. A key finding of our study was the observation that cGAMP enhanced the priming of functionally optimal CD8+ T cells from PLWH on ART. Adjuvant delivery of cGAMP was associated with the production of IFNs but not proinflammatory cytokines and led to induction of antigen-sensitive and highly polyfunctional CD8+ T cells, which have been consistently linked with antiviral efficacy and may even reduce the size of the latent viral reservoir in conjunction with strategies designed to reactivate HIV-1, by eliminating reactivated HIV-infected CD4+ T cells from PLWH.77 As mentioned above, it is difficult to predict the in vivo impact of such interventions from an in vitro model, but the benefit of using cGAMP to induce effective antigen-specific CD8+ T cells has been demonstrated in mice.39,78 Accordingly, cGAMP may be a particularly useful adjuvant to enhance therapeutic CD8+ T cell-mediated control of HIV-1, especially if delivered alongside conserved antigens79 in a single vehicle, exemplified by mRNA-lipid nanoparticles and virus-like particles (VLPs).78,80,81 It is also notable here that cGAMP has been shown to activate latently infected cells in vitro, potentially helping to purge the body reservoir of HIV-1.82
On the basis of these collective observations, we propose that further studies are now warranted to design and test interventions based on the use of the STING agonist cGAMP, either alone or in combination with other factors that may act synergistically, to enhance immune control or even elimination of HIV-1.
Contributors
Conceptualization: N.M., A.S.-C., V.A.
Methodolody: M.P.C-P., L.P., F.P.-C., X.L., E.W., V.M.
Formal Analysis: M.P.C.-P., L.P., F.P.-C., X.L., S.V., N.M., A.S.-C., V.A.
Verification of the underlying data: M.P.C.-P., L.P., V.A.
Investigation: M.P.C-P., L.P., F.P.-C., X.L., N.M., A.S.-C., V.A.
Resources: S.L.-L., R.F., D.A.P., N.C.
Writing Original Draft: M.P.C.-P., F.P.-C., D.A.P., N.M., A.S.-C., V.A.
Writing Review & Editing: M.P.C.-P., F.P.-C., D.A.P., N.M., A.S.-C., V.A.
Supervision: D.A.P., N.M., A.S.-C., V.A.
Funding Acquisition: D.A.P., V.A.
All authors read and approved the final version of the manuscript.
M.P.C.-P. and L.P. contributed equally to this work.
F.P.-C. and X.L. contributed equally to this work.
Data sharing statement
All data reported in this paper will be shared by the lead contact upon request. No data generated during the course of this study were uploaded to external repositories in the public domain.
Declaration of interests
N.M. is a cofounder of Stimunity, a company that develops cGAMP-loaded VLPs. A.S.C has received honoraria from MSD and Gilead for lectures. F.P-C is an inventor on a patent application by the Institut Pasteur (Method for reprogramming CD8+ T cells to enhance their therapeutic potential and applications thereof, US 63/301,532).
Acknowledgements
We thank all blood donors from the Établissement Français du Sang (EFS). Luminex assays were performed at the CYBIO Core Facility (Cochin Institute, France).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104557.
Contributor Information
Nicolas Manel, Email: nicolas.manel@curie.fr.
Asier Saez-Cirion, Email: asier.saez-cirion@pasteur.fr.
Victor Appay, Email: victor.appay@immuconcept.org.
Appendix A. Supplementary data
References
- 1.Appay V., Nixon D.F., Donahoe S.M., et al. HIV-specific CD8+ T cells produce antiviral cytokines but are impaired in cytolytic function. J Exp Med. 2000;192(1):63–75. doi: 10.1084/jem.192.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goulder P.J., Tang Y., Brander C., et al. Functionally inert HIV-specific cytotoxic T lymphocytes do not play a major role in chronically infected adults and children. J Exp Med. 2000;192(12):1819–1832. doi: 10.1084/jem.192.12.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Appay V., Douek D.C., Price D.A. CD8+ T cell efficacy in vaccination and disease. Nat Med. 2008;14(6):623–628. doi: 10.1038/nm.f.1774. [DOI] [PubMed] [Google Scholar]
- 4.Collins D.R., Gaiha G.D., Walker B.D. CD8+ T cells in HIV control, cure and prevention. Nat Rev Immunol. 2020;20(8):471–482. doi: 10.1038/s41577-020-0274-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Betts M.R., Nason M.C., West S.M., et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107(12):4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Almeida J.R., Price D.A., Papagno L., et al. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J Exp Med. 2007;204(10):2473–2485. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hersperger A.R., Martin J.N., Shin L.Y., et al. Increased HIV-specific CD8+ T-cell cytotoxic potential in HIV elite controllers is associated with T-bet expression. Blood. 2011;117(14):3799–3808. doi: 10.1182/blood-2010-12-322727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Migueles S.A., Osborne C.M., Royce C., et al. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity. 2008;29(6):1009–1021. doi: 10.1016/j.immuni.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saez-Cirion A., Lacabaratz C., Lambotte O., et al. HIV controllers exhibit potent CD8 T cell capacity to suppress HIV infection ex vivo and peculiar cytotoxic T lymphocyte activation phenotype. Proc Natl Acad Sci U S A. 2007;104(16):6776–6781. doi: 10.1073/pnas.0611244104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Betts M.R., Price D.A., Brenchley J.M., et al. The functional profile of primary human antiviral CD8+ T cell effector activity is dictated by cognate peptide concentration. J Immunol. 2004;172(10):6407–6417. doi: 10.4049/jimmunol.172.10.6407. [DOI] [PubMed] [Google Scholar]
- 11.Almeida J.R., Sauce D., Price D.A., et al. Antigen sensitivity is a major determinant of CD8+ T-cell polyfunctionality and HIV-suppressive activity. Blood. 2009;113(25):6351–6360. doi: 10.1182/blood-2009-02-206557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trautmann L., Mbitikon-Kobo F.M., Goulet J.P., et al. Profound metabolic, functional, and cytolytic differences characterize HIV-specific CD8 T cells in primary and chronic HIV infection. Blood. 2012;120(17):3466–3477. doi: 10.1182/blood-2012-04-422550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takata H., Buranapraditkun S., Kessing C., et al. Delayed differentiation of potent effector CD8+ T cells reducing viremia and reservoir seeding in acute HIV infection. Sci Transl Med. 2017;9(377) doi: 10.1126/scitranslmed.aag1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goulder P.J., Walker B.D. HIV and HLA class I: an evolving relationship. Immunity. 2012;37(3):426–440. doi: 10.1016/j.immuni.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaslow R.A., Carrington M., Apple R., et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med. 1996;2(4):405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 16.Fellay J., Ge D., Shianna K.V., et al. Common genetic variation and the control of HIV-1 in humans. PLoS Genet. 2009;5(12) doi: 10.1371/journal.pgen.1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nyamweya S., Hegedus A., Jaye A., Rowland-Jones S., Flanagan K.L., Macallan D.C. Comparing HIV-1 and HIV-2 infection: lessons for viral immunopathogenesis. Rev Med Virol. 2013;23(4):221–240. doi: 10.1002/rmv.1739. [DOI] [PubMed] [Google Scholar]
- 18.Jaffar S., Wilkins A., Ngom P.T., et al. Rate of decline of percentage CD4+ cells is faster in HIV-1 than in HIV-2 infection. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;16(5):327–332. doi: 10.1097/00042560-199712150-00003. [DOI] [PubMed] [Google Scholar]
- 19.Esbjornsson J., Mansson F., Kvist A., et al. Long-term follow-up of HIV-2-related AIDS and mortality in Guinea-Bissau: a prospective open cohort study. Lancet HIV. 2019;6(1):E25–E31. doi: 10.1016/S2352-3018(18)30254-6. [DOI] [PubMed] [Google Scholar]
- 20.van der Loeff M.F., Larke N., Kaye S., et al. Undetectable plasma viral load predicts normal survival in HIV-2-infected people in a West African village. Retrovirology. 2010;7:46. doi: 10.1186/1742-4690-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thiebaut R., Matheron S., Taieb A., et al. Long-term nonprogressors and elite controllers in the ANRS CO5 HIV-2 cohort. AIDS. 2011;25(6):865–867. doi: 10.1097/QAD.0b013e328344892e. [DOI] [PubMed] [Google Scholar]
- 22.Esbjornsson J., Jansson M., Jespersen S., et al. HIV-2 as a model to identify a functional HIV cure. AIDS Res Ther. 2019;16(1):24. doi: 10.1186/s12981-019-0239-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leligdowicz A., Onyango C., Yindom L.M., et al. Highly avid, oligoclonal, early-differentiated antigen-specific CD8+ T cells in chronic HIV-2 infection. Eur J Immunol. 2010;40(7):1963–1972. doi: 10.1002/eji.200940295. [DOI] [PubMed] [Google Scholar]
- 24.de Silva T.I., Peng Y., Leligdowicz A., et al. Correlates of T-cell-mediated viral control and phenotype of CD8+ T cells in HIV-2, a naturally contained human retroviral infection. Blood. 2013;121(21):4330–4339. doi: 10.1182/blood-2012-12-472787. [DOI] [PubMed] [Google Scholar]
- 25.Angin M., Wong G., Papagno L., et al. Preservation of lymphopoietic potential and virus suppressive capacity by CD8+ T cells in HIV-2-infected controllers. J Immunol. 2016;197(7):2787–2795. doi: 10.4049/jimmunol.1600693. [DOI] [PubMed] [Google Scholar]
- 26.Lahaye X., Satoh T., Gentili M., et al. The capsids of HIV-1 and HIV-2 determine immune detection of the viral cDNA by the innate sensor cGAS in dendritic cells. Immunity. 2013;39(6):1132–1142. doi: 10.1016/j.immuni.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Coquel F., Silva M.J., Techer H., et al. SAMHD1 acts at stalled replication forks to prevent interferon induction. Nature. 2018;557(7703):57–61. doi: 10.1038/s41586-018-0050-1. [DOI] [PubMed] [Google Scholar]
- 28.Chen S., Bonifati S., Qin Z., et al. SAMHD1 suppresses innate immune responses to viral infections and inflammatory stimuli by inhibiting the NF-κB and interferon pathways. Proc Natl Acad Sci U S A. 2018;115(16):E3798–E3807. doi: 10.1073/pnas.1801213115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng Z., Dai T., He X., et al. The interactions between cGAS-STING pathway and pathogens. Signal Transduct Target Ther. 2020;5(1):91. doi: 10.1038/s41392-020-0198-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lahaye X., Gentili M., Silvin A., et al. NONO detects the nuclear HIV capsid to promote cGAS-mediated innate immune activation. Cell. 2018;175(2):488–501.e22. doi: 10.1016/j.cell.2018.08.062. [DOI] [PubMed] [Google Scholar]
- 31.Zuliani-Alvarez L., Govasli M.L., Rasaiyaah J., et al. Evasion of cGAS and TRIM5 defines pandemic HIV. Nat Microbiol. 2022;7(11):1762–1776. doi: 10.1038/s41564-022-01247-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Motomura K., Chen J., Hu W.S. Genetic recombination between human immunodeficiency virus type 1 (HIV-1) and HIV-2, two distinct human lentiviruses. J Virol. 2008;82(4):1923–1933. doi: 10.1128/JVI.01937-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silvin A., Yu C.I., Lahaye X., et al. Constitutive resistance to viral infection in human CD141+ dendritic cells. Sci Immunol. 2017;2(13) doi: 10.1126/sciimmunol.aai8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berre S., Gaudin R., Cunha de Alencar B., et al. CD36-specific antibodies block release of HIV-1 from infected primary macrophages and its transmission to T cells. J Exp Med. 2013;210(12):2523–2538. doi: 10.1084/jem.20130566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manel N., Hogstad B., Wang Y., Levy D.E., Unutmaz D., Littman D.R. A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature. 2010;467(7312):214–217. doi: 10.1038/nature09337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alanio C., Nicoli F., Sultanik P., et al. Bystander hyperactivation of preimmune CD8+ T cells in chronic HCV patients. Elife. 2015;4 doi: 10.7554/eLife.07916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Briceno O., Lissina A., Wanke K., et al. Reduced naive CD8+ T-cell priming efficacy in elderly adults. Aging Cell. 2016;15(1):14–21. doi: 10.1111/acel.12384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lissina A., Briceno O., Afonso G., et al. Priming of qualitatively superior human effector CD8+ T cells using TLR8 ligand combined with FLT3 ligand. J Immunol. 2016;196(1):256–263. doi: 10.4049/jimmunol.1501140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gutjahr A., Papagno L., Nicoli F., et al. The STING ligand cGAMP potentiates the efficacy of vaccine-induced CD8+ T cells. JCI Insight. 2019;4(7) doi: 10.1172/jci.insight.125107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gutjahr A., Papagno L., Nicoli F., et al. Cutting edge: a dual TLR2 and TLR7 ligand induces highly potent humoral and cell-mediated immune responses. J Immunol. 2017;198(11):4205–4209. doi: 10.4049/jimmunol.1602131. [DOI] [PubMed] [Google Scholar]
- 41.Papagno L., Kuse N., Lissina A., et al. The TLR9 ligand CpG ODN 2006 is a poor adjuvant for the induction of de novo CD8+ T-cell responses in vitro. Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-67704-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gutjahr A., Papagno L., Vernejoul F., et al. New chimeric TLR7/NOD2 agonist is a potent adjuvant to induce mucosal immune responses. eBioMedicine. 2020;58 doi: 10.1016/j.ebiom.2020.102922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price D.A., Brenchley J.M., Ruff L.E., et al. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J Exp Med. 2005;202(10):1349–1361. doi: 10.1084/jem.20051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Angin M., Volant S., Passaes C., et al. Metabolic plasticity of HIV-specific CD8+ T cells is associated with enhanced antiviral potential and natural control of HIV-1 infection. Nat Metab. 2019;1(7):704–716. doi: 10.1038/s42255-019-0081-4. [DOI] [PubMed] [Google Scholar]
- 45.Romero P., Valmori D., Pittet M.J., et al. Antigenicity and immunogenicity of Melan-A/MART-1 derived peptides as targets for tumor reactive CTL in human melanoma. Immunol Rev. 2002;188:81–96. doi: 10.1034/j.1600-065x.2002.18808.x. [DOI] [PubMed] [Google Scholar]
- 46.Pittet M.J., Valmori D., Dunbar P.R., et al. High frequencies of naive Melan-A/MART-1-specific CD8+ T cells in a large proportion of human histocompatibility leukocyte antigen (HLA)-A2 individuals. J Exp Med. 1999;190(5):705–715. doi: 10.1084/jem.190.5.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dutoit V., Guillaume P., Romero P., Cerottini J.C., Valmori D. Functional analysis of HLA-A∗0201/Melan-A peptide multimer+ CD8+ T cells isolated from an HLA-A∗0201- donor: exploring tumor antigen allorestricted recognition. Cancer Immun. 2002;2:7. [PubMed] [Google Scholar]
- 48.Cocchi F., DeVico A.L., Yarchoan R., et al. Higher macrophage inflammatory protein (MIP)-1α and MIP-1β levels from CD8+ T cells are associated with asymptomatic HIV-1 infection. Proc Natl Acad Sci U S A. 2000;97(25):13812–13817. doi: 10.1073/pnas.240469997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xin G., Schauder D.M., Lainez B., et al. A critical role of IL-21-induced BATF in sustaining CD8-T-cell-mediated chronic viral control. Cell Rep. 2015;13(6):1118–1124. doi: 10.1016/j.celrep.2015.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuse N., Sun X., Akahoshi T., et al. Priming of HIV-1-specific CD8+ T cells with strong functional properties from naive T cells. eBioMedicine. 2019;42:109–119. doi: 10.1016/j.ebiom.2019.03.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou X., Xue H.H. Cutting edge: generation of memory precursors and functional memory CD8+ T cells depends on T cell factor-1 and lymphoid enhancer-binding factor-1. J Immunol. 2012;189(6):2722–2726. doi: 10.4049/jimmunol.1201150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim M.V., Ouyang W., Liao W., Zhang M.Q., Li M.O. The transcription factor Foxo 1 controls central-memory CD8+ T cell responses to infection. Immunity. 2013;39(2):286–297. doi: 10.1016/j.immuni.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kawahara H., Minami R., Yokota N. BAG6/BAT3: emerging roles in quality control for nascent polypeptides. J Biochem. 2013;153(2):147–160. doi: 10.1093/jb/mvs149. [DOI] [PubMed] [Google Scholar]
- 54.Bosinger S.E., Utay N.S. Type I interferon: understanding its role in HIV pathogenesis and therapy. Curr HIV/AIDS Rep. 2015;12(1):41–53. doi: 10.1007/s11904-014-0244-6. [DOI] [PubMed] [Google Scholar]
- 55.Favre D., Stoddart C.A., Emu B., et al. HIV disease progression correlates with the generation of dysfunctional naive CD8low T cells. Blood. 2011;117(7):2189–2199. doi: 10.1182/blood-2010-06-288035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pacanowski J., Kahi S., Baillet M., et al. Reduced blood CD123+ (lymphoid) and CD11c+ (myeloid) dendritic cell numbers in primary HIV-1 infection. Blood. 2001;98(10):3016–3021. doi: 10.1182/blood.v98.10.3016. [DOI] [PubMed] [Google Scholar]
- 57.Marlink R., Kanki P., Thior I., et al. Reduced rate of disease development after HIV-2 infection as compared to HIV-1. Science. 1994;265(5178):1587–1590. doi: 10.1126/science.7915856. [DOI] [PubMed] [Google Scholar]
- 58.Duvall M.G., Jaye A., Dong T., et al. Maintenance of HIV-specific CD4+ T cell help distinguishes HIV-2 from HIV-1 infection. J Immunol. 2006;176(11):6973–6981. doi: 10.4049/jimmunol.176.11.6973. [DOI] [PubMed] [Google Scholar]
- 59.Duvall M.G., Precopio M.L., Ambrozak D.A., et al. Polyfunctional T cell responses are a hallmark of HIV-2 infection. Eur J Immunol. 2008;38(2):350–363. doi: 10.1002/eji.200737768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fernandes S.M., Pires A.R., Matoso P., et al. HIV-2 infection is associated with preserved GALT homeostasis and epithelial integrity despite ongoing mucosal viral replication. Mucosal Immunol. 2018;11(1):236–248. doi: 10.1038/mi.2017.44. [DOI] [PubMed] [Google Scholar]
- 61.Makvandi-Nejad S., Rowland-Jones S. How does the humoral response to HIV-2 infection differ from HIV-1 and can this explain the distinct natural history of infection with these two human retroviruses? Immunol Lett. 2015;163(1):69–75. doi: 10.1016/j.imlet.2014.10.028. [DOI] [PubMed] [Google Scholar]
- 62.Lopes A.R., Jaye A., Dorrell L., et al. Greater CD8+ TCR heterogeneity and functional flexibility in HIV-2 compared to HIV-1 infection. J Immunol. 2003;171(1):307–316. doi: 10.4049/jimmunol.171.1.307. [DOI] [PubMed] [Google Scholar]
- 63.Platanias L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5(5):375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 64.Mazewski C., Perez R.E., Fish E.N., Platanias L.C. Type I interferon (IFN)-regulated activation of canonical and non-canonical signaling pathways. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.606456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Noel N., Jacquelin B., Huot N., Goujard C., Lambotte O., Muller-Trutwin M. Interferon-associated therapies toward HIV control: the back and forth. Cytokine Growth Factor Rev. 2018;40:99–112. doi: 10.1016/j.cytogfr.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 66.Jacquelin B., Mayau V., Targat B., et al. Nonpathogenic SIV infection of African green monkeys induces a strong but rapidly controlled type I IFN response. J Clin Invest. 2009;119(12):3544–3555. doi: 10.1172/JCI40093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Su L. Pathogenic role of type I interferons in HIV-induced immune impairments in humanized mice. Curr HIV/AIDS Rep. 2019;16(3):224–229. doi: 10.1007/s11904-019-00444-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sivaraman V., Zhang L., Su L. Type I interferon contributes to CD4+ T cell depletion induced by infection with HIV-1 in the human thymus. J Virol. 2011;85(17):9243–9246. doi: 10.1128/JVI.00457-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saez-Cirion A., Manel N. Immune responses to retroviruses. Annu Rev Immunol. 2018;36:193–220. doi: 10.1146/annurev-immunol-051116-052155. [DOI] [PubMed] [Google Scholar]
- 70.Jennings R.N., Grayson J.M., Barton E.S. Type I interferon signaling enhances CD8+ T cell effector function and differentiation during murine gammaherpesvirus 68 infection. J Virol. 2014;88(24):14040–14049. doi: 10.1128/JVI.02360-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carbonneil C., Aouba A., Burgard M., et al. Dendritic cells generated in the presence of granulocyte-macrophage colony-stimulating factor and IFN-α are potent inducers of HIV-specific CD8 T cells. AIDS. 2003;17(12):1731–1740. doi: 10.1097/00002030-200308150-00002. [DOI] [PubMed] [Google Scholar]
- 72.Martin-Gayo E., Buzon M.J., Ouyang Z., et al. Potent cell-intrinsic immune responses in dendritic cells facilitate HIV-1-specific T cell immunity in HIV-1 elite controllers. PLoS Pathog. 2015;11(6) doi: 10.1371/journal.ppat.1004930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Maelfait J., Bridgeman A., Benlahrech A., Cursi C., Rehwinkel J. Restriction by SAMHD1 limits cGAS/STING-dependent innate and adaptive immune responses to HIV-1. Cell Rep. 2016;16(6):1492–1501. doi: 10.1016/j.celrep.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gaidt M.M., Morrow A., Fairgrieve M.R., Karr J.P., Yosef N., Vance R.E. Self-guarding of MORC3 enables virulence factor-triggered immunity. Nature. 2021;600(7887):138–142. doi: 10.1038/s41586-021-04054-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wagar L.E., Salahudeen A., Constantz C.M., et al. Modeling human adaptive immune responses with tonsil organoids. Nat Med. 2021;27(1):125–135. doi: 10.1038/s41591-020-01145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kwaa A.K.R., Talana C.A.G., Blankson J.N. Interferon alpha enhances NK cell function and the suppressive capacity of HIV-specific CD8+ T cells. J Virol. 2019;93(3):e01541–e01618. doi: 10.1128/JVI.01541-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lima N.S., Takata H., Huang S.H., et al. CTL clonotypes with higher TCR affinity have better ability to reduce the HIV latent reservoir. J Immunol. 2020;205(3):699–707. doi: 10.4049/jimmunol.1900811. [DOI] [PubMed] [Google Scholar]
- 78.Chauveau L., Bridgeman A., Tan T.K., et al. Inclusion of cGAMP within virus-like particle vaccines enhances their immunogenicity. EMBO Rep. 2021;22(8) doi: 10.15252/embr.202152447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hanke T. Aiming for protective T-cell responses: a focus on the first generation conserved-region HIVconsv vaccines in preventive and therapeutic clinical trials. Expert Rev Vaccines. 2019;18(10):1029–1041. doi: 10.1080/14760584.2019.1675518. [DOI] [PubMed] [Google Scholar]
- 80.Gentili M., Kowal J., Tkach M., et al. Transmission of innate immune signaling by packaging of cGAMP in viral particles. Science. 2015;349(6253):1232–1236. doi: 10.1126/science.aab3628. [DOI] [PubMed] [Google Scholar]
- 81.Bridgeman A., Maelfait J., Davenne T., et al. Viruses transfer the antiviral second messenger cGAMP between cells. Science. 2015;349(6253):1228–1232. doi: 10.1126/science.aab3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yamamoto T., Kanuma T., Takahama S., et al. STING agonists activate latently infected cells and enhance SIV-specific responses ex vivo in naturally SIV controlled cynomolgus macaques. Sci Rep. 2019;9(1):5917. doi: 10.1038/s41598-019-42253-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.