Summary
Background
Gut microbial dysbiosis contributes to colorectal cancer (CRC) pathogenesis, possibly mediated in part by increased intestinal permeability to endotoxin lipopolysaccharide (LPS), microbial translocation, and subsequent endotoxemia and inflammation. However, epidemiologic evidence linking circulating markers of microbial translocation with CRC risk is limited.
Methods
We conducted a prospective, nested case–control study of 261 incident CRC cases and 261 controls (matched on age and time of blood draw) among 18,159 men with pre-diagnostic blood specimens in the Health Professionals Follow-Up Study (1993–2009). We examined three complementary markers of microbial translocation and host response to bacteria, including LPS-binding protein (LBP), soluble CD14 (sCD14), and endotoxincore antibody (EndoCAb) immunoglobulin M (IgM), with subsequent risk of CRC. Unconditional logistic regressions were used to estimate odds ratios (ORs) and 95% confidence intervals (CIs).
Findings
Pre-diagnostic circulating levels of sCD14 were associated with a higher risk of incident CRC. Compared to men in the lowest quartile, the multivariable OR was 1.90 (95% CI, 1.13–3.22) for men in the highest quartile (OR per standard deviation [SD] increase, 1.28; 95%CI 1.06–1.53; Ptrend = 0.01). This positive association remained similar after adjusting for C-reactive protein, interleukin-6, and soluble tumor necrosis factor receptor-2, and within strata of putative CRC risk factors. We also observed a suggestive inverse association between EndoCAb IgM and risk of CRC (OR per SD increase, 0.84; 95%CI 0.69–1.02; Ptrend = 0.09).
Interpretation
Microbial translocation and host response to bacteria, as reflected by sCD14, is associated with risk of incident CRC in men.
Funding
US National Institutes of Health.
Keywords: Colorectal cancer, Microbial translocation, Gut dysbiosis
Research in context.
Evidence before this study
Accumulating evidence supports the role of gut microbial dysbiosis in colorectal carcinogenesis, likely mediated in part by increased permeability to endotoxin lipopolysaccharide (LPS) with subsequent endotoxemia and inflammation. However, due to challenges in measuring circulating levels of LPS, epidemiologic studies linking circulating markers of microbial translocation with colorectal cancer (CRC) are limited. LPS-binding protein (LBP), soluble CD14 (sCD14), and endotoxin core antibody (EndoCAb) immunoglobulin M (IgM) have emerged as promising markers to reflect microbial translocation, yet their associations with risk of CRC have not been comprehensively examined. We searched PubMed for articles published between Jan 1, 1990, and January 30, 2022, using the terms “LBP or sCD14 or EndoCAb” and “colorectal cancer”. One nested case–control study from the Multiethnic Cohort reported no association between pre-diagnostic plasma LBP levels and subsequent risk of CRC. However, we did not identify any epidemiologic studies that examined the association between sCD14 or EndoCAb IgM with risk of incident CRC.
Added value of this study
In this prospective, nested case–control study of 261 incident CRC cases and matched controls within the Health Professionals Follow-Up Study, men with plasma sCD14 levels in the highest quartile had a 90% increased risk of incident CRC compared to those in the lowest quartile. The association was independent of CRC putative risk factors and several systemic inflammatory markers. We also observed a suggestive inverse association between EndoCAb IgM and risk of incident CRC.
Implications of all the available evidence
Our findings lend support for the role of microbial translocation, as reflected by higher levels of plasma sCD14 and potentially lower levels of EndoCAb IgM, in colorectal carcinogenesis. Further research is needed to fully uncover the role of this emerging pathway in CRC and its promise in etiology and early detection.
Introduction
Colorectal cancer (CRC) is the 3rd leading cancer type for both incidence and mortality in the United States (US).1 Accumulating evidence supports the role of gut dysbiosis, a persistent imbalance of the microbial community in the gut, in colorectal carcinogenesis.2 Alterations in the intestinal barrier functions resulting either from gut microbiota dysbiosis, environmental, or genetic triggers,3 promote the translocation of bacteria or their components such as the endotoxin lipopolysaccharide (LPS) into the bloodstream, causing endotoxemia, which triggers a pro-inflammatory signaling cascade and immune responses.4
However, human studies linking microbial translocation and host response to bacteria with risk of CRC are limited in part due to challenges in biomarker measurement. LPS is one of the potent virulence factors of Gram-negative bacterial species with a major role in both acute and chronic infections.5 However, LPS has a short half-life and can easily contaminate labware and the environment, reliable measurement of circulating LPS requires LPS-free conditions throughout the collection and measurement processes.6 Such conditions are challenging to achieve, particularly for archived samples collected in cohort studies.
Recently, LPS-binding protein (LBP),7,8 soluble CD14 (sCD14),7, 8, 9, 10 and endotoxin core antibody (EndoCAb) immunoglobulin M (IgM)7,10 have emerged as promising markers to reflect microbial translocation and responses to LPS. LBP specifically binds and transfers bacterial LPS; therefore, increased production of LBP is considered an indicator of the presence of effective LPS in the body.11 sCD14, the soluble form of the LPS co-receptor and primarily produced by macrophages and hepatocytes, could also increase as part of the innate immune response to LPS.12 In LPS-primed monocytes, sCD14 augments Toll-like receptor 4 (TLR-4)13 and inflammatory biomediators. Given the long half-lives of LBP and sCD14 (24–28 h; LPS: 2 min), LBP and sCD14 have been suggested to reflect long-term exposure to endotoxin.14 EndoCAb IgM, inversely associated with LPS levels, is an antibody against LPS core oligosaccharide and another indirect marker of endotoxin exposure and can be measured in healthy human plasma.15,16 Importantly, these three markers can be reliably measured using archived blood.9,10,15,17 Thus far, epidemiologic studies on these markers in relation to subsequent risk of CRC are lacking.
To address these knowledge gaps, we prospectively investigated the associations between circulating pre-diagnostic levels of plasma LBP, sCD14 and EndoCAb IgM, and risk of incident CRC, leveraging high-quality blood biospecimen and rich clinical and lifestyle data collected from a large, prospective US cohort of men, the Health Professionals Follow-Up Study (HPFS).
Methods
Study population
The HPFS is an ongoing prospective cohort study of 51,529 US male health professionals aged 40–75 years at enrolment in 1986. Briefly, participants self-reported information on lifestyle and medical history biennially through follow-up questionnaires with a response rate of approximately 90%. Detailed dietary information was collected using validated food frequency questionnaires (FFQs) every four years. The study protocol was approved by the institutional review boards of the Brigham and Women's Hospital and Harvard T.H. Chan School of Public Health, and those of participating registries as required (IRB protocol number: 2001P001945). All participants provided informed consent upon completion of self-administered questionnaires and return of blood specimens.
Ascertainment of cases and selection of controls
A blood specimen was returned by 18,159 men in the HPFS on ice by overnight courier between 1993 and 1995. More than 95% of samples arrived within 24 h of collection. Procedures for blood collection, handling and storage have been summarized previously.18 Participants who reported a diagnosis of CRC in biennial questionnaires were asked for permission to review their medical records and pathologic reports. We identified unreported lethal CRC cases through the National Death Index and next-of-kin. A study physician blinded to exposure information reviewed records to confirm CRC diagnosis and to extract information on anatomic location. CRC was defined according to the International Classification of Diseases, Ninth Revision (colon cancer [ICD-9 codes 153.0–153.4, 153.6–153.9]; rectal cancer [ICD-9 codes 154.0 or 154.1]). Colon cancer was further classified into proximal colon cancer (ICD-9 codes 153.0, 153.1, 153.4, 153.6, 153.7) and distal colon cancer (ICD-9 codes 153.2 or 153.3). Among all participants with a pre-diagnostic blood specimen, we excluded individuals with a history of inflammatory bowel disease and identified 261 incident CRC cases through 2009.
Using risk set sampling, we randomly selected 1 control for each incident CRC, matching on age (within 2 years) and time of blood draw (within 1 month in the same year) from eligible participants who were alive and free of cancer (except for non-melanoma skin cancer) at the time of diagnosis of the CRC case. A total of 261 incident CRC cases and 261 controls were identified and all were included in this analysis.
Laboratory assays
The archived plasma samples of all cases and controls were extracted in 2021 and shipped to the UCLA AIDS Institute for biomarker measurement at one time. LBP and sCD14 were measured using multiplexed (Luminex platform) assay kits (Fluorokine MAP; R&D Systems); EndoCAb IgM was determined by enzyme-linked immunosorbent assay (ELISA) (Hycult Biotech), as previously described.7 Samples from cases and their matched controls were analyzed in the same batch. Quality control samples were randomly interspersed among the case–control samples. Personnel blinded to quality control and case–control status conducted all the assays. The mean intra-assay coefficients of variation (CVs) from blinded quality control samples were 13.6% for LBP, 8.3% for sCD14, 10.7% for EndoCAb IgM. After considering batch, the CVs were all equal to 4.6%.
Assessment of covariates and other inflammatory markers
Potential confounders, effect modifiers or mediators were identified from the literature19,20 and incorporated into a directed acyclic graph that was used to guide the modeling strategy (Supplementary Fig. S1). Detailed information on putative CRC risk factors was obtained from self-reported questionnaires beginning in 1986 through the time of blood draw (1993–95). This included adult height, current weight, family history of CRC in one or more first-degree relatives, smoking status (never or ever smoked) and pack-years of smoking among ever smokers, physical activity in metabolic equivalent of task (MET)-hours, and regular use (≥2 tablets/week) of aspirin or non-steroidal anti-inflammatory drugs (NSAIDs). We used adult height and current weight information to calculate the body mass index (BMI, kg/m2). Using FFQs, intake of alcohol, red and processed meat, dietary fiber, total folate (from food and supplements), total calcium and total vitamin D was updated every 4 years. Diet quality was assessed by the Alternative Healthy Eating Index (AHEI)-2010 excluding the alcohol component. We used cumulative average BMI, physical activity, aspirin/NSAIDs use, and dietary intake variables by averaging the values from 1986 to the time of blood draw. Biennially, participants reported history of colonoscopy or sigmoidoscopy during the previous 2 years.
We also used extant data on three systemic inflammatory markers measured previously in this nested case–control samples, including C-reactive protein (CRP), interleukin-6 (IL-6), and soluble tumor necrosis factor receptor-2 (sTNFR-2).18 In brief, CRP was measured using a highly sensitive immunoturbidimetric assay (Denka Seiken Co), IL-6 was measured with an ultra-sensitive ELISA (R&D Systems), and sTNFR-2 was measured by ELISA (R&D Systems). The CVs were 7.8% for CRP, 12.1% for IL-6, and 10.1% for sTNFR-2.
Statistical analyses
We first corrected the biomarker concentrations of LBP, sCD14, and EndoCAb IgM using the average batch correction method to minimize the potential influence of laboratory batch-related variability. Generalized extreme studentized deviate (ESD) many-outlier detection was performed to detect outliers and set them as missing in further analyses. Spearman correlation coefficients were calculated between plasma markers of microbial translocation and plasma markers of inflammation among control participants. We also performed factor analysis on these biomarkers to derive three biomarker patterns. We used the iterated principal factor method to extract factors, followed by orthogonal varimax rotation. We retained three factors based on eigenvalues and inspection of scree plots.
To examine the association between LBP, sCD14, EndoCAb IgM and subsequent risk of CRC, we categorized each plasma marker into quartiles based on their distributions among controls. Unconditional logistic regressions were used to estimate the quartile-specific odds ratios (ORs) and 95% confidence intervals (CIs) for CRC, using the lowest quartiles as the respective reference categories. Tests for trend were performed using the median value for each quartile as a continuous variable. We estimated OR (95% CI) per 1 standard deviation (SD) increase. To allow for comparison across different samples or populations,21,22 we also estimated unstandardized regression coefficient and OR per 1000 ng/ml increase in LBP, per 100 ng/ml increase in sCD14, and per 10 MMU/ml increase in EndoCAb IgM. We assessed linearity using Box–Tidwell test and conducted restricted cubic splines23,24 to non-parametrically examine the possible non-linear relation between biomarkers and risk of CRC. We tested for non-linearity using the likelihood ratio test, comparing the model with only the linear term to the model with the linear and the cubic spline terms. As the planned stratified analysis disassembled the matching, unconditional instead of conditional logistic regression was used to estimate OR and 95% CIs to retain samples and allow for direct comparison of effect estimates throughout the analyses.
In the multivariable analyses, we adjusted for the matching factors (age and time of blood draw [continuous]) and for race (white, non-white), BMI (continuous), family history of CRC (yes, no), prior history of sigmoidoscopy/colonoscopy (yes, no), regular use of aspirin (yes, no), regular use of NSAIDs (yes, no), pack-years of smoking (continuous), physical activity (continuous), intake of alcohol, red and processed meat, dietary fiber, total folate (from foods and supplements), total calcium and total vitamin D (all continuous), and AHEI-2010 without alcohol (continuous). To evaluate whether the identified associations were mediated by major systemic inflammatory markers associated with CRC,25, 26, 27 we further adjusted for CRP, IL-6, and sTNFR-2 (all continuous) to estimate the controlled direct effect of our exposures. We also conducted sensitivity analyses after excluding cases diagnosed within 2 years of blood draw and their matched controls to diminish the potential influence of subclinical neoplasia on circulating marker levels.
Furthermore, we performed stratified analyses for the association of per SD increase in sCD14 with risk of CRC according to age at blood draw (<, ≥65 years), time since blood draw (<, ≥6 years), BMI (<25, ≥25 kg/m2), alcohol intake (<15, ≥15 g/day), smoking status (never, ever), prior history of sigmoidoscopy/colonoscopy (yes, no), family history of CRC (yes, no), regular use of aspirin (yes, no) and AHEI-2010 without alcohol (<, ≥ median). Tests for interaction were performed using the Wald test based on a cross-product term of each potential effect modifier and per SD increase in biomarker.
We also assessed whether the association between sCD14 and risk of CRC differed according to age of CRC diagnosis and tumor location, using polytomous logistic regressions (age of CRC diagnosis < median age vs. ≥ median age vs. controls; colon cancer vs. rectal cancer vs. controls). P-values for heterogeneity were estimated based on the Wald test. We used SAS version 9.4 (SAS Institute, Inc) for all statistical analyses. All statistical tests were two-sided, and a P-value less than .05 was considered statistically significant.
Role of the funders
The funders had no role in study design; the collection, analysis, and interpretation of data; writing of the report; or the decision to submit for publication.
Results
Over up to 16 years of follow-up after blood collection, a total of 261 incident CRC cases and 261 matched controls were included in the analysis (Table 1). At the time of blood draw, participants who subsequently developed CRC were more likely than controls to have family history of CRC and less likely to have prior history of sigmoidoscopy/colonoscopy or to use NSAIDs regularly. Also, cases had lower intake of vitamin D and higher circulating levels of CRP and IL-6 than controls.
Table 1.
Characteristics of CRC cases and controls at blood draw, HPFS 1993–1995.a
| Characteristics | Cases (n = 261) | Controls (n = 261) |
|---|---|---|
| Age at blood draw, years | 65.7 (8.4) | 65.6 (8.4) |
| Blood draw to diagnosis, years | 6.9 (4.1) | NA |
| White, n (%) | 245 (93.9) | 253 (96.9) |
| Body mass index, kg/m2b | 26.2 (3.0) | 25.2 (2.6) |
| Family history of CRC, n (%) | 51 (19.5) | 39 (14.9) |
| Prior history of sigmoidoscopy/colonoscopy, n (%) | 149 (57.1) | 167 (64.0) |
| Regular use of aspirin (≥2 tablets/week), n (%)b | 112 (42.9) | 120 (46.0) |
| Regular use of NSAIDs (≥2 tablets/week), n (%)b | 28 (10.7) | 34 (13.0) |
| Ever smokers, n (%) | 144 (55.2) | 144 (55.2) |
| Pack-years among ever smokers, median (IQR) | 15.0 (8.0–27.0) | 15.0 (5.0–26.0) |
| Physical activity, MET-hours/week, median (IQR)b | 25.6 (12.1–40.2) | 25.5 (12.8–37.9) |
| Dietary intakeb | ||
| Alcohol, g/day, median (IQR) | 7.3 (0.9–18.3) | 6.9 (1.9–16.9) |
| Red and processed meats, servings/week, median (IQR) | 5.9 (3.8–9.0) | 6.1 (3.5–8.9) |
| Dietary fiber, g/day | 22.2 (6.3) | 22.7 (6.6) |
| Total folate, μg/day | 496 (201) | 537 (240) |
| Total calcium, mg/day | 909 (328) | 940 (370) |
| Total vitamin D, IU/day, median (IQR) | 403 (243–546) | 410 (262–636) |
| AHEI-2010 without alcohol | 48.1 (10.6) | 48.3 (10.4) |
| Plasma markers of inflammation | ||
| CRP, mg/L, median (IQR) | 1.43 (0.72–2.83) | 1.06 (0.57–2.17) |
| IL-6, pg/ml, median (IQR) | 1.61 (1.00–2.68) | 1.39 (0.93–2.37) |
| sTNFR-2, pg/ml, median (IQR) | 2751 (2356–3273) | 2684 (2323–3273) |
| Plasma markers of microbial translocationc | ||
| LBP, ng/ml, median (IQR) | 4340 (3650–5009) | 4274 (3694–4839) |
| sCD14, ng/ml, median (IQR) | 1212 (1103–1365) | 1191 (1075–1320) |
| EndoCAb IgM, MMU/ml, median (IQR) | 47.5 (32.6–64.4) | 51.8 (35.9–73.2) |
AHEI, Alternative Healthy Eating Index; CRC, colorectal cancer; CRP, C-reactive protein; EndoCAb, endotoxin core antibody HPFS, Health Professionals Follow-up Study (men only); IgM, immunoglobulin M; IL-6, interleukin-6; IQR, interquartile range; LBP, lipopolysaccharide-binding protein; MET, metabolic equivalent of task; NA, not applicable; NSAID, non-steroidal anti-inflammatory drug; sCD14, soluble CD14; SD, standard deviation; sTNFR-2, soluble tumor necrosis factor receptor-2.
Data were presented as mean (SD) unless otherwise indicated.
Cumulative average value was used by averaging the values from baseline to the time of blood draw.
Batch corrected concentrations were used.
A total of 259 cases and 260 controls for LBP (2 missing measurement, 1 outlier), 259 cases and 260 controls for sCD14 (3 outliers), and 253 cases and 259 controls for EndoCAb IgM (10 outliers) were included in the final analyses. LBP and sCD14 levels were higher among cases than controls, while EndoCAb IgM levels were lower. Among controls, most baseline characteristics did not differ systematically by quartile of the three biomarkers (Supplementary Tables S1–S3). sCD14 and LBP were modestly but significantly correlated (Spearman r = 0.28, P < 0.001), while no significant correlation was found between EndoCAb IgM and sCD14 or LBP (Supplementary Table S4). In addition, LBP and sCD14 were modestly correlated with CRP, IL-6, and sTNFR-2 (Spearman r ranged from 0.25 to 0.50), whereas EndoCAb IgM was weakly negatively correlated with sTNFR-2 (r = −0.16) but not correlated with CRP (r = −0.10) and IL-6 (r = −0.02). Using factor analysis, three biomarker patterns were identified and named based on which biomarkers loaded most highly on the pattern, including CRP/IL-6 pattern, sTNFR-2 dominant pattern, and EndoCAb IgM dominant pattern. LBP contributed to CRP/IL-6 pattern, while sCD14 contributed to both the CRP/IL-6 and sTNFR-2 dominant patterns. In contrast, EndoCAb IgM led its own pattern (Supplementary Table S5).
Higher levels of plasma sCD14 were associated with an increased risk of CRC after adjusting for known or putative CRC risk factors (Table 2 and Supplementary Fig. S2B). Compared to men with sCD14 levels in the lowest quartile (Q1), men with levels in the highest quartile (Q4) had a 90% increased risk of CRC (OR, 1.90; 95%CI, 1.13–3.22; Ptrend = 0.01), with a 28% higher risk per SD increase in sCD14 (OR, 1.28; 95%CI 1.06–1.53). Additional adjustment for markers of inflammation, including CRP, IL-6, and sTNFR-2, only slightly attenuated the magnitude of this association. These associations also remained robust after excluding CRC cases diagnosed within 2 years of blood draw (Supplementary Table S6) or using conditional logistic regression (Supplementary Table S7). In contrast, plasma levels of LBP (OR per SD increase, 1.02; 95%CI, 0.86–1.21; Ptrend = 0.88) was not associated with risk of CRC. For EndoCAb IgM, we also identified a suggestive inverse association with subsequent risk of CRC (OR per SD increase, 0.84; 95%CI, 0.69–1.02; Ptrend = 0.09). ORs (95% CI) per 1000 ng/ml increase in LBP, per 100 ng/ml increase in sCD14, and per 10 MMU/ml increase in EndoCAb IgM were reported in Supplementary Table S8. The restricted cubic splines showed a pattern similar to the quartile analysis (Supplementary Fig. S2).
Table 2.
Association of circulating markers of microbial translocation with risk of CRC, HPFS 1993–2009.
| Biomarkers | Quartilea |
Per SD increaseb | Ptrendc | |||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |||
| LBP | ||||||
| No. of cases/controls | 73/65 | 53/65 | 52/65 | 81/65 | ||
| Median, ng/ml | 3103 | 3994 | 4499 | 5308 | ||
| Cut points, ng/ml | 1350–3691 | 3692–4268 | 4269–4832 | 4833–7980 | ||
| Age-adjusted OR (95% CI)d | 1 (referent) | 0.75 (0.46–1.24) | 0.71 (0.43–1.17) | 1.14 (0.71–1.84) | 1.04 (0.88–1.22) | 0.64 |
| MV-adjusted OR (95% CI)e | 1 (referent) | 0.76 (0.46–1.28) | 0.63 (0.37–1.06) | 1.09 (0.66–1.79) | 1.02 (0.86–1.21) | 0.88 |
| MV-adjusted OR (95% CI)f | 1 (referent) | 0.84 (0.48–1.44) | 0.59 (0.34–1.03) | 0.97 (0.56–1.68) | 0.93 (0.77–1.13) | 0.66 |
| sCD14 | ||||||
| No. of cases/controls | 55/65 | 56/65 | 67/65 | 81/65 | ||
| Median, ng/ml | 997 | 1134 | 1249 | 1420 | ||
| Cut points, ng/ml | 649–1074 | 1075–1189 | 1190–1317 | 1318–1949 | ||
| Age-adjusted OR (95% CI)d | 1 (referent) | 1.04 (0.62–1.73) | 1.23 (0.74–2.03) | 1.51 (0.93–2.47) | 1.17 (0.99–1.40) | 0.07 |
| MV-adjusted OR (95% CI)e | 1 (referent) | 1.29 (0.75–2.20) | 1.45 (0.85–2.49) | 1.90 (1.13–3.22) | 1.28 (1.06–1.53) | 0.01 |
| MV-adjusted OR (95% CI)f | 1 (referent) | 1.30 (0.73–2.30) | 1.54 (0.86–2.74) | 1.88 (1.05–3.37) | 1.26 (1.02–1.56) | 0.03 |
| EndoCAb IgM | ||||||
| No. of cases/controls | 77/65 | 69/65 | 60/64 | 47/65 | ||
| Median, MMU/ml | 27.2 | 43.6 | 61.3 | 92.9 | ||
| Cut points, MMU/ml | 10.1–35.8 | 35.9–51.7 | 51.8–73.0 | 73.1–181.9 | ||
| Age-adjusted OR (95% CI)d | 1 (referent) | 0.89 (0.55–1.44) | 0.77 (0.47–1.25) | 0.60 (0.36–1.00) | 0.82 (0.68–0.99) | 0.04 |
| MV-adjusted OR (95% CI)e | 1 (referent) | 0.88 (0.53–1.44) | 0.83 (0.50–1.38) | 0.63 (0.37–1.07) | 0.84 (0.69–1.02) | 0.09 |
| MV-adjusted OR (95% CI)f | 1 (referent) | 0.96 (0.57–1.62) | 0.89 (0.52–1.53) | 0.66 (0.38–1.14) | 0.83 (0.68–1.02) | 0.12 |
CI, confidence interval; CRC, colorectal cancer; CRP, C-reactive protein; EndoCAb, endotoxin core antibody; HPFS, Health Professionals Follow-up Study (men only); IgM, immunoglobulin M; IL-6, interleukin-6; LBP, lipopolysaccharide-binding protein; MV, multivariable; OR, odds ratio; sCD14, soluble CD14; SD, standard deviation; sTNFR-2, soluble tumor necrosis factor receptor-2.
Quartiles of the three biomarkers were defined based on the distribution in the controls.
SD for LBP: 942 ng/ml; SD for sCD14: 194 ng/ml; SD for EndoCAb IgM: 30.6 MMU/ml.
P for trend was calculated using the median of each quartile as a continuous variable.
Adjusted for matching factors (age and time of blood draw [continuous]) and race (white, non-white).
Additionally adjusted for body mass index (continuous), family history of CRC (yes, no), prior history of sigmoidoscopy/colonoscopy (yes, no), regular use of aspirin (yes, no), regular use of non-steroidal anti-inflammatory drugs (yes, no), pack-years of smoking (continuous), physical activity (continuous), alcohol intake, red and processed meat, dietary fiber, total folate (from foods and supplements), calcium, and vitamin D intake (all continuous), and Alternative Healthy Eating Index-2010 score without alcohol (continuous).
Additionally adjusted for CRP, IL-6, and sTNFR-2 (all continuous).
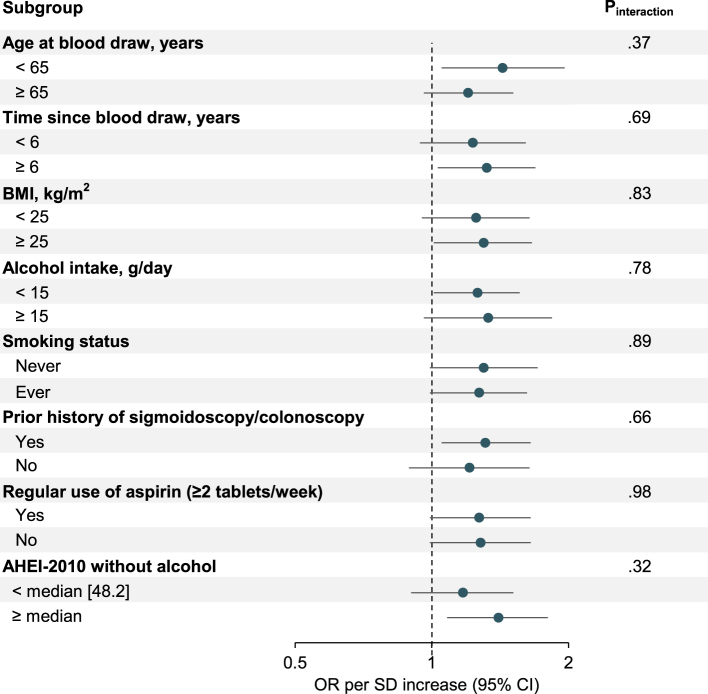
Stratified analyses for sCD14 and CRC according to age at blood draw, time since blood draw, BMI, alcohol intake, smoking status, prior history of sigmoidoscopy/colonoscopy, regular use of aspirin and AHEI-2010 score without alcohol did not reveal any statistically significant interactions (all Pinteraction ≥0.32; Fig. 1 and Supplementary Table S9). Nonetheless, somewhat stronger associations were observed among men who were younger at blood draw (OR per SD increase, 1.43; 95%CI, 1.05–1.96; Ptrend = 0.03) than among those of older age at blood draw (OR per SD increase, 1.20; 95%CI, 0.96–1.51; Ptrend = 0.27), and among individuals with higher AHEI-2010 score without alcohol (above-median [48.2]; OR per SD increase, 1.40; 95%CI, 1.08–1.80; Ptrend = 0.02) compared to those with lower AHEI-2010 score without alcohol (below-median; OR per SD increase, 1.17; 95%CI, 0.90–1.51; Ptrend = 0.19).
Fig. 1.
Associations of sCD14 (per SD increase) with risk of CRC according to age at blood draw, time since blood draw, and putative CRC risk factors, HPFS 1993–2009. AHEI, Alternative Healthy Eating Index; BMI, body mass index; CI, confidence interval; CRC, colorectal cancer; HPFS, Health Professionals Follow-up Study (men only); OR, odds ratio; sCD14, soluble CD14; SD, standard deviation.
Further, plasma sCD14 levels were not differentially associated with risk of CRC according to age of CRC diagnosis or tumor location (all Pheterogeneity ≥0.51) (Table 3), although a somewhat stronger association was observed for cases younger than the median age (73.8 years) (OR per SD increase, 1.42; 95%CI, 1.12–1.82; Ptrend = 0.03), compared to those diagnosed at older ages (OR per SD increase, 1.10; 95%CI, 0.87–1.40; Ptrend = 0.38).
Table 3.
Association of sCD14 with risk of CRC according to age at diagnosis and tumor location, HPFS 1993–2009.
| Quartilea |
Per SD increaseb | Ptrendc | Pheterogeneity | ||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||||
| Age of CRC diagnosis | 0.51 | ||||||
| < median age [73.8 years] | |||||||
| No. of cases/controls | 32/65 | 27/65 | 29/65 | 41/65 | |||
| MV-adjusted OR (95% CI)d | 1 (referent) | 1.08 (0.54–2.14) | 1.31 (0.65–2.63) | 2.03 (1.05–3.92) | 1.42 (1.12–1.82) | 0.03 | |
| ≥ median age | |||||||
| No. of cases/controls | 23/65 | 29/65 | 38/65 | 40/65 | |||
| MV-adjusted OR (95% CI)d | 1 (referent) | 1.61 (0.76–3.41) | 1.49 (0.73–3.07) | 1.51 (0.74–3.10) | 1.10 (0.87–1.40) | 0.38 | |
| Tumor location | 0.79 | ||||||
| Colon | |||||||
| No. of cases/controls | 38/65 | 37/65 | 37/65 | 54/65 | |||
| MV-adjusted OR (95% CI)d | 1 (referent) | 1.23 (0.68–2.24) | 1.13 (0.61–2.07) | 1.81 (1.02–3.22) | 1.25 (1.02–1.53) | 0.05 | |
| Rectal | |||||||
| No. of cases/controls | 11/65 | 17/65 | 17/65 | 19/65 | |||
| MV-adjusted OR (95% CI)d | 1 (referent) | 2.16 (0.88–5.27) | 1.72 (0.70–4.24) | 2.45 (1.01–5.92) | 1.36 (1.02–1.82) | 0.09 | |
CI, confidence interval; CRC, colorectal cancer; HPFS, Health Professionals Follow-up Study (men only); MV, multivariable; OR, odds ratio; sCD14, soluble CD14; SD, standard deviation.
Quartile of sCD14 was defined based on the distribution in the controls.
SD for sCD14: 194 ng/ml.
P for trend was calculated using the median of each quartile as a continuous variable.
Adjusted for matching factors (age and time of blood draw [continuous]), as well as race (white, non-white), body mass index (continuous), family history of CRC (yes, no), prior history of sigmoidoscopy/colonoscopy (yes, no), regular use of aspirin (yes, no), regular use of non-steroidal anti-inflammatory drugs (yes, no), pack-years of smoking (continuous), physical activity (continuous), alcohol intake, red and processed meat, dietary fiber, total folate (from foods and supplements), calcium, and vitamin D intake (all continuous), and Alternative Healthy Eating Index-2010 score without alcohol (continuous).
Discussion
In this prospective, nested case–control study of US men, higher levels of plasma sCD14 were associated with a higher risk of CRC, independent of putative risk factors and systemic inflammatory markers including CRP, IL-6, and sTNFR-2. We also observed a suggestive inverse association between EndoCAb IgM and risk of CRC. However, we found no association between LBP and subsequent risk of CRC. Our findings provide epidemiologic evidence that microbial translocation, as reflected by plasma sCD14 and potentially EndoCAb IgM, may play important roles in colorectal carcinogenesis, and if validated, may have the promise to serve as a pre-diagnostic marker.
Sparse but growing evidence support the link between sCD14 and CRC. For instance, miRNA levels of CD14 were found to be downregulated in 188 microsatellite stable CRC cases compared to normal mucosa.28 In addition, polymorphisms of CD14 such as rs4914 were associated with CRC risk in 479 cases and 486 controls in the Chinese Han population.29 We are not aware of other reports of a positive association between pre-diagnostic plasma sCD14 and subsequent risk of CRC. Notably, the association we observed for sCD14 with CRC was largely independent of systemic inflammatory markers for CRC, including CRP, IL-6, and sTNFR-2,25, 26, 27 indicating that sCD14 or biologic processes captured by sCD14 may independently contribute to colorectal carcinogenesis or operate through inflammatory pathways not captured by the three inflammatory markers. Comprehensive profiling of inflammatory markers is needed to elucidate the independent contribution of sCD14 and related pathways to colorectal carcinogenesis. While validations are warranted, the suggestive stronger associations for participants who are younger or have a healthier diet at blood draw, and for cases diagnosed at younger ages, if confirmed, support the promise of using sCD14 for early CRC detection among individuals not considered at high risk based on other known risk factors. Of particular interest is whether sCD14 is a pre-diagnostic marker for early-onset CRC diagnosed under age 50 or 55, which has been increasing substantially in the US30 and gut dysbiosis has been hypothesized to be the major driver with limited empirical evidence.31
Findings on LBP and CRC have been mixed. Germline polymorphisms in LBP, such as rs223259629 and rs5743533,32 were associated with increased risk of CRC. However, similar to our findings, a recent nested case–control study of 819 CRC from the Multiethnic Cohort (MEC) reported no association between pre-diagnostic plasma LBP levels and risk of CRC.20 It is hypothesized that delayed processing or long-term storage may affect LBP measurements and in part have led to a spurious outcome. Indeed, the range of LBP levels observed in the MEC samples (cases: 2–108 μg/ml; controls: 4–100 μg/ml) stretched notably higher than that typically observed in normal healthy populations (1–24 μg/ml).33 While LBP levels in our study sample had much narrower distributions (cases: 1.8–7.2 μg/ml, controls: 1.7–8.0 μg/ml), we similarly did not identify an association between pre-diagnostic LBP and CRC risk. Additional studies are warranted.
To our knowledge, microbial translocation reflected by suppressed levels of EndoCAb IgM was largely reported among HIV-infected population,10,15,17 and no prior studies have examined the association between EndoCAb IgM and CRC risk. Although not statistically significant after multivariable adjustment, we observed a suggestive inverse association between EndoCAb IgM and CRC risk. While future studies with greater power are needed, the totality of our findings supports the need to further evaluate the role of circulating markers of microbial translocation and host response to bacteria in CRC development. As indirect markers of gut barrier hyperpermeability, plasma levels of LBP, sCD14, and EndoCAb IgM predominately capture responses to LPS, a consequence of increased gut permeability and/or mucosal damage.3 As we increasingly recognized the role of non-bacterial species and the importance of trans kingdom interactions in CRC, studies on markers such as (1 → 3)-β-D-Glucan,34, 35, 36, 37 a fungal translocation marker recently found to be correlated with LPS and sCD14, may be promising. Studies of other recently emerged surrogate markers of gut barrier dysfunction (e.g., zonulin or fatty acid binding protein-23,8), markers that are crucial for the regulation of host-microbiota interplay (e.g., regenerating islet-derived protein 3 A38), and direct measurement of intestinal permeability (e.g., oral probes of 13C-mannitol or rhamnose3), along with deep characterization of the gut microbiome, may contribute further insights on the role of microbial translocation and gut barrier dysfunction in the etiology and prevention of CRC.
Our findings on the association between circulating markers of microbial translocation and risk of CRC are biologically plausible. Some putative CRC risk factors, such as high fat diet and alcohol can damage the intestinal barrier, resulting in either direct induction of epithelial cell damage and intracellular signaling responses or indirect interaction with the intestinal microbiota.3 The impaired gut barrier is partially characterized by increased gut permeability, which permits translocation of LPS and other pathogens into the circulation, activates TLR-4, and promotes endotoxemia and inflammation. More specifically, in the circulation, LPS is first recognized by LBP, which mediates interaction of LPS with CD14 on the cell surface; CD14 then splits LPS aggregates into monomeric molecules and presents them to the TLR4-myeloid differentiation factor 2 (MD-2) complex.13 Once LPS binds the TLR4-MD-2 complex and is recognized, TLR4 is activated, leading to the production of pro-inflammatory cytokines which are necessary to activate immune responses.13 In addition, disruption of the gut barrier contributes to a shift in the composition of the microbiota, with enrichment of deleterious bacterial populations (e.g. Bacteroides39) and depletion or reduction of commensal bacterial species (e.g. Clostridium39). Microbial translocation can further lead to the formation of pro-inflammatory cytokines and release of reactive oxygen species.4 During these processes, chronic inflammation, uncontrolled epithelial cell proliferation, and colorectal neoplasia formation can be potentiated, contributing to the initiation and/or progression of CRC.
The major strength of our study is the nested case–control design embedded with a well-characterized prospective cohort with both high-quality pre-diagnostic blood collection and long-term follow up of disease outcomes. This unparalleled setting allowed us to examine the association between three complementary biomarkers of microbial translocation with subsequent risk of incident CRC. We also implemented a rigorous study design during biomarker measurement. Notably, the CVs for each of the marker were <5% after batch correction, which are considered ideal in epidemiologic studies.40 Furthermore, leveraging previous measurement of well-known systemic inflammatory markers, we reported minimal mediation of the positive association between sCD14 and CRC by CRP, IL-6, and sTNFR-2, highlighting the need of in-depth investigations of both sCD14 related and inflammation related pathways. Finally, the collection of detailed information on lifestyle and medical history also allowed us to adjust for a list of potential covariates to minimize confounding. We have also rigorously adjusted for age throughout the analyses. Nonetheless, several limitations also warrant consideration. First, a single measurement of plasma markers may not represent long-term concentrations. While prior studies reported reasonable stability of sCD14 over a 2-year period,17 studies with repeated blood collection will allow us to evaluate the role of sCD14 trajectories in relation to CRC risk. Measurement error due to categorization of the exposure variables may also exist. However, both continuous and restricted cubic spline analyses support the positive association between sCD14 and risk of CRC. Second, residual confounding due to measurement error in the confounders (such as smoking, physical activity, and dietary intake) or unmeasured confounding cannot be ruled out completely. Third, when testing interaction, additive interaction is preferred in terms of both causality and public health but cannot be assessed in matched case–control studies.41 Forth, the vast majority of our study population comprised white male health professionals, thus limiting the generalizability of the findings to women or other racial/ethnical groups. Studies are also warranted to evaluate the clinical utility of sCD14, in early detection and risk prediction.
In conclusion, in this prospective, nested case–control study of US men, higher levels of plasma sCD14 were associated with an increased risk of incident CRC and we also observed a suggestive inverse association between EndoCAb IgM and subsequent risk of CRC. Our findings support for the role of microbial translocation in colorectal carcinogenesis and point to the value of further investigations of biomarkers of these processes for CRC etiology and prevention.
Contributors
MS and YC accessed and verified the underlying data. YC is responsible for the decision to submit the manuscript.
Concept and design: YC
Acquisition, analysis, or interpretation of data: MS, XZ, JH, BMB, OM, ME, ATC, ELG, YC
Drafting of the manuscript: MS, YC
Critical revision of the manuscript for important intellectual content: MS, XZ, JH, BMB, OM, ME, ATC, ELG, YC
Statistical analysis: MS, XZ, YC
Obtained funding: ATC, ELG, YC
Administrative, technical, or material support: OM, ME, YC
Supervision: YC
All authors have read and approved the final version of the manuscript.
Data sharing statement
Information including the procedures for obtaining and accessing data from the Health Professionals Follow-Up Study is described at https://sites.sph.harvard.edu/hpfs/.
Declaration of interests
ATC has served as an investigator on studies supported by Pfizer, Freenome, and Zoe Ltd. outside the submitted work. ATC also serves as a consultant for Pfizer, Boehringer Ingelheim, Bayer Pharma AG outside the submitted work. YC has served as a consultant for Geneoscopy outside the submitted work.
Acknowledgments
The authors would like to acknowledge the contribution to this study from central cancer registries supported through the Centers for Disease Control and Prevention's National Program of Cancer Registries (NPCR) and/or the National Cancer Institute's Surveillance, Epidemiology, and End Results (SEER) Program. Central registries may also be supported by state agencies, universities, and cancer centers. Participating central cancer registries include the following: Alabama, Alaska, Arizona, Arkansas, California, Colorado, Connecticut, Delaware, Florida, Georgia, Hawaii, Idaho, Indiana, Iowa, Kentucky, Louisiana, Massachusetts, Maine, Maryland, Michigan, Mississippi, Montana, Nebraska, Nevada, New Hampshire, New Jersey, New Mexico, New York, North Carolina, North Dakota, Ohio, Oklahoma, Oregon, Pennsylvania, Puerto Rico, Rhode Island, Seattle SEER Registry, South Carolina, Tennessee, Texas, Utah, Virginia, West Virginia, Wyoming.
Funding/Support: This work was supported by grants U01 CA167552, R01 CA202704 (ATC), R35 CA253185 (ATC), R21 AA027608 (YC), and R37 CA246175 (YC) from the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. ATC is supported as an American Cancer Society Clinical Research Professor and a Massachusetts General Hospital Stuart and Suzanne Steele Research Scholars Award.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104566.
Appendix A. Supplementary data
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Wong S.H., Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol. 2019;16(11):690–704. doi: 10.1038/s41575-019-0209-8. [DOI] [PubMed] [Google Scholar]
- 3.Camilleri M., Vella A. What to do about the leaky gut. Gut. 2022;71(2):424–435. doi: 10.1136/gutjnl-2021-325428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tilg H., Adolph T.E., Gerner R.R., Moschen A.R. The intestinal microbiota in colorectal cancer. Cancer Cell. 2018;33(6):954–964. doi: 10.1016/j.ccell.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Maldonado R.F., Sá-Correia I., Valvano M.A. Lipopolysaccharide modification in Gram-negative bacteria during chronic infection. FEMS Microbiol Rev. 2016;40(4):480–493. doi: 10.1093/femsre/fuw007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marshall J.C., Walker P.M., Foster D.M., et al. Measurement of endotoxin activity in critically ill patients using whole blood neutrophil dependent chemiluminescence. Crit Care. 2002;6(4):342–348. doi: 10.1186/cc1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Epeldegui M., Magpantay L., Guo Y., et al. A prospective study of serum microbial translocation biomarkers and risk of AIDS-related non-Hodgkin lymphoma. Aids. 2018;32(7):945–954. doi: 10.1097/QAD.0000000000001771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shieh A., Epeldegui M., Karlamangla A.S., Greendale G.A. Gut permeability, inflammation, and bone density across the menopause transition. JCI Insight. 2020;5(2) doi: 10.1172/jci.insight.134092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tabung F.K., Birmann B.M., Epstein M.M., et al. Influence of dietary patterns on plasma soluble CD14, a surrogate marker of gut barrier dysfunction. Curr Dev Nutr. 2017;1(11) doi: 10.3945/cdn.117.001396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balagopal A., Philp F.H., Astemborski J., et al. Human immunodeficiency virus-related microbial translocation and progression of hepatitis C. Gastroenterology. 2008;135(1):226–233. doi: 10.1053/j.gastro.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao S., Zhao L. Gut microbiota-based translational biomarkers to prevent metabolic syndrome via nutritional modulation. FEMS Microbiol Ecol. 2014;87(2):303–314. doi: 10.1111/1574-6941.12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landmann R., Knopf H.P., Link S., Sansano S., Schumann R., Zimmerli W. Human monocyte CD14 is upregulated by lipopolysaccharide. Infect Immun. 1996;64(5):1762–1769. doi: 10.1128/iai.64.5.1762-1769.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park B.S., Lee J.O. Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp Mol Med. 2013;45(12):e66. doi: 10.1038/emm.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laugerette F., Furet J.P., Debard C., et al. Oil composition of high-fat diet affects metabolic inflammation differently in connection with endotoxin receptors in mice. Am J Physiol Endocrinol Metab. 2012;302(3):E374–E386. doi: 10.1152/ajpendo.00314.2011. [DOI] [PubMed] [Google Scholar]
- 15.Brenchley J.M., Price D.A., Schacker T.W., et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12(12):1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 16.Barclay G.R. Endogenous endotoxin-core antibody (EndoCAb) as a marker of endotoxin exposure and a prognostic indicator: a review. Prog Clin Biol Res. 1995;392:263–272. [PubMed] [Google Scholar]
- 17.Epstein M.M., Breen E.C., Magpantay L., et al. Temporal stability of serum concentrations of cytokines and soluble receptors measured across two years in low-risk HIV-seronegative men. Cancer Epidemiol Biomarkers Prev. 2013;22(11):2009–2015. doi: 10.1158/1055-9965.EPI-13-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song M., Wu K., Ogino S., Fuchs C.S., Giovannucci E.L., Chan A.T. A prospective study of plasma inflammatory markers and risk of colorectal cancer in men. Br J Cancer. 2013;108(9):1891–1898. doi: 10.1038/bjc.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.GBD 2019 Colorectal Cancer Collaborators Global, regional, and national burden of colorectal cancer and its risk factors, 1990-2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet Gastroenterol Hepatol. 2022;7(7):627–647. doi: 10.1016/S2468-1253(22)00044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Citronberg J.S., Wilkens L.R., Le Marchand L., et al. Plasma lipopolysaccharide-binding protein and colorectal cancer risk: a nested case-control study in the Multiethnic Cohort. Cancer Causes Control. 2018;29(1):115–123. doi: 10.1007/s10552-017-0990-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greenland S., Schlesselman J.J., Criqui M.H. The fallacy of employing standardized regression coefficients and correlations as measures of effect. Am J Epidemiol. 1986;123(2):203–208. doi: 10.1093/oxfordjournals.aje.a114229. [DOI] [PubMed] [Google Scholar]
- 22.Greenland S., Maclure M., Schlesselman J.J., Poole C., Morgenstern H. Standardized regression coefficients: a further critique and review of some alternatives. Epidemiology. 1991;2(5):387–392. [PubMed] [Google Scholar]
- 23.Altman D.G., Royston P. The cost of dichotomising continuous variables. BMJ. 2006;332(7549):1080. doi: 10.1136/bmj.332.7549.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durrleman S., Simon R. Flexible regression models with cubic splines. Stat Med. 1989;8(5):551–561. doi: 10.1002/sim.4780080504. [DOI] [PubMed] [Google Scholar]
- 25.Michels N., van Aart C., Morisse J., Mullee A., Huybrechts I. Chronic inflammation towards cancer incidence: a systematic review and meta-analysis of epidemiological studies. Crit Rev Oncol Hematol. 2021;157 doi: 10.1016/j.critrevonc.2020.103177. [DOI] [PubMed] [Google Scholar]
- 26.Kakourou A., Koutsioumpa C., Lopez D.S., et al. Interleukin-6 and risk of colorectal cancer: results from the CLUE II cohort and a meta-analysis of prospective studies. Cancer Causes Control. 2015;26(10):1449–1460. doi: 10.1007/s10552-015-0641-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan A.T., Ogino S., Giovannucci E.L., Fuchs C.S. Inflammatory markers are associated with risk of colorectal cancer and chemopreventive response to anti-inflammatory drugs. Gastroenterology. 2011;140(3):799–808. doi: 10.1053/j.gastro.2010.11.041. quiz e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slattery M.L., Mullany L.E., Sakoda L., et al. The NF-κB signalling pathway in colorectal cancer: associations between dysregulated gene and miRNA expression. J Cancer Res Clin Oncol. 2018;144(2):269–283. doi: 10.1007/s00432-017-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen R., Luo F.K., Wang Y.L., Tang J.L., Liu Y.S. LBP and CD14 polymorphisms correlate with increased colorectal carcinoma risk in Han Chinese. World J Gastroenterol. 2011;17(18):2326–2331. doi: 10.3748/wjg.v17.i18.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siegel R.L., Fedewa S.A., Anderson W.F., et al. Colorectal cancer incidence patterns in the United States, 1974-2013. J Natl Cancer Inst. 2017;109(8) doi: 10.1093/jnci/djw322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hofseth L.J., Hébert J.R., Chanda A., et al. Early-onset colorectal cancer: initial clues and current views. Nat Rev Gastroenterol Hepatol. 2020;17(6):352–364. doi: 10.1038/s41575-019-0253-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang C.M., Chia V.M., Gunter M.J., et al. Innate immunity gene polymorphisms and the risk of colorectal neoplasia. Carcinogenesis. 2013;34(11):2512–2520. doi: 10.1093/carcin/bgt228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonzalez-Quintela A., Alonso M., Campos J., Vizcaino L., Loidi L., Gude F. Determinants of serum concentrations of lipopolysaccharide-binding protein (LBP) in the adult population: the role of obesity. PLoS One. 2013;8(1) doi: 10.1371/journal.pone.0054600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ellis R.J., Iudicello J.E., Heaton R.K., et al. Markers of gut barrier function and microbial translocation associate with lower gut microbial diversity in people with HIV. Viruses. 2021;13(10) doi: 10.3390/v13101891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramendra R., Isnard S., Mehraj V., et al. Circulating LPS and (1→3)-β-D-Glucan: a Folie à Deux Contributing to HIV-Associated Immune Activation. Front Immunol. 2019;10:465. doi: 10.3389/fimmu.2019.00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mehraj V., Ramendra R., Isnard S., et al. Circulating (1→3)-β-D-glucan is associated with immune activation during human immunodeficiency virus infection. Clin Infect Dis. 2020;70(2):232–241. doi: 10.1093/cid/ciz212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Isnard S., Fombuena B., Sadouni M., et al. Circulating β-d-Glucan as a marker of subclinical coronary plaque in antiretroviral therapy-treated people with human immunodeficiency virus. Open Forum Infect Dis. 2021;8(6):ofab109. doi: 10.1093/ofid/ofab109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang M.Y., Wang J., Guo J. Role of regenerating islet-derived protein 3A in gastrointestinal cancer. Front Oncol. 2019;9:1449. doi: 10.3389/fonc.2019.01449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng Q., Liang S., Jia H., et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun. 2015;6:6528. doi: 10.1038/ncomms7528. [DOI] [PubMed] [Google Scholar]
- 40.Tworoger S.S., Hankinson S.E. Use of biomarkers in epidemiologic studies: minimizing the influence of measurement error in the study design and analysis. Cancer Causes Control. 2006;17(7):889–899. doi: 10.1007/s10552-006-0035-5. [DOI] [PubMed] [Google Scholar]
- 41.Knol M.J., VanderWeele T.J. Recommendations for presenting analyses of effect modification and interaction. Int J Epidemiol. 2012;41(2):514–520. doi: 10.1093/ije/dyr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.