ABSTRACT
Viruses can alter the abundance, evolution, and metabolism of microorganisms in the ocean, playing a key role in water column biogeochemistry and global carbon cycles. Large efforts to measure the contribution of eukaryotic microorganisms (e.g., protists) to the marine food web have been made, yet the in situ activities of the ecologically relevant viruses that infect these organisms are not well characterized. Viruses within the phylum Nucleocytoviricota (“giant viruses”) are known to infect a diverse range of ecologically relevant marine protists, yet how these viruses are influenced by environmental conditions remains under-characterized. By employing metatranscriptomic analyses of in situ microbial communities along a temporal and depth-resolved gradient, we describe the diversity of giant viruses at the Southern Ocean Time Series (SOTS), a site within the subpolar Southern Ocean. Using a phylogeny-guided taxonomic assessment of detected giant virus genomes and metagenome-assembled genomes, we observed depth-dependent structuring of divergent giant virus families mirroring dynamic physicochemical gradients in the stratified euphotic zone. Analyses of transcribed metabolic genes from giant viruses suggest viral metabolic reprogramming of hosts from the surface to a 200-m depth. Lastly, using on-deck incubations reflecting a gradient of iron availability, we show that modulating iron regimes influences the activity of giant viruses in the field. Specifically, we show enhanced infection signatures of giant viruses under both iron-replete and iron-limited conditions. Collectively, these results expand our understanding of how the water column’s vertical biogeography and chemical surroundings affect an important group of viruses within the Southern Ocean.
IMPORTANCE The biology and ecology of marine microbial eukaryotes is known to be constrained by oceanic conditions. In contrast, how viruses that infect this important group of organisms respond to environmental change is less well known, despite viruses being recognized as key microbial community members. Here, we address this gap in our understanding by characterizing the diversity and activity of “giant” viruses within an important region in the sub-Antarctic Southern Ocean. Giant viruses are double-stranded DNA (dsDNA) viruses of the phylum Nucleocytoviricota and are known to infect a wide range of eukaryotic hosts. By employing a metatranscriptomics approach using both in situ samples and microcosm manipulations, we illuminated both the vertical biogeography and how changing iron availability affects this primarily uncultivated group of protist-infecting viruses. These results serve as a foundation for our understanding of how the open ocean water column structures the viral community, which can be used to guide models of the viral impact on marine and global biogeochemical cycling.
KEYWORDS: Monodnaviria, Nucleocytoviricota, Ribovaria, SOTS, iron availability, marine microbiology, metatranscriptomics, phytoplankton, virocells
INTRODUCTION
Viruses are the Earth’s most abundant biological entities and can shape the ecology and biogeochemistry of aquatic ecosystems (1–3). Viruses of protists can drive biogeochemical flux in the water column and have been implicated as key drivers of the efficiency of downward carbon export (4, 5). Further, the “virocell,” or altered metabolic status of a host cell under virus infection, can profoundly impact the metabolism of the host in the environment (6, 7). One of the most widespread groups of protist-infecting double-stranded DNA (dsDNA) viruses in marine systems, the Nucleocytoviricota (“giant viruses”), are known for having relatively large particle and genome sizes (8, 9) and can exceed the diversity of dsDNA bacteriophage in the ocean (10). Thus, large-scale sequencing efforts have illuminated the complex diversity of giant viruses within marine environments, most of which remain uncultivated (11, 12). Furthermore, the potential for giant viruses to alter host metabolism during an infected virocell state has been implicated in sequencing-based studies (11–15) and culture-based experiments (16, 17). Because of their abundance, genetic complexity, and potential for host metabolic alterations in marine ecosystems, the study of giant virus effects on microbial food web dynamics is essential.
The Southern Ocean and its resident microbial community play a significant role in the global carbon cycle, a process shown to be constrained by the availability of the trace metal iron in high-nutrient low-chlorophyll (HNLC) regions (18, 19). The characterization of giant viruses in the Southern Ocean is lacking: most of what we know in this ocean region regarding top-down viral effects relates to dsDNA bacteriophage infecting prokaryotes (20–24). Viral lysis has been shown to be an important loss factor for major eukaryotic phytoplankton communities within the Southern Ocean Antarctic waters (25). One study found that giant viruses of the Phycodnaviridae were the second most dominant group of viruses following dsDNA bacteriophage in Southern Ocean waters (26). Furthermore, understanding depth-dependent and/or temporal drivers of giant virus diversity in the Southern Ocean is necessary to more accurately quantify the biogeochemical feedback of giant virus-protist interactions in the water column. Likewise, limiting nutrients have been shown to dictate marine giant virus-host dynamics in culture-based systems (27, 28); however, there remains limited field-based inference of nutrient-virus interactions in the Southern Ocean.
To address these knowledge gaps, we characterized the diversity of active giant viruses within an important oceanic study site located in the sub-Antarctic zone (SAZ) of the Southern Ocean. Specifically, we document the diversity of active giant viruses at various depths of the upper euphotic zone across a spatiotemporal gradient. Additionally, we tested how iron, a key micronutrient that seasonally limits growth within the SAZ (29), influenced giant virus infection signatures using on-deck incubations which encompassed iron-limited to iron-replete conditions. Based on current and previous results showing a diverse representation of potential giant virus hosts ranging from haptophytes, dinoflagellates, chlorophytes, and pelagophytes, we initially hypothesized that we would detect a wide range of phylogenetically distinct giant virus types with taxon-specific distributions structured by the stratified depth strata observed during the expedition. We also hypothesized that elevated iron availability to SAZ iron-limited communities would result in increased signatures of giant virus infection due to enhanced host physiological health and based on previous culture-based findings in model systems (28).
By screening metatranscriptomic libraries generated from in situ, opportunistic water column sampling, we detected 100 giant virus genomes spanning three phylogenetic families using a database of giant virus metagenome-assembled and cultured isolate genomes as a reference for viral discovery (30). Using this approach, we described patterns in the vertical biogeography of giant viruses, inferring potential hosts as well as contributions to cellular metabolism. We found active viruses of the orders Imitervirales, Algavirales, and Pimascovirales, and that the Mesomimiviridae family made up >80% of all giant virus transcripts across the samples and was prevalent from the surface to a 200-m depth. Giant virus metabolic gene transcripts were abundant throughout the euphotic zone, suggesting they may contribute to key metabolic processes such as photosynthesis and nutrient acquisition. Finally, we found evidence for enhanced infection signatures under iron-limited and iron-replete conditions by leveraging metatranscriptomes generated from incubations across a range of iron availabilities.
RESULTS
Physicochemical status of the water column at the SOTS.
A temporal and depth-resolved metatranscriptomic data set was generated at the Southern Ocean Time Series (SOTS), located alongside the northern edge of the SAZ near the subtropical front (STF). An overview of the cruise track has been previously described by Schallenberg et al. (31), noting frequent intersection during our expedition with an eddy that had surface temperatures of >14°C. Sea surface temperatures during the first three dates ranged from 10.5 to 11.5°C, whereas the final time point sampled was ~13°C, indicating potential interference with this eddy during the March 17 time point. Across all depths sampled, water column temperature ranged between 9 and 13°C and salinity ranged from 34 to 35 up to a 500-m depth (Fig. S2 and Table S1). For only the March 5, 7, and 9 time points, NO3 varied between 5 and 15 μmol L−1, NO2 ranged from 0 to 0.4 μmol L−1, NH4 from 0 to 0.5 μmol L−1, PO4 between 0.6 and 1.2 μmol L−1, and Si from 0.5 to 5 μmol L−1 to 500 m deep (Fig. S2 and Table S1). Dissolved iron concentrations in the upper 50 m ranged between 0.15 and 0.35 nmol L−1, and its variability mirrored salinity and temperature in this stratum. A spike in relative chlorophyll a fluorescence occurred around a 25- to 50-m depth on March 5, 7, and 9; and around a 50- to 100-m depth on March 17 (Fig. S2).
Physicochemical status of the time points sampled during the March 2018 research expedition at the Southern Ocean Time Series (SOTS). Conductivity, temperature, and depth (CTD) data are presented for all four dates sampled, and nutrient data are presented only for March 5, 7, and 9 sampling dates. (A) CTD fluorescence data, expressed as relative fluorescence units. (B) CTD Dissolved oxygen measurements expressed as mg/L. (C) CTD salinity data. (D) CTD temperature data. (E) Nitrate (NO3) measurements. (F) Nitrite (NO2) measurements. (G) Ammonium (NH4) measurements. (H) Phosphate (PO4) measurements. (I) Silicate (Si) measurements. Download FIG S2, PDF file, 0.5 MB (555.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Physicochemical data for time points and depths collected during the March 2018 expedition. No nutrient data were collected for March 17. Download Table S1, DOCX file, 0.02 MB (19.6KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Composition of the microeukaryote community.
Taxonomic distribution of metatranscriptomic reads in the assembly revealed a dominance of Dinophyceae (dinoflagellates), Prymnesiophyceae (haptophytes), and Bacillariophyta (diatoms) in the transcriptionally active pool of microeukaryote protists across the spatiotemporal in situ sampling (Fig. S3). There were also reads assigned to Pelagophyceae (pelagophytes), Spirotichea (ciliate protozoa), and Mamiellophyceae (chlorophytes) which were present but were proportionally lower in transcript abundance throughout the in situ samples (Fig. S3). The proportions of each taxon remained relatively consistent throughout dates and depths; however, we saw decreased proportions of haptophyte reads and/or increased proportions of dinoflagellates and diatom reads with depth (~100 to 200 m, Fig. S3).
Proportions of reads assigned across eukaryotes separated by date and depth sampled. The values represent the summed normalized transcripts (transcripts per million, TPMs) across open reading frames assigned to a given taxonomic rank within the MMTESP database. Download FIG S3, PDF file, 0.6 MB (597.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Detection of the active giant virus community.
We detected 100 unique, transcriptionally active giant virus genomes/metagenome assembled genomes (MAGs) across the sampling set (Fig. S4), contributing 0.0005 to 0.0335% of the total reads. Of these genomes, half had >50% of their total genes detected and four had >80% of their genes detected (Fig. S4). The total normalized reads across these genomes were each proportional to their mean gene count (Fig. S4).
Results of read recruitments to candidate Nucleocytoviricota isolate and metagenome-assembled genomes detected at SOTS. Each dot represents a candidate Nucleocytoviricota genome with transcripts detected. Genes recovered indicate the percentage of genes within each candidate genome that had reads mapped, where the mean genome abundance indicates the total normalized read count (log10 TPM) recruited to that genome. Total number of genes within each candidate genome is represented by dot size. Download FIG S4, PDF file, 0.09 MB (91.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
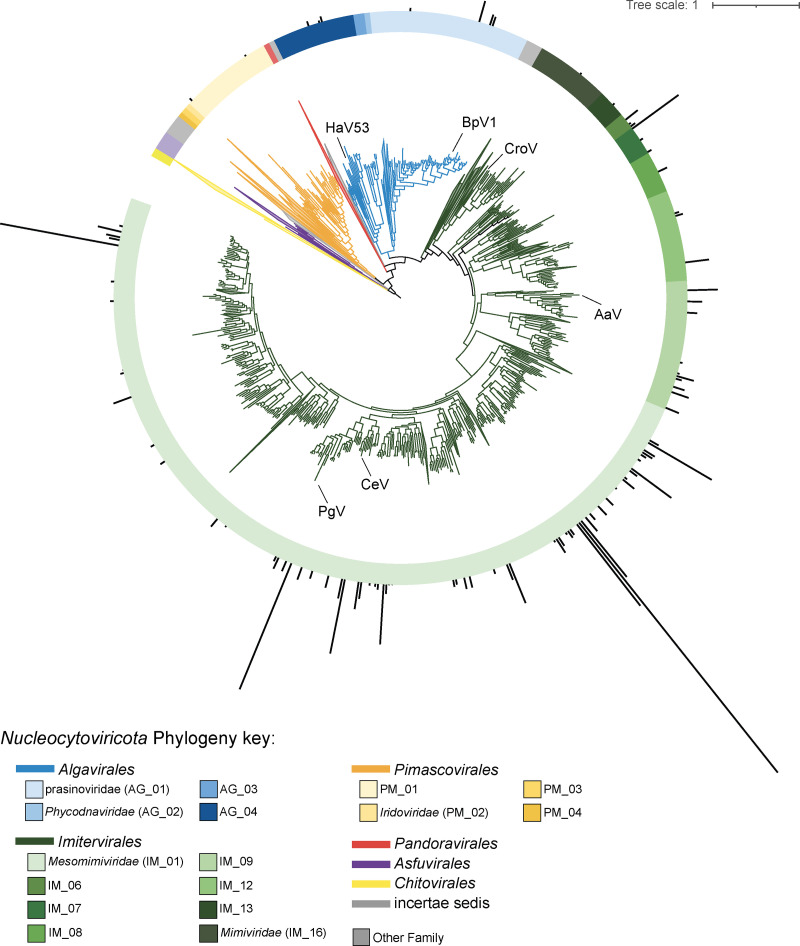
Using a recently established phylogenetic framework for characterizing giant viruses (32), we estimated the phylogenetic assignment of each genome using an analysis of concatenated hallmark proteins. Detected giant virus MAGs fell within the phylogenetically defined orders Algavirales, Imitervirales, and Pimascovirales (Fig. 1 and Table S2).Two of these genomes belong to cultivated isolates: the haptophyte-infecting Chrysochromulina ericina Virus CeV-01B (NC_028094.1, “Mesomimiviridae” or IM_01) and the chlorophyte-infecting Bathycoccus sp. Virus RCC1105 BpV1 (NC_014765.1, “Prasinoviridae” or AG_02; Fig. 1 and Fig. S5). The majority (n = 91) of MAGs/genomes fell within the order Imitervirales (Fig. 1 and Fig. S5). These consisted of 69 Mesomimiviridae (“IM_01”), 10 “IM_09,” 3 “IM_07,” 3 “IM_12,” 3 “IM_06,” 1 “IM_08,” 1 “IM_13,” and 1 Mimiviridae (“IM_16”) assigned families (Fig. S5 and Table S5, available at https://zenodo.org/record/7457861#.Y6BrIuzML0o). The Mesomimiviridae and IM_09 families recruited the most transcripts relative to others (Fig. S5). One of the top three most transcriptionally abundant MAGs in the data set clustered within the same genus (IM_01, g343) as the cultured isolates CeV-01B and Phaeocystis globosa Virus (PgV; Fig. 1 and Fig. S5). The topmost abundant MAG, which belonged to the family Mesomimiviridae, fell within a genus (g329) that does not contain any cultured isolates, contributing ~15% of the total giant virus transcripts (Fig. S5).
FIG 1.
Phylogeny of Nucleocytoviricota genomes and metagenome-assembled-genomes, with detected candidates at the Southern Ocean Time Series (SOTS) shown with their relative transcript abundances (transcripts per million, TPM) in the outermost bar plot. Clades without detected candidates are collapsed. Branches are color-coded by order-level taxonomy. Cultured isolate virus references of interest are labeled in their approximate location along the branches with the following abbreviations: HaV01: Heterosigma akashiwo Virus 01 isolate HaV53 (HaV53), Bathycoccus prasinos Virus 1 (BpV1), Cafeteria roenbergensis Virus (CroV), Aureococcus anophagefferens Virus (AaV), Chrysochromulina ericina Virus (CeV), Phaeocystis globosa Virus (PgV).
Summary of the (A) taxonomic distribution of the 100 genomes. The genome count illustrates the number of genomes detected within a specific family-level assignment. (B) Relative summed expression levels of the family-level genome/metagenomic assembled genome (MAG) assignments across the entire in situ dataset, represented as TPMs. (C) Genome-level contribution towards the total pool of giant-virus transcripts (TPMs), represented as a percent. Each genome is color coded by Family level assignment and the Genus level assignment is shown in text next to each bar. Download FIG S5, PDF file, 1.0 MB (1,007.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Count of giant virus genomes detected, grouped by phylogenetic classification corresponding to Fig. 1. Download Table S2, DOCX file, 0.01 MB (13.7KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Four genomes were assigned to an Algavirales proposed family, the Prasinoviridae (AG_01), which all fell into the genus g177 containing the cultured isolate BpV1 (Fig. 1 and Fig. S5). Three genomes were assigned to the AG_04 family, containing the cultured isolate Heterosigma akashiwo Virus 01 HaV53. Two genomes fell within the PM_01 family of the order Pimascovirales (Fig. 1, Fig. S5).
Defining the vertical biogeography of active giant viruses.
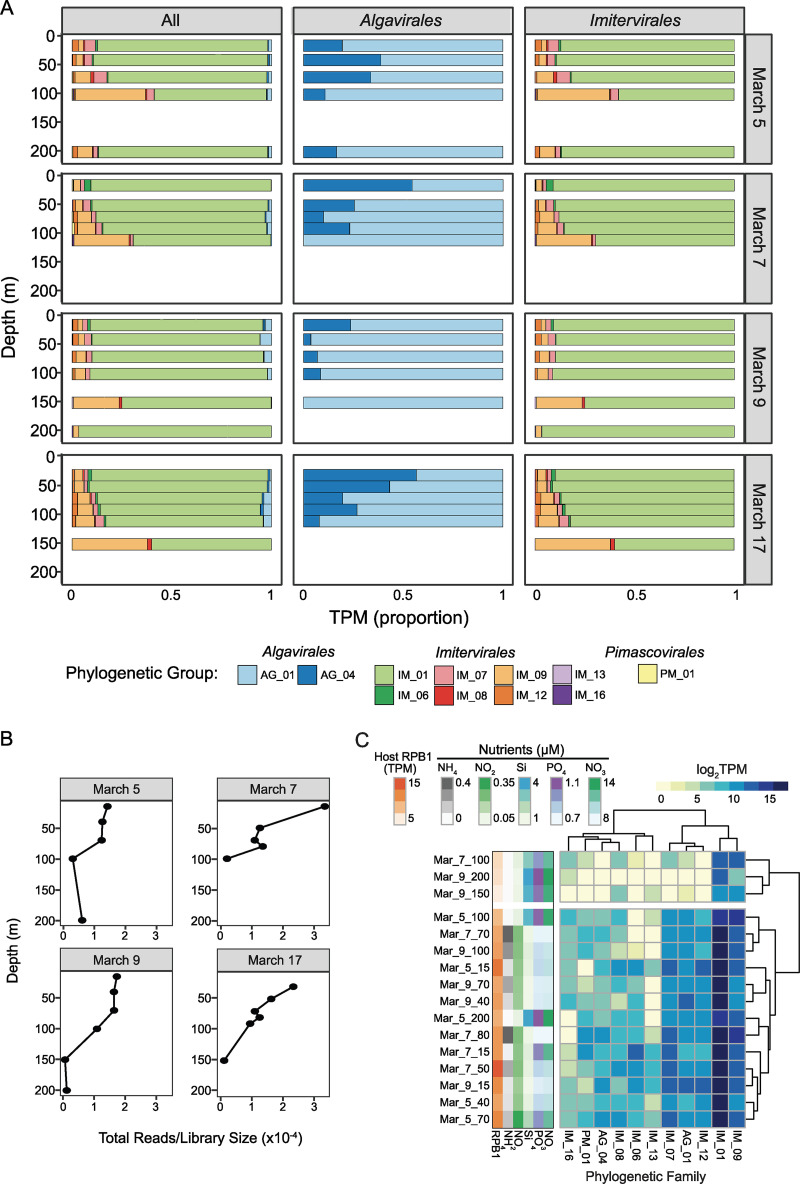
To resolve vertical patterns associated with giant virus phylogeny across depth strata sampled at SOTS, we assessed transcript signatures of the phylogenetically assigned giant virus MAGs/genomes across the spatiotemporal sampling. A principal component analysis (PCA) shows a shift in the summed transcripts per giant virus genome/MAG explained by depth rather than by date sampled (across PC1, 17.62% of the variance explained; Fig. S6). Examining the proportions of transcripts assigned, summarized by family, showed patterns with depth sampled as well (Fig. 2). Generally, transcripts assigned to Mesomimiviridae family genomes dominated the proportion of the transcript pool from the surface (5 to 15 m) to 200-m depth across the sampling dates (Fig. 2A). Transcripts assigned to genomes within the IM_09 family displayed a higher proportion at 100 m (March 5), 125 m (March 7), 150 m (March 9), and 150 m (March 17) compared to their proportions at shallower depths (Fig. 2A). Genomes assigned to the proposed family Prasinoviridae dominated the transcript pool within the order Algavirales, and generally AG_04 transcripts decreased in proportion with depth (Fig. 2A). Depth-specific patterns for transcripts assigned to PM_01 genomes were not as prominent due to their low contribution to the transcript pool across all depths and dates; however, their highest contribution occurred at ~90 m depth on March 7 (Fig. 2A). Summed transcripts across all giant virus MAGs/genomes were highest in the surface layer of the water column and decreased with increasing depth (Fig. 2B). The shifting distribution of nutrients in the upper water column measured during our sampling period correlated with the transcript abundance patterns of giant viruses (Fig. 3C). Where the nutrients nitrate, phosphate, and silicate increased at ~100 to 200 m, there was a decrease in relative transcript abundance across all giant virus families (Fig. 2C). This decrease in giant virus transcripts also coincided with a decreased abundance of total transcripts assigned to the gene encoding the eukaryotic DNA-directed RNA polymerase large subunit marker protein (RPB1, Fig. 2C).
FIG 2.
Taxon-specific transcript abundance patterns of Nucleocytoviricota across a depth-dependent and temporal scale. (A) Shifts in proportions of normalized transcripts assigned to different Nucleocytoviricota genomes by family. (B) Total Nucleocytoviricota transcript counts with depth, normalized by library size. (C) Heatmap of family-level-summed Nucleocytoviricota transcript abundances across depth profiles sampled on March 5, 7, and 9 with corresponding nutrient and total eukaryotic RPB1 transcript abundance data along the left-hand side. Data for March 17 were omitted due to lack of nutrient data. Family-level color codes have been changed (compared to Fig. 1) for visual purposes. Only the short-hand names of the family-level assignments are shown.
FIG 3.
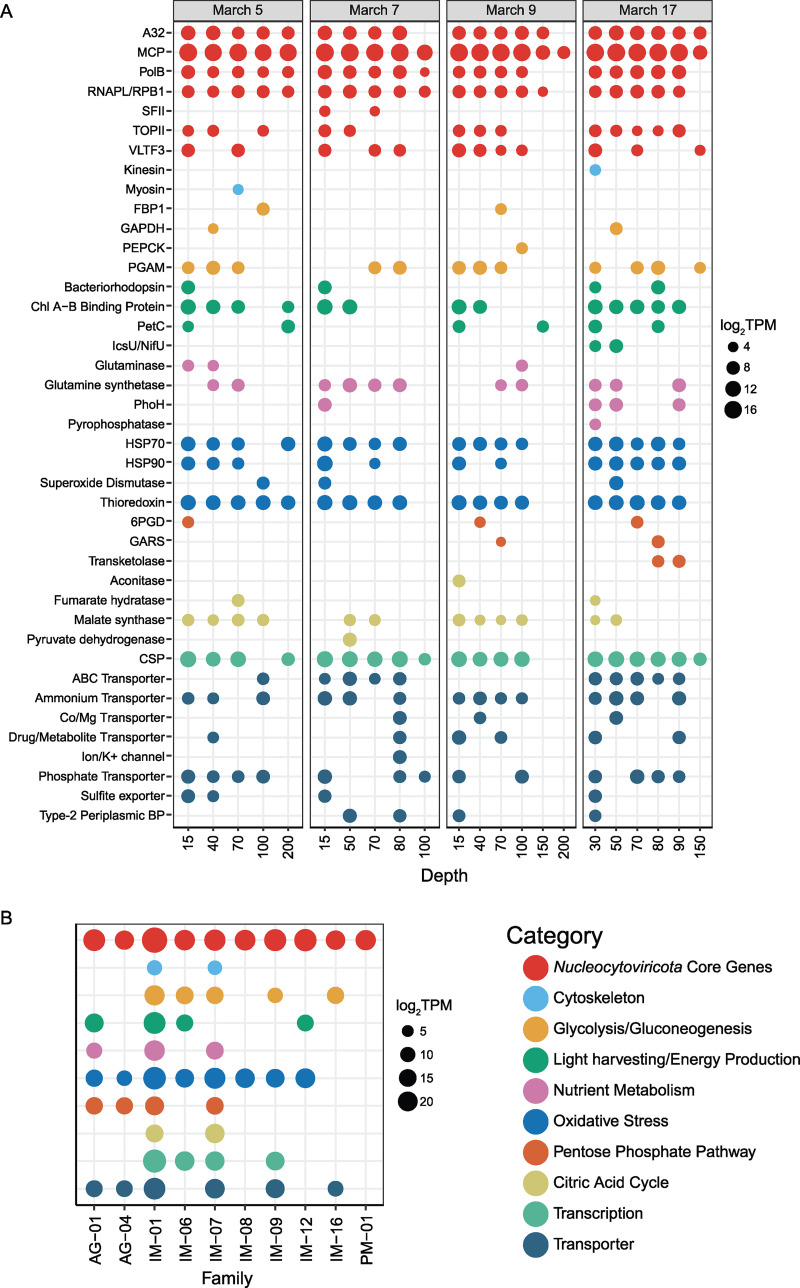
Nucleocytoviricota metabolic gene transcript abundance patterns across a temporal and depth-related scale. (A) Transcript abundances of metabolic genes summed by depth and separated by date sampled. Genes are categorized by broad functional category. (B) Transcript abundances of metabolic genes, summed by functional category and respective genomic phylogenetic assignment to the family level.
Shifts in the transcript abundance patterns of eukaryotic and giant viruses. Each plot shows a principal components (PC) analysis ordination of normalized transcript patterns across (A) Eukaryotic RPB1 candidates colored by date sampled (B) Nucleocytoviricota total genome recruitments colored by date sampled (C) Eukaryotic RPB1 candidates colored by depth sampled (D) Nucleocytoviricota total genome recruitments colored by depth sampled. Download FIG S6, PDF file, 0.5 MB (486.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Five giant virus genomes of the Mesomimiviridae family were positively correlated with chlorophyll a fluorescence (Spearman’s ρ ≥ 0.8, Padj < 0.1), one was positively correlated with NH4 (ρ ≥ 0.8, Padj < 0.1), and two were negatively correlated with NO3 (ρ ≤ 0.8, Padj < 0.1) (Fig. S7). One IM_07 genome was also negatively correlated with NO3 (ρ ≤ 0.8, Padj < 0.1) and one IM_06 genome was positively correlated with chlorophyll a fluorescence (ρ ≥ 0.8, Padj < 0.1) (Fig. S7).
Spearman’s correlations between environmental parameters (CTD and nutrient data) and summed transcript abundances of each giant virus genome/MAG across the in situ dataset. Only correlations with Spearman’s rho (ρ) ±0.8 with an adjusted P value (Holm method) of ≤0.1 are shown. The Family-level assignment of each genome is shown in parentheses next to its ID. Download FIG S7, PDF file, 0.4 MB (454.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Transcribed giant virus structural and metabolic genes with depth.
We assessed depth-specific patterns of the pool of transcribed functional metabolic genes harbored within the giant virus genomes to gain insight into other metabolic consequences of infection across the depth strata. Here, transcripts for the genes encoding “core” giant virus proteins used to generate the concatenated phylogeny (major capsid protein [MCP], DNA polymerase [PolB], A32-like DNA-packaging ATPase [A32], viral late transcription factor 3 [VLFT3], DNA topoisomerase II [TOPII], DNA-directed RNA polymerase beta and alpha subunits [RNAPL/RPB1], and DEAD/SNF2-like helicase [SFII]) were the most abundant throughout the water column across all family members (Fig. 3). Transcripts for the giant virus MCP were present across all depths and dates, and MCP was the most abundant of the core proteins (Fig. 3A).
Transcripts recruited to genes harbored within the giant virus genome categorized as “Cytoskeleton,” “Glycolysis/Gluconeogenesis,” “Light Harvesting/Energy Production,” “Nutrient Metabolism,” “Oxidative Stress,” “Pentose Phosphate Pathway,” “Citric Acid Cycle,” “Transcription,” and “Transport” were queried (Fig. 3). Transcripts of giant virus genes encoding “Light Harvesting/Energy Production” proteins—chlorophyll a binding protein, bacteriorhodopsin, and cytochrome b6-f complex (PetC)—were present throughout all depths and dates (Fig. 3A). Specifically, transcripts for the gene encoding chlorophyll a binding protein were most prevalent at the surface depths (15 to 50 m) but could also be detected at 200 m depth (March 5) and 70 to 90 m depth (March 17, Fig. 3A). Transcripts for genes encoding bacteriorhodopsin-like proteins were consistently present at the surface depths (15 to 40 m, excluding March 9) and for PetC (excluding March 7, Fig. 3A). The phylogenetic assignment of transcripts recruited to genomes with genes encoding “Light Harvesting/Energy Production” proteins belonged to the Mesomimiviridae, IM_06, IM_12, and Prasinoviridae families (Fig. 3B).
Transporters indicating nutrient acquisition were prevalent across the genomes (Fig. 3). Abundant transporters include an ABC transporter detected from 15 to 90 m depth (March 5 and 17), an ammonium transporter detected from 15 to 90 m depth, and a phosphate transporter detected from 15 to 90 m deep (Fig. 3A). Other transporters (Co/Mg transporter, drug/metabolite transporter, sulfite transporter, ion transporter, and a type-2 periplasmic binding transport component) had transcripts detected sporadically across all depths and dates (Fig. 3A). All giant virus families, except IM_06, IM_08, IM_12, and PM_01, had transporters encoded within their genomes that recruited transcripts (Fig. 3B). Genes encoding various nutrient metabolism pathways were also transcribed across the water column (Fig. 3A). The most prominent of these include the genes encoding glutamine synthetase (present from 15- to 90-m depth) and the phosphate starvation-inducible protein (PhoH) (present at 15-m depth on March 7 and 40- to 90-m depth on March 17, Fig. 3A). This pathway was encoded by the Prasinoviridae, Mesomimiviridae, and IM_07 giant virus families (Fig. 3B).
Transcripts encoding genes within central carbon metabolism pathways (citric acid cycle, pentose phosphate pathway, and glycolysis/gluconeogenesis) were less prevalent throughout the water column. Transcripts for phosphoglycerate mutase 2 protein and malate synthase genes had transcripts detected from 15 to 200 m deep across the sampling series (Fig. 3A). Detection of transcripts for other central carbon genes was more sporadic across depth and date (Fig. 3A). The Mesomimiviridae and IM_07 giant virus families harbored genes with detected transcripts belonging to all three central carbon metabolic pathways (Fig. 3B).
Genes encoding heat shock proteins (HSP70, HSP90), thioredoxins, and cold shock proteins (CSP) within the “Oxidative Stress/Transcription” categories were prevalent and abundant throughout depth and dates sampled (15–100 m depth. Figure 3A). All giant virus families, except the Mimiviridae and PM_01, harbored genes related to oxidative stress that had transcripts detected (Fig. 3B). The Mesomimiviridae, IM_06, IM_07, and IM_09 families harbored genes for the CSP protein which had detected transcripts within the data set (Fig. 3B).
Impact of iron availability on giant viruses revealed by on-deck iron incubations.
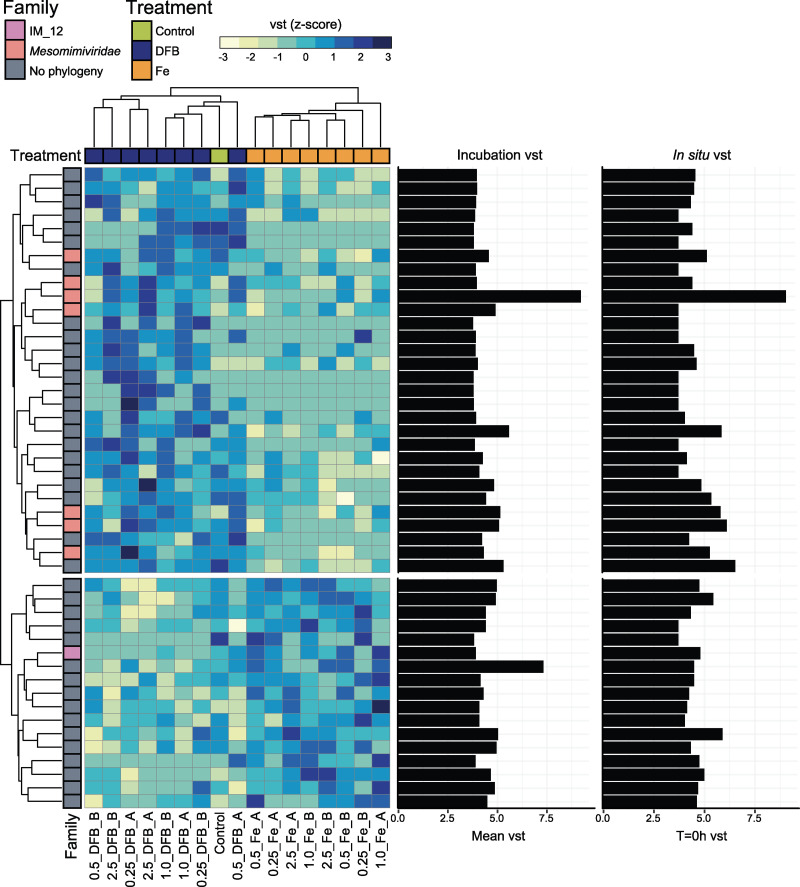
Because the Southern Ocean is known to be seasonally limited by the trace micronutrient iron (29), we investigated the response of active giant viruses across a gradient of iron availability using previously published experimental incubations (33). Out of the 754 total dereplicated MCPs from the co-assembly and the giant virus genomes/MAGs (Fig. 4, Table S3), a total of 47 giant virus MCPs had significantly different (P ≤ 0.05, Mann-Whitney U test) normalized transcript values (variance stabilizing transformation [vst]) when comparing desferrioxamine-B (DFB)-added versus Fe-added incubations. There were 30 MCPs that had statistically significantly (P ≤ 0.05) higher normalized transcript values across the DFB-added incubations versus that in the FeCl3-added incubations (Fig. 4). Out of these, seven MAG MCPs were assigned as Mesomimiviridae, two within the g343 genus that contains the cultured isolate reference genomes, CeV and PgV (Fig. 4, Table S3). The genome MCP with the highest relative transcript values across the incubations and in situ t = 0 h sample was assigned to the Mesomimiviridae g335 genus (no cultured isolates) and was significantly (P ≤ 0.05) higher within the DFB-added condition (Fig. 4, Table S3). There were 17 MCPs that had statistically significantly (P ≤ 0.05) higher vst values across the FeCl3-added incubations versus that in the DFB-added incubations (Fig. 4). Only one phylogenetically assigned giant virus MAG was found within this group and was assigned to the IM_12 g300 genus that contains the chlorophyte-infecting Pyramimonas orientalis Virus (PoV01), Tetraselmis virus 1, and Dishui Lake large algae virus 1 isolates (Fig. 4).
FIG 4.
Expression patterns of Nucleocytoviricota major capsid proteins (MCPs) across an iron availability gradient within an on-deck incubation of the surface microbial community. Only MCPs with significantly different (Mann-Whitney U test, P < 0.05) normalized transcript values (DESeq2’s variance stabilizing transformation [vst]) between the desferrioxamine-B (DFB) added versus the iron chloride (Fe) added incubations are shown in the heatmap. Variability for normalized expression values (z-scores, [observed vst − mean vst]/standard deviation) are standardized across each row (individual MCPs). Each column is annotated by whether DFB, Fe, or nothing (control) was added. Each row is annotated by the phylogenetic assignment of the MCP. Only MCPs originating from the giant virus genomes/metagenome assembled genomes are annotated with phylogeny following Fig. 1. The dendrograms show similarity in expression patterns by incubation (column) and MCP transcripts (row) using Euclidean distance and hierarchical clustering. Bar plots of the averaged vst values for transcripts across the iron incubation metatranscriptomes and the in situ t = 0 h metatranscriptomes are shown to the right of the heatmap.
Detected giant virus major capsid protein hits which had significantly different (Mann-Whitney U test, P < 0.05) vst values between the DFB-added versus Fe-added incubations. Download Table S3, DOCX file, 0.02 MB (22.4KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
DISCUSSION
In this study, we describe the presence and patterns across environmental gradients of active viruses within the phylum Nucleocytoviricota (giant viruses) within an HNLC Southern Ocean system. Samples of opportunity collected across various depths and dates at the SOTS, a region within the SAZ, allowed insight into depth-specific patterns in active giant viruses. Because we found diatoms, dinoflagellates, and haptophytes (Prymnesiophyceae) within our metatranscriptomic libraries, all previously shown to be prevalent within the SOTS surface community (31, 34), we hypothesized that we would detect a rich community of giant virus groups at SOTS as well (34). Indeed, we detected 100 giant virus genomes/MAGs spanning three viral orders, 11 families, and 24 proposed genera (32). This included two cultured isolate genomes: the haptophyte-infecting Chrysochromulina ericina Virus CeV-01B and the chlorophyte-infecting Bathycoccus sp. Virus RCC1105 BpV2. Due to the phylogenetic richness of giant viruses detected in our metatranscriptomes and the large contributions of giant viruses to the overall viral community observed previously (26), we proposed that giant viruses were important contributors to top-down viral predation on the protist community in the Southern Ocean.
As in a study performed within a coastal marine system (30), we found that the phylogenetically rich virus orders Imitervirales and Algavirales were the predominant types within the detected giant viruses at SOTS. We detected only 7 genomes with relatively low transcript abundances in our data set that were assigned to the Algavirales proposed family, the Prasinoviridae, and “AG_04.” However, the proportion of transcripts assigned to Imitervirales, specifically the Mesomimiviridae (IM_01) family, which contains giant viruses of Chrysochromulina (35) and Phaeocystis (36), made up >80% of all giant virus transcripts, whereas a more equal contribution of Algavirales-assigned viruses was observed in the coastal system (30). This suggested that the Mesomimiviridae may contribute primarily to protist-virus infection dynamics within the SAZ during this season, as haptophytes like Phaeocystis spp. have been shown to be the dominant protist taxon at SOTS (34). Further, one giant virus genome within the Mesomimiviridae g329 proposed genus contributed up to 15% of the total giant virus transcripts. Because only one genus within the Mesomimiviridae contains cultured representatives, it was difficult to infer the specific host range of these dominant giant viruses, and thus, further characterization of this diverse family of viruses is necessary. Regardless, we have evidence that a community of active giant viruses, primarily those that infect marine protists, like the Mesomimiviridae, may play a significant role in viral infection dynamics within this HNLC region and across systems of diverging productivity (30). That this family has been detected as the dominant active member in coastal and HNLC marine systems warrants further investigation of the host range of this phylogenetically rich group of viruses.
Using this phylogeny-guided approach, we tracked the depth-dependent patterns of giant viruses to infer taxon-specific lifestyles and putative host ranges. The Mesomimiviridae family dominated the proportion of giant virus transcripts within all surface samples. Furthermore, several genomes within the Mesomimiviridae were positively correlated with chlorophyll a fluorescence. This suggests that some members of the Mesomimiviridae primarily infect hosts residing at the sunlit surface, such as phototrophs. However, we also observed representation of Mesomimiviridae in samples collected up to a 200-m depth, suggesting active infection of hosts past the surface layer. This further illustrates the unknown dynamics of uncultivated Mesomimiviridae-like viruses as it may indicate infection of a wide variety of metabolically diverse hosts (photo-, hetero-, or mixotrophic) that persist with depth. Despite this, the observation of signatures of active infection of giant viruses past the sunlit surface layer is notable because most of our knowledge of virus-host interactions in the environment originates from surface-derived samples.
In contrast to the widespread contribution of active Mesomimiviridae giant viruses at SOTS, we saw taxon-specific localization with depth. Namely, the IM_09 viral family, which contains the isolated pelagophyte-infecting virus Aureococcus anophagefferens Virus (37), had increased transcript representation around the 100- to 150-m zone. This could signify a potential depth “hot spot” of the widespread algal group, Pelagophyceae (38), or related hosts around these depths. It is known that certain pelagophytes are adapted to living at attenuated light levels, specifically within deep chlorophyll maxima (DCM [39]). We did not observe a prominent DCM around this zone; however, the small peaks in relative chlorophyll a fluorescence around these depths could indicate smaller or less abundant fluorescent cells. Because total giant virus transcripts decreased in abundance past ~90 m, it is likely that this host community is present in low abundance around this zone. Indeed, we saw an overall reduction of host signatures concurrent with increased nutrients (e.g., nitrate, phosphate and silicate) with depth (suggesting reduced drawdown by the community). We thus hypothesize that the decreased proportions of active giant viruses with depth is more tightly linked to host abundance and physiological state and that they are most infective within the productive surface layers. This is in contrast to the observation of subsurface viral particle maxima seen frequently in stratified oceanic systems (40), where harmful UV radiation may play a role in modulating bacteriophage distributions in the euphotic zone.
Giant viruses carry genes indicative of cellular metabolic pathways (12, 41) that may allow hijacking of host metabolism for viral replication or even provide an ecological advantage to the host under certain environmental conditions (42). Thus, insight into the presence and contribution of giant virus metabolic genes with depth can provide evidence of the putative host ranges and potential biogeochemical impacts of viral infection in the open ocean environment aside from cellular lysis and death. We observed signatures of cellular metabolism contributed by giant viruses transcribed from 15- to 200-m depth at SOTS. For example, transcripts of genes related to photosynthetic processes were abundant within the Mesomimiviridae, and these transcripts were detected from the surface down to ~90 m depth, where we found signatures of this giant virus family, providing further evidence of a host range that includes phototrophs. Contrastingly, we did not observe transcripts of energy-generating processes encoded in the genomes of IM_09, a family of viruses which putatively infect pelagophyte-like hosts, whereas these transcripts were detected in abundance during the coastal study (30). This could reflect the depth localization of this group of viruses (and/or hosts) in which they persist at attenuated light levels, or it may be due to low levels of viral infection that occur with depth. Likewise, transcripts belonging to central carbon metabolic processes (citric acid cycle, pentose phosphate pathway, glycolysis/gluconeogenesis) were sparsely detected in the depth profiles compared to those in a coastal study where these genes were consistently expressed throughout the diel time series (30). This could be a feature unique to open-ocean environments compared to more productive coastal systems or because virally encoded central carbon metabolic genes are not as readily detected within bulk metatranscriptomic data.
Interestingly, we detected numerous transcripts indicative of nutrient acquisition processes. The most prevalent were assigned to ammonium and phosphate transporters, which may indicate elevated virocell nutrient requirements for viral production or a response to nutrient availability in the water column (43, 44). Additionally, the presence of abundant transcripts assigned to an “ABC-transporter” indicates active transport of some unknown substrate. Although we cannot currently determine the specific substrate, it is possible that giant viruses contribute to the active uptake of ferric iron sources in the water column, as we have shown active uptake of ferric iron and putative iron-siderophores by cyanobacteria, indicating iron-limited conditions during the expedition in the SAZ (33). In addition to nutrient acquisition processes, we saw transcription of genes encoding internal nutrient regulators such as PhoH and glutamine synthetase (45, 46). The detection of these metabolic genes in our data set suggests that giant viruses contribute to host nutrient-cycling processes throughout the water column, and thus raises the question of how an infected virocell contributes differently than an uninfected cell to water column biogeochemical cycles in the open ocean. However, we recognize that we lack a more direct understanding of the actual/realized contributions of giant virus-encoded metabolic genes to their host, so further laboratory experiments confirming the functionality of these genes is needed to interpret these observations.
One of the defining biogeochemical features of the Southern Ocean is the emergence of a seasonally iron-limited microbial community in HNLC regions (29). Indeed, an increase in the photosynthetic health of the phytoplankton community in response to iron additions via on-deck incubations indicated that the community in the SAZ during the time of sampling was partly iron-limited (31, 33). Here, we were also able to stress the community for iron by adding the iron chelator desferrioxamine-B, resulting in profound physiological and transcriptomic responses within the cellular community (33, 47). To date, the impact of the trace metal iron on the virus community has been predominantly characterized for dsDNA bacteriophage, demonstrating the importance of iron to viral reproduction and the regeneration of bioavailable iron (21, 23, 48–51). Iron limitation has been previously shown to negatively impact protist-infecting viruses, such as diatom-infecting RNA viruses (52) and two cultured giant virus-host model systems (28). Therefore, we tested the hypothesis that the availability of iron in the SAZ would negatively influence giant virus infection signatures using the metatranscriptomes generated from the iron incubations. Interestingly, we saw divergent responses to iron availability across the giant virus community, where one subset of the lytic marker gene encoding the structural protein MCP increased in transcript abundance under low-iron conditions (+DFB) and another subset increased under high-iron conditions (+Fe). This “high-iron” subset included an MCP originating from a giant virus MAG within the IM_12 family that clusters with isolated chlorophyte-infecting viruses. This increase in transcripts for MCP under high-iron conditions was expected, because it was hypothesized that viral infection would increase under iron-replete conditions due to the improved physiological state of the hosts, particularly photoautotrophs, which require large amounts of iron for optimal photosynthetic functioning (28, 53). Another study hypothesized that a change in iron redox state and heightened antioxidant production in diatoms undergoing iron limitation was linked to reduced RNA virus infection (52). The mechanisms underlying the elevated signatures of giant virus infection under iron-replete conditions remain unclear, but suggest that at least a subset of the community responds positively to enhanced iron availability in the SAZ region of the Southern Ocean.
The observation of elevated giant virus MCP transcripts under low-iron conditions was unexpected but provides an alternate explanation as to why iron additions trigger bloom formation and sustained abundances of protists living in iron-limited environments (29). Indeed, all of the MCPs elevated under iron-stressed conditions which we were able to phylogenetically classify belonged to the Mesomimiviridae family, whose putative hosts, the haptophytes, dominate the protist community in the SAZ (34). We hypothesize that the physiological history and state of the host during infection play an important role in determining the outcome of virus replication under different iron regimes. For example, a lab-culture-based study which reduced available iron to the host-virus systems Micromonas pusilla-MpV and Phaeocystis globosa-PgV showed a reduction in burst size under iron-stress, although the infectivity of the PgV was unaffected (28). It was hypothesized that P. globosa displayed more efficient virus production during these conditions because it could sustain growth at lower iron concentrations than M. pusilla (28). Because it has been shown that phytoplankton within the SOTS community may be uniquely adapted to chronic low-iron conditions (33, 47), the appearance of active giant viruses in situ and under DFB-added incubations suggests that these viruses may also be adapted to the iron regime in the SAZ. Alternatively, it is possible that the timescale of the lytic cycle was extended due to iron limitation, as proposed during a mesoscale iron fertilization experiment, although those findings were primarily in phage populations (49). Lastly, this observation could be the result of a reduced capacity for the host to defend against viral infection under iron-stressed conditions, leading to increased infection under iron limitation and suppressed infection under iron-rich conditions. However, because host defenses against viral infections are understudied in protist-virus models, it is unclear which systems were affected.
Overall, the patterns in giant virus transcriptional activity with respect to iron availability at SOTS suggest a potentially important role of key nutrients in altering infection dynamics within this seasonally iron-limited system. We provide evidence of enhanced infection signatures under both iron-limited and iron-replete conditions, although we cannot disentangle whether these are indirect effects due to the host cell physiological status or direct effects on the virus itself. Further research evaluating giant virus infection across a natural iron gradient with experimental incubations should be undertaken in areas of differing iron regimes to test whether host adaptation to varying iron availabilities impacts the response to changing conditions. Because our data reflect the short-term responses in gene expression to changes in iron availability (~72 h), we could only capture the early stages of infection and/or multiple stages of infection in the population. The application of more quantitative approaches (such as quantitative PCR), along with high-resolution time series, is needed to better resolve giant virus infection under iron limitation.
Conclusions.
Overall, we identified 100 “active” giant viruses by recruiting metatranscriptomes to giant virus genomes/MAGs collected across a spatiotemporal sampling scheme within an HNLC region in the Southern Ocean. This work highlights the diversity of giant viruses of protists in a nutrient-limited system and provides evidence of active infection and host metabolic rewiring occurring vertically throughout the euphotic zone. Furthermore, we found that iron may play a key role in governing giant virus-host dynamics within this system, which has implications for the response of Southern Ocean microbial communities to changing iron regimes. Since viruses can constrain oceanic biogeochemical cycles and regulate host abundance and ecology, future work should incorporate the isolation and study of giant viruses within the Southern Ocean. Because giant viruses can infect a wide range of protists (e.g., grazers, phototrophs, etc.) that play important roles in the microbial food web, we propose that they should be considered integral components within Southern Ocean microbial communities.
MATERIALS AND METHODS
Water column sample collection and environmental parameters.
Water column samples were collected at SOTS in March 2018, onboard the RV Investigator. Water column structure during the Autumn season is typically stratified with a shallow mixed layer and contains elevated abundances of dominant microeukaryote groups (34). These “samples of opportunity” were collected along a spatiotemporal gradient during the expedition. Samples were collected prior to sunrise using 12-L Niskin bottles on a conductivity, temperature and depth (CTD) rosette at 47°00′01.6″ S, 142°01′14.7″ E (March 5), 46°59′52.6″ S, 141°59'57.6″ E (Mar 7), and 47°00′01.8″ S, 142°00′07.9″ E (March 9), and 46°59′42.5′ S, 142°03′14.0″ E (Mar 17). The following depths were sampled for each date: March 5 (15, 40, 70, 100, and 200 m), March 7 (15, 50, 70, 80, and 100 m), March 9 (15, 40, 70, 100, 150, 200, 300, and 500 m), and March 17 (30, 50, 70, 80, 90, 100, 125, 150, and 200 m). Nutrient concentrations for nitrate (NO3), phosphate (PO4), silicate (Si), nitrite (NO2), and ammonium (NH4) for all sampling dates except for March 17 were determined for unfiltered samples using a Seal AA3 segmented flow system following previous procedures (54). Samples for RNA were collected by filtering approximately 1 L of water on 0.2-μm pore Sterivex filter units and immediately flash-frozen and stored in liquid nitrogen in the field, and then at −80°C until further processing. Filtration was typically completed within a 10- to 15-min time frame.
Iron incubation experimental design.
An on-deck iron incubation experiment was performed (“Growout 1” or GRW1) to elucidate the response of the surface microbial community to a gradient in iron availability. A subset of these data (focusing on cyanobacteria) has been previously published (33). All procedures were performed under trace metal-clean conditions. Briefly, on March 5, unfiltered seawater for GRW1 was collected at 5 m deep from a trace-metal clean pump and poured into 2-L polycarbonate bottles which were cleaned and prepared prior to the incubation by soaking with acid (reagent-grade hydrochloric, 10%) and rinsing three times with seawater. This depth was chosen to capture surface-level community iron limitation, as we assumed this layer would typically be iron-depleted due to decreased inputs from deep waters as well as rapid uptake. These bottles were amended with either iron chloride (FeCl3) or DFB to increase or reduce available iron (Fig. S1) (55, 56). Here, either FeCl3 or DFB was added to the following final concentrations: 0.25, 0.5, 1.0, or 2.5 nM (Fig. S1). All treatments were performed in biological duplicates. We included multiple concentrations of both FeCl3 and DFB to provide a stepwise gradient of iron availability from iron-stressed to iron-replete. The concentrations were chosen in order to sufficiently stress or replenish the community with iron and were based on a previous iron amendment experiment performed within a similar system near New Zealand (56). An unamended treatment (control) was included and a sample for a t = 0 h time point was collected from the water used for the incubations (Fig. S1). Bottles were incubated at ~33% incident irradiance and at ambient surface temperature (~11°C). After 72 h, bottles were destructively sampled to collect cells for RNA by filtering 1 L of water on 0.2-μm pore Sterivex filter units and immediately flash-frozen and stored in liquid nitrogen in the field, and then at −80°C until further processing. The time of incubation was chosen to examine the short-term responses, mainly to reflect changes on the transcriptional level, of the microbial communities. Physiological measurements of the cellular community (chlorophyll a, Fv/Fm) can be found in Gilbert et al. (33).
Schematic of the “Growout-1” (GRW1) iron incubation experimental design. Download FIG S1, PDF file, 0.2 MB (247.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
RNA extraction, library preparation, and metatranscriptome pre-processing.
RNA was extracted using a publicly available phenol-chloroform based protocol (57) and DNA was reduced using the Turbo DNA-free kit (Ambion), performing several rounds if necessary until all DNA was removed. Due to low total RNA mass (<300 ng total RNA), the following samples were pooled: three depths from March 9 (200, 300, and 500 m), three from March 17 (90, 100, and 125 m) and two from Mar 17 (150 and 200 m). Metatranscriptome libraries were prepared by reducing rRNA using the Illumina Ribo-Zero rRNA Removal kit (Bacteria) and sequenced (2 × 151 nt) using the low-input protocol for total RNA on the Illumina Novaseq S4 platform under the DOE Joint Genome Institute (JGI) Community Sequence Proposal ID no. 504140. Raw reads were filtered and trimmed using BBDuk v38.67 and BBMap v38.84 from the BBtools packages (58). Within each set of samples (in situ versus GRW1 incubation), trimmed reads were concatenated and co-assembled (assembling multiple libraries together) using MEGAHIT v1.2.9 (59) with the kmer size parameter set at -k-list 23,43,63,83,103,123. Open reading frames (ORFs) were called from each combined assembly using the gene finding algorithm MetaGeneMark v3.38 (60) using the metagenome style model.
Metatranscriptomic characterization of the microeukaryote community.
The ORFs were taxonomically annotated to classify the active microeukaryote protist community. Here, the ORFs (amino acid) were aligned to the Marine Microbial Eukaryotic Transcriptome Sequencing Project database (61) using the software package EUKulele v.2.0.1 (62) with default parameters. Trimmed reads were recruited to the assembly using BBMap v38.84 (58) with default parameters, and read counts were tabulated using the ORF coordinates using featureCounts (63). Eukaryotic-like reads were normalized using the transcripts-per-million (TPM) method (64).
Nucleocytoviricota (giant viruses) genome database and detection.
To characterize active giant viruses within our data set, we used an approach similar to that of Ha et al. (30) was used. Here, high-quality giant virus MAGs from Schulz et al. (12) (774 genomes), final giant virus MAGs from Moniruzzaman et al. (11) (502 genomes), and genomes from giant virus isolates available within NCBI RefSeq (151 genomes downloaded July 2021) were compiled. We selected the MAGs generated from the Schulz et al. study determined as “high-quality,” where the genome had “low” contamination (hallmark gene copy number had a deviation of >1.2 from the mean of the superclade), ≥90% of core nucleocytoplasmic virus orthologous genes, <30 contigs, minimum assembly size of 100 kb, and at least one contig of >30 kb (12). This approach not only allows characterization of transcripts recruited to full-length giant virus genomes and therefore better phylogenetic resolution, but it also captures giant viruses that are missing hallmark genes (65). The genome set was dereplicated using MASH v2.0 (66) with single-linkage clustering at a MASH distance of ≤0.05 (corresponding to ~95% average nucleotide identity [ANI]), and the genome with the highest N50 contig length was chosen as the representative. The representative genomes were decontaminated using ViralRecall v2.0 (67), whereby contigs with negative scores were removed, resulting in a total of 1,177 genomes in the database. ORFs from each genome were called using Prodigal v2.6.3 (68), and trimmed reads from the in situ profiles were mapped to the resultant ORFs (masking low complexity regions) using CLC Genomics Workbench 10.0 with a 95% similarity fraction and 90% length fraction for more stringent identification of giant virus genomic transcripts (30). In contrast to Ha et al. (30), we mapped reads to the nucleotide ORFs to remain consistent with the 95% ANI genomic dereplication. Next, only genomes which had ≥10% of their genes mapped to transcripts were retained to avoid those with spurious read recruitments (69) resulting in 100 “detected” genomes. These genomes were functionally annotated using eggNOG-mapper v2.1.4 with a DIAMOND alignment E value threshold of 1e-5. Read counts were normalized using the TPM method.
A concatenated protein tree was constructed to assign putative phylogenetic origin of the 100 giant virus genomes, as done previously for phylogenetically benchmarked Giant Virus Orthologous Groups protein markers (32). Here, protein sequences from the 1,177 representative genomes were used to create a concatenated alignment with ncldv_markersearch.py v1.1 (github.com/faylward/ncldv_markersearch). The tree was constructed using IQ-TREE (70) with the LG+F+I+G4 model using 1,000 ultrafast bootstraps to calculate support values (71). Delineation of phylogenetic groups was done according to Aylward et al. (67).
Assessing giant virus activity in relation to iron availability.
The gene encoding the Nucleocytoviricota MCP was queried to assess the magnitude of infection in relation to iron availability, using the GRW1 incubation metatranscriptomes to expand upon the patterns seen in situ with the giant virus genome/MAG approach. We built upon MCPs found within the MAG functional annotations by also targeting giant virus MCP sequences within the GRW1 co-assembly. To do this, a database of MCP protein sequences from cultured giant virus isolates (NCBI RefSeq manually curated from Moniruzzaman et al. [72]) was aligned against the contigs from the GRW1 co-assembly using DIAMOND BLASTx v2.6.0+ with an E value threshold of 1e-10 (73). The resultant hits were aligned to the NCBI nonredundant database (downloaded May 2021) using DIAMOND BLASTx v2.6.0+ with an E value threshold of 1e-10 to retain only those with top hits to a virus. Then, these resultant MCPs were combined with MCPs from the 100 detected giant virus genomes/MAGs and clustered at 95% amino acid identity using CD-HIT (74) to remove redundancy between the MCPs originating from the genomes/MAGs and the assembly. The nucleotide sequences of the final MCPs were re-mapped competitively at 95% nucleotide sequence similarity and 90% read length in CLC Genomics Workbench 10.0. Here, we used the DESeq2 variance stabilizing transformation normalization approach (75) to account for the resulting transcript abundances of one MCP affected by the changes in transcript abundances of others due to changes in iron availability. Because using the MCP protein alone is a weak indicator of phylogenetic origin (32), we did not perform a phylogenetic analysis of the MCP sequences originating from the co-assembly to determine taxonomy, and thus present them as an “unassigned” giant virus.
Statistical analyses.
All statistical analyses and data visualizations were performed using the R statistical software (76). To assess overall trends across dates and depths sampled, a principal-component analysis was performed using the prcomp function in R on log-transformed TPM values of the eukaryotic gene encoding the DNA directed RNA polymerase large subunit protein (RPB1), a gene used as a marker to assess the relative presence of a eukaryotic taxon within metatranscriptomes (72, 77). A PCA was also performed on log-transformed TPMs summarized across each giant virus genome/MAG. To correlate environmental variables to giant virus expression, a Spearman correlation (cor function in R, method = “spearman”) followed by P value adjustment for multiple comparison using the Holm method (“corr.adjust” from the RcmdrMisc package v2.7.1, type = “spearman” [78]) was computed between TPM counts summed across each giant virus genome/MAG and the environmental variables. Because there were no nutrient data for the March 17 sampling date, this date was omitted from nutrient-giant virus comparisons. To identify giant virus MCP transcripts (assembly + genome/MAG) that were significantly different between FeCl3-added and DFB-added incubations in GRW1, a Mann-Whitney U test on the vst-normalized transcripts of each MCP was performed (wilcox.test).
Data availability.
Raw and processed data for the combined assemblies are publicly available through the JGI Data Portal (https://data.jgi.doe.gov) under Project ID no. 1260737 (in situ profiles) and 1260740 (GRW1). To access individual assemblies, see Table S4 at https://zenodo.org/record/7457861#.Y6BrIuzML0o for Project ID numbers. The final set of representative genomes used to for giant viruses read recruitment are available for download on Zenodo (https://zenodo.org/record/6382754#.Y-Pxw-zML0s).
ACKNOWLEDGMENTS
We thank the officers, crew, and research staff of the Marine National Facility and the RV Investigator for their help with sample collection and hydrochemistry data generation.
Financial support for this research was provided by the Australian Research Council’s Discovery program (DP170102108, DP220100289). Ship time was provided via Australia’s Marine National Facility. A National Science Foundation grant (no. OCE-1829641) to S.W.W. supported this work. The work (proposal: 10.46936/10.25585/60001188) conducted by the U.S. Department of Energy Joint Genome Institute (https://ror.org/04xm1d337), a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy under contract no. DE-AC02-05CH11231.
Contributor Information
Steven W. Wilhelm, Email: wilhelm@utk.edu.
Thulani P. Makhalanyane, University of Pretoria
REFERENCES
- 1.Fuhrman JA. 1999. Marine viruses and their biogeochemical and ecological effects. Nature 399:541–548. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- 2.Suttle CA. 2007. Marine viruses: major players in the global ecosystem. Nat Rev Microbiol 5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 3.Breitbart M. 2012. Marine viruses: truth or dare. Annu Rev Mar Sci 4:425–448. doi: 10.1146/annurev-marine-120709-142805. [DOI] [PubMed] [Google Scholar]
- 4.Laber CP, Hunter JE, Carvalho F, Collins JR, Hunter EJ, Schieler BM, Boss E, More K, Frada M, Thamatrakoln K, Brown CM, Haramaty L, Ossolinski J, Fredricks H, Nissimov JI, Vandzura R, Sheyn U, Lehahn Y, Chant RJ, Martins AM, Coolen MJL, Vardi A, DiTullio GR, Van Mooy BAS, Bidle KD. 2018. Coccolithovirus facilitation of carbon export in the North Atlantic. Nat Microbiol 3:537–547. doi: 10.1038/s41564-018-0128-4. [DOI] [PubMed] [Google Scholar]
- 5.Blanc-Mathieu R, Kaneko H, Endo H, Chaffron S, Hernández-Velázquez R, Nguyen CH, Mamitsuka H, Henry N, de Vargas C, Sullivan MB. 2019. Viruses of the eukaryotic plankton are predicted to increase carbon export efficiency in the global sunlit ocean. bioRxiv. 710228.
- 6.Forterre P. 2011. Manipulation of cellular syntheses and the nature of viruses: the virocell concept. Comptes Rendus Chimie 14:392–399. doi: 10.1016/j.crci.2010.06.007. [DOI] [Google Scholar]
- 7.Rosenwasser S, Ziv C, Van Creveld SG, Vardi A. 2016. Virocell metabolism: metabolic innovations during host-virus interactions in the ocean. Trends Microbiol 24:821–832. doi: 10.1016/j.tim.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Fischer MG. 2016. Giant viruses come of age. Curr Opin Microbiol 31:50–57. doi: 10.1016/j.mib.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Koonin EV, Yutin N. 2010. Origin and evolution of eukaryotic large nucleo-cytoplasmic DNA viruses. Intervirology 53:284–292. doi: 10.1159/000312913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mihara T, Koyano H, Hingamp P, Grimsley N, Goto S, Ogata H. 2018. Taxon richness of “Megaviridae” exceeds those of bacteria and archaea in the ocean. Microbes Environ 33:162–171. doi: 10.1264/jsme2.ME17203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moniruzzaman M, Martinez-Gutierrez CA, Weinheimer AR, Aylward FO. 2020. Dynamic genome evolution and complex virocell metabolism of globally-distributed giant viruses. Nat Commun 11:1710. doi: 10.1038/s41467-020-15507-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schulz F, Roux S, Paez-Espino D, Jungbluth S, Walsh DA, Denef VJ, McMahon KD, Konstantinidis KT, Eloe-Fadrosh EA, Kyrpides NC, Woyke T. 2020. Giant virus diversity and host interactions through global metagenomics. Nature 578:432–436. doi: 10.1038/s41586-020-1957-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plugge B, Gazzarrini S, Nelson M, Cerana R, Van J, Etten Derst C, DiFrancesco D, Moroni A, Thiel G. 2000. A potassium channel protein encoded by Chlorella virus PBCV-1. Science 287:1641–1644. doi: 10.1126/science.287.5458.1641. [DOI] [PubMed] [Google Scholar]
- 14.Schvarcz CR, Steward GF. 2018. A giant virus infecting green algae encodes key fermentation genes. Virology 518:423–433. doi: 10.1016/j.virol.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 15.Needham DM, Yoshizawa S, Hosaka T, Poirier C, Choi CJ, Hehenberger E, Irwin NAT, Wilken S, Yung C-M, Bachy C, Kurihara R, Nakajima Y, Kojima K, Kimura-Someya T, Leonard G, Malmstrom RR, Mende DR, Olson DK, Sudo Y, Sudek S, Richards TA, DeLong EF, Keeling PJ, Santoro AE, Shirouzu M, Iwasaki W, Worden AZ. 2019. A distinct lineage of giant viruses brings a rhodopsin photosystem to unicellular marine predators. Proc Natl Acad Sci USA 116:20574–20583. doi: 10.1073/pnas.1907517116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monier A, Chambouvet A, Milner DS, Attah V, Terrado R, Lovejoy C, Moreau H, Santoro AE, Derelle É, Richards TA. 2017. Host-derived viral transporter protein for nitrogen uptake in infected marine phytoplankton. Proc Natl Acad Sci USA 114:E7489–E7498. doi: 10.1073/pnas.1708097114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeAngelis PL, Jing W, Graves MV, Burbank DE, Van Etten JL. 1997. Hyaluronan synthase of Chlorella virus PBCV-1. Science 278:1800–1803. doi: 10.1126/science.278.5344.1800. [DOI] [PubMed] [Google Scholar]
- 18.Boyd P, LaRoche J, Gall M, Frew R, McKay RM. 1999. Role of iron, light, and silicate in controlling algal biomass in subantarctic waters SE of New Zealand. Journal of Geophysical Research: Oceans 104:13395–13408. doi: 10.1029/1999JC900009. [DOI] [Google Scholar]
- 19.Boyd PW, Watson AJ, Law CS, Abraham ER, Trull T, Murdoch R, Bakker DC, Bowie AR, Buesseler KO, Chang H, Charette M. 2000. A mesoscale phytoplankton bloom in the polar Southern Ocean stimulated by iron fertilization. Nature 407:695–702. doi: 10.1038/35037500. [DOI] [PubMed] [Google Scholar]
- 20.Brum JR, Hurwitz BL, Schofield O, Ducklow HW, Sullivan MB. 2017. Seasonal time bombs: dominant temperate viruses affect Southern Ocean microbial dynamics. ISME J 11:588. doi: 10.1038/ismej.2016.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brussaard C, Timmermans K, Uitz J, Veldhuis M. 2008. Virioplankton dynamics and virally induced phytoplankton lysis versus microzooplankton grazing southeast of the Kerguelen (Southern Ocean). Deep Sea Res Part II 55:752–765. doi: 10.1016/j.dsr2.2007.12.034. [DOI] [Google Scholar]
- 22.Evans C, Brussaard CP. 2012. Regional variation in lytic and lysogenic viral infection in the Southern Ocean and its contribution to biogeochemical cycling. Appl Environ Microbiol 78:6741–6748. doi: 10.1128/AEM.01388-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weinbauer MG, Arrieta JM, Griebler C, Herndlb GJ. 2009. Enhanced viral production and infection of bacterioplankton during an iron-induced phytoplankton bloom in the Southern Ocean. Limnol Oceanogr 54:774–784. doi: 10.4319/lo.2009.54.3.0774. [DOI] [Google Scholar]
- 24.Alarcón-Schumacher T, Guajardo-Leiva S, Martinez-Garcia M, Díez B. 2021. Ecogenomics and adaptation strategies of Southern Ocean viral communities. mSystems 6:e00396-21. doi: 10.1128/mSystems.00396-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biggs TEG, Huisman J, Brussaard CPD. 2021. Viral lysis modifies seasonal phytoplankton dynamics and carbon flow in the Southern Ocean. ISME J 15:3615–3622. doi: 10.1038/s41396-021-01033-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flaviani F, Schroeder DC, Lebret K, Balestreri C, Highfield AC, Schroeder JL, Thorpe SE, Moore K, Pasckiewicz K, Pfaff MC. 2018. Distinct oceanic microbiomes from viruses to protists located near the antarctic circumpolar current. Front Microbiol: 9:1474. doi: 10.3389/fmicb.2018.01474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mojica KD, Brussaard CP. 2014. Factors affecting virus dynamics and microbial host-virus interactions in marine environments. FEMS Microbiol Ecol 89:495–515. doi: 10.1111/1574-6941.12343. [DOI] [PubMed] [Google Scholar]
- 28.Slagter HA, Gerringa LJ, Brussaard CP. 2016. Phytoplankton virus production negatively affected by iron limitation. Front Mar Sci 3:156. doi: 10.3389/fmars.2016.00156. [DOI] [Google Scholar]
- 29.Boyd PW, Jickells T, Law CS, Blain S, Boyle EA, Buesseler KO, Coale KH, Cullen JJ, de Baar HJW, Follows M, Harvey M, Lancelot C, Levasseur M, Owens NPJ, Pollard R, Rivkin RB, Sarmiento J, Schoemann V, Smetacek V, Takeda S, Tsuda A, Turner S, Watson AJ. 2007. Mesoscale iron enrichment experiments 1993–2005: synthesis and future directions. Science 315:612–617. doi: 10.1126/science.1131669. [DOI] [PubMed] [Google Scholar]
- 30.Ha AD, Moniruzzaman M, Aylward FO. 2021. High transcriptional activity and diverse functional repertoires of hundreds of giant viruses in a coastal marine system. mSystems 6:e00293-21. doi: 10.1128/mSystems.00293-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schallenberg C, Strzepek RF, Schuback N, Clementson LA, Boyd PW, Trull TW. 2020. Diel quenching of Southern Ocean phytoplankton fluorescence is related to iron limitation. Biogeosciences 17:793–812. doi: 10.5194/bg-17-793-2020. [DOI] [Google Scholar]
- 32.Aylward FO, Moniruzzaman M, Ha AD, Koonin EV. 2021. A phylogenomic framework for charting the diversity and evolution of giant viruses. PLoS Biol 19:e3001430. doi: 10.1371/journal.pbio.3001430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gilbert NE, LeCleir GR, Strzepek RF, Ellwood MJ, Twining BS, Roux S, Pennacchio C, Boyd PW, Wilhelm SW. 2022. Bioavailable iron titrations reveal oceanic Synechococcus ecotypes optimized for different iron availabilities. ISME Commun 2:1–12. doi: 10.1038/s43705-022-00132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eriksen R, Trull TW, Davies D, Jansen P, Davidson AT, Westwood K, van den Enden R. 2018. Seasonal succession of phytoplankton community structure from autonomous sampling at the Australian Southern Ocean Time Series (SOTS) observatory. Marine Ecol Progress Series 589:13–31. doi: 10.3354/meps12420. [DOI] [Google Scholar]
- 35.Gallot-Lavallée L, Pagarete A, Legendre M, Santini S, Sandaa R-A, Himmelbauer H, Ogata H, Bratbak G, Claverie J-M. 2015. The 474-kilobase-pair complete genome sequence of CeV-01B, a virus infecting Haptolina (Chrysochromulina) ericina (Prymnesiophyceae). Genome Announc 3:e01413-15. doi: 10.1128/genomeA.01413-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santini S, Jeudy S, Bartoli J, Poirot O, Lescot M, Abergel C, Barbe V, Wommack KE, Noordeloos AA, Brussaard CP. 2013. Genome of Phaeocystis globosa Virus PgV-16T highlights the common ancestry of the largest known DNA viruses infecting eukaryotes. Proc Natl Acad Sci USA 110:10800–10805. doi: 10.1073/pnas.1303251110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moniruzzaman M, LeCleir GR, Brown CM, Gobler CJ, Bidle KD, Wilson WH, Wilhelm SW. 2014. Genome of brown tide virus (AaV), the little giant of the Megaviridae, elucidates NCLDV genome expansion and host-virus coevolution. Virology 466:60–70. doi: 10.1016/j.virol.2014.06.031. [DOI] [PubMed] [Google Scholar]
- 38.Worden AZ, Janouskovec J, McRose D, Engman A, Welsh RM, Malfatti S, Tringe SG, Keeling PJ. 2012. Global distribution of a wild alga revealed by targeted metagenomics. Curr Biol 22:R675–R677. doi: 10.1016/j.cub.2012.07.054. [DOI] [PubMed] [Google Scholar]
- 39.Latasa M, Cabello AM, Morán XAG, Massana R, Scharek R. 2017. Distribution of phytoplankton groups within the deep chlorophyll maximum. Limnol Oceanogr 62:665–685. doi: 10.1002/lno.10452. [DOI] [Google Scholar]
- 40.Wommack KE, Colwell RR. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol Rev 64:69–114. doi: 10.1128/MMBR.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schulz F, Abergel C, Woyke T. 2022. Giant virus biology and diversity in the era of genome-resolved metagenomics. Nature Rev Microbiol 20:721–736. doi: 10.1038/s41579-022-00754-5. [DOI] [PubMed] [Google Scholar]
- 42.Belhaouari DB, De Souza GAP, Lamb DC, Kelly SL, Goldstone JV, Stegeman JJ, Colson P, La Scola B, Aherfi S. 2022. Metabolic arsenal of giant viruses: host hijack or self-use? Elife 11:e78674. doi: 10.7554/eLife.78674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chung CC, Hwang SP, Chang J. 2003. Identification of a high-affinity phosphate transporter gene in a prasinophyte alga, Tetraselmis chui, and its expression under nutrient limitation. Applied and Environmental Microbiology 69:754–759. doi: 10.1128/AEM.69.2.754-759.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dyhrman ST, Haley ST, Birkeland SR, Wurch LL, Cipriano MJ, McArthur AG. 2006. Long serial analysis of gene expression for gene discovery and transcriptome profiling in the widespread marine coccolithophore Emiliania huxleyi. Applied and Environmental Microbiology 72:252–260. doi: 10.1128/AEM.72.1.252-260.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dyhrman ST. 2016. Nutrients and their acquisition: phosphorus physiology in microalgae. In Borowitzka MA, Beardall J, Raven JA (ed), The physiology of microalgae, p 155–183. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 46.Robertson DL, Smith GJ, Alberte RS. 2001. Glutamine synthetase in marine algae: new surprises from an old enzyme. J Phycology 37:793–795. doi: 10.1046/j.1529-8817.2001.01057.x. [DOI] [Google Scholar]
- 47.Sofen LE, Antipova OA, Ellwood MJ, Gilbert NE, LeCleir GR, Lohan MC, Mahaffey C, Mann EL, Ohnemus DC, Wilhelm SW. 2021. Trace metal contents of autotrophic flagellates from contrasting open-ocean ecosystems. Limnol Oceanogr Lett 7:354–362. doi: 10.1002/lol2.10258. [DOI] [Google Scholar]
- 48.Strzepek RF, Maldonado MT, Higgins JL, Hall J, Safi K, Wilhelm SW, Boyd PW. 2005. Spinning the “ferrous wheel”: the importance of the microbial community in an iron budget during the FeCycle experiment. Global Biogeochem Cycles 19:GB4S26. doi: 10.1029/2005GB002490. [DOI] [Google Scholar]
- 49.Higgins JL, Kudo I, Nishioka J, Tsuda A, Wilhelm SW. 2009. The response of the virus community to a mesoscale iron fertilization in the sub-Arctic Pacific Ocean. Deep Sea Res 56. doi: 10.1016/j.dsr2.2009.06.005. [DOI] [Google Scholar]
- 50.Malits A, Christaki U, Obernosterer I, Weinbauer M. 2014. Enhanced viral production and virus-mediated mortality of bacterioplankton in a natural iron-fertilized bloom event above the Kerguelen Plateau. Biogeosciences 11:6841–6853. doi: 10.5194/bg-11-6841-2014. [DOI] [Google Scholar]
- 51.Poorvin L, Rinta-Kanto JM, Hutchins DA, Wilhelm SW. 2004. Viral release of iron and its bioavailability to marine plankton. Limnol Oceanogr 49:1734–1741. doi: 10.4319/lo.2004.49.5.1734. [DOI] [Google Scholar]
- 52.Kranzler CF, Brzezinski MA, Cohen NR, Lampe RH, Maniscalco M, Till CP, Mack J, Latham JR, Bruland KW, Twining BS. 2021. Impaired viral infection and reduced mortality of diatoms in iron-limited oceanic regions. Nature Geoscience 14:231–237. doi: 10.1038/s41561-021-00711-6. [DOI] [Google Scholar]
- 53.Drakesmith H, Prentice A. 2008. Viral infection and iron metabolism. Nat Rev Microbiol 6:541–552. doi: 10.1038/nrmicro1930. [DOI] [PubMed] [Google Scholar]
- 54.Rees C, Pender L, Sherrin K, Schwanger C, Hughes P, Tibben S, Marouchos A, Rayner M. 2019. Methods for reproducible shipboard SFA nutrient measurement using RMNS and automated data processing. Limnol Oceanogr Methods 17:25–41. doi: 10.1002/lom3.10294. [DOI] [Google Scholar]
- 55.Eldridge ML, Trick CG, Alm MB, DiTullio GR, Rue EL, Bruland KW, Hutchins DA, Wilhelm SW. 2004. Phytoplankton community response to a manipulation of bioavailable iron in HNLC waters of the subtropical Pacific Ocean. Aquatic Microb Ecol 35:79–91. doi: 10.3354/ame035079. [DOI] [Google Scholar]
- 56.Wilhelm SW, King AL, Twining BS, LeCleir GR, DeBruyn JM, Strzepek RF, Breene CL, Pickmere S, Ellwood MJ, Boyd PW. 2013. Elemental quotas and physiology of a southwestern Pacific Ocean plankton community as a function of iron availability. Aquatic Microb Ecol 68:185–194. doi: 10.3354/ame01611. [DOI] [Google Scholar]
- 57.Gilbert N, Pound H, Wilhelm SW. 2020. RNA extraction from Sterivex using phenol:chloroform - cutting off the filter from the Sterivex unit for low biomass samples. Protocolsio doi: 10.17504/protocols.io.bh75j9q6. [DOI] [Google Scholar]
- 58.Bushnell B. 2014. BBMap: BBMap short read aligner, and other bioinformatic tools. Available from https://sourceforge.net/projects/bbmap/.
- 59.Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 60.Zhu W, Lomsadze A, Borodovsky M. 2010. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res 38:e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keeling PJ, Burki F, Wilcox HM, Allam B, Allen EE, Amaral-Zettler LA, Armbrust EV, Archibald JM, Bharti AK, Bell CJ, Beszteri B, Bidle KD, Cameron CT, Campbell L, Caron DA, Cattolico RA, Collier JL, Coyne K, Davy SK, Deschamps P, Dyhrman ST, Edvardsen B, Gates RD, Gobler CJ, Greenwood SJ, Guida SM, Jacobi JL, Jakobsen KS, James ER, Jenkins B, John U, Johnson MD, Juhl AR, Kamp A, Katz LA, Kiene R, Kudryavtsev A, Leander BS, Lin S, Lovejoy C, Lynn D, Marchetti A, McManus G, Nedelcu AM, Menden-Deuer S, Miceli C, Mock T, Montresor M, Moran MA, Murray S, et al. 2014. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol 12:e1001889. doi: 10.1371/journal.pbio.1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krinos AI, Hu SK, Cohen NR, Alexander H. 2020. EUKulele: taxonomic annotation of the unsung eukaryotic microbes. arXiv. doi: 10.48550/arXiv.2011.00089. [DOI]
- 63.Liao Y, Smyth GK, Shi W. 2014. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 64.Wagner GP, Kin K, Lynch VJ. 2012. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci 131:281–285. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- 65.Abergel C, Legendre M, Claverie J-M. 2015. The rapidly expanding universe of giant viruses: Mimivirus, Pandoravirus, Pithovirus and Mollivirus. FEMS Microbiol Rev 39:779–796. doi: 10.1093/femsre/fuv037. [DOI] [PubMed] [Google Scholar]
- 66.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM. 2016. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aylward FO, Moniruzzaman M. 2021. ViralRecall: a flexible command-line tool for the detection of giant virus signatures in ‘omic data. Viruses 13:150. doi: 10.3390/v13020150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tithi SS, Aylward FO, Jensen RV, Zhang L. 2018. FastViromeExplorer: a pipeline for virus and phage identification and abundance profiling in metagenomics data. PeerJ 6:e4227. doi: 10.7717/peerj.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hoang DT, Chernomor O, Von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol 35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moniruzzaman M, Wurch LL, Alexander H, Dyhrman ST, Gobler CJ, Wilhelm SW. 2017. Virus-host relationships of marine single-celled eukaryotes resolved from metatranscriptomics. Nat Commun 8:16054. doi: 10.1038/ncomms16054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 74.Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.R Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 77.Pound HL, Gann ER, Tang X, Krausfeldt LE, Huff M, Staton ME, Talmy D, Wilhelm SW. 2020. The “neglected viruses” of Taihu: abundant transcripts for viruses infecting eukaryotes and their potential role in phytoplankton succession. Front Microbiol 11:338. doi: 10.3389/fmicb.2020.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fox J. 2020. RcmdrMisc: R Commander Miscellaneous functions. R package version 2.7.1. R Foundation for Statistical Computing: Vienna, Austria. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Physicochemical status of the time points sampled during the March 2018 research expedition at the Southern Ocean Time Series (SOTS). Conductivity, temperature, and depth (CTD) data are presented for all four dates sampled, and nutrient data are presented only for March 5, 7, and 9 sampling dates. (A) CTD fluorescence data, expressed as relative fluorescence units. (B) CTD Dissolved oxygen measurements expressed as mg/L. (C) CTD salinity data. (D) CTD temperature data. (E) Nitrate (NO3) measurements. (F) Nitrite (NO2) measurements. (G) Ammonium (NH4) measurements. (H) Phosphate (PO4) measurements. (I) Silicate (Si) measurements. Download FIG S2, PDF file, 0.5 MB (555.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Physicochemical data for time points and depths collected during the March 2018 expedition. No nutrient data were collected for March 17. Download Table S1, DOCX file, 0.02 MB (19.6KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Proportions of reads assigned across eukaryotes separated by date and depth sampled. The values represent the summed normalized transcripts (transcripts per million, TPMs) across open reading frames assigned to a given taxonomic rank within the MMTESP database. Download FIG S3, PDF file, 0.6 MB (597.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Results of read recruitments to candidate Nucleocytoviricota isolate and metagenome-assembled genomes detected at SOTS. Each dot represents a candidate Nucleocytoviricota genome with transcripts detected. Genes recovered indicate the percentage of genes within each candidate genome that had reads mapped, where the mean genome abundance indicates the total normalized read count (log10 TPM) recruited to that genome. Total number of genes within each candidate genome is represented by dot size. Download FIG S4, PDF file, 0.09 MB (91.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Summary of the (A) taxonomic distribution of the 100 genomes. The genome count illustrates the number of genomes detected within a specific family-level assignment. (B) Relative summed expression levels of the family-level genome/metagenomic assembled genome (MAG) assignments across the entire in situ dataset, represented as TPMs. (C) Genome-level contribution towards the total pool of giant-virus transcripts (TPMs), represented as a percent. Each genome is color coded by Family level assignment and the Genus level assignment is shown in text next to each bar. Download FIG S5, PDF file, 1.0 MB (1,007.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Count of giant virus genomes detected, grouped by phylogenetic classification corresponding to Fig. 1. Download Table S2, DOCX file, 0.01 MB (13.7KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Shifts in the transcript abundance patterns of eukaryotic and giant viruses. Each plot shows a principal components (PC) analysis ordination of normalized transcript patterns across (A) Eukaryotic RPB1 candidates colored by date sampled (B) Nucleocytoviricota total genome recruitments colored by date sampled (C) Eukaryotic RPB1 candidates colored by depth sampled (D) Nucleocytoviricota total genome recruitments colored by depth sampled. Download FIG S6, PDF file, 0.5 MB (486.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Spearman’s correlations between environmental parameters (CTD and nutrient data) and summed transcript abundances of each giant virus genome/MAG across the in situ dataset. Only correlations with Spearman’s rho (ρ) ±0.8 with an adjusted P value (Holm method) of ≤0.1 are shown. The Family-level assignment of each genome is shown in parentheses next to its ID. Download FIG S7, PDF file, 0.4 MB (454.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Detected giant virus major capsid protein hits which had significantly different (Mann-Whitney U test, P < 0.05) vst values between the DFB-added versus Fe-added incubations. Download Table S3, DOCX file, 0.02 MB (22.4KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Schematic of the “Growout-1” (GRW1) iron incubation experimental design. Download FIG S1, PDF file, 0.2 MB (247.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
Raw and processed data for the combined assemblies are publicly available through the JGI Data Portal (https://data.jgi.doe.gov) under Project ID no. 1260737 (in situ profiles) and 1260740 (GRW1). To access individual assemblies, see Table S4 at https://zenodo.org/record/7457861#.Y6BrIuzML0o for Project ID numbers. The final set of representative genomes used to for giant viruses read recruitment are available for download on Zenodo (https://zenodo.org/record/6382754#.Y-Pxw-zML0s).