Abstract
The caspase-activated DNase (CAD) is involved in DNA degradation during apoptosis. Chemical modification of murine CAD with the lysine-specific reagent 2,4,6-trinitrobenzenesulphonic acid and the tyrosine-specific reagent N-acetylimidazole leads to inactivation of the nuclease, indicating that lysine and tyrosine residues are important for DNA cleavage by this enzyme. The presence of DNA or the inhibitor ICAD-L protects the enzyme from modification. Amino acid substitution in murine CAD of lysines and tyrosines conserved in CADs from five different species leads to variants with little if any catalytic activity, but unaltered DNA binding (K155Q, K301Q, K310Q, Y247F), with the exception of Y170F, which retains wild-type activity. Similarly, as observed for the previously characterised H242N, H263N, H308N and H313N variants, the newly introduced His→Asp/Glu or Arg exchanges lead to variants with <1% of wild-type activity, with two exceptions: H313R shows wild-type activity, and H308D at pH 5.0 exhibits ∼5% of wild-type activity at this pH. Y170F and H313R produce a specific pattern of fragments, different from wild-type CAD, which degrades DNA non-specifically. The recombinant nuclease variants produced in Escherichia coli were tested for their ability to form nucleolytically active oligomers. They did not show any significant deviation from the wild-type enzyme. Based on these and published data possible roles of the amino acid residues under investigation are discussed.
INTRODUCTION
Apoptosis in its final stages is characterised by nuclear DNA fragmentation (1). The major nucleolytic activity for the cell autonomous apoptotic DNA fragmentation in mammalian cells is the caspase-activated DNase (CAD), also termed DFF40 (DNA fragmentation factor 40 kDa subunit) (2–4). In non-apoptotic cells, this nuclease is bound to an inhibitory protein named ICAD-L/DFF45 (inhibitor of CAD large form/DNA fragmentation factor 45 kDa subunit), forming a heterodimeric complex known as DFF (DNA fragmentation factor). ICAD-L/DFF45 also serves as a specific chaperone for CAD/DFF40 and thus is required for the proper folding of a catalytically competent nuclease (5). Upon activation of caspases in the course of programmed cell death, ICAD-L/DFF45 is cleaved at two specific sites, releasing the nuclease from the DFF complex (2,6). Free CAD/DFF40 oligomerises into its active form, which degrades chromosomal DNA into nucleosomal units (7). Proteins such as histone H1, HMG1, HMG2 and topoisomerase II α bind to CAD/DFF40 and thereby stimulate its activity (7–9).
A deletion analysis of CAD/DFF40 suggested that the catalytic centre is located in the C-terminal part of the enzyme (10,11), whereas structural and mutational studies revealed that the regulatory N-terminal domain (‘CAD-’ or ‘CIDE-N-domain’) is essential for a functional interaction of CAD/DFF40 with its chaperone and inhibitor ICAD-L/DFF45 (12–15).
The C-terminal catalytic domain of CAD/DFF40 contains six conserved histidine residues, some of which, on the basis of chemical modification and site-directed mutagenesis studies, have been shown to be essential for DNA binding and/or cleavage by CAD/DFF40 (16,17). Besides the participation of histidine residues in DNA cleavage and binding by CAD/DFF40, which we have further explored in the present work, very little is known about the composition of the active site of this enzyme. In search of other functionally relevant amino acid residues and in the absence of structural information about the catalytic domain of CAD/DFF40, we have chemically modified recombinant CAD with the lysine-specific reagent 2,4,6-trinitrobenzenesulphonic acid (TNBS) and the tyrosine-specific reagent N-acetylimidazole (NAI). Our experiments show that lysine and tyrosine residues are essential for CAD/DFF40 activity. Guided by an amino acid sequence alignment of five homologous apoptotic nucleases, we have then replaced three conserved lysine residues and two conserved tyrosine residues in the catalytic domain of CAD by glutamine and phenylalanine, respectively. All variants were proficient in DNA binding and, with one exception, almost inactive in DNA cleavage. On the basis of these and previous results, possible roles of the amino acid residues under investigation in the mechanism of DNA binding and cleavage by CAD/DFF40 and the inhibition of CAD/DFF40 by ICAD-L/DFF45 are discussed.
MATERIALS AND METHODS
Expression of GST-mCAD/hICAD-L complex in Escherichia coli and preparation of free GST-mCAD
GST-mCAD/hICAD-L complex and free GST-mCAD were produced as described previously (17). In brief, the GST-mCAD/hICAD-L complex was expressed in E.coli BL21Gold(DE3) cells harbouring plasmids pACET-DFF45 coding for wild-type human ICAD-L and pGEX-2T coding for wild-type GST-tagged murine CAD or the mutant versions, respectively, and purified by affinity chromatography on glutathione–Sepharose 4B beads. To obtain free GST-mCAD, the GST-mCAD/hICAD-L complex bound to the beads was incubated with recombinant caspase-3. After washing with buffer A (20 mM HEPES–NaOH pH 7.4, 100 mM NaCl, 1 mM EDTA) supplemented with 5 mM DTT, 10% glycerol, 0.01% Triton X-100 and 5 mM MgCl2, the free nuclease was eluted from the glutathione–Sepharose 4B beads and dialysed against buffer B (50 mM Na-phosphate, pH 7.5, 100 mM NaCl, 10% glycerol and 1 mM DTT). Protein concentrations were determined by UV spectroscopy, using a molar extinction coefficient calculated according to Pace et al. (18).
Expression of mCAD in mammalian cells
For expression of the mCAD/GST-hICAD-L complex in mammalian cells, the coding regions for wild-type mCAD and the variant Y170F were inserted into the vector pCS2-MT (19,20), and the coding region for a GST-hICAD-L/DFF45 fusion protein was inserted into the vector pCI (Promega). 293T human embryonic kidney cells were cultured in DMEM containing 10% fetal calf serum. Ten micrograms of each expression construct (pCI-GST-hICAD-L/DFF45 and pCS2-MT-mCAD) were used to co-transfect cells cultured in maxi-dishes using TransFast™ (Promega) transfection reagent according to the supplier’s recommendations. Forty-eight hours after transfection, cells were harvested and washed twice with PBS (1.5 ml/maxi-dish). After washing, cells were pelleted, resuspended and lysed by sonication in buffer A supplemented with 5 mM DTT and 10% glycerol. After sonication, Triton X-100 was added to a final concentration of 0.01%. Subsequently, the crude extract was centrifuged and the complex contained in the supernatant was bound to 75 µl of a suspension of glutathione–Sepharose 4B beads. Bound protein was washed twice with the same buffer as described above and resuspended in 175 µl of buffer A supplemented with 5 mM DTT, 10% glycerol and 0.01% Chaps {3-([3-cholamidopropyl]dimethylammonio)-1-propanesulfonate}. Free mCAD was eluted by treating the complex bound to the beads with 5 µl of a caspase-3 preparation for 1 h at ambient temperature. For DNA cleavage, 19 µl of eluted mCAD were incubated with 30 ng/µl assay solution of the plasmid pBSK-VDEX (New England Biolabs) for 15 min at 37°C in a total volume of 25 µl in buffer C (20 mM Tris–HCl, pH 7.4, 100 mM NaCl, 1 mM EDTA, 10 mM DTT, 5% glycerol, 0.01% Chaps) supplemented with 5 mM MgCl2.
Chemical modification of GST-mCAD
Modification of lysine residues of free GST-mCAD and the GST-mCAD/hICAD-L complex was performed using 20 and 60 µM, respectively, TNBS in 10 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM EDTA, 1 mM DTT, 5% glycerol and 0.01% Chaps. For modification of the tyrosine residues 50 and 150 mM, respectively, NAI in 10 mM MES–NaOH pH 7.0 were used. To investigate protection against modification by TNBS or NAI, free GST-mCAD was also modified in the presence of 120 ng/µl assay solution of plasmid DNA (pBSK-VDEX). To analyse the residual activity of the modified nuclease, aliquots of the modification reaction mixtures were transferred into buffer C after defined time intervals, and incubated with caspase-3 (for reasons of standardisation always added) for 10 min at 37°C. In the case of modification of the GST-mCAD/hICAD-L complex and of free GST-mCAD in the absence of DNA, the aliquots were supplemented with plasmid pBSK-VDEX as substrate (25 ng/µl assay solution). To start the DNA cleavage reaction, MgCl2 was added to a final concentration of 5 mM and this reaction mix was then incubated for 10 min at 37°C. The cleavage products were analysed on a 0.8% TBE (100 mM Tris–HCl pH 8.3, 100 mM borate, 2.5 mM EDTA) agarose gel containing 0.05 µg/ml ethidium bromide.
Site-directed mutagenesis
Site-directed mutagenesis of GST-mCAD was performed as described by Kirsch and Joly (21). In brief, a first PCR was performed using a mutagenic primer and an appropriate reverse primer with pGEX-2T-mCAD as template and Pfu DNA polymerase. Then, a second PCR was performed using purified product from the first reaction as megaprimers for an inverse PCR following the instructions of the QuikChange protocol (Stratagene).
In vitro GST-mCAD activity assays
For the in vitro CAD activity assay, aliquots of dialysed free GST-mCAD were incubated in buffer C supplemented with 5 mM MgCl2 for defined time intervals at 37°C, using 25 ng/µl assay solution (10 nM final concentration) plasmid DNA (pBSK-VDEX). Cleavage products were analysed on a 0.8% TBE–agarose gel containing 0.05 µg/ml ethidium bromide.
DNA–cellulose binding assay
DNA binding of GST-mCAD variants was investigated by incubating caspase-3-treated GST-CAD/hICAD-L complex with 50 µl of a DNA–cellulose suspension in buffer B for 25 min at 4°C. Bound protein was washed once with 500 µl of buffer A supplemented with 5 mM DTT, 10% glycerol, 0.01% Triton X-100 and subsequently eluted by incubating the sample for 5 min at 95°C in 10 µl of SDS-gel loading buffer (160 mM Tris–HCl pH 6.8, 2% SDS, 5% β-mercaptoethanol, 40% glycerol and 0.1% bromophenol blue) and analysed by SDS–PAGE.
Size-exclusion chromatography with GST-mCAD variants
Size-exclusion chromatography with GST-mCAD and its variants was performed on a Superose 12 gel filtration column (24 ml bed volume, Pharmacia) equilibrated with 20 mM HEPES–NaOH pH 8.0, 150 mM KCl, 1 mM EDTA and 5 mM MgCl2 using a Merck-Hitachi HPLC system. Twenty micrograms of each variant were loaded onto the column and fractions of 1 ml were collected at a flow rate of 0.5 ml/min. Twenty-microlitre aliquots of the fractions were incubated in buffer C containing 5 mM MgCl2 for 1.75 h at 37°C with 20 ng/µl assay solution plasmid DNA (pBSK-VDEX). Cleavage products were analysed by agarose gel electrophoresis as described above.
RESULTS
Inactivation of CAD by chemical modification with TNBS and NAI and protection from chemical modification by DNA and ICAD-L
Modification of free GST-mCAD with the lysine-specific reagent TNBS and the tyrosine-specific reagent NAI leads to an inhibition of the nucleolytic activity of the enzyme in a time- and dose-dependent manner, suggesting that one or more lysine and tyrosine residues are involved in the DNA cleavage reaction catalysed by CAD (Fig. 1). The presence of the inhibitor hICAD-L or of DNA during the TNBS- and NAI-modification reactions protects the nuclease from being modified, indicating that in the enzyme/substrate and enzyme/inhibitor complexes the essential lysine and tyrosine residues are in close proximity to both, the DNA substrate and the inhibitor, respectively, and therefore, not readily accessible for modification (data not shown).
Figure 1.
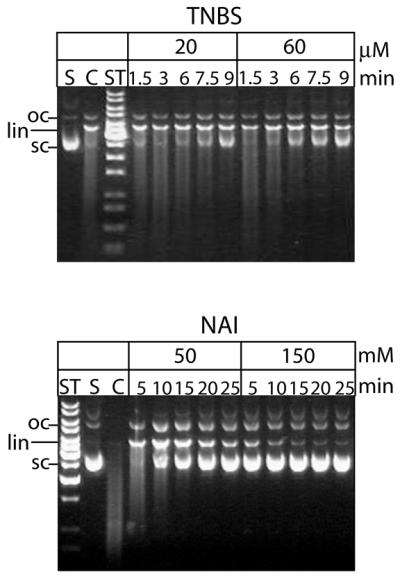
Chemical modification of murine CAD by TNBS and NAI. Free GST-mCAD was modified by TNBS (top) and NAI (bottom) for the indicated time (1.5, 3, 6, 7.5 and 9 min). Subsequently the residual nucleolytic activities of the aliquots withdrawn from the modification reaction mixtures at the indicated time points were measured by a plasmid DNA cleavage assay (10 min). TNBS and NAI inactivate free GST-mCAD in a time- and dose-dependent manner. (ST, length standard; S, substrate DNA; C, control reaction; oc, open circular DNA; lin, linear DNA; sc, supercoiled DNA.)
Effects of the substitution of conserved lysine and tyrosine residues on DNA cleavage by CAD
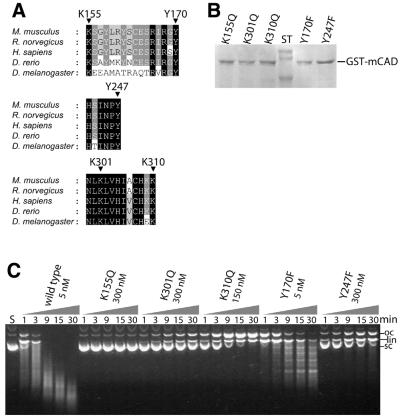
An amino acid sequence alignment reveals that three out of a total of 19 lysine residues (Lys155, Lys301, Lys310) and two out of a total of 13 tyrosine residues (Tyr170, Tyr247) of murine CAD are fully conserved among the apoptotic nucleases from five different species and that all are located in the C-terminal catalytic domain of the enzyme (Fig. 2A). The evolutionary conservation of these residues and the results obtained in the chemical modification experiments prompted us to exchange them by site-directed mutagenesis to glutamine and phenylalanine, respectively. We have chosen these amino acid exchanges in order to test the importance of the functional group (a positive charge and a hydroxy group, respectively) and at the same time to avoid as much as possible any changes in protein integrity caused by an amino acid substitution that would alter the size and polarity of the side chain too much. DNA cleavage experiments with the mCAD variants produced in E.coli and purified to near homogeneity (Fig. 2B) demonstrate that all of the exchanged lysine and tyrosine residues are more or less important for DNA cleavage by CAD (Fig. 2C and Table 1), except for the Y170F variant, which retains wild-type activity. The strongest effect on activity is observed with the variant K155Q, which does not show any detectable DNA cleavage, even after prolonged incubation of the substrate DNA with high amounts of the enzyme. K301Q, K310Q and Y247F exhibit between 1 and 2% of wild-type mCAD activity as measured by the disappearance of supercoiled plasmid DNA. The variant Y170F retains wild-type activity with respect to its initial attack on supercoiled plasmid DNA, but displays a drastically altered cleavage pattern compared with wild-type mCAD by producing defined fragments rather than a ‘smear’.
Figure 2.
Mutational analysis of variants with exchanged conserved lysine and tyrosine residues of GST-mCAD. (A) Amino acid residues Lys155, Lys301 and Lys310 as well as Tyr170 and Tyr247, which were exchanged to glutamine or phenylalanine, respectively, are fully conserved among the apoptotic nucleases known from the five indicated species [Mus musculus (GenBank accession nos AB009377, NM_007859), Rattus norvegicus (GenBank accession no. AF136598), Homo sapiens (GenBank accession nos AF064019, AF039210, AB013918, NM_004402), Danio rerio (GenBank accession no. AF286179) and Drosophila melanogaster (GenBank accession nos AF149797, AB036773)]. Shading is according to the Blosum 62 scoring matrix, with black shading for 100%, dark grey for 80% and light grey for 60% conserved amino acid residues. (B) SDS–PAGE analysis of variants of GST-mCAD that were produced as a complex with hICAD-L in E.coli and activated by treating the complex with recombinant caspase-3. (C) DNA cleavage activity of the GST-mCAD variants were measured by the disappearance of supercoiled plasmid DNA, analysed by agarose gel electrophoresis. GST-mCAD-Y170F is the only variant that retains wild-type activity with respect to the cleavage of supercoiled DNA, whereas all other variants exhibit strongly reduced cleavage activities. GST-mCAD-Y170F produces a pattern of fragments of defined length instead of randomly cut fragments appearing as a ‘smear’ (see wild-type GST-mCAD). Note the different concentrations of the variants as indicated.
Table 1. Properties of variants with substitution of conserved histidine, lysine and tyrosine residues in the C-terminal catalytic domain of the murine CAD.
| Residue | Substitution | ICAD-L bindinga | Relative DNA cleavage activity (%)b | Oligomeric state | DNA bindingc | Remark |
|---|---|---|---|---|---|---|
| Wild-type | – | + | 100 | wt | wt | |
| Lys155 | Gln | + | n.d.c. | wt | wt | |
| Lys301 | Gln | + | 1.4 | wt | wt | |
| Lys310 | Gln | + | 2.3 | wt | wt | |
| Tyr170 | Phe | + | 107.0 | wt | wt | Fragment pattern |
| Tyr247 | Phe | + | 1.2 | wt | wt | |
| His127 | Asnd | + | 25.8 | wt | n.d.e | |
| His242 | Asnd | + | 6.3 | Deviating | n.d.e | Fragment pattern |
| Glu | + | <1.0 | wt | wt | ||
| Arg | + | <1.0 | wt | wt | ||
| His263 | Asnd | + | 0.7 | wt | n.d.e | |
| Asp | + | <1.0 | wt | wt | ||
| Arg | + | <1.0 | wt | wt | ||
| His304 | Asnd | + | 14.5 | wt | n.d.e | |
| His308 | Asnd | + | 9.3 | wt | n.d.e | Fragment pattern |
| Asp | + | ∼1–2 | wt | wt | Residual activity at pH 5.0 | |
| Arg | + | <1.0 | wt | wt | ||
| His313 | Asnd | + | 1.3 | wt | n.d.e | |
| Asp | + | <1.0 | wt | wt | ||
| Arg | + | 109.0 | wt | wt | Fragment pattern |
n.d., not determined; n.d.c., no detectable cleavage; wt, wild-type-like.
aICAD-L binding measured by co-purification of untagged hICAD-L with GST-mCAD.
bMeasured by the disappearance of supercoiled plasmid DNA in steady state cleavage experiments.
cBinding of free GST-mCAD to DNA–cellulose.
dThe properties of these variants were described in Meiss et al. (17).
eSakahira et al. (16) have demonstrated DNA binding for the alanine substitutions at the indicated positions in murine CAD. With the exception of H242A, all other variants bound to DNA–cellulose, similar to wild-type CAD.
Effects of the substitution of conserved histidine residues on DNA cleavage by CAD
In addition to the mutational analysis of conserved lysine and tyrosine residues of CAD, we have produced and analysed new CAD variants with substitutions of conserved histidine residues, which had been shown previously to be important for DNA cleavage (16,17). The residues His242, His263, His308 and His313 were substituted by Asp/Glu and Arg. With the exception of H313R, all variants showed little if any nucleolytic activity (Table 1). His313 of murine CAD, which is conserved in all known caspase−αχτιϖατɛδ DNases from vertebrate organisms, has its counterpart in Arg418 of Drosophila CAD (Drep4) (17), which may explain why the substitution of His313 by Arg in murine CAD does not change the catalytic efficiency of the nuclease. However, like the variants Y170F (this work), H308N and H242N (17), the H313R variant produces a pattern of defined fragments when cleaving DNA (data not shown), pointing towards an involvement of His313 in DNA binding and/or processivity.
CAD variant H308D exhibits residual DNA cleavage activity at low pH
Among all variants with His→Asp/Glu substitutions, H308D is the only one with a residual cleavage activity at pH 5.0. Under these conditions all other variants show little if any activity, and the activity of the wild-type enzyme is much reduced compared with the activity at neutral pH (Fig. 3). At pH 5.0 the carboxy group of the aspartate residue is likely to be partially protonated (assuming a pKa ∼4.7), as is the histidine residue at pH 7.0 (assuming a pKa ∼6.4). Therefore, the protonated carboxy group could take over the putative function of the original histidine residue at this position in acting as the general acid in the mechanism of the CAD-catalysed DNA cleavage by protonating the leaving group after cleavage of a phosphodiester bond.
Figure 3.
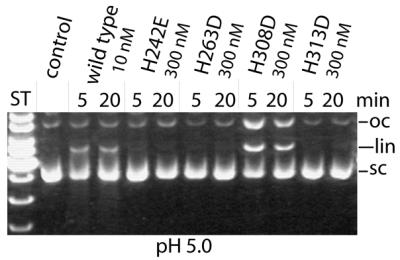
Residual DNA cleavage activities of variants of GST-mCAD with His→Asp/Glu substitutions. Variant H308D is the only variant with a measurable activity at pH 5.0. All other variants, except wild-type GST-mCAD, are virtually inactive at this low pH.
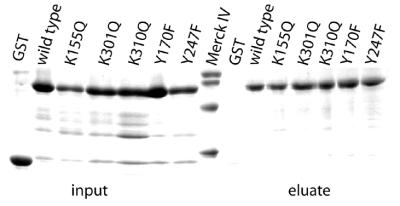
Binding of CAD variants to DNA–cellulose
In order to investigate DNA binding by the murine CAD variants, we have carried out a DNA–cellulose binding assay (16). To this end, GST-mCAD/hICAD-L mutant complexes were treated with caspase-3 and directly applied to the DNA–cellulose resin. Using this method, all GST-mCAD variants were shown to be able to bind to DNA–cellulose (Table 1), whereas the GST-mCAD/hICAD-L complex (data not shown) and GST itself, which were used as controls, did not bind to DNA (Fig. 4), demonstrating that all the GST-mCAD variants produced were not generally affected in DNA binding.
Figure 4.
DNA binding by the GST-mCAD variants. DNA binding of the GST-mCAD variants was investigated using a DNA–cellulose binding assay (16). Caspase-3-treated GST-CAD/hICAD-L complex (2.5 µg) and GST (2.5 µg) were incubated with 50 µl of a DNA–cellulose suspension. After washing, bound proteins were eluted by incubating the suspension for 5 min at 95°C in 10 µl SDS-gel loading buffer and subsequently analysed by SDS–PAGE. As can be seen, all GST-mCAD variants bind to DNA–cellulose, whereas GST alone does not.
CAD variants Y170F and H313R produce a defined DNA cleavage pattern
As already observed for the substitution of His308 and His242 by Asn in mCAD (17), substitution of Tyr170 by Phe and His313 by Arg has a profound effect on the DNA cleavage mode compared with wild-type mCAD. The variants produce a well-defined cleavage pattern when degrading plasmid DNA under conditions where wild-type GST-mCAD produces randomly cut DNA fragments appearing as a ‘smear’. In order to exclude that this pattern was caused by trace amounts of an E.coli protein co-purified with the GST-mCAD variants and to confirm that the altered cleavage behaviour is an intrinsic property of the variant enzymes, we have expressed one variant (Y170F) also in 293-human embryonic kidney cells. The variant expressed in 293 cells, similar to the protein produced in E.coli, after purification produces a defined DNA cleavage pattern, whereas wild-type CAD produces randomly cut DNA fragments (data not shown). Since the activity of the Y170F and H313R variants in attacking supercoiled plasmid DNA is similar to that of wild-type CAD and since their binding to DNA is unaffected, it seems likely that Tyr170 and His313, together with His308 and probably His242, are important for the non-specific cleavage of substrate DNA by mCAD.
Oligomerisation of GST-mCAD variants
After dissociation from the complex with the inhibitor ICAD-L, CAD is known to oligomerise into its active form (7). In order to see if the GST-mCAD variants produced are still able to oligomerise, we have carried out gel filtration experiments with each variant using a Superose 12 column. Under the conditions applied, all variants displayed a similar elution volume (Table 1), with a peak corresponding to the void volume (2 MDa) of the column (data not shown), indicating that the drop in activity measured in the plasmid DNA cleavage assays is not due to impaired oligomerisation of the variant enzymes but rather a consequence of localised changes at or in close proximity to the active site of CAD.
DISCUSSION
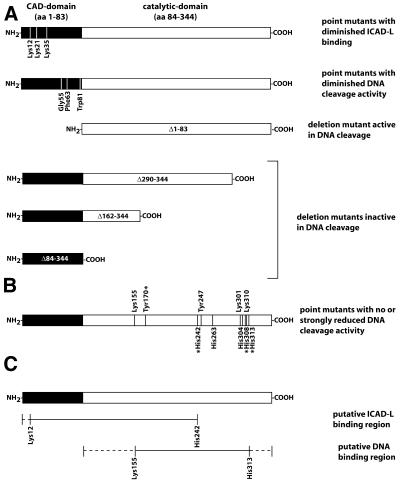
The CAD/DFF40 plays a key role in DNA fragmentation during programmed cell death. Primary sequence analysis suggested that CAD belongs to a new nuclease family, since no sequence similarity with known nuclease families could be detected. To identify functionally important amino acid residues in CAD, DEPC-modification studies had been performed which strongly suggested that histidine residues might be involved in DNA binding and cleavage by this enzyme; not, however, in the interaction with ICAD-L (16,17). In a complementary mutational analysis, histidine residues were identified that might be essential for DNA cleavage by CAD (16,17). We have now extended these studies to find out whether other amino acid residues are essential for DNA cleavage by CAD, and have focused on lysine and tyrosine residues. In addition, we have produced new CAD variants to further analyse the role of histidine residues known to be involved in DNA cleavage by CAD. Our modification experiments demonstrate that lysine and tyrosine residues are required for DNA cleavage and that they are protected from modification by the presence of DNA or the inhibitor ICAD-L. It had been shown previously that the presence of DNA protected CAD from modification by DEPC, indicating that some of the histidine residues are in close proximity to the DNA substrate and inaccessible to the modifying compound. The inhibitory protein ICAD-L, in contrast, did not protect the nuclease from DEPC modification, leading to the conclusion that the substrate binding site and the inhibitor binding site of murine CAD are located on different regions (16). Our finding that TNBS and NAI modification of lysine and tyrosine residues is prevented by the presence of DNA as well as the inhibitory protein hICAD-L, suggests that at least one lysine and one tyrosine residue of CAD, in contrast to histidine residues, contribute directly or indirectly, not only to the substrate binding interface, but also to the inhibitor binding interface of murine CAD. It is known from previous structural and mutational analyses that conserved lysine residues (Lys12, Lys21, Lys35) in the N-terminal regulatory domain of CAD (CAD- or CIDE-N domain) contribute to the interaction with the N-terminal domain of ICAD. Single amino acid substitutions of these lysine residues by alanine had no effect on the nucleolytic activity and complex formation of CAD with ICAD-L; however, the double and triple mutants were proportionally more affected by the amino acid substitutions, with the latter being almost inactive and virtually unable to bind to ICAD-L (11,14). The successive loss of activity is a consequence of the inability of these CAD mutants to bind to the chaperone and inhibitor ICAD-L, which results in the formation of a catalytically incompetent nuclease, and not a consequence of the substitution of a critical active site residue. These results taken together suggest that the N-terminal domain of CAD and the region up to amino acid residue 241 harbours the binding site of CAD for ICAD-L (Fig. 5).
Figure 5.
Variants of CAD with reduced catalytic activity. (A) Inohara et al. (11) and Otomo et al. (14) have identified three conserved lysine residues (Lys12, Lys21 and Lys35) in the N-terminal domain of CAD (amino acids 1–83) that contribute to ICAD-L binding. Inohara et al. (11) have also identified three conserved residues (Gly55, Phe63 and Trp81) in the N-terminal domain of CAD that seem to enhance the nuclease activity of CAD without being essential for catalysis. According to their results, the catalytic nuclease domain is located in the C-terminal region of the protein (amino acids 84–344), since the Δ1–83 variant of CAD exhibits a moderate nucleolytic activity, whereas mutant forms with deletions in the C-terminal domain (Δ290–344, Δ162–344 and Δ84–344) have no nuclease activity. It must be emphasised that Otomo et al. (14) have reported contradictory results, according to which the C-terminal domain of CAD (Δ1–83) does not show any nucleolytic activity. (B) The C-terminal domain of CAD contains several histidine, lysine and tyrosine residues, which have been identified as being catalytically relevant by chemical modification and alignment-guided site-directed mutagenesis (16,17; this work). Variants of CAD with substitutions of the amino acid residues marked with an asterisk (Tyr170, His242, His308 and His313) are active but produce a defined cleavage pattern when hydrolysing DNA. (C) It can be concluded from the structural studies as well as the chemical modification and mutational analyses that the region of CAD interacting with ICAD-L, besides the known interaction of the N-terminal domain (12–15), includes the amino acid residues of CAD up to residue 241, whereas the DNA binding site is between amino acid residues Lys155 and His313.
In many different nucleases, lysine residues are part of the enzyme’s active site (22–31), where they can fulfil several functions. They can serve to correctly position a scissile phosphodiester bond and/or to stabilise the pentacovalent transition state during the enzymatic process, as well as to direct the attacking water molecule for an in-line attack on the phosphodiester bond. For example, in most type II restriction enzymes, a lysine residue is found in the catalytic PD....D/EXK-motif, where it functions mainly to position the attacking water molecule and to stabilise the transition state (reviewed in 32). In murine CAD we have identified by chemical modification, in conjunction with alignment-guided site-directed mutagenesis, three catalytically important lysine residues, Lys155, Lys301 and Lys310, that are located in the C-terminal catalytic domain of the enzyme and are fully conserved in all known CADs from five different species, namely mouse, rat, man, zebrafish and the fruitfly (Fig. 2). CAD variants with the lysine residues substituted by glutamine all exhibit a strongly reduced catalytic activity, with the variant K155Q exhibiting no detectable DNA cleavage activity, indicating that this residue might be particularly important for catalysis, possibly by transition state stabilisation.
Tyrosine residues are also often part of the active site of a nuclease where they are involved in DNA binding by forming stacking interactions with a base or a sugar moiety of the nucleic acid. For example, Tyr76, a key DNA-binding residue of DNase I, which is part of a small loop that fills the minor groove of the DNA, forms a stacking interaction with a deoxyribose and thus plays an important role in the coupling of DNA recognition to phosphodiester bond cleavage by DNase I (33–36). In Serratia nuclease, Tyr76 can only be substituted by phenylalanine to preserve activity, presumably because it is involved in a stacking interaction with a base of the nucleic acid substrate (26,37,38). In the case of topoisomerases Ia and IIa the active site tyrosine residue attacks the scissile phosphodiester bond and forms a covalent intermediate in the mechanism of breakage and rejoining of the DNA (39). The tyrosine residues Tyr170 and Tyr247 of murine CAD when exchanged to phenylalanine show very different effects. The variant Y242F, similar to the variants K301Q and K310Q, shows a strongly reduced cleavage activity, which could mean that Tyr242, together with Lys301 and Lys310, contributes to productive substrate binding by correctly positioning the DNA in the substrate binding site (note that all variants studied here are not deficient in DNA binding as measured by the DNA–cellulose binding assay). The variants Y170F and H313R, in contrast, retain wild-type activity with respect to the cleavage of supercoiled plasmid DNA, but in the course of the cleavage reaction produce a pattern of fragments of defined length instead of randomly cut fragments as observed for wild-type murine CAD. It is likely from these results that Tyr170 and His313, probably together with His308 and His242 (17), for which a similar observation was made, contribute to a more non-specific DNA cleavage mode, which leads to a ‘smear’-like product distribution, but do not participate in catalysis per se.
Murine CAD binds to DNA in the absence of Mg2+, which, however, is needed as the divalent cation cofactor for the DNA cleavage reaction. Using a DNA–cellulose binding assay, it could be demonstrated that the substitution of critical histidine residues abolishes DNA cleavage, not, however, DNA binding of CAD, except for the variant H242A (16). In order to analyse DNA binding by CAD variants with exchanged lysine and tyrosine residues, we have conducted a DNA–cellulose-binding assay with these variants. All of them bind to DNA–cellulose with the same efficiency (as far as this can be measured with such a semi-quantitative assay), indicating that the mutations introduced do not affect the overall DNA-binding capability of CAD. However, this does not exclude that the conserved lysine and tyrosine residues under investigation each contribute to substrate binding by CAD and that their individual substitution leads to a diminished nucleolytic activity. In nucleases, several residues are involved in DNA binding, often by formation of electrostatic interactions between positively charged side chains and the negatively charged phosphodiester backbone, or stacking interactions between aromatic side chains and the bases or the sugar of the nucleic acid substrate. The substitution of one of these residues can lead to a decrease in the catalytic efficiency of the nuclease, but does not necessarily prevent DNA binding. Instead, it could be that in these variants the substrate is not properly positioned, due to the absence of a contact required for a productive interaction.
CONCLUSION
Our investigation of the role of conserved lysine and tyrosine residues of the CAD demonstrates that several lysine and tyrosine residues contribute to DNA binding and cleavage by CAD, and to the inhibition of CAD activity by binding to the inhibitor ICAD-L. Lys155, when substituted by glutamine, shows the most dramatic decrease in nucleolytic activity, emphasising its particular role in the mechanism of DNA cleavage, which could be positioning of the attacking nucleophile or transition state stabilisation. Lys301, Lys310 and Tyr247 also seem to be part of the enzyme/DNA interface, where they could be involved in binding and positioning of the substrate. Tyr170 and His313, like His308 and His242, seem to be involved in DNA binding in such a way as to guarantee a non-specific DNA cleavage mode, which may be associated with the processivity of the enzyme.
Lysine and tyrosine residues co-operate with histidine residues, which we had begun to analyse before (17), in DNA cleavage. Because of the large effects observed with substitution of His263 by asparagine, we had suggested that this histidine residue might act as a general base in the mechanism of DNA cleavage by CAD. We now have evidence that His308 could be the general acid. It is noteworthy that all residues which were shown to affect the DNA cleavage activity when exchanged are located between Lys155 and His313 (Fig. 5), which, therefore, allows one to provisionally define the boundary of the catalytic domain. The function of the essential Mg2+ has not yet been addressed. For this purpose it will be important to identify the amino acid residues involved in Mg2+ binding, which is underway in our laboratory.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Ms Ute Konradi for expert technical assistance. This work has been supported by a grant from the Deutsche Forschungsgemeinschaft (Pi 122/16-1) and the Fonds der Chemischen Industrie. S.R.S. is a member of the Graduiertenkolleg ‘Biochemie von Nukleoproteinkomplexen’. O.G.’s stay at Giessen was supported by the Deutsche Akademische Austauschdienst.
REFERENCES
- 1.Nagata S. (2000) Apoptotic DNA fragmentation. Exp. Cell Res., 256, 12–18. [DOI] [PubMed] [Google Scholar]
- 2.Liu X., Zou,H., Slaughter,C. and Wang,X. (1997) DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis. Cell, 89, 175–184. [DOI] [PubMed] [Google Scholar]
- 3.Enari M., Sakahira,H., Yokoyama,H., Okawa,K., Iwamatsu,A. and Nagata,S. (1998) A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature, 391, 43–50. [DOI] [PubMed] [Google Scholar]
- 4.Halenbeck R., MacDonald,H., Roulston,A., Chen,T.T., Conroy,L. and Williams,L.T. (1998) CPAN, a human nuclease regulated by the caspase-sensitive inhibitor DFF45. Curr. Biol., 8, 537–540. [DOI] [PubMed] [Google Scholar]
- 5.Sakahira H., Iwamatsu,A. and Nagata,S. (2000) Specific chaperone-like activity of inhibitor of caspase-activated DNase for caspase-activated DNase. J. Biol. Chem., 275, 8091–8096. [DOI] [PubMed] [Google Scholar]
- 6.Sakahira H., Enari,M. and Nagata,S. (1998) Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis. Nature, 391, 96–99. [DOI] [PubMed] [Google Scholar]
- 7.Liu X., Zou,H., Widlak,P., Garrard,W. and Wang,X. (1999) Activation of the apoptotic endonuclease DFF40 (caspase-activated DNase or nuclease). Oligomerization and direct interaction with histone H1. J. Biol. Chem., 274, 13836–13840. [DOI] [PubMed] [Google Scholar]
- 8.Toh S.Y., Wang,X. and Li,P. (1998) Identification of the nuclear factor HMG2 as an activator for DFF nuclease activity. Biochem. Biophys. Res. Commun., 250, 598–601. [DOI] [PubMed] [Google Scholar]
- 9.Durrieu F., Samejima,K., Fortune,J.M., Kandels-Lewis,S., Osheroff,N. and Earnshaw,W.C. (2000) DNA topoisomerase IIalpha interacts with CAD nuclease and is involved in chromatin condensation during apoptotic execution. Curr. Biol., 10, 923–926. [DOI] [PubMed] [Google Scholar]
- 10.Inohara N., Koseki,T., Chen,S., Wu,X. and Núñez,G. (1998) CIDE, a novel family of cell death activators with homology to the 45 kDa subunit of the DNA fragmentation factor. EMBO J., 17, 2526–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inohara N., Koseki,T., Chen,S., Benedict,M.A. and Núñez,G. (1999) Identification of regulatory and catalytic domains in the apoptosis nuclease DFF40/CAD. J. Biol. Chem., 274, 270–274. [DOI] [PubMed] [Google Scholar]
- 12.Lugovskoy A.A., Zhou,P., Chou,J.J., McCarty,J.S., Li,P. and Wagner,G. (1999) Solution structure of the CIDE-N domain of CIDE-B and a model for CIDE-N/CIDE-N interactions in the DNA fragmentation pathway of apoptosis. Cell, 99, 747–755. [DOI] [PubMed] [Google Scholar]
- 13.Uegaki K., Otomo,T., Sakahira,H., Shimizu,M., Yumoto,N., Kyogoku,Y., Nagata,S. and Yamazaki,T. (2000) Structure of the CAD domain of caspase-activated DNase and interaction with the CAD domain of its inhibitor. J. Mol. Biol., 297, 1121–1128. [DOI] [PubMed] [Google Scholar]
- 14.Otomo T., Sakahira,H., Uegaki,K., Nagata,S. and Yamazaki,T. (2000) Structure of the heterodimeric complex between CAD domains of CAD and ICAD. Nature Struct. Biol., 7, 658–662. [DOI] [PubMed] [Google Scholar]
- 15.Zhou P., Lugovskoy,A.A., McCarty,J.S., Li,P. and Wagner,G. (2001) Solution structure of DFF40 and DFF45 N-terminal domain complex and mutual chaperone activity of DFF40 and DFF45. Proc. Natl Acad. Sci. USA, 98, 6051–6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakahira H., Takemura,Y. and Nagata,S. (2001) Enzymatic active site of caspase-activated DNase (CAD) and its inhibition by inhibitor of CAD. Arch. Biochem. Biophys., 388, 91–99. [DOI] [PubMed] [Google Scholar]
- 17.Meiss G., Scholz,S.R., Korn,C., Gimadutdinow,O. and Pingoud,A. (2001) Identification of functionally relevant histidine residues in the apoptotic nuclease CAD. Nucleic Acids Res., 29, 3901–3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pace C.N., Vajdos,F., Fee,L., Grimsley,G. and Gray,T. (1995) How to measure and predict the molar absorption coefficient of a protein. Protein Sci., 4, 2411–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rupp R.A.W., Snider,L. and Weintraub,H. (1994) Xenopus embryos regulate the nuclear localization of XMyoD. GenesDev., 8, 1311–1323. [DOI] [PubMed] [Google Scholar]
- 20.Turner D.L. and Weintraub,H. (1994). Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Development, 8, 1434–1447. [DOI] [PubMed] [Google Scholar]
- 21.Kirsch R.D. and Joly,E. (1998) An improved PCR-mutagenesis strategy for two-site mutagenesis or sequence swapping between related genes. Nucleic Acids Res., 26, 1848–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richardson R.M., Pares,X. and Cuchillo,C.M. (1990) Chemical modification by pyridoxal 5′-phosphate and cyclohexane-1,2-dione indicates that Lys-7 and Arg-10 are involved in the p2 phosphate-binding subsite of bovine pancreatic ribonuclease A. Biochem. J., 267, 593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gite S., Reddy,G. and Shankar,V. (1992) Active-site characterization of S1 nuclease. I. Affinity purification and influence of amino-group modification. Biochem. J., 285, 489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Selent U., Ruter,T., Kohler,E., Liedtke,M., Thielking,V., Alves,J., Oelgeschlager,T., Wolfes,H., Peters,F. and Pingoud,A. (1992) A site-directed mutagenesis study to identify amino acid residues involved in the catalytic function of the restriction endonuclease EcoRV. Biochemistry, 31, 4808–4815. [DOI] [PubMed] [Google Scholar]
- 25.Winkler F.K., Banner,D.W., Oefner,C., Tsernoglou,D., Brown,R.S., Heathman,S.P., Bryan,R.K., Martin,P.D., Petratos,K. and Wilson,K.S. (1993) The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J., 12, 1781–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller M.D., Tanner,J., Alpaugh,M., Benedik,M.J. and Krause,K.L. (1994) 2.1 Å structure of Serratia endonuclease suggests a mechanism for binding to double-stranded DNA. Nature Struct. Biol., 1, 461–468. [DOI] [PubMed] [Google Scholar]
- 27.Fisher B.M., Schultz,L.W. and Raines,R.T. (1998) Coulombic effects of remote subsites on the active site of ribonuclease A. Biochemistry, 37, 17386–17401. [DOI] [PubMed] [Google Scholar]
- 28.Fisher B.M., Grilley,J.E. and Raines,R.T. (1998) A new remote subsite in ribonuclease A. J. Biol. Chem., 273, 34134–34138. [DOI] [PubMed] [Google Scholar]
- 29.Garforth S.J., Ceska,T.A., Suck,D. and Sayers,J.R. (1999) Mutagenesis of conserved lysine residues in bacteriophage T5 5′-3′ exonuclease suggests separate mechanisms of endo-and exonucleolytic cleavage. Proc. Natl Acad. Sci. USA, 96, 38–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolt E.L., Sharples,G.J. and Lloyd,R.G. (2000) Analysis of conserved basic residues associated with DNA binding (Arg69) and catalysis (Lys76) by the RusA holliday junction resolvase. J. Mol. Biol., 304, 165–176. [DOI] [PubMed] [Google Scholar]
- 31.Declais A.C., Hadden,J., Phillips,S.E. and Lilley,D.M. (2001) The active site of the junction-resolving enzyme T7 endonuclease I. J. Mol. Biol., 307, 1145–1158. [DOI] [PubMed] [Google Scholar]
- 32.Pingoud A. and Jeltsch,A. (2001) Structure and function of type II restriction endonucleases. Nucleic Acids Res., 29, 3705–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weston S.A., Lahm,A. and Suck,D. (1992) X-ray structure of the DNase I–d(GGTATACC)2 complex at 2.3 Å resolution. J. Mol. Biol., 226, 1237–1256. [DOI] [PubMed] [Google Scholar]
- 34.Doherty A.J., Worrall,A.F. and Connolly,B.A. (1995) The roles of arginine 41 and tyrosine 76 in the coupling of DNA recognition to phosphodiester bond cleavage by DNase I: a study using site-directed mutagenesis. J. Mol. Biol., 251, 366–377. [DOI] [PubMed] [Google Scholar]
- 35.Warren M.A., Evans,S.J. and Connolly,B.A. (1997) Effects of non-conservative changes to tyrosine 76, a key DNA binding residue of DNase I, on phosphodiester bond cleavage and DNA hydrolysis selectivity. Protein Eng., 10, 279–283. [DOI] [PubMed] [Google Scholar]
- 36.Pan C.Q., Ulmer,J.S., Herzka,A. and Lazarus,R.A. (1998) Mutational analysis of human DNase I at the DNA binding interface: implications for DNA recognition, catalysis, and metal ion dependence. Protein Sci., 7, 628–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friedhoff P., Kolmes,B., Gimadutdinow,O., Wende,W., Krause,K.-L. and Pingoud,A. (1996) Analysis of the mechanism of the Serratia nuclease using site-directed mutagenesis. Nucleic Acids Res., 24, 2632–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meiss G., Gast,F.U. and Pingoud,A. (1999) The DNA/RNA non-specific Serratia nuclease prefers double stranded A-form nucleic acids as substrates. J. Mol. Biol., 288, 377–390. [DOI] [PubMed] [Google Scholar]
- 39.Liu Q. and Wang,J.C. (1999) Similarity in the catalysis of DNA breakage and rejoining by type IA and IIA DNA topoisomerases. Proc. Natl Acad. Sci. USA, 96, 881–886. [DOI] [PMC free article] [PubMed] [Google Scholar]