Abstract
In recent years there has been growing interest in the question of how the particular topology of polymeric chains affects their overall dimensions and physical behavior. The majority of relevant studies are based on numerical simulation methods or analytical treatment; however, both these approaches depend on various assumptions and simplifications. Experimental verification is clearly needed but was hampered by practical difficulties in obtaining preparative amounts of knotted or catenated polymers with predefined topology and precisely set chain length. We introduce here an efficient method of production of various single-stranded DNA knots and catenanes that have the same global chain length. We also characterize electrophoretic migration of the produced single-stranded DNA knots and catenanes with increasing complexity.
INTRODUCTION
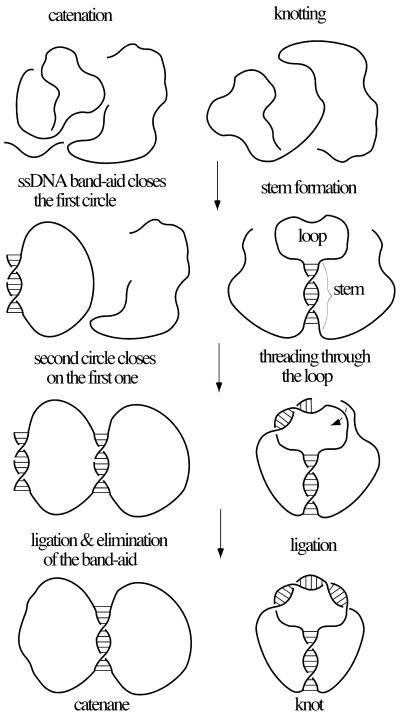
Single-stranded DNA is an ideal nano-engineering material (1,2). Regions of poly(dT) [in the absence of their complement, poly(dA)] are very flexible (3), while regions that are prepared to anneal with each other form a well-defined right-handed, anti-parallel helix with ∼10 bp per turn (4). These two elements, flexible linkers and right-handed helical blocks, permit the design of a great variety of knots and catenanes. Figure 1 illustrates our experimental design for forming different knots and catenanes. The design of catenanes is conceptually simple. We use two different 60mers and a short ‘band-aid’ oligonucleotide. With the exception of regions that can be used for the pairing of two 60mers and the region of complementarity to the band-aid, all the remaining nucleotides are dTs. The band-aid function is to aid one of the 60mers to have its ends brought together for subsequent ligation, while the second 60mer uses the first 60mer as a band-aid (Fig. 1, left). The length of the pairing region between the two 60mers determines the extent of catenation. We have used complementary regions of 12, 20 and 30 bp hoping to obtain singly, doubly and triply linked catenanes, respectively. Upon ligation with T4 DNA ligase, the band-aid is removed during gel electrophoresis performed under DNA-denaturing conditions. The design of knots is conceptually more complex and is a modification of the method originally proposed by Seeman and co-workers (5,6). A 120mer oligonucleotide contains short complementary sequences forming stem and loop regions (Fig. 1, right). Stem-forming regions are both not-divided so that upon annealing they can form relatively stable duplex regions. One of the loop-forming sequences is, however, divided between two ends of the 120mer and therefore the overall stability of the loop region is decreased as compared with the stem region. In addition, formation of the loop is a quasi-third-order type of reaction requiring the simultaneous interaction of three quasi-independent sequence elements, while the stem formation, which is a quasi-second-order type of reaction, requires the interaction of two sequence elements. This sequence design favors a folding process in which the stem should form first and is followed by pairing in the loop. Ligation in the loop region (Fig. 1, right) leads then to the formation of knots. Notice that loop formation preceding formation of the stem would result in the formation of ‘unknots’ (unknotted circles). By varying the length of stem and loop one can direct formation of different knots. We have used stems and loops of 11 or 15 bp.
Figure 1.
General strategy to form catenanes and knots. (Left) Sequence of events leading to the formation of catenanes. Three different oligonucleotides were used in each ligation reaction. The two pairing and ligation events are commutative, i.e. their relative order is not important for the final outcome (formation of catenanes). (Right) Sequence of events leading to formation of knots. Only one type of oligonucleotide is used in the ligation reaction. The sequence of pairing in the stem and loop region is crucial for the final outcome. Knots are only formed when stem is formed first. Unknotted circles are formed when the pairing in the loop precedes this in the stem.
MATERIALS AND METHODS
Oligonucleotides were obtained from Medprobe, Oslo. T4 DNA ligase and exonuclease I, exonuclease III and T4 polynucleotide kinase were supplied by New England Biolabs. Fine chemicals were purchased from Roche and Fluka. The oligonucleotides of 60 or 120 bases, but not the short band-aids or ‘stem splitters’, were phosphorylated by T4 polynucleotide kinase. For the synthesis of knots and catenanes, phosphorylated oligonucleotides and, where appropriate, band-aids or stem splitters were mixed, heated to 80°C for 5 min, annealed at 60°C for 30 min and subsequently allowed to cool to room temperature. The mixtures were ligated overnight at 4°C, at a concentration of 0.4 pmol/ml DNA, with 25 U/ml T4 DNA ligase. If needed, linear molecules left after the ligation were removed by treatment with a mixture of two exonucleases (Escherichia coli exonuclease I and exonuclease III; New England BioLabs and Fermentas, respectively). For the formation of catenanes we used the following oligonucleotides (written in the 5′→3′ direction): ‘first circle’ (Fig. 1, left), GCTCC(T)10GTGTCCTGTCCTGTCCTCTCGTCGTCCCTC(T)10CCTGC; the ‘band-aid’ closing the first circle, AAGGAGCGCAGGAA; the second circle with 12 nt complementary to the first one, GACAGG(T)48CGAGAG; the second circle with 20 nt complementary to the first one, GACAGGACAG(T)40ACGACGAGAG; and the second circle with 30 nt complementary to the first circle, GACAGGACAGGACAC(T)30GAGGGACGACCAGAG. For the formation of knots we have used the following 120mers: constructs with stem and loop regions of 11 bp each (see Fig. 4A), GAGGAG(T)24GTCCTGTCCTG(T)14CTCCTCGTCCC(T)14CAGGACAGGAC(T)24GGGAC; constructs with stem and loop regions of 11 and 15 bp, respectively (see Fig. 4C), GAGGAGAG(T)22GTCCTGTCCTG(T)12CTCTCCTCGTCCCTC(T)12CAGGACAGGAC(T)22GAGGGAC; and constructs with stem and loop regions of 15 bp each (see Fig. 4C), GAGGAGAG(T)20GTGTCCTGTCCTGTC(T)10CTCTCCTCGTCCCTC(T)10GACAGGACAGGACAC(T)20GAGGGAC. Stem-splitting oligonucleotides were complementary to one of the regions forming a stem (see Fig. 3B). To block stem formation in the case of 120mers with a stem of 15 bp we used the following 24mer: AAAAGTGTCCTGTCCTGTCAAAA. The ligation products were separated by denaturing polyacrylamide gel electrophoresis [12% polyacrylamide (PAA), 1× TBE, 8 M urea at 25 V/cm for 2 h] or by alkaline agarose gel electrophoresis [4% NuSieve Agarose (FMC Bioproducts), 50 mM NaOH, 1 mM EDTA at 4 V/cm for 6 h], followed by staining with SYBR Gold (Molecular Probes) and visualization on a UV transilluminator.
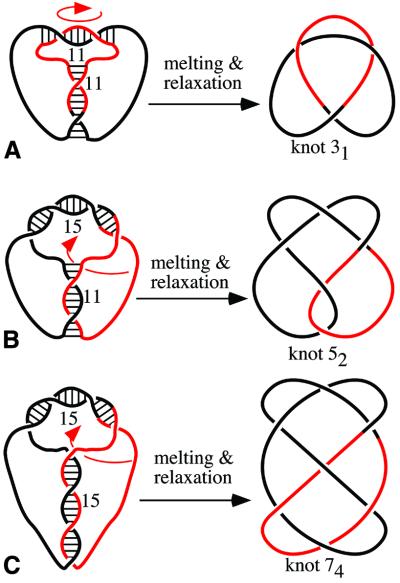
Figure 4.
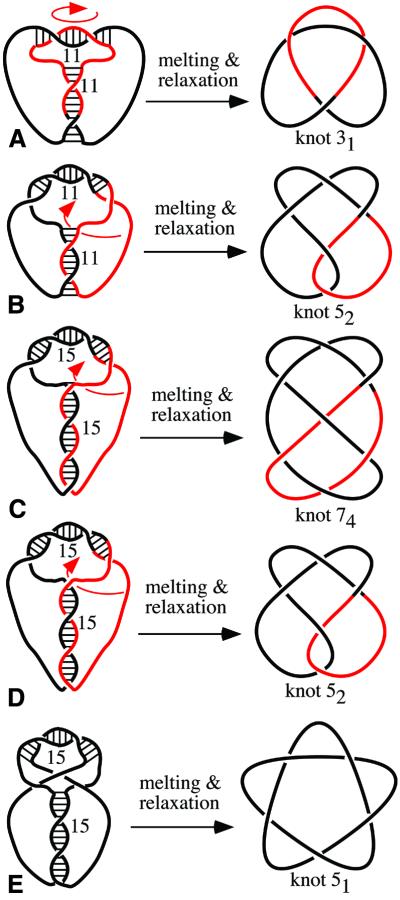
Design of various knots: (A) stem and loop of 11 bp each, (B) stem and loop of 11 and 15 bp, respectively, and (C) stem and loop of 15 bp each. Notice that each pairing region is right-handed and that regions of pairing are anti-parallel. This constraint limits the possible outcomes of the reactions but with oligonucleotides 11/11 and 15/15 we also observed other types of knots (see Fig. 5). It may not be obvious how the ‘constructs’ drawn on the left can be manipulated into standard diagrams of corresponding knots. Arrows show which type of movement of the colored portion needs to be performed to obtain standard representations of knots.
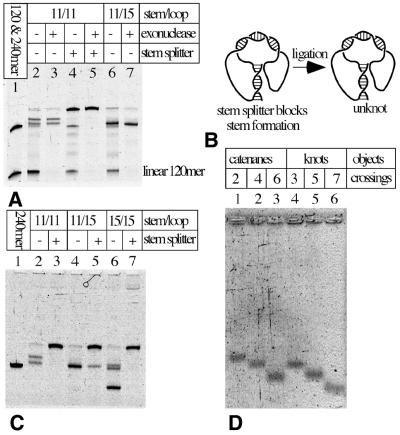
Figure 3.
Gel-electrophoretic analysis of various knots produced upon ligation of knot-forming oligonucleotides. (A) Exonuclease digestion and elimination of knots by the presence of stem splitter allows us to identify knotted DNAs. (B) The principle action of stem splitter. (C) Knots with increasing electrophoretic mobility are formed as pairing regions in stem and loop get longer. (D) Comparison of migration of knots and catenanes with the same total size but with different complexity. (A and C) PAA-denaturing gels with urea and (D) alkaline agarose NuSieve gel 4%.
RESULTS AND DISCUSSION
Formation of catenanes
Ligation reactions generating different catenanes and knots were monitored by denaturing polyacrylamide gel electrophoresis. To distinguish between contaminating linear molecules like unligated substrates or ligated linear multimers, digestion with exonucleases was performed. Figure 2A shows analysis of the ligation reaction with substrates for the generation of catenanes. In the case of the 12 nt-long pairing region between the two 60mers we observed the generation of three major species resisting digestion by exonucleases (Fig. 2A, lane 2). Quickly migrating bands consist of monomeric circular 60mers, as demonstrated by the fact that a band at this position is also obtained when the reaction includes only one of the 60mers and its 14 nt-long band-aid (data not shown). The two slowly migrating species are presumably singly and doubly linked catenanes. DNA species with similar electrophoretic mobilities were also produced in reactions where the pairing regions between two 60mers were 20- and 30-nt long (Fig. 2A lanes 3 and 4, and 5 and 6, respectively). However, each of these reactions produced only one major slowly migrating band consisting presumably of doubly and triply linked catenanes, respectively. We have verified by restriction digestion that these species indeed consist of catenated monomeric circles and not of dimeric rings that could also be produced in these ligation reactions and would then have a similar electrophoretic mobility to catenated circles. The respective DNA species were gel purified and annealed with a band-aid oligonucleotide to create a recognition site for HhaI restriction endonuclease. Upon partial digestion of catenanes composed of two 60mers, equal intensity bands of linear and circular 60mers should appear since only one of the composing circles could be cleaved by HhaI. Figure 2B shows that this is indeed the case. It should be mentioned here that putative dimers composed of two 60mers that are complementary to the band-aid oligonucleotide should produce upon partial digestion linear 120mers and linear 60mers but not circular 60mers. Dimers composed of 60mers that are not complementary to the band-aid oligonucleotide should be resistant to HhaI restriction endonuclease. Mixed dimers are unlikely to form as there is no band-aid for their ligation, but even if they were formed their digestion cannot produce linear and circular 60mers.
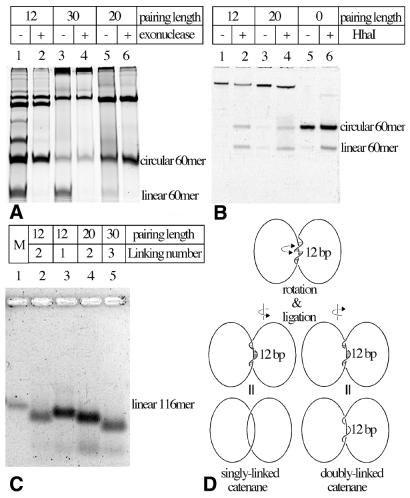
Figure 2.
Gel-electrophoretic analysis of ligation reactions leading to the formation of various catenanes. (A) Exonuclease eliminates linear single-stranded DNA molecules and reveals DNA species that are likely to be catenanes. Notice that 12 nt-long complementary sequences between the two 60mers (annealing of these sequences is required for the circularization of the ‘second’ 60mer, but can also lead to formation of linear multimers composed of these 60mers) result in frequent formation of dimeric and trimeric linear species, which are therefore exonuclease sensitive. Longer complementary sequences eliminate the formation of these dimeric and trimeric linear molecules but promote the formation of species with high molecular weight (see the top of the gel). (B) Partial restriction cleavage of one of the 60mers unambiguously demonstrates the catenated nature of DNA species that we expected to be catenated. In lanes 2 and 4 notice the equal intensity of circular and linear 60mers released from gel-purified slowly migrating species. In lane 6 circular 60mers are partially cut by HhaI and get partially converted to linear 60mers. (C) In alkaline agarose gels catenanes separate according to their expected linking number. M, marker lane containing linear 116mer. (D) Formation of singly and doubly linked catenanes in the case of 12 nt-long complementarity between the two rings. With these substrates the catenanes with linking number 1 (left) and 2 (right) show small energy difference and therefore form with similar probability. (A and B) PAA-denaturing gels with urea and (C) alkaline agarose NuSieve gel 4%.
Electrophoretic migration of catenanes
One of our experimental expectations was to observe ‘natural’ electrophoretic separation of singly, doubly and triply linked catenanes according to their extent of catenation (7). However, under applied electrophoretic conditions (12% PAA, 1× TBE, 8 M urea at 25 V/cm for 2 h) this was apparently not the case. Figure 2A shows, for example, that the catenanes expected to be triply linked migrated at a speed that situated them between catenanes expected to be singly and doubly linked. It is possible, however, that 8 M urea is not sufficient to permanently melt duplex regions, especially when these regions are kept in proximity by catenation and when gels are run at an ambient temperature. In the absence of complete melting, the electrophoretic migration would not only reflect different topology of catenated molecules but would be additionally affected by the different ratio of single- to double-stranded DNA regions in different constructs. To be sure that duplex regions in catenanes are completely melted we decided to analyze gel-purified individual species by electrophoresis on alkaline agarose gels. Figure 2C shows that under such conditions the constructs engineered to produce triply linked catenanes indeed migrate distinctly quicker than constructs intended to produce doubly linked catenanes. Out of the two species produced using 12 bp complementarity between two 60mers, one migrated as expected for singly linked catenanes while the second one co-migrated with a construct expected to produce doubly linked catenanes. Therefore, the migration of completely melted constructs on alkaline agarose gels strongly suggests that our constructs are indeed singly, doubly and triply linked catenanes as predicted in our construction schemes. Figure 2D shows schematically how two different catenanes can be formed in the case of 12 nt complementarity between the two single-stranded DNA oligonucleotides. Upon annealing the two strands are kept together but the nick provides a freedom of rotation. Single-stranded DNA flanking the duplex region is not constrained and this causes a small energy difference between arrangements that upon ligation creates singly linked catenanes (Fig. 2D, left) or doubly linked catenanes (Fig. 2D, right). In the case of constructs with 20 and 30 nt complementarity, the single-stranded DNA flanking the duplex region is already constrained and therefore energy-minimizing topoisomers are produced in the ligation reaction. Probably the mixture of two topoisomers could be obtained by ligation of constructs with 25–26 nt-long complementarity.
Formation of knots
Figure 3A shows an analysis of the ligation reaction with 120mers designed to form knots and having their stem and loop regions set to 11 bp each (see Fig. 4A). Lane 2 (after the marker lane containing 120 and 240 nt-long linear single-stranded oligonucleotides) analyzes the whole ligation reaction and lane 3 contains the species that resisted digestion by exonucleases. Lane 4 analyzes the ligation reaction in the presence of the oligonucleotide that forms a stable duplex with one of the regions required for stem formation. This oligonucleotide should specifically inhibit the formation of knots while not interfering with efficient ligation in the loop region. The presence of the stem-splitting oligonucleotide leads, therefore, to the formation of unknotted circles (Fig. 3B). Lane 5 shows the effect of exonuclease treatment on DNA species present after the ligation performed in the presence of the stem-splitting oligonucleotides and reveals the position of circular 120mer. Comparison of lanes 2, 3, 4 and 5 clearly indicates that in the absence of the stem splitter two types of knots are produced when stem and loop regions were 11 bp each. Notice that upon elimination of linear molecules, knots are almost exclusive products of the ligation reaction and that unknotted circles are hardly ever formed (except when promoted by the stem splitter). Lanes 6 and 7 analyze the ligation reaction with 120mers having their loop and stem regions set to 11 and 15 bp, respectively. Exonuclease treatment (lane 7) reveals that with this construct only one type of knot is formed. This type of knot migrates like the ‘quicker knot’ produced using 120mers with the stem and loop having 11 bp each. Co-migration of these two knotted species is best seen in samples not treated with exonucleases (lanes 2 and 6) and therefore containing small amounts of linear dimers produced during ligations (240 nt long). These linear dimers serve as good internal markers demonstrating that the quicker knot in lane 2 and the only knot in lane 6 show practically the same electrophoretic mobility and therefore are presumably of the same type. Figure 3C shows a comparison of ligation reactions involving 120mers that had their stems and loops set to three different combinations of lengths. All reaction products were, in addition, treated with exonucleases to eliminate linear species. Pairs of corresponding lanes analyze the ligation reactions performed without and with stem splitters. It can be seen again that in the presence of stem splitters ligation reactions are directed to the formation of circular products. In the absence of stem splitters, however, one observes the highly specific formation of knots where the knots seem to get more complex as the length of stem and loop regions increases. Several earlier studies have demonstrated that in the case of knotted DNA molecules of the same length but forming different knots, the more complex knots migrate quicker than simpler knots (8–12). As in Figure 3A, we observe here (Fig. 3C) that 120mers having their loop and stem regions both set to 11 bp produce two different knots (lane 2). The quicker, and therefore more complex, one seems to be identical (with correction for the gel ‘smiling’) to the only knot produced by 120mers with stem and loop regions of 11 and 15 bp, respectively (lane 4). The 120mers having their loop and stem regions both set to 15 bp produce three different knots, the major one shows much higher electrophoretic migration and is presumably significantly more complex than the other two. The slowest knot, which is produced in very small quantities, is most likely of the same type as the one produced by 120mers with stem and loop regions of 11 and 15 bp long, respectively. The third knot is also produced in small amounts and it migrates very closely to the slowest knot suggesting that these two knots have the same minimal number of crossings (11,12).
Electrophoretic migration of knots and catenanes
As already mentioned, it is not certain whether, in PAA gels containing urea but run at an ambient temperature, all duplex regions of DNA are permanently melted, especially when these regions are kept in close proximity by the knotting of short oligonucleotide strands. In the absence of complete melting, the electrophoretic migration would not only be a function of different topologies of knotted molecules but would additionally be affected by the different ratio of single- to double-stranded regions in different constructs. To be sure that duplex regions in knots are completely melted we decided to use harsher denaturing conditions such as in alkaline agarose gels. We gel purified three principal types of knots obtained with different 120mers and analyzed them together with three principal catenanes obtained with pairs of 60mers. Figure 3D (lanes 1–3) reveals an ‘arithmetic progression’ of electrophoretic migration of the catenanes that are most likely singly, doubly and triply linked and, thus, have two, four and six crossings in their standard representation. The three principal knotted species (Fig. 3C, lanes 4–6) also show an ‘arithmetic progression’ of electrophoretic migration with the progression steps practically identical to those observed in catenated species (lanes 1–3). Since the analyzed knots and catenanes have the same overall molecular mass the similarity of the progression steps suggests that the analyzed knots progressively increase their minimal crossing number in steps of two. From the design of three different oligonucleotides used for knotting we expected to obtain knots with three, five and seven crossings (Fig. 4). Therefore, the observed differences in electrophoretic migration of produced knots are consistent with our knotting strategy.
Identification of different knots formed with the same single-stranded DNA constructs
Figure 4 shows the ‘construction plans’ used to generate various knots using oligonucleotides with stem and loop regions of specific length. The figure also shows the expected principal types of knots in their minimal crossing representation. The notation used, 31, 52 and 74, is from the standard tables of knots (13–15), where the main number corresponds to the minimal number of crossings of a given knot and the index number indicates the tabular position of this knot among the knots with a given minimal number of crossings. Presetting stem and loop regions to 11 bp each, we expected efficient formation of trefoil knots having a mathematical notation 31. The stem of 11 bp and the loop of 15 bp were expected to promote the formation of a knot with five crossings known as 52 knot. Oligonucleotides with stem and loop both set to 15 bp were expected to ligate forming knots with seven crossings, which have the mathematical notation 74. The gel analysis of major products is consistent with our expectations, but only in the case of the oligonucleotide with the stem of 11 bp and the loop of 15 bp did the ligation result in formation of unique type of knot. Why were other constructs not so specific? Figure 5A and B shows schematically how an oligonucleotide with stem and loop regions of 11 bp each can form 31 and 52 knots in almost equal proportions. The 11 bp complementarity in the loop can lead, during annealing, to the creation of two principal situations. In the first case there are two right-handed crossings in the loop and the formed knot is the trefoil. In the second case there are three right-handed crossings in the loop and the formed knot is 52. When the complementarity in the loop is of 15 nt the three crossings are almost always created and, therefore, the oligonucleotide with this sequence setting produces almost exclusively 52 knots (see Fig. 3). Figure 5C–E illustrates how an oligonucleotide with stem and loop regions preset to 15 bp each can form two different five crossing knots in addition to 74 knots (Fig. 5C). A 15 bp stem may be at the allowable size limit for this short oligonucleotide and would require high stretching of poly(T) regions (5,6). Such a stretching would oppose completion of the last turn in the stem and this would result then in the formation of 52 knots (Fig. 5D). In a small fraction of the molecules the stress may result in a stem (Fig. 5E). In such a situation the free ends have to cross-back in order to anneal in an anti-parallel way with their complementary sequence. Such a cross-back would lead to the formation of 51 knots (Fig. 5E). The analysis of possible knotting outcomes (Figs 4 and 5) and the observed pattern of electrophoretic migration of formed knots strongly suggests that we have correctly recognized the formed knots. The length of single-stranded DNA (120 nt) was too small to apply the RecA-coating method that was used previously to determine the formed knot types by electron microscopy (16). RecA filaments have the persistence length of 630 nm (17) and therefore could not accommodate the curvature required to follow the knots of the total length of ∼80 nm.
Figure 5.
Formation of two or three different knots from the same substrate. (A and B) Notice that pairing in the loop region extends in both cases over 11 bp and that in both cases the regular right-handed helix is formed in the loop region. The resulting knots are therefore almost equally likely to be formed and, in fact, that was what we observed (see Fig. 3A and C). (C–E) Notice that (C), (D) and (E) differ only in ‘phasing’ of crossings in the 15 bp long stem. In (C) pairing starts after the first crossing, in (D) a base or two pair before the first crossing and in (E) four or five bases pair before the first crossing. These differences determine how the out-coming ends exit the stem to pair within the loop. Notice that in (E) the out-coming ends have to cross-back as otherwise their polarity is not compatible with pairing in the loop. In (A–D) arrows show which type of movement of the colored portion needs to be performed to obtain standard representations of knots. In the case of (E) we invite readers to take a string or rubber tubing and perform the transition from the left to right and contrary.
We would like to stress here that the presented method allows very efficient creation of different knots and catenanes of the same total chain size. The knotting scheme we propose results in almost 100% efficiency of knot formation. Some of the tested substrates produce specifically only one type of knot or catenane while other substrates can result in the formation of two or more species of knots and catenanes. Out of a given type of knot only one ‘enantiomer’ is obtained and these knots with sequence-specific modifications could be used for ‘templating’ chirality specific synthesis reactions. We observed here that, in the denaturated form, single-stranded knots and catenanes of the same total length react very similarly to the increase of minimal crossing number. An increase of crossing number by two causes a similar increase of electrophoretic migration in knots and catenanes. However, knots seem to be shifted in relation to catenanes and the trefoil knot with three crossings migrates like the catenane with four crossings. Likewise, the knot with five crossings migrates like a catenane with six crossings. This result suggests that for the same number of minimal crossings short single-stranded knots are less compact than the catenanes of the same total length. Earlier studies analyzing electrophoretic migration of double-stranded knots and catenanes of the same total length showed that for the same minimal number of crossings knots and catenanes show essentially the same electrophoretic migration (18). However, in these earlier studies, DNA molecules were 6.4 kb long and this meant that their trajectory was essentially not affected by inter-segmental repulsion and would follow random walk trajectory restricted to a given topological space. In our case the dimensions of molecules are of the same order as the Debye length at applied conditions (19) and therefore their trajectory would be non-random and would rather tend to minimize the self-repulsing energy (20).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Jacques Dubochet for his constant support and interest in the project. This work was supported by grants from the Swiss National Science Foundation (31-61636.00 and 31-58841.99) and from the Human Frontiers Science Program.
REFERENCES
- 1.Winfree E., Furong,L., Wenzler,L.A. and Seeman,N.C. (1998) Design and self-assembly of two-dimensional DNA crystals. Nature, 394, 539–544. [DOI] [PubMed] [Google Scholar]
- 2.Braun E., Eichen,Y., Sivan,U. and Ben-Yoseph,G. (1998) DNA-templated assembly and electrode attachement of a conducting silver wire. Nature, 391, 775–778. [DOI] [PubMed] [Google Scholar]
- 3.Libchaber A. and Bar-Ziv,R. (2001) Effects of DNA sequence and structure on binding of RecA to single-stranded DNA. Proc. Natl Acad. Sci. USA, 98, 9068–9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watson J.D. and Crick,F.H.C. (1953) Molecular structure of nucleic acid. A structure for deoxyribonucleic acid. Nature, 171, 373–738. [DOI] [PubMed] [Google Scholar]
- 5.Mueller J.E., Du,S.M. and Seeman,N.C. (1991) Design and synthesis of a knot from single-stranded DNA. J. Am. Chem. Soc., 113, 6306–6308. [Google Scholar]
- 6.Wang H., Du,S.M. and Seeman,N.C. (1993) Tight single-stranded DNA knots. J. Biomol. Struct. Dyn., 10, 853–863. [DOI] [PubMed] [Google Scholar]
- 7.Sundin O. and Varshavsky,A. (1981) Arrest of segregation leads to accumulation of highly intertwined catenated dimers: dissection of final stages of SV40 DNA replication. Cell, 25, 659–669. [DOI] [PubMed] [Google Scholar]
- 8.Dean F.B., Stasiak,A., Koller,T. and Cozzarelli,N.R. (1985) Duplex DNA knots produced by Escherichia coli topoisomerase I, structure and requirements for formation. J. Biol. Chem., 260, 4975–4983. [PubMed] [Google Scholar]
- 9.Stark W.M., Grindley,N.D., Hatfull,G.F. and Boocock,M.R. (1989) Resolvase-catalysed reactions between res sites differeing in the central dinucleotide of subsite I. EMBO J., 10, 3541–3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bates A.D. and Maxwell,A. (1993) In Rickwood,D. and Male,D. (eds), DNA Topology. IRL Press, Oxford.
- 11.Crisona N.J., Kanaar,R., Gonzalez,T.N., Zechiedrich,E.L., Klippel,A. and Cozzarelli,N.R. (1994) Processive recombination by wild-type Gin and an enhancer-independent mutant. Insight into the mechanisms of recombination selectivity and strand exchange. J. Mol. Biol., 243, 437–457. [DOI] [PubMed] [Google Scholar]
- 12.Stasiak A., Katritch,V., Bednar,J., Michoud,D. and Dubochet,J. (1996) Electrophoretic mobility of DNA knots. Nature, 384, 122. [DOI] [PubMed] [Google Scholar]
- 13.Alexander J.W. and Briggs,G.B. (1927) On types of knotted curves. Ann. Math., 28, 562–586. [Google Scholar]
- 14.Rolfsen D. (1976) Knots and Links. Publish or Perish Press, Berkeley, CA.
- 15.Adams C.C. (1994) The Knot Book. W.H. Freeman and Company, New York, NY.
- 16.Krasnow M.A., Stasiak,A., Spengler,S.J., Dean,F., Koller,T. and Cozzarelli,N.R. (1983) Determination of the absolute handedness of knots and catenanes of DNA. Nature, 304, 559–560. [DOI] [PubMed] [Google Scholar]
- 17.Egelman E.H. and Stasiak,A. (1986) Structure of helical RecA-DNA complexes. Complexes formed in the presence of ATP-gamma-S or ATP. J. Mol. Biol., 191, 677–698. [DOI] [PubMed] [Google Scholar]
- 18.Wasserman S.A., Dungan,J.M. and Cozzarelli,N.R. (1985) Discovery of a predicted DNA knot substantiates a model for site-specific recombination. Science, 229, 171–174. [DOI] [PubMed] [Google Scholar]
- 19.Eisenberg D. and Crothers,D. (1979) Physical Chemistry with Applications to the Life Sciences. The Benjamin/Cummings Publishing Company, Inc., Menlo Park, CA.
- 20.O’Hara J. (1998) Energy of knots. In Stasiak,A., Katritch,V. and Kauffman,L.H. (eds), Ideal Knots. World Scientific, Singapore, pp. 288–314.