Abstract
Imaging genetics provides an opportunity to discern associations between genetic variants and brain imaging phenotypes. Historically, the field has focused on adults and adolescents; very few imaging genetics studies have focused on brain development in infancy and early childhood (from birth to age 6 years). This is an important knowledge gap because developmental changes in the brain during the prenatal and early postnatal period are regulated by dynamic gene expression patterns that likely play an important role in establishing an individual’s risk for later psychiatric illness and neurodevelopmental disabilities. In this review, we summarize findings from imaging genetics studies spanning from early infancy to early childhood, with a focus on studies examining genetic risk for neuropsychiatric disorders. We also introduce the Organization for Imaging Genomics in Infancy (ORIGINs), a working group of the ENIGMA (Enhancing NeuroImaging Genetics through Meta-Analysis) consortium, which was established to facilitate large-scale imaging genetics studies in infancy and early childhood.
Keywords: Childhood, Genetics, Imaging, Infant, Magnetic resonance imaging, Pediatric
Imaging genetics reveals considerable information about genetic influences on structural and functional imaging phenotypes (1, 2, 3), but until recently focused largely on the adolescent or adult human brain (4,5). This is an important limitation because the most dynamic phase of human brain development is from embryonic life through early childhood (6) (Figure 1). Disrupted gene expression in this period can produce lifelong changes in brain morphology and function. Even common genetic variations may affect early neurodevelopmental processes, thereby increasing risk for psychiatric conditions later in life (7). These effects may be detectable in early life via neuroimaging, thereby providing opportunities for identifying at-risk populations in infancy for primary prevention and the development of interventions to adjust adverse trajectories earlier in the clinical sequence. In this paper, we review empirical evidence underlying this hypothesis focusing on magnetic resonance imaging (MRI). Studies using ultrasound (8,9) and studies integrating imaging and epigenetic data (10, 11, 12, 13) also provide insights into how genes influence the developing young brain but are beyond the scope of this review. First, we describe the heritability of brain imaging phenotypes in early life. Then, we discuss candidate gene studies of brain structure, function, and connectivity. Next, we review studies characterizing associations between psychiatric risk genes and brain phenotypes in early life using polygenic scores. Then, we discuss genome-wide studies on brain imaging phenotypes in early childhood. Finally, we introduce ORIGINs (the Organization for Imaging Genomics in Infancy), a working group of the ENIGMA (Enhancing NeuroImaging Genetics through Meta-Analysis) consortium, which was established to facilitate large-scale imaging genetics studies in infancy and early childhood.
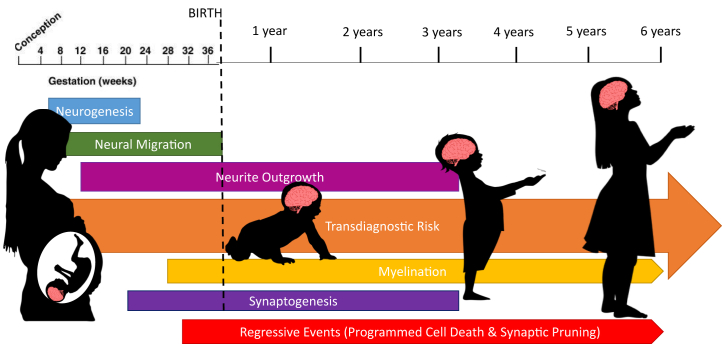
Figure 1.
Early neurodevelopment is a sensitive period for accumulating transdiagnostic risk for psychiatric disorders. Neurodevelopment in the human brain begins from approximately 2 weeks after conception. Neurogenesis and neural migration are primarily prenatal processes, and neurite outgrowth is minimal after 4 years of age. Myelination and synaptic pruning continue beyond 6 years of age (indicated by the small arrows). Genetic influences on these various processes contribute to psychiatric risk, which accumulates across development and may not manifest until later in life (large orange arrow).
Heritability
Twin studies have revealed that many brain phenotypes are heritable in early infancy. Genetic effects explain around 85% of the variance in global white matter volume (WMV) and 56% of the variance in global gray matter volume (GMV), at around 1 month of age (14), while heritability of head size is negligible (15). This contrasts with studies of older children and adults where heritabilities greater than 80% are reported for all 3 phenotypes (global WMV, GMV, and head size) (16, 17, 18). Heritability estimates for global cortical surface area (SA) are high in early infancy (78%), while estimates for global cortical thickness (CT) are lower (29%), with significant genetic overlap between the two (19). This differs from adults, among whom SA and CT are both highly heritable (89% and 81%, respectively), with distinct genetic factors contributing to each measure (20, 21, 22). White matter microstructure is moderately heritable in early life, with 30% to 60% of the variability in mean fractional anisotropy (FA) linked to genetic variation and similar estimates for other diffusivity indices (23,24). In adults, estimates for FA range from 72% to 88% (25). Despite large variations in heritability estimates across individual tracts, a single latent measure of white matter microstructure accounts for a great deal of heritable variation in neonates (50%) (26). Similarly, individual differences in regional CT and SA appear to be driven by a common set of genetic factors influencing cortical structure at the global level (19). In both cases, the pattern of results mirrors temporal changes in gene expression which show strong spatial differences in fetal but not postnatal development (27,28). Finally, genetic effects on resting-state functional MRI phenotypes have been observed during the first 2 years of life. Gao et al. (29) reported modest genetic effects on within-network connectivity in neonates, with 3 visual networks and the right frontoparietal network demonstrating above-average effects. At age 1 year, the most heritable networks were the bilateral frontoparietal networks, the salience network, and 2 visual networks. At age 2 years, genetic effects were the strongest for the auditory network. However, genetic effects were not as strong as those reported in adolescents and adults (30, 31, 32). Genetic effects on between-network connectivity are also minimal in neonates (33). Intergenerational transmission of imaging phenotypes has been reported and likely reflects a combination of genetic, epigenetic, and environmental effects (34, 35, 36). One such study examined the intergenerational transmission from mothers to their 5-year-old children and reported significant effects on sulcal phenotypes in the right frontal and parietal cortices (35).
A recurring theme across studies is that heritability is higher in adulthood than in infancy. This might appear paradoxical because interindividual variation in environmental exposures increases with age, but similar patterns are observed for IQ, where increasing heritability during development is called the Wilson effect (37). The Wilson effect is thought to arise from gene-environment correlations that increase with age. In other words, babies and young children have environments thrust upon them, but as they age, they select, modify, and create environments that are correlated with their genetic predispositions (38). Alternatively, higher heritability observed at later ages could reflect stronger heritability of postnatal processes such as myelination and shifts in proportions of white versus gray matter across development. Determining whether a genetic amplification model applies to neuroimaging phenotypes will require large-scale longitudinal studies that address gene-environment interplay across the life span. The ENIGMA plasticity working group has begun tackling this question. Using 5 longitudinal twin cohorts, they demonstrated that rates of brain change are heritable, and heritability estimates of change rates were higher in adults than in children (39). They subsequently identified variants involved in structural brain changes via a genome-wide association study (GWAS) (40). However, their studies did not include infants or toddlers.
Candidate Gene Approaches
Traditional candidate gene studies test for associations between phenotypic outcomes and variation within specific genes selected for suspected roles in organ development or physiology. In imaging genetics, selection is often based on hypothesized involvement in psychiatric disease. The first candidate gene study of brain imaging phenotypes in neonates focused on global and local brain tissue volumes and several genes with known roles in brain development and putative links to psychiatric disease including DISC1, COMT, NRG1, ESR1, and BDNF (41). Many reported effects mirrored findings in adults; others were unique to infancy. For the BDNF Val/Met polymorphism, Met+ neonates had decreased volumes in regions of the right occipital cortex, left hippocampus, parahippocampus, fusiform gyrus, and inferior temporal gyrus and increased volumes in the motor and somatosensory cortex (41). We highlight this result because a recent study partially replicated the original findings. Specifically, Kawasaki et al. (42) reported that Met+ neonates had significantly smaller relative hippocampal volumes.
The largest traditional candidate gene study to include children under 6 years of age focused on Klotho, a gene linked to age-related decline. A significant interaction between Klotho allele status (rs9536314) and age was observed for total brain volume and total GMV, with KLS-VS heterozygotes having larger volumes in early childhood but not in later childhood/adolescence. Among girls, KL-VS heterozygotes had less WMV than noncarriers, whereas among boys, heterozygotes had greater WMV than noncarriers. No effects were significant in a replication cohort that did not include children younger than 6 years of age (43), supporting the importance of conducting imaging genetics studies in early life to unveil effects that have been found to be absent in older cohorts.
In addition to age, genetic effects on neurodevelopment may vary based on factors such as prematurity and family history. For example, Krishnan et al. (44) hypothesized that polymorphisms in DLG4 would moderate responses to perinatal inflammation and their impact on white matter microstructure based on gene network analysis of the microglial transcriptomic response to injury in mouse models and complementary, data-driven analysis of protein-protein interactions, transcription factors, and human brain gene expression. The team discovered a specific variant in DLG4 (rs17203281) associated with FA in preterm individuals in 2 independent cohorts. Van Steenwinckel et al. (45) adopted a similar approach, identifying key genes and gene networks in animal models of neuroinflammation-induced hypomyelination and then testing for associations in preterm infants. The researchers revealed that Wnt pathway genes were collectively associated with cerebral structural connectivity. In addition, a study of 13 candidate genes revealed that ARVCF, previously linked to schizophrenia, and FADS2, previously linked to intelligence, were associated with white matter FA in preterm infants (46). These studies highlight the importance of considering potential interactions between genetic variation and early-life environmental exposures given that neither DLG4 nor the Wnt pathway genes would be expected to impact diffusion tensor imaging phenotypes in the absence of perinatal inflammation. With regard to family history, Douet et al. (47) reported that the effects of variants in ERBB4 differed in children with and without a family history of schizophrenia and/or bipolar disorder. The TT variant for rs7598440 had more pronounced effects on age-related changes (3–20 years) in CT and SA in children with a family history; these children showed steeper increases in frontal and temporal SA in both early and late childhood.
Several studies explicitly tested for gene-environment interactions using the candidate gene approach. COMT single nucleotide polymorphisms (SNPs) moderated the association between antenatal maternal anxiety and prefrontal and parietal CT in neonates (48). The BDNF genotype (Val66Met) moderates associations between methylation patterns and neonatal hippocampal and amygdala volumes (49). FKBP5, which regulates the hypothalamic-pituitary-adrenal axis, moderates the association between antenatal maternal depressive symptoms and neonatal right hippocampal volume (50). For oxytocin receptor (OXTR) gene variant rs53576, a sex-specific main effect was seen for neonatal hippocampal volume. Left hippocampal volumes were larger in GG-homozygotes than A-allele carriers in boys only. Prenatal maternal anxiety interacted with genotype in both sexes: higher maternal anxiety was associated with larger hippocampal volumes in A-allele carriers (51). Additional details on these studies are found in Table 1. A graphical representation (PhenoGram) (52) of genes and associated phenotypes is given in Figure 2.
Table 1.
Candidate Gene Studies of Imaging Phenotypes in Infancy and Childhood
| Article | Participants, N | Age Group | Ancestry | Gene/SNP-Brain Phenotype Association | Findings |
|---|---|---|---|---|---|
| Studies on Genetic Effects | |||||
| Knickmeyer et al., 2014 (41) | 272 | Neonates (gestational age at MRI: 261–433 days) | Maternal ethnicity—White | ESR1 (rs9340799)—ICV; DISC1 (rs821616), COMT, NRG1, APOE, ESR1 (rs9340799), and BDNF—GMV | Associations in DISC1 and COMT mirrored findings in adults. |
| Dean et al., 2014 (56) | 162 | 2- to 25-month-old infants | Not reported | APOE ε4 allele—↓ MWF and GMV in precuneus, posterior/middle cingulate, lateral temporal, and medial occipitotemporal regions. APOE ε4 allele—↑ MWF and GMV in extensive frontal regions | Infant APOE ε4 allele carriers had lower white matter MWF and GMV measurements than noncarriers in areas preferentially affected by Alzheimer’s disease. |
| Boardman et al., 2014 (46) | 83 preterm infants | Neonates (postmenstrual age 23 + 2 to 32 + 6 weeks) | Multiancestry | ARVCF (rs2518824) and FADS2 (rs174576)—white matter FA | – |
| Douet et al., 2015 (47) | 971 (PING study) | 3–20 years | Multiancestry | ERBB4 (rs7598440)—cortical structures | In the full sample, children with the TT genotype had smaller SA in the occipital and temporal lobes at ages <5 years. When stratifying by family history of schizophrenia and/or bipolar disorder, TT children showed steeper increases in frontal SA in early childhood. |
| Chang et al., 2016 (58) | 1187 (PING study) | 3–20 years | Multiancestry | APOE (ε2ε4—↓ hippocampus; ε4ε4—↓ hippocampal FA; ε3ε4—↑ medial orbitofrontal cortical areas) | The ε4ε4 and ε2ε4 genotypes may negatively influence brain development and brain aging at the extremes of age. |
| Krishnan et al., 2017 (44) | Preterm infants (cohort 1: n = 70; cohort 2 [ePRIME study]: n = 271) | Cohort 1—mean postmenstrual age at scan 40 + 3 weeks; cohort 2—mean postmenstrual age at scan 42 + 4 weeks | Multiancestry | DLG4 (rs17203281)—FA | DLG4 (rs17203281) was associated with structural white matter changes. |
| Van Steenwinckel et al., 2019 (45) | 290 preterm infants (ePRIME study) | Gestational age of 38.29–58.28 weeks | Multiancestry | NFATC4, CSNK1A1, MAPK10, WNT2B, SMAD3, FBXW11, NLK, CSNK1A1L, PLCB2, and WNT5A—white matter structural connectivity | Genomic variation in the Wnt pathway is associated with the levels of connectivity found in their brains. |
| De Vries et al., 2020 (43) | 1387 (PING study) | 3–21 years | Multiancestry | Klotho allele KL-VS; KL-CS × age interaction—TBV, TGMV; Kl-VS × sex—TWMV | A replication in a cohort of 2306 children age 6–12 years (Generation R sample) showed no significant associations. KL-VS’s influence may depend on age and sex. |
| Remer et al., 2020 (57) | 223 | 2–68 months | Multiancestry | APOE ε4 carriers—MWF | ε4 carriers—significant MWF trajectory differences in multiple neuroanatomical locations |
| Kawasaki et al., 2021 (42) | 66 | Newborn infants (37.9–47.6 postmenstrual weeks) | Multiancestry | BDNF-Val66Met variant—hippocampi, amygdalae, TWMV | Met + group—↓ hippocampi, amygdalae, age-dependent declines in % total WMVs, slower age-dependent declines in total brain volumes |
| Cullen et al., 2022 (59) | 208 (dHCP study) | 0–6 weeks | European | rs945270 (intergenic locus downstream of the kinectin 1 [KTN1] gene)—putamen volume | Greater number of C alleles associated with larger volume |
| Studies on Interaction Between Genetic and Environmental Effects | |||||
| Qiu et al., 2015 (48) | 146 (GUSTO cohort) | Neonates (5–17 days) | Asian | COMT—cortical thickness | COMT SNPs (val158met, rs737865 and rs165599)—role in moderating the relationship of antenatal maternal anxiety with dorsolateral prefrontal and parietal cortical thickness in neonates |
| Chen et al., 2015 (49) | 237 (GUSTO) | Neonates (4–17 days) | Asian | BDNF (Val66Met)—hippocampus, amygdala | BDNF (Val66Met)—regulate the sensitivity of the methylome with differential effects on amygdala and hippocampal volume |
| Wang et al., 2017 (50) | 164 Mother-offspring dyads (GUSTO) | Neonates (5–14 days) | Asian | FKBP5—hippocampus | 17 SNPs in the FKBP5 gene showed significant interaction effects with antenatal maternal depressive symptoms on right hippocampal volume. |
| Acosta et al., 2021 (51) | 105 | 11–54 days old | European | OXTR rs53576 × sex—hippocampus | For OXTR SNP rs53576, in boys compared with girls, left hippocampal volumes were significantly larger in GG-homozygotes than A-allele carriers. Higher maternal anxiety was associated with larger hippocampal volumes in A-allele carriers than GG-homozygotes. |
dHCP, developing Human Connectome Project; FA, fractional anisotropy; GMV, gray matter volume; GUSTO, Growing Up in Singapore Toward healthy Outcomes; ICV, intracranial volume; MRI, magnetic resonance imaging; MWF, myelin water fraction; PING, Pediatric Imaging, Neurocognition, and Genetics consortium; SA, surface area; SNP, single nucleotide polymorphism; TBV, total brain volume; TGMV, total GMV; TWMV, total white matter volume.
Figure 2.
PhenoGram of genes associated with brain imaging phenotypes in infants and young children from candidate gene studies and genome-wide association studies. CT, cortical thickness; FA, fractional anisotropy; GMV, gray matter volume; ICV, intracranial volume; MWF, myelin water fraction; PFC, prefrontal cortex; SA, surface area; TBV, total brain volume; WM, white matter; WMV, white matter volume.
Interestingly, imaging genetics studies of infants and young children began at a time when microarray genotyping began making large-scale genotyping practical. The GWAS era quickly highlighted weaknesses in the traditional candidate gene approach. Well-powered GWASs failed to support involvement of many traditional candidate genes in psychiatric disorders. This may partly reflect addressable methodological weaknesses including failure to control for population stratification and thereby increasing the risk of false-positive associations due to differences in ancestry. However, the key disadvantage of the candidate gene approach is likely poor candidate selection, given the inadequacy of current knowledge about underlying biological processes. Subsequent meta-analyses of candidate gene studies relevant to psychiatry, including imaging genetics studies conducted in older populations, revealed poor replicability, false-positive associations, overestimation of effect sizes, and publication bias (53). While the imaging genetics literature for infants and young children is not extensive enough to allow meta-analyses, existing studies likely have similar weaknesses.
One promising approach to addressing these problems is to focus future studies on variants that are robustly associated with mental and neurological disorders or adult brain imaging phenotypes in large-scale GWASs. The ϵ4 allele of the apolipoprotein E (APOE) gene meets this criterion. Not only is ϵ4 the strongest known risk variant for Alzheimer’s disease, it also has well-documented effects on brain structure and cognition in healthy individuals (54,55). In neonates, APOE ϵ3ϵ4 heterozygotes have significantly lower volumes in temporal regions, compared with ϵ3 homozygotes, and lower volume in the frontal and parietal lobes. ϵ3ϵ4 heterozygotes have significantly greater volumes in specific parietal, frontal, and occipital areas (41). Infant ε4 carriers have lower white matter myelin water fraction (MWF) and GMV measurements in the precuneus, posterior/middle cingulate, lateral temporal, and medial occipitotemporal regions, areas which are preferentially affected by Alzheimer’s disease, and greater MWF and GMV measurements in frontal regions (56). Decreased myelin in ε4 carriers in the corticospinal tract, the splenium of the corpus callosum, and frontal white matter, observed in the previous study, was also reported in a longitudinal analysis from the same group in which children were followed from birth to age 5.5 years (57). Regions in which ε4 carriers had greater MWF early on had a decreased rate of MWF development until age 5.5 years, allowing noncarriers to catch up and surpass ε4 carriers at around 3 years of age. In another study, age-related changes in brain structures and cognition were observed to vary depending on genotype, with the smallest hippocampi in ε2ε4 children, the lowest hippocampal FA in younger ε4ε4 children, the largest medial orbitofrontal cortical areas in ε3ε4 children, and age-dependent thinning of entorhinal cortex in ε4ε4 children (58). All these studies suggest that Alzheimer’s disease is a neurodevelopmental disorder as well as a neurodegenerative one.
Six SNPs robustly associated with subcortical volumes in adult GWASs have recently been tested for effects in neonates. An association between rs945270 (an intergenic locus downstream of the kinectin 1 [KTN1] gene) and putamen volume was reported, suggesting that at least some variants have detectable effects across the life span (59).
A significant challenge when performing a candidate gene study informed by existing GWASs is how to prioritize genes for follow-up. Fortunately, an increasing array of in silico tools for searching GWAS literature and performing functional characterization of variants can assist with this task (60, 61, 62). Another challenge is that candidate gene studies informed by GWASs still focus on only a few selected genes/polymorphisms which account for only a fraction of variants involved in psychiatric risk. One approach to overcoming this challenge is to use polygenic risk scores.
Polygenic Risk Score Approaches
Polygenic risk scores (PRSs) estimate an individual’s susceptibility to a complex trait based on prior GWAS summary statistics (63). Efforts such as the Psychiatric Genomics Consortium have produced many well-powered GWASs of psychiatric and neurodevelopmental conditions including schizophrenia, major depressive disorder (MDD), bipolar disorder, attention-deficit/hyperactivity disorder, and autism spectrum disorder (ASD) (64). Summary statistics are often freely available, and researchers can obtain individual data from controlled-access repositories. By examining associations between PRSs and neuroimaging phenotypes measured in infancy and early childhood, researchers are clarifying how genetic risk for these conditions manifests in early life, thereby providing new insights into the etiology of psychiatric and neurodevelopmental disorders. This is a key step in identifying individuals who may benefit from early intervention.
One of the first studies to use this approach in early life was conducted by Xia et al. (65), who found that PRSs for schizophrenia and ASD were not associated with neonatal total brain volume. Like Xia et al. (65), Cullen et al. (59) did not observe associations between neonatal volumes and PRSs for schizophrenia. However, PRSs for schizophrenia were negatively associated with regional GMV and WMV and total WMV in neonates in a different study (66). PRSs for ASD were associated with greater CT and reduced white matter connectivity in children (3 to ∼14 years) (67). In preterm infants, a PRS for 5 conditions (ASD, attention-deficit/hyperactivity disorder, bipolar disorder, MDD, and schizophrenia) predicted reduced volume of the lentiform nucleus, which plays a key role in motor control, cognition, and emotion (68). The authors hypothesized that genetic risk for psychiatric disorders increased vulnerability to abnormal lentiform development in the context of perinatal stress associated with preterm birth but did not include term infants for comparison. Other studies have used PRSs to probe relationships between early-life adversity, genetic risk, and neurodevelopment. Ursini et al. (69) used transcriptomic data to create placental genomic risk scores (PlacGRSs) for schizophrenia. PlacGRSs were calculated like traditional PRSs, but only used markers in genes highly expressed in placenta and differentially expressed in placentae from complicated, compared with normal, pregnancies. PlacGRS was negatively associated with neonatal brain volume in children with perinatal complications, especially in boys. No significant associations were observed for PlacGRSs and nonplacental GRSs for other disorders and traits associated with early-life complications, suggesting that the link between placental biology, genetic risk, perinatal environmental risk, and early brain development outcomes is relatively unique to schizophrenia.
Another important perinatal stressor is maternal depression. Many studies report associations of maternal depressive symptoms with neuroimaging outcomes in early life, but it is unclear whether such associations represent causal effects or arise from genetic confounding. PRSs can be used to test independent effects of maternal depressive symptoms and genetic risk for MDD as well as their interaction. Qiu et al. (70), the first to apply this approach, reported significant interactions between PRSs for MDD and antenatal maternal depressive symptoms for right amygdala volume in Asian (GUSTO [Growing Up in Singapore Towards healthy Outcomes]) and U.S. neonates. However, the direction of the effect differed across cohorts. In Finland, Acosta et al. found patterns similar to those found in the U.S. cohort (71). However, the interaction became nonsignificant after correction for multiple comparisons. The Finnish team also investigated associations of an MDD PRS with infant striatal volumes and found sex-specific effects: the MDD PRS was positively associated with caudate volumes in boys but negatively associated with caudate volumes in girls (72). They did not observe significant interaction effects of the PRS with prenatal maternal depressive symptoms for any dorsal striatal volumes.
PRSs can also be used to understand how molecular pathways shape individual differences in neurodevelopment. For example, a PRS for serum testosterone was recently found to be positively associated with total SA development in female infants (73). Researchers interested in this application of PRSs may implement expression-based PRSs (ePRSs) rather than traditional PRSs. ePRSs integrate genotype data with transcriptomic data to predict expression levels of a particular gene or gene network. Morgunova et al. (74) used this approach to investigate relationships between a coexpression network of the DCC gene, which is robustly associated with multiple psychiatric conditions, and total brain volume in both neonates and older children. Higher ePRSs for the DCC coexpression network were associated with larger brain volumes. A study from GUSTO investigated how genes involved in inflammation interact with maternal depression to shape neonatal brain morphology. They created separate ePRSs for 22 cytokine and chemokine genes expressed in fetal brain and found that ePRSs for TNFRSF19, IL17RB, BMPR1B, IL1RAP, and CXCR4 moderated the impact of maternal depression on specific subcortical volumes and regional CT (75). Using longitudinal data from the same cohort, investigators revealed an age-dependent involvement for transmembrane receptor (TGF-β) variants in moderating effects of prenatal maternal depressive symptoms on amygdala volume (76).
Finally, PRSs have been used to investigate how genetic variants linked to adult and adolescent brain morphology influence early brain development. Xia et al. (65) found that PRSs for WMV and GMV in adolescence showed positive associations with neonatal WMV and GMV, respectively, although the overall proportion of variation explained was low. Morgunova et al. (74) calculated PRSs for brain volume using data from UK Biobank, ENIGMA, CHARGE (Cohorts for Heart and Aging Research in Genomic Epidemiology), and the Early Growth Genetics Consortium. These PRSs did not predict brain volume in their neonate and school-age community cohorts. The first large-scale GWAS of adult intracranial volume tested whether a polygenic score generated from 7 genome-wide significant loci predicted head growth in children of European ancestry who were followed prenatally until 6 years of age (77). The investigators found an age-dependent effect in which the PRS became more predictive in older children, suggesting that adult brain volume is strongly shaped by genetic influences operating in early childhood. Cullen et al. (59) found robust associations between PRSs for adult brainstem, hippocampus, putamen, and thalamus volumes and neonatal volumes, suggesting some stability across the life course.
Reviewed studies highlight the potential of PRS-based investigations of neuroimaging phenotypes in infancy and early childhood (Table 2). They also reveal the importance of considering effects of sex and ancestry. A major limitation of this approach is the lack of sufficiently powered GWASs conducted in non-European populations. European ancestry GWASs do not transfer well to other ancestries and can lead to unpredictable biases (78). Another limitation of PRS studies is that they are based on existing GWASs and hence limited by the power of current datasets. In other words, PRS-based studies, like candidate gene studies, are constrained by current biological knowledge. To fully understand how DNA variants influence brain development, well-powered GWASs of infants and young children encompassing multiple ancestries are needed.
Table 2.
PRS Studies of Imaging Phenotypes in Infancy and Childhood
| Article | Participants, N | Age Group | Ancestry | PRS | Findings |
|---|---|---|---|---|---|
| Studies on Main Genetic Effects | |||||
| Xia et al., 2017 (65) | 561 | 6–161 days | Multiancestry | Polygenic scores for GM and WM from adolescent cohort; polygenic scores for ICV from adolescent and adult cohort | Adolescent WM and GM scores showed positive associations with neonatal WM and GM; adult polygenic scores for ICV did not predict neonatal ICV |
| PRS for schizophrenia and ASD | PRS did not predict global brain volumes. | ||||
| Cullen et al., 2019 (68) | 194 preterm infants | Mean postmenstrual age at scan 42.6 weeks | Multiancestry | PRS from meta-analysis of genome-wide SNP data for 5 psychiatric disorders (ASD, ADHD, bipolar disorder, MDD, and schizophrenia) | ↑PRS—↓ lentiform volume in the mixed ancestral cohort and a European subsample |
| Khundrakpam et al., 2020 (67) | 391 (PING study) | 3–21 years | Multiancestry | PRS for ASD | ↑ PRS for ASD—↑ cortical thickness for a large age span starting from 3 years up to ∼14 years in several cortical regions localized in the bilateral precentral gyri and the left hemispheric postcentral gyrus and precuneus, ↓WM connectivity between the frontal and parietal regions |
| Morgunova et al., 2021 (74) | 142 | Neonates (27 ± 13 days) | Multiancestry | ePRS was created based on the DCC coexpression gene network in the PFC. | ↑ ePRS—↑ total brain volume (GM and WM, adjusted by ICV) |
| Alex et al., 2021 (73) | 430 | Birth–2 years | European | PRS for serum testosterone | ↑ PRS—↑ SA development over time in female infants |
| Cullen et al., 2022 (59) | 208 | 0–6 weeks | European | GPSs for adult subcortical brain volumes | Neonatal volumes of the hippocampus, brainstem, putamen, and thalamus associated with adult GPS |
| GPSs for psychiatric disorders ASD, ADHD, schizophrenia, bipolar disorder, MDD, and cross-disorder (including 8 psychiatric disorders: anorexia nervosa, ADHD, ASD, bipolar disorder, MDD, obsessive-compulsive disorder, schizophrenia, and Tourette syndrome) | None of the neonatal brain volumes showed an association with psychiatric GPS. | ||||
| Le et al., 2022 (66) |
257 |
Postmenstrual age at scan 38–45 weeks |
Preliminary analysis: European; secondary; European and Asian |
PRS for schizophrenia |
↑PRS—↓ right frontal lobe WM, ↓GM and WM superior temporal gyrus volumes and ↓total white matter volume |
| Studies on Interaction Between Genetic and Environmental Risk | |||||
| Qiu et al., 2017 (70) | 168 (GUSTO) and 85 (US) mother-infant dyads | Neonates: GUSTO (5–14 days), US (postconceptual age at the MRI visit 43.02 ± 2.1 weeks) | GUSTO—Asian; US—mixed ancestry | GPRSMDD | A significant interaction was observed between antenatal maternal depressive symptoms and infant GPRSMDD on right hippocampal volume in the Asian cohort and right amygdala volume in both cohorts. A significant interaction was observed between SES and infant GPRSMDD on right amygdala and hippocampal volumes and shapes in the Asian cohort. |
| Wang et al., 2017 (50) | 164 Mother-offspring dyads (GUSTO) | Neonates (5–14 days) | Asian | A genetic risk score was calculated for individual neonates by summing the number of minor alleles of 19 FKBP5 SNPs. | Neonates with a genetic risk score less than or equal to the median showed a positive association between antenatal maternal depressive symptoms and right hippocampal volume. Neonates with a genetic risk score greater than the median showed a negative association between antenatal maternal depressive symptoms and right hippocampal volume. |
| Acosta et al., 2020 (71) | 105 | 11–54 days old | European | PRS-MDD | A nonsignificant interaction effect was observed between PRS-MDD and prenatal maternal depressive symptoms on right amygdala volume. |
| Acosta et al., 2020 (72) | 105 | 11–54 days old | European | PRS-MDD | No significant interaction effects of PRS-MDD with prenatal maternal depressive symptoms were found for infant dorsal striatal volumes. PRS-MDD was more positively associated with caudate volumes in boys compared with girls. |
| Wu et al., 2020 (75) | 161 mother-child dyads (GUSTO) | Neonates (5–14 days) | Asian | A GES was calculated for individuals by summing the number of minor alleles across the SNPs of the gene that were highly correlated with its expression level according to the existing eQTL database. | Positive associations of prenatal maternal depressive symptoms with the hippocampal volume, auditory and prefrontal cortical thickness in neonates high in GESs of the TNF, IL-1, IL-17, chemokine, and TGF family and receptors. |
| Ursini et al., 2021 (69) | 242 | 10–60 days | European | Fractionated genomic risk scores for schizophrenia based on placental gene expression loci (PlacGRSs) | ↑PlacGRSs —↓ neonatal brain volume in singletons and offspring of multiple pregnancies with a history of early-life complications |
| Qiu et al., 2021 (76) | 162 (GUSTO) | Birth–6 years | Asian | A GES was calculated for individuals by summing the number of alleles across the SNPs of the gene that was correlated with TGF-βRI expression level according to the existing eQTL database. | In neonates with a high GES of TGFBR1, higher levels of prenatal maternal depressive symptoms were associated with a smaller right amygdala volume. In children with a low GES of TGFBR1, greater prenatal maternal depressive symptoms predicted greater left and right amygdala volumes at 6 years of age. |
ADHD, attention-deficit/hyperactivity disorder; ASD, autism spectrum disorder; ePRS, expression-based polygenic risk score; eQTL, expression quantitative trait loci; GES, genetic expression score; GM, gray matter; GPRSMDD, genomic profile risk score for major depressive disorder; GPS, genome-wide polygenic score; GUSTO, Growing Up in Singapore Toward healthy Outcomes; ICV, intracranial volume; IL, interleukin; MDD, major depressive disorder; MRI, magnetic resonance imaging; PING, Pediatric Imaging, Neurocognition, and Genetics consortium; PFC, prefrontal cortex; PlacGRS, placental genomic risk score; PRS, polygenic risk score; SA, surface area; SES, socioeconomic status; SNP, single nucleotide polymorphism; US, United States; WM, white matter.
Genome-wide Association Studies
Hypothesis-free GWASs can identify new associations and overturn prior assumptions. However, GWASs of neuroimaging outcomes in infants and young children are very limited. The first GWAS of healthy infants identified several common variants associated with neonatal brain structure (65). An intronic SNP in IGFBP7 was significantly associated with GMV. An intronic SNP in WWOX fell just short of genome-wide significance for WMV. Many top associations tagged transcriptional regulators expressed during brain development (KLF13, LMCD1, TOX3, and TBX4). The investigators also compared their results to large-scale neuroimaging GWASs in adolescents and adults and concluded that genetic determinants of global brain volumes are highly distinct at different ages. In a subsequent GWAS of white matter microstructure in neonates, an intronic SNP in PSMF1 was significantly associated with a tractography-based factor capturing shared variation in FA across 44 white matter bundles (26). Additional loci nearing genome-wide significance were in or near genes with roles in axon growth and guidance, fasciculation, and myelination including B3GAT1, TENM2, NFATC1, and MAP3K13. The above studies are the first of their kind, and replication is crucial. Furthermore, these studies were not large enough to generate stable SNP-wise heritability estimates or evaluate genomic correlations between infant neuroimaging phenotypes and psychiatric disorders.
A smaller study in preterm individuals used genome-wide data and pathway-based and network-based approaches (79). The peroxisome proliferator-activated receptor signaling pathway was found to have a role in white matter development, with 5 genes implicated (AQP7, ME1, PLIN1, SLC27A1, and ACAA1). This inspired the team to examine the peroxisome proliferator-activated receptor pathway in a larger cohort of preterm children. Using machine learning analysis, they uncovered 3 genes associated with cerebral connectivity (PPARG, ITGA6, and FXR1) (80). GWASs are summarized in Table 3 and included in the Figure 2 PhenoGram (52). Gene functions and associated neurological phenotypes/conditions for candidate genes and genes identified via GWASs are provided in Table 4.
Table 3.
Genome-wide Association Studies of Imaging Phenotypes in Infancy and Childhood
| Article | Participants, N | Age Group | Ancestry | Findings |
|---|---|---|---|---|
| Krishnan et al., 2016 (79) | 72 preterm infants | Gestational age 23 + 2 to 32 + 6 weeks | Multiancestry | Identified significant role for lipid pathways and PPAR signaling in influencing development of white matter in preterm infants. Five genes were found to be highly associated with the phenotype: AQP7, ME1, PLIN1, SLC27A1, and ACAA1. |
| Krishnan et al., 2017 (80) | 272 preterm infants | Gestational age 42 weeks + 4 days | Multiancestry | PPARG (6 SNPs), ITGA6 (4 SNPs), FXR1 (2 SNPs) are associated with preterm cerebral endophenotype, particularly insular cortex |
| Xia et al., 2017 (65) | 561 | 6–161 days | Multiancestry | An intronica SNP in IGFBP7 (rs114518130) achieved genome-wide significance for gray matter volume. An intronic SNP in WWOX (rs10514437) neared genome-wide significance for white matter volume. |
| Zhang et al., 2021 (26) | 471 | Neonates (days post conception 293.4 ± 16.6) | Multiancestry | An intronic SNP in the gene PSMF1 was significant for a tractography-based factor that captured shared variation in fractional anisotropy across 44 white matter bundles. |
PPAR, peroxisome proliferator-activated receptor; SNP, single nucleotide polymorphism.
Intronic SNPs are located in a segment of a DNA or RNA molecule which does not code for proteins and interrupts the sequence of genes.
Table 4.
Brain Imaging Phenotype Associated Genes, Their Functions, and Associated Neurologic Phenotypes/Disorders
| Gene | Symbol | Function | Associated Neurologic Phenotype(s)/Disorder(s) |
|---|---|---|---|
| Acetyl-CoA Acyltransferase 1 | ACAA1 | Involved in neuronal growth and myelinogenesis (87) | Alzheimer’s disease (88) |
| Apolipoprotein E | APOE | Facilitates the transfer of cholesterol and phospholipid between cells, key role in neuronal development, brain plasticity, and repair (89) | Alzheimer’s disease, schizophrenia (41) |
| Aquaporin 7 | AQP7 | Allows movement of water, glycerol, and urea across cell membranesa | – |
| Armadillo Repeat Gene Deleted in Velocardiofacial Syndrome | ARVCF | Modulates neural cell-cell adhesion and migration (46) | Schizophrenia (46) |
| Brain-Derived Neurotrophic Factor | BDNF | Regulates cell survival, axonal outgrowth, dendritic growth, and synaptic plasticity (90) | Depression, bipolar disorder, schizophrenia, anxiety, autism, ADHD, substance abuse, eating disorders, Alzheimer’s disease (41) |
| Casein Kinase 1, Alpha 1 | CSNK1A1 | Suppressor of Wnt/β-catenin signalinga | Schizophrenia (91) |
| Casein Kinase 1, Alpha 1-like | CSNK1A1L | Involved in negative regulation of canonical Wnt signaling pathway and peptidyl-serine phosphorylationa | – |
| Catechol-O-Methyltransferase | COMT | Degrades dopamine and other catecholamines (92) | Schizophrenia (41) |
| Discs Large MAGUK Scaffold Protein 4 | DLG4 | Synapse structure and development (44) | Intellectual disability, epilepsy, autism spectrum disorder, schizophrenia (44,93) |
| Disrupted in Schizophrenia 1 | DISC1 | Neural migration, neurite outgrowth, and dendritic arborization (94) | Schizophrenia, bipolar disorder, autism, depression (41) |
| Erb-B2 Receptor Tyrosine Kinase 4 | ERBB4 | Role in neurodevelopment such as glial and neuronal migration, myelination, excitatory neuronal receptor expression, and the onset of puberty (47) | Schizophrenia, bipolar disorder (47) |
| Estrogen Receptor 1 | ESR1 | Mediates estrogen effects on synaptogenesis, growth factor production, and responses to oxidative stress (95) | Anxiety, depression, schizophrenia, Alzheimer’s disease (41) |
| Fatty Acid Desaturase 2 | FADS2 | Essential for neurogenesis, neurotransmission, and protection from oxidative stress (46) | Interact with early dietary exposures to influence childhood IQ (46) |
| F-Box and WD Repeat Domain Containing 11 | FBXW11 | Involved in ubiquitination and proteasomal degradation (96) | Autism spectrum disorder (96) |
| FK506-Binding Protein 5 | FKBP5 | Transcriptional regulation of the HPA axis (50) | Depression, PTSD (50) |
| Fragile X Mental Retardation, Autosomal Homolog 1 | FXR1 | Levels of FXR1 are important for parvalbumin interneurons (97) | Schizophrenia, bipolar disorder (98) |
| Insulin-like Growth Factor-Binding Protein 7 | IGFBP7 | Regulation of availability of IGFsa | Learning and memory (99) |
| Integrin Subunit Alpha 6 | ITGA6 | Involved in insulin-like growth factor 1 signaling (80) | Schizophrenia (100) |
| Kinectin 1 | KTN1 | Encodes the protein kinectin, a receptor that allows vesicle binding to kinesin and is involved in organelle transport (59) | ADHD (59) |
| Klotho | KL | Health and survival (43) | Cognition (43) |
| Malic Enzyme 1 | ME1 | Sex-specific gene regulation in the offspring, key regulator of a T2DM-specific gene expression network (101,102) | – |
| Mitogen-Activated Protein Kinase 10 | MAPK10 | Neuronal proliferation, differentiation, migration, and programmed cell deatha | Cognition (103) |
| Nemo-like Kinase | NLK | Positive effector of the noncanonical Wnt signaling pathway and negative regulator of the canonical Wnt/beta-catenin signaling pathwaya | – |
| Neuregulin 1 | NRG1 | Mediate cell-cell interactions in the brain and other organs, neuronal migration and specification, oligodendrocyte differentiation and myelination, regulation of acetylcholine, and expression of GABA receptors (104) | Schizophrenia, bipolar disorder (41) |
| Nuclear Factor of Activated T-Cells, Cytoplasmic 4 | NFATC4 | Hippocampal plasticity, axonal growth, neuronal survival, and apoptosis (105) | Spatial memorya |
| Oxytocin Receptor | OXTR | Receptor for oxytocin (51) | Depression, autism, eating disorder (51) |
| Perilipin 1 | PLIN1 | Regulates droplet formation in lipopolysaccharide-stimulated microglia (106) | – |
| Peroxisome Proliferator-Activated Receptor Gamma | PPARG | Regulator of adipocyte differentiationa | Schizophrenia (107) |
| Phospholipase C, Beta 2 | PLCB2 | Catalyzes the hydrolysis of phosphatidylinositol 4,5-bisphosphatea | Schizophrenia (108) |
| Proteasome Inhibitor Subunit 1 | PSMF1 | Inhibits activation of the 26S proteasome, a multicatalytic proteinase complex that may play a role in developmental axonal pruning and synaptic plasticity (109) | – |
| SMAD Family Member 3 | SMAD3 | Involved in regulating inflammatory responses (110) | Alzheimer’s disease (110), cognition (111) |
| Solute Carrier Family 27 (Fatty Acid Transporter), Member 1 | SLC27A1 | Involved in fatty acid transport across the blood-brain barrier (112) | – |
| Wingless-Type MMTV Integration Site Family, Member 2B | WNT2B | Regulation of cell growth and differentiationa | Bipolar disorder (113) |
| Wingless-Type MMTV Integration Site Family, Member 5A | WNT5A | Essential role in regulating developmental pathways during embryogenesisa | Schizophrenia (114), memory (115) |
ADHD, attention-deficit/hyperactivity disorder; GABA, gamma-aminobutyric acid; HPA, hypothalamic-pituitary-adrenal; IGF, insulin-like growth factor; PTSD, posttraumatic stress disorder; T2DM, type 2 diabetes mellitus.
GeneCards (116).
General strengths and limitations of GWASs have been reviewed in detail elsewhere (81). In terms of GWASs in infants and young children, the primary limitations of existing studies include their being 1) underpowered due to small samples, 2) mostly cross-sectional rather than longitudinal, 3) only focused on neonates and preterm infants, and 4) the fact that individuals of non-European ancestry constitute such a small proportion of the total samples.
Rigor and Reproducibility
When we consider the rigor and reproducibility of published imaging genetics studies in infants and young children, insufficient power and sample size are significant concerns. Sample sizes for early childhood imaging genetics studies are low, with a mean of 365 for candidate gene studies (median = 216), a mean of 225 for PRS-based studies (median = 168), and a mean of 344 for GWASs (median = 371.5). Consequently, existing studies are powered to detect variants with large effect sizes. It is likely that most variants impacting infant brain imaging phenotypes will explain between 0.1% and 1% of the variance, like other complex traits (82). Furthermore, small samples can produce unstable results, and homogenous sampling can generate statistical inferences that do not represent the overall population. Independent replication is essential to validate results and improve estimation of effects but is currently rare due to difficulties recruiting and scanning large groups of infants.
To improve rigor and reproducibility and fully understand how DNA variants influence brain development in infancy and early childhood across diverse populations and the implications for future research and clinical care, large longitudinal studies are needed. ORIGINs was founded to facilitate such work.
Organization for Imaging Genomics in Infancy
ORIGINs includes investigators from different centers around the world (16 sites, 19 cohorts, 5 countries) who are engaged in neuroimaging research in infancy and early childhood. Our goal is to determine how genetic and environmental factors influence development of brain morphometry, anatomical and functional connectivity, and cognitive and emotional function from birth to age 6 years. In 2020, we received National Institutes of Health funding to create the largest-ever imaging genomics dataset focused on infancy and early childhood. In subsequent sections, we briefly describe who will be included in this dataset, what is being measured, and our data analysis plans.
Participants
Participants will include approximately 6809 children (birth to 6 years of age) participating in neuroimaging studies of early brain development at the University of North Carolina Chapel Hill, University of California Irvine, Max Planck Institute for Human Cognitive and Brain Sciences, Rhode Island Hospital, Northwestern University, University of Denver, University of Rochester, Magee-Womens Hospital of the University of Pittsburgh Medical Center, University of Cape Town, Boston’s Children Hospital/Harvard University, University of Minnesota, University of Washington, Washington University in St. Louis, King’s College London, and National University of Singapore. Eight cohorts have completed initial data collection and 11 are actively scanning. We estimate that 51% of the sample will be female, 49% will be male, 0.05% will be American Indian, Alaska Natives, Native Hawaiians, or Pacific Islanders, 11% will be Asian, 20% will be Black, 6% will be more than one ancestry, and 62% will be White. Individuals identifying as Hispanic/Latinx are expected to make up 15% of the cohort.
Data Measurements
Demographic and Medical History
Health history and demographic information of participants were provided by parents or guardians and/or extracted from medical records. The information includes birth outcomes (gestational age, birth weight), sex, socioeconomic factors (maternal education, total family income), and family history of medical and neuropsychiatric disorders.
Genomic Data
Most participating sites have used/are using saliva samples for DNA extraction. Two cohorts (Drakenstein Child Health Study and GUSTO) used umbilical cord and venous blood specimens. To harmonize data across genotyping platforms, we will impute genomes to a common set of SNPs using the Michigan Imputation Server (83). See the Supplement for details on harmonization and genotyping platforms used by each cohort.
Behavioral Assessments
In a subset of participants (∼3800), we will examine 3 behavioral traits—impulsivity/distractibility, anxiety, and aggressive behavior—that can be reliably measured in very young children and are relevant to multiple psychiatric disorders. Behavioral traits will be measured using age-appropriate versions of the Child Behavior Checklist (84) and the Behavior Assessment System for Children, Second Edition (85,86).
Image Acquisition and Quality Control
3T Siemens scanners (Allegra, Tim Trio, Verio, Skyra, and Prisma) and comparable sequences (Tables S3–S6) were used with all cohorts except for the dHCP (developing Human Connectome Project) and GUSTO. dHCP data were acquired on a Philips Achieva, and newborn T2 structural MRI acquisition for GUSTO was on a 1.5T General Electric scanner. To ensure consistent processing across datasets with the same tools and appropriately standardized parameter settings, all structural, diffusion, and functional connectivity data will be processed at a central site. T1 and T2 structural MRI, diffusion tensor imaging, and resting-state functional MRI acquisition parameters, platform, and harmonization pipeline for each site are detailed in the Supplement.
Data Analysis Plan
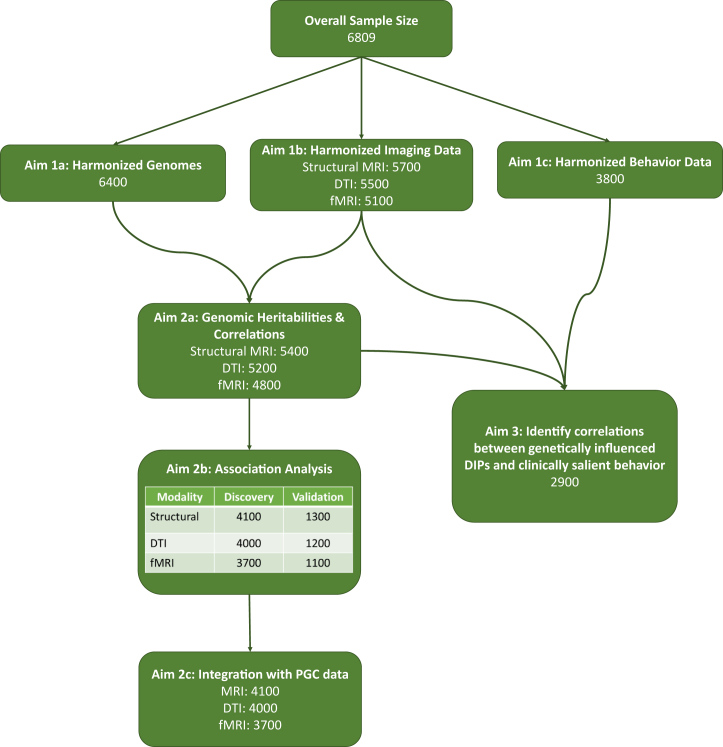
Extracted neuroimaging measures will be analyzed using nonlinear growth models. Growth model parameters, which we refer to as “developmental imaging phenotypes,” will be used to test effects of genetic variants on structural brain development and connectivity using a multivariate GWAS approach. Canonical correlation analysis will be used to identify association patterns between genetically influenced neurodevelopmental traits and clinically salient behaviors. The data analysis and data sharing plan is detailed in the Supplement. A schematic for data analysis is provided as Figure 3.
Figure 3.
Data analysis plan for the ORIGINs (Organization for Imaging Genomics in Infancy). DIP, developmental imaging phenotype; DTI, diffusion tensor imaging; fMRI, functional magnetic resonance imaging; PGC, Psychiatric Genomics Consortium.
Conclusions
Imaging genetics studies of infants and young children have provided evidence that variants associated with psychiatric disorders influence early neurodevelopment both independently and through interactions with environmental factors. In addition, GWASs of neonates and preterm infants have revealed new genes, variants, and molecular pathways implicated in brain development. However, most findings have not been independently replicated. Existing studies, regardless of design, are relatively small and do not encompass diverse ancestries. The ORIGINs initiative is addressing these limitations by creating and harmonizing the largest and most diverse imaging genomics dataset focused on infancy and early childhood to date. This dataset will help reveal how genetic risk for psychiatric disease manifests across infancy and early childhood, in terms of brain structure and function, and assist in early identification of at-risk individuals. Ultimately, identifying genes and molecular pathways associated with early neuroimaging phenotypes could lead to the development of novel prophylactics against complex psychiatric illness.
Acknowledgments and Disclosures
ORIGINs is supported by the National Institute of Mental Health (Grant No. R01MH123716 [to RK]). Cohort 1: The EBDS (Early Brain Development Research) study is supported by the National Institute of Mental Health and National Institute of Child Health and Development (Grant Nos. MH064065, MH070890, and HD053000 [to JHG]), with genotyping funded by the National Institute of Mental Health (Grant No. MH092335 [to RK]). Cohort 2: The GUSTO study was supported by the Singapore National Research Foundation under its Translational and Clinical Research Flagship Programme and administered by the Singapore Ministry of Health’s National Medical Research Council (Grant Nos. NMRC/TCR/004-NUS/2008 and NMRC/TCR/012-NUHS/2014 [to AQ]). Cohort 3: The IBIS study (Infant Brain Imaging Study) was supported by Grant Nos. R01-HD055741, P30-HD003110, and T32-HD040127 from the National Institutes of Health and the Simons Foundation (Grant No. 140209 [to JP]). Cohort 4: The University of North Carolina study was supported through the National Institute on Drug Abuse (Grant No. DA022446 [to KG]). Cohort 5: The University of California Irvine study was supported by the National Institutes of Health (Grant Nos. R01 HD-060628, R01 MH-105538, and UH3 OD-023349) and European Research Council (ERC) (Grant No. ERC-Stg 639766 [to CB]). Cohort 6: The Max Planck Institute for Human Cognitive and Brain Sciences was supported by the Jacobs Foundation and by the German Research Foundation Heisenberg (Grant No. 433758790 [to MAS]). Cohort 7: The University of North Carolina study was supported by the National Institute of Neurological Disease and Stroke (Grant No. NS055754 [to WL]). Cohort 8: The University of North Carolina study was supported by the National Institute of Neurological Disease and Stroke (Grant No. NS055754 [to MS]) and National Institute of Mental Health (Grant No. MH104330 [to RK and MS]). Cohort 9: The Developing Human Connectome Project was funded through a Synergy Grant by the ERC under the European Union’s Seventh Framework Programme (FP/2007-2013)/ERC Grant Agreement No. 319456 (PIs—A. David Edwards, Jo Hajnal, Daniel Rueckert, Stephen Smith). Cohort 10: The Baby Connectome Project was supported by the National Institute of Mental Health (Grant No. U01MH110274 [to WL]). Cohort 11: The DCHS study was funded by the Bill & Melinda Gates Foundation (Grant No. OPP 1017641). Additional support was provided by the Medical Research Council of South Africa. Imaging aspects of the cohort are additionally supported by the National Research Foundation, by the National Institute on Alcohol Abuse and Alcoholism via Grant Nos. R21AA023887 and R01AA026834-01, by the Collaborative Initiative on Fetal Alcohol Spectrum Disorders developmental Grant No. U24 AA014811, and by the United States Brain and Behavior Foundation Independent Investigator Grant No. 24467 and the Wellcome Trust through a Research Training Fellowship (203525/Z/16/Z [to DJS and KAD]). Cohort 12: The University of North Carolina study was supported by the National Institute on Drug Abuse (Grant No. DA379395 [to KG]). Cohort 13 (PHBP) and Cohort 15 (W2W) were funded by Ann & Robert H. Lurie Children’s Hospital of Chicago/Stanley Manne Children’s and by the National Institute of Mental Health (Grant Nos. R01MH121877 and R01MH107652 [to LSW]). Cohort 14: The Boston study was funded by the William Hearst Fund (Harvard University) and by the National Institute of Child Health and Development (Grant No. IH–NICHDR01 HD065762-10) and the Harvard Catalyst/National Institute of Health (Grant No. 5UL1RR025758 [to NG]). Cohort 16: The University of Denver study was supported by Grant Nos. MH109662 and HL155744 (to EPD and BLH) from the National Institute of Mental Health and the National Heart Lung and Blood Institute, respectively. Cohort 17: The University of Denver study was supported by the National Institute of Child Health and Development (Grant No. HD090068 [to PK]). Cohort 18: The Rhode Island Hospital study was supported by the National Institutes of Health (Grant No. UG3OD023313 to Sean C.L. Deoni). Cohort 19: The Rochester-Magee study was supported by the National Institutes of Health (Grant No. 5UG3OD023349 [to TGO’C]).
DS has received research grants and/or consultancy honoraria from Discovery Vitality, Johnson & Johnson, Kanna, L’Oreal, Lundbeck, Orion, Sanofi, Servier, Takeda, and Vistagen. PMT received a research grant from Biogen, Inc. WL is a consultant of and received travel support from Nestlé SA, Switzerland. All other authors report no biomedical financial interests or potential conflicts of interest.
Footnotes
Supplementary material cited in this article is available online at https://doi.org/10.1016/j.biopsych.2023.01.013.
Supplementary Material
References
- 1.Strike L.T., Couvy-Duchesne B., Hansell N.K., Cuellar-Partida G., Medland S.E., Wright M.J. Genetics and brain morphology. Neuropsychol Rev. 2015;25:63–96. doi: 10.1007/s11065-015-9281-1. [DOI] [PubMed] [Google Scholar]
- 2.Gao W., Grewen K., Knickmeyer R.C., Qiu A., Salzwedel A., Lin W., Gilmore J.H. A review on neuroimaging studies of genetic and environmental influences on early brain development. Neuroimage. 2019;185:802–812. doi: 10.1016/j.neuroimage.2018.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson P.M., Jahanshad N., Ching C.R.K., Salminen L.E., Thomopoulos S.I., Bright J., et al. ENIGMA and global neuroscience: A decade of large-scale studies of the brain in health and disease across more than 40 countries. Transl Psychiatry. 2020;10:100. doi: 10.1038/s41398-020-0705-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fatemi S.H., Folsom T.D. The neurodevelopmental hypothesis of schizophrenia, revisited. Schizophr Bull. 2009;35:528–548. doi: 10.1093/schbul/sbn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J.A., Peñagarikano O., Belgard T.G., Swarup V., Geschwind D.H. The emerging picture of autism spectrum disorder: Genetics and pathology. Annu Rev Pathol. 2015;10:111–144. doi: 10.1146/annurev-pathol-012414-040405. [DOI] [PubMed] [Google Scholar]
- 6.Douet V., Chang L., Cloak C., Ernst T. Genetic influences on brain developmental trajectories on neuroimaging studies: From infancy to young adulthood. Brain Imaging Behav. 2014;8:234–250. doi: 10.1007/s11682-013-9260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel Y., Shin J., Abé C., Agartz I., Alloza C., Alnæs D., et al. Virtual ontogeny of cortical growth preceding mental illness. Biol Psychiatry. 2022;92:299–313. doi: 10.1016/j.biopsych.2022.02.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papageorghiou A.T., Sarris I., Ioannou C., Todros T., Carvalho M., Pilu G., et al. Ultrasound methodology used to construct the fetal growth standards in the INTERGROWTH-21st Project. BJOG. 2013;120(suppl 2):27–32. doi: 10.1111/1471-0528.12313. [DOI] [PubMed] [Google Scholar]
- 9.Lamballais S., Jansen P.R., Labrecque J.A., Ikram M.A., White T. Genetic scores for adult subcortical volumes associate with subcortical volumes during infancy and childhood. Hum Brain Mapp. 2021;42:1583–1593. doi: 10.1002/hbm.25292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sparrow S., Manning J.R., Cartier J., Anblagan D., Bastin M.E., Piyasena C., et al. Epigenomic profiling of preterm infants reveals DNA methylation differences at sites associated with neural function. Transl Psychiatry. 2016;6 doi: 10.1038/tp.2015.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fumagalli M., Provenzi L., de Carli P., Dessimone F., Sirgiovanni I., Giorda R., et al. From early stress to 12-month development in very preterm infants: Preliminary findings on epigenetic mechanisms and brain growth. PLoS One. 2018;13 doi: 10.1371/journal.pone.0190602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wheater E.N.W., Galdi P., McCartney D.L., Blesa M., Sullivan G., Stoye D.Q., et al. DNA methylation in relation to gestational age and brain dysmaturation in preterm infants. Brain Commun. 2022;4:fcac056. doi: 10.1093/braincomms/fcac056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hüls A., Wedderburn C.J., Groenewold N.A., Gladish N., Jones M.J., Koen N., et al. Newborn differential DNA methylation and subcortical brain volumes as early signs of severe neurodevelopmental delay in a South African Birth Cohort Study. World J Biol Psychiatry. 2022;23:601–612. doi: 10.1080/15622975.2021.2016955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilmore J.H., Schmitt J.E., Knickmeyer R.C., Smith J.K., Lin W., Styner M., et al. Genetic and environmental contributions to neonatal brain structure: A twin study. Hum Brain Mapp. 2010;31:1174–1182. doi: 10.1002/hbm.20926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smit D.J.A., Luciano M., Bartels M., van Beijsterveldt C.E.M., Wright M.J., Hansell N.K., et al. Heritability of head size in Dutch and Australian twin families at ages 0–50 years. Twin Res Hum Genet. 2010;13:370–380. doi: 10.1375/twin.13.4.370. [DOI] [PubMed] [Google Scholar]
- 16.Gilmore J.H., Knickmeyer R.C., Gao W. Imaging structural and functional brain development in early childhood. Nat Rev Neurosci. 2018;19:123–137. doi: 10.1038/nrn.2018.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jansen A.G., Mous S.E., White T., Posthuma D., Polderman T.J.C. What twin studies tell us about the heritability of brain development, morphology, and function: A review. Neuropsychol Rev. 2015;25:27–46. doi: 10.1007/s11065-015-9278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brouwer R.M., Mandl R.C.W., Schnack H.G., van Soelen I.L.C., van Baal G.C., Peper J.S., et al. White matter development in early puberty: A longitudinal volumetric and diffusion tensor imaging twin study. PLoS One. 2012;7 doi: 10.1371/journal.pone.0032316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jha S.C., Xia K., Schmitt J.E., Ahn M., Girault J.B., Murphy V.A., et al. Genetic influences on neonatal cortical thickness and surface area. Hum Brain Mapp. 2018;39:4998–5013. doi: 10.1002/hbm.24340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panizzon M.S., Fennema-Notestine C., Eyler L.T., Jernigan T.L., Prom-Wormley E., Neale M., et al. Distinct genetic influences on cortical surface area and cortical thickness. Cereb Cortex. 2009;19:2728–2735. doi: 10.1093/cercor/bhp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen C.H., Fiecas M., Gutiérrez E.D., Panizzon M.S., Eyler L.T., Vuoksimaa E., et al. Genetic topography of brain morphology. Proc Natl Acad Sci USA. 2013;110:17089–17094. doi: 10.1073/pnas.1308091110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winkler A.M., Kochunov P., Blangero J., Almasy L., Zilles K., Fox P.T., et al. Cortical thickness or grey matter volume? The importance of selecting the phenotype for imaging genetics studies. Neuroimage. 2010;53:1135–1146. doi: 10.1016/j.neuroimage.2009.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geng X., Prom-Wormley E.C., Perez J., Kubarych T., Styner M., Lin W., et al. White matter heritability using diffusion tensor imaging in neonatal brains. Twin Res Hum Genet. 2012;15:336–350. doi: 10.1017/thg.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee S.J., Steiner R.J., Luo S., Neale M.C., Styner M., Zhu H., Gilmore J.H. Quantitative tract-based white matter heritability in twin neonates. Neuroimage. 2015;111:123–135. doi: 10.1016/j.neuroimage.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vuoksimaa E., Panizzon M.S., Hagler D.J., Hatton S.N., Fennema-Notestine C., Rinker D., et al. Heritability of white matter microstructure in late middle age: A twin study of tract-based fractional anisotropy and absolute diffusivity indices. Hum Brain Mapp. 2017;38:2026–2036. doi: 10.1002/hbm.23502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J., Xia K., Ahn M., Jha S.C., Blanchett R., Crowley J.J., et al. Genome-wide association analysis of neonatal white matter microstructure. Cereb Cortex. 2021;31:933–948. doi: 10.1093/cercor/bhaa266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pletikos M., Sousa A.M.M., Sedmak G., Meyer K.A., Zhu Y., Cheng F., et al. Temporal specification and bilaterality of human neocortical topographic gene expression. Neuron. 2014;81:321–332. doi: 10.1016/j.neuron.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang H.J., Kawasawa Y.I., Cheng F., Zhu Y., Xu X., Li M., et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao W., Elton A., Zhu H., Alcauter S., Smith J.K., Gilmore J.H., Lin W. Intersubject variability of and genetic effects on the brain’s functional connectivity during infancy. J Neurosci. 2014;34:11288–11296. doi: 10.1523/JNEUROSCI.5072-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu J., Yin X., Ge H., Han Y., Pang Z., Liu B., et al. Heritability of the effective connectivity in the resting-state default mode network. Cereb Cortex. 2017;27:5626–5634. doi: 10.1093/cercor/bhw332. [DOI] [PubMed] [Google Scholar]
- 31.Glahn D.C., Winkler A.M., Kochunov P., Almasy L., Duggirala R., Carless M.A., et al. Genetic control over the resting brain. Proc Natl Acad Sci USA. 2010;107:1223–1228. doi: 10.1073/pnas.0909969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu Y., Ma Z., Hamilton C., Liang Z., Hou X., Ma X., et al. Genetic influences on resting-state functional networks: A twin study. Hum Brain Mapp. 2015;36:3959–3972. doi: 10.1002/hbm.22890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blanchett R., Chen Y., Aguate F., Xia K., Cornea E., Burt S.A., et al. Genetic and environmental factors influencing neonatal resting-state functional connectivity [published online Oct 3] Cereb Cortex. 2022 doi: 10.1093/cercor/bhac383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takagi Y., Okada N., Ando S., Yahata N., Morita K., Koshiyama D., et al. Intergenerational transmission of the patterns of functional and structural brain networks. iScience. 2021;24 doi: 10.1016/j.isci.2021.102708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahtam B., Turesky T.K., Zöllei L., Standish J., Grant P.E., Gaab N., Im K. Intergenerational transmission of cortical sulcal patterns from mothers to their children. Cereb Cortex. 2021;31:1888–1897. doi: 10.1093/cercor/bhaa328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fehlbaum L.V., Peters L., Dimanova P., Roell M., Borbás R., Ansari D., Raschle N.M. Mother–child similarity in brain morphology: A comparison of structural characteristics of the brain’s reading network. Dev Cogn Neurosci. 2022;53 doi: 10.1016/j.dcn.2022.101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bouchard T.J. The Wilson effect: The increase in heritability of IQ with age. Twin Res Hum Genet. 2013;16:923–930. doi: 10.1017/thg.2013.54. [DOI] [PubMed] [Google Scholar]
- 38.Martin N.G., Eaves L.J., Heath A.C., Jardine R., Feingold L.M., Eysenck H.J. Transmission of social attitudes. Proc Natl Acad Sci USA. 1986;83:4364–4368. doi: 10.1073/pnas.83.12.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brouwer R.M., Panizzon M.S., Glahn D.C., Hibar D.P., Hua X., Jahanshad N., et al. Genetic influences on individual differences in longitudinal changes in global and subcortical brain volumes: Results of the ENIGMA plasticity working group. Hum Brain Mapp. 2017;38:4444–4458. doi: 10.1002/hbm.23672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brouwer R.M., Klein M., Grasby K.L., Schnack H.G., Jahanshad N., Teeuw J., et al. Genetic variants associated with longitudinal changes in brain structure across the lifespan. Nat Neurosci. 2022;25:421–432. doi: 10.1038/s41593-022-01042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knickmeyer R.C., Wang J., Zhu H., Geng X., Woolson S., Hamer R.M., et al. Common variants in psychiatric risk genes predict brain structure at birth. Cereb Cortex. 2014;24:1230–1246. doi: 10.1093/cercor/bhs401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawasaki Y., Oishi K., Hernandez A., Ernst T., Wu D., Otsuka Y., et al. Brain-derived neurotrophic factor Val66Met variant on brain volumes in infants. Brain Struct Funct. 2021;226:919–925. doi: 10.1007/s00429-020-02207-2. [DOI] [PubMed] [Google Scholar]
- 43.de Vries C.F., Staff R.T., Noble K.G., Muetzel R.L., Vernooij M.W., White T., et al. Klotho gene polymorphism, brain structure and cognition in early-life development. Brain Imaging Behav. 2020;14:213–225. doi: 10.1007/s11682-018-9990-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krishnan M.L., van Steenwinckel J., Schang A.L., Yan J., Arnadottir J., le Charpentier T., et al. Integrative genomics of microglia implicates DLG4 (PSD95) in the white matter development of preterm infant. Nat Commun. 2017;8:428. doi: 10.1038/s41467-017-00422-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Steenwinckel J., Schang A.L., Krishnan M.L., Degos V., Delahaye-Duriez A., Bokobza C., et al. Decreased microglial Wnt/β-catenin signalling drives microglial pro-inflammatory activation in the developing brain. Brain. 2019;142:3806–3833. doi: 10.1093/brain/awz319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boardman J.P., Walley A., Ball G., Takousis P., Krishnan M.L., Hughes-Carre L., et al. Common genetic variants and risk of brain injury after preterm birth. Pediatrics. 2014;133:e1655–e1663. doi: 10.1542/peds.2013-3011. [DOI] [PubMed] [Google Scholar]
- 47.Douet V., Chang L., Lee K., Ernst T., Pediatric Imaging. Neurocognition. and Genetics (PING) Consortium ERBB4 polymorphism and family history of psychiatric disorders on age-related cortical changes in healthy children. Brain Imaging Behav. 2015;9:128–140. doi: 10.1007/s11682-015-9363-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qiu A., Tuan T.A., Ong M.L., Li Y., Chen H., Rifkin-Graboi A., et al. COMT haplotypes modulate associations of antenatal maternal anxiety and neonatal cortical morphology. Am J Psychiatry. 2015;172:163–172. doi: 10.1176/appi.ajp.2014.14030313. [DOI] [PubMed] [Google Scholar]
- 49.Chen L., Pan H., Tuan T.A., Teh A.L., Macisaac J.L., Mah S.M., et al. Brain-derived neurotrophic factor (BDNF) Val66Met polymorphism influences the association of the methylome with maternal anxiety and neonatal brain volumes. Dev Psychopathol. 2015;27:137–150. doi: 10.1017/S0954579414001357. [DOI] [PubMed] [Google Scholar]
- 50.Wang C., Shen M., Guillaume B., Chong Y.S., Chen H., Fortier MV v., et al. FKBP5 moderates the association between antenatal maternal depressive symptoms and neonatal brain morphology. Neuropsychopharmacology. 2018;43:564–570. doi: 10.1038/npp.2017.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Acosta H., Tuulari J.J., Kantojärvi K., Lewis J.D., Hashempour N., Scheinin N.M., et al. A variation in the infant oxytocin receptor gene modulates infant hippocampal volumes in association with sex and prenatal maternal anxiety. Psychiatry Res Neuroimaging. 2021;307 doi: 10.1016/j.pscychresns.2020.111207. [DOI] [PubMed] [Google Scholar]
- 52.Wolfe D., Dudek S., Ritchie M.D., Pendergrass S.A. Visualizing genomic information across chromosomes with PhenoGram. BioData Min. 2013;6:18. doi: 10.1186/1756-0381-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bogdan R., Salmeron B.J., Carey C.E., Agrawal A., Calhoun V.D., Garavan H., et al. Imaging genetics and genomics in psychiatry: A critical review of progress and potential. Biol Psychiatry. 2017;82:165–175. doi: 10.1016/j.biopsych.2016.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shaw P., Lerch J.P., Pruessner J.C., Taylor K.N., Rose A.B., Greenstein D., et al. Cortical morphology in children and adolescents with different apolipoprotein E gene polymorphisms: An observational study. Lancet Neurol. 2007;6:494–500. doi: 10.1016/S1474-4422(07)70106-0. [DOI] [PubMed] [Google Scholar]
- 55.Burggren A.C., Zeineh M.M., Ekstrom A.D., Braskie M.N., Thompson P.M., Small G.W., Bookheimer S.Y. Reduced cortical thickness in hippocampal subregions among cognitively normal apolipoprotein E e4 carriers. Neuroimage. 2008;41:1177–1183. doi: 10.1016/j.neuroimage.2008.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dean D.C., Jerskey B.A., Chen K., Protas H., Thiyyagura P., Roontiva A., et al. Brain differences in infants at differential genetic risk for late-onset Alzheimer disease: A cross-sectional imaging study. JAMA Neurol. 2014;71:11–22. doi: 10.1001/jamaneurol.2013.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Remer J., Dean D.C., Chen K., Reiman R.A., Huentelman M.J., Reiman E.M., Deoni S.C.L. Longitudinal white matter and cognitive development in pediatric carriers of the apolipoprotein ε4 allele. Neuroimage. 2020;222 doi: 10.1016/j.neuroimage.2020.117243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang L., Douet V., Bloss C., Lee K., Pritchett A., Jernigan T.L., et al. Gray matter maturation and cognition in children with different APOE ε genotypes. Neurology. 2016;87:585–594. doi: 10.1212/WNL.0000000000002939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cullen H., Dimitrakopoulou K., Patel H., Curtis C., Batalle D., Gale-Grant O., et al. Common genetic variation important in early subcortical brain development. medRxiv. 2022 doi: 10.1101/2022.08.11.22278677. [DOI] [Google Scholar]
- 60.Patnala R., Clements J., Batra J. Candidate gene association studies: A comprehensive guide to useful in silico tools. BMC Genet. 2013;14:39. doi: 10.1186/1471-2156-14-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.David S. A current guide to candidate gene association studies. Trends Genet. 2021;37:1056–1059. doi: 10.1016/j.tig.2021.07.009. [DOI] [PubMed] [Google Scholar]
- 62.MacArthur D.G., Manolio T.A., Dimmock D.P., Rehm H.L., Shendure J., Abecasis G.R., et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Plomin R., Haworth C.M., Davis O.S. Common disorders are quantitative traits. Nat Rev Genet. 2009;10:872–878. doi: 10.1038/nrg2670. [DOI] [PubMed] [Google Scholar]
- 64.Sullivan P.F., Geschwind D.H. Defining the genetic, genomic, cellular, and diagnostic architectures of psychiatric disorders. Cell. 2019;177:162–183. doi: 10.1016/j.cell.2019.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xia K., Zhang J., Ahn M., Jha S., Crowley J.J., Szatkiewicz J., et al. Genome-wide association analysis identifies common variants influencing infant brain volumes. Transl Psychiatry. 2017;7:e1188. doi: 10.1038/tp.2017.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Le H., Dimitrakopoulou K., Patel H., Curtis C., Cordero-Grande L., Hajnal J., et al. Effect of Schizophrenia common variants on infant brain volumes: cross-sectional study in 207 term neonates in developing human connectome project. Research Square. 2022 doi: 10.21203/rs.3.rs-1950696/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khundrakpam B., Vainik U., Gong J., Al-Sharif N., Bhutani N., Kiar G., et al. Neural correlates of polygenic risk score for autism spectrum disorders in general population. Brain Commun. 2020;2 doi: 10.1093/braincomms/fcaa092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cullen H., Krishnan M.L., Selzam S., Ball G., Visconti A., Saxena A., et al. Polygenic risk for neuropsychiatric disease and vulnerability to abnormal deep grey matter development. Sci Rep. 2019;9:1976. doi: 10.1038/s41598-019-38957-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ursini G., Punzi G., Langworthy B.W., Chen Q., Xia K., Cornea E.A., et al. Placental genomic risk scores and early neurodevelopmental outcomes. Proc Natl Acad Sci USA. 2021;118 doi: 10.1073/pnas.2019789118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qiu A., Shen M., Buss C., Chong Y.S., Kwek K., Saw S.M., et al. Effects of antenatal maternal depressive symptoms and socio-economic status on neonatal brain development are modulated by genetic risk. Cereb Cortex. 2017;27:3080–3092. doi: 10.1093/cercor/bhx065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Acosta H., Kantojärvi K., Hashempour N., Pelto J., Scheinin N.M., Lehtola S.J., et al. Partial support for an interaction between a polygenic risk score for major depressive disorder and prenatal maternal depressive symptoms on infant right amygdalar volumes. Cereb Cortex. 2020;30:6121–6134. doi: 10.1093/cercor/bhaa158. [DOI] [PubMed] [Google Scholar]
- 72.Acosta H., Kantojärvi K., Tuulari J.J., Lewis J.D., Hashempour N., Scheinin N.M., et al. Sex-specific association between infant caudate volumes and a polygenic risk score for major depressive disorder. J Neurosci Res. 2020;98:2529–2540. doi: 10.1002/jnr.24722. [DOI] [PubMed] [Google Scholar]
- 73.Alex A.M., Ruvio T., Xia K., Jha S.C., Girault J.B., Wang L., et al. Influence of gonadal steroids on cortical surface area in infancy. Cereb Cortex. 2022;32:3206–3223. doi: 10.1093/cercor/bhab410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morgunova A., Pokhvisneva I., Nolvi S., Entringer S., Wadhwa P., Gilmore J., et al. DCC gene network in the prefrontal cortex is associated with total brain volume in childhood. J Psychiatry Neurosci. 2021;46:E154–E163. doi: 10.1503/jpn.200081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu Y., Zhang H., Wang C., Broekman B.F.P., Chong Y.S., Shek L.P., et al. Inflammatory modulation of the associations between prenatal maternal depression and neonatal brain. Neuropsychopharmacology. 2021;46:470–477. doi: 10.1038/s41386-020-0774-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Qiu A., Zhang H., Wang C., Chong Y.S., Shek L.P., Gluckman P.D., et al. Canonical TGF-β signaling regulates the relationship between prenatal maternal depression and amygdala development in early life. Transl Psychiatry. 2021;11:170. doi: 10.1038/s41398-021-01292-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Adams H.H.H., Hibar D.P., Chouraki V., Stein J.L., Nyquist P.A., Rentería M.E., et al. Novel genetic loci underlying human intracranial volume identified through genome-wide association. Nat Neurosci. 2016;19:1569–1582. doi: 10.1038/nn.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martin A.R., Gignoux C.R., Walters R.K., Wojcik G.L., Neale B.M., Gravel S., et al. Human demographic history impacts genetic risk prediction across diverse populations. Am J Hum Genet. 2017;100:635–649. doi: 10.1016/j.ajhg.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Krishnan M.L., Wang Z., Silver M., Boardman J.P., Ball G., Counsell S.J., et al. Possible relationship between common genetic variation and white matter development in a pilot study of preterm infants. Brain Behav. 2016;6 doi: 10.1002/brb3.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Krishnan M.L., Wang Z., Aljabar P., Ball G., Mirza G., Saxena A., et al. Machine learning shows association between genetic variability in PPARG and cerebral connectivity in preterm infants. Proc Natl Acad Sci USA. 2017;114:13744–13749. doi: 10.1073/pnas.1704907114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tam V., Patel N., Turcotte M., Bossé Y., Paré G., Meyre D. Benefits and limitations of genome-wide association studies. Nat Rev Genet. 2019;20:467–484. doi: 10.1038/s41576-019-0127-1. [DOI] [PubMed] [Google Scholar]
- 82.Park J.H., Gail M.H., Weinberg C.R., Carroll R.J., Chung C.C., Wang Z., et al. Distribution of allele frequencies and effect sizes and their interrelationships for common genetic susceptibility variants. Proc Natl Acad Sci USA. 2011;108:18026–18031. doi: 10.1073/pnas.1114759108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Das S., Forer L., Schönherr S., Sidore C., Locke A.E., Kwong A., et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Achenbach T.M. In: Encyclopedia of Clinical Neuropsychology. Kreutzer J.S., DeLuca J., Caplan B., editors. Springer; New York: 2011. Child Behavior Checklist. [Google Scholar]
- 85.Reynolds C.R., Kamphaus R.W. 2nd ed. Pearson Assessments; New York: 2004. Behavior Assessment System for Children. [Google Scholar]
- 86.Reynolds C.R., Kamphaus R.W. BASC-2 behavior assessment system for children. Manual Supplement for the Self-Re`port of Personality—Interview. 2nd ed. AGS Publishing; Circle Pines, MN: 2005. [Google Scholar]
- 87.Houdou S., Takashima S., Suzuki Y. Immunohistochemical expression of peroxisomal enzymes in developing human brain. Mol Chem Neuropathol. 1993;19:235–248. doi: 10.1007/BF03160002. [DOI] [PubMed] [Google Scholar]
- 88.Luo R., Fan Y., Yang J., Ye M., Zhang D.F., Guo K., et al. A novel missense variant in ACAA1 contributes to early-onset Alzheimer’s disease, impairs lysosomal function, and facilitates amyloid-β pathology and cognitive decline. Signal Transduct Target Ther. 2021;6:325. doi: 10.1038/s41392-021-00748-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vance J.E., Hayashi H. Formation and function of apolipoprotein E-containing lipoproteins in the nervous system. Biochim Biophys Acta. 2010;1801:806–818. doi: 10.1016/j.bbalip.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 90.Reichardt L.F. Neurotrophin-regulated signalling pathways. Philos Trans R Soc Lond B Biol Sci. 2006;361:1545–1564. doi: 10.1098/rstb.2006.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Uellendahl-Werth F., Maj C., Borisov O., Juzenas S., Wacker E.M., Jørgensen I.F., et al. Cross-tissue transcriptome-wide association studies identify susceptibility genes shared between schizophrenia and inflammatory bowel disease. Commun Biol. 2022;5:80. doi: 10.1038/s42003-022-03031-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yavich L., Forsberg M.M., Karayiorgou M., Gogos J.A., Männistö P.T. Site-specific role of catechol-O-methyltransferase in dopamine overflow within prefrontal cortex and dorsal striatum. J Neurosci. 2007;27:10196–10209. doi: 10.1523/JNEUROSCI.0665-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hormozdiari F., Penn O., Borenstein E., Eichler E.E. The discovery of integrated gene networks for autism and related disorders. Genome Res. 2015;25:142–154. doi: 10.1101/gr.178855.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Porteous D.J., Millar J.K., Brandon N.J., Sawa A. DISC1 at 10: Connecting psychiatric genetics and neuroscience. Trends Mol Med. 2011;17:699–706. doi: 10.1016/j.molmed.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sundermann E.E., Maki P.M., Bishop J.R. A review of estrogen receptor α gene (ESR1) polymorphisms, mood, and cognition. Menopause. 2010;17:874–886. doi: 10.1097/gme.0b013e3181df4a19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Holt R.J., Young R.M., Crespo B., Ceroni F., Curry C.J., Bellacchio E., et al. De novo missense variants in FBXW11 cause diverse developmental phenotypes including brain, eye, and digit anomalies. Am J Hum Genet. 2019;105:640–657. doi: 10.1016/j.ajhg.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shen M., Guo Y., Dong Q., Gao Y., Stockton M.E., Li M., et al. FXR1 regulation of parvalbumin interneurons in the prefrontal cortex is critical for schizophrenia-like behaviors. Mol Psychiatry. 2021;26:6845–6867. doi: 10.1038/s41380-021-01096-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Khlghatyan J., Evstratova A., Chamberland S., Marakhovskaia A., Bahremand A., Toth K., Beaulieu J.M. Mental illnesses-associated Fxr1 and its negative regulator Gsk3β are modulators of anxiety and glutamatergic neurotransmission. Front Mol Neurosci. 2018;11:119. doi: 10.3389/fnmol.2018.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Agis-Balboa R.C., Arcos-Diaz D., Wittnam J., Govindarajan N., Blom K., Burkhardt S., et al. A hippocampal insulin-growth factor 2 pathway regulates the extinction of fear memories. EMBO J. 2011;30:4071–4083. doi: 10.1038/emboj.2011.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ambalavanan A., Girard S.L., Ahn K., Zhou S., Dionne-Laporte A., Spiegelman D., et al. De novo variants in sporadic cases of childhood onset schizophrenia. Eur J Hum Genet. 2016;24:944–948. doi: 10.1038/ejhg.2015.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dahlhoff M., Pfister S., Blutke A., Rozman J., Klingenspor M., Deutsch M.J., et al. Peri-conceptional obesogenic exposure induces sex-specific programming of disease susceptibilities in adult mouse offspring. Biochim Biophys Acta. 2014;1842:304–317. doi: 10.1016/j.bbadis.2013.11.021. [DOI] [PubMed] [Google Scholar]
- 102.Zhong H., Beaulaurier J., Lum P.Y., Molony C., Yang X., MacNeil D.J., et al. Liver and adipose expression associated SNPs are enriched for association to type 2 diabetes. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kunde S.A., Rademacher N., Tzschach A., Wiedersberg E., Ullmann R., Kalscheuer V.M., Shoichet S.A. Characterisation of de novo MAPK10/JNK3 truncation mutations associated with cognitive disorders in two unrelated patients. Hum Genet. 2013;132:461–471. doi: 10.1007/s00439-012-1260-5. [DOI] [PubMed] [Google Scholar]
- 104.Mei L., Xiong W.C. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nat Rev Neurosci. 2008;9:437–452. doi: 10.1038/nrn2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bradley K.C., Groth R.D., Mermelstein P.G. Immunolocalization of NFATc4 in the adult mouse brain. J Neurosci Res. 2005;82:762–770. doi: 10.1002/jnr.20695. [DOI] [PubMed] [Google Scholar]
- 106.Khatchadourian A., Bourque S.D., Richard V.R., Titorenko V.I., Maysinger D. Dynamics and regulation of lipid droplet formation in lipopolysaccharide (LPS)-stimulated microglia. Biochim Biophys Acta. 2012;1821:607–617. doi: 10.1016/j.bbalip.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 107.Sargazi S., Mirani Sargazi F., Moudi M., Heidari Nia M., Saravani R., Mirinejad S., et al. Impact of proliferator-activated receptor γ gene polymorphisms on risk of schizophrenia: A case-control study and computational analyses. Iran J Psychiatry. 2020;15:286–296. doi: 10.18502/ijps.v15i4.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu Y., Qu H.Q., Chang X., Tian L., Glessner J., Sleiman P.A.M., Hakonarson H. Expansion of schizophrenia gene network knowledge using machine learning selected signals from dorsolateral prefrontal cortex and amygdala RNA-seq data. Front Psychiatry. 2022;13 doi: 10.3389/fpsyt.2022.797329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hegde A.N., Upadhya S.C. Role of ubiquitin-proteasome-mediated proteolysis in nervous system disease. Biochim Biophys Acta. 2011;1809:128–140. doi: 10.1016/j.bbagrm.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xu L., Pan C.L., Wu X.H., Song J.J., Meng P., Li L., et al. Inhibition of Smad3 in macrophages promotes Aβ efflux from the brain and thereby ameliorates Alzheimer’s pathology. Brain Behav Immun. 2021;95:154–167. doi: 10.1016/j.bbi.2021.03.013. [DOI] [PubMed] [Google Scholar]
- 111.Muñoz M.D., de la Fuente N., Sánchez-capelo A. TGF-β/Smad3 signalling modulates GABA neurotransmission: Implications in Parkinson’s disease. Int J Mol Sci. 2020;21:590. doi: 10.3390/ijms21020590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mitchell R.W., On N.H., del Bigio M.R., Miller D.W., Hatch G.M. Fatty acid transport protein expression in human brain and potential role in fatty acid transport across human brain microvessel endothelial cells. J Neurochem. 2011;117:735–746. doi: 10.1111/j.1471-4159.2011.07245.x. [DOI] [PubMed] [Google Scholar]
- 113.Zandi P.P., Belmonte P.L., Willour V.L., Goes F.S., Badner J.A., Simpson S.G., et al. Association study of Wnt signaling pathway genes in bipolar disorder. Arch Gen Psychiatry. 2008;65:785–793. doi: 10.1001/archpsyc.65.7.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Evgrafov O.V., Armoskus C., Wrobel B.B., Spitsyna V.N., Souaiaia T., Herstein J.S., et al. Gene expression in patient-derived neural progenitors implicates WNT5A signaling in the etiology of schizophrenia. Biol Psychiatry. 2020;88:236–247. doi: 10.1016/j.biopsych.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chen C.M., Orefice L.L., Chiu S.L., LeGates T.A., Hattar S., Huganir R.L., et al. Wnt5a is essential for hippocampal dendritic maintenance and spatial learning and memory in adult mice. Proc Natl Acad Sci USA. 2017;114:E619–E628. doi: 10.1073/pnas.1615792114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Safran M., Rosen N., Twik M., BarShir R., Stein T.I., Dahary D., et al. In: Practical Guide to Life Science Databases. Abugessaisa I., Kasukawa T., editors. Springer; Singapore: 2021. The GeneCards suite; pp. 27–56. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.