Abstract
We present here an innovative modular and outsourced model of drug research and development for microRNA oligonucleotide therapeutics (miRNA ONTs). This model is being implemented by a biotechnology company, namely AptamiR Therapeutics, in collaboration with Centers of Excellence in Academic Institutions. Our aim is to develop safe, effective and convenient active targeting miRNA ONT agents for the metabolic pandemic of obesity and metabolic-associated fatty liver disease (MAFLD), as well as deadly ovarian cancer.
Keywords: microRNAs, oligonucleotide therapeutics, active targeted delivery, obesity, diabetes, MAFLD, ovarian cancer
1. Introduction
MicroRNAs (miRNAs) are highly conserved small non-coding RNA molecules that post-transcriptionally regulate gene functions through direct degradation of target mRNAs and/or translational repression. The total number of validated human miRNAs was recently estimated to be around 2300 [1,2]. miRNAs are stable, often display cell-type specificity, can be easily quantified from tissues and body fluids, and play a major role in exosomal cell-to-cell communications [3,4,5,6,7].
miRNAs are convenient diagnostic, prognostic and therapeutics markers of various human diseases (e.g., cardiovascular, metabolic, neurologic, infectious) [8,9,10,11,12] and cancers [13,14,15,16]. In addition, miRNAs are increasingly recognized as modulators of disease pathogeneses and are consequently considered bona fide molecular therapeutic targets [17]. Due to miRNAs’ distinct expression patterns and their ability to target numerous transcripts via a one-to-many relationship, miRNA modulators present a unique opportunity to alter the expression of proteins that cannot be feasibly targeted by small-molecule drug-based approaches [18].
Nucleic acid therapeutics (NATs)/oligonucleotide therapeutics (ONTs) represent a new and growing area of drug discovery and development [19,20]. As of 19 December 2022, ONTs (mainly antisense oligonucleotides (ASOs) and small interfering RNAs (siRNAs)) have been initially approved by regulatory authorities in the US and/or the EU. To facilitate R&D process development of NATs/ONTs, the FDA published in June 2022 a draft guidance titled Clinical Pharmacology Considerations for the Development of Oligonucleotide Therapeutics (https://www.fda.gov/regulatory-information/search-fda-guidance-documents/clinical-pharmacology-considerations-development-oligonucleotide-therapeutics (accessed on 22 June 2022)).
Numerous clinical trials are currently evaluating ONTs for rare disease and cancer treatments [21,22]. Some of these should be approved within the coming years. Moreover, several ONTs have been developed for common prevalent diseases, such as cardiovascular and metabolic disorders. Inclisiran (Leqvio®) is an siRNA that targets PCSK9 approved and marketed for the treatment of hypercholesterolemia [23]. Pelacarsen is an ASO that targets lipoprotein(a), a risk factor for cardiovascular diseases (NCT04023552, ClinicalTrials.gov). Fesomersen is a Factor XI ASO, designed to prevent thrombosis ((NCT04534114). ON449 (AZD8233) is an investigational ASO designed to reduce blood cholesterol levels by targeting PCSK9 (NCT04641299). Zilebesiran (ALN-AGT) is an siRNA that targets liver-expressed angiotensinogen to reduce blood pressure in hypertensive patients; it is also currently undergoing phase 2 clinical trials (NCT04936035 and NCT05103332). Additionally, 15 or more additional regulatory approvals are expected in the next 5 years.
The first generation of ONTs to advance into clinical trials incorporated several medicinal chemistry modifications, including the following [24,25,26]:
Phosphorothioate (PS) backbone modifications to reduce nuclease degradation and increase plasma protein binding to facilitate tissues uptake;
Gapmer oligodeoxynucleotides (ODNs) to elicit efficient RNase H cleavage of the target RNA;
Ribose modifications (mainly 2′ position modifications (2′-fluoro, 2′-O-methyl, 2′-O-methoxyethyl) and 2′-4′ locked nucleic acid (LNA)) to enhance stability and specificity.
Although these modifications improved drug potency, they also came with safety issues, such as chirality, hepatotoxicity, renal toxicity, thrombocytopenia, complement activation and immune/hypersensitivity reactions [27,28,29]. Therefore, generation 2 and generation 2.5 ONT drug candidates were designed to avoid toxicity and improve cell penetration and tissue targeting, while also reducing effective therapeutic doses and improving their PK/PD profile, particularly their intra-cellular mean residence time (MRT) [30,31]. To date, at least twelve N-acetylgalactosamine carbohydrate (GalNAc)-conjugated ONTs are in phase II or phase III clinical trials for specific delivery to the liver. Three out of four siRNAs on the market are GalNAc conjugates, including Givorisan/Givlaari®, Lumasiran/Oxlumo® and Inclisiran/Leqvio®.
Although miRNA-based ONTs, including anti-miR compounds, specific miRNA inhibitors, and miRNA mimics, are being tested in pre-clinical studies, phase 1 clinical trials or phase 2 clinical trials [20,32], regulatory authorities have not yet approved their clinical use beyond investigational drugs. miRNAs’ ability to target multiple genes could be advantageous in managing complex diseases that involve the deregulation of multiple signaling pathways. Advances in active targeting delivery of miRNA-based ONTs should accelerate their transition from bench to bedside in the coming years [33]. To improve their PK/PD and safety profile, new chemical modifications have been introduced into the new generations of ONTs. For instance, the novel class of gamma peptide nucleic acid (γPNA) compounds are water-soluble and charge-neutral. Additionally, they neither aggregate nor adhere to surfaces or other macromolecules in a nonspecific manner and they adopt a right-handed helical motif, hybridize to DNA or RNA with an unusually high affinity and sequence specificity [34,35,36]. Each γ modification stabilizes a PNA–RNA duplex by 5 °C. Compared to LNAs and 2′-OMe oligonucleotides, PNAs are shown to be more potent inhibitors of targeted miRNA activity [37].
Although in silico tools can predict the binding of a given miRNA to many target mRNAs, its true biological effects depend on multiple factors, including the tissue expression level of the miRNA, availability of the Argonaute-2 protein (AGO2) and the expression and abundance of the miRNA target genes of interest.
There is mounting experimental evidence that miRNAs can be used as diagnostic/prognostic markers and potential therapeutic targets in various cancers [38,39]. miRNA dysregulation is involved in epithelial–mesenchymal transition (EMT), invasion, proliferation, migration, metastasis, angiogenesis, immune responses and drug resistance of various cancers [40,41,42,43]. Restoring the expression of tumor-suppressor miRNAs or inhibiting overexpressed oncogenic miRNAs (oncomiRs) are promising strategies for targeted cancer therapies. Recently, a peptide nucleic-acid-based antisense has been shown to be a potential new drug candidate for pancreatic cancer [44]. Accumulating evidence suggests that exosomal miRNAs are relevant players in dynamic crosstalk among cancerous, immune, and stromal cells in establishing the tumorigenic microenvironment [45]. In addition, they sustain metastatic niche formation at distant sites.
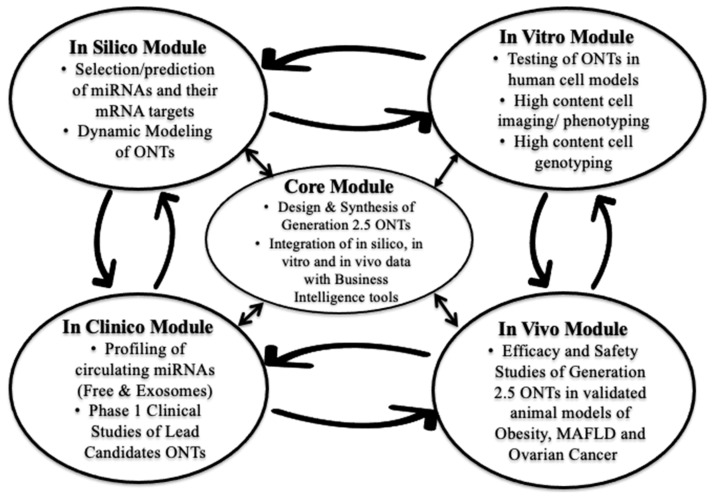
At the inception of AptamiR Therapeutics, we implemented an R&D strategy of small throughput–high yield for the development of miRNA-based ONTs. Instead of relying on brute force drug screening (so called high throughput–low yield screening), we used an innovative model of modular and outsourced drug R&D following the “learn and confirm” strategy championed by Dr. Lewis Sheiner at UCSF [46]. Because of the fast pace of discoveries and publications in the miRNA field and the conveniently expanding commercial availability of tools and reagents required to conduct this kind of R&D activity, AptamiR Therapeutics, Inc. uses a modular parallel and iterative approach, rather than a classical serial one (Figure 1).
Figure 1.
AptamiR Therapeutics’ Modular Parallel and Iterative Strategy of Drug Discovery and Development.
Specifically, we focused on miRNAs as therapeutic agents for the treatment of metabolic pandemics, namely obesity, metabolic (dysfunction)-associated fatty liver disease (MAFLD) and, more recently, ovarian cancer.
2. Results
2.1. Selection of MicroRNAs Involved in Adipose Tissue Functions to Treat Metabolic Pandemics
2.1.1. Background and Goal
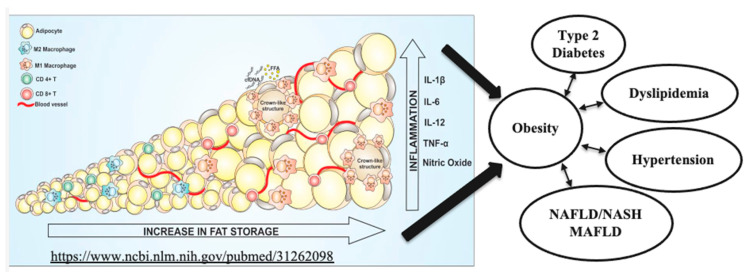
Obesity is a worldwide pandemic, affecting 1/3 of the world population, including children. Its medical, economic and social burdens are significant and growing. Current pharmacotherapy rates for obesity are very low because of limited efficacy, significant side effects, adverse events, restricted access and removal of previously approved drugs. Novel therapies of obesity dramatically improve the lives of millions of subjects, diminish healthcare costs and reducing the death toll linked to obesity-associated cancers and cardiovascular diseases [47,48]. Metabolic (dysfunction)-associated fatty liver disease (MAFLD) is a proposed new terminology that more accurately reflects liver pathogenesis and can help patient stratification for the management of fatty liver disease [49]. MAFLD affects 25% of the global adult population, especially obese patients. There is currently no approved drug to treat MAFLD. MAFLD is the major cause of chronic liver disease, has become the most frequent reason for liver transplantation, and it is associated with substantial morbidity and mortality. The relationships between fat accumulation, inflammation and necrosis resulting in lipotoxicity, dyslipidemia, insulin resistance, diabetes, liver steatosis, inflammation and fibrosis are shown in Figure 2, adapted from [50].
Figure 2.
Medical consequences of adipose tissue hypertrophy, hyperplasia, inflammation and necrosis (adapted from Reference [50], an open-access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/ (https://www.ncbi.nlm.nih.gov/pubmed/31262098 accessed on 20 February 2023)).
Our goal is to develop a novel, safe, effective and convenient therapy for metabolic pandemics, such as obesity and MAFLD. However, obesity and MAFLD are multifactorial diseases that cannot be easily controlled by classical therapeutic agents based on the classical one drug, one target mechanism of action. As microRNAs play diverse roles in obesity and metabolic diseases [51,52,53], as well as regulate gene functions through a one-to-many relationship, our therapeutic strategy is based on the three following innovative principles:
Targeting fat-storing white adipocytes to transform them into fat-burning adipocytes (“browning effect”), instead of altering brain functions, such as appetite/satiety or reducing food intake and absorption (the focus of most pharmacological approaches with significant side effects).
Focusing on miRNA-based ONTs that simultaneously modulate many target genes involved in lipid oxidation, energy expenditure and chronic inflammation (one drug, multiple targets concept) and are well suited for complex and prevalent diseases, such as obesity and MAFLD.
Developing a unique delivery platform to actively target human adipocytes.
Through these efforts, we expect to deliver on our end goal, which is to help patients live longer and healthier lives, while reducing healthcare expenditure.
2.1.2. Strategy
We initiated a search for miRNAs that are unique, conserved across species, universal, abundantly expressed in metabolic tissues/organs and modulate metabolic pathways, especially thermogenesis. This discovery phase of the project was conducted via in silico mining of publicly available datasets, presenting miRNAs as putative regulators of candidate genes.
2.1.3. In Silico Search
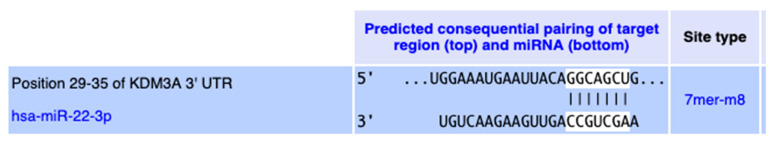
Our initial in silico analysis identified putative miRNAs from a known pool of about 2000 miRNAs and a digitally curated list of 721 genes involved in lipid metabolism, oxidative phosphorylation, mitochondrial functions, respiratory cycle, browning of adipocytes and energy expenditure (AptamiR 721 genes were selected using eight publicly available in silico tools, namely BioCarta; Database for Annotation, Visualization and Integrated Discovery (DAVID); GeneOntology; Gene Set Enrichment Analysis (GSEA); Kyoto Encyclopedia of Genes and Genomes (KEGG); PubGene; Reactome; and STRING). We utilized 34 in silico miRNA target prediction tools and our own proprietary in silico meta-tool (R-AptamiR) to identify 200 miRNAs that potentially bind to these target metabolic genes. A metabolic target of particular interest is the mitochondrial uncoupling protein UCP1, which increases thermogenesis in adipose tissues. The human UCP1 gene structure is notable for a high degree of methylation (“CG islands”) in its promoter region. Methylation of CG islands within gene promoters can lead to their silencing [54]. The human lysine (K)-specific demethylase 3A (KDM3A) is critically important in regulating the expression of metabolic genes and obesity resistance [55]. Using microarray technology, Zhang et al. demonstrated that a significant number of the genes involved in PPAR signaling and fatty acid oxidation (e.g., PPARA, ACADM, ACADL, ACADVL, AQP7) were down-regulated in response to KDM3A knockout [55]. KDM3A directly regulates peroxisome proliferator-activated receptor alpha (PPARA) and UCP1 expression [56]. After a few “learn and confirm” rounds between in silico, in vitro and in vivo experiments, we identified a subset of miRNA targets from the original 200. We found that the human KDM3A 3′ UTR 29–35 region is a conserved target for hsa-miR-22-3p, as shown in Figure 3.
Figure 3.
Conserved interaction between hsa-miR-22-3p and the 3′-UTR region of the KDM3A gene (TargetScanHuman 8.0, www.targetscan.org/vert_80/ (accessed on 20 February 2023)).
The expression of KDM3A is widely distributed across human tissues and organs with low tissue specificity of 0.29 (www.proteinatlas.org/ (accessed on 20 February 2023)).
2.1.4. Validation of miR-22-3p as a Metabolic Target
Using various in silico, in vitro and in vivo tools, we demonstrated that miR-22-3p is an excellent metabolic target that modulates several genes, as illustrated below using the metaMIR tool (http://rna.informatik.uni-freiburg.de (accessed on 20 February 2023)), which ranks miRNAs in relation to gene networks (Table 1).
Table 1.
Prediction and ranking of interactions between various miRNAs and clusters of metabolic genes in humans using the metaMIR Tool (http://rna.informatik.uni-freiburg.de/metaMIR/Input.jsp (accessed on 3 July 2021)).
| miRNA | Final Score | Positive Combo |
|---|---|---|
| hsa-miR-22-3p | 12.11 | AKT1,BDNF,CDKN1A,CREB1,ESR1,HDAC4,HDAC6,KDM3A,KDM6B,KLF6,MAPK14,MECP2,PPARA,PPARGC1B,PRDM16,PTEN,RUNX2,SIRT1,SOD2,SP1,STAT3 |
| hsa-miR-520c-3p | 4.58 | AKT1,CDKN1A,ESR1,HDAC4,KDM6B,KLF6,MECP2,PPARA,PPARGC1B,PRDM16,PTEN,RUNX2,SP1,STAT3 |
| hsa-miR-10b-5p | 4.55 | BDNF,CDKN1A,CREB1,ESR1,HDAC4,KDM3A,PPARA,PPARGC1B,PRDM16,PTEN,SOD2,SP1 |
| hsa-miR-1470 | 4.35 | AKT1,CREB1,HDAC4,KDM6B,KLF6,MAPK14,MECP2,PPARA,PPARGC1B,PTEN,RUNX2,SOD2,STAT3 |
| hsa-miR-5089-3p | 4.29 | AKT1,CREB1,KLF6,MAPK14,MECP2,PRDM16,PTEN,SIRT1,SOD2,STAT3 |
| hsa-miR-7110-5p | 4.12 | CDKN1A,CREB1,KDM6B,KLF6,MECP2,PPARA,PPARGC1B,PRDM16,PTEN,RUNX2,SP1,STAT3 |
| hsa-miR-30a-5p | 4.1 | AKT1,BDNF,CREB1,KDM3A,KDM6B,KLF6,PPARGC1B,PRDM16,PTEN,RUNX2,SIRT1,SOD2 |
| hsa-miR-520a-3p | 3.95 | AKT1,CREB1,ESR1,HDAC4,KDM6B,KLF6,MECP2,PPARGC1B,PRDM16,PTEN,RUNX2,SOD2,STAT3 |
| hsa-miR-520b | 3.83 | AKT1,ESR1,HDAC4,KDM6B,KLF6,MECP2,PPARA,PPARGC1B,PRDM16,PTEN,RUNX2,SOD2,STAT3 |
| hsa-miR-365a-5p | 3.77 | CDKN1A,ESR1,HDAC4,KDM6B,KLF6,MECP2,PPARA,PPARGC1B,PRDM16,SOD2,STAT3 |
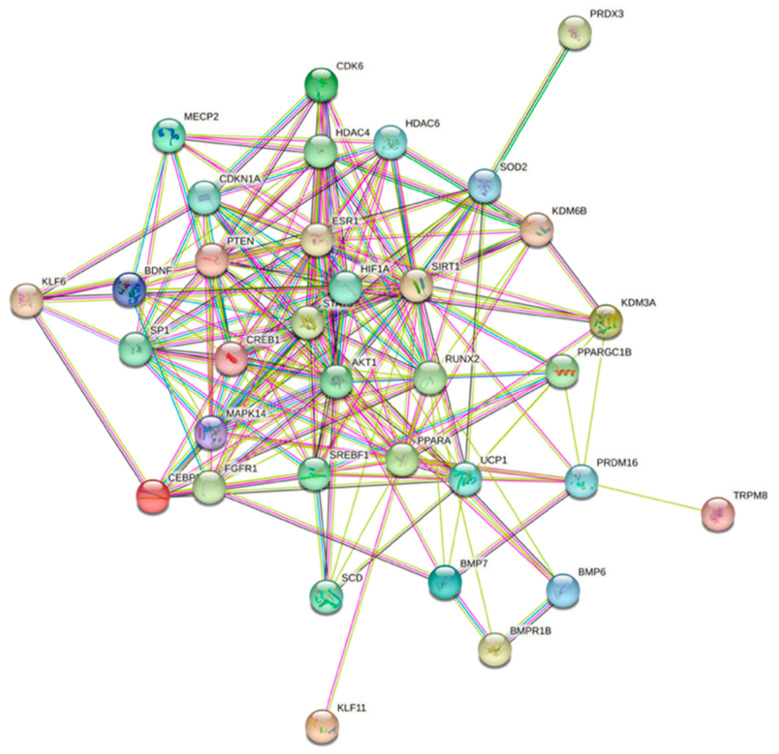
As these genes are required for normal metabolic functions and miR-22-3p is likely to induce degradation or reduce the translation of these genes, a strategy involving antagonizing the functions of miR-22-3p constitutes a viable therapeutic route to the amelioration of metabolic disorders. We further used the protein–protein interaction functional enrichment analysis tool STRING (https://string-db.org (accessed on 3 July 2021)) [57] to illustrate the various interactions within a network of 34 proteins related to miR-22 (Figure 4).
Figure 4.
Prediction of protein–protein interaction networks related to miR-22 using the protein–protein interaction-networks functional enrichment analysis tool String (https://string-db.org (accessed on 3 July 2021)).
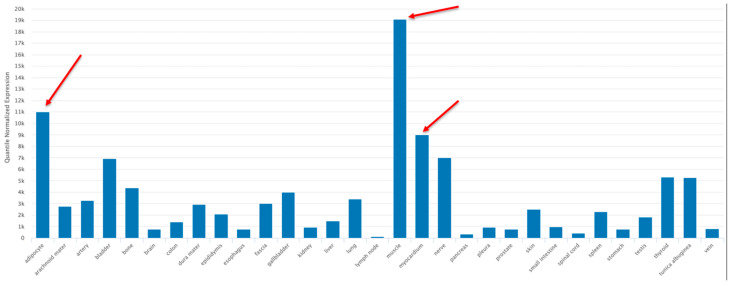
The human tissue/organ distribution of hsa-miR-22-3p was examined using the TissueAtlas2 program (https://www.ccb.uni-saarland.de/tissueatlas2 (accessed on 20 February 2023)) [58]. The tissue specificity index (TSI) gives each specific non-coding RNA molecule a numeric value on a scale from 0 to 1, with 1 meaning that the expression of the molecule was detected in only one specific tissue and 0 meaning that the expression of the molecule was detected in all tissues. The TSI for hsa-miR-22-3p was 0.855, indicating a high tissue-specific expression pattern, especially in the adipocytes, the myocardium and skeletal muscle (red arrows, Figure 5).
Figure 5.
Tissue/organ distribution of miR-22-3p using the TissueAtlas2 program (https://www.ccb.uni-saarland.de/tissueatlas2 (accessed on 20 February 2023)) [58], an Open Access article distributed under the terms of the Creative Commons Attribution-NonCommercial License (https://creativecommons.org/licenses/by-nc/4.0/, accessed on 20 February 2023). Adipocyte, myocardium and skeletal muscle samples are identified by red arrows.
2.1.5. In Vitro and In Vivo Validation of miR-22-3p Antagonism
We then proceeded to explore the metabolic effects of miR-22-3p antagomirs in vitro in primary cultures of human adipocytes and in vivo in mice. The metabolic and energetic benefits of first-generation miR-22-3p antagomirs were summarized in two peer-reviewed articles published in 2020 [59,60]. In vivo proof of concept of miR-22-3p inhibition in mice was performed in the mouse model of diet-induced obesity (DIO) in C57BL/6J male mice. Mice of various ages were allocated to normal chow (10% fat) or a 60% high-fat diet and were treated for up to 12 weeks with a miR-22-3p antagomir or saline. We consistently observed in the presence of miR-22-3p antagomir treatment a reduction in body weight and fat mass without alteration of lean mass; improvement in glucose, insulin sensitivity and lipid profile; increase in thermogenesis; and no modification of food intake or body temperature.
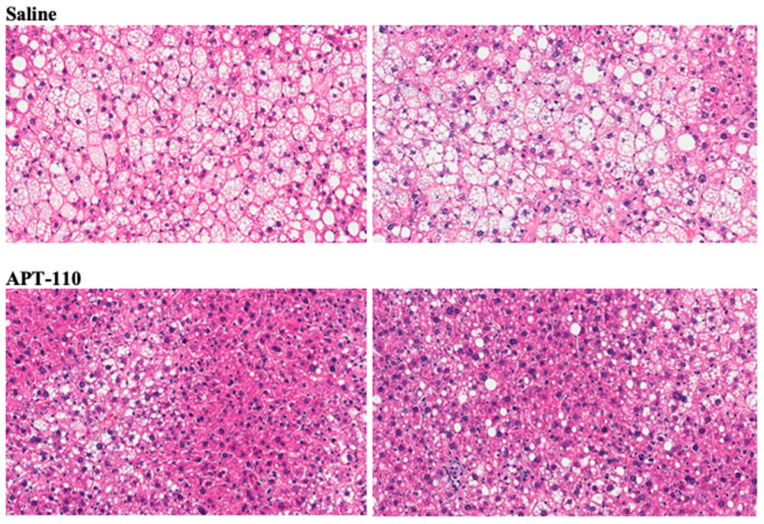
Treatment for 12 weeks with miR-22-3p antagomir APT-110 produced a marked reduction in fatty infiltration of the liver (Figure 6) [60].
Figure 6.
Histologic appearance of livers (H&E staining) at the end of 12 weeks of treatment in mice receiving SC injections of saline or the APT-110 miR-22-3p inhibitor (two samples from each group are shown), reprinted from Reference [60], an open-access article distributed in accordance with the Creative Commons Attribution Non-Commercial (CC BY-NC 4.0) license.
2.1.6. Safety Assessment
Pre-IND toxicology and safety studies of first-generation miR-22-3p antagomir APT-110 (a “naked” single-stranded 18 mer miR-22-3p antagomir containing PS and LNA modifications) were completed in mice and non-human primates according to FDA guidance. In mice, APT-110 administered subcutaneously at 15, 60, and 240 mg/kg/dose on days 1, 3, 5, 8, 15, 22 and 29 was well tolerated and associated with atrophy of adipose tissues at all dose levels. In cynomolgus monkeys, APT-110 was administered subcutaneously at 3.75, 15, 37.5 and 60 mg/kg/dose on days 1, 3, 5, 8, 15, 22 and 29 of the study. Transient activation of blood platelets and the complement pathway was observed right after administration of supra-therapeutic doses. Kidney and liver histologic alterations were also noted. Due to these observations, our first-generation compound was not advanced further.
2.1.7. Design of Generation 2.5 ONTs for Active Targeted Delivery to Metabolic Tissues/Organs
Our new generation 2.5 of targeting ONTs was designed to conduct the following:
Eliminate potential toxicities by replacing PS and LNA modifications with a gamma PNA backbone;
Maintain resistance to nucleases and proteases/peptidases;
Avoid chirality;
Limit binding to serum proteins;
Optimize/simplify chemical synthesis;
Conjugate ONT to a fatty acid or a short peptide for enhanced targeted delivery to adipocytes of a greatly reduced effective dose with an extended duration of action (mean residence time).
2.1.8. Selection of the Membrane Fatty Acid Translocase (FAT) Transporter for Active Targeted Delivery of ONTs to Adipocytes and Metabolic Organs
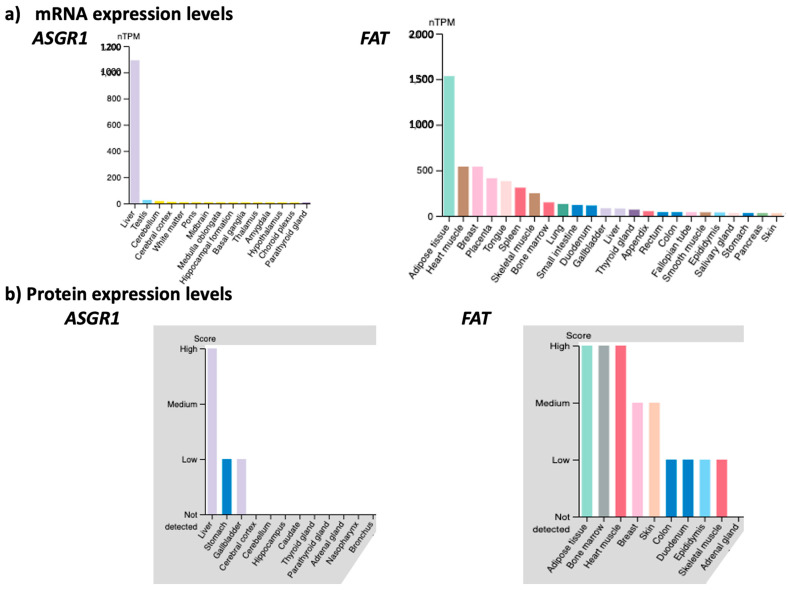
The membrane transporter FAT/CD36/SCARB3 is the main route of uptake by adipose tissues of long-chain fatty acids, as well as short peptides, such as hexarelin, prohibitin and thrombospondin peptide-1 [61,62]. FAT is significantly expressed in cells and tissues sensitive to metabolic dysfunctions, such as adipocytes, hepatocytes, skeletal and cardiac myocytes, pancreatic β-cells, kidney glomeruli and tubules cells, monocytes and macrophages. As the average obese male patient weighs around 200 lbs, of which 40% is adipose tissue, there is a huge amount of FAT available at the surface of the adipose tissues to transport inside the adipocytes our new generation 2.5 of miR-22-3p antagomirs coupled with a fatty acid or peptide. We compared the mRNA and protein level expression of FAT across human tissues to that of asialoglycoprotein receptor 1 (ASGR1), which has been successfully targeted for the preferred delivery of ONTs to the liver (Figure 7). This comparison illustrates the rationale for targeting FAT for the delivery of ONTs to metabolic organs.
Figure 7.
Comparison of mRNA (Panels a) and protein (Panels b) expression levels in various tissues and organs of the asialoglycoprotein receptor 1 (ASGR1) and the fatty acid translocase (FAT) membrane transporter (nTPM = normalized transcript per million) (www.proteinatlas.org (accessed on 27 March 2023)). The figures are shown in “Expression Level” mode in decreasing order from left to right. The tissues/organs with little or no expression are not shown for sake of readability.
2.1.9. In Silico Modeling of Generation 2.5 ONTs
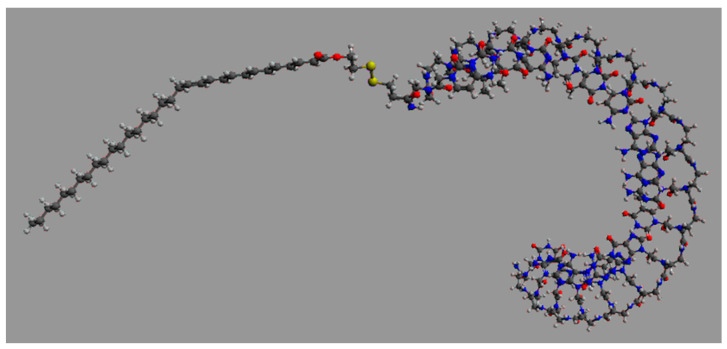
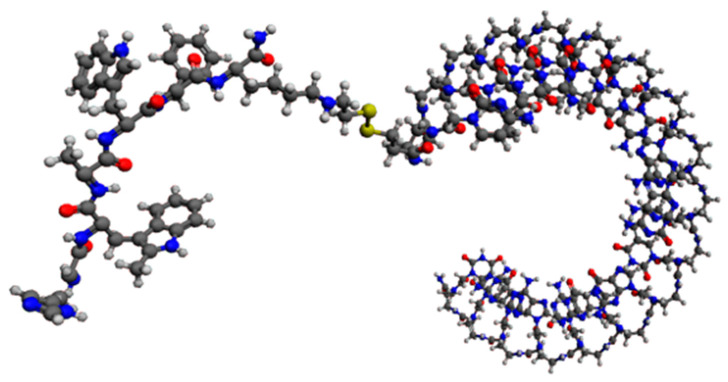
We then focused on significantly reducing the effective dose of ONTs to obtain a greatly improved safety and PK/PD profile, especially in terms of the mean residence time inside the targeted cells. Pr. Pengyu Ren, Ph.D. and his associates at the Department of Biomedical Engineering at the University of Texas at Austin performed high-performance molecular dynamics modeling on graphics processing units of generation 2.5 miR-22-3p antagomirs, coupled with a fatty acid of increasing length from C16 palmitic acid to C22:6 docosahexaenoic acid and C32:6 dotriacontahexaenoic acid (Figure 8) [63,64,65].
Figure 8.
Graphic representation of an 18 mer miR-22-3p antagomir with a PNA (Pna) backbone coupled with C32:6 dotriacontahexaenoic fatty acid (5′-C32-S-S-PnaC-PnaT-PnaT-PnaC-PnaT-PnaT-PnaC-PnaA-PnaA-PnaC-PnaT-PnaG-PnaG-PnaC-PnaA-PnaG-PnaC-PnaT-3′).
To further refine our ability to accurately deliver miRNA antagonists to target tissues, we also studied the coupling of generation 2.5 miR-22-3p antagomirs to the peptide hexarelin. Hexarelin (His-D-2MeTrp-Ala-Trp-D-Phe-Lys-NH2) is a stable analog of growth-hormone-releasing peptide 6 and is a high-affinity ligand for FAT [66]. The interaction of hexarelin with FAT promotes the transcriptional activation of nuclear receptor PPARγ and genes involved in metabolism and thermogenesis [67,68]. Based on these findings, we are also developing generation 2.5 miR-22-3p antagomirs coupled with Hexarelin for active targeting of adipocytes (Figure 9).
Figure 9.
Graphic representation of an 18 mer miR-22-3p antagomir with a PNA backbone coupled with the hexapeptide hexarelin (5′-Hex-S-S-PnaC-PnaT-PnaT-PnaC-PnaT-PnaT-PnaC-PnaA-PnaA-PnaC-PnaT-PnaG-PnaG-PnaC-PnaA-PnaG-PnaC-PnaT-3′).
Together with these optimization studies, we are currently conducting in vitro tests of primary cultures from human adipocytes and in vivo tests of animal models with obesity and MAFLD to understand the metabolic benefits of our generation 2.5 miR-22-3p antagomirs.
2.2. Selection and Targeting of MicroRNAs Involved in Ovarian Cancer Development and Spread
2.2.1. Background and Goal
Cancer epidemiologic and mortality surveys report that ≥300,000 women are diagnosed with ovarian cancer (OC) worldwide and ≥200,000 succumb to OC every year [69,70]. Most patients are diagnosed with advanced OC at stages III or IV. High-grade serous OC (HGSOC) is the most common and deadliest type of OC. Survival of 5 years is only 30% in HGSOC and 18% for patients diagnosed with stage IV tumors. Presently, debulking cytoreductive surgery is the gold standard for the treatment of OC, along with platinum-based chemotherapy regimens. For patients who become platinum resistant, few options are available, and the efficacy of these regimens is limited. Regardless of treatment options, survival rates of OC have barely improved over the years due to lack of specific symptoms and early screening/diagnostic tools, as well as the high rates of drug resistance and cancer relapse. Due to its clinical, biological and molecular complexity, OC is a tumor that is difficult to treat and lacks a clear driver mutation. Targeted therapies are now a strong focus for OC [71,72].
There is now ample evidence that miRNAs play many roles in OC, its tumor microenvironment and resistance to treatments [73,74,75,76,77,78]. Several membrane receptors often overexpressed in OC cells have emerged as potential targets for receptor-mediated therapies [79]. Given the roles of miRNAs in various cellular pathways, including cell survival and differentiation, targeting miRNAs could be a viable approach for the treatment of human cancers, such as OC, through the inhibition and/or stimulation of miRNAs [80]. Our end goal is to develop safe, effective and convenient miRNA-based therapies to cure OC.
2.2.2. Strategy
Building on our knowledge of generation 2.5 miRNA ONTs and targeted delivery to specific cell types, we decided to develop microRNA-based targeting ONTs that can simultaneously modulate several target genes involved in OC and its tumor microenvironment and spreading. We found that our modular parallel and iterative approach could be quickly adapted from one therapeutic area to another.
2.2.3. In Silico Search
In collaboration with the Centre for Computational Biology and Program in Cardiovascular & Metabolic Disorders at Duke-NUS Medical School, Singapore, we used various bioinformatics tools to identify relevant miRNAs involved in OC, some of which are listed in Table 2.
Table 2.
A list of publicly available bioinformatics software used to identify candidate miRNAs and their gene targets, with particular relevance to ovarian cancer.
| Tool | Web Address | Function |
|---|---|---|
| Target Scan Human 8.0 | www.targetscan.org/vert_80 (accessed on 7 November 2022) | Search for predicted miRNA targets |
| metaMIR V 1.1.0 | http://rna.informatik.uni-freiburg.de/metaMIR/Input.jsp (accessed on 7 November 2022) | Predict interactions between miRNAs and clusters of genes in human |
| OncomiR | http://www.oncomir.org/ (accessed on 7 November 2022) | WashU Pan-Cancer miRNome Atlas exploring pan-cancer microRNA dysregulation |
| GeneNet V 3.0.2 | http://www.oncomir.org/ (accessed on 7 November 2022) | R package for learning high-dimensional dependency networks from genomic data |
| Cytoscape V3.7.1 | https://cytoscape.org/ (accessed on 7 November 2022) | Network data integration, analysis and visualization |
| DiffCorr V0.4.2 | https://sourceforge.net/projects/diffcorr/ (accessed on 7 November 2022) | R package to analyze differential correlations biological networks |
| STRING V11.5 | https://string-db.org/ (accessed on 7 November 2022) | Protein–protein interaction networks Functional enrichment analysis |
| Webgestalt V2019 | http://www.webgestalt.org/ (accessed on 7 November 2022) | Gene list over-representation analysis |
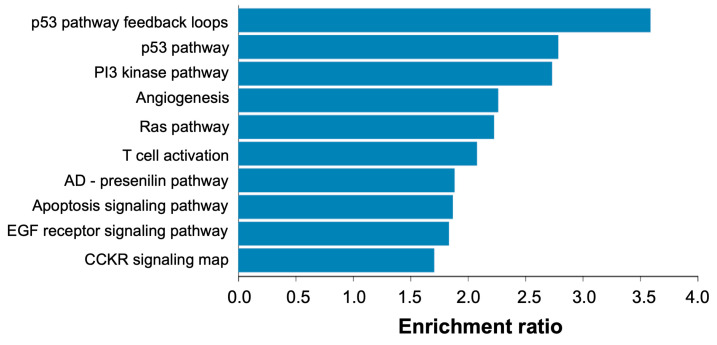
Based on the bioinformatics analyses, we found 441 miRNAs and 1003 potential target genes involved in the proliferation, migration, apoptosis, invasion, metastases, differentiation, epithelial–mesenchymal transition activation and chemoresistance of OC. Of these, 14 miRNAs were associated with >40 target genes, 29 miRNAs with 30–39 genes, 71 miRNAs with 20–29 genes and 244 miRNAs with 5–19 genes. We performed pathway and Gene Ontology biological process over-representation analysis of the 1003 target genes using Webgestalt (http://www.webgestalt.org/ (accessed on 7 November 2022)) and found that genes were over-represented in pathways related to p53, Ras and PI3-kinase signaling, among others (Figure 10).
Figure 10.
Pathway over-representation analysis of candidate miRNA target genes relevant to ovarian cancer. Analysis was performed via Webgestalt (http://www.webgestalt.org/ (accessed on 7 November 2022)) against Panther pathways and using whole human genome as background. The x-axis refers to the enrichment ratio for each pathway (all pathways were significantly enriched at FDR < 5%).
We then built networks of proteins involved in OC as shown in Table 3.
Table 3.
List of 472 proteins related to ovarian cancer with the protein–protein interaction-networks functional enrichment analysis tool String (https://string-db.org (accessed on 7 November 2022)).
| ABCC3 | CALR | CUL4A | FZD2 | KLF9 | NRXN3 | RB1 | STK4 |
| ABL2 | CANX | CXCL1 | FZD6 | KLLN | NSD1 | RBBP8 | STMN2 |
| ACAP2 | CARD18 | CXCL10 | FZD8 | KRAS | NUAK1 | RHOBTB3 | STX17 |
| ACO2 | CASP10 | CXCL11 | GAB2 | LATS2 | OLA1 | RHOC | STXBP4 |
| ACSL4 | CASP8 | CXCL12 | GADD45B | LHX6 | OLFML3 | RNF44 | SUCO |
| ACTC1 | CCL5 | CXCL8 | GALNT1 | LIMK1 | OVOL1 | ROCK1 | SYNCRIP |
| ACTR1A | CCNB1 | CXCL9 | GALNT14 | LOX | P4HA1 | ROCK2 | TAGLN |
| ACTRT3 | CCND1 | CXCR3 | GALR1 | LPIN1 | PA1 | RUNX1 | TAP1 |
| ADAM12 | CCND2 | CYP1B1 | GCNT1 | LRRC15 | PAK2 | RUNX2 | TCF21 |
| ADAM17 | CCNE1 | CYTIP | GCNT2 | LRRK2 | PAPD7 | RUNX3 | TCF4 |
| ADAM19 | CCNG1 | DAAM1 | GCNT4 | LSG1 | PARP1 | S1PR1 | TCF7L1 |
| ADAMDEC1 | CCNG2 | DCN | GCOM1 | LUM | PAX7 | SALL2 | TEX261 |
| ADAMTS17 | CCR2 | DCTN5 | GCSAM | LZTS1 | PCDHA10 | SDC1 | TGFB1 |
| ADAMTS19 | CD1D | DCX | GEMIN4 | MACC1 | PCDHA3 | SEMA4D | THBS2 |
| ADAMTSL1 | CD2 | DDB2 | GFPT2 | MAP2 | PCDHA5 | SEMA6B | TIMM17A |
| AGO1 | CD247 | DICER1 | GM2A | MAP3K1 | PCDHGA10 | SEPTIN6 | TIMMDC1 |
| AKAP13 | CD27 | DKK1 | GNAI3 | MAP3K7 | PCNA | SET | TIMP2 |
| AKR1D1 | CD38 | DLG2 | GPR12 | MAPK1 | PDCD6 | SGCD | TIMP3 |
| AKT1 | CD3D | DLGAP2 | GPR124 | MAPK14 | PDE7A | SHROOM2 | TLN1 |
| AKT2 | CD3E | DNMT1 | GPR83 | MAPK3 | PDGFRA | SIK1 | TLR4 |
| AKT3 | CD44 | DTD2 | GRB7 | MCM2 | PDGFRB | SIK2 | TMEM239 |
| ALG2 | CD55 | DVL3 | HBEGF | MED12L | PDHB | SIRT1 | TMEM45A |
| ANKRD46 | CD68 | E2F2 | HDGF | MET | PDZK1IP1 | SIT1 | TP53 |
| ANXA8L1 | CD74 | E2F3 | HEPHL1 | MLIP | PHEX | SIX2 | TP53I11 |
| APAF1 | CD82 | E2F5 | HEYL | MLLT3 | PHLDB2 | SKAP2 | TRIM2 |
| APC2 | CD8A | EBF1 | HIF1A | MMP10 | PIEZO2 | SLA2 | TRIM27 |
| ARHGAP24 | CD97 | EFEMP1 | HLX | MMP16 | PIGH | SLAMF7 | TRIM31 |
| ARHGAP28 | CDC25A | EGFR | HMGA1 | MMP2 | PIK3CA | SLAMF8 | TRIM52 |
| ARID1A | CDC25B | EIF5A2 | HMGA2 | MMP9 | PKP1 | SLC24A4 | TSC1 |
| ARID3B | CDH1 | ELAVL1 | HMGB1 | MSH5 | PLAG1 | SLC2A3 | TTC14 |
| ARL5B | CDH2 | ELF5 | HNRNPC | MSN | PLAU | SLC31A1 | TUBB3 |
| ASXL3 | CDK1 | ELN | HOXA10 | MT-CO1 | PLD3 | SLC43A2 | TWIST1 |
| ATM | CDK12 | EML1 | HOXA13 | MT-ND2 | PLK1 | SLC4A4 | VAT1L |
| ATP5B | CDK2 | EPAS1 | HOXA9 | MTDH | PLS3 | SLC7A6 | VCAN |
| ATR | CDK4 | EPB41L3 | HOXB2 | MTFR1 | PMAIP1 | SMAD4 | VEGFA |
| AURKB | CDK6 | EPHA2 | ID1 | MTHFD1 | POSTN | SMAD7 | VEGFB |
| AXL | CDKN1A | EPHA4 | ID4 | MTSS1 | POTED | SMTNL2 | VEGFC |
| B3GNT5 | CDKN2A | ERBB2 | IGF1 | MUC1 | POU3F1 | SMURF1 | VIM |
| BAG5 | CEACAM1 | ERBB3 | IGF1R | MUC16 | PPP1R2 | SMYD1 | VTN |
| BAX | CHEK1 | ERBB4 | IGF2BP1 | MYC | PRDM16 | SNAI1 | WDR17 |
| BCL11B | CHEK2 | ESRRG | IGFBPL1 | MYCBP | PRKAA1 | SNAI2 | WNT1 |
| BCL2 | CHI3L1 | FAP | IL1A | MYCN | PROX1 | SOCS1 | WNT5A |
| BCL2L1 | CHST9 | FAR1 | IL2 | MYH9 | PTEN | SOCS2 | WSCD1 |
| BCR | CHSY1 | FBN1 | IL6R | MYO5A | PTGDR | SOD2 | XIAP |
| BIRC5 | COBLL1 | FBXO28 | INHBA | NEFL | PTHLH | SOS2 | XXYLT1 |
| BMF | COL11A1 | FCER1G | INSR | NEFM | PTPN12 | SOX11 | YAP1 |
| BMP3 | COL15A1 | FCRL1 | ITGA5 | NEUROD1 | PTPN4 | SOX12 | YOD1 |
| BMP4 | COL1A2 | FGF1 | ITGB1 | NEUROG1 | PTPRO | SOX4 | YY1 |
| BMP7 | COL3A1 | FGF2 | JAG1 | NF1 | PWWP2A | SOX9 | ZEB1 |
| BNIP3 | COL5A1 | FHL2 | JAG2 | NF2 | R3HDM4 | SPARC | ZEB2 |
| BRAF | COL5A2 | FN1 | JAKMIP2 | NFIX | RAB11FIP3 | SPHK1 | ZNF107 |
| BRCA1 | CPEB3 | FOSL2 | KCNA5 | NFKB1 | RAB22A | SPSB4 | ZNF138 |
| BRCA2 | CPNE3 | FOXA2 | KDR | NHS | RAB30 | SRC | ZNF181 |
| BTLA | CRISPLD2 | FOXD4L1 | KEAP1 | NOB1 | RAB5A | SREBF1 | ZNF346 |
| C10orf128 | CSF1R | FOXF2 | KIAA0101 | NOTCH1 | RACGAP1 | SREBF2 | ZNF423 |
| C11orf58 | CSMD3 | FOXM1 | KIAA0513 | NOTCH2 | RAD51 | SRSF1 | ZNF485 |
| C1orf105 | CTGF | FOXO3 | KLF12 | NOTCH3 | RAP1B | ST7L | ZNF521 |
| CACNA1C | CTNNB1 | FOXP1 | KLF15 | NREP | RARRES1 | STAT3 | ZNF697 |
| CACNG8 | CTSK | FUT4 | KLF4 | NRP1 | RASD1 | STK24 | ZNF706 |
We also built miRNA-mRNA networks. From these networks, 114 miRNAs were shown to interact with 20 to 59 potential targets, as shown in Table 4.
Table 4.
List of miRNAs found to interact with 20 to 59 potential target genes.
| 20 to 29 Target Genes | 30 to 39 Target Genes | 40 to 59 Target Genes | |
|---|---|---|---|
| let-7a-5p | miR-3065-5p | let-7a-3p | miR-1275 |
| let-7b-5p | miR-30a-3p | miR-106a-5p | miR-15a-5p |
| let-7c-5p | miR-30d-3p | miR-106b-5p | miR-15b-5p |
| let-7d-5p | miR-30d-5p | miR-126-5p | miR-16-5p |
| let-7e-5p | miR-30e-3p | miR-129-5p | miR-195-5p |
| let-7f-5p | miR-30e-5p | miR-137 | miR-335-3p |
| let-7g-5p | miR-326 | miR-149-3p | miR-3607-3p |
| let-7i-5p | miR-330-5p | miR-17-5p | miR-373-5p |
| miR-105-5p | miR-340-5p | miR-181a-5p | miR-424-5p |
| miR-1253 | miR-34a-5p | miR-181b-5p | miR-497-5p |
| miR-124-3p | miR-363-3p | miR-181c-5p | miR-548a-5p |
| miR-128-3p | miR-377-3p | miR-181d-5p | miR-548b-5p |
| miR-1290 | miR-381-3p | miR-186-5p | miR-7-1-3p |
| miR-130a-5p | miR-486-3p | miR-200b-3p | miR-548d-3p |
| miR-130b-5p | miR-491-5p | miR-200c-3p | |
| miR-145-5p | miR-494-3p | miR-205-5p | |
| miR-182-5p | miR-506-3p | miR-20b-5p | |
| miR-185-5p | miR-511-5p | miR-30a-5p | |
| miR-1915-3p | miR-513a-3p | miR-30b-5p | |
| miR-200a-3p | miR-519a-3p | miR-30c-5p | |
| miR-204-5p | miR-519d-3p | miR-330-3p | |
| miR-20a-5p | miR-520b | miR-33a-3p | |
| miR-214-3p | miR-539-5p | miR-3688-3p | |
| miR-23a-3p | miR-551b-5p | miR-485-5p | |
| miR-23b-3p | miR-576-5p | miR-526b-3p | |
| miR-25-3p | miR-582-5p | miR-589-3p | |
| miR-26a-5p | miR-603 | miR-590-3p | |
| miR-26b-5p | miR-629-3p | miR-93-5p | |
| miR-27a-3p | miR-661 | miR-940 | |
| miR-27b-3p | miR-664a-3p | ||
| miR-29a-3p | miR-766-3p | ||
| miR-29b-1-5p | miR-92a-3p | ||
| miR-29b-2-5p | miR-92b-3p | ||
| miR-29b-3p | miR-96-5p | ||
| miR-29c-3p | miR-98-5p | ||
| miR-3065-3p | |||
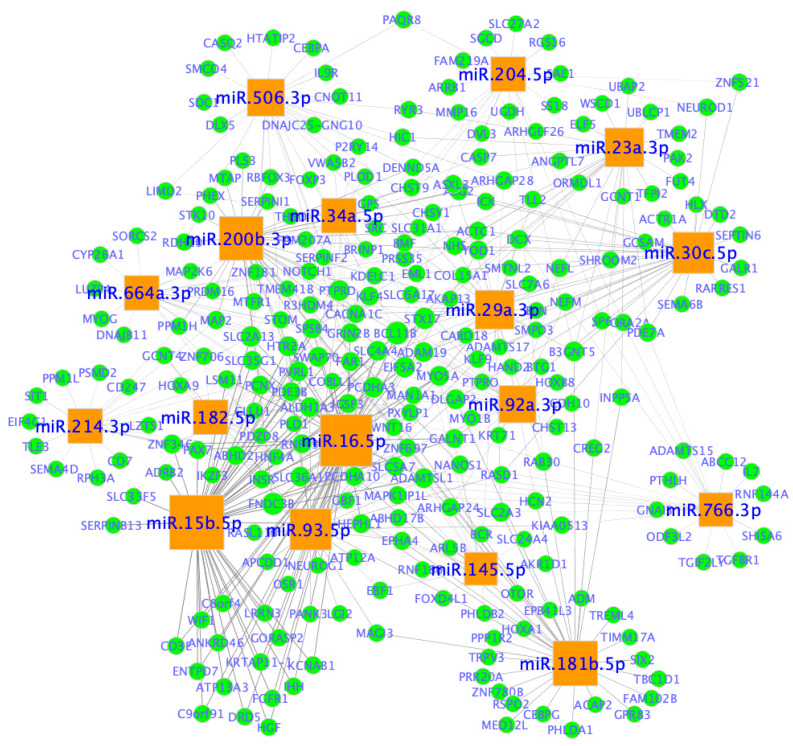
Examples of miRNA-mRNA networks are shown in Figure 11. The two main categories of miRNAs were revealed as follows: one category (e.g., miR-506-3p, miR-204-5p, miR-23a-3p, miR30c-5p, miR-766-3p, miR-181b-5p, miR-214-3p and miR-664a-3p) seems to interact with a specific set of target genes; and the other category (e.g.,miR-34a-5p, miR-29a-3p, miR-92a-3p, miR-145-5p, miR-16-5p, miR-93-5p, miR-15b5p, miR-182-5p and miR-200b-3p) seems to share common target genes.
Figure 11.
miRNA-mRNA networks in ovarian cancer. Network nodes represent miRNAs and genes (mRNAs), whereas edges represent an association between a miRNA and a gene. miRNAs are represented by orange squares and genes are shown in blue circles. miRNA node size is proportional to the number of genes it is predicted to interact with. Network was created in Cytoscape (https://cytoscape.org/ (accessed on 20 January 2023)).
2.2.4. Active Targeted Delivery of Generation 2.5 MiRNA-Based ONTs to Ovarian Cancer Cells via Folic Acid Receptor Alpha (FOLR1)
Building on our knowledge of targeted delivery to specific cell types, as well as learning from the recent development and regulatory approval of antibody drug conjugates (ADCs) and bispecific antibodies (bsAbs) for the treatment of cancers [81,82,83,84,85,86], we looked for a validated target to deliver ONTs to primary OC cells. We selected folic acid receptor alpha (FOLR1), a glycosylphosphatidylinositol (GPI)-anchored cell-surface glycoprotein that is highly expressed at the surface of epithelial ovarian cancer cells [87,88] (Figure 12).
Figure 12.
Tissue/organ distribution of FOLR1 in normal tissues and ovarian cancer, reprinted from [87], an article licensed under Creative Commons Attribution 4.0 International License.
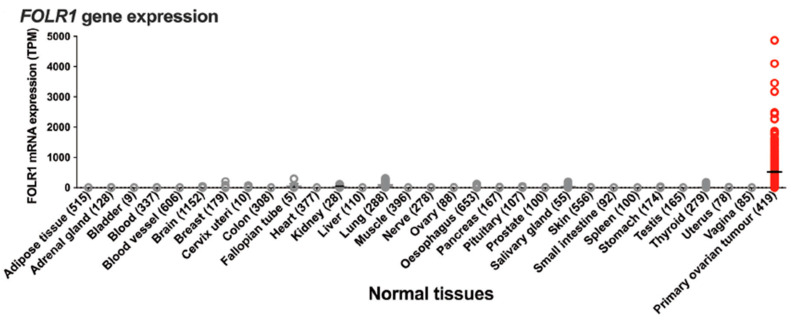
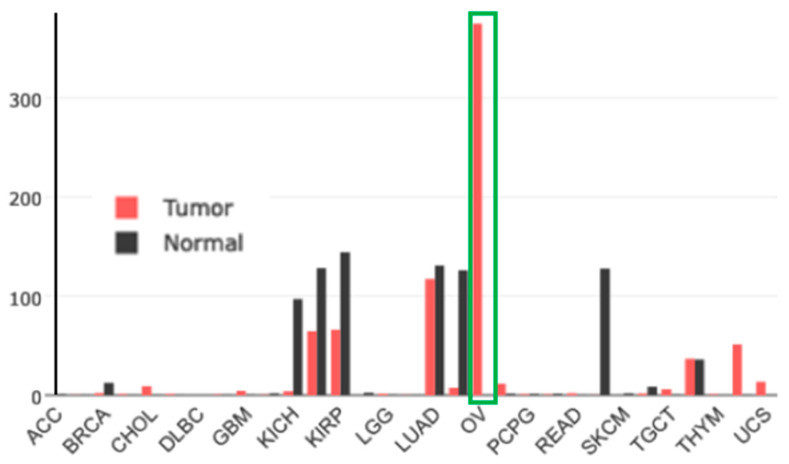
Gene-expression profiling of FOLR1 across tumor samples and paired normal tissues (http://gepia.cancer-pku.cn (accessed on17 February 2023)) shows it to be preferentially expressed in ovarian tumors (Figure 13).
Figure 13.
Gene-expression profile of FOLR1 in human normal and tumor tissue samples using the gene-expression profiling interactive analysis tool (http://gepia.cancer-pku.cn/ (accessed on 17 February 2023)). The cancer and normal ovarian samples are shown in the green rectangle. The height of the bar represents the median expression of the certain tumor type or normal tissue.
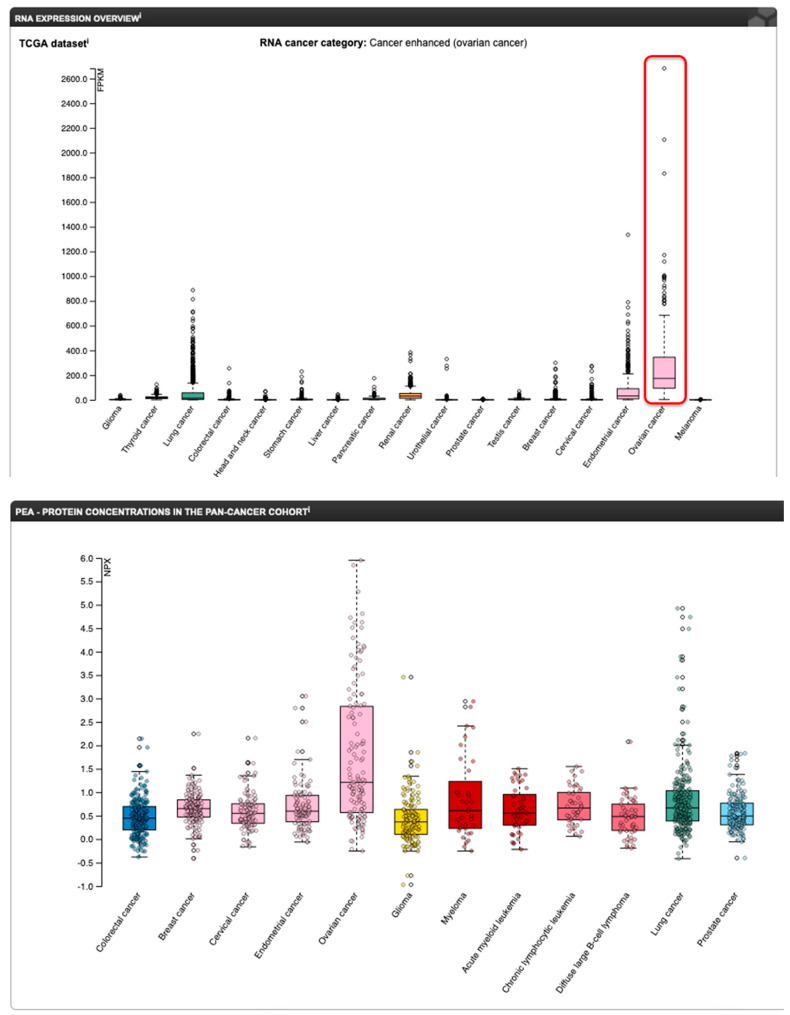
Up to 90% of OC, especially the HGSOC type, overexpress FOLR1 [87,89], and FOLR1 expression is closely associated with the severity of OC (Figure 14) [79,90].
Figure 14.
mRNA (top panel) and protein (bottom panel) expression profile of FOLR1 in various cancers (www.proteinatlas.org (accessed on 17 February 2023)). The red rectangle on the mRNA panel highlights the ovarian cancer samples.
Several FOLR1-targeted therapeutics are currently in late phase clinical trials [91]. Consequently, using our learnings from targeting miRNA therapeutics for obesity and MAFLD, we are now developing ONT candidates conjugated to a fatty acid or a short peptide for enhanced targeted delivery to primary OC cells.
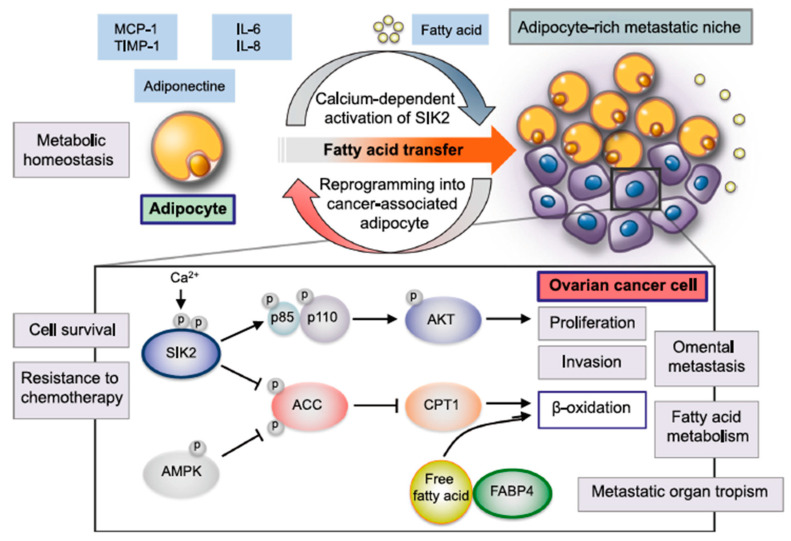
2.2.5. Active Targeted Delivery of Generation 2.5 miRNA-Based ONTs to the Adipocyte-Rich OC Tumor Microenvironment via FAT and FABP4 Transporters
We selected the fatty acid transporters FAT and FABP4, which are highly expressed at the surface of adipocytes (www.proteinatlas.org (accessed on 17 February 2023)), because the adipose-rich omentum microenvironment (cancer-associated adipocytes, CAAs) plays important roles in the spreading and resistance to treatments of ovarian cancer [92]. As recently reviewed by Motohara et al. [92], adipocytes present in the OC tumor microenvironment could augment cancer cell survival, spreading and resistance to chemotherapy via several mechanisms as shown in Figure 15.
Figure 15.
Various roles of adipocytes in the creation of the metabolic tumor microenvironment (TME) in the omentum during ovarian cancer metastasis, reprinted from Reference [92], an open-access article distributed under the terms and conditions of Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/ (accessed on 17 February 2023)).
2.2.6. In Vitro Testing of Generation 2.5 Candidate miRNA Agomirs and Antagomirs Will Be Conducted in Human Cells in Culture including the Following:
Epithelial ovarian cancer cell lines, namely SKOV3, SKOV3/CDDP, PA1, CAOV3, SW626, ES-2 and HO-8910;
Negative control cell lines and other cancer cell lines, namely HepG2 (liver) and A-549 VIM RFP (lung cancer);
Primary cultures of human adipocytes.
Cellular high-content Imaging will be performed using Phenovista (www.phenovista.com (to be performed)). Gene profiling will be conducted using the Nanostring PanCancer IO 360 Gene-Expression Panel (770 unique gene-expression panels) at the single-cell (CosMx SMI) and multi-cellular (GeoMix DSP) levels (www.nanostring.com (to be performed)). Direct identification of miRNA targets will be conducted using ECLIPSEBIO miR-eCLIP technology (www.eclipsebio.com (to be performed)).
Finally, in vivo testing of selected candidate targeting miRNA agomirs and antagomirs will be conducted in orthotopic and patient-derived tumor xenograft (PDX) mouse models of ovarian cancer.
3. Discussion and Conclusions
miRNA-based treatments provide a new frontier in the next generation of therapeutic development and are expected to significantly impact the landscape of disease management. By departing from the traditional “one drug, one target” approach to a “one drug, many targets” paradigm, miRNA therapeutics are expected to reduce polypharmacy and provide significant gains in drug efficacy, safety and patient compliance. Of course, issues surrounding the biophysical properties of miRNA-based treatments, such as in vivo stability, delivery and potential side effects due to immune activation, need to be addressed to enable safer and more efficacious biologics. In our own case, we identified miR-22-3p as a promising candidate for the treatment of metabolic disorders associated with obesity and MAFLD and demonstrated the efficacy of mir-22-3p antagomirs both in vitro in human cells and in vivo pre-clinical animal models. Currently, we are using computational modeling, bioinformatics and experimental approaches to develop generation 2.5 miR-22-3p antagomirs with improved efficacy and reduced side effects for the treatment of the aforementioned metabolic disorders. Furthermore, we are currently exploring the application of miRNA agomirs and antagomirs for the treatment of ovarian cancers where unmet medical needs are quite large.
4. Materials and Methods
4.1. In Vivo Experiments
Diet-induced obesity (DIO) male C57BL/6J mice purchased from the Jackson Laboratory (Bar Harbor, ME, USA) were housed at 24–26 °C with lights turned on at 08:00 and off at 20:00. They were fed at weaning on chow (Beekay rat and mouse diet 1). From six weeks of age, they were started on a 60% high-fat diet (Research Diet D12492). At the age of 12 weeks, the mice were allocated to treatment groups so that the mean and standard deviation for body weight, glucose and insulin were similar across the groups (12 animals per group, 2 animals of the same group per cage). After acclimation for 2 weeks, the mice were administered single subcutaneous injections of saline or the APT-110 miR-22-3p antagomir (15 mg/kg) in the left inguinal fat pad (injections on days 0, 2 and 4 of week 1 of treatment, then once a week for a total of 12 weeks) while they remained on the 60% high-fat diet.
Body weights were measured weekly. Food consumption per cage was measured daily. Blood samples were collected from the cut tip of the tail after the application of lignocaine gel during the in-life phase of the study. For plasma preparation, blood was collected in EDTA-coated microvettes for the measurement of plasma analytes and stored on ice, followed by centrifugation at ~5000× g for 5 min. The resulting plasma was stored at −80 °C until required. Multiple freeze/thaw cycles were avoided. Blood glucose concentration was analyzed as previously reported [93]. Plasma insulin (Cat #: 90080; Crystal Chem, Downers Grove, IL, USA), leptin (Cat #: 90030; Chrystal Chem), adiponectin (Cat # 47-ADPMS-E01, Alpco Diagnostics, Salem, NH, USA), NEFA (Cat # NEFA-HR(2); Wako Diagnostics, Mountain View, CA, USA), ALT (Cat # AL1205, Randox, Crumlin, UK), AST (Cat # AS1202, Randox), total cholesterol (Cat # CH200, Randox) and triglycerides (Cat # TR210, Randox) were measured as per the manufacturers’ recommendations.
Oral glucose tolerance test (OGTT) was performed as follows: six hours prior to the start of the glucose tolerance test (09h00), food was removed, and animals were given clean cages. Mice were dosed with glucose at T = 0 min. Glucose was dosed by oral gavage at a dose of 2.5 g/kg p.o. Blood samples were taken for the analysis of glucose concentration at −30, 0, 30, 60, 120 and 180 min relative to glucose administration. Blood samples were also taken at −30 and +30 min for insulin analysis. Food was returned at the end of the tolerance test.
Mice energy expenditure (EE) was measured by open-circuit calorimetry with the animals in their home cages [93,94]. The physical activity of the mice was recorded while the mice were kept in their original cages. Recordings were taken using an infrared recorder linked to a laptop. Recordings were made on the hour every hour from 7 pm until 8 am. Each recording lasted 10 min. Analysis of activity was conducted by virtually drawing two lines across each cage (thus dividing them into three equal parts). Recordings were analyzed by eye and the number of times a line was broken by a mouse in each 10 min segment was scored.
Body fat and lean content were measured using a Minispec LF90II Nuclear Magnetic Resonance (Bruker Corporation, Fremont, CA, USA). The mice were gently restrained, sufficient to keep them quiescent during this non-invasive technique.
At the end of the study, liver, heart, inguinal, perirenal, epididymal and subscapular fat samples were collected, weighed, then frozen for future gene-expression analysis; otherwise, they were placed in a 10% neutral buffered formalin solution, washed in PBS, pH 7.4 and transferred to 70% ethanol for subsequent processing for histologic analyses. Spleens were weighed and discarded. Blood was collected and processed into serum or plasma aliquots at the time of necropsy by cardiocentesis.
4.2. MiR-22 Antagomir
The miR-22-3p antagomir used in this study was designed by AptamiR Therapeutics, Inc. and custom synthesized (US Patent 62/329,537 on “Inhibition of mir-22 miRNA by APT-110”, initially published on 2 November 2017, WO2017/187426 A1). APT-110 is an 18 mer single-stranded oligonucleotide complementary to nucleotides 2 to 19 of miR-22-3p with phosphorothioate, locked nucleic acid, 2′0-Methyl, DNA and 5-methyl-Cytosine modifications. For subcutaneous dosing in the left inguinal area, APT-110 was prepared in saline (0.9 % NaCl). Control animals received the saline vehicle alone (5 mL/kg).
4.3. Imaging Analyses
Measurement of cells’ perimeter and area utilized the ImageJ image processing program developed by the NIH (https://imagej.nih.gov/ (accessed on 22 June 2020)).
4.4. RNA Sequencing Analyses
RNA sequencing was performed at the Genome Sequencing and Analysis Facility at the University of Texas in Austin on an Illumina HiSeq 4000 system following the manufacturer’s protocol. RNA data analyses were performed by Dr. Sujoy Ghosh at Duke-NUS Medical School, Singapore. Differentially expressed genes from fat (nominal p < 0.01) and liver (nominal p < 0.001) gene-expression data were subjected to pathway over-representation analysis via the Enrichr tool [95] (http://amp.pharm.mssm.edu/Enrichr/ (accessed on 15 November 2019)). A total of 1257 and 1495 genes were analyzed from fat and liver, respectively. Pathway enrichment was further investigated via the KEGG pathway database [96] (www.genome.jp/kegg/pathway.html (accessed on 15 November 2019)) and the WikiPathways database [97] (wikipathways.org). Pathways with a false-discovery rate < 0.1 were considered significantly enriched for differentially expressed genes. A subset of significantly enriched KEGG pathways (adj. p < 1.5 × 10−5) were further visualized via mean-average (MA) plots.
4.5. Statistical Analyses
Results given in the text and data points in the figures are shown as mean ± SEM. Statistical analysis used ANOVA and Student’s t-test, unless non-parametric tests were selected, based on data distribution (GraphPad Prism 8.4).
4.6. Studies Approval
The animal studies were performed according to IACUC-approved protocols and in compliance with the Guide for the Care and Use of Laboratory Animals (National Research Council, 2011) in OLAW-assured and AAALAC-accredited facilities at the Jackson Laboratory, Sacramento, CA (Study ID 40239, 17 April 2015), the University of Buckingham, UK (Study Bu15/030, 30 July 2015), the Drug Dynamics Institute at the University of Texas College of Pharmacy, Austin, TX (IACUC protocol AUP-2015-00125, 10 February 2016) and Aptuit S.r.l., Verona, Italy (Study VPT4074, 22 March 2016).
Acknowledgments
We would like to thank Pengyu Ren, Department of Biomedical Engineering, UT Austin for performing with his team in silico modeling of our ONTs.
Author Contributions
M.T. and S.G. contributed equally to the work. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not Applicable.
Informed Consent Statement
Not Applicable.
Data Availability Statement
Data presented in this article can be accessed in our previously published work [56,57] or by request to the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest. Thibonnier was involved in the design of study protocols, analyses and interpretation of data, the writing of the manuscript and the decision to publish the results. Marc Thibonnier is the founder and a shareholder of AptamiR Therapeutics, Inc.
Patents
Marc Thibonnier is the inventor of several patents assigned to AptamiR Therapeutics, Inc.
Funding Statement
This research received no external funding. Funding for this R&D work came from AptamiR Therapeutics, Inc. Sujoy Ghosh was partially supported by Louisiana Clinical and Translational Science Center grant (NIGMS 2U54GM104940) and by the National Medical Research Council, Singapore.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Alles J., Fehlmann T., Fischer U., Backes C., Galata V., Minet M., Hart M., Abu-Halima M., Grasser F.A., Lenhof H.P., et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019;47:3353–3364. doi: 10.1093/nar/gkz097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kozomara A., Birgaoanu M., Griffiths-Jones S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yedavilli S., Singh A.D., Singh D., Samal R. Nano-Messengers of the Heart: Promising Theranostic Candidates for Cardiovascular Maladies. Front. Physiol. 2022;13:895322. doi: 10.3389/fphys.2022.895322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henning R.J. Cardiovascular Exosomes and MicroRNAs in Cardiovascular Physiology and Pathophysiology. J. Cardiovasc. Transl. Res. 2021;14:195–212. doi: 10.1007/s12265-020-10040-5. [DOI] [PubMed] [Google Scholar]
- 5.Li C., Ni Y.Q., Xu H., Xiang Q.Y., Zhao Y., Zhan J.K., He J.Y., Li S., Liu Y.S. Roles and mechanisms of exosomal non-coding RNAs in human health and diseases. Signal Transduct. Target. Ther. 2021;6:383. doi: 10.1038/s41392-021-00779-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shu Z., Tan J., Miao Y., Zhang Q. The role of microvesicles containing microRNAs in vascular endothelial dysfunction. J. Cell. Mol. Med. 2019;23:7933–7945. doi: 10.1111/jcmm.14716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J., Li S., Li L., Li M., Guo C., Yao J., Mi S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genom. Proteom. Bioinform. 2015;13:17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Assmann T.S., Milagro F.I., Martinez J.A. Crosstalk between microRNAs, the putative target genes and the lncRNA network in metabolic diseases. Mol. Med. Rep. 2019;20:3543–3554. doi: 10.3892/mmr.2019.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennett C.F., Kordasiewicz H.B., Cleveland D.W. Antisense Drugs Make Sense for Neurological Diseases. Annu. Rev. Pharmacol. Toxicol. 2021;61:831–852. doi: 10.1146/annurev-pharmtox-010919-023738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garofalo M., Condorelli G., Croce C.M. MicroRNAs in diseases and drug response. Curr. Opin. Pharmacol. 2008;8:661–667. doi: 10.1016/j.coph.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 11.Jae N., Dimmeler S. Noncoding RNAs in Vascular Diseases. Circ. Res. 2020;126:1127–1145. doi: 10.1161/CIRCRESAHA.119.315938. [DOI] [PubMed] [Google Scholar]
- 12.Chandan K., Gupta M., Sarwat M. Role of Host and Pathogen-Derived MicroRNAs in Immune Regulation During Infectious and Inflammatory Diseases. Front. Immunol. 2019;10:3081. doi: 10.3389/fimmu.2019.03081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shah V., Shah J. Recent trends in targeting miRNAs for cancer therapy. J. Pharm. Pharmacol. 2020;72:1732–1749. doi: 10.1111/jphp.13351. [DOI] [PubMed] [Google Scholar]
- 14.Kousar K., Ahmad T., Abduh M.S., Kanwal B., Shah S.S., Naseer F., Anjum S. miRNAs in Regulation of Tumor Microenvironment, Chemotherapy Resistance, Immunotherapy Modulation and miRNA Therapeutics in Cancer. Int. J. Mol. Sci. 2022;23:13822. doi: 10.3390/ijms232213822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Veiga R.N., Zambalde E.P., Cox L., Jucoski T.S., Kohler A.F., Carvalho T.M., Rodrigues A.C., Ludwig B., Crowley K., de Oliveira J.C., et al. Regulation of Immune Cells by microRNAs and microRNA-Based Cancer Immunotherapy. Adv. Exp. Med. Biol. 2022;1385:75–108. doi: 10.1007/978-3-031-08356-3_3. [DOI] [PubMed] [Google Scholar]
- 16.Otmani K., Rouas R., Lewalle P. OncomiRs as noncoding RNAs having functions in cancer: Their role in immune suppression and clinical implications. Front. Immunol. 2022;13:913951. doi: 10.3389/fimmu.2022.913951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Momin M.Y., Gaddam R.R., Kravitz M., Gupta A., Vikram A. The Challenges and Opportunities in the Development of MicroRNA Therapeutics: A Multidisciplinary Viewpoint. Cells. 2021;10:3097. doi: 10.3390/cells10113097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bajan S., Hutvagner G. RNA-Based Therapeutics: From Antisense Oligonucleotides to miRNAs. Cells. 2020;9:137. doi: 10.3390/cells9010137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corey D.R., Damha M.J., Manoharan M. Challenges and Opportunities for Nucleic Acid Therapeutics. Nucleic Acid. 2022;32:8–13. doi: 10.1089/nat.2021.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chakraborty C., Sharma A.R., Sharma G., Lee S.S. Therapeutic advances of miRNAs: A preclinical and clinical update. J. Adv. Res. 2021;28:127–138. doi: 10.1016/j.jare.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartolucci D., Pession A., Hrelia P., Tonelli R. Precision Anti-Cancer Medicines by Oligonucleotide Therapeutics in Clinical Research Targeting Undruggable Proteins and Non-Coding RNAs. Pharmaceutics. 2022;14:1453. doi: 10.3390/pharmaceutics14071453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taniguchi H., Suzuki Y., Imai K., Adachi Y. Antitumoral RNA-targeted oligonucleotide therapeutics: The third pillar after small molecule inhibitors and antibodies. Cancer Sci. 2022;113:2952–2961. doi: 10.1111/cas.15461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamb Y.N. Inclisiran: First Approval. Drugs. 2021;81:389–395. doi: 10.1007/s40265-021-01473-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bennett C.F., Baker B.F., Pham N., Swayze E., Geary R.S. Pharmacology of Antisense Drugs. Annu. Rev. Pharm. Toxicol. 2017;57:81–105. doi: 10.1146/annurev-pharmtox-010716-104846. [DOI] [PubMed] [Google Scholar]
- 25.Crooke S.T., Baker B.F., Crooke R.M., Liang X.H. Antisense technology: An overview and prospectus. Nat. Rev. Drug Discov. 2021;20:427–453. doi: 10.1038/s41573-021-00162-z. [DOI] [PubMed] [Google Scholar]
- 26.Crooke S.T., Liang X.H., Baker B.F., Crooke R.M. Antisense Technology: A Review. J. Biol. Chem. 2021:100416. doi: 10.1016/j.jbc.2021.100416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alhamadani F., Zhang K., Parikh R., Wu H., Rasmussen T.P., Bahal R., Zhong X.B., Manautou J.E. Adverse Drug Reactions and Toxicity of the Food and Drug Administration-Approved Antisense Oligonucleotide Drugs. Drug Metab. Dispos. 2022;50:879–887. doi: 10.1124/dmd.121.000418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Migliorati J.M., Liu S., Liu A., Gogate A., Nair S., Bahal R., Rasmussen T.P., Manautou J.E., Zhong X.B. Absorption, Distribution, Metabolism, and Excretion of US Food and Drug Administration-Approved Antisense Oligonucleotide Drugs. Drug Metab. Dispos. 2022;50:888–897. doi: 10.1124/dmd.121.000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu A.M., Tu M.J. Deliver the promise: RNAs as a new class of molecular entities for therapy and vaccination. Pharm. Ther. 2022;230:107967. doi: 10.1016/j.pharmthera.2021.107967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moumne L., Marie A.C., Crouvezier N. Oligonucleotide Therapeutics: From Discovery and Development to Patentability. Pharmaceutics. 2022;14:260. doi: 10.3390/pharmaceutics14020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu B., Zhong L., Weng Y., Peng L., Huang Y., Zhao Y., Liang X.J. Therapeutic siRNA: State of the art. Signal Transduct. Target. Ther. 2020;5:101. doi: 10.1038/s41392-020-0207-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanna J., Hossain G.S., Kocerha J. The Potential for microRNA Therapeutics and Clinical Research. Front. Genet. 2019;10:478. doi: 10.3389/fgene.2019.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kara G., Arun B., Calin G.A., Ozpolat B. miRacle of microRNA-Driven Cancer Nanotherapeutics. Cancers. 2022;14:3818. doi: 10.3390/cancers14153818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suparpprom C., Vilaivan T. Perspectives on conformationally constrained peptide nucleic acid (PNA): Insights into the structural design, properties and applications. RSC Chem. Biol. 2022;3:648–697. doi: 10.1039/D2CB00017B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sahu B., Chenna V., Lathrop K.L., Thomas S.M., Zon G., Livak K.J., Ly D.H. Synthesis of conformationally preorganized and cell-permeable guanidine-based gamma-peptide nucleic acids (gammaGPNAs) J. Org. Chem. 2009;74:1509–1516. doi: 10.1021/jo802211n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaddam R.R., Dhuri K., Kim Y.R., Jacobs J.S., Kumar V., Li Q., Irani K., Bahal R., Vikram A. gamma Peptide Nucleic Acid-Based miR-122 Inhibition Rescues Vascular Endothelial Dysfunction in Mice Fed a High-Fat Diet. J. Med. Chem. 2022;65:3332–3342. doi: 10.1021/acs.jmedchem.1c01831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quijano E., Bahal R., Ricciardi A., Saltzman W.M., Glazer P.M. Therapeutic Peptide Nucleic Acids: Principles, Limitations, and Opportunities. Yale J. Biol. Med. 2017;90:583–598. [PMC free article] [PubMed] [Google Scholar]
- 38.Smolarz B., Durczynski A., Romanowicz H., Szyllo K., Hogendorf P. miRNAs in Cancer (Review of Literature) Int. J. Mol. Sci. 2022;23:2805. doi: 10.3390/ijms23052805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Rooij L.A., Mastebroek D.J., Ten Voorde N., van der Wall E., van Diest P.J., Moelans C.B. The microRNA Lifecycle in Health and Cancer. Cancers. 2022;14:5748. doi: 10.3390/cancers14235748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dwivedi S.K.D., Rao G., Dey A., Mukherjee P., Wren J.D., Bhattacharya R. Small Non-Coding-RNA in Gynecological Malignancies. Cancers. 2021;13:1085. doi: 10.3390/cancers13051085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hashemipour M., Boroumand H., Mollazadeh S., Tajiknia V., Nourollahzadeh Z., Rohani Borj M., Pourghadamyari H., Rahimian N., Hamblin M.R., Mirzaei H. Exosomal microRNAs and exosomal long non-coding RNAs in gynecologic cancers. Gynecol. Oncol. 2021;161:314–327. doi: 10.1016/j.ygyno.2021.02.004. [DOI] [PubMed] [Google Scholar]
- 42.Beg A., Parveen R., Fouad H., Yahia M.E., Hassanein A.S. Role of different non-coding RNAs as ovarian cancer biomarkers. J. Ovarian Res. 2022;15:72. doi: 10.1186/s13048-022-01002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pan G., Liu Y., Shang L., Zhou F., Yang S. EMT-associated microRNAs and their roles in cancer stemness and drug resistance. Cancer Commun. 2021;41:199–217. doi: 10.1002/cac2.12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shai A., Galouk E., Miari R., Tareef H., Sammar M., Zeidan M., Rayan A., Falah M. Inhibiting mutant KRAS G12D gene expression using novel peptide nucleic acid-based antisense: A potential new drug candidate for pancreatic cancer. Oncol. Lett. 2022;23:130. doi: 10.3892/ol.2022.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tastan B., Tarakcioglu E., Birinci Y., Park Y., Genc S. Role of Exosomal MicroRNAs in Cell-to-Cell Communication. Methods Mol. Biol. 2022;2257:269–292. doi: 10.1007/978-1-0716-1170-8_14. [DOI] [PubMed] [Google Scholar]
- 46.Sheiner L.B. Learning versus confirming in clinical drug development. Clin. Pharmacol. Ther. 1997;61:275–291. doi: 10.1016/S0009-9236(97)90160-0. [DOI] [PubMed] [Google Scholar]
- 47.Cecchini M. Use of healthcare services and expenditure in the US in 2025: The effect of obesity and morbid obesity. PLoS ONE. 2018;13:e0206703. doi: 10.1371/journal.pone.0206703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okunogbe A., Nugent R., Spencer G., Ralston J., Wilding J. Economic impacts of overweight and obesity: Current and future estimates for eight countries. BMJ Glob. Health. 2021;6:e006351. doi: 10.1136/bmjgh-2021-006351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eslam M., Sanyal A.J., George J., International Consensus P. MAFLD: A Consensus-Driven Proposed Nomenclature for Metabolic Associated Fatty Liver Disease. Gastroenterology. 2020;158:1999–2014 e1991. doi: 10.1053/j.gastro.2019.11.312. [DOI] [PubMed] [Google Scholar]
- 50.Correa L.H., Heyn G.S., Magalhaes K.G. The Impact of the Adipose Organ Plasticity on Inflammation and Cancer Progression. Cells. 2019;8:662. doi: 10.3390/cells8070662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kurylowicz A. microRNAs in Human Adipose Tissue Physiology and Dysfunction. Cells. 2021;10:3342. doi: 10.3390/cells10123342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rakib A., Kiran S., Mandal M., Singh U.P. MicroRNAs: A crossroad that connects obesity to immunity and aging. Immun. Ageing. 2022;19:64. doi: 10.1186/s12979-022-00320-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma L., Gilani A., Yi Q., Tang L. MicroRNAs as Mediators of Adipose Thermogenesis and Potential Therapeutic Targets for Obesity. Biology. 2022;11:1657. doi: 10.3390/biology11111657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Traube F.R., Carell T. The chemistries and consequences of DNA and RNA methylation and demethylation. RNA Biol. 2017;14:1099–1107. doi: 10.1080/15476286.2017.1318241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tateishi K., Okada Y., Kallin E.M., Zhang Y. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature. 2009;458:757–761. doi: 10.1038/nature07777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qian X., Li X., Shi Z., Bai X., Xia Y., Zheng Y., Xu D., Chen F., You Y., Fang J., et al. KDM3A Senses Oxygen Availability to Regulate PGC-1alpha-Mediated Mitochondrial Biogenesis. Mol. Cell. 2019;76:885–895 e887. doi: 10.1016/j.molcel.2019.09.019. [DOI] [PubMed] [Google Scholar]
- 57.Szklarczyk D., Kirsch R., Koutrouli M., Nastou K., Mehryary F., Hachilif R., Gable A.L., Fang T., Doncheva N.T., Pyysalo S., et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51:D638–D646. doi: 10.1093/nar/gkac1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Keller A., Groger L., Tschernig T., Solomon J., Laham O., Schaum N., Wagner V., Kern F., Schmartz G.P., Li Y., et al. miRNATissueAtlas2: An update to the human miRNA tissue atlas. Nucleic Acids Res. 2022;50:D211–D221. doi: 10.1093/nar/gkab808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thibonnier M., Esau C. Metabolic Benefits of MicroRNA-22 Inhibition. Nucleic Acid Ther. 2020;30:104–116. doi: 10.1089/nat.2019.0820. [DOI] [PubMed] [Google Scholar]
- 60.Thibonnier M., Esau C., Ghosh S., Wargent E., Stocker C. Metabolic and energetic benefits of microRNA-22 inhibition. BMJ Open Diabetes Res. Care. 2020;8:e001478. doi: 10.1136/bmjdrc-2020-001478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Glatz J.F.C., Luiken J. Dynamic role of the transmembrane glycoprotein CD36 (SR-B2) in cellular fatty acid uptake and utilization. J. Lipid Res. 2018;59:1084–1093. doi: 10.1194/jlr.R082933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Glatz J.F., Nabben M., Heather L.C., Bonen A., Luiken J.J. Regulation of the subcellular trafficking of CD36, a major determinant of cardiac fatty acid utilization. Biochim. Biophys. Acta. 2016;1861:1461–1471. doi: 10.1016/j.bbalip.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 63.Jing Z., Qi R., Thibonnier M., Ren P. Molecular Dynamics Study of the Hybridization between RNA and Modified Oligonucleotides. J. Chem. Theory Comput. 2019;15:6422–6432. doi: 10.1021/acs.jctc.9b00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jing Z., Ren P. Molecular Dynamics Simulations of Protein RNA Complexes by Using an Advanced Electrostatic Model. J. Phys. Chem. B. 2022;126:7343–7353. doi: 10.1021/acs.jpcb.2c05278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang C., Lu C., Jing Z., Wu C., Piquemal J.P., Ponder J.W., Ren P. AMOEBA Polarizable Atomic Multipole Force Field for Nucleic Acids. J. Chem. Theory Comput. 2018;14:2084–2108. doi: 10.1021/acs.jctc.7b01169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mosa R., Huang L., Wu Y., Fung C., Mallawakankanamalage O., LeRoith D., Chen C. Hexarelin, a Growth Hormone Secretagogue, Improves Lipid Metabolic Aberrations in Nonobese Insulin-Resistant Male MKR Mice. Endocrinology. 2017;158:3174–3187. doi: 10.1210/en.2017-00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marechal L., Laviolette M., Rodrigue-Way A., Sow B., Brochu M., Caron V., Tremblay A. The CD36-PPARgamma Pathway in Metabolic Disorders. Int. J. Mol. Sci. 2018;19:1529. doi: 10.3390/ijms19051529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang Z., Lu X., Huang L., Zhang C., Veldhuis J.D., Cowley M.A., Chen C. Stimulation of endogenous pulsatile growth hormone secretion by activation of growth hormone secretagogue receptor reduces the fat accumulation and improves the insulin sensitivity in obese mice. FASEB J. 2021;35:e21269. doi: 10.1096/fj.202001924RR. [DOI] [PubMed] [Google Scholar]
- 69.Webb P.M., Jordan S.J. Epidemiology of epithelial ovarian cancer. Best Pract. Res. Clin. Obs. Gynaecol. 2017;41:3–14. doi: 10.1016/j.bpobgyn.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 70.Dinkelspiel H.E., Champer M., Hou J., Tergas A., Burke W.M., Huang Y., Neugut A.I., Ananth C.V., Hershman D.L., Wright J.D. Long-term mortality among women with epithelial ovarian cancer. Gynecol. Oncol. 2015;138:421–428. doi: 10.1016/j.ygyno.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Z., Meng F., Zhong Z. Emerging targeted drug delivery strategies toward ovarian cancer. Adv. Drug Deliv. Rev. 2021;178:113969. doi: 10.1016/j.addr.2021.113969. [DOI] [PubMed] [Google Scholar]
- 72.Heh E., Allen J., Ramirez F., Lovasz D., Fernandez L., Hogg T., Riva H., Holland N., Chacon J. Peptide Drug Conjugates and Their Role in Cancer Therapy. Int. J. Mol. Sci. 2023;24:829. doi: 10.3390/ijms24010829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yoshida K., Yokoi A., Kato T., Ochiya T., Yamamoto Y. The clinical impact of intra- and extracellular miRNAs in ovarian cancer. Cancer Sci. 2020;111:3435–3444. doi: 10.1111/cas.14599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zavesky L., Jandakova E., Weinberger V., Hanzikova V., Slanar O., Kohoutova M. Ascites in ovarian cancer: MicroRNA deregulations and their potential roles in ovarian carcinogenesis. Cancer Biomark. 2022;33:1–16. doi: 10.3233/CBM-210219. [DOI] [PubMed] [Google Scholar]
- 75.Strumidlo A., Skiba S., Scott R.J., Lubinski J. The potential role of miRNAs in therapy of breast and ovarian cancers associated with BRCA1 mutation. Hered. Cancer Clin. Pract. 2017;15:15. doi: 10.1186/s13053-017-0076-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Staicu C.E., Predescu D.V., Rusu C.M., Radu B.M., Cretoiu D., Suciu N., Cretoiu S.M., Voinea S.C. Role of microRNAs as Clinical Cancer Biomarkers for Ovarian Cancer: A Short Overview. Cells. 2020;9:169. doi: 10.3390/cells9010169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Segal M., Slack F.J. Challenges identifying efficacious miRNA therapeutics for cancer. Expert Opin. Drug Discov. 2020;15:987–992. doi: 10.1080/17460441.2020.1765770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rattanapan Y., Korkiatsakul V., Kongruang A., Siriboonpiputtana T., Rerkamnuaychoke B., Chareonsirisuthigul T. MicroRNA Expression Profiling of Epithelial Ovarian Cancer Identifies New Markers of Tumor Subtype. Microrna. 2020;9:289–294. doi: 10.2174/2211536609666200722125737. [DOI] [PubMed] [Google Scholar]
- 79.Liang Z., Lu Z., Zhang Y., Shang D., Li R., Liu L., Zhao Z., Zhang P., Lin Q., Feng C., et al. Targeting Membrane Receptors of Ovarian Cancer Cells for Therapy. Curr. Cancer Drug Targets. 2019;19:449–467. doi: 10.2174/1568009618666181010091246. [DOI] [PubMed] [Google Scholar]
- 80.Mirahmadi Y., Nabavi R., Taheri F., Samadian M.M., Ghale-Noie Z.N., Farjami M., Samadi-Khouzani A., Yousefi M., Azhdari S., Salmaninejad A., et al. MicroRNAs as Biomarkers for Early Diagnosis, Prognosis, and Therapeutic Targeting of Ovarian Cancer. J. Oncol. 2021;2021:3408937. doi: 10.1155/2021/3408937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.O’Malley D.M., Matulonis U.A., Birrer M.J., Castro C.M., Gilbert L., Vergote I., Martin L.P., Mantia-Smaldone G.M., Martin A.G., Bratos R., et al. Phase Ib study of mirvetuximab soravtansine, a folate receptor alpha (FRalpha)-targeting antibody-drug conjugate (ADC), in combination with bevacizumab in patients with platinum-resistant ovarian cancer. Gynecol. Oncol. 2020;157:379–385. doi: 10.1016/j.ygyno.2020.01.037. [DOI] [PubMed] [Google Scholar]
- 82.Heo Y.A. Etranacogene Dezaparvovec: First Approval. Drugs. 2023;83:347–352. doi: 10.1007/s40265-023-01845-0. [DOI] [PubMed] [Google Scholar]
- 83.Gilbert L., Oaknin A., Matulonis U.A., Mantia-Smaldone G.M., Lim P.C., Castro C.M., Provencher D., Memarzadeh S., Method M., Wang J., et al. Safety and efficacy of mirvetuximab soravtansine, a folate receptor alpha (FRalpha)-targeting antibody-drug conjugate (ADC), in combination with bevacizumab in patients with platinum-resistant ovarian cancer. Gynecol. Oncol. 2023;170:241–247. doi: 10.1016/j.ygyno.2023.01.020. [DOI] [PubMed] [Google Scholar]
- 84.Thompson S., Dessi J., Self C.H. Preclinical evaluation of light-activatable, bispecific anti-human CD3 antibody conjugates as anti-ovarian cancer therapeutics. MAbs. 2009;1:348–356. doi: 10.4161/mabs.1.4.9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hong Y., Nam S.M., Moon A. Antibody-drug conjugates and bispecific antibodies targeting cancers: Applications of click chemistry. Arch. Pharm. Res. 2023;46:131–148. doi: 10.1007/s12272-023-01433-6. [DOI] [PubMed] [Google Scholar]
- 86.Chen S.H., Dominik P.K., Stanfield J., Ding S., Yang W., Kurd N., Llewellyn R., Heyen J., Wang C., Melton Z., et al. Dual checkpoint blockade of CD47 and PD-L1 using an affinity-tuned bispecific antibody maximizes antitumor immunity. J. Immunother. Cancer. 2021;9:e003464. doi: 10.1136/jitc-2021-003464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bax H.J., Chauhan J., Stavraka C., Santaolalla A., Osborn G., Khiabany A., Grandits M., Lopez-Abente J., Palhares L., Chan Wah Hak C., et al. Folate receptor alpha in ovarian cancer tissue and patient serum is associated with disease burden and treatment outcomes. Br. J. Cancer. 2022;128:342–353. doi: 10.1038/s41416-022-02031-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kobel M., Madore J., Ramus S.J., Clarke B.A., Pharoah P.D., Deen S., Bowtell D.D., Odunsi K., Menon U., Morrison C., et al. Evidence for a time-dependent association between FOLR1 expression and survival from ovarian carcinoma: Implications for clinical testing. An Ovarian Tumour Tissue Analysis consortium study. Br. J. Cancer. 2014;111:2297–2307. doi: 10.1038/bjc.2014.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Markert S., Lassmann S., Gabriel B., Klar M., Werner M., Gitsch G., Kratz F., Hasenburg A. Alpha-folate receptor expression in epithelial ovarian carcinoma and non-neoplastic ovarian tissue. Anticancer Res. 2008;28:3567–3572. [PubMed] [Google Scholar]
- 90.Liu J., Liu L., Antwi P.A., Luo Y., Liang F. Identification and Validation of the Diagnostic Characteristic Genes of Ovarian Cancer by Bioinformatics and Machine Learning. Front. Genet. 2022;13:858466. doi: 10.3389/fgene.2022.858466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Scaranti M., Cojocaru E., Banerjee S., Banerji U. Exploiting the folate receptor alpha in oncology. Nat. Rev. Clin. Oncol. 2020;17:349–359. doi: 10.1038/s41571-020-0339-5. [DOI] [PubMed] [Google Scholar]
- 92.Motohara T., Masuda K., Morotti M., Zheng Y., El-Sahhar S., Chong K.Y., Wietek N., Alsaadi A., Carrami E.M., Hu Z., et al. An evolving story of the metastatic voyage of ovarian cancer cells: Cellular and molecular orchestration of the adipose-rich metastatic microenvironment. Oncogene. 2019;38:2885–2898. doi: 10.1038/s41388-018-0637-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stocker C.J., Wargent E., O’Dowd J., Cornick C., Speakman J.R., Arch J.R., Cawthorne M.A. Prevention of diet-induced obesity and impaired glucose tolerance in rats following administration of leptin to their mothers. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007;292:R1810–R1818. doi: 10.1152/ajpregu.00676.2006. [DOI] [PubMed] [Google Scholar]
- 94.Arch J.R., Trayhurn P. Detection of thermogenesis in rodents in response to anti-obesity drugs and genetic modification. Front. Physiol. 2013;4:64. doi: 10.3389/fphys.2013.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma’ayan A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kanehisa M. KEGG Bioinformatics Resource for Plant Genomics and Metabolomics. Methods Mol. Biol. 2016;1374:55–70. doi: 10.1007/978-1-4939-3167-5_3. [DOI] [PubMed] [Google Scholar]
- 97.Slenter D.N., Kutmon M., Hanspers K., Riutta A., Windsor J., Nunes N., Melius J., Cirillo E., Coort S.L., Digles D., et al. WikiPathways: A multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 2018;46:D661–D667. doi: 10.1093/nar/gkx1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data presented in this article can be accessed in our previously published work [56,57] or by request to the corresponding author.