Summary
Helicobacter suis, hosted by hogs, is the most prevalent gastric non-Helicobacter pylori Helicobacter species found in humans. Recent studies have suggested that H. suis infection has caused many cases of gastric disease, but the transmission route from hogs remains unclear. Diagnostic methods based on H. suis urease activity often yield negative results, and there is no reliable method for diagnosing H. suis infection in clinical practice without gastric biopsy specimens. This study presents the world’s first use of whole-bacterial cell ELISA to simultaneously assess H. suis and H. pylori infections. The ELISAs showed high accuracy, with an area under the ROC curve of 0.96, 100% sensitivity, 92.6% specificity, 76.9% positive predictive value, and 100% negative predictive value for the H. suis test, and an area under the ROC curve of 0.92, 88.2% sensitivity, 87.5% specificity, 65.2% positive predictive value, and 96.6% negative predictive value for the H. pylori test.
Subject areas: Diagnostic procedure, Gastroenterology, Virology
Graphical abstract
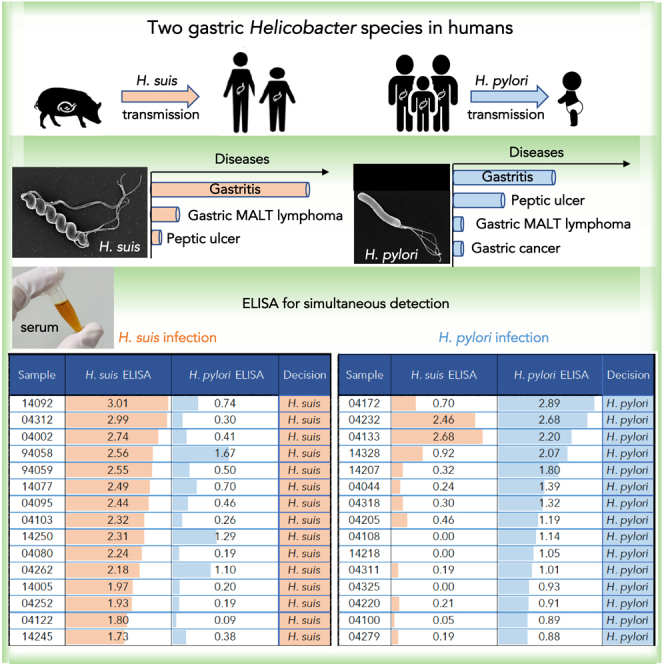
Highlights
-
•
Gastric non-Helicobacter pylori Helicobacter species in humans
-
•
ELISA for the simultaneous diagnosis of Helicobacter suis and H. pylori infection
-
•
Evaluation of the anti-H. suis antibody titers after eradication
-
•
Whole-genome sequences of H. suis strains isolated from humans
Diagnostic procedure; Gastroenterology; Virology
Introduction
Helicobacter suis, which differs from Helicobacter pylori in its bipolar flagella and corkscrew-like spiral morphology, is classified as a member of the gastric non-H. pylori Helicobacter species (NHPH), a group of bacteria that have been known to infect the human stomach since the 1980s.1,2,3,4 The natural hosts of H. suis are macaques and hogs, whereas other NHPH such as Helicobacter heilmannii sensu stricto (s.s.), Helicobacter ailurogastricus, Helicobacter felis, Helicobacter salomonis, Helicobacter baculiformis, Helicobacter cynogastricus, and Helicobacter bizzozeronii infect the stomachs of cats and dogs.5,6,7 Comparative genomic assessment of multilocus sequencing typing (MLST) of H. suis based on seven housekeeping genes (atpA, efp, mutY, ppa, trpC, ureAB, and yphC) revealed high genomic homology between hog- and human-derived isolates, suggesting that H. suis infection in humans is a zoonosis originating from hogs.5,8,9
Because the conventional H. pylori diagnostic tests based on the urease activity, such as the urea breath test (UBT) and rapid urease test (RUT), often yield negative judgments in the case of NHPH infecting the human stomach, PCR or histological examination from gastric biopsy specimens has been performed to diagnose NHPH infection for research purposes. In past reports, the prevalence of NHPH infection was shown to be less than 0.5% by microscopic detection of the NHPH morphology in gastric sections.10,11,12 In more recent reports, Tsukadaira et al. estimated the NHPH infection rate in Japan to be approximately 3.35%.13 When a PCR assay targeting urease genes was performed on the 30 patients with NHPH-associated gastritis, 28 were infected with NHPH as follows (urease genes were not detected in 2 patients): 26 with H. suis and 2 with H. heilmannii s.s. or H. ailurogastricus.13 They concluded that H. suis was the most common NHPH that infected human stomachs.13 The H. suis infection leads to diseases such as gastric mucosa-associated lymphoid tissue (MALT) lymphoma, nodular gastritis, chronic gastritis, and peptic ulcer.14,15,16,17,18,19,20 However, the above reports did not provide proof of H. suis or other NHPH infection because they only used PCR assay and did not isolate the bacteria or analyze the isolated bacteria’s genome to identify the infecting bacteria.
Because most H. pylori infections occur in infancy, usually via transmission from family members, the infection rate of H. pylori has dramatically declined mainly because of improvements in the general hygiene environment in childhood in Japan.21 As a result, the prevalence of H. suis infection could become more significant than the H. pylori infection rate if no measures are taken to prevent H. suis infection.22 Because eradication therapy of H. suis is effective in H. pylori-negative gastric MALT lymphoma,17,23 the H. suis infection could be a predictive marker for the efficacy of eradication therapy for H. pylori-negative gastric MALT lymphoma.24 Therefore, the development of comprehensive methods to diagnose H. suis infection has become a matter of urgency for clinical practice.
Nonetheless, given the current worldwide infection status—i.e., more than half of the world’s population is still infected with H. pylori25—there is a need for rapid diagnostic products that can detect both H. suis and H. pylori infections simultaneously. This is the first report of a serological diagnosis that simultaneously assesses H. suis and H. pylori infection in humans; the study also reports the evaluation of the anti-H. suis antibody titers after eradication.
Results
Classification of the specimens
As shown in Table 1, by using routine methods, PCR, and culture for diagnosing Helicobacter infections, we classified all 101 subjects into the following four groups in advance: an H. suis-infection group (n = 20), an H. pylori-infection group (n = 17), a group with eradication of H. pylori infection (n = 20), and a group with neither H. suis nor H. pylori infection (n = 44). The patients in these groups had gastritis (n = 52), gastric mucosa-associated lymphoid tissue (MALT) lymphoma (n = 23), duodenal MALT lymphoma (n = 1), peptic ulcer (n = 20), autoimmune (metaplastic atrophic) gastritis (n = 4), and endoscopic mucosal resection (EMR) for early gastric cancer (EGC; n = 1). The percentage of men in the H. suis infection group was 85%, which was significantly different from the percentages in the H. suis non-infection groups, i.e., the H. pylori infection group (58.8%), the post-eradication of H. pylori infection group (45.0%), and the non-infection group (45.0%) (all p< 0.05). Table S1 (Individual data of 101 examinees) shows the individual data of the 101 examinees. The partial 23S rRNA gene sequences of the 13 H suis and 16 H pylori strains isolated in this study were compared with those of 21 reference Helicobacter strains, shown in Table S2 (Reference strains of gastric Helicobacter species). The genomic analysis successfully classified every Helicobacter strain with species specificity. All H. suis and H. pylori strains belonged to the same phylogenetic clades of H. suis and H. pylori reference strains, indicating that the genomic identification in this study was correct. We proved that all H. suis strains have more than 99% average nucleotide identity (ANI) values against the H. suis type strain HS1, shown in Figure S1 (Phylogenetic analysis of Helicobacter strains).
Table 1.
Examinee background characteristics
|
H. suis infection (n = 20) |
H. suis non-infection (n = 81) |
|||
|---|---|---|---|---|
| H. pylori infection (n = 17) | Post-eradication of H. pylori infection (n = 20) | Non-infection (n = 44) | ||
| Year of age | ||||
| Average (interquartile range) | 48.5 (44.5–53.8) | 49.0 (40.0–55.0) | 52.0 (40.0–64.5) | 53.5 (46.0–66.5) |
| Sex (%) | ||||
| man | 17 (85.0) | 10 (58.8) | 9 (45.0) | 20 (45.0) |
| woman | 3 (15.0) | 7 (41.2) | 11 (55.0) | 24 (55.0) |
| Disease (%) | ||||
| gastritis | 15 (75.0) | 10 (58.8) | 14 (70.0) | 13 (29.5) |
| gastric MALT lymphoma | 3 (15.0) | 1 (5.9) | 1 (5.0) | 18 (41.0) |
| duodenal MALT lymphoma | 0 (0.0) | 0 (0.0) | 1 (5.0) | 0 (0.0) |
| peptic ulcer | 1 (5.0) | 5 (29.4) | 4 (20.0) | 10 (22.7) |
| autoimmune (metaplastic atrophic) gastritis | 1 (5.0) | 0 (0.0) | 0 (0.0) | 3 (6.8) |
| EGC | 0 (0.0) | 1 (5.9) | 0 (0.0) | 0 (0.0) |
Measurement of the anti-H. suis and anti-H. pylori antibody titers
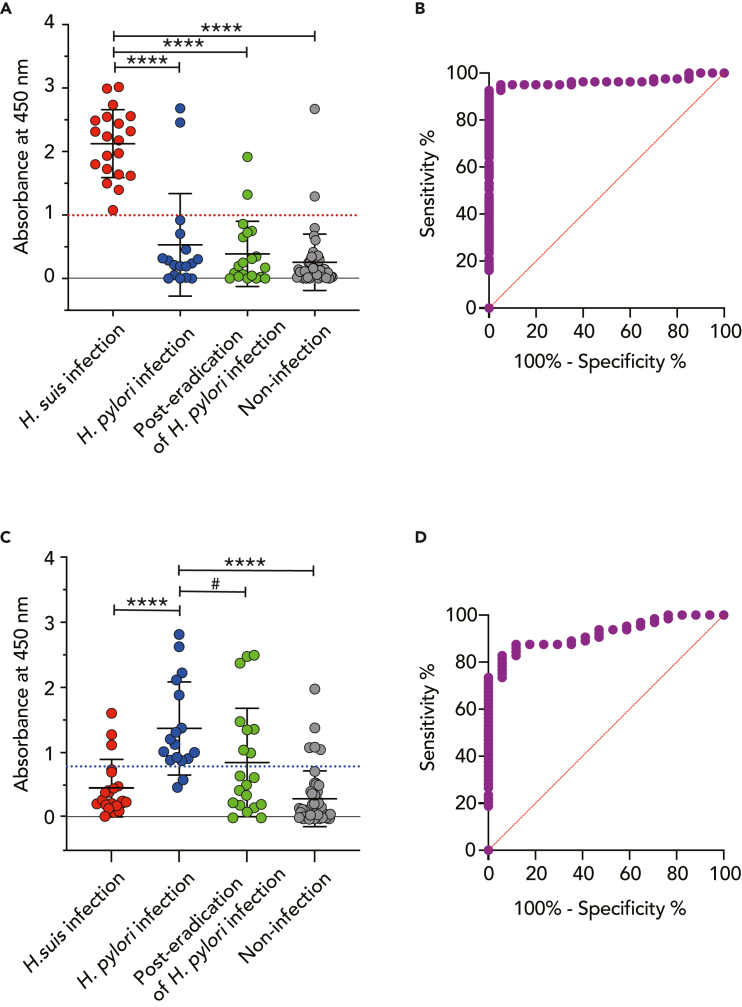
As shown in Figure 1, the mean with standard deviation (SD) of the absorbance at 450 nm of ELISA for identifying H. suis infection in the H. suis infection group was 2.1 ± 0.5, which was significantly higher than those of the H. suis non-infection groups—i.e., the H. pylori infection group (0.5 ± 0.8, p< 0.0001), the post-eradication of H. pylori infection group (0.4 ± 0.5, p< 0.0001), and the non-infection group (0.3 ± 0.4, p< 0.0001) (Figure 1A). The cut-off value (1.0) for the absorbance at 450 nm of H. suis infection was calculated by receiver operating characteristic (ROC) curve analysis between the H. suis infection group (n = 20) and the H. suis non-infection groups (n = 81) —i.e., the H. pylori infection (n = 17), post-eradication of H. pylori infection (n = 20), and non-infection with either H. suis or H. pylori (n = 44) groups (Figure 1B). Meanwhile, the mean with SD of the absorbance at 450 nm of ELISA for identifying H. pylori infection in the H. pylori infection group was 1.4 ± 0.7, which was significantly higher than those of two of the H. pylori non-infection groups—i.e., the H. suis infection group (0.5 ± 0.4, p< 0.0001) and the non-infection group (0.3 ± 0.4, p< 0.0001)—but not the post-eradication of H. pylori infection group (0.9 ± 0.8, p> 0.05) (Figure 1C). Then, the cut-off value (0.8) for the absorbance at 450 nm of H. pylori infection was calculated by ROC curve analysis between the H. pylori infection group (n = 17) and the H. pylori non-infection groups (n = 64), which were the H. suis infection group (n = 20) and the group with non-infection of either H. suis or H. pylori (n = 44) (Figure 1D). The sensitivity, specificity, and positive and negative predictive values (PPV and NPV, respectively) of ELISA for identifying the H. suis infection group against the H. suis non-infection groups were 100% (95% confidence interval [CI]: 83.9–100%), 92.6% (95% CI: 84.8–96.6%), 76.9% (95% CI: 55.9–90.2%), and 100% (95% CI: 93.9–100%), respectively (Table 2). On the other hand, the sensitivity, specificity, PPV, and NPV of ELISA for identifying the H. pylori infection group against the H. pylori non-infection groups were 88.2% (95% CI: 65.7–97.9%), 87.5% (95% CI: 77.2–93.5%), 65.2% (95% CI: 42.8–82.8%) and 96.6% (95% CI: 87–99.4%), respectively (Table 3).
Figure 1.
ELISA for identification of H. suis and H. pylori infection
Serum specimens (n = 101) were divided into four groups: an H. suis-infection group (n = 20), an H. pylori-infection group (n = 17), a group with eradication of H. pylori infection (n = 20), and a group with neither H. suis nor H. pylori infection (n = 44).
(A) ELISA for detecting H. suis infection. ∗∗∗p< 0.0001, the H. suis infection group vs. the H. pylori infection, the post-eradication of H. pylori infection, or the non-infection group. The red dotted line indicates the cut-off value (1.0). The bar represents the mean with SD.
(B) ROC curve constructed from the H. suis-infection group vs. the H. suis non-infection groups, which consisted of the H. pylori infection, the post-eradication of H. pylori infection, and the non-infection groups. The red slanted line is the reference line. The area under the ROC curve (AUC): 0.96 ± 0.02; 95% CI: 0.93 to 1.00; p< 0.0001.
(C) ELISA for detecting H. pylori infection. ∗∗∗∗p< 0.0001, the H. pylori infection group vs. the H. suis infection or non-infection group. #p> 0.05, the H. pylori infection group vs. the post-eradication of H. pylori infection group. The blue dotted line indicates the cut-off value (0.8). The bar represents the mean with SD.
(D) ROC curve constructed from the H. pylori infection group vs. the H. pylori non-infection groups, including the H. suis infection and the non-infection groups. The red slanted line is the reference line. AUC: 0.92 ± 0.03; 95% CI: 0.86 to 0.98; p< 0.0001.
Table 2.
ELISA to identify H. suis infection
| Positive | Negative | Total | ||
|---|---|---|---|---|
| Absorbance | ≥1.0 | 20 | 6 | 26 |
| at 450 nm | <1.0 | 0 | 75 | 75 |
| Total | 20 | 81 | 101 |
Positive includes the H. suis infection group (n = 20). Negative includes the H. suis non-infection groups (n = 81). All specimens (n = 101) were classified into two groups: a group with an H. suis-infection value greater than or equal to the cut-off value (n = 26) and a group with an H. suis-infection value smaller than the cut-off value (n = 75). Cut-off value, 1.0; sensitivity,; specificity, ; PPV, ; NPV, .
Table 3.
ELISA to identify H. pylori infection
| Positive | Negative | Total | ||
|---|---|---|---|---|
| Absorbance | ≥0.8 | 15 | 8 | 23 |
| at 450 nm | <0.8 | 2 | 56 | 58 |
| Total | 17 | 64 | 81 |
Positive includes the H. pylori infection group (n = 17). Negative includes the H. suis infection group and the non-infection group (n = 64). All specimens (n = 81) were classified into two groups: a group with an H. pylori-infection value greater than or equal to the cut-off value (n = 23) and a group with an H. pylori-infection value smaller than the cut-off value (n = 58). Cut-off value, 0.8; sensitivity, ; specificity, ; PPV, ; NPV,.
To validate the test methods, ELISA for identifying H. suis infection was performed at another institute using the same protocol and serum from a separate container. The cut-off value obtained was reproducible at 1.0. Correlation analysis of the data between the two institutes yielded a value of Pearson’s correlation coefficient (R) of 0.99 (95% CI: 0.97 to 0.99; p< 0.0001). Furthermore, the ELISA was always validated with 1800-fold diluted serum in addition to 3600-fold diluted serum, shown in Figure S2 (ELISA for identification of H. suis and H. pylori infection using 1800-fold diluted sera). Cohen’s kappa for the dichotomous results of the positive and negative assessments was 1.0 (complete agreement) between the two ELISAs.
Decline of anti-H. suis antibody titers after eradication
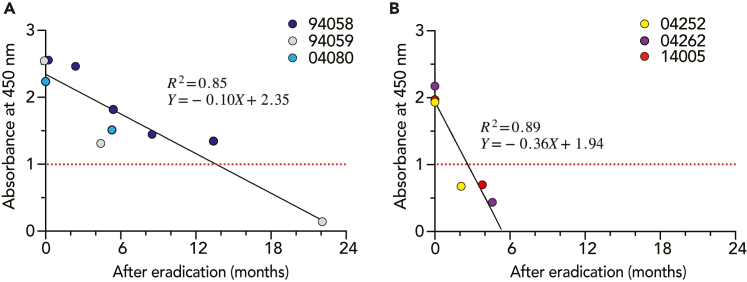
Among 20 individuals belonging to the H. suis infection group, six individuals (nos. 94058, 94059, 04080, 04252, 04262, and 14005) suffering from gastritis were administered the eradication therapy as described in the Methods section. After the eradication therapies of H. suis, the upper gastrointestinal (GI ) endoscopic pathology and PCR for the detection of H. suis were performed, and successful eradication of H. suis infection was judged in all six of these patients. The anti-H. suis antibody titers decreased over time after eradication. The estimated times required for the anti-H. suis antibody titers to decline to 1.0 (the cut-off value for the absorbance at 450 nm) in the 6 specimens were as follows: 15.1 months (no. 94058), 12.3 months (no. 94059), 9.1 months (no. 04080), 1.6 months (no. 04252), 3.1 months (no. 04262), and 2.8 months (no. 14005). Then, the six individuals were divided into the following two groups: a group of individuals exhibiting a slow decline of anti-H. suis antibody titers after eradication (nos. 9405894059, and 04080) and a group exhibiting a fast decline of anti-H. suis antibody titers after eradication (nos. 04252, 04262, and 14005). The time required for the anti-H. suis antibody titers to decline until the cut-off value was re-calculated at 13.6 and 2.6 months after eradication in the slow (Figure 2A) and first (Figure 2B) decline groups, respectively. The difference in eradication efficacy between the slow and fast decline groups seems to be attributed to the difference in antibody titers before eradication (2.4 ± 0.2 and 2.0 ± 0.1 in the slow and fast decline groups, respectively, p< 0.05).
Figure 2.
Measurement of anti-H. suis antibody titers after the eradication of H. suis infection
Six individuals underwent H. suis-eradication treatment, and then were monitored for serum anti-H. suis antibody titers by ELISA. Results for three individuals (nos. 94058, 94059, and 04080) (A) were plotted separately from the other three (nos. 04252, 04262, and 14005)
(B). The red dotted line indicates the cut-off value.
Discussion
The infection source of H. suis
Twenty out of 101 examinees were infected with H. suis, but no other NHPH were identified by the culture and PCR (negative and positive judgments in PCR targeting the hsvA and NHPH 16S rRNA genes, respectively). NHPH other than H. suis, which colonize the stomachs of cats and dogs, are thought to have a lower risk of infection in the human stomach. On the other hand, no H. suis infection has yet been identified in the stomachs of cats and dogs.5,6,7,26,27 Although H. suis colonizes the hog stomach and is the source of zoonotic infection in a high percentage of human cases of H. suis infection,28,29,30 the infection rate among hog farmers is not higher than that of individuals with no contact with animals.31,32 It has been hypothesized that H. suis infects humans via the consumption of contaminated pork.33,34 However, there is no clear evidence regarding the H. suis transmission route. In the present study, sampling was not random, and the population was subject to selection bias. Nevertheless, mixed infections with H. pylori and H. suis were not observed. Most patients infected with H. suis were men (Table 1). Tsukadaira et al. also noted that 25 out of 26 patients infected with H. suis were men.13 However, this study was limited to cases with endoscopic gastritis, so it cannot provide information on infection rates, gender, or other characteristics. To establish a more accurate correlation between infection rates, gender, age, symptoms, endoscopic findings, and disease, large-scale epidemiological studies that cover the general population, including those who undergo serological diagnosis, are necessary.
Comparison of the serological assays for the detection of H. suis and H. pylori infection
Marini et al. have tried detecting anti-Helicobacter antibodies of macaques by ELISA using the H. pylori and H. suis outer membrane protein preparations as antigens.35 In the identification of H. suis infection, the sensitivity and specificity were 70.0% (95% CI: 34.8%–93.3%) and 75.0% (95% CI: 19.4%–99.4%), respectively.35 Meanwhile, in the identification of H. pylori infection, the sensitivity and specificity were 66.7% (95% CI: 9.2%–99.2%) and 63.6% (95% CI: 30.8%–89.1%), respectively.35 Therefore, the ELISA data using the whole bacterial cell lysates as antigens and the 3600-fold diluted sera in this study (Figure 1 and Tables 2 and 3) showed more accuracy than the ELISA data using the outer membrane protein preparations as antigens. When synthetic peptides constructed from each H. suis or H. pylori-specific autotransporter protein sequence were used as antigens, ELISA did not show higher accuracy compared to whole-cell lysates as antigens.
The discrepancy in the judgments made between the serological assay and the PCR or culture
We conducted conventional Giemsa staining and immunohistochemistry (IHC) to identify H. suis and H. pylori in gastric biopsy specimens, shown in Table S1 (Individual data of 101 examinees). To compare the discriminatory ability between the serological assay and PCR or culture using gastric biopsy specimens, we attempted to detect H. suis and H. pylori by Giemsa staining and IHC analysis of the biopsy specimens. Among 17 (H. pylori infection group) and 20 (post-eradication of H. pylori infection group) individuals, four serum specimens (nos. 04232 and 04133 of H. pylori infection group, and nos. 04150 and 04162 of post-eradication of H. pylori infection group) were H. suis-positive by ELISA (Table S1 [Individual data of 101 examinees]). Because the microscopy did not detect H. suis in these specimens, we suggest that the whole-cell lysate of H. suis SNTW101c reacted with anti-H. pylori serum antibodies. For this reason, we advocate simultaneous serological testing for H. suis and H. pylori infections. Among 44 individuals in the non-infection group (PCR and culture negatives), two serum specimens (nos. 04293 and 04018 from patients with gastric MALT lymphoma and peptic ulcer, respectively) were H. suis-positive by ELISA, shown in Table S1 (Individual data of 101 examinees). Histology could not explain the discrepancies between serology and PCR or culture. However, we presume that the individuals had prior infections with H. suis (no. 04293) and H. pylori (no. 04018), because the serum specimens tested strongly positive for H. suis infection (no. 04293) and H. pylori infection (no. 04018) by ELISA, shown in Table S1 (Individual data of 101 examinees). In addition, in the non-infection group, it is assumed that the other four individuals had previously been infected with H. pylori and that the H. pylori were eradicated spontaneously or accidentally because the serum specimens (nos. 04167, 14327, 04157, and 14202) tested positive for H. pylori by ELISA, shown in Table S1 (Individual data of 101 examinees).
Evaluation of the H. suis eradication therapies by serological assay
When H. suis infection becomes apparent, eradication of H. suis is recommended even if the examinees are not suffering from severe illnesses such as gastric MALT lymphoma, nodular gastritis, or intractable peptic ulcer. The regimen for H. pylori eradication has been used for H. suis eradication. In most cases, patients are treated with triple-agent eradication therapy consisting of an inhibitor of gastric acid secretion such as a PPI or a potassium-competitive acid blocker (P-CAB) along with two antibiotics such as amoxicillin and clarithromycin or amoxicillin and metronidazole for 7 to 14 days.14,15,18,36,37,38 In the current study, the time-dependent decline of antibody titers after eradication confirmed the success of the eradication therapies. The serological assay of H. suis infection would be beneficial as a follow-up examination to confirm eradication (Figure 2).
Urgent requirements
Infection with H. pylori in infancy might confer years of protection against H. suis infection. In the present study, none of the H. suis infected examinees had H. pylori infection or prior H. pylori infection that had been eradicated. The H. suis infection rate might have risen with the decline of the H. pylori infection rate. Because there is currently no established method for the clinical diagnosis of H. suis infection, accurate diagnosis of H. suis infection is crucial to avoid wasting precious time on the wrong course of treatment.
Limitations of the study
The PPV of ELISA for discriminating the H. suis infection group from the H. suis non-infection groups was 76.9% (95% CI: 55.9–90.2%) (Table 2). The PPV of ELISA for discriminating the H. pylori infection group from the H. pylori non-infection groups was 65.2% (95% CI: 42.8–82.8%) (Table 3). Concerning diagnostic accuracy, the low values of PPV are the limitation of serological assays. The main reason is that previously eradicated infections may test positive by ELISA, even in cases that tested negative by PCR. Validation of serological tests for diagnosing infectious diseases requires agreement between several independent analytical and measurement results under defined conditions. However, this study represents the first to develop a serological diagnostic method for H. suis infection in humans. Therefore, this assay has only one independent validation group to confirm its efficacy. This is the current limitation of the study.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| HRP-conjugated goat anti-human IgA+IgG+IgM (H+L) antibody | Jackson | Code#109-035-064; RRID: AB_2337583 |
| Rabbit monoclonal anti-H. pylori antibody (clone EP279) | Cell Marque | Cat#AC-0247; RRID: AB_2936230 |
| Rabbit polyclonal anti-H. suis antibody | This paper | N/A |
| HRP-conjugated goat anti-rabbit antibody | Agilent Technologies | Code#P0448; RRID: AB_2617138 |
| Bacterial and virus strains | ||
| H. suis SNTW101c | Rimbara et al.9 | N/A |
| H. pylori TN2GF4 | Matsui et al.39 | N/A |
| H. suis 13 strains isolated from gastric biopsy specimens of subjects | This paper | See Table S1 |
| H. pylori 17 strains isolated from gastric biopsy specimens of subjects | This paper | See Table S1 |
| Biological samples | ||
| Gastric biopsy specimens from subjects | This paper | N/A |
| Serum specimens from subjects | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| H. pylori eradication triple-combination drug VONOSAP Pack 400 (vonoprazan 40 mg, amoxicillin 1500 mg, and clarithromycin 400 mg) | Takeda Pharmaceutical | YJ Code# 6199104X1023 |
| H. pylori eradication triple-combination drug VONOSAP Pack 800 (vonoprazan 40 mg, amoxicillin 1500 mg, and clarithromycin 800 mg) | Takeda Pharmaceutical | YJ Code# 6199104X2020 |
| Critical commercial assays | ||
| DNeasy Blood & Tissue Kits | Qiagen | Cat#69504/69556 |
| Nextera XT DNA Library Prep Kit | Illumina | Illumina advantage product Cat#TG-131-1096 |
| ELISA enzyme substrate KPL SureBlue TMB Microwell Peroxidase Substrate (1-Component) | Sera Care Life Sciences | Material#5120-0075 |
| Nunc-Immuno 96-well microtiter plate | Thermo Fisher Scientific | Cat#439454 |
| NHPH agar plate | This paper | N/A |
| Brucella broth | BD | BD BBLTM -211088 |
| Skirrow | Oxoid | Cat#OXSR0069E |
| Vitox | Oxoid | Cat#R663090 |
| Dent | Oxoid | Cat#OXSR0147E |
| Helicobacter-selective agar plate | Nissui Pharmaceutical | Cat#51035 |
| Histofine antigen-retrieval solution | Nichirei Biosciences | Code#414251F/414252F |
| Master mix | IDT | Code#1055770 |
| Deposited data | ||
| Whole-genome sequences of H. suis 13 strains (GenBank/ENA/DDBJ accession number) | This paper | See Table S1 |
| The 23S-rRNA gene sequences of H. pylori 16 strains (GenBank/ENA/DDBJ accession number) | This paper | See Table S1 |
| The 23S-rRNA gene sequences of Helicobacter 21 type strains (GenBank/ENA/DDBJ accession number) | This paper | See Table S2 |
| Sequence typing (ST) assignment | This paper | PubMLST https://pubmlst.org/organisms/helicobacter-suis |
| Oligonucleotides | ||
|
H. suis hsvA targeting probe-based real-time PCR primer sequence: NHP194003_11930_forward: 5′-CTGGTAATGCATCA TTAGAAGCAAA-3′ |
Rimbara et al.9 | N/A |
| H. suis hsvA targeting probe-based real-time PCR primer sequence: NHP194003_11930_reverse: GATGGGCGCTTCTGGTTTA | Rimbara et al.9 | N/A |
|
H. suis hsvA targeting probe-based real-time PCR probe sequence: NHP194003_11930_probe: 56-FAM/TGTACACAC/ZEN/CAAACAGATG AGCCGT/3IABkFQ |
IDT Rimbara et al.9 |
N/A |
| NHPH 16S rRNA gene targeting probe-based real-time PCR primer sequence: NHPH_16S_F: CAAGTCGAACGATGAAGCCTA | This paper | N/A |
| NHPH 16S rRNA gene targeting probe-based real-time PCR primer sequence: NHPH_16S_R: ATTTGGTATTAATCACCATTTCTAGT |
This paper | N/A |
| NHPH 16S rRNA gene targeting probe-based real-time PCR probe sequence: NHPH_16S_probe: 56-FAM/TTACTCACC/ ZEN/CGTGCGCCACTAATC/3IABkFQ |
IDT This paper |
N/A |
| H. pylori 651 bp DNA fragment of the 23S rRNA gene targeting colony PCR primer sequence: F3: CCGTAGCGAAAGCGAGTCT | This paper | N/A |
| H. pylori 651 bp DNA fragment of the 23S rRNA gene targeting colony PCR primer sequence: R3: CCCGACTAACCCTACGATGA | This paper | N/A |
| Software and algorithms | ||
| ClustalW ver. 2.1 | Open source | http://www.clustal.org/clustal2/ |
| Shovill v1.1.0. | Open source | https://github.com/tseemann/shovill |
| Roary version 3.13.0 | Open source | https://github.com/sanger-pathogens/Roary |
| RAxML-NG v. 1.1 | Open source | https://github.com/amkozlov/raxml-ng |
| pyani 0.2.12 | Open source | https://github.com/widdowquinn/pyani |
| GraphPad Prism 9.4.1 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Other | ||
| H. suis hsvA targeting probe-based real-time PCR protocol | Rimbara et al.9 | N/A |
| NHPH 16S rRNA gene targeting probe-based real-time PCR protocol | This paper | N/A |
| H. pylori 651 bp DNA fragment of the 23S rRNA gene targeting colony PCR protocol | This paper | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Hidenori Matsui (hmatsui@niid.go.jp).
Materials availability
The polyclonal anti-H. suis antibody and ELISA prototype generated by this study are available from the lead contact upon reasonable request. Bacterial strains isolated in this study are also available from the lead contact upon reasonable request.
Experimental model and subject details
Ethics statement
The Research Ethics Committees (REC) of the National Institute of Infectious Diseases (NIID) and Kitasato University approved this study under registration numbers 1284 and 18100, respectively. The ethical review was also approved at the following eight medical institutions: Junpukai Health Maintenance Center, Kyorin University Hospital, Tokai University Hospital, Toyama University Hospital, Takeda Hospital, Aichi Medical University Hospital, Kakogawa Central Municipal Hospital, and Kanazawa Municipal Hospital. We also obtained written informed consent from all examinees, each of whom received an explanation of the study at one of the participating medical institutions.
Collection and transportation of clinical specimens
Serum and gastric biopsy specimens from examinees undergoing medical check-ups or suffering from any gastric disorder who underwent upper GI endoscopy at one of the eight above-named medical institutions were enrolled. However, the examinees with normal stomachs and those with a gastric disease under 20 years of age were excluded from this study. The specimens from participants having healthy stomachs were excluded from this study. All examinees were found to have H. pylori infection by means of two of the following five tests: the UBT, RUT, serum H. pylori antibody test, stool H. pylori antigen test, and histological test. H. pylori, H. suis, and other NHPH infections were finally confirmed by culture of gastric biopsy specimens. Gastric biopsy specimens were collected from the greater curvature of the gastric antrum, the greater curvature of the lower gastric body, and the lesser curvature of the gastric angle, and they were transported to the NIID using the NHPH biopsy transport media for cultivation, PCR, and microscopy. Serum specimens were divided into three containers, transported to the NIID, and stored at -80°C until use. The specimen number assigned by the NIID and the clinical data cannot be used to identify medical institutions or examinees.
Agar plate culture for the detection of Helicobacter species in clinical specimens
The gastric biopsy specimen was homogenized with 300 μL of Brucella broth (BD, Franklin Lakes, NJ, USA) adjusted to pH 5.0 by hydrochloride. A 200 μL aliquot of the tissue homogenates was inoculated onto NHPH agar plates containing 1.5% (wt/vol) agar, Brucella broth, 20% (v) heat-inactivated bovine serum (FBS), Campylobacter selective supplement (Skirrow; Oxoid, Basingstoke, UK), Vitox supplement (Oxoid), and hydrochloric acid to adjust the pH to 5.0, and incubated for more than seven days in a humidified gas mixture (5% O2, 12% CO2, and 83% N2) at 37°C. The grown colonies of primary culture were inoculated onto NHPH agar plates and enriched by modified biphasic culture for 120 h with shaking in a humidified gas mixture at 37°C. The whole-genome sequencing of broth-grown bacteria was carried out as described below. Although both H. pylori and NHPH were grown on the NHPH agar plate as primary cultures, the H. pylori colonies tended to grow faster and to be larger than the small, slow-growing colonies formed by NHPH.9 Therefore, the colonies formed by H. pylori were inoculated onto a Brucella agar plate containing 5% (vol/vol) horse blood for 72 h in a humidified gas mixture at 37°C. NHPH could not grow on this plate.
PCR for the detection of Helicobacter species in clinical specimens
To identify NHPH infections, the DNA was prepared from the remaining 100 μL of homogenates of gastric biopsy specimens using DNeasy Blood & Tissue Kits (Qiagen, Hilden, Germany). Then, the DNA was used as the template for probe-based real-time PCR targeting of the H. suis-specific vacA-type autotransporter protein gene (hsvA) and the NHPH-common region of the 16S rRNA gene. The sequences of the two sets of primers and probes were as follows: NHP194003_11930_forward (5′-CTGGTAATGCATCATTAGAAGCAAA-3′), NHP194003_11930_reverse (5′-GATGGGCGCTTCTGGTTTA-3′), and NHP194003_11930_probe (5’-/56-FAM/TGTACACAC/ZEN/CAAACAGATGAGCCGT/3IABkFQ-3′) for targeting the hsvA gene; NHPH_16S_F (5′-CAAGTCGAACGATGAAGCCTA-3′), NHPH_16S_R (5′- ATTTGGTATTAATCACCATTTCTAGT-3′), and NHPH_16S_probe (5’-/56-FAM/TTACTCACC/ZEN/CGTGCGCCACTAATC/3IABkFQ/-3′) for targeting the NHPH 16S rRNA gene. The diagnosis of H. suis infection was made when both positive results occurred in two types of PCR targeting the hsvA and NHPH 16S rRNA genes.9 The diagnosis of NHPH other than H. suis was made when negative and positive results occurred in PCR targeting the H. suis-specific hsvA and NHPH-common 16S rRNA genes, respectively. We included the culture as a diagnostic criterion to confirm H. pylori infection, whereas we excluded the culture as a diagnostic criterion to confirm H. suis infection. To identify H. pylori infection, the 651 bp DNA fragment of the 23S rRNA gene was amplified by the colony PCR using the following set of primers: F3 (5′- CCGTAGCGAAAGCGAGTCT-3′) and R3 (5′-CCCGACTAACCCTACGATGA-3′). The 23S rRNA gene sequences of Helicobacter pylori strains were deposited at GenBank/EMBL/DDBJ as shown in Table S1. The nucleotide sequences of PCR products were aligned by ClustalW ver. 2.1 (http://www.clustal.org/clustal2/). The 23S rRNA gene sequences of the reference Helicobacter strains were taken from GenBank/EMBL/DDBJ.
Method details
Broth culture of H. pylori and H. suis for the preparation of ELISA antigens
The colonies of H. pylori TN2GF4 grown on a Helicobacter-selective agar plate (Nissui Pharmaceutical, Tokyo) were inoculated into a Brucella broth containing 10% (vol/vol) FBS supplemented with 1% (vol/vol) Vitox supplement and Helicobacter pylori-selective supplement (Dent; Oxoid) for 72 h by shaking in a humidified gas mixture (5% O2, 10% CO2, and 85% N2) at 37°C.39 The frozen stock of H. suis SNTW101c was inoculated onto NHPH agar plates and enriched by modified biphasic culture for 120 h with shaking in a humidified gas mixture at 37°C as described above.
ELISA
Bacterial cells collected by centrifugation were washed twice with phosphate-buffered saline, pH 7.4 (PBS), suspended in distilled water and disrupted by sonication with a Bioruptor II (BMBio, Tokyo) to prepare whole-cell lysates. For the measurement of serum anti-H. suis or anti-H. pylori antibody titer, Nunc-Immuno 96-well microtiter plates (No. 439454; Thermo Fisher Scientific, Waltham, MA, USA) containing 100 μL of whole-cell lysates of H. suis SNTW101c or H. pylori TN2GF4 (4 μg/mL in 0.1 M carbonate/bicarbonate buffer, pH 9.4) were incubated overnight at 4°C. After washing three times with PBS containing 0.05% (vol/vol) Tween 20 (PBS-T), the wells were saturated with 200 μL of blocking buffer (PBS containing 1% BSA). The plates were incubated for 1 h at 37°C while shaking at 500 rpm. After washing three times with PBS-T, the wells were filled with 50 μL of serum samples diluted at 1:1,800 (Figure S2 [ELISA for identification of H. suis and H. pylori infection using 1800-fold diluted sera]) and 1:3,600 (Figure 1) with blocking buffer. The plates were incubated for 1 h at 37°C while shaking at 500 rpm. After washing three times with PBS-T, the wells were filled with 50 μL of horseradish peroxidase (HRP)-conjugated goat anti-human IgA+IgG+IgM (H+L) secondary antibody (Jackson, Bar Harbor, ME, USA) diluted at 1:10,000 with blocking buffer. The plates were incubated for 1 h at 37°C while shaking at 500 rpm. After washing three times with PBS-T, the wells were filled with 50 μL of KPL SureBlue TMB Microwell Peroxidase Substrate (1-Component) (Sera Care Life Sciences, Milford, MA, USA). After 5 min, the color reaction was stopped by adding 50 μL of 1 N hydrochloric acid into the wells. The absorbance at 450 nm (reference wavelength, 630 nm) was measured on a ChroMate-6 microplate reader (Microtec, Chiba, Japan).
Eradication of H. suis
For the treatment of gastric diseases with H. suis infection, each of the following three agents was administered twice daily for a week for eradication of H. suis: vonoprazan (P-CAB: 20 mg), amoxicillin (750 mg), and clarithromycin (200 mg for nos. 04080, 04252, 04262, and 14005 or 400 mg for nos. 94058 and 94059). At least two months after the eradication treatment, an upper GI endoscopy was performed to confirm gastric disease remission and the disinfection of H. suis by PCR.
Quantification and statistical analysis
Differences in the gender ratio among four groups were compared by Fisher's exact test. The anti-H. suis and anti-H. pylori antibody values were compared among four groups using one-way ANOVA, followed by Tukey's post hoc test. Cut-off values, AUCs, sensitivities, and specificities based on the ROC curves were calculated using GraphPad Prism software version 9.5.0 for Mac (GraphPad Software, San Diego, CA, USA). As the diagnostic accuracy by ELIS, the PPV and NPV with 95% CI were calculated using Clinical Calculator 1 (http://vassarstats.net/clin1.html). Pearson’s correlation coefficient (R) was determined using GraphPad Prism software. Cohen's kappa was calculated using the following formula. . Po: observed agreement between the rates. Pe: expected agreement based on chance. Regression lines and R-squares were calculated using GraphPad Prism software. Differences were considered significant when the P-value (two-tailed) was less than 0.05. ∗∗∗∗ and # indicate P<0.0001 and P>0.05, respectively.
Acknowledgments
This work was supported by MEXT/JSPS KAKENHI under grant numbers JP19H03474 (H.M.), JP23H02636 (E.R.), JP20K08365 (T.K.), and JP20H3667 (H.S.), and by AMED under grant numbers JP20fk0108148 and 23fk0108678j0001 (E.R.) and JP20fk0108133 (M.S.).
Author contributions
Conceptualization, H.M., E.R., and K.S.; Methodology – Cultures and PCR, E.R. and S.A.; Methodology – Genomic Analysis, M.S.; Methodology – ELISA, H.M.; Methodology – Microscopy, H.O.; Medical Practice – Eradication, K.M. and K.T.; Medical Practice – Specimen Collection, K.M., K.T., H.S., M.S., T.U., H.T., S.N., A.T., M.S., S.T., and T.S.; Writing, H.M., E.R., and K.M. All authors verified the underlying data and approved the final version of the manuscript for publication.
Declaration of interests
M.H., R.E., and S.M. are inventors of the technology described in this manuscript and are listed as applicants in the international patent application under the PCT (PCT/JP2023/002328, PCT/JP2023/002329, and PCT/JP2023/002330). The authors declare that they have no other competing interests.
Published: March 29, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106522.
Contributor Information
Hidenori Matsui, Email: hmatsui@niid.go.jp.
Katsuhiro Mabe, Email: katsumabe@me.com.
Supplemental information
Data and code availability
-
•
The whole-genome data of H. suis strains have been deposited at GenBank/ENA/DDBJ and will be available from the date of publication.
-
•
The 23S rRNA genome data of H. pylori strains have been deposited at GenBank/ENA/DDBJ and will be available from the date of publication.
-
•
Accession numbers can be found in Table S1 (Individual data of 101 examinees) and Table S2 (Reference strains of gastric Helicobacter species).
-
•
This paper does not report any original code.
-
•
Any additional information about the analysis in this paper is available from the lead contact upon reasonable request.
References
- 1.McNulty C.A., Dent J.C., Curry A., Uff J.S., Ford G.A., Gear M.W., Wilkinson S.P. New spiral bacterium in gastric mucosa. J. Clin. Pathol. 1989;42:585–591. doi: 10.1136/jcp.42.6.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dent J.C., McNulty C.A., Uff J.C., Wilkinson S.P., Gear M.W. Spiral organisms in the gastric antrum. Lancet. 1987;2:96. doi: 10.1016/s0140-6736(87)92754-1. [DOI] [PubMed] [Google Scholar]
- 3.Heilmann K.L. [Spirochete-like bacteria in the human gastric mucosa] Dtsch. Med. Wochenschr. 1988;113:1298–1299. [PubMed] [Google Scholar]
- 4.Heilmann K.L., Borchard F. Gastritis due to spiral shaped bacteria other than Helicobacter pylori: clinical, histological, and ultrastructural findings. Gut. 1991;32:137–140. doi: 10.1136/gut.32.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flahou B., Rossi M., Bakker J., Langermans J.A., Heuvelman E., Solnick J.V., Martin M.E., O'Rourke J., Ngoan L.D., Hoa N.X., et al. Evidence for a primate origin of zoonotic Helicobacter suis colonizing domesticated pigs. ISME J. 2018;12:77–86. doi: 10.1038/ismej.2017.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smet A., Yahara K., Rossi M., Tay A., Backert S., Armin E., Fox J.G., Flahou B., Ducatelle R., Haesebrouck F., Corander J. Macroevolution of gastric Helicobacter species unveils interspecies admixture and time of divergence. ISME J. 2018;12:2518–2531. doi: 10.1038/s41396-018-0199-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taillieu E., Chiers K., Amorim I., Gärtner F., Maes D., Van Steenkiste C., Haesebrouck F. Gastric Helicobacter species associated with dogs, cats and pigs: significance for public and animal health. Vet. Res. 2022;53:42. doi: 10.1186/s13567-022-01059-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang J., Ducatelle R., Pasmans F., Smet A., Haesebrouck F., Flahou B. Multilocus sequence typing of the porcine and human gastric pathogen Helicobacter suis. J. Clin. Microbiol. 2013;51:920–926. doi: 10.1128/JCM.02399-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rimbara E., Suzuki M., Matsui H., Nakamura M., Morimoto M., Sasakawa C., Masuda H., Nomura S., Osaki T., Nagata N., et al. Isolation and characterization of Helicobacter suis from human stomach. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2026337118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morgner A., Lehn N., Andersen L.P., Thiede C., Bennedsen M., Trebesius K., Neubauer B., Neubauer A., Stolte M., Bayerdörffer E. Helicobacter heilmannii-associated primary gastric low-grade MALT lymphoma: complete remission after curing the infection. Gastroenterology. 2000;118:821–828. doi: 10.1016/s0016-5085(00)70167-3. [DOI] [PubMed] [Google Scholar]
- 11.Stolte M., Kroher G., Meining A., Morgner A., Bayerdörffer E., Bethke B. A comparison of Helicobacter pylori and H. heilmannii gastritis. A matched control study involving 404 patients. Scand. J. Gastroenterol. 1997;32:28–33. doi: 10.3109/00365529709025059. [DOI] [PubMed] [Google Scholar]
- 12.Okiyama Y., Matsuzawa K., Hidaka E., Sano K., Akamatsu T., Ota H. Helicobacter heilmannii infection: clinical, endoscopic and histopathological features in Japanese patients. Pathol. Int. 2005;55:398–404. doi: 10.1111/j.1440-1827.2005.01844.x. [DOI] [PubMed] [Google Scholar]
- 13.Tsukadaira T., Hayashi S., Ota H., Kobayashi N., Sekiguchi Y., Kodaira H., Matsumoto T., Horiuchi K., Negishi T., Kurahashi M. Prevalence, clinical features, and esophagogastroduodenoscopy (EGD) findings of non-Helicobacter pylori Helicobacter infection: a study of 50 cases at a single facility in Japan. Helicobacter. 2021;26 doi: 10.1111/hel.12811. [DOI] [PubMed] [Google Scholar]
- 14.Shiratori S., Mabe K., Yoshii S., Takakuwa Y., Sato M., Nakamura M., Kudo T., Kato M., Asaka M., Sakamoto N. Two cases of chronic gastritis with non-Helicobacter pylori Helicobacter infection. Intern. Med. 2016;55:1865–1869. doi: 10.2169/internalmedicine.55.5891. [DOI] [PubMed] [Google Scholar]
- 15.Goji S., Tamura Y., Sasaki M., Nakamura M., Matsui H., Murayama S.Y., Ebi M., Ogasawara N., Funaki Y., Kasugai K. Helicobacter suis-infected nodular gastritis and a review of diagnostic sensitivity for Helicobacter heilmannii-like organisms. Case Rep. Gastroenterol. 2015;9:179–187. doi: 10.1159/000431169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Øverby A., Murayama S.Y., Michimae H., Suzuki H., Suzuki M., Serizawa H., Tamura R., Nakamura S., Takahashi S., Nakamura M. Prevalence of gastric Non-Helicobacter pylori-Helicobacters in Japanese patients with gastric disease. Digestion. 2017;95:61–66. doi: 10.1159/000452400. [DOI] [PubMed] [Google Scholar]
- 17.Kuo S.H., Yeh K.H., Lin C.W., Liou J.M., Wu M.S., Chen L.T., Cheng A.L. Current status of the spectrum and therapeutics of Helicobacter pylori-negative mucosa-associated lymphoid tissue lymphoma. Cancers. 2022;14:1005. doi: 10.3390/cancers14041005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura M., Øverby A., Michimae H., Matsui H., Takahashi S., Mabe K., Shimoyama T., Sasaki M., Terao S., Kamada T., et al. PCR analysis and specific immunohistochemistry revealing a high prevalence of non-Helicobacter pylori Helicobacters in Helicobacter pylori-negative gastric disease patients in Japan: high susceptibility to an Hp eradication regimen. Helicobacter. 2020;25 doi: 10.1111/hel.12700. [DOI] [PubMed] [Google Scholar]
- 19.Iwamuro M., Murayama S.Y., Nakamura M., Hamada K., Tanaka T., Okada H. Helicobacter suis-associated gastritis mimicking conventional H. pylori-associated atrophic gastritis. Case Rep. Gastrointest. Med. 2022;2022 doi: 10.1155/2022/4254605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takigawa H., Masaki S., Naito T., Yuge R., Urabe Y., Tanaka S., Sentani K., Matsuo T., Matsuo K., Chayama K., Kitadai Y. Helicobacter suis infection is associated with nodular gastritis-like appearance of gastric mucosa-associated lymphoid tissue lymphoma. Cancer Med. 2019;8:4370–4379. doi: 10.1002/cam4.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inoue M. Changing epidemiology of Helicobacter pylori in Japan. Gastric Cancer. 2017;20:3–7. doi: 10.1007/s10120-016-0658-5. [DOI] [PubMed] [Google Scholar]
- 22.Øverby A., Yamagata Murayama S., Matsui H., Nakamura M. In the aftermath of Helicobacter pylori: other Helicobacters rising up to become the next gastric epidemic? Digestion. 2016;93:260–265. doi: 10.1159/000445399. [DOI] [PubMed] [Google Scholar]
- 23.Takigawa H., Yuge R., Masaki S., Otani R., Kadota H., Naito T., Hayashi R., Urabe Y., Oka S., Tanaka S., et al. Involvement of non-Helicobacter pylori helicobacter infections in Helicobacter pylori-negative gastric MALT lymphoma pathogenesis and efficacy of eradication therapy. Gastric Cancer. 2021;24:937–945. doi: 10.1007/s10120-021-01172-x. [DOI] [PubMed] [Google Scholar]
- 24.Kadota H., Yuge R., Miyamoto R., Otani R., Takigawa H., Hayashi R., Urabe Y., Oka S., Sentani K., Oue N., et al. Investigation of endoscopic findings in nine cases of Helicobacter suis-infected gastritis complicated by gastric mucosa-associated lymphoid tissue lymphoma. Helicobacter. 2022;27 doi: 10.1111/hel.12887. [DOI] [PubMed] [Google Scholar]
- 25.Hooi J.K.Y., Lai W.Y., Ng W.K., Suen M.M.Y., Underwood F.E., Tanyingoh D., Malfertheiner P., Graham D.Y., Wong V.W.S., Wu J.C.Y., et al. Global prevalence of Helicobacter pylori infection: systematic review and meta-analysis. Gastroenterology. 2017;153:420–429. doi: 10.1053/j.gastro.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 26.Teixeira S., Filipe D., Cerqueira M., Barradas P., Cortez Nunes F., Faria F., Haesebrouck F., Mesquita J.R., Gärtner F., Amorim I. Helicobacter spp. in the stomach of cats: successful colonization and absence of relevant histopathological alterations reveals high adaptation to the host gastric niche. Vet. Sci. 2022;9:228. doi: 10.3390/vetsci9050228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Husnik R., Klimes J., Kovarikova S., Kolorz M. Helicobacter species and their association with gastric pathology in a cohort of dogs with chronic gastrointestinal signs. Animals. 2022;12:1254. doi: 10.3390/ani12101254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joosten M., Flahou B., Meyns T., Smet A., Arts J., De Cooman L., Pasmans F., Ducatelle R., Haesebrouck F. Case report: Helicobacter suis infection in a pig veterinarian. Helicobacter. 2013;18:392–396. doi: 10.1111/hel.12054. [DOI] [PubMed] [Google Scholar]
- 29.De Witte C., Devriendt B., Flahou B., Bosschem I., Ducatelle R., Smet A., Haesebrouck F. Helicobacter suis induces changes in gastric inflammation and acid secretion markers in pigs of different ages. Vet. Res. 2017;48:34. doi: 10.1186/s13567-017-0441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foss D., Kopta L., Paquette J., Bowersock T., Choromanski L., Galvin J., Godbee T., Laurinat R., Sanchez M. Identification of Helicobacter suis in pig-producing regions of the United States. J. Swine Health Prod. 2013;21:242–247. [Google Scholar]
- 31.Matos R., Taillieu E., De Bruyckere S., De Witte C., Rêma A., Santos-Sousa H., Nogueiro J., Reis C.A., Carneiro F., Haesebrouck F., et al. Presence of Helicobacter species in gastric mucosa of human patients and outcome of Helicobacter eradication treatment. J. Personalized Med. 2022;12 doi: 10.3390/jpm12020181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Augustin A.D., Savio A., Nevel A., Ellis R.J., Weller C., Taylor D., Tucker R.M., Ibrahim M.A.A., Bjarnason I., Dobbs S.M., et al. Helicobacter suis is associated with mortality in Parkinson's disease. Front. Med. 2019;6:188. doi: 10.3389/fmed.2019.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Cooman L., Flahou B., Houf K., Smet A., Ducatelle R., Pasmans F., Haesebrouck F. Survival of Helicobacter suis bacteria in retail pig meat. Int. J. Food Microbiol. 2013;166:164–167. doi: 10.1016/j.ijfoodmicro.2013.05.020. [DOI] [PubMed] [Google Scholar]
- 34.De Cooman L., Houf K., Smet A., Flahou B., Ducatelle R., De Bruyne E., Pasmans F., Haesebrouck F. Presence of Helicobacter suis on pork carcasses. Int. J. Food Microbiol. 2014;187:73–76. doi: 10.1016/j.ijfoodmicro.2014.06.016. [DOI] [PubMed] [Google Scholar]
- 35.Marini R.P., Patterson M.M., Muthupalani S., Feng Y., Holcombe H., Swennes A.G., Ducore R., Whary M.M., Shen Z., Fox J.G. Helicobacter suis and Helicobacter pylori infection in a colony of research macaques: characterization and clinical correlates. J. Med. Microbiol. 2021;70 doi: 10.1099/jmm.0.001315. [DOI] [PubMed] [Google Scholar]
- 36.Yoshimura M., Isomoto H., Shikuwa S., Osabe M., Matsunaga K., Omagari K., Mizuta Y., Murase K., Murata I., Kohno S. A case of acute gastric mucosal lesions associated with Helicobacter heilmannii infection. Helicobacter. 2002;7:322–326. doi: 10.1046/j.1523-5378.2002.00103.x. [DOI] [PubMed] [Google Scholar]
- 37.Okamura T., Iwaya Y., Yokosawa S., Suga T., Arakura N., Matsumoto T., Ogiwara N., Higuchi K., Ota H., Tanaka E. A case of Helicobacter heilmannii-associated primary gastric mucosa-associated lymphoid tissue lymphoma achieving complete remission after eradication. Clin. J. Gastroenterol. 2013;6:38–45. doi: 10.1007/s12328-012-0355-9. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura M., Murasato F., Øverby A., Kodama Y., Michimae H., Sasaki K., Flahou B., Haesebrouck F., Murayama S.Y., Takahashi S., et al. Effect of acid suppressants on non-Helicobacter pylori Helicobacters within parietal cells. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.692437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsui H., Takahashi T., Murayama S.Y., Kawaguchi M., Matsuo K., Nakamura M. Protective efficacy of a hydroxy fatty acid against gastric Helicobacter infections. Helicobacter. 2017;22 doi: 10.1111/hel.12430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The whole-genome data of H. suis strains have been deposited at GenBank/ENA/DDBJ and will be available from the date of publication.
-
•
The 23S rRNA genome data of H. pylori strains have been deposited at GenBank/ENA/DDBJ and will be available from the date of publication.
-
•
Accession numbers can be found in Table S1 (Individual data of 101 examinees) and Table S2 (Reference strains of gastric Helicobacter species).
-
•
This paper does not report any original code.
-
•
Any additional information about the analysis in this paper is available from the lead contact upon reasonable request.