Summary
The mammalian circadian clock and glucose metabolism are highly interconnected, and disruption of this coupling is associated with multiple negative health outcomes. Liver is the major source of endogenous glucose production and liver clock is one of the most vital peripheral clock systems. We demonstrate that fatty acid translocase (CD36) is expressed rhythmically in mouse liver and autonomously modulates the diurnal oscillations of liver clock and glucose homeostasis. CD36 knockout in hepatocytes inhibits the relay of insulin signaling and provokes FoxO1 nuclear shuttling, consequently increasing Per1 nuclear expression. Moreover, FoxO1 can activate the central clock gene Per1 at the transcriptional level. These changes lead to a disrupted clock oscillation and behavioral rhythm. Our study first reveal that CD36 is a key regulator of the circadian oscillator and its deficiency may cause liver clock disruption, which aggravates the imbalance of glucose homeostasis and contribute to augmentation and progression of metabolic disease.
Subject areas: Endocrinology, Hepatology, Molecular biology
Graphical abstract
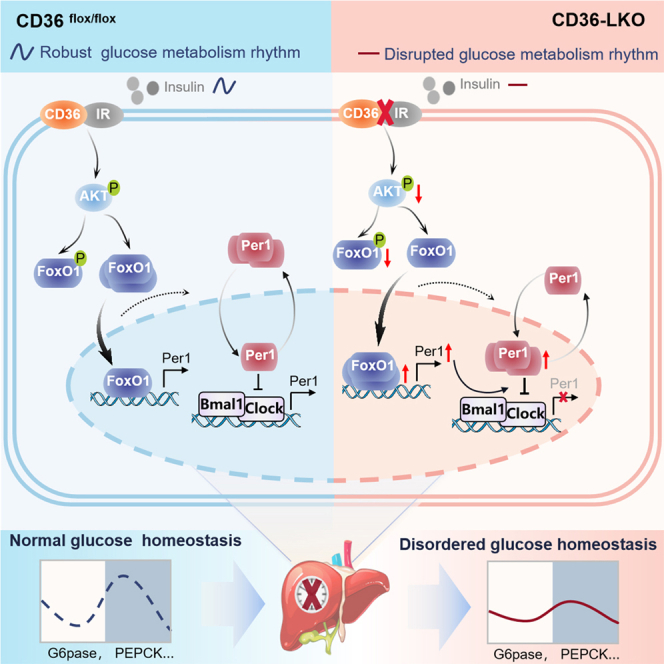
Highlights
-
•
CD36 expresses rhythmically in mouse liver and is a regulator of circadian clock
-
•
CD36 regulates diurnal glucose metabolism via overactivation of FoxO1-Per1 pathway
-
•
CD36 deficiency causes FoxO1 to promote Per1 transcription to reset the liver clock
-
•
Interfering with insulin signaling pathway restores CD36-mediated rhythm disorders
Endocrinology; Hepatology; Molecular biology
Introduction
Circadian rhythms, endogenous 24-h oscillations, are intrinsic to the biology of most multicellular organisms, controlling the temporal organization of many physiological and cellular functions and acting as the main regulator of metabolism.1,2 A surprising number of mammalian peripheral tissues, such as the liver, heart, and kidney, appear to contain the molecular machinery necessary for independent circadian oscillation and regulate the circadian rhythm via neural, hormonal, and metabolic feedback approaches.3 Glucose is a particularly potent entraining cue for peripheral clocks.4 In humans, circadian misalignment regulates glucose and insulin levels. Shift workers often have characteristics related to circadian disruption that may predispose them to a higher risk for diabetes mellitus type 2 (T2DM).5 On the other hand, glucose homeostasis can also regulate and even reset the circadian clock via key signal disruption. Insulin and glucagon have been shown to affect core clock gene expression in peripheral tissues.6,7,8 Patients with diabetes exhibit dampened amplitude of glucose tolerance and insulin secretion rhythms.9 Thus, there is a bidirectional relationship between circadian rhythm and glucose homeostasis. Understanding the link between perturbation of the circadian rhythm and glucose homeostasis is crucial to the development of metabolic syndrome.
As the major source of endogenous glucose production, the liver exhibits strong circadian variation in glucose metabolism, which includes multiple biochemical pathways, such as gluconeogenesis, glycogenesis and glycogenolysis.10,11,12 In addition, many hormones involved in glucose metabolism, such as insulin, glucagon, corticosterone and leptin, exhibit circadian oscillation.4 Glucose transporter activity fluctuates with the circadian cycle, and glucose tolerance and insulin sensitivity also show strong time-of-day variation.13,14 A clear picture of the insulin signaling pathway, which plays a vital role in glucose homeostasis, has emerged.
Fatty acid translocase (CD36) is a scavenger receptor that functions in the uptake of long-chain fatty acids (LCFAs) and is widely expressed on hepatocytes, adipocytes, macrophages and muscle cells.15 CD36 can recognize different ligands and promote different intracellular signaling pathways and thereby plays a significant role in insulin responsiveness, inflammation and lipid metabolism, which accelerates the process of metabolic diseases, such as T2DM, obesity, atherosclerosis and nonalcoholic fatty liver disease.16,17,18 The role of CD36 in glucose homeostasis has been increasingly studied, and clinical studies have illustrated that people with genetic CD36 deficiency are more likely to exhibit glucose and insulin dysfunction and even a propensity to develop metabolic syndrome, which is common in Asians and Africans.17,19,20 Our group previously revealed that loss of CD36 can result in suppression of insulin signal transduction in mice fed a low-fat diet.21 However, no clear evidence has yet shown that CD36 is associated with the circadian rhythm of glucose metabolism.
In mammals, circadian clocks in the suprachiasmatic nucleus (SCN) and peripheral tissues require the alternating actions of activators and repressors of transcription.3 The molecular mechanism underlying circadian clocks is an interlinked feedback loop present in almost all cells that is composed of positive (transcriptional activators BMAL1 and CLOCK) and negative (repressor genes Period1/⅔ and Cryptochrome1/2) arms, which are responsible for generating molecular rhythms. The transcriptional regulators BMAL1 and CLOCK activate the expression of many transcripts, including the Period (PER) and Cryptochrome (CRY) genes, whose protein products inhibit CLOCK and BMAL1, resulting in rhythmic expression.22,23 Another crucial diurnal loop involves the competing nuclear receptors Rev-erbs and Rors, which also modulate Bmal1 expression to perform the physiological function of the clock system.24,25 Notably, insulin signaling has been demonstrated to regulate the translation of Period proteins,8,26 which raises the possibility that CD36 may regulate the circadian clock via insulin signaling.
In this study, we first revealed that CD36 is expressed rhythmically in the mouse liver and can modulate the hepatic circadian clock and that the loss of CD36 in liver disrupts the diurnal variations of clock genes and mouse behavior. Mechanistically, the combination of CD36 and insulin receptor β (IRβ) maintained insulin signaling, thereby maintaining rhythmic oscillations of clock genes and behavior. Loss of CD36 in the liver resets the hepatic circadian clock via the AKT/FoxO1/Per1 pathway, and mistimed insulin signaling disrupts the circadian organization of clock gene expression and mouse behavior, which may lead to a vicious cycle between circadian disruption and glucose homeostasis pathologies and contribute to the augmentation and progression of metabolic disease.
Results
CD36 is indispensable for maintaining the circadian amplitude of Per1
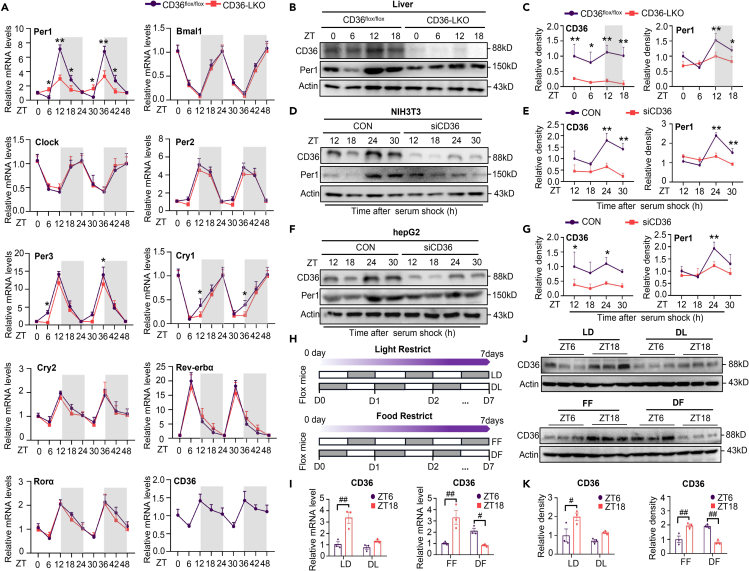
To investigate whether liver CD36 impacts the circadian clock of mice, we examined 48 h mRNA expression profile of several core clock genes in the livers of CD36flox/flox and CD36-LKO mice. Of interest, the amplitude of Per1 oscillation in the liver, especially at ZT12, was notably reduced in CD36-LKO mice compared to CD36flox/flox mice, whereas the rhythmic expression pattern of most other clock genes showed no difference between CD36flox/flox and CD36-LKO mice (Figure 1A), and CD36 knockout in hepatocytes had no effect on the core clock gene expression of the SCN (Figure S1A). Besides, CD36 knockout in hepatocytes also damaged the expression of genes downstream of circadian components in the liver (Figure S1B). Furthermore, hepatic CD36 mRNA expression exhibited a clear diurnal variation and peaked at ZT12, which was consistent with Per1 expression (Figure 1A). To obtain additional evidence that hepatic CD36 modulates the circadian rhythm of Per1, we detected the temporal protein levels of CD36 and Per1 in vivo and in vitro. In accordance with mRNA expression, hepatic CD36 and Per1 protein showed distinct diurnal variation in CD36flox/flox mice, which was not observed in CD36-LKO mice (Figures 1B and 1C). Likewise, CD36 suppression via siRNA strikingly inhibited the rhythmic expression of Per1 protein in NIH3T3 and HepG2 cells after serum shock (Figures 1D–1G). Collectively, these findings suggest that CD36 is a rhythmic-expressing factor and autonomously modulates the circadian amplitude of Per1 in the liver.
Figure 1.
CD36 is a rhythmic expressed factor and necessary to maintain the rhythm of Per1 in vivo and in vitro (see also Figure S1)
(A) mRNA levels of clock genes and CD36 in the livers of CD36flox/flox and CD36-LKO mice over 48 h.
(B and C) Diurnal CD36 and Per1 protein expression levels and relative quantification of western blotting results for livers from CD36flox/flox and CD36-LKO mice.
(D and E) Protein levels of CD36 and Per1 detected over 30 h after CD36 knockdown in NIH3T3 cells.
(F and G) Protein levels of CD36 and Per1 detected over 30 h after CD36 knockdown in HepG2 cells.
(H) Schematic diagram of light and food restriction in CD36flox/flox mice; gray bars represent darkness and feeding.
(I) CD36 mRNA expression in the liver of CD36flox/flox mice subjected to light and food restriction.
(J and K) CD36 protein expression and relative quantification of western blotting results in the liver of CD36flox/flox mice subjected to light restriction and food restriction.
n = 3–5 in each group. Data are shown as the mean ± SEM. Comparison of different groups was carried out using two-way ANOVA. ∗p <0.05, ∗∗p <0.01 versus control groups; #p <0.05, ##p <0.01 compared with ZT6 in CD36flox/flox mice under the LD, DL, FF, and DF conditions. ZT0, the beginning of a subjective circadian period (6:00 a.m.). Gray shading represents the lights-off period from 6:00 p.m. to 6:00 a.m. LD: 12 h/12 h LD cycle; DL: 12 h/12 h DL cycle; FF: free feeding; DF: daytime feeding.
Given the important role of light and food in the synchronization of the central clock and peripheral clock, respectively, we further investigated the effects of these factors on hepatic CD36 rhythmic expression. CD36flox/flox mice were subjected to a reversed LD cycle (dark/light, DL) or daytime feeding (DF) cycle for 7 days (Figure 1H). The flattened diurnal rhythm under the DL cycle indicated that the rhythmic expression of CD36 was light-sensitive. Specifically, under the LD cycle, hepatic CD36 exhibited a nocturnal activation pattern in which the expression of CD36 was notably higher at ZT18 than at ZT6. However, this rhythm was blunted under the DL cycle, with the mRNA and protein expression of CD36 at ZT18 being remarkably suppressed. In addition, daytime feeding reversed the phase of CD36 rhythmic expression, with higher expression at ZT6 than at Z18 under the DF cycle (Figures 1I–1K). Overall, these data suggest that as a component of the peripheral clock, CD36 can respond to light and food signals.
CD36 knockout in hepatocytes disturbs biological behavior rhythms in mice
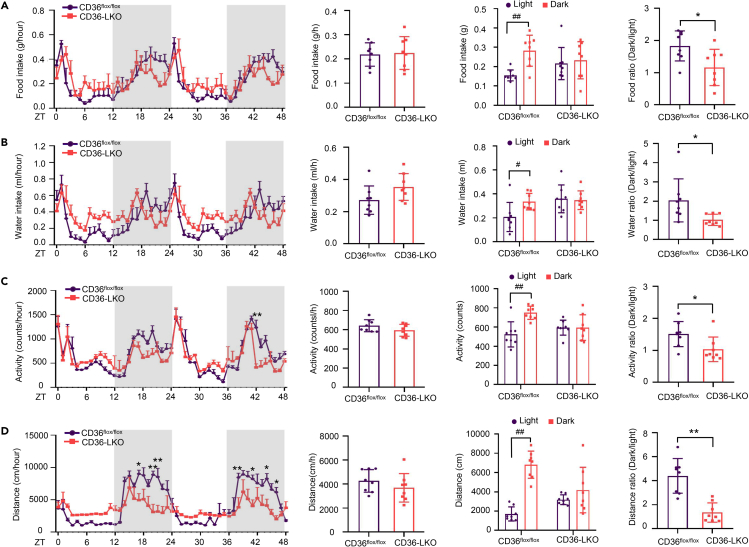
To test whether hepatic CD36 affects behavior and metabolic rhythms in mice, we used TSE metabolic cages to simultaneously monitor diurnal behavioral and metabolic parameters in CD36flox/flox and CD36-LKO mice over 48 h CD36flox/flox mice showed a clear circadian rhythm in the feeding/drinking pattern, whereas CD36-LKO mice consumed less food/water during the dark phase and more food/water during the light phase than CD36flox/flox mice, concomitant with a significantly reduced ratio of dark to light food/water intake (Figures 2A and 2B). Likewise, CD36-LKO mice displayed decreased movement distance/locomotor activity levels during the dark phase and increased levels of those during the light phase, showing a pronounced flattening of locomotor activity rhythm compared with CD36flox/flox mice (Figures 2C and 2D), indicating that loss of hepatic CD36 induced a nearly uniform circadian pattern of attenuation in behavior. CD36 knockout in hepatocytes had no significant effects on metabolic status, including VO2, VCO2, respiratory exchange ratio (RER) and energy expenditure (EE) (Figure S2). Taken together, these results suggest that hepatic CD36 plays a crucial role in maintaining behavioral rhythms in mice.
Figure 2.
CD36 knockout in hepatocytes disorders mouse biological behavior rhythms (see also Figure S2)
Mouse food intake, water intake, locomotor activity and movement distance were monitored for 48 h in 8 to 12-week-old CD36flox/flox and CD36-LKO mice.
(A) Food intake per hour in the light/dark cycle, average food intake per hour, food intake during light and dark phases, and dark/light food ratio in CD36flox/flox and CD36-LKO mice.
(B) Water intake per hour during the light/dark cycle, average water intake per hour, water intake during light and dark phases, and dark/light water ratio in CD36flox/flox and CD36-LKO mice.
(C) Locomotor activity per hour during the light/dark cycle, average locomotor activity per hour, locomotor activity during light and dark phases, and dark/light locomotor activity ratio in CD36flox/flox and CD36-LKO mice.
(D) Movement distance per hour during the light/dark cycle, average movement distance per hour, movement distance during light and dark periods, and dark/light movement distance ratio in CD36flox/flox and CD36-LKO mice.
n = 8 in each group. Data are shown as the mean ± SEM. Comparison of different groups was carried out using two-tailed unpaired Student’s t test or two-way ANOVA. ∗p <0.05, ∗∗p <0.01 compared with the CD36flox/flox group; #p <0.05, ##p <0.01 compared with the light phase in the identical genotype mice. ZT0, the beginning of a subjective circadian period (6:00 a.m.). Gray shading represents the lights-off period from 6:00 p.m. to 6:00 a.m.
CD36 knockout in hepatocytes damages the diurnal fluctuation of serum glucose and insulin sensitivity
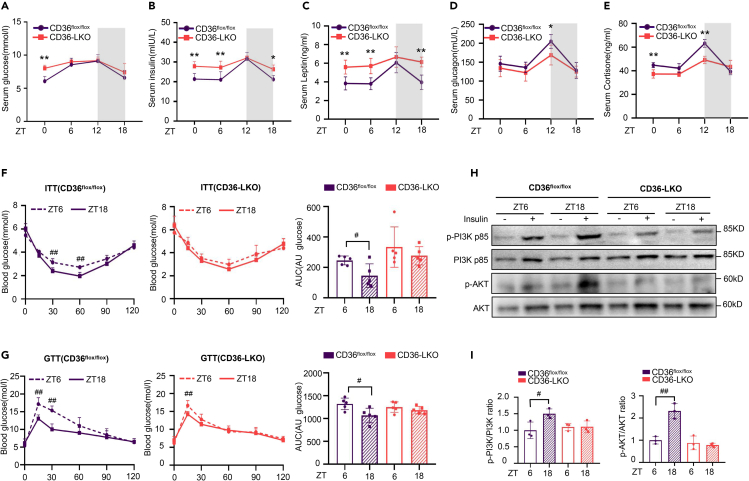
The synthesis and utilization of glucose are involved in the endogenous circadian clock,14,27 and CD36 has been reported to regulate glucose homeostasis.28,29 Accordingly, we wondered whether hepatic CD36 regulates glucose metabolism across the circadian cycle. Temporal analysis of glucose and gluco-regulatory hormones indicated that serum glucose in CD36flox/flox mice displayed an evident diurnal fluctuation, as well as serum insulin, leptin, glucagon and corticosterone. However, these circadian rhythms manifested varying degrees of amplitude reduction in CD36-LKO mice (Figures 3A–3E).
Figure 3.
Hepatic CD36 regulates rhythmic variation of blood glucose and insulin sensitivity
(A–E) Serum glucose (A), serum insulin (B), serum leptin (C), serum glucagon (D), and serum corticosterone (E) were detected across a 24 h cycle in CD36flox/flox and CD36-LKO mice.
(F and G) ITTs and GTTs were performed at ZT6 and ZT18 in CD36flox/flox and CD36-LKO mice. The area under the curve (AUC) was used to quantify the ITT and GTT results.
(H and I) Western blot analysis of AKT, p-AKT (Ser473), PI3K p85, and p-PI3K p85 (Try467) expression in the liver after insulin stimulation. The densitometric quantification of the blots with insulin stimulation is shown below.
n = 3–5 in each group. All data are shown as the mean ± SEM. Comparison of different groups was carried out using two-way ANOVA. ∗p <0.05, ∗∗p <0.01 compared with the CD36flox/flox group; #p <0.05, ##p <0.01 compared with ZT6 in the identical genotype mice. ZT0, the beginning of a subjective circadian period (6:00 a.m.). Gray shading represents the lights-off period from 6:00 p.m. to 6:00 a.m.
To investigate whether CD36 might modulate the circadian variation in glucose tolerance and insulin sensitivity, we performed GTT and ITT tests in CD36flox/flox and CD36-LKO mice at ZT6 and ZT18. Analysis of the area under the curve showed that insulin was more effective in controlling glucose levels at ZT18 than at ZT6, suggesting that insulin sensitivity was ZT dependent in CD36flox/flox mice. However, this diurnal alteration was greatly diminished in CD36-LKO mice (Figure 3F). Consistent with ITT analysis, GTT analysis showed that glucose tolerance also exhibited diurnal variation in CD36flox/flox mice, which was not observed in CD36-LKO mice (Figure 3G). Phosphorylation of serine/threonine kinase (AKT) and phosphoinositide 3-kinase (PI3K) is a marker of insulin signaling pathway activation. Insulin stimulation significantly increased the protein levels of phosphorylated AKT (p-AKT) and phosphorylated PI3K (p-PI3K) at ZT18 compared to ZT6 in CD36flox/flox mice but not in CD36-LKO mice (Figures 3H and 3I), further confirming the attenuated insulin sensitivity rhythm in CD36-LKO mice. Together, these observations suggest that hepatic CD36 is essential for maintaining diurnal variations in serum glucose and insulin sensitivity.
CD36 participates in the diurnal regulation of gluconeogenesis in mice
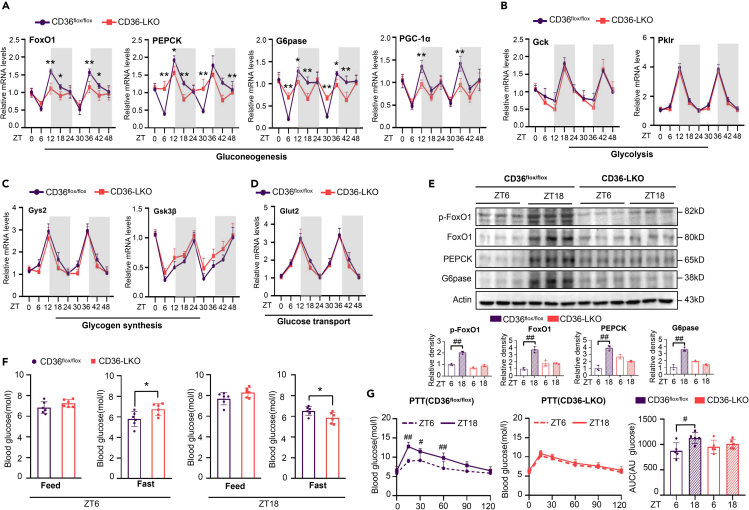
A series of glucose metabolic genes have been reported to be closely controlled by the circadian clock.30,31 To further explore the effect of hepatic CD36 on the glucose metabolism rhythm, we examined the mRNA expression levels of several key genes involved in gluconeogenesis, glycolysis, glucose transport, and glycogen synthesis in the livers of CD36flox/flox and CD36-LKO mice over a time course of 48 h. As shown, most genes displayed potent oscillations in the liver of CD36flox/flox mice, whereas loss of hepatic CD36 markedly dampened the oscillations of key genes involved in gluconeogenesis, such as FoxO1, glucose-6-phosphatase (G6pase), phosphoenolpyruvate carboxykinase 1 (PEPCK), and peroxisome proliferator-activated receptor gamma, coactivator 1 alpha (PGC-1a) (Figure 4A), whereas other genes showed no difference between CD36flox/flox and CD36-LKO mice (Figures 4B–4D). Western blotting analyses revealed that the protein levels of these genes showed a trend similar to that observed with mRNA, and loss of hepatic CD36 decreased FoxO1 phosphorylation, with a blunted circadian expression pattern (Figure 4E).
Figure 4.
CD36 knockout in hepatocytes impairs the circadian rhythm of gluconeogenesis
(A–D) mRNA levels of the key genes involved in hepatic gluconeogenesis (A), glycolysis (B), glycogen synthesis (C) and glucose transport (D) were detected over 48 h in the livers of CD36flox/flox and CD36-LKO mice.
(E) The protein levels of FoxO1, p-FoxO1, PEPCK and G6Pase in livers from the CD36flox/flox and CD36-LKO mice were measured at ZT6 and ZT18 via western blotting.
(F) Blood glucose levels in CD36-LKO and CD36flox/flox mice under feeding conditions or after 12 h of fasting were measured at ZT6 and ZT18.
(G) PTTs were performed at ZT6 and ZT18 in CD36flox/flox and CD36-LKO mice after 16 h of fasting. The area under the curve (AUC) was used to quantify the PTT results.
n = 3–5 in each group. All data are shown as the mean ± SEM. Comparison of different groups was carried out using two-tailed unpaired Student’s t test (F) or two-way ANOVA (A–E, G). ∗p <0.05, ∗∗p <0.01 compared with the CD36flox/flox group; #p <0.05, ##p <0.01 compared with ZT6 in the identical genotype mice. ZT0, the beginning of a subjective circadian period (6:00 a.m.). Gray shading represents the lights-off period from 6:00 p.m. to 6:00 a.m. PEPCK, phosphoenolpyruvate carboxykinase 1; G6pase, glucose-6-phosphatase; FoxO1, Forkhead box O1; PGC-1a, peroxisome proliferator-activated receptor gamma, coactivator 1 alpha; Gck, glucokinase; Pklr, pyruvate kinase L/R; Gys2, glycogen synthase 2; Gsk3β, glycogen synthase kinase 3 beta; Glut2, glucose transporter 2.
To further confirm that hepatic CD36 regulates the circadian rhythm of gluconeogenesis, we challenged CD36-LKO and CD36flox/flox mice with fasting for 12 h and measured blood glucose levels at ZT6 and ZT18. After a 12 h fast, CD36-LKO mice displayed significantly higher blood glucose levels at ZT6 but distinctly lower levels at ZT18 than CD36flox/flox mice (Figure 4F). In addition, pyruvate tolerance test (PTT) results indicated that the gluconeogenesis efficiency was significantly higher at ZT18 than at ZT6 in CD36flox/flox mice but not in CD36-LKO mice (Figure 4G). Therefore, these results, for the first time, demonstrate the pivotal role of CD36 in regulating the circadian rhythm of gluconeogenesis.
Suppression of CD36 increases the nuclear translocation of FoxO1 and Per1
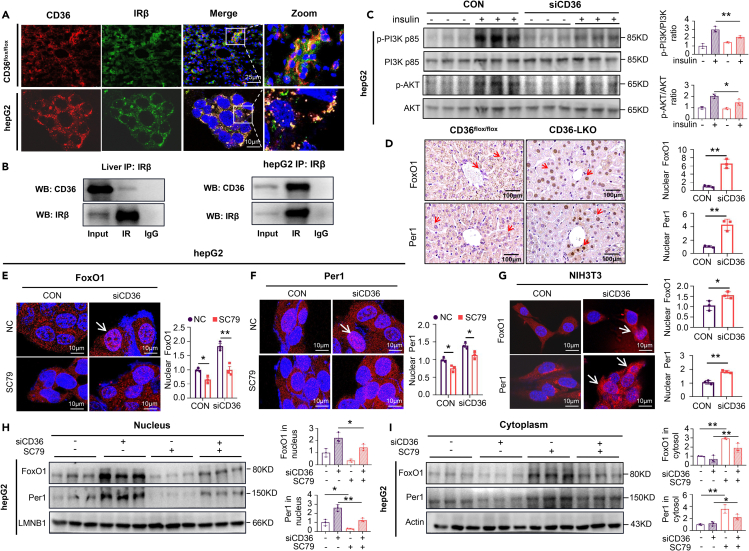
Given that insulin drives Period synthesis to entrain the circadian rhythm,8,32 the interaction between insulin and its receptor initiates a linear signaling cascade, leading to the phosphorylation and nuclear exclusion of FoxO1.33,34 We supposed that activation of the insulin signaling pathway mediated the effects of CD36 on Per1. Consistent with previous studies,21,35 colocalization of CD36 and IRβ could be clearly visualized in liver sections and HepG2 cells via confocal microscopy (Figure 5A), and immunoprecipitation analysis also showed the interaction between CD36 and IRβ (Figure 5B). In addition, insulin was observed to promote the binding of CD36 to Fyn in HepG2 cells (Figure S3A). Furthermore, reduced protein levels of p-AKT and p-PI3K were observed in siCD36 HepG2 cells (Figure 5C), indicating that suppression of CD36 markedly inhibited the activation of the insulin signaling pathway.
Figure 5.
Knockdown of CD36 facilitates nuclear FoxO1 and Per1 expression (see also Figure S3)
(A) Colocalization of CD36 and IRβ was imaged in CD36flox/flox mouse livers and HepG2 cells. Images show the overlay of IRβ (green) and CD36 (red). Yellow punctate structures indicate CD36-IRβ colocalization. Scale bars = 10μm.
(B) Co-IP of CD36 and IRβ in CD36flox/flox mouse liver tissue and HepG2 cells. IP was performed with anti-IRβ antibody, and immunoblotting was used to detect CD36 and IRβ.
(C) Western blot analysis of AKT, p-AKT (Ser473), PI3K p85, and p-PI3K p85 (Try467) expression in HepG2 cells after insulin (100 nmol/L, 5 min) stimulation. The relative quantification is shown on the right.
(D) Immunohistochemistry staining of FoxO1 and Per1 in the livers of CD36flox/flox and CD36-LKO mice. Fifty cells were used for statistical analysis of the number of nuclei positive for FoxO1 and Per1 puncta. Scale bars = 100μm.
(E and F) Effect of CD36 knockdown and the addition of SC79 on the cellular localization of FoxO1 (E) and Per1 (F) in HepG2 cells. Fifty cells were used for statistical analysis of nuclei positive for FoxO1 and Per1 puncta. Scale bars = 10μm.
(G) Cellular localization of FoxO1 and Per1 was imaged in NIH3T3cells after CD36 knockdown. Fifty cells were used for statistical analysis of nuclei positive for FoxO1 and Per1 puncta. Scale bars = 10μm.
(H and I) Immunoblots showing the effect of CD36 knockdown and the addition of SC79 on FoxO1 and Per1 levels in the nucleus (H) and cytoplasm (I) of HepG2 cells.
n = 3 in each group. All data are shown as the mean ± SEM. Comparison of different groups was carried out using two-tailed unpaired Student’s t test (D and G) or two-way ANOVA (C, E, F, H and I). ∗p <0.05, ∗∗p <0.01 versus control groups.
Activation of the insulin signaling pathway inactivates FoxO1 phosphorylation and promotes translocation out of the nucleus.36,37,38 As mentioned above, CD36 knockout in hepatocytes attenuated FoxO1 diurnal variation, and we wondered whether the interaction of CD36, IRβ and Fyn influences the subcellular localization of FoxO1 and Per1. We first examined the effect of CD36 knockdown on the FOXO1 subcellular localization in the presence of insulin, the results showed that insulin promoted nuclear output of FOXO1, whereas siCD36 blocked this effect (Figure S3B). Liver sections from the CD36-LKO mice displayed increased FoxO1 and Per1 staining in cell nuclei compared liver sections from CD36flox/flox mice (Figure 5D). Knockdown of CD36 significantly increased the nuclear translocation of FoxO1 and Per1, whereas these effects were diminished when AKT was activated by its agonist, SC79 (Figures 5E–5G and S3C), illustrating that the subcellular localization of FoxO1 may affect Per1 expression in nuclear and cytoplasmic fractions. Likewise, CD36 knockdown augmented the nuclear protein levels of FoxO1 and Per1, whereas SC79 abolished this effect (Figures 5H and 5I). Accordingly, the above results demonstrate that the insulin signaling pathway mediated the effect of CD36 knockdown on the nuclear shuttling of FoxO1 and Per1.
FoxO1 promotes Per1 transcription to reset the liver clock
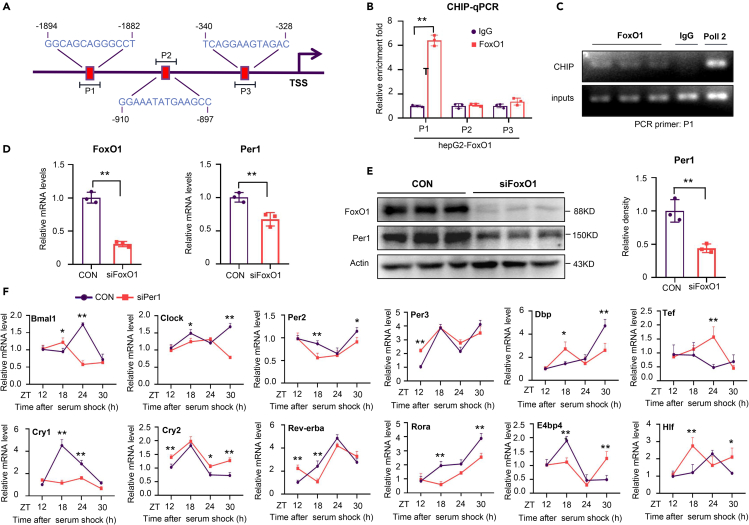
Considering that FoxO1 is a transcription factor that regulates many important biological functions, we performed JASPAR database analysis and predicted that the promoter of Per1 contains multiple FoxO1 binding sites (Figure 6A), suggesting that Per1 transcription may be regulated by FoxO1. To confirm this hypothesis, we conducted a CHIP assay with HepG2 cells to detect FoxO1 enrichment at the promoter of Per1. Indeed, FoxO1 was found to directly associate with a sequence (−1894/-1882) in the Per1 promoter (Figures 6B and 6C). Moreover, FoxO1 suppression via siRNA significantly reduced Per1 mRNA and protein levels in HepG2 cells (Figures 6D and 6E). Overall, these results indicate that FoxO1 can bind to the Per1 promoter to activate Per1 transcription.
Figure 6.
FoxO1 directly targets and positively regulates Per1 to reset the liver clock
(A) Schematic diagrams of the predicted binding sites of FoxO1 in the Per1 promoter.
(B) ChIP assays were performed and followed by real-time PCR.
(C) Recruitment of FoxO1 to the -1894/-1882 region of the Per1 promoter was detected by ChIP assays.
(D and E) HepG2 cells were transfected with siFoxO1. Relative mRNA levels of FoxO1 and Per1 were measured via real-time PCR (D), and the protein levels of FoxO1 and Per1 were detected via western blotting (E).
(F) mRNA levels of clock genes were detected over 30 h in NIH3T3 cells transfected with siPer1.
n = 3 in each group. All data are shown as the mean ± SEM. Comparison of different groups was carried out using two-tailed unpaired Student’s t test (B, D and E) or two-way ANOVA (F). ∗p <0.05, ∗∗p <0.01 versus control groups. ZT0, the beginning of a subjective circadian period (6:00 a.m.). Gray shading represents the lights-off period from 6:00 p.m. to 6:00 a.m.
Per1 knockdown in NIH3T3cells perturbed the circadian oscillation of the core clock genes and clock targeted genes, with an improper increase or decrease in their expression at multiple ZT points (Figure 6F), illustrating that Per1 deficiency induced dysfunction of the clock molecular mechanism. Taken together, these data indicate that FoxO1 modulates the liver clock by directly targeting and stimulating Per1 transcription.
Interfering with the AKT-FoxO1 pathway restored the liver clock and biological behavior rhythm disorders induced by hepatic CD36 deficiency
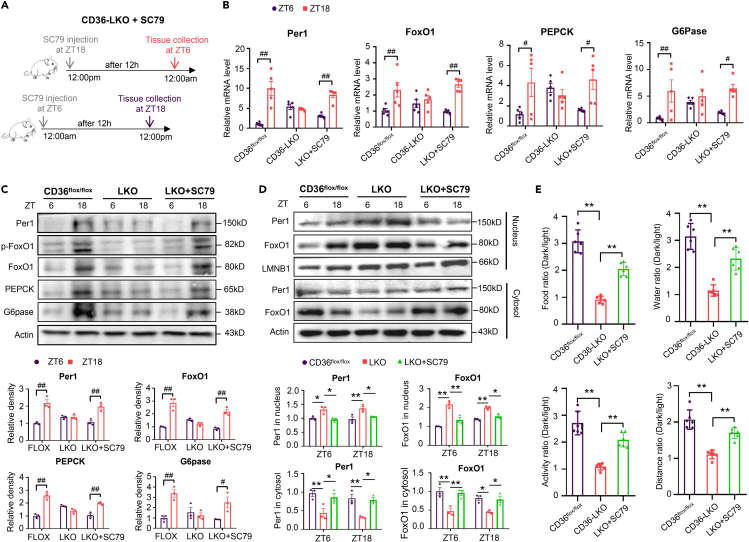
To examine whether inhibition of the insulin signaling pathway is involved in disruption of the liver clock and behavior rhythm in CD36-LKO mice, the in vivo effects of an AKT agonist on circadian rhythm were evaluated. Western blot analyses confirmed that SC79 enhanced Akt activation in CD36flox/flox mice, and this effect was decreased significantly in CD36-LKO mice (Figure S3D). SC79 was intraperitoneally injected into CD36-LKO mice at ZT18 or ZT6, and liver tissues were harvested 12 h after injection (Figure 7A). As expected, SC79 treatment restored the circadian mRNA and protein expression levels of Per1, FoxO1, and the downstream genes in the insulin signaling pathway in CD36-LKO mice (Figures 7B and 7C), as well as the subcellular localization of Per1 and FoxO1 (Figure 7D). We also conducted a series of glucose metabolism-related tolerance tests after SC79 injection. As expected, SC79 restored the reduced diurnal differences in insulin sensitivity, glucose tolerance, and gluconeogenesis caused by CD36 knockout in the liver (Figures S4A–S4C). We further monitored the day-night behavior of CD36flox/flox, CD36-LKO and CD36-LKO+SC79 mice. The circadian rhythm in behavior was robust in CD36flox/flox mice but markedly repressed in CD36-LKO mice, which was restored by SC79 treatment (Figures 7E and S5A–S5D). Consequently, the resetting of the liver clock and behavior rhythm by CD36 is closely linked to the AKT-FoxO1 pathway.
Figure 7.
Intervention of the AKT-FoxO1 pathway restored the dysfunction of the liver clock and biological behavior rhythm in CD36-LKO mice (see also Figures S4 and S5)
(A) Experimental design for examining the effect of SC79 in CD36-LKO mice.
(B) The relative mRNA levels of Per1, FoxO1, G6pase and PEPCK at ZT6 and ZT18 in the livers of CD36-LKO mice after SC79 injection.
(C) The protein expression levels and relative quantification of Per1, FoxO1, p-FoxO1, G6pase and PEPCK at ZT6 and ZT18 in the livers of CD36-LKO mice after SC79 injection.
(D) The protein expression levels and relative quantification of Per1 and FoxO1 at ZT6 and ZT18 in the nuclear fraction and cytosolic fraction of liver extracts from CD36-LKO mice after SC79 injection.
(E) Dark/light ratio of food intake, water intake, locomotor activity and movement distance of CD36-LKO mice after SC79 injection.
n = 5–6 in each group. All data are shown as the mean ± SEM. Comparison of different groups was carried out using two-tailed unpaired Student’s t test (E) or two-way ANOVA (B-D). ∗p <0.05, ∗∗p <0.01 versus control groups; #p <0.05, ##p <0.01 compared with ZT6 mice of the same genotype. ZT, zeitgeber time.
Discussion
The bidirectional relationship between the biological clock and metabolism is well reported, and the balance between them is very important to maintain health and homeostasis in the body.13,39 CD36, a versatile protein, functions as an important regulator of glucose metabolism.35,40 However, no clear evidence has shown that CD36 is implicated in the circadian clock or glucose-related metabolism rhythm. Our most important finding is that CD36 exhibits distinct patterns of diurnal variation in mouse livers and reset the glucose circadian clock in CD36 LKO mice relative to CD36flox/flox mice via a newly discovered FoxO1-Per1 pathway. In this study, we identified CD36 as a potential and powerful circadian regulator of glucose metabolism in the mouse liver and a new link between circadian rhythm and glucose homeostasis. Loss of CD36 in the liver resets the diurnal variations in hepatic clock genes and mouse behaviors as a result to aggravate the imbalance of glucose homeostasis.
The possible regulation of the circadian clock by CD36 is supported by several independent lines of evidence: (1) A 24-h period oscillation of CD36 expression in vivo and in vitro, and CD36 can be a regulator of the peripheral clock. qRT‒PCR and western blotting analyses suggested that CD36 exhibited a distinct diurnal pattern in mouse livers, NIH3T3 and HepG2 cells. In addition, CD36 expression could be changed by restricted light or feeding, suggesting that CD36 responds to food signals and peripheral clocks. (2) A phase relationship with components of the core clock machinery, such as Per1. Impaired amplitude of per1 oscillation was identified in liver-specific knockout mice, NIH3T3 and HepG2 siCD36 cells. (3) A strong association between CD36 and the circadian pattern of metabolism and behavior in mice. Simultaneous monitoring of diurnal metabolic and behavioral parameters using TSE metabolic cages showed that CD36 deficiency induced a significant attenuation of the circadian pattern between food and drink intake, as well as activity. (4) Disruptions of rhythmic serum glucose, insulin sensitivity and gluconeogenesis gene expression in CD36 LKO mice. GTT and ITT analyses suggested that the difference in glucose tolerance and insulin sensitivity between ZT6 and ZT18 disappeared in CD36-LKO mice; furthermore, a dampened oscillation of important key genes involved in gluconeogenesis, such as FoxO1, G6pase, PEPCK, and PGC-1α, in the liver of CD36-LKO mice during most of the circadian cycle was detected. In summary, the crucial role of CD36 in circadian rhythm, even in the diurnal pattern of glucose homeostasis, was well validated.
Given that CD36 is a transmembrane protein, whereas the clock machinery is primarily localized in the nucleus, we further investigated the exact mechanism by which CD36 delivers resetting signals through the cell membrane. CD36 could interact with IRβ and play complex roles in the insulin signaling pathway under several conditions such as multiple nutrient state, different tissue types and mouse models.21,35 Our data showed that the combination of with CD36 and IRβ ensured the circadian rhythm of liver clock through AKT-FoxO1 signaling under a normal chow diet. Exactly, loss of hepatic CD36 inhibited the activation of the insulin signaling pathway and caused misalignment of FoxO1 and p-FoxO1 rhythmic expression, thus dampening the daily rhythm of gluconeogenesis. In addition, CD36 knockdown increased the nuclear localization of FoxO1 and Per1, which could be restored by the AKT agonist SC79. There is growing evidence that metabolic and circadian systems are interconnected at the transcriptional level.41,42,43 Considering that FoxO1 is a transcription factor, we speculated that FoxO1 may mediate liver clock resetting of CD36 by regulating Per1 expression at the transcriptional level. Indeed, the JASPAR database shows that there are several predicted FoxO1 binding sites in the Per1 promoter. ChIP experiments showed that FoxO1 may bind to the Per1 promoter at the −1894/-1882 sites, and FoxO1 knockdown inhibited the expression of Per1. Overall, our data suggested that CD36 resets Per1 oscillation through the AKT-FoxO1 signaling pathway.
The proper phases and amplitudes of endogenous molecular clocks are crucial for the operation of the body’s biological rhythm, which is mainly controlled by Bmal1/Clock activity, and circadian inhibition of CLOCK-BMAL1 transcriptional activity is fundamentally determined by the precisely timed activity as well as nuclear accumulation of Per/Cry, especially Per.44,45,46 We found that FoxO1 activated Per1 transcription, whereas increased nuclear Per may reduce its own expression by inhibiting the CLOCK and BMAL1 heterodimer, which may partly explain the disrupted rhythmic oscillation of Per1 under CD36 deficiency conditions.
We selected hepatocyte-specific knockdown mice to elucidate a promising connection linking CD36, glucose metabolism and the liver clock because the liver is a pivotal metabolic organ with a massive number of metabolic genes manifesting a robust rhythm and which controls glucose uptake, production and metabolism.10,11,12,31 Peripheral oscillators can be uncoupled and reset from the central pacemaker by restricted feeding, and restricted feeding entrains the circadian rhythms in peripheral tissues, predominantly in the liver, but leaves SCN rhythms unaffected.47 In addition, both food availability and the temporal pattern of feeding determine the repertoire, phase, and amplitude of the circadian transcriptome in the mouse liver.48,49 CD36 functions importantly in the uptake of long-chain fatty acids and consequently performs a pivotal role in the metabolic process. We demonstrated that CD36 regulates the circadian rhythm of food intake in mice, which may partly explain why the liver clock is uncoupled from the SCN in our research. This provides the new insight that CD36, in parallel to restricting feeding, may be a powerful and direct factor that integrates peripheral and center clocks.
Although metabolic disorders caused by high expression of CD36 in liver are closely associated with NAFLD,50,51 multiple studies have shown that loss of CD36 inconsistently promotes the progression of NAFLD.52,53 Also, some studies have suggested a favorable effect of CD36 deficiency on obesity-induced insulin resistance, other studies showed a different result.35,54,55,56 These conflicting findings explained that the function of CD36 is complex. Our previous study has found that CD36-LKO mice exhibited attenuated high fat diet (HFD)-induced hepatic steatosis and insulin resistance, and hepatocyte CD36 was identified as a key regulator for de novo lipogenesis in the liver. However, we also proved that CD36 deficiency displayed a slight effect on lipid metabolism under normal chow diet (NCD) condition, suggesting that different dietary states may change the expression of CD36 and the associated metabolic phenotype. Here, we detected the effect of liver CD36 on diurnal variation of insulin sensitivity under the background of NCD from the perspective of circadian rhythm, and we may explore the relationship between CD36 and insulin variation under HFD condition in our later work. In addition, lipid metabolism is also highly related to circadian rhythm, and different dietary states may lead to the disorder of diurnal lipid metabolism.57,58,59 Hence, there exists an explanation of why CD36 showed different effect on lipid metabolism under different dietary states. That is, different dietary states may alter the diurnal variation of CD36, which in turn changes the rhythm of lipid metabolism, ultimately leading to the lipid homeostasis disorder. It will be interesting in the future to explore the relationship between CD36 and diurnal lipid metabolism.
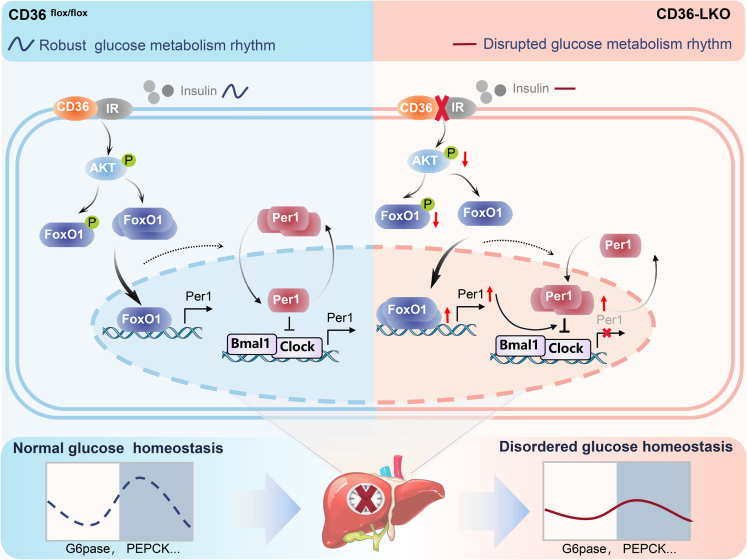
In conclusion, our work defines CD36 as a critical component and regulator of the mammalian clock, playing a crucial role in circadian coordination of the liver clock and glucose homeostasis. In this study, we demonstrated that CD36 acts as a key factor to regulate liver clock function via the AKT-FoxO1-Per1 signaling pathway and that its deficiency causes reduced amplitude of clock output and disruption of behavior rhythm, which may in turn aggravate the imbalance in glucose homeostasis (Figure 8).
Figure 8.
Proposed model
Hepatic CD36 regulates the liver clock via the AKT-FoxO1-Per1 pathway to govern glucose homeostasis. In CD36-LKO mice, inhibition of insulin signaling provokes FoxO1 nuclear shuttling, activates the central clock gene Per1 at the transcriptional level, simultaneously increases Per1 nuclear expression and then disrupts clock loop oscillation and behavioral rhythm, consequently leading to perturbed diurnal glucose homeostasis.
As a key fatty acid transporter, the interaction between the clock component CD36 and rhythmic glucose metabolism suggests that this transmembrane glycoprotein may be a potential mediator in integrating circadian rhythm and metabolism. Future studies will be required to characterize the role of CD36 in integrating energy metabolism and other peripheral clocks, which may pave the way to chronotherapy for metabolic diseases, such as diabetes.
Limitations of the study
One of the limitations of this study is the use of only female animals. It is unclear whether there is a significant sex-dependent dissociation between the phenotype of impaired rhythmic glucose metabolism and liver clock disorder in CD36-LKO mice. Thus, it is necessary to examine possible sex differences of this animal model in the future studies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD36 | Novus | cat# NB400-144; RRID: AB_10003498 |
| Per1 | LifeSpan | cat# LS-C807611 |
| AKT | Cell Signaling Technology | cat# 4691S; RRID: AB_915783 |
| p-AKT | Cell Signaling Technology | cat# 4060S; RRID: AB_2315049 |
| PI3K | Cell Signaling Technology | cat# 4257T; RRID: AB_659889 |
| p-PI3K | GeneTex | cat# GTX132597; RRID: AB_2886688 |
| FoxO1 | Cell Signaling Technology | cat# 2880S; RRID: AB_2106495 |
| p-FoxO1 | Cell Signaling Technology | cat# 9461T; RRID: AB_329831 |
| G6pase | Santa Cruz | cat# sc-167939; RRID: AB_10841606 |
| PEPCK | Santa Cruz | cat# sc-32879; RRID: AB_2160168 |
| β-actin | Bioss | cat# bs-0061R; RRID: AB_10855480 |
| Per1 | Proteintech | cat# 13463-1-AP; RRID: AB_2161536 |
| IRβ | Bioss | cat# bs-0290R; RRID: AB_10856426 |
| IRβ | Cell Signaling Technology | cat# 3025s; RRID: AB_2280448 |
| Fyn | Abcam | cat# ab184276; RRID: AB_2934009 |
| Chemicals, peptides, and recombinant proteins | ||
| SC79 | ApexBio | Cat# B5663 |
| DMEM | HyClone | Cat# SH30243.01 |
| Lipofectamine RNAiMAX Reagent | Invitrogen | Cat# 13778150 |
| HyClone Donor Equine Serum | HyClone | Cat# SH30074.02 |
| RNAiso Plus | TaKaRa | Cat# 9109 |
| protein G magnetic beads | Millipore | Cat# LSKMAGG02 |
| Critical commercial assays | ||
| Mouse Insulin ELISA KIT | Ruixinbio | cat# rx202485m |
| Mouse glucose ELISA KIT | Ruixinbio | cat# rxwb438 |
| Mouse leptin ELISA KIT | Ruixinbio | cat# rx202629m |
| Mouse glucagon ELISA KIT | Ruixinbio | cat# rx202477m |
| Mouse cortisone ELISA KIT | Ruixinbio | cat# rxj202703m |
| PrimeScript RT reagent kit | TaKaRa | cat# RR047A |
| Nuclear and Cytoplasmic Protein Extraction Kit | Beyotime Biotechnology | cat# P0027 |
| EZ-Magna ChIP A/G Chromatin Immunoprecipitation Kit | Millipore | cat# 17-10086 |
| Deposited data | ||
| Original western blot images | This paper | Mendeley Data: https://doi.org/10.17632/zmtm4cvpcb.2 |
| Experimental models: Cell lines | ||
| HepG2 | ATCC | ATCC HB-8065 |
| NIH/3T3 | BNCC | BNCC100843 |
| Experimental models: Organisms/strains | ||
| CD36flox/flox mice | This paper | N/A |
| CD36-LKO mice | This paper | N/A |
| Oligonucleotides | ||
| Primers for qPCR, see Tables S1 and S2 | see Tables S1 and S2 | Primers for qPCR, see Tables S1 and S2 |
| Primers for ChIP-qPCR, see Table S3 | see Table S3 | Primers for ChIP-qPCR, see Table S3 |
| Software and algorithms | ||
| ImageJ | https://imagej.nih.gov/ij/ | |
| GraphPad Prism | GraphPad Software, Inc. | https://www.graphpad.com/scientific-software/prism/ |
| Other | ||
| TSE PhenoMaster metabolic cages | TSE Systems | http://www.tsesystems.com.cn/xdwdxjcxtphenomaster |
| Leica TCS SP8 confocal laser scanning microscope | Leica | https://www.leica-microsystems.com.cn/cn/products/confocal-microscopes/p/sp8-sr/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Yaxi Chen (chenyaxi@cqmu.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Animals and treatments
The CD36flox/flox mice were generated by using a plasmid with Loxp sites flanking CD36 exons 5, to generate CD36-LKO mice, CD36flox/flox mice were crossed with Albumin-cre transgenic mice (Shanghai Model Organisms Center, China) as previously described.60 Female CD36-LKO and CD36flox/flox mice aged 8-12 weeks were used in all experiments. Mice were housed in the specific pathogen-free (SPF) mouse facility of Chongqing Medical University. Animal experiments were conducted in accordance with the guidelines of the Institutional Animal Care and Use Committee of Chongqing Medical University. All mice were maintained under a standard 12 h light/12 h dark cycle at 25°C with free access to water and a regular chow diet. Zeitgeber time zero (ZT0) referred to lights on at 6:00 AM.
Liver samples were harvested from CD36flox/flox and CD36-LKO mice every 6 h over a period of 48 h. Venous blood was collected from the retro-orbital plexuses before decapitation, and tissues were quickly dissected, frozen on dry ice, and stored at −80°C. The serum was separated within 30 minutes and kept at −80°C.
To test the effect of light on hepatic CD36 expression, the CD36flox/flox mice were subjected to the reversed light/dark cycle (dark/light, DL) for 7 days and sacrificed at ZT6 and ZT18 on day 8 under the DL condition. To test the effect of food on hepatic CD36 expression, CD36flox/flox mice were fed exclusively during the light phase (Daytime-restricted feeding, DF) for 7 days and sacrificed at ZT6 and ZT18 on day 8 under DF conditions. For fasting experiments, CD36flox/flox and CD36-LKO mice were subjected to 12 h fasting from ZT18 to ZT6 and blood glucose was measured at ZT6, or 12 h fasting from ZT6 to ZT18 and blood glucose was measured at ZT18. Mice in each group were fasted overnight and intraperitoneally injected with insulin (5 units/kg body weight) 10 minutes before the livers were collected at ZT6 and ZT18 for analysis of insulin signaling. CD36-LKO mice were intraperitoneally injected with vehicle (PBS) at 100 μl or the same dose of the AKT agonist SC79 (ApexBio, Houston, USA) at a concentration of 40 mg/kg for 12 h and then were sacrificed at ZT6 and ZT18 to investigate the effect of insulin signaling on resetting the liver clock.
Cell line and culture
HepG2 (ATCC, Manassas, VA, USA) and NIH3T3 cells (BNCC, Beijing, China) were cultured in DMEM (HyClone, UT, USA) supplemented with 10% fetal bovine serum (FBS) and 100 units/mL penicillin-streptomycin and maintained in a humidified incubator at 37°C with 5% CO2. All cell lines have been authenticated by procell Life Science&Technology Co.,Ltd and tested negative for mycoplasma contamination. The cells were transfected with the indicated constructs using Lipofectamine RNAiMAX Reagent (Invitrogen, CA, USA) and CD36 siRNA (siCD36)/FoxO1 siRNA (siFoxO1)/Per1 siRNA (siPer1) or negative control (-ve CTRi) according to the manufacturer's protocol. SC79 (4 μg/ml, 1 h; ApexBio) was added to the cultures as indicated. For cell serum shock, cells were exposed to 50% horse serum (HyClone, UT, USA) for 2 h. Subsequently, the medium was replaced with recording medium (DMEM supplemented with 1% FBS and 100 units/mL penicillin–streptomycin), and then, the cells were harvested at 6 h intervals from 12 to 30 h.
Method details
Glucose tolerance test (GTT)
Mice were fasted overnight for 12 h. Following fasting, at ZT6 or ZT18, baseline blood glucose was measured using a glucose meter (Roche Diagnostics, Basel, Switzerland), and mice were intraperitoneally injected with glucose (1 g/kg) in isotonic saline. Glucose levels in blood were further measured at 15, 30, 60, 90, and 120 min intervals following injection.
Insulin tolerance test (ITT)
Mice were fasted overnight for 4 h. Following fasting, at ZT6 or ZT18, baseline blood glucose was measured using a glucose meter (Roche Diagnostics, Basel, Switzerland), and mice were intraperitoneally injected with insulin (0.75 U/kg) in isotonic saline. Glucose levels in blood were further measured at 15, 30, 60, 90, and 120 min intervals following injection.
Pyruvate tolerance test (PTT)
Mice were fasted overnight for 16 h. Following fasting, at ZT6 or ZT18, baseline blood glucose was measured using a glucose meter (Roche Diagnostics, Basel, Switzerland), and mice were intraperitoneally injected with pyruvate (2 g/kg) in isotonic saline. Glucose levels in blood were further measured at 15, 30, 60, 90, and 120 min intervals following injection.
Serum biochemistry analysis
The serum levels of insulin, glucose, leptin, glucagon, and cortisone were determined using commercial kits (rx202485m, Ruixinbio, China for insulin; rxwb438, Ruixinbio, China for glucose; rx202629m, Ruixinbio, China for leptin; rx202477m, Ruixinbio, China for glucagon; rxj202703m, Ruixinbio, China for cortisone) according to the manufacturer's instructions.
RNA extraction and RT-qPCR analysis
Total RNA from cells or mouse tissues was isolated using TRIzol reagent (TaKaRa, Japan) and reverse transcribed using a PrimeScript RT reagent kit (TaKaRa, Japan) according to the manufacturers’ protocol. Real-time quantitative PCR (RT-PCR) was performed on a Bio-Rad CFX Connect Real-time PCR system (BioRad, USA) with a SYBR Green PCR Mix kit (TaKaRa, Japan). Gene expression levels were normalized to β-actin, and relative levels were compared to the control using the 2-ΔΔCt method. The specific primer sequences used for real-time PCR are listed in Tables S1 and S2.
Western blotting analysis
Protein extracts from liver tissue or cells were lysed in RIPA buffer, and cytoplasmic and nuclear proteins were extracted from HepG2 cells or liver tissue using a commercial kit (Beyotime Biotechnology, Wuhan, China). The lysates were separated by SDS-PAGE and transferred to PVDF membranes. The membranes were incubated overnight with primary antibodies at 4°C and then incubated with HRP-conjugated secondary antibodies for 2 hours. Immunoreactive protein was detected using an ECL Advance Western Blotting Detection kit (Amersham Bioscience, Piscataway, US). A quantitative analysis was performed using Image J software (V1.53k). The following primary antibodies were purchased: anti-CD36 (1:2000, Novus, cat# NB400-144), anti-Per1 (1:2000, LifeSpan, cat# LS-C807611), anti-AKT (1:1000, CST, cat# 4691S), anti-p-AKT (1:2000, CST, cat# 4060S), anti-PI3K (1:1000, CST, cat# 4257T), anti-p-PI3K (1:500, GeneTex, cat# GTX132597), anti-FoxO1 (1:1000, CST, cat# 2880S), anti-p-FoxO1 (1:1000, CST, cat# 9461T) anti-G6pase (1:2000, Santa Cruz, cat# sc-167939), anti-PEPCK (1:2000, Santa Cruz, cat# sc-32879) and anti-β-actin (1:5000, Bioss, cat# bs-0061R).
Metabolic profiling studies
Experimental mice were housed for 2 weeks in TSE PhenoMaster metabolic cages (TSE Systems; Berlin, Germany) to allow them to acclimate to the new cage environment, and then food and water intake, movement distance, physical activity, oxygen consumption (VO2), carbon dioxide output (VCO2), respiratory exchange ratio (RER) and energy expenditure (EE) were simultaneously measured throughout the 48 h metabolic profiling studies. Food and water were provided ad libitum in the appropriate devices, and all mice were housed under a 12 h light/dark cycle.
Immunohistochemistry
Immunohistochemical studies were performed on sections of 4% paraformaldehyde-fixed and paraffin-embedded liver tissues. Endogenous peroxidases were inactivated using 3% H2O2, followed by blocking with goat serum. Sections were incubated overnight (4°C) with anti-Per1 (1:200; Proteintech, Wuhan, China) and anti-FoxO1 (1:100; CST, Danvers, MA) antibodies. Then, the sections were washed and incubated for 30 min with secondary antibody at 37°C. Histochemical reactions were performed using a diaminobenzidine (DAB) kit, and sections were counterstained with hematoxylin. All images were captured using a Zeiss microscope (Zeiss).
Immunofluorescence
Cells grown in confocal dishes were fixed with 4% paraformaldehyde for 15 min at 37°C, blocked with 3% bovine serum albumin for 30 min at 37°C, and incubated overnight at 4°C with primary antibodies against Per1 (LS-C807611, LifeSpan, US), FoxO1 (2880S, Cell Signaling, US) and IRβ (bs-0290R, Bioss, China). After being washed with PBS three times, the cells were incubated with fluorescence-conjugated secondary antibodies for 1hat 37°C. Cells were washed three times and then stained with DAPI, and images were captured using a Leica TCS SP8 confocal laser scanning microscope (Leica, Germany).
Coimmunoprecipitation (Co-IP)
Proteins from cells and tissues were extracted in NP40 buffer and RIPA buffer, respectively. Endogenous IRβ and Fyn was respectively immunoprecipitated using anti-IRβ antibody (3025s, Cell Signaling, US) and anti-Fyn antibody (ab184276, Abcam, US) coupled with protein G magnetic beads (LSKMAGG02, Millipore, MA, USA). All immunoprecipitated samples were washed three times with cold lysis buffer, and then, the bound proteins were eluted by boiling for 5 min in SDS sample buffer. Rabbit IgG was used as a negative control. The immunoprecipitated protein was further analyzed via western blotting with the respective antibodies.
Chromatin immunoprecipitation (ChIP)
The chromatin immunoprecipitation (ChIP) assay was performed according to the manufacturer’s instructions for an EZ-Magna ChIP A/G Chromatin Immunoprecipitation Kit (17-10086, Millipore, Merck, Bedford, MD). The immunoprecipitated DNA samples were detected via RT-PCR using three primers that were within the −2000 bp to +0 bp region of the Per1 promoter. The ChIP-qPCR primer sequences are provided in Table S3.
Quantification and statistical analysis
Statistical analysis
Statistical analysis was performed with GraphPad Prism 8. Data are shown as the mean with SEM. Comparisons between different groups were performed using two-tailed unpaired Student’s t test or two-way ANOVA. For all analyses, p <0.05 was considered statistically significant.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (32030054, 82170586); the Chongqing Research Program of Basic Research and Frontier Technology (cstc2020jcyj-zdxmX0007); Kuanren Talents Program of the second affiliated hospital of Chongqing Medical University; the 111 Project (No. D20028); Program for Youth Innovation in Future Medicine, Chongqing Medical University.
Author contributions
M.Y.C., Y.Z., and S.Z. performed the experiments, analyzed the data, and wrote the manuscript. D.Y.L., M.Y.Y., Z.Y.W., and M.Y.Z. performed experiments. L.W. analyzed the data. Y.X.C. and X.Z.R. initiated and supervised this work, designed the experiments, and approved the final version of manuscript.
Declaration of interests
The authors declare no competing interests.
Published: March 29, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106524.
Contributor Information
Yaxi Chen, Email: chenyaxi@cqmu.edu.cn.
Xiong Z. Ruan, Email: x.ruan@ucl.ac.uk.
Supplemental information
Data and code availability
-
•
Original western blot images have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Bass J., Lazar M.A. Circadian time signatures of fitness and disease. Science (New York, N.Y.) 2016;354:994–999. doi: 10.1126/science.aah4965. [DOI] [PubMed] [Google Scholar]
- 2.Bass J., Takahashi J.S. Circadian integration of metabolism and energetics. Science (New York, N.Y.) 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dibner C., Schibler U., Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 4.Froy O. The relationship between nutrition and circadian rhythms in mammals. Front. Neuroendocrinol. 2007;28:61–71. doi: 10.1016/j.yfrne.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Poggiogalle E., Jamshed H., Peterson C.M. Circadian regulation of glucose, lipid, and energy metabolism in humans. Metabolism. 2018;84:11–27. doi: 10.1016/j.metabol.2017.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun X., Dang F., Zhang D., Yuan Y., Zhang C., Wu Y., Wang Y., Liu Y. Glucagon-CREB/CRTC2 signaling cascade regulates hepatic BMAL1 protein. J. Biol. Chem. 2015;290:2189–2197. doi: 10.1074/jbc.M114.612358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ikeda Y., Kamagata M., Hirao M., Yasuda S., Iwami S., Sasaki H., Tsubosaka M., Hattori Y., Todoh A., Tamura K., et al. Glucagon and/or IGF-1 production regulates resetting of the liver circadian clock in response to a protein or amino acid-only diet. EBioMedicine. 2018;28:210–224. doi: 10.1016/j.ebiom.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crosby P., Hamnett R., Putker M., Hoyle N.P., Reed M., Karam C.J., Maywood E.S., Stangherlin A., Chesham J.E., Hayter E.A., et al. Insulin/IGF-1 drives PERIOD synthesis to entrain circadian rhythms with feeding time. Cell. 2019;177:896–909.e20. doi: 10.1016/j.cell.2019.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jarrett R.J., Keen H. Diurnal variation of oral glucose tolerance: a possible pointer to the evolution of diabetes mellitus. Br. Med. J. 1969;2:341–344. doi: 10.1136/bmj.2.5653.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang E.E., Liu Y., Dentin R., Pongsawakul P.Y., Liu A.C., Hirota T., Nusinow D.A., Sun X., Landais S., Kodama Y., et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat. Med. 2010;16:1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doi R., Oishi K., Ishida N. CLOCK regulates circadian rhythms of hepatic glycogen synthesis through transcriptional activation of Gys2. J. Biol. Chem. 2010;285:22114–22121. doi: 10.1074/jbc.M110.110361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim H., Zheng Z., Walker P.D., Kapatos G., Zhang K. CREBH maintains circadian glucose homeostasis by regulating hepatic glycogenolysis and gluconeogenesis. Mol. Cell Biol. 2017;37:e00048-17. doi: 10.1128/mcb.00048-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reinke H., Asher G. Crosstalk between metabolism and circadian clocks. Nat. Rev. Mol. Cell Biol. 2019;20:227–241. doi: 10.1038/s41580-018-0096-9. [DOI] [PubMed] [Google Scholar]
- 14.Stenvers D.J., Scheer F.A.J.L., Schrauwen P., la Fleur S.E., Kalsbeek A. Circadian clocks and insulin resistance. Nat. Rev. Endocrinol. 2019;15:75–89. doi: 10.1038/s41574-018-0122-1. [DOI] [PubMed] [Google Scholar]
- 15.Silverstein R.L., Febbraio M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci. Signal. 2009;2:re3. doi: 10.1126/scisignal.272re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glazier A.M., Scott J., Aitman T.J. Molecular basis of the Cd36 chromosomal deletion underlying SHR defects in insulin action and fatty acid metabolism. Mamm. Genome. 2002;13:108–113. doi: 10.1007/s00335-001-2132-9. [DOI] [PubMed] [Google Scholar]
- 17.Miyaoka K., Kuwasako T., Hirano K., Nozaki S., Yamashita S., Matsuzawa Y. CD36 deficiency associated with insulin resistance. Lancet (London, England) 2001;357:686–687. doi: 10.1016/s0140-6736(00)04138-6. [DOI] [PubMed] [Google Scholar]
- 18.Corpeleijn E., van der Kallen C.J.H., Kruijshoop M., Magagnin M.G.P., de Bruin T.W.A., Feskens E.J.M., Saris W.H.M., Blaak E.E. Direct association of a promoter polymorphism in the CD36/FAT fatty acid transporter gene with Type 2 diabetes mellitus and insulin resistance. Diabet. Med. 2006;23:907–911. doi: 10.1111/j.1464-5491.2006.01888.x. [DOI] [PubMed] [Google Scholar]
- 19.Furuhashi M., Ura N., Nakata T., Shimamoto K. Insulin sensitivity and lipid metabolism in human CD36 deficiency. Diabetes Care. 2003;26:471–474. doi: 10.2337/diacare.26.2.471. [DOI] [PubMed] [Google Scholar]
- 20.Hirano K.I., Kuwasako T., Nakagawa-Toyama Y., Janabi M., Yamashita S., Matsuzawa Y. Pathophysiology of human genetic CD36 deficiency. Trends Cardiovasc. Med. 2003;13:136–141. doi: 10.1016/s1050-1738(03)00026-4. [DOI] [PubMed] [Google Scholar]
- 21.Yang P., Zeng H., Tan W., Luo X., Zheng E., Zhao L., Wei L., Ruan X.Z., Chen Y., Chen Y. Loss of CD36 impairs hepatic insulin signaling by enhancing the interaction of PTP1B with IR. FASEB J. 2020;34:5658–5672. doi: 10.1096/fj.201902777RR. [DOI] [PubMed] [Google Scholar]
- 22.Reppert S.M., Weaver D.R. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 23.Partch C.L., Green C.B., Takahashi J.S. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014;24:90–99. doi: 10.1016/j.tcb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sato T.K., Panda S., Miraglia L.J., Reyes T.M., Rudic R.D., McNamara P., Naik K.A., FitzGerald G.A., Kay S.A., Hogenesch J.B. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 25.Preitner N., Damiola F., Lopez-Molina L., Zakany J., Duboule D., Albrecht U., Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 26.Chaves I., van der Horst G.T.J., Schellevis R., Nijman R.M., Koerkamp M.G., Holstege F.C.P., Smidt M.P., Hoekman M.F.M. Insulin-FOXO3 signaling modulates circadian rhythms via regulation of clock transcription. Curr. Biol. 2014;24:1248–1255. doi: 10.1016/j.cub.2014.04.018. [DOI] [PubMed] [Google Scholar]
- 27.Kalsbeek A., la Fleur S., Fliers E. Circadian control of glucose metabolism. Mol. Metab. 2014;3:372–383. doi: 10.1016/j.molmet.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson C.G., Tran J.L., Erion D.M., Vera N.B., Febbraio M., Weiss E.J. Hepatocyte-specific disruption of CD36 attenuates fatty liver and improves insulin sensitivity in HFD-fed mice. Endocrinology. 2016;157:570–585. doi: 10.1210/en.2015-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Son N.H., Basu D., Samovski D., Pietka T.A., Peche V.S., Willecke F., Fang X., Yu S.Q., Scerbo D., Chang H.R., et al. Endothelial cell CD36 optimizes tissue fatty acid uptake. J. Clin. Invest. 2018;128:4329–4342. doi: 10.1172/jci99315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asher G., Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 31.Takeda Y., Kang H.S., Freudenberg J., DeGraff L.M., Jothi R., Jetten A.M. Retinoic acid-related orphan receptor γ (RORγ): a novel participant in the diurnal regulation of hepatic gluconeogenesis and insulin sensitivity. PLoS Genet. 2014;10:e1004331. doi: 10.1371/journal.pgen.1004331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tuvia N., Pivovarova-Ramich O., Murahovschi V., Lück S., Grudziecki A., Ost A.C., Kruse M., Nikiforova V.J., Osterhoff M., Gottmann P., et al. Insulin directly regulates the circadian clock in adipose tissue. Diabetes. 2021;70:1985–1999. doi: 10.2337/db20-0910. [DOI] [PubMed] [Google Scholar]
- 33.Rena G., Guo S., Cichy S.C., Unterman T.G., Cohen P. Phosphorylation of the transcription factor forkhead family member FKHR by protein kinase B. J. Biol. Chem. 1999;274:17179–17183. doi: 10.1074/jbc.274.24.17179. [DOI] [PubMed] [Google Scholar]
- 34.Nakae J., Park B.C., Accili D. Insulin stimulates phosphorylation of the forkhead transcription factor FKHR on serine 253 through a Wortmannin-sensitive pathway. J. Biol. Chem. 1999;274:15982–15985. doi: 10.1074/jbc.274.23.15982. [DOI] [PubMed] [Google Scholar]
- 35.Samovski D., Dhule P., Pietka T., Jacome-Sosa M., Penrose E., Son N.H., Flynn C.R., Shoghi K.I., Hyrc K.L., Goldberg I.J., et al. Regulation of insulin receptor pathway and glucose metabolism by CD36 signaling. Diabetes. 2018;67:1272–1284. doi: 10.2337/db17-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao X., Gan L., Pan H., Kan D., Majeski M., Adam S.A., Unterman T.G. Multiple elements regulate nuclear/cytoplasmic shuttling of FOXO1: characterization of phosphorylation- and 14-3-3-dependent and -independent mechanisms. Biochem. J. 2004;378:839–849. doi: 10.1042/bj20031450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brunet A., Bonni A., Zigmond M.J., Lin M.Z., Juo P., Hu L.S., Anderson M.J., Arden K.C., Blenis J., Greenberg M.E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 38.Lu M., Wan M., Leavens K.F., Chu Q., Monks B.R., Fernandez S., Ahima R.S., Ueki K., Kahn C.R., Birnbaum M.J. Insulin regulates liver metabolism in vivo in the absence of hepatic Akt and Foxo1. Nat. Med. 2012;18:388–395. doi: 10.1038/nm.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Panda S. Circadian physiology of metabolism. Science (New York, N.Y.) 2016;354:1008–1015. doi: 10.1126/science.aah4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maréchal L., Laviolette M., Rodrigue-Way A., Sow B., Brochu M., Caron V., Tremblay A. The CD36-PPARγ pathway in metabolic disorders. Int. J. Mol. Sci. 2018;19:1529. doi: 10.3390/ijms19051529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L., Yang G. PPARs integrate the mammalian clock and energy metabolism. PPAR Res. 2014;2014:653017. doi: 10.1155/2014/653017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adamovich Y., Ladeuix B., Golik M., Koeners M.P., Asher G. Rhythmic oxygen levels reset circadian clocks through HIF1α. Cell Metab. 2017;25:93–101. doi: 10.1016/j.cmet.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 43.Ruberto A.A., Gréchez-Cassiau A., Guérin S., Martin L., Revel J.S., Mehiri M., Subramaniam M., Delaunay F., Teboul M. KLF10 integrates circadian timing and sugar signaling to coordinate hepatic metabolism. Elife. 2021;10:e65574. doi: 10.7554/eLife.65574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen R., Schirmer A., Lee Y., Lee H., Kumar V., Yoo S.H., Takahashi J.S., Lee C. Rhythmic PER abundance defines a critical nodal point for negative feedback within the circadian clock mechanism. Mol. Cell. 2009;36:417–430. doi: 10.1016/j.molcel.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ashimori A., Nakahata Y., Sato T., Fukamizu Y., Matsui T., Yoshitane H., Fukada Y., Shinohara K., Bessho Y. Attenuated SIRT1 activity leads to PER2 cytoplasmic localization and dampens the amplitude of Bmal1 promoter-driven circadian oscillation. Front. Neurosci. 2021;15:647589. doi: 10.3389/fnins.2021.647589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Korge S., Maier B., Brüning F., Ehrhardt L., Korte T., Mann M., Herrmann A., Robles M.S., Kramer A. The non-classical nuclear import carrier Transportin 1 modulates circadian rhythms through its effect on PER1 nuclear localization. PLoS Genet. 2018;14:e1007189. doi: 10.1371/journal.pgen.1007189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Damiola F., Le Minh N., Preitner N., Kornmann B., Fleury-Olela F., Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guan D., Xiong Y., Trinh T.M., Xiao Y., Hu W., Jiang C., Dierickx P., Jang C., Rabinowitz J.D., Lazar M.A. The hepatocyte clock and feeding control chronophysiology of multiple liver cell types. Science (New York, N.Y.) 2020;369:1388–1394. doi: 10.1126/science.aba8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weger B.D., Gobet C., David F.P.A., Atger F., Martin E., Phillips N.E., Charpagne A., Weger M., Naef F., Gachon F. Systematic analysis of differential rhythmic liver gene expression mediated by the circadian clock and feeding rhythms. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2015803118. e2015803118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miquilena-Colina M.E., Lima-Cabello E., Sánchez-Campos S., García-Mediavilla M.V., Fernández-Bermejo M., Lozano-Rodríguez T., Vargas-Castrillón J., Buqué X., Ochoa B., Aspichueta P., et al. Hepatic fatty acid translocase CD36 upregulation is associated with insulin resistance, hyperinsulinaemia and increased steatosis in non-alcoholic steatohepatitis and chronic hepatitis C. Gut. 2011;60:1394–1402. doi: 10.1136/gut.2010.222844. [DOI] [PubMed] [Google Scholar]
- 51.Bieghs V., Wouters K., van Gorp P.J., Gijbels M.J.J., de Winther M.P.J., Binder C.J., Lütjohann D., Febbraio M., Moore K.J., van Bilsen M., et al. Role of scavenger receptor A and CD36 in diet-induced nonalcoholic steatohepatitis in hyperlipidemic mice. Gastroenterology. 2010;138:2477-86–2486.e1-3. doi: 10.1053/j.gastro.2010.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhong S., Zhao L., Wang Y., Zhang C., Liu J., Wang P., Zhou W., Yang P., Varghese Z., Moorhead J.F., et al. Cluster of differentiation 36 deficiency aggravates macrophage infiltration and hepatic inflammation by upregulating monocyte chemotactic protein-1 expression of hepatocytes through histone deacetylase 2-dependent pathway. Antioxid. Redox Signal. 2017;27:201–214. doi: 10.1089/ars.2016.6808. [DOI] [PubMed] [Google Scholar]
- 53.Nassir F., Adewole O.L., Brunt E.M., Abumrad N.A. CD36 deletion reduces VLDL secretion, modulates liver prostaglandins, and exacerbates hepatic steatosis in ob/ob mice. J. Lipid Res. 2013;54:2988–2997. doi: 10.1194/jlr.M037812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hajri T., Hall A.M., Jensen D.R., Pietka T.A., Drover V.A., Tao H., Eckel R., Abumrad N.A. CD36-facilitated fatty acid uptake inhibits leptin production and signaling in adipose tissue. Diabetes. 2007;56:1872–1880. doi: 10.2337/db06-1699. [DOI] [PubMed] [Google Scholar]
- 55.Kennedy D.J., Kuchibhotla S., Westfall K.M., Silverstein R.L., Morton R.E., Febbraio M. A CD36-dependent pathway enhances macrophage and adipose tissue inflammation and impairs insulin signalling. Cardiovasc. Res. 2011;89:604–613. doi: 10.1093/cvr/cvq360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aouadi M., Vangala P., Yawe J.C., Tencerova M., Nicoloro S.M., Cohen J.L., Shen Y., Czech M.P. Lipid storage by adipose tissue macrophages regulates systemic glucose tolerance. Am. J. Physiol. Endocrinol. Metab. 2014;307:E374–E383. doi: 10.1152/ajpendo.00187.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang S., Lin Y., Gao L., Yang Z., Lin J., Ren S., Li F., Chen J., Wang Z., Dong Z., et al. PPAR-γ integrates obesity and adipocyte clock through epigenetic regulation of Bmal1. Theranostics. 2022;12:1589–1606. doi: 10.7150/thno.69054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He Y., B'Nai Taub A., Yu L., Yao Y., Zhang R., Zahr T., Aaron N., LeSauter J., Fan L., Liu L., et al. PPARγ acetylation orchestrates adipose plasticity and metabolic rhythms. Adv. Sci. 2023;10:e2204190. doi: 10.1002/advs.202204190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Le Martelot G., Claudel T., Gatfield D., Schaad O., Kornmann B., Lo Sasso G., Moschetta A., Schibler U. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7:e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zeng H., Qin H., Liao M., Zheng E., Luo X., Xiao A., Li Y., Chen L., Wei L., Zhao L., et al. CD36 promotes de novo lipogenesis in hepatocytes through INSIG2-dependent SREBP1 processing. Mol. Metab. 2022;57:101428. doi: 10.1016/j.molmet.2021.101428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Original western blot images have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.