Summary
Pancreatic ductal adenocarcinoma (PDAC) has been left behind in the evolution of personalized medicine. Predictive markers of response to therapy are lacking in PDAC despite various histological and transcriptional classification schemes. We report an artificial intelligence (AI) approach to histologic feature examination that extracts a signature predictive of disease-specific survival (DSS) in patients with PDAC receiving adjuvant gemcitabine. We demonstrate that this AI-generated histologic signature is associated with outcomes following adjuvant gemcitabine, while three previously developed transcriptomic classification systems are not (n = 47). We externally validate this signature in an independent cohort of patients treated with adjuvant gemcitabine (n = 46). Finally, we demonstrate that the signature does not stratify survival outcomes in a third cohort of untreated patients (n = 161), suggesting that the signature is specifically predictive of treatment-related outcomes but is not generally prognostic. This imaging analysis pipeline has promise in the development of actionable markers in other clinical settings where few biomarkers currently exist.
Keywords: pancreatic cancer, digital pathology, predictive biomarker
Graphical abstract
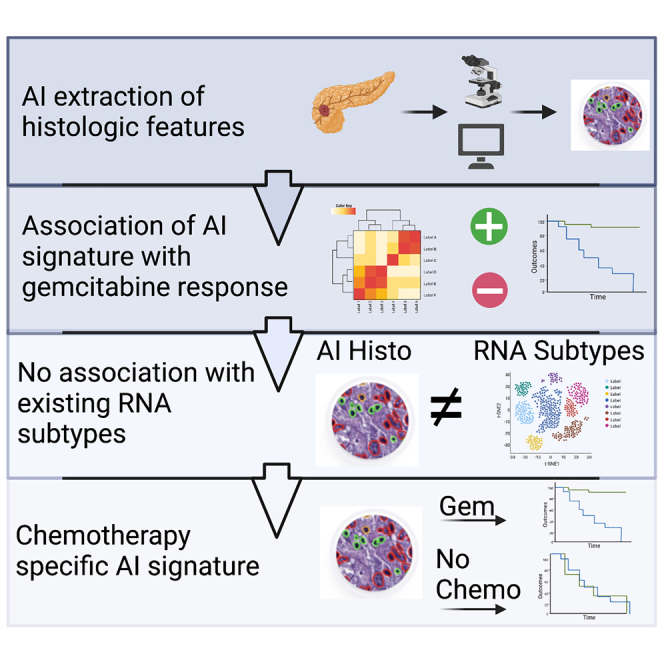
Highlights
-
•
Pancreatic cancer lacks actionable biomarkers for precision therapy
-
•
We report on an AI-derived histologic marker for adjuvant gemcitabine response
-
•
The AI histologic signature is not associated with existing transcriptomic sub-types
-
•
The signature is treatment specific in external cohorts
Nimgaonkar et al. describe the identification of a histologic signature derived using artificial intelligence that stratifies patients receiving adjuvant gemcitabine for pancreatic cancer. The signature is not associated with existing transcriptomic classification schemes and validates specifically to gemcitabine-treated patients in external cohorts.
Introduction
The prognosis for patients diagnosed with localized pancreatic ductal adenocarcinoma (PDAC) remains poor even after successful surgical resection.1 Adjuvant chemotherapy regimens, including modified FOLFIRINOX (5-fluorouracil, irinotecan, and oxaliplatin)-2 and gemcitabine-based regimens,3 have improved overall survival (OS) when compared with observation, but most patients still experience disease recurrence within 2 years. Increasingly, neoadjuvant chemotherapy with or without additional post-operative chemotherapy is being utilized, though the optimal regimen or sequence of regimens remains uncertain.4 Intense study of PDAC tumor genomics has revealed several distinct and reproducible transcriptomic profiles, but to date, there are no validated predictive biomarkers to guide recommendation of one chemotherapy regimen over another in clinical practice.5,6,7,8 There is a need for improved predictive PDAC tumor biomarkers that can prospectively identify patients most likely to benefit from existing chemotherapy regimens using bioanalytes available in the standard-of-care setting.
The advent of digital pathology involving the scanning and computational analysis of digitized whole-slide images has created an opportunity for the discovery and exploitation of novel, sub-visual morphologic biomarkers.9 Quantified morphologic features can identify novel associations to patient outcomes using artificial intelligence (AI).10,11 Morphometric analyses can uncover histologic features associated with response to a particular treatment when a dataset comprises patients treated with a specific agent and outcomes are known.12 More recently, modern deep learning toolkits have enabled rapid segmentation and classification of individual cell types.13 The potential to use deep learning in conjunction with morphometric analysis to identify novel associations between specific cellular compartments in the tumor microenvironment and responses to treatment can enable identification of treatment-specific biomarkers, such as an association between the spatial arrangement of tumor-infiltrating lymphocytes and immune checkpoint inhibitor response.14
In this study, we investigated whether an AI-driven approach to quantitative morphologic feature extraction with modern deep learning toolkits could identify a histologic signature associated with outcomes following administration of a particular adjuvant treatment (gemcitabine) in resected PDAC. We also explored the degree to which an AI-derived histologic signature was associated with adjuvant gemcitabine treatment outcomes and compared its performance with existing transcriptomic sub-types. We then examined the performance of the AI-derived histologic signature in an external cohort of patients who underwent resection of PDAC followed by adjuvant gemcitabine to determine whether our results could be generalized. Finally, we evaluated the signature in another cohort where patients received no adjuvant treatment to ensure that the association with disease-related outcomes was predictive (specific to treatment) and not prognostic (related to the underlying disease process).
Results
Development of a histologic signature known as the visual pancreatic gemcitabine (VPG) signature from TCGA training set
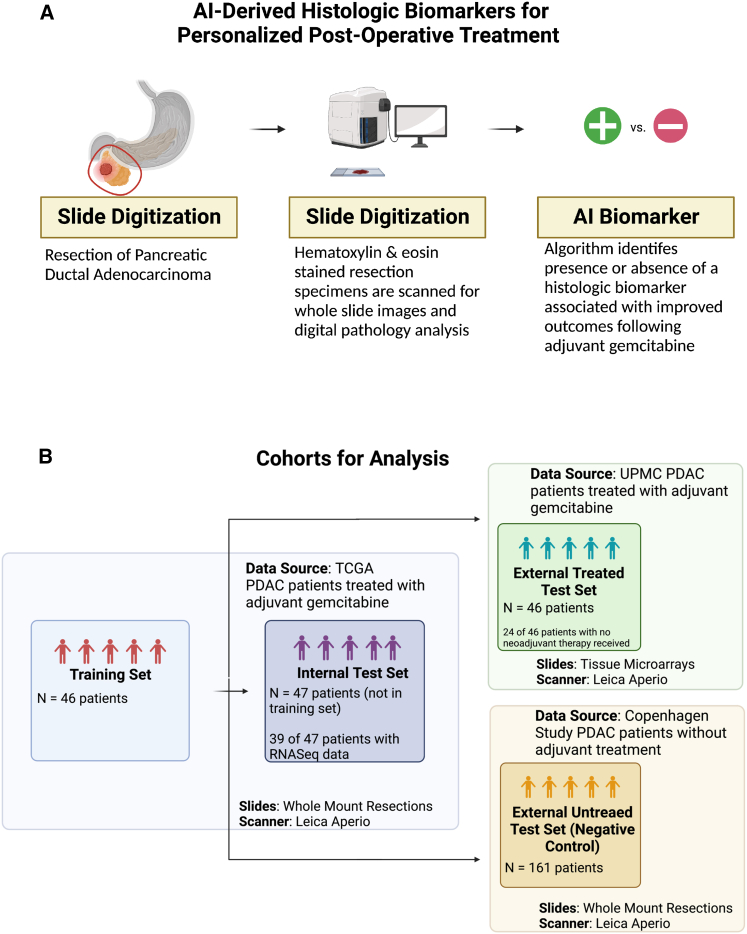
To construct a histologic signature associated with disease-specific survival (DSS) after adjuvant gemcitabine, we analyzed a dataset of scanned whole-slide images and the associated clinical data from a cohort of 93 patients with PDAC treated with adjuvant gemcitabine in The Cancer Genome Atlas (TCGA) (Figure 1A).15 Subsequently, the performance of the signature in stratifying patients was assessed in two external validation cohorts (Figure 1B). One external cohort included 46 patients who underwent PDAC resection followed by gemcitabine treatment at the University of Pittsburgh Medical Center (UPMC) for whom digitally scanned tissue microarrays of tumor specimens were available. The second external cohort included 161 patients from Copenhagen, whose tumors were resected between 1978 and 2008, when adjuvant treatment was not administered as part of the standard of care.16,17,18
Figure 1.
Constructing an experimental approach to identify an AI-derived histologic biomarker associated with outcomes following adjuvant gemcitabine
(A) AI-derived biomarkers could be identified from digitized slides of pancreatic tumor resections, which might guide adjuvant treatment selection.
(B) Data from three patient cohorts were used for this study: (1) TCGA (n = 93 patients), which served as the source for a training set (n = 46) to develop a histologic signature and for a test set (n = 47) to evaluate the performance of the histologic signature; (2) a retrospective cohort from UPMC, which served as a test set external to the data source used for training; and (3) a cohort from a study in Copenhagen, which included patients who received no adjuvant treatment, serving as a negative control.
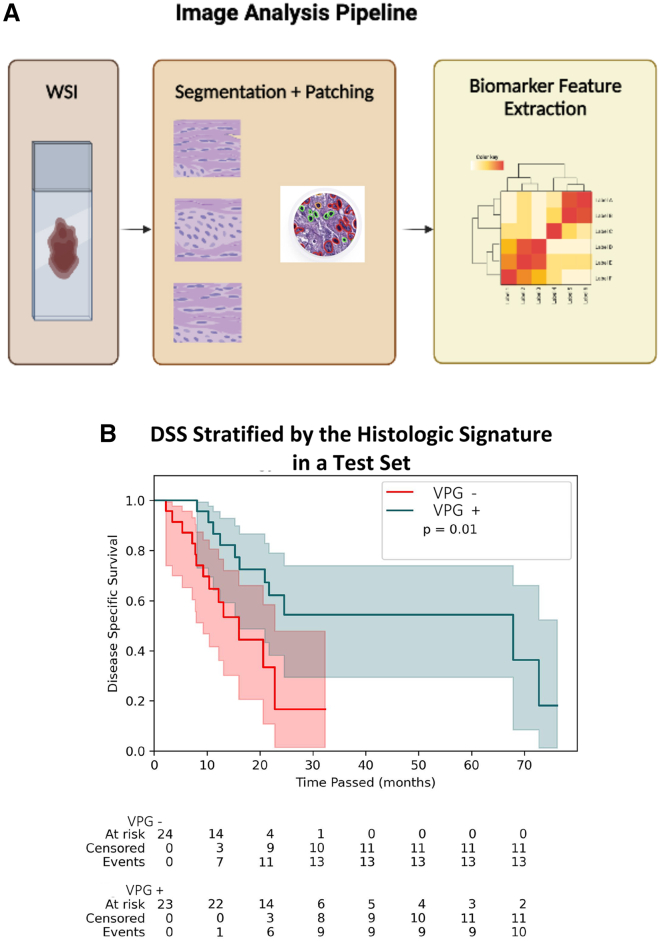
We developed a histologic signature capable of stratifying patients by disease-related outcomes following adjuvant gemcitabine through an image analysis pipeline implemented within TCGA cohort (Figure 2A). Within this dataset, the image analysis pipeline involved nuclei segmentation, extraction of 816 features describing nuclear morphology, feature selection using least absolute shrinkage and selection operator (LASSO) regression, training a cox proportional hazards model incorporating selected features in a training set of 46 patients, and testing the performance of the signature in the test set of the remaining 47 patients. Segmentation of nuclei was performed using a previously published model (HoVer-Net),13,19 and representative images of segmentation in TCGA and all other cohorts are reported in Figures S1–S3. Patients were randomly assigned to the training or test sets, and characteristics were similar between the two groups (Table S1). Through this pipeline, we identified a histologic signature, which we refer to as the VPG signature; the signature incorporates a single feature that describes the variance in nuclear morphology among a tumor’s neoplastic cells. We defined VPG positivity using a threshold determined by the median patient in the training set (positive patients defined by feature quantification greater than the median patient’s feature value of 0.053 units, negative patients defined by feature quantification lower than the median patient). Image examples of VPG+ and VPG− samples are included in Figure S4.
Figure 2.
An image analysis pipeline yields a histologic signature that stratifies disease-specific survival (DSS) following adjuvant gemcitabine
(A) In the image analysis pipeline for this study, whole-slide images (WSIs) from tumor resections were converted to smaller patches before cell-level segmentation and geometric feature extraction describing cellular morphology at the patient level. 816 features are extracted for each individual patient from the available digitized slide. Downstream statistical analysis of these features enables identification of a histologic signature.
(B) Kaplan-Meier curves for the test set from TCGA cohort (n = 47) stratified by the presence or absence of the AI-derived histologic signature. The p value (p = 0.01) corresponds to the log rank test. The median DSS for signature+ patients was 67.9 months (95% CI: [16.2, not reached]), and the median DSS for signature− patients was 16 months (95% CI: [9.3, 22.8]).
The VPG signature stratifies DSS outcomes following adjuvant gemcitabine treatment
We proceeded to assess the performance of this histologic signature in a test set of 47 patients from TCGA cohort. In this test set, VPG+ patients (n = 23) did not differ from VPG− patients (n = 23) in age, gender, or grade of tumor and duration of adjuvant gemcitabine therapy (Table S2). We found that the VPG signature was strongly associated with DSS in the internal validation cohort (log rank p ≤ 0.001) (Figure 2B). The hazard ratio for death for VPG− patients was 2.94 (95% confidence interval [CI]: [1.21, 7.14]). VPG+ patients had a median DSS of 67.9 months (95% CI: [16.2, not reached]), while VPG− patients had a median DSS of 16 months (95% CI: [9.3, 22.8]).
This VPG signature stratifies outcomes following adjuvant gemcitabine where known RNA sub-types cannot
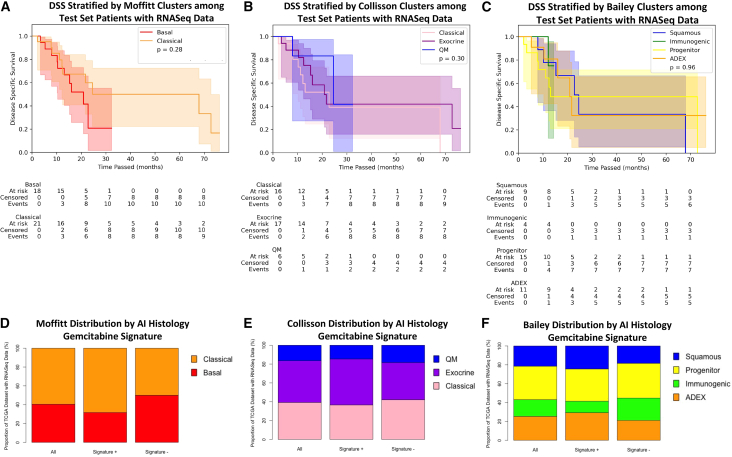
We then evaluated the performance of this signature in the test set relative to three known RNA sequencing (RNA-seq) classification systems described by Moffitt et al.,7 Collisson et al.,6 and Bailey et al.8 To make this comparison, we focused on a sub-group of 39 patients within the test set who had RNA-seq data and classification (performed by TCGA group) available.15 We calculated Kaplan-Meier estimators among VPG+ and VPG− patients, again finding that DSS differed between the two groups (log rank test p value = 0.02, signature + median DSS = 67.9 months, 95% CI [15.3, not reached] vs. signature − median DSS = 16 months [8.0, 22.8]) (Figure S5). There was no difference in DSS between stratifications using any of the three traditional RNA-seq classification approaches (log rank test p values: Moffitt p = 0.28, Collisson: p = 0.30, Bailey: p = 0.96) (Figures 3A–3C). To confirm that the stratification of the RNA-seq clusters was not unique to the patients in the test set, we stratified by the same clusters across the entire gemcitabine-treated TCGA dataset of patients with RNA-seq data available (n = 79) and again failed to observe a difference in survival outcomes across clusters (Figure S6). Of note, the RNA-seq cohorts also did not correlate with DSS among all TCGA patients with RNA-seq data, including those who did not receive adjuvant treatment or who received an adjuvant therapy other than gemcitabine, though there was a trend toward significance among Moffitt sub-sets (Figure S7). We sought to assess whether there was an association between the signature and the RNA-seq clusters by examining the classifications of the signature and RNA-seq clusters among all patients in TCGA cohort with RNA-seq data (n = 79 patients). In this group, the chi-squared values describing the association between the presence of the signature and the Moffitt, Collisson, and Bailey RNA-seq clusters were 2.03 (p = 0.15), 0.71 (p = 0.70), and 2.36 (p = 0.50) respectively, suggesting that the signature is not associated with the RNA-seq clusters (Figures 3D–3F).
Figure 3.
Known RNA-seq sub-types do not stratify patients by outcomes following adjuvant gemcitabine and do not associate with the histologic signature
(A–C) Kaplan-Meier curves describing a sub-group of TCGA test set with RNA-seq data available (n = 39) stratified by (A) RNA-seq clusters previously described by Moffitt et al., (B) RNA-seq clusters previously described by Collisson et al., and (C) RNA-seq clusters previously described by Bailey et al.
(D–F) Among patients with RNA-seq data available across the entire TCGA cohort (n = 79), the proportion of patients falling into the (D) Moffitt clusters, (E) Collisson clusters, and (F) Bailey clusters is graphed among all patients, those who are signature+, and those who are signature−.
The VPG signature validates in an external cohort of gemcitabine-treated patients but not untreated patients
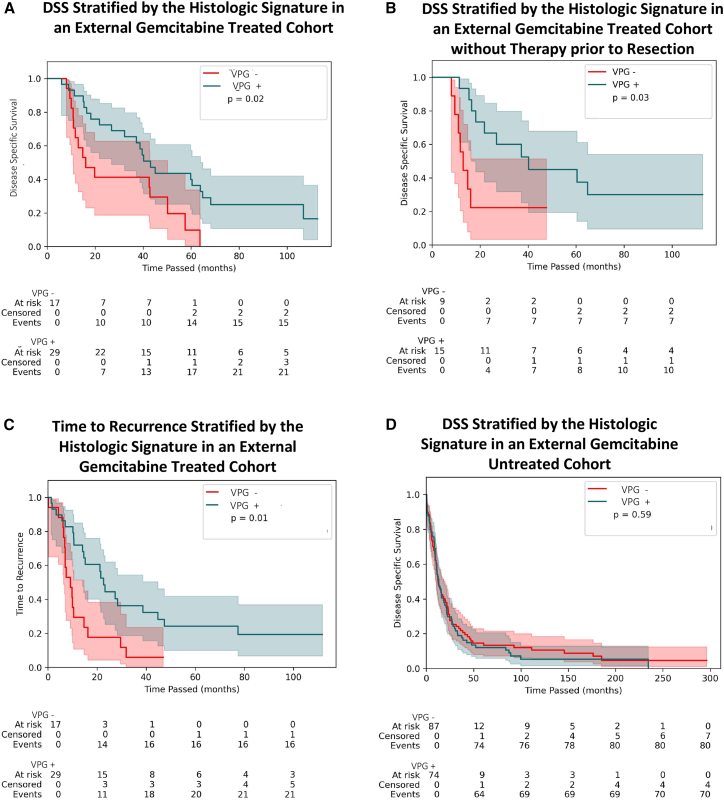
To investigate performance beyond TCGA dataset, we tested our model in a retrospective cohort of adjuvant-gemcitabine-treated patients at UPMC and a retrospective cohort of untreated patients from Copenhagen. The DSS in patients identified as VPG+ was superior to the DSS of those who were VPG− in the UPMC cohort (log rank test p value = 0.02; Figure 4A). The median DSS of VPG+ patients was 43.1 months (95% CI: [26.8, 63.9]), while the median DSS of VPG− patients was 16 months (95% CI: [10.8, 50.1]). Clinical characteristics were similar among patients with and without the signature (see Table S3). 22 of 46 patients in the UPMC cohort had received neoadjuvant chemotherapy prior to resection; in a sub-group analysis of the 24 patients without neoadjuvant chemotherapy, the DSS remained significantly different between VPG+ and VPG− patients (log rank test p = 0.03) (Figure 4B). When performing a similar analysis using the clinically meaningful alternate endpoint of time to recurrence, there was still a difference in the outcomes between VPG+ and VPG− patients (log rank test p = 0.01) (Figure 4C). In order to assess the independent effect of the VPG signature in the UPMC cohort, we constructed a multivariate Cox proportional hazards model of DSS comprising the VPG signature and the covariates of age, performance status as defined by ECOG score, and CA19-9 level before treatment. In the model, the VPG signature was statistically significantly associated with improved DSS (hazard ratio [HR] = 0.41 [0.19, 0.88], p = 0.02), along with the clinical covariate of age. In contrast, in the Copenhagen cohort of untreated patients, the log rank test comparing DSS between VPG+ and VPG− patients showed no association (log rank test p = 0.59), suggesting that the VPG signature is not a prognostic factor for untreated tumors (Figure 4D). The median DSS of VPG+ patients in this cohort was 13.2 months (95% CI: [10.4, 18.6]), and the median DSS of VPG− patients was 12.3 months (95% CI: [10.4, 19.8]). VPG+ (n = 74) and VPG− (n = 87) patients in this cohort were similar in age, gender, and tumor grade (Table S4). It should be noted that the DSS in the untreated Copenhagen cohort did differ from the adjuvant gemcitabine-treated UPMC and TCGA cohorts, as would be expected (Figure S8).
Figure 4.
The signature generalizes to external cohorts of gemcitabine-treated patients but not untreated patients
(A) Kaplan-Meier curves describing DSS among patients receiving adjuvant gemcitabine-based therapy in the UPMC cohort (n = 46) when stratified by the histologic signature. The p value (p = 0.02) corresponds to the log rank test. Median DSS of signature+ patients was 43.1 months (95% CI: [26.8, 63.9]), and the median DSS of signature− patients was 16 months (95% CI: [10.8, 50.1]).
(B) Kaplan-Meier curves describing DSS among patients in the UPMC cohort who had received no therapy prior to surgery (n = 24) (log rank test p = 0.03). Median DSS of signature+ patients was 40.2 months (95% CI: [16.4, not reached]), and the median DSS of signature− patients was 12.9 months (95% CI: [8.1, not reached]).
(C) Kaplan-Meier curves describing time to recurrence among patients who received adjuvant gemcitabine-based therapy (n = 46) (log rank test p = 0.01). Median time to recurrence of signature+ patients was 22.6 months (95% CI: [14.1, 44.8]), and the median time to recurrence among signature− patients of 9.1 months (95% CI: [6.4, 14.7]).
(D) Kaplan-Meier curves describing DSS among patients in the Copenhagen cohort who were untreated (log rank test p = 0.59; signature+ median DSS: 13.2 months [10.4, 19.8], signature− median DSS: 12.3 [10.4, 19.8]).
Discussion
In this study, we applied a modern AI digital pathology approach to identify a histology-based morphological signature associated with treatment outcomes following post-operative treatment with gemcitabine in patients with resected PDAC. The VPG signature developed in this study successfully stratified patients’ DSS following adjuvant gemcitabine and was validated in an external cohort of patients with the additional endpoint of time to recurrence.
The isolation of the VPG signature has several potential clinical implications should the signature be validated in prospective studies. First, given the significant difference in disease-related outcomes between VPG+ and VPG− patients across multiple cohorts tested in this study, this signature may help clinicians identify which patients will benefit from gemcitabine-based therapy after resection. The differences in outcomes between VPG+ and VPG− patients across multiple cohorts tested in this study point toward it being a predictive biomarker in a population where currently none exists. Second, the VPG signature may, on a larger scale, improve the process of designing clinical trials for resected PDAC. Randomizing a large cohort of patients with molecularly heterogeneous tumors to treatment arms without predefined biomarkers compromises results and leads to inefficiency as well as being a waste of precious resources. Third, the VPG signature can be tested and refined for application in other clinical settings, for example in metastatic or borderline resectable PDAC, when FOLFIRINOX and gemcitabine/nab-paclitaxel are both acceptable frontline regimens without a reliable predictive biomarker to help clinicians to recommend one over the other.
Importantly, the VPG signature is derived from images of H&E slides, which are routinely generated for all patients with PDAC. Since no additional tissue or complex molecular testing is required to apply this AI algorithm, both turnaround time and cost are much lower than one would expect for predictive biomarker testing. Validation in the UPMC cohort, in which tissue microarray specimens were used to generate images, shows that feature extraction is feasible across different techniques of tissue preparation.
The favorable performance of this signature, when compared with existing RNA-seq-based clusters, in stratifying disease-related outcomes in patients treated with gemcitabine validates pursuing modalities other than genomics and transcriptomics as potential predictive biomarkers. PDAC RNA-seq sub-types were developed to provide an improved molecular taxonomy of PDAC and, in turn, inform therapeutic development. These sub-types have been correlated with prognosis, including a recent study of the basal and classical sub-types in a multicenter trial,20 but, to our knowledge, their association with prognosis was never previously assessed in TCGA. The prospective COMPASS trial demonstrated that the Moffitt basal and classical sub-types have different outcomes following first-line chemotherapy.21 Subsequent analysis from the same trial revealed that the basal and classical sub-types stratify patients who received modified FOLFIRINOX but not those who received gemcitabine plus nab-paclitaxel.22 While the COMPASS trial featured patients with metastatic disease, our study showed similar results for gemcitabine-treated patients after surgical resection and confirmed that the Bailey and Collisson systems fail to stratify outcomes among gemcitabine-treated patients. Our results paired with those from COMPASS suggest that prevailing molecular taxonomies do not provide adequate predictive stratification for patients treated with gemcitabine-based regimens. Interestingly, our analysis showed that previously designated sub-types did not appear to be prognostic, even when analyzing other treatments. Possible explanations include a smaller proportion of patients in TCGA cohort who received fluoropyrimidine-based therapy or differences between treatment effect in the adjuvant and metastatic settings. Regardless, the performance of the VPG signature in this study validates the capacity for digital pathology approaches to identify biomarkers predictive of treatment response when existing molecular approaches have not been proven to do so. Indeed, published data in other tumor types have shown that digital pathology can predict responses to chemotherapy23,24 and immunotherapy.14 Moreover, the ability to construct such a signature in a training set of fewer than 50 patients illustrates that clinically meaningful tools can be generated from relatively small cohorts of patients and that the same technology can be applied to other clinical contexts.
Results in the untreated Copenhagen cohort suggest that the VPG signature is not a prognostic marker of a tumor’s underlying biology. Instead, the validation of the VPG signature in an external cohort of gemcitabine-treated patients in combination with the data from the Copenhagen cohort suggest that the VPG signature is likely specific to chemotherapy treatment.
Limitations of the study
One limitation of our study is the absence of patients who received adjuvant FOLFIRINOX or other non-gemcitabine regimens. Future evaluation of the VPG signature in such patients is planned and would assess the degree to which the VPG signature associates with outcomes following other chemotherapy regimens as well as the degree to which the signature is specific to gemcitabine alone. In addition, determining the disease-related outcomes of VPG+ patients treated with FOLFIRINOX may help clarify when one regimen (gemcitabine-based therapy or modified FOLFIRINOX) is preferred over the other. Including only patients with resected PDAC, who represent a minority of the PDAC population, is another limitation of our study. Ultimately, a tool addressing the question of whether a patient benefits from gemcitabine-based chemotherapy may have greatest relevance for the needle biopsy specimens used to diagnose more advanced or metastatic disease. Our study also did not account for potential differences in gemcitabine dosing, which may have identified a dose-response relationship between the VPG signature and gemcitabine therapy. The absence of data on dosing and the number of cycles received also limits our ability to draw conclusions about the degree of similarity in treatment received between the internal and external test sets. In addition, the lack of important clinical covariates in our dataset, such as performance status, limits the degree to which we can assess whether the VPG signature is, in fact, an independent predictor of outcomes. It should be noted that differences in the quality of surgical resection represent an important contributor to outcomes that can explain differences between sites but could not be captured with the data available for analysis in this study. Finally, our study does not delve into mechanisms linking the VPG signature and disease-related outcomes. Future investigation into the molecular underpinnings of our VPG signature may facilitate further refinement of the signature while also offering avenues to develop novel therapeutic approaches to improve upon or replace existing therapies.
In summary, this study identifies an AI-based histologic signature that stratifies disease-related outcomes among patients who have received adjuvant gemcitabine after resection of PDAC, where transcriptional profiling-based sub-typing fails to do so. This signature, if validated in prospective cohorts, has the potential to become one of the first clinically applicable predictive biomarkers in PDAC. Finally, if validated in PDAC, the imaging analysis platform underlying this signature may be generalized to other clinical settings, thereby facilitating the emergence of biomarkers to predict treatment response in diseases for which few actionable biomarkers currently exist.
STAR★Methods
Key resources table
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Viswesh Krishna (viswesh@valarlabs.com).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
Analysis of TCGA data for this paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table. Analysis of data reported in this paper from other cohorts will be shared by the lead contact upon request.
-
•
This paper does not report original code. It utilizes previously published models as described below in the model construction section of method details.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Datasets
Data from three cohorts of patients were utilized for this study: a cohort of 93 patients from the TCGA, a cohort of 46 patients from UPMC, and a cohort of 161 patients from Copenhagen. The TCGA cohort of 93 patients was identified by selecting the PDAC patients who had received adjuvant gemcitabine and no 5-fluorouracil. Available histopathologic images, data on disease specific survival, and demographic details were downloaded from the TCGA. The image identified as the diagnostic slide was used for image analysis. RNASeq classifications for the TCGA cohort were obtained from prior work with this cohort.18 The dataset was randomly split in half into a training and test set with no overlap between groups. To be able to make direct comparisons to existing RNA subtypes, 8 patients without RNASeq data were removed from the test set for a sub-group analysis illustrated in Figures 3A–3C and S5. Tests of associations between the histologic signature and RNASeq clusters were made using the 79 patients from the entire TCGA cohort who had RNASeq data available. The UPMC cohort represented a set of 46 patients, who had received gemcitabine after surgical resection; 24 of the 46 patients were treated with resection followed by adjuvant gemcitabine; 22 of the 46 patients were treated with additional neoadjuvant therapy prior to resection and gemcitabine therapy after resection. Clinical data was obtained via manual chart review of the electronic medical record. Digitally scanned tissue microarray specimens were used for image analysis. The cores were 1mm and were obtained from formalin-fixed, paraffin-embedded (FFPE) samples of extra portions of surgical resections. Whole tissue resection specimens were unfortunately not available for analysis for this study. Study of this cohort was approved by the University of Pittsburgh Institutional Review Board. The Copenhagen cohort included 161 patients who underwent pancreaticoduodenectomy between 1978 and 2008 as part of a previously described study.15,16,17 Whole tissue sections stained with hematoxylin and eosin from these patients were scanned using the ImageScope 12.2 (Leica Biosystems, Wetzlar, Germany). Study of this cohort of patients was approved by a regional ethics committee (reference no. KA-20060181).
Method details
Model construction
Scanned histologic images were analyzed through an imaging pipeline that included tissue segmentation, nuclei segmentation, and finally geometric feature extraction. Tissue was segmented via color-based thresholding to remove empty regions of the slide. Patches of size 2132x2132 pixels were extracted from tissue regions and a validated deep learning model developed by Graham et al. was used to segment and classify each nucleus automatically.13 Specifically, the HoVer-Net model extended and trained on the PanNuke Dataset was utilized for this study and classified nuclei into five classes (Neoplastic, Connective, Non-Neoplastic/Epithelial, Necrotic, Inflammatory).19 No modifications were made to the publicly available HoVer-Net segmentation model with reported multi-class Panoptic Quality (mPQ) of 0.4600 and binoptic-class Panoptic Quality (bPQ) of 0.6491 in pancreatic tissue. Descriptive morphometric features were then computed for each nucleus as described in Vrabac et al. (where the HoVer-Net model was also used for segmentation).11 Geometric features were then aggregated first at the patch, and subsequently at the patient level using summary statistics including the mean, standard deviation, skewness and kurtosis to produce the final feature vector for a patient. This feature vector was used as the input to a cox proportional hazards model that used the least absolute shrinkage and selection operator to identify the most correlated features with DSS along with their coefficients on the training set.
Quantification and statistical analysis
Statistical analysis
Feature vectors produced from the imaging analysis pipeline were used as inputs into a cox proportional hazards model that used the least absolute shrinkage and selection operator (with an L1 penalty) to identify features for inclusion in the histologic signature. The median value of the risk scores calculated from the cox model in the TCGA training set was identified and was used as a threshold to label signature positive and negative patients. Kaplan-meier estimators were produced for the different cohorts to compare DSS between signature positive and signature negative patients. The log rank test was used to test for statistically significant differences in DSS with a p value <0.05 deemed significant; all tests were two-sided. Demographic data for patients in each cohort were compiled and compared between signature positive and negative groups using the chi-squared test or Wilcoxon rank-sum test with a p value of <0.05 assessed as statistically significant. Associations between the signature and existing RNA subtypes were tested using the chi-quared test with a p value of <0.05 statistically significant. A multivariate cox proportional hazards model of DSS utilizing age, performance status as defined by ECOG score, and CA19-9 level before treatment in addition to the signature was performed in the UPMC cohort.
Software
Pytorch was utilized for deep learning model inference. Data processing and survival analysis were done with NumPy, pandas, and lifelines packages in Python. Statistical testing of demographic differences and associations with RNA subtypes was performed on R Statistical Software (v4.1.2; R Core Team 2021).
Acknowledgments
The authors thank Anirban Maitra and David Ting for their advice and guidance. Figures 1A, 1B, and 2A were created with BioRender.com. The study was funded by Valar Labs, Inc.
Author contributions
V.N., Viswesh Krishna, A.J., D.V., P.R., and E.A.C. were involved in developing the study, and Viswesh Krishna and P.R. were involved in supervising the study. Vrishab Krishna and E.T. processed the images. Viswesh Krishna and Vrishab Krishna developed the model and ran the statistical tests. Vrishab Krishna and V.N. produced the figures. V.N. and Viswesh Krishna drafted the manuscript. K.S. and A.S. were involved in acquisition of the UPMC samples, and J.S.J. was involved in the acquisition of the Copenhagen samples. All authors critically edited the manuscript.
Declaration of interests
Viswesh Krishna, A.J., D.V., and P.R. are founders of Valar Labs, Inc., and may own stocks. V.N., Vrishab Krishna, E.T., and H.B. are employees of Valar Labs, Inc. E.A.C., D.S., and A.H. are advisors to Valar Labs, Inc.
Published: April 11, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2023.101013.
Supplemental information
References
- 1.Strobel O., Neoptolemos J., Jäger D., Büchler M.W. Optimizing the outcomes of pancreatic cancer surgery. Nat. Rev. Clin. Oncol. 2019;16:11–26. doi: 10.1038/s41571-018-0112-1. [DOI] [PubMed] [Google Scholar]
- 2.Conroy T., Hammel P., Hebbar M., Ben Abdelghani M., Wei A.C., Raoul J.-L., Choné L., Francois E., Artru P., Biagi J.J., et al. FOLFIRINOX or gemcitabine as adjuvant therapy for pancreatic cancer. N. Engl. J. Med. 2018;379:2395–2406. doi: 10.1056/NEJMoa1809775. [DOI] [PubMed] [Google Scholar]
- 3.Oettle H., Neuhaus P., Hochhaus A., Hartmann J.T., Gellert K., Ridwelski K., Niedergethmann M., Zülke C., Fahlke J., Arning M.B., et al. Adjuvant chemotherapy with gemcitabine and long-term outcomes among patients with resected pancreatic cancer: the CONKO-001 randomized trial. JAMA. 2013;310:1473–1481. doi: 10.1001/jama.2013.279201. [DOI] [PubMed] [Google Scholar]
- 4.Oba A., Ho F., Bao Q.R., Al-Musawi M.H., Schulick R.D., Del Chiaro M. Neoadjuvant treatment in pancreatic cancer. Front. Oncol. 2020;10:245. doi: 10.3389/fonc.2020.00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collisson E.A., Bailey P., Chang D.K., Biankin A.V. Molecular subtypes of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2019;16:207–220. doi: 10.1038/s41575-019-0109-y. [DOI] [PubMed] [Google Scholar]
- 6.Collisson E.A., Sadanandam A., Olson P., Gibb W.J., Truitt M., Gu S., Cooc J., Weinkle J., Kim G.E., Jakkula L., et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 2011;17:500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moffitt R.A., Marayati R., Flate E.L., Volmar K.E., Loeza S.G.H., Hoadley K.A., Rashid N.U., Williams L.A., Eaton S.C., Chung A.H., et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 2015;47:1168–1178. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bailey P., Chang D.K., Nones K., Johns A.L., Patch A.-M., Gingras M.-C., Miller D.K., Christ A.N., Bruxner T.J.C., Quinn M.C., et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 9.Bera K., Schalper K.A., Rimm D.L., Velcheti V., Madabhushi A. Artificial intelligence in digital pathology - new tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 2019;16:703–715. doi: 10.1038/s41571-019-0252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beck A.H., Sangoi A.R., Leung S., Marinelli R.J., Nielsen T.O., van de Vijver M.J., West R.B., van de Rijn M., Koller D. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci. Transl. Med. 2011;3:108ra113. doi: 10.1126/scitranslmed.3002564. [DOI] [PubMed] [Google Scholar]
- 11.Vrabac D., Smit A., Rojansky R., Natkunam Y., Advani R.H., Ng A.Y., Fernandez-Pol S., Rajpurkar P. DLBCL-Morph: morphological features computed using deep learning for an annotated digital DLBCL image set. Sci. Data. 2021;8:135. doi: 10.1038/s41597-021-00915-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mi H., Bivalacqua T.J., Kates M., Seiler R., Black P.C., Popel A.S., Baras A.S. Predictive models of response to neoadjuvant chemotherapy in muscle-invasive bladder cancer using nuclear morphology and tissue architecture. Cell Rep. Med. 2021;2:100382. doi: 10.1016/j.xcrm.2021.100382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham S., Vu Q.D., Raza S.E.A., Azam A., Tsang Y.W., Kwak J.T., Rajpoot N. Hover-Net: simultaneous segmentation and classification of nuclei in multi-tissue histology images. Med. Image Anal. 2019;58:101563. doi: 10.1016/j.media.2019.101563. [DOI] [PubMed] [Google Scholar]
- 14.Wang X., Barrera C., Bera K., Viswanathan V.S., Azarianpour-Esfahani S., Koyuncu C., Velu P., Feldman M.D., Yang M., Fu P., et al. Spatial interplay patterns of cancer nuclei and tumor-infiltrating lymphocytes (TILs) predict clinical benefit for immune checkpoint inhibitors. Sci. Adv. 2022;8:eabn3966. doi: 10.1126/sciadv.abn3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas Research Network Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell. 2017;32:185–203.e13. doi: 10.1016/j.ccell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schultz N.A., Werner J., Willenbrock H., Roslind A., Giese N., Horn T., Wøjdemann M., Johansen J.S. MicroRNA expression profiles associated with pancreatic adenocarcinoma and ampullary adenocarcinoma. Mod. Pathol. 2012;25:1609–1622. doi: 10.1038/modpathol.2012.122. [DOI] [PubMed] [Google Scholar]
- 17.Laklai H., Miroshnikova Y.A., Pickup M.W., Collisson E.A., Kim G.E., Barrett A.S., Hill R.C., Lakins J.N., Schlaepfer D.D., Mouw J.K., et al. Genotype tunes pancreatic ductal adenocarcinoma tissue tension to induce matricellular fibrosis and tumor progression. Nat. Med. 2016;22:497–505. doi: 10.1038/nm.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torphy R.J., Wang Z., True-Yasaki A., Volmar K.E., Rashid N., Yeh B., Anderson J.M., Johansen J.S., Hollingsworth M.A., Yeh J.J., Collisson E.A. Stromal content is correlated with tissue site, contrast retention, and survival in pancreatic adenocarcinoma. JCO Precis. Oncol. 2018;2018 doi: 10.1200/PO.17.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gamper J., Koohbanani N.A., Benes K., Graham S., Jahanifar M., Khurram S.A., Azam A., Hewitt K., Rajpoot N. PanNuke dataset extension, insights and baselines. arXiv. 2020 doi: 10.48550/ARXIV.2003.10778. Preprint at. [DOI] [Google Scholar]
- 20.Suurmeijer J.A., Soer E.C., Dings M.P.G., Kim Y., Strijker M., Bonsing B.A., Brosens L.A.A., Busch O.R., Groen J.V., Halfwerk J.B.G., et al. Impact of classical and basal-like molecular subtypes on overall survival in resected pancreatic cancer in the SPACIOUS-2 multicentre study. Br. J. Surg. 2022;109:1150–1155. doi: 10.1093/bjs/znac272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aung K.L., Fischer S.E., Denroche R.E., Jang G.-H., Dodd A., Creighton S., Southwood B., Liang S.-B., Chadwick D., Zhang A., et al. Genomics-driven precision medicine for advanced pancreatic cancer: early results from the COMPASS trial. Clin. Cancer Res. 2018;24:1344–1354. doi: 10.1158/1078-0432.CCR-17-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Kane G.M., Grünwald B.T., Jang G.-H., Masoomian M., Picardo S., Grant R.C., Denroche R.E., Zhang A., Wang Y., Lam B., et al. GATA6 expression distinguishes classical and basal-like subtypes in advanced pancreatic cancer. Clin. Cancer Res. 2020;26:4901–4910. doi: 10.1158/1078-0432.CCR-19-3724. [DOI] [PubMed] [Google Scholar]
- 23.Wang X., Bera K., Barrera C., Zhou Y., Lu C., Vaidya P., Fu P., Yang M., Schmid R.A., Berezowska S., et al. A prognostic and predictive computational pathology image signature for added benefit of adjuvant chemotherapy in early stage non-small-cell lung cancer. EBioMedicine. 2021;69:103481. doi: 10.1016/j.ebiom.2021.103481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saednia K., Lagree A., Alera M.A., Fleshner L., Shiner A., Law E., Law B., Dodington D.W., Lu F.-I., Tran W.T., Sadeghi-Naini A. Quantitative digital histopathology and machine learning to predict pathological complete response to chemotherapy in breast cancer patients using pre-treatment tumor biopsies. Sci. Rep. 2022;12:9690. doi: 10.1038/s41598-022-13917-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Analysis of TCGA data for this paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table. Analysis of data reported in this paper from other cohorts will be shared by the lead contact upon request.
-
•
This paper does not report original code. It utilizes previously published models as described below in the model construction section of method details.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.