Abstract
Background & Aims
Metabolic reprogramming is recognized as a cancer hallmark intimately linked to tumor hypoxia, which supports rapid tumor growth and mitigates the consequential oxidative stress. Phosphofructokinase-fructose bisphosphatase (PFKFB) is a family of bidirectional glycolytic enzymes possessing both kinase and phosphatase functions and has emerged as important oncogene in multiple types of cancer. However, its clinical relevance, functional significance, and underlying mechanistic insights in hepatocellular carcinoma (HCC), the primary malignancy that develops in the most important metabolic organ, has never been addressed.
Methods
PFKFB4 expression was examined by RNA sequencing in The Cancer Genome Atlas and our in-house HCC cohort. The up-regulation of PFKFB4 expression was confirmed further by quantitative polymerase chain reaction in an expanded hepatitis B virus–associated HCC cohort followed by clinicopathologic correlation analysis. Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated protein 9 (Cas9)-mediated PFKFB4 knockout cells were generated for functional characterization in vivo, targeted metabolomic profiling, as well as RNA sequencing analysis to comprehensively examine the impact of PFKFB4 loss in HCC.
Results
PFKFB4 expression was up-regulated significantly in HCC and correlated positively with TP53 and TSC2 loss-of-function mutations. In silico transcriptome-based analysis further revealed PFKFB4 functions as a critical hypoxia-inducible gene. Clinically, PFKFB4 up-regulation was associated with more aggressive tumor behavior. Functionally, CRISPR/Cas9-mediated PFKFB4 knockout significantly impaired in vivo HCC development. Targeted metabolomic profiling revealed that PFKFB4 functions as a phosphatase in HCC and its ablation caused an accumulation of metabolites in downstream glycolysis and the pentose phosphate pathway. In addition, PFKFB4 loss induced hypoxia-responsive genes in glycolysis and reactive oxygen species detoxification. Conversely, ectopic PFKFB4 expression conferred sorafenib resistance.
Conclusions
PFKFB4 up-regulation supports HCC development and shows therapeutic implications.
Keywords: Next-Generation Sequencing, Hypoxia, PFKFB, Glycolysis
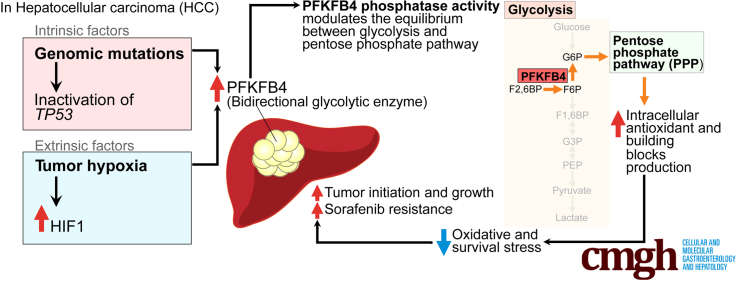
Graphical abstract
Summary.
Intrinsic driver mutations and extrinsic factors could up-regulate phosphofructokinase-fructose bisphosphatase 4 in hepatocellular carcinoma, which leads to aggressive tumor behavior and poor prognosis. Mechanistically, phosphofructokinase-fructose bisphosphatase 4 phosphatase shifted the equilibrium from glycolysis to the pentose phosphate pathway and alleviated cellular stress response in supporting hepatocellular carcinoma progression.
Hepatocellular carcinoma (HCC) is one of the most common yet highly lethal cancers worldwide. Chronic hepatitis B infection, being endemic in East Asia, South East Asia, and sub-Saharan Africa, predisposes to the development of HCC.1 Despite the implementation of surveillance for high-risk patients, HCC still often is diagnosed at advanced stages, resulting in poor prognosis.2 The therapeutic options for advanced HCC are limited as a result of the resilient and aggressive nature of the disease. In particular, extensive interpatient and intratumoral heterogeneities, as well as tumor hypoxia in HCC, together pose significant challenges to the development of effective therapies,3 and this is reflected by the modest survival benefits provided by sorafenib and lenvatinib, the first-line, Food and Drug Administration–approved, molecularly targeted drugs for patients with advanced HCC.4,5 Because the intrinsic and extrinsic factors in HCC often lead to cellular metabolic reprogramming to support the rapid proliferation of cancer cells and to offset the consequentially generated reactive oxygen species (ROS), they might be exploited as cancer’s Achilles’ heel for next-generation cancer therapy development.
Metabolic adaptation is a hallmark of cancer.6 Also known as the Warburg effect, cancer cells switch from oxidative phosphorylation to aerobic glycolysis to produce lactate even under oxygen-sufficient conditions.7 In addition to fulfilling the basal energy requirements, this metabolic switching ensures the generation of sufficient metabolic intermediates, which are served as molecular building blocks to support the high proliferative rate of cancer cells. Conversion of fructose-6-phosphate (F6P) into fructose-1,6-bisphosphate (F1,6BP) is the very first committed step of glycolysis. This critical step is catalyzed by phosphofructokinase 1 (PFK1), an enzyme encoded by Phosphofructokinase, Muscle gene (PFKM) and regulated by four members of phosphofructokinase 2 (PFK2 isoforms 1–4), which also are known as PFK2/fructose-2,6-bisphosphatases (PFKFBs) (Figure 1A).8 Under energy-deprived conditions, PFKFB phosphorylates F6P into fructose-2,6-bisphosphate (F2,6BP) for allosteric activation of PFK1 to drive glycolysis. Conversely, functioning as a bidirectional enzyme with a C-terminal phosphatase domain, PFKFBs also could hydrolyze F2,6BP to inactivate PFK1. The equilibrium between the kinase and phosphatase activities of PFKFBs in positively or negatively modulating the PFK1 activity thus serves as a molecular switch in controlling the directions of the metabolic flux in the glucose metabolism.
Figure 1.
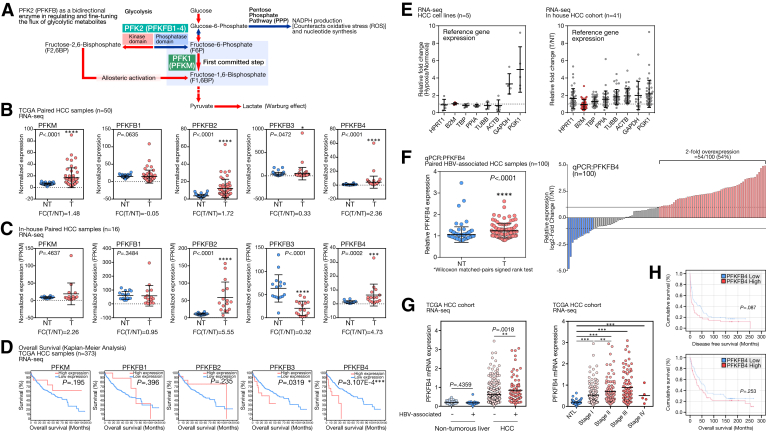
Up-regulation of bidirectional glycolytic enzyme PFKFB4 and its prognostic significance in human HCC. (A) PFKFB4 is one of the PFKFB isoenzymes to allosterically regulate the first committed step of glycolysis by controlling the intracellular ratio of F6P and F2,6BP. Equilibrium of the PFKFB4 kinase and phosphatase activity serves as a molecular switch to fine-tune the flux of metabolites into glycolysis or PPP. (B) Expression of PFK1 (PFKM) and different members of PFKFB family enzymes in the HCC tumors (T) and the nontumorous liver tissues (NT) in the TCGA HCC RNA-seq cohort. (C) Expression of PFK1 (PFKM) and different members of PFKFB family enzymes in the HCC T and NT in our in-house, HBV-associated HCC RNA-seq cohort. (D) The evaluation of the prognostic significance of the expression of PFK1 (PFKM) and different members of PFKFB family enzymes in the TCGA HCC cohort. (E) Change in the expression level of 8 different common reference gene underhypoxia compared with the normoxic condition in a panel of HCC cell lines including Huh7, PLC/PRF5, MHCC97L, Hep3B, and HepG2, and in HCC patient samples compared with the nontumorous liver tissues. (F) Independent qPCR analysis showed a significant up-regulation of PFKFB4 expression in a cohort of 100 paired HBV-associated HCC samples and 54% of them showed 2-fold PFKFB4 overexpression. (G) Expression of PFKFB4 transcript in nontumorous livers and liver tumor samples with or without HBV association, and across different tumor stages of HCC development in TCGA HCC RNA-seq cohort. (H) The differences in overall and disease-free survival rates between the PFKFB4 expression-high and expression-low patients in our in-house HCC cohort. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001. ACTB, Actin beta; B2M, β2 microglobulin; FC(T/NT), relative fold change in the tumor when compared with nontumor; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; FPKM, Fragments Per Kilobase per Million; HPRT1, Hypoxanthine phosphoribosyltransferase 1; NTL, Non-tumorous liver; PGK1, Phosphoglycerate kinase 1; PPIA, Peptidylprolyl isomerase A; TBP, TATA-box binding protein; TUBB, Tubulin beta class I.
The PFKFB proteins have been suggested to play important oncometabolic functions in multiple primary cancers.8 Specifically, several functional genetic screenings have shown that PFKFB isoform 4 (PFKFB4) is functionally unique from other members in supporting cancer cell survival and metastasis. For instance, PFKFB4 was found to be critical in supporting the survival of prostate cancer cells in an unbiased small interfering RNA screening against a panel of metabolic genes.9 Similarly, PFKFB4 also was identified as a key prosurvival gene in glioma cells in another short hairpin RNA kinome and phosphatome library screening.10 Despite both studies identifying PFKFB4 as a critical gene in supporting pro-oncogenic functions, however, the described underlying mechanisms are likely to be context-dependent. In prostate cancer cells, PFKFB4 preferentially diverts the glycolytic flux into the pentose phosphate pathway (PPP) by its phosphatase activity to enhance reduced nicotinamide adenine dinucleotide phosphate (NADPH) production to counteract oxidative stress9; whereas in glioma cells, PFKFB4 mainly directs the glycolytic flux for adenosine triphosphate generation and lactate production in a kinase-dependent manner.10 More recently, PFKFB4 also was shown to drive breast cancer metastasis through the direct phosphorylation and activation of a nonglycolytic substrate, steroid receptor coactivator-3, followed by steroid receptor coactivator-3–induced transcriptional activation of relevant gene expression responsible for supporting the PPP pathway and purine synthesis.11 Although PFKFB4 has been functionally characterized in the aforementioned cancer models, the role of PFKFB4 in liver cancer, a type of cancer that develops in the most important metabolic organ, is still remained to be addressed. In addition, the clinical relevance of PFKFB4 in human tumor tissue samples and its relationship with the background genetic mutations of the tumors has yet to be shown. In this study, we performed an integrative analysis to dissect the undocumented role of PFKFB4 in HCC, covering its clinical relevance in patient samples, functional characterization in vivo, and mechanistic studies at metabolome and transcriptome levels. Our study highlighted the potential association of PFKFB4 with the underlying TP53 mutations in HCC patients and the translation implication of PFKFB4 expression and sorafenib treatment.
Results
PFKFB4 Is the Unique PFKFB Isoform Up-Regulated in HCC and Prognosticates Poor Survival Outcomes
To understand the expression of PFKFB family genes in HCC, the corresponding transcriptome sequencing (RNA sequencing [RNA-seq]) data from the paired HCC samples of The Cancer Genome Atlas (TCGA) database was extracted and examined. Among the PFKFB members, the expression of PFKFB2 (P < .0001), PFKFB3 (P < .0472), and PFKFB4 (P < .0001) was up-regulated significantly in the tumor tissue compared with the nontumorous liver counterparts (Figure 1B). In addition, examination of our in-house RNA-seq of hepatitis B virus (HBV)-associated HCC samples consistently showed that PFKFB2 (P < .0001) and PFKFB4 (P = .0002) were up-regulated significantly in tumor tissue, while PFKFB3 was down-regulated (P < .0001) (Figure 1C). In addition, PFKM, which encodes PFK1, a glycolytic enzyme regulated allosterically by F-2,6-BP, generated by PFKFBs, also only was up-regulated significantly in the TCGA cohort (Figure 1B). Among these differentially expressed PFKFB isoforms, only PFKFB3 and PFKFB4 were associated with significant changes in overall survival in the TCGA cohort (P = .0319 and P = 3.107E-4, respectively) (Figure 1D). Specifically, PFKFB4 was the only PFKFB isozyme that was overexpressed in HCC and correlated with poor prognostic outcomes. Thus, we focused on PFKFB4 for further characterization.
To establish the clinical relevance of PFKFB4 in HCC, real-time quantitative polymerase chain reaction (qPCR) was performed to examine the PFKFB4 expression in an independent expanded patient cohort with 100 pairs of HBV-associated HCCs. Because hypoxia in the tumor microenvironment could profoundly affect gene expression, the expression patterns of 8 commonly used reference genes were first carefully examined in RNA-seq data generated from a panel of hypoxic-treated HCC cell lines and a cohort of paired HCC patient samples (Figure 1E). Among these markers, the β2 microglobulin (B2M) gene was selected as the internal reference gene for subsequent qPCR analysis, considering its relatively stable expression under hypoxia and between the HCC and nontumorous livers. In agreement with RNA-seq, PFKFB4 expression was significantly higher in the tumors (P < .0001) and 54% of these HBV-associated HCC cases showed an increase of at least 2-fold compared with the corresponding nontumorous control (Figure 1F). Similarly, expression of PFKFB4 also was found to be increased significantly in HBV-associated HCC in TCGA data (Figure 1G, left). More importantly, up-regulation of PFKFB4 at a 3-fold cut-off value was associated significantly with more aggressive tumor behavior, including larger tumor size (P = .0288), presence of venous invasion (P = .0025), advanced TNM stage (P = .0355), and younger patient age (P = .0047) (Table 1). Although it did not reach statistical significance, PFKFB4 overexpression showed a trend of association with shorter disease-free survival (P = .087) (Figure 1H). Of note, in the TCGA HCC cohort, PFKFB4 expression displayed a stepwise increase from the early to the advanced stage of HCC, further suggesting its potential association with tumor aggressiveness (Figure 1G, right).
Table 1.
Clinicopathologic Correlation of PFKFB4 in HBV-Associated HCC Patients
| Clinicopathologic parameters | HCC cases |
||
|---|---|---|---|
| PFKFB4 overexpression (at 3-fold cut-off) (n = 46) |
PFKFB4 normal or underexpression (n = 54) | P valuea | |
| Gender | .2584 | ||
| Male | 32 (69.6%) | 43 (79.6%) | |
| Female | 14 (30.4%) | 11 (20.4%) | |
| Average patient age, y (range) | 49.07 (24–71) | 55.69 (29–74) | .0047b |
| Background liver disease | .2146 | ||
| Normal | 3 (6.8%) | 2 (4.0%) | |
| Chronic hepatitis | 22 (50.0%) | 19 (38.0%) | |
| Cirrhosis | 19 (43.2%) | 29 (58.0%) | |
| Tumor size | .0288c | ||
| ≤5 cm | 9 (20.5%) | 21 (42.0%) | |
| >5 cm | 35 (79.5%) | 29 (58.0%) | |
| Tumor encapsulation | .2891 | ||
| Present | 31 (70.5%) | 31 (58.5%) | |
| Absent | 13 (29.5%) | 22 (41.5%) | |
| Venous invasion | .0025b | ||
| Absent | 13 (28.3%) | 32 (59.3%) | |
| Present | 33 (71.7%) | 22 (40.7%) | |
| Tumor microsatellite formation | .3120 | ||
| Absent | 18 (39.1%) | 27 (50.9%) | |
| Present | 28 (60.9%) | 26 (49.1%) | |
| Liver invasion | .0669 | ||
| Absent | 23 (50.0%) | 35 (68.6%) | |
| Present | 23 (50.0%) | 16 (31.4%) | |
| Cellular differentiation | .0945 | ||
| Edmondson grades I–II | 14 (31.8%) | 25 (50.0%) | |
| Edmondson grades III–IV | 30 (68.2%) | 25 (50.0%) | |
| TNM tumor staging | .0355c | ||
| I–II | 11 (20.4%) | 25 (46.3%) | |
| III–IV | 34 (63.0%) | 29 (53.7%) | |
Fisher exact test
P < .05.
P < .01.
Hypoxia Is an Extrinsic Factor Inducing PFKFB4 Expression in HCC in a Hypoxia-Inducible Factor-1–Dependent Manner
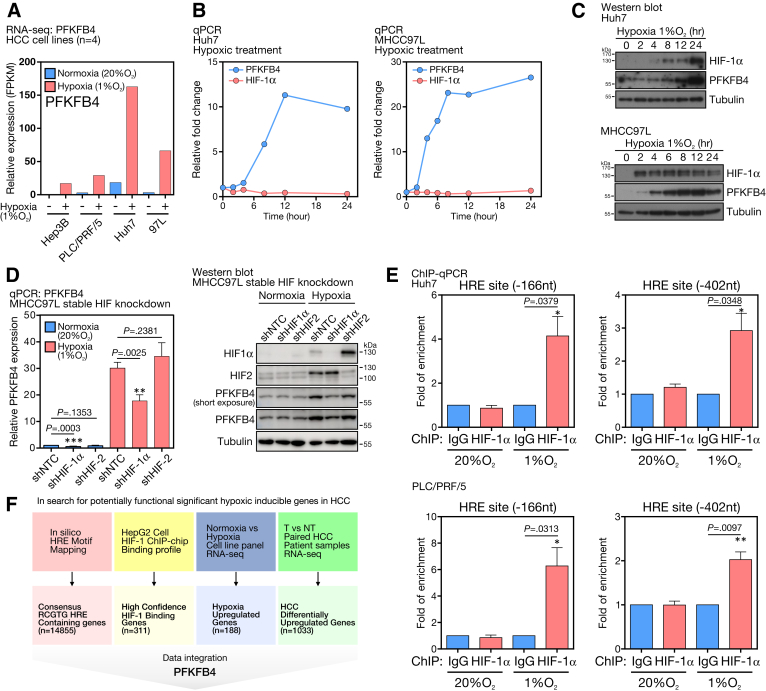
To show that PFKFB4 is hypoxic-inducible in HCC, a panel of HCC cell lines was exposed to hypoxia with 1% oxygen for 24 hours. RNA-seq analysis showed that the expression of PFKFB4 was increased drastically under hypoxia (Figure 2A). To assess the temporal induction pattern of PFKFB4, Human hepatoma 7 (Huh7) and MHCC97L cells were exposed to hypoxia followed by PFKFB4 transcript and protein detection at specified time points. For Huh7 cells, hypoxia-inducible factor (HIF)-1α protein was stabilized after 2 hours and then accumulated throughout the course. There was an induction of the PFKFB4 messenger RNA (mRNA) (Figure 2B, left) and protein starting at 4 hours (Figure 2C, top). The MHCC97L cells showed a very similar induction pattern in which HIF-1α protein was stabilized after 2 hours, accompanied by a 20-fold surge of the PFKFB4 mRNA level from 8 to 24 hours (Figure 2B, right), coupled with a significant increase of PFKFB4 protein expression (Figure 2C, bottom). Furthermore, stable knocking down of HIF-1α, but not HIF-2, in MHCC97L cells, showed a significant reduction in PFKFB4 mRNA and protein levels, suggesting that PFKFB4 was induced by hypoxia via the HIF-1 axis (Figure 2D). To pinpoint the hypoxic responsive elements (HREs) in the PFKFB4 promoter responsible for HIF-1α interaction,12 chromatin-immunoprecipitation (ChIP) assays against the HIF-1α protein were conducted in 2 HCC cells followed by the qPCR detection of specific PCR product covering each individual HRE. Both HRE sites at the -166 and -402 upstream regions from the transcription start site (TSS) of the PFKFB4 promoter were found to be enriched significantly under hypoxic condition (Figure 2E), supporting their positive interactions with HIF-1α, which in turn promotes PFKFB4 expression in HCC cells.
Figure 2.
PFKFB4 expression was extrinsically driven by hypoxia in HCC. (A) RNA-seq analysis for PFKFB4 expression in a panel of 4 HCC cell lines under normoxic and hypoxic conditions. (B) qPCR analysis for PFKFB4 and HIF-1α expression in Huh7 and MHCC97L cells at the indicated time points under hypoxic treatment. (C) Western blot analysis for PFKFB4 and HIF-1α expression in Huh7 and MHCC97L cells at the indicated time points under hypoxic treatment. (D) qPCR and Western blot analysis for PFKFB4 expression in MHCC97L cells with stable HIF-1α and HIF-2 knockdown under normoxic and hypoxic conditions. (E) ChIP-qPCR assay showed the positive interactions between the HIF-1α and the 2 HREs at -166 and -402 upstream positions of the PFKFB4 promoter in Huh7 and PLC/PRF/5 cells under hypoxia. (F) A schematic illustrating the overall in silico data integration strategy for the identification of functionally significant hypoxia-inducible genes in HCC. NT, Non-tumor; shHIF, shRNA against hypoxic-inducible factor; shNTC, non-targeting control shRNA; T, Tumor.
To search for the functionally significant HIF-mediated hypoxia-inducible oncogenes in HCC, a robust bioinformatic analysis pipeline was built to integrate relevant data sets at multiple facets to identify candidate genes that fulfilled the following criteria: (1) contained a consensus RCGTG HRE sequence in their promoter region, (2) showed a positive HIF-binding profile in the HIF-1 ChIP-Chip data,13 (3) showed induced expression in a panel of HCC cell lines under hypoxia compared with normoxia, and (4) were up-regulated significantly in patients’ HCC tumors compared with the corresponding nontumorous liver tissues (Figure 2F). Interestingly, PFKFB4 was identified as the only glycolytic enzyme that fulfilled all the criteria, further suggesting its functional uniqueness and significance in HCC.
p53 Functions as an Upstream Repressor of PFKFB4 and TP53 Mutations Are Associated With PFKFB4 Overexpression in HCC
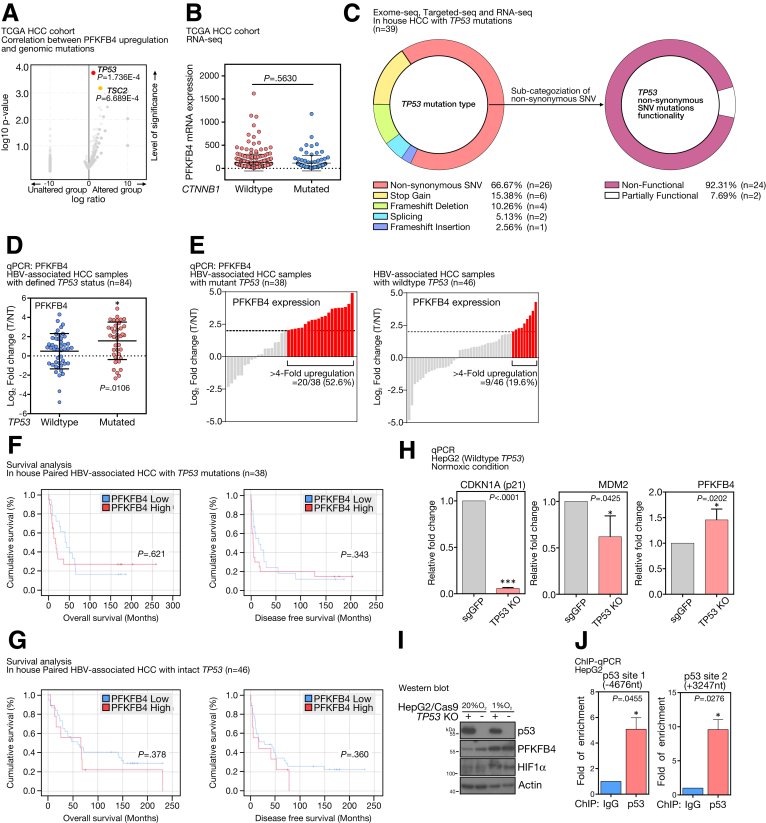
HCCs display a diverse set of driver gene mutations giving rise to intertumoral heterogeneity. To question whether the up-regulated PFKFB4 expression was associated with particular HCC molecular subgroups, we performed an integrative correlation analysis between gene mutations and PFKFB4 expression using TCGA HCC data sets. TP53 (P = 1.736E-4) and TSC2 (P = 6.689E-4) genomic mutations were found to be the top candidates that co-occurred most frequently with PFKFB4 overexpression (Figure 3A). The association between PFKFB4 expression and TP53 mutation tended to be specific because PFKFB4 expression level did not correlate with CTNNB1 mutation, another signature driver gene of HCC (Figure 3B). As validation, we cross-referenced our PFKFB4 qPCR results (Figure 1F) with our in-house databases consisting of exome-sequencing, targeted DNA-sequencing, and RNA-seq and successfully defined the TP53 mutational status in 87 patients (Figure 3C). Of 84 of them with RNA samples available for qPCR, HBV-associated HCCs with TP53 mutations were found to express significantly higher levels of PFKFB4 (P = .0106) (Figure 3D). For instance, PFKFB4 up-regulation of 4-fold or higher was observed in 52.6% of the TP53 mutated cases, while this was present in only 19.6% of the TP53 wild-type cases (Figure 3E).
Figure 3.
PFKFB4 expression was associated with the intrinsic loss-of-function TP53 mutation in HCC. (A) Correlation analysis indicated a significant association between PFKFB4 overexpression with TP53 and TSC2 mutations. (B) Expression of PFKFB4 in the HCC samples with wild-type or mutant CTNNB1 background in TCGA HCC RNA-seq cohort. (C) Categorization of the type of TP53 mutations detected in our in-house HCC cohort. A statistic on the identified nonsynonymous missense TP53 mutations according to their biological functionality. (D) Relative PFKFB4 expression in HBV-associated HCC samples with mutant TP53 (n = 38) and wild-type TP53 (n = 46). (E) qPCR analysis for PFKFB4 expression in HCC samples with wild-type or mutant TP53 background. (F) Comparison of the overall and disease-free survival between the HCC patients with high and low expression of PFKFB4 underlying TP53 mutations. (G) Comparison of the overall and disease-free survival between the HCC patients with high and low expression of PFKFB4 underlying intact TP53. (H) qPCR analysis for PFKFB4, CDKN1A, and MDM2 expression in TP53 KO and control HepG2 cells under normoxic conditions. (I) Western blot analysis for PFKFB4 expression in TP53 KO and control HepG2 cells under normoxic and hypoxic conditions. (J) ChIP-qPCR assay in HepG2 cells showed the positive interactions between the p53 and 2 potential p53 binding sites at -4676 upstream and +3247 downstream positions of the PFKFB4 promoter. ∗P < .05, and ∗∗∗P < .001. NT, Non-tumor; sgGFP,single-guide RNA against Green Fluorescent Protein; SNV, single nucleotide variant; T, Tumor.
To further show the clinical relevance of PFKFB4 overexpression in either TP53 wild-type or mutant background, the PFKFB4 qPCR cohort was divided according to the TP53 status and followed by clinicopathologic correlation analysis. Interestingly, PFKFB4 overexpression was associated significantly with aggressive tumor behavior only in the TP53 mutated cases (Table 2), with younger patient age (P = .019), larger tumor size (P = .003), absence of tumor encapsulation (P = .020), presence of venous invasion (P = .019), and advanced TNM stage (P = .042). In addition, concerning the background liver diseases, PFKFB4 also showed a significant negative correlation with liver cirrhosis (P = .012). However, no association with overall or disease-free survival was found (Figure 3F). In contrast, no association with any pathologic features and prognostic outcomes was found in PFKFB4–up-regulated HCC cases with intact TP53 (Table 2 and Figure 3G). Because TSC2 mutations had a lower incidence rate in HCC, insufficient clinical samples were available to independently verify its correlation with PFKFB4 expression.
Table 2.
Clinicopathologic Correlation of PFKFB4 Overexpression in HBV-Associated HCC Patients With Wild-Type or Mutant TP53
| Clinicopathologic parameters |
TP53 wild-type HCC cases |
TP53 mutated HCC cases |
||||
|---|---|---|---|---|---|---|
| PFKFB4 overexpression (n = 9) | PFKFB4 normal or underexpression (n = 37) | Pa | PFKFB4 overexpression (n = 20) | PFKFB4 normal or underexpression (n = 18) | Pa | |
| Gender | .211 | .485 | ||||
| Male | 5 (55.6%) | 29 (78.4%) | 13 (65.0%) | 14 (77.8%) | ||
| Female | 4 (44.4%) | 8 (21.6%) | 7 (35.0%) | 4 (22.2%) | ||
| Average patient age, y (range) | 46.0 (24–74) | 53.8 (24–66) | .108 | 48.2 (36–71) | 56.5 (29–68) | .019b |
| Background liver disease | 1.000 | .012b | ||||
| Normal | 0 (0.0%) | 3 (8.8%) | 0 (0.0%) | 0 (0.0%) | ||
| Hepatitis | 6 (66.7%) | 17 (50.0%) | 11 (57.9%) | 2 (12.5%) | ||
| Cirrhosis | 3 (33.3%) | 14 (41.2%) | 8 (42.1%) | 14 (87.5%) | ||
| Tumor size | .442 | .003c | ||||
| ≤5 cm | 4 (44.4%) | 10 (29.4%) | 2 (10.5%) | 10 (62.5%) | ||
| >5 cm | 5 (55.6%) | 24 (70.6%) | 17 (89.5%) | 6 (37.5%) | ||
| Tumor encapsulation | .703 | .020b | ||||
| Present | 2 (22.2%) | 11 (31.4%) | 4 (26.7%) | 11 (68.2%) | ||
| Absent | 7 (77.8%) | 24 (68.6%) | 15 (73.3%) | 7 (31.8%) | ||
| Venous invasion | .264 | .019b | ||||
| Absent | 3 (33.3%) | 22 (59.5%) | 4 (20.0%) | 11 (61.1%) | ||
| Present | 6 (66.7%) | 15 (40.5%) | 16 (80.0%) | 7 (38.9%) | ||
| Tumor microsatellite formation | 1.000 | .119 | ||||
| Absent | 4 (44.4%) | 15 (41.7%) | 8 (40.0%) | 12 (66.7%) | ||
| Present | 5 (55.6%) | 21 (58.3%) | 12 (60.0%) | 6 (33.3%) | ||
| Liver invasion | .124 | .745 | ||||
| Absent | 4 (44.4%) | 25 (73.5%) | 12 (60.0%) | 9 (50.0%) | ||
| Present | 5 (55.6%) | 9 (26.5%) | 8 (40.0%) | 9 (50.0%) | ||
| Cellular differentiation | 1.000 | .132 | ||||
| Edmondson grades I–II | 4 (44.4%) | 14 (41.2%) | 3 (15.8%) | 7 (43.8%) | ||
| Edmondson grades III–IV | 5 (55.6%) | 20 (58.8%) | 16 (84.2%) | 9 (56.3%) | ||
| TNM tumor staging | 1.000 | .042b | ||||
| I–II | 3 (33.3%) | 14 (38.9%) | 4 (20.0%) | 10 (55.6%) | ||
| III–IV | 6 (66.7%) | 22 (61.1%) | 16 (80.0%) | 8 (44.4%) | ||
Fisher exact test.
P < .05.
P < .01.
The association of TP53 mutation with PFKFB4 up-regulation suggested that p53 might function as an upstream negative regulator of PFKFB4. By using Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated protein 9 (Cas9)-mediated gene knockout (KO), TP53 KO cells were generated to investigate their impact on PFKFB4 transcription. TP53 KO was performed successfully on Hepatoma G2 (HepG2) cells and the loss of function of p53 in the KO clone was confirmed by the significant reduction of p21 (P < .0001) and MDM2 levels (P = .0425) (Figure 3H, left and middle), both being well-established downstream target genes of p53. TP53 KO was sufficient to promote significant PFKFB4 mRNA up-regulation (P = .0202) in normoxic conditions (Figure 3H, right). Similarly, up-regulation of PFKFB4 protein in TP53 KO cells was observed compared with control cells (Figure 3I). A previous study predicted 2 potential p53 binding sites in the PFKFB4 gene, with 1 located at the -4676 upstream of the TSS and another positioned at the +3247 downstream of the TSS.14 To show their roles as bona fide p53 binding sites, ChIP assays against p53 were performed in parental HepG2 cells, followed by the qPCR detection of PCR product covering each p53 binding site. Interestingly, both predicted p53 binding sites were enriched significantly, suggesting their positive interactions with p53 and their involvement in p53-mediated PFKFB4 down-regulation (Figure 3J).
Loss of PFKFB4 Attenuates HCC Cell Growth and Tumor Incidence In Vivo
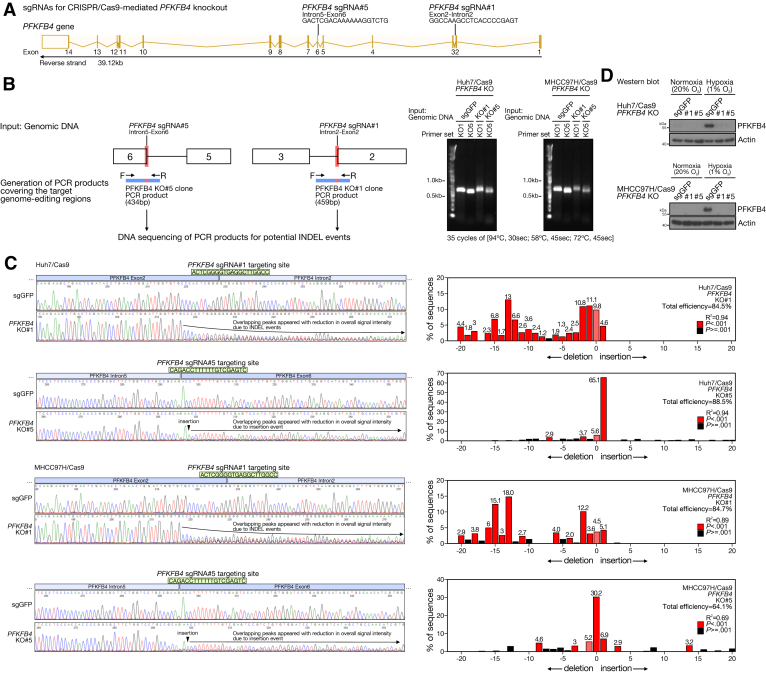
To functionally characterize PFKFB4 in HCC cells, 2 stable CRISPR/Cas9-mediated PFKFB4 KO cells were generated (Figure 4A). Huh7 and MHCC97H cells were selected because both cells showed a minimal level of PFKFB4 expression under normoxia, but a great extent of PFKFB4 induction upon hypoxia. Successful PFKFB4 KO in Huh7 and MHCC97H cells was confirmed at the genomic DNA level (Figure 4B and C), and also by Western blot, with no PFKFB4 protein observed under hypoxia when compared with the negative single-guide RNA against Green Fluorescent Protein (sgGFP) control (Figure 4D). To characterize the role of PFKFB4 in regulating tumor growth in vivo, PFKFB4 KO and control Huh7 cells were injected subcutaneously into nude mice. Subcutaneous tumors without PFKFB4 showed a significant reduction in growth rate and tumor mass and size (Figure 5A and B). Similarly, subcutaneous tumors derived from PFKFB4 KO MHCC97H cells also showed a slower initial tumor growth rate compared with the control tumors (Figure 5C).
Figure 4.
Generation and validation of CRISPR/Cas9-mediated PFKFB4 KO and control HCC cell lines. (A) Schematic illustrating the specifically designed sgRNAs that guide the Cas9-mediated gene editing at the indicated regions of the PFKFB4 genes for the generation of PFKFB4 KO HCC cells. (B) To examine the CRISPR/Cas9-mediated gene-editing events in PFKFB4 KO cells, genomic DNA was first extracted from PFKFB4 KO and control Huh7 and MHCC97H cells. The extracted DNA then served as a template for the PCR amplification of the specific PCR product covering the sgRNA targeting regions across the exon 2–intron 2 junction and the intron 5–exon 5 junction of the PFKFB4 gene, followed by direct Sanger sequencing. The amplification of PCR products covering the 2 sgRNA target regions in PFKFB4 genes in control and PFKFB4 KO Huh7 and MHCC97H cells was confirmed by agarose gel electrophoresis. (C) PCR products generated from control and PFKFB4 KO Huh7 as well as MHCC97H cells were individually subjected to direct Sanger sequencing. Sequencing analysis showed a significant reduction of sequencing signals across the predicted PFKFB4 sgRNA targeting sites in both PFKFB4 KO#1 and KO#5 cells owing to the CRISPR/Cas9-mediated INDEL events (right panel). Similar findings were not observed around the 2 PFKFB4 sgRNA targeting sites in the control cells. Paired sequencing data generated from the PFKFB4 KO and control cells covering individual PFKFB4 sgRNA target sites were subjected to in silico Tracking of Indels by Decomposition (TIDE) analysis for further examination of the underlying INDEL events. PFKFB4 sgRNA#1 mainly introduced deletions around the sgPFKFB4#1 target region, while PFKFB4 sgRNA#5 primarily introduced a single base insertion in the sgPFKFB4#5 target region (left panel). (D) Western blot analysis for PFKFB4 expression in the PFKFB4 KO and control Huh7 and MHCC97H cells.
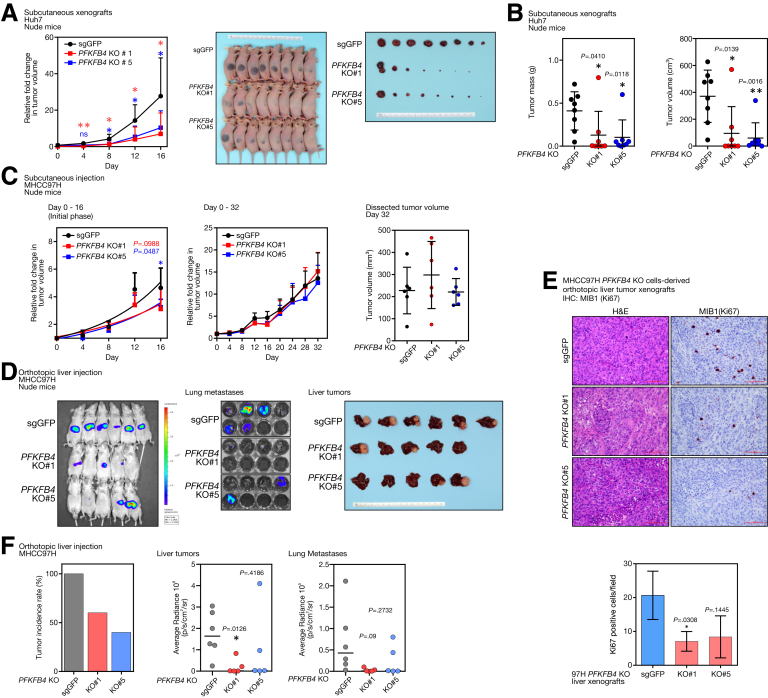
Figure 5.
CRISPR/Cas9-mediated PFKFB4 KO suppressed tumorigenicity of HCC cells in vivo. (A) A comparison of the growth rates between the PFKFB4 KO and control Huh7 cell–derived subcutaneous tumor xenografts. The volume of the dissected tumors among different experimental groups was compared at the end point. (B) The tumor mass and tumor volume of the PFKFB4 KO and control Huh7 cell–derived subcutaneous tumor xenografts. (C) Comparison of the growth rates between the PFKFB4 KO and control MHCC97H cell–derived subcutaneous tumor xenografts at the initial phase, and up to 32 days after subcutaneous injection. The tumor volume between the PFKFB4 KO and control MHCC97H tumor xenografts on day 32 after subcutaneous injection was shown. (D) Bioluminescence imaging of the MHCC97H cell–derived liver orthotopic xenografts and lung tissues. A macroscopic view of the dissected liver tumors was shown. (E) PFKFB4 KO and control HCC cell–derived orthotopic liver xenografts were sectioned and processed for H&E staining and MIB1 (Ki67) staining. The number of MIB1 (Ki67)-positive cells in the PFKFB4 KO and control MHCC97H cell–derived orthoptic liver tumor xenografts were counted and compared. (F) The overall tumor incidence rate of the orthotopic liver tumor formation and the corresponding bioluminescence signals detected from the dissected liver and lung tissues. ∗P < .05, ∗∗P < .01. IHC, Immunohistochemistry; sgGFP, single-guide RNA against Green Fluorescent Protein.
To further show the role of PFKFB4 in vivo, luciferase-labeled MHCC-97H PFKFB4 KO and control cells were injected orthotopically into the left lobe of the liver and incubated for 8 weeks (Figure 5D). Interestingly, both PFKFB4 KO groups showed a marked reduction in tumor incidence, which was 60% (3 of 5) and 40% (2 of 5) in the PFKFB4 KO#1 and PFKFB4 KO#5 groups, respectively, as compared with a 100% (6 of 6) tumor incidence rate in the control group. With immunohistochemistry, a reduction of Ki67-positive cells was observed in the PFKFB4 KO orthotoptic liver tumors (Figure 5E). Furthermore, the abolishment of PFKFB4 caused a reduction in tumor size and lung metastasis according to bioluminescence imaging (Figure 5F). Taken together, our results suggested that PFKFB4 plays a pro-oncogenic role in promoting HCC incidence and supporting HCC growth.
Ablation of PFKFB4 Causes an Increase of PFK1 Downstream Metabolites in Glycolysis Under Hypoxia in HCC Cells
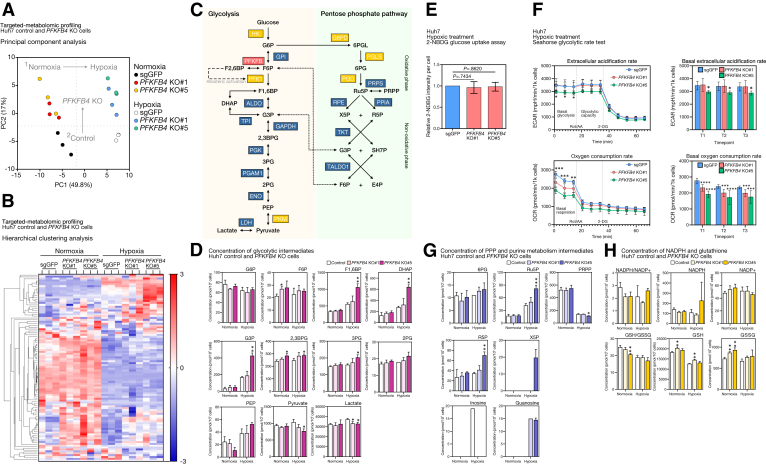
To examine the PFKFB4-mediated metabolic reprogramming in HCC, Huh7 PFKFB4 KO and control cells were subjected to targeted metabolomics analysis with capillary electrophoresis–time-of-flight mass spectrometry (CE-TOFMS) and triple quadrupole mass spectrometry. The triplicate samples for each of the control and 2 PFKFB4 KO clones exposed to either normoxia or hypoxia for 24 hours were included and the intracellular concentrations of 116 metabolites from 12 major human metabolic pathways involved in energy metabolism subsequently were quantified. The principal component analysis was performed to compare the overall metabolic profiles of the 18 samples (Figure 6A). It was found that the triplicates of each clone clustered together, suggesting their consistent clonality. The clusters of the 2 PFKFB4 clones were in close proximity, suggesting their high similarities, but at the same time were distinctly separated from the control samples. In addition, samples exposed to hypoxia were noticeably separated from the normoxic samples. In addition, hierarchical clustering analysis generated a heat map that depicted the distinct metabolic profile of the PFKFB4 KO cells, in which a considerable number of metabolites were up-regulated significantly (Figure 6B, upper right).
Figure 6.
PFKFB4 functions as a phosphatase in HCC cells and its loss resulted in the accumulation of glycolytic metabolites and distorted PPP. (A) Principal component analysis illustrating the overall differences between the metabolomic profiles between the PFKFB4 KO and control Huh7 cells under normoxic and hypoxic conditions. (B) Unsupervised hierarchical clustering analysis of the metabolomic profiles between the PFKFB4 KO and control Huh7 cells under normoxic and hypoxic conditions. (C) Schematic diagram showing the key metabolic enzymes in glycolysis and PPP and their metabolic substrates and products. (D) The concentration of glycolytic intermediates in PFKFB4 KO and control Huh7 cells under normoxic and hypoxic conditions. (E) PFKFB4 KO and control cells were cultured with glucose-free medium and subjected to hypoxic treatment for 24 hours, followed by incubation with 2-NDBG, the fluorescently labeled glucose analog, for 10 minutes, followed by flow cytometry. The relative 2-NDBG intensities per cell between the PFKFB4 KO and control cells are shown. (F) The overall glycolytic rate and oxidative phosphorylation rate between PFKFB4 KO and control cells was assessed by the Seahorse glycolytic rate test kit on a Seahorse bioanalyzer. The relative glycolytic rate as a function of the extracellular acidification rate (ECAR) (top panel) and the oxidative phosphorylation rate as a function of the oxygen consumption rate (OCR) (bottom panel) are shown. (G) The concentration of PPP and purine metabolism intermediates in PFKFB4 KO and control Huh7 cells under normoxic and hypoxic conditions. (H) The concentration of NADPH and glutathione in PFKFB4 KO and control Huh7 cells under normoxic and hypoxic conditions. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001. DHAP, Dihydroxyacetone phosphate; PC, Principal component; PEP, Phosphoenolpyruvate.
Because PFKFB4 primarily catalyzes the reaction between F6P and F2,6BP, the metabolic changes along the glycolytic pathway in the PFKFB4 KO cells were first specifically inspected (Figure 6C, left). In normoxic conditions, loss of PFKFB4 did not produce any drastic dysregulation in glycolysis, except for an increase in F6P and 2,3-diphosphoglyceric acid (2,3DPG) within 10%–30%. However, in hypoxic conditions, up-regulation was observed in multiple PFK1 downstream metabolites, including F1,6BP, glyceraldehyde-3-phosphate (G3P), dihydroxyacetone phosphate, 2,3DPG, and 3-phosphoglyceric acid (3PG). The up-regulation was particularly significant in the KO#5 clone, which reached a 1.9-fold increase for F1,6BP, a 2.5-fold increase for G3P, and also a 1.2-fold and a 1.3-fold increase for 2,3DPG and 3PG, respectively. Intriguingly, this increase in the level of glycolytic intermediates did not result in the complete promotion of glycolysis in both KO clones because the concentrations of both end products, pyruvate and lactate, were counterintuitively down-regulated by 10%–30% and 10%, respectively (Figure 6D). No significant changes in the overall glycolysis rate were confirmed further by independent metabolic assays (Figure 6E and F).
Dysregulation in the PPP and Purine Metabolism in PFKFB4 KO Cells
Apart from glycolysis, the activity of PFKFB4 also is believed to indirectly influence the PPP through regulation of the PFK1-mediated committed step in glycolysis (Figure 6C, right). Indeed, an increase of PPP metabolite levels was detected in the PFKFB4 KO cells under hypoxia. For instance, most of the intermediates were increased collectively, including 6-phosphogluconic acid and ribulose 5-phosphate of the oxidative arm, as well as ribose-5-phosphate and xylulose 5-phosphate of the nonoxidative arm (Figure 6G, top). Similarly, the up-regulation was particularly significant in the KO#5 clone, which showed a 2-fold increase for both ribulose 5-phosphate and ribose-5-phosphate, and the level of xylulose 5-phosphate also increased from the undetectable range to a detectable concentration of 20 pmol/106 cells. Moreover, because purine metabolism is coupled to the PPP, some dysregulation was observed under hypoxia after PFKFB4 abolishment, including the most drastic changes in inosine and guanosine concentrations (Figure 6G, bottom). Because PPP is indispensable for the production of NADPH, an anabolic cofactor, its generation and consequential level of antioxidants were examined further. However, the alterations in hypoxic PPP did not influence the NADPH/oxidized nicotinamide adenine dinucleotide phosphate (NADP+) ratio (Figure 6H, top); but this ratio was reduced under normoxic conditions in both PFKFB4 KO cells by 30%. The absolute concentration of the oxidized form NADP+ was increased by 10%–20%, accompanied by a reduction of NADPH by 10%–20%. A similar pattern also was observed in the Glutathione/Glutathione disulfide (GSH/GSSG) ratio, which was decreased by 10%–20%, but with the total concentration of glutathione increased by 10% (Figure 6H, bottom).
Transcriptomic Analysis Identifies a Series of Differentially Expressed Metabolic and Stress-Related Genes in PFKFB4 KO Xenografts
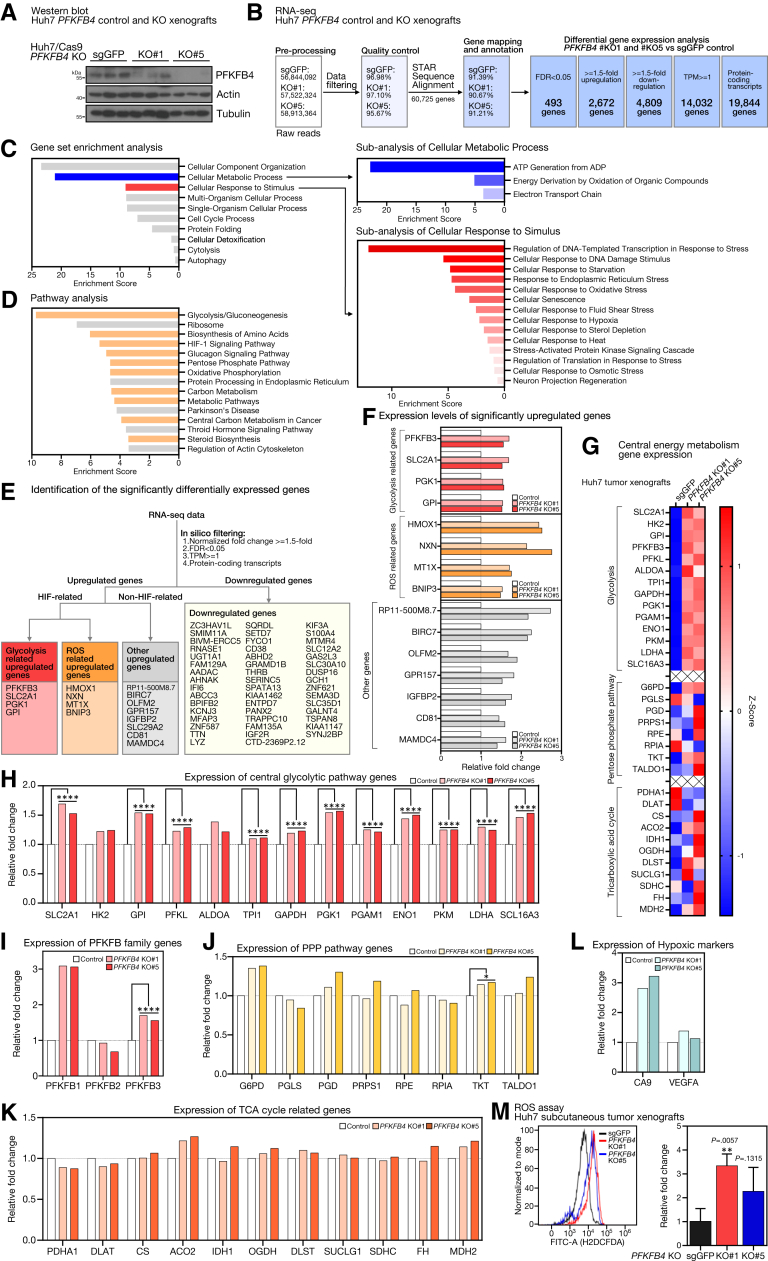
To further show the global gene expression changes after PFKFB4 KO, unbiased RNA sequencing was performed on the Huh7 PFKFB4 KO and the control subcutaneous xenografts. Successful depletion of PFKFB4 in the xenografts was first confirmed by Western blot before RNA-seq (Figure 7A). The workflow of the raw RNA-seq data processing and the subsequent differential gene expression analysis are shown (Figure 7B). In brief, by comparing both PFKFB4 KO groups with the control, 493 genes and 3048 transcripts were identified as differentially expressed, with a false discovery rate (FDR) value of <0.05. In addition, 2672 genes showed a ≥1.5-fold up-regulation and 4809 genes showed a ≥1.5-fold down-regulation.
Figure 7.
PFKFB4 loss resulted in the increase of genes related to glycolysis, hypoxic response, and intracellular ROS modulation in vivo. (A) Western blot analysis for PFKFB4 expression in PFKFB4 KO and control Huh7 cell–derived subcutaneous tumor xenografts. (B) An overview of the bioinformatic pipeline for the identification of the significantly differentially expressed genes between the PFKFB4 KO and the control tumor xenografts. (C) Gene set enrichment analysis of the differentially expressed genes between the PFKFB4 KO and control tumor xenografts. (D) Subanalysis of the cellular metabolic process and cellular response to stimulus revealed significant enrichment with energy metabolism and cellular stress responses including hypoxia. (E) Identification of the significantly differentially expressed genes between the PFKFB4 KO and control tumor xenografts. The significantly up-regulated genes were classified further into HIF-related and non–HIF-related. (F) The expression changes of the significantly up-regulated genes in PFKFB4 KO tumor xenografts compared with control are shown. (G) Heat map showing the expression changes of the enzymes that are involved in the central energy metabolism including glycolysis, PPP, and TCA cycle in PFKFB4 KO tumor xenografts compared with control. (H) The expression of the central glycolytic pathway genes in the PFKFB4 KO tumor xenografts compared with control. (I) The expression of other PFKFB family genes in PFKFB4 KO tumor xenografts compared with control. (J) The expression of the PPP genes in the PFKFB4 KO tumor xenografts compared with control. (K) The expression of the TCA cycle-related genes in the PFKFB4 KO tumor xenografts compared with control. (L) The expression of the hypoxic markers in the PFKFB4 KO tumor xenografts compared with control. (M) Quantification of the intracellular ROS levels by flow cytometry–based H2DCFDA staining in the PFKFB4 KO tumor xenografts compared with control. ∗∗P < .01, Student t test. ∗FDR, <0.05, ∗∗∗∗FDR, <0.0001. ADP, adenosine diphosphate; ATP, adenosine triphosphate; CA9, Carbonic anhydrase 9; FITC, Fluorescein isothiocyanate; sgGFP, single-guide RNA against Green Fluorescent Protein; TPM, Transcript Per Million; VEGFA, Vascular endothelial growth factor A.
In silico ontology analyses were performed for the biological interpretation of the 493 significant differentially expressed genes. First, gene set enrichment analysis showed that genes involved in cellular component organization, cellular metabolic process, and cellular response to stimulus were most affected (Figure 7C, left). Subanalysis showed the vast majority of cellular metabolic process–related genes were responsible for adenosine triphosphate generation, while most cellular stimulus response–related genes were implicated in the regulation of transcription in response to wide-ranging cellular stress, including starvation, endoplasmic reticulum, oxidative stress, and hypoxia (Figure 7C, right). Second, pathway enrichment analysis showed glycolysis and gluconeogenesis as the top dysregulated pathways followed by ribosome, biosynthesis of amino acids, HIF-1 signaling, glucagon signaling, and PPP (Figure 7D). Third, for a more in-depth characterization of these differentially expressed genes, 4 major criteria including (1) FDR <0.05, (2) normalized fold change ≥1.5-fold compared with control, (3) basal expression with transcripts per kilobase million (TPM) >1, and (4) only considering protein-coding transcript, were set to ensure the high confidence screening for the most critically altered genes with actual expression (Figure 7E, top). Of the 63 genes identified, 16 were up-regulated (Figure 7E, left) and 47 were down-regulated (Figure 7E, right). Of note, among the up-regulated genes, 50% (8 of 16) were known to be hypoxia-responsive genes, of which 25% (4 of 16) were involved in the mitigation of oxidative stress (HMOX1, BNIP3, MT1X, and NXN), whereas the other 25% (4 of 16) were canonical glycolytic enzymes and transporters (PFKFB4, GPI, PGK2, and SLC2A1) (Figure 7F). However, no distinctive pattern could be observed for the down-regulated genes.
Because the aforementioned analyses consistently suggested that cellular metabolism was the most dysregulated biological process upon the loss of PFKFB4, the gene expression patterns of the central glucose metabolic pathways, including glycolysis, PPP, and tricarboxylic acid (TCA) cycle, were examined further. Because each metabolic intermediate reaction often is catalyzed by multiple enzymatic isoforms, for clarity, only the dominant isoforms with the highest TPM were considered. Intriguingly, the expression levels of glycolytic enzymes showed an overall up-regulation, but those of the PPP and TCA cycles remained relatively stable (Figure 7G). Specifically, the increase in glycolytic gene expression reached statistical significance for all enzymes, except HK2 and ALDOA, and the degree of up-regulation was between 10% and 60% (Figure 7H). Apart from the central axis of glycolysis, the expression of the PFKFB family members also was examined. Although PFKFB1 showed 3-fold up-regulation in the PFKFB4 KO xenograft, its expression was far too low (TPM, <0.5), indicating relatively low confidence of actual gene expression, while the PFKFB2 expression did not show any significant changes (FDR, 0.973). In contrast, PFKFB3, the most proglycolytic PFKFB isoform, was increased significantly by 60% (FDR, <0.0001) (Figure 7I). The overall PPP and TCA cycle gene expression was not notably altered (Figure 7J and K) except for transketolase (TKT), which showed a significant, 15% increase in expression. The expression of hypoxic markers carbonic anhydrase 9 (CA9) and vascular endothelial growth factor A (VEGFA) showed a general increasing trend in PFKFB4 KO xenografts (Figure 7L). Up-regulation of multiple hypoxic-responsive genes and ROS-related genes in the RNA-seq led us to postulate that loss of PFKFB4 might increase the level of intracellular ROS during tumor formation. To test this hypothesis, a flow cytometry–based 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) assay was performed to evaluate the ROS levels in the Huh7 PFKFB4 KO subcutaneous xenografts. Indeed, the PFKFB4-ablated xenografts showed a 2- to 3-fold increase in the intracellular ROS level, whereas the percentages of cell death in different experimental groups were not affected (Figure 7M).
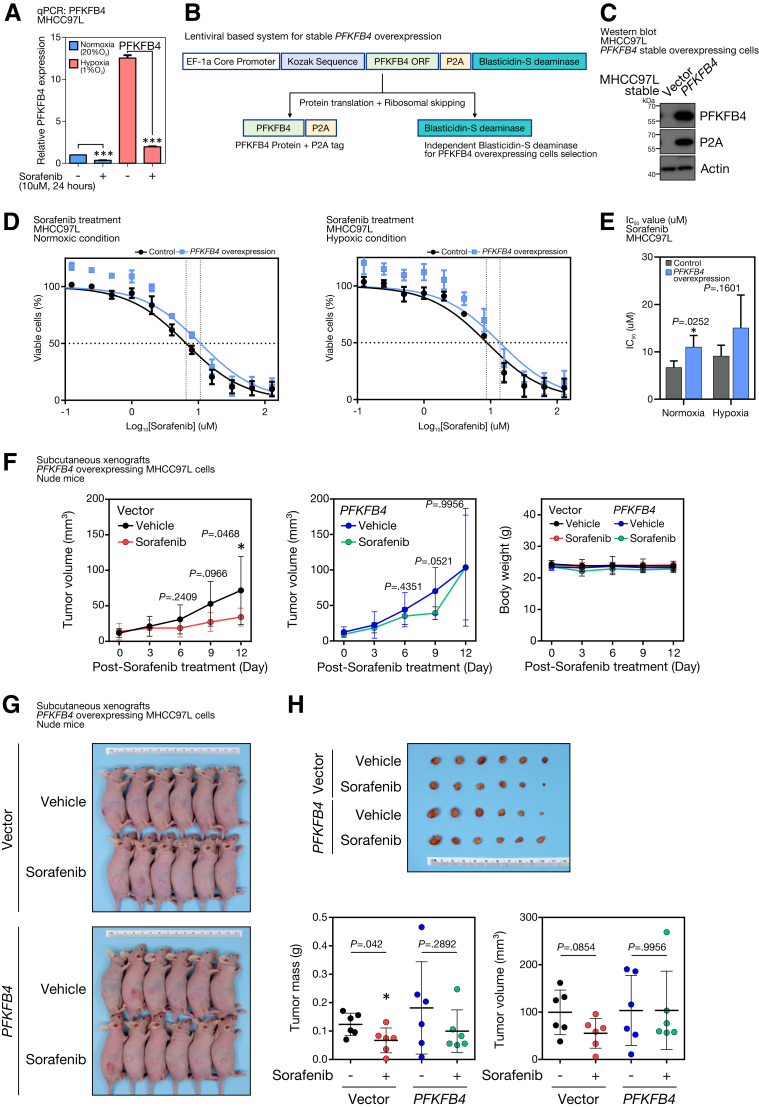
Gain of PFKFB4 Increases Sorafenib Resistance in HCC Cells
Sorafenib is the most widely multiple tyrosine kinase inhibitor for treating advanced HCC. However, drug resistance usually develops shortly and eventually renders the tumor refractory to sorafenib treatment. Interestingly, sorafenib treatment significantly could suppress the PFKFB4 levels in HCC cells under both normoxic and hypoxic conditions (Figure 8A) and we questioned whether PFKFB4 could function as a sorafenib target and serve as a marker to reflect the sensitivity of sorafenib treatment. Because sorafenib could suppress the HIF-1α level15 and hence indirectly may down-regulate PFKFB4 expression, control and PFKFB4-overexpressing cells were established by a bicistronic lentiviral system (Figure 8B) and used to address this postulation. Successful stable overexpression of PFKFB4 in MHCC97L cells was confirmed by Western blot (Figure 8C). The PFKFB4-overexpressing cells were subjected to 24 hours of normoxia or hypoxia pretreatment and then challenged with 12 different concentrations of sorafenib for an additional 48 hours in the corresponding oxygen level–regulated conditions. Compared with the vector control cells, the dose-response curve of the PFKFB4 overexpressing group showed a horizontal shift to the right, reflecting a higher tolerance toward sorafenib treatment (Figure 8D). Such observation was found consistently in both normoxic and hypoxic conditions, with 63.5% and 52.7% increases in the average half-maximal inhibitory concentration, respectively (Figure 8E). Similarly, PFKFB4 overexpressing MHCC97L cell–derived subcutaneous tumors did not show a significant reduction in the tumor volume (Figure 8F) as well as tumor mass (Figure 8G and H) compared with the vector control tumors.
Figure 8.
Ectopic PFKFB4 expression promoted sorafenib resistance to HCC cells. (A) PFKFB4 transcript expression in sorafenib-treated MHCC97L cells under normoxic (20% O2) and hypoxic (1% O2) conditions. (B) Schematic diagram illustrating the lentiviral-based bicistronic system that drives PFKFB4 overexpression. (C) Western blot analysis for PFKFB4 in PFKFB4-overexpressing and control MHCC97L cells. (D) Dosage response curves of sorafenib in PFKFB4-overexpressing and control MHCC97L cells under normoxic and hypoxic conditions. (E) Half-maximal inhibitory concentration (IC50) values of sorafenib (umol/L ± SD) in PFKFB4-overexpressing and control MHCC97L cells. (F) Vector and PFKFB4-overexpressing MHCC97L cells, respectively, were injected subcutaneously into immunodeficient nude mice. At 2 weeks postinjection, the tumor-bearing mice were subjected to vehicle or sorafenib (5 mg/kg) treatment by daily oral gavage. The tumor volume and body weight were monitored and compared as indicated. (G) On day 12 post-treatment, the mice were killed, and the tumors in all the test groups were dissected. (H) The dissected tumor volume and tumor mass between the vehicle and sorafenib treatment groups were measured and compared. ∗P < .05 and ∗∗∗P < .001. EF-1a, Elongation factor 1-alpha.
Discussion
Several studies have shown the significance of PFKFB4 in human cancers, however, its association with HCC, the dominant form of primary liver cancer affecting the most important metabolic organ, remains elusive. Our study highlighted the uniqueness of PFKFB4 among the PFKFB family in HCC, in which it not only frequently was overexpressed, but its up-regulation also was associated with aggressive tumor behavior, metastatic features, and poor survival outcome. PFKFB4 expression can be influenced dynamically by both intrinsic genomic mutations and extrinsic microenvironmental stress. KO of TP53 in relevant HCC cells could increase PFKFB4 transcript and protein levels, while blockade of HIF-1α could cause a reduction. These findings suggest that p53 likely functions as an upstream suppressor of PFKFB4 and mutations of these critical tumor-suppressor genes in HCC development might cause PFKFB4 dysregulation. On the contrary, hyperactivation of HIF-1α in responding to cellular stress functions as an upstream activator to drive PFKFB4 expression. Of note, a previous study suggested that another transcription factor, peroxisome proliferator- activated receptor gamma (PPARγ), also could drive PFKFB4 transcription in HepG2 cells under the normoxic condition.16
Ros et al14 showed that PFKFB4 expression was increased in cancer cells with p53 deficiency and PFKFB4 was essential to support biosynthetic activities and redox homeostasis via the PPP pathway in a p53 null background.9 Our findings not only complement their hypothesis but also further show the clinical relevance of such a relationship in an HCC patient cohort with loss-of-function TP53 mutations and showed that PFKFB4 overexpression correlated significantly with aggressive tumor behavior in this specific HCC molecular subgroup. Our previous large-scale mutational screening identified a novel molecular subset of HBV-associated HCC underlying recurrent TSC2 mutations, which is associated with mammalian target of rapamycin (mTOR) activation.17 The PFKFB4 up-regulation in this distinct class of HCC further suggested a potential novel regulatory mechanism of PFKFB4 expression by Tuberous Sclerosis Complex 2 (TSC2)-inactivation, loss-driven mTOR activation. Intriguingly, Feng and Wu et al18 reported that in acute myeloid leukemia cells, the knockdown of TSC2 only boosted the expression of PFKFB3, but not PFKFB4 or other PFKFB isoforms. Moreover, the mTOR-induced PFKFB3 was claimed to be mediated through the stabilization of HIF-1a, while in our unpublished data, TSC2 KO did not impact the HIF-1a protein level in HCC cells. Such difference suggests that the TSC2/mTOR regulation of PFKFB4 might be a specific feature in HCC and independent of the HIF-1 pathway, which would warrant further investigation.
The positive relationship between hypoxia, HIF-1α, and PFKFB4 was established previously, however, their functional importance in the context of human HCC still has not been examined systematically. We have shown that hypoxia-stabilized HIF-1a but not HIF-2 was responsible to drive PFKFB4 transcript expression and the accumulation of PFKFB4 protein was presented in a sequential and time-dependent manner. Of note, instead of using different RNA interference techniques to inhibit the expression of PFKFB4 on the post-transcriptional level, our study adopted the CRISPR/Cas9 technology to eradicate PFKFB4 expression at the genome level. PFKFB4 KO cell models avoid the effect of inadequate suppression of PFKFB4 expression, especially when the drastic effect of hypoxia induction of PFKFB4 transcript overcame that of the short hairpin RNA inhibition. By using xenotransplantation mouse models, PFKFB4 was shown to play oncogenic functions in HCC cells. For instance, subcutaneous tumors showed significant growth impairment in the absence of PFKFB4. Consistently, orthotopic liver xenotransplantation showed that the abolishment of PFKFB4 led to a remarkable decrease in tumor incidence. It appears that in the liver tumor microenvironment, PFKFB4 plays a pivotal role in tumor initiation, where PFKFB4 KO cells possibly failed to go through tumorigenesis and therefore no tumors were formed.
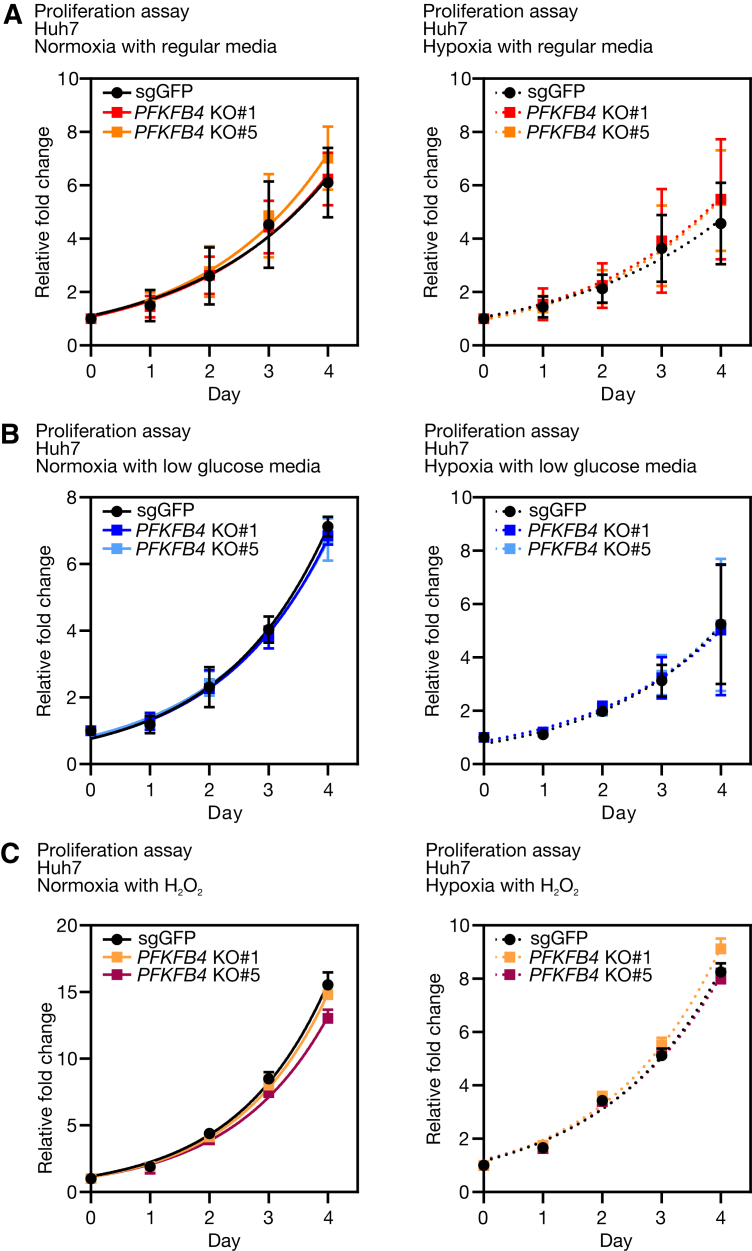
Most of the protumorigenic phenotypic changes of PFKFB4 only appeared to be noticeable in vivo but not in vitro (Figure 9). Such discrepancy might be attributed to variations in microenvironmental factors. Because the PFKFB isoenzymes are responsible for metabolic homeostasis and stress adaptation, they most likely are sensitive to exogenous cues. One speculation is that the in vivo culture media did not fully recapitulate the nutrient composition of HCC or the liver microenvironment. Indeed, several recent studies accentuated the significance of nutrient content in determining the metabolic fidelity of cancer cells.19,20 Another speculation is that the formation of tumor hypoxia in the in vivo and in vitro systems might be different. In the in vitro settings, hypoxia was achieved by immediate exposure of cultured cells to 1% oxygen for hours to days, whereas in the in vivo settings, it was more likely a gradual process that lasted for weeks to months. In other words, the former condition resembled acute hypoxia, while the latter imitated chronic hypoxia, similar to the progressive expansion growth of tumor but with insufficient neovascularization. It is known that the pathophysiologic consequences vary with hypoxic subtypes, in which distinct mechanisms would take place in response to exogenous stress.21 PFKFB4 might preferentially facilitate cellular adaptation to a chronic hypoxic microenvironment.
Figure 9.
The cell proliferation rate of PFKFB4 KO and control Huh7 cells in different culturing conditions in vitro. (A) Comparison of the in vitro proliferation rate between PFKFB4 KO and control Huh7 cells under normoxic and hypoxic conditions with regular culture media. (B) Comparison of the in vitro proliferation rate between PFKFB4 KO and control Huh7 cells under normoxic and hypoxic conditions with low glucose–supplemented media. (C) Comparison of the in vitro proliferation rate between PFKFB4 KO and control Huh7 cells under normoxic and hypoxic conditions in the presence of hydrogen peroxide. sgGFP, single-guide RNA against Green Fluorescent Protein.
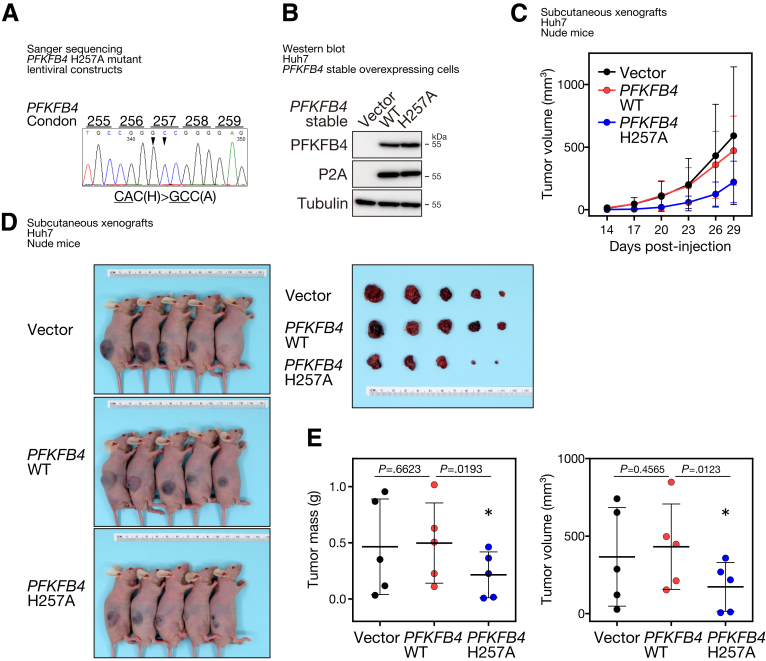
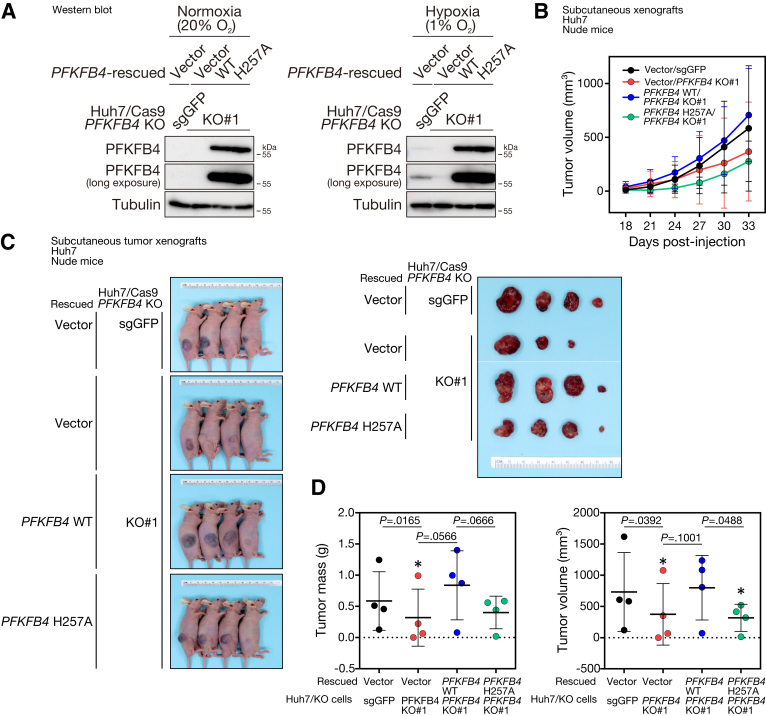
Loss of p53, hyperactivation of mTOR, and hypoxia do share similarities and are known to be linked to up-regulation of glycolysis. For instance, p53 retrains glycolysis by up-regulating the expression of antiglycolytic regulators, such as TP53 induced glycolysis regulatory phosphatase (TIGAR), as well as down-regulating the expression of multiple glycolytic enzymes and the glucose transporter protein type 1 (GLUT1).22 mTOR hyperactivation increases hexokinase 2 (HK2) translation and activation, and it also up-regulates the expression of multiple glycolytic enzymes via Myc.23 Stabilization of the HIF-1 complex under hypoxia induces the transcription of the majority of the glycolytic enzymes, including GLUT1, PFKFB3, and lactate dehydrogenase A (LDHA).24 All circumstances suggest that the expression of PFKFB4 may be triggered upon metabolic reprogramming that favors glycolysis. However, whether PFKFB4 functions to further boost the canonical glycolysis as a positive feedback loop, or to channel the excess glucose into alternative pathways to support certain metabolic requirements, has remained controversial and appeared to be context-dependent in different cancer types. By targeted metabolomic analysis in HCC cell lines, abolishment of PFKFB4 in hypoxia was found to cause an increase of PFK1 downstream intermediates, including F1,6BP, G3P, dihydroxyacetone phosphate, 2,3BPG, and 3PG. This shift in glycolytic flux suggested that the activity of PFK1 might be promoted in the absence of PFKFB4. In other words, PFKFB4 normally functions as a phosphatase and favors the dephosphorylation of F2,6BP into F6P in HCC cells. The phosphatase activity of PFKFB4 indeed is important in supporting the pro-oncogenic functions in the context of HCC. For instance, our extended investigation has provided additional evidence that ectopic expression of a well-characterized, phosphatase-defective PFKFB4 mutant, H257A,25,26 significantly could suppress the growth of the subcutaneous tumor xenografts derived from Huh7 HCC cells in mice, which resulted in a significant decrease of the tumor volume and tumor mass at the end point (Figure 10). Consistently, stable overexpression of the exogenous PFKFB4 H257A also was found to be less efficient in rescuing the PFKFB4 KO–mediated growth suppression on tumor xenografts compared with the wild-type PFKFB4 (Figure 11). Of note, the increase of glycolytic metabolites upon PFKFB4 loss was limited to only a few PFK1 downstream reactions, but the full activation of the entire glycolytic pathway was not achieved, as indicated by the decrease of both glycolytic end products: pyruvate and lactate. This observation further indicated the specialized regulatory role of PFKFB4 in fine-tuning the first committed step of glycolysis. Although the alterations in glycolysis suggested that bisphosphatase is the dominant functional domain of PFKFB4, the direct impact on the level of F2,6BP in PFKFB4 KO remains to be assessed because no mass spectrometry–based method is currently available for its detection.
Figure 10.
Stable overexpression of PFKFB4 phosphatase defective mutant could suppress subcutaneous tumor growth in vivo. (A) The lentiviral construct for driving stable expression of PFKFB4 phosphatase defective mutant, H257A was prepared by site-directed mutagenesis and its identity was confirmed by Sanger sequencing. (B) Western blot analysis for PFKFB4 protein expression in control, PFKFB4 wild-type (WT), and PFKFB4 H257A stable–overexpressing Huh7 cells. (C) Comparison of the growth rates among the control, PFKFB4 WT, and PFKFB4 H257 stable–overexpressing Huh7 cell–derived subcutaneous tumor xenografts. (D) The volume of the dissected tumors among different experimental groups was compared at the end point. (E) The tumor mass and tumor volume of the dissected control, PFKFB4 WT, and PFKFB4 H257A mutant stable–overexpressing Huh7 cell–derived subcutaneous tumor xenografts. ∗P < .05.
Figure 11.
Stable overexpression of wild-type (WTO but not the phosphatase defective mutant form of PFKFB4 could rescue the PFKFB4 KO–induced suppression of subcutaneous tumor growth in vivo. (A) Western blot analysis for PFKFB4 protein expression in vector-rescued control KO cells and vector, wild-type PFKFB4, and phosphatase defective PFKFB4-rescued PFKFB4 KO Huh7 cells subjected to normoxic and hypoxic conditions for 24 hours. (B) Comparison of the growth rates among the vector-rescued control KO cells and vector, wild-type PFKFB4, and phosphatase defective PFKFB4-rescued PFKFB4 KO cell–derived subcutaneous tumor xenografts. (C) The tumor volume of the dissected tumors among different experimental groups was compared at the end point. (D) The tumor mass and tumor volume of the control, PFKFB4 WT, and PFKFB4 H257A mutant stable overexpressing Huh7 cell–derived subcutaneous tumor xenografts after dissection. ∗P < .05. sgGFP, single-guide RNA against Green Fluorescent Protein.
In addition, a general increase in PPP metabolite level also was observed in PFKFB4 KO cells under hypoxia. The comparatively lower extent of the increase of PPP metabolites compared with those in glycolysis might suggest this has an indirect impact. One plausible explanation is that the build up of glycolytic intermediates may interfere with the activity of the PPP enzymes because some intermediates are commonly shared between both pathways. For instance, F6P and G3P are also the substrates and products of transaldolase and transketolase, respectively. Thus, the distorted balance of these glycolytic intermediates could interfere with the substrate-to-product ratios and redirect the orientation of the metabolic reactions that are readily reversible in the nonoxidative phase of PPP. The consequential accumulation of the nonoxidative PPP metabolites might eventually block the oxidative phase of PPP, leading to the observed overall increase in levels of PPP metabolites. The PFKFB4 KO–induced metabolic dysregulation resembled the effect of TKT stable knockdown in HCC cells as we reported previously, in which the suppression of a single reaction in the nonoxidative PPP caused the accumulation of metabolic intermediates from glycolysis, oxidative PPP, and nonoxidative PPP.27 Moreover, a decrease in NADPH/NADP+ and GSH/GSSG ratio was observed in PFKFB4 KO groups. These findings indicated a comparatively higher oxidative stress status in the PFKFB4 KO cells. NADPH is an essential cofactor produced by the PPP that supplies the reducing power of the recovery of oxidized antioxidants. This explains the co–down-regulation of the GSH/GSSG ratio. In addition, the total glutathione level was increased, suggesting the activity of glutathione synthetase might be promoted in attempting to counteract oxidative stress. Intriguingly, these alterations were present only in normoxia, but not in hypoxia, implying that HIF-mediated reprogramming might rescue such phenotypes in the in vitro culture condition.
RNA-seq analysis suggested that xenografts with PFKFB4 abolishment showed highly glycolytic phenotypes, which echoed our metabolomic results. For instance, more than 80% of the enzymes in the central glycolytic pathway showed a significant increase in gene expression, and the glucose influx transporter as well as the lactate efflux transporter also showed the same induction upon PFKFB4 loss. HIF-1–mediated transcriptional reprogramming appeared to be a major driver of these changes because hypoxia response was among the most affected pathways in the gene enrichment analysis. In line with this, other nonglycolytic hypoxia-inducible genes also were found to be up-regulated, including HMOX1, BNIP3, MT1X, and NXN. HMOX1 encodes for heme oxygenase, which is an essential regulator of ferroptosis and is regarded as a protective mechanism against oxidative damage.28,29 BNIP3 belongs to the B-cell lymphoma 2 (Bcl-2) protein family, and, apart from its apoptotic regulatory role, it also facilitates autophagy and mitophagy under cellular stress.30 MT1X and NXN encode for metallothionein 1X and nucleoredoxin, and both are involved in ROS detoxification. Taken together, the up-regulation of glycolysis-related and oxidative stress–related hypoxia-responsive genes suggested that tumor cells in the PFKFB4 KO xenografts appeared to be experiencing a higher level of oxidative stress, which then might hyperactivate the HIF-1 pathway to promote the Warburg effect and ROS elimination to facilitate cell survival.
Hypoxia-induced therapeutic resistance has been a major obstacle to solid cancer treatment. Sorafenib is the first approved systemic therapeutic agent for the treatment of patients with advanced HCC, but the survival benefit it provides is quite modest, largely because of the development of drug resistance. It has been reported that sorafenib-induced cell death in HCC was dependent on the generation of ROS.31 Because PFKFB4 is essential in suppressing ROS accumulation through the reprogramming of glucose metabolism, the action of PFKFB4 possibly could eliminate the sorafenib-induced ROS accumulation and eventually attenuate the therapeutic efficacy. PFKFB4 up-regulation may be considered a biomarker of sorafenib resistance after further validation of its protein expression in HCC tissues and patient-derived liquid biopsies. Although PFKFB4 inhibition as adjuvant therapy with sorafenib sounds like an attractive approach to overcome sorafenib resistance, the mechanistic action of the PFKFB4 inhibitor regarding its specificity toward the kinase or phosphatase has to be considered carefully. For instance, a first-in-class PFKFB4 selective inhibitor, 5-(n-(8-methoxy-4-quinolyl)amino)pentyl nitrate (5MPN), has been suggested to exert cytostatic property on lung cancer and multiple myeloma models by exclusively inhibiting the F6P binding site of the PFKFB4 kinase domain.32 However, because PFKFB4 is more likely to function as a phosphatase in the context of HCC, administration of 5MPN for HCC might further tilt the reactions toward the dephosphorylation of F2,6BP, which might counterintuitively enhance the oncogenic property of PFKFB4. Hence, it is imperative to explore alternative PFKFB4 inhibitors for selective blockade of its phosphatase activity, which eventually may be useful to serve as an alternative therapy for HCC patients with TP53 mutations.
Materials and Methods
Patient Samples
The use of human samples was approved by the Institutional Review Board of the University of Hong Kong/Hospital Authority Hong Kong West Cluster (UW09-185, UW17-056, and UW19-781). HCC patient samples were obtained immediately after surgical resection from the operation room, snap-frozen in liquid nitrogen, and stored at -80ºC. All patients had chronic HBV infection and tested positive for hepatitis B surface antigen in their sera. Frozen sections were cut from tumors and stained for histologic examination to ensure a homogenous cell population of tissues. The clinical parameters of the patients, where applicable and available, including tumor size (in centimeters), background liver disease, metastatic features of HCC (liver invasion, tumor microsatellite formation, tumor encapsulation, and venous invasion), cellular differentiation grading, and TNM staging were collected and subjected to clinicopathologic correlation analysis (SPSS Statistics V25, IBM).
Tissue Culture and In Vitro Sorafenib Treatment
All cells were cultured in a humidified incubator at 37ºC with 5% carbon dioxide. The 293FT cells were purchased from Life Technologies; Hep3B, HepG2, and PLC/PRF/5 cells were purchased from ATCC; Huh7 cells were obtained from the JCRB Cell Bank (Japan); MHCC97L and MHCC97H cells were obtained from Fudan University (China); and MIHA cells were a generous gift from the Marion Bessin Liver Research Center. The 293FT, Huh7, MHCC97L, MHCC97H, and MIHA cells were maintained in Dulbecco’s modified Eagle medium (DMEM)–high glucose medium (Gibco) supplemented with 10% fetal bovine serum (v/v) and 1% streptomycin and penicillin (v/v). Similar culture supplements were provided to other cell lines, instead MEM medium (Gibco) was used for culturing Hep3B, HepG2, and PLC/PRF/5 cells. In addition, 1 mmol/L sodium pyruvate also was provided to Hep3B, HepG2, MHCC97L, MHCC97H, and MIHA cells.
For in vitro sorafenib treatment, 10 mmol/L sorafenib p-toluenesulfonate salt (LC Laboratories) stock solution in dimethyl sulfoxide was prepared. The control and PFKFB4-overexpressing MHCC97H cells then were incubated with the indicated concentrations of sorafenib for 24 hours followed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay for quantifying the percentage of viable cells after the sorafenib treatment compared with the vehicle-treated control.
Mouse Models and In Vivo Sorafenib Treatment
All animal experiments were approved by the Committee on the Use of Live Animals in Teaching and Research (5256-19) (Li Ka Shing Faculty of Medicine, the University of Hong Kong). All of the mice were supplied and maintained by the Centre for Comparative Medicine Research (Li Ka Shing Faculty of Medicine, the University of Hong Kong). For subcutaneous xenotransplantation, cells to be injected were trypsinized and counted. For each injection, 1 × 106 Huh7 cells, 5 × 105 97H cells, and 5 × 105 97L cells were resuspended in a 100-uL injection vehicle solution consisting of ice-cold serum-free medium and Matrigel matrix (Discovery Labware) (1:1; v/v). Cells were injected subcutaneously to the right flank of the 6- to 8-week-old male immunodeficient BALB/c nude mice with a 27G needle. The tumor size was calculated by the following formula: 1/2 × length × width × depth was recorded every 3–4 days after the tumor incidence. The mice were killed after 4 time points when the humane end point was reached.
For liver orthotopic xenotransplantation, cells to be injected into the liver of the 6- to 8-week-old male immunodeficient BALB/c nude mice were trypsinized and counted. For each injection, 1 × 106 luciferase-labeled MHCC97H cells were resuspended in a 15-uL injection vehicle solution consisting of ice-cold serum-free medium and Matrigel matrix (Corning) (1:1; v/v). Each mouse was anesthetized with 200 uL of a 1% ketamine and 0.1% xylazine mixture. A midline incision was performed on the mouse to expose the liver and the cells were injected orthotopically into the right lobe with a microliter syringe. At 6 weeks after surgery, the mice were subjected to bioluminescence imaging using an IVIS Spectrum imaging system (PerkinElmer), and the tumor size and degree of lung metastases were quantified by the emitted luciferase signals.
For in vivo sorafenib treatment, the vehicle solution was prepared by mixing 75% ethanol and Kolliphor EL (Sigma) (1:1, v/v) at 65°C. To achieve a final dosage of 5 mg/kg per mouse, 200 mg sorafenib p-toluenesulfonate salt was first dissolved into 15 mL vehicle solution at 65°C, aliquoted, and stored at -80°C. Starting from day 14 postsubcutaneous injection, a 5-fold dilution of the vehicle and sorafenib stock solution in sterile water was freshly prepared, and 100 uL of the working vehicle and sorafenib solution was administered by oral gavage daily. The tumor volume and body weight of the mice were monitored every 3 days from the start date of sorafenib treatment. At the experimental end point, the tumors from different experimental groups were dissected and their volume and mass were measured and compared.
Hypoxic Treatment
The tissue culture plates were placed inside a modular incubator chamber (Billups-Rothenberg). The chamber was sealed to ensure no leakage and was purged with hypoxic gas (94% N2, 5% CO2, and 1% O2) at a rate of 40 L/min for 2 minutes. The chamber then was placed inside the tissue culture incubator for the indicated duration.
RNA Extraction and Real-Time Reverse-Transcription qPCR Analysis
Cells (5 × 105) per well were seeded onto a 6-well plate 1 day before RNA collection. To lyse the cells, 1 mL TRIzol reagent (Life Technologies) was added to each well and incubated for 5 minutes on an orbital shaking platform at 150 rpm at room temperature and collected in the microfuge tubes. The RNA was extracted by standard chloroform extraction, isopropanol precipitation followed by centrifugation and washing with 70% ethanol. The dried RNA eventually was dissolved in 20–50 uL diethylpyrocarbonate (DEPC) water, with concentration being determined.
For reverse-transcription qPCR analysis, 1 ug RNA was subjected to reverse-transcription and the resultant complementary DNA (cDNA) product was diluted 5-fold with UltraPure Distilled water (Invitrogen) before the qPCR reaction. PowerSYBR Green Master Mix (Applied Biosystems) was used in the presence of the primer pair for the corresponding target gene and the cDNA. The detailed primer sequences are listed in Table 3. qPCR reactions were run on the StepOnePlus Real-Time PCR system or the QuantStudio 5 System (Applied Biosystems), and the comparative cycle threshold (ΔΔCT) method was used to compare the changes in gene expression between the tested samples and the control.
Table 3.
Primers for qPCR Analysis
| Primer | Sequence, 5’>3’ |
|---|---|
| PFKFB4 F | GGCAGGGAGTTTGCCAAGA |
| PFKFB4 R | TCGTTGAGGACCTTCCACTG |
| HIF-1α F | CGTTCCTTCGATCAGTTGTC |
| HIF-1α R | TCAGTGGTGGCAGTGGTAGT |
| HPRT F | CTTTGCTGACCTGCTGGATT |
| HPRT R | CTGCATTGTTTTGCCAGTGT |
| B2M F | CCACTGAAAAAGATGAGTATGCCT |
| B2M R | CCAATCCAAATGCGGCATCTTCA |
| CDKN1A (p21) F | GAGGCCGGGATGAGTTGGGAGGAG |
| CDKN1A (p21) R | CAGCCGGCGTTTGGAGTGGTAGAA |
| MDM2 F | TAGTATTTCCCTTTCCTTTGATGA |
| MDM2 R | CACTCTCCCCTGCCTGATAC |
B2M, β2 microglobulin; F, forward primer; R, reverse primer.
Protein Extraction, Antibodies, and Western Blot
Cells (1 × 106) were seeded onto a 60-mm plate 1 day before protein collection. Cells were washed twice with ice-cold phosphate-buffered saline (PBS) before harvest. Cells were scraped with RIPA buffer (50 mmol/L Tris-HCl pH = 6.8, 150 mmol/L NaCl, 1 mmol/L EDTA pH = 7.4, 1% NP40, 0.05% sodium dodecyl sulfate [SDS]) supplemented with Complete Protease Inhibitor Cocktail (Roche) and lysed on ice for 20 minutes followed by centrifugation at 12,000 rpm for 15 minutes at 4°C. The lysate supernatant was collected and subjected to Bradford assay for concentration determination. A similar protein extraction protocol also was applied to the tumor xenograft sample, except approximately 0.1 g tumor tissue was first cut and transferred to a 2-mL screw cap tube containing 450 uL RIPA buffer for homogenization with a Precellys Evolution Homogenizer (Bertin Technologies) at 9000 rpm for 30 seconds twice. A total of 20 ug per lane of protein samples was separated on 10% SDS–polyacrylamide gel electrophoresis. The following primary antibodies were used for Western blot at the indicated dilution in 4% milk/Tris-buffered saline with 0.1% Tween 20 detergent (TBST) solution: PFKFB4 (ab137785, 1:500; Abcam), p53 (sc-126, 1:1000; Santa Cruz), porcine teschovirus-1 2A (P2A) peptide (ABS31, 1:5000; Merck Millipore), tubulin (T9026, 1:5000; Sigma), actin (A5316, 1:5000; Sigma), and HIF-2α antibodies (ab199, 1:500; Abcam); while HIF-1α antibody (3716, 1:500; Cell Signaling Technologies) was diluted in 4% bovine serum albumin/TBST. After protein transfer, the polyvinylidene difluoride membrane was blocked with 4% milk/TBST for 1 hour and incubated with the primary antibodies overnight. The membrane was washed 3 times with TBST for 5 minutes, followed by incubation with the horseradish peroxidase–conjugated secondary antibodies for 2 hours. The washed membrane eventually was subjected to image capture using the Amersham Imager 600 (GE) in the presence of WesternBright ECL solution (Advansta). For MIB1 (Ki67) immunohistochemistry, sections of formalin-fixed, paraffin-embedded PFKFB4 KO and control orthotopic liver tumor xenograft tissues were subjected to boiling for 15 minutes in a Tris-based antigen unmasking solution (pH = 9.0) for antibody retrieval followed by immunostaining with a Ki67-specific antibody (RM-9106, 1:100; Thermo Fisher).
ChIP Assay
Cells subjected to normoxic or hypoxic treatment (1% O2) for 6 hours were fixed with 1% formaldehyde at room temperature for 10 minutes. The fixation then was quenched by adding glycine solution to a final concentration of 125 mmol/L. Formaldehyde cross-linked cells were collected by centrifugation and then resuspended in membrane lysis buffer (5 mmol/L KOH, pH = 8.0, 85 mmol/L KCl, 0.5% NP-40, and 0.5% SDS) supplemented with 1× Complete Protease Inhibitors (Roche) and incubated on ice for 30 minutes. Cell nuclei were collected by centrifugation and the cross-linked DNA was digested by Micrococcal nuclease (New England Biolabs) for 5 minutes. Digested DNA was released from the nuclei by 3 freeze-thaw cycles and then subjected to the ChIP assay using the EZ-ChIP assay kit (Millipore) according to the manufacturer’s protocol. HIF-1α antibody (ab1; Abcam) and p53 antibody (2527; Cell Signaling Technologies) were used for the ChIP pulldown, while normal rabbit IgG (sc-2027; Santa Cruz) served as the negative control. With the Power SYBR GREEN master mix (Applied Biosystems) and AmpliTaq Gold 360 master mix (Applied Biosystems) mixed at a 1:1 ratio (v/v), the purified ChIP products served as a template for qPCR analysis in the presence of a 360 GC enhancer solution and the corresponding qPCR primer sets. All the experiments were performed 3 times independently. Primer sets that target specific regions upstream and downstream of the PFKFB4 gene in the ChIP-qPCR analysis are listed in Table 4.
Table 4.
Primers for ChIP-qPCR Analysis
| Primer | Sequence, 5’>3’ |
|---|---|
|
PFKFB4 HRE (-166) ChIP F (chromsome 3: 48557099) |
GCCTGCACAATCCGGGCCTTGAA |
|
PFKFB4 HRE (-166) ChIP R (chromsome 3: 48556879) |
GCCGCGACAGCCCATGTCTATCTT |
|
PFKFB4 HRE (-402) ChIP F (chromsome 3: 48557278) |
TCGTCCCACTGCATGAAAACCCC |
|
PFKFB4 HRE (-402) ChIP R (chromsome 3: 48557140) |
GCGCTGATTTGCATAGGCCCCAC |
|
PFKFB4 p53 site (-4676) ChIP F (chromsome 3: 48553565) |
AAAGGAACCCATGAGGGAAGTT |
|
PFKFB4 p53 site (-4676) ChIP R (chromsome 3: 48553505) |
TGAGCATGTGCGGAAGGA |
|
PFKFB4 p53 site (+3247) ChIP R (chromsome 3: 48561435) |
CGTCCACACTGCCTGGAA |
|
PFKFB4 p53 site (+3247) ChIP F (chromsome 3: 48561493) |
CACAGCCCAACTCCATTGC |
F, forward primer; R, reverse primer.
Generation of CRISPR/Cas9-Mediated PFKFB4 KO Cells
Specific single-guide RNAs (sgRNAs) against PFKFB4 and TP53 were designed using the E-CRISP prediction program from the German Cancer Research Center (DKFZ) and were cloned individually into the lenti-Guide-Puro vector, a gift from Zhang Feng, Massachusetts Institute of Technology (MIT).33 sgRNAs against PFKFB4 with high specificity and efficiency scores were designed and 2 sequences each targeting exon 2 to intron 2 (sgRNA#1) and intron 5 to exon 6 (sgRNA#5) junctions of the PFKFB4 gene were adopted for the subsequent experiments. The details of the sgRNA sequence are listed in Table 5. The cloned vectors were co-transfected with packaging plasmids consisting of group-specific antigen (GAG), regulator of expression of virion proteins (REV), and vesicular stomatitis virus glycoprotein (VSV-G) into 293FT cells for lentiviral packaging. Viral supernatant containing the lentivirus was collected at 48 hours and 72 hours post-transfection. To generate the CRISPR/Cas9 KO cells, the target HCC cells with stable Cas9 overexpression driven by the lenti–Cas9–blast system (a gift from Zhang Feng, MIT33) were transduced with viral supernatant of the sgRNAs followed by puromycin selection (Huh7 cells, 0.75 ug/mL; MHCC97L and H cells, 0.5 ug/mL) for 2 weeks to enrich the positively transduced cells. For comparison, a control sgRNA against green fluorescent protein (GFP) also was transduced and served as a control. The successful KO of PFKFB4 was evaluated by PCR and Sanger sequencing at the genomic DNA level using specific primer sets in Table 6 and by Western blot at the protein level using PFKFB4-specific antibodies. Paired sequencing data were evaluated further for potential insertion and deletion (INDEL) events introduced around the sgRNA target regions in PFKFB4 genes in the KO cells by in silico Tracking of Indels by Decomposition (TIDE) analysis.34
Table 5.
Primers for Cloning the Corresponding sgRNA Expression Constructs for CRISPR/Cas9 KO in HCC Cells
| Primer | Sequence, 5’>3’ |
|---|---|
| PFKFB4 sgRNA#1 S | CACCGGCCAAGCCTCACCCCGAGT |
| PFKFB4 sgRNA#1 AS | AAACACTCGGGGTGAGGCTTGGCC |
| PFKFB4 sgRNA#5 S | CACCGACTCGACAAAAAAGGTCTG |
| PFKFB4 sgRNA#5 AS | AAACCAGACCTTTTTTGTCGAGTC |
| TP53 sgRNA S | CACCGATCCACTCACAGTTTCCAT |
| TP53 sgRNA AS | AAACATGGAAACTGTGAGTGGATC |
AS, antisense; S, sense.
Table 6.
Primers for Amplification and Sequencing of PCR Products Specifically Covering the Two sgRNA Target Regions in the PFKFB4 Gene
| Primer | Sequence, 5’>3’ |
|---|---|
| PFKFB4-KO1-DNA-1F | TGTGTGGGGATGCTAGGAAG |
| PFKFB4-KO1-DNA-1R | TGTAGGTCTTGACCACGTCC |
| PFKFB4-KO5-DNA-1F | CCTTTCACCGCCTGCTTATG |
| PFKFB4-KO5-DNA-1R | TCACACACGACCTTCTGACA |
F, forward primer; R, reverse primer.
Generation of PFKFB4-Overexpressing Cells
The open reading frame (ORF) of PFKFB4 was amplified with pfu polymerase (Promega) from cDNA samples of a hypoxic-treated HCC cell line using 2 specific sets of primers listed in Table 7. The PFKFB4 ORF then was cloned into a bicistronic lentiviral vector derived from the lenti–Cas9–blast system (a gift from Zhang Feng, MIT33) using 2-step molecular cloning involving the restriction enzymes AgeI (Takara Bio) and BamHI (NEB). This vector drives the simultaneous expression of PFKFB4 and the downstream blasticidin as a single transcript, which ensures blasticidin selection could effectively enrich PFKFB4-overexpressing cells. The sequence of the final assembled PFKFB4 ORF and the integrity of the ribosomal skipping sequence P2A plus the downstream blasticidin-S-deaminase was confirmed by Sanger sequencing. PFKFB4 H257A mutant was derived from the PFKFB4 wild-type construct by site-directed mutagenesis using the 2-step PCR method with the primer sets listed in Table 7. The wild-type and mutant lenti–PFKFB4–blast vectors were transfected into 293FT cells for viral packaging. The viral supernatant was collected to transduce the MHCC97H cells, followed by blasticidin selection at a concentration of 4 ug/mL. For comparison, empty vector control also was transduced and served as a negative control. The successful overexpression of PFKFB4 was evaluated by Western blot using the PFKFB4 and P2A-peptide antibodies.
Table 7.
Primers for Cloning PFKFB4 Overexpression Construct
| Primer | Sequence, 5’>3’ |
|---|---|
| PFKFB4 AgeI F | GGACCGGTGCCACCATGGCGTCCCCACGGGAATT |
| PFKFB4 480 BamHI R | CAGGATCCACACAGATGGAC |
| PFKFB4 480 BamHI F | GTGGATCCTGAGGTCATAGC |
| PFKFB4 stop-1 BamHI R | CCGGATCCCTGGTGAGCAGGCACCGTGA |
| PFKFB4 H257A F | CTCTGCCGGGCCGGGGAGAGC |
| PFKFB4 H257A R | GCTCTCCCCGGCCCGGCAGAG |
F, forward primer; R, reverse primer.
Targeted Metabolomics Analysis
The preparation of samples was based strictly on the Sample Preparation Protocol of Metabolite Extraction for Adherent cells (Human Metabolome Technologies, Japan). In brief, for each clone, a triplicate of at least 5 × 106 cells was collected after 24-hour hypoxic treatment or normoxic culture conditions. To extract the metabolites, the cells were washed twice with 5% mannitol solution (FUJIFILM Wako) and 800 uL methanol (FUJIFILM Wako) was added to each dish for a 30-second incubation. A total of 550 uL of the provided internal standard was added to each dish and mixed by pipetting. A total of 1 mL metabolite extract was transferred to 1.5-mL microfuge tubes and centrifuged at 2300 × g at 4°C for 5 minutes. The supernatant was transferred to the provided Ultrafree centrifugal filter units for further purification by centrifugation at 9100 × g at 4°C for 2 hours and stored at -80ºC.
The metabolomics analysis was performed by Human Metabolome Technologies, with CE-TOFMS (Agilent Technologies) and triple quadrupole mass spectrometry (Agilent Technologies) for the analyses of cationic and anionic metabolites, respectively, as previously described.35, 36, 37 The CE-TOFMS data were analyzed with Masterhands version 2.17.1.11 (Keio University, Japan) and the CE triple quadrupole mass spectrometry data were analyzed with MassHunter Quantitative Analysis B.0600 (Agilent Technologies).
Glucose Uptake Assay and Glycolytic Rate Test
To evaluate the glucose uptake rate, control and PFKFB4 KO Huh7 cells were subjected to hypoxic treatment for 24 hours followed by incubation with 100 μg/mL 2-NDBG (Invitrogen), a fluorescent glucose analog, in glucose-free DMEM for 10 minutes. The cells then immediately were washed with ice-cold PBS once, followed by resuspending in 300 μL for flow cytometry analysis.
To compare the glycolytic rates between the control and PFKFB4 KO Huh7 cells under hypoxia, the glycolytic rate test kit was used on Seahorse XFe96 Analyzer (Agilent Technologies), following the manufacturer’s protocol. In brief, a triplicate of control and PFKFB4 KO Huh7 cells were seeded on the Seahorse XF cell culture microplate at a density of 4 × 105 cells per well overnight followed by hypoxic treatment at 1% O2 for 24 hours. The cells then were washed once with the XF assay medium supplemented with 1 mmol/L pyruvate, 2 mmol/L glutamate, and 10 mmol/L glycose, and incubated with the same fresh assay medium in a 37°C incubator without CO2 for 1 hour before the glycolytic rate test assay. Standard assay mode was chosen in which the rotenone plus antimycin A, both being inhibitors of the mitochondrial electron transport chain, and 2-deoxy-D-glucose, an inhibitor of glycolysis, were auto-injected into the test plate during the glycolytic rate test. The real-time metabolic data were acquired and visualized by Seahorse Wave controller software.
Whole Transcriptome (RNA-seq) Analysis of Tumor Xenografts
For each experimental group, approximately 0.1 g tissue was cut from the tumor xenografts and transferred to a 2-mL screw cap tube containing 1 mL TRIzol reagent. The tissue then was lysed and homogenized with a Precellys Evolution Homogenizer (Bertin Technologies) at 9000 rpm for 30 seconds for 2 cycles separated by a 30-second interval. The RNA then was extracted from the homogenate. The RNA-seq analysis was conducted by the Center for PanorOmic Sciences at the University of Hong Kong. In brief, the RNA quality was confirmed with the 2100 Bioanalyzer (Agilent). All samples had an A260/280 ratio between 1.5 and 2.0 and an RNA integrity number higher than 7.5. The RNA-seq was performed on the NovaSeq 6000 Sequencing System (Illumina). To ensure the reliability of the results, the analysis excluded reads with more than 5% unknown bases and reads with more than 50% of bases with a quality value of 11 or lower. The GENCODE Human Genome GRCh38 was used as the reference genome and reads were aligned with STAR (Cold Spring Harbor Laboratory).38 The transcript expression was quantified by RSEM (University of Wisconsin-Madison).39 Differential expression analysis and FDR scoring were performed using EBSeq (University of Wisconsin-Madison).40 Gene set enrichment and pathway enrichment analyses were performed using the Partek software package (Partek Incorporated).
In Vivo Quantification of Intracellular ROS Level
The tumor xenograft to be tested was first diced into small pieces of approximately 1 mm3 with surgical blades and dissociated into a single-cell suspension by the gentleMACS dissociator with a series of 3 human tumor dissociation programs (h-tumor_01.01, h-tumor_02.01, and h-tumor_03.01) in the presence of DMEM/F12 culture medium with 20 mmol/L ROCK inhibitor (Sigma), 2.5 mg/mL liberase (Sigma), and 100 mg/mL DNAse I (Roche). Each step was performed twice and a 5-minute incubation at 37ºC was introduced between the 3 different programs. The cell suspension was centrifuged at 900 rpm for 4 minutes, and the cell pellets were washed with DMEM/F12 twice to remove the red blood cells and unwanted cell debris. The number of viable cells was evaluated with Trypan blue staining using a Countess II automated cell counter (Life Technologies). To evaluate the intracellular ROS level, cell pellets consisting of 1 × 105 live cells were resuspended in 1 mL ice-cold PBS with 2 umol/L CM-H2DCFDA (Invitrogen). The fluorescence signal was acquired by the Novocyte Advanteon Flow Cytometer (Agilent) and analyzed with FlowJo v10.7 (BD).
Statistical Analysis
GraphPad Prism 6.0 software (GraphPad Software) was used for statistical analyses. The Student t test was used for most experiments unless stated otherwise. Data on the graph are representative of 3 independent experiments (n = 3). Data are expressed as means ± SD. P < .05 was considered statistically significant.
All authors had access to the study data and reviewed and approved the final manuscript.
Acknowledgments
The authors are grateful for the assistance and technical support provided by the Genomics Core, Bioinformatics Core, and the Imaging and Flow Cytometry Core of the Center for PanorOmic Sciences at The University of Hong Kong. The authors also thank Dr H. T. Ma for his advice on the use of Tracking of Indels by Decomposition (TIDE) analysis in examining the genome editing in knockout cells.
CRediT Authorship Contributions
Charles Shing Kam (Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Writing – original draft: Equal; Writing – review & editing: Supporting)
Daniel Wai-Hung Ho (Conceptualization: Equal; Data curation: Equal; Formal analysis: Equal; Funding acquisition: Equal; Investigation: Equal)
Vanessa Sheung-In Ming (Data curation: Equal; Investigation: Equal)
Lu Tian (Data curation: Equal; Investigation: Equal)
Karen Man-Fong Sze (Investigation: Equal; Methodology: Equal)
Vanilla Xin Zhang (Investigation: Equal; Methodology: Equal)
Yu-Man Tsui (Investigation: Equal; Methodology: Equal)
Abdullah Husain (Investigation: Supporting; Methodology: Equal)
Joyce Man-Fong Lee (Methodology: Equal)
Carmen Chak-Lui Wong (Methodology: Equal)
Albert Chi-Yan Chan (Resources: Equal)
Tan-To Cheung (Resources: Equal)
Lo-Kong Chan (Conceptualization: Equal; Data curation: Equal; Formal analysis: Equal; Funding acquisition: Equal; Investigation: Equal; Supervision: Equal; Writing – original draft: Equal; Writing – review & editing: Equal)
Irene O.L. Ng, MD, PhD (Conceptualization: Equal; Funding acquisition: Equal; Resources: Equal; Supervision: Equal; Writing – original draft: Equal; Writing – review & editing: Equal)
Data Availability Statement
Specific profiling data and materials described in this study will be made available upon request.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This study was funded and supported by the Health and Medical Research Fund (07182546) of the Food and Health Bureau of the Hong Kong Special Administrative Region government and the Hong Kong Research Grant Council Theme-based Research Scheme (T12-704/16-R and T12-716/22-R). Irene Oi-Lin Ng is Loke Yew Professor in Pathology.
Contributor Information
Lo-Kong Chan, Email: lokongchan@gmail.com.
Irene Oi-Lin Ng, Email: iolng@hku.hk.
References
- 1.Bray F., Ferlay J., Soerjomataram I., et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Yang J.D., Hainaut P., Gores G.J., et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16:589–604. doi: 10.1038/s41575-019-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan L.K., Tsui Y.M., Ho D.W., Ng I.O. Cellular heterogeneity and plasticity in liver cancer. Semin Cancer Biol. 2022;82:134–149. doi: 10.1016/j.semcancer.2021.02.015. [DOI] [PubMed] [Google Scholar]
- 4.Llovet J.M., Ricci S., Mazzaferro V., et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 5.Kudo M., Finn R.S., Qin S., et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet. 2018;391:1163–1173. doi: 10.1016/S0140-6736(18)30207-1. [DOI] [PubMed] [Google Scholar]
- 6.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 8.Ros S., Schulze A. Balancing glycolytic flux: the role of 6-phosphofructo-2-kinase/fructose 2,6-bisphosphatases in cancer metabolism. Cancer Metab. 2013;1:8. doi: 10.1186/2049-3002-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ros S., Santos C.R., Moco S., et al. Functional metabolic screen identifies 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 as an important regulator of prostate cancer cell survival. Cancer Discov. 2012;2:328–343. doi: 10.1158/2159-8290.CD-11-0234. [DOI] [PubMed] [Google Scholar]
- 10.Goidts V., Bageritz J., Puccio L., et al. RNAi screening in glioma stem-like cells identifies PFKFB4 as a key molecule important for cancer cell survival. Oncogene. 2012;31:3235–3243. doi: 10.1038/onc.2011.490. [DOI] [PubMed] [Google Scholar]
- 11.Dasgupta S., Rajapakshe K., Zhu B., et al. Metabolic enzyme PFKFB4 activates transcriptional coactivator SRC-3 to drive breast cancer. Nature. 2018;556:249–254. doi: 10.1038/s41586-018-0018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H., Lu C., Fang M., et al. HIF-1alpha activates hypoxia-induced PFKFB4 expression in human bladder cancer cells. Biochem Biophys Res Commun. 2016;476:146–152. doi: 10.1016/j.bbrc.2016.05.026. [DOI] [PubMed] [Google Scholar]
- 13.Xia X., Lemieux M.E., Li W., et al. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc Natl Acad Sci U S A. 2009;106:4260–4265. doi: 10.1073/pnas.0810067106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ros S., Floter J., Kaymak I., et al. 6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 is essential for p53-null cancer cells. Oncogene. 2017;36:3287–3299. doi: 10.1038/onc.2016.477. [DOI] [PubMed] [Google Scholar]
- 15.Liu L.P., Ho R.L., Chen G.G., Lai P.B. Sorafenib inhibits hypoxia-inducible factor-1alpha synthesis: implications for antiangiogenic activity in hepatocellular carcinoma. Clin Cancer Res. 2012;18:5662–5671. doi: 10.1158/1078-0432.CCR-12-0552. [DOI] [PubMed] [Google Scholar]
- 16.Shu Y., Lu Y., Pang X., et al. Phosphorylation of PPARgamma at Ser84 promotes glycolysis and cell proliferation in hepatocellular carcinoma by targeting PFKFB4. Oncotarget. 2016;7:76984–76994. doi: 10.18632/oncotarget.12764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho D.W.H., Chan L.K., Chiu Y.T., et al. TSC1/2 mutations define a molecular subset of HCC with aggressive behaviour and treatment implication. Gut. 2017;66:1496–1506. doi: 10.1136/gutjnl-2016-312734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng Y., Wu L. mTOR up-regulation of PFKFB3 is essential for acute myeloid leukemia cell survival. Biochem Biophys Res Commun. 2017;483:897–903. doi: 10.1016/j.bbrc.2017.01.031. [DOI] [PubMed] [Google Scholar]
- 19.Cantor J.R., Abu-Remaileh M., Kanarek N., et al. Physiologic medium rewires cellular metabolism and reveals uric acid as an endogenous inhibitor of UMP synthase. Cell. 2017;169:258–272. e17. doi: 10.1016/j.cell.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vande Voorde J., Ackermann T., Pfetzer N., et al. Improving the metabolic fidelity of cancer models with a physiological cell culture medium. Sci Adv. 2019;5:eaau7314. doi: 10.1126/sciadv.aau7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bayer C., Shi K., Astner S.T., et al. Acute versus chronic hypoxia: why a simplified classification is simply not enough. Int J Radiat Oncol Biol Phys. 2011;80:965–968. doi: 10.1016/j.ijrobp.2011.02.049. [DOI] [PubMed] [Google Scholar]
- 22.Shen L., Sun X., Fu Z., et al. The fundamental role of the p53 pathway in tumor metabolism and its implication in tumor therapy. Clin Cancer Res. 2012;18:1561–1567. doi: 10.1158/1078-0432.CCR-11-3040. [DOI] [PubMed] [Google Scholar]
- 23.Masui K., Tanaka K., Akhavan D., et al. mTOR complex 2 controls glycolytic metabolism in glioblastoma through FoxO acetylation and upregulation of c-Myc. Cell Metab. 2013;18:726–739. doi: 10.1016/j.cmet.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denko N.C. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat Rev Cancer. 2008;8:705–713. doi: 10.1038/nrc2468. [DOI] [PubMed] [Google Scholar]
- 25.Hasemann C.A., Istvan E.S., Uyeda K., Deisenhofer J. The crystal structure of the bifunctional enzyme 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase reveals distinct domain homologies. Structure. 1996;4:1017–1029. doi: 10.1016/s0969-2126(96)00109-8. [DOI] [PubMed] [Google Scholar]
- 26.Yuen M.H., Mizuguchi H., Lee Y.H., et al. Crystal structure of the H256A mutant of rat testis fructose-6-phosphate,2-kinase/fructose-2,6-bisphosphatase. Fructose 6-phosphate in the active site leads to mechanisms for both mutant and wild type bisphosphatase activities. J Biol Chem. 1999;274:2176–2184. doi: 10.1074/jbc.274.4.2176. [DOI] [PubMed] [Google Scholar]
- 27.Xu I.M., Lai R.K., Lin S.H., et al. Transketolase counteracts oxidative stress to drive cancer development. Proc Natl Acad Sci U S A. 2016;113:E725–E734. doi: 10.1073/pnas.1508779113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryter S.W., Choi A.M. Heme oxygenase-1: redox regulation of a stress protein in lung and cell culture models. Antioxid Redox Signal. 2005;7:80–91. doi: 10.1089/ars.2005.7.80. [DOI] [PubMed] [Google Scholar]
- 29.Sun X., Ou Z., Chen R., et al. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology. 2016;63:173–184. doi: 10.1002/hep.28251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J., Ney P.A. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–946. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coriat R., Nicco C., Chereau C., et al. Sorafenib-induced hepatocellular carcinoma cell death depends on reactive oxygen species production in vitro and in vivo. Mol Cancer Ther. 2012;11:2284–2293. doi: 10.1158/1535-7163.MCT-12-0093. [DOI] [PubMed] [Google Scholar]
- 32.Chesney J., Clark J., Lanceta L., et al. Targeting the sugar metabolism of tumors with a first-in-class 6-phosphofructo-2-kinase (PFKFB4) inhibitor. Oncotarget. 2015;6:18001–18011. doi: 10.18632/oncotarget.4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brinkman E.K., Chen T., Amendola M., van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soga T., Heiger D.N. Amino acid analysis by capillary electrophoresis electrospray ionization mass spectrometry. Anal Chem. 2000;72:1236–1241. doi: 10.1021/ac990976y. [DOI] [PubMed] [Google Scholar]
- 36.Soga T., Ohashi Y., Ueno Y., et al. Quantitative metabolome analysis using capillary electrophoresis mass spectrometry. J Proteome Res. 2003;2:488–494. doi: 10.1021/pr034020m. [DOI] [PubMed] [Google Scholar]
- 37.Soga T., Ueno Y., Naraoka H., et al. Simultaneous determination of anionic intermediates for Bacillus subtilis metabolic pathways by capillary electrophoresis electrospray ionization mass spectrometry. Anal Chem. 2002;74:2233–2239. doi: 10.1021/ac020064n. [DOI] [PubMed] [Google Scholar]
- 38.Dobin A., Davis C.A., Schlesinger F., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leng N., Dawson J.A., Thomson J.A., et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29:1035–1043. doi: 10.1093/bioinformatics/btt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Specific profiling data and materials described in this study will be made available upon request.