Abstract
Background
SARS-CoV-2 ribonucleic acid (RNA) has been detected in feces, but RNA is not infectious. This systematic review aims to answer if fecal SARS-CoV-2 is experimentally infectious and if evidence of human fecal-oral SARS-CoV-2 transmission exists.
Methods
On September 19, 2022, we searched PubMed, Embase, Web of Science, medRxiv, and bioRxiv. Biomedical studies inoculating SARS-CoV-2 from feces, rectal, or anal swabs in cells, tissue, organoids, or animals were included. Epidemiological studies of groups differing in exposure to fecal SARS-CoV-2 were included. Risk of bias was assessed using standardized tools. Results were summarized by vote counting, tabulation, and a harvest plot. PROSPERO registration no. CRD42020221719.
Results
A total of 4,874 studies were screened; 26 studies were included; and 13 out of 23 biomedical studies (56.5%) succeeded in infection. Two (66.7%) epidemiological studies found limited evidence suggesting fecal-oral transmission. All studies had concerns about the risk of bias.
Conclusions
It is possible to experimentally infect cell cultures, organoids, and animals with fecal SARS-CoV-2. No strong epidemiologic evidence was found to support human fecal-oral transmission. We advise future research to study fecal infectivity at different time points during infection, apply appropriate controls, use in vivo models, and study fecal exposure as a risk factor of transmission in human populations.
Key Words: COVID-19, Feces, Stool, Infection, Gastrointestinal disease, Anal canal
Background
The pandemic coronavirus disease 2019 (COVID-19)1 was caused by the newly emerged severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2; family Coronaviridae, genus Betacoronavirus).2, 3 SARS-CoV-2 is recognized to be transmitted primarily by droplets and aerosols during close contact, and secondarily by fomites.4
Yet, as SARS-CoV-2 ribonucleic acid (RNA) has been detected in the feces of COVID-19 patients, a fecal-oral route of transmission is also considered. A meta-analysis of symptomatic COVID-19 patients reported the prevalence of SARS-CoV-2 RNA reliably detected in fecal samples, rectal, or anal swabs to be 47% (95% confidence interval 38-55).5 The discovery of in-host genomic variability in fecal SARS-CoV-2 RNA suggests ongoing viral evolution depending on active infection of the gastrointestinal tract.6
Direct evidence of enterocyte infection is available, as SARS-CoV-2 RNA and particles were discovered in the biopsies of esophageal, gastric, small intestinal, colonic, and rectal tissues from COVID-19 patients.7, 8, 9, 10, 11 However, the evidence of active infection of intestinal cells along with fecal shedding of SARS-CoV-2 RNA does not necessarily imply fecal-oral transmission to be possible. Reverse transcription polymerase chain reaction (RT-PCR) cannot distinguish transmissible virus particles from fragments with no infectious potential.
Experimentally, human small intestinal and colonic organoids and human colon cell lines have also been successfully infected with SARS-CoV-2.12, 13, 14 Further, intranasal, intragastric, and intratracheal inoculation with SARS-CoV-2 of nonhuman primates led to infection of intestinal segments from the stomach to rectum.15, 16, 17
On this basis, SARS-CoV-2 is now appreciated as an enteric pathogen. During the COVID-19 pandemic, the World Health Organization (WHO) made recommendations for safe sanitation, for example, to limit exposure to feces from infected individuals.18 Medical societies around the world also advised endoscopy units to postpone nonurgent cases and reorganize their workflow to mitigate the risk of virus transmission.19 Though the fecal-oral route today is not considered a main route of infection,4 the possibility of fecal-oral transmission has not yet been ruled out.
The aim of this systematic review was to investigate if SARS-CoV-2 has a potential for fecal-oral transmission by answering if any epidemiologic evidence for or against human fecal-oral transmission exists and if fecally shed SARS-CoV-2 is infectious to culture cells, tissues, organoids, or live animals.
Methods
This systematic review was performed in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses 2020 guidelines.20 The methods were prespecified in a protocol available on PROSPERO (registration no. CRD42020221719).21
Two reviewers identified studies for inclusion by title and abstract screening and full-text screening and then performed data extraction, risk of bias assessment, and certainty of evidence assessment. These activities were undertaken independently, with each reviewer blinded to the other’s decisions. Disagreements were resolved by discussion and consensus. A third reviewer was available for consultation in case of disagreements, though all discrepancies were resolved by consensus.
Literature search
We designed a search strategy using subject headings and free-text words synonymous with or related to “SARS-CoV-2” and “fecal-oral transmission” combined by Boolean operators. It was developed in cooperation with a health care librarian with systematic review expertise. The reproducible search strings are available in Supplementary Appendix 1.
Five electronic databases were searched (platforms in parentheses): Embase (embase.com), PubMed (pubmed.gov), Web of Science Core Collection (webofknowledge.com), bioRxiv (medrxiv.org), and medRxiv (medrxiv.org). Preprint databases were included to minimize the publication bias.
All searches were run on September 19, 2022. Reference lists of the included articles were manually screened to identify additional studies.
The searches were limited to dates of publication ranging from December 31, 2019, onwards. On that day, the WHO China Country Office was for the first time notified of an outbreak of the disease that would later be known as COVID-19.22 No language or study design limits were applied.
Study records
The search results were exported to EndNote version X9.3.3 (Clarivate Analytics). Duplicates were identified using EndNote’s duplicate identification tool. The records were then uploaded to Covidence (Veritas Health Innovation) for study selection and data extraction.
Study selection
Studies fulfilling the following criteria were included:
-
–
Epidemiologic studies of groups differing in exposure to fecally shed SARS-CoV-2. An outcome should be RT-PCR–confirmed SARS-CoV-2 infection.
-
–
Biomedical studies of cell, tissue, organoid, or live animal inoculation with SARS-CoV-2 obtained from human or animal feces, rectal, or anal swabs. The outcome of infection should be identified by use of electron microscopy or change in viral titer by quantitative RT-PCR.
When the results of the outcome of interest were not sufficiently reported, the study was still eligible, and the corresponding authors were contacted via e-mail (maximum 2 e-mail attempts, time limit of clarification was 1 week).
Published and preprint reports were included.
We excluded abstracts, consensus documents, editorials, exercises, features, forums, guidance, highlights, images, insights, interviews, news, opinions, panels, personal views, perspectives, podiums, policies, practices, protocols, seminars, rapid reviews, reviews, symposia, and previous systematic reviews.
We also excluded comments or commentaries, correspondences, rapid and short reports, and letters to the editor presenting no original results.
Reports in languages other than English were excluded, but relevant titles are listed in the results.
Data extraction
Items recorded from the included studies are listed in Supplementary Table S1.
Risk of bias assessment for individual studies
Two different tools were used. The tool for epidemiologic studies was developed with inspiration from tools of the Joanna Briggs Institute to be suited for different study designs.23 A new tool for biomedical studies was developed. Both are available in Supplementary Appendix 2.
To assess potential outcome-reporting biases, analyses specified in the methods sections of the included studies, and protocols if available, were compared to the outcomes.
The risk of bias assessment did not affect the final inclusion.
Data synthesis
We used vote counting for data summary. Effect estimates were categorized into binary variables (evidence for or against fecal-oral transmission) and reported as proportions. The included studies were tabulated and summarized in a harvest plot.24
Certainty of evidence
Certainty of evidence was assessed by a Grading of Recommendations Assessment, Development, and Evaluation–inspired approach.25 Four levels of certainty were used: high, moderate, low, and very low. Beginning from high certainty, we voted the level a step down when 1 of the following criteria were met: risk of bias assessments (if more than half of the included studies were assessed medium or high); imprecision (if either epidemiologic or biomedical evidence came from only 1 or 2 small studies); inconsistency (if not a large proportion of the included studies, ie, more than 85%, agreed on the risk of fecal-oral transmission); and indirectness (if most outcomes were surrogates for fecal-oral transmission). As publication bias was not possible to estimate because of the binary effect estimate, this was not included.
Results
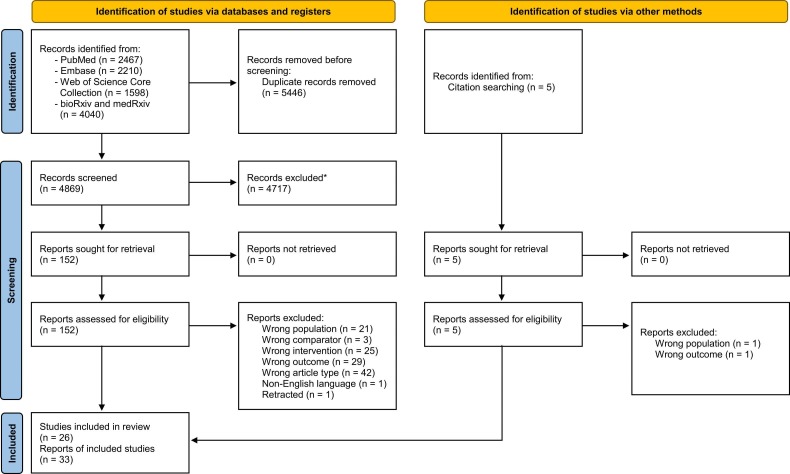
The systematic search identified 10,315 records ( Fig 1). After duplicates removal we screened 4,869 records, from which we reviewed 152 full text documents and finally included 30 reports of 24 studies.10, 14, 15, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52 A list of studies excluded during full-text screening is available in Supplementary Appendix 3 along with the reasons for exclusion. One possible eligible record in Chinese was identified, but was excluded because of non-English language.53 Later, we searched the references of the included reports and other reviews and thus identified 3 reports of 2 additional studies eligible for inclusion.54, 55, 56
Fig. 1.
Flowchart from record identification to study inclusion. *Duplicates were removed using the duplicate identification tool of EndNote version X9.3.3 (Clarivate Analytics) and then manually. Figure setup from Page et al.20
Across all study designs, 15 of 26 included studies (57.7%) provided any evidence favoring possible fecal-oral transmission of SARS-CoV-2.
Epidemiologic studies of fecal-oral SARS-CoV-2 transmission
Three epidemiologic studies were eligible for inclusion ( Table 1).
Table 1.
Characteristics of the included epidemiologic studies
| Source | Publication date* | Site (city or province, country) | Study design | Exposed |
Unexposed |
Group definition | Group comparability | Support of fecal-oral hypothesis† | Risk of bias | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Infected | Uninfected | Infected | Uninfected | ||||||||
| Al Mayahi ZK et al26 | 05/15/2021 | Rustaq, Oman | Case series | 4 | 0 | 34 | 2 | Exposed group cleaned up diarrhea from a COVID-19-positive patient. | Health workers in the same hospital ward | Yes | Medium |
| Isanovic M et al38 | 07/04/2022 | Columbia, SC, USA | Cohort study | 1 | 42 | 10,915 | 352,799 | Exposed group worked at a wastewater treatment plant having ½ 12 h daily on facility grounds. | Same area | No | Medium |
| Kang M et al40 | 09/01/2020 | Guangzhou, China | Cross-sectional | 4 | 57 | 0 | 136 | Exposed group lived vertically aligned to the index patient in flats connected by bathroom drainage pipes. | Same apartment complex with similar restrictive measures | Yes | Medium |
MM/DD/YYYY.
Conclusion by the study authors.
In 2 out of 3 studies (66.7%), the authors concluded fecal-oral SARS-CoV-2 transmission to be a possible explanation of the described spread of infection. Al Mayahi et al described a cluster of COVID-19 patients in a hospital.26 After a patient developed diarrhea, all 4 health care workers who cleaned up a large spill of loose stool were subsequently infected. Of the remaining health care workers exposed to SARS-CoV-2–positive patients, 2 out of 36 became infected. Kang et al described a COVID-19 dissemination in a high-rise building; all infected persons lived in flats connected by dried-out traps to a shared drainage pipe.40 In all flats not connected to this drainage pipe, no one became infected.
In 1 study, the authors describe the risk of infection from wastewater as minimal. Isanovic et al followed a group of wastewater treatment plant workers for 6 months, testing them for SARS-CoV-2 every 2 weeks.38 They found no significant difference between the worker case rate and that of the surrounding population.
Biomedical studies of fecal-oral SARS-CoV-2 transmission
23 studies were eligible for inclusion, inoculating cells, tissues, organoids, or animals in vivo with fecal SARS-CoV-2 ( Table 2).
Table 2.
Characteristics of the included biomedical studies
| Source | Publication date* | Site (city or province, country) | Sample type | Sample origin | Sample note | Donors | Samples | Sample Ct-range | Inoculation test system | Outcome measure | Infection success rate† | Risk of bias |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Albert S et al27, 28 | 05/11/2021 | Valencia, Spain | Feces | Human | In Sigma Virocult at 4 °C for a maximum of 24 h before processing | 5 | 8 | 31.8-41.3 | Cells, Vero E6 | qPCR | 0/8 | Medium |
| Audsley JM et al29 | 05/10/2021 | Melbourne, Australia | Feces | Human | Placed in viral transport medium | 1 | 2 | 23-36 | Cells, Vero/hSLAM | qPCR | 1/2 | Medium |
| Bailie CR et al30 | 02/10/2021 | Melbourne, Australia | Rectal swab | Human | N/A | 2 | 2 | 37-38 | Cells, Vero/hSLAM | qPCR, TEM | 0/2 | High |
| Barroso-Arévalo S et al31 | 07/15/2021 | Madrid, Spain | Rectal swab | Dog | Placed in viral transport medium | 1 | 1 | 26.34 | Cells, Vero E6 | qPCR | 1/1 | High |
| Binder RA et al32 | 09/09/2020 | Durham, NC, USA | Rectal swab | Human | Self-collected. Placed in viral transport medium | 3 | 3 | 33.3-38.6 | Cells, Vero E6 | qPCR | 0/3 | Medium |
| Cerrada-Romero C al33 | 05/05/2022 | Madrid, Spain | Feces | Human | N/A | 27 | 31 | 24.3-39.6 | Cells, Vero E6 | qPCR | 0/31 | High |
| Das Adhikari U et al10 | 07/10/2021 | Boston, MA, USA | Feces | Human | N/A | 5 | 5 | 4.5 × 103-7.2 × 107 (RNA copies/mL) | Cells, Vero E6 | qPCR | 3/5 | Medium |
| Dergham J et al34 | 06/18/2021 | Marseilles, France | Feces | Human | Stored at −80 °C | 1 | 2 | 16.22-17.29 | Cells, Vero E6 | qPCR | 2/2† | Medium |
| Fumian TM et al35 | 01/09/2021 | Rio de Janeiro, Brazil | Feces | Human | Stored at −80 °C for up to 7 months | 4 | 4 | 29.8-32.4 | Cells, Vero E6 | qPCR | 0/4 | High |
| Gortázar C et al36, 37 | 06/12/2021 | Castilla-La Mancha, Spain | Rectal swab | Ferret | N/A | 1 | 1 | 34.5 | Cells, Vero E6 | qPCR | 1/1 | High |
| Jeong HW et al39 | 07/23/2020 | Cheongju, Korea | Feces | Human | N/A | 1 | 1 | 2.2 (log10 RNA copies/mL) | In vivo, Ferrets | qPCR | 1/1‡ | High |
| Jiao, L et al15 | 12/08/2020 | Yunnan, China | Feces | Rhesus monkey | N/A | N/A | N/A | N/A | Cells, Vero E6 | qPCR, TEM | 1/N/A | High |
| Kim JM et al41 | 05/08/2020 | N/A, Korea | Feces | Human | N/A | 8 | 13 | 27-27,310 (RNA copies/µL) | Cells, CaCo-2 | qPCR | 0/13 | High |
| McAloose D et al42, 43, 44, 45 | 10/13/2020 | Bronx, NY, USA | Feces | Tiger, lion§ | Stored at −80 °C | 7 | 14 | 22.3-36.3** | Cells, Vero (CCL-81) Vero 76 Vero E6 |
qPCR | 2/14 | High |
| Pedersen RM et al46 | 11/09/2021 | Odense, Denmark | Rectal swab | Human | N/A | N/A | 12, 13†† | 25-39‡‡ | Cells, CaCo-2 Vero E6 |
qPCR | 0/12 0/13 |
Medium |
| Wang W et al47 | 03/11/2020 | Hubei, Shandong and Beijing, China | Feces | Human | N/A | N/A | 4 | 22.3-38.4§§ | N/A | TEM | 2/4 | High |
| Wölfel R et al48 | 04/01/2020 | Munich, Germany | Feces | Human | Shipped in native conditions | 4 | 13 | 3.5-7.7 (log10 RNA copies/g)‡‡ | Cells, Vero E6 | qPCR | 0/13 | High |
| Wurtzer S et al49 | 08/15/2022 | Ivry-sur-Seine, France | Feces | Golden Syrian hamster | Inoculation within 30 min of collection | 2 | 2 | 4 × 106-6 × 107 (RNA copies/mg) | In vivo, Golden Syrian Hamster | qPCR | 0/2 | Medium |
| Xiao F et al50 | 05/18/2020 | Guangzhou, China | Feces | Human | N/A | 1 | N/A | 23.34 | Cells, Vero E6 | TEM | 1/1 | High |
| Yao H et al54, 55 | 10/29/2020 | Zhejiang, China | Feces | Human | Viral isolation within 4 h from sampling | 3 | 3 | <28 | Cells, Vero (CCL-81) | qPCR | 3/3 | High |
| Young BE et al56 | 08/28/2020 | Singapore | Feces | Human | Shipped in native conditions | 7 | 7 | N/A | Cells, Vero E6 | qPCR | 0/7 | High |
| Zhang Y et al51, 52 | 11/25/2020 | Beijing, China | Feces | Human | N/A | 1/1 | 1/1 | 24 | Cells, Vero | TEM | 1/1 | High |
| Zhou J et al14 | 05/13/2020 | Hong Kong, China | Feces | Human | From an archive | 1 | 1 | 33.6 | Organoids, Human enteric | qPCR | 1/1 | Medium |
Ct-range, cycle threshold-range; N/A, not available; qPCR, reverse transcription quantitative polymerase chain reaction; TEM, transmission electron microscopy.
MM/DD/YYYY. Publication date of the first peer-reviewed report.
Number of samples resulting in infection out of the total samples inoculated. Except for the lack of clarity in Dergham et al,34 in all studies doing replicates, all replicates of each sample had the same outcome.
Data from correspondence with the study authors.
1 Malayan tiger (2 samples), 3 Amur tigers (6 samples), and 3 African lions (6 samples).
Only available for 7 of 14 samples.
12 samples inoculating Caco-2 cells and 13 samples inoculating Vero E6 cells. Not clear if some of these are the same samples.
Read from the figure.
Range for all 44 positive fecal samples. Only 4 samples with low Ct were used for inoculation.
In 13 out of 23 studies (56.5%), SARS-CoV-2 from at least 1 fecal sample or rectal swab succeeded in infection (20 of 145 samples, 13.8%).
The most frequent method was inoculation of different Vero lineage cells (18 studies); using this method, 10 studies succeeded in infection (16 out of 112 samples, 14.3%). The most frequent sample type was feces (18 studies); using this sample type, 11 studies succeeded in infection (18 out of 113 samples, 15.9%). 18 studies used human samples for inoculation, with 9 of these (50.0%) succeeding infection (15 out of 126 samples, 11.9%).
Only the studies by Kim et al and Pedersen et al included infections of cell cultures from other than the Vero-lineage; in total, 25 inoculations of CaCo-2 cells led to zero infections.41, 46 Zhou et al successfully made the only inoculation of organoids.14 Two studies made inoculations in vivo; Jeong et al used live ferrets with a successful outcome39; Wurtzer et al inoculated live golden Syrian hamsters unsuccessfully.49
Four studies mentioned doing replicates, all of which had the same outcome for every individual sample. 15, 39, 46, 49
Risk of bias assessment
There were concerns about the overall risk of bias for all included studies (26/26), with 15 of these assessed as having a high risk of bias (Supplementary Table S2 and Supplementary Appendix 2).
In all included studies the outcomes reported in methods and results were without major discrepancies. All studies were reported in peer-reviewed journals, though some were identified also in preprint reports.28, 37, 43, 45, 54
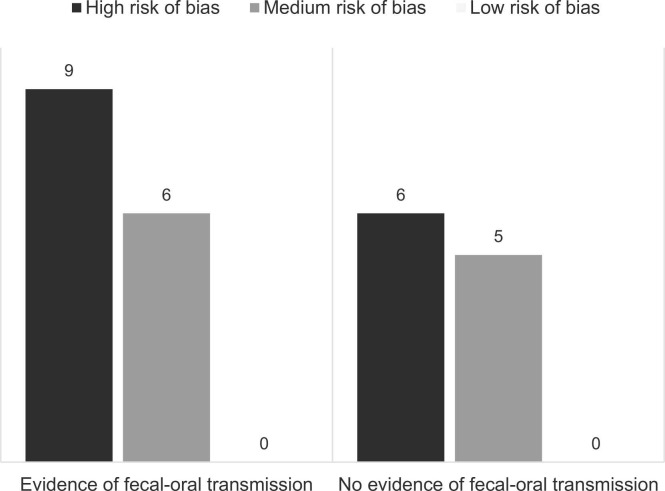
For a combined overview of the reviewed studies providing evidence for and against the possibility of fecal-oral transmission of SARS-CoV-2, see Figure 2.24 The concern of risk of bias seems to be equally distributed between evidence for and against fecal-oral SARS-CoV-2 transmission.
Fig. 2.
Harvest plot of the included studies. A combined overview of the evidence of fecal-oral SARS-CoV-2 transmission from all included studies. Inspired by Ogilvie et al.24 Left panel: either biomedical studies with successful infection of cells, tissues, organoids, or animals by fecal SARS-CoV-2 (success rate ≥ 1/n) or epidemiologic studies concluding their results are supporting the hypothesis of fecal-oral SARS-CoV-2 transmission. Right panel: the opposite.
Certainty of evidence
The certainty of the evidence for fecal-oral transmission of SARS-CoV-2 was judged low. The evidence was downgraded 1 step each for risk of bias assessment, inconsistency, and indirectness.
Discussion
This study set out to review the available biomedical and epidemiologic evidence of fecal-oral SARS-CoV-2 transmission. We found that fecally shed SARS-CoV-2 can infect culture cells and organoids in vitro and ferrets and golden Syrian hamsters in vivo, and that fecal-oral transmission is backed up by limited epidemiologic evidence (Table 1, Table 2).
The epidemiologic evidence for fecal-oral SARS-CoV-2 transmission can be described as circumstantial. Kang et al reported probable fecal-oral transmission of SARS-CoV-2 by bioaerosols through a drainage pipe in a high-rise building in China.40 Although the authors described the circumstances in great detail, including tracer gas testing, the possibility of other routes of transmission still exists. As a study reported isolation of infectious virus from plastic and steel surfaces for up to 72 hours,57 undetected fomite transmission might still be rendered probable. The retrospective study design further implies the risk of unreported meetings between the affected families.
Al Mayahi et al reported how a group of health care workers with high exposure to feces from COVID-19 patients were infected. The authors emphasize other routes of infection to be equally possible. As Kang et al and Al Mayahi et al only described situations and did not apply any statistics, it is difficult to quantitate the outcome and estimate the likelihood of observing the reported situation if fecal-oral SARS-CoV-2 transmission is not possible. Also considering our risk of bias assessment of both studies to be medium, we find it difficult to conclude anything from these 2 studies.
Isanovic et al reported how there was no difference between SARS-CoV-2 infection in a wastewater treatment plant and the surrounding community. A recent study found a higher prevalence of certain gastrointestinal pathogens in fecal samples from wastewater treatment plant workers than from the more distant surrounding population.58 This may be explained by the increased exposure to droplets, hand-to-mouth contact, or inhalation of aerosols. Infectious SARS-CoV-2 has been isolated from aerosols for up to 3 hours, which renders the fecal bioaerosol transmission hypothesis possible.57 In their study, Isanovic et al detected a fecal indicator virus in all air samples collected on the treatment plant grounds, but no SARS-CoV-2 RNA.
Most of the included biomedical studies used Vero E6 cell assays or other Vero cell lines to evaluate infectivity. Vero E6 cells have been used for SARS-CoV-1 research by many laboratories.59 Accordingly, the cell line was a natural choice for SARS-CoV-2 research, although it has been suggested that Vero E6 cells are less susceptible to SARS-CoV-2 infection than enteroids14 or human primary airway epithelial cells.2
It is interesting how Jeong et al succeeded in infection of ferrets but were not able to determine the outcome from infection of Vero cells because of cell toxicity.39 Ferrets and golden Syrian hamsters have been found to be excellent animal models of SARS-CoV-2 infection and transmission.60, 61 Generally, animals may be more relevant models of disease transmission between humans compared to cell cultures.
The quantity of SARS-CoV-2 in the swabs or samples is most likely important for the probability of isolation. A significant negative relation between the RT-PCR cycle threshold (Ct) value and culture positivity of Vero E6 cells has been discovered elsewhere,62 and 2 studies found that only respiratory samples with an RT-PCR Ct value below 24 or 30 were capable of successfully infecting Vero CCL-81 cells63 or Vero E6 cells,56 respectively. The relation between Ct value and culture positivity is supported by our findings, as only 2 included studies succeeded in infection with a sample Ct greater than 30.14, 36 In this context, it should be noted that Ct values of different samples from COVID-19 patients vary over the course of the disease.64
Studies of respiratory samples have found isolation of infectious SARS-CoV-2 to be possible for significantly fewer days than SARS-CoV-2 RNA detection.65 This supports the hypothesis that the timing of sampling may be important. Because of inconsistent reporting of symptom onset and no systematic sampling across the included studies, no specific conclusions can be made on the temporal distribution of the probability of detecting infectious fecal SARS-CoV-2.
This study had several strengths. A wide search strategy helped in retrieving a high number of results. Searching several databases including preprint servers limited the risk of publication bias. To further limit the bias, 2 researchers did all the screening, data extraction, bias assessments, and certainty of evidence assessment separately and blinded to each other’s decisions. Our results contribute to the existing knowledge by giving a comprehensive summary of the current research on SARS-CoV-2 fecal-oral transmission.
Also, this study had some limitations. First, the evidence presented in this manuscript neither excludes nor proves fecal-oral transmission of SARS-CoV-2 between humans. However, several biomedical studies have confirmed the hypothetical possibility. Second, in the screening process, the reviewers were not blinded to journal titles, study authors, or their institutions. Third, vote counting provides no information on the magnitude of effects, and it does not account for differences in the relative sizes of included studies.66 However, vote counting was assessed the best method, as there was no consistent effect measure. Fourth, to increase the certainty in diagnosis, only epidemiologic studies of RT-PCR-confirmed participants were included. This resulted in exclusion of studies based on seropositivity as a marker of previous infection (see Supplementary Appendix 3). It may therefore be possible to locate more epidemiologic evidence, although the risk of bias from reduced diagnostic accuracy will then be present.
Implications for practice and policy
Generally, findings from this systematic review support the biomedical hypothesis of infectious SARS-CoV-2 in feces but found only limited circumstantial evidence of documented fecal-oral transmission between humans. As the overall certainty of evidence was judged low, we emphasize that fecal-oral SARS-CoV-2 transmission between humans is still only hypothetical. It could be discussed if, for example, the WHO recommendations on sanitation18 and the medical society recommendations on endoscopy workflow19 are necessary as precautionary measures to fecal-oral transmission.
Implications for research
There are a couple of important questions that should be addressed in future research:
-
1)
We propose larger and more systematic cell culture infection studies assessing the timeframe from symptom onset to when it is possible to isolate infectious SARS-CoV-2 from feces.
-
2)
As all but 2 included cell inoculation studies did not apply positive and negative controls, we emphasize the necessity of appropriate controls to ensure the reliability of cell assays.
-
3)
We recommend testing the fecal-oral hypothesis by natural exposure of animals to feces from infected individuals. This will provide stronger evidence on whether exposure to feces containing infectious viral particles actually may lead to infection of organisms more complex than culture cells.
-
4)
Also, we propose controlled epidemiologic studies examining shared toilets and direct exposure to human feces as possible risk factors of SARS-CoV-2 transmission. This may provide the most transferable knowledge on the effect of efforts taken to limit fecal-oral transmission.
Conclusions
In conclusion, the present systematic review has found 3 epidemiologic studies, of which 2 studies based on circumstantial evidence supported the hypothesis of possible fecal-oral SARS-CoV-2 transmission and 1 cohort study did not. Further, we found 23 studies experimentally inoculating culture cells, organoids, and live ferrets and hamsters with fecally shed SARS-CoV-2, leading to successful infection in 13 studies (56.5%).
Although this systematic review found fecal SARS-CoV-2 to be infectious, the study found no strong direct evidence for fecal-oral transmission of SARS-CoV-2 between humans. Because of comprehensive concerns of the risk of bias in the included studies and the majority of evidence being experimental infection of culture cells, this study, only with low certainty, supports that SARS-CoV-2 has a potential for fecal-oral transmission between humans.
Ethical approval statement
Ethical approval was not required because this study retrieved and synthesized data from already published studies.
Acknowledgments
We thank Professor Henrik Dimke of the Department of Molecular Medicine, University of Southern Denmark, for making his expertise available to resolve disagreements in the title and abstract screening, data extraction, risk of bias assessment, and certainty of evidence assessment though this was never necessary. We also thank librarian Helene Sognstrup of Aarhus University Library Health Sciences for providing her expertise on designing the search strategy.
Footnotes
Funding/support: This work was supported by Aarhus University Research Foundation (grant number 30714).
Conflicts of interest: None to report.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.ajic.2023.04.170.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
References
- 1.World Health Organization [internet]. WHO Director-General's opening remarks at the media briefing on COVID-19; 2020. Accessed December 18, 2022. https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020.
- 2.Zhu N., Zhang D., Wang W., et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization [internet]. Technical guidance: Naming the coronavirus disease (COVID-19) and the virus that causes it; 2020. Accessed December 18, 2022. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease.
- 4.Center for Disease Control and Prevention [internet]. Scientific Brief: SARS-CoV-2 Transmission; 2021. Accessed December 18, 2022. https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/sars-cov-2-transmission.html. [PubMed]
- 5.Zhou J.Q., Liu G.X., Huang X.L., Gan H.T. The importance of fecal nucleic acid detection in patients with coronavirus disease (COVID-19): a systematic review and meta-analysis. J Med Virol. 2022;94:2317–2330. doi: 10.1002/jmv.27652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu Y., Kang L., Shen Z., et al. Dynamics of severe acute respiratory syndrome coronavirus 2 genome variants in the feces during convalescence. J Genet Genom. 2020;47:610–617. doi: 10.1016/j.jgg.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin L., Jiang X., Zhang Z., et al. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 2020;69:997–1001. doi: 10.1136/gutjnl-2020-321013. [DOI] [PubMed] [Google Scholar]
- 8.Qian Q., Fan L., Liu W., et al. Direct evidence of active SARS-CoV-2 replication in the intestine. Clin Infect Dis. 2021;73:361–366. doi: 10.1093/cid/ciaa925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das Adhikari U., Eng G., Farcasanu M., et al. Fecal severe acute respiratory syndrome coronavirus 2 (SARS-Cov-2) RNA is associated with decreased coronavirus disease 2019 (COVID-19) survival. Clin Infect Dis. 2022;74:1081–1084. doi: 10.1093/cid/ciab623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Livanos A.E., Jha D., Cossarini F., et al. Intestinal host response to SARS-CoV-2 infection and COVID-19 outcomes in patients with gastrointestinal symptoms. Gastroenterology. 2021;160:2435–2450. doi: 10.1053/j.gastro.2021.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamers M.M., Beumer J., van der Vaart J., et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stanifer M.L., Kee C., Cortese M., et al. Critical role of type III interferon in controlling SARS-CoV-2 infection in human intestinal epithelial cells. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.107863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou J., Li C., Liu X., et al. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat Med. 2020;26:1077–1083. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
- 15.Jiao L., Li H., Xu J., et al. The gastrointestinal tract is an alternative route for SARS-CoV-2 infection in a nonhuman primate model. Gastroenterology. 2021;160:1647–1661. doi: 10.1053/j.gastro.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng W., Bao L., Liu J., et al. Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Science. 2020;369:818–823. doi: 10.1126/science.abc5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munster V.J., Feldmann F., Williamson B.N., et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585:268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.World Health Organization [internet]. Water, sanitation, hygiene, and waste management for SARS-CoV-2, the virus that causes COVID-19; 2020. Accessed December 18, 2022. https://www.who.int/publications/i/item/WHO-2019-nCoV-IPC-WASH-2020.4.
- 19.Lui R.N., Wong S.H., Sánchez-Luna S.A., et al. Overview of guidance for endoscopy during the coronavirus disease 2019 pandemic. J Gastroenterol Hepatol. 2020;35:749–759. doi: 10.1111/jgh.15053. [DOI] [PubMed] [Google Scholar]
- 20.Page M.J., McKenzie J.E., Bossuyt P.M., et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:n71. doi: 10.1136/bmj.n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.PROSPERO: International prospective register of systematic reviews [internet]. Frische S, Termansen MB, Dimke H. Fecal-oral transmission of SARS-CoV-2? A systematic review of studies of epidemiology and viral infectivity; 2020. Accessed December 18, 2022. https://www.crd.york.ac.uk/prospero/display_record.php?ID=CRD42020221719.
- 22.World Health Organization [internet]. Pneumonia of unknown caus—China; 2020. Accessed December 18, 2022. https://www.who.int/csr/don/05-january-2020-pneumonia-of-unkown-cause-china/en/.
- 23.Joanna Briggs Institute: Faculty of health and medical sciences at University of Adelaide [internet]. Critical appraisal tools. Accessed December 18, 2022. https://jbi.global/critical-appraisal-tools.
- 24.Ogilvie D., Fayter D., Petticrew M., et al. The harvest plot: a method for synthesising evidence about the differential effects of interventions. BMC Med Res Methodol. 2008;8:8. doi: 10.1186/1471-2288-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guyatt G.H., Oxman A.D., Vist G.E., et al. GRADE: an emerging consensus on rating quality of evidence and strength of recommendations. BMJ. 2008;336:924–926. doi: 10.1136/bmj.39489.470347.AD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al Mayahi Z.K., Al Kindi N., Al Shaqsi N., et al. Non-respiratory droplet transmission of COVID-19 in the isolation ward of a secondary hospital in Oman: a return to isolation basics. Infect Dis Clin Pract. 2021;29:e371–e375. doi: 10.1097/IPC.0000000000001022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Albert S., Ruíz A., Pemán J., Salavert M., Domingo-Calap P. Lack of evidence for infectious SARS-CoV-2 in feces and sewage. Eur J Clin Microbiol Infect Dis. 2021;40:2665–2667. doi: 10.1007/s10096-021-04304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Albert S., Ruíz A., Pemán J., Salavert M., Domingo-Calap P. Lack of evidence for infectious SARS-CoV-2 in feces and sewage. medRxiv. 2021 doi: 10.1101/2021.05.11.21256886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Audsley J., Holmes N.E., Mordant F.L., et al. Temporal differences in culturable severe acute respiratory coronavirus virus 2 (SARS-CoV-2) from the respiratory and gastrointestinal tracts in a patient with moderate coronavirus disease 2019 (COVID-19) Infect Control Hosp Epidemiol. 2021;43:1286–1288. doi: 10.1017/ice.2021.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bailie C.R., Franklin L., Nicholson S., et al. Symptoms and laboratory manifestations of mild COVID-19 in a repatriated cruise ship cohort. Epidemiol Infect. 2021;149 doi: 10.1017/S0950268821000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barroso-Arévalo S., Rivera B., Domínguez L., Sánchez-Vizcaíno J.M. First detection of SARS-CoV-2 B.1.1.7 variant of concern in an asymptomatic dog in Spain. Viruses. 2021;13:1379. doi: 10.3390/v13071379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Binder R.A., Alarja N.A., Robie E.R., et al. Environmental and aerosolized severe acute respiratory syndrome coronavirus 2 among hospitalized coronavirus disease 2019 patients. J Infect Dis. 2020;222:1798–1806. doi: 10.1093/infdis/jiaa575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cerrada-Romero C., Berastegui-Cabrera J., Camacho-Martínez P., et al. Excretion and viability of SARS-CoV-2 in feces and its association with the clinical outcome of COVID-19. Sci Rep. 2022;12:7397. doi: 10.1038/s41598-022-11439-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dergham J., Delerce J., Bedotto M., La Scola B., Moal V. Isolation of viable SARS-CoV-2 virus from feces of an immunocompromised patient suggesting a possible fecal mode of transmission. J Clin Med. 2021;10:2696. doi: 10.3390/jcm10122696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fumian T.M., Malta F.C., Dos Santos D.R.L., et al. SARS-CoV-2 RNA detection in stool samples from acute gastroenteritis cases, Brazil. J Med Virol. 2021;93:2543–2547. doi: 10.1002/jmv.26786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gortázar C., Barroso-Arévalo S., Ferreras-Colino E., et al. Natural SARS-CoV-2 infection in kept ferrets, Spain. Emerg Infect Dis. 2021;27:1994–1996. doi: 10.3201/eid2707.210096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gortázar C., Barroso-Arévalo S., Ferreras-Colino E., et al. Natural SARS-CoV-2 infection in kept ferrets, Spain. bioRxiv. 2021 doi: 10.3201/eid2707.210096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Isanovic M., Correa Velez K.E., Norman R.S. Dispersion of SARS-CoV-2 RNA across a wastewater treatment plant and its workers. Water Environ J. 2022;36:713–722. doi: 10.1111/wej.12812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeong H.W., Kim S.M., Kim H.S., et al. Viable SARS-CoV-2 in various specimens from COVID-19 patients. Clin Microbiol Infect. 2020;26:1520–1524. doi: 10.1016/j.cmi.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang M., Wei J., Yuan J., et al. Probable evidence of fecal aerosol transmission of SARS-CoV-2 in a high-rise building. Ann Intern Med. 2020;173:974–980. doi: 10.7326/M20-0928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim J.M., Kim H.M., Lee E.J., et al. Detection and isolation of SARS-CoV-2 in serum, urine, and stool specimens of COVID-19 patients from the Republic of Korea. Osong Public Health Res Perspect. 2020;11:112–117. doi: 10.24171/j.phrp.2020.11.3.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartlett S.L., Diel D.G., Wang L., et al. SARS-CoV-2 infection and longitudinal fecal screening in Malayan Tigers (Panthera tigris jacksoni), Amur Tigers (Panthera tigris altaica), and African Lions (Panthera leo krugeri) at the Bronc Zoo, New York, USA. J Zoo Wildl Med. 2021;51:733–744. doi: 10.1638/2020-0171. [DOI] [PubMed] [Google Scholar]
- 43.Bartlett S.L., Diel D.G., Wang L., et al. SARS-CoV-2 infection and longitudinal fecal screening in Malayan Tigers (Panthera tigris jacksoni), Amur Tigers (Panthera tigris altaica), and African Lions (Panthera leo krugeri) at the Bronc Zoo, New York, USA. bioRxiv. 2020 doi: 10.1638/2020-0171. [DOI] [PubMed] [Google Scholar]
- 44.McAloose D., Laverack M., Wang L., et al. From people to Panthera: natural SARS-CoV-2 infection in Tigers and Lions at the Bronx Zoo. mBio. 2020;11 doi: 10.1128/mBio.02220-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McAloose D., Laverack M., Wang L., et al. From people to Panthera: natural SARS-CoV-2 infection in Tigers and Lions at the Bronx Zoo. bioRxiv. 2020 doi: 10.1128/mBio.02220-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pedersen R.M., Tornby D.S., Bang L.L., et al. Rectally shed SARS-CoV-2 in COVID-19 inpatients is consistently lower than respiratory shedding and lacks infectivity. Clin Microbiol Infect. 2022;28(2):304.E1–304.E3. doi: 10.1016/j.cmi.2021.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang W., Xu Y., Gao R., et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wölfel R., Corman V.M., Guggemos W., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 49.Wurtzer S., Lacote S., Murri S., et al. Reduction in SARS-CoV-2 virus infectivity in human and hamster feces. Viruses. 2022;14:1777. doi: 10.3390/v14081777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao F., Sun J., Xu Y., et al. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg Infect Dis. 2020;26:1920–1922. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y., Chen C., Song Y., et al. Excretion of SARS-CoV-2 through faecal specimens. Emerg Microbes Infect. 2020;9:2501–2508. doi: 10.1080/22221751.2020.1844551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y., Chen C., Zhu S., et al. Isolation of 2019-nCoV from a stool specimen of a laboratory-confirmed case of the coronavirus disease 2019 (COVID-19) China CDC Wkly. 2020;2:123–124. [PMC free article] [PubMed] [Google Scholar]
- 53.Xu K., Cai H., Shen Y., et al. Management of COVID-19: the Zhejiang experience. J Zhejiang Univ. 2020;49:147–157. doi: 10.3785/j.issn.1008-9292.2020.02.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yao H., Lu X., Chen Q., et al. Patient-derived mutations impact pathogenicity of SARS-CoV-2. medRxiv. 2020 [Google Scholar]
- 55.Yao H., Lu X., Chen Q., et al. Patient-derived SARS-CoV-2 mutations impact viral replication dynamics and infectivity in vitro and with clinical implications in vivo. Cell Discov. 2020;6:76. doi: 10.1038/s41421-020-00226-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Young B.E., Ong S.W.X., Ng L.F.P., et al. Viral dynamics and immune correlates of coronavirus disease 2019 (COVID-19) severity. Clin Infect Dis. 2021;73:e2932–e2942. doi: 10.1093/cid/ciaa1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Doremalen N., Bushmaker T., Morris D.H., et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodríguez-Molina D., Berglund F., Blaak H., et al. Carriage of ESBL‑producing Enterobacterales in wastewater treatment plant workers and surrounding residents—the AWARE Study. Eur J Clin Microbiol Infect Dis. 2021 doi: 10.1007/s10096-021-04387-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ogando N.S., Dalebout T.J., Zevenhoven-Dobbe J.C., et al. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J Gen Virol. 2020;101:925–940. doi: 10.1099/jgv.0.001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim Y.I., Kim S.G., Kim S.M., et al. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe. 2020;27:704–709. doi: 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sia S.F., Yan L.M., Chin A.W.H., et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Singanayagam A., Patel M., Charlett A., et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bullard J., Dust K., Funk D., et al. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin Infect Dis. 2020;71:2663–2666. doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zheng S., Fan J., Yu F., et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369:m1443. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Kampen J.J.A., van de Vijver D.A.M.C., Fraaij P.L.A., et al. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19) Nat Commun. 2021;12:267. doi: 10.1038/s41467-020-20568-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Borenstein M., Rothstein H., Hedges L., Higgins J. In: Introduction to Meta-Analysis. 1st ed. Borenstein M., Rothstein H., Hedges L., Higgins J., editors. John Wiley & Sons, Ltd.; 2009. Vote counting—a new name for an old problem; pp. 251–255. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material