Abstract
An essential function of the epidermis is to provide a physical barrier that prevents the loss of water. Essential mediators of this barrier function include ceramides, cholesterol, and very long chain fatty acids, and their alteration causes human pathologies, including psoriasis and atopic dermatitis. A frameshift mutation in the human ZNF750 gene, which encodes a zinc finger transcription factor, has been shown to cause a seborrhea-like dermatitis. Here, we show that genetic deletion of the mouse homolog ZFP750 results in loss of epidermal barrier function, which is associated with a substantial reduction of ceramides, nonpolar lipids. The alteration of epidermal lipid homeostasis is directly linked to the transcriptional activity of ZFP750. ZFP750 directly and/or indirectly regulates the expression of crucial enzymes primarily involved in the biosynthesis of ceramides. Overall, our study identifies the transcription factor ZFP750 as a master regulator epidermal homeostasis through lipid biosynthesis and thus contributing to our understanding of the pathogenesis of several human skin diseases.
The transcription factor ZFP750 is crucial for skin homeostasis, acting as a master regulator of lipid synthesis.
INTRODUCTION
The epidermis is a multilayered tissue that has a crucial function in protecting the organism from water loss, xenobiotics, ultraviolet light, and harmful pathogens (1–4). In particular, the outermost layer of the epidermis, the stratum corneum (SC), provides a permeability barrier that prevents loss of water and electrolytes. The SC is composed of enucleated fully differentiated keratinocytes (corneocytes), which are embedded in an extracellular matrix of neutral lipids. This matrix is mainly composed of ceramides, cholesterol, and free fatty acids (FAs) (5–9). Alteration of the epidermal permeability barrier function results in several human epidermal pathologies such as psoriasis (10), atopic dermatitis (11), and enhanced susceptibility to infections (12). Therefore, unveiling the cellular processes that regulate the formation of the SC and lipid synthesis is of fundamental importance for understanding the pathogenesis of several human skin disorders.
The human zinc finger (C2H2-type) protein ZNF750 is a transcription factor involved in terminal epidermal differentiation and cancer (13–17). In vitro experiments in human keratinocytes indicate that ZNF750, together with p63 (13, 18), belongs to a complex network of transcription factors, which, in combination with different transcriptional regulators, allows the expression of late terminal differentiation genes. In particular, ZNF750, by interacting with the chromatin modifiers REST corepressor 1 (RCOR1), lysine demethylase 1A (KDM1A), and C-terminal binding protein 1/2 (CTBP1/2), represses the expression of progenitor genes including retinoblastoma binding protein 8 (RBBP8), Homer Scaffold protein 3 (HOMER3), Laminin subunit gamma 1 (LAMC1), and collagen type V alpha 2 chain (COL5A2). In addition, by interacting with RCOR1, Kruppel-like factor 4 (KLF4), and CTBP1/2, ZNF750 activates differentiation genes such as Periplakin (PPL) and Plakophilin 1 (PKP1) (19). Mutations of ZNF750 have been associated with the development of skin diseases including seborrhea-like dermatitis with psoriasiform elements (20, 21). In particular, the frameshift mutation 56_57dupCC in the Znf750 gene, identified in psoriatic patients, results in the production of a truncated protein, lacking the zinc-finger domain. ZNF750 down-regulation results in the reduction of genes implicated in epidermal differentiation and skin barrier formation, including Filaggrin (FLG); Loricrin (LOR); serine protease inhibitor Kazal-type 5 (SPINK5); Arachidonate 12-Lipoxygenase, 12R type (ALOX12B); and Desmoglein1 (DSG1) (22).
To gain insight into the in vivo role of ZNF750 (mouse homolog ZFP750), we have generated constitutive knockout (KO) mice by deleting exon 2. Here, we show that ZFP750 is essential for epidermal development and homeostasis by acting as a critical regulator of skin lipid metabolism. ZFP750 by directly and/or indirectly regulating the expression of crucial enzymes in the biosynthesis of ceramides plays an essential role in the regulation of skin barrier permeability. Mice with Zfp750 ablation die shortly after birth and show defects in cornified envelope assembly. Overall, our study identifies ZFP750 as a crucial transcription factor for skin development, acting as a master regulator of genes involved in epidermal lipid synthesis.
RESULTS
Zfp750−/− mice show alteration of skin barrier function and perinatal lethality
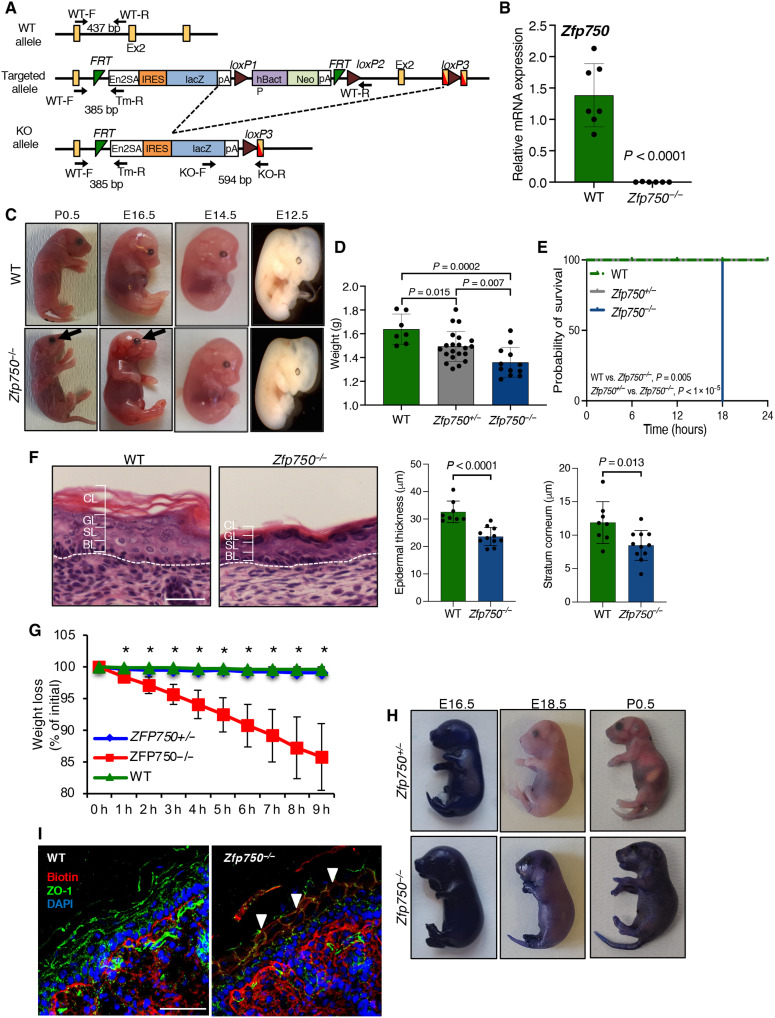
To gain insight into the in vivo role of the transcription factor ZFP750, we generated constitutive KO mice using the strategy illustrated in Fig. 1A. We deleted exon 2 by crossing mice carrying the loxP site (targeted allele) with a transgenic mouse line expressing Cre recombinase under the control of the cytomegalovirus (CMV) promoter. Polymerase chain reaction (PCR) analysis of genomic DNA extracted from newborn mice confirmed the Cre-mediated recombination of Zfp750-floxed alleles (KO allele) (fig. S1A). It resulted in an effective reduction of Zfp750 mRNA (Fig. 1B). Pups from heterozygous backcrosses were born at the (23) expected mendelian ratio (fig. S1B), although a slight reduction in the percentage of Zfp750−/− mice was observed. After birth, Zfp750−/− mice had shiny, wrinkled, and sticky skin, and the eyes were already opened. The lack of eyelids was readily evident on day 16.5 of gestation (E16.5) (Fig. 1C, black arrows). In addition, Zfp750−/− mice showed a reduction in body weight when compared to their control littermates (Fig. 1D) and died within 20 hours (Fig. 1E). Milk was only observed in the stomach of the wild-type (WT) mice, suggesting that newborn Zfp750−/− pups were neglected by the mother. The heterozygous mice (Zfp750+/−) did not show any obvious phenotype and were similar to the WT mice. Next, taking advantage of the lacZ reporter gene under the control of ZFP750 promoter (Fig. 1A), we monitored the expression of ZFP750 protein during in vivo skin development. At E14.5, ZFP750 is mainly expressed in the suprabasal layer [keratin-14 (Krt-14)–negative cells] (fig. S1C). During the following stages of embryonic development [embryonic day 17.5 (E17.5) and postnatal day 0.5 (P0.5)] Krt14-positive cells are still negative for ZFP750, while Krt10- and loricrin-positive cells show high levels of ZFP750 expression (fig. S1C).
Fig. 1. Alteration of skin barrier function in Zfp750−/− mice.
(A) Schematic representation of the strategy used for generating the KO mouse. Primers for genotyping are indicated with arrows. bp, base pair. (B) Real-Time PCR showing the absence of Zfp750−/− mRNA in the epidermis of Zfp750−/− mice. Data are normalized to β-actin and relative to WT. Bars represent the mean ± SD (n = 6 mice per genotype). (C) Gross phenotype of Zfp750−/− mice. Pictures of the WT and Zfp750−/− mice showing shiny and wrinkled skin and lack of eyelids (black arrow). (D) Weight at the birth of the indicated mice. (E) Kaplan Meier analysis of the post-natal death of Zfp750−/−mice. Zfp750−/−mice die within 20 hrs (WT, n = 2; ZFP750+/−, n = 9; ZFP750−/−, n = 5). (F) Hematoxylin and eosin staining of skin from WT and Zfp750−/−− mice showing the reduction of the epidermal thickness. Scale bar, 20 μm. CL, cornified layer; GL, granular layer; SL, spinous layer; BL, basal layer. (G) Zfp750−/− mice loss weight within 9 hours (h) after birth (*P < 0.05). (H) Skin permeability assay. Mice were stained with toluidine blue. E17.5, embryonic day 17.5; P0.5, postnatal day 0.5. (I) Inside-out skin barrier was assessed using EZ-Link sulfo-NHS-LC-biotin. Mice were injected intradermally into the skin of the back with EZ-Link sulfo-NHS-LC-biotin (10 mg/ml). Scale bar, 50 μm. ZO-1, zonula occludens-1; DAPI, 4′,6-diamidino-2-phenylindole.
The perinatal lethality observed in Zfp750−/− mice led us to hypothesize that the barrier function of the skin was altered. To investigate this, we first performed histological analysis of newborn skin and monitored the body weight of Zfp750−/− mice. Zfp750−/− mice were characterized by a reduction in epidermal thickness and a tightly compacted SC when compared with control mice (Fig. 1F and fig. S1D), and Zfp750−/− mice lost up to 15% of their initial body weight within 9 hours after birth (Fig. 1G). The loss of weight was due to dehydration as shown by impaired skin permeability barrier function. Using the toluidine blue exclusion assay to assess the outside-in permeability barrier, control mice excluded the dye, while Zfp750−/− mice showed notable dye penetration into the skin (Fig. 1H). The alteration of the skin barrier function was also confirmed by an inside-out tight junction barrier assay (Fig. 1I). Together, these results demonstrate that Zfp750−/− mice die shortly after birth and show a severe alteration of both the “outside-in” and the “inside-out” skin barrier function that results in water loss, indicating a crucial role of ZFP750 in the skin permeability barrier.
Zfp750−/− mice show impaired skin development
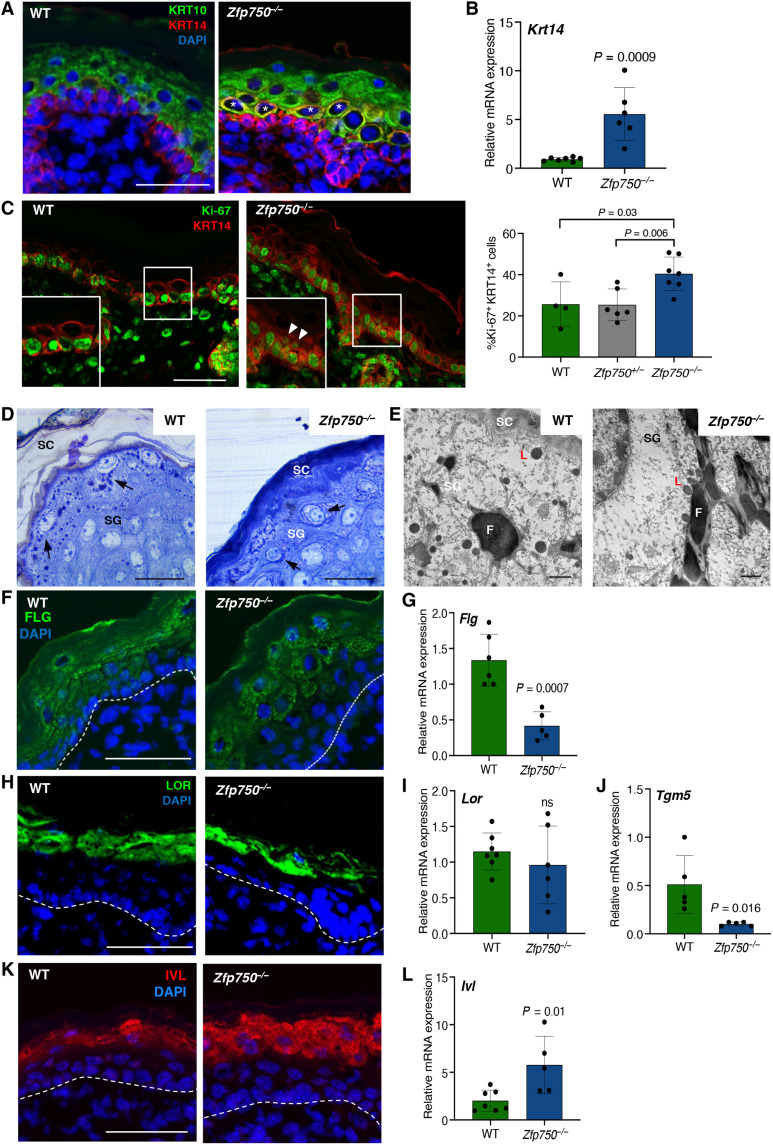
In human keratinocytes, ZNF750 acts both as a repressor of progenitor genes and an activator of differentiation genes (19). To evaluate the role of ZFP750 in development, we analyzed the effect of its deletion on keratinocyte proliferation, as well as on the shift of basal (Krt14 positive cells) versus spinous layer (Krt10 positive cells). Deletion of Zfp750 did not grossly affect the structure of both basal and suprabasal cell layers (Fig. 2A). However, in the epidermis of Zfp750−/− mice, we observed several cells that were positive both for Krt14 and Krt10 (Fig. 2A, white asterisks), suggesting that loss of ZFP750 delays/impairs the keratin switch at the early differentiation stage, as also indicated by the increase of Krt14 mRNA expression (Fig. 2B). This led us to investigate whether the deletion of ZFP750 could affect proliferation in the epidermis by assessing the expression of the proliferation marker Ki-67. Our analysis revealed that the proliferation rate was significantly increased in the epidermis of Zfp750−/− mice (Fig. 2C). The deregulation of epidermal differentiation was already present at E17.5 in both dorsal and ventral skin, confirming the role of ZFP750 in early skin development during embryogenesis (fig. S2, A and B). To evaluate whether the regulatory axis described in human keratinocytes (13) is also conserved in mouse skin, we evaluated p63, ZFP750, and KLF4 expression in E14.5 and P0.5 stages. As expected, p63 was expressed in the basal cells both at E14.5 and P0.5. At E14.5, ZFP750 is coexpressed with p63, although its expression diverged from p63 at P0.5 (fig. S3A), being restricted to the upper basal layers. Chromatin immunoprecipitation experiments showed that p63 binds to the ZFP750 promoter (fig. S3B), indicating that p63 regulates ZFP750 expression in the early phases of epidermal development, although at later stages additional, factors must be responsible for its transcriptional activation. Furthermore, in the epidermis of Zfp750−/− newborn mice, the Klf4 and p63 mRNA levels (fig. S3, C and D) were comparable with those of control mice, suggesting that Klf4 is not regulated by ZFP750. These results indicate that p63 and ZFP750 cooperate during epidermal development and that the regulatory axis involving p63-ZFP750 and KLF4 is not completely conserved in mouse skin development.
Fig. 2. Epidermal terminal differentiation is altered in ZFP750−/− mice.
(A) Immunofluorescence analysis of the proliferating basal layer (Krt14) and differentiated spinous layer (Krt10) in newborn WT and Zfp750−/− mice. Zfp750−/− mice shows the presence of cells expressing simultaneously Krt14 and Krt10 (asterisks). Scale bar, 50 μm. (B) Krt14 mRNA levels in the epidermis of the indicated mice. Data were normalized to β-actin and relative to WT (n = 6 mice per genotype). (C) Zfp750−/− mice displayed increased cell proliferation. Immunofluorescence analysis for the assessment of proliferation (Ki-67) in newborn WT and Zfp750−/− mice. The staining shows an increase of the Ki67+Krt14+ cells. Scale bar, 50 μm. (D) Representative micrographs of semithin sections of skin from WT and ZFP750−/− mice, stained with toluidine blue. ZFP750−/− image reveals a more densely packed SC and perturbation of filaggrin (F) granule distribution. (E) Representative transmission electron micrograph of WT and Zfp750−/− skin. L, Loricrin; SG, stratum granulosum; SC, stratum corneum. Scale bars, 50 μm (D) and 500 nm (E). (F) Immunofluorescence and (G) mRNA expression of filaggrin (FLG) in dorsal skin of the indicated mice. (H) Immunofluorescence and (I) mRNA expression of loricrin (LOR) in dorsal skin isolated from WT and ZFP750−/− newborn mice. Scale bar, 50 μm. (J) mRNA expression of transglutaminase 5 (Tgm5) in the epidermis isolated from WT and Zfp750−/− newborn mice. (K) Immunofluorescence and (L) mRNA expression of involucrin (IVL) in dorsal skin isolated from WT and ZFP750−/− newborn mice. Scale bar, 50 μm. For (A) to (K), a representative micrograph is shown (n = 4 mice per genotype). Data are normalized to β-actin and relative to WT (n = 5 mice per genotype). Bars represent the means ± SD.
ZNF750 has also been implicated in controlling the expression of late epidermal differentiation genes (19, 22). We therefore investigated whether the barrier defects observed in the KO mice were due to altered expression of the cornified envelope protein components. We first focused our attention on filaggrin and loricrin expression, as these two proteins have been reported to be regulated by ZNF750. Transmission electron microscopy (TEM) highlighted that keratohyalin granules containing pro-filaggrin (F) were smaller in the KO mice when compared with the control littermates (Fig. 2D, black arrows). A substantial reduction was also observed in loricrin-containing L-granules (Fig. 2D). We also observed a more densely packed SC in Zfp750−/− mice (Fig. 2D). In addition, perturbation of filaggrin granule distribution (arrows), which appears marginated in the cytoplasm, forming a circular edge in the cells of the stratum granulosum (SG) was observed in Zfp750−/− mice compared with a regular distribution in WT mice (Fig. 2D). Notably, transmission electron micrograph of WT skin showed the presence of large and irregular filaggrin (F) in the SG. On the contrary, ultrastructural analysis of a granular cell in Zfp750−/− skin reveals smaller and elongated morphology of filaggrin (F) granules (Fig. 2E).
In line with this observation, ZFP750 mutant mice showed reduced levels of filaggrin (FLG) protein and mRNA (Fig. 2, F and G, and fig. S4A). Filaggrin granules were almost absent in the Zfp750−/− SC (Fig. 2F), and their distribution was altered, being arranged around the nucleus, and not linearly distributed along the SG, as shown in WT epidermis (Fig. 2F). Loricrin levels were unchanged both at protein and mRNA level (Fig. 2, H and I, and fig. S4B) in Zfp750−/− mice despite the reduction observed in L-granule size (Fig. 2E).
Transglutaminases (TGMs) are calcium-dependent enzymes that catalyze the posttranslational modification of proteins via formation of isopeptide bonds. Four different TGM named as TGM1, TGM2, TGM3, and TGM5 are mainly involved in the formation of the cholesteryl ester (CE) (24). The expression of transglutaminase enzymes was also significantly reduced (Tgm5; Fig. 2J) or had a tendency to decrease (Tgm1 and Tgm3; fig. S4C) after the deletion of ZFP750.
On the contrary, the expression of involucrin (IVL) was significantly up-regulated both at protein and mRNA levels in Zfp750−/− newborn mice when compared with WT mice (Fig. 2, K and L, and fig. S4D); this is probably due to an attempt of the tissue to rescue the barrier function, as already described in other KO mice models presenting with barrier defects (25).
Lipid barrier composition is altered in Zfp750−/− mice
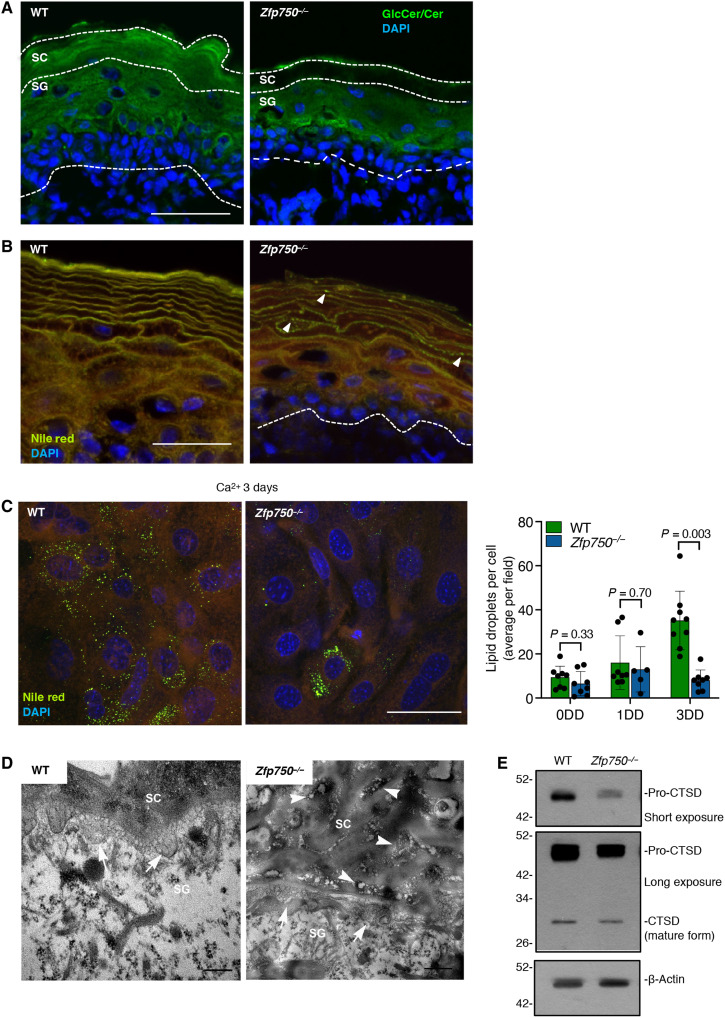
To test whether deletion of ZFP750 also affects the lipid cornified envelope composition resulting in compromised skin barrier function, we first assessed by immunofluorescence analysis the profile of ceramides and glucosylceramide (GlcCer), the major glycosphingolipid of the epidermis and the precursor of ceramides (26). Skin from newborn mice was stained with antibody that recognized both ceramides and GlcCer (27). In control mice, GlcCer/Cer was mainly localized in both the SC and granular layer (Fig. 3A). On the contrary, GlcCer/Cer was completely absent in the SC of the skin sampled from Zfp750−/− mice. Next, we used Nile red dye to analyze the lipid composition of the SC. In the SC isolated from control mice, we observed a bright yellow-gold color indicating the presence of both polar and nonpolar lipids (Fig. 3B). In contrast, in the SC of Zfp750−/− mice, the presence of both lipids was notably reduced, and this reduction was more evident for the nonpolar lipids (ceramides, green fluorescence). Reduction in nonpolar lipids was also confirmed in keratinocytes derived from Zfp750−/− mice after 3 days of differentiation (Fig. 3C). Moreover, while in WT mice lipids were distributed in a weave-like manner, lipids in the SC of Zfp750−/− mice showed a pearl-like distribution (Fig. 3B, white arrowheads). Dot-like distribution of lipids in the SC was also confirmed by TEM analysis (Fig. 3D, white arrowheads). The ultrastructural analysis of the WT mice showed that the interface between the granular layer and the SC was filled with lamellar bodies (LBs) that appeared as oval-shaped vesicles (Fig. 3D, white arrows and fig. S4E). In contrast, Zfp750−/− mice showed an interface almost devoid of LBs (Fig. 3D, white arrows and fig. S4E). To further corroborate the reduction of LBs in ZFP750−/− mice, we used cathepsin D to trace LBs distribution and amounts in the epidermis (28). We observed a weak staining of cathepsin D in the epidermis of KO mice (fig. S4F). This reduction was also confirmed by Western blot analysis. Both the proenzyme and the enzymatically active forms were substantially reduced in the epidermis of Zfp750−/− mice (Fig. 3E). These results indicate that loss of ZFP750 alters epidermal lipid composition, therefore contributing to the dysfunction of the cutaneous permeability barrier.
Fig. 3. Aberrant lipid composition in Zfp750−/−epidermis.
(A) Immunofluorescence analysis of glucosylceramide and ceramide (GlcCer/Cer) localization in the epidermis. Scale bars, 50 μm. (B) Nile red staining of the epidermal lipids showing a substantial decrease of the green fluorescence (neutral lipid species) in Zfp750−/− mice. Mutant mice showed a dot-like pattern (white arrowheads). Scale bars, 50 μm. (C) The intracellular content of lipid droplets is reduced in in vitro differentiated keratinocytes derived from Zfp750−/− mice. A representative micrograph of three independent experiments is shown. Scale bar, 50 μm. DD, Day of Differentiation. (D) Transmission electron micrographs showing abnormal lipid secretion (arrows) in ZFP750−/− mice (ii and iv) at the apical surface of the granular layer; persisting vesicle aggregates are found in the spaces among cells of SC (arrowheads). Scale bars, 200 nm (WT) and 500 nm (Zfp750−/−). A representative micrograph is shown (n = 4). (E) Western blot of CTSD showing a marked decrease in the precursor pro-CTSD in ZFP750−/− (β-actin, loading control). A representative immunoblot is shown (n = 2).
ZFP750 regulates the expression of genes involved both in the protein and lipid components of the cornified envelope
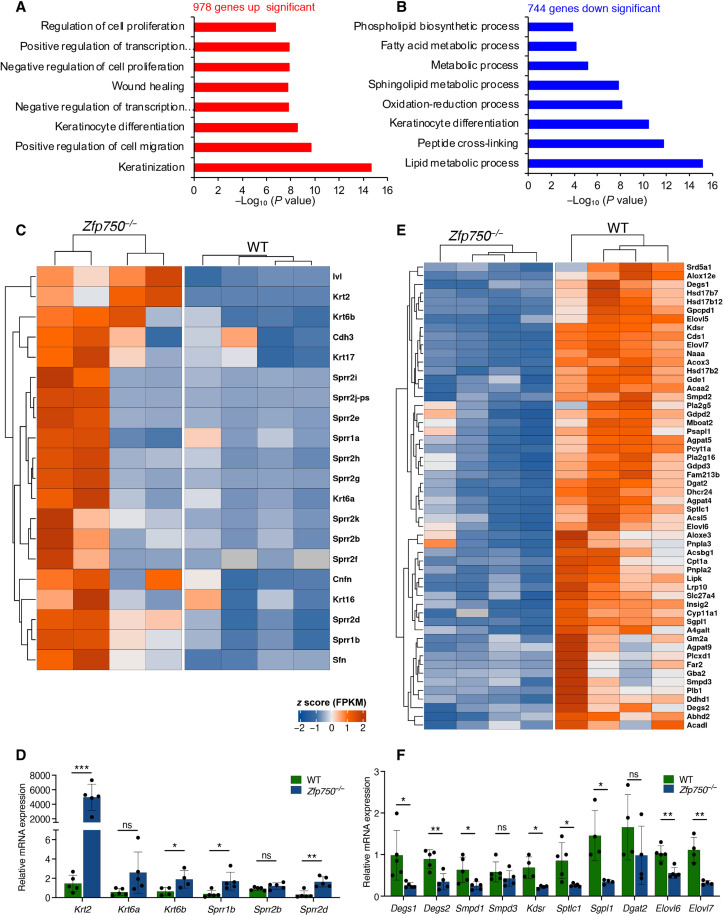
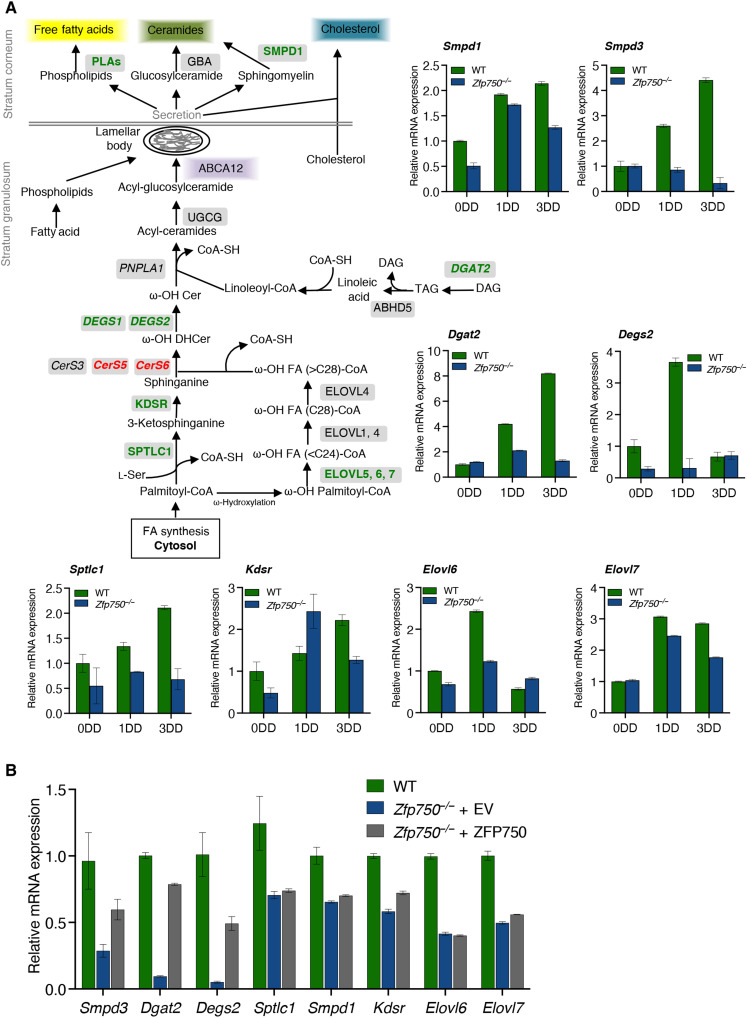
To gain insight into the molecular mechanisms underlying the phenotype observed in Zfp750−/− mice, total RNA was extracted from the epidermis of control and mutant mice and subjected to sequencing. Genetic deletion of ZFP750 resulted in the alteration of the expression of 1722 genes (fig. S5A). Nine hundred and seventy-eight genes responsible for the process of keratinization, regulation of cell migration, and proliferation were up-regulated (Fig. 4A). In particular, deletion of ZFP750 significantly up-regulated the expression of genes involved in the process of keratinization such as Krt2, Krt6b, Sprr1b, and Sprr2d (Fig. 4, C and D). The up-regulation of these genes could be due to a deregulation of the differentiation program after ZFP750 deletion. Genes encoding the small proline rich family (i.e., Sprr1b and Sprr2d) have also been shown to be up-regulated in Zfp750−/− mice with skin barrier defects in an attempt to rescue the abnormalities of the tissue (25, 29, 30). In addition, in agreement with the defects in CE development, deletion of ZFP750 also resulted in the down-regulation of several genes coding for late differentiation markers and precursors of the CE including Lce1i, Lce1h, Lce1l, and Lce1m. On the contrary, 744 genes were down-regulated in the Zfp750−/− epidermis, and Gene Ontology analysis revealed enrichment in pathways involved in lipid metabolism and keratinocyte differentiation (Fig. 4B). In particular, the expression of 55 genes involved in lipid metabolism (Degs1, Degs2, Sgpl1, Elovl6, Elovl7, and Kdsr) were repressed in skin derived from Zfp750−/− mice, suggesting that ZFP750 transcriptionally regulates their expression (Fig. 4, E and F). These results indicate that the transcription factor ZFP750 regulates epidermal development by controlling the expression of genes involved in the assembly of both the protein and the lipid components of the cornified cell envelope.
Fig. 4. Genetic deletion of Zfp750 deregulates genes involved in lipid metabolism, keratinocyte differentiation and keratinization.
(A and B) Gene Ontology terms of significantly up-regulated (red bars) and down-regulated (Blue bars) genes obtained from RNA seq. (C) Heatmap of the 20 genes belonging to the keratinization process deregulated in the epidermis of Zfp750−/− mice. (D) Validation of RNA seq by qPCR. Keratinization genes are up-regulated in Zfp750−/− mice. (E) Heatmap of the 55 genes involved in lipid metabolism and deregulated in the epidermis of Zfp750−/− mice. Bars represent the means ± SD (n = 4 mice per genotype). Data are normalized to β-actin and relative to WT. (ns, not significant; *P < 0.05, **P < 0.01, and ***P < 0.001). (F) Validation of RNA-seq by qPCR. Lipid metabolic genes are down-regulated in Zfp750−/− mice. Bars represent the means ± SD (n ≥ 4 mice per genotype). For (D) and (F), data are normalized to β-actin and relative to WT. (ns, not significant; *P < 0.05, **P < 0.01, and ***P < 0.001).
ZFP750 is a critical regulator of ceramide metabolism in the epidermis
Epidermal ceramides and their corresponding metabolites have a crucial role in the formation and maintenance of skin barrier integrity (26). The de novo biosynthesis of ceramides containing very-long-chain (VLC) and ultralong-chain (ULC) FAs starts at the endoplasmic reticulum (ER) of the spino/granular layer of the epidermis where the first step is the condensation of palmitoyl-CoA and l-serine by the enzyme serine palmitoyl transferase (SPT; SPTLC1 and SPTLC2 subunits) (Fig. 5A); these biosynthetic pathways are skin specific (31, 32). Although the RNA sequencing approach does not give information to identify the direct ZFP750 transcriptional targets, it provides interesting information about the deregulated pathways. The expression of the subunit Sptlc1 was substantially reduced in the epidermis of Zfp750−/− mice and did not increase during terminal differentiation of Zfp750−/− keratinocytes (Fig. 5A). In addition, the expression of the 3-ketodihydrosphingosine reductase (Kdsr), which subsequently reduces the product of SPT in sphinganine, was notably reduced in the epidermis of Zfp750−/− mice (Fig. 5A). We also found that the expression of the enzymes ceramide synthase (CerS) and desaturase (Degs), which catalyze the production of dihydroceramide and ceramide, respectively, was deregulated by the deletion of ZFP750. In particular, Degs2 expression increased during terminal differentiation of WT keratinocytes, while in the absence of ZFP750, the up-regulation of Degs2 expression was completely abolished (Fig. 5A). In contrast, CerS5 and CerS6 mRNA expression was up-regulated.
Fig. 5. ZFP750 controls the expression of genes implicated in ceramide biosynthesis.
(A) Schematic representation of the ceramide biosynthetic pathway and analysis of the mRNA levels by qPCR of the indicated enzymes during the differentiation of murine WT and Zfp750−/−keratinocytes. Enzymes down-regulated and up-regulated are highlighted in green and red, respectively. Data are normalized to TATA-binding protein (TBP) and relative to WT. One of three independent experiments is shown. Data represent the mean of three technical replicates (n = 3, PCR runs) ± SD. (B) Re-expression of ZFP750 in keratinocytes derived from Zfp750−/− mice partially restores the mRNA levels of Smpd3, Dgat2, and Degs2. Data are normalized to TBP and relative to WT. One of two independent experiments is shown (n = 2). Data represent the mean of three technical replicates (n = 3, PCR runs) ± SD. PLAs, acid phospholipases; GBA, glucocerebrosidase; SMPD1, acid sphingomyelinase 1; ABCA12, ATP-binding cassette 12; UGCG, UDP-glucose:ceramide glucosyltransferase; PNPLA1, patatin-like phospholipase domain-containing protein 1; DEGS1-2, Delta 4-Desaturase, sphingolipid 1-2; CerS, Ceramide synthase; KDSR, 3-ketosphinganine reductase; SPT, serine palmitoyl transferase; ELOVL, elongase of very FAs; DGAT2, diacylglycerol acetyltransferase 2; ABHD5; abhydrolase domain containing 5.
The FA component of ceramides, VLC and ULC FAs, are synthesized in the ER of keratinocytes, and their synthesis is mediated by a family of seven enzymes called elongation of VLC FAs 1 to 7 (Elovl1 to Elovl7) that catalyzes the first and rate-limiting reaction of the four reactions that constitute the long-chain FA elongation cycle (33). We observed an increase in Elovl6 expression during WT keratinocyte differentiation, although this up-regulation was not observed during the differentiation of keratinocytes derived from Zfp750−/− mice (Fig. 5A). Moreover, Elovl7 expression was partially reduced during Zfp750−/− keratinocyte differentiation. Diacylglycerol acyltransferase (DGAT) regulates the final step of triglyceride synthesis and loss of DGAT2 in mice results in the impairment of permeability barrier function (34): Loss of ZFP750 completely abolished the up-regulation of Dgat2 during keratinocyte differentiation (Fig. 5A).
Proper formation of the lipid barrier depends also on the processing of the lipids stored in the LB and by their release at the interface between SG and SC. The key lysosomal proteins that regulate the extracellular processing of the pro-barrier lipids are β-glucocerebrosidase (GBA), acid sphingomyelinase 1 (SMPD1), and prosaposin (35). As shown in Fig. 5A, the expression of Smpd1 mRNA increased during the differentiation of both WT and ZFP750−/− keratinocytes. However, the increase was notably less in Zfp750−/− keratinocytes. On the other hand, the up-regulation of neutral sphingomyelinase (Smpd3) expression during keratinocyte differentiation (36) was abolished in Zfp750−/− keratinocytes (Fig. 5A). To further confirm that ZFP750 is a key regulator of enzymes involved in epidermal ceramide metabolism, we re-expressed ZFP750 in keratinocytes isolated from ZFP750−/− mice. Reintroduction of ZFP750 substantially recovered the mRNA levels of Smpd3, Dgat2, and Degs2, suggesting a direct role of ZFP750 in regulating the transcription of these genes (Fig. 5B). Together, these results demonstrated that ZFP750 is a key transcription factor for controlling the expression of crucial enzymes involved in the formation of the lipid cornified envelope.
Epidermal lipid composition is deregulated in Zfp750−/− mice
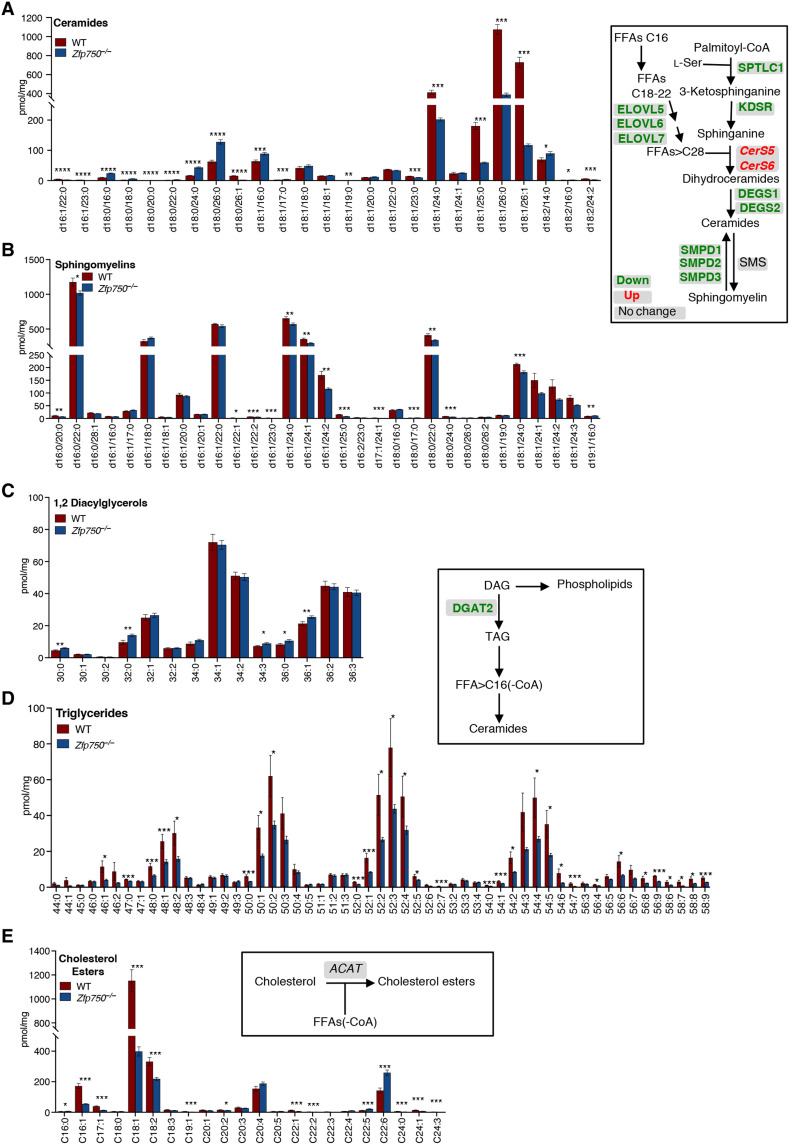
To explore whether the deregulation in the expression of several genes involved in lipid metabolism was also associated with a deregulation of the endogenous epidermal lipid composition, we performed lipidomic analysis by UPLC-MS/MS on the epidermis isolated from newborn Zfp750−/− and control mice. Our analysis revealed that the number of ceramide species, which play a critical role in skin barrier permeability, was profoundly deregulated (Fig. 6A). In particular, the following ceramide species (including the saturated ceramides (d18:0/16:0), (d18:1/16:0), (d18:0/24:0), (d18:0/26:0) showed significant accumulation. On the contrary, in the epidermis of Zfp750−/− mice, there was a significant decrease of monounsaturated ceramides with C24-26, such as (d18:1/24:0), (d18:1/25:0), (d18:1/26:0), and (d18:1/26:1). This ceramide profile is likely a consequence of the reduction in the mRNA levels of the key enzymes involved in ceramide biosynthesis including Sptlc1, Kdsr, Degs1, Degs2, Smpd1-3, Elovl6, and Elovl7 (Fig. 6). In mammalian cells, most of the ceramide is also converted to sphingomyelin by sphingomyelin synthase. As a consequence of the reduction in the amount of several ceramide species, we also observed a significant reduction of sphingomyelins in the epidermis of the mutant mice (Fig. 6B).
Fig. 6. Zfp750 genetic deletion results in the impairment of epidermal lipid composition.
(A to E) MS analysis of the indicated lipid species extracted from the epidermis of newborn WT and Zfp750−/− mice. Bars represent the means ± SE (n = 13 mice per genotype). FFAs, free FAs; SMS, sphingomyelin synthase; DAG, diacyglycerols; TAG, triglycerides; ACAT, acyl-CoA:cholesterol acyltransferase. (*P < 0.05, **P < 0.01, and ***P < 0.001).
The epidermal neutral lipid metabolism also plays a critical role in the maintenance of a proper permeability barrier in the skin (37). Of relevance, our analysis revealed that the amount of triacylglycerol (TAG) is remarkably reduced in epidermis isolated from Zfp750−/− mice (Fig. 6D), and this was associated with a significant increase of several 1,2-diacyglycerol species in the epidermis of the mutant mice (Fig. 6C).
Cholesterol is an essential component of the epidermis; however, CEs, which are less polar than free cholesterol, enhance skin hydrophobicity for barrier function (38). In particular, epidermis isolated from newborn mice mainly contains CE with C16-18 FA. The levels of cholesterol esters (C16-18) were significantly reduced in the epidermis of mutant mice (Fig. 6E).
Phospholipids contained in the LB are metabolized in the SC into free FAs, which, in turn, contribute to the acidification of the corneum (38). Our observations also indicate that ZFP750 is required for the proper biosynthesis of phospholipids. Specifically, the amount of phospholipids, namely, phosphatidylethanolamine, phosphatidylserine, phosphatidylcholine, and phosphatidylinositol, was found significantly deregulated in the epidermis of Zfp750−/− mice (fig. S6). Specifically, several phosphatidylserine species including ether bonds showed marked accumulation in the epidermis of Zfp750−/− mice. On the other hand, only few phosphatidylinositol species were found reduced in the epidermis of Zfp750−/− mice. No significant changes were found, instead, in the phosphatidylethanolamine group.
This lipidomic analysis confirmed the reduction of key keratinocytes-specific lipids, demonstrating the relevance of ZFP750 in allowing the synthesis of a proper lipid barrier in the epidermis.
Human ZNF750 controls the expression of genes involved in ceramide biosynthesis
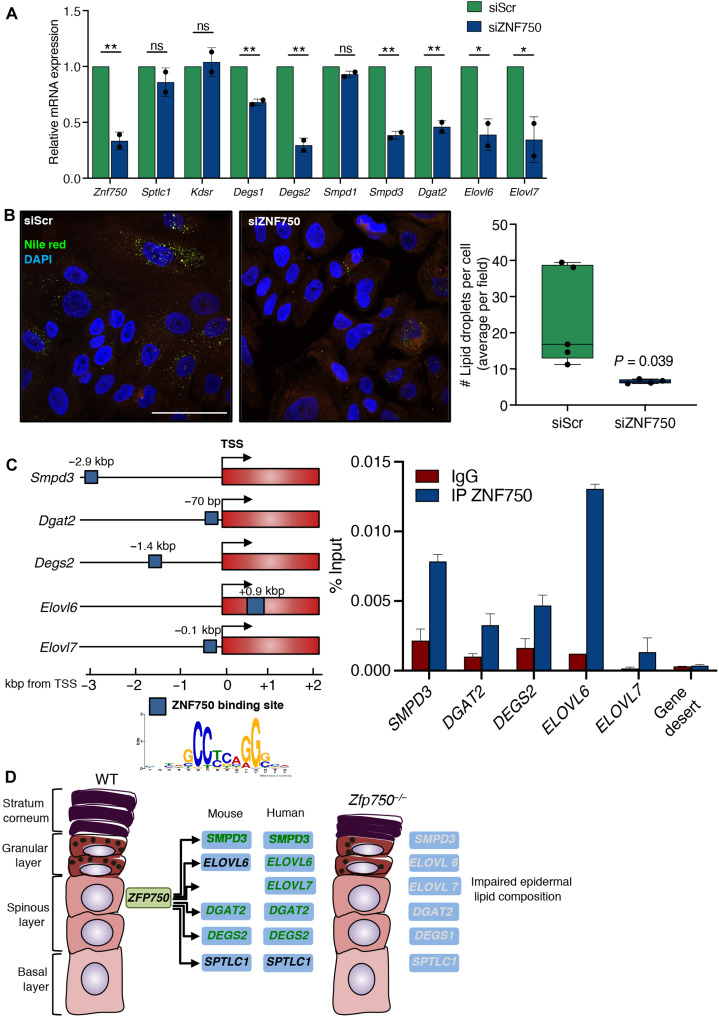
To assess whether the regulation of epidermal lipid metabolism by ZFP750 is also conserved in humans, we inhibited the expression of ZNF750 in human keratinocytes by small interfering RNA (siRNA). Inhibition of ZNF750 expression resulted in a significant reduction in the mRNA levels of Degs1, Degs2, Dgat2, Smpd3, Elovl6, and Elovl7 (Fig. 7A). The deregulation of these genes was associated with a significant reduction in the levels of nonpolar lipids (Fig. 7B). To gain insight into how ZNF750 controls the expression of these genes during keratinocyte differentiation, we examined the promoter region −5 kilo base pair (kbp)/+1 kbp from the transcription start site (TSS) of the indicated genes for putative binding sites for ZNF750. Sequences matching the annotated ZNF750 consensus sequence (19) were found in the promoter region of the following genes: Degs2, Dgat2, Smpd3, Elovl6, and Elovl7. To confirm the bioinformatic analysis, we subsequently performed chromatin immunoprecipitation assays. We observed that endogenous ZNF750 directly binds the promoter region of Degs2, Dgat2, Smpd3, Elovl6, and Elovl7 (Fig. 7C). Overall, these results suggest that ZFP750 directly and/or indirectly regulates the expression of key enzymes implicated in synthesizing epidermal lipids.
Fig. 7. Down-regulation of human ZNF750 expression recapitulates the defects in lipid biosynthesis.
(A) Analysis of mRNA levels of lipid biosynthesis enzymes by qPCR following ZNF750 knockdown in differentiated human epidermal keratinocytes (HEKn) (*P < 0.05 and **P < 0.01). Bars represent the means ± SD of two independent experiments (n = 2). Data are normalized to human TBP and relative to the scramble condition. siScr, scrambled siRNAcontrol; siZNF750, silencer ZNF750. (B) Znf750 knockdown in HEKn results in the reduction of intracellular lipid droplet content. Bars represent the means ± SD. Scale bar, 50 μm. (C) ChIP-qPCR analysis showing the binding of endogenous ZNF750 to the promoter region of the indicated genes. The ZNF750 binding site position is shown. Gene desert is used as a negative control region. One of two independent experiments is shown (n = 2). Data represent the mean of three technical replicates (n = 3, PCR runs) ± SD. (D) Proposed model of ZFP750 as master regulator of the epidermal lipid biosynthesis. The direct target genes of both ZFP750 and ZNF750 are shown in green.
DISCUSSION
The results shown in the exon 2 deleted ZFP750 mice support the hypothesis that ZFP750 is a master regulator of epidermal barrier function by controlling the expression of several enzymes involved in the synthesis of epidermal lipids and, consequently, the epidermal lipid composition (Fig. 7D). The data provide a mechanistic explanation for the reported frameshift mutation in the human Znf750 gene causing a seborrhea-like dermatitis with psoriasiform elements (20).
Different proteins and transcription factors have been described as playing an important role in regulating keratinocyte differentiation and epidermal lipid synthesis, providing a progressively more complex and coherent picture on the regulation at the transcriptional level of lipid synthesis. Peroxisome proliferator–activated receptor isoforms (PPARs) (PPARα, PPARβ/δ, and PPARγ) are activated by lipids such as FAs and ceramide, and this activation results in the transcription of a subset of key epidermal genes including GBA, Abca12, Tgm1, loricrin, and involucrin (39, 40). Nonetheless, loss of PPARα or PPARγ genes in mice does not result in obvious morphological changes in the epidermis (41, 42). Conversely, PPARβ/δ null mice show a delay in skin barrier recovery after acute perturbation. This is associated with reduced numbers of LBs and extracellular lipid in the SC (43). In addition, vitamin D receptor and steroid receptor coactivator participate in the regulation of epidermal sphingolipid and ceramide synthesis in cultured keratinocytes by regulating the expression of ELOVL3, ELOVL4, ceramide glucosyltransferase, and ABCA12 (44). Moreover, the expression of ELOVL3 and stearoyl-CoA desaturases is controlled by T cell factor, and its in vivo disruption leads to a major change of lipid composition, compromising the water permeability barrier (45). In this context, our results indicate that deletion of ZFP750 extensively deregulates the expression of a substantial number of enzymes involved in the regulation of lipid, sphingolipid, and ceramide synthesis, which results in an impaired epidermal lipid composition and macroscopic morphological changes of the epidermis associated with a perturbation of skin barrier function. Although alteration of skin barrier function often is incompatible with survival, we cannot rule out that additional developmental defects are the cause of the severe phenotype observed.
Three main lipid metabolic pathways are regulated by ZFP750. The first is the pathway of ceramide biosynthesis. Specifically, Zfp750−/− epidermis displays a significant reduction in the amount of ceramides with long chain FAs (C>22), whereas ceramide species with short-chain FAs (C16-22) are increased. Although most of the key enzymes involved in ceramide biosynthesis are down-regulated in ZFP750−/− mice, the observed ceramide species profile is most likely caused by a deficit in the expression of ELOVL6 and ELOVL7, which are the key enzymes in regulating the elongation of FA, elovanoids, from C16 to C22 (46). Second, synthesis of triglycerides is impaired. We propose that the molecular mechanism underlying the reduction in triglycerides is caused by defective expression of DGAT2. This is further confirmed by the reduction in lipid droplet content in keratinocytes isolated from Zfp750−/− mice since DGAT2 is required for their formation (47). As a result, in vivo deletion of DGAT2 results in the alteration of the epidermal barrier integrity. In particular, Dgat2 KO mice show lipopoenia and die shortly after birth, partly due to impaired epidermal permeability barrier function (34). Here, we show that DGAT2 is a key target of ZFP750. While this may explain, at the molecular level, the phenotype observed in our mouse model, we cannot exclude the possibility that other enzymes deregulated in the epidermis of ZFP750−/− mice also contribute to the observed phenotype. Mice with conditional deletion of the Sptlc2 gene in keratinocytes show reduced levels of ceramides, which also impair the water barrier function of the epidermis (47). In addition, by the age of 2 weeks and older, Sptlc2 KO mice develop psoriasis-like skin lesions. Last, Zfp750−/− mice show substantially reduced cholesterol esters, which plays an important role in the formation of the epidermal barrier (38).
Epidermal lipids are not only essential for the formation of the physical barrier of the skin but are also able to regulate epidermal differentiation (48, 49). Lipids can act as signaling molecules by activating PPARs and liver X receptors and, hence, regulate gene expression. Moreover, ceramides stimulate keratinocyte differentiation and inhibit proliferation in part by increasing Activator Protein 1 (AP-1) activity (50). Therefore, ZFP750 not only regulates terminal epidermal differentiation by directly controlling the expression of differentiation genes including PPL and PKP1 but also partly by regulating epidermal lipid composition.
The p63/ZNF750/KLF4 axis regulates in vitro differentiation of human keratinocytes (13). In our mouse model, genetic deletion of Zfp750 results in an impairment of keratinocyte differentiation associated with a defect in the transition from proliferating to differentiating keratinocytes together with an increase of basal cells proliferation. Although ZFP750 appears not to be expressed in the epidermal basal layer, the increased proliferation observed may in part caused by compensatory mechanisms. Several studies report that when the epidermal barrier is compromised, increased proliferation is frequently observed as a secondary response (50). Animal deficient for essential FA or inhibition of cholesterol synthesis shows an increase in DNA synthesis, which is limited to the epidermal basal layer (51). Moreover, perturbations in canonical Notch signaling compromise epidermal barrier function, which, in turn, leads to postnatal hyperproliferation as an indirect secondary reaction (52). A second possible hypothesis is that Krt10-positive cells expressing ZFP750 regulate cell proliferation of the basal cell layer. It has been shown that microtubule disruption results in increased actomyosin contractility, which is both necessary and sufficient to induce stem cell hyperproliferation (53).
Defects in keratinocyte terminal differentiation are also observed in p63-null mice, indicating that these transcription factors cooperate in the regulation of epidermal differentiation (18). However, it should also be noted that mice lacking p63 exhibit severe developmental anomalies, including limb truncations, craniofacial malformations, and the lack of an intact epidermis, anomalies that are not displayed by Zfp750−/− mice. In addition, our data contribute to dissect the relative contribution of each member of this axis. In murine skin, this axis is mostly conserved in Zfp750−/− mice. While p63 directly regulates the expression of ZFP750 in the early epidermal differentiation stage (Krt14-positive cells), in the late stage of differentiation (loricrin-positive cells), ZFP750 is not under the control of p63. Therefore, additional factors may work together with p63 to turn on ZFP750 expression. Moreover, KLF4 expression is not reduced in the keratinocytes and epidermis derived from Zfp750−/− mice, indicating that KLF4 expression is not controlled by ZFP750. Hence, our findings indicate that, at least in mice, ZFP750 regulates keratinocyte differentiation independently from KLF4 and, to some degree, from p63. Overall, our findings suggest that ZFP750 is not required for differentiation because the stratification process properly take place. Therefore, we can conclude that ZFP750 does not participate to the development of the epidermis, which take place during embryogenesis. On the contrary, our results indicate that ZFP750 is mainly required for the maintenance of skin homeostasis in postnatal mice. The thinner epidermis may be the results of the following two possible mechanisms: (i) defect in delamination and migration of the differentiated keratinocytes (54, 55) and (ii) reduced lipid content in the SC, which make it more compacted. However, further studies are needed to assess the contribution of delamination in our model. Time lapse experiment and the generation of inducible skin-specific ZFP750 KO mice may provide the answer to the involvement of delamination in the observed phenotype.
The ZNF750 gene is located on the PSORS2 locus, one of the nine loci with statistically significant linkage to psoriasis (56). Accordingly, ZNF750 has been found mutated in a family affected by seborrhea-like dermatitis with psoriasiform elements and point mutations, both in the promoter region and in the first untranslated exon have been described (20, 21). The disease is characterized by a diminished or altered content of ceramides in the epidermis with an impairment of epidermal barrier function (31). Therefore, it is tempting to speculate that deregulation of the ZNF750 transcriptome might be involved in the pathogenesis of human skin disorders including psoriasis, and the involvement of ZNF750 in patients with psoriasis needs further investigation. To further support the relevance of ZNF750 in human skin disease, recently, it has been shown that keratinocytes isolated from patients with ectodermal dysplasia syndrome (AEC) displayed a ZNF750-reduced nuclear expression that it is associated with a delayed or altered epidermal differentiation. ZNF750 expression was rescued by treatment with the P53-reactivating compounds called PRIMA-1MET (57) and topical treatment of two patients with AEC allowed re-epithelialization of the eroded skin and a notable loss of pain after few weeks (58). This suggest that ZNF750 and its downstream pathway may be pharmacologically modulated, paving the way for novel therapeutic approaches for human skin disorders. Therefore, we would conclude that ZFP750 is one of the missing pieces in regulating lipid, sphingolipid, and ceramide synthesis in the epidermis. Moreover, ZFP750/ZNF750 is a key transcription factor that plays a crucial role in the regulation of epithelial tissue homeostasis. One limitation of our study is that the constitutive deletion of ZFP750 cannot allow us to discriminate whether those defects are mainly due to ZFP750 expression in the skin or additional factors may contribute. Therefore, it would be interesting to assess whether those defects are intrinsic to skin by generating mice lacking ZFP750 selectively in epidermal keratinocytes by using Tg(KRT14-Cre/ERT2) transgenic mice. Moreover, this strategy will allow to study the role of ZFP750 also in adult skin both in physiological and pathological condition by possibly overcoming the lethal phenotype of the constitutive Zfp750−/− mice. Overall, our findings potentially have profound implications for understanding the pathophysiology of several human skin diseases.
MATERIALS AND METHODS
Experimental design
The aim of this study is to investigate the in vivo role of the transcription factor ZFP750, which has been shown to play an important role in epidermal differentiation and function.
All studies were conducted in accordance with the ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines. Heterozygous mice were backcrossed onto C57BL/6J for seven generation, then male and female heterozygous mice were intercrossed to obtain homozygous null mice, and then littermate WT mice were used as controls. Males/females were equally distributed. All animal experiments were carried out under procedural guidelines, severity protocols, and within the United Kingdom with ethical permission from the Home Office [United Kingdom and Italy; number license 817/2016 and 147/2021 (Italy)]. No available data were excluded from the analysis. Each mouse represented one experimental unit.
For cell culture experiments, a minimum of two independent biological replicates were performed. Western blots and micrograph are representative images of at least two independent experiments. Real-time PCR assays were performed in technical triplicates of multiple biological replicates. The numbers of biological replicates for each experiment are stated in the figure legends.
Mice
The embryonic stem cell carrying the target mutation a “KO-first” allele targeted to the Zfp750 genomic locus named Zfp750tm1a(EUCOMM)Wtsi (Mouse Genome Informatics allele ID: 2442210) were obtained from European Mouse Mutant Cell Repository (23). Briefly, mouse embryonic stem cells were injected into blastocysts and transplanted in pseudopregnant mice to generate chimera mice. Germline transmission of the targeted allele was confirmed by PCR. Constitutive KO mice were generated by crossing heterozygous mice carrying tm1a allele with transgenic mice for CMV-Cre. Heterozygous mice were backcrossed onto C57BL/6J for seven generation, then male and female heterozygous mice were intercrossed to obtain homozygous null mice, and then littermate WT mice were used as controls. Mice were subjected to listed procedures under the project license released from the Home Office [number license 817/2016 (Italy) and 70/8654 (United Kingdom)].
DNA extraction and genotyping
Genomic DNA was isolated from ear biopsies using the REDExtract-N-Amp Tissue PCR Kit according to the manufacturer’s protocol (Sigma-Aldrich). For PCR analysis, the extracted DNA was used as template to amplify specific regions of WT and targeted alleles. PCR conditions were as follows: 95°C for 5′, 30 cycles (95°C for 30″, 62°C for 30″, 72°C for 30″), 72°C for 1′. Primers sequences for the amplification are reported in table S1.
Outside-in SC barrier assay
Skin permeability was tested by the diffusion of toluidine blue dye as previously described. Briefly, euthanized embryos and newborn pups were subjected to dehydration and rehydration with a methanol gradient, washed in phosphate-buffered saline (PBS) for 1 min, and stained for 2 min with 0.1% toluidine blue (Sigma-Aldrich) in water. After destaining with PBS, mice were photographed. The SC barrier function was evaluated on the basis of the degree of dye penetration.
Inside-out tight junction barrier assay
To test the tight junction barrier in mice, EZ-Link sulfo-NHS-LC-biotin (10 mg/ml; Pierce Chemical Co.) in PBS containing 1 mM CaCl2 was injected intradermally (50 μl) into the back skin of newborn mice. After 30 min of incubation, mice were euthanized, skin was isolated, embedded in OCT, and then frozen. Cryosections with a thickness of 10 μm were cut and fixed in 95% cold ethanol at 4°C for 20 min and then in 100% acetone at room temperature for 5 min. Sections were blocked with 10% goat serum in PBS and incubated with an anti-ZO1 antibody (1:200; Invitrogen, #402200) overnight at 4°C. After washes in PBS, sections were incubated for 1 hour with a mixture of goat anti-rabbit Alexa Fluor 488 nm (Invitrogen), streptavidin-conjugated Texas Red (Calbiochem), and 4′,6-diamidino-2-phenylindole (DAPI) (1 μg/ml; Sigma-Aldrich). The slides were mounted with ProLong Gold Antifade Mountan (Invitrogen) and analyzed by a Nikon Confocal microscope (Nikon Eclipse Ti).
Epidermis isolation
Skin from newborn mice was isolated and incubated dermis side down in 0.25% Trypsin-EDTA (Gibco) overnight at 4°C. Then, epidermis was peeled from dermis, washed in PBS, and snap frozen.
RNA extraction and quantitative real-time PCR
For RNA extraction, epidermis previously isolated was lysed in RLT buffer (QIAGEN) using steel beads and the TissueLyser LT homogenizer (QIAGEN). For RNA extraction from mouse and human primary keratinocytes, they were directly lysed in RLT buffer. Total RNA was extracted using the RNeasy Mini Kit (QIAGEN) following the manufacturer’s protocol. RNA (500 ng) were reverse-transcribed by GoScript Reverse Transcription System (Promega); Qualitative PCR (qPCR) analyses were performed with the GoTaq qPCR Master Mix (Promega). Relative mRNA levels were calculated with the 2-ΔΔCt method after normalization to β-actin or TATA-binding protein. Primers sequences used for qPCR are reported in table S1.
Cell culture and transfection
Mouse primary keratinocytes were isolated from P0.5 mice as previously described (59). Briefly, neonatal mouse skin was isolated and incubated dermis side down in Trypsin-EDTA 0.25% (Gibco) overnight at 4°C. The following day, the epidermis was peeled from the dermis to collect keratinocytes. The cells were cultured in Eagle’s minimum essential medium, supplemented with 8% chelated fetal bovine serum (FBS), 1% (v/v) antimycotic, and with 50 μM calcium to sustain proliferation. For re-expression of ZFP750 in keratinocytes derived from Zfp750−/− mice, cells were plated into a type I collagen-coated six-well plate. ZFP750−/− keratinocytes were transfected either with a green fluorescent protein (GFP) empty vector as control or with pCMV6-entry-ZFP750 (OriGene, MR224624) with Lipofectamine 3000 according to the manufacturer’s protocol. Twenty-four hours after transfection, cells were induced to differentiate for 24 hours and then collected for downstream analysis.
Neonatal human epidermal keratinocytes (HEKns; Life Technologies) were cultured in EpiLife medium with human keratinocyte growth supplements (Life Technologies).
For silencing experiments, HEKns were transfected with 20 nM of either scramble (Scr) or ZNF750-specific siRNA with Lipofectamine RNAiMAX according to the manufacturer’s protocol. Forty-eight hours after transfection, HEKn were induced to differentiate for 3 days before harvesting. Both human and mouse keratinocytes differentiation was induced by raising the calcium concentration to 1.2 mM.
Protein extraction and Western blot analysis
Epidermis was separated from the dermis using PBS with 10 mM EDTA and heating at 65°C for 30 s. Then, the epidermis was lysed in SDS buffer [100 mM Tris-HCl, 20 mM dithiothreitol, 5 mM EDTA, 2% SDS, and 2X protease inhibitor cocktail (PIC) (Roche)] with T25 ULTRA-TURRAX and left on a rotator overnight at 4°C. The following day, the samples were centrifuged to remove debris and the supernatant was collected for Western blot analysis.
Proteins were resolved on a polyacrylamide gel and blotted onto a nitrocellulose membrane (G&E Healthcare). The membranes were blocked with 10% milk in 0.1% PBS-Tween for 1 hour at 4°C. Membranes were then incubated with primary antibody for overnight at 4°C, washed, and then hybridized with the appropriate horseradish peroxidase–conjugated secondary antibody (rabbit and mouse; Bio-Rad, Hercules, CA, USA). Detection was performed with the ECL Chemiluminescence Kit (PerkinElmer, Waltham, MA, USA). The following antibodies were used: anti-KRT14 (1:50,000; BioLegend, #905301), anti-IVL (1:5000; BioLegend, #924401), anti-LOR (1:1000; BioLegend, #905101), anti-FLG (1:20,000; BioLegend, #905801), anti-cathepsin D (1:1000; Abcam), and anti–β-actin (1:50,000; Sigma-Aldrich, A-5441). Uncropped Western blot scans are provided in the Supplementary Materials.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assay was performed according to the Myers Lab ChIP-seq protocol (Huntsville, HudsonAlpha Institute for Biotechnology). Briefly, 3,000,000 of neonatal human keratinocytes were cross-linked in a solution containing 1% formaldehyde (Sigma-Aldrich) for 10′ at room temperature (RT). The reaction was stopped by the addition of 0.0125 M glycine (Sigma-Aldrich) for 5′ at RT. After lysis, chromatin was sonicated into 200- to 500-bp fragments using a Diagenode Bioruptor. Chromatin was precleared overnight with Protein G Sepharose beads (G&E Healthcare). Immunoprecipitation was performed overnight at 4°C using 2 μm of either nonspecific immunoglobulin G (IgG) (Invitrogen) or specific ZNF750 antibody (Sigma-Aldrich, HPA023012). The following day, the chromatin was washed and eluted, and purified DNA was used for qPCR analysis.
For ChIP assay on mouse keratinocytes, newborn WT mice were killed and skin was incubated in 0.25% trypsin (Gibco) for 4 hours at 37°C. Subsequently, primary mouse keratinocytes were extracted as previously described (59). Cells in suspension were fixed in solution containing 1% formaldehyde (Sigma-Aldrich) for 10′ at room temperature (RT) and then processed as described above. Immunoprecipitation was performed overnight at 4°C using one microgram of either nonspecific IgG (Invitrogen) or specific P63 antibody (Cell Signaling Technology, #13109). A negative control region was used to further confirm the specificity of the immunoprecipitation. Primer sequences used for ChIP-qPCR are listed in table S1.
Histological and immunofluorescence analysis
Hematoxylin and eosin staining was performed on paraffin-embedded tissue. Briefly, tissue sections (5 μm) were dewaxed and rehydrated, then stained for 4 min with hematoxylin (Merck), shortly washed in distilled water, and incubated for 1 min in eosin solution (Merck). Slides were then mounted using a mounting medium (Bio Mount HM, Bio Optica).
For immunofluorescence analysis, the skin was isolated from euthanized newborn pups, embedded in OCT (Bio Optica), and frozen at −80°C. Cryosections were cut at 10 μm thickness, thawed at RT for 5 min, and subsequently fixed in formalin (Bio Optica) for 10 min. After rinses in PBS, they were permeabilized with 0.1% Triton X-100 (Sigma-Aldrich) in PBS for 10 min and then flushed with PBS. Blocking was performed for 1 hour at RT with 10% goat serum after which primary antibodies were incubated either for 2 hours at RT or overnight at 4°C in blocking buffer. After removal of the primary antibody, the sections were washed in PBS and subsequently incubated for 1 hour at RT with fluorescent secondary antibody and DAPI (1 μg/ml; Sigma-Aldrich). Last, the slides were mounted with ProLong Gold Antifade Mountant (Invitrogen) and analyzed by a Nikon Confocal microscope (Nikon Eclipse Ti). For immunofluorescence studies on embryos, they were euthanized and fixed in formalin for a minimum of 24 hours after which they were processed for paraffin embedding. In this case, sections were cut at 4 μm thickness, deparaffinized, rehydrated, and subjected to antigen retrieval in 0.01 M sodium citrate buffer (pH 6.0; Sigma-Aldrich) for 15′ and then processed as described above.
The primary antibodies were as follows: p63 (1:1000; Cell Signaling Technology, #13109), KRT14-LL02 (1:1000; Abcam, ab7800), KRT10 (1:1000; Covance, PRB-159P), IVL (1:500; BioLegend, #924401), Filaggrin (1:1000; BioLegend, #905801), Loricrin (1:1000; BioLegend, #905101), β-galactosidase (1:500; Abcam, ab9361), Ki-67 (1:400; Cell Signaling Technology, #12202), zonula occludens-1 (1:200; Invitrogen, #402200), GlcCer/Cer (1:300; GlycoBiotech, RAS0011), and anti-CTSD (1:100; Abcam, ab75852,). All secondary antibodies were Alexa Fluor–conjugated (Invitrogen).
Electron microscopy
Mouse skin was fixed in 2% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) at 4°C overnight. Fixation was performed with 1% OsO4. Samples were then dehydrated in graded ethanol and embedded in Epon resin. Ultrathin sections were stained with 2% uranyl acetate and examined under a transmission electron microscope (JEOL JEM-2100Plus, Japan Electron Optics Laboratory). Images were captured with a digital camera (Tietz Video and Image Processing Systems).
Lipid analysis by Nile red
A stock solution of Nile red (1 mg/ml; Sigma-Aldrich) in acetone was diluted to a working solution of 1 μg/ml in 75:25 glycerol:water followed by a rapid vortexing. Mouse frozen skin sections were thawed at room temperature for 5 min, rapidly rinsed in PBS, and stained in the dark with the working solution for 30 min, after which the coverslip was added. For Nile red staining on primary human and mouse keratinocytes, the cells were grown on a coverslip. They were washed once in PBS and subsequently stained as described for skin sections. Images were acquired with a Nikon confocal microscope (Nikon Eclipse Ti). Lipid droplets stained by Nile red were quantified using the ImageJ software (https://imagej.nih.gov/ij/).
Whole transcriptome analysis by RNA-seq
Total RNA was extracted from mouse epidermis using QIAzol (QIAGEN, IT) according to the manufacturer’s instruction. RNA libraries for sequencing were generated using the same amount of RNA for each sample as previously described (60).
RNA-seq analysis
RNA-seq data were analyzed using the RNA-seq analysis cloud pipeline (Tuxedo Suite-based) (61). In particular, quality checks and filtering were performed with FastQC and NGS QC Toolkit, respectively, while reads mapping was carried out with the TopHat aligner against mm10 reference genome (62). Next, transcript assembly and abundance estimation were performed using Cufflinks and FeatureCounts softwares. We tested for differential gene expression between WT and KO mice using CuffDiff. Only genes with adjusted P value (q value) < 0.05 were considered significant. Ontology enrichment were realized with the Auto-GO software through the Enrichr API (63).
UPLC-MS/MS lipid profiling
Approximately 10 mg of each tissue sample was accurately weighed and then homogenized with 200 μl of water (Millipore ultrapure) using Bullet Blender tissue homogenizer (Next Advance, Averill Park, NY, USA). Lipids were extracted with 1 ml of chloroform/methanol [2:1 (v/v)] with the internal standard mixture containing phosphatidylcholine 15:0-18:1(d7), phosphatidylethanolamine 15:0-18:1(d7), phosphatidylserine 15:0-18:1(d7), phosphatidylinositol 15:0-18:1(d7), lysophosphatidylcholine 18:1(d7), lysophosphatidylethanolamine 18:1(d7), cholesterol ester 18:1(d7), diacylglycerols 15:0-18:1(d7), triglycerides 15:0-18:1(d7)-15:0, sphingomyelin 18:1(d9), and ceramides 16:0-d18:1(d7) (Avanti Polar Lipids, Alabaster, AL, USA) based on the modification of method published (64). After centrifugation at 13,500g and 4°C for 10 min, the organic phase was collected and dried under nitrogen. The dried residue was subsequently dissolved in 1 ml of methanol containing 5 mM ammonium acetate, pending ultraperformance liquid chromatography–tandem mass spectrometry (UPLC-MS/MS) analysis.
Lipid profiling was performed on a Waters ACQUITY UPLC system coupled with a Waters XEVO TQ-S MS controlled by MassLynx 4.1 software (Waters, Milford, MA). Chromatographic separation was performed with an ACQUITY BEH C18 column (1.7 μm, 100 mm by 2.1 mm internal dimensions) (Waters, Milford, MA). Gradient elution was started at a flow rate of 0.4 ml/min, with the injected volume being 5 μl and the column temperature at 40°C. Both mobile phases A and B contained 5 mM ammonium formate in a solution of A [acetonitrile/H2O (95:5, v/v) and B [acetonitrile/isopropanol (10:90, v/v)], respectively. The lipids were eluted using the following gradients: 0 to 2 min, 60% B; 2 to 8 min, 60 to 100% B; 8 to 10 min, 100% B; and 10 to 12 min, 60% B. The capillary voltage of the ESI+ mode was 3.2 kV. Desolvation gas flow (liter/hour) was set as 1000. The electrospray ion source temperature and desolvation temperature were maintained at 150° and 500°C, respectively. Selected lipids were detected with multiple reaction monitoring mode with specified precursor and product ions as described previously (65–67). Peak integration and quantitation of raw data of lipids generated by UPLC-Triple Quadrupole Mass Spectroscopy was performed using the TargetLynx application manager (v4.1, Waters, Milford, MA, USA).
Bioinformatic analysis
Bioinformatic analysis was carried out interrogating the DAVID database (https://david.ncifcrf.gov). For promoter analysis, JASPAR and FIMO websites were interrogated (68, 69). The analysis spanned from 5-kbp upstream the TSS up to 1-kbp downstream the TSS. Only putative binding sites with higher degree of conservation between human and mouse and with P value < 0.05 and q value < 0.01 were considered.
Statistical analysis
A minimum of four mice were included per experimental group. For western blots and cell culture experiments, a minimum of two biological replicates were performed. For parametric data, data significance was analyzed using a two-tailed unpaired Student’s t test. Prism software version 9 (GraphPad Software Inc.) was used for all statistical analyses. All results are expressed as means ± SD. P < 0.05 was considered significant. n refers to biological replicates that are reported in each figure legend.
Acknowledgments
We would like to thank G. Raschellà for helpful comments and suggestions. We thank the staff members of the animal facility at the University of Rome Tor Vergata (Italy).
Funding: This work has been supported primarily by the MUR-PNRR (M4C2I1.3 PE6 project PE00000019 Heal Italia CUP: E83C22004670001) (to E.C., G.M., and M.A.). This work has also been supported by Associazione Italiana per la Ricerca contro il Cancro (AIRC) IG#20473 (2018–2022) and IG 2022 ID 27366 (2023–2027) (to G.M.), AIRC IG#22206 (2019–2023) (to E.C.), Fondazione Luigi Maria Monti IDI-IRCCS ricerca corrente (to E.C.), Ministry of Health & MAECI Italy-China Science and Technology Cooperation PGR00961 (to E.C.), The National Key R&D Program of China 2022YFA0807300 (to Y.W.), Regione Lazio through LazioInnova Progetto Gruppo di Ricerca n 85-2017-14986 (n 33 & n 55-2021-T0002E0001; to G.M. and E.C.), Progetti di Ricerca Scientifica d’Ateneo CUP:E83C22001850005 (to M.A.), and CAMS Innovation Fund for Medical Sciences 2021-I2M-5-008 (to Q.S.).
Author contributions: Conceptualization: G.M. and E.C. Methodology: A.B., M.A., M.C., F.D.N., M.F., L.D.A., L.F., M.P., R.N., L.W., W.J., Q.S., Y.S., and Y.W. Investigation: A.B., M.A., M.C., F.D.N., L.F., R.N., and L.W. Visualization: A.B., M.A., L.D.A., L.F., R.N., and L.W. Supervision: M.A., G.M., E.C., R.A.K. Writing—original draft: M.A., G.M., E.C., and R.A.K. Writing–review and editing: M.A., A.B., G.M., and E.C.
Competing interests: G.M., E.C., and M.A. are members of the Editorial Board of CDDpress/Springer Nature. The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Gene expression data of RNA-seq are deposited in GEO repository (accession number GSE220813).
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Tables S1 to S6
Supplementary Materials (Uncropped Western blot scans)
References
REFERENCES AND NOTES
- 1.E. Candi, R. Schmidt, G. Melino, The cornified envelope: A model of cell death in the skin. Nat. Rev. Mol. Cell Biol. 6, 328–340 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Y.-C. Hsu, E. Fuchs, Building and maintaining the skin. Cold Spring Harb. Perspect. Biol. 14, a040840 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.C. Blanpain, E. Fuchs, Epidermal homeostasis: A balancing act of stem cells in the skin. Nat. Rev. Mol. Cell Biol. 10, 207–217 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.V. Frezza, A. Terrinoni, C. Pitolli, A. Mauriello, G. Melino, E. Candi, Transglutaminase 3 protects against photodamage. J. Invest. Dermatol. 137, 1590–1594 (2017). [DOI] [PubMed] [Google Scholar]
- 5.K. R. Feingold, P. M. Elias, Role of lipids in the formation and maintenance of the cutaneous permeability barrier. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1841, 280–294 (2014). [DOI] [PubMed] [Google Scholar]
- 6.P. Shamaprasad, C. O. Frame, T. C. Moore, A. Yang, C. R. Iacovella, J. A. Bouwstra, A. L. Bunge, C. McCabe, Using molecular simulation to understand the skin barrier. Prog. Lipid Res. 88, 101184 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.M. Blaess, L. Kaiser, O. Sommerfeld, R. Csuk, H.-P. Deigner, Drug triggered pruritus, rash, papules, and blisters - is AGEP a clash of an altered sphingolipid-metabolism and lysosomotropism of drugs accumulating in the skin? Lipids Health Dis. 20, 156 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.T. Kono, Y. Miyachi, M. Kawashima, Clinical significance of the water retention and barrier function-improving capabilities of ceramide-containing formulations: A qualitative review. J. Dermatol. 48, 1807–1816 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.M. Szántó, R. Gupte, W. L. Kraus, P. Pacher, P. Bai, PARPs in lipid metabolism and related diseases. Prog. Lipid Res. 84, 101117 (2021). [DOI] [PubMed] [Google Scholar]
- 10.H. Takahashi, H. Tsuji, M. Minami-Hori, Y. Miyauchi, H. Iizuka, Defective barrier function accompanied by structural changes of psoriatic stratum corneum. J. Dermatol. 41, 144–148 (2014). [DOI] [PubMed] [Google Scholar]
- 11.H.-J. Lee, S.-H. Lee, Epidermal permeability barrier defects and barrier repair therapy in atopic dermatitis. Allergy Asthma Immunol. Res. 6, 276–287 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.P. W. Wertz, Lipids and the permeability and antimicrobial barriers of the skin. J Lipids. 2018, 1–7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.G. L. Sen, L. D. Boxer, D. E. Webster, R. T. Bussat, K. Qu, B. J. Zarnegar, D. Johnston, Z. Siprashvili, P. A. Khavari, ZNF750 is a p63 target gene that induces KLF4 to drive terminal epidermal differentiation. Dev. Cell 22, 669–677 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.M. Hazawa, D.-C. Lin, H. Handral, L. Xu, Y. Chen, Y.-Y. Jiang, A. Mayakonda, L.-W. Ding, X. Meng, A. Sharma, S. Samuel, M. M. Movahednia, R. W. Wong, H. Yang, C. Tong, H. P. Koeffler, ZNF750 is a lineage-specific tumour suppressor in squamous cell carcinoma. Oncogene 36, 2243–2254 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.M. Cassandri, A. Butera, I. Amelio, A. M. Lena, M. Montanaro, A. Mauriello, L. Anemona, E. Candi, R. A. Knight, M. Agostini, G. Melino, ZNF750 represses breast cancer invasion via epigenetic control of prometastatic genes. Oncogene 39, 4331–4343 (2020). [DOI] [PubMed] [Google Scholar]
- 16.A. Butera, M. Cassandri, F. Rugolo, M. Agostini, G. Melino, The ZNF750–RAC1 axis as potential prognostic factor for breast cancer. Cell Death Discov. 6, 135 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D.-C. Lin, J.-J. Hao, Y. Nagata, L. Xu, L. Shang, X. Meng, Y. Sato, Y. Okuno, A. M. Varela, L.-W. Ding, M. Garg, L.-Z. Liu, H. Yang, D. Yin, Z.-Z. Shi, Y.-Y. Jiang, W.-Y. Gu, T. Gong, Y. Zhang, X. Xu, O. Kalid, S. Shacham, S. Ogawa, M.-R. Wang, H. P. Koeffler, Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat. Genet. 46, 467–473 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.A. Yang, R. Schweitzer, D. Sun, M. Kaghad, N. Walker, R. T. Bronson, C. Tabin, A. Sharpe, D. Caput, C. Crum, F. McKeon, p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398, 714–718 (1999). [DOI] [PubMed] [Google Scholar]
- 19.L. D. Boxer, B. Barajas, S. Tao, J. Zhang, P. A. Khavari, ZNF750 interacts with KLF4 and RCOR1, KDM1A, and CTBP1/2 chromatin regulators to repress epidermal progenitor genes and induce differentiation genes. Genes Dev. 28, 2013–2026 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.R. Y. Birnbaum, A. Zvulunov, D. Hallel-Halevy, E. Cagnano, G. Finer, R. Ofir, D. Geiger, E. Silberstein, Y. Feferman, O. S. Birk, Seborrhea-like dermatitis with psoriasiform elements caused by a mutation in ZNF750, encoding a putative C2H2 zinc finger protein. Nat. Genet. 38, 749–751 (2006). [DOI] [PubMed] [Google Scholar]
- 21.C.-F. Yang, W.-L. Hwu, L.-C. Yang, W.-H. Chung, Y.-H. Chien, C.-F. Hung, H.-C. Chen, P.-J. Tsai, C. S. J. Fann, F. Liao, Y.-T. Chen, A promoter sequence variant of ZNF750 is linked with familial psoriasis. J. Invest. Dermatol. 128, 1662–1668 (2008). [DOI] [PubMed] [Google Scholar]
- 22.I. Cohen, R. Y. Birnbaum, K. Leibson, R. Taube, S. Sivan, O. S. Birk, ZNF750 is expressed in differentiated keratinocytes and regulates epidermal late differentiation genes. PLOS ONE 7, e42628 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.W. C. Skarnes, B. Rosen, A. P. West, M. Koutsourakis, W. Bushell, V. Iyer, A. O. Mujica, M. Thomas, J. Harrow, T. Cox, D. Jackson, J. Severin, P. Biggs, J. Fu, M. Nefedov, P. J. de Jong, A. F. Stewart, A. Bradley, A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474, 337–342 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.R. L. Eckert, M. T. Sturniolo, A.-M. Broome, M. Ruse, E. A. Rorke, Transglutaminase function in epidermis. J. Invest. Dermatol. 124, 481–492 (2005). [DOI] [PubMed] [Google Scholar]
- 25.P. J. Koch, P. A. de Viragh, E. Scharer, D. Bundman, M. A. Longley, J. Bickenbach, Y. Kawachi, Y. Suga, Z. Zhou, M. Huber, D. Hohl, T. Kartasova, M. Jarnik, A. C. Steven, D. R. Roop, Lessons from loricrin-deficient mice. J. Cell Biol. 151, 389–400 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.M. Rabionet, K. Gorgas, R. Sandhoff, Ceramide synthesis in the epidermis. Biochim. Biophys. Acta 1841, 422–434 (2014). [DOI] [PubMed] [Google Scholar]
- 27.T. Yanagi, M. Akiyama, H. Nishihara, J. Ishikawa, K. Sakai, Y. Miyamura, A. Naoe, T. Kitahara, S. Tanaka, H. Shimizu, Self-improvement of keratinocyte differentiation defects during skin maturation in ABCA12-deficient harlequin ichthyosis model mice. Am. J. Pathol. 177, 106–118 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.F. Egberts, M. Heinrich, J.-M. Jensen, S. Winoto-Morbach, S. Pfeiffer, M. Wickel, M. Schunck, J. Steude, P. Saftig, E. Proksch, S. Schütze, Cathepsin D is involved in the regulation of transglutaminase 1 and epidermal differentiation. J. Cell Sci. 117, 2295–2307 (2004). [DOI] [PubMed] [Google Scholar]
- 29.P. Djian, K. Easley, H. Green, Targeted ablation of the murine involucrin gene. J. Cell Biol. 151, 381–388 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.A. Sandilands, C. Sutherland, A. D. Irvine, W. H. I. McLean, Filaggrin in the frontline: Role in skin barrier function and disease. J. Cell Sci. 122, 1285–1294 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.K.-K. Hong, H.-R. Cho, W.-C. Ju, Y. Cho, N.-I. Kim, A study on altered expression of serine palmitoyltransferase and ceramidase in psoriatic skin lesion. J. Korean Med. Sci. 22, 862–867 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.T. Hornemann, Y. Wei, A. von Eckardstein, Is the mammalian serine palmitoyltransferase a high-molecular-mass complex? Biochem. J. 405, 157–164 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.S. J. Stone, H. M. Myers, S. M. Watkins, B. E. Brown, K. R. Feingold, P. M. Elias, R. v. Farese, Lipopenia and skin barrier abnormalities in DGAT2-deficient mice. J. Biol. Chem. 279, 11767–11776 (2004). [DOI] [PubMed] [Google Scholar]
- 34.H. Gallala, O. Macheleidt, T. Doering, V. Schreiner, K. Sandhoff, Nitric oxide regulates synthesis of gene products involved in keratinocyte differentiation and ceramide metabolism. Eur. J. Cell Biol. 83, 667–679 (2004). [DOI] [PubMed] [Google Scholar]
- 35.L. Bin, L. Deng, H. Yang, L. Zhu, X. Wang, M. G. Edwards, B. Richers, D. Y. M. Leung, Forkhead box C1 regulates human primary keratinocyte terminal differentiation. PLOS ONE 11, e0167392 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.F. P. W. Radner, J. Fischer, The important role of epidermal triacylglycerol metabolism for maintenance of the skin permeability barrier function. Biochim. Biophys. Acta 1841, 409–415 (2014). [DOI] [PubMed] [Google Scholar]
- 37.M. Tachi, M. Iwamori, Mass spectrometric characterization of cholesterol esters and wax esters in epidermis of fetal, adult and keloidal human skin. Exp. Dermatol. 17, 318–323 (2008). [DOI] [PubMed] [Google Scholar]
- 38.J. W. Fluhr, J. Kao, S. K. Ahn, K. R. Feingold, P. M. Elias, M. Jain, Generation of free fatty acids from phospholipids regulates stratum corneum acidification and integrity. J. Invest. Dermatol. 117, 44–51 (2001). [DOI] [PubMed] [Google Scholar]
- 39.M. Schmuth, V. Moosbrugger-Martinz, S. Blunder, S. Dubrac, Role of PPAR, LXR, and PXR in epidermal homeostasis and inflammation. Lipids 1841, 463–473 (2014). [DOI] [PubMed] [Google Scholar]
- 40.L. G. Kömüves, K. Hanley, A.-M. Lefebvre, M.-Q. Man, D. C. Ng, D. D. Bikle, M. L. Williams, P. M. Elias, J. Auwerx, K. R. Feingold, Stimulation of PPARα promotes epidermal keratinocyte differentiation in vivo. J. Invest. Dermatol. 115, 353–360 (2000). [DOI] [PubMed] [Google Scholar]
- 41.D. Cipolletta, M. Feuerer, A. Li, N. Kamei, J. Lee, S. E. Shoelson, C. Benoist, D. Mathis, PPAR-γ is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature 486, 549–553 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.M.-Q. Man, G. D. Barish, M. Schmuth, D. Crumrine, Y. Barak, S. Chang, Y. Jiang, R. M. Evans, P. M. Elias, K. R. Feingold, Deficiency of PPARβ/δ in the epidermis results in defective cutaneous permeability barrier homeostasis and increased inflammation. J. Invest. Dermatol. 128, 370–377 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Y. Oda, Y. Uchida, S. Moradian, D. Crumrine, P. M. Elias, D. D. Bikle, Vitamin D receptor and coactivators SRC2 and 3 regulate epidermis-specific sphingolipid production and permeability barrier formation. J. Invest. Dermatol. 129, 1367–1378 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D. Fehrenschild, U. Galli, B. Breiden, W. Bloch, P. Schettina, S. Brodesser, C. Michels, C. Günschmann, K. Sandhoff, C. M. Niessen, C. Niemann, TCF/lef1-mediated control of lipid metabolism regulates skin barrier function. J. Invest. Dermatol. 132, 337–345 (2012). [DOI] [PubMed] [Google Scholar]
- 45.A. Kihara, Very long-chain fatty acids: Elongation, physiology and related disorders. J. Biochem. 152, 387–395 (2012). [DOI] [PubMed] [Google Scholar]
- 46.J. A. Olzmann, P. Carvalho, Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 20, 137–155 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.K. Nakajima, M. Terao, M. Takaishi, S. Kataoka, N. Goto-Inoue, M. Setou, K. Horie, F. Sakamoto, M. Ito, H. Azukizawa, S. Kitaba, H. Murota, S. Itami, I. Katayama, J. Takeda, S. Sano, Barrier abnormality due to ceramide deficiency leads to psoriasiform inflammation in a mouse model. J. Invest. Dermatol. 133, 2555–2565 (2013). [DOI] [PubMed] [Google Scholar]
- 48.W. M. Holleran, Y. Takagi, Y. Uchida, Epidermal sphingolipids: Metabolism, function, and roles in skin disorders. FEBS Lett. 580, 5456–5466 (2006). [DOI] [PubMed] [Google Scholar]
- 49.S. Kim, I. Hong, J. S. Hwang, J. K. Choi, H. S. Rho, D. H. Kim, I. Chang, S. H. Lee, M.-O. Lee, J. S. Hwang, Phytosphingosine stimulates the differentiation of human keratinocytes and inhibits TPA-induced inflammatory epidermal hyperplasia in hairless mouse skin. Mol. Med. 12, 17–24 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.J. A. Segre, Epidermal barrier formation and recovery in skin disorders. J. Clin. Investig. 116, 1150–1158 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.E. Proksch, W. M. Holleran, G. K. Menon, P. M. Elias, K. R. Feingold, Barrier function regulates epidermal lipid and DNA synthesis. Br. J. Dermatol. 128, 473–482 (1993). [DOI] [PubMed] [Google Scholar]
- 52.C. Blanpain, W. E. Lowry, H. A. Pasolli, E. Fuchs, Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes Dev. 20, 3022–3035 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.W. Ning, A. Muroyama, H. Li, T. Lechler, Differentiated daughter cells regulate stem cell proliferation and fate through intra-tissue tension. Cell Stem Cell 28, 436–452.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Y. A. Miroshnikova, H. Q. Le, D. Schneider, T. Thalheim, M. Rübsam, N. Bremicker, J. Polleux, N. Kamprad, M. Tarantola, I. Wang, M. Balland, C. M. Niessen, J. Galle, S. A. Wickström, Adhesion forces and cortical tension couple cell proliferation and differentiation to drive epidermal stratification. Nat. Cell Biol. 20, 69–80 (2018). [DOI] [PubMed] [Google Scholar]
- 55.K. R. Mesa, K. Kawaguchi, K. Cockburn, D. Gonzalez, J. Boucher, T. Xin, A. M. Klein, V. Greco, Homeostatic epidermal stem cell self-renewal is driven by local differentiation. Cell Stem Cell 23, 677–686.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.J. Tomfohrde, A. Silverman, R. Barnes, M. A. Fernandez-Vina, M. Young, D. Lory, L. Morris, K. D. Wuepper, P. Stastny, A. Menter, A. Bowcock, Gene for familial psoriasis susceptibility mapped to the distal end of human chromosome 17q. Science 264, 1141–1145 (1994). [DOI] [PubMed] [Google Scholar]
- 57.R. Shalom-Feuerstein, L. Serror, E. Aberdam, F.-J. Müller, H. van Bokhoven, K. G. Wiman, H. Zhou, D. Aberdam, I. Petit, Impaired epithelial differentiation of induced pluripotent stem cells from ectodermal dysplasia-related patients is rescued by the small compound APR-246/PRIMA-1MET. Proc. Natl. Acad. Sci. U.S.A. 110, 2152–2156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.E. Aberdam, L. N. Roux, P.-H. Secrétan, F. Boralevi, J. Schlatter, F. Morice-Picard, S. Sol, C. Bodemer, C. Missero, S. Cisternino, D. Aberdam, S. Hadj-Rabia, Improvement of epidermal covering on AEC patients with severe skin erosions by PRIMA-1MET/APR-246. Cell Death Dis. 11, 30 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.U. Lichti, J. Anders, S. H. Yuspa, Isolation and short-term culture of primary keratinocytes, hair follicle populations and dermal cells from newborn mice and keratinocytes from adult mice for in vitro analysis and for grafting to immunodeficient mice. Nat. Protoc. 3, 799–810 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.C. Contadini, A. Ferri, M. Di Martile, C. Cirotti, D. Del Bufalo, F. De Nicola, M. Pallocca, M. Fanciulli, F. Sacco, G. Donninelli, A. Capone, E. Volpe, N. Keller, S. Miki, D. Kawauchi, D. Stupack, F. Furnari, D. Barilà, Caspase-8 as a novel mediator linking Src kinase signaling to enhanced glioblastoma malignancy. Cell Death Differ. 30, 417–428 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.M. D’Antonio, P. D’Onorio De Meo, M. Pallocca, E. Picardi, A. M. D’Erchia, R. A. Calogero, T. Castrignanò, G. Pesole, RAP: RNA-Seq analysis pipeline, a new cloud-based NGS web application. BMC Genomics 16, S3 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.C. Trapnell, A. Roberts, L. Goff, G. Pertea, D. Kim, D. R. Kelley, H. Pimentel, S. L. Salzberg, J. L. Rinn, L. Pachter, Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.E. Y. Chen, C. M. Tan, Y. Kou, Q. Duan, Z. Wang, G. V. Meirelles, N. R. Clark, A. Ma’ayan, Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.J. Folch, M. Lees, G. H. S. Stanley, A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509 (1957). [PubMed] [Google Scholar]
- 65.J. Franco, B. Rajwa, C. R. Ferreira, J. P. Sundberg, H. HogenEsch, Lipidomic profiling of the epidermis in a mouse model of dermatitis reveals sexual dimorphism and changes in lipid composition before the onset of clinical disease. Metabolites 10, 299 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.A. K. Nilsson, U. Sjöbom, K. Christenson, A. Hellström, Lipid profiling of suction blister fluid: Comparison of lipids in interstitial fluid and plasma. Lipids Health Dis. 18, 164 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.T. Zhang, S. Chen, X. Liang, H. Zhang, Development of a mass-spectrometry-based lipidomics platform for the profiling of phospholipids and sphingolipids in brain tissues. Anal. Bioanal. Chem. 407, 6543–6555 (2015). [DOI] [PubMed] [Google Scholar]
- 68.C. E. Grant, T. L. Bailey, W. S. Noble, FIMO: Scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.A. Sandelin, JASPAR: An open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 32, D91–D94 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Tables S1 to S6
Supplementary Materials (Uncropped Western blot scans)
References