Abstract
A plate confrontation experiment is commonly used to study the mechanism by which Trichoderma spp. antagonize and parasitize other fungi. Previous work with chitinase gene expression (ech42) during the precontact period of this process in which cellophane and dialysis membranes separated Trichoderma harzianum and its host Rhizoctonia solani resulted in essentially opposite results. Here, we show that cellophane membranes are permeable to proteins up to at least 90 kDa in size but that dialysis membranes are not. ech42 was expressed during the precontact stage of the confrontation between Trichoderma atroviride and its host only if the cellophane was placed between the two fungi. These results are consistent with enzyme diffusion from T. atroviride to R. solani generating the trigger of ech42 gene expression.
The ability of some species of Trichoderma to antagonize and parasitize other fungi has made them effective biocontrol agents against a range of plant pathogens (4, 8, 13). The mechanism responsible for biocontrol is unknown, although both hydrolytic enzymes (chitinases, glucanases, and proteases) and antibiotics play an important role (5, 9, 11, 14, 15). In particular, two chitinases (ech42 and chit33) and a protease (prb1) appear important, even though the events triggering the expression of these genes during antagonistic interaction of Trichoderma with other fungi are not well understood.
Recently (6, 16), ech42 expression has been shown to be triggered by diffusable factors whose formation does not require contact between Trichoderma and Rhizoctonia solani. Cortes et al. (6) used Northern blot analysis and detected ech42 gene expression even though Trichoderma and R. solani were separated by a cellophane membrane. In contrast, Zeilinger et al. (16) separated the two fungi with a dialysis membrane but found no ech42 expression with a green fluorescent protein (GFP) reporter system. Resolving this apparent discrepancy is critical to understanding how cell wall degradation products, which both groups (6, 16) reported to elicit ech42 gene expression, are formed.
In our laboratories, we frequently cultivate Trichoderma spp. on agar plates covered with a cellophane membrane, since this facilitates the removal of the fungus from the plate for subsequent analysis. Since this mode of cultivation also works well with macromolecular substrates such as xylan (unpublished observations), we suspected that the cellophane membrane may not be completely impermeable to proteins. We therefore compared the use of cellophane and dialysis membranes as tools to separate the two colonies during confrontation experiments.
To determine if macromolecules can permeate the cellophane and dialysis membranes used in the confrontation experiment, circular pieces of the cellophane membrane or the dialysis tubing (2-cm diameter) were wetted, wrapped around the bottom of an ISCO cup, and fixed with a tight rubber band. The cup was then filled with 1 ml of a solution of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) low-molecular-weight standard proteins (2 mg/ml) and placed in a 30-mm-diameter Petri dish containing 2 ml of distilled water. After incubation for 24 h at room temperature, 1 ml of water was withdrawn, mixed with 2 ml of 96% ethanol, allowed to stand for 2 h at −20°C, and centrifuged (13,000 × g, 4°C, 10 min). The precipitate was resuspended in 1 ml of SDS-PAGE sample buffer (1), boiled (3 min, 95°C), and subjected to SDS-PAGE in 7.5% separation gels (1). Marker protein controls were also dissolved in SDS-PAGE sample buffer, boiled as described above, and run on the same gel.
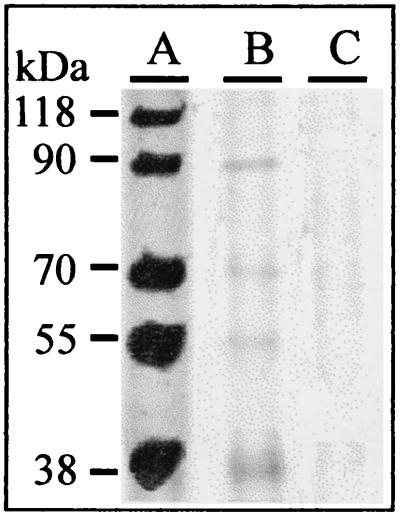
We incubated membranes with Trichoderma atroviride strain P1 (ATCC 74058) by growing the fungus on potato dextrose agar (Merck, Darmstadt, Germany), covered with the membrane, for 4 days at 30°C. By this time, the fungus had covered the entire plate as a fine, hairy mycelium. We scraped the fungus from the plate with a spatula and rinsed the membrane with 200 ml of distilled water. Circular pieces (2-cm diameter) were cut from portions of the membrane that were free of fungal hyphae, as determined microscopically. We found a small, but clearly detectable, amount of all but the largest marker protein (118 kDa) in the water (Fig. 1, lane B). The control (membrane not incubated with T. atroviride [data not shown]) also showed the same result, indicating that the cellophane membrane is permeable to proteins up to 90 kDa and that cultivation of Trichoderma on it does not further alter this property. In contrast, no proteins diffused through the dialysis membrane (Fig. 1, lane C). From this experiment we concluded that the cellophane membrane is not completely impermeable to proteins and that its use in the confrontation assay would not prevent proteins and macromolecules of similar sizes from diffusing from one fungus to the other.
FIG. 1.
SDS-PAGE of marker proteins applied onto the cellophane and into closed dialysis membrane tubes (lane A) and of material leaking through the cellophane (lane B) and the dialysis membrane (lane C). Forty micrograms of marker proteins (lane A) and 20 μl of samples (lanes B and C) were applied, and gels were stained with Coomassie blue. Samples in lanes B and C were concentrated such that they were directly comparable to that in lane A (see the text for details).
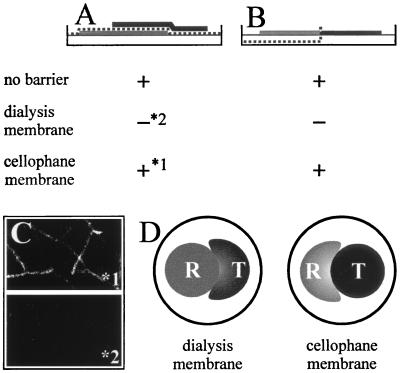
This result suggests that the reason for the different findings may be attributed to the type of membrane used; it also implies that experiments performed with a dialysis membrane would not show ech42 gene expression but that those performed with a cellophane membrane would. We tested these hypotheses by using T. atroviride ZEGA 2#6, carrying six copies of the ech42::GFP reporter construct (16), and R. solani strain 1450 (Institute of Plant Pathology, Naples, Italy) as the plant-pathogenic host, as previously described (6, 16). Plate confrontation assays were carried out with agar plates covered with cellophane and incubated in the dark (12). For confrontation assays that avoid physical contact, the host and mycoparasite were separated by dialysis membranes (Sigma, Deisenhofen, Germany; cutoff size, 12 kDa) or cellophane, as previously described (6, 16) (see Fig. 2). Microscopic analyses of ech42::GFP expression were performed using a fluorescence microscope (DMRE/HC; Leica, Solms, Germany) fitted with a Leica filter set (L4 band-pass, 450- to 490-nm excitation filter, 515- to 560-nm emission filter). Small pieces (5 to 10 mm2) of the cellophane were cut out at the interaction zone, and hyphae were removed from the cellophane with a drop of sterile water. Digitized pictures (Fig. 2) were obtained using a video capture system with an attached Sony DXC/950P camera. From these experiments we concluded that the membranes used, cellophane and dialysis, were responsible for the differences in ech42 gene expression and not other differences in the two experimental setups.
FIG. 2.
Confrontation assay between T. atroviride (T) ech42::GFP (ZEGA 2#6; black areas) and R. solani (R; gray areas). (A) Arrangement of the membranes according to the work of Cortes et al. (6). The membrane is shown by a dotted line. (B) Arrangement of the membranes according to the work of Zeilinger et al. (16). + indicates ech42::GFP expression, and − indicates no expression. 1 and 2 refer to the two photographs shown in panel C. (C) Fluorescence microscopy photographs of the effects of using the membrane arrangement of Cortes et al. (6) with a cellophane (1*) and a dialysis (2*) membrane on ech42::GFP expression. (D) Patterns of colony growth of Trichoderma and Rhizoctonia under the experimental conditions of Cortes et al. (6) when a dialysis or a cellophane membrane was used to separate the two fungi.
Interestingly, when the dialysis membrane was used in the experimental setup of Cortes et al. (6), R. solani inhibited Trichoderma (Fig. 2D). This result suggests that macromolecule diffusion is required for T. atroviride to attack R. solani and to overcome the competitive action (e.g., for nutrients) by the host. Whether Trichoderma prevents the formation of or inactivates a host-derived inhibitory compound remains to be clarified, but to the best of our knowledge, no inhibitory compounds from R. solani have been described.
These results led us to conclude that the expression of ech42 during the precontact period of a mycoparasitic interaction requires the diffusion of a macromolecule of 12 to 90 kDa (i.e., larger than the 12-kDa cutoff size of the dialysis membrane but not larger than the largest marker that penetrated the cellophane membrane) from one fungus to the other. This confirms that our previous hypothesis (16) is correct and resolves the apparent inconsistency between our results and those of Cortes et al. (6).
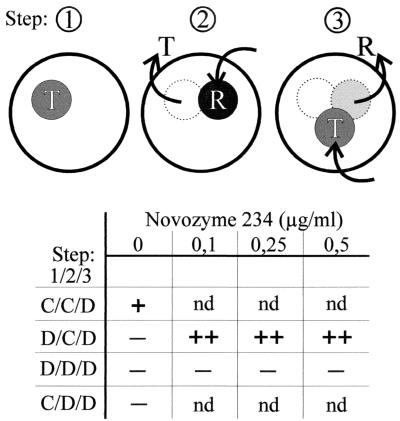
Having two types of membranes with different levels of permeability for fungal extracellular macromolecules available further provided us with a tool to learn whether this triggering macromolecule is produced by Trichoderma or Rhizoctonia. To this end, we grew T. atroviride on synthetic medium (16) with 0.1% (wt/vol) glucose on plates that had been covered with one of the two membranes (see Fig. 3) as described above (step 1). Thereafter, the membrane plus the fungus was removed and the plate was covered with a new membrane and inoculated with Rhizoctonia (step 2). Finally, Rhizoctonia was removed, a dialysis membrane was placed on the plate, and the plate was inoculated with T. atroviride ZEGA 2#6 (step 3). In these experiments, GFP formation was observed only when a cellophane membrane was used both in the first and in the second step (Fig. 3), suggesting that the production of a macromolecule by Trichoderma and its action on Rhizoctonia are crucial for ech42 expression.
FIG. 3.
Schematic drawing of the consecutive steps of the plate confrontation assay. Disks represent plates with colonies of the fungi T. atroviride ZEGA 2#6 (T) and R. solani (R). Arrows pointing from the plates indicate removal of the fungus; arrows pointing into the plate indicate inoculation of agar disks with the respective fungus. The membranes used in the four different experiments are indicated in the numerical order of the three steps, with C meaning cellophane, and D meaning dialysis membrane. Novozyme 234 was applied as a solution on top of the agar, distributed equally, and allowed to diffuse into the agar for 2 h at room temperature before a membrane was added. − indicates no fluorescence, and + indicates fluorescence when samples were examined under a fluorescence microscope. nd, not determined.
The blockage of macromolecule penetration into the agar in step 1 could be fully compensated for by inclusion of a low concentration (0.1 to 0.5 μg/ml) of a lytic enzyme preparation of T. harzianum in the second plate (Fig. 2) (we chose Novozyme 234 [Sigma, St. Louis, Mo.] for this experiment, as this preparation is readily available and the experiment is thus easily repeatable by other workers). However, this addition did not compensate for the use of a dialysis membrane in step 2, indicating that contact between the macromolecule and Rhizoctonia was required. The final triggering of ech42 expression (step 3) was observed even when a dialysis membrane was used in this step, indicating that the inducer is of low molecular weight. These data support a model in which an attack on Rhizoctonia by a macromolecule of Trichoderma releases a low-molecular-weight inducer of ech42 expression. As this macromolecule releases a compound from Rhizoctonia and because a similar effect could be obtained with Novozyme 234, we hypothesize that this macromolecule is an enzyme, most likely a chitinase as its action can be inhibited by allosamidine (16). The system used here will be useful in the purification of this enzyme from Novozyme 234 and other extracellular culture fluids of Trichoderma biocontrol strains.
Acknowledgments
The first two authors contributed equally to the manuscript and are listed in alphabetical order.
This study was supported by a grant from the FWF (Austrian Science Foundation, P12748-MOB).
Thanks are due to E. M. Kubicek-Pranz for critically discussing the experimental approach used in this paper.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Wiley-Interscience; 1990. [Google Scholar]
- 2.Baek J-M, Howell C R, Kenerley C M. The role of an extracellular chitinase from Trichoderma virens Gv29-8 in the biocontrol of Rhizoctonia solani. Curr Genet. 1999;35:41–50. doi: 10.1007/s002940050431. [DOI] [PubMed] [Google Scholar]
- 3.Carsolio C, Benhamou N, Haran S, Cortes C, Gutierrez A, Herrera-Estrella A. Role of the Trichoderma harzianum endochitinase gene, ech42, in mycoparasitism. Appl Environ Microbiol. 1999;65:929–935. doi: 10.1128/aem.65.3.929-935.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chet I. Trichoderma: application, mode of action and potential as a biocontrol agent of soilborne plant pathogenic fungi. In: Chet I, editor. Innovative approaches to plant disease control. New York, N.Y: Wiley; 1987. pp. 137–160. [Google Scholar]
- 5.Chet I, Benhamou N, Haran S. Mycoparasitism and lytic enzymes. In: Harman G E, Kubicek C P, editors. Trichoderma and Gliocladium. Vol. 2. 1998. pp. 153–171. Enzymes, biological control and commercial application. Taylor and Francis Ltd., London, United Kingdom. [Google Scholar]
- 6.Cortes C, Gutierrez A, Olmedo V, Inbar J, Chet I, Herrera-Estrella A. The expression of genes involved in parasitism by Trichoderma harzianum is triggered by a diffusible factor. Mol Gen Genet. 1998;260:218–225. doi: 10.1007/s004380050889. [DOI] [PubMed] [Google Scholar]
- 7.Flores A, Chet I, Herrera-Estrella A. Improved biocontrol activity of Trichoderma harzianum by overexpression of the proteinase-encoding gene prb1. Curr Genet. 1997;31:30–37. doi: 10.1007/s002940050173. [DOI] [PubMed] [Google Scholar]
- 8.Hjeljord L, Tronsmo A. Trichoderma and Gliocladium in biological control: an overview. In: Harman G E, Kubicek C P, editors. Trichoderma and Gliocladium. Vol. 2. 1998. pp. 129–151. Enzymes, biological control and commercial application. Taylor and Francis Ltd., London, United Kingdom. [Google Scholar]
- 9.Howell C R. The role of antibiosis in biocontrol. In: Harman G E, Kubicek C P, editors. Trichoderma and Gliocladium. Vol. 2. 1998. pp. 173–183. Enzymes, biological control and commercial application. Taylor and Francis Ltd., London, United Kingdom. [Google Scholar]
- 10.Limon M C, Pintor-Toro J A, Benitez T. Increased antifungal activity of Trichoderma harzianum transformants that overexpress a 33-kDa chitinase. Phytopathology. 1999;89:254–261. doi: 10.1094/PHYTO.1999.89.3.254. [DOI] [PubMed] [Google Scholar]
- 11.Lorito M. Chitinolytic enzymes and their genes. In: Harman G E, Kubicek C P, editors. Trichoderma and Gliocladium. Vol. 2. 1998. pp. 73–99. Enzymes, biological control and commercial application. Taylor and Francis Ltd., London, United Kingdom. [Google Scholar]
- 12.Lorito M, Mach R L, Sposato P, Strauss J, Peterbauer C K, Kubicek C P. Mycoparasitic interaction relieves binding of Cre1 carbon catabolite repressor protein to promoter sequence of ech-42 (endochitinase-encoding) gene of Trichoderma harzianum. Proc Natl Acad Sci USA. 1996;93:14868–14872. doi: 10.1073/pnas.93.25.14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papavizas G C. Trichoderma and Gliocladium: biology, ecology and potential for biocontrol. Annu Rev Phytopathol. 1985;23:23–54. [Google Scholar]
- 14.Schirmböck M, Lorito M, Wang Y L, Hayes C K, Arisan-Atac I, Scala F, Harman G E, Kubicek C P. Parallel formation and synergism of hydrolytic enzymes and peptaibol antibiotics: molecular mechanisms involved in the antagonistic action of Trichoderma harzianum against phytopathogenic fungi. Appl Environ Microbiol. 1994;60:4364–4370. doi: 10.1128/aem.60.12.4364-4370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woo S L, Donzelli B, Scala F, Mach R L, Harman G E, Kubicek C P, Del Sorbo G, Lorito M. Disruption of ech42 (endochitinase-encoding) gene affects biocontrol activity in Trichoderma harzianum strain P1. Mol Plant-Microbe Interact. 1998;12:419–429. [Google Scholar]
- 16.Zeilinger S, Galhaup C, Payer K, Woo S L, Mach R L, Fekete C, Lorito M, Kubicek C P. Chitinase gene expression during mycoparasitic interaction of Trichoderma harzianum with its host. Fungal Genet Biol. 1999;26:131–140. doi: 10.1006/fgbi.1998.1111. [DOI] [PubMed] [Google Scholar]