Summary
Background
Ebola virus disease (EVD) outbreaks have emerged in Central and West Africa. EVD diagnosis relies principally on RT-PCR testing with GeneXpert®, which has logistical and cost restrictions at the peripheral level of the health system. Rapid diagnostic tests (RDTs) would offer a valuable alternative at the point-of-care to reduce the turn-around time, if they show good performance characteristics. We evaluated the performance of four EVD RDTs against the reference standard GeneXpert® on stored EVD positive and negative blood samples collected between 2018 and 2021 from outbreaks in eastern Democratic Republic of the Congo (DRC).
Methods
We conducted a prospective and observational study in the laboratory on QuickNavi-Ebola™, OraQuick® Ebola Rapid Antigen, Coris® EBOLA Ag K-SeT, and Standard® Q Ebola Zaïre Ag RDTs using left-over archived frozen EDTA whole blood samples. We randomly selected 450 positive and 450 negative samples from the EVD biorepositories in DRC, across a range of GeneXpert® cycle threshold values (Ct-values). RDT results were read by three persons and we considered an RDT result as “positive”, when it was flagged as positive by at least two out of the three readers. We estimated the sensitivity and specificity through two independent generalized (logistic) linear mixed models (GLMM).
Findings
476 (53%) of 900 samples had a positive GeneXpert Ebola result when retested. The QuickNavi-Ebola™ showed a sensitivity of 56.8% (95% CI 53.6–60.0) and a specificity of 97.5% (95% CI 96.2–98.4), the OraQuick® Ebola Rapid Antigen test displayed 61.6% (95% CI 57.0–65.9) sensitivity and 98.1% (95% CI 96.2–99.1) specificity, the Coris® EBOLA Ag K-SeT showed 25.0% (95% CI 22.3–27.9) sensitivity and 95.9% (95% CI 94.2–97.1) specificity, and the Standard® Q Ebola Zaïre Ag displayed 21.6% (95% CI 18.1–25.7) sensitivity and 99.1% (95% CI 97.4–99.7) specificity.
Interpretation
None of the RDTs evaluated approached the "desired or acceptable levels" for sensitivity set out in the WHO target product profile, while all of the tests met the "desired level" for specificity. Nevertheless, the QuickNavi-Ebola™ and OraQuick® Ebola Rapid Antigen Test demonstrated the most favorable profiles, and may be used as frontline tests for triage of suspected-cases while waiting for RT-qPCR confirmatory testing.
Funding
Institute of Tropical Medicine Antwerp/EDCTP PEAU-EBOV-RDC project.
Keywords: EVD, EBOV, GeneXpert, RDT sensitivity, Specificity
Research in context.
Evidence before this study
Several Ebola rapid diagnostic tests (RDTs) received emergency use authorization from the US Food and Drug Administration or World Health Organization (WHO) such as the OraQuick® Ebola Rapid Antigen Test (OraSure Technologies), the ReEBOV® Antigen Rapid Test (Coregenix), the Standard® Q Line Ebola Zaire Ag test (SD Biosensor) and the DPP Ebola Antigen System (Chembio Diagnostics).
We searched PubMed for articles published in English between Jan 1, 2014, and August 31, 2022, with terms “Ebola Virus Disease diagnosis” and “Ebola Rapid Diagnostic Test”. When combining both search terms, we found 68 papers of which 28 described the implementation and/or the performance of Ebola rapid tests. Among all evaluations performed, none of the RDTs studied achieved the desired (sensitivity >98%, specificity >99%) levels of both sensitivity and specificity as set out in the WHO target product profile for Ebola RDTs. In outbreak settings where a large number of blood samples were tested with a wide range of Ebola viral load levels, the QuickNavi™-Ebola RDT (Denka, Niigata, Japan) showed better performance than OraQuick® Ebola Rapid Antigen Test (FDA/WHO approved) and Coris® EBOLA Ag K-SeT rapid test, as it achieved the WHO-desired clinical specificity of at least 99%.
Added value of this study
In this study, we used the left-over samples from 2018 to 2021 Ebola virus disease (EVD) outbreaks in eastern Democratic Republic of Congo (DRC), which were stored for long-term at the Institut National de Recherche Biomédicale (INRB) biorepositories. Here, we conducted a laboratory prospective study to evaluate, the performance of four Ebola RDTs run side by side on the same set of archived whole blood samples against the GeneXpert® as the reference standard, as three independent operators could read RDTs results.
The added value of this study can be summarized as follows: 1) identical biological specimens submitted to the same testing conditions (RDTs brand, lab-operators, work conditions and environment), 2) same sample size to be tested, 3) each RDT result was read by three independent operators to give the final result, 4) individual and combined performance of RDTs determined in order to support the ranking of RDTs, 5) the possibility of using the screening panel at the point-of-care for triage and isolation of EVD suspect-cases while waiting for RT-qPCR.
Implications of all the available evidence
In this study, we determined individual and combined performance of RDTs run in the same conditions. This method strongly reduced variations associated with lab-operators technique and interpretation, work and environment conditions. The performance obtained, helped in raising the opportunity to use RDTs at the point-of-care to triage and isolate Ebola suspect-cases while waiting for confirmatory RT-qPCR. Throughout this study, the results obtained allowed 1) ranking RDTs evaluated according to their sensitivity and specificity, 2) proposing a screening panel combining the two most sensitive RDTs with an “OR” criterion for inclusion in order to detect at least one viral target protein. The implementation of RDTs is expected to be less-resource intensive, although it will require training for their correct use in the field, accurate results interpretation and reporting, correct sample shipment conditions to the laboratory for confirmation, and additional clinical and epidemiological data to support quick decision-making on the ground.
Introduction
Ebola virus disease (EVD) outbreaks have emerged in Central and West Africa where they posed major threats to global health security.1, 2, 3 Over the last ten years (2012–2022), twelve EVD outbreaks were reported in the world among which the two largest and deadliest caused 11,310 and 2287 deaths, in West Africa (2013–2016) and in eastern Democratic Republic of the Congo (DRC) (2018–2020), respectively.2,4, 5, 6 Early diagnosis is key to control EVD outbreak as it allows triaging and isolation of cases, case finding and follow-up of their contacts, investigation of the cause of death and clinical management of cases, implementation of therapeutics and vaccines clinical trials, post-epidemic surveillance and survivor's follow-up.7, 8, 9, 10, 11, 12, 13
During EVD outbreaks, diagnosis currently relies principally on the GeneXpert®, a semi-automated Reverse Transcription quantitative Polymerase Chain Reaction (RT-qPCR) closed system. This technique offers a lower-risk of contamination, high sensitivity (>99%) and specificity (>95%), ease of the use (minimal technical knowledge and few steps required), and short turn-around time (<2 h).14,15 However, the use of GeneXpert® is costly and requires skilled personnel, infrastructure and equipment, reagents and uninterrupted power supply to be continuously run at the peripheral healthcare of the health system. As most of these requirements are not usually available at the peripheral level, the use of the GeneXpert® is strongly hampered, calling out for reliable rapid diagnostic tests (RDTs) which are suitable for field settings.14, 15, 16, 17, 18
RDTs are easy-to-use and less-resource intensive assays detecting viral antigens in the blood and other bodily fluids without requiring any power supply, cold chain, sophisticated equipment or highly trained-personnel compared to the GeneXpert®. Thus, RDTs can complement the GeneXpert® in the detection of EVD cases at the point-of-care. RDTs offer several advantages as they 1) considerably reduce the turn-around time, 2) allow a quick decision-making on case management and death screening during or after outbreaks, 3) can help patients with reactive RDT to be transferred to the adequate health facility, and 4) can increase the access and acceptability of EVD testing at the point-of-care.7,11,12,14,16,17,19, 20, 21 However, RDTs need to be highly sensitive and specific to allow early detection of viremic patients during the course of the disease and reduce the consequences of false-positive/negative results during the public health interventions.10,22,23
Most of the previous Ebola virus (EBOV) RDTs evaluated (including outbreaks in the DRC) displayed variable performance characteristics, without reaching the desired (sensitivity >98%, specificity >99%) or acceptable (sensitivity >95%, specificity >99%) level of sensitivity and specificity as set out in the World Health Organization target product profile (TPP) for EBOV tests.11,17,18,24 Therefore, some concerns with RDTs still unresolved such as 1) the consistency in the performance characteristics in suboptimal or controlled settings, 2) the necessity to use RDTs to triage and rule out EVD in symptomatic patients at the peripheral care level, and 3) the development of algorithms to be used at the point-of-care in low resource settings.17
The Institut National de Recherche Biomédicale (INRB) as the National Reference Laboratory of the DRC has long-term storage for left-over samples from different EVD outbreaks, including those from eastern DRC (2018–2021). As a large and well-characterized EVD number of negative and positive samples were available at INRB biorepositories, a unique opportunity emerged to establish a head-to-head comparison of several RDTs with the same batch of samples. In this study, we estimated the performance characteristics of four EVD RDTs against the GeneXpert® as the reference standard on a set of stored EVD positive and negative whole blood samples from eastern DRC outbreaks.
Methods
Study design, population and setting
We conducted a prospective laboratory study to evaluate two lots of QuickNavi-Ebola™ and Coris® EBOLA Ag K-SeT, and one lot of OraQuick® Ebola Rapid Antigen and Standard® Q Ebola Zaïre Ag RDTs (Supplemental Table S1). We used archived frozen whole blood samples from North Kivu (Beni, Butembo, Mangina and Katwa) and Ituri (Komanda, Mambasa, Tchomia, Bunia and Biakato) provinces, during DRC EVD outbreaks (2018–2021). Those samples managed and tested within INRB field laboratories were collected in patients admitted in Ebola Treatment Units for diagnosis. The left-over samples following primary testing were temporarily stored across sites, and later shipped to INRB biorepositories in Kinshasa or Goma for long-term storage.
Sample selection, sample processing and data collection
The sequences of the study procedures are summarized in Supplemental Fig. S1.
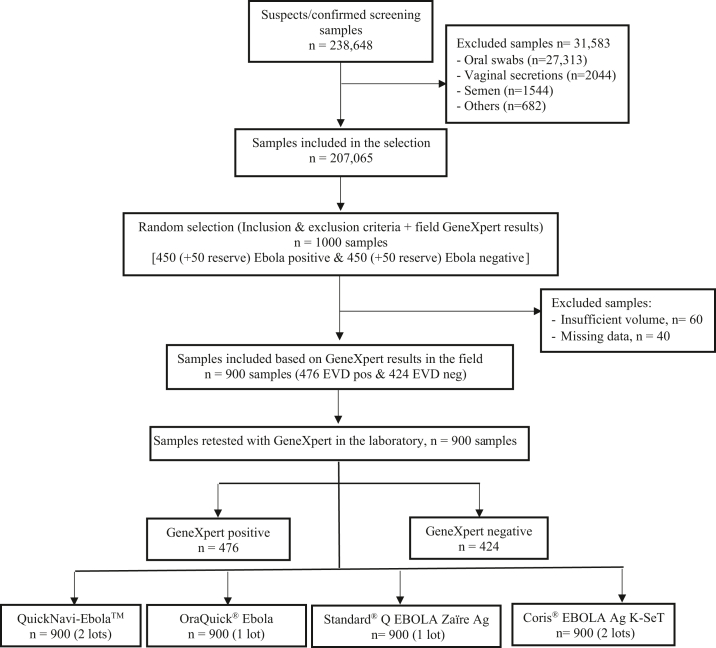
We initially used field laboratory datasets to randomly select 450 positive and 450 negative samples (plus 50 additional per category to foresee any replacement) based on GeneXpert® results at the time of outbreak response. This sample size was chosen to obtain sufficiently precise estimates of the sensitivity and specificity, and be able to find clinically meaningful differences in them with sufficient power.
Thereafter, we sorted out the selected samples from biorepositories in Kinshasa and Goma to check for their corresponding volumes. For EVD positive patients, we included one aliquot from the first diagnostic sample or the first positive sample of the patient's follow-up, in case of insufficient volume of the initial sample. Finally, we included 476 positive and 424 negative samples having a volume >500 μl and complete information in the ‘study dataset’ (ID, type of specimen, timing of sampling, Xpert Ebola results, Cycle threshold value, availability of epidemiological data).
The lab testing (GeneXpert® and RDTs) was carried out at the Biosafety level-3 of Rodolphe Mérieux INRB Laboratory in Goma (North Kivu Province, DRC) by experienced lab-technicians. The final set of samples was randomly mixed, relabeled, aliquoted (one copy per RDT brand and per lot, and one for the GeneXpert® retesting), and stored in identified cryoboxes (one aliquot specimen per cryobox). The study personnel completed a training on the protocol, Good Clinical and Laboratory Practices, GeneXpert® and RDTs proficiency prior to the study initiation. For standardization purposes between operators, we used the package insert of each kit to develop practical bench-aids for RDTs and the GeneXpert® assay. All the samples were re-run with the GeneXpert®, their results recorded into the ‘study database’ and double-blinded to operators while running the RDTs.
Based on the cycle threshold values (Ct-values) of Ebola virus targets glycoprotein (GP) and nucleoprotein (NP), sample results were categorized as follows: EVD negative 1) GP not detected/NP not detected, or 2) GP not detected/Ct of NP ≥ 40, or 3) GP detected/NP not detected; EVD positive 1) GP detected/NP detected and Ct < 40, or 2) GP not detected/NP detected and Ct < 40.
Samples with invalid results were re-run from the original tube at the end of all the GeneXpert® testing. Samples were processed per RDT brand and per lot: only one brand was run at a time by two operators, one operator per lot assisted by his/her buddy. As there was a risk that operators got used to the reactivity patterns of the tests while processing aliquots in the same sequence, we randomly allocated the samples boxes to be processed for the same lot while changing also the operators and buddies.
RDTs results were directly read from the device by three persons following this order: the buddy of the operator, the buddy of the other operator and the operator themself. The first two readers independently wrote the results on their individual reporting form then, asked for the operator themself to give their result which was noted on another form by the buddy. We considered an RDT result as “positive”, when it was flagged as positive by at least two out of the three readers. After daily sample processing, the results were entered into an Excel sheet which automatically displayed the final result based on the reports of two out of three readers.
Data analysis
Data were analyzed using R version 4.0.3 (R Foundation for Statistical Computing, Vienna, Austria). Basic characteristics of participants and samples were summarized using descriptive analyses. The diagnostic performance of each of the RDTs was evaluated against the GeneXpert® reference standard for the primary outcome. Sensitivity was defined as the probability that patients with a positive GeneXpert® Ebola assay result had a positive RDT result. Specificity was defined as the probability that patients with a negative GeneXpert® Ebola assay result had a negative RDT result. Sensitivity and specificity were estimated directly using proportions and associated Wilson confidence intervals (CIs), and through two independents generalized (logistic) linear mixed models (GLMM) using the lme4 package in R25 when adjusting for different viral loads (measured through the proxy variable Ct-value) and for sources of technical variation, as well as to compare the diagnostic accuracy between the RDTs.
We modelled the binary RDT outcome (positive or negative) as a function of test type and Ct-value (for sensitivity). We considered including sample, lab technician and RDT lot as a random effect. Inclusion of interaction (between RDT and Ct-value) and random slopes (i.e. per RDT rather than jointly) were tested at the 5% significance level when relevant. CIs were constructed using the profile likelihood when possible and with the asymptotic Wald approximation otherwise. Equality of sensitivity and specificity of different RDTs was tested with Tukey-style pairwise comparisons using the multcomp package in R.26
Ethical issues
This study received the approval of Kinshasa School of Public Health Ethics committee (Approval Number: ESP/CE/06/2022).
Role of funding source
This study was funded by the Institute of Tropical Medicine-Antwerp, the EDCTP PEAU-EBOV-RDC project under grant agreement RIA2018EF-2087, and through the FA5 DRC Program funded by the Directorate General for Development Cooperation and Humanitarian Aid (DGD) of the Belgian government.
The authors had full access to the study datasets and had the final responsibility for the decision to submit for publication. The funders have not played any role in the study design, data collection, data analyses, interpretation and writing of report.
Results
We tested a total 900 whole blood samples in the laboratory (Fig. 1). Out of 900 samples re-tested on the GeneXpert®, 476 (53%) were EBOV positive and 424 (47%) EBOV negative. For EBOV positive samples, the median NP Ct-value was 27.7 (IQR 22.5–35.0), with minimum of 15.9 and maximum of 39.9. The distribution of NP Ct-values in EBOV positive samples followed a bimodal pattern, with large number of samples in the range of 19–26 and 34–39 (Supplemental Fig. S2).
Fig. 1.
Study flow diagram.
Sensitivities and specificities of the four RDTs evaluated versus the reference standard GeneXpert® are shown in Table 1.
Table 1.
Rapid diagnostic test performance compared with GeneXpert Ebola assay as reference standard.
| RDT | Individual | Positive by 1/3 reader | Positive by 2/3 readers | Positive by 3/3 readers |
|---|---|---|---|---|
| Coris | ||||
| Sensitivity | 24.9% (23.3–26.5) | 27.8% (25.0–30.8) | 25.0% (22.3–27.9) | 21.8% (19.3–24.6) |
| Specificity | 95.6% (94.8–96.4) | 94.8% (93.0–96.2) | 95.9% (94.2–97.1) | 96.2% (94.7–97.4) |
| OraQuick | ||||
| Sensitivity | 61.4% (58.8–63.9) | 64.1% (59.6–68.4) | 61.6% (57.0–65.9) | 58.6% (54.0–63.1) |
| Specificity | 97.8% (96.8–98.5) | 96.0% (93.5–97.6) | 98.1% (96.2–99.1) | 99.3% (97.8–99.8) |
| QuickNavi | ||||
| Sensitivity | 56.4% (54.5–58.2) | 58.2% (55.0–61.3) | 56.8% (53.6–60.0) | 54.1% (50.9–57.3) |
| Specificity | 97.1% (96.3–97.7) | 96.1% (94.5–97.3) | 97.5% (96.2–98.4) | 97.6% (96.3–98.5) |
| Standard Q line | ||||
| Sensitivity | 21.2% (19.1–23.5) | 22.5% (18.9–26.6) | 21.6% (18.1–25.7) | 19.5% (16.1–23.4) |
| Specificity | 98.7% (97.9–99.3) | 98.1% (96.2–99.1) | 99.1% (97.4–99.7) | 99.1% (97.4–99.7) |
Note: estimate (95% confidence interval).
Table 1 provides sensitivity and specificity while interpreting RDT positive results compared to the GeneXpert® as the reference-standard by 1/3, 2/3, 3/3 readers, as well as individual reads (all reads were considered as independent observations).
When we considered 1/3, 2/3, 3/3 or individual reads (all reads taken as independent observations) flagging a test as positive, the sensitivity and the specificity were similar for each RDT (Table 1). The overall performance of all RDTs observed showed that QuickNavi-Ebola™ and OraQuick® Ebola Rapid Antigen were the most sensitive tests whereas the Standard® Q Ebola Zaïre Ag was the most specific (98.7%) (Table 1). We also noted a good agreement between the different lab-operators, as for all six lots we found at least 95% agreement between all three readers (all positive or all negative) (Supplemental Table S2).
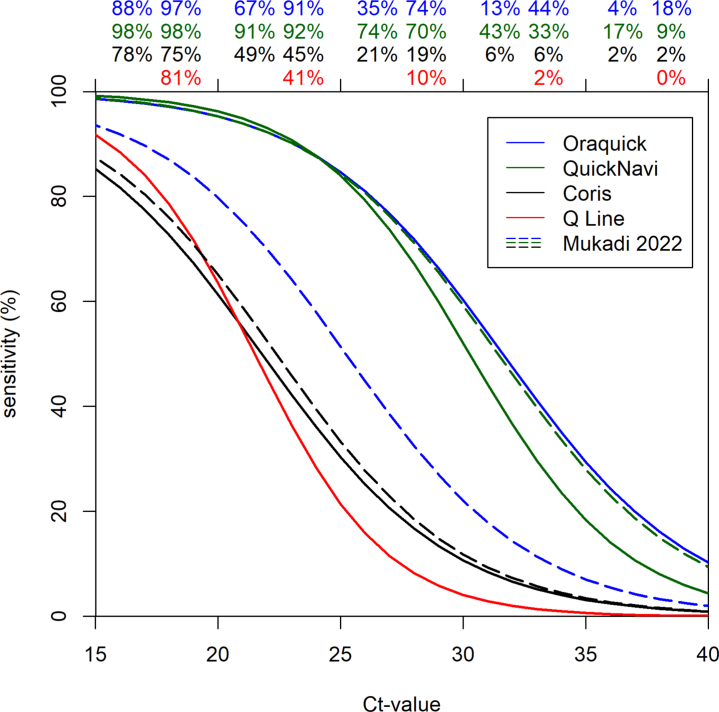
The estimated sensitivity at different observed Ct-values is displayed in Fig. 2 (continuous lines). The very steep curves indicate the large impact of Ct-value (output of a model with different slopes). The sensitivity of the four RDTs declined as the Ct-values increased. The QuickNavi-Ebola™ and OraQuick® Ebola Rapid Antigen tests showed respectively 98% and 97% sensitivity in samples at 17.5 NP Ct-values compared to the Coris® EBOLA Ag K-SeT (sensitivity: 75%) and Standard® Q Ebola Zaïre Ag (sensitivity: 81%) tests. At an NP Ct-values of 22.5, the sensitivity of the QuickNavi-Ebola™ and OraQuick® Ebola Rapid Antigen was maintained above 90%, whereas it has significantly dropped to under 50% for Coris® EBOLA Ag K-SeT and Standard® Q Ebola Zaïre Ag. We observed a drop in sensitivity under 20% at NP Ct-values >35.0 for the QuickNavi-Ebola™ and OraQuick® Ebola Rapid Antigen, whereas the threshold of <20% sensitivity was already reached with the NP Ct-values >27.5 for the Coris® EBOLA Ag K-SeT and Standard® Q Ebola Zaïre Ag tests (Fig. 2: continuous lines).
Fig. 2.
Sensitivity of the rapid diagnostic tests as a function of GeneXpert Ct-value. Continuous lines indicate the sensitivity of RDTs in the current study, whereas the dashed lines indicate the sensitivity of RDTs in the previous study (Mukadi et al.). The estimates of sensitivity on top of the graph are for Mukadi et al. (left) (Lancet Infect Dis. 2022 Mar 14:S1473-3099(21)00675-7) and the current study (right), respectively. The very steep curves indicate the large impact of Ct-value (output of a model with different slopes). The percentages on the figure do not show the estimated sensitivity in the bins, but instead give the modelled sensitivity in the middle of the bin (i.e. at 17.5, 22.5, 27.5 etc.) which is an estimate for the sensitivity conditional on a sample with an observed concentration that corresponds with a Ct-value of such a number.
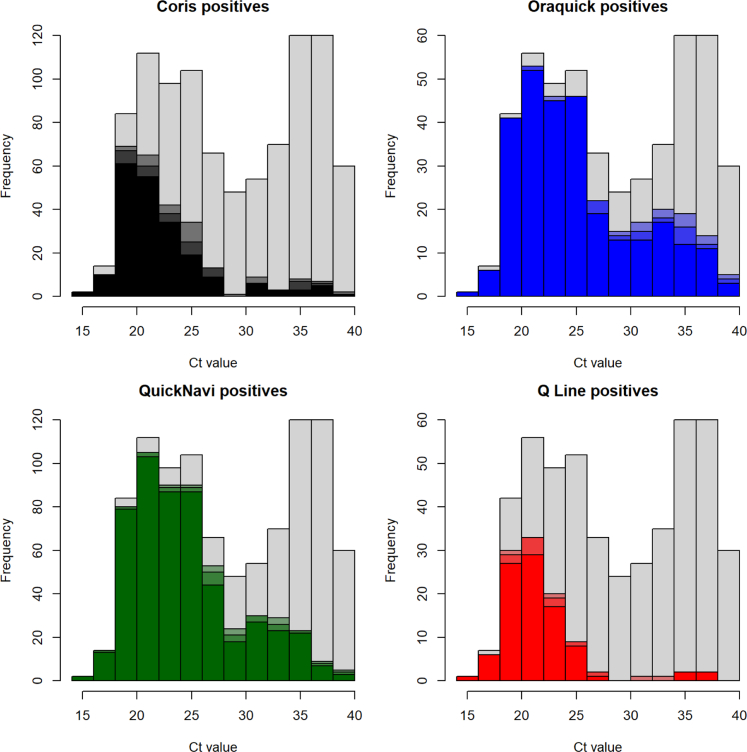
Fig. 3 shows the RDT positivity for GeneXpert® positive samples per category of Ct-value. The QuickNavi-Ebola™ and OraQuick® Ebola Rapid Antigen detected more positives in samples with low and medium Ct-values (high and medium viral loads), but they detected lower positives in specimens with high Ct-values (low viral load). The Coris® EBOLA Ag K-SeT and Standard® Q Ebola Zaïre Ag had low sensitivity around the median Ct-values. Among 103 true positives samples with the Standard® Q Ebola Zaïre Ag test, 99 detected VP40 protein (89 by three readers); 54 detected GP protein (46 by three readers) including 4 samples that were VP40 negative; and 9 detected NP protein (data not shown).
Fig. 3.
RDT positivity for GeneXpert positive samples by Ct-value. This histogram describes the frequency of RDT positive (coloured) and negative (grey) samples compared by type of RDTs per Ct-value among Xpert positive samples. The grey bars indicate the negative results as flagged by three readers. Colours indicate the positivity by RDT and by Ct-value, the darkest colour indicates a test being positive for all 3 readers, and the lighter colours for 2 and 1 reader positive (Each colour represents RDT brand).
The lot effect was small and not significant for both the sensitivity (p = 0.92) and specificity models (p = 0.89). The random sample ID effect was highly significant for both sensitivity and specificity (p < 0.0001), even after adjusting for Ct-value.
The final model for sensitivity included the type of RDT, Ct-value, and sample ID as random effect. All differences in sensitivity between the four RDTs were significant except for the Coris® EBOLA Ag K-SeT versus Standard® Q Ebola Zaïre Ag (p = 0.16). The p-value between QuickNavi-Ebola™ and OraQuick® Ebola Rapid Antigen was 0.03, all others were <0.0001. Despite the significant difference, OraQuick® Ebola Rapid Antigen Test and QuickNavi-Ebola™ performed similarly, and considerably better than Coris® EBOLA Ag K-SeT and Standard® Q Ebola Zaïre Ag. The significant difference between QuickNavi-Ebola™ and OraQuick® Ebola Rapid Antigen seems to be explained by the OraQuick® Ebola Rapid Antigen test performing slightly better than QuickNavi-Ebola™ in samples with high Ct-values especially (Fig. 2, Fig. 3). All the tests showed high specificity, except for the Coris® EBOLA Ag K-SeT which performed significantly worse than Standard® Q Ebola Zaïre Ag (p = 0.005). We combined overall sensitivities and specificities by Ct-values for different RDTs to get testing strategies. Combining OraQuick® Ebola Rapid Antigen and QuickNavi-Ebola™ tests as a panel would lead to a modest increase of sensitivity compared to a single test, with only a small reduction of specificity. Adding either Coris® EBOLA Ag K-SeT or Standard® Q Ebola Zaïre Ag to the panel was not likely to increase the sensitivity much and would in case of Coris® risk a considerable reduction in specificity. Combining OraQuick® Ebola Rapid Antigen Test and QuickNavi-Ebola™ with an “AND” criterion would further reduce the sensitivity to undesirable levels (Table 2).
Table 2.
Raw sensitivity and specificity of testing strategies of 1 or more RDTs.
| Positive (+) if | sensitivity | specificity |
|---|---|---|
| Oraquick + | 61.6% (57.0–65.9) | 98.1% (96.2–99.1) |
| QuickNavi + | 56.8% (53.6–60.0) | 97.5% (96.2–98.4) |
| OraQuick + OR QuickNavi + | 68.8% (65.7–71.7) | 96.0% (94.4–97.2) |
| OraQuick + OR QuickNavi + OR Coris + | 69.6% (66.6–72.5) | 92.6% (90.5–94.2) |
| OraQuick + OR QuickNavi + OR Q Line + | 68.8% (65.7–71.7) | 95.3% (93.6–96.6) |
| OraQuick + AND QuickNavi + | 49.6% (46.4–52.8) | 99.6% (98.9–99.9) |
Discussion
Throughout this study, the QuickNavi-Ebola™ and OraQuick® Ebola RDTs did not reach the “desired or acceptable level” of sensitivity following the WHO TPP for EBOV tests. However, both tests showed specificities close to the “desired or acceptable level” of specificity (>99%) with 97.5% and 98.1%, respectively.
A previous evaluation of plasma samples with Coris® EBOLA Ag K-SeT showed sensitivities of 98.7% (samples with low Ct-values), 62.1% (samples with high Ct-values), and 88.6% (overall sensitivity).27 However, the sample size was small (n = 210), the majority of samples had low Ct-values (<34.0), and the reference standard used was less sensitive.27 During the 2018–2021 EVD outbreaks in DRC, the Coris® EBOLA Ag K-SeT was run on a large number of blood samples (n = 819 and n = 900), but it displayed poor performances (38.9% and 25.0% sensitivity, 97.4% and 95.9% specificity) compared to GeneXpert®.17 QuickNavi-Ebola™ was reported to have a low limit of detection compared to OraQuick® Ebola Rapid Antigen Test.14 In outbreak settings, the QuickNavi-Ebola™ showed good performances in two studies,14,17 while less impressive performance was observed in the current study (56.8% sensitivity versus 97.5% specificity). However, as shown previously,17 test performance is strongly dependent on the Ct-values of the tested samples. We therefore compared not only the overall performance, but also taking into account the Ct-value (Fig. 2).
Since our study has more samples with higher Ct-values than previously,17 we could expect results for overall sensitivity to be worse. However, when the results are analyzed by Ct-value for QuickNavi-Ebola™ and Coris® EBOLA Ag K-SeT, the differences are generally small and its performance is in line with the previous study.17 In summary, the QuickNavi-Ebola™ test showed good performance characteristics in samples with low Ct-values, but performs poorly in samples with high Ct-values. Conversely, the OraQuick® Ebola Rapid Antigen test overall estimates are in line with the previous study (57.4% sensitivity and 98.3% specificity),17 which masks that it performed better in this study taking into account the Ct-value, with performance especially better for the middle and lower Ct-values, compared to our previous study (Fig. 2). These findings are also consistent with previous reports (manufacturer: 84% sensitivity, 98% specificity at mainly low Ct-values).13,20,28 The Standard® Q Ebola Zaïre Ag (designed to detect both VP40 and NP) showed a very low sensitivity (21.6%) for a good specificity (99.1%). This finding is in line with a previous study,11 that showed high specificity for low sensitivity compared to other RDTs. Additionally, the Standard® Q Ebola Zaïre Ag showed excess pooling on the sample reception pad, resulting in failure to flow on the membrane,11 as we noticed too while using whole blood samples.
A few technical conditions could be pointed out as potential source for the suboptimal performance of the RDTs evaluated here. First, the storage and transportation conditions could have modified the quality of the biological specimens used. During the outbreak, samples were subjected to multiple shipments and variable cold chain conditions from primary sites of testing to the temporary and long-term storage sites. Technical requirements in the implementation of this study such as freeze-thaw process and aliquoting could have changed the quality of the specimens. In order to compensate possible sample quality loss due to the material storage over time, we kept one aliquot to be re-tested with the GeneXpert®. Decay of antigens in the samples could have partially affected the detection of these antigens in the RDTs. Viral proteins such as VP40 and NP are usually abundantly expressed in infected organisms and thus represent ideal targets for RDTs. We consequently expected our RDTs to detect more positives as those abundant viral proteins were targeted.
Using archived whole blood samples instead of plasma could have slowed the migration over the membrane. However, during EBOV outbreaks the laboratory procedures do not allow specimen centrifugation, in order to mitigate the risk of environmental contamination. Wonderly et al. observed a reduced specificity of RDTs using plasma versus EDTA whole blood, but no effect on the sensitivity was found.11 This observation warrants for separate performance evaluation with whole blood and plasma specimens. In our study, we only had EDTA whole blood available for testing. Therefore, care should be taken before extrapolating these results to plasma or fresh samples tested in the field.11 In samples with low Ct-values, the high concentration of antigens could have formed antibody–antigen complexes sticking to the membrane and reducing flow across the sample pad (prozone effect).
In our previous study,17 we evaluated QuickNavi-Ebola™, OraQuick® Ebola Rapid Antigen, Coris® EBOLA Ag K-SeT, and added Standard® Q Ebola Zaïre Ag test in the current study. The QuickNavi-Ebola™ and OraQuick® Ebola Rapid Antigen tests did not achieve the “acceptable level” of performance as stated by the WHO TPP for EBOV tests,11,17,18,24 although their respective specificities had almost reached 99% (97.5% and 98.1%). Nonetheless, we can propose their use as frontline diagnosis in remote areas to triage and isolate suspected-cases. All individuals will be isolated separately in the triage ward while waiting for RT-qPCR results i.e. those with at least one positive RDT in the high-risk area and those with negative results in low-risk area. To use RDTs in the field, we should take into consideration 1) the performance characteristics of the most sensitive tests, 2) the conditions of biosafety, and 3) the ease of use.
In the current study, RDTs performance was ranked as follows (sensitivity and specificity): 1) OraQuick® Ebola Rapid Antigen Test (61.6% and 98.1%) and QuickNavi-Ebola™ (56.8% and 97.5%), 2) Coris® EBOLA Ag K-SeT (25.0% and 95.9%) and Standard® Q Ebola Zaïre Ag (21.6% and 99.1%). In our previous study, RDTs were categorized in the following manner (sensitivity and specificity): 1) QuickNavi-Ebola™ (87.4% and 99.6%), 2) OraQuick® Ebola Rapid Antigen Test (57.4% and 98.3%) and 3) Coris® EBOLA Ag K-SeT (38.9% and 97.4%).17 At the point-of-care, we propose a screening panel consisting of the QuickNavi-Ebola™ and OraQuick® Ebola Rapid Antigen Test (high combined sensitivity and specificity) using finger prick or venous blood with an “OR” criterion for inclusion (Table 2). To our knowledge, Ebola RDTs and GeneXpert® evaluated in this study can only be applicable to EBOV species. Therefore, it will be useful to test those diagnostic tools against other Ebola species in further studies. Using this panel of two RDTs at a time will allow the detection of at least one target viral protein (NP or VP40), as the presence of antigens may vary in positive cases according to the stage of the infection. At this stage, RDT use will not generate cost savings for EVD testing as RT-qPCR will still be needed to discriminate positives from negatives among suspected-cases. From our studies, we learned that both QuickNavi-Ebola™ and OraQuick® Ebola Rapid Antigen tests can be easily run and interpreted in field conditions. However, their implementation will require 1) adequate training on their correct use, 2) accurate reporting of their results, 3) correct shipment of clinical specimens to the laboratory for confirmatory RT-qPCR with venous blood collected after RDTs analysis, in order to exclude any false positives and false negatives.5,14,17 In the meantime, stringent and adequate measures should be taken to carefully triage and manage isolation of suspect-cases in care units, in order to mitigate the risk of EVD nosocomial transmission. Furthermore, the implementation of these RDTs should be supported by a clear at-risk communication plan, an excellent psychosocial environment and messaging, and detailed clinical and epidemiological data.
Contributors
DM-B, ADW, BKJ, JvG, KKA, SA-M: conceived the study, the methodology and wrote the original draft. DM-B, JB-P, FE-A, FM-M, NMM, EMM, MAK-M, ET-T, EI-N: performed investigation and data curation. DM-B, BKJ, EIN, JB-P, FE-A, FM-M and ADW: accessed and verified all underlying data. BKJ, JvG, DM-B, and ADW: performed formal data analysis. JDK, HK-M, BKJ, PM-K, SM-M, MK, AT, PF, JMM, JJM-T, MEM, PNF, SR, AL, AN-N: edited the manuscript. DM-B, ADW, BKJ, JDK, PF, JMM, MK, AT, PNF, JJM-T, AL, AN-N, SR: visualized the study documents. JMM, KKA, JvG, SA-M: validated the study documents and the manuscript.
All authors read and approved the final version of the manuscript, and ensure it is the case.
Data sharing statement
The data statement is shared separately in the supplementary documents. The data sharing agreement will be available three months after official paper release.
Declaration of interests
US CDC provided the Xpert® Ebola cartridges. FIND purchased OraQuick Ebola Rapid Antigen tests and donated to INRB through US CDC partnership. Institute of Tropical Medicine-Antwerp purchased Coris® Ag K-SeT and Standard® Q line Zaïre Ebola Rapid diagnostic tests with the financial support of the EDCTP PEAU- EBOV-RDC project under grant agreement RIA2018EF-2087, and through the FA5 DRC Program funded by the Directorate General for Development Cooperation and Humanitarian Aid (DGD) of the Belgian government.
Hokkaido University provided QuickNavi Ebola rapid tests to INRB via Japanese International Cooperation Agency (JICA). Rodolphe Mérieux INRB-Goma Laboratory supported the study with laboratory supplies.
DMB is a PhD fellow supported by the Belgian Directorate-general Development Cooperation and Humanitarian Aid.
JB-P, ADW, FE-A, BKJ, FM-M, JDK, HK-M, EI-N, PM-K, PM-K, SM-M, MK, AT, PF, NMM, EMM, MAK-M, ET-T, PNF, SR, AL, AN-N, MEM, JMM, JJM-T, KKA, JvG, SA-M declare no competing interests.
Acknowledgements
During the tenth EVD outbreak in DRC, INRB received technical, material and logistical support through its different partners to operate the laboratory response: World Health Organization Headquarter (WHO, Geneva) and Regional Office (Afro/WHO, Brazzaville), Africa Centre for Disease Control and Prevention (Africa-CDC), US National Institute of Allergy and Infectious Diseases/National Institutes of Health (NIAID/NIH), US Centers for Diseases Control and Prevention, Japanese International Cooperation Agency (JICA), The Science and Technology Research Partnership for Sustainable Development (SATREPS), Institut de Recherche pour le Développement (IRD, Montpellier), Institute of Tropical Medicine-Antwerp (ITM Antwerp), Institut Pasteur de Dakar (IPD), United States Army Medical Research Institute of Infectious Diseases (USAMRIID, USA), United States Agency for International development (USAID), USAID Predict Metabiota DRC, UCLA DRC Health Research and Training Program.
We are very grateful to our colleagues from INRB Headquarter in Kinshasa, Rodolphe Mérieux INRB-Goma Laboratory, INRB field laboratories (Beni, Butembo, Mangina, Katwa, Komanda, Mambasa, Tchomia, Bunia and Biakato cities), and partners from different organizations through which support was provided to this study.
Study Group: Hugo Kavunga-Membo, PhD1,2, Elie Ishara-Nshombo, DVM2, Stijn Roge, PhD4, Noella Mulopo-Mukanya, MSc2, Espérance Tsiwedi-Tsilabia, MSc2, Emile Muhindo-Milonde, MSc2, Marie-Anne Kavira-Muhindo, MSc2, Maria E. Morales-Betoulle6, Antoine Nkuba-Ndaye, PhD1,3
Affiliations
1. Institut National de Recherche Biomédicale, INRB, Kinshasa, DRC
2. Rodolphe Mérieux INRB-Goma Laboratory, Goma, North Kivu, DRC
3. Service de Microbiologie, Departement de Biologie Médicale, Cliniques Universitaires de Kinshasa, Université de Kinshasa, DRC
4. Institute of Tropical Medicine, Antwerp, Belgium
5. Health Emergencies program, World Health Organization, Geneva, Switzerland
6. US Centers for Disease Control and Prevention, Atlanta, Georgia, United States of America
7. International Institute for Zoonosis Control, Hokkaido University, Sapporo, Japan
8. University of Antwerp, Antwerp, Belgium
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104568.
Contributor Information
Daniel Mukadi-Bamuleka, Email: daniel.mukadi@inrb.net.
Study Group:
Hgo Kavunga-Membo, Elie Ishara-Nshombo, Stijn Roge, Noella Mulopo-Mukanya, Espérance Tsiwedi-Tsilabia, Emile Muhindo-Milonde, Marie-Anne Kavira-Muhindo, Maria E. Morales-Betoulle, and Antoine Nkuba-Ndaye
Appendix A. Supplementary data
References
- 1.Jacob S.T., Crozier I., Fischer W.A., 2nd, et al. Ebola virus disease. Nat Rev Dis Prim. 2020;6(1):13. doi: 10.1038/s41572-020-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moran Z., Rodriguez W., Ahmadou D., et al. Comparative performance study of three Ebola rapid diagnostic tests in Guinea. BMC Infect Dis. 2020;20:670. doi: 10.1186/s12879-020-05339-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malvy D., McElroy A.K., de Clerck H., Günther S., van Griensven J. Ebola virus disease. Lancet. 2019;393(10174):936–948. doi: 10.1016/S0140-6736(18)33132-5. [DOI] [PubMed] [Google Scholar]
- 4.World Health organization 10th Ebola outbreak in the democratic republic of the Congo declared over; vigilance against flare-ups and support for survivors must continue. 2020. https://www.who.int/news/item/25-06-2020-10th-ebola-outbreak-in-the-democratic-republic-of-the-congo-declared-over-vigilance-against-flare-ups-and-support-for-survivors-must-continue Available online:
- 5.Mukadi-Bamuleka D., Sanogo O.Y., Bulabula-Penge J., et al. Postmortem surveillance for Ebola virus using OraQuick Ebola rapid diagnostic tests, eastern democratic republic of the Congo, 2019–2020. Emerg Infect Dis. 2022;28(No. 2) doi: 10.3201/eid2802.210981. www.cdc.gov/eid [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mulangu S., Dodd L.E., Davey R.T., Jr., et al. A randomized, controlled trial of Ebola virus disease therapeutics. N Engl J Med. 2019;381(24):2293–2303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Couturier C., Wada A., Louis K., et al. Characterization and analytical validation of a new antigenic rapid diagnostic test for Ebola virus disease detection. PLoS Negl Trop Dis. 2020;14(1):e0007965. doi: 10.1371/journal.pntd.0007965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cnops L., De Smet B., Mbala-Kingebeni P., van Griensven J., Ahuka-Mundeke S., Ariën K.K. Where are the Ebola diagnostics from last time? Nature. 2019;565(7740):419–421. doi: 10.1038/d41586-019-00212-y. [DOI] [PubMed] [Google Scholar]
- 9.Perkins M.D., Kessel M. What Ebola tells us about outbreak diagnostic readiness. Nat Biotechnol. 2015;33(5):464–469. doi: 10.1038/nbt.3215. [DOI] [PubMed] [Google Scholar]
- 10.Fleck F. Rapid Ebola tests hold promise. Bull World Health Organ. 2015;93(4):215–216. doi: 10.2471/BLT.15.020415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wonderly B., Jones S., Gatton M.L., et al. Comparative performance of four rapid Ebola antigen-detection lateral flow immunoassays during the 2014-2016 Ebola epidemic in West Africa. PLoS One. 2019;14(3):e0212113. doi: 10.1371/journal.pone.0212113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shorten R.J., Brown C.S., Jacobs M., Rattenbury S., Simpson A.J., Mepham S. Diagnostics in Ebola Virus Disease in resource-rich and resource-limited settings. PLoS Negl Trop Dis. 2016;10(10):e0004948. doi: 10.1371/journal.pntd.0004948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broadhurst M.J., Brooks T.J., Pollock N.R. Diagnosis of Ebola virus disease: past, present, and future. Clin Microbiol Rev. 2016;29(4):773–793. doi: 10.1128/CMR.00003-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makiala S., Mukadi D., De Weggheleire A., et al. Clinical evaluation of QuickNaviTM-ebola in the 2018 outbreak of Ebola virus disease in the democratic republic of the Congo. Viruses. 2019;11:589. doi: 10.3390/v11070589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katawera V., Kohar H., Mahmoud N., et al. Enhancing laboratory capacity during Ebola virus disease (EVD) heightened surveillance in Liberia: lessons learned and recommendations. Pan Afr Med J. 2019;33(Suppl 2):8. doi: 10.11604/pamj.supp.2019.33.2.17366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhillon R.S., Srikrishna D., Kelly J.D. Deploying RDTs in the DRC Ebola outbreak. Lancet. 2018;391(10139):2499–2500. doi: 10.1016/S0140-6736(18)31315-1. [DOI] [PubMed] [Google Scholar]
- 17.Mukadi-Bamuleka D., Bulabula-Penge J., De Weggheleire A., et al. Field performance of three Ebola rapid diagnostic tests used during the 2018-20 outbreak in the eastern Democratic Republic of the Congo: a retrospective, multicentre observational study. Lancet Infect Dis. 2022;22(6):891–900. doi: 10.1016/S1473-3099(21)00675-7. [DOI] [PubMed] [Google Scholar]
- 18.Emperador D.M., Mazzola L.T. Wonderly Trainor B, Chua A, Kelly-Cirino C. Diagnostics for filovirus detection: impact of recent outbreaks on the diagnostic landscape. BMJ Glob Health. 2019;4(Suppl 2) doi: 10.1136/bmjgh-2018-001112. e001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stone C., Mahony J.B. Point-of-care (POC) tests for infectious diseases– the next generation! Ann Infect Dis Epidemiol. 2018;3(1) 1025. [Google Scholar]
- 20.VanSteelandt A., Aho J., Franklin K., et al. Operational evaluation of rapid diagnostic testing for Ebola Virus Disease in Guinean laboratories. PLoS One. 2017;12(11):e0188047. doi: 10.1371/journal.pone.0188047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeMers H.L., He S., Pandit S.G., et al. Development of an antigen detection assay for early point-of-care diagnosis of Zaire ebolavirus. PLoS Negl Trop Dis. 2020;14(11):e0008817. doi: 10.1371/journal.pntd.0008817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chua A.C., Cunningham J., Moussy F., Perkins M.D., Formenty P. The case for improved diagnostic tools to control Ebola Virus Disease in West Africa and how to get there. PLoS Negl Trop Dis. 2015;9(6):e0003734. doi: 10.1371/journal.pntd.0003734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tembo J., Simulundu E., Changula K., et al. Recent advances in the development and evaluation of molecular diagnostics for Ebola virus disease. Expert Rev Mol Diagn. 2019;19(4):325–340. doi: 10.1080/14737159.2019.1595592. [DOI] [PubMed] [Google Scholar]
- 24.World health Organization Target product profile for Zaïre ebolavirus rapid, simple test to be used in the control of the Ebola outbreak in West Africa. https://www.who.int/tools/target-product-profile-database/item/the-case-for-improved-diagnostic-tools-to-control-ebola-virus-disease-in-west-africa-and-how-to-get-there-br-br-ebola-diagnostic-plos-neglected-tropical-diseases-7283
- 25.Bates D., Maechler M., Bolker B., Walker S. Fitting linear mixed-effects models using lme 4. J Stat Software. 2015;67(1):1–48. doi: 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- 26.Hothorn T., Bretz F., Westfall P. Simultaneous inference in general parametric models. Biom J. 2008;50(3):346–363. doi: 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
- 27.Colavita F., Biava M., Mertens P., et al. EBOLA Ag K-SeT rapid test: field evaluation in Sierra Leone. Clin Microbiol Infect. 2018;24(6):653–657. doi: 10.1016/j.cmi.2017.10.019. [DOI] [PubMed] [Google Scholar]
- 28.US Food and Drug Administration Evaluation of automatic class III designation for OraQuick Ebola rapid antigen test decision summary. 2020. https://www.accessdata.fda.gov/cdrh_docs/reviews/DEN190025.pdf
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.