Abstract
When Salmonella enterica invades mammalian cells, it activates signals leading to increased expression of inflammatory mediators. One such mediator is nitric oxide (NO), which is produced under control of the enzyme inducible NO synthase (iNOS). Induction of iNOS in response to Salmonella infection has been demonstrated, but the bacterial effector molecules that regulate expression of the enzyme have not been identified. In the study reported here, an analysis of Salmonella-dependent iNOS expression in macrophages was carried out. Wild-type Salmonella strains increased the levels of both iNOS protein and mRNA in murine macrophage cell lines in an invasion-independent fashion. Mutant strains lacking a functional pathogenicity island 1-encoded type III secretion system, as well as strains lacking the invasins SipB, SipC, and SipD, were impaired in iNOS induction. Complementation experiments indicated that all three of the invasins were required for induction of iNOS expression. These results suggested that an effector protein, translocated into macrophages via the type III secretion system in a SipB-, SipC-, and SipD-dependent manner, might be the ultimate mediator of iNOS induction. In keeping with this idea, a mutant strain deficient in SopE2, a recently described homolog of SopE, was found to be impaired in the induction of iNOS expression. These observations suggest that iNOS expression is regulated by signals activated by SopE2 (and possibly SopE) and that the role of SipB, SipC, and SipD in this process is to facilitate translocation of the relevant effector.
Infections with Salmonella enterica (most commonly serovar Typhimurium) constitute a significant public health problem in most countries. In the United States, it is estimated that between 800,000 and 4 million cases occur every year, and of these about 500 are fatal (13). The organism usually causes a self-limited, acute intestinal inflammation, manifesting clinically with diarrhea and vomiting, but sometimes it can lead to disseminated, potentially serious infections. One of the most important characteristics of Salmonella virulence is the ability of the organism to invade mammalian cells. After it is ingested in contaminated food, Salmonella first invades the epithelial cells of the intestine, and then, once it has crossed the epithelial barrier, the macrophages of the lamina propria and Peyer's patches (23). A crucial bacterial structure required for the invasion process is a specialized secretory apparatus, the type III secretion system, which is encoded at a locus known as Salmonella pathogenicity island 1 (SPI1) (4). The SPI1 type III secretion system is involved in the signal-peptide-independent export of a set of proteins (referred to here as effector proteins) that are ultimately translocated into the cytosol of the host cells. The translocated effectors interact with specific cellular proteins to induce dramatic cytoskeletal and membrane rearrangements that lead to the active uptake of the bacteria by the cell (14). Other effects include the induction of apoptosis, the opening of chloride channels, and the activation of cellular signal transduction pathways leading to the increased expression of proinflammatory cytokines (4, 18, 20, 21, 31). Salmonella mutants that lack either a functional SPI1 type III secretion system or certain of the effector proteins that are secreted through it are defective in invasion and have reduced virulence in animal models of disease (10, 21).
The effector proteins SipB, SipC, and SipD have been of particular interest because of their involvement in invasion. They are secreted into bacterial supernatants individually but are mutually interdependent for translocation into the host cell (4). Mutant strains lacking any one of these proteins fail to translocate the others, resulting in an invasion-defective phenotype (4, 21). In addition, SipB, SipC, and SipD are also involved in the translocation of several other effector proteins, including SopB, SopE, SptP, and AvrA (9, 11, 15, 16, 40). While the roles of SipB and SipD in invasion may be largely to act as chaperones during translocation, SipC has been shown to have direct effects on actin polymerization and bundling (17). Together with SipA, an actin-binding effector, and SopE, a guanine nucleotide exchange factor for Rho GTPases, SipC plays an important part in inducing the cytoskeletal rearrangements required for invasion (14, 43).
During the course of cell invasion, bacterial lipopolysaccharide (LPS) and some of the Salmonella effectors activate cellular signaling pathways, leading to changes in the expression of various genes, including those for inflammatory mediators such as interleukin-8 and tumor necrosis factor alpha (20). The increased production of these cytokines is involved in the generation of an acute inflammatory response that, on the one hand, helps in the clearance of the organism and the induction of adaptive immunity and, on the other, contributes to tissue damage. Another factor that plays a similar double-edged role in the response to Salmonella is nitric oxide (NO), a labile gas with antimicrobial, proinflammatory, and immunomodulatory effects (28). NO is produced from arginine in a reaction catalyzed by the enzyme nitric oxide synthase (NOS). There are three isoforms of NOS, which differ in patterns of expression and dependence on calcium. The main isoform expressed in macrophages is NOS2, also known as iNOS because of its inducible expression and its calcium-independent activity. This isoform is active even at the trace levels of calcium found within resting cells and, once expressed, is capable of sustained, high-output NO production.
Increased expression of iNOS following infection with Salmonella was recently demonstrated in intestinal epithelial cell lines (33, 39). Since the organism is susceptible to NO and its reaction products (6, 22), this response could constitute an important innate defense mechanism. On the other hand, the proinflammatory effects of NO could contribute to pathological tissue damage. Elucidating how Salmonella induces the expression of iNOS would thus shed light on an important aspect of the host reaction to the organism. In the study reported here, we have examined the induction of iNOS in macrophages following infection with Salmonella in order to test the hypothesis that one or more of the bacterial effector proteins may be involved in regulating expression of the enzyme.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The wild-type S. enterica serovar Typhimurium strain SL1344, and the isogenic mutant strains VV341, EE633, and EE638 have been described previously (21, 41). The strains SB201 (invG::cam), and SB136 (invA::aphT), also isogenic with SL1344, were provided by Jorgé Galan, Yale University. The SL1344 derivative GG5 (sopE::pMG1) was constructed by integrating an ampicillin-resistant suicide plasmid, pMG1, into the sopE locus of SL1344 (L. Guogas and C. Lee, unpublished results), and was provided by Catherine Lee, Harvard Medical School. The wild-type serovar Dublin strain 2229 and the isogenic mutant strains A1, AV1, B1, SB2, and SE1 have been characterized elsewhere (11, 35, 40) and were provided by Edouard E. Galyov, Institute for Animal Health, Compton, United Kingdom, who also kindly gave us the SopE-deficient, wild-type serovar Typhimurium strain F98 and its isogenic, SopE2-deficient derivative, SE2.1, both of which have been described previously (2), as well the serovar Dublin strain SC2, in which the entire gene encoding SptP was removed (E. Galyov, unpublished data). All bacteria were grown in Luria-Bertani (LB) medium, with ampicillin added to 100 μg/ml for strains transformed with β-lactamase-encoding plasmids. To prepare bacteria for macrophage infection, one to two colonies of the appropriate strains were inoculated into 2 ml of LB medium and shaken at 37°C for several hours until the culture reached saturation. Then, 10 μl of the saturated culture was diluted into 10 ml of fresh medium, followed by incubation overnight at 37°C without shaking in tightly capped, 12-ml tubes.
Cell culture.
The murine macrophage cell lines RAW264.7 and J774 were maintained in Dulbecco modified Eagle medium (DMEM) containing 10% heat-inactivated fetal calf serum, 50 U of penicillin per ml, and 50 μg of streptomycin per ml at 37°C in an atmosphere of 5% carbon dioxide. To prepare the cells for infection, they were detached from the tissue culture plates by incubating them for 10 min in phosphate-buffered saline (PBS) containing 1 mM EGTA and 4 mM EDTA. The cells were counted and then resuspended in medium without antibiotics, and 0.5-ml aliquots (1.5 × 106 cells) were seeded into the wells of a 24-well tissue culture plate. The cells were allowed to adhere for 2 h before infecting them with Salmonella strains.
Infections.
Aliquots (1 ml) of the overnight standing cultures of Salmonella were centrifuged at 12,000 × g for 5 min, and the bacteria were washed twice with sterile PBS. The final pellet was resuspended in antibiotic-free DMEM at a density of ca. 2 × 108 bacteria/ml. Then, 50-μl aliquots of the suspension (ca. 107 bacteria) were added to the wells containing the RAW264.7 or J774 cells. The multiplicity of infection (MOI) was therefore 5 to 10:1. After incubation at 37°C for 1 h, the culture supernatants were aspirated, and the cells were washed twice with sterile PBS before being overlaid with medium containing 100 μg of gentamicin per ml. The cells were incubated at 37°C for different times before preparing protein extracts or RNA.
Since wild-type strains of Salmonella invaded the cells more efficiently than the mutant strains (see Results), there was a significant difference in the numbers of the two types of bacteria surviving after overnight incubation. To minimize this difference, tetracycline was added to a final concentration of 10 μg/ml to cells infected with wild-type strains of Salmonella 2 h after the end of the infection period. The addition of the antibiotic inhibited the multiplication of intracellular bacteria, as found by others (5). In our experiments, addition of the drug reduced the number of wild-type bacteria surviving at the end of the experiment from 0.36 ± 0.06 × 106 to 0.28 ± 0.02 × 104, thereby ensuring that the numbers of surviving wild-type and mutant Salmonella were comparable. Tetracycline was always added to uninfected cells to control for any effects the drug might have on iNOS expression.
Quantitation of bacterial invasiveness.
Invasiveness was determined by means of a standard gentamicin protection assay (21). In brief, after the completion of the infection period, the cells were incubated in gentamicin-containing medium in order to kill extracellular bacteria. Two hours later, the supernatant medium was aspirated from the cultures, and the cells were washed twice with sterile PBS. The cells were then lysed in 0.2 ml of sterile 1% Triton X-100. Serial dilutions of the lysates were made, and aliquots plated onto LB agar to determine the number of viable, gentamicin-protected (intracellular) bacteria. The mean number and standard deviation of intracellular bacteria, expressed as a percentage of the original inoculum, were calculated from triplicate wells.
To quantitate bacterial attachment, the cells were lysed immediately after completion of the infection period, and serial dilutions of the lysates were plated to obtain the number of “cell-associated” bacteria. The number of attached bacteria was then calculated by subtracting the number of internalized bacteria (as determined above) from the cell-associated bacteria (12).
Western blotting.
At different times after completion of infection, the cells were washed with ice-cold Tris-buffered saline (TBS; 10 mM Tris-Cl, pH 8.0; 150 mM NaCl) and then lysed in 0.2 ml of 1% NP-40 in TBS containing 10 μg of aprotinin and leupeptin per ml and 2 mM phenylmethylsulfonyl fluoride. After incubation on ice for 15 min, the lysates were cleared by centrifugation at 12,000 × g for 15 min at 4°C. The protein concentrations of the cleared lysates were estimated by the DC protein assay (Bio-Rad, Hercules, Calif.) according to the manufacturer's instructions. Equal amounts of protein, along with prestained molecular weight markers (Gibco-BRL, Grand Island, N.Y.), were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 9% acrylamide gels and then transferred to a nitrocellulose membrane by semidry blotting as previously described (3). The membrane was blocked with 5% nonfat dry milk in TBS-Tween 20 (TBST; TBS containing 0.05% Tween 20) for 30 min at room temperature. The filter was then divided in two, using the prestained markers as a guide. The upper half of the filter (68 kDa and above) was probed with an antibody specific for iNOS (NOS2; Santa Cruz Biotechnology, Santa Cruz, Calif.). The lower half of the filter was probed with an antibody to the housekeeping protein elongation factor 1α (anti-EF1α; Upstate Biotechnology, Lake Placid, N.Y.) to confirm equal loading of lanes. The blot was then developed with horseradish peroxidase-conjugated second antibodies and enhanced chemiluminescence. Bands were quantitated with NIH Image software after scanning the autoradiograms.
Northern blotting.
Total RNA was prepared from infected cells using Trizol reagent (Gibco-BRL, Grand Island, N.Y.), according to the manufacturer's instructions. Equal amounts of total RNA (15 to 20 μg per lane) were electrophoresed on a 1.2% formaldehyde-agarose gel, photographed under UV transillumination, and then blotted to nitrocellulose membrane according to a standard protocol (34). After baking the membrane at 80°C for 1 h, it was hybridized overnight at 65°C to an [α-32P]dCTP-labeled probe specific for murine iNOS, washed, and exposed for autoradiography.
Construction of Sip expression plasmids and transformation into Salmonella.
The reading frames encoding SipC and SipD were amplified by PCR from the genomic DNA of wild-type Salmonella (SL1344) using the following primers: SipC-5′, 5′-CGCCGAAGCTTAATATGTTAATTAGTAATG-3′; SipC-3′, 5′-GCGAATTCTTTCAGATTAAGCGCG-3′; SipD-5′, 5′-GCGAATTCATCTATACGCCATCATG-3′; and SipD-3′, 5′-AGAAGAGTGCGGCCGCCATTATTAATATCCTC-3′. Restriction sites incorporated into the primers were used to clone the PCR products into the bacterial expression vector pBH (Roche, Nutley, N.J.) downstream of a trp-lac combination promoter and a ribosome binding site. The SipC reading frame was cloned into the HindIII and EcoRI sites of the vector to give the plasmid pBHSipC, while the SipD reading frame was cloned into the EcoRI and NotI sites to give pBHSipD. To construct a plasmid expressing both SipC and SipD, the SipD-encoding fragment was cloned into the EcoRI and NotI sites of pBHSipC, downstream of the SipC reading frame. The sequences of the SipC-3′ and SipD-5′ primers were such that the resultant plasmid (pBHSipCD) reproduced the sequence of the SipC and SipD encoding region of the wild-type genomic locus, except for the introduction of the EcoRI site (24). pBHSipC, pBHSipD, and pBHSipCD were individually transformed into appropriate Salmonella strains by electroporation (32), and transformants were selected on ampicillin-containing medium.
Analysis of Sip proteins in bacterial supernatants.
Salmonella strains (wild-type, mutant, and transformed) were grown exactly as described above for the infections. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the overnight standing cultures to a final concentration of 0.5 mM. Incubation at 37°C was continued for a further 3 h, after which the bacteria were spun down and the supernatants were filtered through 0.45-μm-pore-size filters. The filtered supernatants were mixed with trichloroacetic acid (final concentration, 10%) and incubated on ice for 30 min. The precipitated proteins were collected by centrifugation at 15,000 × g for 15 min, washed with acetone, dried, and dissolved in SDS-PAGE sample buffer. After separation on 9% acrylamide gels, the proteins were stained with Gel Code Blue (Pierce Chemical Co., Rockford, Ill.) according to the manufacturer's recommendations.
RESULTS
Induction of iNOS by Salmonella is sensitive to heat, chemical fixation, and inhibition of protein synthesis.
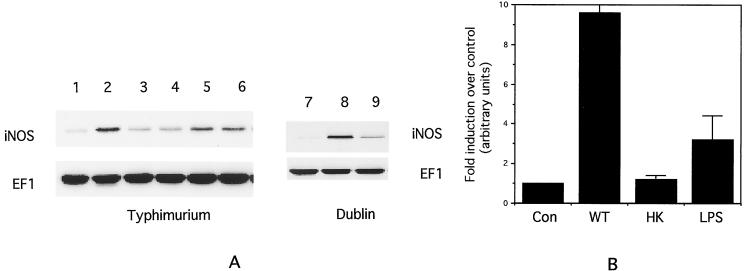
RAW264.7 cells were infected at an MOI of 5 to 10:1 with the wild-type serovar Typhimurium strain SL1344. Cell lysates were prepared about 18 h after infection, and Western blotting was carried out to analyze iNOS expression as well as expression of the housekeeping protein EF1α. As shown in Fig. 1A, lanes 1 and 2, infection with SL1344 resulted in a significant increase in iNOS protein expression (an average of ca. ninefold over control, as determined by quantitation of band intensities; Fig. 1B). This increase could be detected as early as 4 h after infection (data not shown) but was maximal at 18 h. The level of EF1α did not change appreciably, confirming the equal loading of lanes.
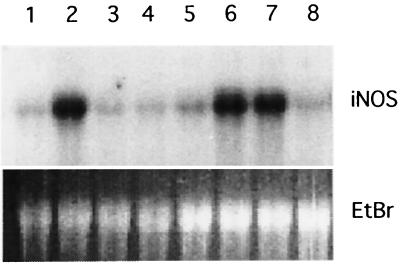
FIG. 1.
(A) Western blot of iNOS and EF1 expression in RAW264.7 cells infected with Salmonella. Lane 1, control; lane 2, SL1344; lane 3, heat-killed SL1344; lane 4, formaldehyde-fixed SL1344; lane 5, 100 ng of LPS per ml; lane 6, heated LPS; lane 7, control; lane 8, 2229; lane 9, chloramphenicol-treated 2229. (B) Quantitation of mean iNOS band intensities from three separate experiments similar to that shown in panel A. Con, control; WT, wild-type Salmonella; HK, heat-killed Salmonella; LPS, 100 ng of LPS per ml. Error bars indicate the standard deviations.
To determine if bacterial viability was required for iNOS induction, the experiment was carried out with an equal number of SL1344 organisms which had been either heated to 80°C for 10 min or fixed with 2% formaldehyde for 20 min. Both of these treatments significantly reduced the level of iNOS induction (Fig. 1A, lanes 3 and 4, and Fig. 1B). Treating the cells with 100 ng of LPS per ml for 18 h resulted in an increase in iNOS expression that was appreciably lower than that induced by Salmonella (ca. fourfold over that for the control; Fig. 1, lane 5, and Fig. 1B). The LPS-induced increase was unaffected by heating the LPS to 80°C for 10 min (Fig. 1, lane 6). These results indicate that the Salmonella had to be viable to induce iNOS expression. They also suggest that it is unlikely that the LPS in the bacterial cell wall is sufficient to explain the induction.
Similar results were obtained with a second wild-type Salmonella strain (strain 2229, serovar Dublin; Fig. 1, lanes 7 and 8), as well as with a second murine macrophage cell line, J774 (data not shown).
To determine if bacterial protein synthesis was required for iNOS induction, the Salmonella strains were treated for 1 h at 37°C with 100 μg of chloramphenicol per ml before they were added to the cells. Chloramphenicol was also added to the cell culture medium during the 1-h infection period. As shown in Fig. 1, lanes 7 to 9, for the Dublin strain 2229, this treatment significantly reduced the level of iNOS that was induced, suggesting that the bacteria had to be actively synthesizing protein for proper induction. The same effect was observed with the wild-type Typhimurium strain SL1344 (data not shown).
Hersh et al. have reported that infection of RAW264.7 with wild-type Salmonella leads to apoptosis and that the mechanism of cell death involves binding of SipB to caspase 1 (18). We did not observe any evidence of Salmonella-induced apoptosis in our studies, either by trypan blue exclusion or by examination of nuclear morphology (data not shown). This difference may be attributable to the much lower MOIs used in our experiments (5 to 10:1 versus 100:1 in the studies of Hersh et al.) and to the fact that we used tetracycline to inhibit intracellular multiplication of wild-type Salmonella.
Induction of iNOS by Salmonella requires an intact SPI1-encoded type III secretion system and specific effector proteins.
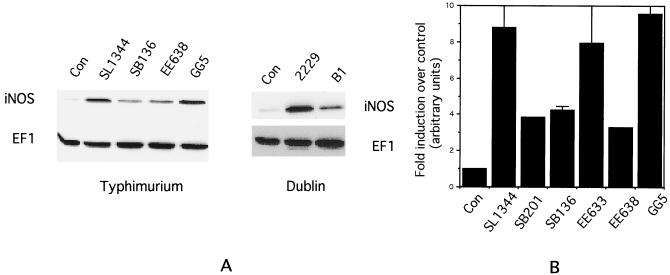
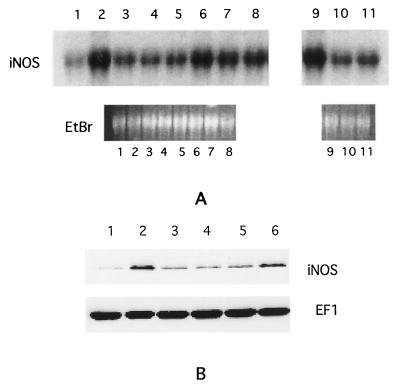
The requirement for bacterial viability and protein synthesis led us to inquire whether the secretion of the Salmonella effector proteins via the SPI1 type III system might be involved in iNOS induction. To evaluate this possibility, we examined mutant strains of Salmonella that were deficient in components of this type III secretion system or various effector proteins for their ability to induce iNOS in RAW264.7 cells. The results of such an experiment are shown in the Western blot of Fig. 2A, which indicates that the strains SB136, which lacks InvA (a structural component of the type III apparatus), and EE638, which lacks the effectors SipC, SipD, and SipA, are impaired in iNOS induction, while strain GG5, which lacks the effector protein SopE, induces iNOS to levels comparable to the wild-type SL1344 strain (lanes 1 to 5). Similarly, the serovar Dublin strain B1, which lacks Sips B, C, D, and A, is impaired in iNOS induction compared to the wild-type 2229 strain (lanes 6 to 8). Quantitation of iNOS band intensities from several such experiments is shown in Fig. 2B, which indicates that the mutant strains induce iNOS protein to levels which are about 50% of that induced by the wild-type strain. Plating of serial dilutions of the bacterial suspensions used to infect the cells confirmed that the numbers of input bacteria were comparable for all of the strains.
FIG. 2.
(A) Western blot of iNOS and EF1 expression in RAW264.7 cells infected with different strains of Salmonella. Strain designations are indicated above the lanes. (B) Quantitation of mean iNOS band intensities from five separate experiments similar to that shown in panel A. Strain designations are indicated below the columns. Error bars indicate the standard deviations.
Similar experiments were carried out with a large number of mutant strains of both the Typhimurium and the Dublin serovars. The results of these experiments are summarized in Table 1. Inspection of the results shown in the table indicates that strains that lack a functional SPI1 type III secretion system (VV341, SB201, and SB136) are unable to induce iNOS to wild-type levels. EE638, which lacks Sips C, D, and A, is also impaired in iNOS induction, but EE633, which lacks SipA, behaves like the wild-type strain. The results from the latter two strains indicate that Sips C and D, but not SipA, are required for inducing iNOS expression. The same conclusion can be drawn from the phenotypes of the Dublin strains A1 and B1, i.e., that SipA is not required for induction of iNOS. The results also indicate that SipB by itself is not sufficient to induce iNOS expression since strain EE638 (which expresses SipB, but not Sips C, D, and A) is deficient in this function.
TABLE 1.
Induction of iNOS protein in RAW264.7 cells by various Salmonella strains
| S. enterica strain | Deficient protein(s) | % iNOS expression |
|---|---|---|
| Serovar Typhimurium | ||
| SL1344 | Wild type | 100 |
| VV341 | HilA | 34 |
| SB201 | InvG | 44 |
| SB136 | InvA | 48 |
| EE633 | SipA | 90 |
| EE638 | Sips C, D, and A | 37 |
| GG5 | SopE | 108 |
| Serovar Dublin | ||
| 2229 | Wild type | 100 |
| A1 | SipA | 96 |
| B1 | Sips B, C, D, and A | 54 |
| AV1 | AvrA | 97 |
| SB2 | SopB | 76 |
| SE1 | SopE | 116 |
| SC2 | SptP | 103 |
Induction of iNOS by Salmonella does not require invasion of cells.
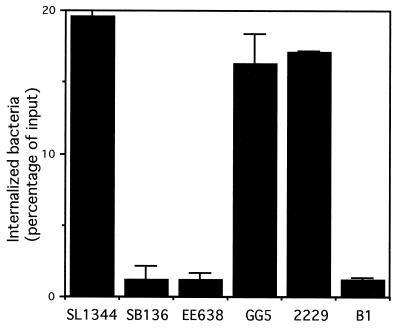
Studies in epithelial cell lines have indicated that structural components of the SPI1-encoded type III secretion system such as InvG and InvA, as well as Sips C and D, are essential for invasion (21). SipB is also involved in this process since Sips B, C, and D are mutually interdependent for translocation into host cells (4). Since macrophages are professional phagocytic cells and may not behave like the nonphagocytic epithelial cells, we addressed the invasiveness of the various Salmonella strains in RAW264.7 by means of a standard gentamicin protection assay. The results of such an experiment are shown in Fig. 3. It is clear that the strains SB136 (deficient in the InvA structural component of the type III secretion system), EE638 (deficient in Sips C, D, and A), and B1 (deficient in Sips B, C, D, and A) are significantly defective in invasion compared to the two wild-type strains SL1344 and 2229. The strain EE633 (deficient in SipA) had wild-type levels of invasion (data not shown). Similar, although less dramatic, differences in internalization between wild-type and mutant bacteria were observed when bacteria were opsonized with 20% normal mouse serum (ca. 15% internalized for strain SL1344 versus ca. 3% internalized for strain EE638). These results indicate that even in the phagocytic RAW264.7 cells, internalization of Salmonella is facilitated by an intact type III secretion system and by Sips B, C, and D.
FIG. 3.
Invasiveness of different Salmonella strains based on the mean numbers of internalized bacteria (percent input) 2 h after infection of RAW264.7 cells. Error bars indicate the standard deviations.
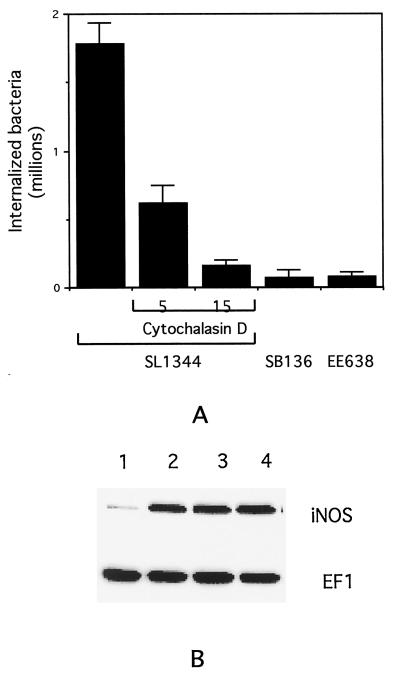
Based on these studies of invasiveness, it seemed possible that the effects of Sips B, C, and D on iNOS induction described in the previous section could be secondary to decreased entry of the corresponding mutant bacteria into the cells. To address this possibility, we inhibited the internalization of the wild-type Salmonella strain SL1344 by treating the RAW264.7 cells with the actin depolymerizing agent cytochalasin D for 1 h before and during the course of infection (12). The number of internalized bacteria was determined, and representative results are displayed in Fig. 4A. The results indicate clearly that cytochalasin D treatment produces a dose-dependent decrease in the number of internalized wild-type Salmonella. At 15 μg of cytochalasin D per ml, the number of internalized wild-type bacteria is comparable to the number of internalized mutant strains SB136 and EE638.
FIG. 4.
(A) Effect of cytochalasin D (at 5 or 15 μg/ml) on internalization of wild-type Salmonella (SL1344) into RAW264.7 cells compared to the number of internalized SB136 and EE638 mutant strains. Error bars indicate the standard deviations. (B) Western blot indicating the effect of cytochalasin D treatment on iNOS and EF1 expression in RAW264.7 cells. Lane 1, uninfected cells; lane 2, untreated cells infected with SL1344; lane 3, cytochalasin D-treated (5 μg/ml) cells infected with SL1344; lane 4, cytochalasin D-treated (15 μg/ml) cells infected with SL1344.
We then tested the effect of cytochalasin D on the ability of wild-type Salmonella to induce expression of iNOS. As shown in Fig. 4B, in contrast to the effects on internalization, cytochalasin D had little or no effect on iNOS expression, even at a dose of 15 μg/ml. Cytochalasin D treatment alone did not alter iNOS protein expression from control levels (data not shown). These observations indicate that cell invasion is not required for iNOS induction by Salmonella, making it unlikely that the requirement for Sips B, C, and D in this process is secondary to their role in invasiveness.
There were no significant differences between wild-type and mutant bacterial strains in their attachment to RAW264.7 cells (data not shown), indicating that the differences in iNOS induction could not be attributed to variations in adherence of the bacteria to the cells.
Effect of Salmonella on iNOS mRNA.
To examine the effect of Salmonella infection on iNOS mRNA, Northern blots were carried out on total RNA extracted from infected RAW264.7 cells. In initial experiments, induction of iNOS mRNA was found to be maximal at about 18 h after infection and required bacterial viability, as observed for the iNOS protein (data not shown). We then went on to examine the mutant strains of Salmonella for their ability to induce iNOS mRNA. As shown in Fig. 5, the wild-type strain SL1344 (lane 2) induced a dramatic increase in iNOS mRNA level over control (lane 1), but the mutants SB201 (deficient in InvG; lane 3), SB136 (deficient in InvA; lane 4), and EE638 (deficient in the effectors Sips C, D, and A; lane 5) did not. A strain deficient in the effector SopE (GG5; lane 6) showed wild-type levels of iNOS induction, as did the strain EE633 (deficient in the effector SipA; data not shown). Similar results were obtained with the Dublin serovar; the wild-type strain 2229 showed a marked increase in iNOS mRNA, but the mutant B1 (deficient in the effectors Sips B, C, D, and A) showed very little increase over uninfected cells (data not shown and also see the Northern blot of Fig. 7). Confirming the results obtained with iNOS protein, treatment of the cells with 15 μg of cytochalasin D per ml did not inhibit iNOS mRNA induction by wild-type Salmonella (Fig. 5, lane 7), while cytochalasin D by itself had no effect (lane 8). Thus, the results of the Northern blot analysis substantiate the iNOS protein data and suggest that the regulation of iNOS expression by Salmonella occurs at the level of mRNA.
FIG. 5.
Northern blot of iNOS mRNA expression in RAW264.7 cells 18 h after infection with different strains of Salmonella. Lane 1, control; lane 2, SL1344; lane 3, SB201; lane 4, SB136; lane 5, EE638; lane 6, GG5; lane 7, cytochalasin D-treated (15 μg/ml) cells infected with SL1344; lane 8, cytochalasin D-treated (15 μg/ml) cells. The ethidium bromide-stained 28S rRNA band (EtBr) is shown to indicate the loading of lanes.
FIG. 7.
(A) Northern blot of iNOS expression in RAW264.7 cells infected with wild-type, mutant, and transformed strains of Salmonella. Lane 1, uninfected cells; lane 2, SL1344; lane 3, EE638; lane 4, EE638/C; lane 5, EE638/D; lane 6, EE638/CD; lane 7, EE638/CD, cells treated with 15 μg of cytochalsin D per ml; lane 8, EE638/CD, no IPTG; lane 9, 2229; lane 10, B1; lane 11, B1/CD. EtBr, ethidium bromide-stained 28S rRNA band. (B) Western blot of iNOS and EF1 expression in RAW264.7 cells infected with wild-type, mutant, and transformed strains of Salmonella. Lane 1, uninfected cells; lane 2, SL1344; lane 3, EE638; lane 4, EE638/C; lane 5, EE638/D; lane 6, EE638/CD.
Complementation of mutant Salmonella strains with Sip expression plasmids.
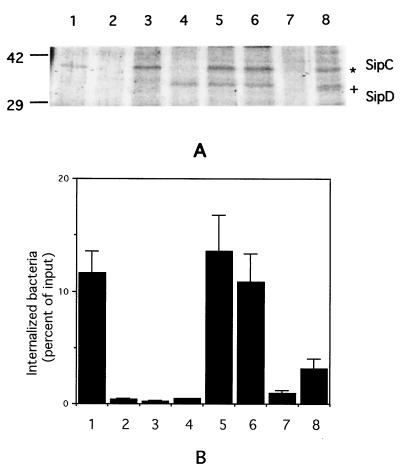
The data presented above implicate SipB, SipC, and SipD in the regulation of iNOS expression by Salmonella. To confirm this idea and to determine which of these proteins is actually required for iNOS induction, we created expression plasmids expressing SipC and SipD, either individually or in combination, and transformed them into strains EE638 (which expresses SipB) and B1 (which does not express SipB). Transformants were selected and grown overnight in standing cultures, and expression of the plasmid-encoded Sips was examined by analyzing the proteins present in the bacterial supernatants. The results of such an experiment are shown in Fig. 6A, which indicates that proteins of the appropriate size (SipC, ca. 42 kDa; SipD, ca. 36 kDa) are present at similar levels in the supernatants of the relevant transformed strains. The plasmid-encoded proteins were expressed even when the bacteria were not induced with IPTG (compare lanes 5 and 6 in Fig. 6A).
FIG. 6.
(A) Expression of Sips C and D in wild-type, mutant, and transformed strains of Salmonella. Lane 1, SL1344; lane 2, EE638; lane 3, EE638/C; lane 4, EE638/D; lane 5, EE638/CD; lane 6, EE638/CD, no IPTG; lane 7, B1; lane 8, B1/CD. The numbers to the left of the gel indicate molecular mass in kilodaltons. (B) Invasiveness of wild-type, mutant, and transformed strains of Salmonella. The column numbers refer to the same strains and conditions as the lane numbers in panel A. Error bars indicate the standard deviations.
To confirm the functionality of the introduced proteins, we carried out invasion assays on the wild-type, mutant, and transformed strains. The results are shown in Fig. 6B and indicate that strain EE638/CD (the transformant expressing Sips B, C, and D) displays invasiveness comparable to wild-type Salmonella, with or without IPTG induction, while EE638/C (the transformant expressing Sips B and C but not SipD), EE638/D (the transformant expressing Sips B and D but not SipC), and B1/CD (the transformant expressing Sips C and D but not SipB) are significantly impaired in invasiveness. These findings are consistent with previously reported observations that Sips B, C, and D are all required for invasion (4, 21) and confirm that the plasmid-encoded SipC and SipD are capable of carrying out at least one of the functions ascribed to them.
We then went on to examine the ability of the transformed strains to induce iNOS expression 18 h after infection of RAW264.7 cells. As shown in the Northern blot of Fig. 7A, strain EE638/CD (the transformant expressing Sips B, C, and D, lane 6) induced iNOS mRNA expression comparable to wild-type Salmonella (lane 2). In contrast, strains EE638/C (the transformant expressing Sips B and C but not SipD; lane 4), EE638/D (the transformant expressing Sips B and D but not SipC; lane 5), and B1/CD (the transformant expressing Sips C and D but not SipB; lane 11) are as defective as the parental mutant strains (EE638, lane 3; B1, lane 10) in the induction of iNOS mRNA. Treating the cells with cytochalasin D did not significantly reduce the ability of EE638/CD to induce iNOS (lane 7), a result consistent with our observations on the wild-type strain. In keeping with the observation that IPTG induction was not required for expression of plasmid-encoded Sips C and D (Fig. 6), iNOS induction occurred in response to EE638/CD even without IPTG treatment of the bacteria (lane 8). Similar results were obtained when Western blotting was used to analyze iNOS protein expression (Fig. 7B). These observations confirm the involvement of Sips B, C, and D in iNOS induction by Salmonella and further indicate that all three of these proteins are required for this function.
Involvement of Sip-dependent effectors in iNOS induction.
The analysis of the mutant strains is consistent with the idea that an effector protein or proteins whose translocation is dependent on both the type III secretion system and the Sips B, C, and D might be involved in inducing iNOS expression. Four such effectors, SopB, SopE, SptP, and AvrA (9, 11, 15, 16, 40) have been considered, and the results shown in Table 1 indicate that they are not required for iNOS induction. Recently, a structural homolog of SopE, SopE2, has been identified (2). SopE2 is expressed in all Salmonella strains tested, in contrast to SopE, which is encoded in a temperate bacteriophage found only in some strains (16, 29). Like SopE, SopE2 is found in protein aggregates in culture supernatants of SipB-deficient strains, suggesting that the translocation of the latter protein is also likely to be dependent on Sips B, C, and D (2).
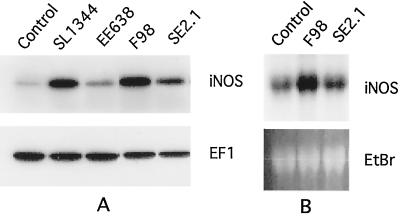
We tested a SopE-deficient, wild-type serovar Typhimurium strain, F98, as well as its isogenic, SopE2-deficient derivative, SE2.1, for the ability to induce iNOS expression in RAW264.7 cells. The results shown in Fig. 8 indicate that F98 induced iNOS expression as well as the wild-type strain SL1344, a finding consistent with the results obtained with two other SopE-deficient strains that we have tested (GG5 and SE1). However, iNOS induction by strain SE2.1 was reduced, suggesting that the absence of SopE2 (in a SopE-deficient background) impaired this function. These results indicate that SopE2 is involved in inducing iNOS expression.
FIG. 8.
(A) Western blot of iNOS and EF1 expression in RAW264.7 cells infected with wild-type and mutant strains of Salmonella. (B) Northern blot of iNOS expression in RAW264.7 cells infected with wild-type and mutant strains of Salmonella. EtBr, ethidium bromide-stained 28S rRNA band. Strain designations are indicated above the lanes.
DISCUSSION
The production of NO is an important antimicrobial defense mechanism, but when uncontrolled it can lead to pathological inflammatory states (28). Indeed, elevated NO levels and increased expression of iNOS have been implicated in clinical problems such as rheumatoid arthritis and inflammatory bowel disease (1, 7, 19, 27). Elucidating the mechanisms that regulate iNOS expression is thus relevant to understanding both innate immunity, and the pathogenesis and treatment of chronic inflammatory conditions.
Like several other microorganisms, Salmonella has been noted to increase expression of iNOS in mammalian cells, although the mechanism of induction has not been worked out in detail. Two recent studies have shown that infection of intestinal epithelial cell lines by Salmonella is associated with increased iNOS expression, and a correlation between invasiveness and iNOS induction was observed (33, 39). Our findings extend these studies to macrophages and also demonstrate the involvement of SipB, SipC, and SipD in the regulation of iNOS expression. These effectors have been previously shown to play an important role in invasion, thereby providing an explanation for the link between invasiveness and iNOS induction noted in the earlier studies (33, 39). However, our results show clearly that the two processes can be dissociated based on the effects of cytochalasin D treatment (Fig. 4 and 5). This uncoupling from invasion has been noted for other effects induced by Salmonella, i.e., the activation of NF-κB, as well as transepithelial signaling for neutrophil recruitment (8, 12), and is consistent with the observation that Sips can be translocated into mammalian cells by extracellularly situated bacteria (4).
One of the functions of Sips B, C, and D is the formation of a translocation apparatus for the delivery of certain effector proteins (AvrA, SopB, SopE, and SptP) into the mammalian cytosol (9, 11, 15, 16, 40). The involvement of one of these effectors in iNOS induction seemed likely based on our observation that Sips B, C, and D were all required for the function. However, our initial series of experiments ruled out a role for AvrA, SopB, SopE, and SptP (Table 1), suggesting that yet another Sip-dependent effector might be the ultimate mediator of iNOS induction. Indeed, we found that a strain deficient in SopE2, a recently identified homolog of SopE that is also likely to be dependent on Sips B, C, and D for translocation (2), was clearly impaired in its ability to induce iNOS expression (Fig. 8).
Our findings thus suggest a simple model in which SopE2, translocated into the mammalian cell via the SPI1 type III secretion system and the SipB, -C, and -D complex, initiates signals leading to increased iNOS expression. SopE2 is likely to have the same function as SopE in relation to the activation of Rho GTPases (14), thus providing an obvious mechanism for the initiation of signaling. The close structural, and presumed functional, homology between the two molecules raises the possibility that SopE may also have the ability to induce iNOS expression. Although strains GG5 and SE1 (both deficient in SopE) behaved like the corresponding wild-type strains with respect to iNOS expression (Table 1), the presence of SopE2 in these strains probably compensated for the lack of SopE. A strain expressing SopE but deficient in SopE2 will be required to test the involvement of the former effector in iNOS induction. Finally, it is worth mentioning here that since Rho GTPases have been implicated in the Salmonella-activated signaling pathways leading to the expression of proinflammatory cytokines in epithelial cells (20), SopE and SopE2 are likely to have a broad role in inducing inflammation that extends beyond the production of NO in macrophages.
Complementation of the SopE2-deficient strain SE2.1 with a plasmid-encoded SopE2 gene (2) did not restore iNOS induction to wild-type levels (data not shown). Although this failure of rescue raises the possibility that some other gene might be involved in the regulation of iNOS expression, we think it unlikely since (i) the sopE2 mutation does not cause obvious effects on the expression or secretion of any other Salmonella secretory protein (2) and (ii) our analysis of the SipB-, SipC-, and SipD-deficient mutants and their transcomplemented derivatives clearly indicates a role for a Sip-dependent effector in iNOS induction. The failure of complementation may reflect a requirement for specifically regulated expression of SopE2 (expression at a specific time during invasion, or at a specific site within the macrophage) that cannot be achieved by the plasmid-encoded effector.
Although our findings have highlighted the involvement of Sips B, C, and D and SopE2 in iNOS induction, they do not exclude a role for LPS in this process. LPS is an important inducer of iNOS expression in macrophages, especially in conjunction with cytokines such as interferon gamma (42). Furthermore, a mutant of Salmonella with altered LPS structure has been shown to be impaired in inducing NO production, both in vitro and in vivo (26). Sip-deficient, as well as SopE2-deficient, mutants of Salmonella have residual iNOS inducing ability (Fig. 2 and 8, for example), which could be attributable to the effects of LPS acting through the recently described Toll-like receptors (30). It is likely that the final level of iNOS expression is determined by signals activated by both LPS and SopE2.
We have not directly addressed the in vivo significance of our observations. However, evidence from the published literature suggests that our findings are likely to be relevant to Salmonella pathogenicity. B1, the mutant strain of serovar Dublin deficient in Sips A, B, C, and D, has been found to be markedly impaired in inducing intestinal fluid secretion and neutrophil transmigration in bovine ligated ileal loops (11). Similar findings were obtained when serovar Typhimurium mutant strains deficient in SipB, SipC, or SipD were fed to calves (37). Although the decreased pathogenicity of the Sip-deficient strains in these studies could be related to their poor invasiveness, at least some aspects of virulence (neutrophil transmigration, NF-κB activation) have been shown to be independent of cell invasion (8, 12). An alternative possibility, therefore, is that one of the effectors dependent on Sips B, C, and D for translocation might be involved in inducing intestinal pathology. The involvement of several such effectors has been examined in bovine models. Strains deficient in SopB, SipA, or SptP all behave like wild-type Salmonella after being fed to calves (36, 37). In the bovine ligated ileal loop model, strains deficient in SptP or AvrA are unimpaired in enteropathogenesis, and strains deficient in SopB or SopE are only modestly impaired (11, 31, 35, 38). Thus, these in vivo observations have an excellent correlation with our in vitro findings: Sips B, C, and D are required for inducing both intestinal pathology and iNOS expression; conversely, SipA, SopB, SopE, AvrA, and SptP are not required for either of these functions. Together with the known involvement of NO in inflammatory conditions of the bowel (7, 19, 27), our results suggest that the SopE2-dependent induction of iNOS in epithelial cells and tissue macrophages of the gut will have an important role in the intestinal pathology associated with Salmonella infection. Experiments to address this possibility will be a fruitful avenue of further research.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants T-32-DK-07477 (B.J.C.) and DK-50989 (B.A.M.).
We thank Jorgé Galan, Edouard Galyov, and Catherine Lee for kindly providing bacterial strains; Isabel Fernandez and Milton Silva for assisting with some of the experiments; Catherine Lee, Cathryn Nagler-Anderson, and Shiv Pillai for helpful comments on the manuscript; and W. Allan Walker for his continued support.
REFERENCES
- 1.Amin A R, Attur M, Abramson S B. Nitric oxide synthase and cyclooxygenases: distribution, regulation and intervention in arthritis. Curr Opin Rheumatol. 1999;11:202–209. doi: 10.1097/00002281-199905000-00009. [DOI] [PubMed] [Google Scholar]
- 2.Bakshi C S, Singh V P, Wood M W, Jones P W, Wallis T S, Galyov E E. Identification of SopE2, a Salmonella secreted protein which is highly homologous to SopE and involved in bacterial invasion of epithelial cells. J Bacteriol. 2000;182:2341–2344. doi: 10.1128/jb.182.8.2341-2344.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaudhuri A, Orme S, Vo T, Wang W, Cherayil B J. CD40-mediated signals inhibit the binding of TRAF2 to the CD40 cytoplasmic domain. J Immunol. 1997;159:4244–4251. [PubMed] [Google Scholar]
- 4.Collazo C M, Galan J E. The invasion-associated type III secretion system of Salmonella typhimurium directs the translocation of Sip proteins into the host cell. Mol Microbiol. 1997;24:747–756. doi: 10.1046/j.1365-2958.1997.3781740.x. [DOI] [PubMed] [Google Scholar]
- 5.Darji A, Guzman C A, Gerstel B, Wachholz P, Timmis K N, Wehland J, Chakraborty T, Weiss S. Oral somatic transgene vaccination using attenuated S. typhimurium. Cell. 1997;91:765–775. doi: 10.1016/s0092-8674(00)80465-1. [DOI] [PubMed] [Google Scholar]
- 6.DeGroote M A, Granger D, Xu Y, Campbell G, Prince R, Fang F C. Genetic and redox determinants of nitric oxide cytotoxicity in a Salmonella typhimurium model. Proc Natl Acad Sci USA. 1995;92:6399–6403. doi: 10.1073/pnas.92.14.6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dijkstra G, Moshage H, van Dulleman H M, de Jager-Krikken A, Tieboshc A T, Kleibeuker J H, Jansen P L, van Goor H. Expression of nitric oxide synthases and formation of nitrotyrosine and reactive oxygen species in inflammatory bowel disease. J Pathol. 1998;186:416–421. doi: 10.1002/(SICI)1096-9896(199812)186:4<416::AID-PATH201>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 8.Eaves-Pyles T, Szabo C, Salzmann A L. Bacterial invasion is not required for activation of NF-κB in enterocytes. Infect Immun. 1999;67:800–804. doi: 10.1128/iai.67.2.800-804.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu Y, Galan J E. The Salmonella typhimurium tyrosine phosphatase SptP is translocated into host cells and disrupts the actin cytoskeleton. Mol Microbiol. 1998;27:359–368. doi: 10.1046/j.1365-2958.1998.00684.x. [DOI] [PubMed] [Google Scholar]
- 10.Galan J E, Curtiss R., III Cloning and molecular characterization of genes whose protein products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci USA. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galyov E E, Wood M W, Rosqvist R, Mullan P B, Watson P R, Hedges S, Wallis T S. A secreted effector protein of Salmonella dublin is translocated into eukaryotic cells and mediates inflammation and fluid secretion in infected ileal mucosa. Mol Microbiol. 1997;25:903–912. doi: 10.1111/j.1365-2958.1997.mmi525.x. [DOI] [PubMed] [Google Scholar]
- 12.Gewirtz A T, Siber A M, Madara J L, McCormick B A. Orchestration of neutrophil movement by intestinal epithelial cells in response to Salmonella typhimurium can be uncoupled from bacterial internalization. Infect Immun. 1999;67:608–617. doi: 10.1128/iai.67.2.608-617.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glynn M K, Bopp C, Dewitt W, Dabney P, Mokhtar M, Angulo F J. Emergence of multidrug resistant Salmonella enterica serotype Typhimurium DT104 infections in the United States. N Engl J Med. 1998;338:1331–1338. doi: 10.1056/NEJM199805073381901. [DOI] [PubMed] [Google Scholar]
- 14.Hardt W D, Chen L M, Schuebel K E, Bustelo X R, Galan J E. S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell. 1998;93:815–826. doi: 10.1016/s0092-8674(00)81442-7. [DOI] [PubMed] [Google Scholar]
- 15.Hardt W D, Galan J E. A secreted Salmonella protein with homology to an avirulence determinant of plant pathologic bacteria. Proc Natl Acad Sci USA. 1997;94:9887–9892. doi: 10.1073/pnas.94.18.9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardt W D, Urlaub H, Galan J E. A target of the centisome 63 type III secretion system of Salmonella typhimurium is encoded by a cryptic bacteriophage. Proc Natl Acad Sci USA. 1998;95:2574–2579. doi: 10.1073/pnas.95.5.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayward R D, Koronakis V. Direct nucleation and bundling of actin by the SipC protein of invasive Salmonella. EMBO J. 1999;18:4926–4934. doi: 10.1093/emboj/18.18.4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hersh D, Monack D M, Smith M R, Ghori N, Falkow S, Zychinsky A. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc Natl Acad Sci USA. 1999;96:2396–2401. doi: 10.1073/pnas.96.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herulf M, Ljung T, Hellstrom P M, Wietzburg E, Lundberg J O. Increased luminal nitric oxide in inflammatory bowel disease as shown with a novel minimally invasive method. Scand J Gastroenterol. 1998;33:164–169. doi: 10.1080/00365529850166897. [DOI] [PubMed] [Google Scholar]
- 20.Hobbie S, Chen L M, Davis R J, Galan J E. Involvement of mitogen-activated protein kinase pathways in the nuclear responses and cytokine production induced by Salmonella typhimurium in cultured intestinal epithelial cells. J Immunol. 1997;159:5550–5559. [PubMed] [Google Scholar]
- 21.Hueck C J, Hantman M J, Bajaj V, Johnston C, Lee C A, Miller S I. Salmonella typhimurium secreted invasion determinants are homologous to Shigella Ipa proteins. Mol Microbiol. 1995;18:479–490. doi: 10.1111/j.1365-2958.1995.mmi_18030479.x. [DOI] [PubMed] [Google Scholar]
- 22.Incze K, Farkas J, Mihalyi V, Zukal E. Antibacterial effect of cytoseine-nitrosothiol and possible precursors thereof. Appl Microbiol. 1974;27:202–205. doi: 10.1128/am.27.1.202-205.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen V B, Harty J T, Jones B D. Interactions of the invasive pathogens Salmonella typhimurium, Listeria monocytogenes, and Shigella flexneri with M cells and murine Peyer's patches. Infect Immun. 1998;66:3758–3766. doi: 10.1128/iai.66.8.3758-3766.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaniga K, Trollinger D, Galan J E. Identification of two targets of the type III protein secretion system encoded by the inv and spa loci of Salmonella typhimurium that have homology to the Shigella IpaD and IpaA proteins. J Bacteriol. 1995;177:7078–7085. doi: 10.1128/jb.177.24.7078-7085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaniga K, Uralil J, Bliska J B, Galan J E. A secreted protein tyrosine phosphatase with modular effector domains in the bacterial pathogen Salmonella typhimurium. Mol Microbiol. 1996;21:633–641. doi: 10.1111/j.1365-2958.1996.tb02571.x. [DOI] [PubMed] [Google Scholar]
- 26.Khan S A, Everest P, Servos S, Foxwell N, Zahringer U, Brade H, Rietschel E T, Dougan G, Charles I G, Maskell D J. A lethal role for lipid A in Salmonella infections. Mol Microbiol. 1998;29:571–579. doi: 10.1046/j.1365-2958.1998.00952.x. [DOI] [PubMed] [Google Scholar]
- 27.Kimura H, Hokari R, Miura S, Shigematsu T, Hirokawa M, Akiba Y, Kurose I, Higuchi H, Fujimori H, Tsuzuki Y, Serizawa H, Ishii H. Increased expression of an inducible isoform of nitric oxide synthase and the formation of peroxynitrite in colonic mucosa of patients with active ulcerative colitis. Gut. 1998;42:180–187. doi: 10.1136/gut.42.2.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacMicking J, Xie Q W, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 29.Mirold S, Ratsch W, Rohde M, Stender S, Tschape H, Russmann H, Igwe E, Hardt W D. Isolation of a temperate bacteriophage encoding the type III effector protein SopE from epidemic S. typhimurium strains. Proc Natl Acad Sci USA. 1999;96:9845–9850. doi: 10.1073/pnas.96.17.9845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Modlin R L, Brightbill H D, Godowski P J. The Toll of innate immunity on microbial pathogens. N Engl J Med. 1999;340:1834–1835. doi: 10.1056/NEJM199906103402312. [DOI] [PubMed] [Google Scholar]
- 31.Norris F A, Wilson M P, Wallis T S, Galyov E E, Majerus P W. SopB, a protein required for virulence of Salmonella dublin, is an inositol phosphate phosphatase. Proc Natl Acad Sci USA. 1998;95:14057–14059. doi: 10.1073/pnas.95.24.14057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Callaghan D, Charbit A. High efficiency transformation of Salmonella typhimurium and Salmonella typhi by electroporation. Mol Gen Genet. 1990;223:156–158. doi: 10.1007/BF00315809. [DOI] [PubMed] [Google Scholar]
- 33.Salzman A L, Eaves-Pyles T, Linn S C, Denenberg A G, Szabo C. Bacterial induction of inducible nitric oxide synthase in cultured human intestinal epithelial cells. Gastroenterology. 1998;114:93–102. doi: 10.1016/s0016-5085(98)70637-7. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 35.Schesser K, Dukuzumuremyi J M, Cilio C, Borg S, Wallis T S, Patterson S, Galyov E E. The Salmonella YopJ homolog AvrA does not possess YopJ-like activity. Microb Pathog. 2000;28:59–70. doi: 10.1006/mpat.1999.0324. [DOI] [PubMed] [Google Scholar]
- 36.Tsolis R M, Adams L G, Ficht T A, Baumler A J. Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect Immun. 1999;67:4879–4885. doi: 10.1128/iai.67.9.4879-4885.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsolis R M, Adams L G, Hantman M J, Scherer C A, Kimbrough T, Kingsley R A, Ficht T A, Miller S I, Baumler A J. SspA is required for lethal Salmonella enterica serovar Typhimurium infections in calves but is not essential for diarrhea. Infect Immun. 2000;68:3158–3163. doi: 10.1128/iai.68.6.3158-3163.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wallis T S, Galyov E E. Molecular basis of Salmonella-induced enteritis. Mol Microbiol. 2000;36:997–1005. doi: 10.1046/j.1365-2958.2000.01892.x. [DOI] [PubMed] [Google Scholar]
- 39.Witthoft T, Eckmann L, Kim J M, Kagnoff M F. Enteroinvasive bacteria directly activate expression of iNOS and NO production in human colon epithelial cells. Am J Physiol. 1998;275:G564–G571. doi: 10.1152/ajpgi.1998.275.3.G564. [DOI] [PubMed] [Google Scholar]
- 40.Wood M W, Rosqvist R, Mullan P B, Edwards M H, Galyov E E. SopE, a secreted protein of Salmonella dublin is translocated into the target eukaryotic cell via a sip-dependent mechanism and promotes bacterial entry. Mol Microbiol. 1996;22:327–338. doi: 10.1046/j.1365-2958.1996.00116.x. [DOI] [PubMed] [Google Scholar]
- 41.Wray C, Sojka W J. Experimental Salmonella typhimurium infection in calves. Res Vet Sci. 1978;25:139–143. [PubMed] [Google Scholar]
- 42.Xie Q W, Cho H, Calaycay J, Mumford R A, Swiderek K M, Lee T D, Ding A, Troso T, Nathan C. Cloning and characterization of inducible nitric oxide synthase from mouse macrophages. Science. 1992;256:225–228. doi: 10.1126/science.1373522. [DOI] [PubMed] [Google Scholar]
- 43.Zhou D, Moosekar M, Galan J E. Role of the S. typhimurium actin-binding protein SipA in bacterial internalization. Science. 1999;283:2092–2096. doi: 10.1126/science.283.5410.2092. [DOI] [PubMed] [Google Scholar]