Abstract
Charcot–Marie–Tooth (CMT) disease is one of the most common inherited neurological disorders, affecting either axons from the motor and/or sensory neurons or Schwann cells of the peripheral nervous system (PNS) and caused by more than 100 genes. We previously identified mutations in FGD4 as responsible for CMT4H, an autosomal recessive demyelinating form of CMT disease. FGD4 encodes FRABIN, a GDP/GTP nucleotide exchange factor, particularly for the small GTPase Cdc42. Remarkably, nerves from patients with CMT4H display excessive redundant myelin figures called outfoldings that arise from focal hypermyelination, suggesting that FRABIN could play a role in the control of PNS myelination. To gain insights into the role of FGD4/FRABIN in Schwann cell myelination, we generated a knockout mouse model (Fgd4SC–/–), with conditional ablation of Fgd4 in Schwann cells. We show that the specific deletion of FRABIN in Schwann cells leads to aberrant myelination in vitro, in dorsal root ganglia neuron/Schwann cell co-cultures, as well as in vivo, in distal sciatic nerves from Fgd4SC–/– mice. We observed that those myelination defects are related to an upregulation of some interactors of the NRG1 type III/ERBB2/3 signalling pathway, which is known to ensure a proper level of myelination in the PNS. Based on a yeast two-hybrid screen, we identified SNX3 as a new partner of FRABIN, which is involved in the regulation of endocytic trafficking. Interestingly, we showed that the loss of FRABIN impairs endocytic trafficking, which may contribute to the defective NRG1 type III/ERBB2/3 signalling and myelination. Using RNA-Seq, in vitro, we identified new potential effectors of the deregulated pathways, such as ERBIN, RAB11FIP2 and MAF, thereby providing cues to understand how FRABIN contributes to proper ERBB2 trafficking or even myelin membrane addition through cholesterol synthesis. Finally, we showed that the re-establishment of proper levels of the NRG1 type III/ERBB2/3 pathway using niacin treatment reduces myelin outfoldings in nerves of CMT4H mice.
Overall, our work reveals a new role of FRABIN in the regulation of NRG1 type III/ERBB2/3 NRG1signalling and myelination and opens future therapeutic strategies based on the modulation of the NRG1 type III/ERBB2/3 pathway to reduce CMT4H pathology and more generally other demyelinating types of CMT disease.
Keywords: Charcot–Marie–Tooth, myelin outfoldings, endocytic trafficking, NRG1/ERBB2/3, niacin treatment
Charcot–Marie–Tooth type 4H is associated with aberrant myelination of peripheral nerves. El-Bazzal et al. show that myelin defects are related to impaired NRG1/ERBB2/3 signalling and endocytic trafficking, and that pharmacological treatments targeting these defective molecular pathways reduce pathology in CMT4H mice.
Introduction
Charcot–Marie–Tooth (CMT) disease, also known as hereditary motor and sensory neuropathy (HMSN), is one of the most common inherited group of neurological diseases, with an overall prevalence of about 1/2500.1 Clinically, CMT diseases are characterized by progressive muscular weakness starting at the distal extremities, pes cavus deformity, loss of deep tendon reflexes, associated with mild to moderate distal sensory loss.2 Two main subgroups can be defined based on electrophysiological and histopathological characteristics: the demyelinating form (CMT1) resulting from primary damage of myelinating Schwann cells (SCs) and the axonal form (CMT2), affecting the axons from motor (MNs) and/or sensory neurons (SNs). Patients affected with CMT1 have reduced nerve-conduction velocities (NCVs; ≤ 38 m/s), whereas patients affected with CMT2 show slightly reduced to normal NCVs but reduced amplitudes (≥38 m/s).3 In addition, CMT disease is characterized by extensive clinical and genetic heterogeneity, with around 100 genes identified to date and mutations segregating following all modes of inheritance.4
In 2007, we and others identified FGD4/FRABIN as the causative gene of CMT4H,5 a rare autosomal recessive demyelinating form of CMT.6,7 The disease is characterized by typical findings of CMT, such as distal amyotrophy and foot deformities, with early-onset and slow progression. Motor and sensory NCVs are strongly reduced in all patients, but there is a strong inter- and intra-familial variability in the severity of the disease. The diagnosis is established by the presence of biallelic pathogenic variants in the FGD4 gene, encoding the protein FRABIN. More than 30 mutations are described to date in FGD4, most of them resulting in a loss of the protein or a non-functional truncated protein.6,8,9 Remarkably, nerves from patients affected with CMT4H display characteristic myelin abnormalities defined as myelin outfoldings, which arise from aberrant focal hypermyelination. In the peripheral nervous system (PNS), the myelination process is mainly regulated by the NRG1 type III/ERBB2/3 pathway: notably, the amount of axonal NRG1 type III and its downstream signalling, via the activation of the ERBB2/3 receptors, determines the thickness of the myelin sheath.10,11 In consequence, this pathway has to be tightly regulated to avoid improper myelination. Previous work has reported that myelin outfoldings might arise from an enhanced NRG1 type III/ERBB2/3 signalling, as well as the overactivation of the downstream effectors AKT/mTOR.12–14 A growing body of work suggests that the dysregulation of ERBB receptor trafficking may be the cause of an impairment of NRG1 type III/ERBB2/3 signalling in SCs and a common pathogenic mechanism for several CMT4 subtypes, such as CMT4B1 (OMIM #601382), CMT4B2 (OMIM #604563), CMT4C (OMIM #601596)15,16 and CMT4D (OMIM #601455).17 However, in CMT4H, the implication of FRABIN in the regulation of PNS myelination and the above-mentioned signalling pathways remains poorly studied.
FRABIN is a ubiquitously expressed protein containing five functional domains: a Dbl domain, responsible for the guanine exchange factor (GEF) activity toward CDC42 and RAC1, an N-terminal F-actin binding domain and three domains [two pleckstrin homology (PH) and one FYVE domain] implicated in binding to phosphoinositides (PIs).18,19 PIs are known regulators of endocytic trafficking20 and previous work by Horn et al.21 has shown an impairment of endocytosis in the absence of FRABIN, without providing a link to the aberrant hypermyelination in CMT4H.
We hereby provide evidence that FRABIN is required for proper myelination of the PNS by regulating NRG1 type III/ERBB2/3 signalling in both in vitro and in vivo models of CMT4H. In particular, we show that the specific deletion of FRABIN in SCs leads to aberrant myelination in vitro, in dorsal root ganglia (DRG) neurons/SCs co-cultures and in vivo, in distal sciatic nerves of conditional knock-out mice. We demonstrate that the myelination defects in CMT4H are related to impaired NRG-1 type III/ERBB2/3 signalling and defective endocytic trafficking. Finally, we show that the re-establishment of proper levels of the different actors of the NRG1 type III/ERBB2/3 pathway, using niacin treatment, reduces the proportion of myelin outfoldings in nerves of our CMT4H mouse model.
Material and methods
Animals
To generate a conditional Fgd4 null allele, we first generated a mouse with a floxed allele by flanking Fgd4 exon 4 with lox-P sites. Its excision, by crossing with a transgenic mouse expressing the Cre-recombinase, introduces a frameshift and is predicted to generate a premature stop codon in exon 5. Mice heterozygous for the Fgd4 floxed allele (Fgd4fl/+) have been developed on a pure C57BL/6N background in collaboration with ‘La Clinique de la Souris’ (ICS; Strasbourg; http://www.ics-mci.fr) by performing homologous recombination in embryonic stem cells (ES) derived from C57BL/6N mouse. The Fgd4fl/+ line was maintained on a pure C57BL/6N background (Taconic Biosciences A/S, Denmark). Genotyping of Fgd4fl/+ mice was performed by PCR on DNA isolated from tail biopsies using direct PCR lysis reagent (#VI-102-T, Viagen Biotech Inc.), following the manufacturer’s instructions, using the primers flox-LF (5′-CGAACCCTTAGCGATCTGTT-3′) and flox-LR (5′-TTTTCCTAGCTGGCGTGTTT-3′).
To obtain an allele with specific deletion of Fgd4 in SCs, we crossed Fgd4fl/+ mice with transgenic mice expressing the Cre-recombinase under a promoter-specific for SCs [B6N.FVB-Tg(Mpz-cre)26Mes/J], ‘so-called P0-Cre mice’, available at the Jackson Laboratories (#017927).22 We obtained Fgd4fl/+; P0-Cre, so-called Fgd4SC–/+ that we backcrossed for at least 10 generations on C57BL/6N (Taconic) background. The P0-Cre transgene was detected by PCR using the following primers: P0Cre-F (5′-CCACCACCTCTCCATTGCAC-3′) and P0Cre-R (5′-GCTGGCCCAAATGTTGCTGG-3′). For ubiquitous deletion of Fgd4, Fgd4fl/+; CMV-Cre, mice, so-called Fgd4–/+, were generated by crossing the floxed Fgd4fl/+ line with transgenic mice expressing the Cre-recombinase under the CMV promoter [B6.C-Tg (CMV-cre)1Cgn/J], available at the Jackson Laboratories (#006054). The CMV-Cre transgene was detected by PCR using the following primers: CMVCre-F (5′- AGGTTCGTTCACTCATGGA-3′) and CMVCre-R (5′-TCGACCAGTTTAGTTACCC-3′). The excision of the exon 4 in the Fgd4–/+ mice was detected using the primers flox-LF (5′-CGAACCCTTAGCGATCTGTT-3′) and flox-ER (5′-CAAGCCTCAGCTTCACTTCC-3′). Animals were housed in an animal facility with a 12 h light/12 h dark environment and ad libitum access to water and a normal diet. All experiments were done in accordance with a national appointed ethical committee for animal experimentation (Ministère de l’Education Nationale, de l’Enseignement Supérieur et de la Recherche; Authorization No. 2019062110352453 v4).
Niacin treatment
One-month-old Fgd4SC–/– mice were injected daily with saline solution or niacin (60 mg/kg) for 8 weeks (n = 4 saline-treated animals and 5 niacin-treated animals). Nicotinic acid (#PHR1276, Niacin, Sigma-Aldrich) was diluted in sterile saline solution and was daily injected intraperitoneally at a dose of 60 mg/kg.
Primary cell culture
Schwann cells culture
Primary rat SCs were prepared from the sciatic nerves of postnatal Day 3 pups. SCs were maintained in Dulbecco's Modified Eagle's Medium (#41965039 ThermoFisher Scientific), 2 mM L-glutamine (#25030024, ThermoFisher Scientific), 10% foetal bovine serum (FBS; #10270098, ThermoFisher Scientific), 2 μM forskolin (#344270, Merck) and 2 ng/ml recombinant human NRG1-β 1 (#396-HB, R&D Systems) as described in Poitelon et al.23 SCs were not used beyond the fourth passage.
DRG neurons/SCs co-cultures
Mouse DRG neurons were isolated after dissecting the spinal cord of embryonic Day 13.5 (E13.5) embryos and seeded on 12 mm glass coverslips, as described.23 In detail, after isolation, DRGs were dissociated and then incubated in 0.25% trypsin solution (#25200056, ThermoFisher Scientific) for 45 min at 37°C. DRGs were mechanically dissociated and ∼40 000 cells were seeded on matrigel- (#356234, Corning) coated coverslips in C-medium, composed of minimum essential medium (MEM) (#11090081, ThermoFisher Scientific), 2 mM L-glutamine (#25030024, ThermoFisher Scientific), 10% FBS (#10270098, ThermoFisher Scientific), 4 mg/ml D-glucose (#G5146, Sigma-Aldrich), 50 ng/ml nerve growth factor (NGF) (#N6009, Sigma-Aldrich). After 24 h, the C-medium was replaced by NB medium, composed as follows: neurobasal medium (#21103049, ThermoFisher Scientific), 4 g/l D-glucose (#G5146, Sigma-Aldrich), 2 mM L-glutamine (#25030024, ThermoFisher Scientific), 50 ng/ml NGF (#N6009, Sigma-Aldrich) and B27 supplement 1× (50×, #17504044, ThermoFisher Scientific) and changed every 2 days. After 5 days of NB medium, myelination was induced by adding 50 µg/ml ascorbic acid (#A0278, Sigma-Aldrich) to the C-medium for 12 days.
Drugs
Co-cultures were treated using nicotinic acid (niacin, #PHR1276, Sigma-Aldrich) diluted in MEM (#11090081, ThermoFisher Scientific) at a final concentration of 5 mM. Endosidin-2 (ES2; #SML168, 5 mg, Sigma-Aldrich) was diluted in MEM at a final concentration of 1 µM. Niacin or ES2 were added concomitantly to ascorbic acid at Day 6 of the co-culture.
Analysis of myelination
Around 10 fields per coverslip were randomly acquired with a Zeiss ApoTome.2 microscope (Zeiss) using a ×20 objective. Myelin abnormalities were defined as an excessive redundant or abnormal thickening of myelin visualized along with myelin basic protein (MBP) positive segments and quantified in ∼150–160 myelinated segments for each co-culture and at least three co-cultures. Results were expressed as a percentage of MBP-positive segments showing myelin abnormalities on the total of MBP-positive fibres.
RNA sequencing
RNA-sequencing and bioinformatics analysis was performed by the Genomics and Bioinformatics facility (GBiM) from the U1251/Marseille Medical Genetics laboratory. mRNA sequencing (mRNA-Seq) was performed, in duplicate, on total RNA samples extracted from DRG/SC co-cultures derived from either conditional knockout (Fgd4SC–/–) or wild-type (WT) littermates’ embryos (four samples in total). Total RNA was extracted from each condition (two coverslips of control Fgd4fl/fl and Fgd4SC–/– DRG/SC co-cultures, in two replicates), using Purelink silica membrane, anion exchange resin, spin-column kits, following the manufacturer’s recommendations (Purelink RNA Minikit, #12183018A, ThermoFisher Scientific). Before sequencing, the quality of total RNA samples was assessed using the Agilent Bioanalyzer (Agilent Technologies): only RNAs with RNA Integrity Numbers (RIN) above 8 were deemed suitable for sequencing and used for library preparation. For each sample, a library of stranded mRNA was prepared from 500 ng of total RNA after capture of RNA species with poly-A tail poly(A), using the Kapa mRNA HyperPrep kit (Roche), following the manufacturer’s instructions. The quality and profile of individual libraries have been quantified and visualized using Qubit™ and the Agilent Bioanalyzer dsDNA High Sensibility Kit (Agilent Technologies), respectively. Indexed libraries were pooled and sequenced (2 × 75 bp paired-end sequencing) on an Illumina NextSeq 500 platform (Illumina).
Data processing and differential gene expression analysis
The quality of sequencing reads was assessed using fastQC v0.11.5 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Raw sequencing reads were mapped to the mouse reference genome (Mus musculus) genome assembly GRCm38 (mm10) using STAR v2.5.3a24 and bam files were indexed and sorted using Sambamba v0.6.6.25 After mapping, the number of reads per feature (GENCODE v34 annotations) was determined using Stringtie v1.3.1c.26 Differential gene expression analysis was performed using a Wald test thanks to the DESeq2 package.27P-values were adjusted for multiple testing using the method described by Benjamini and Hochberg (BH).28 Only transcripts with an adjusted P-value (false discovery rate, FDR) below 0.05 were considered as significantly differentially expressed. Relative expressions of the most variable features between samples have been plotted as heat maps using the Pheatmap R package. Differentially expressed genes (DEGs) were visualized in the form of volcano plots, representing the Log2FC (log2 of the expression fold change) and the adjusted P-value, prepared using the EnhancedVolcano R package. Enrichment (gene set enrichment Analysis, GSEA) and overrepresentation (singular enrichment analysis, SEA) of Gene Ontology (GO) term annotation in the DEGs were performed using a Mann–Whitney test and hypergeometric test, respectively, using enrichGO from the R-package clusterProfiler (v3.10.15). For functional GO annotation, we selected the DEGs with a statistical significance of Padj value < 0.05 (SEA). Pathway analysis was conducted, using Reactome, which uses a hypergeometric test with an adjustment of the P-value (BH) to detect pathway, enriched in the data set of DEGs from the top10 GO biological process (BP) terms.
Statistical analyses
The applied statistical tests, as well as the number of replicates, are indicated in the figure legends. For all experiments based on primary co-cultures, results were obtained from at least three independent cell cultures.
Animals were matched by gender, genotype and age. Mice were randomly assigned to experimental groups and gait experiments, as well as processing and analysis of the tissues, were performed in blind. The significance of the results for electron microscopy, primary cultures and in vivo niacin treatment was evaluated with one- or two-way ANOVA or two-tailed Student’s t-test according to experimental design. Statistical analyses were performed using GraphPad Prism. All data are presented as mean ± SEM.
For gait analysis, each mouse was characterized by the median score of the left and right paw for motor function studies. The scores were analysed according to the ANCOVA statistics with weight at the corresponding age as a covariate, using29 and SPSS Statistics 19.30 We calculated the effect size and its confidence interval. Results were expressed as means and SEM.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and/or its Supplementary material. These data are available from the corresponding author upon reasonable request.
Results
Specific loss of FRABIN in Schwann cells induces myelin abnormalities in vitro and in vivo
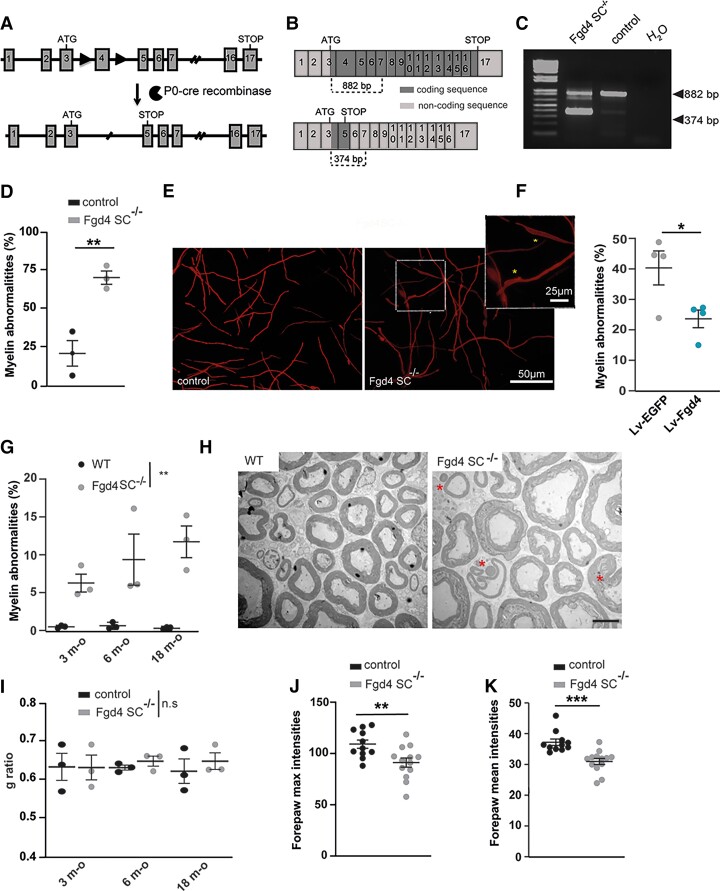
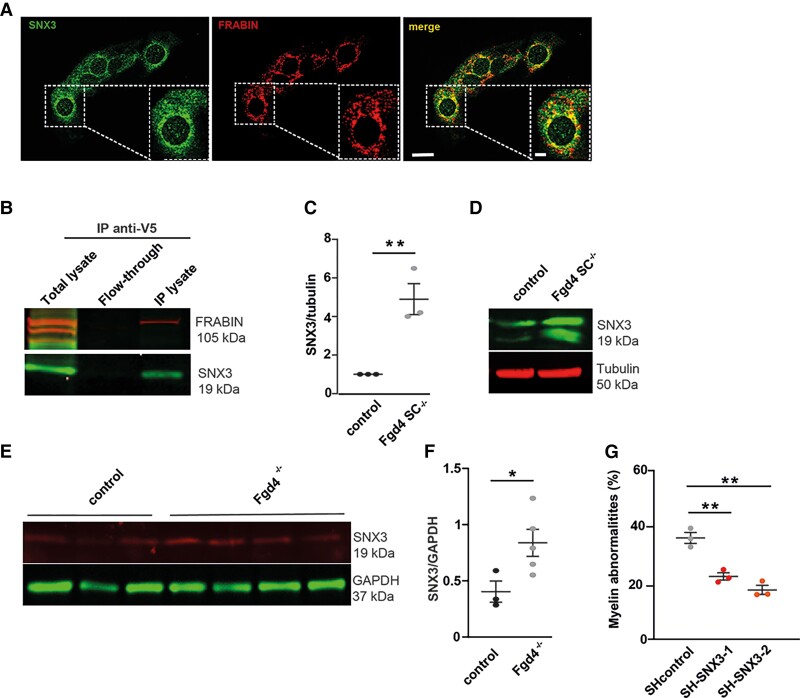
To determine the pathomechanisms underlying CMT4H disease, we generated mice with an Fgd4 floxed allele, in order to study the effect of Fgd4 ablation in the different cell types constitutive of the PNS (i.e. motor and DRG neurons and Schwann cells), by crossing them with transgenic mice expressing Cre-recombinase under the requested cell-specific promoter (Fig. 1A). To this aim, the exon 4 of Fgd4 was flanked with LoxP sites by homologous recombination. Because previous work from Horn et al.21 showed that the clinical phenotype and the molecular basis of CMT4H relied on the loss of function of FGD4/FRABIN in SCs, without detectable primary contributions from neurons, we generated a conditional null allele mouse model, referred to as Fgd4SC–/–, by abolishing FRABIN’s expression specifically in SCs. Conditional ablation of Fgd4 in SCs was obtained by mating mice homozygous for the floxed allele (Fgd4fl/fl) with transgenic P0-Cre mice to drive expression of the Cre-recombinase in SCs at E13.5.22 We demonstrated the effective deletion of the floxed exon 4 in this conditional knockout Fgd4SC–/– mouse model, by RT-PCR, using total RNA extracted from sciatic nerves of Fgd4SC–/– and control mice (Fig. 1B and C). As expected, we show that the deletion of exon 4 leads to a shorter fragment amplified between exons 3 and 7, in the mutant nerves (374 bp in Fgd4SC–/– versus 882 bp in the WT). Due to the lack of antibodies recognizing specifically mouse FRABIN, we were, unfortunately, unable to assess the deletion by western blot.
Figure 1.
Generation and phenotypic characterization of a new mouse model of CMT4H. (A–C) Generation of a conditional knockout mouse model for CMT4H. (A) The floxed allele (Fgd4fl/+) was generated by flanking exon 4 with loxP sites (black arrowheads) in the Fgd4 gene leading to a frameshift by generation of a premature stop codon in exon 5. The conditional deletion of Fgd4 in SCs is induced by crossing Fgd4fl/fl mice with mice expressing the cre recombinase under the Mpz/P0 promoter. For simplification the conditional knockout Fgd4fl/fl; P0cre mice are referred to as Fgd4SC–/–. (B) Fgd4 transcript before (top) or after (bottom) excision of exon 4 following cre-recombinase expression. (C) Excision of the exon 4 from Fgd4 transcript was verified by RT-PCR using RNA extracted from the sciatic nerves of Fgd4SC–/– and control mice. Due to exon 4 excision, the size of the amplified amplicon was 882 bp and 374 bp in WT and Fgd4SC–/– sciatic nerves, respectively. (D and E) Loss of Fgd4/FRABIN alters myelination in vitro. (D) Number of myelin anomalies quantified in WT and Fgd4SC–/– co-cultures. The presence of focal hypermyelination defects was evaluated in 150–160 myelinated segments per coverslip and three independent cultures. Myelinated segments were visualized by immunolabelling of MBP. Data are expressed as mean ± SEM. Statistical analysis: two-way ANOVA with Sidak post hoc test. (E) Illustration of myelinated segments in control and Fgd4SC–/– co-cultures. Examples of myelin abnormalities that can be observed in the enlarged image and identified by asterisks. (F) Overexpression of Fgd4/FRABIN in Fgd4SC–/– co-cultures reduces the proportion of abnormally myelinated fibres. Fgd4SC–/– co-cultures were infected with Lv-EGFP or Lv-Fgd4, 1 day after seeding. Data are expressed as mean percentage ± SEM (n = 4 independent cultures). Statistical analysis: unpaired Student’s t-test. (G and H) Loss of Fgd4/FRABIN alters PNS myelination in vivo. (G) Proportion of abnormal myelinated fibres in the distal part of the sciatic nerve of 3-, 6- and 18-month-old WT and Fgd4SC–/– mice (n = 3 animals per genotype). Data are expressed as mean percentage ± SEM. Statistical analysis: two-way repeated-measures ANOVA (group × time) with Sidak post hoc test. (H) Electron microscopy pictures illustrating myelination of sciatic nerves of WT (left) and Fgd4SC–/– (right) mouse. Asterisks indicate outfoldings. Scale bar = 5 µm. (I) g-Ratio analysis revealed no statistical difference in myelin thickness in the sciatic nerve of at 3-, 6- and 18-month-old WT and Fgd4SC–/– mice. A total of 500–1000 axons between 0.5 and 6 µm for animals (n = 3 per genotype) were analysed. Data are presented as mean ± SEM (n = 3 animals). (J and K) Maximum and mean forepaw intensities are significantly decreased in Fgd4SC–/– mice compared to WT mice. Data are expressed as mean ± SEM (n = 11 WT and n = 13 Fgd4SC–/– mice). *P < 0.05, **P < 0.01, ***P < 0.001.
We previously observed that the loss of FRABIN leads to aberrant myelin structures called outfoldings in nerves of patients affected with CMT4H.5,6,9 Here, we wanted to assess whether such abnormalities could be reproduced in our Fgd4SC–/– mouse model both in vitro and in vivo. In vitro, we established DRG/SC co-cultures from E13.5 Fgd4SC–/– embryos, in which myelination was induced for 15 days using ascorbic acid. Myelinated segments were visualized by MBP immunostaining (Fig. 1D and E). Indeed, we noticed irregularly shaped MBP-positive fibres reflecting focal hypermyelination (Fig. 1E), similar to those previously described in a co-culture model derived from the mouse model for CMT4B1.13,31 In Fgd4SC–/– DRG/SC co-cultures, the proportion of these abnormal MBP+ fibres represented 71.4 ± 4.3% of total MBP+ fibres, as compared to control co-cultures, where they represent 21.6 ± 8.3% (Fig. 1D). To determine whether the presence of those myelin defects was related to a loss of Fgd4/FRABIN, we overexpressed Fgd4/FRABIN in our mutant Fgd4SC–/– co-culture model using lentivirus (Lv). Overexpressing Fgd4/FRABIN led to a significant decrease in the proportion of abnormal MBP+ segments in mutant Fgd4SC–/– co-cultures (39.8 ± 5.6%) to a level comparable to control conditions (23.1 ± 2.9%; Fig. 1F). This finding confirms that the loss of Fgd4/FRABIN in SCs is a major contributor to the altered myelin phenotype.
Altered myelination was in parallel evaluated, over-time, in vivo, in the sciatic nerves of both Fgd4SC–/– and control mice. We observed out- and infoldings, as described in the nerves of CMT4H patients5,7–9 (Fig. 1H) and confirming the observations by Horn et al.21 in their CMT4H mouse model, where a Dhh promoter was used to drive Cre recombinase expression, leading to Fgd4 ablation specifically in Schwann cells The proportion of abnormal myelinated fibres was higher in Fgd4SC–/– nerves as compared to controls, as soon as 3 months old, and increased significantly over time (3 months old: control 0 ± 0,1% and Fgd4SC–/– 4.9 ± 1.9%; 6 months old: control 0.46 ± 0.32% and Fgd4SC–/– 9.1 ± 5.1%; 18 months old: control 0.44 ± 0.26% and Fgd4SC–/– 11.7 ± 2.1%; Fig. 1G and H). The presence of altered myelin was, however, not correlated to a defect of myelin thickness, as attested by similar g-ratios in mutant versus control sciatic nerves at all studied time points (Fig. 1I and Supplementary Fig. 1A). Finally, in addition to myelination defects, 12-month-old Fgd4SC–/– mice display significant denervation of the neuromuscular junctions (NMJs) of the gastrocnemius muscle (% of fully innervated NMJs: WT 75.3 ± 3.1% versus Fgd4SC–/– 50.3±7%; % of intermediate NMJs: WT 14.5 ± 1.9% versus Fgd4SC–/– 27.06±6.6% and % of denervated NMJs: WT 10.1 ± 1.4% versus Fgd4SC–/– 22.6±1.6%), suggesting late distal axonal degeneration (Supplementary Fig. 1B and C).
To assess the locomotor performance of our CMT4H model, we monitored footprint analyses using the gait test at 6, 12 and 18 months. For measures of intensity parameters on which the body mass could have an impact, an ANCOVA was performed, using the genotype as a fixed factor (Fgd4SC–/– versus respective control) and body mass as covariate, at the three ages. In 18-month-old mice, we detected impaired motor functions, as evidenced by a difference in parameters reflecting the pressure exerted by each paw (i.e. intensity parameters). Indeed, the maximal forepaw intensity was lower in Fgd4SC–/– mice (92.4 ± 3.9) compared to controls (110.6 ± 4.8) [F(1,24) = 8.37, P < 0.008] as well as the mean forepaw intensity: 31.08 ± 0.7 for Fgd4SC–/– versus 37.7 ± 1.6 for WT [F(1,24) = 17.28, P < 0.001]. These differences exceed the limits between pathology and typicality as shown by the large effect’s sizes (η2 = 0.28, CI from 0.11 to 0.57 and η2 = 0.43, CI from 0.19 to 0.53, respectively; Fig. 1J and K).
Specific loss of FRABIN in SCs leads to a deregulation of the neuregulin1 type III-ERBB2/3 myelination pathway
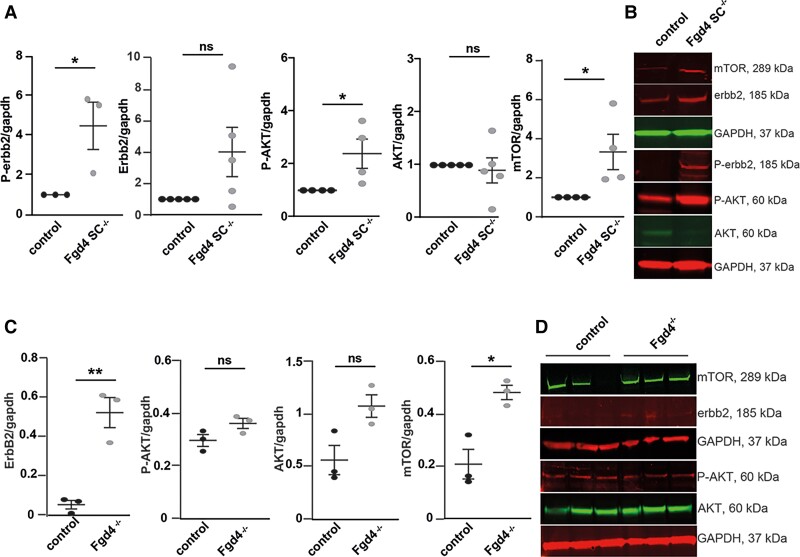
Previous studies reported that myelin outfoldings might arise from an enhanced NRG1 type III/ERBB2/3/AKT/mTOR pathway activation.12,13 Therefore, we assessed, by western blot, the level of expression of key components of this pathway, both in vitro and in vivo (Fig. 2 and Supplementary Fig. 5). In vitro, we noticed a significant increase in the levels of the phosphorylated Erbb2 receptor (P-ERBB2), phosphorylated AKT (P-Akt) and mTOR, in Fgd4SC–/– co-cultures as compared to control (Fig. 2A and B and Supplementary Fig. 5A–G). The level of full-length and cleaved forms of NRG-1 type III was not significantly different in vitro (Supplementary Fig. 1D and E and Supplementary Fig. 6I and J). Abnormal regulation of the NRG1 type III-ERBB2/3 pathway was confirmed in vivo, in the sciatic nerves of full knockout mice (Fgd4–/–). We used full knockout animals (Fgd4–/–), rather than conditional knockout animals (Fgd4SC–/–) because, in the latter, the normal expression of Frabin in the axons (which are not knockout) may compensate for any change in the level of expression of the NRG1 type III/ErbB2/3 pathway due to the knockout in Schwann cells. In Fgd4–/– sciatic nerves, we observed, by western blot, a significant upregulation of ERBB2 and mTOR, as compared to control nerves, while levels of AKT and its phosphorylated form were not different (Fig. 2C and D and Supplementary Fig. 5H–M). We also noticed a significant decrease of the full-length NRG1 type III (i.e. the pro-protein) and a concomitant increase of the levels of the C-terminal NRG1 fragment,32 which indicates an increase in the cleavage of the NRG1 type III pro-protein (Supplementary Fig. 1F and G and Supplementary Fig. 6K and L). Overall, those observations demonstrate an enhanced activation of the NRG1 type III/ERBB2/3/AKT/mTOR pathway.
Figure 2.
Neuregulin-1 type III/ERBB2/3/AKT/mTOR pathway is upregulated in CMT4H models. (A and B) Phosphorylated ERBB2 (P-ERBB2) and AKT (P-AKT) as well as mTOR expression are significantly increased in Fgd4SC–/– co-cultures compared to control. (A) Levels of expression of P-ERBB2, ERBB2, P-AKT, AKT and mTOR were assessed by western blot analysis. Data are expressed as mean ± SEM (n = 3–4 co-cultures) and normalized to the control values. Statistical analysis: unpaired Student’s t-test. (B) Western blot pictures illustrating the expression of the markers described in A. (C and D) Levels of expression of P-ERBB2, ERBB2, P-AKT, AKT and mTOR were assessed by western blot analysis in the sciatic nerves of Fgd4–/– mice compared to WT mice. (C) Data are expressed as mean ± SEM (n = 3 animals per genotype). Statistical analysis: unpaired Student’s t-test. (D) Western blot pictures illustrating the expression of the markers described in C. *P < 0.05, **P < 0.01, ***P < 0.001.
Loss of FRABIN does not affect the myelination-related gene expression programme
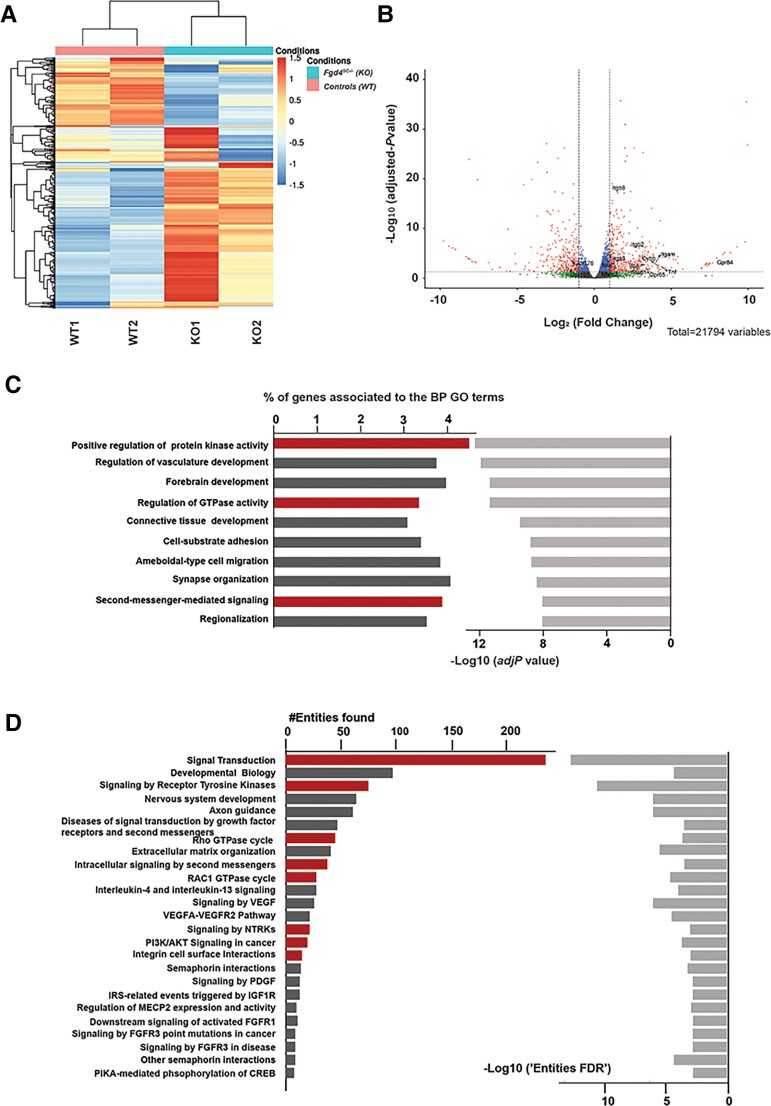
To examine global gene expression involved in the myelination abnormalities linked to the absence of FRABIN in SCs, we performed bulk mRNA-sequencing on duplicates from Fgd4SC–/– and WT co-cultures. The heat map of unsupervised hierarchical clustering of the four samples (two Fgd4SC–/– and two WT) showed that the Fgd4SC–/– samples clustered separately from the controls (Fig. 3A). GSEA on the entire set of expression data highlighted a total of 2451 genes DEGs between Fgd4SC–/– and WT co-cultures. Among them, 1679 were overexpressed and 772 underexpressed (Fig. 3B).
Figure 3.
Transcriptional profiles of in vitro myelin samples (DRG/SC co-cultures) from conditional knockout (Fgd4SC–/–) and control (WT) mice. (A) Full heat map of unsupervised hierarchical clustering of the four samples (n = 2 replicates for Fgd4SC–/– and 2 replicates for WT). The scale bar unit is obtained applying a variance stabilizing transformation to the count data (DESeq2: VarianceStabilizingTransformation) before normalization. (B) Volcano plots showing the distribution of gene expression fold changes and adjusted P-values between the two conditions. A total number of 21 794 genes were tested. Padj < 0.05 was used as the threshold to reject the null hypothesis and consider the difference in gene expression. Red plots represent significantly deregulated genes [adjusted P-value < 0.05, with fold change (FC) > 2 or <−2]. Genes with significant deregulation (adjusted P-value < 0.05) but with small FC (−2 < FC < 2) are indicated in blue. Green and grey dots represent genes with non-significant fold changes (adjusted P > 0.05). (C) Top 10 BP GO terms enriched in the DEGs. We identified BPs with an adjusted P-value lower than 0.05. The bars on the left represent the percentage of DEGs determined for each represented GO term from the total number of DEGs. Light grey bars on the right represent the enrichment score (–Log10 of adjusted P-value) for each GO term. GO terms with particularly interesting functions, regarding the NRG1 type III pathway and FRABIN, are highlighted in red. (D) Top 25 reactome pathways enriched for the 536 genes found in the top 10 significantly enriched GO terms from C (adj. P-value < 0.001). The bars represent the number of DEGs found in each reactome pathway. Pathways with particularly interesting functions regarding the NRG1 type III pathway and FRABIN are highlighted in red.
Functional GO annotation of the DEGs, with statistical significance of Padj value < 0.05, showed enrichment in several BPs. Interestingly, in the top enriched BP GO terms, we found ‘positive regulation of protein kinase activity’ (GO:0045860), ‘regulation of GTPase activity’ (GO:0043087), ‘second-messenger-mediated signalling’ (GO:0019932) or ‘cell–substrate adhesion’ (Fig. 3C), which are related to the known functions of FRABIN (regulation of GTPase, binding to PIs) and important processes in myelination. Enrichment analysis of the reactome pathways using the list of the 536 DEGs from the top 10 enriched GO terms confirmed major deregulations in signal transduction in Fgd4SC–/– co-cultures. As expected, considering the known function of FRABIN as a GEF for small RhoGTPases, we observed enrichment of genes encoding proteins involved in the RAC1 GTPase cycle (#R-HSA-9013149), Rho GTPase cycle (#R-HSA-9012999) and intracellular signalling by second messengers (#R-HSA-9006925; Fig. 3D). In addition, this analysis of pathway enrichment highlights new signalling transduction pathways, which might be important for proper myelination, such as integrin cell surface interactions (Fig. 3D).
In-depth analysis of DEGs from the top enriched GO terms revealed several deregulated genes, such as members of the integrin family (in particular Itgb8), members of the G protein-coupled receptor (Gpcr) family, such as Gpr65, or other signalling proteins (Nfat5, Tnf, Cybb, stab1; Supplementary Table 1). These genes are highlighted in Fig. 3B. We did not reveal any changes of expression in NRG1 type III/ERBB2/3 pathway and associated canonical downstream pathways, nor in myelin structural genes (Supplementary Table 1). However, very interestingly, we observed a significant change of expression of Maf (FC = 1.57, Padj = 8.2 × 10−5) in Fgd4SC–/– co-cultures, a transcription factor acting downstream of the neuregulin signalling and regulating cholesterol biosynthesis in Schwann cells33 (Supplementary Table 1). Also, there was a significant upregulation of Erbin (FC = 1.65, Padj = 0.000341), a protein interacting with ERBB2, which regulates ERBB2’s function and localization at the membrane.34 Finally, we noticed an upregulation of Rab11fip2 (FC = 1.69, Padj = 0.000378), a regulator of membrane trafficking.35,36
Interestingly, we were able to confirm that the protein levels of ERBIN are significantly increased in the sciatic nerves of 1-year-old Fgd4–/– animals in comparison to control animals (Supplementary Fig. 2A and B).
Specific loss of FRABIN in Schwann cells impairs endosomal trafficking
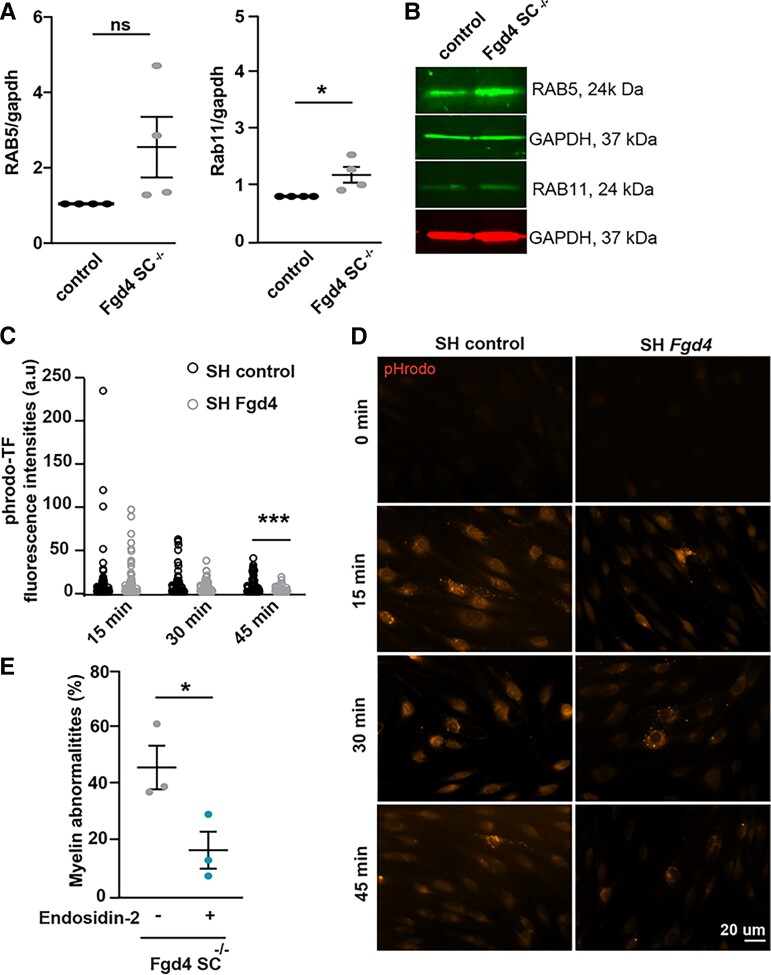
Emerging data propose that dysregulation of ERBB receptor trafficking may be the cause of the impairment of NRG type III/ERBB2/3 signalling in SCs and represent a common pathogenic mechanism in several CMT4 subtypes.15 FRABIN is a GEF responsible for GDP/GTP exchange on the small RhoGTPases CDC42 and RAC1, and also contains two PH and one FYVE domains, which are implicated in interactions with PIs. PIs are lipid regulators of endocytic trafficking.20 Previous work by Horn et al.21 has shown an impairment of endocytosis in the absence of FRABIN, but its impact on myelination defects has not been studied. To determine whether the myelination defects observed in the Fgd4SC–/– mice are connected to defective endocytic trafficking, we looked at the expression of the early endosomal marker RAB5 and the recycling endosomal marker RAB11, in vitro. In Fgd4SC–/– co-cultures, RAB5 expression levels are increased compared to controls but without reaching statistical significance. In contrast, RAB11 is significantly upregulated in Fgd4SC–/– co-cultures compared to control (Fig. 4A and B and Supplementary Fig. 5N–Q). To follow more precisely the endocytic trafficking following FRABIN’s loss, we monitored the trafficking of pHrodo-transferrin (pHrodo-TF), over time, in primary SCs knocked-down (KD) for Fgd4/FRABIN, as well as in control cells. We induced Fgd4/FRABIN KD using a shRNA targeting Fgd4 (SHFgd4), previously described by Horn et al.,21 delivered in rat primary SCs, using a Lv (Supplementary Fig. 3C). SCs were then pulse-labelled, 48 h post-infection, with fluorescent pHrodo-TF for 30 min at 4°C to prevent uptake and then chased by incubating at 37°C to allow uptake (15 min) and recycling (30 and 45 min). pHrodo-TF has the property to become fluorescent once internalized in the endosomes. By quantifying the levels of fluorescence of pHrodo-TF, we noticed similar levels of internalized TF in SCs expressing a control shRNA (SHcontrol) or SHFgd4, after 15 min of incubation, suggesting a similar capacity of TF-endocytosis between the control and Fgd4 KD conditions (Fig. 4C and D). In both control and Fgd4 KD conditions, the level of fluorescence of phrodo-TF decreases over time due to TF recycling back to the membrane. Interestingly, in comparison to the control conditions, the levels of fluorescence of pHrodo-TF are significantly lower after 45 min of incubation in Fgd4 KD conditions (SHcontrol: 7.7 ± 1.3 a.u. and SHFgd4: 2.07 ± 0.4 a.u.; Fig. 4C and D). This decrease in TF-retention in the endosomes suggested an increase of pHrodo-TF recycling back to the membrane in conditions of loss of Fgd4/FRABIN.
Figure 4.
Altered endocytic trafficking contribute to the abnormal myelination observed in Fgd4SC–/– conditions. (A and B) Expression of two endosomal markers (i.e. RAB5: early endosome and RAB11: recycling endosome) in Fgd4SC–/– co-cultures: RAB11 is significantly upregulated in in Fgd4SC–/– co-cultures as compared to control (A). Quantification of RAB5 and RAB11 expression levels in western blot observed in B. Data are expressed as mean ± SEM (n = 3 co-cultures) and normalized to the control values. Statistical analysis: unpaired Student’s t-test. (B) Illustration of RAB5 and RAB11 expression patterns observed by western blot in control and Fgd4SC–/– co-cultures. (C and D) pHrodo-Transferrin (pHrodo-TF) trafficking is altered in primary SCs infected with a Lv expressing a shRNA control or directed against Fgd4. After Lv infection (48 h), SCs were incubated with pHrodo-TF during 15, 30 or 45 min and immediately fixed in PFA 4%. pHrodo-TF is weakly fluorescent at a neutral pH, but brightly fluorescent in acidic compartments such as endosomes. (C) pHrodo-TF fluorescence was quantified in infected cells (n = 60–80 cells for each condition, from three independent cultures) and data are represented as individual values for each time point. Statistical analysis: unpaired Student’s t-test for each time point. (D) Examples of pHrodo-TF labelling after 0, 15, 30 or 45 min in control (shRNA-control) and knocked-down (shRNA-Fgd4) conditions. (E) The blocking of endosomal recycling using Endosidin-2 (1 µM) reduces significantly the proportion of abnormal myelin in Fgd4SC–/– co-cultures compared to non-treated conditions. Data are expressed as mean percentage ± SEM (n = 3 co-cultures). Statistical analysis: unpaired Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001.
To assess whether the observed endocytic trafficking defect could participate in the improper myelination observed in the Fgd4SC–/– co-cultures, we treated the co-cultures with ES2 (1 µM), a compound known to reduce exocytosis and endosomal recycling.37 Interestingly, treating Fgd4SC–/– co-cultures with ES2 decreases significantly the number of myelin abnormalities observed in conditions of loss of FRABIN (non-treated Fgd4SC–/–: 47 ± 6.8%, ES2-treated Fgd4SC–/–: 17.5 ± 5.7%; Fig. 4E). Altogether, these data support a defect in endosomal trafficking following Fgd4/FRABIN loss, which may compromise proper myelination signalling and contribute to the abnormal myelination observed in vitro (Supplementary Fig. 4).
Sorting nexin 3, a new partner of FRABIN, contributes to CMT4H pathogenesis
As the function of FRABIN in vesicle trafficking remains poorly studied, we searched for FRABIN interactors that could be involved in this process. To achieve this, we performed a yeast two-hybrid (Y2H) screening analysis in a human foetal brain library, using Fgd4 cDNA as bait. Among the partners identified, we focused on Sorting nexin 3 (SNX3). SNX3 belongs to the Sorting nexin protein family, implicated in membrane trafficking. SNX3 is associated with the early endosomes through its PX domain and interacts, as FRABIN, with the polyphosphoinositide PI3P.38 In addition, SNX3 is known to facilitate the recycling of the transferrin receptor.39 We first assessed the localization of FRABIN and SNX3 in the S16 rat SC line by immunofluorescence. We noticed vesicular staining of the endosomal marker SNX3, as previously described by Xu et al.,38 and colocalization with FRABIN, especially at the perinuclear region (Fig. 5A). In parallel, we evaluated their potential interaction by co-immunoprecipitation. HEK293 cells previously transfected with FRABIN-His-V5 were subjected to immunoprecipitation using an anti-V5 antibody. Western blot analysis revealed that SNX3 was co-immunoprecipitated in the lysate fraction (Fig. 5B and Supplementary Fig. 6A).
Figure 5.
SNX3, a new partner of FRABIN, contribute to abnormal myelination in vitro. (A) FRABIN (red) colocalizes with the endosomal marker SNX3 (green) in the immortalized S16 rat SC line. Colocalization areas are visualized in yellow in the merge picture. Scale bar of the whole picture = 20 µm, scale bar of the enlarged picture = 10 µm. (B) Co-immunoprecipitation (IP) of SNX3 and FRABIN in HEK293 cells overexpressing V5-tagged FRABIN.SNX3 was detected using an anti-SNX3 antibody after immunoprecipitation of V5-FRABIN using an anti-V5 antibody. Note that both FRABIN and SNX3 are found only in the IP lysate, and not in the flow-through fraction. (C and D) SNX3 levels are increased in Fgd4SC–/–co-cultures. SNX3 levels of expression were evaluated by western blot in both control and Fgd4SC–/– co-cultures. (C) Quantification of proteins levels of SNX3 observed by western blot in co-cultures. Data are expressed as mean ± SEM (n = 3 co-cultures) and normalized to the control values. Statistical analysis: unpaired Student’s t-test. (D) Western blot illustration of SNX3 expression profile, normalized to tubulin. (E and F) SNX3 levels are increased in sciatic nerves from knockout CMT4H animals (Fgd4–/–). SNX3 levels of expression were evaluated by western blot in sciatic nerves from 1-year-old Fgd4–/– and control mice. (E) Quantification of protein levels of SNX3 observed by western blot in 1-year-old Fgd4–/– and control’s sciatic nerves. Data are expressed as mean ± SEM (n = 3 controls and n = 5 Fgd4–/–). Statistical analysis: unpaired Student’s t-test. (F) Western blot illustration of SNX3 expression profile, normalized to GAPDH. (G) Knockdown of Snx3 reduces significantly the number of myelin abnormalities in Fgd4SC–/– co-cultures. Fgd4SC–/– co-cultures were infected at Day 1 after plating with Lv expressing either SH control, or SH-SNX3 (1 and 2). Myelination was then induced by ascorbic acid (50 µM) treatment over 12 days. Data are expressed as mean ± SEM (n = 3 co-cultures). Statistical analysis: one-way ANOVA, with Sidak post hoc test (multiple comparison to SH control). *P < 0.05, **P < 0.01, ***P < 0.001.
We then examined the levels of SNX3 in control and knockout conditions, by western blot, in vitro and in vivo. We observed a significant increase in the levels of expression of SNX3 in Fgd4SC–/– co-cultures as well as in the sciatic nerves of Fgd4–/– animals in comparison to controls (Fig. 5C–F and Supplementary Fig. 6D and E). To determine whether the upregulation of SNX3 was contributing to CMT4H pathology, we downregulated the expression of SNX3 using RNA interference in Fgd4SC–/– co-cultures. Fgd4SC–/– co-cultures were infected with Lv expressing either a control shRNA or two different shRNAs targeting Snx3. The efficiency of SNX3 KD was evaluated 1 week post-infection by western blot (Supplementary Fig. 3A and B and Supplementary Fig. 6M). By quantifying the number of MBP+ segments harbouring myelination abnormalities, we noticed a significant reduction in their percentage following Snx3 KD in Fgd4SC–/– co-cultures with a KD of Snx3: 36.5% ± 1.9 in Fgd4SC–/– co-cultures + Lv-SH control, 22.7% ± 1.3 in Fgd4SC–/– co-cultures + Lv-SH SNX3-1, or 17.8% ± 1.6 in Fgd4SC–/– co-cultures + Lv-SH SNX3-2 (Fig. 5G). We conclude that SNX3 is a partner of FRABIN in SCs and that an increase in SNX3 levels is likely to contribute to the focal hypermyelination events in CMT4H (Supplementary Fig. 4).
Niacin reduces myelin abnormalities in both in vitro and in vivo models of CMT4H
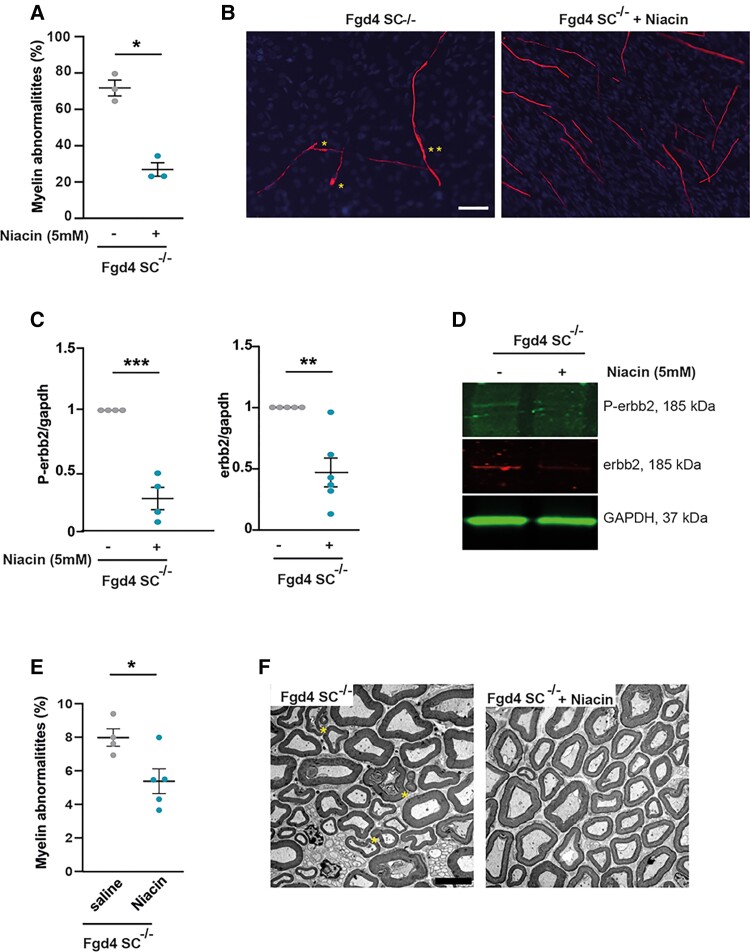
It is known that the amount of axonal NRG1-type III, and its downstream signalling via the activation of the ERBB2/3 receptors, determines the thickness of myelin sheath.10,11 Also, nerves from patients affected with CMT4H, due to loss of function mutations in FRABIN, display abnormalities characteristic of aberrant hypermyelination, such as outfoldings.5,7–9 Here we showed that this abnormal myelination observed in the absence of FRABIN is due to abnormal regulation of the NRG1 type III/ERBB2/3 pathway (Fig. 2 and Supplementary Fig. 4). We therefore aimed at lowering myelination by reducing the NRG1 type III/ERBB2/3 signalling using nicotinic acid (niacin). Indeed, niacin has been shown to promote the activation of the α-secretase TACE, which leads to a shorter cleaved form of NRG1-type III unable to link its ERBB2/3 receptors.40 Therefore, cleavage of NRG1 type III by TACE negatively regulates PNS myelination. We first tested niacin treatment in vitro by adding 5 mM of niacin, concomitantly to ascorbic acid, every 2 days over 10 days, in Fgd4SC–/– co-cultures. We observed that niacin treatment reduces by ∼60% the number of myelin defects (Fgd4SC–/–: 71.4% ± 4.4 versus Fgd4SC–/– + niacin: 28.4% ± 4.6; Fig. 6A and B). We then checked, by western blot, whether the benefit of niacin treatment was due to a downregulation of NRG1 type III/ErbB2/3 signalling. After niacin treatment, we observed a significant reduction of the levels of expression of P-ErbB2 and ErbB2 (Fig. 6C and D and Supplementary Fig. 6I and J).
Figure 6.
Downregulating NRG1-type III/ERBB2/3 signalling using niacin reduces myelination defects both in vitro and in vivo. (A and B) Niacin treatment reduces significantly myelin abnormalities in Fgd4SC–/– co-cultures. Fgd4SC–/– co-cultures were treated every 2 days with niacin (5mM) concomitantly to ascorbic acid addition (50 µM) over 12 days. (A) Quantification of the myelin abnormalities. Data are expressed as mean ± SEM (n = 3 co-cultures) and normalized to the control values. Statistical analysis: unpaired Student’s t-test. (B) Examples of MBP immunostaining in Fgd4SC–/– co-cultures treated (right) or non-treated (left) with niacin. Examples of myelin abnormalities are indicated by asterisks (*outfolding, **tomacula). (C and D) Niacin treatment leads to a downregulation of the NRG1-type III/ERBB2/3 signalling pathway. P-ERBB2 and ERBB2 expression levels were assessed by western blot in non-treated and niacin-treated Fgd4SC–/– co-cultures. (C) Quantification of P-ERBB2 and ERBB2 protein levels. Data are expressed as mean ± SEM (n = 3 co-cultures). Statistical analysis: unpaired Student’s t-test. (D) Western blot pictures illustrating the expression of the markers described in C. (E and F) Niacin treatment reduces the proportion of abnormal myelin fibres in vivo. One-month-old Fgd4SC–/– mice were daily injected with saline solution or niacin (60 mg/kg) over 8 weeks. Myelin abnormalities were quantified in the distal part of the sciatic nerves of the mice. (E) Quantification of the myelin abnormalities in sciatic nerves of treated and non-treated Fgd4SC–/– mice. Data are expressed as mean ± SEM (n = 4 saline-treated animals and 5 niacin-treated animals). Statistical analysis: unpaired Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001 (F) Electron microscopy pictures illustrating myelination of sciatic nerves of saline-treated (left) and niacin-treated Fgd4SC–/– (right) mouse. Asterisks indicate outfoldings. Scale bar = 5 µm.
We then evaluated the myelination status of the sciatic nerves of niacin-treated Fgd4SC–/– mice compared with saline-injected Fgd4SC–/– mice. As we observed a significant proportion of myelin defects as soon as 3 months of age in Fgd4SC–/– mice, mice were treated via IP injections for 8 weeks starting from 4 weeks of age (before the onset of myelin defects). Importantly, we noticed a significant reduction of the number of defective myelin fibres in treated versus non-treated animals (Fgd4SC–/– + niacin: 5% ± 0.7 versus non-treated Fgd4SC–/–: 7.4% ± 0.5; Fig. 6E and F).
These observations demonstrate that the overactivation of the NRG1 type III/ERBB2/3 pathway underlies the myelination anomalies linked to CMT4H pathogenesis. Consequently, reducing NRG1 type III/ERBB2/3 NRG1 activation via niacin represents an interesting treatment perspective for CMT4H.
Discussion
CMT4H is an autosomal recessive demyelinating subtype of CMT disease, which we first described in 2005,5 and for which we, and others, described FGD4 as the culprit gene in 2007.6,7 One distinctive feature of CMT4H is the presence in the nerves of patients of recurrent loops of myelin, called outfoldings.5,7–9,41,42 These rare myelination abnormalities have also been reported in other autosomal recessive demyelinating subtypes of CMT, i.e. CMT4B1 (OMIM #601382), CMT4B2 (OMIM #604563) and CMT4F (OMIM #614895), linked to mutations, respectively, in MTMR2 (OMIM #603557), SBF2/MTMR13 (OMIM #607697) and PRX (OMIM #605725), suggesting common pathophysiological mechanisms originating from an initial SC defect.31,43,44 Here, we sought to investigate these mechanisms by using both in vitro and in vivo models of CMT4H disease. Indeed, we generated a conditional knockout mouse model, with specific ablation of Fgd4/FRABIN in SCs, that we call Fgd4SC–/– and from which we derived our in vitro myelin model based on DRG/SC co-cultures. We showed that the loss of Fgd4/FRABIN in SCs was sufficient to reproduce, both in vitro and in vivo, the characteristic myelin defects (i.e. outfoldings) observed in the nerves of CMT4H patients. In vivo, we detected the presence of those myelin abnormalities in the sciatic nerves of Fgd4SC–/– animals, as early as 3 months of age, and showed that their proportion progressively increase over time. Our results agree with previous data observed in another CMT4H mouse model, using a different promoter (Dhh) to drive Cre recombinase expression in SCs.21 In our Fgd4SC–/– model, we also provided, for the first time, evidence of secondary axonal degeneration reflected by late muscle denervation of the gastrocnemius of 12-month-old mutant mice, a phenomenon previously described in various de-/dysmyelinating CMT.45,46 Gait tests revealed that Fgd4SC–/– mice display late motor impairment (i.e. at 18 months), resulting in the decrease of the pressure exerted by forepaws. These defects may reflect foot misplacement and deformation, as well as distal muscle weakness and amyotrophy observed in CMT4H patients.5 Overall, we showed that our Fgd4SC–/– mouse model mimics most of the pathogenic features of CMT4H and is a reliable tool to study the molecular mechanisms underlying CMT4H-related aberrant myelination.
In this model and the derived in vitro myelin model, we showed that the observed myelination defects were connected to a dysregulation of the axo-glial NRG1 type-III/ERBB2/3 pathway and its downstream effectors, P-AKT and mTOR. Notably, we noticed a strong upregulation of P-ERBB2, P-AKT and mTOR in the Fgd4SC–/– co-cultures and of ERBB2, and mTOR in the sciatic nerves of the full knockout Fgd4–/– mice. In the PNS, it is known that the amount of axonal NRG1 type III, and its downstream signalling via the activation of the ERBB2/3 receptors and downstream effectors AKT and mTOR,34 determines the thickness of the myelin sheath.10,11 Previous work from Bolino et al.13 in models of aberrant focal hypermyelination suggested that the upregulation of this signalling pathway is likely to contribute to the hypermyelination process, including the generation of aberrant loops of myelin. Also, several studies reported that myelin outfoldings might arise from the overactivation of PI3K, AKT and mTOR kinases.12–14,47 Indeed, constitutive activation of AKT or mTORC1 in SCs results in increased thickness and aberrant myelin14,48,49 and the use of the mTORC1 inhibitor, rapamycin, ameliorates myelin outfoldings observed in a DRG/SC co-culture model of CMT4B1.50
Intriguingly, here, the upregulation of the NRG1 type III/ERBB2/3/AKT/mTOR pathway and aberrant focal hypermyelination in Fgd4SC–/– co-cultures is not correlated to the upregulation of myelin proteins. Indeed, neither the genes encoding structural myelin proteins, from compact (Mpz, Pmp22, Mbp, Pmp2) and non-compact (Gjb1, Mag, Prx) myelin, nor the transcription factors regulating their expression (Egr2, Sox10) were found upregulated in Fgd4SC–/– co-cultures. Our results are in accordance with other recent studies,51,52 describing, in different mouse models, an increase in myelin thickness, due to an activation of the NRG1 type III/ERBB2/3 pathway, independently of a change in the expression in the most common myelin genes or the master Egr2 transcription factor. Although NRG1 type III/ERBB2/3 interaction promotes myelination by stimulating three canonical signalling pathways (PI3K/AKT/mTOR, MAPK/ERK and CaN/NFATc4),48,51 which are thought to converge on the activation of EGR2/KROX20, our results provide further evidence that the activation of the NRG1type III/ERBB2/3 pathway and its downstream effectors (here PI3K/AKT/mTOR) can lead to enhanced production of myelin independently of the master myelin gene transcription factor Egr2.
Other downstream NRG1 type III/ERBB-dependent mechanisms might cause the increase in myelin thickness in CMT4H, such as the spatiotemporal regulation of small GTPases regulating actin dynamics (e.g. RAC1 or CDC42) to increase membrane spreading and motility around the axons53 or else the regulation of cholesterol biosynthesis.54 Interestingly, our RNA-Seq data show increased transcript levels of the transcription factor Maf in Fgd4SC–/– conditions. Kim et al.33 showed that Maf is activated following NRG1/ERBB2 signalling and has a crucial role in PNS myelination through the regulation of cholesterol biosynthesis,33 a major constituent of myelin membranes. We were not able to confirm this upregulation, at the protein level, in sciatic nerves from 1-year-old Fgd4–/– mice, which may due to the late myelination stage at which we tested its expression. Indeed, Maf expression is strongly induced during the early phase of SC myelination and its ablation leads to hypomyelination. Our results suggest that Frabin has a role in the transduction of signals regulating cholesterol biosynthesis in SCs and might help to understand how NRG1 type III/ERBB2/3 coordinates myelin protein production and cholesterol synthesis in these cells.
Moreover, our transcriptomic data did not reveal any changes of expression in the NRG1 type III/ERBB2/3 signalling pathway in Fgd4SC–/– co-cultures, suggesting a regulation of those proteins at the protein level. In our case, we noticed a significant increase in the protein levels of ERBB2 and P-ERBB2 but not of NRG1 type III, showing that the loss of Fgd4/FRABIN in SCs may upregulate specifically ERBB2 levels rather than NRG1 type III itself. Indeed, emerging evidence indicates that ERBB receptor signalling is regulated by endocytic trafficking that controls its degradation or recycling towards the plasma membrane.15,55–57 Interestingly, previous work by Horn and colleagues21 has shown that the downregulation of Fgd4/FRABIN in a rat SC line impairs the endocytosis of the transferrin receptor (Tfrc). Here, by performing transferrin uptake assays, we demonstrate that the downregulation of Fgd4/FRABIN in primary SC do not affect the basal level of Tfrc’s endocytosis but rather promotes its recycling back to the membrane. In Fgd4SC–/– co-cultures, we also observed the upregulation of the small GTPase RAB11, which regulates the trafficking of cargoes to the endocytic recycling compartment for direct recycling to the plasma membrane.58
In our RNA-Seq data, we also noticed an increase in the levels of the Rab11fip2 transcript in Fgd4SC–/– co-cultures. Rab11fip2 encodes an effector of Rab11, member of the Rab11-family interacting proteins,59,60 which link endocytic vesicles with cytoskeletal motors for transport of cargoes along actin and microtubules and regulate the balance of transport on these two filament types.36 Although we were not able to confirm this deregulation at the protein level due to a lack of specificity of the antibodies used to detect Rab11FIP2, this result further supports the fact that altered endosomal recycling is a key mechanism underlying CMT4H physiopathology. Indeed, in Fgd4SC–/– co-cultures, we showed, for the first time, that slowing down endosomal recycling using ES2 decreases significantly the proportion of abnormal myelinated segments.
Our data support the fact that FRABIN, through binding to PIs, regulates the endocytic trafficking of cargo proteins, such as ERBB receptors. Consequently, the enhancement of NRG1 type III/ERBB2/3 signalling observed in our models could be due to accelerated recycling of the ERBB2 receptor towards the SC plasma membrane, rather than to increased ERBB2 production. The upregulation of Erbin, observed in our RNA-Seq data in Fgd4SC–/– co-cultures and confirmed in sciatic nerves from 1-year-old Fgd4–/– mice, further strengthens this hypothesis of altered ERBB2 trafficking and recycling in CMT4H. Indeed, ERBIN, through interaction with ERBB2, is implicated in the stabilization and internalization of the ERBB2 protein.34 Importantly, deletion of Erbin enhances ERBB2 internalization and degradation and leads to hypomyelination of mouse peripheral nerves. Conversely, the upregulation of Erbin observed here in Fgd4SC–/– co-cultures and Fgd4–/– sciatic nerves, is likely to reduce ERBB2 degradation and increase its levels at the membrane, hence causing hypermyelination.
Finally, by Y2H experiments, we identified for the first time the Sorting Nexin 3 (SNX3) protein, as a partner of FRABIN in the PNS. SNX3 belongs to the Sorting Nexins (SNXs) family of PI-binding proteins61 and is associated with early endosomes through its PX domain. Most interestingly, we observed that SNX3 is strongly upregulated in conditions of loss of Fgd4/FRABIN and conversely, that downregulation of Snx3 using shRNA, in Fgd4SC–/– co-cultures, reduces the aberrant myelination, typical of the disease. Accumulating evidence suggests that SNX proteins, in addition to binding to membranes play a critical role in cargo selection.62 Interestingly, modulation of SNX3 expression has been reported to alter the delivery of Tfrc to recycling endosomes.38,39 Moreover, SNX3 has been recently identified as a novel regulator, independent of the retromer complex, mediating tubular endosomal recycling of clathrin-independent endocytosis,63 the mechanism by which ERBB2 and ERBB364 are internalized,65 and which is likely to implicate actin remodelling and RhoGTPases.66
The fate and sorting of cargoes packaged in vesicles of the endo-lysosomal system rely greatly on the heterogeneity of vesicles’ membranes, variously enriched in different PIs that help to shape compartmental identity.20 Importantly, FGD4/FRABIN, in addition to one F-actin binding domain and the RhoGEF domain, has two PH domains with binding specificity to PI(34,5)P3, PI(4,5)P2 and PI(3,4)P2 and one FYVE domain with binding specificity to PI(3)P.6,67 This specific protein structure, together with the demonstrated interaction with SNX3, strongly suggests that FRABIN could play a major role in orchestrating ERBB2 trafficking and hence the activated signalling cascades downstream from NRG1 type III/ERBB2/3 interaction. Although additional experiments are required to identify all actors and steps involved in the pathomechanisms underlying CMT4H, the results of our RNA-Seq experiments point out integrin signalling as an important downstream deregulated pathway. Interestingly, in CMT4C, SH3TC2 associates with the laminin receptor, integrin-α6 and aberrant Rab11-dependent endocytic trafficking of this integrin is thought to be involved in the demyelination seen in patients’nerves.68 Like RAB11FIP2, SH3TC2 is a Rab11 effector.69 It also interacts with ERBB2/3 after its activation at the plasma membrane70 and plays a role in ERBB2/3 receptor internalization and/or intracellular trafficking. Therefore, it is tempting to speculate that SH3TC2 could be implicated in the defective ERBB receptor trafficking observed here in CMT4H and might act synergistically or antagonistically with FRABIN to regulate the activation of NRG1 type III/ERBB2/3 downstream effectors by targeting them to particular endocytosis compartments.
Overall, our results point to defective endosomal recycling as one main cause of the aberrant myelination associated with the loss of Fgd4/FRABIN. Among other genes responsible for autosomal recessive demyelinating CMT, beyond SH3TC2 (CMT4C, OMIM #601596), MTMR2 (CMT4B1, OMIM #601382) has also been reported to regulate endocytic trafficking. All three genes (FGD4, SH3TC2, MTMR2) encode proteins which are connected to defects in endocytic signalling.13,70–72 This suggests that defective endocytic trafficking and NRG1 type III/ERBB2/3 pathway is a key common pathogenic mechanism for these forms of CMT and a potential common treatment for those CMTs.
Interestingly, previous work in models of CMT4B1 showed that treatment with niacin efficiently restored proper myelination. Nicotinic acid/niacin is a compound known to enhance TACE activity73 and hence negatively regulates NRG1 type-III/ERBB2/3 signalling.13 Indeed, BACE-1 and TACE are known to cleave NRG1 type III at distinct cleavage sites, thereby regulating myelination either positively for BACE-174 or negatively for TACE.75 Because we showed that the dysregulation of the NRG1 type-III/ERBB2/3 signalling pathway is a major contributor to aberrant myelination in CMT4H, we used niacin to assess its capacity to rescue myelin outfoldings in our models. Remarkably, the use of niacin improved myelination of both Fgd4SC–/– co-cultures and animals’ sciatic nerves.
Overall, our study provides new key insights into the pathophysiological mechanisms underlying CMT4H. We demonstrate that FRABIN plays a major role in PNS myelination by regulating the levels of NRG1 type III/ERBB2/3 signalling through accelerated recycling of the ERBB2 receptor towards the SC plasma membrane. By identifying SNX3 as a new interactor of FRABIN or new potential effectors of the deregulated pathways, such as ERBIN and MAF, we provide cues to understand how FRABIN contributes to proper ERBB2 trafficking or even myelin membrane addition through cholesterol synthesis.
Our results open new avenues for a better understanding and treatment of CMT4H as well as for other hypermyelinating neuropathies.
Supplementary Material
Acknowledgements
We thank the PiCSL-FBI core facility (IBDM, AMU-Marseille), member of the France BioImaging national research infrastructure, in particular Nicolas Brouilly, who performed the electron microscopy experiments. We are grateful to the Genomics and Bioinformatics facility (GBiM), namely Camille Humbert, Christel Castro, Nathalie Trévisiol and Catherine Aubert, from the U 1251/Marseille Medical Genetics laboratory, for performing RNA-Seq experiments and analysis. We thank Thomas Marissal for his help with the statistical analysis.
Contributor Information
Lara El-Bazzal, Aix Marseille Univ, INSERM, MMG, U 1251, Marseille, France.
Adeline Ghata, Aix Marseille Univ, INSERM, MMG, U 1251, Marseille, France.
Clothilde Estève, Aix Marseille Univ, INSERM, MMG, U 1251, Marseille, France.
Jihane Gadacha, Aix Marseille Univ, INSERM, MMG, U 1251, Marseille, France.
Patrice Quintana, Aix Marseille Univ, INSERM, MMG, U 1251, Marseille, France.
Christel Castro, Aix Marseille Univ, INSERM, MMG, U 1251, Marseille, France.
Nathalie Roeckel-Trévisiol, Aix Marseille Univ, INSERM, MMG, U 1251, Marseille, France.
Frédérique Lembo, Aix Marseille Univ, INSERM, CNRS, CRCM, Institut Paoli-Calmettes, Marseille, France.
Nicolas Lenfant, Aix Marseille Univ, INSERM, MMG, U 1251, Marseille, France.
André Mégarbané, Department of Human Genetics, Gilbert and Rose-Marie Chagoury School of Medicine, Lebanese American University, Beirut, Lebanon.
Jean-Paul Borg, Aix Marseille Univ, INSERM, CNRS, CRCM, Institut Paoli-Calmettes, Marseille, France.
Nicolas Lévy, Aix Marseille Univ, INSERM, MMG, U 1251, Marseille, France.
Marc Bartoli, Aix Marseille Univ, INSERM, MMG, U 1251, Marseille, France.
Yannick Poitelon, Department of Neuroscience and Experimental Therapeutics, Albany Medical College, Albany, NY, USA.
Pierre L Roubertoux, Aix Marseille Univ, INSERM, MMG, U 1251, Marseille, France.
Valérie Delague, Aix Marseille Univ, INSERM, MMG, U 1251, Marseille, France.
Nathalie Bernard-Marissal, Aix Marseille Univ, INSERM, MMG, U 1251, Marseille, France.
Funding
This research work was supported by the French Association against Myopathies (Association Française contre les Myopathies) grants: # MNH-Decrypt, # TRIM-RD and # MoTharD).
Competing interests
The authors declare that there is no conflict of interest.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Skre H. Genetic and clinical aspects of Charcot–Marie–Tooth’s disease. Clin Genet. 1974;6:98–118. [DOI] [PubMed] [Google Scholar]
- 2. Harding AE, Thomas PK. The clinical features of hereditary motor and sensory neuropathy types I and II. Brain. 1980;103:259–280. [DOI] [PubMed] [Google Scholar]
- 3. Pareyson D, Scaioli V, Laura M. Clinical and electrophysiological aspects of Charcot–Marie–Tooth disease. Historical article review. Neuromolecular Med. 2006;8(1–2):3–22. [DOI] [PubMed] [Google Scholar]
- 4. Pipis M, Rossor AM, Laura M, Reilly MM. Next-generation sequencing in Charcot–Marie–Tooth disease: Opportunities and challenges. Nat Rev Neurol. 2019;15:644–656. [DOI] [PubMed] [Google Scholar]
- 5. De Sandre-Giovannoli A, Delague V, Hamadouche T, et al. Homozygosity mapping of autosomal recessive demyelinating Charcot–Marie–Tooth neuropathy (CMT4H) to a novel locus on chromosome 12p11.21-q13.11. J Med Genet. 2005;42:260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Delague V, Jacquier A, Hamadouche T, et al. Mutations in FGD4 encoding the Rho GDP/GTP exchange factor FRABIN cause autosomal recessive Charcot–Marie–Tooth type 4H. Am J Hum Genet. 2007;81:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stendel C, Roos A, Deconinck T, et al. Peripheral nerve demyelination caused by a mutant Rho GTPase guanine nucleotide exchange factor, frabin/FGD4. Am J Hum Genet. 2007;81:158–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baudot C, Esteve C, Castro C, et al. Two novel missense mutations in FGD4/FRABIN cause Charcot–Marie–Tooth type 4H (CMT4H). J Peripher Nerv Syst. 2012;17:141–146. [DOI] [PubMed] [Google Scholar]
- 9. Boubaker C, Hsairi-Guidara I, Castro C, et al. A novel mutation in FGD4/FRABIN causes Charcot Marie Tooth disease type 4H in patients from a consanguineous Tunisian family. Ann Hum Genet. 2013;77:336–343. [DOI] [PubMed] [Google Scholar]
- 10. Michailov GV, Sereda MW, Brinkmann BG, et al. Axonal neuregulin-1 regulates myelin sheath thickness. Science.. 2004;304:700–703. [DOI] [PubMed] [Google Scholar]
- 11. Taveggia C, Zanazzi G, Petrylak A, et al. Neuregulin-1 type III determines the ensheathment fate of axons. Neuron. 2005;47:681–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goebbels S, Oltrogge JH, Wolfer S, et al. Genetic disruption of Pten in a novel mouse model of tomaculous neuropathy. EMBO Mol Med. 2012;4:486–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bolino A, Piguet F, Alberizzi V, et al. Niacin-mediated Tace activation ameliorates CMT neuropathies with focal hypermyelination. EMBO Mol Med. 2016;8:1438–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Domenech-Estevez E, Baloui H, Meng X, et al. Akt regulates axon wrapping and myelin sheath thickness in the PNS. J Neurosci. 2016;36:4506–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee SM, Chin LS, Li L. Dysregulation of ErbB receptor trafficking and signaling in demyelinating Charcot–Marie–Tooth disease. Mol Neurobiol. 2017;54:87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Markworth R, Bähr M, Burk K. Held up in traffic-defects in the trafficking machinery in Charcot–Marie–Tooth disease. Front Mol Neurosci. 2021;14:695294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jiang LT, Chen YH, Huang JH, Tong WF, Jin LJ, Li LX. Aberrant neuregulin 1/ErbB signaling in Charcot–Marie–Tooth type 4D disease. Mol Cell Biol. 2022;42:e0055921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ono Y, Nakanishi H, Nishimura M, et al. Two actions of frabin: Direct activation of Cdc42 and indirect activation of Rac. Oncogene. 2000;19:3050–3058. [DOI] [PubMed] [Google Scholar]
- 19. Umikawa M, Obaishi H, Nakanishi H, et al. Association of frabin with the actin cytoskeleton is essential for microspike formation through activation of Cdc42 small G protein. J Biol Chem. 1999;274:25197–25200. [DOI] [PubMed] [Google Scholar]
- 20. Cullen PJ, Carlton JG. Phosphoinositides in the mammalian endo-lysosomal network. Subcell Biochem. 2012;59:65–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Horn M, Baumann R, Pereira JA, et al. Myelin is dependent on the Charcot–Marie–Tooth Type 4H disease culprit protein FRABIN/FGD4 in Schwann cells. Brain. 2012;135:3567–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Feltri ML, D'Antonio M, Previtali S, Fasolini M, Messing A, Wrabetz L. P0-Cre transgenic mice for inactivation of adhesion molecules in Schwann cells. Ann N Y Acad Sci. 1999;883:116–123. [PubMed] [Google Scholar]
- 23. Poitelon Y, Feltri ML. The pseudopod system for axon–glia interactions: Stimulation and isolation of Schwann cell protrusions that form in response to axonal membranes. Methods Mol Biol. 2018;1739:233–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dobin A, Gingeras TR. Mapping RNA-seq reads with STAR. Curr Protoc Bioinformatics. 2015;51:11.14.1–11.14.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P. Sambamba: Fast processing of NGS alignment formats. Bioinformatics. 2015;31:2032–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. Stringtie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300. [Google Scholar]
- 29. Cohen J. Statistical power analysis for the behavioral sciences. 2nd ed. Routledge;1988; [Google Scholar]
- 30. Field AP. Discovering statistics using SPSS. Sage Publications; 2005. [Google Scholar]
- 31. Bolis A, Coviello S, Bussini S, et al. Loss of Mtmr2 phosphatase in Schwann cells but not in motor neurons causes Charcot–Marie–Tooth type 4B1 neuropathy with myelin outfoldings. J Neurosci. 2005;25:8567–8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Falls DL. Neuregulins: Functions, forms, and signaling strategies. Exp Cell Res. 2003;284:14–30. [DOI] [PubMed] [Google Scholar]
- 33. Kim M, Wende H, Walcher J, et al. Maf links Neuregulin1 signaling to cholesterol synthesis in myelinating Schwann cells. Genes Dev. 2018;32:645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Newbern J, Birchmeier C. Nrg1/ErbB signaling networks in Schwann cell development and myelination. Semin Cell Dev Biol. 2010;21:922–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Prekeris R. Rabs, Rips, FIPs, and endocytic membrane traffic. Sci World J. 2003;3:870–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Machesky LM. Rab11FIP proteins link endocytic recycling vesicles for cytoskeletal transport and tethering. Biosci Rep. 2019;39:BSR20182219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang C, Brown MQ, van de Ven W, et al. Endosidin2 targets conserved exocyst complex subunit EXO70 to inhibit exocytosis. Proc Natl Acad Sci U S A. 2016;113:E41–E50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xu Y, Hortsman H, Seet L, Wong SH, Hong W. SNX3 regulates endosomal function through its PX-domain-mediated interaction with PtdIns(3)P. Nat Cell Biol. 2001;3:658–666. [DOI] [PubMed] [Google Scholar]
- 39. Chen C, Garcia-Santos D, Ishikawa Y, et al. Snx3 regulates recycling of the transferrin receptor and iron assimilation. Cell Metab. 2013;17:343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen J, Cui X, Zacharek A, et al. Niaspan treatment increases tumor necrosis factor-alpha-converting enzyme and promotes arteriogenesis after stroke. J Cereb Blood Flow Metab. 2009;29:911–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fabrizi GM, Taioli F, Cavallaro T, et al. Further evidence that mutations in FGD4/frabin cause Charcot–Marie–Tooth disease type 4H. Neurology. 2009;72:1160–1164. [DOI] [PubMed] [Google Scholar]
- 42. Arai H, Hayashi M, Hayasaka K, Kanda T, Tanabe Y. The first Japanese case of Charcot–Marie–Tooth disease type 4H with a novel FGD4 c.837-1G>A mutation. Neuromuscul Disord. 2013; 23:652–655. [DOI] [PubMed] [Google Scholar]
- 43. Guilbot A, Delague V. De la souris à l'homme: La périaxine responsable d'une forme autosomale récessive de la maladie de Charcot–Marie–Tooth. Medecine/sciences. 2001;17:663–665. [Google Scholar]
- 44. Robinson FL, Niesman IR, Beiswenger KK, Dixon JE. Loss of the inactive myotubularin-related phosphatase Mtmr13 leads to a Charcot–Marie–Tooth 4B2-like peripheral neuropathy in mice. Proc Natl Acad Sci U S A. 2008;105:4916–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moss KR, Bopp TS, Johnson AE, Höke A. New evidence for secondary axonal degeneration in demyelinating neuropathies. Neurosci Lett. 2021;744:135595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Previtali SC, Quattrini A, Bolino A. Charcot–Marie–Tooth type 4B demyelinating neuropathy: Deciphering the role of MTMR phosphatases. Expert Rev Mol Med. 2007;9:1–16. [DOI] [PubMed] [Google Scholar]
- 47. Figlia G, Norrmen C, Pereira JA, Gerber D, Suter U. Dual function of the PI3K–Akt–mTORC1 axis in myelination of the peripheral nervous system. eLife. 2017;6:e29241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Figlia G, Gerber D, Suter U. Myelination and mTOR. Glia. 2018;66:693–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Beirowski B, Wong KM, Babetto E, Milbrandt J. mTORC1 promotes proliferation of immature Schwann cells and myelin growth of differentiated Schwann cells. Proc Natl Acad Sci U S A. 2017;114:E4261–E4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Guerrero-Valero M, Grandi F, Cipriani S, et al. Dysregulation of myelin synthesis and actomyosin function underlies aberrant myelin in CMT4B1 neuropathy. Proc Natl Acad Sci U S A. 2021;118:e2009469118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Belin S, Ornaghi F, Shackleford G, et al. Neuregulin 1 type III improves peripheral nerve myelination in a mouse model of congenital hypomyelinating neuropathy. Hum Mol Genet. 2019;28:1260–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Scapin C, Ferri C, Pettinato E, et al. Enhanced axonal neuregulin-1 type-III signaling ameliorates neurophysiology and hypomyelination in a Charcot–Marie–Tooth type 1B mouse model. Hum Mol Genet. 2019;28:992–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sparrow N, Manetti ME, Bott M, et al. The actin-severing protein cofilin is downstream of neuregulin signaling and is essential for Schwann cell myelination. J Neurosci. 2012;32:5284–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Quintes S, Goebbels S, Saher G, Schwab MH, Nave KA. Neuron–glia signaling and the protection of axon function by Schwann cells. J Peripher Nerv Syst. 2010;15:10–16. [DOI] [PubMed] [Google Scholar]
- 55. Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Exp Cell Res. 2008;314:3093–3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tomas A, Futter CE, Eden ER. EGF Receptor trafficking: Consequences for signaling and cancer. Trends Cell Biol. 2014;24:26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. McLean JW, Wilson JA, Tian T, et al. Disruption of endosomal sorting in Schwann cells leads to defective myelination and endosomal abnormalities observed in Charcot–Marie–Tooth disease. J Neurosci. 2022;42:5085–5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Redpath GMI, Betzler VM, Rossatti P, Rossy J. Membrane heterogeneity controls cellular endocytic trafficking. Front Cell Dev Biol. 2020;8:757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hales CM, Griner R, Hobdy-Henderson KC, et al. Identification and characterization of a family of Rab11-interacting proteins. J Biol Chem. 2001;276:39067–39075. [DOI] [PubMed] [Google Scholar]
- 60. Baetz NW, Goldenring JR. Rab11-family interacting proteins define spatially and temporally distinct regions within the dynamic Rab11a-dependent recycling system. Mol Biol Cell. 2013;24:643–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Teasdale RD, Collins BM. Insights into the PX (phox-homology) domain and SNX (sorting nexin) protein families: Structures, functions and roles in disease. Biochem J. 2012;441:39–59. [DOI] [PubMed] [Google Scholar]
- 62. Wang J, Fedoseienko A, Chen B, Burstein E, Jia D, Billadeau DD. Endosomal receptor trafficking: Retromer and beyond. Traffic. 2018;19:578–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tian Y, Kang Q, Shi X, et al. SNX-3 mediates retromer-independent tubular endosomal recycling by opposing EEA-1-facilitated trafficking. PLoS Genet. 2021;17:e1009607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Reif R, Adawy A, Vartak N, et al. Activated ErbB3 translocates to the nucleus via clathrin-independent endocytosis, which is associated with proliferating cells. J Biol Chem. 2016;291:3837–3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Barr DJ, Ostermeyer-Fay AG, Matundan RA, Brown DA. Clathrin-independent endocytosis of ErbB2 in geldanamycin-treated human breast cancer cells. J Cell Sci. 2008;121:3155–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sandvig K, Kavaliauskiene S, Skotland T. Clathrin-independent endocytosis: An increasing degree of complexity. Histochem Cell Biol. 2018;150:107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kim Y, Ikeda W, Nakanishi H, et al. Association of frabin with specific actin and membrane structures. Genes Cells. 2002;7:413–420. [DOI] [PubMed] [Google Scholar]
- 68. Vijay S, Chiu M, Dacks JB, Roberts RC. Exclusive expression of the Rab11 effector SH3TC2 in Schwann cells links integrin-α6 and myelin maintenance to Charcot–Marie–Tooth disease type 4C. Biochim Biophys Acta. 2016;1862:1279–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Stendel C, Roos A, Kleine H, et al. SH3TC2, a protein mutant in Charcot–Marie–Tooth neuropathy, links peripheral nerve myelination to endosomal recycling. Brain. 2010;133:2462–2474. [DOI] [PubMed] [Google Scholar]
- 70. Gouttenoire EA, Lupo V, Calpena E, et al. Sh3tc2 deficiency affects neuregulin-1/ErbB signaling. Glia. 2013;61:1041–1051. [DOI] [PubMed] [Google Scholar]
- 71. Bolis A, Coviello S, Visigalli I, et al. Dlg1, Sec8, and Mtmr2 regulate membrane homeostasis in Schwann cell myelination. J Neurosci. 2009;29:8858–8870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lee HW, Kim Y, Han K, Kim H, Kim E. The phosphoinositide 3-phosphatase MTMR2 interacts with PSD-95 and maintains excitatory synapses by modulating endosomal traffic. J Neurosci. 2010;30:5508–5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chen J, Cui X, Zacharek A, et al. Niaspan increases angiogenesis and improves functional recovery after stroke. Ann Neurol. 2007;62:49–58. [DOI] [PubMed] [Google Scholar]
- 74. Hu X, Hicks CW, He W, et al. Bace1 modulates myelination in the central and peripheral nervous system. Nat Neurosci. 2006;9:1520–1525. [DOI] [PubMed] [Google Scholar]
- 75. La Marca R, Cerri F, Horiuchi K, et al. TACE (ADAM17) inhibits Schwann cell myelination. Nat Neurosci. 2011;14:857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and/or its Supplementary material. These data are available from the corresponding author upon reasonable request.