Rheumatology key message.
Heritability estimates of clinically defined gout and its subtypes were first determined by genome-wide meta-analyses.
Dear Editor, Gout is the most common form of inflammatory arthritis associated with elevated serum urate (hyperuricaemia). It has three major subtypes: renal underexcretion (RUE) type, renal overload (ROL) type and the combined (RUE + ROL) type (Fig. 1A). Historically associated with overindulgence in meats, seafood and alcohol, gout has been called the ‘disease of kings’. However, literature reveals gout to have been understood as a hereditary trait as early as the second century [1]. Nevertheless, unlike with serum urate [2], the heritability of gout, i.e. the percentage variance of phenotype that is explained by inherited genetic variants, remains inconclusive.
Figure 1.
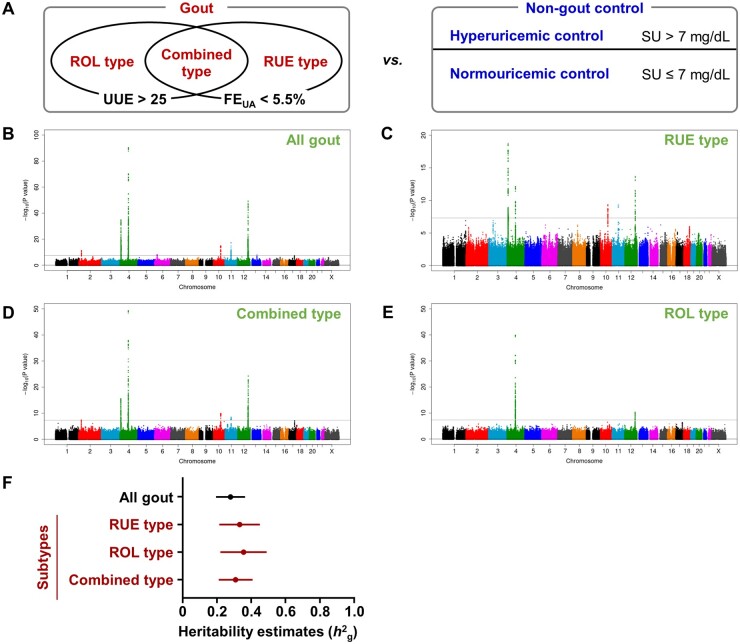
Overview of the results of meta-analyses and SNP-based heritability estimates of gout. (A) Design of GWASs (gout cases vs non-gout controls) conducted in this study. (Left) Subtype classification of gout. UUE, urinary urate excretion (mg/h/1.73 m2); FEUA, fractional excretion of uric acid (%). FEUA was calculated using the following equation: ([urine urate]/[serum urate]) × ([serum creatinine]/[urine creatinine]) × 100. (Right) Classification of non-gout controls. In addition to normouricaemic subjects without gout characterized by serum urate (SU) ≤7 mg/dL (normouricaemic control), hyperuricaemic subjects without gout characterized by SU >7 mg/dL (hyperuricaemic control) were employed. (B–E) Manhattan plots of genome-wide meta-analyses for genetic loci that influence the risk of gout: (B) all gout, (C) RUE type, (D) combined (RUE + ROL) type and (E) ROL type. SNPs with a minor allele frequency ≥1% were analysed. The horizontal axis represents chromosomal positions; the vertical axis indicates the −log10(P-value) for assessment of the association. Horizontal lines represent the genome-wide significance threshold (α = 5 × 10−8). Information on significant loci identified in the present genome-wide meta-analysis is summarized in Supplementary Table S5, available at Rheumatology online. (F) Estimation of the SNP-based heritability of gout and its subtypes using clinically defined gout patients in Japanese populations (vs non-gout controls). Horizontal bars indicate the standard error of heritability. Further information, including the prevalence of gout and each subtype, and a comparison with the SNP-based heritability estimates of gout (vs normouricaemic controls) are shown in Supplementary Table S1, available at Rheumatology online
A classic twin study conducted in the USA reported that despite confirmation of the strong heritability of hyperuricaemia, individual variability in gout was substantially influenced by environmental factors shared between co-twins, not by genetic factors [3]. In contrast, a cross-sectional study of a Taiwanese population suggested a genetic predisposition to gout [4]. Another study using the UK Biobank Resource, which calculated heritability estimates of gout (proportion of variance in gout explained by common inherited genetic variants based on an additive model of inheritance), suggested that in European individuals, the common genetic variant-mediated heritabilities of high serum urate and gout were similar [5]. However, these previous studies analysed gout patients that included self-reported cases. No information is available on the heritability of gout subtypes, as it has hitherto been unclear as to whether a genetic association of this type may apply to other populations. To address these issues, we herein investigated heritability estimates of gout, including its subtypes, in Japanese populations.
Via genome-wide association study (GWAS) meta-analyses of clinically defined gout with its subtypes in Japanese gout cases and normouricaemic controls (serum urate ≤7 mg/dl; without a past history of gout) of Japanese males, we previously identified genomic loci that influence gout susceptibility [6]. To extend the range covered for serum urate concentrations in controls and create a more generalized population, we herein employed ‘non-gout controls’ by adding asymptomatic hyperuricaemic controls comprising Japanese males (serum urate >7 mg/dl; without gout), an excluded population in our previous studies [6], to the normouricaemic controls. This allowed us to estimate the heritability of clinically defined gout by reconducting GWAS meta-analyses (3053 gout cases vs 5637 non-gout controls; Fig. 1B–E). Details of the results and related methods are described in the supplementary material, available at Rheumatology online. From the results [i.e. summary statistics of 1,029,593 single nucleotide polymorphisms (SNPs), which have a minor allele frequency (MAF) ≥1% and were not palindromic SNPs], we estimated the SNP-based heritability of gout (Fig. 1F, Supplementary Table S1, available at Rheumatology online) by the use of linkage disequilibrium score regression, as described previously [7], with minor modifications. After adjusting for the effects of covariates, the heritability for all gout was estimated at 27.9% when subjects without gout were selected as controls. These results indicate that genetic factors are significant contributors to individual variability in gout.
We next investigated the SNP-based heritability of three major subtypes of gout: the RUE type (654 cases), the ROL type (486 cases) and the combined type (905 cases) (Fig. 1F). The heritability estimates were 33.2% for the RUE type, 35.5% for the ROL type and 30.9% for the combined type, confirming genetic influence. Strong genetic correlations were also detected among the major subtypes (Supplementary Table S2, available at Rheumatology online). Although heritability estimates for the normal type (the minor subtype of gout) could not be obtained owing to the small sample size (92 cases), our findings will improve the understanding of the genetic basis of the risk of gout.
To estimate polygenic contributions to the heritability of gout, single nucleotide variations (SNVs) were then categorized into 53 overlapping functional categories based on annotation data according to the full baseline model, as previously described [8]. When focused on only the SNPs, no significant enrichments were observed; however, expanding the analysis targets to SNVs with MAF ≥0.1% enabled us to detect statistically significant enrichments in four functional categories (Supplementary Table S3, available at Rheumatology online). These results suggest the important contributions to heritability of somewhat rare genetic variations in non-coding regions. Further results of meta-analyses using SNVs and the SNV-based heritability of gout, including major subtypes, are summarized in Supplementary Fig. S1 and Table S4, available at Rheumatology online.
We acknowledge a limitation to this study. There seems to be a divergence between SNP-based heritability estimates for gout uncovered in this study (Fig. 1F) and those for serum urate (∼14%) previously reported in similar Japanese populations [7]; however, the reasons for this remain unclear. One possibility is the previously noted theoretical underestimation of heritabilities in this type of SNP-based survey, especially for serum urate [2].
In summary, we uncovered the relative contributions of heritability to phenotypic variation of gout and several of its subtypes. This study is the first to provide heritability estimates for clinically defined gout and to quantitatively show the existence of genetic predisposition to its three major subtypes.
Supplementary data
Supplementary data are available at Rheumatology online.
Supplementary Material
Acknowledgements
The authors express our sincere thanks to all the participants in this study. Our heartfelt gratitude goes to the members of the Japan Multi-Institutional Collaborative Cohort (J-MICC) Study for their support. We also specially acknowledge N. Hamajima and H. Tanaka for sample collection and their continuous encouragement. We also thank K. Morichika, M. Miyazawa, M. Seki, M. Someya, R. Kikuchi, M. Horie, S. Fujiwara and Y. Mori (National Defence Medical College) for their technical assistance. Y.T. is an Excellent Young Researcher in the MEXT Leading Initiative for Excellent Young Researchers. Data and sample collection for the cohorts participating in the present study were approved by the respective research ethics committees (National Defence Medical College and Nagoya University). All the studies were performed according to the guidelines of the Declaration of Helsinki. All participants provided their written informed consent.
Collaborators: The members of the Japan Gout Genomics Consortium (Japan Gout) are Kimiyoshi Ichida, Yukinori Okada, Tappei Takada, Seiko Shimizu, Yuya Shirai, Ken Yamamoto, Ituro Inoue, the J-MICC Study Group (principal investigator Keitaro Matsuo, Kenji Wakai, Yohko Nakamura, Hiroto Narimatsu, Kiyonori Kuriki, Sadao Suzuki, Hidemi Ito, Asahi Hishida, Takashi Tamura, Yoshikuni Kita, Teruhide Koyama, Kokichi Arisawa, Hiroaki Ikezaki, Keitaro Tanaka and Chihaya Koriyama), Toru Shimizu, Hiroshi Ooyama, Keiko Ooyama, Mitsuo Nagase, Yuji Hidaka, Miho Toba, Risa Tanaka, Yuzo Takada, Takahiro Nakamura, Satoko Iwasawa, Satoko Suzuki, Yuka Miyoshi, Hiroshi Nakashima, Yutaka Sakurai, Masashi Tsunoda and Nariyoshi Shinomiya.
Contributor Information
Yu Toyoda, Department of Integrative Physiology and Bio-Nano Medicine, National Defense Medical College, Saitama, Japan.
Masahiro Nakatochi, Public Health Informatics Unit, Department of Integrated Health Sciences, Nagoya University Graduate School of Medicine, Aichi, Japan.
Akiyoshi Nakayama, Department of Integrative Physiology and Bio-Nano Medicine, National Defense Medical College, Saitama, Japan.
Yusuke Kawamura, Department of Integrative Physiology and Bio-Nano Medicine, National Defense Medical College, Saitama, Japan.
Hirofumi Nakaoka, Human Genetics Laboratory, National Institute of Genetics, Shizuoka, Japan; Department of Cancer Genome Research, Sasaki Institute, Sasaki Foundation, Tokyo, Japan.
Kenji Wakai, Department of Preventive Medicine, Nagoya University Graduate School of Medicine, Aichi, Japan.
Keitaro Matsuo, Division of Cancer Epidemiology and Prevention, Aichi Cancer Center, Aichi, Japan; Division of Cancer Epidemiology, Nagoya University Graduate School of Medicine, Aichi, Japan.
Hirotaka Matsuo, Department of Integrative Physiology and Bio-Nano Medicine, National Defense Medical College, Saitama, Japan.
for the Japan Gout Genomics Consortium:
Kimiyoshi Ichida, Yukinori Okada, Tappei Takada, Seiko Shimizu, Yuya Shirai, Ken Yamamoto, and Ituro Inoue
Data availability statement
Data are available upon reasonable request to the corresponding author.
Authors’ contributions
H.M. conceived and designed the study with the assistance of Y.T., M.N., A.N. and Y.K. Y.T., M.N., A.N., Y.K., H.N. and H.M. analysed the data. M.N. performed the statistical analysis. K.W., K.M. and H.M. were involved in sample collection. H.M. organized this collaborative study. A.N. and Y.K. provided intellectual input and assisted with the preparation of the manuscript. Y.T., M.N., and H.M. wrote the manuscript. Y.T., M.N., and A.N. contributed equally to this work.
Funding
The research conducted by the National Defence Medical College was supported by JSPS KAKENHI [16H06279 (PAGS), 221S0002, 17H04128, 20H00566, 20K23152 and 21H03350], the Ministry of Defence of Japan, the Nakajima Foundation, the Kawano Masanori Memorial Foundation for Promotion of Pediatrics and the Gout and Uric Acid Foundation of Japan. The research conducted by the Nagoya University Graduate School of Medicine was also supported by JSPS KAKENHI [16H06277 (CoBiA) and 22H04923] and Grants-in-Aid for Scientific Research on Innovative Areas (221S0001) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Conflicts of interest: The authors have declared no conflicts of interest.
References
- 1. Nuki G, Simkin PA.. A concise history of gout and hyperuricemia and their treatment. Arthritis Res Ther 2006;8(Suppl 1):S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Major TJ, Dalbeth N, Stahl EA, Merriman TR.. An update on the genetics of hyperuricaemia and gout. Nat Rev Rheumatol 2018;14:341–53. [DOI] [PubMed] [Google Scholar]
- 3. Krishnan E, Lessov-Schlaggar CN, Krasnow RE, Swan GE.. Nature versus nurture in gout: a twin study. Am J Med 2012;125:499–504. [DOI] [PubMed] [Google Scholar]
- 4. Kuo CF, Grainge MJ, See LC. et al. Familial aggregation of gout and relative genetic and environmental contributions: a nationwide population study in Taiwan. Ann Rheum Dis 2015;74:369–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cadzow M, Merriman TR, Dalbeth N.. Performance of gout definitions for genetic epidemiological studies: analysis of UK Biobank. Arthritis Res Ther 2017;19:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nakayama A, Nakatochi M, Kawamura Y. et al. Subtype-specific gout susceptibility loci and enrichment of selection pressure on ABCG2 and ALDH2 identified by subtype genome-wide meta-analyses of clinically defined gout patients. Ann Rheum Dis 2020;79:657–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nakatochi M, Kanai M, Nakayama A. et al. Genome-wide meta-analysis identifies multiple novel loci associated with serum uric acid levels in Japanese individuals. Commun Biol 2019;2:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Finucane HK, Bulik-Sullivan B, Gusev A. et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet 2015;47:1228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon reasonable request to the corresponding author.