Abstract
The targeted screening and sequencing approaches for COVID-19 surveillance need to be adjusted to fit the evolving surveillance objectives which necessarily change over time. We present the development of variant screening assays that can be applied to new targets in a timely manner and enable multiplexing of targets for efficient implementation in the laboratory. By targeting the HV69/70 deletion for Alpha, K417N for Beta, K417T for Gamma, and HV69/70 deletion plus K417N for sub-variants BA.1, BA.3, BA.4, and BA.5 of Omicron, we achieved simultaneous detection and differentiation of Alpha, Beta, Gamma, and Omicron in a single assay. Targeting both T478K and P681R mutations enabled specific detection of the Delta variant. The multiplex assays used in combination, targeting K417N and T478K, specifically detected the Omicron sub-variant BA.2. The limits of detection for the five variants of concern were 4–16 copies of the viral RNA per reaction. Both assays achieved 100% clinical sensitivity and 100% specificity. Analyses of 377 clinical samples and 24 wastewater samples revealed the Delta variant in 100 clinical samples (nasopharyngeal and throat swab) collected in November 2021. Omicron BA.1 was detected in 79 nasopharyngeal swab samples collected in January 2022. Alpha, Beta, and Gamma variants were detected in 24 wastewater samples collected in May–June 2021 from two major cities of Alberta (Canada), and the results were consistent with the clinical cases of multiple variants reported in the community.
Keywords: SARS-CoV-2, variants of concern, multiplex RT-qPCR assays, single nucleotide mutation, clinical analysis of viral RNA, wastewater surveillance
Introduction
The SARS-CoV-2 virus continues to mutate and evolve rapidly, resulting in many variants and sub-variants.1−20 These variants carry various mutations that confer increased infectivity and evasion of immune response. Many of the mutations are located in the spike (S) protein (Table S1).2,3 The World Health Organization (WHO) has designated five variants of concern (VOCs): Alpha (B.1.1.7 and descendant lineage), Beta (B.1.351 and descendant lineage), Gamma (P.1 and descendant lineage), Delta (B.1.617.2 and descendant lineage), and Omicron (B.1.1.529 and sub-variants and descendant lineages).5 Continuing evolution of Omicron has resulted in a family of sub-variants: BA.1, BA.2, BA.3, BA.4, BA.5, XBB.1.5, and XBB.1.x.5,6 Due to its high vaccine-breakthrough capability, the Omicron variant is prevailing in areas with high vaccination rates.2 In contrast, in regions where populations are not vaccinated, the variants with less vaccine-breakthrough capability, such as Alpha, Beta, and Gamma, could coexist.
The detection of SARS-CoV-2 variants is an important component of COVID-19 surveillance. Depending on the stage of the pandemic, detection of SARS-CoV-2 variants has been used for (i) guiding public health contact-tracing investigations with the objective to contain newly detected variants of concern in a particular geographic area; (ii) the identification of variants to determine likely susceptibility or resistance to monoclonal antibody treatment; (iii) linking with clinical and epidemiological data to better understand disease severity, breakthrough infections, and reinfections associated with particular variants; and (iv) surveillance information as one of the many factors in public health policy and strategy decisions.
Sequencing approaches provide the highest resolution of information and are essential for detecting new emerging variants but suffer from timeliness and capacity issues, while complementary “targeted screening” assays can be much more timely and enable screening of a higher proportion of COVID-19 positive cases in a community or jurisdiction. The challenge with targeted screening tests is the need to constantly update and optimize targets included in the assays. Targeted screening and sequencing approaches need to be adjusted to fit the evolving surveillance objectives throughout the pandemic.
In this study, we present an approach for the development of variant screening assays that can be applied to new targets and new variants in a timely manner. We chose target mutations that confer increased infectivity and evasion of immune response, which favor the survival of new variants. These mutations are naturally selected and may appear in future SARS-CoV-2 variants.2,3 Therefore, these targets used for identification of the current variants have the potential to be used for detection of future newer variants. We recognize that SARS-CoV-2 strains and variants continue to evolve, but our approach is adaptable in a timely manner to newer targets for newer variants.
Materials and Methods
Reagents
The following reagents and materials were purchased from the respective suppliers: Proteinase K and RLT Lysis buffer from QIAgen (Germantown, MD, USA), TaqPath 1-Step RT-qPCR Master Mix, CG, and RNA-grade glycogen from Thermo Fisher Scientific (Carlsbad, CA, USA), NEB Luna Universal Probe One-Step RT-qPCR Kit from New England BioLabs (Whitby, Ontario, Canada), CDC N1 primer-probes 2019-nCoV RUO kit from Integrated DNA Technologies (Coralville, IA, USA), Proteinase K Inhibitor from Millipore Sigma (Oakville, Ontario, Canada), RNasin Plus RNase Inhibitor from Promega (Madison, WI, USA), SPRIselect magnetic beads from Beckman Coulter (Brea, CA, USA), polyethylene glycol Bio Ultra 8000 from Millipore Sigma, and 2-mercaptoethanol of biotechnology grade from BioShop Canada (Burlington, Ontario, Canada).
Purified RNA from the wild-type SARS-CoV-2 and variants Alpha, Beta, Gamma, and Delta was used as positive controls and was provided by our colleagues in the Li Ka Shing Institute of Virology, University of Alberta. The pure RNA was diluted in THE RNA Storage Solution (Thermo Fisher Scientific) containing 1.2 U/μL of RNasin Plus RNase Inhibitor. The dilutions were kept at −80 °C.
TaqMan minor groove binder (MGB) probes were obtained from Applied Biosystems (Waltham, CA, USA). Other oligonucleotides were obtained from Integrated DNA Technologies. Primers and probes were designed using Primer Express 3.0.1 software. The sequences of primers, probes, and amplicons are summarized in Table 1.
Table 1. Primer and Probe Combinations Developed for the Detection and Discrimination of SARS-CoV-2 Variants of Concerna.
| amplicon &; primer name | Oligo sequence (from 5′ to 3′) |
|---|---|
| K417N amplicon (89 bp) (EPI_ISL_16540372) | 5′ GAGGTGATGAAGTCAGACAAATCGCTCCAGGGCAAACTGGAAATATTGCTGATTATAATTATAAATTACCAGATGATTTTACAGGCTGC |
| K417T amplicon (89 bp) (EPI_ISL_16645710) | 5′ GAGGTGATGAAGTCAGACAAATCGCTCCAGGGCAAACTGGAACGATTGCTGATTATAATTATAAATTACCAGATGATTTTACAGGCTGC |
| K417N/T F- primer | 5′ GAGGTGATGAAGTCAGACAAATCG |
| K417N/T R-primer | 5′ GCAGCCTGTAAAATCATCTGGTAA |
| K417N probe (G22813T) | 5′ FAM- CAAACTGGAAAtATTGCTGATT-NFQ-MGB |
| K417T probe (A22812C) | 5′ NED- AACTGGAAcGATTGCT-NFQ-MGB |
| HV69/70 deletion amplicon (74 bp) (EPI_ISL_601443) | 5′CCTTTCTTTTCCAATGTTACTTGGTTCCATGCTATCTCTGGGACCAATGGTACTAAGAGGTTTGATAACCCTGT |
| HV69/70 deletion F-primer | 5′ CCTTTCTTTTCCAATGTTACTTGGTT |
| HV69/70 deletion R-primer | 5′ ACAGGGTTATCAAACCTCTTAGTACCA |
| HV69/70 deletion Probe (21765–21770 deletion) | 5′ Yakima-ATGCTATCT/ZEN/CTGGGACCAA- IABkFQ |
| T478K amplicon (87 bp) (EPI_ISL_16647223) | 5′ CCTTTTGAGAGAGATATTTCAACTGAAATCTATCAGGCCGGTAGCAAACCTTGTAATGGTGTTGAAGGTTTTAATTGTTACTTTCCT |
| T478K F-primer | 5′ CCTTTTGAGAGAGATATTTCAACTGAAAT |
| T478K R-primer | 5′ AGGAAAGTAACAATTAAAACCTTCAACAC |
| T478K probe (C22995A) | 5′ FAM- CCGGTAGCAaACCTTGTA-NFQ-MGB |
| P681R amplicon (89 bp) (EPI_ISL_16647223) | 5′ CCCATTGGTGCAGGTATATGCGCTAGTTATCAGACTCAGACTAATTCTCGTCGGCGGGCACGTAGTGTAGCTAGTCAATCCATCATTGC |
| P681P F-primer | 5′ CCCATTGGTGCAGGTATATGC |
| P681R R-primer | 5′ GCAATGATGGATTGACTAGCTACACT |
| P681R probe (C23604G) | 5′ VIC- AGACTAATTCTCgTCGGCG-NFQ-MGB |
Genome position according to the GISAID reference strain: hCoV-19/Wuhan/WIV04/2019 (EPI_ISL_402124 in GISAID). The single nucleotide mutations associated with variants of concern are indicated with lower case letters and in red color. F-primer represents forward-primer. R-primer indicates reverse-primer. IABkFQ denotes Iowa black fluorescence quencher. ZEN indicates internal ZEN black quencher. NFQ-MGB is the nonfluorescent quencher-minor groove binder.
Clinical Specimens
Two hundred and twenty nasopharyngeal and throat swab samples were collected at a COVID Assessment Site in Alberta, Canada, in November 2021 and another 157 nasopharyngeal swab samples were collected in Alberta in January 2022. RNA from these nasopharyngeal and throat swab samples was extracted by our colleagues in the Provincial Laboratory for Public Health, Alberta Precision Laboratories. These are accredited clinical laboratories, responsible for province-wide clinical testing and diagnosis of COVID-19 using the MagMAX Viral/Pathogen Nucleic Acid Kit in combination with the KingFisher Flex System. The extracted RNA was kept at −80 °C.
Wastewater (Sewage) Samples and Concentration of Virus on Electronegative Membranes
Wastewater sewage samples were collected in May and June of 2021 from two wastewater treatment plants located in Calgary and Edmonton, two major cities in the province of Alberta, Canada. Raw sewage from a post grit chamber located between the influent and primary treatment was collected from a 24 h composite sampler. The collected samples were labeled with the date, time, and sampling location. All the samples were stored at 4 °C after collection and immediately shipped to the laboratory for analysis.
Both the liquid and solid phases of wastewater sewage samples were processed to capture viral materials for the subsequent analysis.21 An 80 mL aliquot of each wastewater sewage sample was centrifuged at 3000 rpm for 30 min. The resulting supernatant was transferred into a separate tube. The remaining solid pellet was re-suspended in beef extract solution (3% w/v beef extract in 0.05 M glycine, pH 9.0) at a volume ratio of 1:5. The pellet suspension was agitated at 800 rpm for 30 min at room temperature on a shaker followed by centrifugation at 10,000g for 10 min, and the resulting supernatant was transferred into a new tube and neutralized with HCl. The neutralized supernatant was combined with the supernatant from the initial centrifugation step. Then, MgCl2 (1 M) was added to the combined supernatant to reach the final concentration of 25 mM MgCl2. This mixture was then filtered through an electronegative membrane filter, which was made of nitrocellulose mixed ester and had a diameter of 47 mm and a pore size of 0.45 μm (Millipore Sigma). The membrane with the captured viruses was subsequently used to extract viral RNA.
Extraction of Viral RNA from Wastewater Samples Followed by RNA Detection with the CDC SARS-CoV-2 N1 Assay
The RNA of viruses captured on the electronegative membrane was extracted utilizing the RNA extraction method that we previously developed.22 A viral inactivation and RNA preservation (VIP) buffer was used to break the viral capsule and preserve the released viral RNA. The released viral RNA was concentrated on magnetic beads. The magnetic beads were resuspended in 30 μL of RNase-free water containing 90 ng/μL of Proteinase K inhibitor and 40 units of RNasin Plus RNase Inhibitor.
Then, 5 μL of the resultant bead solution was used as the input sample for the subsequent analysis, including the detection of total SARS-Cov-2 RNA using the N1 assay (CDC) as described previously.22
Singleplex RT-qPCR Assays, Separately Targeting the HV69/70 Deletion and K417N, K417T, T478K, or P681R Mutation
The NEB Luna Universal Probe One-Step RT-qPCR Kit was used to perform all assays. Primers-probe sets for each of the K417N, K417T, T478K, and P681R mutations and the HV69/70 deletion targets were designed using Primer Express 3.0.1 software, and their sequences are summarized in Table 1. Annealing temperatures and concentrations of primers and probes were optimized for each of the single target assays: HV69/70 deletion and K417N, K417T, T478K, and P681R mutations. For optimization of the primer concentrations, a range of concentrations (100 to 800 nM) of each forward primer were paired with different concentrations of its corresponding reverse primer. Probe concentrations from 50 to 500 nM were tested in the optimization experiments. Each singleplex assay was conducted to test variant RNA containing specific mutations and the wild-type SARS-CoV-2 RNA in parallel at annealing temperatures ranging from 55 to 65 °C to find the optimal temperature capable of differentiating mutations from the wild-type sequence. The optimized conditions for each assay are shown in Supporting Information Table S2.
RT-qPCRs (20 μL of total reaction volume) for each singleplex assay contained 10 μL of Luna Universal One–Step Reaction Mix (2×), 1 μL of Luna WarmStart RT Enzyme Mix (20×), optimal concentrations of the corresponding forward primer, reverse primer, and probe for each assay (shown in Table S2), and 5 μL of the template (sample). RT-qPCR thermal cycling conditions were as follows: 55 °C for 10 min, 95 °C for 1 min, and 45 cycles of 95 °C for 10 s and 60–64 °C annealing temperature (specified in Table S2) for 1 min. All the RT-qPCR assays were performed on a QuantStudio 3 Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific) using the QuantStudio Design & Analysis Software v1.5.1.
ABG Multiplex RT-qPCR Assay Targeting the H69/V70 Deletion and K417N and K417T Mutations
The ABG multiplex assay conditions were optimized on the basis of three singleplex assays targeting the HV69/70 deletion and K417N or K417 T mutation (Table S2). The ABG multiplex RT-qPCRs (20 μL of total reaction volume) contained 10 μL of Luna Universal One–Step Reaction Mix (2×), 1 μL of Luna WarmStart RT Enzyme Mix (20×), 100 nM of each forward primer, 800 nM of each reverse primer, 250 nM of probe for the HV69/70 deletion target, 500 nM of each probe for the K417N and K417T mutation targets, and 5 μL of sample (template). The multiplex RT-qPCR thermal cycling conditions were as follows: 55 °C for 10 min, 95 °C for 1 min, and 45 cycles of 95 °C for 10 s and 61 °C annealing temperature for 1 min. The ABG multiplex RT-qPCR assays were performed on a QuantStudio 3 Real-Time PCR System and data analyzed using the QuantStudio Design & Analysis Software v1.5.1 (Applied Biosystems, Thermo Fisher Scientific). This instrument has four spectral channels, with one channel used for the reference and a maximum of three channels for a multiplex assay in a single run.
Delta Multiplex RT-qPCR Assay Targeting the T478K and P681R Mutations
The Delta assay targeting both T478K and P681R mutations was optimized on the basis of the two singleplex assays for these targets. The Delta assay multiplex RT-qPCRs (20 μL of total reaction volume) contained 10 μL of Luna Universal One–Step Reaction Mix (2×), 1 μL of Luna WarmStart RT Enzyme Mix (20×), 400 nM of forward primer for the T478K target, 100 nM of forward primer for the P681R target, 800 nM of each reverse primer, 250 nM of each probe for the T478K and P681R mutation targets, and 5 μL of sample (template). The multiplex RT-qPCR thermal cycling conditions were as follows: 55 °C for 10 min, 95 °C for 1 min, and 45 cycles of 95 °C for 10 s and 64 °C for 1 min. The Delta assay was conducted on the same instrument as the ABG assay.
Dynamic Range, Efficiency, Analytical Sensitivity, Specificity, and Reproducibility of the ABG and Delta Multiplex RT-qPCR Assays
The dynamic range for these five singleplex RT-qPCR and two multiplex RT-qPCR assays was determined by testing 10-fold serial dilutions of quantified pure RNA in triplicate. The log values of the copies of pure SARS-CoV-2 variant RNA were plotted against the corresponding Ct values to generate standard curves (Figures S1 and S2). The slope of each standard curve was used to calculate the RT-qPCR efficiency of each assay using the following equation: E = −1 + 10(−1/slope), where E represents PCR efficiency.
The analytical sensitivity of the two assays was determined by testing twofold serial dilutions of variant RNA. The limit of detection (LOD) for the ABG multiplex RT-qPCR assay was determined by testing Alpha, Beta, and Gamma RNA ranging from 4 to 250 copies/reaction. The LOD of the Delta multiplex RT-qPCR assay was determined by testing Delta RNA ranging from 8 to 250 copies/reaction. In the ABG assay, reactions containing 16–250 copies of RNA were conducted in six replicates, while reactions containing 4 and 8 copies of RNA were performed in 10 replicates. In the Delta assay, reactions containing 32–250 copies of RNA were conducted in six replicates, while reactions containing 8 and 16 copies of RNA were performed in 10 replicates. The LOD was defined as the lowest RNA concentration detected in all of 10 replicates. Ct values over 40 were considered as a non-detectable threshold.
The analytical specificity of the ABG and Delta assays was determined by testing ∼105 copies of wild-type SARS-CoV-2, Alpha, Beta, Gamma, and Delta RNA individually using the ABG and Delta multiplex RT-qPCR assays.
The variability of multiple analyses between days and within the same day of the ABG and Delta assays was determined using samples containing high (Ct value of ∼20) and low (Ct value greater than ∼34) amount of Alpha, Beta, and Gamma RNA (for the ABG assay) and Delta RNA (for the Delta assay), respectively. All samples were tested in five replicates, each set on three different days.
Application of the ABG and Delta Assays
Using the ABG assay, we tested 79 samples of Omicron sub-variant BA.1 positive and 78 samples of SARS-CoV-2 negative. These samples were provided by the Provincial Laboratory for Public Health, Alberta Precision Laboratories (APL), Alberta, Canada. Omicron sub-variant BA.1 positive samples were confirmed by APL using an in-house multiplex RT-qPCR assay targeting the E gene, HV69/70 deletion, and N501Y mutation. We also applied the ABG assay to test RNA samples extracted from wastewater samples collected from Edmonton and Calgary, Alberta, Canada, in May and June of 2021.
Using the Delta assay, we tested 100 samples of Delta variant positive and 120 samples of SARS-CoV-2 negative. These samples were provided by APL. The Delta variant was confirmed by APL using genome sequencing.
Statistical Analysis
The variability of the ABG and Delta assays was determined by calculating percent coefficients of variation (%CV) for high and low viral load samples. Clinical sensitivity (true positives/[true positives + false negatives] × 100%), clinical specificity (true negatives/[true negatives + false positives] × 100%), positive predictive value (PPV) (true positives/[true positives + false positives] × 100%), and negative predictive value (NPV) (true negatives/[true negatives + false negatives] × 100%) of the ABG and Delta assays were calculated in comparison with the reference methods. The results of the ABG assay were compared with those obtained by our reference lab (APL). The results of the Delta assay were compared with those obtained by APL using genome sequencing.
Results
Design of Multiplex RT-qPCR Assays for Simultaneous Detection and Identification of Alpha, Beta, Gamma, Delta, and Omicron Variants
The five SARS-CoV-2 variants of concern carry many mutations in the spike (S) protein of the virus (Table S1). Identification of each variant usually requires sequencing of the spike protein gene (S gene) within the SARS-CoV-2 RNA. Alternatively, the S gene with specific mutations or combination of mutations unique to each variant could be analyzed and used as a marker for the variant.
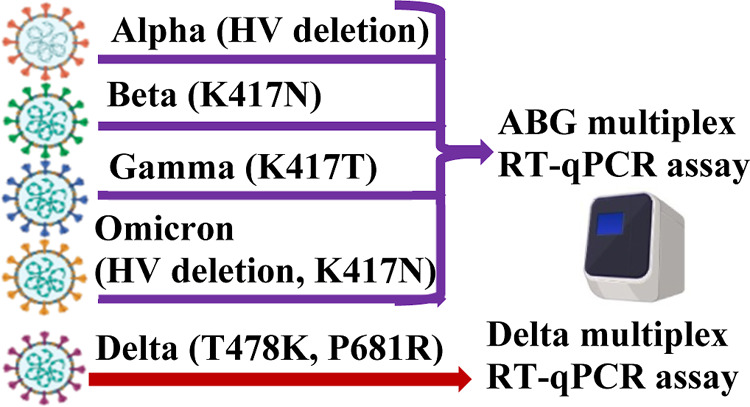
We considered the important S gene mutations that are known to play critical roles in the viral infection. As briefly described below, we compared and identified a combination of mutations that can be used to detect the five variants and differentiate among them. Specific mutations included the HV69/70 deletion and the K417N, K417T, T478K, and P681R mutations. The HV69/70 deletion, which is the loss of two amino acids at positions 69 and 70 within the spike protein, results from the deletion of 6 nucleotides (21765–21770) in the S gene. The amino acid changes at positions K417N, K417T, T478K, and P681R of the spike protein each result from a single nucleotide mutation. Our analysis and comparison of these common or unique mutations in the five variants led us to design two multiplex RT-qPCR assays. The ABG assay, targeting the HV69/70 deletion and the K417N and K417T mutations, detects and differentiates the Alpha, Beta, and Gamma variants. The Delta assay, targeting the T478K and P681R mutations, detects and identifies the Delta variant. Omicron, including sub-variants BA.1, BA.3, BA.4, and BA.5, is detected using the ABG assay on the basis of both the HV69/70 deletion and the K417N mutation. The BA.2 sub-variant of Omicron can be identified on the basis of K417N and T478K mutations.
To improve discrimination between targets with a single nucleotide difference, we incorporated the TaqMan MGB probes for fluorescence detection. The MGB at the 3′ end of probes can stabilize probe-target hybrids and provide better discrimination of a single nucleotide mismatch.23,24
Development of the ABG and Delta Assays for the Identification of Variants of Concern: Alpha, Beta, Gamma, Delta, and Omicron
Prior to conducting the multiplex RT-qPCR assay, we first optimized the concentration of the primers-probe set for each target in each single target RT-qPCR assay. The optimal concentrations of the primers-probe sets are listed in Table S2. We also determined the optimal annealing temperatures, 61 °C for the HV69/70 deletion assay and 60 °C for the K417N and K417T assays, which enabled the discrimination of each mutation from the wild-type sequence of SARS-CoV-2 (Table S2). Under the optimal conditions, the PCR efficiency for the three targets involving HV69/70 deletion and K417N and K417T mutations was 96, 97, and 96%, respectively (Figure S1). Efficiency in the range of 90–110% is considered excellent performance.25,26
To perform the ABG multiplex RT-qPCR assay in a single tube, we combined the primers and probes of the three targets at the optimized concentrations. We further fine-tuned the overall annealing temperature (61 °C) of the ABG assay for maximum discrimination of all the three targets from the wild-type sequence. To evaluate whether the amplification efficiency of three targets in the ABG assay was compromised, we conducted the ABG multiplex assay and three single target assays in parallel. We found that for a wide range of concentrations (102, 104, or 106 copies) of Alpha, Beta, and Gamma RNA, the ΔCt value between singleplex and ABG multiplex assays for each target was less than 1 (Table S3). The ABG multiplex assay achieved RT-qPCR efficiency of 94–95% for all three targets in the same reaction tube (Table 2, Figure S1). These results indicate that having similar amplification efficiencies for all three targets in both singleplex and multiplex assays meets the criteria of optimal performance.26
Table 2. RT-qPCR Efficiencies and LODs of ABG Assay and Delta Assay for the Detection of SARS-CoV-2 Variants of Concern.
| multiplex RT-qPCR assay | target | RT-qPCR efficiency (%)a | limit of detection (copies per reaction)b |
|---|---|---|---|
| ABG | HV69/70 deletion | 95 | 4 |
| K417N | 94 | 8 | |
| K417T | 95 | 8 | |
| Delta | T478K | 99 | 16 |
| P681R | 105 | 16 |
RT-qPCR efficiency = −1 + 10(−1/slope), where the slope refers to the slope of standard curves (Figures S1 and S2).
The detailed information regarding the LOD of the two assays for the five targets is shown in Table S4 and Table S5.
Using the same process, we optimized primer-probe sets and annealing temperature for the targets involving the T478K and P681R mutations (Table S2). The PCR amplification efficiency was 95% for the T478K target and 90% for the P681R target in the singleplex format (Figure S2). We compared the performance of the Delta multiplex assay for both targets with the corresponding single target assay. We found that the ΔCt value between singleplex and multiplex assays for each target at identical concentrations (102, 104, or 106 copies) was less than 1 (Table S3). The efficiency of the Delta multiplex assay for targets T478K and P681R was 99 and 105%, respectively (Table 2, Figure S2). These results indicate that the performance and conditions of the Delta multiplex assay were optimal.25,26
Assessment of the Multiplex RT-qPCR Assay Performance: Analytical Sensitivity, Specificity, and Reproducibility
We determined the LOD of both ABG and Delta multiplex assays by analyzing viral RNA of Alpha, Beta, Gamma, and Delta variants. We performed 6–10 replicate analyses using viral RNA, with each reaction containing 4–250 copies of the viral RNA. Our results show that as few as 4 copies of RNA of Alpha variant (HV69/70 deletion) were consistently detected in 10 replicate analyses using the ABG assay (Table S4). Beta (K417N target) and Gamma (K417T target) were consistently detected in all 10 replicate analyses when there were 8 or more copies of viral RNA. Similarly, the Delta multiplex assay for both T478K and P681R targets provided consistent positive detection in all 10 replicates when there were 16 or more copies of viral RNA (Table S5). These results represent LOD of 4, 8, 8, and 16 copies of the viral RNA of Alpha, Beta, Gamma, and Delta variants.
We evaluated the specificity of the ABG multiplex and Delta assays by analyzing RNA of the variants (Alpha, Beta, Gamma, and Delta) and wild-type SARS-CoV-2. The positive signal from the ABG assay was observed only when the specific target corresponding to the Alpha, Beta, or Gamma variant was present (Figure 1). Similarly, the positive signals from the Delta assay for both T478K and P681R mutations were observed only when the RNA of Delta variant was present (Figure 1). These results demonstrate good analytical specificity of both the ABG and Delta multiplex RT-qPCR assays.
Figure 1.
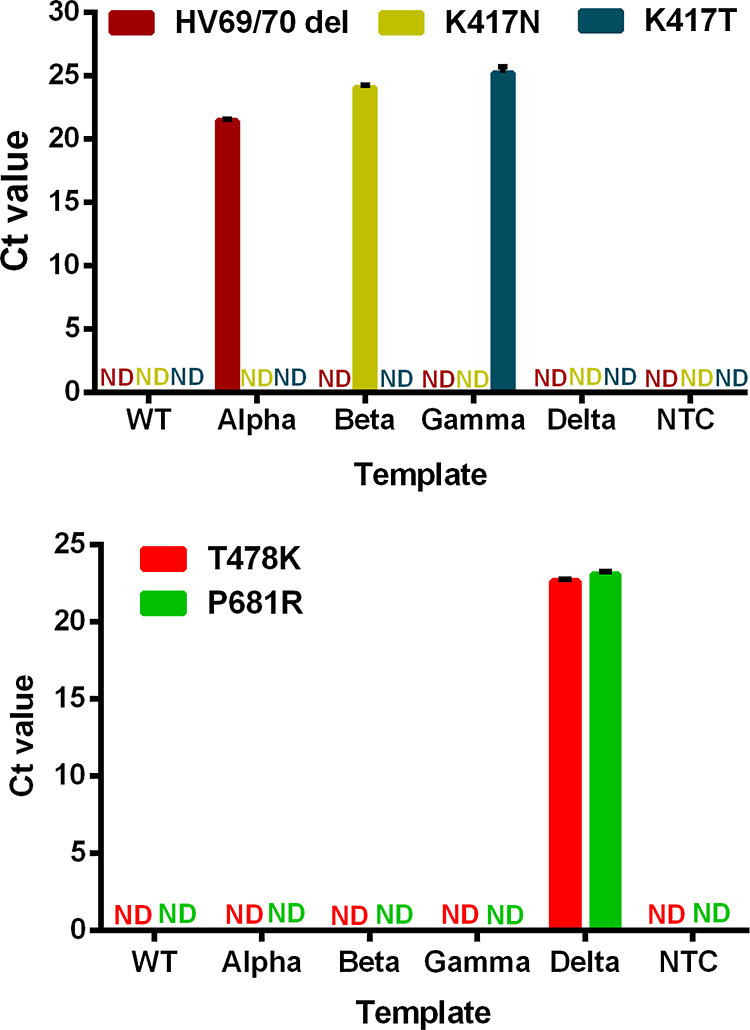
Specificity of ABG (top) and Delta (bottom) multiplex RT-qPCR assays on testing the wild-type SARS-CoV-2 and its four variants: Alpha, Beta, Gamma, and Delta. WT denotes wild-type. NTC denotes no target control. ND denotes not detectable.
We evaluated the reproducibility of both assays by performing five replicate analyses each on three different days. Using the ABG assay, we analyzed samples containing 8 or 50,000 copies of viral RNA of Alpha, Beta, and Gamma in equal numbers. The coefficient of variation (CV) within the same day ranged from 0.3 to 1.8%, and the CV ranged from 1.1 to 2.3% between days (Table S6). We also analyzed samples containing 16 or 50,000 copies of RNA of the Delta variant using the Delta assay. The intra- and inter-day variations were 0.5–1.7% and 0.9–2.7%, respectively (Table S6).
Detection of Omicron Sub-variant BA.1 in Clinical Samples Using the ABG Multiplex RT-qPCR Assay
To demonstrate the application of the new assays, we analyzed 157 nasopharyngeal swab samples collected in Alberta, Canada, in January 2022. The ABG assay correctly detected 79 positives for Omicron sub-variant BA.1 (Table S7) and 78 negatives for SARS-CoV-2, which corresponds to a clinical sensitivity of 100% and a clinical specificity of 100% (Table 3).
Table 3. Clinical Sensitivity, Specificity, PPV, and NPV of Delta and ABG Multiplex RT-qPCR Assaysa.
| genome
sequencing results for Delta and multiplex RT-qPCR assay results for Omicron sub-variant BA.1 provided
by APL |
|||||||
|---|---|---|---|---|---|---|---|
| multiplex assay | multiplex assay result | positive | negative | sensitivity | specificity | PPV | NPV |
| Delta assay (T478K and P681R) | positive | 100 | 0 | 100/100 (100%) | 100/100 (100%) | ||
| negative | 0 | 120 | 120/120 (100%) | 120/120 (100%) | |||
| ABG assay (HV 69/70 deletion and K417N) | positive | 79 | 0 | 79/79 (100%) | 79/79 (100%) | ||
| negative | 0 | 78 | 78/78 (100%) | 78/78 (100%) | |||
Nasopharyngeal and throat swab samples (220) were collected in November 2021 and were analyzed for the Delta variant. Nasopharyngeal swab samples (157) were collected in January 2022 and were analyzed for Omicron sub-variant BA.1. APL is Alberta Precision Laboratories. PPV, positive predictive value; NPV, negative predictive value. Samples positive for both T478K and P681 R targets were determined as positive for Delta variant. Samples positive for both the HV69/70 deletion and K417N mutation targets were deemed as positive for Omicron sub-variant BA.1.
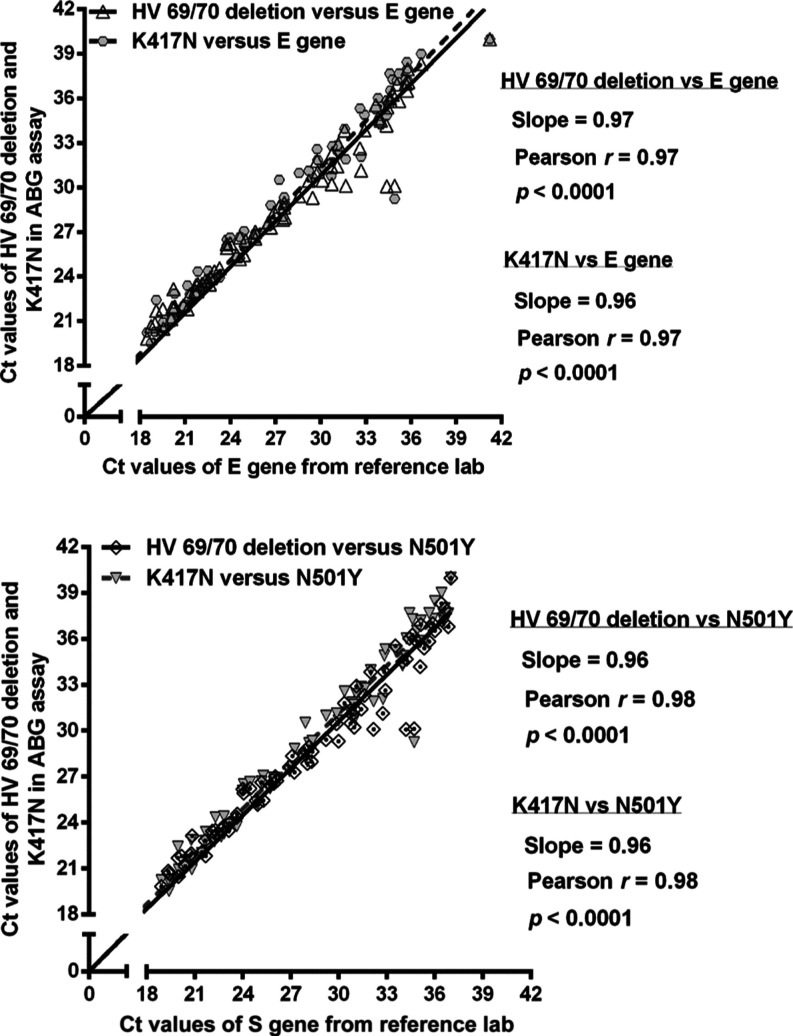
We compared the Ct values obtained from the ABG assay for both the HV69/70 deletion and K417N mutation with the Ct values obtained by our reference lab (APL) using RT-qPCR for the E gene (Figure 2). The three sets of Ct values from the analysis of the same 79 NPS samples were significantly correlated (p < 0.0001), with Pearson correlation coefficients of 0.96–0.97 (Figure 2). According to the clinical diagnostic methodology conducted in January 2022 when Omicron BA.1 was the predominant variant in the tested community, samples testing positive for both the HV69/70 deletion and N501Y mutation were considered positive for Omicron sub-variant BA.1. Thus, we compared the Ct values obtained from the ABG assay with the Ct values of the assay that targeted N501Y. Significant correlations (p < 0.0001), with a Pearson correlation coefficient of 0.96, indicate agreement of the ABG multiplex assay results with the results of the reference lab (Figure 2).
Figure 2.
Correlation between Ct values obtained using the ABG assay for the HV69/70 deletion and K417N mutation targets in the S gene and Ct values provided by the reference lab for the E gene (top) and the N501Y target in the S gene (bottom). The nasopharyngeal swab samples were collected in Alberta, Canada, in January 2022.
Detection of the Delta Variant in Clinical Samples Using the Delta Multiplex RT-qPCR Assay
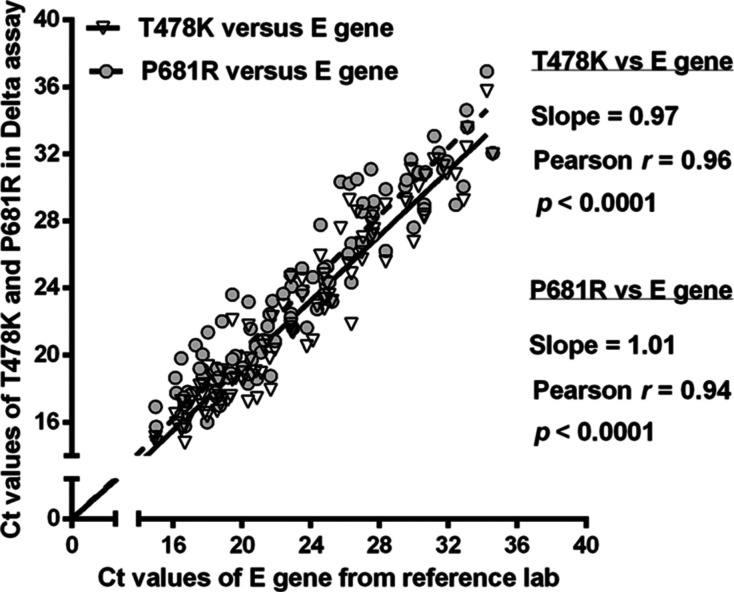
To evaluate the performance of our Delta assay for clinical samples, we analyzed a total of 220 nasopharyngeal and throat swab samples collected at a COVID Assessment Site in Alberta, Canada, in November 2021. These samples were previously analyzed by APL using their in-house singleplex RT-qPCR targeting the E gene,27 and positive samples were confirmed to be Delta using genome sequencing. Among these samples, 100 samples were Delta variant positive (Table S8) and 120 samples were SARS-CoV-2 negative. The overall results of our Delta multiplex RT-qPCR assay showed 100 positives and 120 negatives, fully consistent with the results of APL. Furthermore, the Ct values obtained using our Delta assay for both the T478K and P681R targets in the S gene correlated with the Ct values obtained by APL for the analysis of the E gene. The Pearson correlation coefficients were 0.94–0.97, and correlations were significant (p < 0.0001) (Figure 3).
Figure 3.
Correlation between Ct values obtained using the Delta assay for the T478K and P681R mutation targets in the S gene and Ct values provided by the reference lab for the E gene. Nasopharyngeal and throat swab samples were collected in Alberta, Canada, in November 2021.
ABG Multiplex RT-qPCR Assay for Wastewater Surveillance of Alpha, Beta, and Gamma Variants
To demonstrate the application of the assays for wastewater surveillance, we applied the ABG multiplex assay to analyze wastewater samples for detection of variants Alpha, Beta, and Gamma. We analyzed the wastewater samples collected from Calgary and Edmonton, Alberta, Canada, from May 16 to June 22, 2021. We found that Alpha was the dominant variant in wastewater during this sampling period. With the exception of only one sample collected from Edmonton, all other 23 wastewater samples contained detectable RNA of the Alpha variant. The concentrations were 14–862 copies per 100 mL of wastewater samples from Calgary and 5–366 copies per 100 mL of wastewater samples from Edmonton (Figure 4; Table S9). Beta was detected in 2 of 12 Calgary wastewater samples and 4 of 12 Edmonton wastewater samples. The concentrations were 2–28 copies per 100 mL of wastewater. Gamma was detected in 5 of 12 Calgary samples and 6 of 12 Edmonton samples. The relative concentrations of Alpha, Beta, and Gamma detected in wastewater are in concordance with the number of clinical cases caused by these three VOCs during the period of wastewater sampling, from May 16 to June 22, 2021. As reported by Alberta Health Services (Figure S3), Alpha was the variant leading to the most clinical COVID-19 cases during that sampling period. The cases caused by Gamma were much fewer than those caused by Alpha and more than those caused by Beta.28
Figure 4.
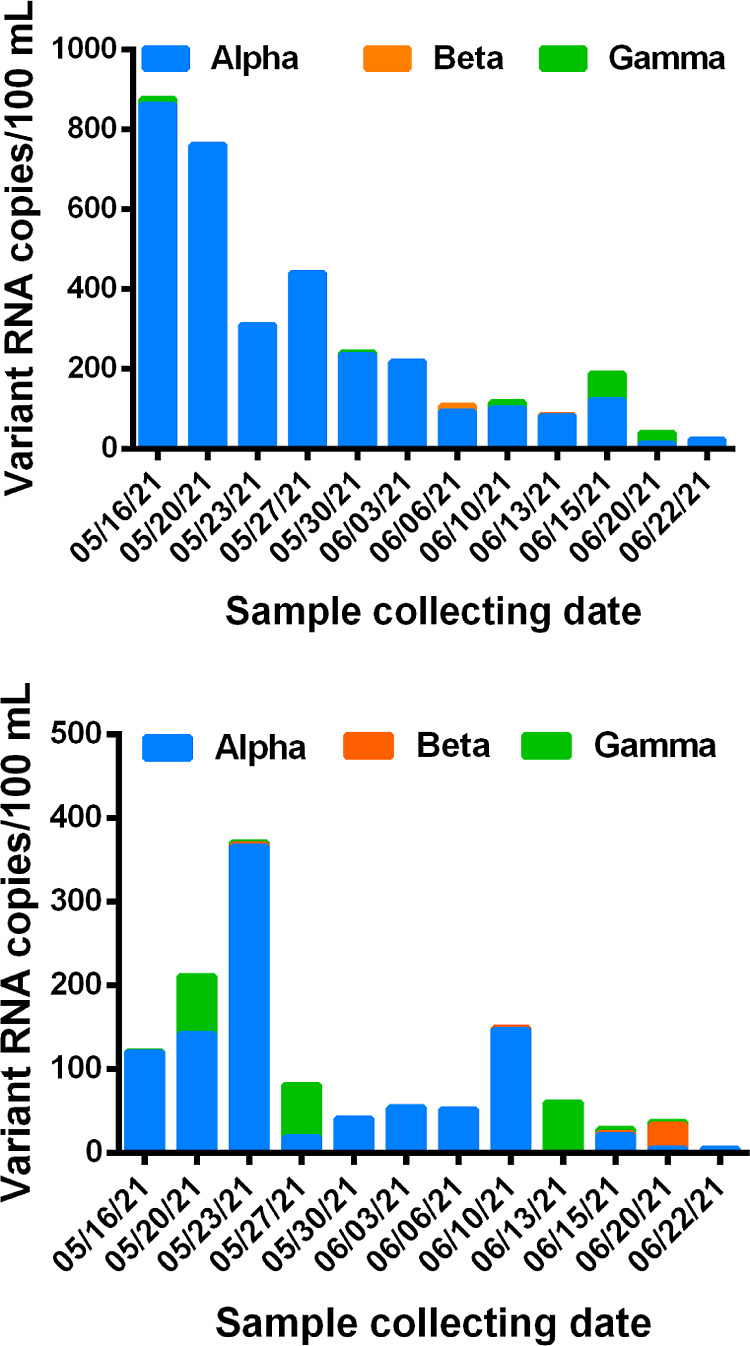
Detection of Alpha, Beta, and Gamma in wastewater samples collected from two major cities, Calgary (top) and Edmonton (bottom), of Alberta, Canada, from May 16 to June 22, 2021.
The total copies of Alpha, Beta, and Gamma in each sample were lower than the total number of SARS-CoV-2 RNA determined using the CDC SARS-CoV-2 N1 assay (Table S9), indicating that the majority of SARS-CoV-2 RNA present in wastewater during the sampling period was from the wild-type SARS-CoV-2. These results suggest extended virus shedding from patients infected by the wild-type SARS-CoV-2, and the new variants emerging in the community. These findings are consistent with those reported in other studies.29,30 Hokajärvi et al.30 reported that the SARS-CoV-2 viral particle and RNA were stable over 1 month in wastewater at 4 °C, which is the typical water temperature in Alberta during the time of sampling.
Discussion
Monitoring SARS-CoV-2 variants is an important component of COVID-19 surveillance throughout the pandemic. We recognize that as assays targeting unique mutations are developed and evaluated, SARS-CoV-2 is undergoing further mutations and evolving into new variants and sub-lineages. Therefore, it is challenging to keep up with the continuing mutations of SARS-CoV-2 and the changing surveillance needs. For assays to have the potential to detect new variants, it is important to choose naturally selected mutations. Many studies have confirmed that natural selection is the dominant mechanism of SARS-CoV-2 evolution. Viral evolution selects mutations that are able to strengthen viral infectivity and enhance immune escape.1,2 Thus, mutations in the current variants of concern may appear in future SARS-CoV-2 variants.15 Using naturally selected mutations as targets, through the examples of the multiplex assays for the five variants of concern, we show an approach that can be applied to assays for new variants in a timely manner and enables multiplexing of targets for efficient implementation in the laboratory.
A number of mutations in the dominant variants are located in the spike (S) protein of SARS-CoV-2 (Table S1).2,3 Many of these mutations are naturally selected, conferring increased infectivity and evasion of immune response.1−3 For example, mutation N501Y, located in the receptor-binding domain (RBD) of the S protein, confers elevated affinity for host receptors.9 Mutation E484K enhances binding affinity for host receptors while diminishing antibody binding.10 Mutations K417N and K417T moderately decrease the affinity of the S protein to host receptors while facilitating immune evasion.10 Mutation D614G confers a moderate advantage for transmissibility and infectivity.7,8 The HV69/70 deletion confers a twofold higher level of infectivity as well as higher disease severity based on hospitalizations and fatalities.11,12 Mutation T478K, located in the region of interaction with the host receptor, diminishes the neutralizing activity of antibodies.2,13 Mutation P681R enhances the cleavage of the full-length spike protein to S1 and S2, leading to increased virial replication.14
As a consequence of the diverse combination of the mutations associated with the increased infectivity and capacity of immune evasion in different variants of concern,16 Beta and Gamma variants are more lethal than Alpha. The Delta variant caused a significantly increased vaccine breakthrough compared with Alpha, Beta, and Gamma.17 Patients infected with the Delta variant had higher hospital admission and fatality rates than patients infected with the Alpha variant.18 Patients infected with the Omicron variant had higher vaccine breakthrough albeit lower hospitalization rates and less disease severity.15,17,19,20 Omicron sub-variants BA.4 and BA.5 are about 36% more infectious than BA.2.15 The lower hospitalization rates and seemingly less disease severity with Omicron sub-variants may at least be partially attributable to the prior exposure of the population through infections and/or multiple immunizations, as well as to the virus-specific characteristics.
Sequencing of SARS-CoV-2 strains has provided crucial information about the mutations within its genome. While extremely useful, genome sequencing has a long turnaround time and high costs and is not available in many places due to a lack of resources and expertise. By targeting unique mutations present in variants, RT-qPCR offers a faster and less expensive approach to detect a specific variant for routine surveillance. For example, on the basis of the HV69/70 deletion in the S gene of the Alpha variant, many jurisdictions used S gene target failure (SGTF) to indicate the occurrence of Alpha. The SGTF method can only indicate Alpha and Omicron (BA.1, BA.3, BA.4, and BA.5) containing the HV69/70 deletion, whereas Beta, Gamma, Delta, and Omicron BA.2 lacking the HV69/70 deletion cannot be detected. The SGTF method cannot discriminate a specific variant among the five variants of concern.31
To detect and identify variants of concern, a few studies reported multiple RT-qPCR assays.32−37 Most studies identified a partial list of variants such as Alpha and Delta,32 Delta,33 or Omicron.34 Only one study35 reported a two-reaction multiplex RT-qPCR genotyping assay to differentiate five variants by examining six different mutations simultaneously. However, this genotyping assay is expensive because the primers and probes targeting all six mutations are required at the same time even when only a single variant is being detected. In one reaction, mutations L452R, E484K, and N501Y were targeted. In the second reaction, HV69/70 deletion and K417N and T478K mutations were used as targets. HV69/70 deletion and N501Y mutation positive were interpreted as Alpha. K417N, E484K, and N501Y mutations positive were interpreted as Beta. E484K and N501Y mutations positive were interpreted as Gamma. Thus, two reactions involving all six pairs of primers and six probes must be used at the same time to discriminate Alpha, Beta, and Gamma. Similarly, two multiplex RT-PCRs were required to target L452R and T478K mutations for the detection of the Delta variant.
In our study, we first aligned the S gene sequences and identified unique and naturally selected mutations as the targets of analysis. We chose the minimum combination of the targets so that all five variants can be detected simultaneously (Table S1). Among the five variants of concern, Alpha contains the HV69/70 deletion, Beta contains the K417N mutation, Gamma contains the K417T mutation, Delta contains the T478K and P681R mutations, Omicron BA.2 contains the K417N and T478K mutations, and Omicron BA.1, BA.3, BA.4, and BA.5 contain the combination of the HV69/70 deletion and K417N and T478K mutations. To specifically identify the five variants of concern with minimum targets, we chose the HV69/70 deletion for Alpha, K417N for Beta, K417T for Gamma, the combination of the T478K and P681R mutations for Delta, the combination of the K417N and T478K mutations for Omicron BA.2, and the combination of the HV69/70 deletion and K417N mutation for Omicron BA.1, BA.3, BA.4, and BA.5.
By targeting five mutations in variants of concern, our multiplex RT-qPCR assays enable simultaneous detection and identification of five variants of concern as well as the discrimination of Omicron sub-variant BA.2 from other Omicron sub-variants. We chose five naturally selected mutations as multiplex RT-qPCR targets and incorporated the MGB probes to improve the discrimination of single nucleotide mutations. By targeting the HV69/70 deletion and the K417N and K417T mutations in the S gene, our first multiplex RT-qPCR assay (the ABG assay) enabled detection and differentiation of Alpha (HV69/70 deletion), Beta (K417N mutation), and Gamma (K417T mutation). When the Delta variant emerged, we were able to develop the Delta assay within a week using the same strategy that we used to develop the ABG assay. By targeting both T478K and P681R mutations in the S gene, our multiplex RT-qPCR assay enabled the detection and identification of the Delta variant. As the virus continues to evolve into Omicron and its sub-variants, naturally selected mutations including HV69/70 deletion, K417N, and T478K reappear in the new variants. Our already developed ABG assay, targeting a combination of the HV69/70 deletion and K417N mutation, enabled the detection of sub-variants BA.1, BA.3, BA.4, and BA.5. The use of both ABG and Delta assays, targeting the K417N and T478K mutations, enabled the discrimination of the BA.2 sub-variant of Omicron. As of March 15, 2023, the currently circulating variant of interest is XBB.1.5, which is a combination of Omicron BA.2.10.1 and BA.2.75 sublineages, according to the information from WHO.5 This variant contains the K417N and T478K mutations, which can be detected and discriminated from the previous dominant variants Omicron BA.4 and BA.5 using our two assays. Therefore, our multiplex RT-qPCR assays used in combination achieved our objective of simultaneous detection and differentiation of the five variants of concern and have the potential to be used for detection of newer variants. The strategy and similar considerations should be useful for designing multiplex assays for newer variants (strains, lineages) in a timely manner.
Applications of the multiplex assays to the analysis of clinical and wastewater samples demonstrated the detection of all five variants of concern. Alpha, Beta, and Gamma were detected in wastewater samples collected in May–June 2021; Delta was detected in nasopharyngeal and throat swab samples collected in November 2021; whereas Omicron BA.1 was detected in nasopharyngeal swab samples collected in January 2022. The LODs of all the targets in the ABG and Delta assays ranged from 4 to 16 copies per reaction. The multiplex assays and the strategy involved in their development can be adapted for the detection of naturally selected mutations in future dominant variants. This strategy is also suitable for developing an assay for the detection of other viruses and their variants.
Acknowledgments
We thank LeeAnn Turnbull (Provincial Laboratory for Public Health, Alberta Precision Laboratories, Edmonton, Alberta, Canada) for coordinating RNA extracts of the clinical specimens and Dr. Judy Yuanyuan Qiu (Provincial Laboratory for Public Health, Edmonton, Alberta, Canada) for providing wastewater samples. This work was supported by the New Frontiers in Research Fund, the Natural Sciences and Engineering Research Council of Canada, the Canadian Institutes of Health Research, the Social Sciences and Humanities Research Council of Canada, the Canada Research Chairs Program, Alberta Innovates, and Alberta Health.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmeasuresciau.3c00005.
Amino acid changes in the spike protein of variants of concern, optimal RT-qPCR conditions of singleplex and multiplex assays, performance comparison of five targets between singleplex and multiplex RT-qPCR assays, analytical sensitivities of two multiplex assays, reproducibility of two assays, detection of the Omicron variant in clinical samples, detection of the Delta variant in clinical samples, detection of the Alpha, Beta, and Gamma variants in wastewater samples, PCR standard curves, and variants of concern COVID-19 cases in Alberta (PDF)
Author Contributions
CRediT: Yanming Liu conceptualization (lead), data curation (lead), formal analysis (lead), investigation (lead), methodology (lead), validation (lead), visualization (lead), writing-original draft (lead), writing-review & editing (lead); Teresa Kumblathan conceptualization (supporting), data curation (supporting), formal analysis (supporting), investigation (supporting), methodology (supporting), validation (supporting), visualization (supporting), writing-original draft (supporting), writing-review & editing (supporting); Michael A. Joyce resources (equal), validation (supporting), writing-review & editing (supporting); D. Lorne Tyrrell funding acquisition (supporting), project administration (supporting), resources (supporting), writing-review & editing (supporting); Graham Tipples resources (equal), validation (equal), writing-review & editing (supporting); Xiaoli Pang resources (equal), validation (equal), writing-review & editing (supporting); Xing-Fang Li conceptualization (equal), formal analysis (supporting), funding acquisition (supporting), investigation (supporting), methodology (supporting), project administration (equal), resources (equal), supervision (equal), validation (equal), visualization (supporting), writing-original draft (supporting), writing-review & editing (equal); X. Chris Le conceptualization (lead), data curation (supporting), formal analysis (supporting), funding acquisition (lead), investigation (supporting), methodology (supporting), project administration (lead), resources (lead), supervision (equal), validation (supporting), visualization (supporting), writing-original draft (supporting), writing-review & editing (lead).
The authors declare no competing financial interest.
Supplementary Material
References
- Chen J.; Wang R.; Wang M.; Wei G. W. Mutations strengthened SARS-CoV-2 infectivity. J. Mol. Biol. 2020, 432, 5212–5226. 10.1016/j.jmb.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R.; Chen J.; Wei G. W. Mechanisms of SARS-CoV-2 evolution revealing vaccine-resistant mutations in Europe and America. J Phys Chem Lett 2021, 12, 11850–11857. 10.1021/acs.jpclett.1c03380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay V.; Lucas A.; Panja S.; Miyauchi R.; Mallela K. M. Receptor binding, immune escape, and protein stability direct the natural selection of SARS-CoV-2 variants. J. Biol. Chem. 2021, 297, 101208. 10.1016/j.jbc.2021.101208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A.; Holmes E. C.; O’Toole Á.; Hill V.; McCrone J. T.; Ruis C.; du Plessis L.; Pybus O. G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Int. Microbiol. 2020, 5, 1403–1407. 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO) , 2022. Tracking SARS-CoV-2 Variants. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/(accessed 21 March 2023).
- Majumdar S.; Sarkar R. Mutational and phylogenetic analyses of the two lineages of the Omicron variant. J. Med. Virol. 2022, 94, 1777–1779. 10.1002/jmv.27558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora P.; Pöhlmann S.; Hoffmann M. Mutation D614G increases SARS-CoV-2 transmission. Signal Transduction Targeted Ther. 2021, 6, 101–102. 10.1038/s41392-021-00502-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante J. A.; Liu Y.; Liu J.; Xia H.; Johnson B. A.; Lokugamage K. G.; Zhang X.; Muruato A. E.; Zou J.; Fontes-Garfias C. R.; Mirchandani D.; et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2021, 592, 116–121. 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H.; Chen Q.; Yang G.; He L.; Fan H.; Deng Y. Q.; Wang Y.; Teng Y.; Zhao Z.; Cui Y.; Li Y.; et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science 2020, 369, 1603–1607. 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton M. I.; MacGowan S. A.; Kutuzov M. A.; Dushek O.; Barton G. J.; van der Merwe P. A. Effects of common mutations in the SARS-CoV-2 Spike RBD and its ligand, the human ACE2 receptor on binding affinity and kinetics. Elife 2021, 10, e70658 10.7554/eLife.70658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K. A.; Gubbay J.; Hopkins J.; Patel S.; Buchan S. A.; Daneman N.; Goneau L. W. S-gene target failure as a marker of variant B.1.1.7 among SARS-CoV-2 isolates in the greater Toronto area, December 2020 to March 2021. JAMA J. Am. Med. Assoc. 2021, 325, 2115–2116. 10.1001/jama.2021.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng B.; Kemp S. A.; Papa G.; Datir R.; Ferreira I. A.; Marelli S.; Harvey W. T.; Lytras S.; Mohamed A.; Gallo G.; Thakur N.; et al. Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B. 1.1. 7. Cell Rep 2021, 35, 109292. 10.1016/j.celrep.2021.109292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm A.; Toptan T.; Pallas C.; Wolf T.; Goetsch U.; Gottschalk R.; Vehreschild M. J.; Ciesek S.; Widera M. Antibody-mediated neutralization of authentic SARS-CoV-2 B.1.617 variants harboring L452R and T478K/E484Q. Viruses 2021, 13, 1693. 10.3390/v13091693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Liu J.; Johnson B. A.; Xia H.; Ku Z.; Schindewolf C.; Widen S. G.; An Z.; Weaver S. C.; Menachery V. D.; Xie X.; et al. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. Cell Rep 2022, 39, 110829. 10.1016/j.celrep.2022.110829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.; Qiu Y.; Wang R.; Wei G. W. Persistent Laplacian projected Omicron BA.4 and BA.5 to become new dominating variants. Comput. Biol. Med. 2022, 151, 106262. 10.1016/j.compbiomed.2022.106262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A.; Zia T.; Suleman M.; Khan T.; Ali S. S.; Abbasi A. A.; Mohammad A.; Wei D. Q. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: an insight from structural data. J. Cell. Physiol. 2021, 236, 7045–7057. 10.1002/jcp.30367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen P. A.; Olsen R. J.; Long S. W.; Subedi S.; Davis J. J.; Hodjat P.; Walley D. R.; Kinskey J. C.; Ojeda Saavedra M.; Pruitt L.; Reppond K.; et al. Delta variants of SARS-CoV-2 cause significantly increased vaccine breakthrough COVID-19 cases in Houston, Texas. Am. J. Clin. Pathol. 2022, 192, 320–331. 10.1016/j.ajpath.2021.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twohig K. A.; Nyberg T.; Zaidi A.; Thelwall S.; Sinnathamby M. A.; Aliabadi S.; Seaman S. R.; Harris R. J.; Hope R.; Lopez-Bernal J.; Gallagher E.; et al. Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: a cohort study. Lancet Infect. Dis. 2022, 22, 35–42. 10.1016/S1473-3099(21)00475-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed A. M.; Ciling A.; Taha T. Y.; Chen I. P.; Khalid M. M.; Sreekumar B.; Chen P. Y.; Kumar G. R.; Suryawanshi R.; Silva I.; Milbes B.; et al. Omicron mutations enhance infectivity and reduce antibody neutralization of SARS-CoV-2 virus-like particles. Proc. Natl. Acad. Sci. U.S.A. 2022, 119, e2200592119 10.1073/pnas.2200592119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Wu S.; Wu B.; Yang Q.; Chen A.; Li Y.; Zhang Y.; Pan T.; Zhang H.; He X. SARS-CoV-2 Omicron strain exhibits potent capabilities for immune evasion and viral entrance. Sig. Transduct. Target. Ther. 2021, 6, 430–433. 10.1038/s41392-021-00852-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumblathan T.; Liu Y.; Qiu Y.; Pang L.; Hrudey S. E.; Le X. C.; Li X. F. An efficient method to enhance recovery and detection of SARS-CoV-2 RNA in wastewater. J. Environ. Sci. 2023, 130, 139–148. 10.1016/j.jes.2022.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Kumblathan T.; Feng W.; Pang B.; Tao J.; Xu J.; Xiao H.; Joyce M. A.; Tyrrell D. L.; Zhang H.; Li X. F.; Le X. C. On-site viral inactivation and RNA preservation of gargle and saliva samples combined with direct analysis of SARS-CoV-2 RNA on magnetic beads. ACS Meas. Sci. Au 2022, 2, 224–232. 10.1021/acsmeasuresciau.1c00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y.; Nellåker C.; Karlsson H. Evaluation of minor groove binding probe and Taqman probe PCR assays: Influence of mismatches and template complexity on quantification. Mol. Cell. Probes 2006, 20, 311–316. 10.1016/j.mcp.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Kutyavin I. V.; Afonina I. A.; Mills A.; Gorn V. V.; Lukhtanov E. A.; Belousov E. S.; Singer M. J.; Walburger D. K.; Lokhov S. G.; Gall A. A.; Dempcy R.; Reed M. W.; Meyer R. B.; Hedgpeth J. 3’-minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 2000, 28, 655–661. 10.1093/nar/28.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson G.; Nolan T.; Bustin S. A.. Real-time quantitative PCR, pathogen detection and MIQE. In PCR Detection of Microbial Pathogens; Totowa N. J., Ed.; Humana Press, 2013, pp 1–16. [DOI] [PubMed] [Google Scholar]
- TaqMan Assay Multiplex PCR Optimization Application Guide, 2022. https://assets.thermofisher.com/TFS-Assets/LSG/manuals/taqman_optimization_man.pdf (accessed 30 March, 2023).
- Pabbaraju K.; Wong A. A.; Douesnard M.; Ma R.; Gill K.; Dieu P.; Fonseca K.; Zelyas N.; Tipples G. A. Development and validation of RT-PCR assays for testing for SARS-CoV-2. JAMMI 2021, 6, 16–22. 10.3138/jammi-2020-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberta Health Services (AHS) , 2022. COVID-19 Alberta Statistics - Interactive Aggregate Data on COVID-19 Cases in Alberta. https://www.alberta.ca/stats/covid-19-alberta-statistics.htm#variants-of-concern (accessed 21 March 2023).
- Karthikeyan S.; Levy J. I.; De Hoff P.; Humphrey G.; Birmingham A.; Jepsen K.; Farmer S.; Tubb H. M.; Valles T.; Tribelhorn C. E.; Tsai R.; et al. Wastewater sequencing reveals early cryptic SARS-CoV-2 variant transmission. Nature 2022, 609, 101–108. 10.1038/s41586-022-05049-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokajärvi A. M.; Rytkönen A.; Tiwari A.; Kauppinen A.; Oikarinen S.; Lehto K. M.; Kankaanpää A.; Gunnar T.; Al-Hello H.; Blomqvist S.; Miettinen I. T.; et al. The detection and stability of the SARS-CoV-2 RNA biomarkers in wastewater influent in Helsinki, Finland. Sci. Total Environ. 2021, 770, 145274. 10.1016/j.scitotenv.2021.145274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger C. M.; Lienhard R.; Seth-Smith H. M.; Roloff T.; Wegner F.; Sieber J.; Bel M.; Greub G.; Egli A. PCR performance in the SARS-CoV-2 Omicron variant of concern?. Swiss Med. Wkly. 2021, 151, w30120. 10.4414/smw.2021.w30120. [DOI] [PubMed] [Google Scholar]
- Erster O.; Mendelson E.; Kabat A.; Levy V.; Mannasse B.; Assraf H.; Azar R.; Ali Y.; Bucris E.; Bar-Ilan D.; Mor O.; et al. Specific detection of SARS-CoV-2 variants B.1.1.7 (Alpha) and B.1.617.2 (Delta) using a one-step quantitative PCR assay. Microbiol. Spectr. 2022, 10, 021766–e2221. 10.1128/spectrum.02176-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garson J. A.; Badru S.; Parker E.; Tedder R. S.; McClure M. O. Highly sensitive and specific detection of the SARS-CoV-2 Delta variant by double-mismatch allele-specific real time reverse transcription PCR. J. Clin.Virol. 2022, 146, 105049. 10.1016/j.jcv.2021.105049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibai M.; Wang H.; Yeung P. S.; Sahoo M. K.; Solis D.; Mfuh K. O.; Huang C.; Yamamoto F.; Pinsky B. A. Development and evaluation of an RT-qPCR for the identification of the SARS-CoV-2 Omicron variant. J. Clin. Virol. 2022, 148, 105101. 10.1016/j.jcv.2022.105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung P. S. W.; Wang H.; Sibai M.; Solis D.; Yamamoto F.; Iwai N.; Jiang B.; Hammond N.; Truong B.; Bihon S.; Santos S.; et al. Evaluation of a rapid and accessible reverse transcription-quantitative PCR approach for SARS-CoV-2 variant of concern identification. J. Clin. Microbiol. 2022, 60, 001788–e222. 10.1128/jcm.00178-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabbaraju K.; Zelyas N.; Wong A.; Croxen M. A.; Lynch T.; Buss E.; Murphy S.; Shokoples S.; Kanji J.; Tipples G. Evolving strategy for an evolving virus: development of real-time PCR assays for detecting all SARS-CoV-2 variants of concern. J. Virol. Meth. 2022, 307, 114553. 10.1016/j.jviromet.2022.114553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelyas N.; Pabbaraju K.; Croxen M. A.; Lynch T.; Buss E.; Murphy S. A.; Shokoples S.; Wong A.; Kanji J. N.; Tipples G. Precision response to the rise of the SARS-CoV-2 B.1.1.7 variant of concern by combining novel PCR assays and genome sequencing for rapid variant detection and surveillance. Microbiol. Spectr. 2021, 9, e00315–e00321. 10.1128/Spectrum.00315-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.