Abstract
Identifying novel molecular mechanisms of exhausted CD8 T cells (Tex) is a key goal of improving immunotherapy of cancer and other diseases. However, high-throughput interrogation of in vivo Tex can be costly and inefficient. In vitro models of Tex are easily customizable and quickly generate high cellular yield, offering an opportunity to perform CRISPR screening and other high-throughput assays. We established an in vitro model of chronic stimulation and benchmarked key phenotypic, functional, transcriptional, and epigenetic features against bona fide in vivo Tex. We leveraged this model of in vitro chronic stimulation in combination with pooled CRISPR screening to uncover transcriptional regulators of T cell exhaustion. This approach identified several transcription factors, including BHLHE40. In vitro and in vivo validation defined a role for BHLHE40 in regulating a key differentiation checkpoint between progenitor and intermediate subsets of Tex. By developing and benchmarking an in vitro model of Tex, we demonstrate the utility of mechanistically annotated in vitro models of Tex, in combination with high-throughput approaches, as a discovery pipeline to uncover novel Tex biology.
INTRODUCTION
T cell exhaustion is a distinct cell state that arises during chronic viral infection and cancer. Exhausted CD8 T cells (Tex) are defined by reduced proliferative potential, high and sustained expression of inhibitory receptors (IRs), decreased production of effector cytokines, and a distinct transcriptional and epigenetic program (1). The inability to durably reverse exhaustion represents a major barrier to treatment of cancers and chronic viral infections. Efforts to develop more effective therapies for these diseases are inhibited by a limited understanding of the molecular mechanisms that underlie Tex biology. Despite a substantial body of work analyzing Tex biology in search of novel therapeutic targets, most of these efforts have been relatively low-throughput due to limitations of in vivo mouse models. Defining scenarios in which key aspects of Tex biology can be modulated and even screened in a high-throughput fashion could reveal new therapeutic opportunities.
Mouse models of chronic viral infection, such as lymphocytic choriomeningitis virus (LCMV), and cancer have been instrumental in establishing our understanding of CD8 T cell exhaustion, including discoveries with therapeutic relevance such as checkpoint pathway blockade (2–4). Despite their utility, in vivo models have limitations in efficiency and cellular yield. Mouse models of T cell exhaustion often generate relatively small numbers of Tex, limiting approaches that require large cell numbers. As a result, high-throughput experimental assays are challenging due to differences in scale between cells required and cells available. For example, shRNA or CRISPR screens in in vivo models require pooling of large numbers of mice to obtain sufficiently high cell/guide numbers to achieve robust screening results. Increasing the input cell number in the interest of maximizing yield can perturb pathogenesis and alter cellular differentiation. Adoptive transfer of a higher number of LCMV-specific CD8 T cells results in faster control of viral replication and/or increased immunopathology (5–7) and can influence CD8 T cell differentiation (8–10), circumventing the very biology of T cell exhaustion that is the intent of the study. Thus, although in vivo CRISPR screening models have uncovered valuable new insights into Tex and other T cell states, overcoming limitations in scale would allow additional layers of biology to be interrogated.
In vitro models are an attractive alternative to in vivo mouse models for screening and other discovery platforms because cellular output can be generated quickly and efficiently. In vitro models can be scaled up to maximize cellular yield and/or large amounts of cellular material for high-throughput assays (e.g. proteomics, metabolomics). However, modeling complex in vivo phenomena in vitro also has inherent challenges. In vitro models cannot fully recapitulate the complexities and nuance of in vivo Tex biology: for example, 3D tissue architecture and cell homing/migration, are challenging to model in vitro. Yet the reductionist nature of in vitro models provides a unique opportunity to dissect specific pathways. In vitro cultures can be easily customized to interrogate the causal effects of a diverse range of stimuli or conditions, such as cytokines and other secreted molecules, hypoxia, metabolites, pharmacological small molecules, or even genetic manipulations. Regardless, the utility of any model depends on an accurate evaluation of which features of in vivo biology are effectively modeled in vitro and which are not. Detailed benchmarking and analysis of in vitro models of Tex could have considerable value given the importance of this cell type in human disease.
Sustained TCR engagement is a central driver of CD8 T cell exhaustion in vivo (11–13). Repeated or continuous TCR stimulation in vitro can also successfully induce phenotypic features of Tex, such as loss of proliferative potential (14), expression of PD-1 and other IRs (15), reduced effector function (16), or a combination of these properties (17–21). Collectively, these studies demonstrate the utility of in vitro models approximating T cell exhaustion. However, defining how much of the overall program of in vivo Tex biology is accurately captured in these settings remains a challenge. A more detailed understanding of which aspects of Tex biology are and are not capable of being modeled in vitro will enable more successful downstream application of in vitro models and screens to identify underlying mechanisms and therapeutic opportunities.
To address these issues, we developed an in vitro model that approximates Tex via chronic administration of cognate peptide in a manner intended to achieve continuous antigenic stimulation. We benchmarked these in vitro chronically stimulated CD8 T cells against in vivo Tex generated during chronic LCMV infection to confirm that phenotypic, functional, transcriptional, and epigenetic features of Tex were appropriately recapitulated in vitro. We then leveraged this in vitro culture model in combination with CRISPR/Cas9 screening of transcriptional pathways to identify a role for the transcription factor (TF) BHLHE40 in T cell exhaustion. We returned to mouse models of chronic viral infection to interrogate the function of BHLHE40 in in vivo Tex and found that BHLHE40 regulates a differentiation checkpoint between progenitor Tex and intermediate and terminally differentiated subsets of Tex. Our data suggest that biology targeted by BHLHE40 may be relevant for therapeutically manipulating these Tex subsets involved in response to checkpoint blockade.
Here we demonstrate the utility of mechanistically annotated in vitro models of T cell exhaustion. In combination with CRISPR technology, this in vitro model of chronic stimulation enables high-throughput screening of transcriptional regulators of Tex, after which individual hits can be further interrogated in vitro and in vivo. This foundation of in vitro modeling of Tex will allow scaling of other discovery approaches to probe deeper into the mechanisms of CD8 T cell exhaustion.
RESULTS
Chronic antigenic stimulation in vitro induces the molecular phenotype of CD8 T cell exhaustion
Many in vitro models recapitulate some of the known features of Tex (14, 17–19, 21). However, it is often unclear which features or biological modules of in vivo biology can and cannot be recreated in vitro. To address this question, we aimed first to develop an in vitro model of Tex and subsequently to deeply interrogate and benchmark the phenotypic, functional, transcriptional, and epigenetic features of this model in comparison to in vivo Tex generated during chronic LCMV infection.
We established an in vitro model of Tex, building on previous work from Beltra et al (15). Naïve CD8 T cells were isolated from mice transgenic for a TCR that recognizes the DbGP33−41 epitope of LCMV (P14 mice). These P14 cells were co-cultured with dendritic cells from WT C57BL/6 mice that were pulsed with DbGP33−41 peptide. To mimic chronic antigenic stimulation characteristic of that which drives Tex differentiation in vivo, P14 cells were further stimulated with DbGP33−41 peptide and IL-2 every 2 days thereafter (Fig. 1A). This timing was chosen based on previous work showing that peptide stimulation under in vitro conditions lasts ~2 days or less (22). We also generated an in vitro acutely stimulated condition, in which P14 cells received DbGP33−41 peptide only at day 0. At day 7 of in vitro culture, P14 cells were harvested and analyzed for activation and differentiation by flow cytometry. As a benchmark for bona fide Tex biology, in vivo CD8 Tex were generated via LCMV clone 13 (Cl13) infection and analyzed in parallel.
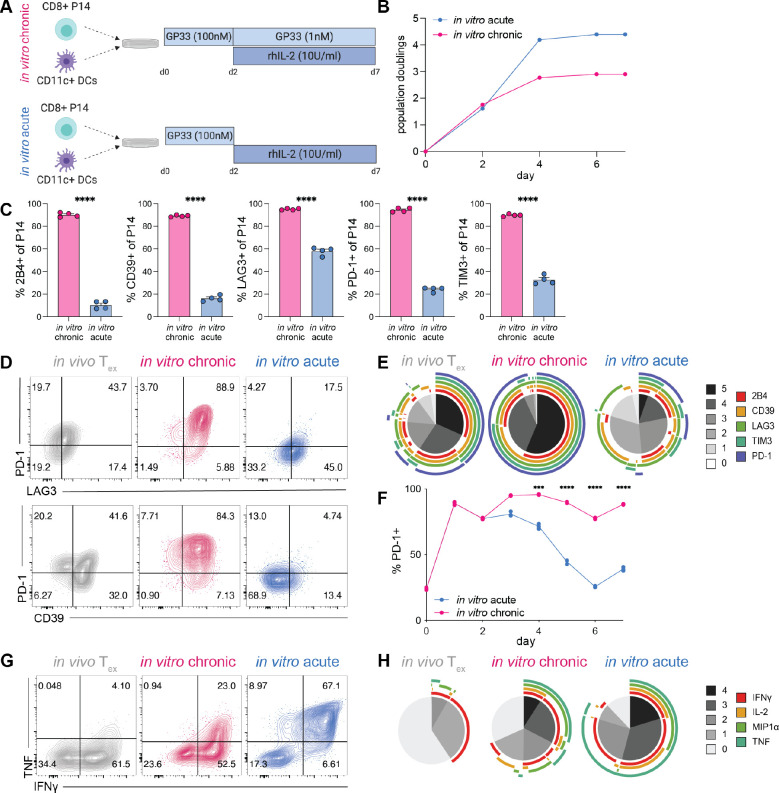
Fig. 1. Chronic antigenic stimulation in vitro induces key features of Tex.
(A) Experiment schematic of chronic and acute stimulation of P14 cells in vitro. (B) Cell expansion during chronic and acute stimulation in vitro. (C) Percent expression of IRs by chronically and acutely stimulated P14 cells (gated on CD44hi CD8+ live singlets); two technical replicates of two biological replicates shown. Significance calculated by unpaired two-tailed t test; ****p<0.0001. (D) Representative flow cytometry data and (E) SPICE analysis of IR co-expression by in vivo Tex (LCMV-Cl13 30dpi), in vitro chronically stimulated P14 cells, and in vitro acutely stimulated P14 cells (gated on CD44hi CD8+ live singlets). (F) Longitudinal PD-1 expression on in vitro chronically and acutely stimulated P14 cells. Representative of 2 experiments; significance calculated by paired two-tailed t test; *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. (G) Representative flow cytometry data and (H) SPICE analysis of co-production of effector cytokines in in vivo Tex (LCMV-Cl13 30dpi), in vitro chronically stimulated P14 cells, and in vitro acutely stimulated P14 cells. (B-E, G-H) Representative of >3 experiments. (D,G) Numbers in flow cytometry plots indicate percentage of parent population within each gate. (E,H) Grayscale sections in SPICE plots indicate number of IRs/cytokines co-expressed; colored bands indicate individual IRs/cytokines.
In vivo Tex are phenotypically distinct from naïve (Tn), effector (Teff) and memory (Tmem) CD8 T cells and are characterized by: 1) decreased proliferative potential, 2) high and stable expression of inhibitory receptors (IRs), and 3) decreased production of effector cytokines (11, 12, 23). We benchmarked our in vitro model against these known hallmarks of in vivo Tex, beginning with proliferation and expansion. Both in vitro conditions induced proliferation and expansion between days 0 and 4 of culture, after which proliferation slowed and eventually plateaued. However, whereas acutely stimulated P14 cells reached 4 population doublings, chronically stimulated P14 cells were unable to maintain similar levels of expansion and plateaued at less than 3 population doublings, despite continuous exposure to antigenic stimulation (Fig. 1B). These data suggest that persistent antigenic stimulation is sufficient to reduce proliferative expansion and/or potential. To interrogate the contribution of proliferation and cell death to this overall decreased expansion, we assessed expression of the cell cycle protein KI67 and pro- and anti-apoptotic proteins BIM and BCL-2, respectively. KI67 expression was maintained between days 4–7 of in vitro culture; however, this ongoing cell cycle was accompanied by an increasing BIM/BCL-2 ratio, indicating that chronic stimulation results in sustained proliferation but also increased sensitivity to cell death, leading to high cell turnover and a net overall lack of expansion in cell numbers (Fig. S1A). These results are consistent with observations in vivo Tex, for which ongoing stimulation is associated with continued cell cycle and proliferation, but no net increase in cell numbers (24–31).
Chronic antigen stimulation in vitro was sufficient to induce high expression of multiple IRs, characteristic of in vivo Tex (12, 32). Whereas acutely stimulated P14 cells demonstrated significantly lower expression of IRs, such as PD-1 and LAG3, than in vivo Tex, chronic stimulation induced high expression of IRs as a percentage (Fig. 1C) and on a per cell level, as quantified by MFI (Fig. S1B). Furthermore, a higher proportion of chronically stimulated P14 cells co-expressed combinations of IRs, including PD-1 and LAG3 or CD39 (Fig. 1D), than acutely stimulated P14 cells or even in vivo Tex. Because increased diversity and co-expression of IRs is associated with progression toward terminal exhaustion (26, 32, 33), we performed SPICE analysis to assess IR co-expression. More than 50% of chronically stimulated P14 cells co-expressed all 5 IRs analyzed, an even higher proportion of IR co-expression than observed on in vivo Tex (~30%) (Fig. 1E). In contrast, less than 10% of acutely stimulated P14 cells co-expressed 5 or more IRs. Furthermore, whereas acute stimulation induced IR expression shortly after activation that waned over time, high expression of IRs was maintained on P14 cells that had undergone chronic stimulation in vitro (Fig. 1F), consistent with stable maintenance of IR expression by in vivo Tex. We compared this antigen-specific in vitro model of chronic stimulation to a previously published model of chronic stimulation using non-antigen-specific TCR engagement (18). Although IR expression was similar between both in vitro models, chronic stimulation via αCD3/αCD28 resulted in substantially decreased expansion and cellular yield (Fig. S1C). Also, to assess whether additional rounds of antigen administration would further enhance features of Tex, we extended our in vitro chronic stimulation by an additional 3 days. Chronically stimulated P14 cells at d10 of in vitro culture had comparable IR expression to those at day 7, suggesting little increase in exhaustion phenotype; however, extending chronic stimulation in vitro to 10 days resulted in poorer cell recovery (Fig. S1D).
We next evaluated our in vitro generated P14 populations for reduced and/or altered effector cytokine production characteristic of Tex (11, 12, 23). Whereas in vitro acutely stimulated P14 cells produced multiple effector cytokines in response to peptide restimulation, chronically stimulated P14 cells had a reduced capacity to produce TNF and IFNγ, though the extent of loss of cytokine production was not as severe as that of in vivo Tex (Fig. 1G). Because hierarchical loss of effector cytokine production is associated with increased severity of exhaustion (11, 12), we assessed cytokine co-production via SPICE analysis. Approximately 20% of P14 cells were able to co-produce all 4 effector cytokines analyzed after acute stimulation in vitro. This multi-functional population was substantially reduced in in vitro chronically stimulated P14 cells to approximately 10% and nearly absent in in vivo Tex (Fig. 1H). Collectively, these data suggest that chronic stimulation in vitro was sufficient to recapitulate known cellular hallmarks of Tex biology, including reduced proliferative potential, sustained high expression and co-expression of multiple IRs, and decreased production of effector cytokines.
In vitro chronically stimulated CD8 T cells develop a transcriptional signature of Tex
In addition to phenotypic and functional properties, in vivo generated Tex are defined by unique transcriptional and epigenetic landscapes distinct from those of Tn, Teff, and Tmem (1, 12, 34–38). To assess global transcriptional changes induced by acute versus chronic stimulation in vitro, we analyzed P14 cells by RNA-seq on days 0, 4, and 7 of in vitro culture. By number of differentially expressed genes (DEGs), the most substantial differences were observed between naïve (d0) and in vitro stimulated P14 cells at d4 regardless of stimulation condition (Fig. 2A), indicating that the most robust transcriptional changes occurred upon the transition from naïve to activated CD8 T cell. Whereas acute and chronically stimulated P14 cells only differed by 634 DEGs at d4 of in vitro culture, this number grew to 2844 by d7, suggesting that acute and chronic stimulation induced similar early transcriptional programs that diverged significantly by d7 (Fig. 2A). Furthermore, unbiased DEG analysis identified a cluster of genes uniquely expressed at d7 of chronic stimulation, including Havcr2, Ccl3, and Xcl1 (Fig. 2B). Consistent with these observations, principal component analysis (PCA) revealed that acutely and chronically stimulated P14 cells clustered together at d4 (Fig. S2A). By d7, however, acutely and chronically stimulated P14 cells occupied distinct principal component space from all other in vitro generated populations. Collectively, these data indicate that, despite some overlap in transcriptional programs shortly after activation, acute and chronic stimulation in vitro generate unique and divergent transcriptional states by d7. Furthermore, this transcriptional divergence between d4 and d7 of in vitro culture suggests coordinated temporal regulation of gene expression in response to both acute and chronic stimulation in vitro.
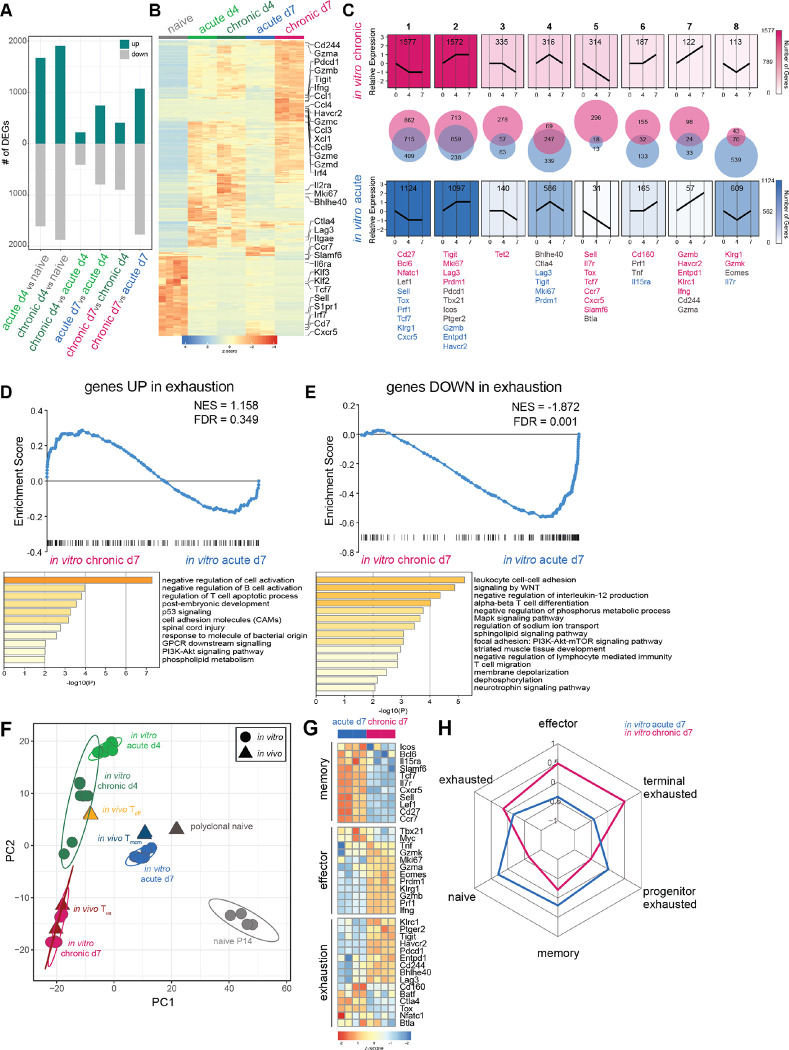
Fig. 2. In vitro chronically stimulated P14 cells develop a transcriptional signature of Tex.
(A) Number of differentially expressed genes (DEGs; filtered on log2 fold change (lfc)>1 and p<0.05) between pairwise comparisons as indicated. (B) Top DEGs (filtered on lfc>6; p<0.01) from all pairwise comparisons between in vitro generated CD8 T cell subsets. (C) Trajectory analysis (see Methods) of gene expression patterns during chronic or acute stimulation in vitro. (D-E) [top] Gene set enrichment analysis (GSEA) of genes (D) upregulated and (E) downregulated in in vivo Tex (41) after chronic or acute stimulation in vitro. [bottom] Gene ontology (GO) analysis of leading edge genes. (F) Principal component analysis (PCA) of RNA-seq data from in vitro chronically and acutely stimulated P14 cells and previously published in vivo CD8 T cell subsets (37). (G) Gene expression of manually curated lists of genes associated with Tmem, Teff, or Tex in in vitro chronically and acutely stimulated P14 cells. (H) Gene set variation analysis (GSVA) indicating comparative enrichment for various gene signatures (28, 41) after chronic or acute stimulation in vitro.
To further interrogate temporal transcriptional trajectories induced by acute and chronic stimulation in vitro, we manually binned genes into patterns of relative expression at days 0, 4, and 7 of in vitro culture (Fig. 2C). The two most common temporal patterns of gene expression in both acutely and chronically stimulated P14 were a decrease or increase from d0 to d4 followed by stable maintenance of expression from d4 to d7 (patterns 1 and 2, respectively). Whereas some genes within these patterns biased either toward chronic or acute stimulation, most genes were shared between both conditions. These two most common temporal gene patterns confirm that the greatest magnitude of change in gene expression occur upon the transition from naïve to activated CD8 T cell and highlight common biology found in both in vitro conditions.
To investigate transcriptional trajectories preferentially associated with acute stimulation, we examined genes in patterns 4 (transient upregulation at d4) and 8 (transient downregulation at d4) (Fig. 2C). Pattern 4 contained 586 genes that transiently increased during acute stimulation. Of these, less than half overlapped between acute and chronic stimulation; 339 of these changes were found only after acute stimulation. Genes in pattern 4 preferentially responsive to acute stimulation included Lag3, Tigit, and Mki67, consistent with acutely stimulated P14 cells becoming transiently activated before returning to a relative state of quiescence. Conversely, gene expression of Lag3, Tigit, and Mki67 remained elevated in chronically stimulated P14 cells at d7 (pattern 2), consistent with a continued role for these genes in Tex. Pattern 8, also biased toward acute stimulation conditions, displayed an inverse pattern, with genes decreasing in expression at d4 then returning to baseline by d7. Of these 600 transcriptional changes biased toward acute stimulation, less than 100 were shared with chronic stimulation. For example, expression of Il7r, which encodes the IL-7 receptor crucial for homeostatic proliferation in Tn and Tmem, followed this pattern of transient decrease. Thus, acute stimulation in vitro recapitulated key temporal changes in gene expression reminiscent of in vivo T cell activation and transition to a post-effector state.
To interrogate transcriptional trajectories preferentially associated with chronic stimulation, we examined genes that decreased (pattern 5) or increased (pattern 7) in expression throughout the course of in vitro culture (Fig. 2C). Pattern 5 was dominated by 296 genes biased toward chronic stimulation, with only 18 genes shared with acute stimulation. Genes in this pattern included Sell, Il7r, Tcf7, Cxcr5, and Slamf6, genes typically associated with stemness and quiescence. Pattern 7 included only 24 genes shared by both conditions and 98 genes biased toward chronic stimulation, including genes encoding IRs, such as Havcr2 (TIM3) and Entpd1 (CD39), as well as genes related to effector function, such as Ifng and Gzmb. Increased transcription despite decreased protein expression of effector molecules is a pattern consistent with early transcriptional studies of Tex that first noted this disconnect between effector function and transcription of effector genes (12). Collectively, these patterns suggest differential regulation of transcriptional trajectories in response to chronic or acute stimulation in vitro. Furthermore, genes within temporal patterns preferentially responsive to chronic stimulation in vitro are associated with transcriptional evidence of persistent activation as well as robust downregulation of genes associated with durability, stemness, and other programs associated with Tmem development.
To quantify how gene expression induced by chronic stimulation in vitro compared to that of in vivo Tex, we employed gene set enrichment analysis (GSEA) (39, 40) of a previously published transcriptional signature of exhaustion (41). Genes upregulated in in vivo Tex trended toward enrichment in in vitro chronically stimulated P14 cells (Fig. 2D), although this enrichment did not reach statistical significance. One possible interpretation of this result is that chronic stimulation in vitro recapitulates some, but not all, features of in vivo Tex. It is also possible that a strong transcriptional signature of activation in this in vitro model obscures significant enrichment of this exhaustion signature. To investigate which transcriptional pathways from this in vivo Tex gene signature were and were not captured by chronic stimulation in vitro, we focused on the leading edge of enriched genes and applied gene ontology (GO) analysis. Leading edge genes included known exhaustion-related genes, such as Pdcd1, Lag3, Tigit, and Prdm1 (Fig. S2B). GO analysis of these leading edge genes highlighted multiple pathways involved in negative regulation of T cell activation (Fig. 2D). These data were supported by the observation that a gene signature of in vitro activation was more highly enriched in chronically stimulated cells at d4 compared to d7 (Fig. S2C). Conversely, the in vivo Tex gene signature enriched in chronically stimulated cells at d7 compared to d4 (Fig. S2D). Collectively, these GSEA analyses indicate an overall transcriptional profile characterized by progression towards exhaustion and waning signature of cell activation during chronic stimulation in vitro.
Some genes upregulated in in vivo Tex were not robustly enriched after chronic stimulation in vitro, such as Ctla4 and Tox (Fig. S2E). GO analysis of genes absent from the leading edge highlighted pathways related to cell activation as well as cytokine signaling and inflammation (Fig. S2F), further emphasizing antigen-specific TCR stimulation as the basis of our in vitro model of chronic stimulation, rather than cytokine/chemokine signaling, inflammation, or other aspects that contribute to development of in vivo Tex. These observations were supported by low expression of Tox mRNA observed after chronic stimulation in vitro (Fig. S2G). Furthermore, although TOX protein expression increased marginally after in vitro chronic stimulation compared to acute stimulation, induction of TOX in in vitro chronically stimulated P14 cells was modest compared to that of in vivo Tex (Fig. S2H). Low expression of TOX in this and other previously published in vitro models of Tex (17, 18) indicates that, although TOX may be required for Tex to form and persist in vivo, there are likely features of Tex are less dependent on high and sustained TOX expression that can be captured using in vitro models. Thus, these GSEA and GO analyses suggest that chronic stimulation in vitro was sufficient to recreate some aspects of transcriptional programs distinctly upregulated in in vivo Tex, such as IR expression and negative regulation of cell activation, but not others, such as inflammatory signaling and high and sustained expression of TOX.
GSEA also revealed a pattern of genes downregulated in Tex that were likewise significantly negatively enriched in in vitro chronically stimulated P14 cells (Fig. 2E). Leading edge genes included Tcf7, Il7r, Sell, Lef1, Ccr7, and other known Tn- and Tmem-associated genes (Fig. S2I). GO analysis of these leading edge genes revealed pathways involved in cell-cell adhesion, WNT, MAPK, ion, and sphingolipid signaling, confirming that in vitro chronic stimulation significantly recapitulates transcriptional downregulation of programs associated with quiescence and memory. In total, these GSEA results suggest that, whereas this in vitro model of chronic stimulation partially captures transcriptional programming upregulated in in vivo Tex, this model appeared to broadly capture the transcriptional programs distinctly downregulated in Tex.
To compare transcriptional profiles of in vitro chronically and acutely stimulated CD8 T cells to those of in vivo Tn, Teff, Tmem, and Tex, we computationally merged our RNA-seq data set with a previously published data set of in vivo generated antigen-specific CD8 T cells (37) and visualized the results by PCA. (Note that differences between Tn populations represent naïve P14 cells versus polyclonal naïve CD8 T cells.) PCA revealed co-localization of acutely and chronically stimulated P14 cells at d4 of in vitro culture (Fig. 2F), consistent with earlier data (Fig. S2A). Both P14 populations at d4 also co-localized with in vivo Teff (Fig. 2F), consistent with a dominant signature of T cell activation at this early time point in vitro. By day 7, however, acutely and chronically stimulated P14 cells transcriptionally diverged to occupy distinct principal component space. Whereas in vitro acutely stimulated P14 cells at d7 co-localized with in vivo Tmem, in vitro chronically stimulated P14 cells at d7 overlapped nearly completely with in vivo Tex (Fig. 2F). These data suggest that, during the first 4 days of in vitro culture, acute and chronic stimulation drove similar transcriptional programming consistent with programs of in vivo Teff. However, the absence of further stimulation in acute conditions in vitro allowed P14 cells to adopt Tmem-like transcriptional programming. In contrast, continued chronic antigenic stimulation in vitro induced Tex-like transcriptional changes.
We next sought to evaluate transcriptional programs induced by acute and chronic stimulation in vitro in the context of known Teff, Tmem, and Tex biology. We therefore evaluated expression of key effector-, memory- and exhaustion-associated genes in in vitro chronic and acutely stimulated P14 cells (Fig. 2G). By d7, in vitro acutely stimulated P14 cells highly expressed many genes associated with Tmem (Bcl6, Tcf7, Sell, Lef1, Slamf6, Cxcr5, Cd27, and Ccr7), reflecting the overlap in principal component space described above (Fig. 2F). In contrast, by d7, chronically stimulated P14 cells upregulated some Tex-associated genes (Pdcd1, Havcr2, Entpd1, Lag3, Cd244, and Tigit) but lacked robust expression of others (Cd160, Batf, Ctla4, Tox, Nfatc1). Chronic stimulation also induced expression of some Teff-associated genes (Klrg1, Tnf, Ifng, Eomes, Prf1, Gzmb), consistent with increased transcription of Teff-related genes despite reduced protein expression by in vivo Tex. Overall, these data indicate that acute stimulation in vitro was sufficient to induce expression of many known Tmem-related genes, whereas chronic stimulation in vitro induced expression of known activation- and Tex-related genes. Furthermore, these results suggest that chronic antigenic stimulation in vitro was sufficient to induce many Tex-associated transcriptional changes but additional stimuli may be necessary to fully induce and/or sustain certain pathways, including the TOX-NFAT axis.
In vivo Tex are heterogeneous and consist of biologically distinct subsets, including progenitor (or stem-like), intermediate (Teff-like), and terminal populations (24, 26, 33, 42, 43). Progenitor Tex express the TF TCF1 and provide the long-term proliferative reserve needed to give rise to intermediate and terminally differentiated subsets of Tex (24, 27–29). Progenitor Tex also provide the proliferative burst necessary for response to checkpoint blockade (24, 27, 29). Intermediate Tex re-acquire some effector activity and give rise to terminal Tex that are often found in non-lymphoid tissues and tumors (33, 44). To investigate whether in vitro chronically stimulated P14 cells recapitulated the biology of any of these Tex subsets, we used gene set variation analysis (GSVA) (45) to quantify enrichment of transcriptional signatures of progenitor and terminal Tex subsets (28) in in vitro chronically and acutely stimulated P14 cells. We also included signatures from major CD8 T cell states, such as Tn, Teff, and Tmem, as well as a signature of bulk Tex which contained progenitor, intermediate, and terminal populations (41). Whereas in vitro chronically stimulated P14 cells enriched for signatures of Tex and Teff (Fig. 2H), in vitro acutely stimulated P14 cells enriched for Tn and Tmem gene signatures. Furthermore, in vitro chronically stimulated P14 cells enriched for a signature of terminal Tex, even more strongly than for the signature of bulk Tex, and lacked enrichment for a signature of progenitor Tex. These data are also consistent with the absence of genes involved in stem and progenitor biology in in vitro chronically stimulated P14 cells, such as Tcf7, Lef1, Slamf6, and Cxcr5 (Fig. 2G). This GSVA data suggests that chronic stimulation in vitro may induce a transcriptional profile more representative of terminally differentiated Tex.
Progenitor and terminal subsets of Tex are defined not only by differential transcriptional signatures but also by differences in phenotype and function. For example, progenitor and terminal subsets of Tex are associated with discrete TF hallmarks: whereas TCF1 expression is associated with progenitor Tex (27, 28, 46, 47), expression of Eomes is associated with terminally differentiated Tex subsets (26). We therefore sought to evaluate whether the terminally differentiated transcriptional signature of in vitro chronically stimulated P14 cells was associated with phenotypic or functional differences. Indeed, in vitro chronically stimulated P14 cells and in vivo Tex, but not in vitro acutely stimulated P14 cells, displayed high co-expression of PD-1 and Eomes (Fig. S2J), consistent with terminal Tex differentiation (26). Similarly, whereas bulk in vivo Tex had both TCF1+ progenitor and TCF1− terminal populations, TCF1 expression was almost entirely absent in P14 cells after in vitro chronic stimulation (Fig. S2K), in line with prior work linking chronic stimulation in vitro to Tcf7 promoter methylation and gene silencing (17). Additionally, chronic stimulation in vitro led to increased production of Granzyme B compared to bulk in vivo Tex (Fig. S2L). This observation aligns with recent studies documenting increased cytotoxic potential in terminal Tex compared to other Tex subsets (33, 44). Taken together, these TF expression patterns and increased cytotoxic potential confirm earlier transcriptional data indicating that chronic stimulation in vitro induces a cell state with similarities to the terminal subset of in vivo Tex.
In vitro chronically stimulated CD8 T cells develop epigenetic signatures of Teff and Tex
Recent studies have shown that Tex develop an epigenetic landscape distinct from Teff or Tmem (37, 38, 48). To assess global epigenetic changes induced by acute versus chronic stimulation in vitro, we analyzed P14 cells by ATAC-seq on days 0, 4, and 7 of in vitro culture. By number of differentially accessible chromatin regions (DACRs), the most substantial differences were observed between naïve (d0) and in vitro stimulated P14 cells at d4 (~30,000 DACRs) regardless of stimulation condition (Fig. S3A), indicating that the most robust epigenetic changes occurred upon the transition from naïve to activated CD8 T cell. Whereas acute and chronically stimulated P14 cells only differed by 547 DACRs at d4 of in vitro culture, this number grew to 12160 by d7 (Fig. S3A), suggesting that acute and chronic stimulation induced similar early chromatin accessibility programs that diverged significantly by d7. By PCA, acutely and chronically stimulated P14 cells initially co-localized at d4 but later diverged to occupy distinct principal component space by d7 (Fig. S3B). Collectively, these epigenetic data reveal distinct overall trajectories of differentiation associated with chronic or acute stimulation in vitro, consistent with the transcriptional trajectories observed earlier.
To understand how the global epigenetic programs of in vitro chronically and acutely stimulated P14 cells compare to those of in vivo Teff, Tmem, and Tex, we computationally merged our in vitro ATAC-seq data set with a previously published ATAC-seq data set of in vivo generated CD8 T cell populations (48) and visualized the results by PCA (Fig. 3A). P14 cells at d4 of in vitro chronic and acute stimulation clustered closely with in vivo Tex in principal component space, perhaps reflecting activation-associated chromatin accessibility. However, by d7, acutely stimulated P14 cells co-localized with in vivo Tmem. Chronically stimulated P14 cells at d7 instead remained co-localized near in vivo Tex in principal component space, indicating chromatin accessibility at many Tex-specific loci after chronic stimulation in vitro. Indeed, accessibility at the −23.8kb enhancer in the Pdcd1 locus, previously described as uniquely accessible in in vivo Tex with downstream functional effects on PD-1 expression (38), was observed only in in vitro chronically stimulated P14 cells and in vivo Tex (Fig. 3B) but not in in vitro acutely stimulated P14 cells. Chronically stimulated P14 cells also co-localized in principal component space with in vivo Teff (Fig. 3A), suggesting that chronic stimulation in vitro generated a chromatin accessibility profile with characteristics of both Teff and Tex.
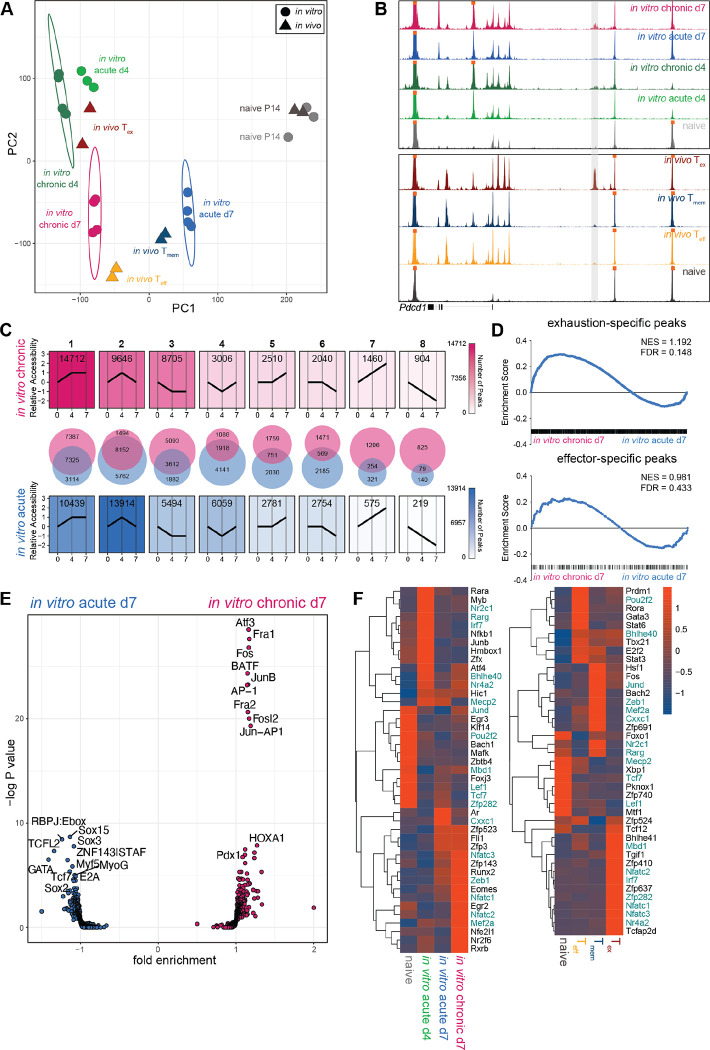
Fig. 3. In vitro chronically stimulated P14 cells develop epigenetic signatures of Teff and Tex.
(A) PCA of ATAC-seq data from in vitro chronically and acutely stimulated P14 cells and previously published in vivo CD8 T cell subsets (48). (B) ATAC-seq signal tracks for in vitro chronically and acutely stimulated P14 cells and previously published in vivo CD8 T cell subsets (48); −23.1kb enhancer of Pdcd1 locus highlighted in gray. (C) Trajectory analysis (see Methods) of chromatin accessibility patterns during in vitro chronic or acute stimulation. (D) Peak set enrichment analysis (PSEA) of Tex- and Teff-specific ACRs in in vitro chronically and acutely stimulated P14 cells. (E) Transcription factor (TF) motif accessibility in differential ACRs between in vitro chronically and acutely stimulated P14 cells. (F) Rank calculated via Taiji analysis in [left] in vitro chronically and acutely stimulated P14 cells and [right] previously published in vivo CD8 T cell subsets (37), filtered by mean>0.0001 and fold change>5. Heat scale indicates z-score of rank as calculated by PageRank algorithm. Colored text indicates shared TFs between in vitro and in vivo analyses.
To define the temporal epigenetic changes induced by chronic and acute stimulation in vitro, we manually binned individual accessible chromatin regions (ACRs) into patterns based on relative accessibility over the course of in vitro culture (Fig. 3C). During chronic stimulation in vitro, the most frequent pattern was an increase in chromatin accessibility between d0 and d4, followed by stable maintenance from d4 to d7 (pattern 1). This most frequent pattern indicates that chronic stimulation in vitro preserved a majority of ACRs opened during the transition from naïve to activated cell. In contrast, the most common pattern during in vitro acute stimulation was a transient increase from d0 to d4 followed by a return to baseline by d7 (pattern 2). This observation is consistent with the notion that, in the absence of further TCR engagement in vitro, P14 cells preferentially close many ACRs that became transiently accessible during activation, returning to a state of relative epigenetic inaccessibility. Many ACRs binned into pattern 2 were shared between chronic and acute stimulation in vitro (8152 shared ACRs), suggesting some common epigenetic patterning of transient accessibility associated with initial activation. ACRs that progressively decreased in accessibility over in vitro culture (pattern 8) represented the least common pattern in both acute and chronic stimulation, followed by ACRs that progressively increased in accessibility (pattern 7). Patterns 7 and 8 were also characterized by the least amount of overlap between in vitro chronic and acute stimulation (254 and 79 shared ACRs, respectively), indicating divergent epigenetic landscapes and biology preferentially associated with either chronic or acute stimulation in vitro. Overall, these data suggest that, unlike acutely stimulated cells, P14 cells undergoing in vitro chronic stimulation induced epigenetic changes consistent with some aspects of Tex, including a prominent pattern of sustained changes in accessibility.
To compare chromatin accessibility profiles of in vitro chronically and acutely stimulated P14 cells to in vivo Tn, Teff, Tmem, and Tex, we performed peak set enrichment analysis (PSEA) (49) using signatures of subset-specific ACRs, or “peaks,” curated from a previously published data set (48). PSEA revealed enrichment for Tmem-specific peaks after acute stimulation in vitro (Fig. S3C), consistent with shared epigenetic programming of quiescence. Conversely, PSEA of in vitro chronically stimulated P14 cells revealed enrichment for Tex-specific ACRs (Fig. 3D). Enrichment for Tn- (Fig. S3C) and Teff-specific peaks (Fig. 3D) were not significant, suggesting that both in vitro acutely and chronically stimulated P14 cells share some ACRs with in vivo Tn and Teff. Collectively, these PSEA results indicate that acute stimulation in vitro induced a chromatin accessibility landscape similar to that of Tmem whereas chronic stimulation in vitro generated an epigenetic profile similar to that of Tex.
To investigate how the epigenetic landscape induced by chronic stimulation in vitro might affect downstream TF binding, we performed TF motif enrichment analysis in DACRs between in vitro chronically stimulated P14 cells and in vivo Tex. Whereas ACRs unique to in vitro chronic stimulation enriched for binding motifs of Fli1 and RUNX family members, NFAT binding motifs predominated ACRs unique to in vivo Tex (Fig. S3D). ACRs unique to in vivo Tex also enriched for T-box family motifs and NFAT:AP1 composite motifs. Prior work demonstrated that constitutive expression of partnerless NFAT (engineered to be unable to interact with AP1) can induce expression of TOX and other hallmarks of Tex (50) and that NFAT is necessary for induction and stabilization of TOX in vivo (49). The relative absence of monomeric NFAT binding sites in in vitro chronically stimulated P14 cells suggests that antigenic stimulation in vitro may not be sufficient to remodel chromatin in a manner that accommodates partnerless NFAT binding to the extent of in vivo Tex. Furthermore, these results suggest that a comparative lack of monomeric NFAT sites in the chromatin landscape induced by chronic stimulation in vitro may be associated with only modest induction of TOX expression (Fig. S2H). This analysis identified differential modules of TF binding potential between in vitro chronically stimulated P14 cells and in vivo Tex, providing context for which transcriptional circuits might be accurately modeled in vitro.
To further examine TF binding potential downstream of differential chromatin accessibility between in vitro acutely and chronically stimulated P14 cells, we conducted TF motif enrichment analysis in DACRs between these two conditions (Fig. 3E). ACRs unique to in vitro acutely stimulated P14 cells enriched for TCF family binding motifs, consistent with the role of these TFs in restoring stemness and quiescence following acute stimulation, as well as Sox family binding motifs. Conversely, ACRs unique to chronically stimulated P14 cells enriched for AP1 family binding motifs, including BATF, Fos, Fosl2, AP1, and Jun-AP1 dimer motifs. Strong enrichment for AP1 family member motifs could potentially indicate disruption of canonical Fos/Jun signaling via formation of heterodimers, leading to a reduction in AP1 available to partner with NFAT, as has been implicated by studies describing the role of BATF in Tex (51).
To predict TF networks central to differential biology in these in vitro models of chronic and acute stimulation, we computationally integrated chromatin accessibility and TF motif data (from ATAC-seq) with gene expression data (from RNA-seq) and applied Taiji PageRank analysis (52, 53). Taiji analysis constructs networks of interactions between TFs to infer importance, or rank, based on connectivity within these networks. Taiji scans motif binding sites in ACRs from ATAC-seq, then assigns TF motifs found in enhancer or promoter regions to target genes. Once TFs have been linked to gene targets, Taiji constructs putative TF networks then weights TF nodes in these networks based on gene expression data from RNA-seq. The PageRank algorithm is then applied to determine rank, or importance, based on node weight and connectivity. Highly ranked TFs will have high gene expression as well as binding motifs present in ACRs at loci of multiple other TFs, indicating that their influence extends to multiple transcriptional networks. To contextualize analysis of our in vitro data sets within known CD8 T cell biology, we applied Taiji analysis in parallel to a previously published in vivo RNA- and ATAC-seq data set (37). Taiji analysis identified many of the same ranked TFs in acute and chronic stimulation in vitro and in vivo (Fig. 3F). For example, Lef1 and Tcf1, TFs associated with stemness and quiescence, ranked highly in Tn as well as in vitro acutely stimulated P14 at d7 and in vivo Tmem. Zeb1 ranked highly in in vitro acutely stimulated P14 cells at d7 and in vivo Tmem, confirming a role for this TF in coordinating memory-associated transcriptional networks (54). Zeb1 also ranked highly in in vitro chronically stimulated P14 cells at d7, indicating potential activity in Tex biology. Similarly, effector-related TFs, such as Myb, Junb, Nfkb1, and Fli1, as well as TFs without previously defined roles in T cell differentiation, such as Mecp2, ranked highly in in vitro acutely stimulated P14 cells at d4 and in vivo Teff. Exhaustion-related TFs, such as Eomes, Nr4a2, and multiple NFAT family members, ranked highly in in vitro chronically stimulated P14 cells and in vivo Tex. Despite the relative lack of enrichment of NFAT binding motifs observed in ACRs of in vitro chronically stimulated P14 cells compared to in vivo Tex (Fig. S3D), Nfatc1 and Nfatc2 emerged as highly ranked TFs via Taiji analysis. This observation suggests that these transcriptional networks have activity and influence during chronic stimulation in vitro, though perhaps to a lesser extent than in in vivo Tex. These data are also consistent with a key role for NFAT circuitry in CD8 T cell exhaustion but suggest that in vitro settings may access this biology less efficiently than in vivo Tex.
Many transcription factors have divergent roles in different cellular and epigenetic contexts. For example, NR4A family members operate downstream of TCR activation but also have non-redundant functions in transcriptional regulation of Tex (55). Taiji analysis of in vitro generated P14 cells reflected these dual roles in both acute and chronic activation, with Nr4a2 ranking highly in in vitro acutely stimulated P14 cells at d4 and in vitro chronically stimulated P14 cells at d7 (Fig. 3F). This observation suggests persistent or preferential use of Nr4a2 activity in the trajectory toward Tex, consistent with in vivo observations (55). Taiji analysis revealed another key transcriptional circuit centered around Bhlhe40. Bhlhe40 was predicted to have divergent functions in both effector- and exhaustion-associated biology due to rank in in vitro acutely stimulated P14 cells at d4 and chronically stimulated P14 cells at d7; Bhlhe40 also ranked highly in both in vivo Teff and Tex. As such, Bhlhe40 could represent a point of convergence for transcriptional control of both Teff and Tex biology. Identification of these TFs suggests that Taiji analysis can reveal TFs with non-overlapping roles in these distinct cellular contexts.
Pooled CRISPR screening identifies novel transcriptional regulators of CD8 T cell differentiation
To leverage this in vitro model as a discovery tool and interrogate regulators of Tex, we used a pooled CRISPR screening approach (56). We applied the same sgRNA library previously used in in vivo CRISPR screening of Tex and Tmem (56, 57) to facilitate comparison of results from in vitro versus in vivo CRISPR screening. Using P14 cells from Cas9+/− GFP P14 mice, in vitro chronic and acute stimulation proceeded as described above. At ~24 hours post-activation, Cas9+/− GFP P14 cells were transduced with a pooled sgRNA VEX-expressing retrovirus (RV) library targeting 150 TFs (56, 57). Transduced P14 cells were then returned to co-culture under acute or chronic stimulation conditions in vitro. VEX+GFP+ P14 cells were sorted and sgRNA cassettes were sequenced at day 2 (baseline) and day 7 of in vitro culture (Fig. 4A). Positive and negative selection at the population level were quantified via sequencing.
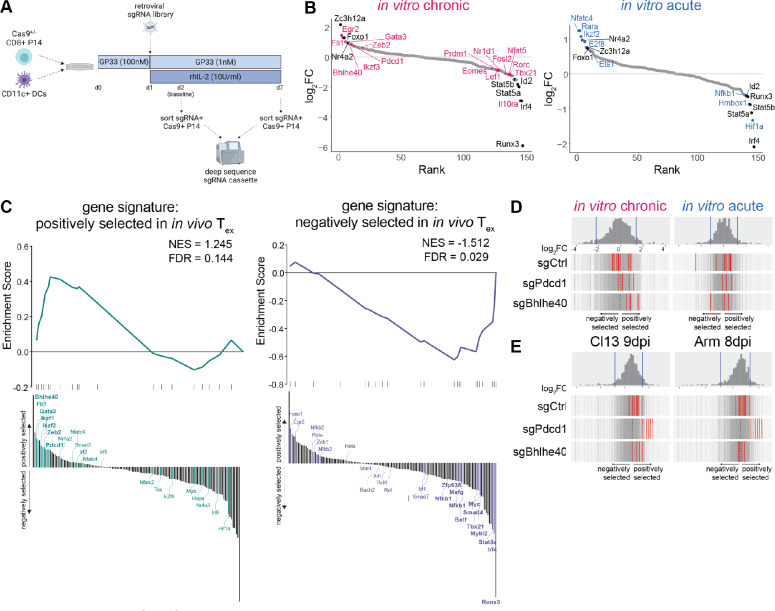
Fig. 4. Pooled CRISPR screening in in vitro chronically stimulated P14 cells identifies novel transcriptional regulators of CD8 T cell differentiation.
(A) Experiment schematic of pooled CRISPR screening in in vitro chronically stimulated P14 cells. (B) Genes targeted by pooled CRISPR screen ranked by magnitude of selection (as quantified by lfc) after acute and chronic stimulation in vitro. Colored text indicates hits unique to each condition; black text indicates hits common to both. (C) GSEA of gene sets constructed from [left] positively selected and [right] negatively selected sgRNAs from previously published pooled in vivo CRISPR screening (57). Waterfall plot illustrates rank order of hits in in vitro chronically stimulated P14 cells; bold lettering indicates leading edge genes. (D-E) Selection of individual sgRNAs against negative control genes, Pdcd1, and Bhlhe40 (D) after acute and chronic stimulation in vitro and (E) from in vivo data set (57) in PBMC at 9dpi of LCMV-Cl13 and 8dpi of LCMV-Arm. Histogram and vertical gray bars indicate distribution of all sgRNAs; vertical red bars represent indicated sgRNAs. (B-D) Representative of two experiments (two technical replicates and 2 biological replicates each).
Hits from in vitro pooled CRISPR screening included many TFs with known roles in CD8 T cell biology. For example, sgRNAs negatively selected after both in vitro acute and chronic stimulation included TFs involved in CD8 T cell activation and proliferation, such as Id2, Irf4, Runx3, Stat5a, and Stat5b (Fig. 4B), identifying these as required factors for optimal CD8 T cell differentiation in vitro. sgRNAs positively selected after both in vitro acute and chronic stimulation included Foxo1 and Nr4a2, consistent with prior literature describing these TFs in restraining initial T cell activation and effector development (55, 58–60). This screen also identified targets preferentially associated with chronic stimulation in vitro, including several TFs involved in transcriptional regulation of Tex. As a positive control for Tex-specific biology, sgRNAs against Pdcd1 were included; as expected, these sgRNAs were among the top positively selected hits after chronic stimulation in vitro. Other sgRNAs positively selected after chronic stimulation in vitro included those targeting TFs with known roles in activation and effector function, such as Fli1 and Zeb2 (54, 57, 61, 62), as well as TFs with recently identified roles in regulation of stability and/or maintenance of Tex, such as Gata3 and Egr2 (63–65) (Fig. 4B). sgRNAs negatively selected after chronic stimulation in vitro, such as Eomes, Tbx21, Prdm1, and Nfat5, reflected transcriptional targets previously implicated in Tex (26, 49, 50, 66–69). Identification of these TFs with known roles in CD8 T cell differentiation and function confirms that these in vitro models of acute and chronic stimulation can not only accommodate high-throughput assays such as pooled CRISPR screening, but also recapitulate known biology for cell types of interest.
To benchmark this in vitro CRISPR screening approach against in vivo Tex, we next compared in vitro CRISPR screen data to a previously published CRISPR screen in in vivo Tex that used the same sgRNA library (57). To quantify overlap between hits from CRISPR screening in vitro and in vivo, we generated gene sets from positively or negatively selected hits from CRISPR screening of in vivo Tex and used GSEA to evaluate enrichment of these gene sets in vitro. GSEA revealed significant enrichment of the positively selected in vivo Tex gene set in hits positively selected after chronic stimulation in vitro (Fig. 4C). Leading edge genes included TFs strongly implicated in Tex biology in vitro and in vivo, such as Bhlhe40, Fli1, Gata3, Ikzf1/2, and Zeb2. Conversely, positively selected hits in vivo that did not appear in the leading edge of overlap with in vitro Tex included Tox, Nfatc2, and Nfatc4, highlighting the relative dearth of the TOX/NFAT signaling axis in vitro. Hif1a, though strongly positively selected in in vivo Tex, was negatively selected after chronic stimulation in vitro, pointing to differential regulation of hypoxia-related transcriptional signaling, potentially due to non-hypoxic conditions in vitro. GSEA also showed significant enrichment of the negatively selected in vivo Tex gene set in hits negatively selected after chronic stimulation in vitro (Fig. 4C). Leading edge genes included TFs associated with activation and effector programs, such as Runx3, Irf4, Tbx21, Myc, Batf (Fig. 4C). Negatively selected hits in vivo that were not replicated in vitro included TFs such as Foxo1, Egr2, and Bcl6, reflecting the relative absence of transcriptional networks associated with stem-like Tex progenitors during chronic stimulation in vitro. This analysis comparing results from CRISPR screening in both chronic stimulation in vitro and in vivo Tex highlight the ability of this in vitro model of chronic stimulation to recapitulate transcriptional networks that coordinate key aspects of Tex biology. Conversely, analysis of differentially selected TFs between these two models could be useful for parsing pathways responsive to other environmental factors unique to in vivo Tex, such as NFAT signaling, inflammation, or hypoxia.
After benchmarking pooled CRISPR screening in our in vitro model of chronic stimulation to previously published in vivo CRISPR screening with the same library, we probed our in vitro dataset further to identify novel regulators of Tex. BHLHE40 was of particular interest because it was previously identified via Taiji analysis as a highly ranked TF in both in vitro chronically stimulated P14 cells and in vivo Tex (Fig. 3F). Bhlhe40 was also among the most robustly enriched leading edge genes positively selected in CRISPR screening of chronic stimulation in vitro and in vivo Tex (Fig. 4C). All sgRNAs against Bhlhe40 were positively selected after chronic stimulation in vitro, with some displaying stronger selection than guides against Pdcd1 (Fig. 4D). Conversely, sgRNAs against Bhlhe40 were not positively selected after acute stimulation in vitro, indicating biology biased toward chronic stimulation. Bhlhe40 also displayed positive selection in in vivo CRISPR screening in chronic (Cl13) but not acute infection (Arm) (Fig. 4E), confirming Tex-specific biology in vivo.
BHLHE40 is a transcriptional regulator of Tex differentiation
Pooled CRISPR screening and Taiji analysis of the in vitro model of chronic stimulation both identified BHLHE40 as a potential regulator of Tex and Teff biology. BHLHE40 has been previously implicated in several aspects of T cell biology, including regulation of CD4 T cell lineage commitment and differentiation (70, 71) and maintenance of stemness in CD4 and CD8 tumor-infiltrating lymphocytes (TIL) as well as tissue-resident memory CD8 T cells (72, 73). Moreover, BHLHE40 has recently been implicated in the response of CD8 TIL to checkpoint blockade therapy (74). However, it is unclear whether BHLHE40 functions differently in the formation of Tex versus Teff and, if so, how BHLHE40 might regulate establishment and/or maintenance of Tex. Furthermore, Tex biology includes a hierarchy of subsets linked to developmental lineage and differential response to checkpoint blockade; it is unclear where in this hierarchy BHLHE40 impacts Tex.
To determine whether BHLHE40 has the potential to differentially regulate Teff and Tex, we first used Taiji analysis to construct TF networks downstream of BHLHE40 in in vitro acutely stimulated P14 cells at d4 versus in vitro chronically stimulated P14 cells at d7. Some TFs were common to both settings; however, most TFs in downstream networks regulated by BHLHE40 were found in only one condition (Fig. S4A). For example, Bcl6, Rxra, Runx1, and several KLF family members preferentially connected downstream of BHLHE40 after acute stimulation in vitro. In contrast, after chronic stimulation in vitro, BHLHE40 connected to a set of Ets family TFs. These observations suggest cell context-specific transcriptional regulation by BHLHE40. We also performed TF co-occurrence analysis to identify TFs that might bind chromatin in tandem with BHLHE40. We found many TFs for which binding motifs co-occur within <100bp proximity of binding motifs of BHLHE40, indicating potential of these TFs to co-bind and co-regulate transcription with BHLHE40 (Fig. S4B); for example, binding motifs for BHLHE40 and BCL6 occur in close proximity at the Runx1 promoter (Fig. S4C). Because RUNX1 is capable of partially antagonizing RUNX3, which, in turn, promotes effector responses (57), our data suggests that co-repression of RUNX1 by BHLHE40 and BCL6 could allow more effective RUNX3-driven Teff circuitry in intermediate Tex. Taken together, these results highlight ways in which BHLHE40 may cooperate with other known TFs to regulate CD8 T cell differentiation.
To interrogate the role of BHLHE40 in CD8 T cell differentiation, we employed a loss-of-function approach using shRNA knockdown (KD). We first tested this approach in our in vitro model of chronic stimulation (Fig. S5A). shRNA KD led to an 82% reduction in gene expression of Bhlhe40 (Fig. S5B). In in vitro chronically stimulated P14 cells, Bhlhe40 KD resulted in a decrease in percent expression and MFI of multiple IRs, including PD-1, TIM3, and 2B4 (Fig. S5C), suggesting that BHLHE40 may promote IR expression and providing further support for a potential role for BHLHE40 in CD8 T cells during chronic stimulation.
Whereas our in vitro model of chronic stimulation allowed us to screen many TFs in parallel and identify genes of interest, including Bhlhe40, we next used the LCMV model to test if and how BHLHE40 regulates CD8 T cell differentiation and function in vivo. We thus employed shRNA KD of Bhlhe40 during viral infection with the acutely-resolving Armstrong strain (Arm) or chronic clone 13 strain (Cl13) of LCMV (Fig. 5A). Congenically distinct P14 cells were transduced with an RV expressing either a control shRNA or shRNA against Bhlhe40 and adoptively co-transferred at a 1:1 ratio (Fig. S5D) into Arm- or Cl13-infected recipient mice as described (75). At 8 days post infection (dpi), Bhlhe40 KD resulted in a competitive numerical advantage in Cl13 infection (Fig. S5E) but not in Arm (Fig. S5F). However, by 22–31dpi of Cl13, this advantage of Bhlhe40 KD increased to ~70–80% of transferred P14 cells (Fig. 5B). These data confirm results from CRISPR screening in the in vitro model in which absence of Bhlhe40 led to a proliferation and/or survival advantage in settings of chronic but not acute stimulation (Fig. 4B,D).
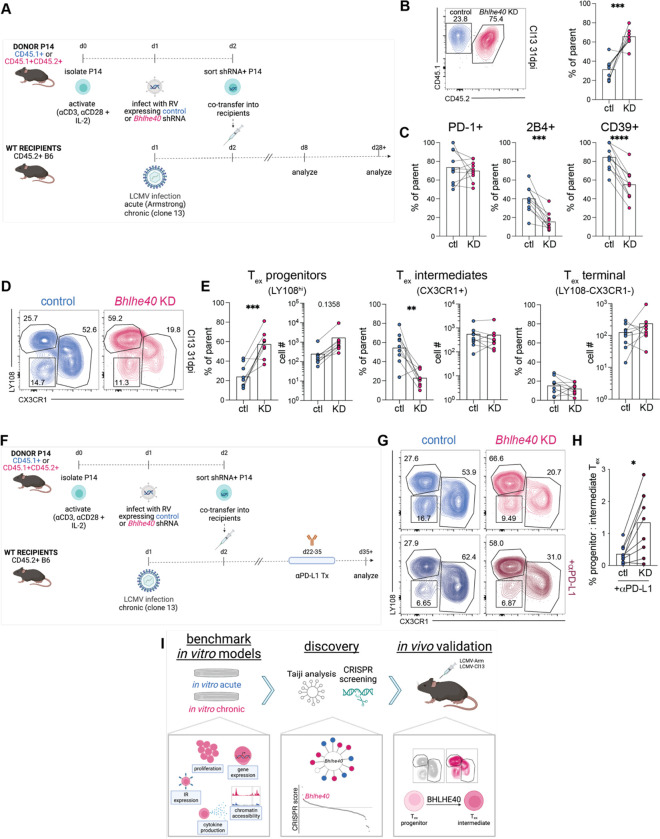
Fig. 5. BHLHE40 is a transcriptional regulator of Tex differentiation.
(A) Experiment schematic of adoptive co-transfer of control and Bhlhe40 knockdown (KD) P14 cells into LCMV Arm- or Cl13-infected recipient mice. (B) [left] Concatenated (n=10) flow cytometry plot and [right] summary data of frequency of control and Bhlhe40 KD P14 cells (gated on GFP+ CD44hi CD8+ live singlets) at 31dpi of LCMV-Cl13. (C) Percent expression of IRs in control and Bhlhe40 KD P14 cells at 31dpi of LCMV-Cl13. (D) Concatenated (n=10) flow cytometry plots of frequency of Tex subsets in control and Bhlhe40 KD P14 cells at 31dpi of LCMV-Cl13. (E) Frequency and total number of progenitor, intermediate, and terminal Tex in control and Bhlhe40 KD P14 cells at 31dpi of LCMV-Cl13. (F) Experiment schematic of adoptive co-transfer of control and Bhlhe40 KD P14 cells into LCMV Cl13-infected recipient mice, with αPD-L1 treatment or vehicle control between 22–35dpi. (G) Concatenated (n=10) flow cytometry plots of frequency of Tex subsets in control and Bhlhe40 KD P14 cells at 37dpi of LCMV-Cl13, after treatment with vehicle control [top] or αPD-L1 [bottom] from 22–35dpi. (H) Ratio of progenitor to intermediate Tex in control and Bhlhe40 KD P14 cells after αPD-L1 treatment. (I) Schematic detailing discovery pipeline. (B-G) n=10 mice, representative of 3 experiments. Significance calculated by paired two-tailed t test; *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. (B,D,G) Numbers in flow cytometry plots indicate percentage of parent population within each gate.
Next, to investigate whether BHLHE40 impacts phenotype and function early after infection, we analyzed subset distribution and differentiation states of Teff in control or Bhlhe40 shRNA-transduced P14 cells in Arm and Cl13. At 7dpi of Arm infection, shRNA KD of Bhlhe40 led to significantly decreased proportions and numbers of KLRG1+ Teff (Fig. S5G). Furthermore, Bhlhe40 KD significantly reduced per-cell protein expression (MFI) of KLRG1 (Fig. S5H). At 7dpi of Cl13 infection, before the formation of mature Tex, Bhlhe40 KD also resulted in decreased proportions of KLRG1+ Teff-like cells (Fig. S5I). These data support the idea that Bhlhe40 regulates terminal Teff differentiation during the first week of infection. However, production of TNF and IFNγ was not impacted by Bhlhe40 KD (Fig. S5J), suggesting that BHLHE40 controls differentiation state rather than effector function.
We then assessed impact of loss of Bhlhe40 on Tex differentiation at a late time point in Cl13 infection after which exhaustion had been established (49, 76). By 31dpi of Cl13, shRNA KD of Bhlhe40 reduced percent expression and MFI of some IRs, including 2B4 and CD39, but not others, such as PD-1 (Fig. 5C, Fig. S5K,L), confirming earlier observations from the in vitro model of chronic stimulation (Fig. S5C). Furthermore, because IR co-expression is associated with progression toward terminal Tex (26, 32, 33), these results suggest that Bhlhe40 may promote terminal differentiation of Tex.
We next examined the effect of Bhlhe40 KD on the differentiation hierarchy between subsets of Tex using expression of LY108 and CX3CR1: high expression of LY108 (a surrogate of TCF1 expression) (33) distinguishes the progenitor Tex subset, CX3CR1 expression identifies the intermediate and more effector-like Tex subset, and the LY108-CX3CR1- population forms the terminally differentiated Tex subset (42, 43). shRNA KD of Bhlhe40 resulted in an increase of the Tex progenitor subset: whereas roughly 25% of control-transduced P14 cells were progenitor Tex, this population increased to almost 60% after Bhlhe40 KD (Fig. 5D). This increase was seen by percent and absolute number (Fig. 5E). In contrast, loss of Bhlhe40 led to decreased proportions of the Tex intermediate subset, which represented ~50% of control-transduced P14 cells but only ~20% after Bhlhe40 KD (Fig. 5D). However, the total number of intermediate Tex was similar due to the overall numerical advantage of Bhlhe40 KD P14 cells (Fig. 5E). Bhlhe40 KD did not affect subset distribution of terminal Tex (Fig. 5D,E). Because Bhlhe40 KD resulted in accumulation of Tex progenitors at the expense of Tex intermediates, these data point to a role for Bhlhe40 in controlling conversion from Tex progenitors into effector-like Tex intermediates. We also examined the impact of Bhlhe40 gain-of-function via retroviral overexpression (Fig. S5M) on Tex differentiation and found the opposite effect: overexpression of Bhlhe40 promotes differentiation of Tex intermediates at the expense of both progenitor and terminal Tex (Fig. S5N,O).
Response to immunotherapy, such as PD-1 blockade, hinges on differentiation of Tex progenitors into Tex intermediates (24, 27, 29, 42, 43); thus, these results also implicate BHLHE40 in regulating this crucial checkpoint in the response to immunotherapy. To evaluate whether the accumulation of Tex progenitors associated with Bhlhe40 KD would prove advantageous in synergy with PD-1 pathway blockade, we deployed Bhlhe40 KD in combination with αPD-L1 treatment during Cl13 infection (Fig. 5F). Regardless of αPD-L1 treatment, Bhlhe40 KD led to increased numbers and percentages of Tex progenitors (Fig. 5G, S5P). In combination with Bhlhe40 KD, αPD-L1 treatment resulted in increased percentages of CX3CR1+ intermediates (Fig. 5G, S5P) but this increase was not significant for absolute number of cells. Although PD-1 pathway blockade increased the proportion of Tex intermediates, the accumulation of Tex progenitors created by Bhlhe40 KD remained intact: whereas the ratio of progenitor to intermediate Tex in control shRNA plus αPD-L1 was roughly 0.5:1, this ratio was inverted to approximately 1.5:1 in Bhlhe40 KD plus αPD-L1 conditions (Fig. 5H). Overall, these data suggest that, despite the ability of PD-1 pathway blockade to induce a burst of proliferation and differentiation of Tex progenitors into Tex intermediate cells, blocking PD-1 signaling is largely insufficient to override transcriptional reprogramming of this differentiation checkpoint maintained by shRNA KD of Bhlhe40.
Overall, these data suggest that in vitro screening using a model that captures a subset of Tex biology can identify targets that have specific roles at a distinct stage of in vivo Tex differentiation. This approach of chronic stimulation in vitro, from benchmarking against known Tex biology to incorporation of pooled CRISPR screening to in vivo validation, represents a discovery pipeline that can be applied to identifying additional novel regulators of Tex biology (Fig. 5I).
DISCUSSION
The inability to durably reverse CD8 T cell exhaustion remains a major barrier to the treatment of cancer and chronic viral infection. A better understanding of the molecular cues that induce and maintain exhaustion will enable development of more effective therapeutics to prevent or reverse this state of CD8 T cell differentiation. In vivo models have engendered foundational knowledge about Tex. However, in vivo models generate low numbers of Tex and are time-consuming, costly, and difficult to scale. These systems are thus less than ideal for generating large amounts of biological material for high-throughput screening techniques or approaches such as proteomics and/or metabolomics. In vitro models that can recapitulate many or even some features of Tex are an attractive alternative to in vivo models due to their scalability, speed, efficiency, and ability to be customized. Here, we established a model of in vitro chronic antigenic stimulation and comprehensively defined the functional, phenotypic, transcriptional, and epigenetic features of this model in comparison to known in vivo Tex biology. Understanding which features of exhaustion can be recapitulated and interrogated in vitro allowed us to then use this in vitro model to perform high-throughput CRISPR-based screening to identify novel transcriptional regulators of Tex.
To induce features of Tex in vitro, we used chronic antigenic stimulation as the primary driver of exhaustion biology. We opted to mimic physiological TCR engagement by stimulating P14 cells with cognate peptide presented by professional antigen-presenting cells (APCs). Our goal was to induce high antigenic stimulation with peptide presented by a professional APC, then reduce stimulation strength ~100-fold, consistent with the physiological dynamics of viral or antigen load in chronic infections (11). We also aimed to limit the amount of rest between stimulations to achieve continuous rather than intermittent stimulation. We observed, as have other studies using repeated TCR stimulation to mimic exhaustion in vitro (14, 15, 17–20), that CD8 T cells displayed decreased proliferative potential, high expression of IRs, and reduced production of effector cytokines after chronic stimulation in vitro. Benchmarking these phenotypic, transcriptional, and epigenetic features of our in vitro model of chronic stimulation against in vivo CD8 Tex revealed where this model recapitulated known features of Tex and where this in vitro approach diverged from in vivo biology. Many aspects of exhaustion biology were robustly captured by chronic stimulation in vitro: for example, in vitro chronically stimulated P14 expressed IRs at population and per-cell levels comparable to or even higher than in vivo Tex. Yet other aspects of in vivo biology were incompletely captured in vitro, including the NFAT/TOX signaling axis. However, defining which aspects of exhaustion biology were captured using this in vitro model provided a foundation to leverage this approach for discovery-based assays downstream.
A notable feature of our in vitro model of chronic stimulation is the generation of multiple phenotypic, functional, transcriptional, and epigenetic features of Tex despite relatively low gene and protein expression of TOX. Previous studies have identified TOX as a key transcriptional and epigenetic regulator essential for the formation of Tex (49, 77–79). However, because Tex fail to develop in vivo in the absence of TOX, interrogating which exhaustion-related pathways function dependently and independently of TOX has been difficult. The failure of Tex to persist in the absence of TOX likely reflects a TOX-dependent Tex program of survival and durability, in addition to epigenetic effects, in the setting of chronic antigen stimulation (49, 77, 78). However, lack of knowledge about TOX-independent aspects of the Tex program suggests that such pathways may only be apparent using in vitro models. Our in vitro model of chronic stimulation, in which many phenotypic, functional, transcriptional, and epigenetic features of Tex are generated in the absence of high and sustained expression of TOX, may represent an opportunity to mechanistically interrogate these Tex-related pathways to reveal novel therapeutic targets.
In vivo Tex form a proliferative hierarchy consisting of progenitor subsets that differentiate into downstream intermediate and terminal subsets. Chronic stimulation in vitro largely induced transcriptional, phenotypic, and functional features that overlapped robustly with those of in vivo terminal Tex, based on enrichment of a gene signature of terminally differentiated Tex, high co-expression of IRs, terminally differentiated TF signature, and increased cytotoxic potential after chronic stimulation in vitro. It will be interesting to interrogate whether P14 cells subjected to chronic stimulation in vitro first pass through progenitor and intermediate states before undergoing accelerated terminal differentiation or if these cells bypass upstream differentiation steps in favor of directly inducing features of terminal Tex. If chronic stimulation in vitro is indeed insufficient for establishment and maintenance of progenitor Tex, it is likely that the generation of cells with features of progenitor Tex in vitro may require soluble cytokine/chemokine cues or even periods of rest or “rescue” to allow for upregulation of stem- and memory-like transcriptional pathways (20, 80). Although TCF1 signaling is known to be requisite for development of progenitor Tex in vivo (81), the cell-extrinsic cues that drive this transcriptional programming remain poorly understood. Future studies could apply gain-of-function screens to in vitro modeling of Tex to address this question.
The in vitro Tex model described here has generated many insights into CD8 T cell exhaustion; however, no single in vitro model will completely capture the complex in vivo biology of Tex. An advantage of the current work was benchmarking against bona fide in vivo Tex generated via the LCMV model of chronic viral infection in which CD8 T cell exhaustion was first defined (82, 83). Comparison to in vivo Tex identified many ways in which this in vitro model recapitulates known exhaustion biology; this benchmarking also identifies ways it does not (e.g. TOX/NFAT signatures). Furthermore, other models of in vitro Tex (14, 19, 20) may capture slightly different aspects of exhaustion biology. In vitro approaches can be tuned to address different features of known in vivo biology: for example, by introducing hypoxia or known inflammatory mediators. As the potential use of in vitro models of Tex, including variations of engineered cell therapies, expands, benchmarking to define the bounds of biology accurately captured will prove useful. In vitro Tex models may also have utility in medium- to high-throughput drug or chemical screening, approaches not feasible in vivo. Thus, well-annotated in vitro Tex models should have considerable applicability due to scalability, ease of use, and ability to enable novel types of investigation into cellular mechanisms of exhaustion.
Another advantage of this and other in vitro models of T cell differentiation is the ability to apply high-throughput discovery assays due to increased efficiency, flexibility, and cell yield. We illustrated the utility of this in vitro model in combination with pooled CRISPR screening toward the discovery of novel biology. Compared to recently published in vivo CRISPR screens (56, 57, 84), we performed CRISPR screening in our in vitro model of chronic stimulation with relative logistical ease. For example, due to sgRNA library size and cell recovery requirements, in vivo CRISPR screening in an adoptive transfer model required pooling of 4–6 donor P14 mice and upwards of 30 recipient mice per time point per infection (i.e. over 120 total mice) (57). Using the same sgRNA library, we were able to perform our experiments with 2 mice each and used each P14 mouse as a biological replicate. Additionally, whereas the in vivo adoptive transfer model required 12+ hours of flow cytometric sorting (as well as proportional amounts of requisite reagents) to isolate CRISPR-edited cells, we performed sorting for these cells in under 2 hours.
Comparative analysis of transcriptional and epigenetic data from this in vitro model of chronic stimulation and in vivo Tex enabled interpretation of results from CRISPR screening within the context of which aspects of Tex biology were accurately recapitulated in vitro. For example, because the TOX/NFAT signaling axis was transcriptionally underrepresented in our in vitro model of chronic stimulation, we anticipated a lack of selection for these TFs during CRISPR screening. Similarly, because we did not incorporate modeling of hypoxia in vitro, Hif1a did not emerge as a positively selected hit, even though this TF was positively selected during in vivo CRISPR screening. However, CRISPR screening in our in vitro model of chronic stimulation identified many TFs with known roles in general CD8 T cell biology, including hits relating to T cell activation, such as Fli1 and Zeb2, as well as TFs with known roles in Tex, such as Eomes, Tbx21, and Prdm1. CRISPR screening also uncovered recently described TFs or TFs with previously undescribed roles in Tex biology, such as Egr2, Gata3, and Bhlhe40.
Both Taiji analysis and pooled CRISPR screening in our model of in vitro chronic stimulation identified BHLHE40, a novel transcriptional regulator of CD8 T cell differentiation. Prior work has shown that BHLHE40 has a role in anti-tumor CD8 T cell immunity (73). However, the mechanism of action of BHLHE40 has remained incompletely understood. In this study, we demonstrated via in vivo loss- and gain-of-function experiments that BHLHE40 controls a differentiation checkpoint between progenitor and intermediate Tex subsets. Progenitor Tex have garnered attention because of their role in seeding proliferation and differentiation events underlying response to PD-1 pathway blockade (24, 27, 29). However, intermediate Tex, due to their numerical expansion and upregulation of effector-associated genes, are postulated to be the functional cellular currency necessary to exert therapeutic control over infection or tumor burden upon checkpoint blockade (42, 43). Our finding that decreased BHLHE40 activity results in accumulation of progenitor Tex at the expense of intermediate Tex supports a key role for this TF in the transition from progenitor to intermediate Tex and indicates that intact BHLHE40 activity is likely required for formation of intermediate Tex. Notably, PD-1 pathway blockade was insufficient to override transcriptional reprogramming by Bhlhe40 knockdown. These data suggest that unimpaired function of BHLHE40 is also likely required for robust cellular responses to checkpoint blockade by facilitating increased conversion of progenitor to intermediate Tex. This observation is consistent with recent work describing decreased efficacy of checkpoint blockade therapy in genetic deficiency of Bhlhe40 in a transplantable tumor model (74). An appropriate balance between progenitor and intermediate subsets is likely required for optimal Tex function, both at steady state in established exhaustion and in response to checkpoint blockade therapy. Our finding that BHLHE40 and, possibly, other TFs can be leveraged to manipulate this balance has powerful therapeutic implications.
From benchmarking our in vitro model of chronic stimulation against known in vivo Tex to perturbing this model and identifying functionally relevant TFs that regulate Tex biology, we have demonstrated that in vitro models can increase our understanding of the molecular mechanisms that underlie Tex. This discovery pipeline can ultimately be used to uncover novel actionable pathways that can be exploited towards the therapeutic reversal of T cell exhaustion in chronic viral infection and cancer.
MATERIALS AND METHODS
Mice
Mice were maintained in a specific-pathogen-free facility at the University of Pennsylvania (UPenn). Experiments and procedures were performed in accordance with the Institutional Animal Care and Use Committee (IACUC) of UPenn. P14 transgenic mice expressing a TCR specific for the LCMV peptide DbGP33−41 were bred in-house and backcrossed onto the C57BL/6 background (Charles River); recipient WT C57BL/6 mice were purchased from Charles River. LSL-Cas9-GFP mice were purchased from Jackson Laboratory (JAX) and bred to CD4CRE and P14 mice on the JAX C57BL/6 background (referred to as C9P14). For in vitro experiments, mice were between 8–20 weeks of age; for in vivo experiments, mice were between 6–8 weeks of age. For all experiments, mice were age- and sex-matched and randomly assigned to experimental groups.
Adoptive T cell transfer
CD8 T cells were isolated from peripheral blood of donor P14 mice via gradient centrifugation with Histopaque-1083 (Sigma-Aldrich). 500 naïve P14 cells were adoptively transferred intravenously (i.v.) into 6–8-week-old recipient mice 1 day prior to infection. Recipients were of a distinct congenic background to allow for identification of donor populations. For experiments with in vivo Tex, mice were sacrificed between 22–31dpi with LCMV-Cl13.
Infections
LCMV strains Armstrong (Arm) and clone 13 (Cl13) were propagated and titers were determined as previously described (6). C57BL/6 mice were infected intraperitoneally (i.p.) with 2×105 plaque-forming units (PFU) of LCMV-Arm or i.v. with 4×106 PFU LCMV-Cl13.
In vitro chronic and acute stimulation
CD8 T cells were isolated from spleens of naïve P14 transgenic mice by homogenizing against a 70μm cell strainer. The cell suspension was then washed and passed through a 70μm cell strainer an additional time. CD8 T cells were isolated via negative selection kit (StemCell) per manufacturer’s instructions. Dendritic cells (DCs) were isolated from spleens of naïve WT C57BL/6 mice by cutting samples into small pieces and incubating at 37°C for 45 minutes with 1U/ml DNase I (Roche) and 10U/ml collagenase D (Roche) in complete RPMI (cRPMI): RPMI 1640 (Corning Cellgro) supplemented with 10% FBS, 50μM β-mercaptoethanol, 2mM glutamine (Gibco), 100U/ml penicillin (Sigma), 100μg/ml streptomycin (Sigma), 100μM non-essential amino acids (Invitrogen), 1mM sodium pyruvate (Invitrogen), and 20mM HEPES buffer (Invitrogen). Cells were then homogenized against a 70μm cell strainer and red blood cells were lysed in ACK lysis buffer (Gibco) for 5 minutes. The cell suspension was quenched and washed with PBS then passed through a 70μm cell strainer an additional time. DCs were isolated with CD11c+ Microbeads (Miltenyi) per manufacturer’s instructions then pulsed with DbGP33−41 peptide (0.1μM, GenScript) in cRPMI for 30 minutes at 37°C. CD8 P14 cells and DCs were then co-cultured in cRPMI at a 1:1 ratio (105 DCs + 105 P14 cells per well) in a flat-bottom 96-well plate as previously described (15). At days 2, 4, and 6 of co-culture, P14 cells were counted via BD Accuri, normalized to a concentration of 0.5×106 P14 cells/ml (105 P14 cells per well), and re-plated into in vitro culture. Chronically stimulated P14 cells received DbGP33−41 peptide (1nM) at days 2, 4, and 6; acutely stimulated P14 cells received initial peptide stimulation but no subsequent doses of peptide at days 2, 4, and 6. Both in vitro conditions received 10U/ml recombinant human IL-2 (Peprotech) at days 2, 4, and 6.
In vitro chronic and acute stimulation via aCD3/aCD28 was performed as previously described (18).
Flow cytometry
Ex vivo single cell suspensions were prepared by homogenizing against a 70μm cell strainer followed by red blood cell lysis with ACK lysis buffer (Gibco). In vitro single cell suspensions were prepared by resuspending cells in well with multichannel pipette. Cells were washed with staining buffer (PBS with 2% FCS) and surface staining was performed for 1 hour at 4°C in staining buffer. For intranuclear staining, permeabilization was performed using the Foxp3/Transcription Factor Fixation/Permeabilization kit (eBioscience) for 30 minutes at 4°C, following which TF staining was performed for 2 hours at 4°C. For intracellular cytokine staining (ICS), permeabilization was performed using the BD Cytofix/Cytoperm™ Fixation/Permeabilization Solution Kit (BD) for 30 minutes at 4°C, following which ICS was performed for 1 hour at 4°C. Samples were run on a BD LSR II. Voltages on the machine were standardized using fluorescent targets and rainbow beads (Spherotech). Data were analyzed with FlowJo software (v10.7.1, TreeStar). Cell sorting was performed on a BD FACSAria with a 100μm nozzle; a circulating temperature system was used at 4°C when sorting for sequencing and 37°C when sorting for in vivo transfer. A small aliquot of all sorted samples was run as a purity check.
Preparation of RNA- and ATAC-seq libraries
At day 7 of in vitro culture, live CD8 T cells were sorted to a purity of >95% for each replicate. Naïve CD8 T cells were also sorted from P14 mice. To extract RNA, 5×104 cells were resuspended in buffer RLT supplemented with β-mercaptoethanol and processed with an RNeasy Micro Kit (Qiagen) per manufacturer’s instructions. mRNA libraries were prepared using the SMART-Seq v4 Ultra Low Input RNA Kit (Takara) per manufacturer’s instructions. Extracted RNA and libraries were assessed for quality on a TapeStation 2200 instrument (Agilent). ATAC libraries were generated as described with minor changes (85). Briefly, nuclei from 5×105 cells were isolated using a lysis solution composed of 10mM Tris-HCl, 10mM NaCl, 3mM MgCl2, and 0.1% IGEPAL CA-630. Immediately following cell lysis, nuclei were pelleted in DNA Lo-Bind 1.5ml tubes (Eppendorf) and resuspended in TD Buffer with Tn5 transposase (Illumina). Transposition reaction was performed at 37°C for 45 minutes. DNA fragments were purified from enzyme solution using MinElute Enzyme Reaction Cleanup Kit (Qiagen). Libraries were barcoded (Nextera Index Kit, Illumina) and amplified with NEBNext High Fidelity PCR Mix (New England Biolabs). Library quality was assessed using a TapeStation instrument. RNA and ATAC libraries were quantified using a KAPA Library Quantification Kit and sequenced on an Illumina NextSeq 550 instrument (150bp, paired-end) on high-output flow cells.
RNA-seq data processing and analysis
FASTQ files were aligned using STAR (v2.5.2a) against the mm10 mouse reference genome. The aligned files were processed using PORT gene-based normalization (https://github.com/itmat/Normalization). Computational merging of in vitro and in vivo data sets was performed using the Combat function in the sva R package (v3.40.0). Differentially expressed genes (DEGs) were identified with DESeq2 (v1.32.0, DESeq function) using log2 fold change > 1 and adjusted p value < 0.05; genes were first filtered on minimum expression (median 20 reads per group). GSEA (39, 40) and GSVA (45) were used to calculate enrichment scores; an FDR of 0.25 was used to determine statistical significance in GSEA.
ATAC-seq data processing and analysis
The script used for processing raw ATAC-seq FASTQ data is available at the following GitHub repository: https://github.com/wherrylab/jogiles_ATAC/blob/master/Giles _Wherry_ATAC_pipeline_mm10_UPennCluster. In brief, samples were aligned to mm10 reference genome with Bowtie2 (v2.1.0). Unmapped, unpaired, and mitochondrial reads were removed using samtools (v1.1). ENCODE Blacklist regions were removed (https://sites.google.com/site/anshulkundaje/projects/blacklists). PCR duplicates were removed using Picard (v1.141). Peak calling was performed with MACS2 (v2.1.1) with an FDR q-value = 0.01. A union peak list for each experiment was created by combining all peaks in all samples; peaks had to be present in 2 of 4 technical replicates in order to be called. Overlapping peaks were merged using bedtools (v2.29.2) merge. The number of reads in each peak was determined with bedtools (v2.29.2) coverage. Peaks were annotated using HOMER (v4.11.1). Computational merging of in vitro and in vivo data sets was performed using the Combat function in the sva R package (v3.40.0). Differentially accessible chromatin regions (DACRs) were identified with the R package DESeq2 (v1.32.0, DESeq function) using log2 fold change > 1 and adjusted p value < 0.05. ATAC-seq signal tracks were generated with the gviz R package (v1.36.1). ATAC signal tracks are generated using gviz with bigwigs normalized for library size and group scaled across all samples within each dataset; peaks out of range are indicated with a contrast color on top. Scripts for peak set enrichment are available at: https://github.com/wherrylab/jogiles_ATAC/blob/master/PSEA.Rmd. In brief, bedtools (v2.29.2) intersect was used to find overlapping peaks between the experiment and peak set of interest. Peak names between the experiment and peak set of interest were unified using custom R scripts. GSEA (39, 40) was used to calculate enrichment scores; an FDR of 0.25 was used to determine statistical significance.
Transcriptional and epigenetic trajectory analysis
To evaluate transcriptional and epigenetic trajectories, genes were manually binned into patterns based on changes in gene expression and chromatin accessibility between days 0 and 4, and days 4 and 7 of in vitro culture. A log2 fold change > 1 between two timepoints was scored as 1; a log2 fold change < −1 was scored as −1; a log2 fold change between 1 and −1 was scored as 0. The relative gene expression and chromatin accessibility was determined by starting all genes at a baseline of 0, then adding these scores over time.
Taiji analysis
Taiji (v1.3) analysis was implemented as previously described (http://wanglab.ucsd.edu/star/taiji) (52, 53). Normalized ranks were compared across conditions and top TFs were chosen using cut-offs of mean > 0.0001 and fold change of 5 above the mean. TFs were visualized using the pheatmap R package (v1.0.12).
TF co-occurrence analysis
The TF-COMB python package (https://tf-comb.readthedocs.io/en/latest/) (86) was used to find TF binding sites in ACRs using the CIS-BP motif library of TFs (cisbp.ccbr.utoronto.ca) for mm10. Market basket analysis was done on these TFBSs to identify and count co-occurring transcription factors using the default window of 100bp and no allowed overlap. TFs were then filtered on minimum gene expression from RNAseq (median 20 reads per group).
Pooled CRISPR screening
Vector and library construction
SpCas9 sgRNA was expressed using pSL21-VEX (U6-sgRNA-EFS-VEX, available through Addgene). 4–5 sgRNA were designed against functional domains of each TF (56), synthesized by Integrated DNA Technologies (IDT), pooled in equal molarity, PCR-amplified, and cloned into BsmBI-digested SL21 vector as previously described (57).
In vitro retroviral transduction
CD8 T cells were isolated from spleens of naïve C9P14 transgenic mice as described above. Total splenocytes from naïve WT C57BL/6J mice were isolated via collagenase/DNase digestion as described above. Total splenocytes were pulsed with DbGP33−41 peptide (0.1μM, GenScript) in cRPMI for 30 minutes at 37°C and then co-cultured with C9P14 cells at a 10:1 ratio (106 splenocytes + 105 C9P14 cells per well) in a flat-bottom 96-well plate. 24 hours after initial stimulation, C9P14 cells were enriched using an EasySep magnetic CD8 negative selection kit (StemCell) per manufacturer’s instructions and spin-transduced for 60 minutes at 2000×g 37°C with RV supernatant containing polybrene (4mg/mL) and IL-2 (100U/mL) (75). 6 hours later, C9P14 cells were returned to in vitro co-culture with either DbGP33−41 pulsed or unpulsed splenocytes for in vitro chronic and acute stimulation conditions, respectively. Chronic and acute stimulation in vitro proceeded as described above, with cells being counted, normalized to a concentration of 0.5×106 C9P14 cells/ml (105 C9P14 cells per well), and put back into culture either with or without DbGP33−41 peptide at days 3 and 5. A portion of transduced C9P14 cells were kept in in vitro culture without splenocyte co-culture; 24 hours after transduction, sgRNA+Cas9+ cells were sorted from this population for baseline sequencing. sgRNA+Cas9+ cells were sorted from in vitro chronically and acutely stimulated P14 populations for sequencing at day 7.
Isolated library construction, sequencing, and data processing
Library preparation proceeded as previously described (57). Read counts of each individual sgRNA were calculated and compared to the sequence of reference sgRNA as previously described (56). Significant hits were calculated via MAGeCK (v0.5.8) (87) with p < 0.05 and log2 fold change < 0.5.
GSEA comparing in vitro and in vivo CRISPR screening
Gene signatures were created from significantly enriched and depleted hits (significance calculated via MAGeCK (v0.5.8) (87) with p < 0.1) from previously published in vivo CRISPR screening (57) in spleen at 14dpi with LCMV-Cl13. GSEA (39, 40) was used to calculate enrichment of these gene signatures in the total ranked order gene set from CRISPR screening in in vitro chronically stimulated P14 cells.
shRNA and overexpression vector generation
shRNA vector against Bhlhe40 was generated as previously described (88, 89) using guide sequence TAGGATACACTTGAAATCCGTG. Overexpression plasmid encoding Bhlhe40 was kindly gifted by M. M. Gubin (University of Texas MD Anderson Cancer Center). RV was generated for each construct as previously described (75) using 293T cells (ATCC).
In vivo retroviral transduction
Single cell suspensions were prepared by mechanically disrupting spleens from congenically distinct P14 mice by homogenizing against a 70μm cell strainer. CD8 T cells were enriched using an EasySep magnetic negative selection kit (StemCell) per manufacturer’s instructions. P14 cells were stimulated with αCD3 (1mg/mL; Invitrogen), αCD28 (0.5mg/mL; Invitrogen), and recombinant human IL-2 (100U/ml; Peprotech) in cRPMI. 24 hours post activation, P14 cells were spin-transduced for 60 minutes at 2000×g 37°C with RV supernatant containing polybrene (4mg/mL) and IL-2 (100U/mL) (75). Approximately 24 hours later, GFP+ cells were sorted on a BD FACSAria II. Sorted cells were washed with and resuspended in warmed PBS. P14 cells transduced with control (Krt8 shRNA or empty OE vector) or experimental RV (Bhlhe40 shRNA or Bhlhe40 OE vector) were pooled at a 1:1 ratio (2.5×104 control + 2.5×104 Bhlhe40 KD/OE P14 cells per mouse) and transferred i.v. into congenically distinct recipient mice infected with LCMV-Arm or -Cl13 one day prior.
αPD-L1 treatment
αPD-L1 (clone 10F.9G2; BioXcell) treatment (200ug/mouse) was administered i.p. every 3 days from 22dpi-35dpi for a total of 5 treatments (2); PBS was used as a vehicle control.
Immunoblots
2×106 cells per condition were sorted. Whole-cell lysates were prepared in RIPA buffer and separated on a 4–15% gel by SDS-PAGE, transferred to a PVDF membrane and blocked in 5% non-fat milk in TBST. Primary antibodies against the following proteins were used: GAPDH (1:1000; CST) and Bhlhe40 (1:500; Novus Biologicals) in 1% non-fat milk in TBST. HRP-conjugated anti-rabbit secondary antibodies were used (CST). Signal was detected using SuperSignal West Femto ECL substrate (Thermo) and imaged on a BioRad ChemiDoc Touch imaging system.
Statistical analyses
GraphPad Prism software was used for all statistical analyses. Statistics were calculated by paired two-tailed t test. The statistical details for each experiment are provided in the associated figure legends.
Supplementary Material
Acknowledgements
We thank all members of the Wherry laboratory for helpful discussions and critical analysis of the manuscript. Figure schematics were created with Biorender.com.
Funding
This work was supported by National Institutes of Health grant T32 AR007442 (JEW), Parker Institute for Cancer Immunotherapy Scholar Award (JEW), National Institutes of Health grant T32 CA009140 (JRG), Cancer Research Institute-Mark Foundation Fellowship (JRG), Australia NHMRC C.J. Martin Fellowship GNT1111469 (SFN), Mark Foundation Momentum Fellowship (SFN), National Institutes of Health grant NCI-CA234842 (ZC), National Institutes of Health F30 fellowship F30AI129263 (OK), National Science Foundation Graduate Research Fellowship (YJH), and National Institutes of Health grants AI155577 (EJW), AI115712 (EJW), AI117950 (EJW), AI108545 (EJW), AI082630 (EJW), and CA210944 (EJW).
Footnotes
Competing Interests
EJW is a member of the Parker Institute for Cancer Immunotherapy that supports research in the Wherry lab. EJW is an advisor for Danger Bio, Marengo, Janssen, NewLimit, Pluto Immunotherapeutics Related Sciences, Santa Ana Bio, Synthekine, and Surface Oncology. EJW is a founder of and holds stock in Surface Oncology, Danger Bio, and Arsenal Biosciences. OK holds equity in Arsenal Biosciences and is an employee of Orange Grove Bio. RPS is an employee of Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA
Code and data availability
RNA- and ATAC-seq data have been deposited in the NCBI Gene Expression Omnibus (GEO) database and are accessible through the GEO SuperSeries accession number GSE211015. All other relevant data are available from the corresponding author upon reasonable request.
References
- 1.McLane L. M., Abdel-Hakeem M. S., Wherry E. J., CD8 T Cell Exhaustion During Chronic Viral Infection and Cancer. Annu Rev Immunol 37, 457–495 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Barber D. L. et al. , Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 439, 682–687 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Krummel M. F., Allison J. P., CTLA-4 engagement inhibits IL-2 accumulation and cell cycle progression upon activation of resting T cells. J Exp Med 183, 2533–2540 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leach D. R., Krummel M. F., Allison J. P., Enhancement of antitumor immunity by CTLA-4 blockade. Science 271, 1734–1736 (1996). [DOI] [PubMed] [Google Scholar]
- 5.Blattman J. N., Wherry E. J., Ha S. J., van der Most R. G., Ahmed R., Impact of epitope escape on PD-1 expression and CD8 T-cell exhaustion during chronic infection. J Virol 83, 4386–4394 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Odorizzi P. M., Pauken K. E., Paley M. A., Sharpe A., Wherry E. J., Genetic absence of PD-1 promotes accumulation of terminally differentiated exhausted CD8+ T cells. J Exp Med 212, 1125–1137 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeitziner S. M., Walton S. M., Torti N., Oxenius A., Adoptive transfer of cytomegalovirus-specific effector CD4+ T cells provides antiviral protection from murine CMV infection. Eur J Immunol 43, 2886–2895 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Marzo A. L. et al. , Initial T cell frequency dictates memory CD8+ T cell lineage commitment. Nat Immunol 6, 793–799 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Badovinac V. P., Haring J. S., Harty J. T., Initial T cell receptor transgenic cell precursor frequency dictates critical aspects of the CD8(+) T cell response to infection. Immunity 26, 827–841 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wirth T. C., Pham N. L., Harty J. T., Badovinac V. P., High initial frequency of TCR-transgenic CD8 T cells alters inflammation and pathogen clearance without affecting memory T cell function. Mol Immunol 47, 71–78 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Wherry E. J., Blattman J. N., Murali-Krishna K., van der Most R., Ahmed R., Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J Virol 77, 4911–4927 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wherry E. J. et al. , Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 27, 670–684 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Schietinger A. et al. , Tumor-Specific T Cell Dysfunction Is a Dynamic Antigen-Driven Differentiation Program Initiated Early during Tumorigenesis. Immunity 45, 389–401 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKinney E. F., Lee J. C., Jayne D. R., Lyons P. A., Smith K. G., T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature 523, 612–616 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beltra J. C. et al. , IL2Rbeta-dependent signals drive terminal exhaustion and suppress memory development during chronic viral infection. Proc Natl Acad Sci U S A 113, E5444–5453 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Briceno O. et al. , Reduced naive CD8(+) T-cell priming efficacy in elderly adults. Aging Cell 15, 14–21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao M. et al. , Rapid in vitro generation of bona fide exhausted CD8+ T cells is accompanied by Tcf7 promotor methylation. PLoS Pathog 16, e1008555 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vardhana S. A. et al. , Impaired mitochondrial oxidative phosphorylation limits the self-renewal of T cells exposed to persistent antigen. Nat Immunol 21, 1022–1033 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lynn R. C. et al. , c-Jun overexpression in CAR T cells induces exhaustion resistance. Nature 576, 293–300 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber E. W. et al. , Transient rest restores functionality in exhausted CAR-T cells through epigenetic remodeling. Science 372, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scharping N. E. et al. , Mitochondrial stress induced by continuous stimulation under hypoxia rapidly drives T cell exhaustion. Nat Immunol 22, 205–215 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaech S. M., Ahmed R., Memory CD8+ T cell differentiation: initial antigen encounter triggers a developmental program in naïve cells. Nat Immunol 2, 415–422 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wherry E. J., T cell exhaustion. Nat Immunol 12, 492–499 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Blackburn S. D., Shin H., Freeman G. J., Wherry E. J., Selective expansion of a subset of exhausted CD8 T cells by alphaPD-L1 blockade. Proc Natl Acad Sci U S A 105, 15016–15021 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shin H., Blackburn S. D., Blattman J. N., Wherry E. J., Viral antigen and extensive division maintain virus-specific CD8 T cells during chronic infection. J Exp Med 204, 941–949 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paley M. A. et al. , Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science 338, 1220–1225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Im S. J. et al. , Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu T. et al. , The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci Immunol 1, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Utzschneider D. T. et al. , T Cell Factor 1-Expressing Memory-like CD8(+) T Cells Sustain the Immune Response to Chronic Viral Infections. Immunity 45, 415–427 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Li H. et al. , Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell 176, 775–789.e718 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giles J. R. et al. , Human epigenetic and transcriptional T cell differentiation atlas for identifying functional T cell-specific enhancers. Immunity 55, 557–574.e557 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blackburn S. D. et al. , Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol 10, 29–37 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beltra J. C. et al. , Developmental Relationships of Four Exhausted CD8(+) T Cell Subsets Reveals Underlying Transcriptional and Epigenetic Landscape Control Mechanisms. Immunity 52, 825–841.e828 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doering T. A. et al. , Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity 37, 1130–1144 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pauken K. E., Wherry E. J., Overcoming T cell exhaustion in infection and cancer. Trends Immunol 36, 265–276 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pereira R. M., Hogan P. G., Rao A., Martinez G. J., Transcriptional and epigenetic regulation of T cell hyporesponsiveness. J Leukoc Biol 102, 601–615 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scott-Browne J. P. et al. , Dynamic Changes in Chromatin Accessibility Occur in CD8(+) T Cells Responding to Viral Infection. Immunity 45, 1327–1340 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sen D. R. et al. , The epigenetic landscape of T cell exhaustion. Science 354, 1165–1169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subramanian A. et al. , Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mootha V. K. et al. , PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Bengsch B. et al. , Epigenomic-Guided Mass Cytometry Profiling Reveals Disease-Specific Features of Exhausted CD8 T Cells. Immunity 48, 1029–1045.e1025 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zander R. et al. , CD4(+) T Cell Help Is Required for the Formation of a Cytolytic CD8(+) T Cell Subset that Protects against Chronic Infection and Cancer. Immunity 51, 1028–1042 e1024 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hudson W. H. et al. , Proliferating Transitory T Cells with an Effector-like Transcriptional Signature Emerge from PD-1(+) Stem-like CD8(+) T Cells during Chronic Infection. Immunity 51, 1043–1058.e1044 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller B. C. et al. , Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol 20, 326–336 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hänzelmann S., Castelo R., Guinney J., GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14, 7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jadhav R. R. et al. , Epigenetic signature of PD-1+ TCF1+ CD8 T cells that act as resource cells during chronic viral infection and respond to PD-1 blockade. Proc Natl Acad Sci U S A 116, 14113–14118 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leong Y. A. et al. , CXCR5(+) follicular cytotoxic T cells control viral infection in B cell follicles. Nat Immunol 17, 1187–1196 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Pauken K. E. et al. , Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354, 1160–1165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khan O. et al. , TOX transcriptionally and epigenetically programs CD8(+) T cell exhaustion. Nature 571, 211–218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinez G. J. et al. , The transcription factor NFAT promotes exhaustion of activated CD8(+) T cells. Immunity 42, 265–278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quigley M. et al. , Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nat Med 16, 1147–1151 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu B. et al. , Epigenetic landscapes reveal transcription factors that regulate CD8(+) T cell differentiation. Nat Immunol 18, 573–582 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang K., Wang M., Zhao Y., Wang W., Taiji: System-level identification of key transcription factors reveals transcriptional waves in mouse embryonic development. Sci Adv 5, eaav3262 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guan T. et al. , ZEB1, ZEB2, and the miR-200 family form a counterregulatory network to regulate CD8(+) T cell fates. J Exp Med 215, 1153–1168 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen J. et al. , NR4A transcription factors limit CAR T cell function in solid tumours. Nature 567, 530–534 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi J. et al. , Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol 33, 661–667 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Z. et al. , In vivo CD8(+) T cell CRISPR screening reveals control by Fli1 in infection and cancer. Cell 184, 1262–1280 e1222 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Delpoux A. et al. , FOXO1 constrains activation and regulates senescence in CD8 T cells. Cell Rep 34, 108674 (2021). [DOI] [PubMed] [Google Scholar]
- 59.Hess Michelini R., Doedens A. L., Goldrath A. W., Hedrick S. M., Differentiation of CD8 memory T cells depends on Foxo1. J Exp Med 210, 1189–1200 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Staron M. M. et al. , The transcription factor FoxO1 sustains expression of the inhibitory receptor PD-1 and survival of antiviral CD8(+) T cells during chronic infection. Immunity 41, 802–814 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dominguez C. X. et al. , The transcription factors ZEB2 and T-bet cooperate to program cytotoxic T cell terminal differentiation in response to LCMV viral infection. J Exp Med 212, 2041–2056 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Omilusik K. D. et al. , Transcriptional repressor ZEB2 promotes terminal differentiation of CD8+ effector and memory T cell populations during infection. J Exp Med 212, 2027–2039 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wagle M. V. et al. , Antigen-driven EGR2 expression is required for exhausted CD8(+) T cell stability and maintenance. Nat Commun 12, 2782 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang R. et al. , Distinct epigenetic features of tumor-reactive CD8+ T cells in colorectal cancer patients revealed by genome-wide DNA methylation analysis. Genome Biol 21, 2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singer M. et al. , A Distinct Gene Module for Dysfunction Uncoupled from Activation in Tumor-Infiltrating T Cells. Cell 166, 1500–1511.e1509 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Intlekofer A. M. et al. , Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol 6, 1236–1244 (2005). [DOI] [PubMed] [Google Scholar]
- 67.McLane L. M. et al. , Role of nuclear localization in the regulation and function of T-bet and Eomes in exhausted CD8 T cells. Cell Rep 35, 109120 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shin H. et al. , A role for the transcriptional repressor Blimp-1 in CD8(+) T cell exhaustion during chronic viral infection. Immunity 31, 309–320 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Agnellini P. et al. , Impaired NFAT nuclear translocation results in split exhaustion of virus-specific CD8+ T cell functions during chronic viral infection. Proc Natl Acad Sci U S A 104, 4565–4570 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang X. O. et al. , Requirement for the basic helix-loop-helix transcription factor Dec2 in initial TH2 lineage commitment. Nat Immunol 10, 1260–1266 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu F. et al. , The transcription factor Bhlhe40 is a switch of inflammatory versus antiinflammatory Th1 cell fate determination. J Exp Med 215, 1813–1821 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li C. et al. , The Transcription Factor Bhlhe40 Programs Mitochondrial Regulation of Resident CD8(+) T Cell Fitness and Functionality. Immunity 51, 491–507 e497 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang L. et al. , Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 564, 268–272 (2018). [DOI] [PubMed] [Google Scholar]
- 74.Salmon A. J. et al. , BHLHE40 Regulates the T-Cell Effector Function Required for Tumor Microenvironment Remodeling and Immune Checkpoint-Therapy Efficacy. Cancer Immunol Res, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kurachi M. et al. , Optimized retroviral transduction of mouse T cells for in vivo assessment of gene function. Nat Protoc 12, 1980–1998 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Angelosanto J. M., Blackburn S. D., Crawford A., Wherry E. J., Progressive loss of memory T cell potential and commitment to exhaustion during chronic viral infection. J Virol 86, 8161–8170 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Alfei F. et al. , TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature 571, 265–269 (2019). [DOI] [PubMed] [Google Scholar]
- 78.Scott A. C. et al. , TOX is a critical regulator of tumour-specific T cell differentiation. Nature 571, 270–274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Seo H. et al. , TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8(+) T cell exhaustion. Proc Natl Acad Sci U S A 116, 12410–12415 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Abdel-Hakeem M. S. et al. , Epigenetic scarring of exhausted T cells hinders memory differentiation upon eliminating chronic antigenic stimulation. Nat Immunol 22, 1008–1019 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen Z. et al. , TCF-1-Centered Transcriptional Network Drives an Effector versus Exhausted CD8 T Cell-Fate Decision. Immunity 51, 840–855 e845 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zajac A. J. et al. , Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med 188, 2205–2213 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gallimore A., Dumrese T., Hengartner H., Zinkernagel R. M., Rammensee H. G., Protective immunity does not correlate with the hierarchy of virus-specific cytotoxic T cell responses to naturally processed peptides. J Exp Med 187, 1647–1657 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dong M. B. et al. , Systematic Immunotherapy Target Discovery Using Genome-Scale In Vivo CRISPR Screens in CD8 T Cells. Cell 178, 1189–1204.e1123 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bentsen M., Heger V., Schultheis H., Kuenne C., Looso M., TF-COMB - Discovering grammar of transcription factor binding sites. Comput Struct Biotechnol J 20, 4040–4051 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li W. et al. , MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 15, 554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen R. et al. , In vivo RNA interference screens identify regulators of antiviral CD4(+) and CD8(+) T cell differentiation. Immunity 41, 325–338 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Giles J. R. et al. , Shared and distinct biological circuits in effector, memory and exhausted CD8(+) T cells revealed by temporal single-cell transcriptomics and epigenetics. Nat Immunol 23, 1600–1613 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA- and ATAC-seq data have been deposited in the NCBI Gene Expression Omnibus (GEO) database and are accessible through the GEO SuperSeries accession number GSE211015. All other relevant data are available from the corresponding author upon reasonable request.