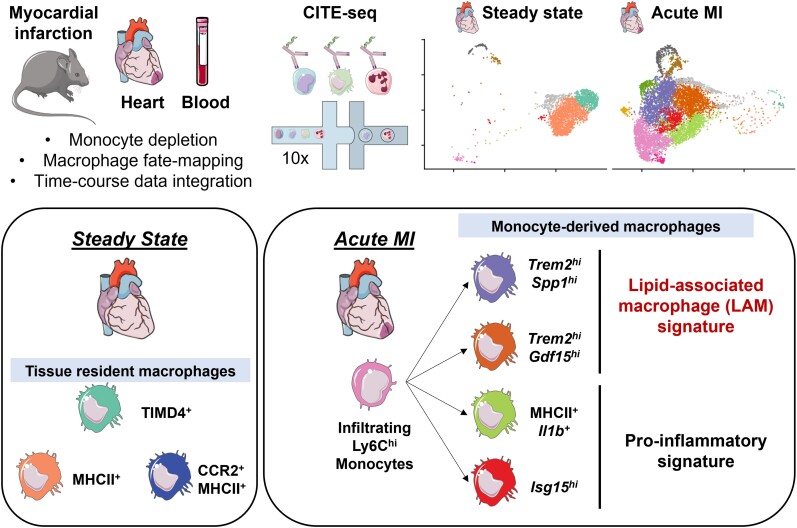
Abstract
Aims
Macrophages have a critical and dual role in post-ischaemic cardiac repair, as they can foster both tissue healing and damage. Multiple subsets of tissue resident and monocyte-derived macrophages coexist in the infarcted heart, but their precise identity, temporal dynamics, and the mechanisms regulating their acquisition of discrete states are not fully understood. To address this, we used multi-modal single-cell immune profiling, combined with targeted cell depletion and macrophage fate mapping, to precisely map monocyte/macrophage transitions after experimental myocardial infarction.
Methods and results
We performed single-cell transcriptomic and cell-surface marker profiling of circulating and cardiac immune cells in mice challenged with acute myocardial infarction, and integrated single-cell transcriptomes obtained before and at 1, 3, 5, 7, and 11 days after infarction. Using complementary strategies of CCR2+ monocyte depletion and fate mapping of tissue resident macrophages, we determined the origin of cardiac macrophage populations. The macrophage landscape of the infarcted heart was dominated by monocyte-derived cells comprising two pro-inflammatory populations defined as Isg15hi and MHCII+Il1b+, alongside non-inflammatory Trem2hi cells. Trem2hi macrophages were observed in the ischaemic area, but not in the remote viable myocardium, and comprised two subpopulations sequentially populating the heart defined as Trem2hiSpp1hi monocyte-to-macrophage intermediates, and fully differentiated Trem2hiGdf15hi macrophages. Cardiac Trem2hi macrophages showed similarities to ‘lipid-associated macrophages’ found in mouse models of metabolic diseases and were observed in the human heart, indicating conserved features of this macrophage state across diseases and species. Ischaemic injury induced a shift of circulating Ly6Chi monocytes towards a Chil3hi state with granulocyte-like features, but the acquisition of the Trem2hi macrophage signature occurred in the ischaemic tissue. In vitro, macrophages acquired features of the Trem2hi signature following apoptotic-cell efferocytosis.
Conclusion
Our work provides a comprehensive map of monocyte/macrophage transitions in the ischaemic heart, constituting a valuable resource for further investigating how these cells may be harnessed and modulated to promote post-ischaemic heart repair.
Keywords: Monocyte, Macrophage, Inflammation, Myocardial infarction, Single-cell RNA-seq
Graphical Abstract
Graphical abstract.
Time for primary review: 20 days
1. Introduction
Macrophages are critically involved in cardiac repair after myocardial infarction (MI), where they have a dual role as they can either promote tissue repair or precipitate myocardial damage.1 In the infarcted heart, cardiac macrophages form a large group of cells with different ontogenies,2 temporal dynamics and states, and delineating the contribution of each macrophage subtype to post-MI cardiac repair remains a major challenge.3 On one hand, resident tissue macrophages (RTMs) that self-renew independently of circulating monocyte input have cardioprotective functions.4,5 On the other hand, recruited monocyte-derived macrophages that numerically predominate during the acute post-MI phase adopt more heterogeneous phenotypes, and can either drive tissue repair or damage.3 Fundamental work investigating the kinetics of monocyte/macrophage transitions in the infarcted heart based on flow cytometry or bulk transcriptomics has established the notion that functionally distinct subsets of monocytes and monocyte-derived cells are present in the heart with specific temporal dynamics.6–8 More recent advances based on single-cell transcriptomic profiling have highlighted a substantial heterogeneity of monocyte-derived macrophages in various disease contexts.9 Although single-cell analysis has provided new insights into cardiac resident macrophage diversity4 or the Type I interferon (IFN) response in macrophages after ischaemic injury,10,11 a detailed analysis of the monocyte-to-macrophage transition after acute MI has not been performed previously, and the identity of monocyte-derived macrophage subsets, their dynamics, and the mechanisms regulating their acquisition of discrete states associated with specific functional capacities remain to be precisely characterized. The recent introduction of multi-modal single-cell analysis, and in particular, the simultaneous measurement of transcript expression and cell-surface markers in single cells via cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq),12 offers a unique opportunity to refine our understanding of monocyte-derived macrophage heterogeneity in the infarcted myocardium.
Here, we used CITE-seq,12 combined with targeted depletion of circulating CCR2+ monocytes and fate-mapping analysis of tissue resident macrophages, to precisely characterize circulating and cardiac monocyte-derived cell states after experimental MI. We identified monocyte-derived macrophage subsets with discrete gene expression signatures and analysed their dynamics in the infarcted heart. Notably, we identified macrophages presenting a Trem2hi lipid-associated gene expression signature acquired in the ischaemic heart, and reminiscent of a lipid-associated macrophage (LAM) signature found in other tissues in metabolic disease contexts. Altogether, our work presents a refined time-resolved map of monocyte/macrophage transitions after experimental MI, providing a valuable resource for further understanding the multifaceted roles of monocytes and macrophages in ischaemic heart repair.
2. Methods
Detailed experimental methods are available with Supplementary material online. Single-cell RNA-seq and CITE-seq data shown in this report can be browsed in a web-accessible interface: https://infection-atlas.org/Rizzo2022/.
All animal studies and numbers of animals used conform to the Directive 2010/63/EU of the European Parliament and have been approved by the appropriate local authorities (Regierung von Unterfranken, Würzburg, Germany, Akt.-Z. 55.2-DMS-2532-2-743 and 2-865). Illustrations were created using Servier Medical Art (smart.servier.com). In all experiments, animals were killed by cervical dislocation under isoflurane anaesthesia (induced by isoflurane inhalation 4.0%).
3. Results
3.1. Single-cell CITE-seq analysis of macrophages in the healthy and infarcted heart
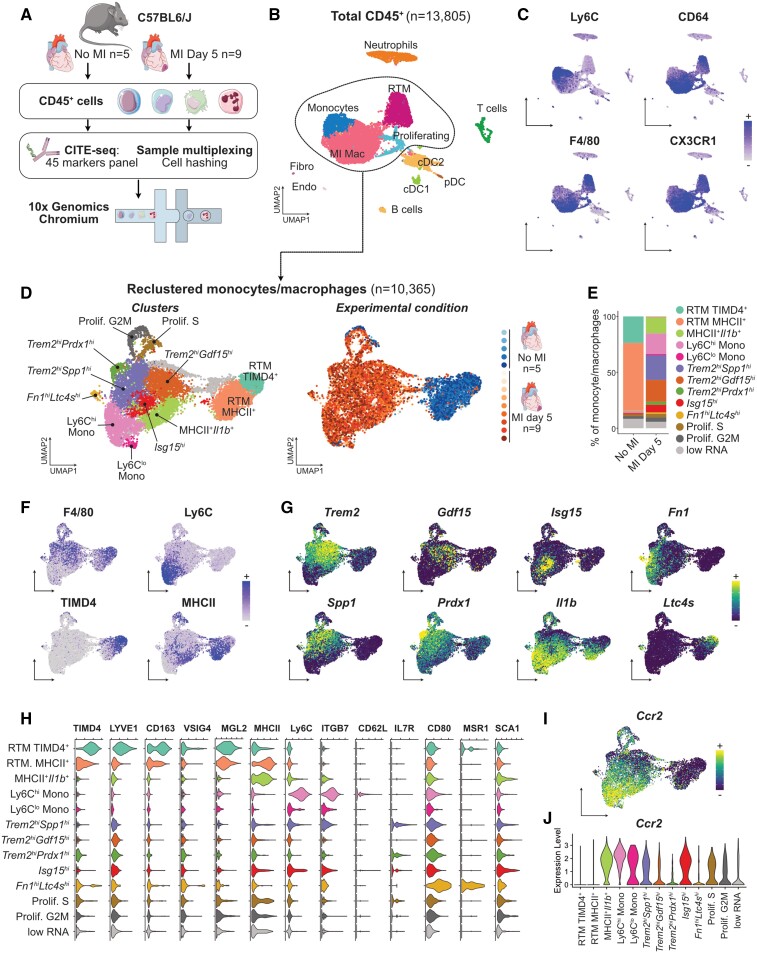
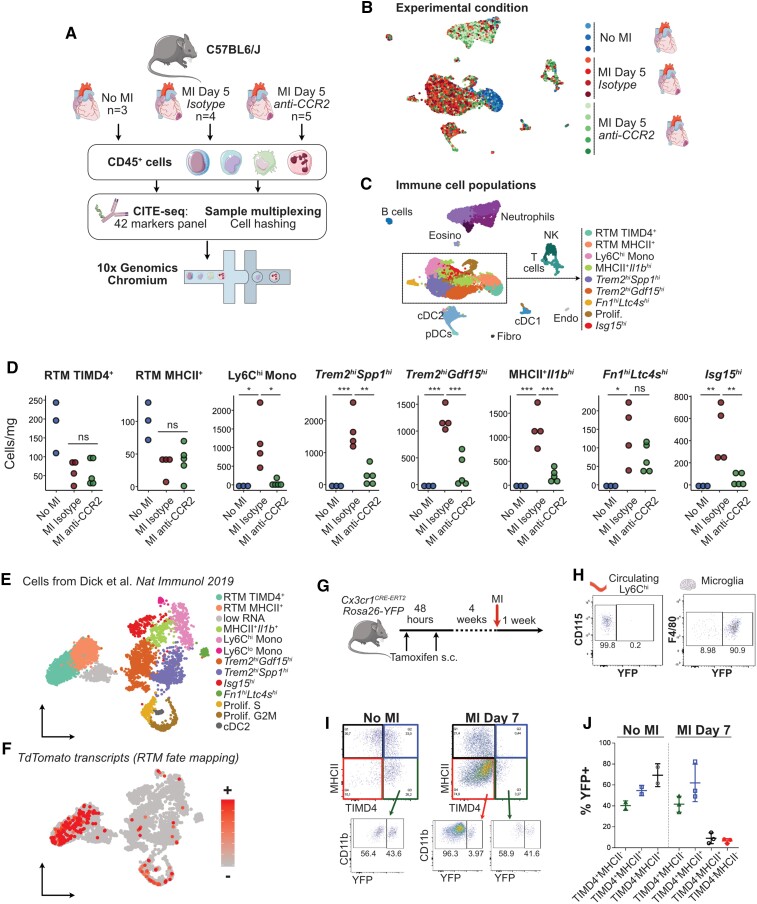
Myocardial infarction is characterized by rapid and massive infiltration of inflammatory monocytes and a shift in the cardiac monocyte/macrophage landscape.6,7 To obtain a high-dimensional characterization of monocyte/macrophage shifts in the ischaemic heart based on transcriptomic profiling and analysis of cell-surface marker expression, we performed CITE-seq analysis of cardiac CD45+ cells encompassing 45 epitopes defining all the main immune lineages and discrete subsets of innate and adaptive immune cells (see Section 2). We analysed CD45+ cells extracted from the heart of male wild-type C57BL6/J mice in the steady state (n = 5) and at 5 days after MI (n = 9), comprising a total of 13 805 cells (Figure 1A and B). We performed our main analysis at Day 5 after MI, as it represents a time point where differentiated macrophages are abundant, but active Ly6Chi monocyte recruitment is still ongoing,7 which allows capturing unsynchronized cells at various stages of the monocyte-to-macrophage transition in a single data set. Immune cell lineages were identified using canonical surface markers of T cells (CD3), NK cells (NK1.1), γδT cells (γδTCR), B cells (CD19), dendritic cells (DCs: MHCII, CD11c) including XCR1+ cDC1, plasmacytoid DCs (SiglecH), neutrophils (Ly6G), F4/80+CD64+CX3CR1+Ly6Clo macrophages comprising steady-state RTMs and macrophages appearing after MI (MI Mac), and monocytes (Ly6C+; Figure 1C, see Supplementary material online, Figure S1A). Cellular identities, including small clusters of contaminating endothelial cells and fibroblasts, were confirmed by examination of enriched transcripts (see Supplementary material online, Figure S1B). Monocytes/macrophages (n = 10 365 cells) were extracted and reclustered in silico to precisely delineate subpopulations and their distribution in the healthy and infarcted heart (Figure 1D and E). Based on differentially expressed cell-surface markers and transcripts (Figure 1F–H, see Supplementary material online, Figure S1C and D), we annotated 13 populations of monocytes/macrophages and their relative abundance in the steady-state and ischaemic heart (Figure 1D and E, see Supplementary material online, Figure S1E and F).
Figure 1.
CITE-seq analysis of the monocyte/macrophage landscape in the steady-state and infarcted heart. For all graphs in this figure, cells were obtained from n = 5 mice without MI, and n = 9 mice with MI, pooled from two independent experiments (see Section 2). (A) Experimental design summary; (B) UMAP representation of transcriptome-based clustering of n = 13 805 total cardiac CD45+ cells; (C) CITE-seq signal for the indicated monocyte/macrophage surface markers projected onto the total CD45+ cells UMAP plot; (D) n = 10 365 cells corresponding to monocytes and macrophages (including proliferating macrophages) were extracted for clustering and UMAP dimensional reduction analysis with annotated cell clusters (left) and sample of origin colour coded on the UMAP plot (right); (E) average proportions of each cluster according to experimental condition; (F) surface markers and (G) transcript expression projected onto the UMAP plot for selected markers used to identify and annotate clusters; (H) expression of the indicated surface markers in each monocyte/macrophage cluster; (I and J) expression of Ccr2 projected on the UMAP plot of monocyte/macrophages (I) and shown across clusters (J). RTM, resident tissue macrophages; MI Mac, MI-associated macrophages; (p)DC, (plasmacytoid) dendritic cell; Endo, endothelial cells; Fibro, fibroblasts.
At the steady state, we found two RTM populations, the first one represented 23.40 ± 2.28% of all steady-state macrophages and was defined as TIMD4+LYVE1+MHCIIlowFolr2hi, corresponding to previously described cardiac RTM defined as Lyve1+,13 TIMD4+,4 or more recently ‘TLF’ (TIMD4+LYVE1+Folr2+).14 Here, we termed this population RTM-TIMD4 (Figure 1D–F). The second RTM population was MHCII+ with low expression of RTM-TIMD4 surface markers and transcripts, called RTM-MHCII (60.33 ± 1.52% of all steady-state macrophages) (Figure 1D–H and see Supplementary material online, Figure S1C, E and F). We identified surface markers differentially expressed by RTM subsets: compared with MI-associated macrophages, RTM-TIMD4 and RTM-MHCII expressed higher surface MGL2 (Figure 1H). RTM-TIMD4 additionally expressed high CD163, and part of this cluster expressed VSIG4 (Figure 1H). While RTM-TIMD4 and RTM-MHCII were mostly negative for Ccr2 expression (Figure 1I and J), previous reports have identified a third cardiac RTM subset defined as CCR2+MHCII+,4 and it was recently proposed that three RTM subsets populate mouse tissues, namely ‘TLF’, MHCII+, and CCR2+ macrophages, each presenting a specific core gene expression signature conserved across organs.14 To better resolve steady-state populations of cardiac monocyte/macrophages, we reclustered cells from the steady state alone, which recovered similar populations (see Supplementary material online, Figure S2A–C). Ccr2 expression was observed in Ly6Chi monocytes (which represent monocytes within the tissue, as ensured by exclusion of intravenous leucocytes during sorting, see Supplementary Methods). Ccr2 was also detected within the RTM-MHCII+ cluster, which is in line with the existence of resident CCR2+MHCII+ macrophages in the heart.4,14 However, in our hands, CCR2+MHCII+ RTM did not form a separate cluster as proposed in Dick et al.4,14 We manually gated Ccr2+ cells within the RTM-MHCII+ population (see Supplementary material online, Figure S2D and E) and analysed expression of ‘core signature genes’ of the three RTMs conserved across organs (including the heart) as defined in Dick et al.14 Within these three bona fide RTM alone, relative expression of the core signature genes appeared mostly consistent with the distribution recently proposed in Dick et al.14 When considering all steady-state cardiac populations, relative expression of the genes of the ‘CCR2+ RTM’ signature was highest in monocytes, excepted for MHCII encoding transcripts (see Supplementary material online, Figure S2F and G).
At Day 5 post-MI, the monocyte/macrophage compartment was dominated by Ly6Chi monocytes (17.56 ± 6.58% of monocytes/macrophages at Day 5 post-MI), pro-inflammatory macrophages expressing MHCII and enriched in Il1b, Tnip3, Tlr2, or Tnfsf9 (MHCII+Il1b+; 14.51 ± 3.70% of monocytes/macrophages), and Type I IFN signature cells enriched for Isg15 (previously termed IFNIC: IFN-inducible cells10; Isg15hi, 6.25 ± 1.91%; Figure 1D–G). We also delineated three clusters sharing enrichment for Trem2 (Trem2hi; Figure 1D and G). A first Trem2hi population had high expression of Spp1 encoding osteopontin (Trem2hiSpp1hi, 21.20 ± 4.63% of monocytes/macrophages), a second cluster was enriched for Gdf15 (Trem2hiGdf15hi, 20.74 ± 5.01% of monocytes/macrophages), while a third cluster was characterized by enrichment for Prdx1 (Trem2hiPrdx1hi; 2.51 ± 1.24% of monocytes/macrophages; Figure 1D and G). Low levels of Ly6Clow monocytes and a population defined as Fn1hiLtc4shi were observed. Finally, we recovered a minor population characterized by mixed expression of tissue resident and MI-associated macrophage markers, found at similar levels in both experimental conditions, showing low transcript variety and a low RNA content (‘low RNA’ cluster, see Supplementary material online, Figure S1C and D). We interpret these cells as low-quality cells damaged during sample preparation, and did not consider them further in subsequent analyses.
As the data presented in Figure 1, results from computational integration of two independent CITE-seq experiments where samples were pooled using cell hashing15 (see Section 2), we ensured reproducibility of our observations by analysing both experiments independently with similar analysis parameters as those used initially in Figure 1. The result of the analysis is presented in Supplementary material online, Figure S3. In each separate experiment, similar populations of monocytes/macrophages were recovered, although some small populations (Trem2hiPrdx1hi and Ly6Clo monocytes enriched in, e.g. Ace or Ear2) were not resolved at the clustering resolution used (see Supplementary material online, Figure S3A–J). We also observed variability in distribution of RTM to the RTM-TIMD4+ or RTM-MHCII+ clusters, which we attribute to ‘intermediate’ TIMD4intMHCIIint cells, also observed by flow cytometry (see Supplementary material online, Figure S3L), distributing preferentially to the RTM-MHCII+ (Experiment 1) or RTM-TIMD4+ (Experiment 2) clusters in different experiments (see Supplementary material online, Figure S3B, G and K). Clustering analysis of each biological replicate independently also provided consistent results, although the analysis of much lower cell numbers in each single replicate overall yielded a lower clustering granularity (see Supplementary Analysis R Notebook).
3.2. Trem2hi macrophage subsets have a LAM transcriptional signature conserved in humans
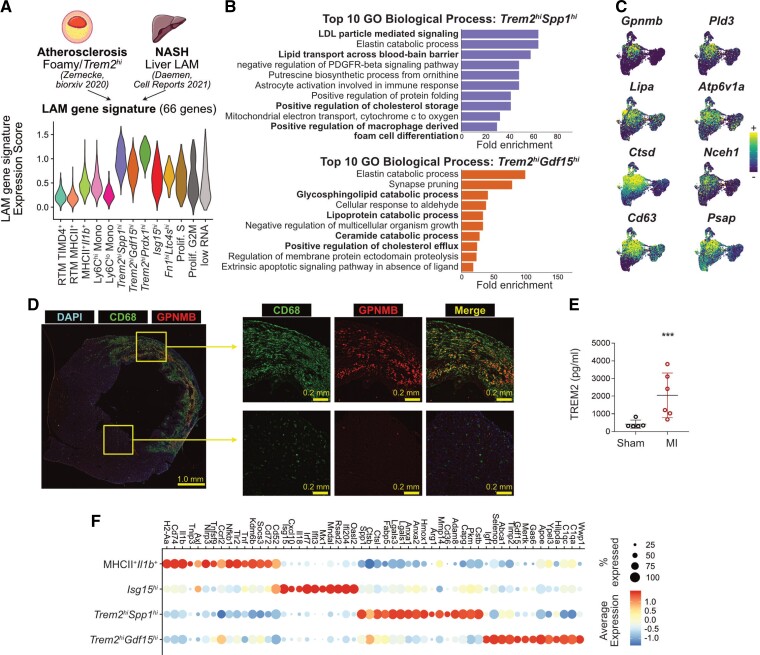
Trem2 expression is characteristic of a LAM (lipid-associated macrophage) signature previously defined by single-cell RNA-seq in metabolic disease contexts, such as in the obese adipose tissue,16 in atherosclerotic lesions,17,18 and in the liver in experimental models of NAFLD19 and NASH.20 At first glance, cardiac Trem2hi macrophage populations appeared enriched for some characteristic LAM transcripts (e.g. Gpnmb, Spp1, Ctsd, Cd63, Psap; see Supplementary material online, Figure S1C). To further explore gene expression similarities across tissues, we first extracted a list of 66 commonly enriched transcripts (see Section 2) in Foamy/Trem2hi macrophages in atherosclerotic lesions,18 and LAMs in the NASH liver,20 two lipid driven pathologies, and used these to calculate a ‘LAM gene signature’ score in cardiac macrophages. The LAM signature was highest in Trem2hiSpp1hi and Trem2hiPrdx1hi macrophages, followed by the Trem2hiGdf15hi cluster (Figure 2A). Gene ontology analysis revealed putatively enriched biological processes relevant to a LAM state, for example, ‘positive regulation of macrophage foam cell formation’ in Trem2hiSpp1hi, and ‘positive regulation of cholesterol efflux’ in Trem2hiGdf15hi macrophages (Figure 2B). LAM-signature transcripts (e.g. Gpnmb, Pld3, Nceh1, Psap) showed a clear and specific enrichment in Trem2hi macrophage populations (Figure 2C). When directly integrating scRNA-seq data obtained from the infarcted heart, NASH liver, and atherosclerotic aortas, Trem2hi MI-associated cardiac macrophage populations clustered together with Foamy/Trem2hi macrophages from atherosclerotic vessels18 and LAMs from NASH liver20 (see Supplementary material online, Figure S4A–C). Gpnmb has previously been associated with the LAM signature19–21 and appeared specifically enriched in Trem2hi MI-associated cardiac macrophages with a LAM signature (Figure 2C). At Day 5 after MI, GPNMB expressing CD68+ macrophages were observed specifically in the infarcted area of the myocardium (Figure 2D, see Supplementary material online, Figure S5), indicating that LAM-signature macrophages accumulate in the infarcted area. TREM2 protein levels were increased in the heart of infarcted mice compared with sham controls at Day 5 (Figure 2E), further confirming cardiac accumulation of TREM2 expressing cells after MI.
Figure 2.
Identification of Trem2hi macrophages with a LAM signature in the ischaemic myocardium. (A) Sixty-six LAM-signature genes were extracted from the indicated data sets and a gene expression score was applied to cardiac macrophages (LAM-signature expression score), represented here as a violin plot. The atherosclerosis data set consists of a pool of n = 12 scRNA-seq data sets (see Zernecke et al.18); the NASH data set of one scRNA-seq library (Daemen et al.20); the LAM-signature expression score was applied to cardiac macrophages (n = 10 365 cells; cells were obtained from n = 5 mice without MI, and n = 9 mice with MI, pooled from two independent experiments, see Figure 1, see Section 2). (B) Fold enrichment for the top 10 enriched Gene Ontology (GO) Biological Processes in the indicated cardiac macrophage cluster (all with adjusted P-value <0.05); macrophage clusters were determined in analysis shown in Figure 1D from n = 10 365 cells; cells were obtained from n = 5 mice without MI, and n = 9 mice with MI, pooled from two independent experiments. (C) Expression of selected LAM-signature transcripts projected on the UMAP plot of monocytes/macrophages as detailed in Figure 1D (n = 10 365 cells; cells were obtained from n = 5 mice without MI, and n = 9 mice with MI, pooled from two independent experiments). (D) Immunofluorescence labelling of CD68 and GPNMB in Day 5 infarcts with overview of a full myocardial section and high magnification images of the infarcted area and remote non-ischaemic myocardium (representative pictures shown for n = 1 heart). (E) Total levels of TREM2 detected by ELISA in extracts from the hearts of mice after sham operation (n = 5 mice) or permanent MI (Day 5; n = 6 mice). (F) Dot plot showing expression of the selected transcripts in the indicated cardiac macrophage clusters (all the transcripts shown significantly enriched in the relevant clusters with adjusted P-value <0.05); macrophage clusters were determined in analysis shown in Figure 1D from n = 10 365 cells; cells were obtained from n = 5 mice without MI, and n = 9 mice with MI, pooled from two independent experiments. NASH, non-alcoholic steatohepatitis.
The macrophage LAM signature is conserved across species in various disease contexts.16,18,22 To evaluate whether the LAM signature was also found in the diseased human heart, we analysed recently published data from human ischaemic cardiomyopathy patients (n = 3 samples ischaemic left ventricle, 1 sample remote non-ischaemic left ventricle, and 1 sample control myocardium).23 A population of macrophages enriched for characteristic genes of the LAM signature (e.g. TREM2, SPP1, CTSD, FABP5) was readily observed (see Supplementary material online, Figure S4D and E).
We further analysed the gene expression signature of the major Trem2hiSpp1hi and Trem2hiGdf15hi populations, relative to other abundant MI-associated macrophages, that is MHCII+Il1b+ and Isg15hi clusters, with a focus on experimentally validated effectors (Figure 2F). MHCII+Il1b+ macrophages were enriched in transcripts associated with a pro-inflammatory and pathogenic profile (e.g. Axl,24Nlrp325), while Isg15hi highly expressed Il1826 in addition to their typical Type I IFN response signature (Isg15, Irf7, Cxcl10).10,11 In contrast, Trem2-enriched populations highly expressed genes involved in immune modulation, tissue repair and efferocytosis (Trem2hiSpp1hi: Hmox1,27Arg1,28Anxa129; Trem2hiGdf15hi: Igf1,30Gdf15,31Mertk,32Timp2,33Apoe34). Both Trem2hi populations were enriched for expression of the profibrotic, LAM-signature transcript19Spp1 (with highest levels observed in Trem2hiSpp1hi). Trem2hiSpp1hi were additionally enriched for profibrotic Mmp14 (encoding MT1-MMP)35 (Figure 2F). Altogether, our data show the accumulation of two populations of Trem2hi macrophages in the ischaemic heart harbouring a LAM signature also identified in other organs and disease contexts and in the diseased human heart.
3.3. Dynamics of monocyte/macrophage populations in the infarcted heart
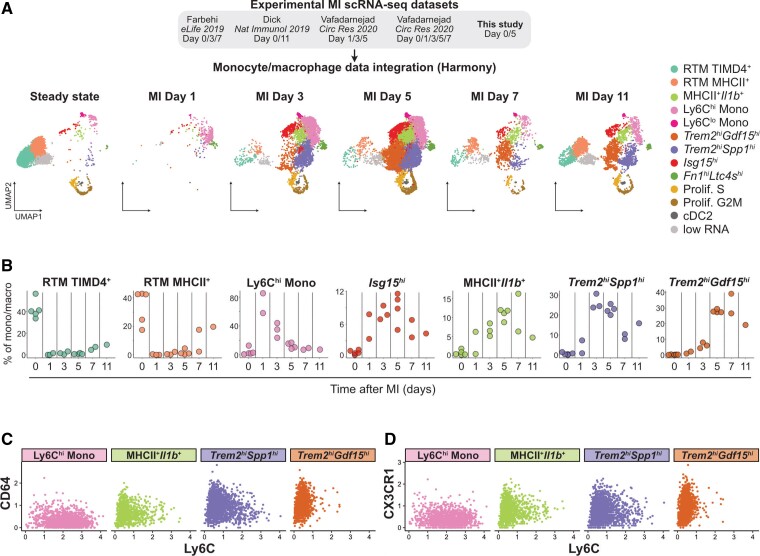
To resolve the kinetics of MI-associated macrophage accumulation, we integrated our data with existing scRNA-seq data sets of cardiac macrophages from the steady state and at 1, 3, 5, 7, and 11 days after MI4,36,37 (Figure 3A and see Supplementary material online, Figure S6A and B), representing a total of 24 637 cells. Similar MI-associated populations were recovered, although the minute Trem2hiPrdx1hi population grouped with Trem2hiSpp1hi macrophages at this clustering resolution (see Supplementary material online, Figure S6C). We observed dynamic transitions within the monocyte/macrophage compartment (Figure 3A and B). Trem2hiSpp1hi cells peaked at Days 3–5 post-MI, while Trem2hiGdf15hi cells peaked at Days 5–7 (Figure 3A and B). Infiltration of MHCII+Ilb1+ and Isg15hi macrophages also peaked at Days 3–7. At Days 7 and 11, partial recovery of RTM subsets was observed (Figure 3A and B). To better resituate our observations relative to previous analyses of monocyte/macrophage dynamics in the infarcted heart performed using flow cytometry,6 we analysed the expression of Ly6C, a monocyte surface marker, vs. expression of macrophage differentiation markers (CD64, CX3CR1, and F4/80) on monocytes and the major MI-associated macrophage populations (Figure 3C and D, see Supplementary material online, Figure S6D). Ly6Chi monocytes where Ly6ChiCD64lowCX3CR1lowF4/80low and Trem2hiGdf15hi cells appeared as fully differentiated macrophages (Ly6ClowCD64hiCX3CR1hiF4/80hi), while Trem2hiSpp1hi had a Ly6CintCD64intCX3CR1intF4/80int profile suggestive of a monocyte-to-macrophage intermediate state (Figure 3C and D, see Supplementary material online, Figure S6D). Altogether, these data suggest that the Trem2hiSpp1hi population represents a monocyte-to-macrophage intermediate state while Trem2hiGdf15hi cells are differentiated macrophages, and that these populations sequentially peak in the infarcted heart.
Figure 3.
Monocyte/macrophage dynamics in the infarcted heart. (A) The indicated MI data sets were integrated using Harmony and monocyte/macrophage population identified; the resulting clustering analysis and UMAP plot are shown split according to time point after MI with monocyte/macrophage clusters colour coded; n = 24 637 total cells from n = 6 independent scRNA-seq data sets. (B) Proportion of the indicated clusters within total monocyte/macrophage over the post-MI time continuum, calculated from the integrated data set. Each data point represents proportion of the indicated cluster at the indicated time point in independent scRNA-seq libraries (see also Supplementary material online, Figure S5). (C) CITE-seq signal for Ly6C vs. CD64 and (D) Ly6C vs. CX3CR1 in the indicated cell populations (analysis in C and D performed on monocyte/macrophage clusters determined in analysis shown in Figure 1D from n = 10 365 cells; cells were obtained from n = 5 mice without MI, and n = 9 mice with MI, pooled from two independent experiments).
3.4. Trem2hi macrophage populations originate from recruited monocytes
Macrophage origin (i.e. self-renewing tissue resident macrophage vs. monocyte-derived macrophage) has been proposed as a major driver of cardiac macrophage function.3 We thus investigated the origin of the major MI-associated macrophage populations. High Ccr2 expression in MHCII+Ilb1+ and Isg15hi clusters (Figure 1I and J), clearly indicates a monocytic origin. Trem2hiSpp1hi and Trem2hiGdf15hi macrophages expressed intermediate and low levels of Ccr2, respectively (Figure 1I), and lacked expression of surface markers associated with RTM subsets (CD163, TIMD4, LYVE1, VSIG4, MHCII; Figure 1), suggesting a monocytic origin. We hypothesized that Trem2hi macrophages originate from Ly6ChiCcr2+ monocytes that lose Ccr2 expression upon differentiation in the heart.
To test this hypothesis, we first depleted circulating Ly6Chi monocytes in mice using the anti-CCR2 monoclonal antibody MC-21.38 Twenty-four hours after a single injection, Ly6Chi monocytes were efficiently depleted from the bloodstream as previously described38 (see Supplementary material online, Figure S7A), and MI was induced. Monocyte depletion was maintained by daily i.p. injections of anti-CCR2 until Day 5 after MI (see Supplementary material online, Figure S7B), where cardiac CD45+ cells were sampled for CITE-seq analysis (Figure 4A–C). Analysis of CITE-seq data identified the main CD45+ cell populations and monocyte/macrophage subsets (Figure 4C) similar to those described above (Figure 1). While levels of RTM subsets were not affected by CCR2+ cell depletion, known circulating monocyte-derived populations (Ly6Chi monocytes, MHCII+Il1b+ macrophages, Isg15hi) were strongly reduced in the infarcted heart at Day 5. The Fn1hiLtc4shi population was not affected, indicating that it is independent of CCR2+ monocyte recruitment. We furthermore noted a drastic reduction of Trem2hiSpp1hi and Trem2hiGdf15hi macrophages counts, indicating that accumulation of these cells is dependent on circulating monocyte infiltration (Figure 4D). Additionally, cDC139 and cDC2, proposed to be Ccr2 dependent in inflammatory contexts40 were reduced (see Supplementary material online, Figure S7D). Cardiac neutrophils, T cells, NK cells, and pDC numbers were not affected by anti-CCR2 (see Supplementary material online, Figure S7C).
Figure 4.
MI-associated macrophage populations originate from recruited CCR2+ monocytes. (A) Schematic representation of the experimental design; (B and C) UMAP representation of scRNA-seq analysis (n = 10 831 cells) with (B) sample of origin and (C) biological identity of cell clusters colour coded on the UMAP plot; (D) absolute counts of the indicated cell clusters (per mg of cardiac tissue); data shown in A–D were obtained from one experiment with n = 3 mice without MI, n = 4 mice at Day 5 after MI treated with isotype control; n = 5 mice at Day 5 after MI treated with anti-CCR2; (E) annotated UMAP plot of cells from Dick et al.4 (n = 5802 cells from n = 1 scRNA-seq data set from mice without MI and n = 1 scRNA-seq data sets from mice at 11 days after MI) extracted from integrated data analysis shown in Figure 3 and (F) identification of TdTomato+ fate mapped RTMs, cells ordered according to transcript detection, that is, cells with detectable transcripts moved to front of the plot; (G) experimental setup for CX3CR1-based fate mapping of tissue resident macrophages; (H) recombination controls in Ly6Chi monocytes and microglia after the 4 weeks washout period; (I and J) fate mapping of cardiac macrophages before and at 7 days post-MI, pre-gated on live CD45+CD11b+F4/80hiLy6Clow. Data shown in G–J were obtained in one experiment with n = 2 mice without MI and n = 3 mice at Day 7 after MI.
To further confirm that Trem2hi macrophage populations derive from monocytes, we then analysed the expression of TdTomato transcripts that mark RTM in pulse-labelled Cx3cr1CreERT2 mice in Dick et al.4 from our integrated analysis (Figure 4E and F). TdTomato transcripts mapped preferentially to tissue resident and proliferating macrophages, while only scattered cells were observed in Trem2hi subsets and MHCII+Il1b+ cells (Figure 4E and F). We additionally performed fate-mapping analysis of RTM and MI-associated macrophages by flow cytometry in tamoxifen-pulsed Cx3cr1creERT2-Rosa26EYFP mice41 after a 4-week washout period (Figure 4G), when circulating Ly6Chi monocytes had lost YFP expression while microglia were >90% YFP+ (Figure 4H). Only partial recombination was observed in long living macrophages such as TIMD4+MHCII− RTM (40.35 ± 4.45%) in the steady state, which remained stable after MI (41.63 ± 7.65% YFP+ in TIMD4+MHCII− RTM; Figure 4I and J). At 7 days after MI TIMD4−MHCII− macrophages, most MI-associated macrophage populations including Trem2hi subsets, as defined by CITE-seq (Figure 1), were overwhelmingly YFPneg (93.47 ± 2.25%; Figure 4I and J). Although partial tamoxifen-induced recombination in RTM represents a limitation of our analysis, these data nevertheless suggest that pre-existing RTM do not represent a major source of MI-associated TIMD4−MHCII− macrophages (Figure 4I and J). Altogether, our complementary approaches of circulating CCR2+ monocyte depletion, fate mapping of RTM in previously published scRNA-seq data, and fate mapping of RTM by flow cytometry indicate that Trem2-enriched, MI-associated macrophage subsets originate from recruited monocytes.
3.5. The macrophage Trem2hi/LAM signature is acquired in the ischaemic heart
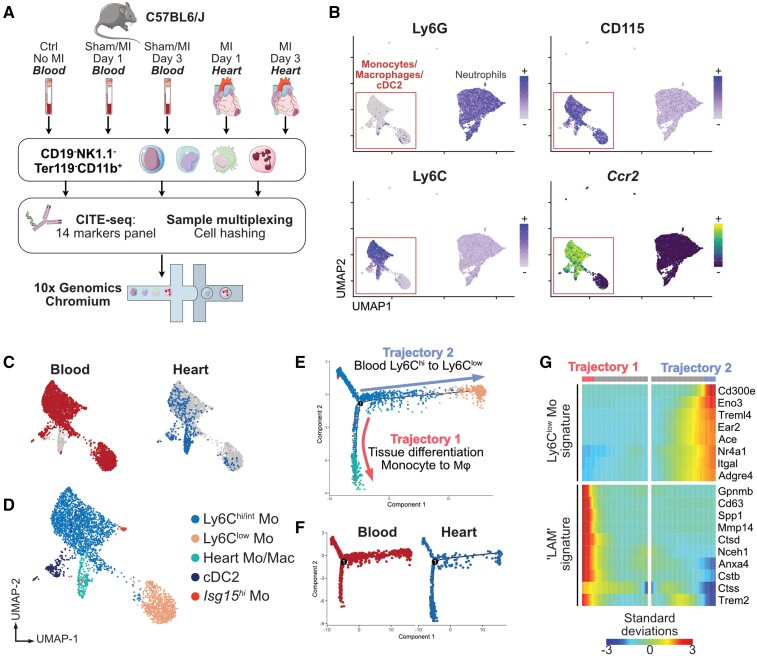
We next investigated whether MI-associated cardiac monocyte/macrophage states could be acquired remotely, notably via the production of ischaemic injury-associated atypical monocytes arising from emergency myelopoiesis,42 that would transit in the bloodstream before infiltrating the myocardium. We performed CITE-seq analysis of CD19−NK1.1−Ter119−CD11b+ cells from the blood of control mice, sham-operated animals at 1 and 3 days post-surgery, and mice with MI at 1 and 3 days. Cardiac cells from the ischaemic heart at 1 and 3 days post-MI were also included37 (Figure 5A).
Figure 5.
Monocytes/macrophages acquire the Trem2hi LAM signature in the ischaemic heart. (A) Experimental design overview; (B) Ccr2 transcript expression and CITE-seq signal for the indicated surface markers in blood and heart CD19−NK1.1−Ter119−CD11b+ cells projected on the UMAP plot (n = 9848 cells); (C) tissue of origin of single cells corresponding to monocytes/macrophages/cDC2 (n = 3378 cells) projected on the UMAP plot; (D) clustering analysis and annotation of cell clusters; (E) pseudotime analysis of monocytes/macrophages in Monocle, cell identity colour-code identical to D (n = 3378 cells); (F) pseudotime analysis split according to tissue origin; (G) heatmap of pseudotime gene expression variation for selected genes on branches of the pseudotime tree (as indicated on E; n = 3378 cells; all indicated genes show statistically significant variation). All data shown in Figure 5 were obtained from n = 1 pooled cell preparation per experimental condition, grouped in a single (n = 1) multiplexed scRNA-seq data set.
We first investigated remote priming of blood monocytes by MI by separately reclustering cells obtained from the peripheral blood. This revealed expansion of injury-associated Ly6Chi monocytes enriched for Chil3 (encoding Chitinase-like protein 3 also known as Ym1) and granulocyte marker genes (Lcn2, Prtn3) induced by MI, and also sham surgery at 1 day after MI (Chil3hi monocytes, see Supplementary material online, Figure S8A–E). Bulk-sorted bone marrow Ly6Chi monocytes also showed increased levels of Chil3, Lcn2, and Prtn3 at 3 days after MI (see Supplementary material online, Figure S8F). These results are consistent with the emergence of Ym1+ monocytes enriched in granulocyte marker transcripts induced by tissue injury43 or during emergency myelopoiesis.44 Although we noted a slight increase of the LAM-signature expression score in Chil3hi monocytes compared with baseline Ly6Chi monocytes, most LAM-signature transcripts were expressed at low levels and only two genes (Gapdh, Lpl) were significantly enriched in Chil3hi monocytes compared with baseline Ly6Chi monocytes (see Supplementary material online, Figure S8G).
To analyse the acquisition of the LAM signature by monocytes infiltrating the ischaemic heart, we analysed blood and cardiac CD115+Ly6G− cells together (3378 cells; Figure 5). One cell cluster contained almost only cells extracted from the ischaemic heart and expressed Ccr2 indicating an infiltrating monocyte origin (Heart Mo/Mac, Figure 5A–D). Pseudotime ordering of monocytes/macrophages in Monocle45 delineated two major trajectories (Figure 5E and F), with blood cells expectedly following a Ly6Chi to Ly6Clow trajectory with the acquisition of Ly6Clow monocyte marker genes (Ace, Treml4, Nr4a1, Itgal; Figure 5G), while cells infiltrated in the heart acquired expression of Trem2 and transcripts associated with the LAM signature (Figure 5G). These data indicate that MI induces a peripheral shift of Ly6Chi monocytes towards a Chil3hi granulocyte-like state in the blood, while in the ischaemic heart monocyte-derived macrophages acquire the full Trem2hi LAM signature.
3.6. Monocyte-to-Trem2hi macrophage transition in the heart and potential inducers of the macrophage LAM signature
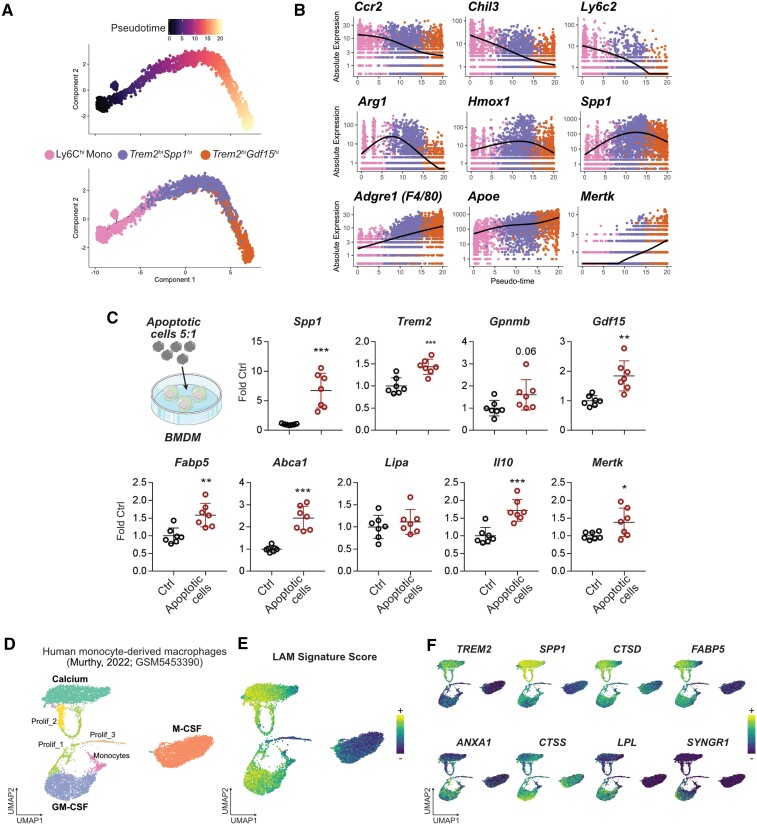
Based on our observations that (i) Trem2hiSpp1hi and Trem2hiGdf15hi macrophages sequentially peak in the heart, (ii) Trem2hiSpp1hi likely represents monocyte-to-macrophage differentiation intermediates, while Trem2hiGdf15hi is differentiated macrophages, (iii) Trem2hiSpp1hi and Trem2hiGdf15hi macrophages share features of a LAM gene expression signature, (iv) cells corresponding to Trem2hiSpp1hi and Trem2hiGdf15hi macrophages are not observed in the bloodstream (i.e. they acquire their gene expression signature in the injured tissue), and (v) Trem2hiSpp1hi and Trem2hiGdf15hi macrophages are lost upon depletion of circulating monocytes, we performed pseudotime analysis in Monocle,45 following the assumption that Trem2hiSpp1hi and Trem2hiGdf15hi macrophages originate from Ly6Chi monocytes, and might represent different states along a monocyte-to-macrophage differentiation trajectory (Figure 6A). This analysis yielded a putative trajectory with Ly6Chi monocytes at its beginning, and Trem2hiGdf15hi macrophages at its end, with progressive loss of monocyte marker genes (Ccr2, Chil3, Ly6c2) and acquisition of macrophage differentiation markers (Adgre1, Mertk, Apoe), while Spp1 and genes involved in the differentiation of monocytes to alternatively activated macrophages (e.g. Hmox1, Arg1) were transiently increased (Figure 6B). These results suggest that Trem2hiSpp1hi cells are monocyte/macrophage intermediates that may represent a transitional state towards fully differentiated Trem2hiGdf15hi macrophages.
Figure 6.
Monocyte transition to Trem2hi macrophages and potential inducers of the LAM signature. (A) Pseudotime trajectory analysis of Ly6Chi monocytes, Trem2hiSpp1hi and Trem2hiGdf15hi macrophages. (B) Expression of the indicated transcripts according to pseudotime and colour coded by cell population of origin. In A and B, cells belonging to the indicated clusters (n = 4633 total cells) were extracted from the analysis shown in Figure 1D (n = 10 365 cells obtained from n = 5 mice without MI, and n = 9 mice with MI, pooled from two independent experiments). (C) Expression of the indicated transcripts in mouse bone marrow–derived macrophages (BMDM) in control condition or after overnight exposure to apoptotic cells (Apo = apoptotic thymocyte at a 5:1 apoptotic cell:macrophage ratio). Each data point represents macrophages from one mouse assayed in technical duplicates, total n = 7 per condition, pooled from two independent experiments (*P < 0.05; **P < 0.01; ***P < 0.001). (D) Reanalysis of data from Murthy et al.48 with UMAP of in vitro human monocyte-derived macrophages differentiated in the presence of calcium, GM-CSF or M-CSF. (E) LAM-signature score projected on the UMAP plot. (F) Expression of the indicated LAM-signature transcripts induced by calcium (top) or GM-CSF (bottom).
We next investigated potential inducers of the LAM signature. Based on the gene expression profile of cardiac Trem2hi macrophages enriched for phagocytic genes and with low expression of pro-inflammatory cytokines, we hypothesized that efferocytosis, a major regulator of macrophage anti-inflammatory gene expression in the ischaemic heart,46 might be involved in acquisition of the LAM signature. Expression of Spp1, Trem2, Gdf15, and Fabp5 was increased in mouse bone marrow–derived macrophages after exposure to apoptotic cells, alongside previously documented efferocytosis-induced genes such as Mertk, Abca1, or Il1046,47 (Figure 6C). A recent report48 showed that exposing human monocyte to calcium, which is released from necrotic cells to activate macrophages,49 leads to their differentiation into macrophages producing high levels of SPP1, a marker of Trem2hi macrophages. Reanalysing data from Murthy et al.,48 we observed that exposure to calcium was associated with high expression of not only SPP1, but also many characteristic genes of the LAM signature such as TREM2, CTSD, and FABP5, while monocyte-derived macrophages exposed to GM-CSF also acquired some markers of the LAM signature (Figure 6D–F). Altogether, these data suggest that phagocytosis of dead cells, and exposure to damage associated molecular patterns released by dead cells, might be involved in macrophage acquisition of the Trem2hi LAM signature.
4. Discussion
Using single-cell CITE-seq analysis of circulating and cardiac monocytes/macrophages, combined with fate mapping and CCR2+ monocyte depletion, we mapped monocyte/macrophage dynamics and characterized monocyte-derived macrophage heterogeneity in experimental MI. This allowed us to precisely delineate MI-associated monocyte-derived macrophage populations in the ischaemic heart, including Trem2hi macrophage subsets with a tissue-acquired LAM gene expression signature.
We identified previously described populations of TIMD4+, MHCII+, and CCR2+ RTMs,4,14 and could confirm CD16350 or identify novel surface markers (MGL2, VSIG4) of these populations, that may be useful for cardiac macrophage characterization by flow cytometry or targeting of specific resident macrophage subsets. Consistent with previous reports, these populations almost entirely vanished from the infarcted heart immediately after MI.4,51
Rapid shifts in monocyte/macrophage populations after acute MI had previously been characterized by flow cytometric analyses (e.g. Nahrendorf et al.6 or Weinberger et al.52). Our CITE-seq and time-dependent scRNA-seq characterization of cardiac monocytes/macrophages provide a refined and accurate picture of these shifts and identify previously unrecognized MI-associated macrophage states. Notably, we confirm that bona fide Ly6Clow monocytes only infiltrate the heart in low numbers,53 and identified a population of Fn1hiLtc4shi macrophages with a discrete gene expression signature and surface expression of some RTM markers (TIMD4 and VSIG4). The origin, localization, and function of these macrophages remain to be determined. The major populations of MI-associated macrophages comprised two pro-inflammatory populations (Isg15hi and MHCII+Il1b+), and three Trem2hi macrophage populations with low inflammatory gene expression. All the major MI-associated macrophage populations originate from infiltrating monocytes as determined by CCR2+ cell depletion and fate-mapping analysis.
Trem2hi macrophages comprised three subpopulations (two large populations termed Trem2hiSpp1hi and Trem2hiGdf15hi, and a smaller Trem2hiPrdx1hi subset), and showed gene expression features reminiscent of a LAM signature previously observed in obese adipose tissue,16 atherosclerotic lesions,17,18 and the liver in models of NAFLD19 and NASH.20 Cardiac Trem2hi LAM-signature macrophages were derived from monocytes, similar to previous observations in the liver,19,20 and adipose tissue.16 However, the LAM signature is also shared by TREM2-dependent disease-associated microglia,54 indicating that its acquisition does not depend on macrophage origin and is mostly driven by the local tissue environment. Features of the LAM transcriptional signature may not only be induced by pathological lipid loading but rather more generally induced in contexts of tissue damage. In line, our data indicate that Trem2hi LAM-signature macrophage populations likely differentiate from monocytes in injured areas of the myocardium, as GPNMB, a highly specific marker of the LAM signature, was exclusively seen on CD68+ macrophages within the infarcted area of the heart, but not in the infarct border zone or remote non-affected tissue. Major transcriptional hubs involved in the regulation of lipid homeostasis are activated also in response to efferocytosis of dead cells, such as the liver-X-receptor pathway47 which was recently implicated in microglial acquisition of the LAM gene expression in response to chronic phagocytic challenge.55 This raises the possibility that macrophages with high efferocytosis activity, and lipid loading from dead cell engulfment, acquire a LAM signature. In line, exposure of bone marrow–derived macrophages to apoptotic cells in vitro induced expression of several characteristic LAM transcripts. More recently, the LAM signature was also observed in steady-state bile-duct macrophages, and LAM genes were induced by lipids during in vitro monocyte-to-macrophage differentiation,21 indicating that also homeostatic exposure to high lipid loads may drive acquisition of a LAM signature. Acquisition of the LAM signature might not only depend on lipid-related pathways, but also be driven by other microenvironmental cues. Exposure to calcium, a damage associated molecular pattern released by necrotic cells,48 strongly induced expression of characteristic LAM markers in human monocyte-derived macrophages. Specific inflammatory cytokines might dictate the monocyte-to-macrophage transition in the heart, as recently shown for IFN-γ and GM-CSF in the context of neuro-inflammation.56In vitro, our data indicate that some features of the LAM signature can be driven by GM-CSF. Importantly, macrophages with a LAM signature were observed in human ischaemic cardiomyopathy samples. While this represents a different pathological context compared with acute cardiac ischaemia in our mouse model, this provides proof-of-concept that the TREM2+ LAM signature can be observed in the disease human heart, consistent with the LAM signature also being observed in humans in atherosclerosis,18 adipose tissue,16 and in the steatotic human liver,21 indicating conservation of this macrophage state across species and disease contexts.
In the ischaemic mouse heart, features of the Trem2hi LAM signature were shared by three macrophage subpopulations. A small population was highly enriched for Prdx1 and genes involved in iron handling (Ftl1, Fth1, Slc40a1, Slc48a1). Two other Trem2hi populations represented the most abundant MI-associated macrophages and were characterized as Trem2hiSpp1hi and Trem2hiGdf15hi. Their sequential presence in the ischaemic heart, as well as expression patterns of Ccr2 and monocyte/macrophage differentiation markers, suggest that Trem2hiSpp1hi cells are a monocyte/macrophage intermediate giving rise to fully differentiated Trem2hiGdf15hi macrophages. However, our data do not exclude that pro-inflammatory macrophage populations (Isg15hi and MHCII+Il1b+) could also shift towards a non-inflammatory Trem2hiGdf15hi state overtime after MI.
Our analysis of circulating monocytes indicates that ischaemic injury induces a shift towards monocytes enriched for the expression of Chil3 (encoding Ym1) and several granulocyte-associated genes (e.g. Prtn3, Lcn2). Recent reports proposed that mature monocytes arise from two distinct pathways in the steady state, with monocyte-dendritic progenitors and granulocyte-monocyte progenitors differentiating to monocytes with a ‘DC-like’ state or a ‘neutrophil-like’ state, respectively.57 Our results are thus consistent with an MI-induced shift towards the production of ‘granulocyte-like’ monocytes, which appear similar to Ym1+Ly6Chi monocytes that emerge after tissue injury.43 Bulk transcriptome analysis of human monocytes sampled 48 h after acute MI showed upregulation of LCN2, a prototypical marker of the granulocyte-like signature, as well as other granulocyte-associated transcripts (IL1RN, CXCR1),58 indicating that a similar shift in circulating monocyte transcriptome might occur in patients with MI. While our data indicate that the macrophage LAM signature is acquired in the ischaemic heart, and not directly induced in blood monocytes by MI, it remains to be determined whether injury-associated Chil3hi monocytes are primed towards acquiring the LAM signature once infiltrated in the infarcted myocardium.
5. Conclusion
In conclusion, our work provides a novel high-resolution view of the heterogeneity and dynamics of monocyte/macrophage transitions during the acute post-MI inflammation phase and constitutes a valuable resource for further investigating how these cells may be harnessed and modulated to promote post-ischaemic heart repair.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Authors’ contributions
Performed experiments: G.R., J.G., M.Pi, E.V., A.R., S.R.B., P.A., T.K., N.D., A.P.A.L., K.S., C.C.; Analysed and interpreted data: G.R., J.G., M.Pi, E.V., A.R., S.R.B., P.A., T.K., N.D., A.P.A.L., K.S., C.H., J.S.S., A.Z., A.E.S., C.C.; Created the web-based interface: O.D.; Provided critical mouse lines, assays, and reagents: M.Pr., M.M., K.S., C.H.; Revised and edited the manuscript: J.S.S., M.Pr., M.M., C.H., A.Z.; Designed the study: G.R., A.E.S., C.C.; Wrote the manuscript: G.R., A.E.S., C.C.
Supplementary Material
Contributor Information
Giuseppe Rizzo, Comprehensive Heart Failure Center, University Hospital Würzburg, Am Schwarzenberg 15, A15, 97078 Würzburg, Germany; Institute of Experimental Biomedicine, University Hospital Würzburg, Josef-Schneider-Str. 2, D16, 97080 Würzburg, Germany.
Julius Gropper, Comprehensive Heart Failure Center, University Hospital Würzburg, Am Schwarzenberg 15, A15, 97078 Würzburg, Germany; Institute of Experimental Biomedicine, University Hospital Würzburg, Josef-Schneider-Str. 2, D16, 97080 Würzburg, Germany.
Marie Piollet, Comprehensive Heart Failure Center, University Hospital Würzburg, Am Schwarzenberg 15, A15, 97078 Würzburg, Germany; Institute of Experimental Biomedicine, University Hospital Würzburg, Josef-Schneider-Str. 2, D16, 97080 Würzburg, Germany.
Ehsan Vafadarnejad, Helmholtz Institute for RNA-based Infection Research (HIRI), Helmholtz-Center for Infection Research (HZI), Josef-Schneider-Str. 2, D15, 97080 Würzburg, Germany.
Anna Rizakou, Institute of Experimental Biomedicine, University Hospital Würzburg, Josef-Schneider-Str. 2, D16, 97080 Würzburg, Germany.
Sourish Reddy Bandi, Comprehensive Heart Failure Center, University Hospital Würzburg, Am Schwarzenberg 15, A15, 97078 Würzburg, Germany; Institute of Experimental Biomedicine, University Hospital Würzburg, Josef-Schneider-Str. 2, D16, 97080 Würzburg, Germany.
Panagiota Arampatzi, Core Unit Systems Medicine, University Hospital Würzburg, Josef-Schneider-Str. 2, D15, 97080 Würzburg, Germany.
Tobias Krammer, Helmholtz Institute for RNA-based Infection Research (HIRI), Helmholtz-Center for Infection Research (HZI), Josef-Schneider-Str. 2, D15, 97080 Würzburg, Germany.
Nina DiFabion, Helmholtz Institute for RNA-based Infection Research (HIRI), Helmholtz-Center for Infection Research (HZI), Josef-Schneider-Str. 2, D15, 97080 Würzburg, Germany.
Oliver Dietrich, Helmholtz Institute for RNA-based Infection Research (HIRI), Helmholtz-Center for Infection Research (HZI), Josef-Schneider-Str. 2, D15, 97080 Würzburg, Germany.
Anahi-Paula Arias-Loza, Comprehensive Heart Failure Center, University Hospital Würzburg, Am Schwarzenberg 15, A15, 97078 Würzburg, Germany.
Marco Prinz, Institute of Neuropathology, Faculty of Medicine, University of Freiburg, Freiburg, Germany; Signalling Research Centres BIOSS and CIBSS, University of Freiburg, Freiburg, Germany; Center for Basics in NeuroModulation (NeuroModulBasics), Faculty of Medicine, University of Freiburg, Freiburg, Germany.
Matthias Mack, Department of Internal Medicine II, Nephrology, Franz-Josef-Strauss Allee 11, University Hospital Regensburg, 93053 Regensburg, Germany.
Kai Schlepckow, German Center for Neurodegenerative Diseases (DZNE) Munich, 81377 Munich, Germany.
Christian Haass, German Center for Neurodegenerative Diseases (DZNE) Munich, 81377 Munich, Germany; Division of Metabolic Biochemistry, Faculty of Medicine, Biomedical Center (BMC), Ludwig-Maximilians-Universität München, 81377 Munich, Germany; Munich Cluster for Systems Neurology (SyNergy), 81377 Munich, Germany.
Jean-Sébastien Silvestre, Université de Paris, PARCC, INSERM, F-75015 Paris, France.
Alma Zernecke, Institute of Experimental Biomedicine, University Hospital Würzburg, Josef-Schneider-Str. 2, D16, 97080 Würzburg, Germany.
Antoine-Emmanuel Saliba, Helmholtz Institute for RNA-based Infection Research (HIRI), Helmholtz-Center for Infection Research (HZI), Josef-Schneider-Str. 2, D15, 97080 Würzburg, Germany.
Clément Cochain, Comprehensive Heart Failure Center, University Hospital Würzburg, Am Schwarzenberg 15, A15, 97078 Würzburg, Germany; Institute of Experimental Biomedicine, University Hospital Würzburg, Josef-Schneider-Str. 2, D16, 97080 Würzburg, Germany.
Funding
This work was supported by the Interdisciplinary Center for Clinical Research (Interdisziplinäres Zentrum für Klinische Forschung, IZKF), University Hospital Würzburg (E-352 and A-384 to A.Z., E-353 to C.C.), the Deutsche Forschungsgemeinschaft (DFG; German Research Foundation, Projects 374031971-TRR 240, 324392634-TR221, ZE827/13-1, 14-1, project numbers 396923792 to A.Z., 432915089 to A.Z. and C.C., 458539578 and 471705758 to C.C.), the SFB1525 project number 453989101 (projects A1 and B3 to A.Z., project PS2 to A.E.S. and C.C., project A6 to C.C.), and the BMBF within the Comprehensive Heart Failure Centre Würzburg (BMBF 01EO1504 to C.C. and A.E.S.). A.E.S. is supported by the EMBO Young Investigator Program. This work was supported by grants from the Deutsche Forschungsgemeinschaft under Germany’s Excellence Strategy within the framework of the Munich Cluster for Systems Neurology (EXC 2145 SyNergy, ID 390857198 to C.H.) and the Helmholtz-Gemeinschaft Zukunftsthema ‘Immunology and Inflammation’ (ZT-0027 to C.H.).
Data availability
Single-cell RNA-sequencing data generated for this report has been deposited in Gene Expression Omnibus (GSE135310, GSE197441, GSE197853).
References
- 1. Frodermann V, Nahrendorf M. Macrophages and cardiovascular health. Physiol Rev 2018;98:2523–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Molawi K, Wolf Y, Kandalla PK, Favret J, Hagemeyer N, Frenzel K, Pinto AR, Klapproth K, Henri S, Malissen B, Rodewald HR, Rosenthal NA, Bajenoff M, Prinz M, Jung S, Sieweke MH. Progressive replacement of embryo-derived cardiac macrophages with age. J Exp Med 2014;211:2151–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zaman R, Hamidzada H, Epelman S. Exploring cardiac macrophage heterogeneity in the healthy and diseased myocardium. Curr Opin Immunol 2020;68:54–63. [DOI] [PubMed] [Google Scholar]
- 4. Dick SA, Macklin JA, Nejat S, Momen A, Clemente-Casares X, Althagafi MG, Chen J, Kantores C, Hosseinzadeh S, Aronoff L, Wong A, Zaman R, Barbu I, Besla R, Lavine KJ, Razani B, Ginhoux F, Husain M, Cybulsky MI, Robbins CS, Epelman S. Self-renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nat Immunol 2019;20:29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bajpai G, Bredemeyer A, Li W, Zaitsev K, Koenig AL, Lokshina I, Mohan J, Ivey B, Hsiao HM, Weinheimer C, Kovacs A, Epelman S, Artyomov M, Kreisel D, Lavine KJ. Tissue resident CCR2− and CCR2+ cardiac macrophages differentially orchestrate monocyte recruitment and fate specification following myocardial injury. Circ Res 2019;124:263–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nahrendorf M, Swirski FK, Aikawa E, Stangenberg L, Wurdinger T, Figueiredo JL, Libby P, Weissleder R, Pittet MJ. The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. J Exp Med 2007;204:3037–3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Leuschner F, Rauch PJ, Ueno T, Gorbatov R, Marinelli B, Lee WW, Dutta P, Wei Y, Robbins C, Iwamoto Y, Sena B, Chudnovskiy A, Panizzi P, Keliher E, Higgins JM, Libby P, Moskowitz MA, Pittet MJ, Swirski FK, Weissleder R, Nahrendorf M. Rapid monocyte kinetics in acute myocardial infarction are sustained by extramedullary monocytopoiesis. J Exp Med 2012;209:123–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mouton AJ, DeLeon-Pennell KY, Rivera Gonzalez OJ, Flynn ER, Freeman TC, Saucerman JJ, Garrett MR, Ma Y, Harmancey R, Lindsey ML. Mapping macrophage polarization over the myocardial infarction time continuum. Basic Res Cardiol 2018;113:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gessain G, Bleriot C, Ginhoux F. Non-genetic heterogeneity of macrophages in diseases—a medical perspective. Front Cell Dev Biol 2020;8:613116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. King KR, Aguirre AD, Ye YX, Sun Y, Roh JD, Ng RP Jr, Kohler RH, Arlauckas SP, Iwamoto Y, Savol A, Sadreyev RI, Kelly M, Fitzgibbons TP, Fitzgerald KA, Mitchison T, Libby P, Nahrendorf M, Weissleder R. IRF3 and type I interferons fuel a fatal response to myocardial infarction. Nat Med 2017;23:1481–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Calcagno DM, NgRP, Jr., Toomu A, Zhang C, Huang K, Aguirre AD, Weissleder R, Daniels LB, Fu Z, King KR. The myeloid type I interferon response to myocardial infarction begins in bone marrow and is regulated by Nrf2-activated macrophages. Sci Immunol 2020;5:eaaz1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R, Smibert P. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 2017;14:865–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chakarov S, Lim HY, Tan L, Lim SY, See P, Lum J, Zhang XM, Foo S, Nakamizo S, Duan K, Kong WT, Gentek R, Balachander A, Carbajo D, Bleriot C, Malleret B, Tam JKC, Baig S, Shabeer M, Toh SES, Schlitzer A, Larbi A, Marichal T, Malissen B, Chen J, Poidinger M, Kabashima K, Bajenoff M, Ng LG, Angeli V, Ginhoux F. Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science 2019;363:eaau0964. [DOI] [PubMed] [Google Scholar]
- 14. Dick SA, Wong A, Hamidzada H, Nejat S, Nechanitzky R, Vohra S, Mueller B, Zaman R, Kantores C, Aronoff L, Momen A, Nechanitzky D, Li WY, Ramachandran P, Crome SQ, Becher B, Cybulsky MI, Billia F, Keshavjee S, Mital S, Robbins CS, Mak TW, Epelman S. Three tissue resident macrophage subsets coexist across organs with conserved origins and life cycles. Sci Immunol 2022;7:eabf7777. [DOI] [PubMed] [Google Scholar]
- 15. Stoeckius M, Zheng S, Houck-Loomis B, Hao S, Yeung BZ, Mauck WM 3rd, Smibert P, Satija R. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol 2018;19:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jaitin DA, Adlung L, Thaiss CA, Weiner A, Li B, Descamps H, Lundgren P, Bleriot C, Liu Z, Deczkowska A, Keren-Shaul H, David E, Zmora N, Eldar SM, Lubezky N, Shibolet O, Hill DA, Lazar MA, Colonna M, Ginhoux F, Shapiro H, Elinav E, Amit I. Lipid-associated macrophages control metabolic homeostasis in a Trem2-dependent manner. Cell 2019;178:686–98.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cochain C, Vafadarnejad E, Arampatzi P, Pelisek J, Winkels H, Ley K, Wolf D, Saliba AE, Zernecke A. Single-cell RNA-Seq reveals the transcriptional landscape and heterogeneity of aortic macrophages in murine atherosclerosis. Circ Res 2018;122:1661–1674. [DOI] [PubMed] [Google Scholar]
- 18. Zernecke A, Erhard F, Weinberger T, Schulz C, Ley K, Saliba A-E, Cochain C. Integrated scRNA-seq analysis identifies conserved transcriptomic features of mononuclear phagocytes in mouse and human atherosclerosis. bioRxiv 2020. doi: 10.1101/2020.12.09.417535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Remmerie A, Martens L, Thone T, Castoldi A, Seurinck R, Pavie B, Roels J, Vanneste B, De Prijck S, Vanhockerhout M, Binte Abdul Latib M, Devisscher L, Hoorens A, Bonnardel J, Vandamme N, Kremer A, Borghgraef P, Van Vlierberghe H, Lippens S, Pearce E, Saeys Y, Scott CL. Osteopontin expression identifies a subset of recruited macrophages distinct from Kupffer cells in the fatty liver. Immunity 2020;53:641–57.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Daemen S, Gainullina A, Kalugotla G, He L, Chan MM, Beals JW, Liss KH, Klein S, Feldstein AE, Finck BN, Artyomov MN, Schilling JD. Dynamic shifts in the composition of resident and recruited macrophages influence tissue remodeling in NASH. Cell Rep 2021;34:108626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guilliams M, Bonnardel J, Haest B, Vanderborght B, Bujko A, Martens L, Thoné T, Browaeys R, De Ponti FF, Remmerie A, Wagner C, Vanneste B, Zwicker C, Vanhalewyn T, Gonçalves A, Lippens S, Devriendt B, Cox E, Ferrero G, Wittamer V, Willaert A, Kaptein SJF, Neyts J, Dallmeier K, Geldhof P, Casaert S, Deplancke B, ten Dijke P, Hoorens A, Vanlander A, Berrevoet F, Van Nieuwenhove Y, Saeys Y, Saelens W, Van Vlierberghe H, Devisscher L, Scott CL. Spatial proteogenomics reveals distinct and evolutionarily-conserved hepatic macrophage niches. Cell 2022;185:379–396.e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ramachandran P, Dobie R, Wilson-Kanamori JR, Dora EF, Henderson BEP, Luu NT, Portman JR, Matchett KP, Brice M, Marwick JA, Taylor RS, Efremova M, Vento-Tormo R, Carragher NO, Kendall TJ, Fallowfield JA, Harrison EM, Mole DJ, Wigmore SJ, Newsome PN, Weston CJ, Iredale JP, Tacke F, Pollard JW, Ponting CP, Marioni JC, Teichmann SA, Henderson NC. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 2019;575:512–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rao M, Wang X, Guo G, Wang L, Chen S, Yin P, Chen K, Chen L, Zhang Z, Chen X, Hu X, Hu S, Song J. Resolving the intertwining of inflammation and fibrosis in human heart failure at single-cell level. Basic Res Cardiol 2021;116:55. [DOI] [PubMed] [Google Scholar]
- 24. DeBerge M, Glinton K, Subramanian M, Wilsbacher LD, Rothlin CV, Tabas I, Thorp EB. Macrophage AXL receptor tyrosine kinase inflames the heart after reperfused myocardial infarction. J Clin Invest 2021;131:e139576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Louwe MC, Olsen MB, Kaasboll OJ, Yang K, Fosshaug LE, Alfsnes K, Ogaard JDS, Rashidi A, Skulberg VM, Yang M, de Miranda Fonseca D, Sharma A, Aronsen JM, Schrumpf E, Ahmed MS, Dahl CP, Nyman TA, Ueland T, Melum E, Halvorsen BE, Bjoras M, Attramadal H, Sjaastad I, Aukrust P, Yndestad A. Absence of NLRP3 inflammasome in hematopoietic cells reduces adverse remodeling after experimental myocardial infarction. JACC Basic Transl Sci 2020;5:1210–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xiao H, Li H, Wang J-J, Zhang J-S, Shen J, An X-B, Zhang C-C, Wu J-M, Song Y, Wang X-Y, Yu H-Y, Deng X-N, Li Z-J, Xu M, Lu Z-Z, Du J, Gao W, Zhang A-H, Feng Y, Zhang Y-Y. IL-18 cleavage triggers cardiac inflammation and fibrosis upon beta-adrenergic insult. Eur Heart J 2018;39:60–69. [DOI] [PubMed] [Google Scholar]
- 27. Vijayan V, Wagener F, Immenschuh S. The macrophage heme-heme oxygenase-1 system and its role in inflammation. Biochem Pharmacol 2018;153:159–167. [DOI] [PubMed] [Google Scholar]
- 28. Yang Z, Ming XF. Functions of arginase isoforms in macrophage inflammatory responses: impact on cardiovascular diseases and metabolic disorders. Front Immunol 2014;5:533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ferraro B, Leoni G, Hinkel R, Ormanns S, Paulin N, Ortega-Gomez A, Viola JR, de Jong R, Bongiovanni D, Bozoglu T, Maas SL, D'Amico M, Kessler T, Zeller T, Hristov M, Reutelingsperger C, Sager HB, Doring Y, Nahrendorf M, Kupatt C, Soehnlein O. Pro-angiogenic macrophage phenotype to promote myocardial repair. J Am Coll Cardiol 2019;73:2990–3002. [DOI] [PubMed] [Google Scholar]
- 30. Zaman R, Hamidzada H, Kantores C, Wong A, Dick SA, Wang Y, Momen A, Aronoff L, Lin J, Razani B, Mital S, Billia F, Lavine KJ, Nejat S, Epelman S. Selective loss of resident macrophage-derived insulin-like growth factor-1 abolishes adaptive cardiac growth to stress. Immunity 2021;54:2057–2071.e6. [DOI] [PubMed] [Google Scholar]
- 31. Kempf T, Zarbock A, Widera C, Butz S, Stadtmann A, Rossaint J, Bolomini-Vittori M, Korf-Klingebiel M, Napp LC, Hansen B, Kanwischer A, Bavendiek U, Beutel G, Hapke M, Sauer MG, Laudanna C, Hogg N, Vestweber D, Wollert KC. GDF-15 is an inhibitor of leukocyte integrin activation required for survival after myocardial infarction in mice. Nat Med 2011;17:581–588. [DOI] [PubMed] [Google Scholar]
- 32. Wan E, Yeap XY, Dehn S, Terry R, Novak M, Zhang S, Iwata S, Han X, Homma S, Drosatos K, Lomasney J, Engman DM, Miller SD, Vaughan DE, Morrow JP, Kishore R, Thorp EB. Enhanced efferocytosis of apoptotic cardiomyocytes through myeloid-epithelial-reproductive tyrosine kinase links acute inflammation resolution to cardiac repair after infarction. Circ Res 2013;113:1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kandalam V, Basu R, Abraham T, Wang X, Soloway PD, Jaworski DM, Oudit GY, Kassiri Z. TIMP2 deficiency accelerates adverse post-myocardial infarction remodeling because of enhanced MT1-MMP activity despite lack of MMP2 activation. Circ Res 2010;106:796–808. [DOI] [PubMed] [Google Scholar]
- 34. Ali K, Middleton M, Pure E, Rader DJ. Apolipoprotein E suppresses the type I inflammatory response in vivo. Circ Res 2005;97:922–927. [DOI] [PubMed] [Google Scholar]
- 35. Alonso-Herranz L, Sahun-Espanol A, Paredes A, Gonzalo P, Gkontra P, Nunez V, Clemente C, Cedenilla M, Villalba-Orero M, Inserte J, Garcia-Dorado D, Arroyo AG, Ricote M. Macrophages promote endothelial-to-mesenchymal transition via MT1-MMP/TGFbeta1 after myocardial infarction. Elife 2020;9:e57920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Farbehi N, Patrick R, Dorison A, Xaymardan M, Janbandhu V, Wystub-Lis K, Ho JW, Nordon RE, Harvey RP. Single-cell expression profiling reveals dynamic flux of cardiac stromal, vascular and immune cells in health and injury. Elife 2019;8:e43882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vafadarnejad E, Rizzo G, Krampert L, Arampatzi P, Arias-Loza PA, Nazzal Y, Rizakou A, Knochenhauer T, Reddy Bandi S, Nugroho VA, Schulz DJ, Roesch M, Alayrac P, Vilar J, Silvestre JS, Zernecke A, Saliba AE, Cochain C. Dynamics of cardiac neutrophil diversity in murine myocardial infarction. Circ Res 2020;127:e232–e249. [DOI] [PubMed] [Google Scholar]
- 38. Mack M, Cihak J, Simonis C, Luckow B, Proudfoot AE, Plachy J, Bruhl H, Frink M, Anders HJ, Vielhauer V, Pfirstinger J, Stangassinger M, Schlondorff D. Expression and characterization of the chemokine receptors CCR2 and CCR5 in mice. J Immunol 2001;166:4697–4704. [DOI] [PubMed] [Google Scholar]
- 39. Schlitzer A, Sivakamasundari V, Chen J, Sumatoh HR, Schreuder J, Lum J, Malleret B, Zhang S, Larbi A, Zolezzi F, Renia L, Poidinger M, Naik S, Newell EW, Robson P, Ginhoux F. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat Immunol 2015;16:718–728. [DOI] [PubMed] [Google Scholar]
- 40. Bosteels C, Neyt K, Vanheerswynghels M, van Helden MJ, Sichien D, Debeuf N, De Prijck S, Bosteels V, Vandamme N, Martens L, Saeys Y, Louagie E, Lesage M, Williams DL, Tang SC, Mayer JU, Ronchese F, Scott CL, Hammad H, Guilliams M, Lambrecht BN. Inflammatory type 2 cDCs acquire features of cDC1s and macrophages to orchestrate immunity to respiratory virus infection. Immunity 2020;52:1039–1056.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Goldmann T, Wieghofer P, Muller PF, Wolf Y, Varol D, Yona S, Brendecke SM, Kierdorf K, Staszewski O, Datta M, Luedde T, Heikenwalder M, Jung S, Prinz M. A new type of microglia gene targeting shows TAK1 to be pivotal in CNS autoimmune inflammation. Nat Neurosci 2013;16:1618–1626. [DOI] [PubMed] [Google Scholar]
- 42. Schultze JL, Mass E, Schlitzer A. Emerging principles in myelopoiesis at homeostasis and during infection and inflammation. Immunity 2019;50:288–301. [DOI] [PubMed] [Google Scholar]
- 43. Ikeda N, Asano K, Kikuchi K, Uchida Y, Ikegami H, Takagi R, Yotsumoto S, Shibuya T, Makino-Okamura C, Fukuyama H, Watanabe T, Ohmuraya M, Araki K, Nishitai G, Tanaka M. Emergence of immunoregulatory Ym1(+)Ly6C(hi) monocytes during recovery phase of tissue injury. Sci Immunol 2018;3:eaat0207. [DOI] [PubMed] [Google Scholar]
- 44. Yanez A, Coetzee SG, Olsson A, Muench DE, Berman BP, Hazelett DJ, Salomonis N, Grimes HL, Goodridge HS. Granulocyte-monocyte progenitors and monocyte-dendritic cell progenitors independently produce functionally distinct monocytes. Immunity 2017;47:890–902.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 2014;32:381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang S, Weinberg S, DeBerge M, Gainullina A, Schipma M, Kinchen JM, Ben-Sahra I, Gius DR, Yvan-Charvet L, Chandel NS, Schumacker PT, Thorp EB. Efferocytosis fuels requirements of fatty acid oxidation and the electron transport chain to polarize macrophages for tissue repair. Cell Metab 2019;29:443–456.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. A-Gonzalez N, Bensinger SJ, Hong C, Beceiro S, Bradley MN, Zelcer N, Deniz J, Ramirez C, Diaz M, Gallardo G, de Galarreta CR, Salazar J, Lopez F, Edwards P, Parks J, Andujar M, Tontonoz P, Castrillo A. Apoptotic cells promote their own clearance and immune tolerance through activation of the nuclear receptor LXR. Immunity 2009;31:245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Murthy S, Karkossa I, Schmidt C, Hoffmann A, Hagemann T, Rothe K, Seifert O, Anderegg U, von Bergen M, Schubert K, Rossol M. Danger signal extracellular calcium initiates differentiation of monocytes into SPP1/osteopontin-producing macrophages. Cell Death Dis 2022;13:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rossol M, Pierer M, Raulien N, Quandt D, Meusch U, Rothe K, Schubert K, Schoneberg T, Schaefer M, Krugel U, Smajilovic S, Brauner-Osborne H, Baerwald C, Wagner U. Extracellular Ca2 + is a danger signal activating the NLRP3 inflammasome through G protein-coupled calcium sensing receptors. Nat Commun 2012;3:1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pinto AR, Paolicelli R, Salimova E, Gospocic J, Slonimsky E, Bilbao-Cortes D, Godwin JW, Rosenthal NA. An abundant tissue macrophage population in the adult murine heart with a distinct alternatively-activated macrophage profile. PLoS One 2012;7:e36814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Heidt T, Courties G, Dutta P, Sager HB, Sebas M, Iwamoto Y, Sun Y, Da Silva N, Panizzi P, van der Laan AM, Swirski FK, Weissleder R, Nahrendorf M. Differential contribution of monocytes to heart macrophages in steady-state and after myocardial infarction. Circ Res 2014;115:284–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Weinberger T, Rauber S, Schneider V, Messerer D, Fischer M, Rudi WS, Kobayashi Y, Schulz C. Differential MHC-II expression and phagocytic functions of embryo-derived cardiac macrophages in the course of myocardial infarction in mice. Eur J Immunol 2021;51:250–252. [DOI] [PubMed] [Google Scholar]
- 53. Hilgendorf I, Gerhardt LM, Tan TC, Winter C, Holderried TA, Chousterman BG, Iwamoto Y, Liao R, Zirlik A, Scherer-Crosbie M, Hedrick CC, Libby P, Nahrendorf M, Weissleder R, Swirski FK. Ly-6Chigh monocytes depend on Nr4a1 to balance both inflammatory and reparative phases in the infarcted myocardium. Circ Res 2014;114:1611–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, David E, Baruch K, Lara-Astaiso D, Toth B, Itzkovitz S, Colonna M, Schwartz M, Amit I. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 2017;169:1276–1290.e17. [DOI] [PubMed] [Google Scholar]
- 55. Nugent AA, Lin K, van Lengerich B, Lianoglou S, Przybyla L, Davis SS, Llapashtica C, Wang J, Kim DJ, Xia D, Lucas A, Baskaran S, Haddick PCG, Lenser M, Earr TK, Shi J, Dugas JC, Andreone BJ, Logan T, Solanoy HO, Chen H, Srivastava A, Poda SB, Sanchez PE, Watts RJ, Sandmann T, Astarita G, Lewcock JW, Monroe KM, Di Paolo G. TREM2 regulates microglial cholesterol metabolism upon chronic phagocytic challenge. Neuron 2020;105:837–854.e9. [DOI] [PubMed] [Google Scholar]
- 56. Amorim A, De Feo D, Friebel E, Ingelfinger F, Anderfuhren CD, Krishnarajah S, Andreadou M, Welsh CA, Liu Z, Ginhoux F, Greter M, Becher B. IFNgamma and GM-CSF control complementary differentiation programs in the monocyte-to-phagocyte transition during neuroinflammation. Nat Immunol 2022;23:217–228. [DOI] [PubMed] [Google Scholar]
- 57. Weinreb C, Rodriguez-Fraticelli A, Camargo FD, Klein AM. Lineage tracing on transcriptional landscapes links state to fate during differentiation. Science 2020;367:eaaw3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ruparelia N, Godec J, Lee R, Chai JT, Dall'Armellina E, McAndrew D, Digby JE, Forfar JC, Prendergast BD, Kharbanda RK, Banning AP, Neubauer S, Lygate CA, Channon KM, Haining NW, Choudhury RP. Acute myocardial infarction activates distinct inflammation and proliferation pathways in circulating monocytes, prior to recruitment, and identified through conserved transcriptional responses in mice and humans. Eur Heart J 2015;36:1923–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell RNA-sequencing data generated for this report has been deposited in Gene Expression Omnibus (GSE135310, GSE197441, GSE197853).