Abstract
The DNAJB1-PRKACA fusion is the signature genetic event of fibrolamellar hepatocellular carcinoma (FL-HCC), a rare but lethal liver cancer that primarily affects adolescents and young adults. A deletion fuses the first exon of the HSP40 gene (DNAJB1), with exons 2–10 of protein kinase A (PRKACA), producing the chimeric kinase DNAJB1-PKAca (J-PKAca). The HSP40 portion’s scaffolding/chaperone function has been implicated in redirecting substrate recognition to upregulate oncogenic pathways, but the direct substrates of this fusion are not fully known. We integrated cell-based and in vitro phosphoproteomics to identify substrates targeted directly by PKA and J-PKAca, comparing phosphoproteome profiles from cells with in vitro rephosphorylation of peptides and proteins from lysates using recombinant enzymes. We identified a subset of phosphorylation sites in both cell-based and in vitro experiments, as well as altered pathways and proteins consistent with observations from related studies. We also treated cells with PKA inhibitors that function by two different mechanisms (rpcAMPs and PKI) and examined phosphoproteome profiles, finding some substrates that persisted in the presence of inhibitors and revealing differences between WT and chimera. Overall, these results provide potential insights into J-PKAca’s oncogenic activity in a complex cellular system and may provide candidate targets for therapeutic follow-up.
Keywords: fibrolamellar hepatocellular carcinoma, FL-HCC, protein kinase A, PKA, DNAJB1-PRKACA, phosphoproteomics, kinase inhibitors, kinase—substrate identification
Graphical Abstract
INTRODUCTION
Fibrolamellar hepatocellular carcinoma (FL-HCC) is a primary liver cancer that occurs in young people without a gender bias or underlying liver diseases. It accounts for less than 1% of all primary liver cancers but represents the majority of HCCs in patients younger than 30 years of age.1,2 Surgery is the mainstay of treatment in FL-HCC. No chemotherapeutic agents have been shown to have significant efficacy in HCC.3 The gross anatomic feature of FL-HCC is an expanding, heterogeneous tumor mass with areas of increased vascularity and necrosis’ including areas of fibrosis similar to focal nodular hyperplasia, a benign vascular-fibrotic lesion in the liver.4
Due to the rarity of FL-HCC and a lack of representative experimental systems, the molecular basis for carcinogenesis in these tumors has been elusive and difficult to study. Recently, however, there has been a breakthrough in understanding the pathogenesis and expression profile of the FL-HCC. Honeyman and colleagues discovered a novel, chimeric transcript that is present in all studied samples of FL-HCC.5 Detection of a single, consistent genetic deletion in one copy of chromosome 195,6 results in the formation of a chimeric gene, DNAJB1 —PRKACA, which combines the first exon of DNAJB1, the heat shock protein 40 (HSP40), with exons 2 through 10 of PRKACA, the catalytic subunit of protein kinase A (PKAca). This gives rise to a chimeric construct in which the N-terminal helix of PKAca is fused to the J-domain of DNAJB1 (J-PKAca). This chimera has been found to be the main driver of FL-HCC,7 and while some of the changes downstream of its expression have been characterized, the detailed molecular pathogenesis producing those changes remains poorly understood.
PKAca is one of the best understood human kinases, and there is ample evidence to support the hypothesis that its kinase activity plays a role in FL-HCC.5,6 Although protein kinases represent a significant class of potential drug targets, with more than 60 FDA-approved small-molecule kinase inhibitors and many more in development or in clinical trials8,9 PKA itself seems to be a challenging target, likely due to its function in many normal cell types leading to dose-limiting toxicities and thus it may suffer from a narrow therapeutic window.10 Furthermore, previous work has suggested that the catalytic activity of the chimeric fusion is not significantly different from that of the native PKAca enzyme,5,7 yet the phenotypic differences observed in FL-HCC suggest that increased activity on native PKAca substrates is not the only contributor to oncogenic transformation; additional mechanisms may be activated by the chimera.7 DNAJB1 (otherwise known as HSP40) is a chaperone protein with numerous protein—protein interaction partners, and a recent study suggests that J-PKAca functions as a scaffold via its J (HSP40) component to assemble signaling elements that are then aberrantly phosphorylated by the catalytic portion of the chimera to contribute to the pathogenicity of FL-HCC.11 Other recent work has characterized the allosteric effects of the J-domain on important aspects of the PKA activation mechanism, observing changes to the dynamics of the holoenzyme bound to regulatory subunits that impact important factors like cAMP binding, surfaces accessible for interaction with binding partners, and potentially localization.12 Consequently, understanding the downstream pathways activated by excess WT PKAca activity vs aberrant J-PKAca chimera signaling may help to find new viable drug targets for FL-HCC. However, both PKA and HSP40 are upstream of a very broad set of cellular pathways, and their downstream effects are governed by particular regulatory interactions that determine substrates under a given set of biological conditions. Therefore, identifying particular direct substrates of both WT PKAca and the J-PKAca chimera in cells would be valuable for follow-up on candidate pathways for therapeutic intervention.
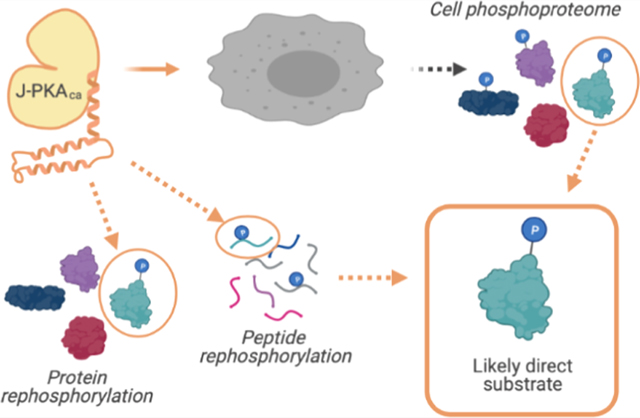
Many high-throughput approaches for the identification of phosphopeptides with mass spectrometry-based proteomics have been described (some reviewed in ref 13), but these do not typically enable clear assignment of direct substrates relative to those that may have been phosphorylated by other kinases downstream of the target kinase’s activation. We seek to understand whether this chimeric fusion between HSP40 and PKA results in direct phosphorylation of a different set of substrates that could lead to its activation of unique pathways. Therefore, in the present study, we have employed an integrated strategy termed kinase assay linked with phosphoproteomics (KALIP)14–17 (similar to other approaches employing cell lysates as a substrate pool for in vitro kinase reactions18,19 ) for comparing the specificity and identifying direct substrates of WT PKAca and the chimeric mutant J-PKAca (Figure 1). We identified phosphopeptides from cells overexpressing WT PKAca or J-PKAca (Figure 1A), as well as from in vitro kinase reactions with recombinant, purified WT PKAca or J-PKAca using either digested peptides (Figure 1B) or whole proteins as (Figure 1C) the substrate pool. These experiments identified some phosphosites associated uniquely with the chimeric fusion enzyme and enabled system analyses that revealed similarities and differences in the phosphosite motifs and pathways activated in the presence of the WT PKAca vs J-PKAca. We also examined the differential WT PKAca and J-PKAca phosphoproteomes in response to two different PKA inhibitors in cells, identifying pathways and substrates that seemed to persist despite the apparent inhibition of other substrates and pathways. Overall, these findings of direct substrates, and substrates with differential inhibitor responses, could assist in prioritizing candidates for next-generation treatment of FL-HCC.
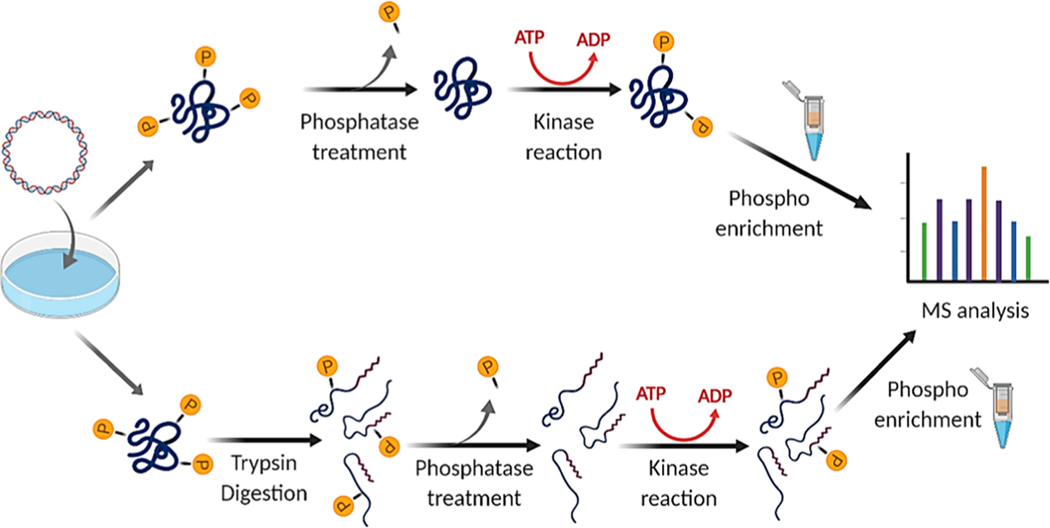
Figure 1.
Summary of workflow for protein-level (top) and peptide-level (bottom) KALIP rephosphorylation experiments (image created with BioRender).
MATERIALS AND METHODS
Recombinant Enzyme Expression and Purification
High-level expression of human PKAca and J-PKAca in Escherichia coli was achieved by the construction of pET28a (+) vector that contained the protein gene subcloned prior to a phage T7 RNA polymerase promoter. Each species was expressed in the E. coli cell line BL21(DE3) by growing the bacteria in LB medium at 37 °C. Protein expression was induced by the addition of 0.4 mM IPTG and carried out overnight at 24 °C before harvesting the cells. Affinity purification was carried out using a Ni-NTA affinity resin (Thermo Scientific HisPur Ni-NTA Resin, catalogue number: 88221). Protein molecular weight was confirmed by SDS-PAGE (Figure S1). The activity of purified enzymes was confirmed by the PepTag nonradioactive protein kinase assay (Promega, catalogue number: V5340) (Figure S2).
Human Cell Culture and Transfection
Human embryonic kidney 293 cells (HEK293-T) were maintained in modified Dulbecco’s medium (DMEM) supplemented with 10% fetal bovine serum, 100 μg/mL of streptomycin, and 100 IU/mL of penicillin in 5% CO2 at 37 °C. Cells were grown to 80—90% confluency and then transiently transfected using the Lipofectamine 3000 transfection kit (Thermo Fisher Scientific) with 10 μg of plasmid containing either human PRKACA gene (pcrDNA3.1-PRKACA, Addgene plasmid #100890), DNAJB1—PRKACA gene (pcrDNA3.1-Chimera, Addgene plasmid #100891), or empty pcrDNA3.1 vector. Both plasmids were gifts from Sanford Simon.5 Cells were incubated with plasmids for 36 h at 37 °C and 5% carbon dioxide in transfection media (DMEM/10% FBS) and then either harvested directly or treated with 0.1 mM sodium pervanadate for 15 min at 4 °C prior to harvesting. The cells were then washed twice with PBS, and the pellets were frozen at −80 °C until further processing as described in the Sample Preparation from Cell Lysates section. Sample aliquots were used to verify kinase expression via Western blotting, as described in the Supporting Information.
Inhibitor Treatment
Cells were maintained in modified Dulbecco’s medium (DMEM) supplemented with 10% fetal bovine serum, 100 μg/mL of streptomycin, and 100 IU/mL of penicillin in 5% CO2 at 37 °C and grown to ∼80% confluency before transfection with plasmids as described above. Small-molecule inhibitor rpcAMPs (cAMPs-Rp triethylammonium salt, Tocris, Cat. #1337) (370 μM) was added with transfection (36 h of total treatment) or after 12 h of transfection (24 h of total treatment); cell-permeable peptide inhibitor PKI (myristoylated PKI14–22 amide, Tocris, Cat. #2546) (6.4 μM) was added 12 h after transfection (24 h of total treatment). The cells were harvested after a total of 36 h in transfection media (DMEM/10% FBS). Cells were briefly pulsed with 0.1 mM sodium pervanadate for 15 min at 4 °C prior to harvesting the cells to inhibit phosphatase activity and prevent peptide dephosphorylation to “lock” phosphorylation sites in place for downstream analysis. The cells were then washed twice with PBS, and pellets were frozen at −80 until further processing, as described in the Sample Preparation from Cell Lysates section.
Sample Preparation from Cell Lysates
For all experiments except the native protein rephosphorylation, peptides were prepared as previously described:20 cell pellets that had been snap-frozen at −80 °C were solubilized in lysis buffer (20% acetonitrile, 7 M urea, 2 M thiourea, 0.4 M TEAB, 4 mM DTT, phosphatase inhibitor cocktail (Pierce Phosphatase Inhibitor Mini Tablets, catalogue number: A32957), and Roche cOmplete, Mini EDTA-free Protease Inhibitor Cocktail 04693159001) and probe-sonicated to shear DNA. The cell debris was cleared by centrifugation at 16 000g for 30 min at 4 °C, and the supernatant containing the soluble proteins was collected. Protein concentrations were measured using the Bradford assay, and the lysate containing ∼1 mg of total protein was reduced using TCEP (5 mM) and alkylated with iodoacetamide (40 mM) for 1 h at room temperature in the dark. Proteins were then digested with proteomics-grade trypsin (Pierce Trypsin Protease, MS grade-catalogue number 90058) at a ratio of 1:50 by mass (trypsin/total protein in lysate) overnight at 37 °C. On the next day, samples were acidified with 10% trifluoroacetic acid (TFA), desalted with Waters Oasis HLB cartridges, and concentrated to dryness using a Speed-Vac.
For the native protein rephosphorylation experiments, cell lysates were prepared in lysis buffer containing 50 mM Tris·HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% Nonidet P-40, and protease inhibitor cocktail for 20 min on ice. Cell debris was cleared using centrifugation at 16 000g and 4 °C for 15 min. The supernatant containing soluble proteins was collected and measured for protein concentration and used in native protein rephosphorylation experiments as described below.
Phosphopeptide Enrichment
Sequential enrichment by metal oxide affinity chromatography (SMOAC) (Thermo Fisher Scientific) was used to enrich phosphopeptides according to the manufacturer’s instructions. This method uses a two-step consecutive enrichment of phosphopeptides using TiO2 and Fe-NTA spin columns.21 The recovered phosphopeptides from this step were concentrated to dryness using a Speed-Vac.
Kinase Reactions for Rephosphorylation
For Peptide Rephosphorylation.
Peptide digests were prepared as described above. Samples were then resuspended in 100 μL of lambda phosphatase reaction buffer (50 mM Hepes, 100 mM NaCl, 2 mM DTT, 1 mM MnCl2, pH 7.5). Lambda phosphatase (New England Biolabs P0753S) was added to each sample (2800–3000 units) and incubated overnight at 30 °C. The phosphatase was deactivated by heating at 75°C for 10 min. Samples were incubated in a kinase assay reaction buffer (100 mM Tris–HCl pH 7.8, 10 mM MgCl2, 5 mM DTT, 2 mM ATP) containing the purified recombinant kinase (approximately 5000 units; estimated by in vitro kinase reaction with the recombinant, purified enzyme and relative quantification of bands via PepTag Nonradioactive protein kinase assays; Figure S2) at 30 °C for 90 min. Reactions were quenched by the addition of 0.5% TFA to a pH below 3. The samples were then desalted using Waters HLB cartridges and concentrated to dryness using a Speed-Vac.
For Native Protein Rephosphorylation.
Samples (2 mg of total protein in 300 μL of lysis buffer each) were treated with lambda phosphatase (2800–3000 units) in phosphatase buffer (500 mM Tris–HCl, pH 7.5, 1 mM Na2 EDTA, 50 mM dithiothreitol) and 2 mM Mn ion (MnCl2) and incubated at 37 °C for 90 min. Since it was preferred to keep the proteins in their native, folded state, heat inactivation was not desirable; therefore, to stop the reaction, a cocktail of two tablets of phosphatase inhibitor (Pierce Phosphatase Inhibitor Mini Tablets, catalogue number: A32957) and two tablets of protease inhibitor (Roche cOmplete, Mini, EDTA-free Protease Inhibitor Cocktail #04693159001) dissolved in 5 ml of kinase reaction buffer was prepared and 150 μL of this cocktail was added to each sample, stirring gently with a small magnetic stirrer while holding in a water bath for 10 min. A fraction of the reaction was kept as control for MS analysis, and for rephosphorylation reaction, the purified recombinant kinases (∼5000 units, estimated as described above for peptide rephosphorylation) were added to each sample in kinase buffer (20 mM potassium phosphate, 180 mM KCl, 20 mM MgCl2, 5 mM DTT, and 200 μM ATP) while gently stirring (as above) for 60 min at 30 °C. The kinase reaction was quenched by heat inactivation (75 °C for 10 min) and dialyzed overnight against trypsin digestion buffer (100 mM Tris–HCl, 10 mM CaCl2, 5 mM DTT, pH 7.8) in Pur-a-lyzer midi dialysis tubes (Sigma-Aldrich PURD10005) for buffer exchange, resulting in a minimal amount of protein precipitation. Samples were then resolubilized using up to 2 M urea, alkylated with iodoacetamide, digested using trypsin, desalted and concentrated to dryness, and phosphoenriched using the SMOAC strategy as described for other samples above.
Mass Spectrometry Data Acquisition and Data Analysis
The phosphopeptide samples were then analyzed by LC-MS/MS as previously20 using a Thermo Scientific Easy NanoLC LC 1000 system coupled to a high-resolution Orbitrap Fusion Tribrid Mass Spectrometer using a reverse phase C18 column with a 60 min linear gradient of 2–30% B (solvent A: water +.01% formic acid; solvent B: acetonitrile+.01% formic acid) at a flow rate of 200 nL/min. The mass spectrometer was operated in a data-dependent mode with a resolution of 60 000 and a scan range of 300–1500 m/z. The top 12 most abundant ions were selected with a dynamic exclusion time of 15 s for high collision dissociation (HCD), and fragments were analyzed.
Raw data files were processed for peptide, protein, and phosphosite identification using PEAKS Studio XPro software (Bioinformatics Solutions Inc.) against the complete UniProt human reference proteome (containing both reviewed and unreviewed entries). Details of phosphorylation sites per protein were extracted from the exported PEAKS PTM search result tables for further comparisons and analyses using a custom-built PHP script we named PEAKS-ModExtractor (https://gitlab.com/jackbrennan07/peaks-modextractor). Instructions for using Peaks-ModExtractor are available at the link provided.
Qualitative Data Analysis to Identify Shared and Unique Sites (Cell-Based Phosphoproteomic Profiling, In Vitro Rephosphorylation, and Cell-Based Inhibitor Experiments).
For all experiments, lists of phosphopeptides/phosphorylation sites identified were analyzed qualitatively to extract a final list of sequences/sites that were identified robustly and compared to each other to determine which were shared and which were uniquely present in a given experiment: first, during the PEAKS Studio Xpro export process, a false discovery rate (FDR) cutoff for peptide identification was applied and only peptides with FDR ≤ 1% were included. Next, results from PEAKS-ModExtractor outputs were filtered to include only those with Ascore values ≤30. This value was selected as a cutoff based on the previously published evaluation of the Ascore metric performance,22,23 which demonstrated that phosphopeptides with Ascores above this cutoff are reliably high confidence for identifying and localizing the modification. Then, data sets from each experiment were compared using BioVenn (http://www.biovenn.nl/index.php)24 and additionally filtered to focus on more robust observations by only including phosphopeptides observed in both replicates. The resulting lists were then qualitatively background-subtracted (also using BioVenn) by removing any sites that were also identified in the experiment’s respective negative control (the empty vector-only control for the cell-based experiments and phosphatase-treated inputs for the in vitro rephosphorylation experiments). As a further approach to ensure interpretation of the identified peptides/proteins in context, we included a reference table from the CRAPome,25 a repository of proteins commonly identified across many affinity purification mass spectrometry (AP-MS) experiments compiled at and downloaded from https://reprint-apms.org, and provided information about how often a given protein has been observed (via average spectral counts across CRAPome experiments) with our results listed in Supporting Table S1. It should be noted that this CRAPome reference is not from phosphopeptide enrichment by SMOAC and therefore may not fully represent the type of background contaminants relevant to this experiment —however, it may still be a useful reference for considering the cellular context of different protein abundances and the nature of different types of binding interactions relevant to our result list.
Comparing Tryptic Peptides vs Sites in Their Protein Context.
Peptide sequences pulled from the PEAKS result tables by PEAKS-ModExtractor were used to compare results from the peptide rephosphorylation experiment; however, because phosphorylation at the protein level (i.e., in cells and in the native protein rephosphorylation experiment) occurred before protease digestion, the identified peptide lists were not sufficient to analyze substrate motifs. Thus, phosphorylation site(s) per protein identifier were used to retrieve the sequences surrounding the identified phosphosites (−7 to +7) by retrieving protein region sequence information from UniProt via the Retrieve/ID Mapping function, downloading the results as noncompressed “source list”.fasta files. The resulting consolidated lists of 15 AA sequences −7 to +7 from each phosphorylation site were used for comparison analyses. Motif visualization was performed using PTM-Logo.26 Sequences of <15 or >15 amino acids in length (arising from, e.g. isoform conflicts, redundant partial sequences in unreviewed entries, etc.) were excluded from PTM-logo motif analysis due to limitations of the motif tool. In cases where nonuniform sequences needed to be evaluated (e.g. the peptide rephosphorylation experiment), the Weblogo resource was used.27
Raw data, associated PEAKS result files, and PEAKS-ModExtractor outputs are available for download at the MassIVE repository (https://massive.ucsd.edu, accession #MSV000086707).
RESULTS
Cell-Based Phosphoproteomics Profiles and Signaling Pathway Comparison
To compare the signaling profiles of WT PKAca and its oncogenic mutant J-PKAca, constructs containing each protein were transiently expressed in HEK293 cells in duplicate. Cells transfected with empty vectors were used as a control. Expression levels were verified by Western blot against the human cAMP-dependent protein kinase A catalytic subunit, which recognizes both WT PKAca and J-PKAca (Figure S1). It should be noted that the expression of both WT- and J-PKAca were relatively high in the cell line model and may have resulted in differences in ratios of catalytic to regulatory subunits that would affect the physiological behavior of the kinase activation. Cells were treated briefly with the phosphatase inhibitor pervanadate to enhance phosphorylation site stability for downstream analysis and lysed in denaturing lysis buffer. Phosphopeptides were enriched using sequential metal oxide affinity chromatography (SMOAC), as described in the Materials and Methods section, and analyzed on an Orbitrap Fusion LC-MS/MS. Mass spectrometry data were processed using PEAKS Studio XPro, and modified peptides were extracted from the PTM search export using PEAKS-ModExtractor. The outputs were filtered to ignore modifications other than phosphorylation of STY and Ascore values below 30. To control for background phosphorylation, phosphosite identifications from the vector-only control were filtered from the sites identified in WT PKAca- and J-PKAca-expressing cells. These background-subtracted lists were used to make phosphosite comparisons between replicates and conditions.
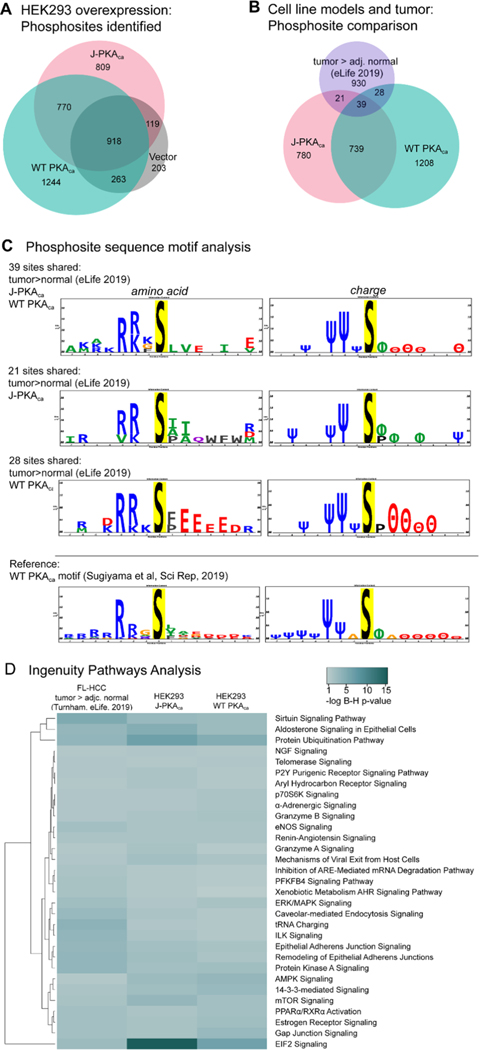
Overall, 3195 phosphosites were confidently identified in both replicates from WT PKAca-expressing cells and 2616 from J-PKAca-expressing cells. After filtering out those that were also observed in the vector control, 1244 were unique to WT PKAca, 809 to J-PKAca, and 770 were common between the two (Figure 2A). Pathway activation was evaluated using ingenuity pathways analysis (IPA) in the phosphorylation analysis mode for each phosphosite list from WT PKAca- and J-PKAca-expressing cells and also for phosphosites identified as higher abundance in tumor vs adjacent normal tissues (>1 log2-fold change or only in tumors) from a recent phosphoproteomics study of FL-HCC patient samples.11 Full pathway analysis tables are available in the Supporting Information. Some of the pathways observed as significantly enriched in the overexpression cell lines were likely cell-type-specific, as they were not observed as activated from the patient sample data. However, a number of substrates and key pathways enriched in the FL-HCC patient sample data were also observed in WT PKAca- and J-PKAca-expressing cells (Figure 2B,D), in particular, the ERK/MAPK signaling pathway that was identified as important in the patient sample study by Turnham et al. These results support that despite the caveat related to the ratio of catalytic to regulatory subunit expression levels, it seems reasonable to use HEK293 cells overexpressing the kinase constructs as model systems for examining direct substrates of these kinases in this context. However, they also suggest that most of the pathway activation may arise broadly from overactive PKA and not necessarily from substrate interactions unique to the DNAJB1/HSP40 chaperone interactions, given that pathways activated in WT PKAca-expressing cells were overall the same as in J-PKAca-expressing cells. The only pathways with stronger enrichment in J-PKAca-expressing cells vs WT PKAca included EIF2 signaling, protein ubiquitination, and aldosterone signaling in epithelial cells (Figure 2D), none of which were very highly enriched in the patient sample data. With respect to specific phosphorylation sites, direct comparisons of the HEK293 cell overexpression models with the patient FL-HCC data are subject to the caveat that the phosphoenrichment technique, instrumentation, peptide identification software, and human proteome databases were not the same; however, there were nevertheless still some sites observed in common (Figure 2B).
Figure 2.
Characteristics of phosphoprotein signaling and sites. (A) Venn diagram representing phosphosites confidently observed from PKAca- , J-PKAca-, and vector-only expressing cells (created using BioVenn https://www.biovenn.nl/). (B) Phosphosites observed (same lists used as input for IPA) were compared between the HEK293 overexpression models and FL-HCC tumor tissues (tumor > adj. normal, eLife, 201911). Venn diagrams for comparisons of individual sites and for protein identifiers were created with BioVenn. (C) Sequence motif analysis for the overlapping phosphosite observations was performed using PTM-Logo,23 also including motif analysis for the sites observed as phosphorylated by WT PKAca in a previously reported study by Sugiyama et al. in 2019, performing whole-protein in vitro phosphorylation.25 Only significantly over-represented amino acids and charge characteristics at positions −7 to +7 are shown in sequence logos. Charge logo legend: Ψ = basic residue, e.g., K/R; θ = acidic residue, e.g., D/E; ϕ = hydrophobic residue; P = proline; Δ = other. (D) Heatmap generated with https://heatmapper.ca, illustrating selected pathway activation enrichment scores from IPA (Supporting Table S3), comparing phosphorylation sites observed as higher in tumor vs normal FL-HCC tissue in the Turnham et al. study vs HEK293 cells overexpressing J-PKAca or WT PKAca (after filtering each to remove sites observed in vector-only control). Multiple comparison-corrected Benjamini–Hochberg p-values were determined in IPA, with the significance cutoff set at p = 0.05, and only pathways with significant B–H p-value enrichment scores from the Turnham data and the J-PKAca data were included in the heatmap.
The sequences surrounding the overlapping sites (Table 1) were further examined. The amino acid motif around most of the 39 phosphorylation sites observed in common among the FL-HCC tumor>normal, J-PKAca, and WT PKAca data sets showed similar characteristics to that seen for direct phosphorylation of proteins by recombinant WT PKAca in experiments reported by Sugiyama et al.28 (Figure 2C), suggesting that many of these sites are likely to be direct substrates in FL-HCC that arise from abnormal levels of PKA activity. Intriguingly, the 21 sites that were observed in common only in FL-HCC tumor > normal/J-PKAca exhibited the expected enrichment of hydrophobic amino acids at +1, whereas the 28 sites shared only in FL-HCC tumor>normal/WT PKAca did not. This suggests that the 21 FL-HCC tumor > normal/J-PKAca sites may also represent unique direct substrates of the J-PKAca mutant, while those observed as unique in the WT PKAca data may be more likely to be downstream and not direct. This was also supported by direct phosphorylation experiments (as described below and in Table 2).
Table 1.
Comparing Sites Observed in FL-HCC Tissue (Higher in Tumor vs Adjacent Normal; Turnham et al., eLife, 2019)11 and the HEK293 J-PKAca and/or WT PKAca Overexpression Models
| all three | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| uniprot ID | gene name | entry name | protein name | site | sequence | directa |
| P12956 | XRCC6 | XRCC6_HUMAN | X-ray repair cross-complementing protein 6 (Ku70) | 520 | AMNKRLGSLVDEFKE | xxxx |
| P39023 | RPL3 | RL3_HUMAN | 60S ribosomal protein L3 | 13 | FSAPRHGSLGFLPRK | xxxx |
| Q02790 | FKBP4 | FKBP4_HUMAN | peptidyl-prolyl cis-trans isomerase FKBP4 | 78 | LDRKDKFSFDLGKGE | xx |
| Q08211 | DHX9 | DHX9_HUMAN | ATP-dependent RNA helicase A (DEAH box protein 9) | 449 | VTQPRRISAVSVAER | xx |
| Q04917 | YWHAH | 1433F_HUMAN | 14–3-3 protein eta (protein AS1) | 145 | ASGEKKNSVVEASEA | xx |
| Q14690 | PDCD11 | RRP5_HUMAN | protein RRP5 homologue (NF-kappa-B-binding protein) | 926 | ARKLRKGSEHQAIVQ | x |
| P11940 | PABPC1 | PABP1_HUMAN | polyadenylate-binding protein 1 (PABP-1) | 96 | RDPSLRKSGVGNIFI | x |
| P48147 | PREP | PPCE_HUMAN | prolyl endopeptidase (PE) | 667 | VGRSRKQSNPLLIHV | x |
| P62888 | RPL30 | RL30_HUMAN | 60S ribosomal protein L30 | 10 | AAKKTKKSLESINSR | x |
| Q9Y2W1 | THRAP3 | TR150_HUMAN | thyroid hormone receptor-associated protein 3 | 377 | EKIKEKGSFSDTGLG | x |
| Q9H0D6 | XRN2 | XRN2_HUMAN | 5′– 3′ exoribonuclease 2 | 678 | PEETRRNSLGGDVLF | x |
| P14625 | HSP90B1 | ENPL_HUMAN | endoplasmin (94 kDa glucose-regulated protein) (GRP-94) | 306 | AKEEKEESDDEAAVE | x |
| P48444 | ARCN1 | C0PD_HUMAN | coatomer subunit delta (delta-coat protein) | 437 | RHDSRRNTLEWCLPV | |
| Q5T5Y3 | CAMSAP1 | CAMP1_HUMAN | calmodulin-regulated spectrin-associated protein 1 | 629 | SIVSRRPSEGPQPLV | |
| 060716 | CTNND1 | CTND1_HUMAN | catenin delta-1 (cadherin-associated Src substrate) | 268 | PQVRVGGSSVDLHRF | |
| P12081 | HARS1 | HARS1_HUMAN | histidine-tRNA ligase, cytoplasmic | 27 | GLKQQKASAELIEEE | |
| P08670 | VIM | VIME_HUMAN | vimentin | 72 | SSAVRLRSSVPGVRL | |
| Q8WUI4 | HDAC7 | HDAC7_HUMAN | histone deacetylase 7 | 573 | CLRGRKASLEELQSV | |
| Q6P2E9 | EDC4 | EDC4_HUMAN | enhancer of mRNA-decapping protein 4 | 1389 | GKAARRLSLMLHGLV | |
| P53618 | C0PB1 | C0PB_HUMAN | coatomer subunit β (β-coat protein) | 933 | HIRIRAKSQGMALSL | |
| 095757 | HSPA4L | HS74L_HUMAN | heat shock 70 kDa protein 4L | 579 | LKKGKVKSIDLPIQS | |
| Q15149 | PLEC | PLEC_HUMAN | plectin | 2886 | PVRNRRLTVNEAVKE | |
| Q09666 | AHNAK | AHNK_HUMAN | neuroblast differentiation-associated protein AHNAK | 332 | PKAGLRVSAPEVSVG | |
| 000560 | SDCBP | SDCB1_HUMAN | syntenin-1 | 131 | KIGLRLKSIDNGIFV | |
| Q13501 | SQSTM1 | SQSTM_HUMAN | sequestosome-1 | 272 | RSRLTPVSPESSSTE | |
| Q15149 | PLEC | PLEC_HUMAN | plectin | 3785 | DYVRRRLTAEDLFEA | |
| P22626 | HNRNPA2B1 | R0A2_HUMAN | heterogeneous nuclear ribonucleoproteins A2/B1 | 225 | NFGPGPGSNFRGGSD | |
| P11586 | MTHFD1 | C1TC_HUMAN | C1-tetrahydrofolate synthase, cytoplasmic (C1-THF synthase) | 490 | VNGVRRFSDIQIRRL | |
| Q7Z406 | MYH14 | MYH14_HUMAN | myosin-14 (myosin heavy chain 14) | 105 | RMNPPKFSKAEDMAE | |
| 095613 | PCNT | PCNT_HUMAN | pericentrin | 1814 | ADQERRHSQALEALQ | |
| P35579 | MYH9 | MYH9_HUMAN | myosin-9 | 81 | KMNPPKFSKVEDMAE | |
| Q14980 | NUMA1 | NUMA1_HUMAN | nuclear mitotic apparatus protein 1 | 2047 | KQADRRQSMAFSILN | |
| P27816 | MAP-4 | MAP4_HUMAN | microtubule-associated protein 4 (MAP-4) | 636 | TGTGKKCSLPAEEDS | |
| P13861 | PRKAR2A | KAP2_HUMAN | cAMP-dependent protein kinase type II-α regulatory subunit | 58 | PAATPRQSLGHPPPE | |
| P98174 | FGD1 | FGD1_HUMAN | FYVE, RhoGEF, and PH domain-containing protein 1 | 135 | EGPQRLRSDPGPPTE | |
| P47897 | QARS1 | SYQ_HUMAN | glutamine-tRNA ligase | 70 | LRDTRRLSFLVSYIA | |
| Q8NDT2 | RBM15B | RB15B_HUMAN | putative RNA-binding protein 15B | 532 | ADLVRDRTPPHLLYS | |
| 095140 | MFN2 | MFN2_HUMAN | mitofusin-2 | 442 | AEEIRRLSVLVDDYQ | |
| 060832 | DKC1 | DKC1_HUMAN | H/ACA ribonucleoprotein complex subunit DKC1 | 21 | KKKKERKSLPEEDVA | |
| J-PKAca cells and FL-HCC | ||||||
|
| ||||||
| uniprot ID | gene name | entry name | protein name | site | sequence | directa |
| 043707 | ACTN4 | ACTN4_HUMAN | α-actinin-4 (nonmuscle α-actinin-4) | 423 | EKFRQKASIHEAWTD | xxx |
| 014579 | C0PE | C0PE_HUMAN | coatomer subunit epsilon | 99 | AHESRRDSIVAELDR | xx |
| P10809 | HSPD1 | CH60_HUMAN | 60 kDa heat shock protein, mitochondrial (Hsp60) | 488 | KNAGVEGSLIVEKIM | xx |
| Q14204 | DYNC1H1 | DYHC1_HUMAN | cytoplasmic dynein 1 heavy chain 1 | 1230 | DIMRRKDSAIQQQVA | xx |
| P07900 | HSP90AA1 | HS90A_HUMAN | heat shock protein HSP 90-α | 641 | LEINPDHSIIETLRQ | x |
| P21333 | FLN-A | FLNA_HUMAN | filamin-A (FLN-A) | 1084 | LQGGSAGSPARFTID | |
| Q16637 | SMN1 | SMN_HUMAN | survival motor neuron protein (Gemin-1) | 28 | FRRGTGQSDDSDIWD | |
| P61604 | HSPE1 | CH10_HUMAN | 10 kDa heat shock protein, mitochondrial (Hsp10) | 21 | DRVLVERSAAETVTK | |
| 060825 | PFKFB2 | F262_HUMAN | 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 2 | 466 | PVRMRRNSFTPLSSS | |
| Q12955 | ANK3 | ANK3_HUMAN | ankyrin-3 | 1459 | EKTDRRQSFASLALR | |
| P02545 | LMNA | LMNA_HUMAN | prelamin-A/C [cleaved into: lamin-A/C (70 kDa lamin)] | 392 | ERLRLSPSPTSQRSR | |
| Q9UI14 | RABAC1 | PRAF1_HUMAN | prenylated Rab acceptor protein 1 | 41 | WLERRRATIRPWSTF | |
| Q08211 | DHX9 | DHX9_HUMAN | ATP-dependent RNA helicase A | 1142 | IRQISRPSAAGINLM | |
| P83731 | RPL24 | RL24_HUMAN | 60S ribosomal protein L24 | 149 | IVKPVKVSAPRVGGK | |
| Q15126 | PMVK | PMVK_HUMAN | phosphomevalonate kinase (PMKase) | 113 | VSDTRRVSDIQWFRE | |
| J-PKAca cells and FL-HCC | ||||||
|
| ||||||
| uniprot ID | gene name | entry name | protein name | site | sequence | directa |
| Q9NZU5 | LMCD1 | LMCD1_HUMAN | LIM and cysteine-rich domains protein 1 | 16 | NPGVKKMSLGQLQSA | |
| Q8IVT5 | KSR1 | KSR1_HUMAN | kinase suppressor of Ras 1 | 888 | PKLNRRLSHPGHFWK | |
| 043707 | ACTN4 | ACTN4_HUMAN | α-actinin-4 (nonmuscle α-actinin-4) | 367 | LQTKLRLSNRPAFMP | |
| Q14789 | G0LGB1 | G0GB1_HUMAN | golgin subfamily B member 1 | 2489 | QLEERHLSIILEKDQ | |
| Q06210 | GFPT1 | GFPT1_HUMAN | glutamine-fructose-6-phosphate aminotransferase 1 | 205 | AVGTRRGSPLLIGVR | |
| 060488 | ACSL4 | ACSL4_HUMAN | long-chain-fatty-acid-CoA ligase 4 | 674 | IPIKVRLSPEPWTPE | |
| WT PKAca cells and FL-HCC | ||||||
|
| ||||||
| uniprot ID | gene name | entry name | protein name | site | sequence | directa |
| P11142 | HSPA8 | HSP7C_HUMAN | heat shock cognate 71 kDa protein | 329 | RDAKLDKSQIHDIVL | xx |
| P11142 | HSPA8 | HSP7C_HUMAN | heat shock cognate 71 kDa protein | 511 | TNDKGRLSKEDIERM | x |
| P78559 | MAP-1A | MAP1A_HUMAN | microtubule-associated protein 1A (MAP-1A) | 667 | EMEEVHPSDEEEEDA | |
| P17096 | HMGA1 | HMGA1_HUMAN | high mobility group protein HMG-I/HMG-Y | 102 | EEGISQESSEEEQ__ | |
| Q8NDT2 | RBM15B | RB15B_HUMAN | putative RNA-binding protein 15B | 609 | DRGRTTHSPYEERSR | |
| Q8WWI1 | LM0–7 | LM07_HUMAN | LIM domain only protein 7 (LMO-7) | 246 | RHHKREDSFESLDSL | |
| P27708 | CAD | PYR1_HUMAN | CAD protein (pyrimidine biosynthesis fusion complex) | 1406 | GAGGRRLSSFVTKGY | |
| Q16643 | DBN1 | DREB_HUMAN | drebrin | 142 | NGLARLSSPVLHRLR | |
| P49736 | MCM2 | MCM2_HUMAN | DNA replication licensing factor MCM2 | 139 | RRGLLYDSDEEDEER | |
| Q00610 | CLTC | CLH1_HUMAN | clathrin heavy chain 1 | 191 | YSVDRKVSQPIEGHA | |
| Q9NZB2 | FAM120A | F120A_HUMAN | constitutive coactivator of PPAR-γ-like protein 1 | 30 | LQKLARGSLVGGGRQ | |
| Q96S38 | RPS6KC1 | KS6C1_HUMAN | ribosomal protein S6 kinase delta-1 (S6K-delta-1) | 427 | LNRSPEESFDIKEVK | |
| Q9NNW5 | WDR6 | WDR6_HUMAN | WD repeat-containing protein 6 | 514 | VCGDRRGSVLLFPSR | |
| P02545 | LMNA | LMNA_HUMAN | prelamin-A/C [cleaved into: lamin-A/C (70 kDa lamin)] | 51 | VYIDRVRSLETENAG | |
| Q9UK76 | JPT1 | JUPI1_HUMAN | Jupiter microtubule-associated homologue 1 | 87 | SGLQRRNSSEASSGD | |
| Q8WX93 | PALLD | PALLD_HUMAN | palladin | 1121 | PRSRSRDSGDENEPI | |
| P46821 | MAP-1B | MAP1B_HUMAN | microtubule-associated protein 1B (MAP-1B) | 832 | AERSLMSSPEDLTKD | |
| Q5JRA6 | MIA3 | TG01_HUMAN | transport and Golgi organization protein 1 homologue (TANGO1) | 1539 | MALQKKLSQEEYERQ | |
| Q92905 | C0PS5 | CSN5_HUMAN | COP9 signalosome complex subunit 5 | 284 | EAQLGRGSFMLGLET | |
| Q02952 | AKAP12 | AKA12_HUMAN | A-kinase anchor protein 12 (AKAP12) | 627 | KKRVRRPSESDKEDE | |
| P21359 | NF-1 | NF1_HUMAN | neurofibromin (neurofibromatosis-related protein NF-1) | 2543 | KLLGTRKSFDHLISD | |
| Q9C0C9 | UBE20 | UBE20_HUMAN | (E3-independent) E2 ubiquitin-conjugating enzyme | 515 | SGTSRKKSIPLSIKN | |
| Q96S38 | RPS6KC1 | KS6C1_HUMAN | ribosomal protein S6 kinase delta-1 (S6K-delta-1) | 423 | ISKFLNRSPEESFDI | |
| Q6UX71 | PLXDC2 | PXDC2_HUMAN | plexin domain-containing protein 2 | 506 | AMKFRRGSGHPAYAE | |
| Q86UU1 | PHLDB1 | PHLB1_HUMAN | pleckstrin homology-like domain family B member 1 | 324 | GGHERPPSPGLRGLL | |
| 094806 | PRKD3 | KPCD3_HUMAN | serine/threonine-protein kinase D3 (protein kinase C nutype) | 731 | ARIIGEKSFRRSVVG | |
| A6NHR9 | SMCHD1 | SMHD1_HUMAN | structural maintenance of chromosomes flexible hinge domain-containing protein 1 | 1697 | ALLKRKLSEQEELKK | |
| Q15084 | PDIA6 | PDIA6_HUMAN | protein disulfide-isomerase A6 | 428 | VEDDIDLSDVELDDL | |
Number of direct phosphorylation observations at the protein or peptide level (see Supporting Table S1).
Table 2.
Summary of Phosphosites Seen in FL-HCC Tumor > Adj. Normal (Turnham)11 and as Direct Substrates in Rephosphorylation Reactions by J-PKAca and/or WT PKAca In Vitro
| all three | |||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| uniprot ID | gene name | entry name | protein name | site | sequence | peptide | protein |
| P12956 | XRCC6 | XRCC6_HUMAN | X-ray repair cross-complementing protein 6 (Ku70) | 520 | AMNKRLGSLVDEFKE | x | x |
| P39023 | RPL3 | RL3_HUMAN | 60S ribosomal protein L3 | 13 | FSAPRHGSLGFLPRK | x | x |
| P60174 | TPI1 | TPIS_HUMAN | triosephosphate isomerase | 21 | KMNGRKQSLGELIGT | x | x |
| 043707 | ACTN4 | ACTN4_HUMAN | α-actinin-4 | 423 | EKFRQKASIHEAWTD | x | xa |
| Q02790 | FKBP4 | FKBP4_HUMAN | peptidyl-prolyl cis-trans isomerase FKBP4 | 78 | LDRKDKFSFDLGKGE | xa | xa |
| 014579 | COPE | C0PE_HUMAN | coatomer subunit epsilon | 99 | AHESRRDSIVAELDR | x | |
| P10809 | HSPD1 | CH60_HUMAN | 60 kDa heat shock protein, mitochondrial | 488 | KNAGVEGSLIVEKIM | x | |
| P11142 | HSPA8 | HSP7C_HUMAN | heat shock cognate 71 kDa protein | 329 | RDAKLDKSQIHDIVL | x | |
| P04075 | ALDOA | ALD0A_HUMAN | fructose-bisphosphate aldolase A | 46 | SIAKRLQSIGTENTE | x | |
| Q08211 | DHX9 | DHX9_HUMAN | ATP-dependent RNA helicase A | 449 | VTQPRRISAVSVAER | x | |
| Q04917 | YWHAH | 1433F_HUMAN | 14–3-3 protein eta | 145 | ASGEKKNSVVEASEA | x | |
| Q14204 | DYNC1H1 | DYHC1_HUMAN | cytoplasmic dynein 1 heavy chain 1 | 1230 | DIMRRKDSAIQQQVA | x | |
| 095819 | MAP4K4 | M4K4_HUMAN | mitogen-activated protein kinase kinase kinase kinase 4 | 900 | QDPTRKGSVVNVNPT | x | |
| P16949 | STMN1 | STMN1_HUMAN | stathmin | 16 | KELEKRASGQAFELI | x | |
| P78527 | PRKDC | PRKDC_HUMAN | DNA-dependent protein kinase catalytic subunit | 893 | WDREKRLSFAVPFRE | x | |
| J-PKAca rephosphorylation and FL-HCC | |||||||
|
| |||||||
| uniprot ID | gene name | entry name | protein name | site | sequence | peptide | protein |
| Q9Y2W1 | THRAP3 | TR150_HUMAN | thyroid hormone receptor-associated protein 3 | 377 | EKIKEKGSFSDTGLG | x | |
| Q9H0D6 | XRN2 | XRN2_HUMAN | 5′–3′ exoribonuclease 2 (DHM1-like protein) | 678 | PEETRRNSLGGDVLF | x | |
| P07900 | HSP90AA1 | HS90A_HUMAN | heat shock protein HSP 90-α | 641 | LEINPDHSIIETLRQ | x | |
| P62987 | UBA52 | RL40_HUMAN | ubiquitin-60S ribosomal protein L40 | 57 | LEDGRTLSDYNIQKE | x | |
| P31040 | SDHA | SDHA_HUMAN | succinate dehydrogenase flavoprotein subunit, mitochondrial | 530 | AAVFRVGSVLQEGCG | x | |
| P14625 | HSP90B1 | ENPL_HUMAN | endoplasmin (94 kDa glucose-regulated protein) | 306 | AKEEKEESDDEAAVE | x | |
| 060749 | SNX2 | SNX2_HUMAN | sorting nexin-2 | 185 | FSVKRRFSDFLGLHS | x | |
| P49419 | ALDH7A1 | AL7A1_HUMAN | α-aminoadipic semialdehyde dehydrogenase | 84 | IARVRQASVADYEET | x | |
| Q9Y230 | RUVBL2 | RUVB2_HUMAN | RuvB-like 2 | 419 | VCRKRKGTEVQVDDI | x | |
| Q9NR30 | DDX21 | DDX21_HUMAN | nucleolar RNA helicase 2 | 121 | VTKNEEPSEEEIDAP | x | |
| Q02543 | RPL18A | RL18A_HUMAN | 60S ribosomal protein L18a | 123 | RHRARAHSIQIMKVE | x | |
| P27708 | CAD | PYR1_HUMAN | CAD protein (pyrimidine biosynthesis fusion complex) | 1859 | PPRIHRASDPGLPAE | x | |
| WT PKAca rephosphorylation and FL-HCC | |||||||
|
| |||||||
| uniprot ID | gene name | entry name | protein name | site | sequence | peptide | protein |
| P34932 | HSPA4 | HSP74_HUMAN | heat shock 70 kDa protein 4 | 76 | RFHGRAFSDPFVEAE | x | |
| Q14690 | PDCD11 | RRP5_HUMAN | protein RRP5 homologue (programmed cell death protein 11) | 926 | ARKLRKGSEHQAIVQ | x | |
| P11940 | PABPC1 | PABP1_HUMAN | polyadenylate-binding protein 1 | 96 | RDPSLRKSGVGNIFI | x | |
| P48147 | PREP | PPCE_HUMAN | prolyl endopeptidase | 667 | VGRSRKQSNPLLIHV | x | |
| P62888 | RPL30 | RL30_HUMAN | 60S ribosomal protein L30 | 10 | AAKKTKKSLESINSR | x | |
| P11142 | HSPA8 | HSP7C_HUMAN | heat shock cognate 71 kDa protein | 511 | TNDKGRLSKEDIERM | x | |
Seen in either J-PKAca or WT PKAca rephosphorylation but not both; see Supporting Table S1 for details.
Identification of Direct In Vitro Substrates of PKAca and J-PKAca Using HEK293 Cell Line Models
We were interested in determining which of the phosphorylation sites observed in our experiments were most likely direct substrates of either J-PKAca and/or WT PKAca. PKA is known to phosphorylate a number of motifs that have variations on the basic R/K-R/K-X-S/T motif, including R-X-R-R-X-S-Φ (where Φ is a hydrophobic residue), R-R-X-S-Φ, and R-R-X-S/T;29 however, these motifs are very simplistic and generic, with insufficient information about the full range of amino acids,especially C-terminal to the phosphosite, to adequately facilitate identification of direct substrates from cell-based phosphosite profiles. To explore this, we chose the Kinase Assay LInked with Phosphoproteomics (KALIP) approach,14,16,20 in which cell lysates from the HEK293 kinase construct overexpression models were used as substrate pools for rephosphorylation by purified kinase in vitro (as illustrated in Figure 1). This provided a more relevant comparison to the phosphosite profiles we observed from cell-based phosphoproteomics described in the previous section. Previously, in the work by Sugiyama et al., PKA was one of >360 kinases profiled for phosphorylation of whole-protein substrates in vitro, using HeLa cell lysate as the substrate pool.28 In that work, cell lysates prepared in a nondenaturing buffer were treated with phosphatase to remove pre-existing phosphate groups and then heated to 75 °C to inactivate the phosphatase, which also could have denatured many of the proteins.30 We were most interested in maintaining the native structural states of the proteins and avoiding heat treatment, so we used whole-protein lysate from the cells overexpressing J-PKAca or WT PKAca as the substrate pool after inactivating the phosphatase using phosphatase inhibitors. However, heat treatment would have also denatured endogenous kinases that may produce background phosphorylation; since we did not heat-treat, those kinases may have still been active. This led to the caveat that some observations may not result from direct phosphorylation. Accordingly, we also performed KALIP rephosphorylation on the trypsin digest of the lysate from cells overexpressing J-PKAca or WT PKAca, which was subject to its own caveat that trypsin digest would disrupt the N-terminal R/K motifs typically present in PKA substrates. However, by combining the information from these two experiments, we were able to balance the caveats of each to learn more about the C-terminal substrate motifs independent of the optimal N-terminal motif, confirm the amino acid patterns seen for +1 to +4 in the work from Sugiyama et al., compare substrate profiles for J-PKAca and WT PKAca, and cross-reference for sites also observed in FL-HCC tumor tissues by Turnham et al.11
Peptide Rephosphorylation.
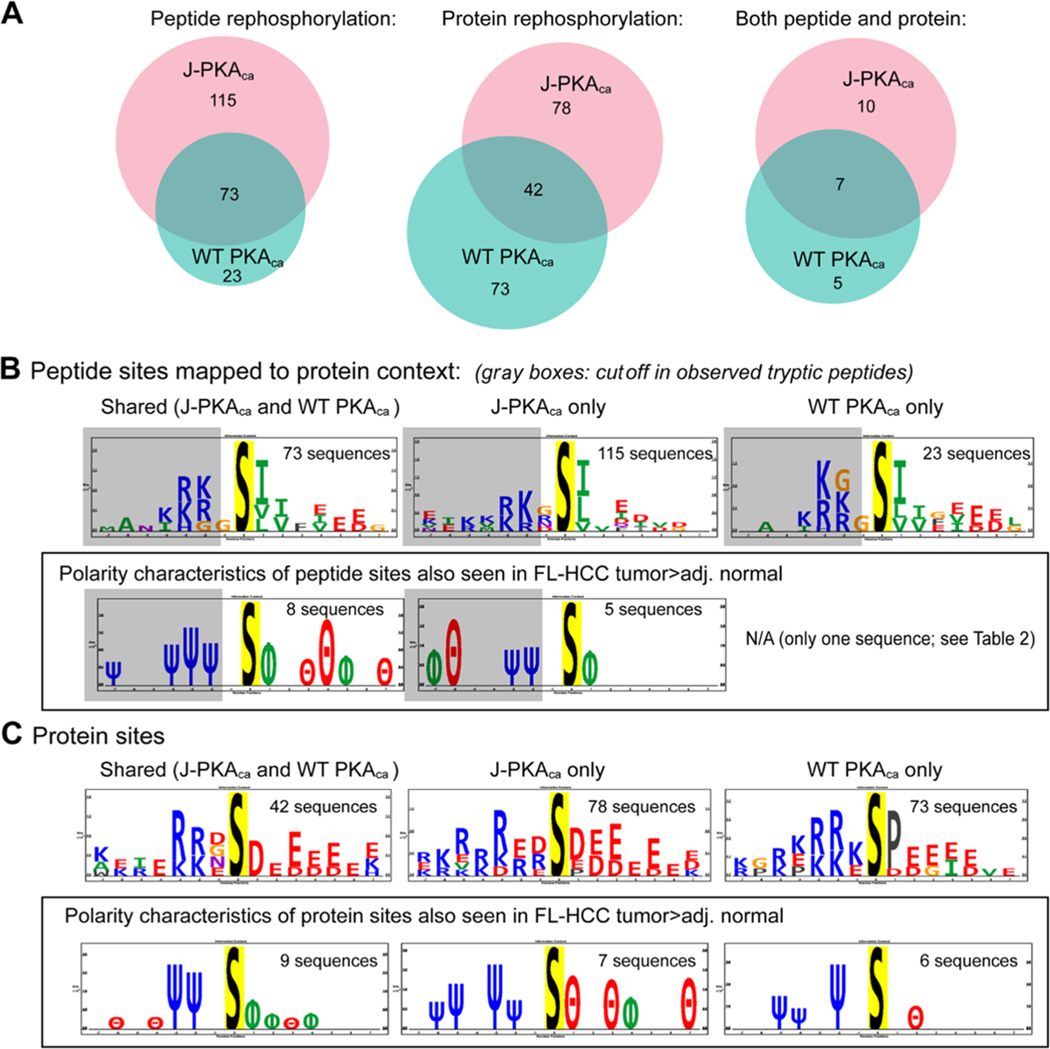
Considering only the peptides robustly identified in both kinase reaction replicates, 188 phosphopeptides were observed for the J-PKAca vs 96 for the WT PKAca in the peptide rephosphorylation experiments and 120 phosphorylation sites on 96 proteins for J-PKAca vs 116 sites on 99 proteins for WT PKAca in the protein rephosphorylation experiments. In general, the phosphorylation motifs observed in the peptide experiment matched the known hydrophobic motif-S-Φ-Φ-C-terminal to the phosphorylation site, which several studies (including an expressed oriented peptide library consisting of S-centered sequences from human proteins reported by the Rinehart and Turk, as well as the protein rephosphorylation study by Sugiyama et al.) have previously reported.19,28 The vast majority of the sequences surrounding the sites were truncated in the tryptic digest that was used for peptide rephosphorylation, with very little missed cleavage; thus, the peptides themselves in the kinase reaction did NOT have the common N-terminal -(R/K)-(R/K)-X-S-motif. However, interestingly, when mapped back to the context of their full proteins, the sites phosphorylated still largely contained the N-terminal positively charged residues (R/K) at −2 and −3 (Figure 3B). We speculate that perhaps this reflects the evolution of direct PKA substrates as containing both N- and C-terminal components of the motif and that the motif on the C-terminal side of the phosphosite is to some degree sufficient to direct PKA phosphorylation as seen in our peptide rephosphorylation experiment. Further, a much larger number of unique phosphopeptides were seen for the J-PKAca peptide reaction, and the sequence motif for those was slightly different from that for the shared and WT PKAca-unique sequences, showing less enrichment of hydrophobic amino acids at +2 and more acidic amino acids at +3. These differences suggest some intrinsic distinctions in substrate recognition of J-PKAca vs WT PKAca— for example, this may indicate that J-PKAca substrate recognition is less reliant on the N-terminal basic amino acid motif, with the C-terminal motif being sufficient for phosphorylation by the mutant. It could also mean that the J-domain alters catalytic dynamics in a way that depends more on the C-terminal motif and leads to increased turnover for more substrates, which may not be reflected in the Km or Vmax values previously characterized using a limited set of substrates that all contained the N-terminal basic motif.31,32
Figure 3.
Peptide and protein rephosphorylation experiments. (A) Venn diagrams illustrating the number of phosphopeptides robustly observed in both replicates of the peptide and/or protein rephosphorylation experiment using recombinant J-PKAca or WT PKAca, showing common and unique peptides for each. (B) Sequence motif analysis using PTM-logo23 (sites mapped to their protein context) for peptide rephosphorylation experiment, overall (top) and highlighting amino acid polarity characteristics for those also seen as higher in FL-HCC tumors by Turnham et al.11 (in a box). Almost all observed phosphopeptides were truncated within 1–2 amino acid N-terminals of the phosphosite (illustrated by gray boxes in sequence logos). Number of sequences used to generate motifs provided for context. (C) Sequence motifs for sites observed by protein rephosphorylation, including all sites observed (top; likely contains upstream/downstream sites as well as direct) and those also seen as more abundant in FL-HCC tumor tissues11 (as for peptide rephosphorylation in a box). Additional detailed information (UniProt identifiers, phosphosites, tc.) provided in Table 2 and Supporting Table S1. Charge logo legend: Ψ = basic residue, e.g., K/R; θ = acidic residue, e.g., D/E; ϕ = hydrophobic residue; P = proline; Δ = other.
Protein Rephosphorylation.
For the protein rephosphorylation experiments, the background phosphorylation by nondenatured kinases in the cell lysate is evident from the sequence motifs observed (Figure 3C); however, several were also identified by Sugiyama et al.28 (see Supporting Table S2) and/or as higher in FL-HCC tumor than in normal tissue by Turnham et al.11 (Supporting Table S1), which did exhibit the expected PKA N- and/or C-terminal motifs. These observations suggest that those sites also identified in Turnham et al.11 are likely to be bona fide direct substrates of J-PKAca and/or WT PKAca in FL-HCC tumor tissues (summary of these proteins is shown in Table 2).
Both Peptide and Protein Rephosphorylation.
Twenty-two phosphosites were identified in common in both the peptide and protein experiments (17 sites for J-PKAca and 12 sites for WT PKAca). Of those, 10 were unique to J-PKAca, 5 to WT PKAca, and 7 were common between both (Figure 3A). Three of the sites in common sites were also seen as higher in tumor than normal in FL-HCC tissues11 (Table 2): the DNA repair protein XRCC6/Ku70 (Ku70 S520), 60S ribosomal protein L3 (RPL3 S13), and triosephosphate isomerase (TPIS S21). All three of these sites have been reported to be phosphorylated in many prior studies (as curated in the PhosphoSitePlus database, https://phosphosite.org); RPL3 S13 and TPIS S21 were also observed in the PKA protein rephosphorylation experiment performed by Sugiyama et al.; however, they have not been studied in depth as PKA substrates, and Ku70 S520 has not been previously associated with PKA.
Phosphoproteomics of Inhibitor-Treated Cells Expressing J-PKAca or WT PKAca
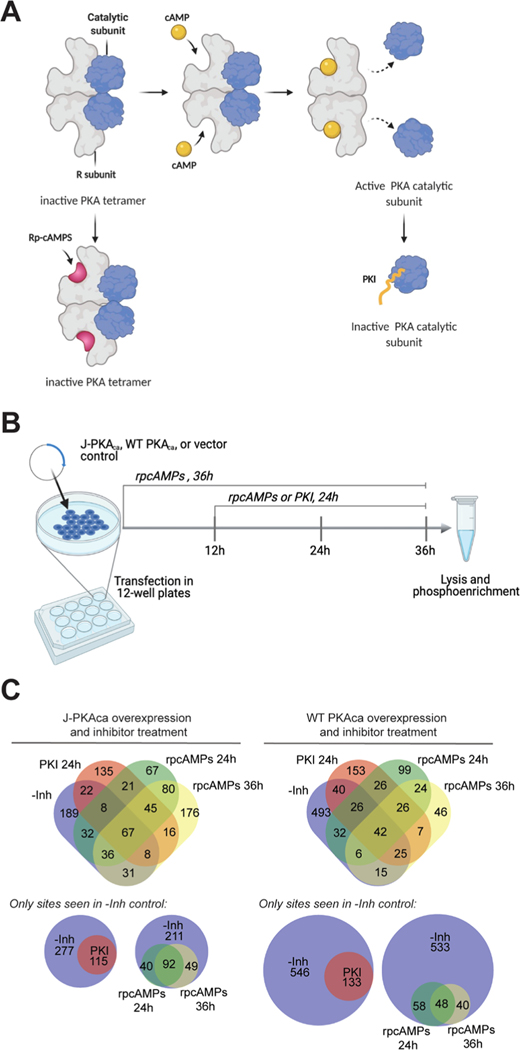
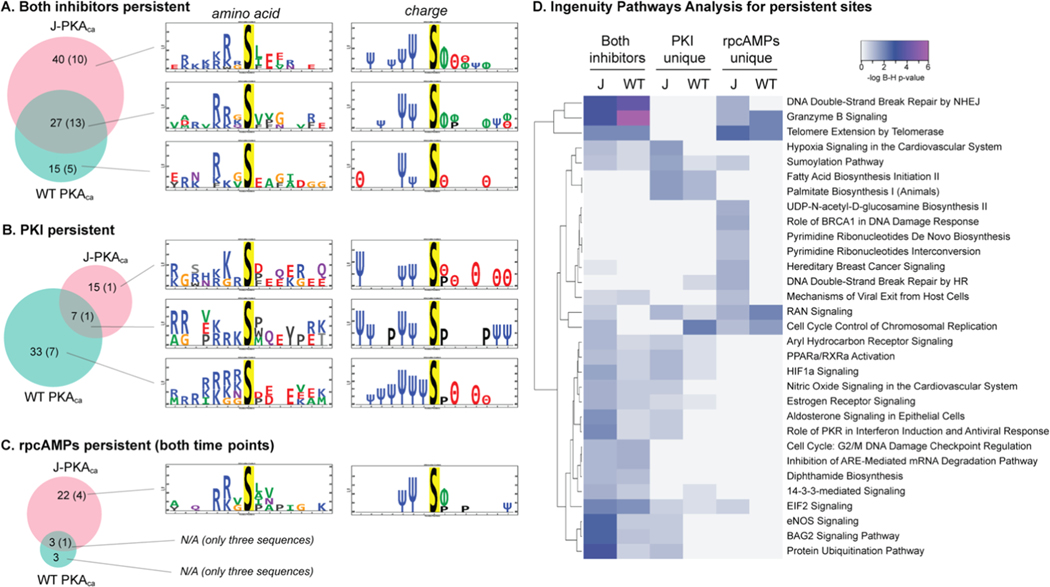
We were also interested in how PKA inhibitors would affect the phosphorylation profile in cells expressing chimeric J-PKAca vs the WT PKAca. We applied two PKA inhibitors with different inhibition mechanisms, rpcAMPs and a truncated, myristoylated form of protein kinase A inhibitor peptide (PKI14–22) (Figure 4). rpcAMPs competitively inhibit the cAMP-induced activation of the kinase by binding to the regulatory subunit (best characterized for RIα) and stabilizing the inactive conformation of the PKAca catalytic subunit in the holoenzyme,33,34 while PKI is an endogenous peptide that competitively inhibits substrate binding to the catalytic subunit.31,35 Cells overexpressing each version of the kinase were treated with each inhibitor in culture across different points in the time line between transfection and harvesting (Figure 4B); lysates were harvested in the presence of phosphatase inhibitor, and phosphopeptides were enriched and identified by LC-MS/MS. Two incubation time span experiments were performed for rpcAMPs (36 h coincubation for the entire transfection time line and addition after 12 h from transfection for 24 h inhibitor treatment). PKI was added after 12 h for a 24 h inhibitor treatment to minimize any potential effects of peptide biostability on the results; myristoylated PKI14–22 has been used in cell-based experiments as long as 24–96 h;36,37 therefore, 24 h should be well within its biostability window. Both inhibitors resulted in a noticeable reduction of phosphopeptides for PKAca and J-PKAca (Figure 4C); however, full 36 h cotreatment with rpcAMPs during the course of J-PKAca overexpression was also associated with observation of a substantial number of new phosphopeptides not seen in the -inh control. Further, a subset of certain phosphopeptides seemed to persist in cells expressing each version of the kinase even in the presence of each inhibitor (Figure 4C and Table 3), with additional subsets persisting in PKI-treated cells or in rpcAMP-treated cells through both time points (see Supporting Table S1 for full lists).
Figure 4.
Phosphoproteomics of inhibitor-treated cells. (A) Different mechanisms of inhibition for two cAMP-PKA specific inhibitors PKI and rpcAMPs. rpcAMP binds to the PKA regulatory subunit and prevents its dissociation from the catalytic subunit and is also resistant toward cyclic nucleotide phosphodiesterases. PKI is a substrate-competitive inhibitor of the catalytic subunit. (B) Experimental design: cells cultured in 12-well plates were transfected in duplicate with J-PKAca, WT PKAca, or empty vector and incubated for 36 h. Inhibitor was added at t = 0 h (rpcAMPs) or t = 12 h (PKI or rpcAMPs) for 36 or 24 h treatment, followed by lysis, trypsin digest, and phosphoenrichment for LC-MS/MS (created with BioRender.com). (C) Venn diagrams illustrating the outcomes from experiments by kinase construct and inhibitor. Top: four-way Venn diagrams (created with http://bioinformatics.psb.ugent.be/webtools/Venn/) showing numbers of phosphosites identified in each sample but not the corresponding vector-only control. Bottom: subsetted, simplified proportional Venn diagrams (created with http://biovenn.nl) showing only phosphosites observed in the -Inh controls and the various inhibitor treatments.
Table 3.
Persistent Phosphorylation Sites Observed
| persistently observed: both kinase constructs, each inhibitor, all time points | |||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| uniprot ID | gene name | entry name | protein name | site | sequence | in Turnham tumor > normal? | direct evidence? |
| P08670 | VIM | VIME_HUMAN | vimentin | 72 | SSAVRLRSSVPGVRL | x | |
| P11940 | PABPCi | PABP1_HUMAN | polyadenylate-binding protein 1 (PABP-1) | 96 | RDPSLRKSGVGNIFI | x | x |
| P12956 | XRCC6 | XRCC6_HUMAN | X-ray repair cross-complementing protein 6 (Ku70) | 520 | AMNKRLGSLVDEFKE | x | xxxx |
| P13861 | PRKAR2A | KAP2_HUMAN | cAMP-dependent protein kinase type II-α regulatory subunit | 58 | PAATPRQSLGHPPPE | x | |
| P48147 | PREP | PPCE_HUMAN | prolyl endopeptidase | 667 | VGRSRKQSNPLLIHV | x | x |
| Q02790 | FKBP4 | FKBP4_HUMAN | peptidyl-prolyl cis—trans isomerase FKBP4 | 78 | LDRKDKFSFDLGKGE | x | xx |
| Q04917 | YWHAH | 1433F_HUMAN | 14–3-3 protein eta | 145 | ASGEKKNSVVEASEA | x | xx |
| P78527 | PRKDC | PRKDC_HUMAN | DNA-dependent protein kinase catalytic subunit | 893 | WDREKRLSFAVPFRE | x | xx |
| P55010 | EIF5 | IF5_HUMAN | eukaryotic translation initiation factor 5 | 410 | VVYSKAASVPKVETV | ||
| O00231 | PSMD11 | PSD11_HUMAN | 26S proteasome non-ATPase regulatory subunit 11 | 14 | VEFQRAQSLLSTDRE | x | |
| P26373 | RPL13 | RL13_HUMAN | 60S ribosomal protein L13 | 77 | VRAGRGFSLEELRVA | x | |
| P08670 | VIM | VIME_HUMAN | vimentin | 73 | SAVRLRSSVPGVRLL | ||
| Q9P2K5 | MYEF2 | MYEF2_HUMAN | myelin expression factor 2 (MEF-2) | 520 | GMRERIGSKGNQIFV | ||
| P52272 | HNRNPM | HNRPM_HUMAN | heterogeneous nuclear ribonucleoprotein M | 365 | ENMGRFGSGMNMGRI | - | |
| P23588 | EIF4B | IF4B_HUMAN | eukaryotic translation initiation factor 4B (eIF-4B) | 359 | SQSTRAASIFGGAKP | ||
| Q12872 | SFSWAP | SFSWA_HUMAN | splicing factor, suppressor of white-apricot homologue | 238 | KAKQARNSQFDFLRF | ||
| Q14980 | NUMA1 | NUMA1_HUMAN | nuclear mitotic apparatus protein 1 | 1969 | QETLRRASMQPIQIA | ||
| Q9NR30 | DDX21 | DDX21_HUMAN | nucleolar RNA helicase 2 | 487 | LKGFRNGSFGVLVAT | ||
| P52272 | HNRNPM | HNRPM_HUMAN | heterogeneous nuclear ribonucleoprotein M | 397 | GGGGGGGSVPGIERM | x | |
| Q13310 | PABPC4 | PABP4_HUMAN | polyadenylate-binding protein 4 (PABP-4) | 96 | RDPSLRKSGVGNVFI | ||
| P07900 | HSP90AAi | HS90A_HUMAN | heat shock protein HSP 90-α | 330 | HLAVKHFSVEGQLEF | xx | |
| Q9UHXi | PUF60 | PUF60_HUMAN | poly(U)-binding-splicing factor PUF60 | 206 | NIKVGRPSNIGQAQP | x | |
| Q14320 | FAM50A | FA50A_HUMAN | protein FAM50A | 276 | VTKARGKSGPLFNFD | x | |
| Q6P2Q9 | PRPF8 | PRP8_HUMAN | pre-mRNA-processing-splicing factor 8 | 2079 | EWRVRAISAANLHLR | ||
| P50502 | ST13 | F10A1_HUMAN | Hsc70-interacting protein (Hip) | 156 | ILYAKRASVFVKLQK | ||
| P20700 | LMNBi | LMNBi_HUMAN | lamin-B1 | 158 | TALGDKKSLEGDLED | ||
| Pi3639 | EEF2 | EF2_HUMAN | elongation factor 2 (EF-2) | 23 | KANIRNMSVIAHVDH | xx | |
It is difficult to confidently evaluate the motifs for phosphosites that were not observed in the presence of inhibitor vs the control since the absence of evidence is not necessarily evidence of absence (particularly for mass spectrometry of peptides). However, it is useful to evaluate the sequence characteristics for the sites that were consistently observed across conditions. We found some interesting differences in the motifs observed for phosphosites that persisted in the presence of different inhibitors with each kinase construct (Figure 5). Generally, the sites that persisted in the presence of both inhibitors that were either unique for J-PKAca or shared for both kinase constructs exhibited motifs that were more similar to the known substrate preference motif (K/R at −2 and/or −3 and hydrophobic residues at +1 and/or +2) and were different from those persisting only in the WT PKAca inhibitor experiments (Figure 5A). Also, a larger number of persistent sites were seen uniquely for J-PKAca than for WT PKAca. This might indicate that J-PKAca is more resistant to inhibition via either mode (substrate-competitive and regulatory subunit stabilization, as illustrated in Figure 4A). Differences were seen for which sites persisted in the presence of PKI vs rpcAMPs, as well. The number of sites that persisted through PKI treatment was greater for the WT PKAca-expressing cells than for J-PKAca, although the motifs were not very different (when generalized to polarity) and were not very similar to PKA substrate preference motifs; also, very few sites persisted in the presence of rpcAMPs but not PKI for WT PKAca. On the other hand, a much larger number of sites persisted through rpcAMP treatment in J-PKAca-overexpressing cells than for WT PKAca, and the sequences around those sites closely resembled the PKA substrate motif. This may suggest that rpcAMP, in particular, is less effective at J-PKAca inhibition, while PKI inhibition of the chimeric form is not as impacted.
Figure 5.
Motifs observed for persistent phosphosites. Venn diagrams for number of sites that persisted in the presence of inhibitors for each kinase construct (number of those that also had evidence that they may be direct substrates indicated in parentheses) and amino acid/polarity motifs for the sets of sequences associated with each group. (A) Both inhibitors, all time points. (B) Persisting through PKI but not rpcAMPs. (C) Persisting through both time courses of rpcAMPs but not PKI (Venn diagrams created with http://biovenn.nl; motif logos created with PTM-logo23). Charge logo legend: Ψ = basic residue, e.g., K/R; θ = acidic residue, e.g., D/E; ϕ = hydrophobic residue; P = proline. (D) Heatmap generated using https://heatmapper.ca illustrating ingenuity pathways analysis (Supporting Table S4) showing the log10 Benjamini–Hochberg p-values for enrichment of canonical pathways represented by the sites observed as persistent in the presence of both inhibitors or uniquely for each inhibitor, per kinase construct. J = K-PKAca; WT = WT PKAca.
DISCUSSION
PKA signaling is involved in the control of a wide variety of cellular processes from metabolism to ion channel activation, cell growth and differentiation, gene expression, and apoptosis. The J-PKAca hallmark mutation in FL-HCC seems to affect these processes in complex ways that go beyond straightforward amplification of normal PKA pathways, including novel activation of Wnt/β-catenin, Ras–Raf–Erk pathways, and others,7,11 and also seems to evade the regulatory control of endogenous inhibitors like the PKI peptides, despite the expression of those inhibitors in tumor cells and a lack of difference in biochemical inhibition by various forms of PKI in vitro for J-PKAca vs WT PKAca.31 So far, although pathways have been characterized and some upstream/downstream kinase activation has been predicted,11 the direct kinase–substrate relationships that lead to these signaling outcomes have not been identified, and the molecular mechanisms by which J-PKAca promotes oncogenesis in FL-HCC are still not fully understood. While the work described here does not comprehensively map or prove these mechanisms, it provides evidence for potential direct substrates that may be important to pursue in the future.
Disrupted Regulatory Interactions vs Unique Downstream Substrate/Pathway Activation
Turnham et al. proposed that the J-domain/HSP40 promotes aberrant interactions with novel substrates through its chaperone functions.11 Other recent work has proposed that changes to the allosteric connectivity in the kinase resulting from the J-domain fusion affect the ability of the chimeric version of the kinase to interact with its regulatory domains, leading to aberrant phosphorylation and activation of established PKA signaling pathways.12,32 Our data contain evidence that supports both of these proposals, and our experiments are particularly helpful for comparing J-PKAca with the WT kinase head-to-head. On the one hand, we observed overall that the pathways induced by expression of the J-PKAca mutant were not substantially different from those induced by WT PKAca, and most of the phosphorylation sites we detected in J-PKAca-overexpressing cells that were shared with tumor vs normal FL-HCC tissue (Turnham et al.11) were also detected in WT PKAca-over-expressing cells. This suggests that for the most part, J-PKAca overexpression produces very similar downstream outcomes as just overexpressing/overactivating PKA activity more generally, supporting the idea that the key differences lie in the regulation of PKA kinase activity and not necessarily in new substrates phosphorylated by the mutant. On the other hand, some specific phosphorylation sites we detected in J-PKAca-expressing cells, but not in WT, also support that there may be particular events related to the J-domain chimera, e.g., phosphorylation of KSR1 at S888, which is homologous to a PKA-phosphorylated site in murine KSR1 that has been linked to cAMP/PKA activation of the ERK1/2 cascade38 (a pathway that was suggested as key to the J-PKAca mechanism by Turnham et al.), and/or additional heat shock proteins by J-PKAca (Tables 1 and 2). However, most of the J-PKAca-“unique” sites from each given experiment were also seen across various versions of the WT experiments, suggesting that they are not truly unique to the mutant. S423 of actinin-4 (ACTN4 S23) was the one apparently direct site that was seen in all three J-PKAca experiments, but only in WT PKAca peptide rephosphorylation and not the corresponding WT protein rephosphorylation and cell experiments; this site was also higher in tumor vs normal in FL-HCC and may be worth additional study, given the ACTN4’s role in malignancy and metastasis.39
We also identified sites and pathways that were differentially affected by the PKA inhibitors rpcAMPs and PKI(14–22), leading to a set of phosphorylation sites that persisted in the presence of each inhibitor (Figures 4 and 5). While we cannot rule out the potential that overexpression of the catalytic domain to levels higher than can be sufficiently modulated by the regulatory subunit available in the cell, and many of the persistent substrates and pathways were common to both forms of the enzyme (Table 3), intriguingly some were not (Figure 5). The J-PKAca chimera seemed to be particularly resistant to inhibition by rpcAMPs, which is consistent with the mechanism proposed by Lu et al. from cryo-EM structures of the holoenzyme,12 in which the J-domain seems to affect the dynamics and allostery of the regulatory subunit interactions, resulting in destabilization such that J-PKAca is more easily activated by cAMP. It is plausible that destabilization of those regulatory subunit interactions could also destabilize interaction of the mutant holoenzyme with rpcAMPs and enable cAMP to compete more effectively for J-PKAca binding, which would allow the mutant to retain higher activity in the presence of rpcAMPs at the concentrations used in our experiments. Also, the persistence of signaling pathways associated with pyrimidine biosynthesis and DNA damage response in J-PKAca-expressing cells in the presence of rpcAMPs (Figure 5D) further suggests that some unique mechanisms are present downstream of the mutant. Overall, our data should be useful to others for the study of the specific mechanisms that may govern aberrant activation resulting from amplification of PKA activity due to the J-domain fusion’s disruption of regulatory complexes, as well as potential unique downstream pathways.
Candidate Direct Substrates That May Govern Pathogenesis in FL-HCC
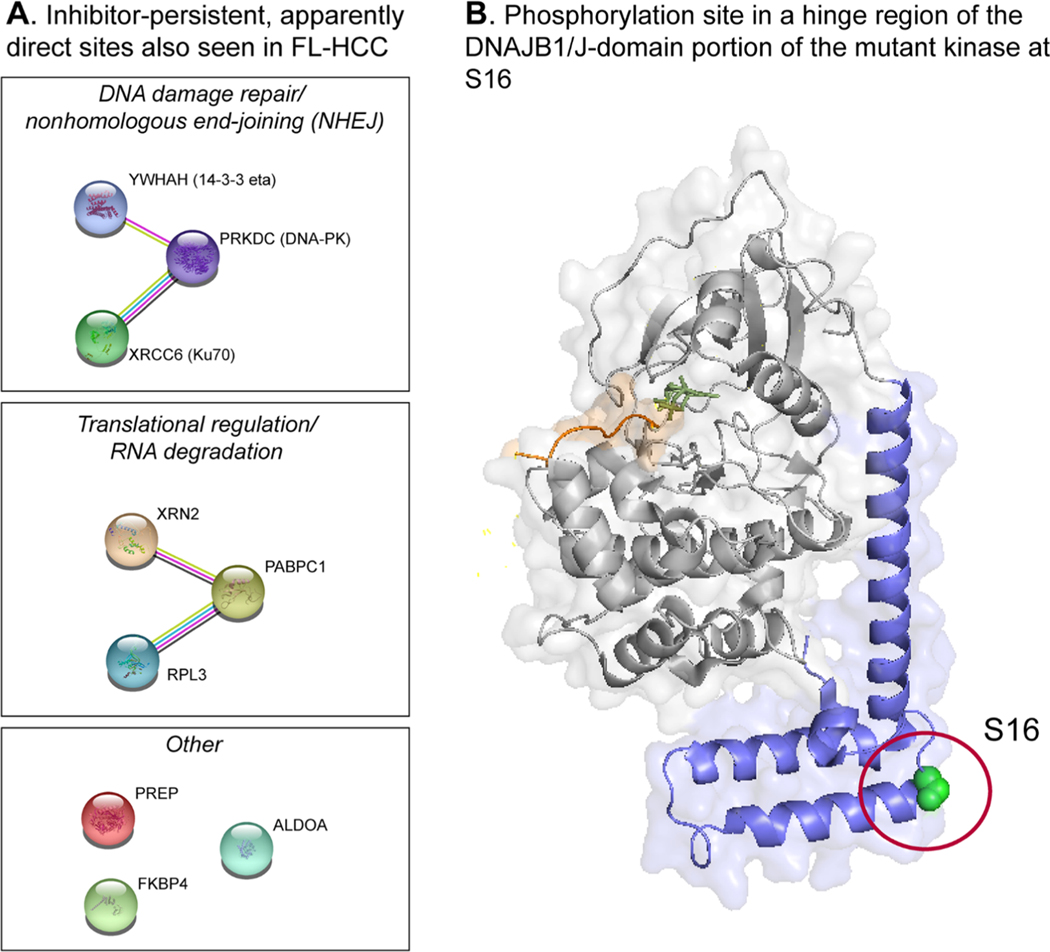
A key goal of this study was to identify direct substrates of J-PKAca that could be contributing to pathogenesis in FL-HCC. The KALIP workflow enabled us to determine a number of apparently direct substrates of both WT PKAca and J-PKAca, including some that were also observed by others in FL-HCC tumor tissues11 and that connect to pathways that are well supported by other studies. Two specific sites on two proteins were identified across all experiments (for mutant and WT kinase): S520 of XRCC6 (the DNA damage repair/nonhomologous end-joining-related protein Ku70) and S13 of RPL3 (a component of the 60S ribosome), strongly suggesting that they are direct substrates of both forms of the kinase. These sites were also persistent in the presence of inhibitors PKI and rpcAMPs. A number of other sites that were present in FL-HCC tissues were also apparently persistent in the presence of inhibitors in our experiments, and we saw evidence for direct phosphorylation by J-PKAca/WT PKAca for some of these (Table 3). One potentially notable site observed as persistent to inhibitors for both J-PKAca and WT was S58 on the regulatory subunit RIIα. This site is in a disordered region of the protein not visible in the published structures, but it has previously been observed in other phosphoproteomics studies40–43 and is close to a CDK2 phosphorylation site (T54) that is important for regulation of PKA localization to the centrosome during mitosis.41 According to STRING analysis,44 several of the apparently direct sites are on proteins associated with either RPL3 or Ku70 in complexes involved in translational regulation (RPL3) or nonhomologous end-joining (NHEJ) (Ku70) (Figure 6A). This raises the interesting possibility that deregulated PKA signaling via these particular complexes may be important to FL-HCC pathogenesis but also difficult to target via PKA directly, so other potential druggable targets in those networks (e.g., DNA-PK or the ribosome) may be valuable for future FL-HCC therapeutic development.
Figure 6. Specific sites of interest.
(A) Potential functional relationships between the proteins with sites we observed in direct phosphorylation experiments that were also seen in FL-HCC by Turnham et al.10 and to persist across conditions in our inhibitor experiments (J-PKAca and WT PKAca). (B) Illustration showing the location of the phosphosite observed for the J-domain (adapted from PDB entry 4WB7).
Phosphorylation of the J-Domain at S16
In all of our experiments with J-PKAca, we observed that S16 of the J-domain was phosphorylated, and we also observed this in the rephosphorylation experiments. However, because we were introducing recombinant J-PKAca in the kinase reaction, we cannot confirm that this is a direct autophosphorylation substrate of the mutant kinase, and this site was not reported in studies by Turnham et al.11 Further, it was also detected in one run of the WT control experiment (albeit in a single replicate and at a level close to the noise, far lower than in any J-PKAca experiment and likely an artifact of MS analysis), so we are cautious about overinterpreting this phosphorylation site. Nevertheless, this site is located on the hinge region of the J-domain (Figure 6B), and it was observed as persistent in all of the J-PKAca inhibitor experiments; given the importance of J-domain dynamics on the regulatory interactions described by other studies,12,32 it is possible that this phosphorylation event is significant, and it may be worthwhile for future structural studies to take it into account.
CONCLUSIONS
Overall, we identified a number of phosphorylation sites that are associated with deregulated PKA activity in these overexpression model systems that also were rephosphorylated in vitro by recombinant enzyme, suggesting that they are direct substrates of either J-PKAca, WT PKAca, or both. Even though we used HEK293 cells as our model and FL-HCC is a liver cancer, a substantial number of the sites we detected were consistent with sites detected at higher levels in FL-HCC tissues vs normal, lending confidence to the relevance of these phosphorylation events to this cancer. While not comprehensive examinations of the pathophysiology of this disease, the data presented here provide potential lead mechanisms for further evaluation as targets. Of those we identified, some of the key players (DNA-PK and the ribosome) are currently being pursued as drug targets in preclinical and clinical studies, and candidate compounds may be available for testing sensitivity of this cancer type. Since safely inhibiting PKA activity itself is a challenging prospect, the potential targets presented here may represent more viable opportunities for therapeutic intervention in FL-HCC.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health/National Cancer Institute through R01CA183571 (L.L.P.). The authors thank Sanford Simon for the expression plasmids used in this study. The table of contents graphic was created with BioRender.com. The authors also acknowledge John Blankenhorn for early data analysis iterations for the cell-based experiments in Protein Pilot 5.0 that were replaced by PEAKS Studio Xpro and not used in the final version of this manuscript.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.1c00500.
SDS-PAGE gel showing recombinant PKAca and J-PKAca expression and purification (Figure S1); kinase activity assay confirming active recombinant PKAca and J-PKAca (Figure S2); Western blot confirming PKAca and J-PKAca in HEK293 cells (Figure S3) (PDF)
Outputs from PEAKS results processed through the PEAKS-ModExtractor script; compare all with tumor > normal summary.xlsx (Table S1); list of PKA direct sites from Sugiyama Scientific Repts 2019.xlsx (Table S2); IPA comparison table, Turnham, J-PKAca–WT PKA.xlsx (Table S3); IPA comparison table inhibitor-persistent sites.xlsx (Table S4) (ZIP)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.jproteome.1c00500
The authors declare no competing financial interest.
Contributor Information
Adak Karamafrooz, Department of Biochemistry, Molecular Biology, and Biophysics, University of Minnesota, Minneapolis, Minnesota 55455, United States.
Jack Brennan, Independent Technology Consultant, LIC, Boston, Massachusetts 02129, United States.
David D. Thomas, Department of Biochemistry, Molecular Biology, and Biophysics, University of Minnesota, Minneapolis, Minnesota 55455, United States
Laurie L. Parker, Department of Biochemistry, Molecular Biology, and Biophysics, University of Minnesota, Minneapolis, Minnesota 55455, United States
REFERENCES
- (1).Eggert T; McGlynn KA; Duffy A; Manns MP; Greten TF; Altekruse SF Fibrolamellar hepatocellular carcinoma in the USA, 2000–2010: A detailed report on frequency, treatment and outcome based on the Surveillance, Epidemiology, and End Results database. United Eur. Gastroenterol. J 2013, 1, 351–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Torbenson M. Fibrolamellar carcinoma: 2012 update. Scientifica 2012, 2012, No. 743790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Ang CS; Kelley RK; Choti MA; Cosgrove DP; Chou JF; Klimstra D; Torbenson MS; Ferrell L; Pawlik TM; Fong Y; O’Reilly EM; Ma J; McGuire J; Vallarapu GP; Griffin A; Stipa F; Capanu M; Dematteo RP; Venook AP; Abou-Alfa GK Clinicopathologic characteristics and survival outcomes of patients with fibrolamellar carcinoma: data from the fibrolamellar carcinoma consortium. Gastrointest. Cancer Res 2013, 6, 3–9. [PMC free article] [PubMed] [Google Scholar]
- (4).Andersen JB Fibrolamellar hepatocellular carcinoma: a rare but distinct type of liver cancer. Gastroenterology 2015, 148, 707–710. [DOI] [PubMed] [Google Scholar]
- (5).Honeyman JN; Simon EP; Robine N; Chiaroni-Clarke R; Darcy DG; Lim II; Gleason CE; Murphy JM; Rosenberg BR; Teegan L; Takacs CN; Botero S; Belote R; Germer S; Emde AK; Vacic V; Bhanot U; LaQuaglia MP; Simon SM Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science 2014, 343, 1010–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Darcy DG; Chiaroni-Clarke R; Murphy JM; Honeyman JN; Bhanot U; LaQuaglia MP; Simon SM The genomic landscape of fibrolamellar hepatocellular carcinoma: whole genome sequencing of ten patients. Oncotarget 2015, 6, 755–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Kastenhuber ER; Lalazar G; Houlihan SL; Tschaharganeh DF; Baslan T; Chen CC; Requena D; Tian S; Bosbach B; Wilkinson JE; Simon SM; Lowe SW DNAJB1-PRKACA fusion kinase interacts with beta-catenin and the liver regenerative response to drive fibrolamellar hepatocellular carcinoma. Proc. Natl. Acad. Sci. U.S.A 2017, 114, 13076–13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Ferguson FM; Gray NS Kinase inhibitors: the road ahead. Nat. Rev. Drug Discovery 2018, 17, 353–377. [DOI] [PubMed] [Google Scholar]
- (9).Roskoski R. FDA-approved small molecule protein kinase inhibitor compilation. http://www.brimr.org/PKI/PKIs.htm. [DOI] [PubMed] [Google Scholar]
- (10).Propper DJ; Saunders MP; Salisbury AJ; Long L; O’Byrne KJ; Braybrooke JP; Dowsett M; Taylor M; Talbot DC; Ganesan TS; Harris AL Phase I study of the novel cyclic AMP (cAMP) analogue 8-chloro-cAMP in patients with cancer: toXicity, hormonal, and immunological effects. Clin. Cancer Res 1999, 5, 1682–1689. [PubMed] [Google Scholar]
- (11).Turnham RE; Smith FD; Kenerson HL; Omar MH; Golkowski M; Garcia I; Bauer R; Lau HT; Sullivan KM; Langeberg LK; Ong SE; Riehle KJ; Yeung RS; Scott JD An acquired scaffolding function of the DNAJ-PKAc fusion contributes to oncogenic signaling in fibrolamellar carcinoma. eLife 2019, 8, No. e44187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Lu TW; Aoto PC; Weng JH; Nielsen C; Cash JN; Hall J; Zhang P; Simon SM; Cianfrocco MA; Taylor SS Structural analyses of the PKA RIIbeta holoenzyme containing the oncogenic DnaJB1-PKAc fusion protein reveal protomer asymmetry and fusion-induced allosteric perturbations in fibrolamellar hepatocellular carcinoma. PLoS Biol. 2020, 18, No. e3001018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Bodenmiller B; Aebersold R. Quantitative analysis of protein phosphorylation on a system-wide scale by mass spectrometry-based proteomics. Methods Enzymol. 2010, 470, 317–334. [DOI] [PubMed] [Google Scholar]
- (14).Xue L; Arrington JV; Tao WA Identification of Direct Kinase Substrates via Kinase Assay-Linked Phosphoproteomics. Methods Mol. Biol 2016, 1355, 263–273. [DOI] [PubMed] [Google Scholar]
- (15).Xue L; Geahlen RL; Tao WA Identification of direct tyrosine kinase substrates based on protein kinase assay-linked phosphoproteomics. Mol. Cell. Proteomics 2013, 12, 2969–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Arrington J; Xue L; Wang WH; Geahlen RL; Tao WA Identification of the Direct Substrates of the ABL Kinase via Kinase Assay Linked Phosphoproteomics with Multiple Drug Treatments. J. Proteome Res 2019, 18, 1679–1690. [DOI] [PubMed] [Google Scholar]
- (17).Xue L; Wang WH; Iliuk A; Hu L; Galan JA; Yu S; Hans M; Geahlen RL; Tao WA Sensitive kinase assay linked with phosphoproteomics for identifying direct kinase substrates. Proc. Natl. Acad. Sci. U.S.A 2012, 109, 5615–5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Kettenbach AN; Wang T; Faherty BK; Madden DR; Knapp S; Bailey-Kellogg C; Gerber SA Rapid determination of multiple linear kinase substrate motifs by mass spectrometry. Chem. Biol 2012, 19, 608–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Barber KW; Miller CJ; Jun JW; Lou HJ; Turk BE; Rinehart J. Kinase Substrate Profiling Using a Proteome-wide Serine-Oriented Human Peptide Library. Biochemistry 2018, 57, 4717–4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Perez M; Blankenhorn J; Murray KJ; Parker LL High-throughput Identification of FLT3 Wild-type and Mutant Kinase Substrate Preferences and Application to Design of Sensitive In Vitro Kinase Assay Substrates. Mol. Cell. Proteomics 2019, 18, 477–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Choi J; Bomgarden R; Patel B; Foster L; Snovida S; Rogers J.c. Sequential Enrichment Using Metal Oxide Affinity Chromatography (SMOAC) to Enhance Phosphoproteome Coverage for Quantitative Proteomic Analysis; ThermoScientific, 2018. [Google Scholar]
- (22).Suni V; Imanishi SY; Maiolica A; Aebersold R; Corthals GL Confident site localization using a simulated phosphopeptide spectral library. J. Proteome Res 2015, 14, 2348–2359. [DOI] [PubMed] [Google Scholar]
- (23).Beausoleil SA; Villen J; Gerber SA; Rush J; Gygi SP A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol 2006, 24, 1285–1292. [DOI] [PubMed] [Google Scholar]
- (24).Hulsen T; de Vlieg J; Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 2008, 9, No. 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Mellacheruvu D; Wright Z; Couzens AL; Lambert JP; St-Denis NA; Li T; Miteva YV; Hauri S; Sardiu ME; Low TY; Halim VA; Bagshaw RD; Hubner NC; Al-Hakim A; Bouchard A; Faubert D; Fermin D; Dunham WH; Goudreault M; Lin ZY; Badillo BG; Pawson T; Durocher D; Coulombe B; Aebersold R; Superti-Furga G; Colinge J; Heck AJ; Choi H; Gstaiger M; Mohammed S; Cristea IM; Bennett KL; Washburn MP; Raught B; Ewing RM; Gingras AC; Nesvizhskii AI The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 2013, 10, 730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Saethang T; Hodge K; Yang CR; Zhao Y; Kimkong I; Knepper MA; Pisitkun T. PTM-Logo: a program for generation of sequence logos based on position-specific background amino-acid probabilities. Bioinformatics 2019, 35, 5313–5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Crooks GE; Hon G; Chandonia JM; Brenner SE WebLogo: a sequence logo generator. Genome Res. 2004, 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Sugiyama N; Imamura H; Ishihama Y. Large-scale Discovery of Substrates of the Human Kinome. Sci. Rep 2019, 9, No. 10503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Rust HL; Thompson PR Kinase consensus sequences: a breeding ground for crosstalk. ACS Chem. Biol 2011, 6, 881–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Rawat S; Raman Suri C; Sahoo DK Molecular mechanism of polyethylene glycol mediated stabilization of protein. Biochem. Biophys. Res. Commun 2010, 392, 561–566. [DOI] [PubMed] [Google Scholar]
- (31).Averill AM; Rehman HT; Charles JW; Dinh TA; Danyal K; Verschraegen CF; Stein GS; Dostmann WR; Ramsey JE Inhibition of the chimeric DnaJ-PKAc enzyme by endogenous inhibitor proteins. J. Cell. Biochem 2019, 120, 13783–13791. [DOI] [PubMed] [Google Scholar]
- (32).Olivieri C; Walker C; Karamafrooz A; Wang Y; Manu VS; Porcelli F; Blumenthal DK; Thomas DD; Bernlohr DA; Simon SM; Taylor SS; Veglia G. Defective internal allosteric network imparts dysfunctional ATP/substrate-binding cooperativity in oncogenic chimera of protein kinase A. Commun. Biol. 2021, 4, No. 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Badireddy S; Yunfeng G; Ritchie M; Akamine P; Wu J; Kim CW; Taylor SS; Qingsong L; Swaminathan K; Anand GS Cyclic AMP analog blocks kinase activation by stabilizing inactive conformation: conformational selection highlights a new concept in allosteric inhibitor design. Mol. Cell. Proteomics 2011, 10, No. M110004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Dostmann WR (RP)-cAMPS inhibits the cAMP-dependent protein kinase by blocking the cAMP-induced conformational transition. FEBS Lett. 1995, 375, 231–234. [DOI] [PubMed] [Google Scholar]
- (35).Wang Z; Cole PA Catalytic mechanisms and regulation of protein kinases. Methods Enzymol. 2014, 548, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Zheng XL; Matsubara S; Diao C; Hollenberg MD; Wong NC Activation of apolipoprotein AI gene expression by protein kinase A and kinase C through transcription factor, Sp1. J. Biol. Chem 2000, 275, 31747–31754. [DOI] [PubMed] [Google Scholar]
- (37).Farrow B; Rychahou P; Murillo C; O’Connor KL; Iwamura T; Evers BM Inhibition of pancreatic cancer cell growth and induction of apoptosis with novel therapies directed against protein kinase A. Surgery 2003, 134, 197–205. [DOI] [PubMed] [Google Scholar]
- (38).Smith FD; Langeberg LK; Cellurale C; Pawson T; Morrison DK; Davis RJ; Scott JD AKAP-Lbc enhances cyclic AMP control of the ERK1/2 cascade. Nat. Cell Biol 2010, 12, 1242–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Honda K. The biological role of actinin-4 (ACTN4) in malignant phenotypes of cancer. Cell Biosci. 2015, 5, No. 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Bian Y; Song C; Cheng K; Dong M; Wang F; Huang J; Sun D; Wang L; Ye M; Zou H. An enzyme assisted RP-RPLC approach for in-depth analysis of human liver phosphoproteome. J. Proteomics 2014, 96, 253–262. [DOI] [PubMed] [Google Scholar]
- (41).Carlson CR; Witczak O; Vossebein L; Labbe JC; Skalhegg BS; Keryer G; Herberg FW; Collas P; Tasken K. CDK1-mediated phosphorylation of the RIIalpha regulatory subunit of PKA works as a molecular switch that promotes dissociation of RIIalpha from centrosomes at mitosis. J. Cell Sci 2001, 114, 3243–3254. [DOI] [PubMed] [Google Scholar]
- (42).Dephoure N; Zhou C; Villen J; Beausoleil SA; Bakalarski CE; Elledge SJ; Gygi SP A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.A 2008, 105, 10762–10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Olsen JV; Vermeulen M; Santamaria A; Kumar C; Miller ML; Jensen LJ; Gnad F; CoX J; Jensen TS; Nigg EA; Brunak S; Mann M. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signaling 2010, 3, No. ra3. [DOI] [PubMed] [Google Scholar]
- (44).Szklarczyk D; Gable AL; Lyon D; Junge A; Wyder S; Huerta-Cepas J; Simonovic M; Doncheva NT; Morris JH; Bork P; Jensen LJ; Mering CV STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.