Abstract
The ability of Candida albicans to respond to diverse environments is critical for its success as a pathogen. The RIM101 pathway controls gene expression and the yeast-to-hyphal transition in C. albicans in response to changes in environmental pH in vitro. In this study, we found that the RIM101 pathway is necessary in vivo for pathogenesis. First, we show that rim101−/rim101− and rim8−/rim8− mutants have a significant reduction in virulence using the mouse model of hematogenously disseminated systemic candidiasis. Second, these mutants show a marked reduction in kidney pathology. Third, the rim101−/rim101− and rim8−/rim8− mutants show defects in the ability to damage endothelial cells in situ. Finally, we show that an activated allele of RIM101, RIM101-405, is a suppressor of the rim8− mutation in vivo as it rescues the virulence, histological, and endothelial damage defects of the rim8−/rim8− mutant. These results demonstrate that the RIM101 pathway is required for C. albicans virulence in vivo and that the function of Rim8p in pathogenesis is to activate Rim101p.
Candida albicans is a human pathogen of significant medical importance. It can cause life-threatening systemic infections in susceptible individuals, such as those with suppressed immune systems (18). In fact, C. albicans now ranks fourth on the list of most frequently acquired nosocomial bloodstream infections (20).
Hematogenously disseminated C. albicans colonizes diverse organs during infection (18). Several properties have been identified that are likely to be required for successful dissemination and to cause disease, including secretion of degradative enzymes and the ability to switch between the yeast and hyphal growth forms (3, 10, 17). The ability to undergo the yeast-to-hyphal transition appears to be critical for pathogenesis, as mutants unable to form hyphae are less virulent in the hematogenously disseminated mouse model (8, 14, 16). This transition is regulated by signal transduction pathways that respond to environmental signals and stimulate changes in gene expression. One environmental signal that regulates the yeast-to-hyphal transition in vitro is extracellular pH. The ability to respond to extracellular pH appears to be important in vivo. For example, De Bernardis et al. have shown that mutants which are unable to grow at a given pH in vitro are limited in their sites of infection in vivo (5).
We identified two homologs of the RIM101 pathway in C. albicans, RIM101 and RIM8, also referred to as PRR2 and PRR1, respectively (4, 21, 22). In Saccharomyces cerevisiae and Aspergillus nidulans, the zinc finger transcription factor Rim101p (PacCp) is regulated by proteolytic processing in response to environmental pH (15, 19). At alkaline pH, several proteins, including Rim8p (PalFp), stimulate the processing of Rim101p to the active form which governs changes in gene expression (15, 19). In C. albicans, the RIM101 pathway regulates several alkaline responses, including stimulation of alkaline response genes, repression of acidic response genes, and stimulation of the yeast-to-hyphal transition (4, 21, 22). However, the RIM101 pathway is not required for growth at either alkaline or acidic pH in vitro. Because the RIM101 pathway plays an important role in responding to the environment in vitro, we examined the requirement for this pathway in pathogenesis. Here, we show that the RIM101 pathway is required for several host-pathogen interactions and thus pathogenesis. Further, our use of a suppressor mutation has allowed us to narrow the range of RIM101 pathway functions that are required for pathogenesis.
MATERIALS AND METHODS
Strains and plasmids.
The C. albicans strains used in this study are derivatives of SC5314 and are described in Table 1. Our parent strain, BWP17, is a triply marked auxotrophic strain which we used to generate the rim101−/rim101− and rim8−/rim8− mutants by PCR directed gene knockout (4, 26). All strains were reverted to prototrophy using plasmid pGEM-HIS1 (26) to create His+ strains and plasmid pRS-ARG-URA-BN (see below) to create Arg+ and Ura+ strains as described below. A prototrophic revertant of our parent strain, BWP17 (26), was generated as follows. First, BWP17 was transformed with NotI-digested pRS-ARG-URA-BN to generate an Arg+ Ura+ strain. This strain was then transformed with NruI-digested pGEM-HIS1 (26) to generate the prototrophic strain DAY185. DAY44, DAY62, and DAY114 were described previously (4). To generate the prototrophic rim101−/rim101− and rim8−/rim8− strains, DAY5 and DAY61 (4, 26) were transformed with NruI-digested pGEM-HIS1 to generate the prototrophic strains DAY25 and DAY117, respectively. To generate prototrophic RIM101/rim101− and RIM8/rim8− heterozygous strains, we made DAY2 and BWP35 (4, 26) prototrophic as follows. DAY2 was transformed with NruI-digested pGEM-HIS1 to generate the His+ derivative DAY176. DAY176 was then transformed with NotI-digested pRS-ARG-URA-BN to generate the prototrophic strain DAY203. BWP35 was transformed with NotI-digested pRS-ARG-URA-BN to generate the Ura+ derivative DAY191. DAY191 was then transformed with NruI-digested pGEM-HIS1 to generate the prototrophic strain DAY205.
TABLE 1.
C. albicans strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| DAY25 | ura3Δ::λimm434 HIS1::his1::hisG arg4::hisG rim101::ARG4 | This study |
| ura3Δ::λimm434 his1::hisG arg4::hisG rim101::URA3 | ||
| DAY44 | ura3Δ::λimm434 his1::hisG arg4::hisG rim101::ARG4::pRIM101::HIS1 | 4 |
| ura3Δ::λimm434 his1::hisG arg4::hisG rim101::URA3 | ||
| DAY62 | ura3Δ::λimm434 his1::hisG arg4::hisG rim8::ARG4 pRIM101-405::HIS1 | 4 |
| ura3Δ::λimm434 his1::hisG arg4::hisG rim8::URA3 RIM101 | ||
| DAY106 | ura3Δ::λimm434 his1::hisG arg4::hisG rim8::ARG4::pRIM8::HIS1 | This study |
| ura3Δ::λimm434 his1::hisG arg4::hisG rim8::URA3 | ||
| DAY114 | ura3Δ::λimm434 his1::hisG arg4::hisG rim8::ARG4 pRIM101::HIS1 | 4 |
| ura3Δ::λimm434 his1::hisG arg4::hisG rim8::URA3 RIM101 | ||
| DAY117 | ura3Δ::λimm434 HIS1::his1::hisG arg4::hisG rim8::ARG4 | This study |
| ura3Δ::λimm434 his1::hisG arg4::hisG rim8::URA3 | ||
| DAY185 | ura3Δ::λimm434 HIS1::his1::hisG ARG4::URA3::arg4::hisG | This study |
| ura3Δ::λimm434 his1::hisG arg4::hisG | ||
| DAY203 | ura3Δ::λimm434 HIS1::his1::hisG ARG4::URA3::arg4::hisG rim101::ARG4 | This study |
| ura3Δ::λimm434 his1::hisG arg4::hisGRIM101 | ||
| DAY205 | ura3Δ::λimm434 HIS1::his1::hisG ARG4::URA3::arg4::hisG rim8::ARG4 | This study |
| ura3Δ::λimm434 his1::hisG arg4::hisGRIM101 |
The rim8−/rim8− mutant was complemented as follows. First wild-type RIM8 was amplified from genomic DNA in a PCR with primers RIM8 −500 clone (5′-gggtcgacCCATTGTCTGTGGTTCGCTCTACC) and seq3c-rim8 (5′-GTTCCTGGACAAATCGTCATCC) using standard reaction conditions (26). This fragment was ligated into pGEMT (Promega) to generate pGEMT-RIM8. pGEMT-RIM8 was digested with SalI, and the RIM8-containing band was ligated into SalI-digested pGEM-HIS1, generating pDDB88. pDDB88 was digested with NruI and transformed into DAY61 (4) to generate DAY117.
pRS-ARG-URA-BN is a derivative of pRS-ARG-URA-BH (26). pRS-ARG-URA-BH was digested with ClaI and religated with the Cla-Not linker CGATGCGGCCGCAT. This linker introduces a NotI site into the ClaI site, generating pRS-ARG-URA-BN.
Media and growth conditions.
C. albicans was routinely grown in YPD (2% Bacto Peptone, 1% yeast extract, 2% dextrose). Selection following transformation was done on synthetic medium containing 6.7% yeast nitrogen base with ammonium sulfate and without amino acids, 2% dextrose, and uridine at 80 μg/ml, except when selecting for URA3, and supplemented with the necessary auxotrophic requirements of the cells (1). Cell densities were determined by dilution and counting on a hemacytometer.
Virulence assays.
C. albicans strains were passaged overnight at room temperature in YPD at least three times prior to injection. Cells were harvested, washed, counted using a hemacytometer, and resuspended at a density of 2 × 106/ml in sterile phosphate-buffered saline (Irvine Scientific, Irvine, Calif.). BALB/c mice (≥23 g, male) (B and K Universal, Fremont, Calif.) were infected through the lateral tail vein with 500 μl (106 organisms). Mouse survival was monitored at least twice daily, and moribund mice were euthantized by cervical dislocation. Following injections, dilutions of the fungal suspension were plated on YPD to ensure that similar numbers of viable cells had been injected into the mice. We used C. albicans strain DAY25 to infect 9 mice; DAY44, -62, -106, -203, and -205 to infect 10 mice each; DAY185 and -117 to infect 20 mice each; and DAY114 to infect 21 mice. Statistically significant differences between curves were determined using the Wilcoxon rank sum test. For these analyses, P values of <0.05 were considered to be significant.
Fungal burden.
Fungal burdens were determined by injecting 106 C. albicans organisms into the tail veins of ≥23-g male BALB/c mice (Harlan). C. albicans were prepared and injected as described above. At 20 and 40 h postinfection, the kidneys and livers were removed aseptically from five mice per strain per time point. The organs were weighed, homogenized, diluted in sterile saline, and plated in Sabouraud dextrose agar (Difco). Colonies were counted after incubation of the plates at 37°C for 24 to 48 h, and results were expressed as log CFU per gram of infected organ.
Histopathology.
Concomitant with the fungal burden experiment, kidneys were removed aseptically from two mice per strain per time point. Kidneys were fixed for at least 4 h in 100% ethanol at room temperature. They were rinsed and stored in 10% buffered Formalin prior to embedding in paraffin. Thin sections were prepared and stained with hematoxylin and eosin, followed by Gomori methenamine silver, and examined by light microscopy. We found that using both stains allowed visualization of both C. albicans cells and neutrophils within the same section.
Endothelial damage.
Candida-stimulated endothelial damage was measured using a chromium release assay described previously (7). The inoculum size was 105 cells/well of a 24-well tissue culture plate. C. albicans and endothelial cells were incubated together for 3 h. Specific release of chromium was calculated using the formula [(2 × experimental release) − (2 × spontaneous release)]/[total incorporation − (2 × spontaneous release)] as described previously (7). All experiments were performed in triplicate and repeated twice.
RESULTS
Virulence and the RIM101 pathway.
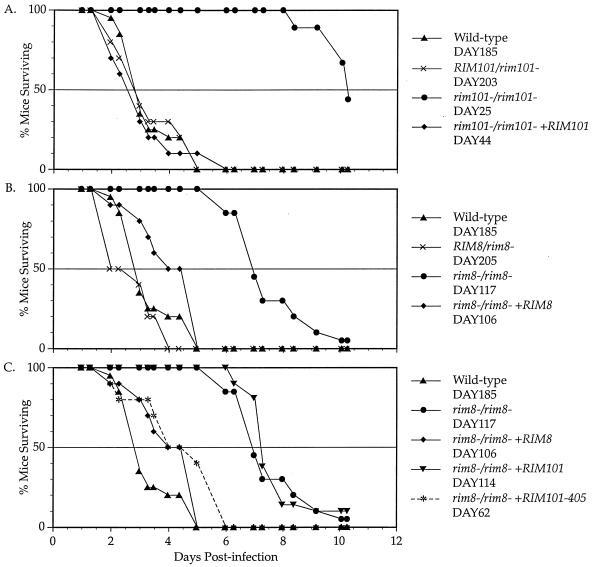
To determine if the RIM101 pathway has a role in virulence, we analyzed the effect of RIM101 pathway mutants in the hematogenously disseminated murine model. Wild-type and mutant strains were injected into mice via the tail vein, and survival of the mice was monitored (Fig. 1A to C). The wild-type and heterozygous RIM101/rim101− strains (DAY185 and DAY203) had similar degrees of virulence, with mice having a 3-day median survival time (Fig. 1A). However, the homozygous rim101−/rim101− strain (DAY25) had markedly diminished virulence compared to the wild-type strain, with mice having a 10-day median survival time (P < 0.0001). The complemented rim101−/rim101− +RM101 strain (DAY44) was as virulent as the wild type, demonstrating that the virulence defect of the rim101−/rim101− mutant is due to loss of Rim101p. Thus, Rim101p is required for virulence in this animal model.
FIG. 1.
Survival curves of mice following injection with C. albicans. Experiments were done simultaneously but broken up to simplify viewing. Survival was monitored in mice infected with RIM101 mutants (A), RIM8 mutants (B), and RIM8 suppressor strains (C). No mice died between the last time point (day 10) and the end of the experiment (day 14).
We also examined the rim8−/rim8− mutant (Fig. 1B). The heterozygous RIM8/rim8− strain (DAY205) maintained wild-type levels of virulence. However, the rim8−/rim8− strain (DAY117) had markedly diminished virulence, with mice having a 7-day median survival time (P < 0.0001). We noted that the rim8−/rim8− mutant was significantly more virulent than the rim101−/rim101− mutant (P < 0.0005). Mice infected with the complemented rim8−/rim8− +RIM8 strain (DAY106) had a median survival time of 4.5 days, which was not statistically separable from that of the wild type (P > 0.10). Thus, the rim8−/rim8− mutant virulence defect is due to loss of Rim8p. These results suggest that Rim8p is required for virulence in this animal model. Because the complemented rim8−/rim8− strain behaved identically to the suppressed rim8−/rim8− +RIM101-405 strain (see below), we focused only on the suppressed strain.
To demonstrate that we had successfully infected mice with viable C. albicans, we analyzed the fungal burdens in the livers and kidneys of the RIM101 pathway mutants (Table 2). The wild-type, rim101−/rim101−, rim101−/rim101− +RIM101, and rim8−/rim8− strain livers had similar fungal burdens after both 20 and 40 h of infection. Thus, the rim101−/rim101− and rim8−/rim8− strains were able to colonize the liver as well as the wild-type strain was during the first 40 h of infection. The kidneys of these strains had similar fungal burdens 20 h postinfection as well. However, both the rim101−/rim101− and rim8−/rim8− mutants failed to show the dramatic fungal burden increase seen 40 h postinfection in the kidneys of mice infected with the wild-type strain and, to a lesser extent, in those infected with the rim101−/rim101− +RIM101 strain. These results demonstrate that the rim101−/rim101− and rim8−/rim8− mutants were able to colonize the kidneys as well as the wild type did during the first 20 h of infection. However, the rim101−/rim101− and rim8−/rim8− mutants grow less well than the wild type in the kidneys during prolonged infection.
TABLE 2.
Fungal burdens
| Strain | Relevant genotype | 20 h
|
40 h
|
||
|---|---|---|---|---|---|
| Kidneys | Livers | Kidneys | Livers | ||
| DAY185 | Wild type | 5.85 ± 0.40a | 3.90 ± 0.09 | 7.41 ± 0.67 | 3.57 ± 0.48 |
| DAY25 | rim101−/rim101− | 5.21 ± 0.19 | 3.78 ± 0.26 | 4.89 ± 0.32b | 3.00 ± 0.15 |
| DAY44 | rim101−/rim101− +RIM101 | 5.53 ± 0.51 | 3.35 ± 0.34 | 6.03 ± 0.91 | 3.27 ± 0.60 |
| DAY117 | rim8−/rim8− | 5.76 ± 0.43 | 3.88 ± 0.07 | 5.30 ± 0.48b | 3.29 ± 0.40 |
| DAY62 | rim8−/rim8− +RIM101-405 | 5.75 ± 0.46 | 3.66 ± 0.26 | 5.61 ± 0.40b | 2.66 ± 0.13 |
Units are log CFU per gram of tissue.
Statistically significantly different from the wild type (P < 0.001) by analysis of variance.
Histopathology.
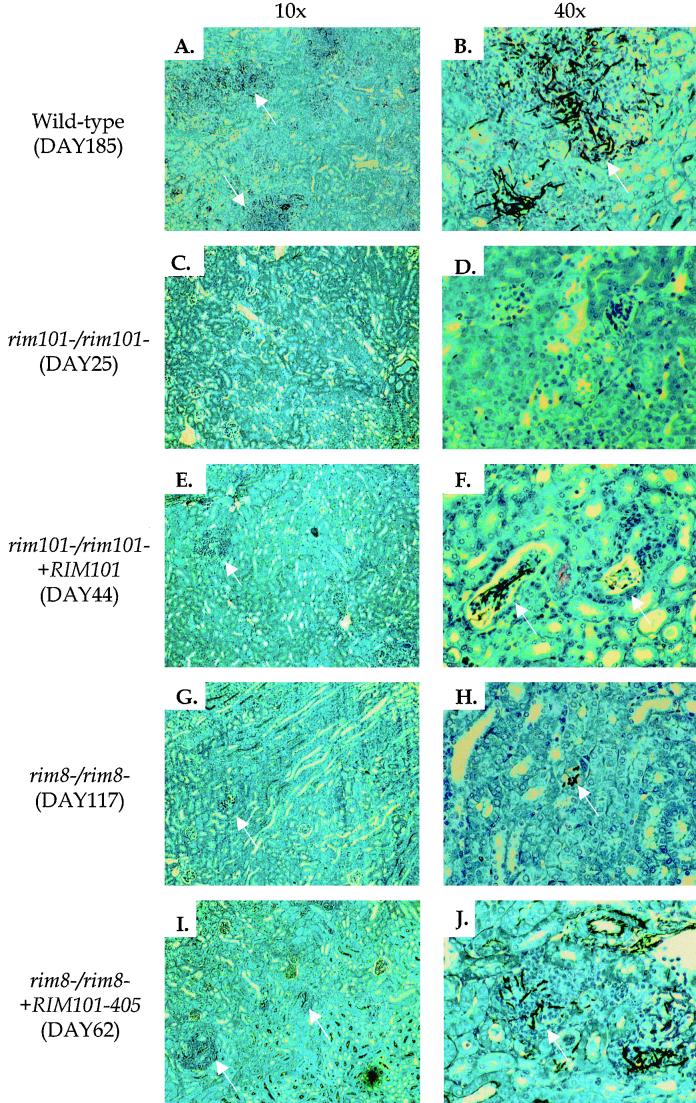
Several features of a pathogenic infection are observed in murine kidneys infected with C. albicans (18). To determine whether the RIM101 pathway is required for these responses, we removed kidneys infected with the wild-type strain and RIM101 pathway mutants for histological examination. Forty hours postinfection, three features of a successful infection were seen in kidneys colonized with wild-type and rim101−/rim101− +RIM101 cells. First, most wild-type cells had germinated, forming long hyphae within the kidney (Fig. 2B). Second, wild-type cells elicited a strong host immune response, as seen by the number of neutrophils surrounding the C. albicans organisms (Fig. 2A). Third, microabscesses, the sites of concentrated C. albicans and neutrophils, were disseminated throughout the kidney (Fig. 2A). The rim101−/rim101− and rim8−/rim8− strains showed defects in these assays: there were few organisms, and those that were apparent were often in the yeast form (Fig. 2D and H). Further, there was poor stimulation of the immune response and there were few microabscesses (Fig. 2C and G). Similar results were seen 20 h postinfection (data not shown). We noted that the rim101−/rim101− mutant was more defective than the rim8−/rim8− mutant by these three criteria (compare panels C and D with G and H in Fig. 2). Thus, both Rim101p and Rim8p are required for host-pathogen interactions leading to normal germination and the development of lesions in the murine kidney.
FIG. 2.
Histological samples of kidneys infected with C. albicans for 40 h. Mice were injected with the wild-type (A and B), rim101−/rim101− (C and D), rim101−/rim101− +RM101 (E and F), rim8−/rim8− (G and H), and rim8−/rim8− +RIM101-405 (I and J) strains. Sections were visualized using either a 10× (A, C, E, G, and I) or a 40× (B, D, F, H, and J) objective. Arrows denote microabscesses in the 10× panels and representative C. albicans cells in the 40× panels. Note that arrows are absent in panels C and D due to the absence of visible microabscesses and C. albicans cells, respectively.
Endothelial damage.
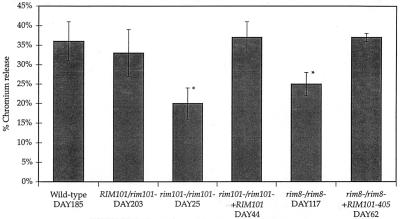
We analyzed the ability of RIM101 pathway mutants to cause damage to endothelial cells in situ. Filler et al. proposed that Candida-stimulated endothelial damage occurs shortly after intravenous infection and is important for entry into the underlying tissues (7). We assayed endothelial cell damage by monitoring 51Cr release into the medium from lysed endothelial cells in the presence and absence of C. albicans (7). The wild-type, RIM101/rim101−, and rim101−/rim101− +RM101 strains caused ∼35% damage after 3 h of incubation with endothelial cells and were not statistically significantly different from each other (Fig. 3). The rim101−/rim101− and rim8−/rim8− strains caused 20 and 25% damage to endothelial cells, respectively, a significant reduction compared to the wild type (P < 0.001). Further, the rim101−/rim101− strain was more defective than the rim8−/rim8− strain (P < 0.05). Endothelial cell damage requires C. albicans germination (9), but we found that all strains, including the rim101−/rim101− and rim8−/rim8− mutants, germinated equally well (data not shown). Since the RIM101 pathway mutant germinated normally in this assay, these results suggest that the damage defect is due to a downstream event, such as reduced expression of proteases or other factors required for damage (12). Thus, both Rim101p and Rim8p are required to induce wild-type levels of damage to endothelial cells.
FIG. 3.
Endothelial damage. Damage to endothelial cells was determined by a chromium release assay. Wild-type and RIM101 pathway mutants were incubated with endothelial cells for 3 h, and release of 51Cr from lysed endothelial cells was measured. Asterisks indicate values that statistically significantly differ from wild-type values (P < 0.001) by analysis of variance.
Suppression of virulence defects.
Suppression analysis is a powerful tool with which to identify the cause of a mutant phenotype. The RIM101-405 allele expresses a C-terminally truncated Rim101p which should not require processing for activity and restores filamentation to the rim8−/rim8− mutant in vitro (4). We used this allele to elucidate the nature of the rim8−/rim8− defect in virulence by genetic suppression studies. First, the rim8−/rim8− +RIM101-405 strain was more virulent than the rim8−/rim8− strain (P < 0.0001) and was statistically indistinguishable from the wild-type strain or the complemented rim8−/rim8− +RIM8 strain (Fig. 1C). Introduction of full-length RIM101 into the rim8−/rim8− strain did not have this effect, which demonstrated that this effect is an attribute of the RIM101-405 mutation and not due to increased RIM101 gene dosage. Although the RIM101-405 allele rescued the virulence defect, it did not rescue the fungal burden defect of the rim8−/rim8− mutant in kidneys 40 h postinfection (Table 2).
Second, we checked for rescue of the rim8−/rim8− mutant phenotype by histological examination. In kidneys 20 h postinfection, the rim8−/rim8− +RIM101-405 strain mimicked the rim8−/rim8− strain: few cells had germinated, there was little immune response, and few microabscesses were apparent (data not shown). Forty hours postinfection, the results mimicked the wild-type strain: many cells had germinated to form hyphae (Fig. 2J), there was a pronounced immune response, and microabscesses were apparent throughout the kidneys (Fig. 2I). Third, we looked for rescue in the endothelial damage assay (Fig. 3). The rim8−/rim8− +RIM101-405 strain caused 37% damage, a marked increase over the rim8−/rim8− strain (P < 0.0001). Because the RIM101-405 allele, which expresses a C-terminally truncated protein, rescues many phenotypes of the rim8−/rim8− mutant, these results argue that the function of Rim8p in virulence is to promote activation of Rim101p.
DISCUSSION
Rim101p and Rim8p are required for host-pathogen interactions based upon three assays. First, rim101−/rim101− and rim8−/rim8− mutants have a severe virulence defect in a murine model. Second, by histological examination, these mutants germinate poorly, fail to stimulate a strong immune response, and fail to form microabscesses disseminated throughout the kidney. Third, these mutants do not stimulate endothelial cell damage to wild-type levels. Thus, the RIM101 pathway is required for the host-pathogen interactions leading to virulence.
Previous in vitro studies of RIM101 pathway mutants did not distinguish the rim101−/rim101− and rim8−/rim8− mutants (4, 21, 22). Here, we found that although both mutants were defective for all host-pathogen interactions, they do show differences in the three assays. First, the rim8−/rim8− mutant is more virulent than the rim101−/rim101− mutant. Second, the rim8−/rim8− mutant causes more hallmarks of pathology in kidneys than the rim101−/rim101− mutant. Third, the rim8−/rim8− mutant stimulates more endothelial cell damage than the rim101−/rim101− mutant. Thus, we found that both Rim8p and Rim101p are critical for responses in vitro and in vivo but that the assays used in these studies are more sensitive to perturbations of the RIM101 pathway.
Why is the rim101−/rim101− mutant less virulent than the rim8−/rim8− mutant? Our genetic suppression analyses argue that the function of Rim8p in vivo is to stimulate processing of Rim101p. Studies of both A. nidulans and S. cerevisiae argue that, in the rim8−/rim8− mutant, only unprocessed Rim101p is present (15, 19). However, in the rim101−/rim101− mutant, no Rim101p is present. Thus, one simple model to explain the difference in virulence is that both unprocessed and processed Rim101p function during infection. The rim8−/rim8− mutant has a virulence defect because of the absence of processed Rim101p. The rim101−/rim101− mutant has a more severe phenotype due to the absence of both processed and unprocessed Rim101p. We predict that molecular analysis of Rim101p (PacC) pathways in C. albicans and other fungi will support this new hypothesis that unprocessed Rim101p has a functional role in target gene regulation.
In kidneys 20 and 40 h postinfection, the rim8−/rim8− and rim8−/rim8− +RIM101-405 strains produced similar fungal burdens. However, these strains varied markedly in their virulence. Similar results were reported for other mutants, including the hwp1−/hwp1− and chs3−/chs3− mutants (2, 13, 25). These results indicate that the fungal burden does not necessarily predict the virulence phenotype. However, these results do suggest that the RIM101 pathway is not required for colonization of the kidney but is required for maintenance of infection.
We did find a strong correlation among the damage, histological, and virulence results. In fact, the histological and damage results appear to predict the virulence result of RIM101 pathway mutants. Similar results are seen for mutations affecting HWP1 and SAP2 (11, 12, 24, 25). Thus, we found that the histological and damage phenotypes are good predictors of the virulence phenotype.
One useful finding to come from these studies is that our wild-type strain appears to maintain normal virulence. We had previously created a triply marked auxotrophic strain to allow for rapid PCR-directed gene knockouts (two markers) and complementation (one marker). Here, we have analyzed host interactions with a prototrophic derivative of BWP17, DAY185. Mice infected with DAY185 appear to have survival times similar to those of mice infected with SC5314 and a related strain, CAI12 (23, 25). DAY185 stimulates endothelial damage to levels similar to those reported for SC5314 (12). Further, DAY185 grows well in kidneys and elicits a strong immune response. Thus, the BWP17 strain is useful to generate mutants rapidly and for consequent in vivo analyses as well.
What is the function of the RIM101 pathway in pathogenesis? Our results argue that the RIM101 pathway is not simply required for growth in vivo. We see that RIM101 pathway mutants have a defect in fungal burden compared to the wild type. However, if we compare the rim8−/rim8− and rim8−/rim8− +RIM101-405 mutants, which produce similar fungal burdens, we find that the rim8−/rim8− +RIM101-405 strain behaves like the wild type in the other assays. Thus, a defect in growth is not sufficient to explain the role of the RIM101 pathway in pathogenesis. The rim8−/rim8− +RIM101-405 strain has an activity lacking in the rim8−/rim8− strain. We suggest that stimulation of host cell damage may be a candidate for this activity. Filler et al. have suggested that endothelial cell damage may be required to stimulate expression of cytokines and leukocyte adhesion molecules (6). If similar events occur in the kidney, then we predict that mutants that inflict less cell damage would elicit a weaker immune response. This is exactly what we see. Thus, we propose that one function of the RIM101 pathway in pathogenesis is to regulate the expression of genes that stimulate host cell damage.
ACKNOWLEDGMENTS
We thank the nurses at Harbor-UCLA Medical Center for collecting umbilical cords; Quynh-Trang Phan and Angela Sanchez for preparing endothelial cells; S. French for help with the histology; H. K. Lee, S. Klein, and D. Shepard for technical support; and Jim Ericson for help with photography of the histological sections. We are indebted to T. Lamb and H. Shuman for helpful criticism of the manuscript and to Scott Filler for numerous helpful and stimulating discussions. D.D. thanks Debra and Fia McWilliam for continued support throughout the course of this work.
This research was supported by a Mycology Scholar Award from the Burroughs Wellcome Fund to A.P.M. and by Public Health Service grants PO1AI-37194 and RO1AI-19990. A.S.I. is supported by a grant-in-aid from the American Heart Association, Western States Affiliate 9960030Y.
REFERENCES
- 1.Adams A, Gotschling D E, Kaiser C A, Stearns T. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- 2.Bulawa C E, Miller D W, Henry L K, Becker J M. Attenuated virulence of chitin-deficient mutants of Candida albicans. Proc Natl Acad Sci USA. 1995;92:10570–10574. doi: 10.1073/pnas.92.23.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cutler J E. Putative virulence factors of Candida albicans. Annu Rev Microbiol. 1991;45:187–218. doi: 10.1146/annurev.mi.45.100191.001155. [DOI] [PubMed] [Google Scholar]
- 4.Davis D, Wilson R B, Mitchell A P. RIM101-dependent and -independent pathways govern pH responses in Candida albicans. Mol Cell Biol. 2000;20:971–978. doi: 10.1128/mcb.20.3.971-978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Bernardis F, Mühlchlegel F A, Cassone A, Fonzi W A. The pH of the host niche controls gene expression in and virulence of Candida albicans. Infect Immun. 1998;66:3317–3325. doi: 10.1128/iai.66.7.3317-3325.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filler S G, Pfunder A S, Spellberg B J, Spellberg J P, Edwards J E., Jr Candida albicans stimulates cytokine production and leukocyte adhesion molecule expression by endothelial cells. Infect Immun. 1996;64:2609–2617. doi: 10.1128/iai.64.7.2609-2617.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filler S G, Swedloff J N, Hobbs C, Luckett P M. Penetration and damage of endothelial cells by Candida albicans. Infect Immun. 1995;63:976–983. doi: 10.1128/iai.63.3.976-983.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gale C A, Bendel C M, McClellan M, Hauser M, Becker J M, Berman J, Hostetter M K. Linkage of adhesion, filamentous growth, and virulence in Candida albicans to a single gene, INT1. Science. 1998;279:1355–1358. doi: 10.1126/science.279.5355.1355. [DOI] [PubMed] [Google Scholar]
- 9.Ghannoum M A, Filler S G, Ibrahim A S, Fu Y, Edwards J E., Jr Modulation of interactions of Candida albicans and endothelial cells by fluconazole and amphotericin B. Antimicrob Agents Chemother. 1992;36:2239–2244. doi: 10.1128/aac.36.10.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghannoum M A. Potential role of phospholipases in virulence and fungal pathogenesis. Clin Microbiol Rev. 2000;13:122–143. doi: 10.1128/cmr.13.1.122-143.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hube B, Sanglard D, Odds F C, Hess D, Monod M, Schäfer W, Brown A J P, Gow N A R. Disruption of each of the secreted aspartyl proteinase genes SAP1, SAP2, and SAP3 of Candida albicans attenuates virulence. Infect Immun. 1997;65:3529–3538. doi: 10.1128/iai.65.9.3529-3538.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ibrahim A S, Filler S G, Sanglard D, Edwards J E, Jr, Hube B. Secreted aspartyl proteinases and interactions of Candida albicans with human endothelial cells. Infect Immun. 1998;66:3003–3005. doi: 10.1128/iai.66.6.3003-3005.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohler J R, Fink G R. Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc Natl Acad Sci USA. 1996;93:13223–13228. doi: 10.1073/pnas.93.23.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leberer E, Ziegelbauer K, Schmidt A, Harcus D, Dignard D, Ash J, Johnson L, Thomas D Y. Virulence and hyphal formation of Candida albicans require the Ste20-like protein kinase CaCla4p. Curr Biol. 1997;7:539–546. doi: 10.1016/s0960-9822(06)00252-1. [DOI] [PubMed] [Google Scholar]
- 15.Li W, Mitchell A P. Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics. 1997;145:63–73. doi: 10.1093/genetics/145.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lo H J, Kohler J R, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink G R. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 17.Mitchell A P. Dimorphism and virulence in Candida albicans. Curr Opin Microbiol. 1998;1:687–692. doi: 10.1016/s1369-5274(98)80116-1. [DOI] [PubMed] [Google Scholar]
- 18.Odds F C. Candida and candidosis. 2nd ed. 1988. Bailiere Tindall, London, England. [Google Scholar]
- 19.Orejas M, Espeso E A, Tilburn J, Sarkar S, Arnst H N, Penalva M A. Activation of the Aspergillus PacC transcription factor in response to alkaline pH requires proteolysis of the carboxy-terminal moiety. Genes Dev. 1995;9:1622–1632. doi: 10.1101/gad.9.13.1622. [DOI] [PubMed] [Google Scholar]
- 20.Pfaller M A, Jones R N, Messer S A, Edmond M B, Wenzel R P. National surveillance of nosocomial blood stream infection due to Candida albicans: frequency of occurrence and antifungal susceptibility inthe SCOPE Program. Diagn Microbiol Infect Dis. 1998;31:327–332. doi: 10.1016/s0732-8893(97)00240-x. [DOI] [PubMed] [Google Scholar]
- 21.Porta A, Ramon A M, Fonzi W A. PRR1, a homolog of Aspergillus nidulans palF, controls pH-dependent gene expression and filamentation in Candida albicans. J Bacteriol. 1999;181:7516–7523. doi: 10.1128/jb.181.24.7516-7523.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramon A M, Porta A, Fonzi W A. Effect of environmental pH on morphological development of Candida albicans is mediated via the PacC-related transcription factor encoded by PRR2. J Bacteriol. 1999;181:7524–7530. doi: 10.1128/jb.181.24.7524-7530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rieg G, Fu Y, Ibrahim A S, Zhou X, Filler S G, Edwards J E., Jr Unanticipated heterogeneity in growth rate and virulence among Candida albicans AAF1 null mutants. Infect Immun. 1999;67:3193–3198. doi: 10.1128/iai.67.7.3193-3198.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharkey L L, McNemar M D, Saporito-Irwin S M, Sypherd P S, Fonzi W A. HWP1 functions in the morphological development of Candida albicans downstream of EFG1, TUP1, and RBF1. J Bacteriol. 1999;181:5273–5279. doi: 10.1128/jb.181.17.5273-5279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsuchimori N, Sharkey L L, Fonzi W A, French S W, Edwards J E, Jr, Filler S G. Reduced virulence of HWP1-deficient mutants of Candida albicans and their interactions with host cells. Infect Immun. 2000;68:1997–2002. doi: 10.1128/iai.68.4.1997-2002.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson R B, Davis D, Mitchell A P. Rapid hypothesis testing in Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]