Summary
Cortical circuits are composed predominantly of pyramidal-to-pyramidal neuron connections, yet their assembly during embryonic development is not well understood. We show that mouse embryonic Rbp4-Cre cortical neurons, transcriptomically closest to layer 5 pyramidal neurons, display two phases of circuit assembly in vivo. At E14.5, they form a multi-layered circuit motif, composed of only embryonic near-projecting-type neurons. By E17.5, this transitions to a second motif involving all three embryonic types, analogous to the three adult layer 5 types. In vivo patch clamp recordings and two-photon calcium imaging of embryonic Rbp4-Cre neurons reveal active somas and neurites, tetrodotoxin-sensitive voltage-gated conductances, and functional glutamatergic synapses, from E14.5 onwards. Embryonic Rbp4-Cre neurons strongly express autism-associated genes and perturbing these genes interferes with the switch between the two motifs. Hence, pyramidal neurons form active, transient, multi-layered pyramidal-to-pyramidal circuits at the inception of neocortex, and studying these circuits could yield insights into the etiology of autism.
Keywords: embryonic development, layer 5, pyramidal neurons, neuronal activity, in vivo imaging, transient circuits, in vivo patch clamp, synapses, single cell sequencing, autism
Graphical abstract
Highlights
-
•
Mouse embryonic pyramidal neurons display two phases of circuit assembly in vivo
-
•
Pyramidal neurons first form a multi-layered circuit before cortical lamination begins
-
•
This circuit is transiently active with functional synapses and active conductances
-
•
Perturbing autism-associated genes interferes with the switch between the two phases
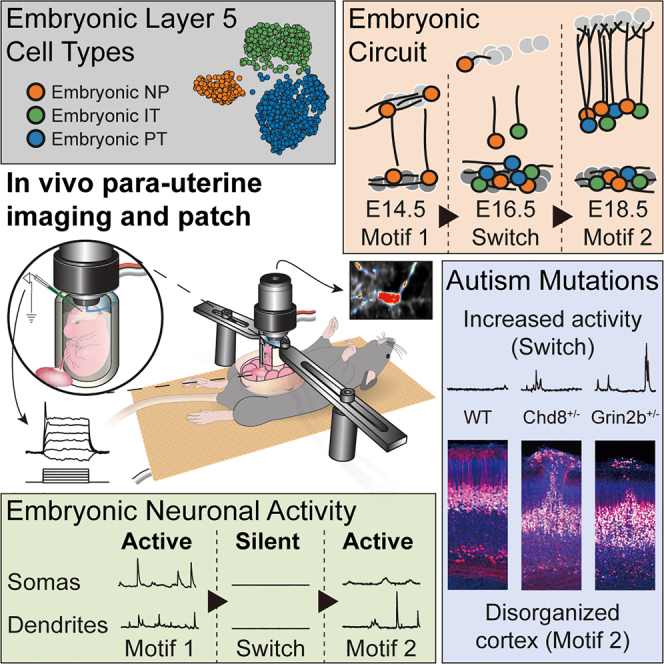
During mouse embryonic development, cortical layer 5 pyramidal neurons switch between two distinct active circuit motifs, the first of which constitutes a multi-layered circuit motif at the inception of the neocortex. Further, the switch is perturbed by mutations in autism-associated genes.
Introduction
The neocortex contains neuronal circuits that are highly interconnected, with the most common class of neurons, pyramidal neurons (PN),1 receiving a majority of their connections from other PNs.2,3,4,5,6 PNs are of different types with distinct transcriptomic identities and are organized into distinct layers.1,7,8 While the activity and communication within PN-to-PN circuits have been intensively investigated, in vivo, in the adult,4,9,10,11,12,13,14,15 when and how the first active PN circuits assemble in vivo are not known. Answers to these questions are central to understanding cortical circuit development. Moreover, since neurodevelopmental disorders are associated with defects within cortical circuits,16,17,18,19,20 insights into pyramidal circuit formation may also be relevant for understanding the mechanisms of diseases such as autism spectrum disorder.
During cortical development, PNs migrate to their final locations in neocortex, with layers forming in an inside-out fashion.21,22,23,24,25,26 PNs that will populate layers 5 and 6 (L5-PNs and L6-PNs) are born first. In mice, these neurons appear between embryonic day (E) 11.5 and E14.5 and start to migrate into the developing cortex from E12.5 onwards.21,23,27,28 Among the early-born PNs, L6-PNs receive the majority of their inputs from upper layer neurons,5 which arrive in cortex later, from E15.5 onwards. Therefore, most of the L6-PN circuitry likely forms only after E15.5. In contrast, L5-PNs—which are composed of three cell types: near-projecting neurons (NP), intratelencephalic neurons (IT), and pyramidal tract neurons (PT, also referred to as extratelencephalic neurons)—receive 50%–70% of their inputs, recurrently, from other L5-PNs.2,6 Therefore, the majority of L5-PN connectivity could form early in embryonic cortical development.
Neuronal activity starts during embryonic development.29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44 Cajal-Retzius cells, as early as E14.5,32,33 and PNs, from E15.5 onwards, show calcium transients in their somas, in vivo.34,35 Moreover, at E14.5, thalamic neurons display correlated activity in vitro, and this correlated activity can reach cortex by E16.5, when thalamic axons arrive in the cortex.45,46 However, the time when cortical PNs first assemble into circuits with other cortical PNs in vivo and the point at which activity appears and becomes correlated within these circuits remain unknown.
Neurons within circuits transmit activity using active conductances via neurites across synapses. Therefore, to understand if early born PNs form active circuits, it is necessary to measure activity within their individual neurites, to find active conductances, and to detect functional synapses in vivo. While in vivo two-photon imaging in embryos has been performed,32,33,34,47,48,49,50 these recordings did not resolve activity within individual neurites and were not targeted to cell types. Further, thus far, no patch clamp recordings, capable of revealing active conductances, have been performed in embryonic neurons in vivo.
Insight into the timing and assembly of the first active PN circuits is also important in the context of neurodevelopmental disorders such as autism spectrum disorder and schizophrenia.51 Indeed, common circuit dysfunctions have been hypothesized to underlie the phenotypic similarities across the heterogeneous mixture of genetic abnormalities associated with these neurodevelopmental disorders.16,17,18,19,20,52,53 Further, patches of disorganized cortical tissue have been observed in brains of both children with autism as well as in mouse models of the disorder, around the time of birth.54,55,56,57,58,59,60,61 However, it remains unknown if mutations in autism-related genes may perturb the development of PN-to-PN circuits during embryonic development.
Here, we developed a method with sufficient mechanical stability to perform both two-photon imaging from individual neurites and two-photon targeted patch clamp recordings, in healthy living mouse embryos connected to the dam, to investigate these questions. We found that embryonic cortical PNs form two distinct active circuit motifs during embryonic development that switch after E15.5, the first of which constitutes a new multi-layered circuit motif at the inception of the neocortex. Further, the perturbation of autism-associated genes interferes with the switch.
Results
Embryonic Rbp4-Cre cortical neurons have L5-PN identity
Rbp4-Cre KL100 mice express Cre in adult L5-PNs.62,63 During embryonic development, cells with neuronal morphology were labeled close to the surface of the neocortex in Rbp4-Cre KL10062,63 mice, when crossed with a fluorescent Cre-reporter line, encoding either tdTomato64 or GCaMP6s65 (Figure S1). The first Rbp4-Cre neurons appeared at E13.5, a time when L5-PNs first enter the preplate, the precursor to the cortical plate.21 From here on, we refer to embryonic Rbp4-Cre cortical neurons as “Rbp4-Cre neurons”.
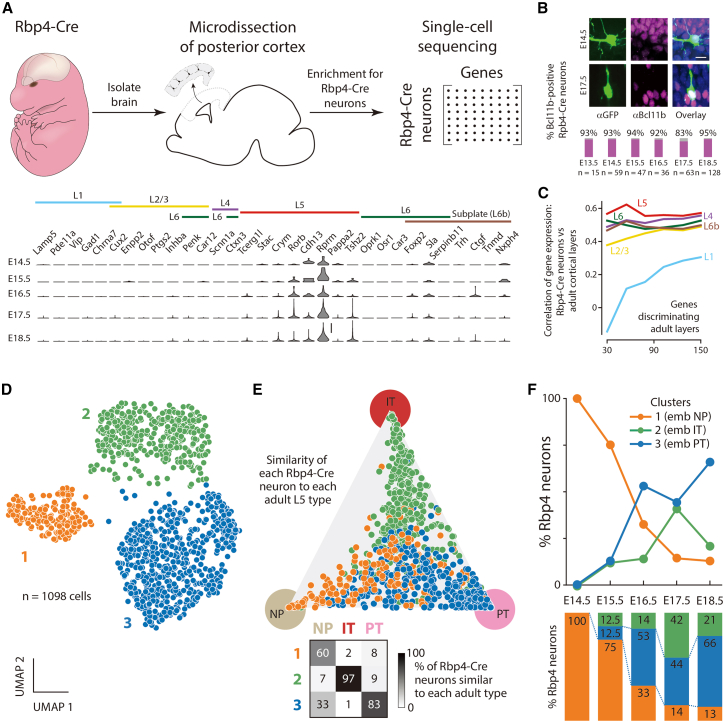
Because neuronal identity is most comprehensively defined by transcriptomic identity, assessed through single cell sequencing,7,8,66,67,68 we performed single-cell RNA sequencing of cells from the developing cortex every day from E14.5 to E18.5 and identified Rbp4-Cre neurons via tdTomato expression (Figures 1A and S1). To compare the identity of Rbp4-Cre neurons with adult cortical neurons, we used a panel of 26 marker genes with layer-specific expression.7,8 On all studied embryonic days, Rbp4-Cre neurons expressed marker genes associated with adult cortical L5-PNs. In particular, from E13.5 onwards, Rbp4-Cre neurons expressed Bcl11b (Citip2), the expression of which is restricted in the adult cortex to L5-PNs69 (Figure 1B). Moreover, using increasing numbers of genes that best distinguished gene expression in different adult cortical layers,7,8 the gene expression of Rbp4-Cre neurons remained most highly correlated with that of adult L5-PNs (Figure 1C). These experiments suggest that, during embryonic development, Rbp4-Cre neurons are transcriptomically closest to adult L5-PNs.
Figure 1.
Rbp4-Cre neurons have L5-PN identity
(A) Top: Single-cell RNA sequencing workflow (Figure S1). Bottom: Expression profile of layer-specific genes7,8,70 in Rbp4-Cre neurons.
(B) Top: Rbp4-Cre neurons (green), Bcl11b (magenta), Hoechst (blue). Bottom: fraction of Rbp4-Cre neurons expressing Bcl11b. n = number of Rbp4-Cre neurons.
(C) Correlation of cortical layer-specific neuronal genes’ expression between Rbp4-Cre neurons and adult cortical layers for up to 150 genes.7,8
(D) UMAP embedding of Rbp4-Cre neurons’ single cell transcriptomes.71 Color: Leiden clusters.72
(E) Top: Rbp4-Cre types (colored as in D) embedded in a triangle representing the similarity between each cell’s expression profile from the three adult L5-PN types (NP, IT, and PT)8 (Figure S1). Bottom: For each adult type, percent of Rbp4-Cre neurons of each embryonic type associated with that adult type.
(F) Percent of neurons from each type on each embryonic day (colored as in D and labels derived from E).
Scale bars: 20 transcripts (A), 10 μm (B).
See also Figure S1.
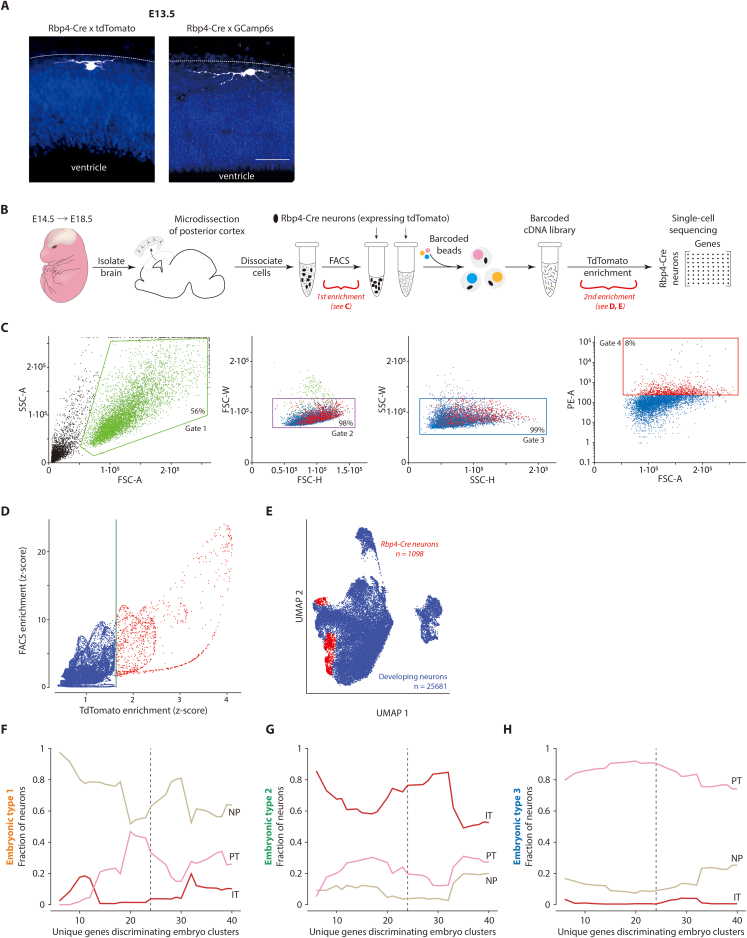
Figure S1.
Rbp4-Cre neurons appear in cortex from E13.5 onwards, and divide into three embryonic types, related to Figure 1
(A) Rbp4-Cre neurons at E13.5, near surface of developing cortex (dotted line), immunostained against either tdTomato (left) or GCaMP6s (right) (white), counterstained with Hoechst (blue).
(B) Single cell RNA sequencing workflow overview demonstrating two distinct stages of enrichment for tdTomato-expressing Rbp4-Cre neurons, from dissociated embryonic cortical tissue (red brackets).
(C) Initial enrichment of Rbp4-Cre neurons was performed by isolating cells positive for the tdTomato marker from the dissociated cortical tissue using FACS. Gates were selected based on the size and granularity of sorted events. Gate 1 was chosen to exclude debris, while gates 2 and 3 were chosen to exclude doublets. Gate 4 was chosen to select cells with higher tdTomato fluorescence. Box: events filtered at each gate.
(D) Additional enrichment for positively identified tdTomato-expressing Rbp4-Cre neurons. Relative enrichment of cells over a negatively FACS sorted population (blue) was used to define a baseline tdTomato expression level. Cells with significantly greater tdTomato expression than in the baseline were selected as positively identified Rbp4-Cre neurons (red). Green line: 6σ threshold of tdTomato expression.
(E) UMAP embedding of all 25681 excitatory neuron transcriptomes (blue) demonstrating the location of the subset of 1098 positively identified Rbp4-Cre neurons (red).
(F–H) Rbp4-Cre neuron divide into three embryonic types, each associated with one adult layer 5 type, stably across an increasing number of genes. Fraction of neurons within each Rbp4-Cre neuron type (1 (F), 2 (G), and 3 (H)) with a significant conditional probability of being sampled from the expression profile of one of the three adult cell types. Adult types: near-projecting neurons (NP) (beige), intratelencephalic neurons (IT) (red), and pyramidal tract neurons (PT) (pink) (as sequenced and identified in8 from VISp). Genes were selected to best discriminate the embryonic clusters, while still being differentially expressed across the three adult types. Dotted line: 24 differentially expressed genes used to generate Figure 1E. Scale bars: 50 μm (A).
Adult L5-PNs are classified into IT, PT, and NP neuron types.2,73 To understand if Rbp4-Cre neurons also divide into cell types, we clustered the transcriptomes of individual Rbp4-Cre neurons.71,72 We found three distinct clusters (Figure 1D). In each cluster, most of the neurons had gene expression profiles closest to one and only one of the three adult L5-PN types, with conditional probability >99% (Figures 1E and S1). Hence, we named each cluster through this correspondence as embryonic-NP, embryonic-IT, and embryonic-PT. The three Rbp4-Cre neuron types showed distinct relative proportions on each embryonic day (Figure 1F). At E14.5, all Rbp4-Cre neurons were embryonic-NP. From E15.5 onwards, the fraction of embryonic-IT and embryonic-PT neurons gradually increased. These results suggest that Rbp4-Cre neurons divide into three cell types and that this cell type composition is developmentally regulated.
Rbp4-Cre neurons form transient layers populated by different cell types
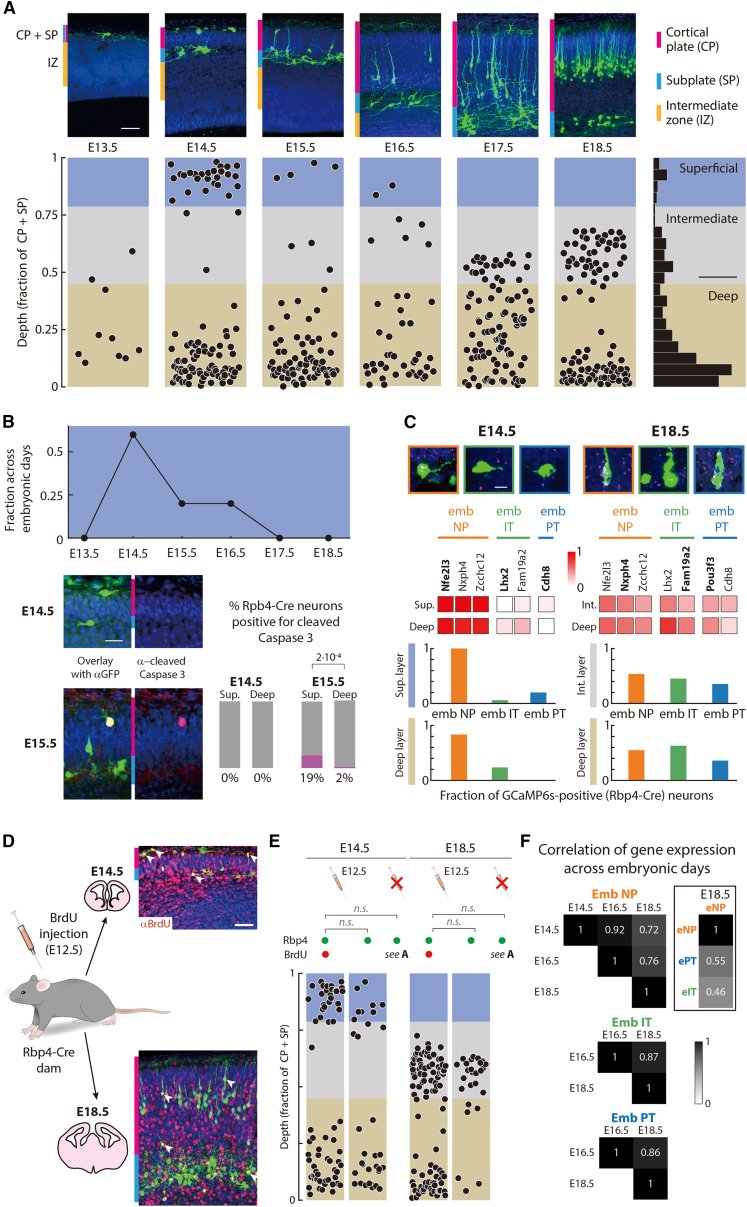
Given that the cell type composition of Rbp4-Cre neurons changed during embryonic development, we asked if the spatial organization of Rbp4-Cre neurons also varied across embryonic days. To reveal the location of GCaMP6s-expressing Rbp4-Cre neurons, we collected brain sections every day from E13.5 to E18.5 and stained them with a GFP antibody (Figure 2A). Rbp4-Cre neurons were distributed throughout the depth of the developing preplate at E13.5. By E14.5, Rbp4-Cre neurons had organized into two distinct layers: a superficial layer near the surface of cortex and a deep layer below the cortical plate. The number of Rbp4-Cre neurons within the superficial layer decreased with time, with the maximum decrease occurring from E14.5 to E15.5 and all superficial layer neurons disappearing before E17.5 (Figure 2B). This decrease was not due to GCaMP6s expression, since we found a similar decrease in Rbp4-Cre neurons expressing tdTomato (Figure S2). Further, a greater fraction of superficial layer neurons stained positive for cleaved Caspase-3, a marker of apoptosis, compared to neurons in the deep layer (Figure 2B). From E15.5 onwards, an intermediate layer appeared between the superficial and deep layers. By E18.5, neurons in the intermediate layer adopted a morphology akin to adult L5-PNs, with neurites extending to the surface of cortex (Figure 2A). Therefore, Rbp4-Cre neurons exist in two different spatial configurations during the embryonic development of cortex: a deep layer and a transient superficial layer around E14.5, and the same deep layer and an intermediate layer from E17.5 to E18.5. Neurons in the superficial layer undergo apoptosis, resulting in a transition between the two spatial configurations from E15.5 to E16.5.
Figure 2.
Rbp4-Cre neurons distribute into superficial, intermediate, and deep layers
(A) Top: Rbp4-Cre neurons (green), Hoechst (blue). Bottom left: Rbp4-Cre neuron depths normalized to the cortical plate and subplate thickness, (Figure S2). Bottom right: distribution of Rbp4-Cre neuron depths across days (blue: superficial layer; gray: intermediate layer; beige: deep layer).
(B) Top: Fraction of all superficial layer Rbp4-Cre neurons found on each embryonic day. Bottom left: Immunostaining Rbp4-Cre neurons (green), cleaved Caspase 3 (magenta), Hoechst (blue). Bottom right: Fraction of Rbp4-Cre neurons expressing cleaved Caspase 3. Fisher’s exact test.
(C) Top: Example in situ hybridizations of type-specific markers (magenta) (Figure S2), Rbp4-Cre neurons (green), Hoechst (blue). Middle: Fraction of Rbp4-Cre neurons containing embryonic-NP, IT, and PT-specific in situ hybridization markers; bold: genes shown in examples, above. Bottom: fractions of embryonic-NP, IT, and PT Rbp4-Cre neurons (based on in situ hybridizations).
(D) Immunostaining Rbp4-Cre neurons (green), BrdU (red), Hoechst (blue). Arrowheads: example Rbp4-Cre neurons incorporating BrdU.
(E) Normalized depth of Rbp4-Cre neurons incorporating BrdU (left), compared to Rbp4-Cre neurons without BrdU in the same embryos (center) and control embryos without BrdU injection (as in A). χ2 test.
(F) Correlation of gene expression: comparing across time vs. type (on E18.5).
Scale bars: 50 μm (top, A), 10% (bottom right, A), 20 μm (bottom left, B), 10 μm (top, C), 25 μm (D).
See also Figure S2.
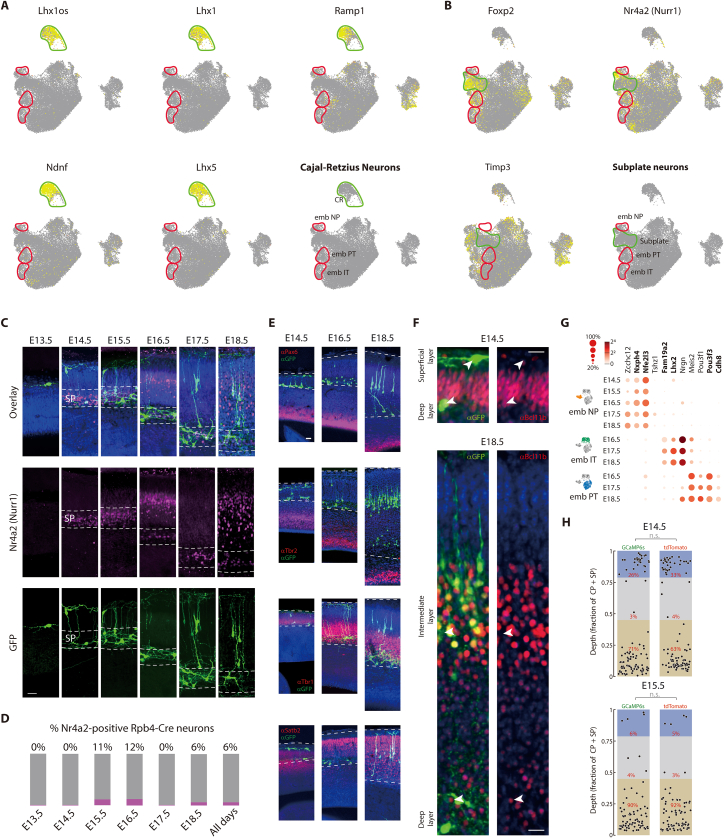
Figure S2.
Rbp4-Cre neurons are located in the region from the subplate to the surface of cortex, are distinct from Cajal-Retzius cells and subplate neurons, and express layer 5 markers, related to Figure 2
(A) Within the UMAP embedding of all 25681 sequenced excitatory neuron transcriptomes (gray), cells expressing a number of genes previously associated with Cajal-Retzius cells66,68,79 overlap in a location (green outline) distinct from positively identified Rbp4-Cre neurons (as in Figure S1) (red outline; Rbp4-Cre neurons) (labeled in bottom right). Clusters of Rbp4-Cre neurons are additionally labeled based on the cluster identification from Figure 1E.
(B) Within the UMAP embedding of all 25681 sequenced excitatory neuron transcriptomes (gray), cells expressing genes previously associated with subplate neurons overlap in a location (green outline) distinct from positively-identified Rbp4-Cre neurons (as in Figure S1) (red outline; Cre neurons) (labeled in the bottom right). Clusters of Rbp4-Cre neurons are additionally labeled based on the cluster identification from Figure 1E.
(C) Rbp4-Cre neurons (green), counterstained with Hoechst (blue), show that neurons in the deep layer lie physically within the subplate (SP; dotted line), as localized by the expression of a common subplate marker, Nr4a2 (Nurr1) (magenta).
(D) Only a small fraction of Rbp4-Cre neurons express Nr4a2 from E13.5 to E18.5.
(E) Rbp4-Cre neurons are located in the region from the subplate to the surface of cortex. Rbp4-Cre neurons (stained using GFP antibody labeling GCaMP6s, green) in both spatial configurations (on E14.5, E16.5, and E18.5) co-labeled with antibodies labeling different zones in the developing cortical wall (Pax6 (first row), Tbr2 (second row), Tbr1 (third row), Satb2 (fourth row), red), counterstained with Hoechst (blue). Dotted line outlines the area from subplate to surface of cortex.
(F) Rbp4-Cre neurons (stained using GFP antibody, green), counterstained with Hoechst (blue), show that neurons within both the superficial and deep layers at E14.5, as well as neurons within both the intermediate and deep layers at E18.5, all colocalize with Bcl11b (red), the expression of which is restricted in the adult cortex to layer 5.69 Arrows: example Rbp4-Cre neurons.
(G) Genes selective for each embryonic layer 5 type. Transcript counts for each gene shown as colored circles in all types at all ages. Radius of circle: fraction of cells expressing the gene; color of circle: mean normalized transcripts per cell (log2). Bold text: genes used for in situ hybridization, shown in Figure 2C.
(H) Spatial distribution of Rbp4-Cre neurons into layers is indistinguishable when expressing GCaMP6s compared to tdTomato on both E14.5 and E18.5, and Rbp4-Cre neurons. Quantifying the distribution of Rbp4-Cre neurons into layers at E14.5 (top) and E15.5 (bottom) in embryos generated by using the GCaMP6s-tTA2 reporter line, or a tdTomato reporter line. Probability: χ2 test comparing the fraction of Rbp4-Cre neurons in each layer for the two different reporter lines, p = 0.05. Scale bars: 20 μm (C, E, F).
During development, the cortical plate is bordered above by the marginal zone, containing Cajal-Retzius cells,74,75,76 and below by the subplate, containing subplate neurons.77,78 From E14.5 onwards, Rbp4-Cre neurons were located entirely within the cortical plate and subplate (Figure S2). Since superficial layer Rbp4-Cre neurons were close to the surface of the cortical plate, while neurons within the deep layer were within the subplate, it is possible that some Rbp4-Cre neurons have Cajal-Retzius or subplate identity. However, genes previously identified as markers of Cajal-Retzius cells66,68,79 were expressed in neurons with distinct gene expression profiles from Rbp4-Cre neurons. Similarly, genes identified as markers of subplate neurons7,66 were expressed in neurons distinct from Rbp4-Cre neurons, and Rbp4-Cre neurons did not express the common subplate marker, Nr4a2. Instead, at E14.5, in both superficial and deep layers, Rbp4-Cre neurons expressed Bcl11b, an L5-PN marker. Similarly, at E18.5, Rbp4-Cre neurons in both the intermediate and deep layers expressed Bcl11b. Therefore, all Rbp4-Cre neurons, independent of neocortical depth, have L5-PN but not Cajal-Retzius or subplate identity.
To reveal how the three Rbp4-Cre cell types divide into the three layers, we performed in situ hybridization against genes that are preferentially expressed in each Rbp4-Cre cell type (Figures 2C and S2). At E14.5, both superficial and deep layers were composed predominantly of embryonic-NP type. This confirmed the previous finding that, at E14.5, all Rbp4-Cre neurons formed a single cluster corresponding to the embryonic-NP type. At E18.5, both intermediate and deep layers were composed of all three types. Therefore, superficial layer Rbp4-Cre neurons are always embryonic-NP type. At E14.5, Rbp4-Cre neurons in the deep layer were also embryonic-NP type, but the cell type composition gradually changed to include all three types by E18.5. Similarly, the intermediate layer was a mixture of all three types.
The morphology of embryonic-NP Rbp4-Cre neurons changed during the development of the neocortex. At E14.5, they had a tangential orientation, with neurites extending horizontally, parallel to the surface of cortex. In contrast, at E18.5, embryonic-NP neurons in the intermediate layer had a vertical morphology, extending neurites primarily perpendicular to the surface of cortex. This change in morphology and location could be due to the presence of distinct subtypes of embryonic-NP neurons or due to a change in morphology within the embryonic-NP type. Different subtypes could appear because of differences in neuronal birth dates80,81,82 or by changes in the pattern of gene expression during development. Injecting dams with bromodeoxyuridine (BrdU) on embryonic day E12.5, we observed overlap with Rbp4-Cre neurons in both the superficial and deep layers at E14.5, as well as in both the intermediate and deep layers at E18.5 (Figure 2D). Further, this distribution was statistically indistinguishable from that observed across all Rbp4-Cre neurons (Figure 2E). We then compared the gene expression profiles of embryonic-NP neurons from E14.5 to E18.5, on every second embryonic day, using correlation as a measure of similarity (Figure 2F). The correlations of embryonic-NP neurons across the three time points were similar to those observed within embryonic-PT and IT types, from E16.5 to E18.5. Further, compared to the correlations between embryonic-NP and both embryonic-PT and IT neurons on a single embryonic day (E18.5), embryonic-NP neurons were more correlated across different embryonic days. Therefore, embryonic-NP neurons across embryonic days overlap in their birth date and remain more similar in gene expression to each other, than compared to the other Rbp4-Cre neuron types on a single embryonic day. These observations are consistent with a single embryonic-NP type of Rbp4-Cre neurons across different embryonic days, despite the changes in their morphology and location.
Para-uterine imaging enables recordings of activity from neurites of Rbp4-Cre neurons in living embryos
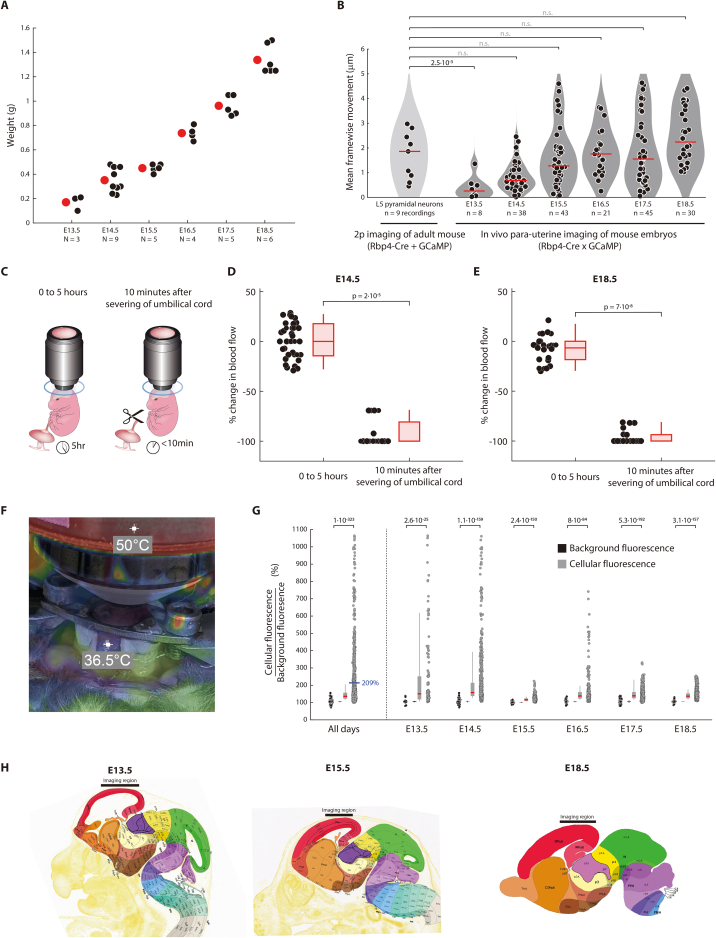
To understand if Rbp4-Cre neurons form active circuits in vivo, we developed a method that provides enhanced mechanical stability to the embryo, and thereby the resolution necessary to image individual neurites in vivo. To increase mechanical stability, we placed the embryo within a holder matched to the embryo’s size, filled the holder with agar, and placed a cover glass on top (Figure 3A). Additionally, to isolate the embryo from the anesthetized dam’s movements due to breathing and beating of the dam’s heart, the holder was externally supported. In this way, embryos, ranging in weight from 0.17 g at E13.5 to 1.3 g at E18.5, were stable enough such that the amount of movement between two-photon imaging frames was similar to that observed in adult mice (Figure S3). Moreover, to keep the embryo healthy during imaging, we held the embryo in the holder within the dam’s abdominal cavity adjacent to the uterus (“para-uterine”) (Figure 3A), decreasing strain on the umbilical cord. Blood flow within the embryonic cortex was constant for at least 5 h, as opposed to deteriorating within 10 min upon disruption of the umbilical cord (Video S1; Figure S3). Further, the para-uterine location maintained the embryo at physiological temperature.83,84 These improvements allowed the imaging of Rbp4-Cre neurons for at least 5 h, with a resolution sufficient to resolve individual neurites.
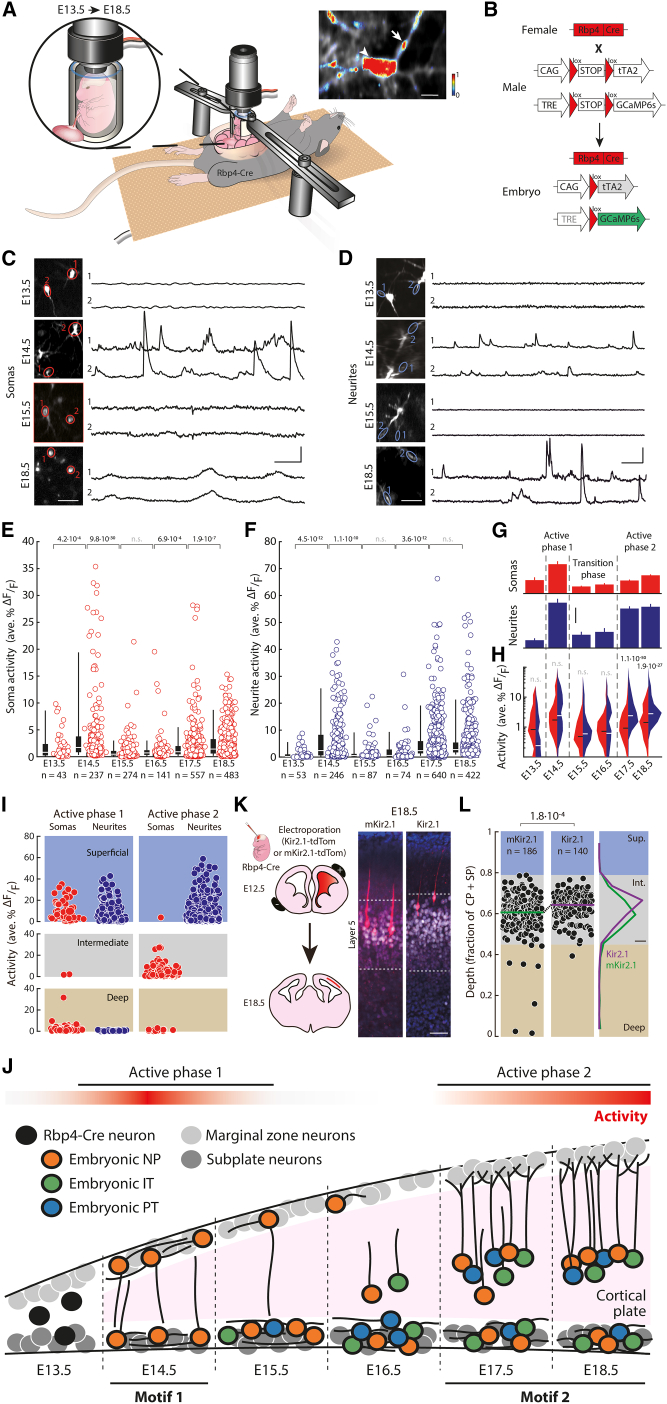
Figure 3.
Rbp4-Cre neurons show two phases of increased spontaneous activity
(A) Schematic diagram of in vivo para-uterine two-photon calcium imaging (Figure S3). Top right: single embryonic neuron; arrowhead: soma activity; arrow: neurite activity; color: normalized calcium activity.
(B) Mating strategy to drive GCaMP6s expression in Rbp4-Cre neurons.
(C and D) Two-photon imaging of somas (red, C) and neurites (blue, D) of Rbp4-Cre neurons. Two regions of interest (ROIs) (left) and their recorded activity traces (right).
(E and F) Activity of individual somas (E) and neurites (F) of Rbp4-Cre neurons. Circles: activity of each ROI; box (25–75 percentile) and whisker (5–95 percentile); white line: median; n = number of somas or neurites. Recordings from 3 (E13.5), 9 (E14.5), 5 (E15.5), 4 (E16.5), 5 (E17.5), and 6 (E18.5) embryos (Figures S4 and S5).
(G) Activity (mean ± SEM) (data from E [soma, red] and F [neurite, blue]). Dotted line: separation of active phases and transition phase.
(H) Distribution of activity (data from E [soma, red] and F [neurite, blue]) in log-scale. Horizontal lines: median (black: soma; white: neurite).
(I) Activity in the two active phases across the three layers.
(J) Schematic of Rbp4-Cre neuron development, highlighting the two circuit motifs and phases of activity.
(K) Left: schematic of electroporations. Right: Immunostaining of electroporated Rbp4-Cre neurons (red), Bcl11b (white), Hoechst (blue).
(L) Distribution of mKir2.1- or Kir2.1-positive Rbp4-Cre neurons’ normalized depths (as in Figure 2A) (10 mKir2.1-tdTomato and 10 Kir2.1-tdTomato electroporated embryos). Colored lines: medians. n = number of neurons.
(E, F, H, L) Wilcoxon rank-sum test.
Scale bars: 10 μm (inset, A), 40 μm (left, C, D), 25 s and 25 %ΔF/F (right, C), 25 s and 50 %ΔF/F (right, D), 2 ave. %ΔF/F (G), 20 μm (K), 10% (L).
See also Figures S3, S4, and S5.
Figure S3.
Characterizing in vivo para-uterine method for imaging cortical neurons, related to Figure 3
(A) Mean (red dot) of embryonic weights on each day (black dots) ranges from 0.17 g at E13.5 to 1.3 g at E18.5. N = number of embryos.
(B) Stability of in vivo para-uterine two-photon imaging of cortical neurons in embryos is similar to stability of two-photon imaging of cortical neurons in adult mice. Movement per frame (recorded from 5 to 10 Hz), averaged per recording, computed via rigid motion correction between frames.145In vivo embryonic recordings were made as schematized in Figure 3A. Adult recordings were made in head-fixed Rbp4-Cre mice, injected with AAV expressing Cre-dependent GCaMP. Probability: Wilcoxon rank-sum test; n = number of recordings from 3 (E13.5), 9 (E14.5), 5 (E15.5), 4 (E16.5), 5 (E17.5), and 6 (E18.5) embryos and 3 adult mice.
(C–E) Embryonic blood flow does not change following 5 h of imaging, but degrades rapidly following severing of the umbilical cord. Blood flow at the surface of the brain was imaged in visible light prior to and following 5 h of imaging (C). The difference in blood flow is quantified (left), at E14.5 (D) and E18.5 (E). Blood flow at the surface of the brain was also imaged immediately prior to, and 10 min following the severing of the umbilical cord. The difference in blood flow is quantified (right), at E14.5 (D) and E18.5 (E). Box-and-whiskers: distribution of changes in blood flow across each time window, as box (25–75 percentile) and whisker (5–95 percentile); red lines: median. Probability: Wilcoxon rank-sum test.
(F) Temperature of embryo stabilized para-uterine under the two-photon microscope. Infrared image is aligned with a visible light image, where the embryo can be observed within the holder (as schematized in Figure 3A). The 36.5°C marker labels the embryo,83,84 which is visible through the opening in the holder allowing for the exit of the umbilical cord. The second 50°C marker labels the objective heater, providing a secondary source of heat during imaging. Image was taken following 5 h of imaging. (G) Fluorescence of Rbp4-Cre neurons is significantly increased over background fluorescence from E13.5 to E18.5. Mean cellular fluorescence, normalized by the pixel size of each cell, compared against the mean pixel fluorescence within three background regions selected within the imaging window, collected across all neurons, across all embryonic days (left) and on each embryonic day (right). Black circles: ratio of fluorescence within individual background regions in each imaging plane compared to each other; gray circles: ratio of fluorescence within individual neurons compared to each background region in the same imaging plane; box-and-whiskers: distributions across background fluorescence ratios (black) and cellular fluorescence ratios (gray) as box (25–75 percentile) and whisker (10–90 percentile); black and red line: median; blue line and text: mean. Probability: Wilcoxon rank-sum test comparing cellular fluorescence ratios to background fluorescence ratios. Recordings from 3 (E13.5), 9 (E14.5), 5 (E15.5), 4 (E16.5), 5 (E17.5), and 6 (E18.5) embryos.
(H) Imaging region shown with respect to the embryonic brain at E13.5 (left), E15.5 (middle), and E18.5 (right). Imaging region was centered over the posterior dorsal pallium. Images taken from Allen Developing Brain Atlas (http://atlas.brain-map.org/).159
Video was taken after 5 h of para-uterine imaging and 10 min after the disruption of the umbilical cord
We tested several systems for driving expression of calcium sensors in Rbp4-Cre neurons: adeno-associated viruses, herpes simplex viruses, electroporation, and mouse lines. Amongst these systems, GCaMP6s-tTA2 reporter mice, when crossed with Rbp4-Cre mice, generated GCaMP6s expression in Rbp4-Cre neurons throughout cortex, with sufficient signal-to-noise ratio for two-photon imaging during embryonic cortical development (Figures 3A, 3B, and S3). GCaMP6s-tTA2 mice were double transgenics that we created by crossing mice expressing tetracycline-controlled transactivator protein (tTA2)85 with mice expressing tTA2-dependent GCaMP6s,65,86 both under the control of Cre. Rbp4-Cre neurons showed an average fluorescence that was 109% above the background, from E13.5 to E18.5 (Figure S3). Therefore, the combination of “para-uterine imaging” and the use of GCaMP6s-tTA2 mice enabled the in vivo imaging of activity in both somas and neurites of Rbp4-Cre neurons, across the embryonic development of the cortical plate (Figures 3C and 3D).
Rbp4-Cre neurons show two phases of increased activity
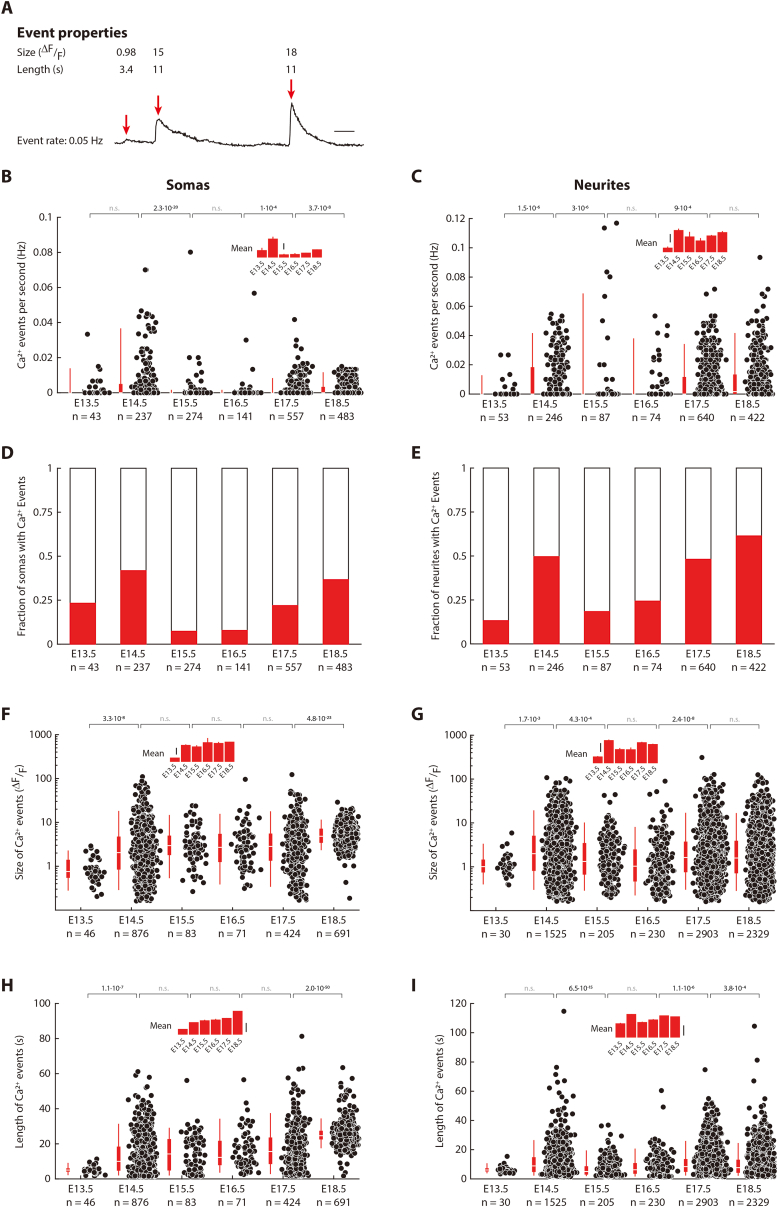
At E14.5 and from E17.5 onwards, we observed spontaneous calcium events in both Rbp4-Cre somas and neurites (Figures 3C and 3D; Video S2) in the posterior part of cortex (posterior dorsal pallium; Figure S3). Calcium event properties, such as the length and size, differed across embryonic days (Figure S4). Therefore, to assess the overall change of activity, we quantified the total change of GCaMP6s fluorescence (%ΔF/F) across each recording, normalized per second (Figures 3E, 3F, and S5). In both Rbp4-Cre somas and neurites, spontaneous calcium activity significantly increased from E13.5 to E14.5. This was followed by a significant decrease in activity from E14.5 to E15.5 (Video S2). Activity stayed at a low level from E15.5 to E16.5. From E16.5 to E17.5, activity in both Rbp4-Cre somas and neurites significantly increased. In somas, the activity then increased further from E17.5 to E18.5 while, in neurites, the increased activity plateaued from E17.5 to E18.5. Although the dam was anesthetized during imaging, different anesthetics did not change the overall amplitude of spontaneous calcium activity in either active phase (Figure S5), despite their distinct modes of action.87,88 Therefore, we found two periods of time with increased spontaneous calcium activity, independent of the dam’s anesthesia: the first at E14.5, and the second starting at E17.5, separated by a transition phase from E15.5 to E16.5, where Rbp4-Cre neurons showed reduced activity (Figure 3G). These two phases were also reflected in the fraction of active neurons (Figure S4).
Figure S4.
Fraction of Rbp4-Cre ROIs with calcium events, as well as the frequency, length, and size of calcium events all vary across embryonic days, related to Figure 3
(A) Spontaneous calcium activity recorded from a single Rbp4-Cre neuron, showing three detected events, with event properties quantified for each event.
(B–I) Quantification of event properties for Rbp4-Cre somas (B, D, F, and H) and neurites (C, E, G, and I). On each embryonic day from E13.5 to E18.5, these event statistics include the event rate (B and C), fraction of Rbp4-Cre ROIs showing spontaneous calcium events in each 10-min recording (D and E) and, for each detected calcium event, the size (F and G) and length (H and I) of the event. Circles: event property of each ROI; box (25–75 percentile) and whisker (5–95 percentile); white line: median. Inset: mean ± SEM. (B – E) n = number of Rbp4-Cre ROIs (somas or neurites) recorded from 3 (E13.5), 9 (E14.5), 5 (E15.5), 4 (E16.5), 5 (E17.5), and 6 (E18.5) embryos. (F – I) n = number of detected calcium events. Probability: Wilcoxon rank-sum test. Scale bar: 5s (A), 2⋅10−3 Hz (inset, B), 4⋅10−3 Hz (inset, C), 2 ΔF/F (inset, F), 2 ΔF/F (inset, G), 10s (inset, H), 10s (inset, I).
Figure S5.
Characterizing calcium activity and migration in embryonic Rbp4-Cre neurons, related to Figure 3
(A and B) Activity in the somas (A) and neurites (B) of Rbp4-Cre neurons (data from Figures 3E and 3F) with neurons recorded in the same embryo colored identically.
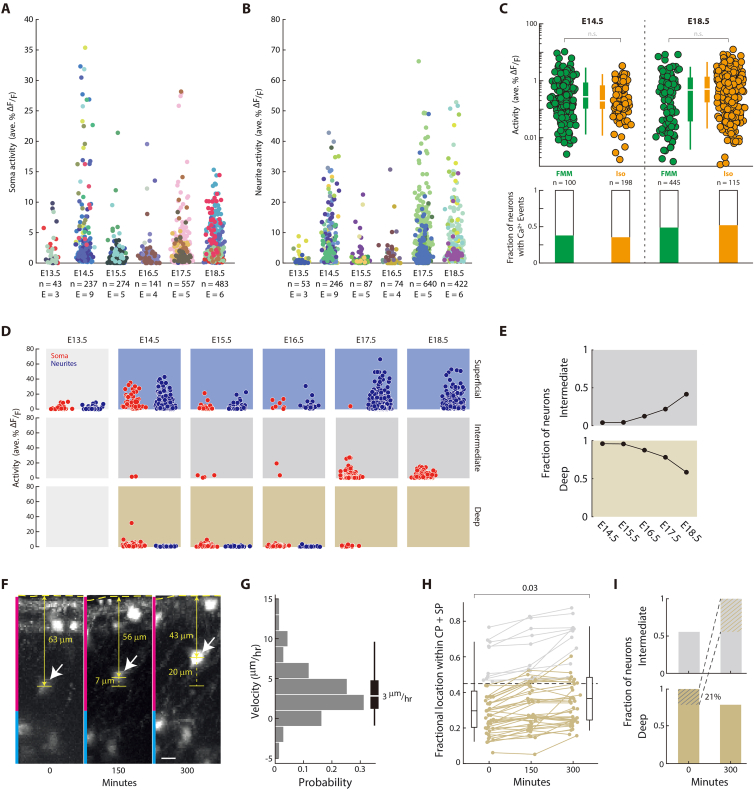
(C) Changing the anesthetic used in the dam does not change the overall calcium activity in Rbp4-Cre neurons, during both embryonic phases of increased activity. In vivo para-uterine two-photon imaging of Rbp4-Cre neurons was performed at E14.5 (left) and E18.5 (right), to characterize the activity during both active phases, with the dam anesthetized using two different anesthetics: a mixture of Fentanyl-Medetomidine-Midazolam (FMM), and 1.75% Isoflurane. Probability: Wilcoxon rank-sum test. n = number of Rbp4-Cre neurons recorded from 8 (E14.5) and 6 (E18.5) embryos.
(D) Activity in Rbp4-Cre neurons’ somas (red) and neurites (blue), in each layer, on each embryonic day. Spatial layers colored as in Figure 2A (blue: superficial layer; gray: intermediate layer; beige: deep layer). Somas and neurites were not assigned to layers at E13.5 (light gray) because of the limited thickness of cortex combined with the limited depth resolution of 2p imaging. Deep layer neurons were poorly sampled >E17.5, because the increasing depth made them difficult to image from with 2p imaging.
(E–I) Rbp4-Cre neurons migrate from the deep layer to the intermediate layer.
(E) Fraction of all neurons within the intermediate and deep layers, from E14.5 to E18.5, found in each of the two layers. Layers colored as in Figure 2A (gray: intermediate layer; beige: deep layer).
(F) In vivo para-uterine time lapse imaging of a population of Rbp4-Cre neurons labeled with tdTomato, at E16.5, over 5 h, in the cortical plate (magenta) and subplate (cyan). White arrow: migrating cell; horizontal dotted line: surface of cortex; yellow line: distance of cell to surface; vertical dotted line: distance migrated.
(G) Distribution of migration velocities for all recorded migrating neurons, averaged across 5 h of time lapse imaging. n = 48 neurons from 2 E16.5 embryos. Box (25–75 percentile) and whisker (5–95 percentile); white line (median).
(H) Locations of layer 5 neurons in intermediate (gray) and deep (beige) layers (colored based on position at t = 0), followed using para-uterine time lapse imaging for 5 h. Shown as a fraction of the combined cortical plate (CP) and subplate (SP) thickness. Box-and-whiskers: distribution of neuronal locations at the start of imaging (left) and after 300 min (right) as box (25–75 percentile) and whisker (5–95 percentile); black line: median; dotted line: boundary between deep and intermediate layer at t = 0; p value: Wilcoxon rank-sum test; n = 48 neurons from 2 E16.5 embryos.
(I) Fraction of Rbp4-Cre neurons within each layer at the start of imaging (left) and after 300 min (right), normalized to the maximum within each layer (data from (H)). 21% of neurons in the deep layer move to the intermediate layer within the imaging period (colored shading), and no neurons move in the reverse direction. Scale bar: 10 μm (F).
Sequentially showing imaging of GCaMP6s in somas of Rbp4-Cre neurons at E14.5 (10X real-time), neurites of Rbp4-Cre neurons at E14.5 (15X real-time), somas and neurites of Rbp4-Cre neurons at E15.5 (15X real-time), somas of Rbp4-Cre neurons at E18.5 (30X real-time), and neurites of Rbp4-Cre neurons at E18.5 (15X real-time). Motion correction was applied to the video, and the fire colormap in FIJI was used to visualize changes. Arrowheads: calcium events
In the first phase of increased activity, somas and neurites of Rbp4-Cre neurons showed no significant difference in activity from each other (Figure 3H). Further, neurons in both the superficial and deep layers were active (Figures 3I and S5). In contrast, in the second phase, neurites showed significantly higher activity than somas. In this phase, we recorded primarily from the intermediate layer, and active Rbp4-Cre neurons were found in this layer. Therefore, active somas and neurites were present in all imaged cortical layers, in both phases of increased activity, but there was a qualitative change between the relative activity of somas and neurites from the first phase to the second.
Taken together, during embryonic development, Rbp4-Cre neurons switch their activity, spatial organization, and cell type identity in a way that is coordinated in time (Figure 3J). Initially, Rbp4-Cre neurons are present in a highly active deep-and-superficial layer configuration composed of a single embryonic-NP type. This constitutes the first organizational motif, which persists until E15.5. From E15.5 to E16.5, activity significantly decreases, coinciding with a decrease in superficial layer neurons and the formation of an intermediate layer. By E17.5, Rbp4-Cre neurons switch to a new, deep-and-intermediate layer configuration, which persists until birth. This second motif again shows increased activity but contains all three Rbp4-Cre neuron types.
Rbp4-Cre neurons migrate from the deep layer to the intermediate layer between the active phases
From E16.5 onwards, the number of neurons in the intermediate layer increased as the number of neurons in the deep layer decreased (Figures 2A and S5). To test if this change in proportion is due to neurons in the deep layer migrating to the intermediate layer, we performed in vivo time-lapse imaging over 5 h at E16.5. We observed neurons moving from the deep layer toward the intermediate layer, with an average speed of 3 μm/h, a similar speed to what has been reported before.89,90,91 Each hour, 4% of neurons within the deep layer moved into the intermediate layer. This suggests that neurons in the deep layer migrate to form the intermediate layer, during the time interval when the Rbp4-Cre neurons show decreased activity.
Perturbation of activity alters the layered organization of Rbp4-Cre neurons
Since both the activity and spatial organization of Rbp4-Cre neurons changed in a coordinated way across embryonic days, we asked if perturbing the activity of Rbp4-Cre neurons may change their spatial localization. We performed in utero electroporation of embryos with a plasmid expressing, in a Cre-dependent manner, either the inward rectifier potassium channel Kir2.192 or a non-conducting Kir2.1 channel (mKir2.1) as a control,93,94 both fused to tdTomato (Figure 3K). To ensure that we targeted Rbp4-Cre neurons already at E14.5, we performed electroporation at E12.5. The electroporated plasmids enter progenitor neurons at E12.5 and, once Cre becomes active in postmitotic Rbp4-Cre neurons, should result in the hyperpolarization of the Cre-expressing postmitotic neurons.21,22,23,24,25,26 Compared to mKir2.1-tdTomato expressing Rbp4-Cre neurons, Kir2.1-tdTomato expressing Rbp4-Cre neurons localized significantly closer to the surface of cortex (Figure 3L). Therefore, hyperpolarization of postmitotic Rbp4-Cre neurons affects their location within the neocortex at E18.5.
Rbp4-Cre neurons have active conductances
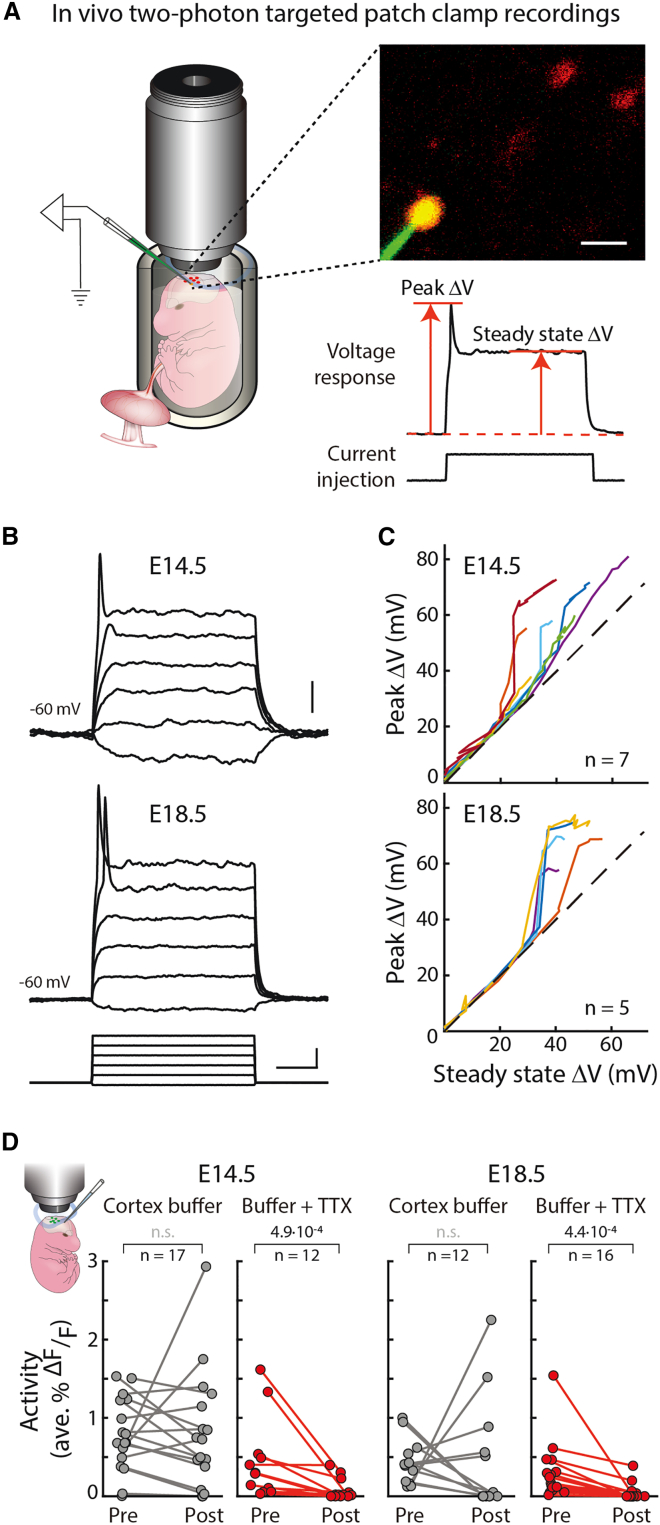
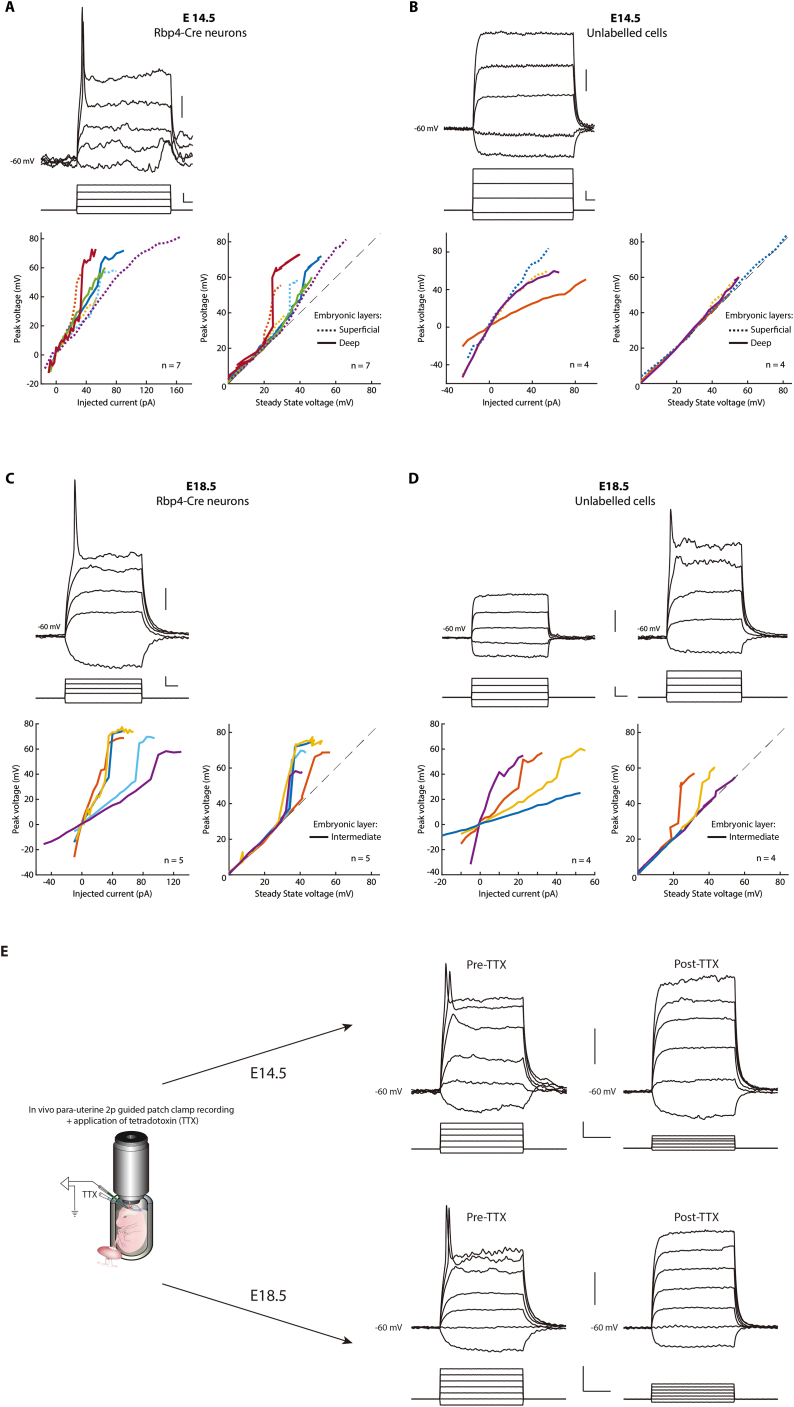
To determine if neurons that are active in the two phases have active conductances, we performed in vivo two-photon targeted patch clamp recordings from Rbp4-Cre neurons labeled with tdTomato (Figure 4A). Interestingly, all recorded Rbp4-Cre neurons, at E14.5 and E18.5, displayed active conductances as indicated by a nonlinear increase in peak voltage, with increasing current steps (Figures 4B and 4C). This included neurons within both the superficial and deep layers, at E14.5, and within the intermediate layer, at E18.5 (Figure S6). These active conductances disappeared upon application of a blocker of voltage-gated sodium channels, TTX. In contrast, none of the recorded unlabeled cells in the vicinity of labeled Rbp4-Cre neurons, independent of depth, displayed active conductances at E14.5. Even at E18.5, only half of nearby unlabeled cells had active conductances. Therefore, during both phases of increased calcium activity, Rbp4-Cre neurons display TTX-sensitive active conductances, suggesting that voltage-gated sodium channels may contribute to the increase in calcium activity.
Figure 4.
Rbp4-Cre neurons display active conductances during both active phases of embryonic development
(A) Left: schematic of in vivo two-photon targeted patch clamp recordings from Rbp4-Cre neurons. Right top: imaging field. Rbp4-Cre neurons (red), Alexa Fluor 488 filled pipette (green) and patched neuron (yellow). Right bottom: voltage change with peak and steady state voltage labeled, in response to current injection.
(B) Rbp4-Cre neurons’ voltage responses to graded current injections (bottom).
(C) Peak versus steady state voltage. n = number of neurons (Figure S6).
(D) Calcium activity following TTX application. Wilcoxon signed rank test. n = number of neurons recorded in 3 E14.5 and 3 E18.5 embryos.
Scale bars: 20 μm (inset, A), 10 mV (top, B), 50 ms and 40 pA (bottom, B).
See also Figure S6.
Figure S6.
All Rbp4-Cre neurons display voltage gated sodium channels dependent active conductances, at both E14.5 and E18.5, related to Figure 4
(A–D) All Rbp4-Cre neurons (A, C), but not all nearby unlabeled cells (B, D), display active conductances, at both E14.5, and E18.5. Voltage responses (top) of an Rbp4-Cre neuron and a nearby unlabeled cell at E14.5 (A and B) and E18.5 (C and D) to graded intracellular current injections (middle), recorded in current clamp mode. The voltage responses to current are quantified in both current-voltage curves and the peak voltage compared to the steady-state voltage (as in Figure 4C) for both Rbp4-Cre neurons (C) and nearby unlabeled neurons (D), to visualize any nonlinearity in the peak voltage response. The line type labels the embryonic layer in which each recorded neuron was found (E14.5: Dotted: superficial layer, solid: deep layer; E18.5: Solid: intermediate layer). Due to visibility at E18.5, it was not possible to record from deep layer neurons. n = number of neurons.
(E) Left: schematic of para-uterine imaging combined with patch clamp recording to perform in vivo two-photon targeted electrophysiology from Rbp4-Cre neurons together with application of TTX. Right: Voltage responses of an Rbp4-Cre neuron (at E14.5 (top) and E18.5 (bottom)) to graded intracellular current injections, recorded in current clamp mode before (left, pre) and after (right, post) application of TTX. Scale bar: 10 mV (top, A), 50 ms and 20 pA (middle, A), 10 mV (top, B), 50 ms and 20 pA (middle, B), 10 mV (top, C), 50 ms and 20 pA (middle, C), 10 mV (top, D), 50 ms and 20 pA (middle, D), 20 mV (top, E), 10 ms and 40 pA (bottom, E).
To test this, we imaged the change in spontaneous calcium activity in Rbp4-Cre neurons, before and after the application of TTX. At both E14.5 and E18.5, calcium activity significantly decreased following the application of TTX (Figure 4D). In contrast, applying cortex buffer alone resulted in no significant change in the spontaneous activity on either embryonic day. This suggests that voltage-gated sodium channels contribute to the recorded calcium activity during embryonic development.
Rbp4-Cre neurons form synapses
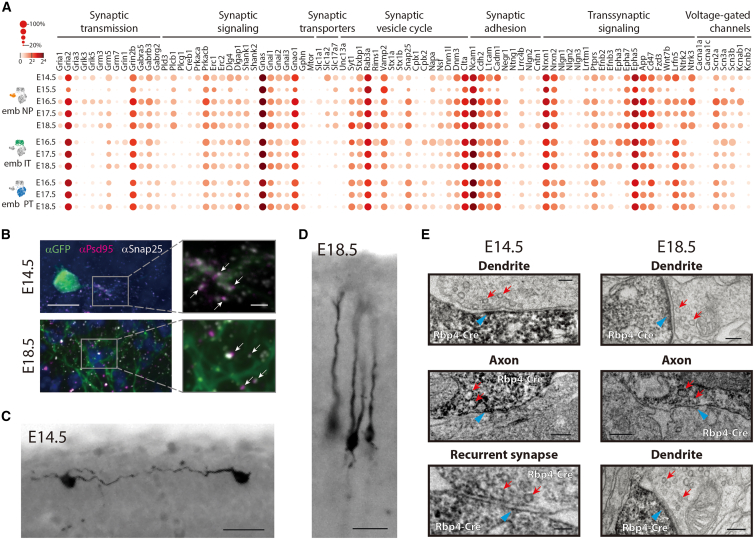
To identify whether Rbp4-Cre neurons could form cortical circuits with other Rbp4-Cre neurons, we explored their expression of genes underlying neuronal communication. Many genes associated with synaptic function and active neuronal membrane properties were expressed within all three Rbp4-Cre neuron types across developmental days E14.5 to E18.5 (Figure 5A, Data S1). This included genes involved in chemical synaptic transmission, such as AMPA and NMDA receptor subunits; genes involved in electrical synaptic transmission, such as Connexin 45 (Gjc1); genes involved in pre- and postsynaptic signaling, such as Erc1 and PSD-95 (Dlg4); genes associated with the synaptic vesicle cycle, such as Snap25 and Vamp2; and genes necessary for active neuronal membrane properties, such as voltage-gated calcium, sodium, and potassium channels.95,96 Therefore, the molecular components underlying neuronal communication are present within all three types of Rbp4-Cre neurons from E14.5 to E18.5.
Figure 5.
Rbp4-Cre neurons have synapses already at E14.5
(A) Expression (circles) of selected genes (Data S1) related to neuronal communication.95,96 Radius of circles: fraction of cells expressing the gene; color of circles: mean normalized transcripts per cell (log2).
(B) Immunostaining of Rbp4-Cre neurons (green), Snap25 (presynaptic; gray), PSD-95 (postsynaptic; magenta), Hoechst (blue). Right: Zoom (arrows: colocalization of Snap25 and PSD-95).
(C and D) DAB staining of Rbp4-Cre neurons in tissue prepared for EM.97
(E) Synaptic contacts involving Rbp4-Cre neurons (DAB, darker cells), presynaptic vesicles (red arrow) and postsynaptic densities (blue arrowhead).
Scale bars: 10 μm (left, B), 2 μm (right, B), 25 μm (C), 25 μm (D), 100 nm (E).
We then explored whether Rbp4-Cre neurons form synapses. First, we performed immunostaining against a presynaptic marker, Snap25, and a postsynaptic marker, PSD-95. Both markers showed punctate labeling on the neurites of Rbp4-Cre neurons at E14.5 and at E18.5 (Figure 5B). Additionally, Snap25 and PSD-95 colocalized in many puncta. Next, we visualized Rbp4-Cre neurons with electron microscopy (EM) using DAB97 to identify the presence of synaptic specializations onto or from Rbp4-Cre neurons (Figures 5C–5E). At both E14.5 and E18.5, we observed close junctional contacts involving Rbp4-Cre neurons, with synaptic vesicles marking the presynaptic side and postsynaptic densities marking the postsynaptic side. Some of these synapses had Rbp4-Cre neurons presynaptically, while others had Rbp4-Cre neurons postsynaptically. Additionally, at E14.5, we observed synapses with Rbp4-Cre neurons on both sides, i.e. recurrent synapses between two Rbp4-Cre neurons. This suggests that Rbp4-Cre neurons both form and receive synaptic contacts and, already at E14.5, that pairs of Rbp4-Cre neurons have recurrent synapses.
Rbp4-Cre neurons are sensitive to AMPA and NMDA and show excitatory synaptic potentials
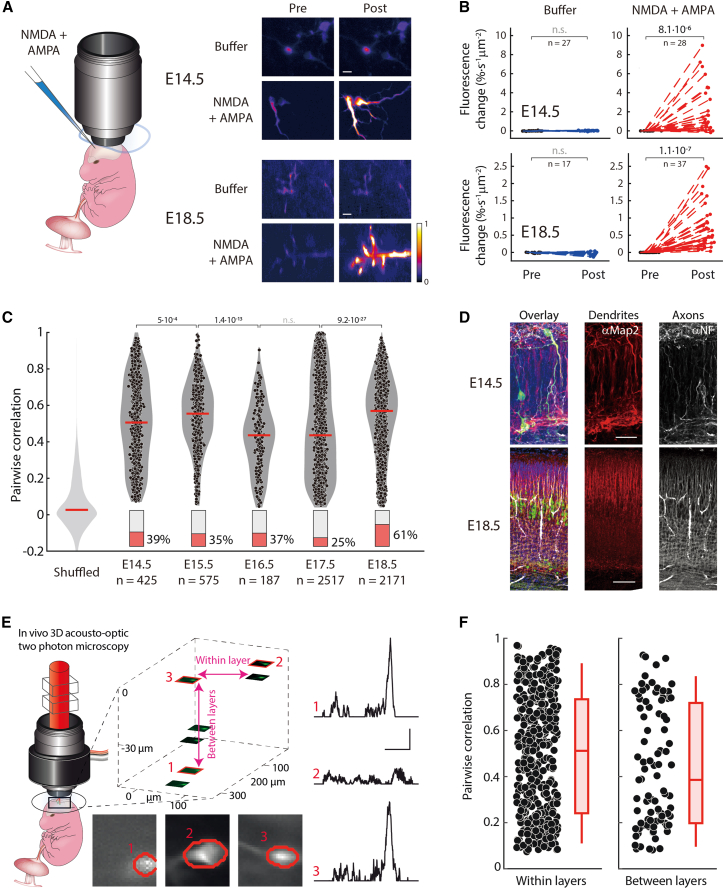
To determine if the synapses during embryonic development are functional, we applied agonists of glutamatergic synaptic transmission while imaging from Rbp4-Cre neurons (Figure 6A). On both E14.5 and E18.5, applying a mixture of AMPA and NMDA onto cortex was closely followed by a significant increase in calcium in all imaged Rbp4-Cre neurons (Figure 6B). In contrast, cortex buffer alone led to no significant change in calcium. Additionally, following application of TTX, in vivo two-photon targeted patch clamp recordings from Rbp4-Cre neurons revealed spontaneous excitatory synaptic potentials at both E14.5 and E18.5 (Figure S7). The response to glutamatergic agonists and the presence of spontaneous excitatory synaptic potentials suggest that Rbp4-Cre neurons form functional synapses during embryonic development.
Figure 6.
Rbp4-Cre neurons form active circuits already at E14.5
(A) Left: schematic of NMDA+AMPA injection during in vivo embryonic two-photon imaging. Right: Rbp4-Cre neurons; color: normalized calcium activity).
(B) Change in fluorescence before (Pre) and after (Post) application of either cortex buffer (blue) or NMDA+AMPA (red). Wilcoxon signed rank test. n = number of Rbp4-Cre neuron ROIs from 3 E14.5 and 3 E18.5 embryos (Figure S7).
(C) Pairwise correlations of Rbp4-Cre neurons’ calcium activity, that are significantly greater than random (Figure S7). Shuffled data is on the left. Filled circles: correlations; gray shading: distribution; red line: median. Bars: percent of neuron pairs with correlations significantly greater than random (red). Wilcoxon rank-sum test. n = pairs of Rbp4-Cre neurons recorded from 3 (E13.5), 9 (E14.5), 5 (E15.5), 4 (E16.5), 5 (E17.5), and 6 (E18.5) embryos.
(D) Immunostaining of Rbp4-Cre neurons (green), Map2 (dendrites, red), NF (axons, white), Hoechst (blue).
(E) Left: schematic in vivo para-uterine imaging using 3D acousto-optic two-photon microscope. Top middle: mean projections around each soma. Bottom middle: Three zoomed examples (red outline, top middle). Right: Δf/f activity from examples. Cells 1 and 3 have high correlation.
(F) Pairwise correlations of E14.5 Rbp4-Cre neurons’ activity that are significantly greater than random, within and across layers. Dots: pairwise correlations; box (25–75 percentile) and whisker (5–95 percentile); line: median.
Scale bars: 10 μm (A), 30 μm (top, D), 100 μm (bottom, D), 20s and 5 %ΔF/F (E).
See also Figure S7.
Figure S7.
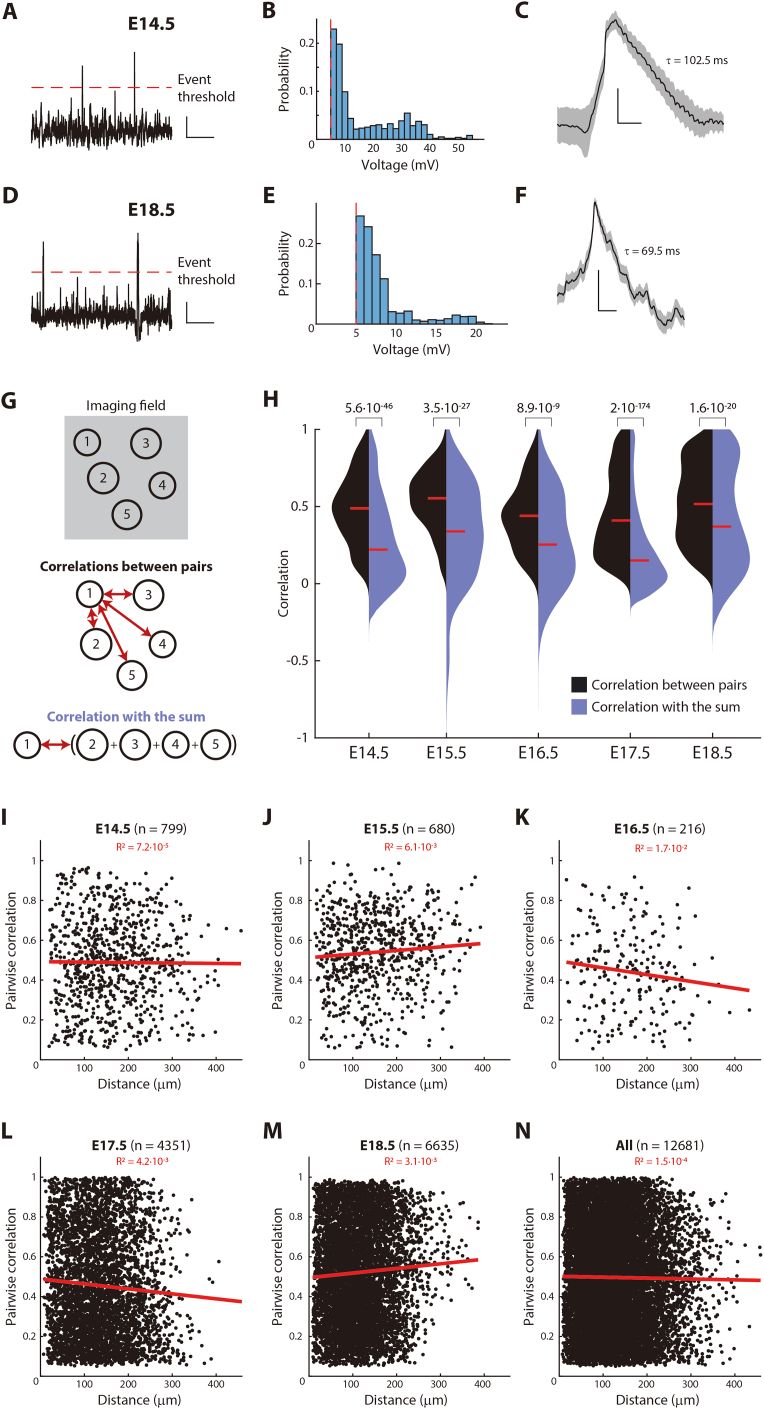
Rbp4-Cre neurons display spontaneous excitatory synaptic potentials, and correlated activity which does not extend across the population, related to Figure 6
(A–F) Spontaneous excitatory synaptic potentials are found in both active phases of embryonic development, at E14.5 (A – C) and E18.5 (D – F).
(A and D) Traces recorded in current clamp mode during in vivo two-photon targeted patch clamp recordings from Rbp4-Cre neurons, following the application of TTX to the surface of cortex. Synaptic potentials were defined as deflections greater than 5 mV (dotted line: event threshold).
(B and E) Distribution of amplitudes greater than the event threshold.
(C and F) Average across events, normalized to the peak voltage of each event, showing the characteristic shape of a synaptic potential. Decay time (τ) shown in ms. Black line: mean; gray shading: SEM.
(G and H) Population activity across Rbp4-Cre neurons is less correlated than activity across pairs of Rbp4-Cre neurons. (G) Schematic comparing the “correlation between pairs” of neurons (black), and “correlation with the sum” of neuronal activity (blue) within all neurons recorded in an imaging field (top). Pairwise correlations of neuron 1 with each other neuron among all imaging field (middle). Correlation of the activity of neuron 1 with the sum of activity in all other neurons in the imaging window (bottom).
(H) From E14.5 to E18.5, the distribution of correlations between pairs of neurons (that are significantly greater than random) (black shaded area) is compared to the distribution of correlations with the sum (for all recorded neurons) (blue shaded area). Probability: Wilcoxon rank-sum test. Correlations computed from recordings in 9 (E14.5), 5 (E15.5), 4 (E16.5), 5 (E17.5), and 6 (E18.5) embryos.
(I–M) Pairwise correlations of spontaneous calcium activity, that are significantly greater than random, between Rbp4-Cre neurons, on each embryonic day from E14.5 to E18.5, do not decrease with distance.
(N) Combining all pairwise correlations, significantly greater than random, across all embryonic days.
(I–N) Red line: best fit trend of correlations across distance (red: R2 quality of fit); n = number of pairs from recordings in 9 (E14.5), 5 (E15.5), 4 (E16.5), 5 (E17.5), and 6 (E18.5) embryos.
Scale bars: 5s and 2.5 mV (A), 50 ms and 25% of peak (C), 5s and 2.5 mV (D), 50 ms and 25% of peak (F).
Activity in Rbp4-Cre neurons is correlated
Communication between neurons can lead to correlated activity. From E14.5 to E18.5, we observed many pairs of Rbp4-Cre neurons with correlations significantly greater than expected at random (Figure 6C). This correlated activity did not extend across the whole population, either through synchronous activity98 or through propagating waves of activity (Figure S7), but was apparent in pairs of Rbp4-Cre neurons. At E14.5, both superficial and deep layers had active neurons (Figure S5). Further, Rbp4-Cre neurons extended axons and dendrites both within and between the layers (Figure 6D). Hence, we asked if neurons communicate only within their own layer, or across layers. Using a 3D acousto-optic two-photon microscope, we recorded simultaneously from neurons in both layers (Figure 6E). Pairs of neurons were not only significantly correlated within the same layer but also across layers, in both cases showing correlation coefficients as high as 0.9 (Figure 6F). Taken together, the significant pairwise correlations suggest that, from E14.5 onwards, Rbp4-Cre neurons communicate with each other.
Perturbing autism-associated genes in Rbp4-Cre neurons changes circuit organization and activity
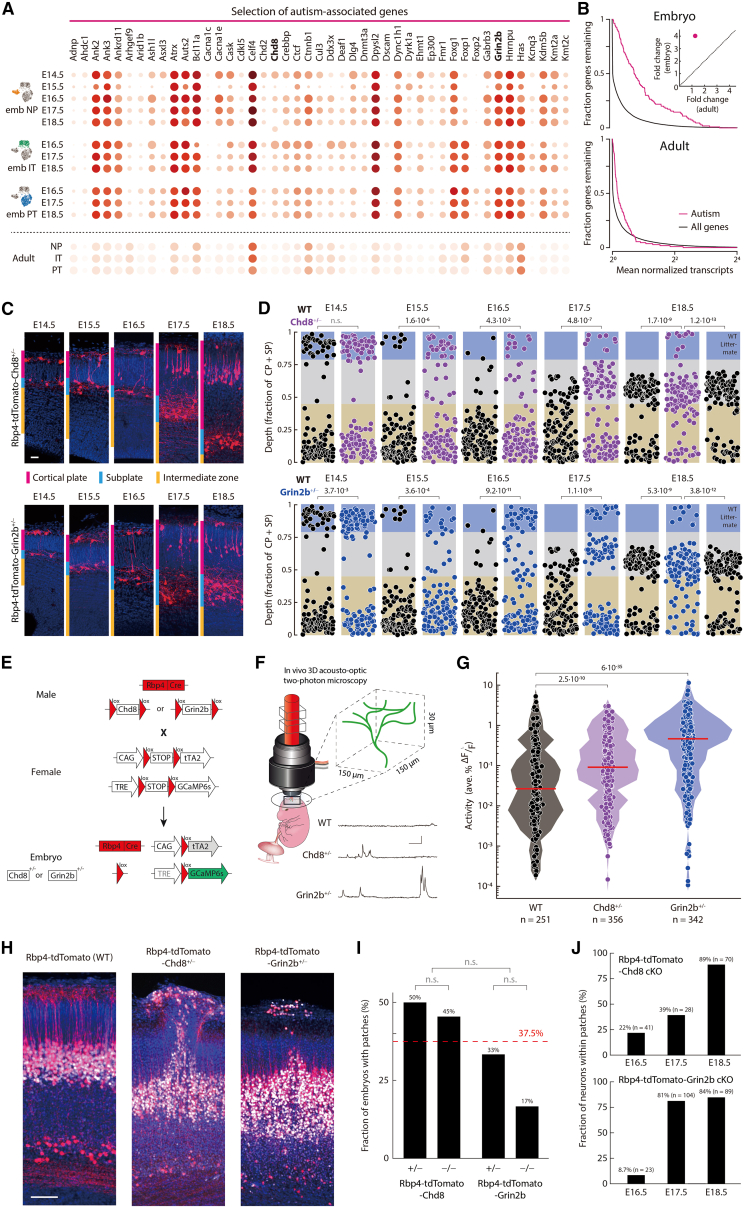
Because neurodevelopmental disorders have been associated with cortical circuit dysfunction,16,17,26,54,99,100,101,102,103 we analyzed the expression of neurodevelopmental disease-associated genes in Rbp4-Cre neurons between E14.5 and E18.5 and compared this with expression in adult L5-PNs.8 A number of genes associated with schizophrenia104,105,106 and autism spectrum disorder107 were expressed across all three types of Rbp4-Cre neurons (Figure 7A; Data S2). Gene expression associated with both neurodevelopmental diseases was significantly higher than the average expression across all genes, with schizophrenia-associated genes showing about 3-fold higher expression and autism-associated genes showing 4-fold higher expression (Figure 7B; Data S2). Importantly, while schizophrenia-associated genes were expressed similarly, autism-associated genes showed a 4-fold increase in expression in Rbp4-Cre neurons compared to adult L5-PNs.
Figure 7.
Perturbing autism-associated genes selectively in Rbp4-Cre neurons disrupts circuit organization and activity during embryonic development
(A) Expression (circles) of selected genes (Data S2) associated with autism spectrum disorder107 in the three Rbp4-Cre neuron types and adult L5-PN types.8 Radius of circles: fraction of cells expressing the gene; color of circles: mean normalized transcripts per cell (log2).
(B) Fraction of genes with a mean transcript count greater than the number of transcripts shown on the x axis, for all genes (black), and genes associated with autism spectrum disorder (magenta) in Rbp4-Cre neurons (top) and adult L5-PNs (bottom). Inset: Fold change of autism-associated gene expression compared to all genes in embryos and adult.
(C) Immunostaining of cortex of Rbp4-tdTomato-Chd8+/− (top) and Rbp4-tdTomato-Grin2b+/− (bottom) mice (Figure S8). Rbp4-Cre neurons (red), Bcl11b, (white), Hoechst (blue).
(D) Normalized depths of Rbp4-Cre neurons (as in Figure 2A) in Rbp4-tdTomato (WT) and the two mutant (top, Chd8+/−; bottom, Grin2b+/−) embryos (Figure S9). 125 neurons from each mouse line, sampled at random. χ2 test.
(E) Mating strategy to generate Rbp4-GCaMP6s-tTA2-Chd8+/− (Chd8+/−) and Rbp4-GCaMP6s-tTA2-Grin2b+/− (Grin2b+/−) embryos.
(F) Example recordings from Rbp4-Cre neurons’ dendrites in Rbp4-GCaMP6s-tTA2 (WT), Grin2b+/−, and Chd8+/− embryos at E16.5 using 3D acousto-optic two-photon microscope.
(G) Distribution of activity in E16.5 embryos, shown in log-scale, for WT and two mutant genotypes. Circles: activity of each neurite; red line: median; shading: distribution. Wilcoxon rank-sum test. n = number of neurites.
(H) Immunostaining of local patches of cortical disorganization in Rbp4-tdTomato-Chd8+/− and Rbp4-tdTomato-Grin2b+/− mice, at E18.5. Rbp4-Cre neurons (red), Bcl11b, (white), Hoechst (blue).
(I) Fraction of mutant mice, of each genotype, showing at least one patch, summed across E16.5 to E18.5. Red line: Average across all four genotypes. Data from 14 (Rbp4-tdTomato-Chd8+/−), 11 (Rbp4-tdTomato-Chd8−/−), 9 (Rbp4-tdTomato-Grin2b+/−), and 6 (Rbp4-tdTomato-Grin2b−/−) embryos. Fisher’s exact test (p = 0.05, prior to Bonferroni correction).
(J) Fraction of neurons within the superficial layer that are located within patches of disorganization, in embryos with at least one patch. n = number of superficial layer neurons on each embryonic day.
Scale bars: 20 μm (C), 25s and 25 %ΔF/F (F), 50 μm (H).
See also Figures S8 and S9.
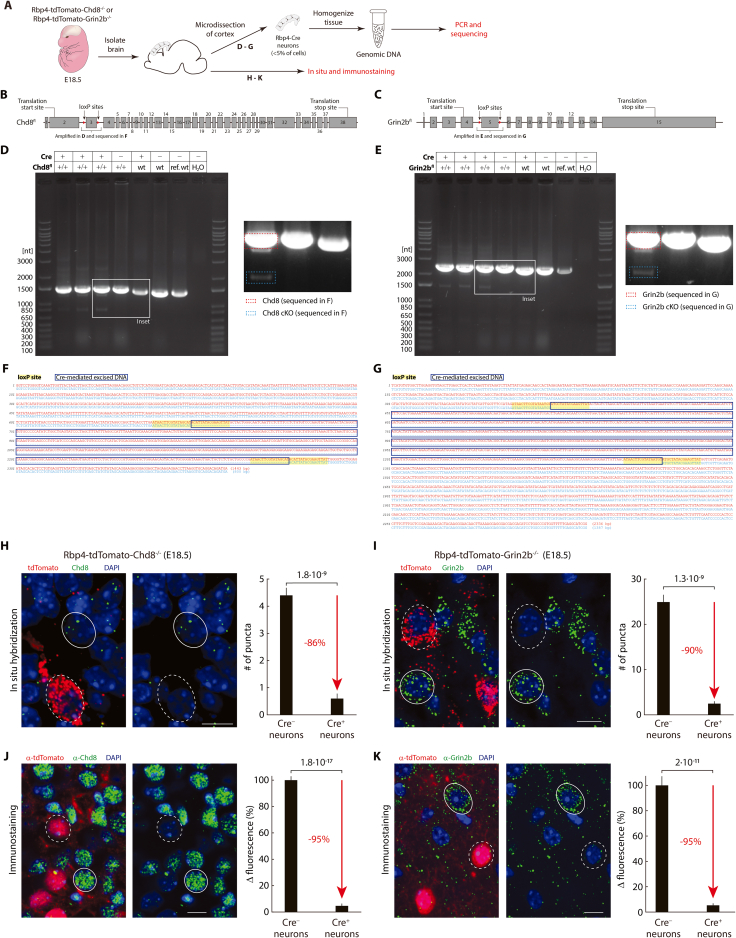
Therefore, we perturbed two known autism-associated genes, Chd8 and Grin2b,107,108,109 specifically in Rbp4-Cre neurons and investigated their effect on the spatial organization of these neurons during embryonic development. Since autism can be associated with both heterozygous and homozygous mutations,110,111,112,113,114 we bred both heterozygous and homozygous knockout mice for the gene Chd8 (Rbp4-tdTomato-Chd8+/− and Rbp4-tdTomato-Chd8−/− mice) or Grin2b (Rbp4-tdTomato-Grin2b+/− and Rbp4-tdTomato-Grin2b−/− mice) and confirmed the loss of RNA and protein expression in homozygous knockouts (Figure S8). Control mice had tdTomato in Rbp4-Cre neurons but were wild type for Chd8 and Grin2b.
Figure S8.
Crossing floxed Chd8 and Grin2b mouse lines with Rbp4-Cre results in the conditional knockout of Chd8 and Grin2b in Rbp4-Cre neurons, related to Figure 7
(A) Schematic of strategy to demonstrate Cre-mediated excision and resulting selective knockout of Chd8 and Grin2b in E18.5 Rbp4-tdTomato-Chd8−/− or Rbp4-tdTomato-Grin2b−/− embryos.
(B and C) Schematic diagram of Chd8 (B) and Grin2b (C) transcripts (Chd8: Ensembl 212; Grin2b: Ensembl 202), with approximate location of inserted loxP sites, region amplified in D and E (and sequenced in F and G), translational start sites, and translation end sites. Introns not shown to scale.
(D and E) Regions within Chd8 (D) and Grin2b (E) genes, including the loxP sites, were PCR amplified and electrophoretically separated. Above: genotype of embryo for Cre and either floxed Chd8 or Grin2b; ref. wt: commercial mouse genomic DNA. Inset: Zoom in (with increased contrast) showing one example of each genotype and examples of longer (red) and shorter (blue) bands, with relative intensity in agreement with the presence of a small fraction of Cre-expressing cells (i.e. weaker band of the shorter fragment) and a large fraction of cells not expressing Cre (i.e. stronger band of the longer fragment) in the cortical samples.
(F and G) Aligned sequences of excised bands from D and E. Red: sequence of Chd8 (F) and Grin2b (G) bands from longer fragment (derived from cells not expressing Cre) (yellow overlay: loxP sites); blue: sequence of bands from shorter fragment (derived from cells with Cre expression) (blue box: excised region).
(H and I) Left: In situ hybridization of probes against Chd8 (H) or Grin2b (I) (green) and tdTomato (red: marking Rbp4-Cre neurons) in E18.5 Rbp4-tdTomato-Chd8−/− (H) or Rbp4-tdTomato-Grin2b−/− (I) embryos. Example Cre-positive (dashed circles: tdTomato-expressing) and Cre-negative cells (solid circles: lacking tdTomato expression). Right: Number of puncta per cell in Cre-positive compared to Cre-negative cells (mean ± sem). n = 25 cells of each type.
(J and K) Left: Immunostaining against Chd8 (J) or Grin2b (K) (green) and tdTomato (red: marking Rbp4-Cre neurons) in E18.5 Rbp4-tdTomato-Chd8−/− (J) or Rbp4-tdTomato-Grin2b−/− (K) embryos. Example Cre-positive (dashed circles: tdTomato-expressing) and Cre-negative cells (solid circles: lacking tdTomato expression). Right: Quantification of change in fluorescence between Cre-positive cells compared to Cre-negative cells (mean ± sem). Fluorescence was normalized by mean fluorescence within Cre-negative neurons in each slice. n = 50 cells (J) and 30 cells (K) of each type. (H–K) Probability: Wilcoxon rank-sum test. Scale bars: 10 μm (H–K).
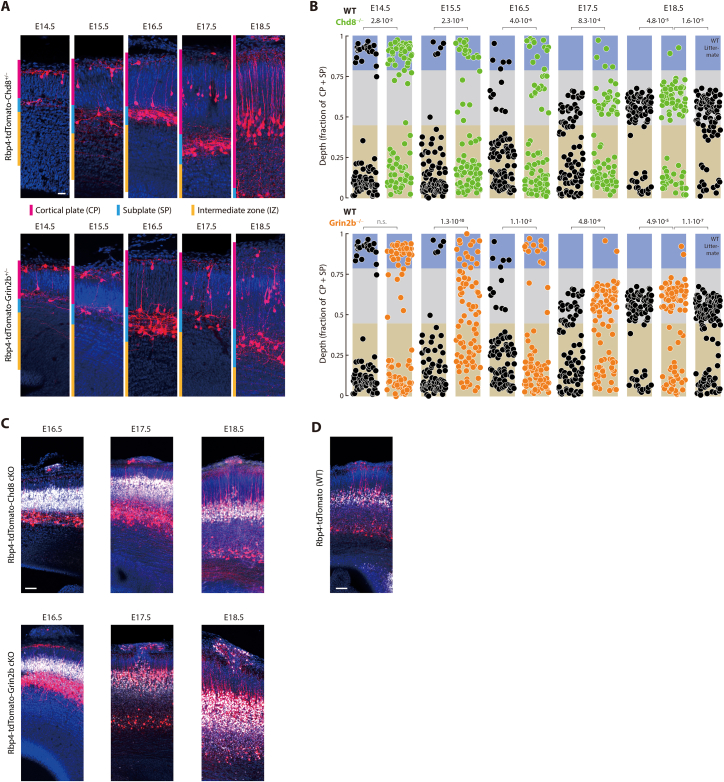
To reveal the spatial distribution of Rbp4-Cre neurons in the four different mutant mouse lines and control, we collected brain sections from E14.5 to E18.5 and stained them with a tdTomato antibody (Figures 7C and S9). Remarkably, in all four mutant mouse lines but not in the control, we observed Rbp4-Cre neurons near the surface of the neocortex until E18.5. The number of superficial layer neurons from E15.5 onwards on each embryonic day was significantly higher in all four conditional mouse lines compared to control (Figures 7D and S9). Therefore, a common effect of perturbing two different and unrelated autism-associated genes in Rbp4-Cre neurons was the abolishment of the transience of the superficial layer. In control mice, the deep-and-superficial layer configuration persisted only until E15.5. Following that day, the circuit switched to a deep-and-intermediate layer configuration. In contrast, in all four mutant mice, the superficial layer persisted and therefore, following E15.5, Rbp4-Cre neurons were found in all three layers.
Figure S9.
Perturbing autism-associated genes selectively in Rbp4-Cre neurons disrupts organization of layer 5 during embryonic development, related to Figure 7
(A) Rbp4-Cre neurons (stained using tdTomato antibody, red) in Rbp4-tdTomato-Chd8−/− (top) and Rbp4-tdTomato-Grin2b−/− (blue) mice, from E14.5 to E18.5, within the cortical plate (magenta), subplate (cyan), and intermediate zone (yellow), counterstained with Hoechst (blue).
(B) Distribution of Rbp4-Cre neuronal locations as a fraction of the cortical plate and subplate thickness, from E14.5 to E18.5, in control (WT, black), Rbp4-tdTomato-Chd8−/− (top, green), and Rbp4-tdTomato-Grin2b−/− (bottom, orange) mice. 85 neurons from each mouse line, sampled at random, displayed on each embryonic day. Layer boundaries derived from Figure 2A (blue: superficial layer; gray: intermediate layer; beige: deep layer). Probability: χ2 test comparing the fraction of Rbp4-Cre neurons in each layer between the conditional knockout mouse and control mice, on each embryonic day; p = 0.05.
(C) Local patches of disorganization in Rbp4-tdTomato-Chd8 and Rbp4-tdTomato-Grin2b conditional knockout (cKO) mice (examples from each embryonic day from E16.5 to E18.5) show Rbp4-Cre neurons (stained using tdTomato antibody, red) at the surface and disrupted intermediate layer, including both neurons expressing Cre (red) and not expressing Cre (stained using Bcl11b antibody, white), counterstained with Hoechst (blue).
(D) Superficial layer Rbp4-Cre neurons in E18.5 control mouse, without disorganization of the underlying intermediate layer. Scale bar: Scale bar: 20 μm (A, C, D).
We then asked if there was a change in the activity of Rbp4-Cre neurons in the Chd8 or Grin2b knockout mice. We bred heterozygous Rbp4-GCaMP6s-tTA2-Chd8+/− and Rbp4-GCaMP6s-tTA2-Grin2b+/− mice together with littermate controls, Rbp4-GCaMP6s-tTA2-Chd8+/+ and Rbp4-GCaMP6s-tTA2-Grin2b+/+ mice, and imaged Rbp4-Cre neurons during the transition phase at E16.5 (Figure 7E). We recorded activity within the superficial layer, including both somas and apical dendrites of Rbp4-Cre neurons. Here, we again used a 3D acousto-optic two-photon microscope (Figure 7F). Note that due to differences in the scan rate, dwell time, and laser power, the amplitude of activity recorded in this imaging mode was lower and not directly comparable to that recorded earlier (Figures 3C–3F). By comparison to littermate controls, there was a significant increase in the activity of Rbp4-Cre neurons, in both Chd8 and Grin2b heterozygous knockout mice (Figure 7G). Therefore, the monoallelic deletion of autism-associated genes in Rbp4-Cre neurons interferes with the transition phase of the biphasic activity during embryonic development.
In addition to the presence of Rbp4-Cre neurons in the superficial layer after E15.5 and their increased activity at E16.5, in a subset of mutant animals, we also observed the presence of non-uniform patches of disorganized structure involving Rbp4-Cre neurons. These patches were characterized by a cluster of closely spaced Rbp4-Cre neurons at the surface of the neocortex together with a distortion of the intermediate layer directly under the cluster of surface neurons, such that the intermediate layer extended closer to the neurons at the surface. We found disorganized patches in all four mutant mouse lines, on all embryonic days, from E16.5 onwards (Figures 7H and S9). In control mice, we occasionally observed several Rbp4-Cre neurons at the surface of cortex (3/31 embryos), but no associated disorganization of the underlying intermediate layer. Averaging across all four lines, 37.5% of embryos showed patches of disorganization within cortex from E16.5 onwards (Figure 7I). Further, within the subset of mice that showed disorganized patches at E18.5, greater than 80% of neurons at the surface of cortex were located within patches, whereas at E16.5, less than 20% of neurons were located within patches (Figure 7J). Notably, this form of local patches of disorganization is reminiscent of patches of disorganization observed in the cortices of children with autism.54
Discussion
When and how the first active PN circuits form and whether these circuits are perturbed in neurodevelopmental disorders are central questions in understanding both neocortical development and the etiology of neurodevelopmental disorders.115,116,117,118,119,120 Since L5-PNs are born early in embryonic development and have the highest degree of recurrent connectivity in the adult, we argued that these can form one of the earliest PN-to-PN circuits. Using mouse genetics and confirmed by single cell transcriptomics, we found that cortical neurons expressing Cre in Rbp4-Cre mice were transcriptionally closest to L5-PNs. By investigating the spatial distribution and in vivo activity of this population of neurons during the embryonic development of neocortex in wild-type and autism-associated gene mutant mice, we made two sets of observations. The first set relates to the question of when and how the first active PN circuits develop; the second set relates to the question of how mutations in autism-associated genes affect the formation of embryonic PN circuits.
Pyramidal neuron circuit formation
First, we found that cortical PNs form active, multi-layered, transient circuits at the inception of the neocortex. The current view on PN development is that they migrate into the cortical plate layer-by-layer, in an inside-out configuration.22,81 We showed that Rbp4-Cre neurons form into active, transient two-layered circuits that span the neocortex from subplate to marginal zone, prior to their migration to form layer 5 in the cortical plate. The evidence for active circuits includes the presence of chemical synapses, with recurrent synapses between Rbp4-Cre neurons; the TTX-sensitive active conductances; the sensitivity of activity to TTX; the neurons’ sensitivity to AMPA and NMDA; the recorded spontaneous excitatory postsynaptic potentials; and the correlated activity within and across the two layers. The evidence that these circuits are transient includes the apoptosis of Rbp4-Cre neurons in the superficial layer and the migration of Rbp4-Cre neurons from the deep layer to form layer 5. The existence of a multi-layered PN circuit at E14.5 suggests a new, early developmental step in the formation of neocortex.
The spatial organization of the transient PN-to-PN circuit at E14.5 is reminiscent of previously identified subplate to Cajal-Retzius circuits.77,121,122,123,124 However, based on several lines of evidence—single-cell transcriptomic identity of Rbp4-Cre neurons, in situ hybridization of marker mRNA, and antibody staining of marker proteins—the neurons described in this study had neither subplate nor Cajal-Retzius cell identity. Rather, at E14.5, the Rbp4-Cre neurons were closest to NP type L5-PNs. Additionally, previous work has found that PNs participate in transient synaptic circuits with subplate neurons during their migration to their final location in neocortex31 and that the organization of cortical circuits is instructed through subcortical inputs after E16.5.35,45 However, transient circuits between embryonic PNs have not been previously described. We showed such circuits prior to the thalamic innervation of the cortical plate.45,125,126
Second, we found that activity in cortical PNs can show complex patterns during embryonic development. Instead of a monotonic increase in activity across development as has been previously described,36,37,123 Rbp4-Cre neurons showed a biphasic activity pattern: two phases of increased activity, separated by a transition phase with decreased activity. The first phase of increased activity occurred at E14.5, at a time when no active PN circuits have been previously described. The second phase of increased activity occurred from E17.5 to E18.5, when we detected increased activity within dendrites compared to the somas, potentially reflecting inputs driven, directly or indirectly, by the incoming axons of thalamic principal neurons.35,45,46
Further, we showed that perturbing embryonic cortical activity in postmitotic developing PNs can fine-tune their laminar position within cortex. It has previously been shown that hyperpolarizing cortical neuronal progenitor cells changes the laminar position of their postmitotic daughter cells,127,128 while hyperpolarizing L2/3-PNs postmitotically did not result in a change in laminar position.127 In contrast, our results show that hyperpolarizing Rbp4-Cre neurons postmitotically shifted their laminar positions toward the surface of the brain. Therefore, the effect of activity perturbations on laminar position in postmitotic PNs is cell type specific.
Third, we found that the composition of PN types within a cortical layer is tightly regulated during embryonic development. Whereas types of PNs within the same layer have been shown to have distinct functional roles in the adult, their establishment during embryonic development has not been well understood. We showed that Rbp4-Cre neurons divide into three distinct types (embryonic-NP, IT, and PT), each of which is closest to a single adult L5-PN type. The two-layered, transient Rbp4-Cre neuron circuit was entirely composed of embryonic NP neurons, while embryonic-IT and PT neurons are only incorporated into circuits later.
Altogether, our findings reveal a switch in pyramidal circuits during early development: Rbp4-Cre neurons show a switch in activity, spatial organization, and cell type identity, all of which are coordinated in time (Figure 3J).
Effect of autism-associated gene mutations on pyramidal circuit formation
Neurodevelopmental disorders, such as autism spectrum disorder, have been associated with defects in cortical circuits.16,17,26,54,99,100,101,102,103,129,130 Notably, a recent genetic study of autistic patients suggested that, during development, autism-associated genes are strongly enriched in maturing excitatory neurons, compared to immature progenitors.131 Further, in human cortical organoids, mutations in autism-associated genes perturb the maturation of lower layer PNs.120 However, how and when mutant autism associated genes affect embryonic circuit development and activity in vivo are not well understood. Using perturbations of two different autism-associated genes selectively in Rbp4-Cre neurons, we made two observations.
First, we found that these genetic manipulations interfere with the transition phase of the biphasic PN activity pattern during embryonic development. We recorded an increased activity in mutant Rbp4-Cre neurons at E16.5, relative to activity in wild-type conditions.
Second, we found that the genetic manipulations resulted in the perturbation of the multi-layered transient circuits. Superficial layer Rbp4-Cre neurons persisted through embryonic development until E18.5, one day prior to birth. Further, an increasing fraction of these superficial layer neurons clustered together, at the surface, as the embryos developed. These clusters were associated with a disorganization of the developing neocortex directly under each cluster, such that Rbp4-Cre neurons were displaced toward the surface. Intriguingly, this disorganization also involved neurons that did not express Cre, suggesting communication between the Cre-expressing subset of neurons and other L5-PNs. Further, the patchy disorganization of mouse cortex that we report is reminiscent of the patchy disorganization of cortical tissue observed in children with autism.54,56
Our work identifying embryonic PN-to-PN active circuit motifs, together with the in vivo imaging and recording methods that we developed, provide an opportunity to study the effects of genes associated with neurodevelopmental disorders, and in particular with autism spectrum disorder, on identified circuits in the living embryo.
Cortical development in mice and humans
Our experiments were performed in mice. However, the cytoarchitecture of the six layered neocortex132 as well as the major cortical cell types are conserved between mice and human, with changes primarily in the relative proportions of each cell type.133,134 The cortex is thinner in mice compared to humans. The expansion in humans is most pronounced in the upper layer neurons, with the upper layer constituting only a fifth of the cortical plate thickness in adult mice, compared to more than half the cortical plate thickness in adult humans.135,136 The increase is thought to result from an increase in the proliferation of precursor cells during the development of the human neocortex.136,137 The timelines of mouse and human development also differ. Mouse gestation is 18.5 days, while human gestation is 280 days. This difference is due not only to mouse development being faster than human development, but also because mice are born earlier in development, compared to humans, at a stage equivalent to gestational day (GD) 116 in humans, i.e. less than half way through the normal human gestational period.138,139 In mice, most cortical neurons are generated between E11.5 and E17.5. In humans, the proliferation of cortical precursors commences around GD 35 (E10.5 days in mice) and extends beyond GD 14027 (corresponding to the early postnatal period in mice). Further, during mouse embryonic development, the subplate is thinner than the cortical plate, but in human development, the subplate is thicker than the cortical plate.124,140 The inception of the human neocortex occurs around GD 45. The E14.5 to E18.5 period that we studied in mouse development corresponds to approximately GD 64 to GD 105 in human development. The first synaptic structures have been described in human cortex in the first trimester, around GD 40141 and the first active membrane properties after GD 112,142 around the time of birth (E18.5) in mice. Spontaneous activity in humans occur after GD 140, equivalent to postnatal development in the mouse.143 Given these differences in timing,139 relating the development of active PN-to-PN circuits in different mammalian species to a common reference point is useful and, in this paper, we used the inception of neocortex as this reference point.
Limitations of the study
In this study, we focused on the embryonic development of Rbp4-Cre neurons. Using genetically identified neurons, we could not be certain of the lineage of these neurons. Instead, we could only identify gene expression profiles of the labeled cells and measure the similarity of the genetically defined population across time. Additionally, because a proportion of Rbp4-Cre neurons in all layers were born on the same day, we were unable to specifically target Rbp4-Cre neurons within a single layer via electroporation. Further, we were restricted in the depth of our recordings, by the physical limitations of two-photon microscopy. Hence, at E18.5, a time when the cortex was thicker, we were not able to record the activity of neurons within the deep layer. Finally, while previous work has argued that the transcriptome can be used to identify the cell type of a given neuron,7,8,66 it is a legitimate concern how to put this in the context of neurons previously classified by their morphology, birth date, or spatial location. Given this, we have referred to our cell population as Rbp4-Cre neurons and we associate them with L5-PN identity only with respect to their transcriptomic correlation with adult L5-PNs.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-GFP | Invitrogen | Cat# A-11122; RRID: AB_221569 |
| Rat monoclonal anti-GFP | Nacalai | Cat# 04404-84; RRID: AB_10013361 |
| Goat polyclonal anti Nurr1/NGFI-Bbeta/NR4A2 | R&D Systems | Cat# AF2156; RRID: AB_2153894 |
| Rat monoclonal anti CTIP2/BCL11B | Merck | Cat# MABE1045 |
| Rabbit anti-PSD95 | Abcam | Cat# ab269863 |
| Goat polyclonal anti-snap25 | Antibody Online | Cat# ABIN1742235 |
| Chicken polyclonal anti-Map2 | Encor Biotechnology | Cat# CPCA-MAP2; RRID: AB_2138173 |
| Mouse monoclonal anti-Neurofilament H | BioLegend | Cat# 801602; RRID: AB_2715851 |
| Rabbit anti-cleaved caspase-3 | Abcam | Cat# ab2302; RRID: AB_302962 |
| Rabbit polyclonal RFP antibody (anti-tdTomato) | Rockland | Cat# 600-401-379; RRID: AB_2209751 |
| Rabbit polyclonal anti-GFP | Chemicon | Cat# AB3080; RRID: AB_91337 |
| Rat monoclonal anti-BrdU | Abcam | Cat# ab6236 |
| Rabbit polyclonal anti-Chd8 | Bethyl | Cat# a301-225a; RRID: AB_890577 |
| Rabbit polyclonal anti-NMDAR2B | Invitrogen | Cat# 71-8600; RRID: AB_2534001 |
| Rabbit polyclonal anti-Satb2 | Abcam | Cat# ab34735; RRID: AB_2301417 |
| Rabbit polyclonal anti-Tbr2 | Abcam | Cat# ab23345; RRID: AB_778267 |
| Rabbit polyclonal anti-Tbr1 | Abcam | Cat# ab31940; RRID: AB_2200219 |
| Rabbit polyclonal anti-Pax6 | Millipore | Cat# AB2237; RRID: AB_1587367 |
| Alexa Fluor 488 donkey anti-rabbit IgG (H + L) | ThermoFisher | Cat# A-21206; RRID: AB_2535792 |
| Alexa Fluor 633 donkey anti-goat IgG (H + L) | ThermoFisher | Cat# A-21082; RRID: AB_2535739 |
| Cy3-AffiniPure donkey anti-rat IgG (H + L) | Jackson ImmunoResearch Laboratories | Cat# 712-165-153; RRID: AB_2340667 |
| Biotin-SP-affinipure goat anti-rabbit IgG, F(ab’)2 | Jackson Immuno Research Laboratories | Cat# 111-065-047; RRID: AB_2337964 |
| Bacterial and virus strains | ||
| PHP.eB AAV-CAG-FLEX-GCaMP7s | Bharioke et al., 202298 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Fentanyl | Janssen | N/A |
| Medetomidine | Virbac AG | Cat#QN05CM91 |
| Midazolam | Sintetica | N/A |
| Isoflurane (Attane) | Provet AG | Cat#QN01AB06 |
| Opal 520 dye | Akoya Bio | Cat#FP1487001KT |
| Opal 570 dye | Akoya Bio | Cat#FP1488001KT |
| Opal 690 dye | Akoya Bio | Cat#FP1497001KT |
| AMPA | Tocris | Cat#0254 |
| NMDA | Tocris | Cat#0114 |
| TTX | Tocris | Cat#1069 |
| 0.2% glutaraldehyde | Electron Microscopy Sciences | Cat#16200 |
| 1.0% osmium tetroxide | Electron Microscopy Sciences | Cat#19110 |
| 3,3-diaminobenzidine (DAB) | Sigma-Aldrich | Cat#D800′ |
| BrdU (5-bromo-2′-deoxyuridine), Thymidine analog | Abcam | Cat#ab142567 |
| Critical commercial assays | ||
| RNAscope Multiplex Fluorescent Reagent Kit v2 Assay | ACD Bio-techne | Cat#323100 |
| Chromium Gene Expression Kit | 10x Genomics | v3.1 |
| Vectastain Elite ABC kit | Vector Laboratories | Cat#PK-6100 |
| AllPrep DNA/RNA Micro Kit | Qiagen | Cat#1053393 |
| GoTaq® Long PCR Master Mix | Promega | Cat#M4021 |
| NucleoSpin® Gel and PCR Clean-Up Kit | Mackerey-Nagel | Cat#740609.250 |
| Deposited data | ||
| Raw single cell sequencing data | This paper, deposited at NCBI Sequence Read Archive (SRA) | SRA: PRJNA924256 |
| Processed single cell sequencing count tables | This paper, deposited at Mendeley Data | https://doi.org/10.17632/6cw35fj7z4.1 |
| Experimental models: Organisms/strains | ||
| Rbp4-Cre KL100 | MMRRC | MMRRC: 031125-UCD |
| Ai94(TITL-GCaMP6s) | Jackson | RRID: IMSR_JAX:024104 |
| CAG-stop-tTA2 | Jackson | RRID: IMSR_JAX:014092 |
| Chd8em3Lutzy/J | Jackson | RRID: IMSR_JAX:031555 |
| Grin2bem5Lutzy/J | Jackson | RRID: IMSR_JAX:032664 |
| Ai9(RCL-tdT) | Jackson | RRID: IMSR_JAX:007909 |
| Oligonucleotides | ||
| RNAscope 3-plex Positive Control Probe-Mm | ACD Bio-techne | Cat#320881 |
| RNAscope 3-Plex Negative Control Probe | ACD Bio-techne | Cat#320871 |
| Mm-GCamP6s-01 | ACD Bio-techne | Cat#557091 |
| Mm-Lhx2 | ACD Bio-techne | Cat#485791 |
| Mm-Pou3f3 | ACD Bio-techne | Cat#441521 |
| Mm-Reln | ACD Bio-techne | Cat#405981 |
| Mm-Nxph4 | ACD Bio-techne | Cat#489641 |
| Mm-Crym | ACD Bio-techne | Cat#466131 |
| Mm-Nfe2I3 | ACD Bio-techne | Cat#486201 |
| Mm-Fam19a2 | ACD Bio-techne | Cat#452631 |
| Mm-Cdh8 | ACD Bio-techne | Cat#485461 |
| Mm-Zcchc12 | ACD Bio-techne | Cat#524441 |
| tdTomato | ACD Bio-techne | Cat#317041 |
| Mm-Chd8-E3 | ACD Bio-techne | Cat#457771 |
| Mm-Grin2b-O1 | ACD Bio-techne | Cat#1223851 |
| Chd8 Forward: GGTCCTGGGGTCAAATTGGT | Microsynth | N/A |
| Chd8 Reverse: TCATCTGTCCTGCACCTTAAGG | Microsynth | N/A |
| Grin2b Forward: TCATGTGTGGCTTGGAGGTG | Microsynth | N/A |
| Grin2b Reverse: CCGATGCCTCAAAAACCACG | Microsynth | N/A |
| Recombinant DNA | ||
| pGP-AAV-CAG-FLEX-jGCaMP7s-WPRE | Dana et al., 2019144 | Addgene # 104495 |
| CAG-FLEX-Kir2.1-T2A-tdTomato | This paper | N/A |
| CAG-FLEX-Kir2.1Mut-T2A-TdTomato | This paper | N/A |
| Software and algorithms | ||
| NoRMCorre | Pnevmatikakis and Giovannucci, 2017145 | https://github.com/flatironinstitute/NoRMCorre |
| MATLAB R2020a | Mathworks | RRID: SCR_001622 |
| Custom code (MATLAB) to analyze data (not including single cell RNA sequencing) and Custom code (Python) to analyze single cell RNA sequencing data | This paper | https://doi.org/10.5281/zenodo.7689306 |
| Other | ||
| Chromium Single Cell Instrument | 10x Genomics | N/A |
| FACSAria Fusion Cell Sorter | BD Biosciences | N/A |
| Ultramicrotome | Leica | EM UC7 |
| Tecnai Spirit Transmission Electron Microscope | FEI | N/A |
| Galvo-galvo scanning two-photon microscope | Femtonics | N/A |
| FemtoSMART resonant-galvo scanning microscope | Femtonics | N/A |
| Temperature controller | CWE | Cat#TC1000 |
| Acousto-optic microscope | Femtonics | N/A |
| Epifluorescence microscope | Olympus | SZX16 |
| ORCA-Flash4.0 V3 CMOS camera | Hamamatsu | N/A |
| Datex Ohmeda Isotec 5 continuous flow vaporizer | Groppler Medizintechnik | N/A |
| MouseSTAT Jr with Paw Sensor (Pulse oximeter) | Kent Scientific | N/A |
| Stereotactic micromanipulator | Narishige | SM-15R |
| Microinjector | Narishige | IM-9B |
| Multiclamp amplifier | Molecular Devices | 700B |
| IXplore Spin Microscope | Olympus | N/A |
| FV3000-BX63L upright confocal laser scanning microscope | Olympus | N/A |
| 25× water immersion objective | Nikon | APO LWD 25X/1.10W |
| P-97 micropipette puller | Sutter Instrument Company | N/A |
| Nano-injector | WPI | Nanoliter 2020 |
| Electroporator | BTX Apparatus | ECM 830 |
| IR camera | Imaging source | DMK 22BUC03 |
| Mastercycler X50s thermal cycler | Eppendorf | N/A |
| iBright FL1000 imaging system | Thermo Fisher | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Botond Roska (botond.roska@iob.ch).
Materials availability
All unique reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Animals
Animal experiments were performed in accordance with standard ethical guidelines (European Communities Guidelines on the Care and Use of Laboratory Animals, 86/609/EEC) and were approved by the Veterinary Department of the Canton of Basel-Stadt. The following transgenic mouse lines were used: Rbp4-Cre KL100,62,63 Ai94(TITL-GCaMP6s)65 crossed with CAG-stop-tTA285 (GCaMP6s x tTA2), Chd8em3Lutzy/J, Grin2bem5Lutzy/J and Ai9(RCL-tdT).64 All mice were of C57BL/6 background, 55–120 days old, and maintained on a normal 12-h light/dark cycle in a pathogen-free environment with ad libitum access to food and drinking water. Embryos were randomly assigned to all experiments, thereby sampling across both male and female embryos. For adult mouse imaging, both male and female mice were used.
Method details
Timed pregnancies
To determine embryonic age, we bred GCaMP6ski/ki x tTA2ki/ki or Ai9ki/ki males to Rbp4-Cretg/tg females on a strict schedule where males were placed in the females’ cage in the evening and plugs were checked the following morning. Pregnant females (dams) were monitored, and pregnancy was confirmed visually 10 days after the detection of the plug. Experiments were then performed from E13.5 - E18.5.
For generating mice that are heterozygote and homozygote conditional knockouts for Chd8 or Grin2b as well as littermate controls, we bred, conditional Chd8+/− x Rbp4tg/- mice to conditional Chd8+/− x Ai9ki/ki mice and conditional Grin2b+/− x Rbp4tg/- mice to conditional Grin2b+/− x Ai9ki/ki mice. In addition, we also bred, conditional Chd8+/− x Rbp4tg/- mice to conditional Chd8−/− x Ai9ki/ki mice and conditional Grin2b+/− x Rbp4tg/- to conditional Grin2b−/− x Ai9ki/ki mice to increase the number of embryos homozygous for a conditional knockout of Chd8 or Grin2b. To control for germline mutations,146 we also used heterozygous embryos from crosses between conditional Chd8−/− x Ai9ki/ki mice or conditional Grin2b−/− x Ai9ki/ki mice to Rbp4tg/tg mice.
To record activity from Rbp4-Cre neurons in the context of heterozygous conditional knockouts for Chd8 and Grin2b, as well as littermate controls, we bred conditional Chd8+/− x Rbp4tg/tg males or Grin2b−/− x Rbp4tg/tg males to GCaMP6ski/ki x tTA2ki/ki females, using the same strict schedule used to determine embryonic age, introduced above.
Para-uterine embryonic stabilization
Dams, pregnant with embryos between E13.5 and E18.5, were anesthetized with a mixture of Fentanyl-Medetomidine-Midazolam (Fentanyl (Janssen, 0.05 mg/kg); Medetomidine (Virbac, 0.5 mg/kg); Midazolam (Sintetica, 5 mg/kg)) through the initial surgery to prepare each embryo and the subsequent imaging session. A nasal mask was used to deliver humidified oxygen to the dam, at a flow rate of 0.8 L/min (Respironics EverFlo OPI pump, Phillips). For experiments where recordings were performed under Isoflurane anesthesia, dams were anesthetized via 1.75% Isoflurane (Provet) delivered using a Respironics EverFlo OPI pump, Phillips, UniVet Porta Anesthetic machine (using a Datex Ohmeda Isotec 5 continuous flow vaporizer), Groppler Medizintechnik). The desired mixture of isoflurane with humidified oxygen was generated by the vaporizer and applied to the mouse through a nasal mask (Groppler Medizintechnik), with a flow rate of 0.8 L/min.
Dams were placed on their back on a heating pad (TC1000, CWE), centered on a movable metal plate (MB1515/M, Thorlabs), and the hair on the abdomen was removed. A 1.5 cm long incision was placed along the midline of the abdomen. A heat reflective foil was placed above the abdomen to decrease heat loss. The abdominal skin and wall were retracted using the CD + Labs System (retractor tip blunt (ACD-012); latex elastomer (ACD-011); magnetic fixator (magnet replaced by metal pin to fit plate) (ACD-002) (Southmedic Inc.)). A single uterine horn was isolated within the abdominal cavity, and the uterus surgically opened. A single embryo, within its yolk sac, was then isolated, and the yolk sac opened. The embryo was then placed in a custom 3D-printed holder. Low-melting–point agar at 37–39°C was applied around the embryo to fill the space between the embryo and the holder. A titanium headplate was glued to the embryo holder using UV glue (NOA 68, Norland Optical Adhesive), with the circular opening centered above the head of the embryo. The titanium headplate was tightly screwed to custom metal connectors, which were in turn attached to the metal plate through posts (TR30 V/M, Thorlabs), thereby holding the embryo holder securely in position within the abdominal cavity of the dam, with the embryo’s body mostly immersed. HBSS (14175046, Gibco) warmed to 37°C was used to fill the abdominal cavity of the dam. To improve optical access, the skin above the developing cortex was then removed, prior to imaging, creating an imaging region (Figure S3). In addition, for embryos older than E14.5, the cranial bone was removed. A glass coverslip (10 mm round, 631–0665, VWR (Marienfeld)) was glued to the holder such that the glass covered the embryonic brain, applying minimal pressure on the brain. The metal plate supporting the dam and the immobilized embryo was then transferred to a two-photon microscope for imaging.
In-vivo para-uterine two-photon imaging
Dams were anesthetized and maintained at a surgical depth, as assessed by the lack of spontaneous movement, as well as the absence of a paw withdrawal reflex, in response to a toe pinch. To maintain the anesthesia, top-up doses of FMM anesthesia (10–30% of the initial bolus) were injected into the dam, between recordings, approximately every 30 min. Throughout the experiment, a nasal mask was used to deliver humidified oxygen to the dam, at a flow rate of 0.8 L/min (Respironics EverFlo OPI pump, Phillips). For experiments where recordings were performed under Isoflurane anesthesia, dams were maintained under anesthesia via 1.75% Isoflurane (Provet) delivered using a Respironics EverFlo OPI pump, Phillips, UniVet Porta Anesthetic machine (using a Datex Ohmeda Isotec 5 continuous flow vaporizer), Groppler Medizintechnik). The desired mixture of isoflurane with humidified oxygen was generated by the vaporizer and applied to the mouse through a nasal mask (Groppler Medizintechnik), with a flow rate of 0.8 L/min.
Dams were monitored throughout all recording sessions using an IR camera within the recording setup (DMK 22BUC03, Imaging Source). The dam’s breathing remained rapid and shallow, without gasping, a hallmark of stable anesthesia at surgical depth.147 In addition, a pulse oximeter attached to a sensor applied to the rear paw (MouseSTAT Jr and Paw Sensor, Kent Scientific) was used to monitor the heart rate and blood oxygenation of each mouse during experiments. For all dams, the heart rate was maintained at less than 400 bpm,148 and the blood oxygenation never dropped below 90%, when averaged over a minute.
GCaMP6s expressing neurons were imaged using three different two-photon laser scanning microscopes.149 During two-photon imaging, signal-to-noise ratio decreases with depth, due to scattering within the tissue. Due to imaging being performed within a few days of Cre expression, Rbp4-Cre neurons generally had low levels of GCaMP6s expression, restricting the overall signal-to-noise ratio. Therefore, recording depth was limited. This restriction was particularly severe for deep layer neurons in the second phase of increased activity, as deep layer neurons are found at depths >250 microns, at E18.5, compared to only <100 microns, at E14.5. Therefore, in the second phase of increased activity, we recorded primarily from Rbp4-Cre neurons in the intermediate layer.
The first microscope was a Femtonics galvo-galvo scanning microscope equipped with a 25× water immersion objective (APO LWD 25×/1.10W, Nikon). Imaging w–s performed at 5–10 Hz within the developing embryonic cortex, at 920 nm. Each recording of spontaneous calcium activity, from E13.5 to E18.5, was 10 min in length.
The second microscope used was an acousto-optic (AO) microscope150,151 at 920 nm, also equipped with a 25× water immersion objective (APO LWD 25×/1.10W, Nikon). This microscope was used for simultaneous imaging across a three-dimensional (3D) cortical volume. Multi-cube scanning mode150 (Figures 6E and 6F) as well as a volume scanning mode (high-speed arbitrary frame scan) (Figures 7F and 7G) were used. For multi-cube scanning, 3D imaging regions composed of multiple imaging planes, centered around individual neurons or neurites, were selected manually within the imaged embryonic cortical volume. Each imaging plane was chosen to be approximately twice the size of the selected object in the horizontal dimensions. 5–7 imaging planes, generating a cube, were selected in the vertical dimension, to account for any remaining movement within the imaging volume. Recording speed varied with the number of recorded objects. Each recording was 5 min in length. For high-speed arbitrary frame scan mode, 9–11 planes were selected with a distance of 3–5 microns between each plane. All recordings were 10 min in length.
The third two-photon microscope used was a FemtoSMART resonant-galvo scanning microscope. It was equipped with an Olympus 16× water immersion objective (0.8 NA). This microscope was used for simultaneous imaging and electrophysiology.
The temperature of the embryo was supported at 37.5°C using a temperature controller (TC1000, CWE), with the temperature probe held within the abdominal cavity of the dam. In addition, the objective was heated using an objective heater set at 50°C (OWS-1, Warner Instruments) to further decrease the rate of heat loss from the embryo (Figure S3).
For the in vivo imaging of Rbp4-Cre anatomy, 3D volumes within the cortex of Rbp4-Cre x Ai9 embryos at E16.5 were imaged for 5 h, plane-by-plane, using the galvo-galvo two-photon microscope at 950 nm.
Electrophysiology in combination with in vivo para-uterine imaging
For electrophysiological recordings we used whole-cell patch pipettes, pulled from borosilicate glass with filament (O.D.: 1.5 mm, I.D.: 0.86 mm) using a P-97 micropipette puller (Sutter Instrument Company) and filled with intracellular solution containing: 0.2 mM EGTA, 130 mM K-gluconate, 4 mM KCl, 2 mM NaCl, 10 mM HEPES, 4 mM ATP-Mg, 0.3 mM GTP-Tris, 14 mM phosphocreatine-Tris, 0.050 mM Alexa 488 and brought to pH 7.25 (with dilute NaOH or HCl) and 292 mOsm (by addition of H2O).
Embryos were prepared, as described for para-uterine imaging, above, with the exception of the coverslip which was cut to allow access of the patch pipette, and the opening of the dura. A ground wire was positioned in the immersion solution, near the surface of the brain (cortex buffer: 125 mM NaCl, 5 mM KCl, 10 mM glucose, 10 mM HEPES, 2 mM MgSO4 and 2 mM CaCl2). Signals (sampled at 50 kHz and low pass filtered at 10 kHz) were recorded using a National Instruments Board connected to a Multiclamp 700B amplifier (Molecular Devices).
The whole-cell patch pipette, filled with green fluorescent dye, was then lowered through the opening in the dura under positive pressure (100 mbar). Dye from the pipette filled the extracellular space, the pressure was lowered (25–35 mbar) and the pipette was advanced diagonally. Rbp4-Cre positive neurons were visualized by their expression of tdTomato. Adjacent non-Rbp4-Cre neurons were identified by the absence of red fluorescence, and by the injection of dye into the extracellular space.
The pressure on the pipette was further reduced just before touching the membrane of the target neuron, and finally released to form a gigaohm seal under visual guidance. Slow and fast pipette capacitances were compensated, and whole-cell access was achieved by applying a negative pulse of pressure on the pipette.
Current steps were applied in current clamp mode to the patched cell, and the resulting voltage change was recorded, both in the presence and absence of TTX. Additionally, subthreshold membrane voltage was recorded in current clamp mode following the application of TTX.
In utero electroporation
CAG-FLEX-Kir2.1-T2A-tdTomato and CAG-FLEX-Kir2.1Mut-T2A-TdTomato constructs were generated by amplifying Kir2.1-T2A-tdTomato from pCAG-Kir2.1-T2A-tdTomato (Plasmid #60598, Addgene) and mKir2.1-T2A-tdTomato from pCAG-Kir2.1Mut-T2A-tdTomato (Plasmid #60644, Addgene).93 Using Gibson cloning, the fragments were inserted into pAAV-CAG-Flex-mRuby2-GSG-P2A-GCaMP6s-WPRE-pA backbone (Plasmid #68717, Addgene)152 from which mRuby2-GSG-P2A-GCaMP6s-WPRE was removed.
In utero electroporations were performed in embryos of females with timed pregnancies, on E12.5. The mother was deeply anesthetized with isoflurane and buprenorphine (Temgesic, 0.05 mg/kg) was injected subcutaneously to the mother for analgesia. A 2 cm incision was performed at the abdominal midline to expose the embryos and 0.1 mL of Ritodrine (Sigma-Aldrich, 12 mg/mL) was injected to avoid premature abortion. 500 nL of Kir or mKir DNA (1–3 mg/mL) mixed with 1% Fast Green (Sigma-Aldrich) was injected into the lateral ventricle of the brain with a nano-injector (Nanoliter 2020, WPI). For the electroporation, five square pulses of 35V were delivered through the placenta at 950 ms intervals using a 3 mm tweezertrode driven by an electroporator (BTX Apparatus, ECM 830). Following electroporation, the embryos were place back within the abdomen of the dam, the surgical incision was sutured. Embryos were extracted for immunohistological staining at E18.5, following the protocol described under “immunohistological staining of embryonic tissue” below.
Pharmacological manipulations in combination with in vivo para-uterine imaging
For the application of pharmacological agents during either calcium imaging or electrophysiological recordings (concurrent with imaging), a pipette, pulled from borosilicate glass with filament (O.D.: 1.0 mm, I.D.: 0.5 mm) using a P-97 micropipette puller (Sutter Instrument Company), was used.
Pipette were filled with a pharmacological agent, diluted with cortex buffer (as above). The two agents used were a mixture of AMPA (5 mM) (Cat no. 0254, Tocris) and NMDA (10 mM) (Cat no. 0114, Tocris), and TTX (1 mM) (Cat no. 1069, Tocris). In control applications, only cortex buffer was added to the pipette, without any pharmacological agent.
Embryos were prepared, as described for electrophysiological recordings, above. The cut coverslip allowed access of the pipette to the surface of cortex. The pipette was advanced diagonally until the tip was close to but not touching the surface of the brain, as monitored visually via brightfield imaging through the two-photon microscope.
Calcium imaging and electrophysiology were performed from Rbp4-Cre neurons under the coverslip, within 250 μm of the application site.
The pipette was connected to a syringe via silicone tubing, to allow the application of pressure to the fluid in the pipette. The meniscus of the fluid within the pipette was monitored visually during the application of pressure, to ensure that the fluid was delivered to the embryo (DMK 22BUC03, Imaging Source). In total, 500 nL of fluid was added into the buffer between the coverslip and the brain. In calcium imaging experiments using TTX, we made recordings immediately prior to and following the application of TTX (or cortex buffer, in control experiments) to the surface of cortex. In electrophysiological experiments using TTX, we patched a cell prior to TTX application, and took recording both before and after TTX application. In calcium imaging experiments using AMPA+NMDA, we performed 10-min recordings, and applied AMPA+NMDA (or cortex buffer, in control experiments) to the surface of cortex, 3 min into each recording.
Birth dating of embryonic cortical neurons
Dams, pregnant with embryos at E12.5, were injected with BrdU (5-bromo-2′-deoxyuridine) (Abcam, ab142567) intraperitoneally. Dams were then returned to their home. Embryos were extracted for immunohistological staining at E14.5 and E18.5, following the protocol described under “immunohistological staining of embryonic tissue” below.
Surgical isolation of embryonic cortex for single cell RNA sequencing
Dams, pregnant with embryos between E14.5 and E18.5, were anesthetized with a mixture of Fentanyl-Medetomidine-Midazolam (Fentanyl (Janssen, 0.05 mg/kg), Medetomidine (Virbac, 0.5 mg/kg); Midazolam (Sintetica, 5 mg/kg)). Dams were placed on their back and the hair on the abdomen was shaved. A 1.5 cm long incision was placed along the midline of the abdomen. The uterine wall was surgically opened longitudinally. A single embryo, within its yolk sac, was then isolated and the yolk sac was opened. The embryo was removed and placed in ice-cold oxygenated brain slice solution (sucrose (10 mM), trehalose (125 mM), KCL (3 mM), NaH2PO4 (1.25 mM), HEPES (26 mM), Dextrose (20mM), adjusted to pH 7.3). The brain was isolated, and the embryonic cortex removed. The posterior two-thirds of both cortical hemispheres were dissociated. 10 mL of papain activation solution was prepared (0.5 M EDTA, 100 mM L-Cysteine, 100 mM 2Me-EtOH) 30 min before the dissociation. 20–40 μL of papain solution (LS003126, Worthington) was then activated with 100–200 μL of papain activation solution, as per the manufacturer’s instructions. Each sample of dissected embryonic cortex was transferred into 1.5 mL Eppendorf tubes, cut into smaller square pieces of 1 mm2, and incubated in 1 mL of activated papain solution for 40–45 min at 37°C. Dissociated cortical cells were spun down at 1300 rpm for 2 min to remove as much papain solution as possible. Cells were then washed three times with DMEM (D4947, Sigma-Aldrich), supplemented with GlutaMAX (35050061, Gibco) and DNase (2000 U/ml; D4263, Sigma). Cells were spun down, again, at 1300 rpm for 2 min, and then r–suspended in 0.5 - 1mL DMEM GlutaMAX, to which B27 (12587-010, Gibco) was added. To remove debris, cells were filtered through a cell strainer (352235, Falcon).
Sorting was performed on a FACSAria Fusion Cell Sorter (BD Biosciences) using a 100 μm nozzle and a sheath pressure of 20 psi. After recording several thousand events, the gating strategy was set using the FACSDiva software (v8.0.2, BD Biosciences). Size and granularity of the population of events was determined using the forward and side scatter values, and gates were set to exclude cell debris, doublets, and clumps. A final gate was set to select cells with high tdTomato fluorescence using a 582/15 nm bandpass filter (example gating strategy shown in Figure S1). Cells were sorted in single-cell drop mode into a tube containing DMEM without phenol red (D4947, Sigma-Aldrich), supplemented with GlutaMAX (35050061, Gibco) and B27 (12587-010, Gibco). Sorted cells were immediately processed for single cell RNA sequencing (as described below). In some cases, tdTomato-negative cells were also sorted, in order to aid in isolating Rbp4-Cre neurons (detailed below).
Single cell RNA sequencing
Cellular suspensions (8,000 cells per lane) were loaded on a 10x Genomics Chromium Single Cell instrument to generate single-cell Gel Beads in Emulsion (GEMs). Single-cell RNA-Seq libraries were prepared using GemCode Single Cell 3′ Gel Bead and Library Kit according to the manufacturer’s manual (version CG000204_Rev_C). The Chromium Gene Expression version 3.1 (v3.1) kit was used for all samples. Reverse transcription of GEMs was performed in a Eppendorf Mastercycler X50s thermal cycler using 0.2 mL PCR tube strips (E0030124286, Eppendorf): 53°C for 45 min, 85°C for 5 min, held at 4°C. After reverse transcription, the GEMs emulsion was broken and the single-stranded cDNA was cleaned up with DynaBeads MyOne Silane Beads (37002D, Life Technologies). cDNA was amplified using a Eppendorf Mastercycler X50s thermal cycler in 0.2 mL PCR tube strips using the following program: 98°C for 3 min, cycled 12 ×: 98°C for 15 s, 63°C for 20 s, 72°C for 1 min; then 72°C for 1 min, hold at 4°C. Amplified cDNA product was cleaned up with the SPRIselect Reagent Kit (0.6 × SPRI; B23317, Beckman Coulter). Indexed sequencing libraries were constructed using the reagents in the Chromium Single Cell 3′ library kit v3.1 (PN-1000121, 10x Genomics), following these steps: (i) fragmentation, end repair and A-tailing, (ii) post fragmentation, end repair and A-tailing; double-sided size selection with SPRIselect Reagent Kit (0.6 × SPRI and 0.8 × SPRI), (iii) adaptor ligation, (iv) post-ligation cleanups with SPRIselect (0.8 × SPRI), (v) sample index PCR using the Chromium multiplex kit (PN-120262, 10x Genomics), and (vi) post-sample index double-sided size selection with SPRIselect Reagent Kit (0.6 × SPRI and 0.8 × SPRI). The barcode sequencing libraries were measured using a Qubit 2.0 with a Qubit dsDNA HS assay kit (Q32854, Invitrogen) and the quality of the libraries assessed on a 2100 Bioanalyzer (Agilent) using a high-sensitivity DNA kit (5067-4626, Agilent). Libraries were sent to the EMBL Genomics Core Facility (Heidelberg) for sequencing. Libraries were loaded at 240 p.m. on an Illumina HiSeq4000 with 150 cycle kits using the following read length: 28 cycles Read 1, 8 cycles i7 index and 91 cycles Read 2.
Immunohistological staining of embryonic tissue
Dams, pregnant with embryos between E13.5 and E18.5, were anesthetized with a mixture of Fentanyl-Medetomidine-Midazolam (Fentanyl (Janssen, 0.05 mg/kg), Medetomidine (Virbac, 0.5 mg/kg); Midazolam (Sintetica, 5 mg/kg)). Dams were placed on their back and the hair on the abdomen was shaved. A 1.5 cm long incision was placed along the midline of the abdomen. The uterine wall was surgically opened longitudinally. A single embryo, within its yolk sac, was then isolated and the yolk sac was opened. The embryo was removed from the yolk sac, decapitated and the brain was removed, while immersed in ice-cold cortex buffer. Brains were immediately transferred into 4% (w/v) paraformaldehyde (diluted in phosphate buffered saline (PBS)). Brains were kept at 4°C, for 12 h, and then washed for 24 h in PBS at 4°C. To improve antibody penetration, the tissue was then washed with PBS and cryopreserved by incubation in 30% (wt/vol) sucrose at 4°C until the tissue sunk to the bottom of the tube. Three sequential freeze-thaw cycles (using dry ice) were applied, and the tissue was then stored at −80°C until further processing.
After washing in PBS, brains were embedded in 3% agarose (SeaKem LE Agarose, Lonza) and 150 μm thick vibratome sections were cut (VT1000S vibratome, Leica Biosystems). For preparation for staining against BrdU, antigen retrieval was performed by incubating the sections in 2M HCl (J/4270/17, Fisher Chemical) at 37° for 20 min, followed by 3 washes of a solution of borax (composed of 0.6% (w/v) Boric Acid (B7901, Sigma) and 12.7% (w/v) Sodium tetraborate decahydrate (B3545, Sigma), adjusted to a pH of 8.6, using 2M HCl (J/4270/17, Fisher Chemical)) at room temperature for 5 min each. Sections were then washed in PBS at room temperature for 5 min, before being transferred to blocking solution, as follows. For all tissues, sections were incubated for 24 h in blocking buffer containing 10% (v/v) normal donkey serum (NDS) (S30-100ML, Merck), 0.5% (v/v) Triton X-100 (93443-100ML, Sigma), 1% (w/v) BSA (A4161-1G, Sigma) and 0.01% (w/v) sodium azide (S2002-25G, Sigma) in PBS. Primary antibodies were diluted in a buffer containing 3% (v/v) NDS, 1% (w/v) BSA, 0.01% (w/v) sodium azide and 0.5% Triton X-100 in PBS and incubated for 3–7 days at room temperature. The primary antibodies used in this study were: rabbit polyclonal anti-GFP (A-11122, Invitrogen), rat monoclonal anti-GFP (04404-84, Nacalai), goat polyclonal anti Nurr1/NGFI-Bbeta/NR4A2 (AF2156, R&D Systems), rat monoclonal anti CTIP2/BCL11B (MABE1045, Merck), rabbit anti-PSD95 (ab269863, Abcam); goat polyclonal anti-snap25 (ABIN1742235, antibody online); chicken polyclonal anti-Map2 (CPCA-MAP2, Encor Biotechnology Inc); mouse monoclonal anti-Neurofilament H (801602, BioLegend); rabbit anti-cleaved caspase-3 (ab2302, Abcam); rabbit polyclonal RFP antibody (anti-tdTomato) (600-401-379, Rockland); rat monoclonal anti-BrdU (ab6236, Abcam); rabbit polyclonal anti-Chd8 (a301-225a, Bethyl); rabbit polyclonal anti-NMDAR2B (71–8600, Invitrogen); rabbit polyclonal anti-Satb2 (ab34735, Abcam); rabbit polyclonal anti-Tbr2 (ab23345, Abcam); rabbit polyclonal anti-Tbr1 (ab31940, Abcam); rabbit polyclonal anti-Pax6 (AB2237, Millipore). Secondary antibody incubation was performed for 24 h at room temperature in the same buffer as the primary. The secondary antibodies used in this study were all from ThermoFisher unless otherwise stated: Alexa Fluor 488 donkey anti-rabbit IgG (heavy and light chains (H + L), A-21206); Alexa Fluor 568 donkey anti-rabbit IgG (H + L, A10042); Alexa Fluor 647 donkey anti-rabbit IgG (H + L, A-31573); Alexa Fluor 488 donkey anti-rat IgG (H + L, A-21208); Alexa Fluor 568 donkey anti-goat IgG (H + L, A-11057); Alexa Fluor 633 donkey anti-goat IgG (H + L, A-21082); Cy3-AffiniPure donkey anti-rat IgG (H + L, 12-165-153, Jackson ImmunoResearch Laboratories). The secondary antibody solution was replaced with a solution of Hoechst (10 μg/mL) (33342, ThermoFisher) diluted to 1:10000 in PBS for 10 min. Slides were then washed 3 times in PBS. Finally, coverslips (631–0974, VWR) were added onto brain slices embedded using Pro-Long Gold Antifade Mountant (P36982, ThermoFisher).
In situ hybridization
Whole embryonic brains were dissected at E14.5 and E18.5 and then fixed in 4% (wt/vol) paraformaldehyde (28908, ThermoFisher) in PBS at 4°C for 8 to 12 h. The tissue was then washed with PBS and cryopreserved by sequential incubations in 10%, 20% and 30% (wt/vol) sucrose at 4°C until the tissue sunk to the bottom of the tube. Embryonic brains were then embedded in Optimal Cutting Temperature (OCT) Compound (25608-930, VWR), frozen on dry ice and stored at −80°C. 35 μm coronal sections were cut (CryoStar NX70 Cryostat 957000, ThermoFisher), excluding the olfactory cortex, and placed on Super-Frost Ultra Plus GOLD slides (11976299, ThermoFisher). Slides were placed on a 60°C hot plate for 1 to 2 h for increased tissue adhesion and were then air dried at −20°C for 2 to 4 h, before being stored at −80°C until use.
The RNAscope Multiplex Fluorescent Reagent Kit v2 Assay (323100, ACD Bio-techne) was used to detect RNA in embryonic cortical cells according to the user manual (Section: “Fixed-frozen tissue samples”), with the exception of a shorter incubation period with Protease III. Slides were incubated with 5 drops of Protease III for 4 min at 40°C to facilitate probe penetration into the cells while minimizing damage to the tissue. Combinations of 3 probes per slide were used in their respective channels (C1, C2 and C3). Probes used were RNAscope 3-plex Positive Control Probe-Mm (320881), RNAscope 3-Plex Negative Control Probe (320871), Mm-GCamP6s-01 (557091), Mm-Lhx2 (485791), Mm-Pou3f3 (441521), Mm-Reln (405981), Mm-Nxph4 (489641), Mm-Crym (466131), Mm-Nfe2I3 (486201), Mm-Fam19a2 (452631), Mm-Cdh8 (485461), Mm-Zcchc12 (524441), tdTomato (317041), Mm-Chd8-E3 (457771), and Mm-Grin2b-O1 (1223851). Slides were stored overnight at room temperature after hybridization in 5x Saline Sodium Citrate (SSC; 20X stock solution containing 3 M sodium chloride and 300 mM trisodium citrate, pH 7.0). Opal 520 dye (FP1487001KT, Akoya Bio), Opal 570 dye (FP1488001KT, Akoya Bio) or Opal 690 dye (FP1497001KT, Akoya Bio) diluted 1:1500 in TSA buffer (provided with the Multiplex Fluorescent Reagent Kit v2) were used as fluorescent labels. Before mounting the slides, DAPI solution (provided with the kit) was added to the slides for 1 min. Coverslips (631–0974, VWR) were added onto brain slices embedded using Pro-Long Gold Antifade Mountant (P36982, ThermoFisher).
Image acquisition for immunohistology and in situ hybridization
A spinning disc microscope (Axio Imager M2 upright microscope, Yokogawa CSU W1 dual camera T2 spinning disk confocal scanning unit, Visitron VS-Homogenizer on an Olympus IXplore Spin confocal spinning disc microscope system) was used to image immunohistologically stained slides, using 20X (UPLSAPO20X, Olympus) and 40X (UPLSAPO40×S, Olympus) objectives. In situ hybridization slides were imaged using an Olympus FV3000-BX63L upright confocal laser scanning microscope, using 20X (UPLSAPO20X, Olympus) and 40X oil (UPLANFL N40×, Olympus) objectives. The image acquisition settings were adjusted to maximally fill the available imaging dynamic range. Data collection and analysis were not blind to the conditions of the experiments.
Whole mouse brain fixation and slice preparation for electron microscopy
Dams, pregnant with embryos at either E14.5 or E18.5, were anesthetized with a mixture of Fentanyl-Medetomidine-Midazolam (Fentanyl (Janssen, 0.05 mg/kg), Medetomidine (Virbac, 0.5 mg/kg); Midazolam (Sintetica, 5 mg/kg)). Dams were placed on their back and the hair on the abdomen was shaved. A 1.5 cm long incision was placed along the midline of the abdomen. The uterine wall was surgically opened longitudinally. A single embryo, within its yolk sac, was then isolated and the yolk sac was opened. A single embryo, within its yolk sac, was then isolated and the yolk sac was opened. The embryo was removed from the yolk sac, decapitated and the brain was removed, while immersed in ice-cold cortex buffer. Brains were immediately transferred into 4% (w/v) paraformaldehyde (diluted in phosphate buffered saline (PBS)). Brains were kept at 4°C, for 12 h, and then washed for 24 h in PBS at 4°C. Brains were fixed overnight at 4°C in freshly prepared phosphate buffer (0.1 M, pH 7.4) containing 4% paraformaldehyde (15700, Electron Microscopy Sciences) and 0.2% glutaraldehyde (16200, Electron Microscopy Sciences). After fixation, brains were embedded in 3% agarose (SeaKem LE Agarose, Lonza) and vibratome-sectioned (VT1000S vibratome, Leica Biosystems) into 60 μm thick slices. Slices were placed into multi-well culture dishes containing phosphate buffer saline (PBS) or phosphate buffer (0.1 M pH 7.4) containing 1% paraformaldehyde for longer storage.
Immunostaining for immunoelectron microscopy
As in,97 to increase penetration of the antibody, 2 freeze/thaw cycles were applied (using liquid nitrogen). Slices were thawed in a cryoprotectant solution containing 2% v/v glycerol (G5516, Sigma-Aldrich) and 20% v/v dimethyl sulfoxide (DMSO) (D8418, Sigma-Aldrich) diluted in PBS. Slices were then agitated in a solution of 0.3% peroxide (107209, Merck) diluted in PBS for 10 min, and then washed three times in PBS/BSA-c (900.022, Aurion). Slices were incubated overnight, at 4°C, with an anti-GFP primary antibody (AB3080, Chemicon) diluted 1:500 with PBS/BSA-c. Slices were then washed three times in PBS/BSA-c for 5 min before incubation for 90 min with a secondary antibody (Biotin-SP-affinipure goat anti-rabbit IgG, F(ab’)2 (111-065-047, Jackson Immuno Research Laboratories)) diluted 1:300 in PBS/BSA-c. After three washes, 5 min each, in tris-buffered saline (TBS: 0.1 M, 0.9% NaCl, pH 8) slices were treated with a peroxidase-based enzymatic detection system (Vectastain Elite ABC kit (PK-6100, Vector Laboratories)). Slices were then washed three times in TBS (0.05 M, 0.9% NaCl, pH 7.6) (T5030, Sigma-Aldrich), before incubation for 20 min, at room temperature, with a mixture of 0.04% (w/v) 3,3-diaminobenzidine (DAB) (D8001, Sigma-Aldrich) and 0.015% (v/v) hydrogen peroxidase (107209, Merck) diluted in TBS. Staining was terminated by rinsing the slices three times in TBS for 5 min each.
Transmission electron microscopy (TEM)
As in,97 slices were transferred into glass scintillation vials containing cacodylate buffer (0.1 M, pH 7.4) (C0250, Sigma-Aldrich) and washed three times before post-fixation for 40 min with 1.0% osmium tetroxide (19110, Electron Microscopy Sciences) diluted in cacodylate buffer (0.1 M, pH 7.4). After a further three washes in ddH2O, slices were incubated in 1.0% (w/v) uranyl acetate dissolved in ddH2O. The slices were then rinsed three times in ddH2O and dehydrated through a graded ethanol series. After the final dehydration in 100% ethanol, slices were incubated twice in 100% propylene oxide (82320, Sigma-Aldrich). Slices were further infiltrated with a 1: 1 mixture of Durcupan resin (Resin A/M (44611, Sigma-Aldrich); B hardener (44612, Sigma-Aldrich); D hardener (44614, Sigma-Aldrich); DMP 30 (13600, Electron Microscopy Sciences)) and 100% propylene oxide, before being infiltrated with pure Durcupan resin. Finally, slices were flat embedded and cured overnight at 60°C. Thin sections of 50 nm each were cut using a Leica EM UC7 ultramicrotome and transferred onto formvar support film copper slot grids (FF2010-CU, Electron Microscopy Sciences) for TEM imaging.
Images were recorded at a magnification of between 8200× and 9900× for overviews and at a magnification of 16500× for imaging from immunolabeled synapses (corresponding to a pixel size of 2.8 nm) using a Tecnai Spirit TEM (FEI, Eindhoven Company) operated at 120 kV using a side-mounted 2 K × 2 K CCD camera (Veleta, Olympus).
Imaging in adult L5-PNs
PHP.eB AAV-CAG-FLEX-GCaMP7s, generated from the plasmid pGP-AAV-CAG-FLEX-jGCaMP7s-WPRE (Plasmid #104495, Addgene)144 was produced as previously described in.153 For systemic administration of AAVs, Rbp4-Cre mice were anesthetized with 1.75% isoflurane. 0.5–20 μL of purified AAV, with the volume adjusted to 50 μL by adding saline solution (0.9%), was injected retro-orbitally into the sinus using a 30-gauge micro-fine insulin syringe.154 A minimum of 2 × 1010 genome copies (GC) of virus were injected per gram of mouse weight. Animals were imaged at least 3 weeks following AAV infection.
For implantation of cranial windows, mice were anesthetized with Fentanyl-Medetomidine-Midazolam (FMM) (Fentanyl (Janssen, 0.05 mg/kg), Medetomidine (Virbac AG, 0.5 mg/kg), Midazolam (Sintetica, 5 mg/kg)). To prevent dehydration of the cornea during surgery, we applied Coliquifilm (Allergan) to the eyes. The skin was removed, the skull cleared of tissue, and a thin titanium holder was attached to the skull with dental cement (Superbond C&B) allowing for head fixation during calcium imaging.155 A 4 mm diameter craniotomy was made over the left hemisphere, exposing the visual cortices and surrounding areas. After removal of the bone, dehydration of the cortical surface was minimized by repeatedly applying cortex buffer to the surface. The cortical surface was covered with a 4 mm diameter glass coverslip and sealed with UV glue (NOA 68, Norland), stabilized by an additional layer of dental cement. Buprenorphine (Temgesic, 0.05–0.1 mg/kg) was injected 20 min before the end of the surgery to provide extended pain relief during the immediate recovery from surgery. At the end of surgery, mice were woken using an antagonist mixture to counteract FMM anesthesia (Atipamezol (Virbac, 2.5 mg/kg), Flumazenil (Sintetica, 0.5 mg/kg)). Mice were monitored daily following surgery and allowed to recover for at least 10 days before any recordings.
Mice were head-fixed, in darkness, within the Femtonics galvo-galvo scanning two-photon setup, and Rbp4-Cre neurons were imaged under the Nikon 25× water immersion objective (1.1 NA). Imaging was performed at 5–10 Hz. The resulting recordings (used to generate Figure S3) were then analyzed as with embryonic recordings, as detailed below.
In vivo para-uterine blood flow imaging
Dams were anesthetized and maintained at a surgical depth, as assessed by the lack of spontaneous movement, as well as the absence of a paw withdrawal reflex, in response to a toe pinch. To maintain the anesthesia, top-up doses of FMM anesthesia (10–30% of the initial bolus) were injected into the dam, between recordings, approximately every 30 min. Throughout the experiment, a nasal mask was used to deliver humidified oxygen to the dam, at a flow rate of 0.8 L/min (Respironics EverFlo OPI pump, Phillips). Dams were monitored visually throughout blood flow imaging sessions. In addition, a pulse oximeter attached to a sensor applied to the rear paw (MouseSTAT Jr and Paw Sensor, Kent Scientific) was used to monitor the heart rate and blood oxygenation of each mouse during experiments. For all dams, the heart rate was maintained at less than 400 bpm,148 and the blood oxygenation never dropped below 90%, when averaged over a minute.
Embryos were imaged under a widefield microscope (SZX16, Olympus) in the brightfield, to observe blood flow throughout the imaging region at the beginning of the experiment, after 5 h and, 10 min following disruption of the umbilical cord (severing of the cord). Recordings were collected using a CMOS camera (ORCA-Flash4.0 V3, Hamamatsu). Data was recorded via the CameraLink interface, using the HCImage Live software (Hamamatsu).
Extracting and analyzing genomic DNA
Samples from the frontal cortex of Rbp4tg/-Grin2b+/+, Rbp4tg/-Chd8+/+, Rbp4−/−Grin2b+/+, Rbp4−/−Chd8+/+, Rbp4+/−Chd8−/−Grin2b−/−, and wild-type mice were collected and homogenized with a pellet pestle (SCERSP749540-0000, VWR), in RLT Plus Lysis Buffer (1053393, Qiagen). Total genomic DNA was extracted using the AllPrep DNA/RNA Micro Kit (80284, Qiagen), following the manufacturer’s instruction. The genomic deletion of exon 3 of the CHD8 gene and exon 4/5 of the GRIN2B gene was assessed by PCR amplification of their respective genomic locations, including the flanking intronic regions, using the GoTaq Long PCR Master Mix (Cat. #M4021, Promega) and the following primers: chd8Ex3F (5’ – ggtcctggggtcaaattggt), chd8Ex3R (5’ – tcatctgtcctgcaccttaagg), grin2bEx4/5F (5’ – tcatgtgtggcttggaggtg) and grin2bEx4/5R (5’ – ccgatgcctcaaaaaccacg) (synthesized by Microsynth). For each reaction, the optimal annealing temperature was determined by a PCR gradient. PCR products were electrophoretically separated on 1% agarose gels (SeaKem LE Agarose: 50004, Lonza) and visualized on an iBright FL1000 imaging system (Thermo Fisher Scientific). As one control, a sample of reference mouse genomic DNA (labeled as ref. WT in Figure S8) (69239-3, Sigma) was prepared identically, and also run on the gel. Sizes of the bands were estimated using a 1 kb Plus DNA Ladder (10787018, Thermo Fisher Scientific). In samples from Rbp4tg/-Grin2b+/+ and Rbp4tg/-Chd8+/+ mice, several PCR products were identified. One had a length similar to that of the single PCR product identified in the ref. wt and wild-type samples, and the rest were shorter (Figure S8). For all Rbp4tg/-Grin2b+/+, Rbp4tg/-Chd8+/+ mice, several bands were excised, extracted and cleaned up using the NucleoSpin Gel and PCR Clean-Up Kit (740609.250, Mackerey-Nagel) and sequenced by Next-Generation Sequencing (NGS) at the MGH CCIB DNA Core (Harvard Medical School, Boston, USA). The reads were assembled to sequence contigs, which were aligned against the Mus musculus C57BL/6J genome (GRCm39, GCA_000001635.9). In this way, the band labeled by the red box in Figure S8 was identified as the uncut sequence of either Chd8 or Grin2b, while the band labeled by the blue box in Figure S8 was identified as the same sequence following excision of the central section, by Cre. The sequence for one of the replicates is shown in Figure S8.
Quantification and statistical analysis
Filtering of calcium traces
On the galvo-galvo scanning microscope, two-photon recordings were imported as 3D matrices of recorded activity within a spatial field of view over time. Similarly, recordings taken with high -speed arbitrary frame scan mode on the acousto-optic two-photon microscope were also imported as 3D matrices across time, by taking the mean projection across all imaging planes, at each time point. Following image stabilization,145 custom code (MATLAB R2020a, Mathworks) was used for manual selection of elliptical regions of interest (ROIs) within the imaging field of view containing individual cells or dendrites. The distance between elliptical ROIs was quantified as the Euclidean distance between the centers of each of the two ellipses. The average of the pixels within the ROI at each time point was defined as the raw activity trace for each ROI. Three separate regions of background expression were also selected within each imaging field of view.
To estimate the per pixel fluorescence within individual neuronal somas as a function of the background activity in the embryonic tissue (as in Figure S3), the raw activity traces were integrated over time and normalized by the number of pixels within each ROI, to obtain a raw calcium activity per pixel over the recording interval. The same raw pixel fluorescence value was computed for the three background regions selected within each imaging window. The amplitude of the cellular fluorescence as a function of the per pixel background fluorescence, for each background region, was computed within each imaging window, and compared against the distribution of background fluorescence (as a function of other background ROIs).
For recordings taken on the acousto-optic two-photon microscope in multi-cube scanning mode, the mean projection across imaging planes at each time point was computed to generate a 3D matrix for each ROI across time. Following image stabilization,145 custom code (MATLAB R2020a, Mathworks) was used for manually selecting a single ROI, containing the imaged soma or neurite, within each mean imaging field. To identify a responsive object within the manually selected ROI, custom code was used to identify pixels which responded with similar temporal profiles. The mean pairwise correlation of each pixel with its surrounding pixels over the recorded time interval was computed (2 concentric rings of surrounding pixels; MATLAB R2020a, Mathworks). The correlation of each pixel within the ROI was compared against the distribution of pairwise correlations for pixels outside the manually selected ROI, within the same mean projected imaging plane. Pixels within the manually selected ROI that had a significantly higher mean pairwise correlation were identified as part of the recorded object (>2σ). If the fraction of significant pixels within the manually selected ROI was greater than 20%, the mean response of all such pixels was defined as the raw activity trace for each recorded object.
To denoise the raw traces, recordings were first converted to Fourier domain. A threshold, set at 150% of the mean amplitude component of the trace, was then applied to the amplitude component across the entire frequency range. This filter reduces white noise within the frequency spectrum, without a bias toward any specific frequency component. For recordings on the galvo-galvo microscope, the total power within the filtered frequency spectrum of the three background ROIs, was used as a secondary threshold. If an ROI contained a power less than the mean over the background ROIs’, the filtered trace was set to zero. In contrast, if the power of an ROI was greater than the background ROIs’ power, the filtered amplitude component was inverted back to obtain a denoised trace in time. To appropriately account for spontaneous activity, a %ΔF/F trace was then computed by utilizing the mean amplitude across the entire recording as the estimate of F. For all embryonic days following E13.5, a rolling mean with a time constant of 62.5 s was then subtracted away from the filtered activity trace, to ensure that the baseline was stable across the recording length. This trace was then used for all further analyses. For recordings on E13.5, a shorter rolling mean with a time constant of 12.5 s was subtracted away from the filtered activity trace. Finally, to obtain an estimate of the total activity within each ROI, the average of the %ΔF/F activity across time was used, yielding a unit of %(ΔF/F)/s.
While event-based analyses were also performed (see below), %(ΔF/F)/s was selected for most analyses in the paper since it provides an estimate of the overall calcium activity, and is less dependent on assumptions about the calcium kinetics of each individual event (as required to detect calcium events). Through the use of this ΔF/F, we normalized for the difference in amplitude across different neurons. However, even with this normalization, neurons within different layers showed a wide range of amplitudes due to the increased scattering with increased depth of recordings. Therefore, we did not compare calcium activity between neurons in different layers.
For recordings made to assess the effect of TTX application, recordings were filtered as detailed above. The total activity within each ROI was determined, in a 10-min recording immediately prior to, and a 10-min recording immediately following the injection of TTX.
For recordings made to assess the effect of AMPA+NMDA application, the 3D matrix was image stabilized145 and the same custom code as above (MATLAB R2020a, Mathworks) was used for manual selection of elliptical ROIs within the imaging field of view. Calcium activity in each ROI, per unit area, was calculated by summing across all pixels within each ROI and then dividing by the size of the ROI. The baseline fluorescence was defined as the average fluorescence within each ROI prior to the application of AMPA+NMDA, i.e. across the first 3 min of each recording. The fluorescence change, as a fraction of the baseline, was then computed and the change in the first minute of each recording (i.e. 2 min before the application of AMPA+NMDA) was compared to the change in the fifth minute of the same recording (i.e. 2 min following the application of AMPA+NMDA) (as in Figure 6B).
Computing event-based statistics
Calcium events were defined as fluctuations of ΔF/F above baseline with a length greater than 1.5s and a minimum average activity of at least 10%(ΔF/F)/s. The size of each event is defined as the total ΔF/F recorded from start to end. The frequency of events in each ROI was computed by dividing the total number of events by the total length of the recording. The properties of the calcium events recorded are examined in Figure S4.
Quantifying voltage changes in response to current steps
Every embryonic neuron, independent of recording day, responded with a maximum of a single nonlinear depolarization, at each current step, which was always located close to the initiation of the current injection. For the voltage curve recorded in response to a single current injection, the peak voltage in the first half of the current injection was defined as the peak voltage, and the mean voltage across the second half of the current injection was defined as the steady state voltage. Any active conductance was thereby represented by a peak voltage that was higher than the steady state voltage.
Quantifying excitatory synaptic potentials
Excitatory synaptic potentials were computed from subthreshold membrane voltage recorded in the presence of TTX. Recordings, taken at 50000 Hz, were downsampled by averaging recorded values in 50 frames bins, to generate traces with a frequency of 1000 Hz. A rolling mean with a time constant of 1s in length was subtracted away from the downsampled traces, to ensure that the baseline was stable across the recording length. We termed this trace the membrane voltage with adjusted baseline, and it was used for all further analyses.
Visual inspection of the amplitude distribution of the membrane voltage with adjusted baseline demonstrated a number of peaks within a long tail of amplitude values, suggesting the presence of events of quantal size. A threshold of 5 mV was chosen, and for each event crossing this threshold, a window of 100 ms prior and 400 ms following the event was selected. This 500 ms interval was then averaged across events to obtain the average shape of an excitatory event within Rbp4-Cre neurons (separately at both E14.5 and E18.5).
Location of cells within immunohistological sections
The bottom edge of the subplate (defined by the lower border of staining with CTIP2 (Bcll1b) and Nurr1 (Nr4a2) antibodies), the top edge of the cortical plate (defined by the outer edge of Hoechst staining), as well as the center of individual Rbp4-Cre neurons (defined by a GFP antibody) within coronal sections of the embryonic brain were manually annotated. A custom macro for Fiji156 (modified from the points_to_curve_distance macro) was used to compute the perpendicular distance of each cell to the two edges. The fractional distance of Rbp4-Cre neurons from the bottom of the subplate, normalized by the total distance within the cortical plate and subplate was computed, on all embryonic days. The fractional locations for the set of Rbp4-Cre neurons across all embryonic days was clustered into three clusters by hierarchical clustering of the mean of the pairwise Euclidean norms. The boundaries of these clusters were used to define the fractional edges of each of the three layers of Rbp4-Cre neurons, within the embryonic cortex.
Quantifying in vivo para-uterine imaging of anatomy
Due to movement between the imaging of each plane, custom code was used to register images using a rigid transformation, within each 3D stack, to a reference plane, and the registered stacks output as TIFF stacks (MATLAB R2020a, Mathworks). Fiji156 was then used to rotate the TIFF stacks into a projection onto the xy plane, thereby showing the location of cells within the cortical plate and subplate. Individual cells were selected that were identifiable across all time points. As with the location of cells within immunohistological sections, the bottom edge of the subplate, the top edge of the cortical plate, as well as the center of individual Rbp4-Cre neurons within coronal sections of the embryonic brain were manually annotated. A custom macro for Fiji156 (modified from the points_to_curve_distance macro) was used to compute the perpendicular distance to the two edges. The fractional distance of Rbp4-Cre neurons from the bottom of the subplate, normalized by the total distance within the cortical plate and subplate was computed, and neurons classified into the individual layers using the quantitative boundaries defined from immunohistological staining. The average location of neurons within different anatomical layers could then be quantified across time.
Quantification for in situ hybridization
All GCaMP6s-positive cells per image in cortex were manually identified. GCaMP6s-positive cells were then assessed for colocalization of markers for embryonic-NP (Nfe2I3, Nxph4, Zcchc12, and Reln (on E14.5 only)), embryonic-IT (Lhx2 and Fam19a2) and embryonic-PT (Cdh8 and Pou3f3 (on E18.5 only)). GCaMP6s-positive neurons were identified by their anatomical location as being members of a specific layer (superficial and deep at E14.5; intermediate and deep at E18.5). The fraction of GCamP6s-positive cells in each layer that were positive for each marker was then calculated on both embryonic days. Finally, the fraction of all GCaMP6s-positive cells positive for at least one of the markers for each type was computed.
Quantification of blood flow rate
Brightfield recordings of blood vessels, at the surface of the embryonic cortex, were obtained at 100 Hz and were imported as 3D matrices of the 2D spatial field of view over time. 3D matrices were stabilized,145 and identical blood vessels identified by visual inspection of the pattern of blood vessels, at the start of imaging, after 5 h of imaging, and following the disruption of the umbilical cord. Custom code (MATLAB R2020a, Mathworks) was used to draw cross-sections across a subset of the blood vessels. The amplitude of activity averaged across the pixels within the cross-section of the blood vessel was computed. This amplitude within each blood vessel was filtered by the application of a rolling mean with a time constant of 25 ms, followed by the subtraction of a rolling mean with a time constant of 3s, to denoise the recorded signal and ensure that the baseline was stable across the recording length.
The blood flow within each blood vessel results in changes in amplitude as blood cells pass the cross-section. Therefore, we quantify the number of fluctuations in amplitude across the recorded interval. We computed the fractional change in the number of fluctuations from the recording at the start of imaging to the recording after 5 h, and the fractional change in the number of fluctuations from the recording after 5 h to the recording 10 min following the disruption of the umbilical cord, per blood vessel.
Correlation analyses
The correlation of the spontaneous activity within Rbp4-Cre neurons in each recording was computed as the pairwise Pearson correlation of the denoised activity in each pair of neurons. To determine that the observed correlations were greater than that expected from a random pair of neurons with the same overall activity, the spontaneous activity traces for each pair of neurons were reordered, 1000 times. In detail, individual events within each spontaneous activity trace (defined as deviations of the ΔF/F > 0, as introduced above) were shuffled in time, to maintain both the overall spontaneous activity level as well as the time constant of individual events (which differed across embryonic days). The distribution of pairwise correlations of both neurons’ activity across the 1000 randomized pairs defines the random distribution for each pair of neurons, and neuron pairs showing significant pairwise correlations were those with correlations significantly higher than this distribution (>3σ). Note that this distribution is specific to the level of activity within each pair of neurons. To obtain a model of the random correlations across all pairs, the distribution of correlations across all pairs of neurons that were not significantly different from their specific random distribution was used.
To calculate the synchrony across a population of Rbp4-Cre neurons within a recording (as for Figure S7), we computed the pairwise Pearson correlation between activity within each soma (filtered as described above) and the sum of the activity in all other somas within a given recording, i.e. excluding the region of interest for which the synchrony is being computed (MATLAB 2020a, Mathworks).98
Single-cell RNA sequencing alignment and pre-processing
Analysis was performed in Python (v3.7.7). The workflow for the analysis was managed using Snakemake (v5.19.3). Sequencing results were aligned to a reference and barcodes identified using StarSolo (v2.7.5a). The reference was generated from the mouse genome (GRCm38) and annotation (GRCm38.93) and modified to include the sequences for tdTomato (Addgene plasmid # 22799)64 and the Cre recombinase present in the mouse strain (personal communication with S. Gong).62,63
Raw count matrices for each sample were loaded into and primarily handled within ScanPy (v1.5.1). To filter cell barcodes associated with empty droplets, count matrices were temporarily transferred to R (v3.6.1) using rpy2 (v3.3.2) and EmptyDrops157 was applied using DropletUtils (v1.6.1). Droplets with a false-discovery rate less than 0.001 were classified as non-ambient and retained for subsequent steps.
To identify multiplets (cell barcodes associated with multiple cells), we used Scrublet (v0.2.1) to simulate the creation of multiplets from our data and score our observed cells in comparison.158 The threshold separating likely multiplets from individual cells was estimated using a mixture of two Gaussians fit to the simulated scores, visual inspection confirmed that one Gaussian captured the singlet distribution and the other Gaussian captured the multiplet distribution. The point where these two distributions crossed was used as the threshold beyond which cells were excluded as being more likely a multiplet than a singlet. The process of simulation, identifying the threshold, and excluding cells beyond the threshold was repeated for each sample individually.
Cells were filtered as low-quality if they expressed fewer than 500 distinct genes or if the fraction of their reads originating from mitochondria exceeded 3 standard deviations from the median fraction of mitochondrial reads within the sample. To reduce the influence of outliers, median absolute deviation was used to estimate the standard deviation. Mitochondrial and ribosomal genes were filtered out of most subsequent analyses.
Count matrices across all samples were aggregated, and a Leiden clustering (leidenalg, v0.8.0) was performed.72 Unless otherwise specified, gene expression is presented as log2 pseudocounts of transcripts which were first normalized to a total library size of 10,000 in each cell. Three individual cells were selected at random from every cluster and manually annotated by a trained observer according to their individual transcriptomes. Additional cells were annotated when necessary. In total, 106 cells were annotated in this manner. Throughout the process, an interactive cell browser was used to assist experimenters in exploring the data (UCSC Cell Browser v0.7.11).
Isolation of Rbp4-Cre neurons
When enriching for tdTomato-labeled cells (Rbp4-Cre neurons) with FACS, a relatively permissive gate was used to help ensure a representative set of labeled cells. Additional enrichment of the sequenced single cells was performed to further isolate Rbp4-Cre neurons (Figure S1). First, clusters annotated as excitatory neurons or their progenitors were selected (a total of 25681 cells). Second, from these excitatory neurons, communities of cells with high tdTomato were located and isolated. Community detection was performed by applying a weighted kernel density smoothing to a UMAP embedding (60-nearest-neighbor graph, top 400 overdispersed genes)71 where the weights are the tdTomato transcript count in each cell and the kernel bandwidth is half of the bandwidth determined by Scott’s rule. This approach was selected because (i) it is more robust to dropout and ambient RNA than applying a threshold at the single-cell level (ii) it may provide a more-principled cutoff for inclusion for the lineages contained in our data than clustering.
To validate this method we used the same kernel and UMAP projection to compare communities of cells identified as having high tdTomato expression with communities of cells which FACS selected as possible Rbp4-Cre neurons. The degree of FACS enrichment was estimated by comparing the position on the UMAP plot of cells which were positive- and negative-selected by FACS and sequenced separately by dividing the kernel density of the positive-sorted cells by the density of the negative-sorted cells at each cell position. The highest FACS enrichment was observed in communities of cells containing the highest tdTomato expression (Figure S1).
The threshold for a cell to be included as tdTomato-labeled was 6 estimated standard deviations above the median kernel-smoothed tdTomato expression (Figure S1). To avoid the influence of outliers, this threshold was determined using cells which had a FACS enrichment ratio of less than 1 and by estimating the standard deviation using the median absolute deviation. Following this enrichment, we obtained a population of 1098 positive-identified Rbp4-Cre neurons, which were used for all further analyses.
Embedding and clustering of Rbp4-Cre neurons
Embeddings of Rbp4-Cre neurons were created by applying UMAP to a 60-nearest neighbor graph created directly from the top 400 overdispersed gene representation.71 Leiden clusters were calculated at a resolution of 0.2.72
Statistical analysis of single cell RNA sequencing in Rbp4-Cre neurons in relation to the adult cortex
RNA sequencing data from single cells of the adult cortex were loaded as counts of reads mapping to exons (from8), normalized by the effective gene length to approximate the transcript abundance, and then library-normalized to a total of 10000 normalized counts per cell. Subsequent comparisons were performed in log2 space using pseudocounts.
To compare gene expression of Rbp4-Cre neurons to that of cells from each adult layer, we identified the top genes discriminating the different adult layers using Wilcoxon rank sum. The number of genes selected was varied to verify that results were robust. Spearman correlations of gene expression were calculated between the Rbp4-Cre neurons and each of the adult layers.
Individual Rbp4-Cre neurons were compared to types of adult layer 5 cells using a Bayesian approach. Starting with a uniform prior, the posterior probability of each single cell was updated based on the genetic identity of the transcripts observed inside it and the conditional probability of observing transcripts with those genetic identities in each of the three adult layer 5 cell types. The joint distribution between genes and cell types in the adult was pre-processed in two ways: (i) additive smoothing was applied by adding 1 to the number of transcripts observed in each combination of adult cell type and gene before normalizing to create the joint probability distribution and (ii) the joint probability distribution was normalized such that each gene had the same marginal probability of being observed. The gene space employed for these comparisons was the intersection of the top 600 genes differentially expressed between the adult types and different numbers of genes differentially expressed by the clusters of Rbp4-Cre neurons. The fraction of cells which had a posterior probability greater than 99% (Bonferroni-adjusted for multiple comparisons) of being sampled from one of the three adult layer 5 cell types revealed that the cell type association of clusters was not sensitive to the number of genes selected within a range of 7–40 unique genes.
The ternary plot was created using the posterior probability of each Rbp4-Cre neuron being one of the three adult cell types, using the set of 24 unique genes, chosen as discussed above. To prevent the majority of cells from overlapping each other at the extreme corners of the triangles, the values presented on the ternary plot were curved by taking the 15th root of each cells’ probabilities followed by normalization to a total of one.
Distribution of gene expression in Rbp4-Cre neurons
Genes associated with synaptic transmission, signaling, the synaptic vesicle cycle, and active membrane properties were identified from appropriate Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways including Glutamatergic synapse, GABAergic synapse (postsynaptic), and Synaptic vesicle cycle.96 Genes associated with transsynaptic signaling were identified from a review of synaptic development.95 Spearman correlation was used for comparing correlations of transcripts across days, within each of the embryonic cell types.
The list of schizophrenia associated genes was chosen from a genome wide association study of the disease,104 the OMIM database,105 and supplemented with genes found to be associated with schizophrenia in a genome wide association study of multiple neurodevelopmental diseases.106 To ensure a lack of bias in the representation, the list was not revised further, except for the removal of genetic loci and genes not annotated in our dataset (primarily because of the absence of a mouse ortholog), including: Apol2, Apol4, Cacna1l, Chi3l1, ChrnaB4, Daoa, Disc2, Fam5b, Nlgn4x, Sczd1, Sczd11, Sczd12, Sczd2, Sczd3, Sczd5, Sczd6, Sczd7, and Sczd8.
The list of autism spectrum disorder associated genes was chosen by ordering genes by their SFARI gene score (from 1 to 3) and, then secondarily, by the number of reports107 (accessed as of August 29, 2020). In this ordering, all genes with greater than 10 reports were chosen. To ensure a lack of bias in the representation, the list was not revised further, except for the removal of genes that were not expressed in the adult dataset8: Kmt5b, Nexmif, Scn2a.
For each gene, the mean number of transcripts across (i) the population of Rbp4-Cre neurons and (ii) the population of adult layer 5 neurons (combining NP, IT, and PT types in VISp)8 was quantified. Expression values for each gene were presented as a circle plot, separated by embryonic type and age. To compare expression of disease-associated genes to the set of all genes, in both Rbp4-Cre neurons and adult layer 5 neurons, the distribution of mean expression was separately computed for the subset of genes associated with each disease and for the full set of genes. Distributions of disease gene expression were compared to the distribution of the full set of genes using Wilcoxon rank-sum, with a p value below 0.05 (Bonferroni corrected for 6 comparisons). A significant difference indicated that the set of genes associated with a given disease had higher than expected expression compared to the full set. For each of the diseases, the fold difference in gene expression was compared between Rbp4-Cre neurons and adult layer 5 neurons. In each case, fold difference was computed by dividing the mean expression across genes associated with the disease by the mean expression across the full set of genes.
Statistical analyses
All statistical comparisons of distributions, except for the statistical significance of pairwise correlations (discussed under correlations, above), the statistical significance in the analysis of gene expression profiles (discussed under distributions of gene expression, above), and the analysis of distribution of neurons into layers (for Figures 2E, 7D, S2, and S9) were performed using either the Wilcoxon rank-sum test, for unpaired data, or the Wilcoxon signed rank test, for paired data. The significance threshold used for both tests was 3σ. The comparison of the spatial distribution of neurons into layers (Figures 2E, 7D, S2, and S9) was performed using a chi-squared test (χ2 test).
Acknowledgments
We thank D. Jothimani, E. Mace, A. Herrero-Navarro, R. Morikawa, E.S. Ruthazer, N. Verma for comments; S. Curtoni, D. Rey, and T. Marzolla for mouse work; S. Gong for Rbp4-Cre BAC sequence; V. Juvin (SciArtWork) for graphic design; and Roska lab for discussions. Support: M.M.: EMBO Long Term Fellowship (ALTF 1552-2015) Marie Curie Fellowship, SFARI Bridge to Independence Award, and HFSP Postdoctoral Fellowship (LT000447/2016); B. Roska: SNSF Synergia grant (CRSII3_141801), ERC advanced grant (RETMUS N°669157, HURET N°883781), Louis-Jeantet Foundation award, Körber Foundation award, SNSF grant (31003A_182523), and the NCCR ‘Molecular Systems Engineering’ grant.
Author contributions
Conceptualization: M.M., A. Bharioke, B. Roska; Methodology: M.M., A. Bharioke; Investigation: M.M., A. Bharioke, G.K., V.M.-J., A. Brignall, T.R., A.G.-M., T.U., S.H., D.P., J.K., S.P.; Data curation: M.M., A. Bharioke, A. Brignall, C.S.C.; Formal analysis: M.M., A. Bharioke, C.S.C.; Resources: N.L., B.G.-S., B. Rózsa; Software: M.M., A. Bharioke, C.S.C.; Supervision, B. Roska; Writing: M.M., A. Bharioke, B. Roska.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: April 17, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2023.03.025.
Supplemental information
Data and code availability
-
•
Single-cell RNA-sequencing data have been deposited at the NCBI Sequence Read Archive (with processed count tables in Mendeley Data), and both datasets are publicly available as of the date of publication. Accession numbers are listed in the key resources table. All microscopy data and other recordings reported in this paper will be shared by the lead contact upon request.
-
•
All original code has been deposited at Zenodo and is publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Harris K.D., Shepherd G.M.G. The neocortical circuit: themes and variations. Nat. Neurosci. 2015;18:170–181. doi: 10.1038/nn.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim E.J., Juavinett A.L., Kyubwa E.M., Jacobs M.W., Callaway E.M. Three types of cortical layer 5 neurons that differ in brain-wide connectivity and function. Neuron. 2015;88:1253–1267. doi: 10.1016/j.neuron.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown A.P.Y., Cossell L., Strom M., Tyson A.L., Vélez-Fort M., Margrie T.W. Analysis of segmentation ontology reveals the similarities and differences in connectivity onto L2/3 neurons in mouse V1. Sci. Rep. 2021;11:4983. doi: 10.1038/s41598-021-82353-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wertz A., Trenholm S., Yonehara K., Hillier D., Raics Z., Leinweber M., Szalay G., Ghanem A., Keller G., Rózsa B., et al. Single-cell–initiated monosynaptic tracing reveals layer-specific cortical network modules. Science. 2015;349:70–74. doi: 10.1126/science.aab1687. [DOI] [PubMed] [Google Scholar]
- 5.Vélez-Fort M., Rousseau C.V., Niedworok C.J., Wickersham I.R., Rancz E.A., Brown A.P.Y., Strom M., Margrie T.W. The stimulus selectivity and connectivity of layer six principal cells reveals cortical microcircuits underlying visual processing. Neuron. 2014;83:1431–1443. doi: 10.1016/j.neuron.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young H., Belbut B., Baeta M., Petreanu L. Laminar-specific cortico-cortical loops in mouse visual cortex. Elife. 2021;10:e59551. doi: 10.7554/eLife.59551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tasic B., Menon V., Nguyen T.N., Kim T.K., Jarsky T., Yao Z., Levi B., Gray L.T., Sorensen S.A., Dolbeare T., et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 2016;19:335–346. doi: 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tasic B., Yao Z., Graybuck L.T., Smith K.A., Nguyen T.N., Bertagnolli D., Goldy J., Garren E., Economo M.N., Viswanathan S., et al. Shared and distinct transcriptomic cell types across neocortical areas. Nature. 2018;563:72–78. doi: 10.1038/s41586-018-0654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morishima M., Kawaguchi Y. Recurrent connection patterns of corticostriatal pyramidal cells in frontal cortex. J. Neurosci. 2006;26:4394–4405. doi: 10.1523/JNEUROSCI.0252-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cossell L., Iacaruso M.F., Muir D.R., Houlton R., Sader E.N., Ko H., Hofer S.B., Mrsic-Flogel T.D. Functional organization of excitatory synaptic strength in primary visual cortex. Nature. 2015;518:399–403. doi: 10.1038/nature14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petreanu L., Mao T., Sternson S.M., Svoboda K. The subcellular organization of neocortical excitatory connections. Nature. 2009;457:1142–1145. doi: 10.1038/nature07709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kondo S., Yoshida T., Ohki K. Mixed functional microarchitectures for orientation selectivity in the mouse primary visual cortex. Nat. Commun. 2016;7:13210. doi: 10.1038/ncomms13210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee W.-C.A., Bonin V., Reed M., Graham B.J., Hood G., Glattfelder K., Reid R.C. Anatomy and function of an excitatory network in the visual cortex. Nature. 2016;532:370–374. doi: 10.1038/nature17192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keller A.J., Roth M.M., Scanziani M. Feedback generates a second receptive field in neurons of the visual cortex. Nature. 2020;582:545–549. doi: 10.1038/s41586-020-2319-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han Y., Kebschull J.M., Campbell R.A.A., Cowan D., Imhof F., Zador A.M., Mrsic-Flogel T.D. The logic of single-cell projections from visual cortex. Nature. 2018;556:51–56. doi: 10.1038/nature26159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mullins C., Fishell G., Tsien R.W. Unifying views of autism spectrum disorders: a consideration of autoregulatory feedback loops. Neuron. 2016;89:1131–1156. doi: 10.1016/j.neuron.2016.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Del Pino I., Rico B., Marín O. Neural circuit dysfunction in mouse models of neurodevelopmental disorders. Curr. Opin. Neurobiol. 2018;48:174–182. doi: 10.1016/j.conb.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 18.Golden C.E., Buxbaum J.D., De Rubeis S. Disrupted circuits in mouse models of autism spectrum disorder and intellectual disability. Curr. Opin. Neurobiol. 2018;48:106–112. doi: 10.1016/j.conb.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foss-Feig J.H., Adkinson B.D., Ji J.L., Yang G., Srihari V.H., McPartland J.C., Krystal J.H., Murray J.D., Anticevic A. Searching for cross-diagnostic convergence: neural mechanisms governing excitation and inhibition balance in schizophrenia and autism spectrum disorders. Biol. Psychiatry. 2017;81:848–861. doi: 10.1016/j.biopsych.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robertson C.E., Baron-Cohen S. Sensory perception in autism. Nat. Rev. Neurosci. 2017;18:671–684. doi: 10.1038/nrn.2017.112. [DOI] [PubMed] [Google Scholar]
- 21.Greig L.C., Woodworth M.B., Galazo M.J., Padmanabhan H., Macklis J.D. Molecular logic of neocortical projection neuron specification, development and diversity. Nat. Rev. Neurosci. 2013;14:755–769. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Angevine J.B., Sidman R.L. Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature. 1961;192:766–768. doi: 10.1038/192766b0. [DOI] [PubMed] [Google Scholar]
- 23.Molyneaux B.J., Arlotta P., Menezes J.R.L., Macklis J.D. Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 24.Rakic P. Mode of cell migration to the superficial layers of fetal monkey neocortex. J. Comp. Neurol. 1972;145:61–83. doi: 10.1002/cne.901450105. [DOI] [PubMed] [Google Scholar]
- 25.Berry M., Rogers A.W. The migration of neuroblasts in the developing cerebral cortex. J. Anat. 1965;99:691–709. [PMC free article] [PubMed] [Google Scholar]
- 26.Jabaudon D. Fate and freedom in developing neocortical circuits. Nat. Commun. 2017;8:16042. doi: 10.1038/ncomms16042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cadwell C.R., Bhaduri A., Mostajo-Radji M.A., Keefe M.G., Nowakowski T.J. Development and arealization of the cerebral cortex. Neuron. 2019;103:980–1004. doi: 10.1016/j.neuron.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kriegstein A.R., Noctor S.C. Patterns of neuronal migration in the embryonic cortex. Trends Neurosci. 2004;27:392–399. doi: 10.1016/j.tins.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 29.Galli L., Maffei L. Spontaneous impulse activity of rat retinal ganglion cells in prenatal life. Science. 1988;242:90–91. doi: 10.1126/science.3175637. [DOI] [PubMed] [Google Scholar]
- 30.Maffei L., Galli-Resta L. Correlation in the discharges of neighboring rat retinal ganglion cells during prenatal life. Proc. Natl. Acad. Sci. USA. 1990;87:2861–2864. doi: 10.1073/pnas.87.7.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohtaka-Maruyama C., Okamoto M., Endo K., Oshima M., Kaneko N., Yura K., Okado H., Miyata T., Maeda N. Synaptic transmission from subplate neurons controls radial migration of neocortical neurons. Science. 2018;360:313–317. doi: 10.1126/science.aar2866. [DOI] [PubMed] [Google Scholar]
- 32.Yuryev M., Andriichuk L., Leiwe M., Jokinen V., Carabalona A., Rivera C. In vivo two-photon imaging of the embryonic cortex reveals spontaneous ketamine-sensitive calcium activity. Sci. Rep. 2018;8:16059. doi: 10.1038/s41598-018-34410-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuryev M., Pellegrino C., Jokinen V., Andriichuk L., Khirug S., Khiroug L., Rivera C. In vivo calcium imaging of evoked calcium waves in the embryonic cortex. Front. Cell. Neurosci. 2015;9:500. doi: 10.3389/fncel.2015.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Q., Cohen M.A., Alsina F.C., Devlin G., Garrett A., McKey J., Havlik P., Rakhilin N., Wang E., Xiang K., et al. Intravital imaging of mouse embryos. Science. 2020;368:181–186. doi: 10.1126/science.aba0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Antón-Bolaños N., Sempere-Ferràndez A., Guillamón-Vivancos T., Martini F.J., Pérez-Saiz L., Gezelius H., Filipchuk A., Valdeolmillos M., López-Bendito G. Prenatal activity from thalamic neurons governs the emergence of functional cortical maps in mice. Science. 2019;364:987–990. doi: 10.1126/science.aav7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allene C., Cossart R. Early NMDA receptor-driven waves of activity in the developing neocortex: physiological or pathological network oscillations? J. Physiol. 2010;588:83–91. doi: 10.1113/jphysiol.2009.178798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corlew R., Bosma M.M., Moody W.J. Spontaneous, synchronous electrical activity in neonatal mouse cortical neurones. J. Physiol. 2004;560:377–390. doi: 10.1113/jphysiol.2004.071621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCabe A.K., Chisholm S.L., Picken-Bahrey H.L., Moody W.J. The self-regulating nature of spontaneous synchronized activity in developing mouse cortical neurones. J. Physiol. 2006;577:155–167. doi: 10.1113/jphysiol.2006.117523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luhmann H.J., Sinning A., Yang J.-W., Reyes-Puerta V., Stüttgen M.C., Kirischuk S., Kilb W. Spontaneous neuronal activity in developing neocortical networks: from single cells to large-scale interactions. Front. Neural Circuits. 2016;10:40. doi: 10.3389/fncir.2016.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allène C., Cattani A., Ackman J.B., Bonifazi P., Aniksztejn L., Ben-Ari Y., Cossart R. Sequential generation of two distinct synapse-driven network patterns in developing neocortex. J. Neurosci. 2008;28:12851–12863. doi: 10.1523/JNEUROSCI.3733-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Owens D.F., Boyce L.H., Davis M.B., Kriegstein A.R. Excitatory GABA responses in embryonic and neonatal cortical slices demonstrated by gramicidin perforated-patch recordings and calcium imaging. J. Neurosci. 1996;16:6414–6423. doi: 10.1523/JNEUROSCI.16-20-06414.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bortone D., Polleux F. KCC2 expression promotes the termination of cortical interneuron migration in a voltage-sensitive calcium-dependent manner. Neuron. 2009;62:53–71. doi: 10.1016/j.neuron.2009.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Owens D.F., Kriegstein A.R. Patterns of intracellular calcium fluctuation in precursor cells of the neocortical ventricular zone. J. Neurosci. 1998;18:5374–5388. doi: 10.1523/JNEUROSCI.18-14-05374.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mayer S., Chen J., Velmeshev D., Mayer A., Eze U.C., Bhaduri A., Cunha C.E., Jung D., Arjun A., Li E., et al. Multimodal single-cell analysis reveals physiological maturation in the developing human neocortex. Neuron. 2019;102:143–158.e7. doi: 10.1016/j.neuron.2019.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moreno-Juan V., Filipchuk A., Antón-Bolaños N., Mezzera C., Gezelius H., Andrés B., Rodríguez-Malmierca L., Susín R., Schaad O., Iwasato T., et al. Prenatal thalamic waves regulate cortical area size prior to sensory processing. Nat. Commun. 2017;8 doi: 10.1038/ncomms14172. ncomms14172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guillamón-Vivancos T., Aníbal-Martínez M., Puche-Aroca L., Moreno-Bravo J.A., Valdeolmillos M., Martini F.J., López-Bendito G. Input-dependent segregation of visual and somatosensory circuits in the mouse superior colliculus. Science. 2022;377:845–850. doi: 10.1126/science.abq2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kawasoe R., Shinoda T., Hattori Y., Nakagawa M., Pham T.Q., Tanaka Y., Sagou K., Saito K., Katsuki S., Kotani T., et al. Two-photon microscopic observation of cell-production dynamics in the developing mammalian neocortex in utero. Dev. Growth Differ. 2020;62:118–128. doi: 10.1111/dgd.12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Higuchi Y., Kita Y., Murakami F. In vivo imaging of cortical interneurons migrating in the intermediate/subventricular zones. Neurosci. Res. 2016;110:68–71. doi: 10.1016/j.neures.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 49.Yanagida M., Miyoshi R., Toyokuni R., Zhu Y., Murakami F. Dynamics of the leading process, nucleus, and Golgi apparatus of migrating cortical interneurons in living mouse embryos. Proc. Natl. Acad. Sci. USA. 2012;109:16737–16742. doi: 10.1073/pnas.1209166109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ang E.S.B.C., Haydar T.F., Gluncic V., Rakic P. Four-dimensional migratory coordinates of GABAergic interneurons in the developing mouse cortex. J. Neurosci. 2003;23:5805–5815. doi: 10.1523/JNEUROSCI.23-13-05805.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marín O. Developmental timing and critical windows for the treatment of psychiatric disorders. Nat. Med. 2016;22:1229–1238. doi: 10.1038/nm.4225. [DOI] [PubMed] [Google Scholar]
- 52.Kaiser T., Feng G. Modeling psychiatric disorders for developing effective treatments. Nat. Med. 2015;21:979–988. doi: 10.1038/nm.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Courchesne E., Pierce K. Why the frontal cortex in autism might be talking only to itself: local over-connectivity but long-distance disconnection. Curr. Opin. Neurobiol. 2005;15:225–230. doi: 10.1016/j.conb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 54.Stoner R., Chow M.L., Boyle M.P., Sunkin S.M., Mouton P.R., Roy S., Wynshaw-Boris A., Colamarino S.A., Lein E.S., Courchesne E. Patches of disorganization in the neocortex of children with autism. N. Engl. J. Med. 2014;370:1209–1219. doi: 10.1056/NEJMoa1307491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dufour A., Dumon C., Gouty-Colomer L.-A., Eftekhari S., Ferrari D.C., Ben-Ari Y. Prenatal reduction of E14.5 embryonically fate-mapped pyramidal neurons in a mouse model of autism. Eur. J. Neurosci. 2022;56:3875–3888. doi: 10.1111/ejn.15724. [DOI] [PubMed] [Google Scholar]
- 56.Rabelo L.N., Queiroz J.P.G., Castro C.C.M., Silva S.P., Campos L.D., Silva L.C., Nascimento E.B., Martínez-Cerdeño V., Fiuza F.P. Layer-specific changes in the prefrontal glia/neuron ratio characterizes patches of gene expression disorganization in children with autism. J. Autism Dev. Disord. 2022 doi: 10.1007/s10803-022-05626-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shin Yim Y., Park A., Berrios J., Lafourcade M., Pascual L.M., Soares N., Yeon Kim J., Kim S., Kim H., Waisman A., et al. Reversing behavioral abnormalities in mice exposed to maternal inflammation. Nature. 2017;549:482–487. doi: 10.1038/nature23909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim S., Kim H., Yim Y.S., Ha S., Atarashi K., Tan T.G., Longman R.S., Honda K., Littman D.R., Choi G.B., Huh J.R. Maternal gut bacteria promote neurodevelopmental abnormalities in mouse offspring. Nature. 2017;549:528–532. doi: 10.1038/nature23910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choi G.B., Yim Y.S., Wong H., Kim S., Kim H., Kim S.V., Hoeffer C.A., Littman D.R., Huh J.R. The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science. 2016;351:933–939. doi: 10.1126/science.aad0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Orosco L.A., Ross A.P., Cates S.L., Scott S.E., Wu D., Sohn J., Pleasure D., Pleasure S.J., Adamopoulos I.E., Zarbalis K.S. Loss of Wdfy3 in mice alters cerebral cortical neurogenesis reflecting aspects of the autism pathology. Nat. Commun. 2014;5:4692. doi: 10.1038/ncomms5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peñagarikano O., Abrahams B.S., Herman E.I., Winden K.D., Gdalyahu A., Dong H., Sonnenblick L.I., Gruver R., Almajano J., Bragin A., et al. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell. 2011;147:235–246. doi: 10.1016/j.cell.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gong S., Doughty M., Harbaugh C.R., Cummins A., Hatten M.E., Heintz N., Gerfen C.R. Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J. Neurosci. 2007;27:9817–9823. doi: 10.1523/JNEUROSCI.2707-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gerfen C.R., Paletzki R., Heintz N. GENSAT BAC Cre-recombinase driver lines to study the functional organization of cerebral cortical and basal ganglia circuits. Neuron. 2013;80:1368–1383. doi: 10.1016/j.neuron.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R., et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Madisen L., Garner A.R., Shimaoka D., Chuong A.S., Klapoetke N.C., Li L., van der Bourg A., Niino Y., Egolf L., Monetti C., et al. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron. 2015;85:942–958. doi: 10.1016/j.neuron.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Di Bella D.J., Habibi E., Stickels R.R., Scalia G., Brown J., Yadollahpour P., Yang S.M., Abbate C., Biancalani T., Macosko E.Z., et al. Molecular logic of cellular diversification in the mouse cerebral cortex. Nature. 2021;595:554–559. doi: 10.1038/s41586-021-03670-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Loo L., Simon J.M., Xing L., McCoy E.S., Niehaus J.K., Guo J., Anton E.S., Zylka M.J. Single-cell transcriptomic analysis of mouse neocortical development. Nat. Commun. 2019;10:134. doi: 10.1038/s41467-018-08079-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bandler R.C., Vitali I., Delgado R.N., Ho M.C., Dvoretskova E., Ibarra Molinas J.S., Frazel P.W., Mohammadkhani M., Machold R., Maedler S., et al. Single-cell delineation of lineage and genetic identity in the mouse brain. Nature. 2022;601:404–409. doi: 10.1038/s41586-021-04237-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arlotta P., Molyneaux B.J., Chen J., Inoue J., Kominami R., Macklis J.D. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 70.Schuman B., Machold R.P., Hashikawa Y., Fuzik J., Fishell G.J., Rudy B. Four Unique Interneuron Populations Reside in Neocortical Layer 1. J. Neurosci. 2019;39:125–139. doi: 10.1523/JNEUROSCI.1613-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Becht E., McInnes L., Healy J., Dutertre C.-A., Kwok I.W.H., Ng L.G., Ginhoux F., Newell E.W. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2018;37:38–44. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 72.Traag V.A., Waltman L., van Eck N.J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 2019;9:5233. doi: 10.1038/s41598-019-41695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Groh A., Meyer H.S., Schmidt E.F., Heintz N., Sakmann B., Krieger P. Cell-type specific properties of pyramidal neurons in neocortex underlying a layout that is modifiable depending on the cortical area. Cereb. Cortex. 2010;20:826–836. doi: 10.1093/cercor/bhp152. [DOI] [PubMed] [Google Scholar]
- 74.Sekine K., Kubo K.i., Nakajima K. How does Reelin control neuronal migration and layer formation in the developing mammalian neocortex? Neurosci. Res. 2014;86:50–58. doi: 10.1016/j.neures.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 75.Kilb W., Frotscher M. Cajal-Retzius cells: organizers of cortical development. e-Neuroforum. 2016;7:82–88. doi: 10.1007/s13295-016-0031-5. [DOI] [Google Scholar]
- 76.Marín-Padilla M. Cajal–Retzius cells and the development of the neocortex. Trends Neurosci. 1998;21:64–71. doi: 10.1016/S0166-2236(97)01164-8. [DOI] [PubMed] [Google Scholar]
- 77.Allendoerfer K.L., Shatz C.J. The Subplate, a transient neocortical structure: its role in the development of connections between thalamus and cortex. Annu. Rev. Neurosci. 1994;17:185–218. doi: 10.1146/annurev.ne.17.030194.001153. [DOI] [PubMed] [Google Scholar]
- 78.Ohtaka-Maruyama C. Subplate neurons as an organizer of mammalian neocortical development. Front. Neuroanat. 2020;14:8. doi: 10.3389/fnana.2020.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kuang X.-L., Zhao X.-M., Xu H.-F., Shi Y.-Y., Deng J.-B., Sun G.-T. Spatio-temporal expression of a novel neuron-derived neurotrophic factor (NDNF) in mouse brains during development. BMC Neurosci. 2010;11:137. doi: 10.1186/1471-2202-11-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Oberst P., Fièvre S., Baumann N., Concetti C., Bartolini G., Jabaudon D. Temporal plasticity of apical progenitors in the developing mouse neocortex. Nature. 2019;573:370–374. doi: 10.1038/s41586-019-1515-6. [DOI] [PubMed] [Google Scholar]
- 81.Bayer S.A., Altman J., others . Raven Press; 1991. Neocortical Development. [Google Scholar]
- 82.Yoshinaga S., Shin M., Kitazawa A., Ishii K., Tanuma M., Kasai A., Hashimoto H., Kubo K.I., Nakajima K. Comprehensive characterization of migration profiles of murine cerebral cortical neurons during development using FlashTag labeling. iScience. 2021;24:102277. doi: 10.1016/j.isci.2021.102277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Refinetti R. The circadian rhythm of body temperature. Front. Biosci. 2010;15:564–594. doi: 10.2741/3634. [DOI] [PubMed] [Google Scholar]
- 84.Reitman M.L. Of mice and men – environmental temperature, body temperature, and treatment of obesity. FEBS Lett. 2018;592:2098–2107. doi: 10.1002/1873-3468.13070. [DOI] [PubMed] [Google Scholar]
- 85.Miyamichi K., Amat F., Moussavi F., Wang C., Wickersham I., Wall N.R., Taniguchi H., Tasic B., Huang Z.J., He Z., et al. Cortical representations of olfactory input by trans-synaptic tracing. Nature. 2011;472:191–196. doi: 10.1038/nature09714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen T.-W., Wardill T.J., Sun Y., Pulver S.R., Renninger S.L., Baohan A., Schreiter E.R., Kerr R.A., Orger M.B., Jayaraman V., et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hemmings H.C., Riegelhaupt P.M., Kelz M.B., Solt K., Eckenhoff R.G., Orser B.A., Goldstein P.A. Towards a comprehensive understanding of anesthetic mechanisms of action: a decade of discovery. Trends Pharmacol. Sci. 2019;40:464–481. doi: 10.1016/j.tips.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pavel M.A., Petersen E.N., Wang H., Lerner R.A., Hansen S.B. Studies on the mechanism of general anesthesia. Proc. Natl. Acad. Sci. USA. 2020;117:13757–13766. doi: 10.1073/pnas.2004259117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Simó S., Jossin Y., Cooper J.A. Cullin 5 regulates cortical layering by modulating the speed and duration of Dab1-dependent neuronal migration. J. Neurosci. 2010;30:5668–5676. doi: 10.1523/JNEUROSCI.0035-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Boitard M., Bocchi R., Egervari K., Petrenko V., Viale B., Gremaud S., Zgraggen E., Salmon P., Kiss J.Z. Wnt signaling regulates multipolar-to-bipolar transition of migrating neurons in the cerebral cortex. Cell Rep. 2015;10:1349–1361. doi: 10.1016/j.celrep.2015.01.061. [DOI] [PubMed] [Google Scholar]
- 91.Noda M., Iwamoto I., Tabata H., Yamagata T., Ito H., Nagata K.I. Role of Per3 , a circadian clock gene, in embryonic development of mouse cerebral cortex. Sci. Rep. 2019;9:5874. doi: 10.1038/s41598-019-42390-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Arcangeli A., Bianchi L., Becchetti A., Faravelli L., Coronnello M., Mini E., Olivotto M., Wanke E. A novel inward-rectifying K+ current with a cell-cycle dependence governs the resting potential of mammalian neuroblastoma cells. J. Physiol. 1995;489:455–471. doi: 10.1113/jphysiol.1995.sp021065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xue M., Atallah B.V., Scanziani M. Equalizing excitation–inhibition ratios across visual cortical neurons. Nature. 2014;511:596–600. doi: 10.1038/nature13321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tinker A., Jan Y.N., Jan L.Y. Regions responsible for the assembly of inwardly rectifying potassium channels. Cell. 1996;87:857–868. doi: 10.1016/S0092-8674(00)81993-5. [DOI] [PubMed] [Google Scholar]
- 95.Siddiqui T.J., Craig A.M. Synaptic organizing complexes. Curr. Opin. Neurobiol. 2011;21:132–143. doi: 10.1016/j.conb.2010.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Knott G.W., Holtmaat A., Trachtenberg J.T., Svoboda K., Welker E. A protocol for preparing GFP-labeled neurons previously imaged in vivo and in slice preparations for light and electron microscopic analysis. Nat. Protoc. 2009;4:1145–1156. doi: 10.1038/nprot.2009.114. [DOI] [PubMed] [Google Scholar]
- 98.Bharioke A., Munz M., Brignall A., Kosche G., Eizinger M.F., Ledergerber N., Hillier D., Gross-Scherf B., Conzelmann K.-K., Macé E., Roska B. General anesthesia globally synchronizes activity selectively in layer 5 cortical pyramidal neurons. Neuron. 2022;110:2024–2040.e10. doi: 10.1016/j.neuron.2022.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gilman S.R., Iossifov I., Levy D., Ronemus M., Wigler M., Vitkup D. Rare De Novo Variants Associated with Autism Implicate a Large Functional Network of Genes Involved in Formation and Function of Synapses. Neuron. 2011;70:898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Forsyth J.K., Nachun D., Gandal M.J., Geschwind D.H., Anderson A.E., Coppola G., Bearden C.E. Synaptic and gene regulatory mechanisms in schizophrenia, autism, and 22q11.2 copy number variant–mediated risk for neuropsychiatric disorders. Biol. Psychiatry. 2020;87:150–163. doi: 10.1016/j.biopsych.2019.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Birnbaum R., Weinberger D.R. Genetic insights into the neurodevelopmental origins of schizophrenia. Nat. Rev. Neurosci. 2017;18:727–740. doi: 10.1038/nrn.2017.125. [DOI] [PubMed] [Google Scholar]
- 102.Ebert D.H., Greenberg M.E. Activity-dependent neuronal signalling and autism spectrum disorder. Nature. 2013;493:327–337. doi: 10.1038/nature11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Forrest M.P., Parnell E., Penzes P. Dendritic structural plasticity and neuropsychiatric disease. Nat. Rev. Neurosci. 2018;19:215–234. doi: 10.1038/nrn.2018.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Neale B.M., Corvin A., Walters J.T.R., Farh K.-H., Holmans P.A., Lee P., Bulik-Sullivan B., Collier D.A., Huang H., et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schizophrenia (MIM Number: 181500) Online Mendelian Inheritance in Man, OMIM. Johns Hopkins University, Baltimore, MD. https://omim.org/
- 106.Cross-Disorder Group of the Psychiatric Genomics Consortium Electronic address plee0@mghharvardedu. Cross-Disorder Group of the Psychiatric Genomics Consortium. Won H., Feng Y.-C.A., Rosenthal J., Zhu Z., Tucker-Drob E.M., Nivard M.G., Grotzinger A.D., Posthuma D., et al. Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell. 2019;179:1469–1482.e11. doi: 10.1016/j.cell.2019.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Abrahams B.S., Arking D.E., Campbell D.B., Mefford H.C., Morrow E.M., Weiss L.A., Menashe I., Wadkins T., Banerjee-Basu S., Packer A. SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs) Mol. Autism. 2013;4:36. doi: 10.1186/2040-2392-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Cicek A.E., Kou Y., Liu L., Fromer M., Walker S., et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Iossifov I., Levy D., Allen J., Ye K., Ronemus M., Lee Y.-H., Yamrom B., Wigler M. Low load for disruptive mutations in autism genes and their biased transmission. Proc. Natl. Acad. Sci. USA. 2015;112:E5600–E5607. doi: 10.1073/pnas.1516376112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yin J., Schaaf C.P. Autism genetics – an overview. Prenat. Diagn. 2017;37:14–30. doi: 10.1002/pd.4942. [DOI] [PubMed] [Google Scholar]
- 111.De Rubeis S., Buxbaum J.D. Genetics and genomics of autism spectrum disorder: embracing complexity. Hum. Mol. Genet. 2015;24:R24–R31. doi: 10.1093/hmg/ddv273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schaaf C.P., Sabo A., Sakai Y., Crosby J., Muzny D., Hawes A., Lewis L., Akbar H., Varghese R., Boerwinkle E., et al. Oligogenic heterozygosity in individuals with high-functioning autism spectrum disorders. Hum. Mol. Genet. 2011;20:3366–3375. doi: 10.1093/hmg/ddr243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Frazier T.W. Autism spectrum disorder associated with germline heterozygous PTEN mutations. Cold Spring Harb. Perspect. Med. 2019;9:a037002. doi: 10.1101/cshperspect.a037002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Busch R.M., Srivastava S., Hogue O., Frazier T.W., Klaas P., Hardan A., Martinez-Agosto J.A., Sahin M., Eng C., Developmental Synaptopathies Consortium Neurobehavioral phenotype of autism spectrum disorder associated with germline heterozygous mutations in PTEN. Transl. Psychiatry. 2019;9:253. doi: 10.1038/s41398-019-0588-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Katz L.C., Shatz C.J. Synaptic activity and the construction of cortical circuits. Science. 1996;274:1133–1138. doi: 10.1126/science.274.5290.1133. [DOI] [PubMed] [Google Scholar]
- 116.Verhage M., Maia A.S., Plomp J.J., Brussaard A.B., Heeroma J.H., Vermeer H., Toonen R.F., Hammer R.E., van den Berg T.K., Missler M., et al. Synaptic assembly of the brain in the absence of neurotransmitter secretion. Science. 2000;287:864–869. doi: 10.1126/science.287.5454.864. [DOI] [PubMed] [Google Scholar]
- 117.Li M., Cui Z., Niu Y., Liu B., Fan W., Yu D., Deng J. Synaptogenesis in the developing mouse visual cortex. Brain Res. Bull. 2010;81:107–113. doi: 10.1016/j.brainresbull.2009.08.028. [DOI] [PubMed] [Google Scholar]
- 118.Boyce W.T., Levitt P., Martinez F.D., McEwen B.S., Shonkoff J.P. Genes, environments, and time: the biology of adversity and resilience. Pediatrics. 2021;147:e20201651. doi: 10.1542/peds.2020-1651. [DOI] [PubMed] [Google Scholar]
- 119.Eyring K.W., Geschwind D.H. Three decades of ASD genetics: building a foundation for neurobiological understanding and treatment. Hum. Mol. Genet. 2021;30:R236–R244. doi: 10.1093/hmg/ddab176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Paulsen B., Velasco S., Kedaigle A.J., Pigoni M., Quadrato G., Deo A.J., Adiconis X., Uzquiano A., Sartore R., Yang S.M., et al. Autism genes converge on asynchronous development of shared neuron classes. Nature. 2022;602:268–273. doi: 10.1038/s41586-021-04358-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kanold P.O. The first cortical circuits: subplate neurons lead the way and shape cortical organization. Neuroform. 2019;25:15–23. doi: 10.1515/nf-2018-0010. [DOI] [Google Scholar]
- 122.Riva M., Genescu I., Habermacher C., Orduz D., Ledonne F., Rijli F.M., López-Bendito G., Coppola E., Garel S., Angulo M.C., Pierani A. Activity-dependent death of transient Cajal-Retzius neurons is required for functional cortical wiring. Elife. 2019;8:e50503. doi: 10.7554/eLife.50503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Molnár Z., Luhmann H.J., Kanold P.O. Transient cortical circuits match spontaneous and sensory-driven activity during development. Science. 2020;370:eabb2153. doi: 10.1126/science.abb2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kostović I. The enigmatic fetal subplate compartment forms an early tangential cortical nexus and provides the framework for construction of cortical connectivity. Prog. Neurobiol. 2020;194:101883. doi: 10.1016/j.pneurobio.2020.101883. [DOI] [PubMed] [Google Scholar]
- 125.López-Bendito G., Molnár Z. Thalamocortical development: how are we going to get there? Nat. Rev. Neurosci. 2003;4:276–289. doi: 10.1038/nrn1075. [DOI] [PubMed] [Google Scholar]
- 126.Molnár Z., Adams R., Goffinet A.M., Blakemore C. The role of the first postmitotic cortical cells in the development of thalamocortical innervation in the reeler mouse. J. Neurosci. 1998;18:5746–5765. doi: 10.1523/JNEUROSCI.18-15-05746.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vitali I., Fièvre S., Telley L., Oberst P., Bariselli S., Frangeul L., Baumann N., McMahon J.J., Klingler E., Bocchi R., et al. Progenitor hyperpolarization regulates the sequential generation of neuronal subtypes in the developing neocortex. Cell. 2018;174:1264–1276.e15. doi: 10.1016/j.cell.2018.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hurni N., Kolodziejczak M., Tomasello U., Badia J., Jacobshagen M., Prados J., Dayer A. Transient cell-intrinsic activity regulates the migration and laminar positioning of cortical projection neurons. Cereb. Cortex. 2017;27:3052–3063. doi: 10.1093/cercor/bhx059. [DOI] [PubMed] [Google Scholar]
- 129.Writing Committee for the Attention-Deficit/Hyperactivity Disorder. Autism Spectrum Disorder. Bipolar Disorder. Major Depressive Disorder. Obsessive-Compulsive Disorder. and Schizophrenia ENIGMA Working Groups. Patel Y., Parker N., Shin J., Howard D., et al. Virtual histology of cortical thickness and shared neurobiology in 6 psychiatric disorders. JAMA Psychiatr. 2021;78:47–63. doi: 10.1001/jamapsychiatry.2020.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fu J.M., Satterstrom F.K., Peng M., Brand H., Collins R.L., Dong S., Wamsley B., Klei L., Wang L., Hao S.P., et al. Rare coding variation provides insight into the genetic architecture and phenotypic context of autism. Nat. Genet. 2022;54:1320–1331. doi: 10.1038/s41588-022-01104-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Velmeshev D., Perez Y., Yan Z., Valencia J.E., Castaneda-Castellanos D.R., Schirmer L., Mayer S., Wick B., Wang S., Nowakowski T.J., et al. Single-cell analysis of prenatal and postnatal human cortical development. bioRxiv. 2022 doi: 10.1101/2022.10.24.513555. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mountcastle V.B. The columnar organization of the neocortex. Brain. 1997;120:701–722. doi: 10.1093/brain/120.4.701. [DOI] [PubMed] [Google Scholar]
- 133.Hodge R.D., Bakken T.E., Miller J.A., Smith K.A., Barkan E.R., Graybuck L.T., Close J.L., Long B., Johansen N., Penn O., et al. Conserved cell types with divergent features in human versus mouse cortex. Nature. 2019;573:61–68. doi: 10.1038/s41586-019-1506-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Polioudakis D., de la Torre-Ubieta L., Langerman J., Elkins A.G., Shi X., Stein J.L., Vuong C.K., Nichterwitz S., Gevorgian M., Opland C.K., et al. A Single-Cell Transcriptomic Atlas of Human Neocortical Development during Mid-gestation. Neuron. 2019;103:785–801.e8. doi: 10.1016/j.neuron.2019.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.DeFelipe J., Alonso-Nanclares L., Arellano J.I. Microstructure of the neocortex: Comparative aspects. J. Neurocytol. 2002;31:299–316. doi: 10.1023/A:1024130211265. [DOI] [PubMed] [Google Scholar]
- 136.Kriegstein A., Noctor S., Martínez-Cerdeño V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat. Rev. Neurosci. 2006;7:883–890. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
- 137.Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat. Rev. Neurosci. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Semple B.D., Blomgren K., Gimlin K., Ferriero D.M., Noble-Haeusslein L.J. Brain development in rodents and humans: identifying benchmarks of maturation and vulnerability to injury across species. Prog. Neurobiol. 2013;106–107:1–16. doi: 10.1016/j.pneurobio.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Workman A.D., Charvet C.J., Clancy B., Darlington R.B., Finlay B.L. Modeling transformations of neurodevelopmental sequences across mammalian species. J. Neurosci. 2013;33:7368–7383. doi: 10.1523/JNEUROSCI.5746-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kostović I., Išasegi I.Ž., Krsnik Ž. Sublaminar organization of the human subplate: developmental changes in the distribution of neurons, glia, growing axons and extracellular matrix. J. Anat. 2019;235:481–506. doi: 10.1111/joa.12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zecevic N. Synaptogenesis in layer I of the human cerebral cortex in the first half of gestation. Cereb. Cortex. 1998;8:245–252. doi: 10.1093/cercor/8.3.245. [DOI] [PubMed] [Google Scholar]
- 142.Moore A.R., Filipovic R., Mo Z., Rasband M.N., Zecevic N., Antic S.D. Electrical excitability of early neurons in the human cerebral cortex during the second trimester of gestation. Cereb. Cortex. 2009;19:1795–1805. doi: 10.1093/cercor/bhn206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Moore A.R., Zhou W.-L., Jakovcevski I., Zecevic N., Antic S.D. Spontaneous electrical activity in the human fetal cortex in vitro. J. Neurosci. 2011;31:2391–2398. doi: 10.1523/JNEUROSCI.3886-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dana H., Sun Y., Mohar B., Hulse B.K., Kerlin A.M., Hasseman J.P., Tsegaye G., Tsang A., Wong A., Patel R., et al. High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nat. Methods. 2019;16:649–657. doi: 10.1038/s41592-019-0435-6. [DOI] [PubMed] [Google Scholar]
- 145.Pnevmatikakis E.A., Giovannucci A. NoRMCorre: An online algorithm for piecewise rigid motion correction of calcium imaging data. J. Neurosci. Methods. 2017;291:83–94. doi: 10.1016/j.jneumeth.2017.07.031. [DOI] [PubMed] [Google Scholar]
- 146.Steinmetz N.A., Buetfering C., Lecoq J., Lee C.R., Peters A.J., Jacobs E.A.K., Coen P., Ollerenshaw D.R., Valley M.T., de Vries S.E.J., et al. Aberrant cortical activity in multiple GCaMP6-expressing transgenic mouse lines. eNeuro. 2017;4 doi: 10.1523/ENEURO.0207-17.2017. ENEURO.0207-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Adams S., Pacharinsak C. Mouse anesthesia and analgesia. Curr. Protoc. Mouse Biol. 2015;5:51–63. doi: 10.1002/9780470942390.mo140179. [DOI] [PubMed] [Google Scholar]
- 148.Fleischmann T., Jirkof P., Henke J., Arras M., Cesarovic N. Injection anaesthesia with fentanyl-midazolam-medetomidine in adult female mice: importance of antagonization and perioperative care. Lab. Anim. 2016;50:264–274. doi: 10.1177/0023677216631458. [DOI] [PubMed] [Google Scholar]
- 149.Trachtenberg J.T., Chen B.E., Knott G.W., Feng G., Sanes J.R., Welker E., Svoboda K. Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature. 2002;420:788–794. doi: 10.1038/nature01273. [DOI] [PubMed] [Google Scholar]
- 150.Szalay G., Judák L., Katona G., Ócsai K., Juhász G., Veress M., Szadai Z., Fehér A., Tompa T., Chiovini B., et al. Fast 3D imaging of spine, dendritic, and neuronal assemblies in behaving animals. Neuron. 2016;92:723–738. doi: 10.1016/j.neuron.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Katona G., Szalay G., Maák P., Kaszás A., Veress M., Hillier D., Chiovini B., Vizi E.S., Roska B., Rózsa B. Fast two-photon in vivo imaging with three-dimensional random-access scanning in large tissue volumes. Nat. Methods. 2012;9:201–208. doi: 10.1038/nmeth.1851. [DOI] [PubMed] [Google Scholar]
- 152.Rose T., Jaepel J., Hübener M., Bonhoeffer T. Cell-specific restoration of stimulus preference after monocular deprivation in the visual cortex. Science. 2016;352:1319–1322. doi: 10.1126/science.aad3358. [DOI] [PubMed] [Google Scholar]
- 153.Jüttner J., Szabo A., Gross-Scherf B., Morikawa R.K., Rompani S.B., Hantz P., Szikra T., Esposti F., Cowan C.S., Bharioke A., et al. Targeting neuronal and glial cell types with synthetic promoter AAVs in mice, non-human primates and humans. Nat. Neurosci. 2019;22:1345–1356. doi: 10.1038/s41593-019-0431-2. [DOI] [PubMed] [Google Scholar]
- 154.Yardeni T., Eckhaus M., Morris H.D., Huizing M., Hoogstraten-Miller S. Retro-orbital injections in mice. Lab Anim. 2011;40:155–160. doi: 10.1038/laban0511-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Holtmaat A., Bonhoeffer T., Chow D.K., Chuckowree J., De Paola V., Hofer S.B., Hübener M., Keck T., Knott G., Lee W.-C.A., et al. Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nat. Protoc. 2009;4:1128–1144. doi: 10.1038/nprot.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lun A.T.L., Riesenfeld S., Andrews T., Dao T.P., Gomes T., participants in the 1st Human Cell Atlas Jamboree. Marioni J.C. EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 2019;20:63. doi: 10.1186/s13059-019-1662-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wolock S.L., Lopez R., Klein A.M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 2019;8:281–291.e9. doi: 10.1016/j.cels.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Allen Reference Atlas - Developing Mouse Brain [brain atlas] http://atlas.brain-map.org/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video was taken after 5 h of para-uterine imaging and 10 min after the disruption of the umbilical cord
Sequentially showing imaging of GCaMP6s in somas of Rbp4-Cre neurons at E14.5 (10X real-time), neurites of Rbp4-Cre neurons at E14.5 (15X real-time), somas and neurites of Rbp4-Cre neurons at E15.5 (15X real-time), somas of Rbp4-Cre neurons at E18.5 (30X real-time), and neurites of Rbp4-Cre neurons at E18.5 (15X real-time). Motion correction was applied to the video, and the fire colormap in FIJI was used to visualize changes. Arrowheads: calcium events
Data Availability Statement
-
•
Single-cell RNA-sequencing data have been deposited at the NCBI Sequence Read Archive (with processed count tables in Mendeley Data), and both datasets are publicly available as of the date of publication. Accession numbers are listed in the key resources table. All microscopy data and other recordings reported in this paper will be shared by the lead contact upon request.
-
•
All original code has been deposited at Zenodo and is publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.