Abstract
Eukaryotic mRNAs are modified at the 5′ end with a methylated guanosine (m7G) that is attached to the transcription start site (TSS) nucleotide. The TSS nucleotide is 2′-O-methylated (Nm) by CMTR1 in organisms ranging from insects to human. In mammals, the TSS adenosine can be further N6-methylated by RNA polymerase II phosphorylated CTD-interacting factor 1 (PCIF1) to create m6Am. Curiously, the fly ortholog of mammalian PCIF1 is demonstrated to be catalytic-dead, and its functions are not known. Here, we show that Pcif1 mutant flies display a reduced fertility which is particularly marked in females. Deep sequencing analysis of Pcif1 mutant ovaries revealed transcriptome changes with a notable increase in expression of genes belonging to the mitochondrial ATP synthetase complex. Furthermore, the Pcif1 protein is distributed along euchromatic regions of polytene chromosomes, and the Pcif1 mutation behaved as a modifier of position-effect-variegation (PEV) suppressing the heterochromatin-dependent silencing of the white gene. Similar or stronger changes in the transcriptome and PEV phenotype were observed in flies that expressed a cytosolic version of Pcif1. These results point to a nuclear cotranscriptional gene regulatory role for the catalytic-dead fly Pcif1 that is probably based on its conserved ability to interact with the RNA polymerase II carboxy-terminal domain.
Keywords: Pcif1, m6Am, m6A, N6-methyladenosine, position-effect variegation (PEV), transcription
INTRODUCTION
RNA polymerase II transcripts are capped at the 5′ end with a 7-methylguanosine (m7G) moiety which is attached via an inverted 5′–5′ triphosphate bridge to the transcription start site (TSS) nucleotide (Shatkin 1976). This cap0 structure is seen in lower eukaryotes like yeast. In organisms ranging from insects (Haussmann et al. 2022) to human (Belanger et al. 2010), the TSS nucleotide is 2′-O-methylated (Nm) on the ribose by CMTR1 to create the cap1 structure (Inesta-Vaquera and Cowling 2017). In viral and mammalian RNA caps, the TSS adenosine (Am) can be further methylated at the N6 position to form the dimethylated m6Am (Wei et al. 1975; Moyer and Banerjee 1976). The activity responsible for this m6A methylation was partially purified from human HeLa cell extracts (Keith et al. 1978), and 40 yr later identified to be the RNA polymerase II phosphorylated CTD-interacting factor 1 (PCIF1) (Akichika et al. 2019).
PCIF1 was originally identified as a nuclear protein that uses its amino-terminal WW domain to interact with the phosphorylated carboxy-terminal domain (CTD) of RNA polymerase II (Fan et al. 2003). PCIF1 interacts with the Ser5-phosphorylated CTD of the RNA Pol II, suggesting a recruitment to the nascent RNA early during transcription (Akichika et al. 2019; Boulias et al. 2019; Sendinc et al. 2019). Presence of the m6Am modification on the RNA cap is associated with RNA stability in human cells (Boulias et al. 2019) and mouse testes (Pandey et al. 2020). Additionally, it is shown to also regulate cap-dependent translation (Akichika et al. 2019; Sendinc et al. 2019). We previously reported that mice lacking Pcif1 are viable and fertile, with no dramatic phenotypes, except a slightly reduced body weight (Pandey et al. 2020). Interestingly, insect Pcif1 orthologs have mutations in the catalytic motif that are predicted to render them inactive as RNA methylases. We demonstrated that purified Drosophila Pcif1 is inactive as a methylase in vitro, but it retained a WW domain capable of specifically interacting with Ser5-phosphorylated CTD of RNA polymerase II (Pandey et al. 2020). This makes the fly system a unique model to investigate the noncatalytic role of Pcif1.
In this study, we generated and analyzed two independent fly mutants of Pcif1 (CG11399) to examine the in vivo role of Drosophila melanogaster Pcif1. Our results show that the protein acts in the nucleus to regulate levels of hundreds of transcripts and is required for female fertility and to a lesser extent for male fertility. We also observed that Pcif1 localized to polytene chromosome interbands known to be transcriptionally active sites and open euchromatin regions. Moreover, we observed that loss of the nuclear Pcif1 protein modified position-effect variegation (PEV) by suppressing the chromatin-dependent silencing of the white (w) gene in wm4h mutants. We propose a model where nuclear catalytic-dead fly Pcif1 associates with RNA Pol II CTD to regulate gene expression.
RESULTS
Fly Pcif1 mutants display reduced body weight and decreased fertility
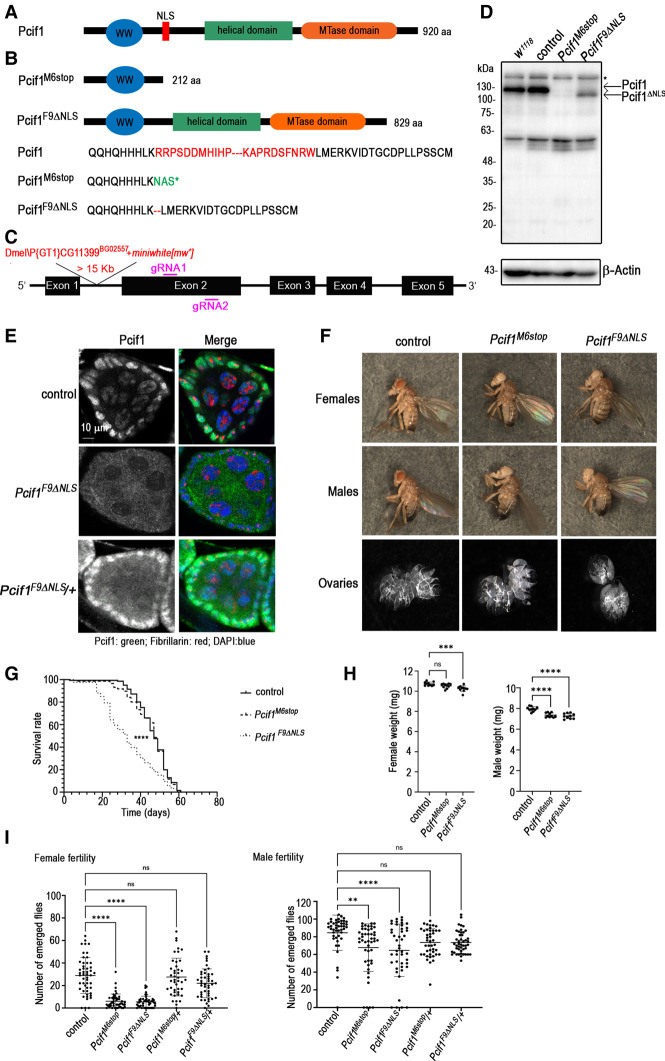
Drosophila Pcif1 is a multidomain protein of 920 amino acids with a predicted amino-terminal WW domain and a carboxy-terminal methyltransferase domain (Fig. 1A). A central helical domain homologous to an alpha helix-rich region in the crystal structure of the human PCIF1 (Akichika et al. 2019) is indicated. A putative nuclear localization signal (NLS) is also identified between the WW and helical domains.
FIGURE 1.
Molecular characterization of Pcif1 in Drosophila melanogaster. (A) Structural organization of the wild-type Pcif1 protein. The amino-terminal WW domain, the nuclear localization signal (NLS), the helical domain, and the carboxy-terminal methyl-transferase (MTase) domain are shown. (B) The CRISPR–Cas9 mutagenesis generated the Pcif1M6stop allele, which causes a frameshift ending in a premature stop codon generating a truncated protein and the Pcif1F9ΔNLS allele which has an in-frame deletion of the NLS. (C) Illustration of the Pcif1 locus with the indication of the P element insertion site in the Pcif1BG02557 line (Dmel\PCG11399BG02557), here used as control, and of the position of the sgRNAs, gRNA1, and gRNA2 designed for the CRISPR–Cas9 mutagenesis (purple lines). (D) Western blot analysis of head protein extracts from Pcif1BG02557 control, or Pcif1M6stop, and Pcif1F9ΔNLS mutants as indicated. Wild-type Pcif1 and Pcif1ΔNLS protein sizes are indicated by the black arrows. β-Actin was used as loading control. The asterisks indicate unspecific bands. (E) Immunostaining of ovaries dissected from Pcif1BG02557 controls, Pcif1F9ΔNLS homozygous, or Pcif1F9ΔNLS/+ heterozygous females. The egg chamber, formed by an external layer of follicle cells (somatic cells) that surround the nurse cells (germ cells), is shown. Pcif1: green, DNA: blue, Fibrillarin (nucleolar marker): red. (F) Pictures of 5-d-old females, males, or dissected ovaries of either Pcif1BG02557 (control) or Pcif1M6stop and Pcif1F9ΔNLS mutants. (G) Longevity analysis of Pcif1BG02557 control (n = 150) or Pcif1M6stop (n = 150) and Pcif1F9ΔNLS (n = 130) mutants. (H) Weight analysis of Pcif1BG02557 control or Pcif1M6stop and Pcif1F9ΔNLS mutants: each dot represents the weight of a group of 10 adult flies per genotype for either males or females as indicated. Ten replicates were performed. (I) Fertility analysis of females or males expressed as the number of emerging flies per cross of one individual of the indicated genotype with three wild-type flies of the opposite sex. Log-rank (longevity) and one-way ANOVA (weight and fertility) tests were performed using GraphPad Prism9. P-value: (***) P < 0.001, (****) P < 0.0001. ns: nonsignificant.
To investigate the physiological role of Drosophila Pcif1, we used the CRISPR–Cas9 genome engineering tool to generate a genomic deletion in the region encoding for the NLS in the w1118 background. Two mutated alleles were identified: Pcif1M6stop and Pcif1F9ΔNLS. The deletion in the Pcif1M6stop allele induces a frameshift ending in a premature stop codon (Fig. 1B). On the other hand, the Pcif1F9ΔNLS allele has an in-frame deletion of the NLS, generating a protein of 829 amino acids (Fig. 1B). To clean up any potential off-target mutations, the selected alleles were backcrossed for up to 10 generations with a fly line carrying a P-element inserted in the first intron of Pcif1 (P{GT1}CG11399BG02557), also referred to as the Pcif1BG02557 line (Fig. 1C). This P-element contains a miniwhite [mw+] gene that confers a red eye color to the flies in a w1118 context, while the Pcif1 CRISPR/Cas9 mutants generated in a w1118 background have white eyes, thus allowing to trace the presence of the Pcif1 mutations along generations (see Materials and Methods). Thus, the two Pcif1 mutant alleles used for our studies are in the w1118; Pcif1BG02557 background, with the original Pcif1BG02557 line hereafter referred to as control. Western blot analysis of fly head lysates indicated that the control flies expressed the Pcif1 protein at similar levels as the w1118 strain, while the absence of detectable signal in the homozygous Pcif1M6stop flies confirmed these to be null mutants (Fig. 1D). In contrast, flies homozygous for the Pcif1F9ΔNLS allele expressed a slightly smaller protein, which is consistent with the deletion of the NLS sequence. Immunofluorescence analysis showed that the wild-type protein was nuclear in the control fly ovaries, while the Pcif1ΔNLS protein failed to enter the nucleus and was consequently stranded in the cytoplasm (Fig. 1E). In heterozygous, Pcif1ΔNLS/+ ovaries, immunostaining revealed the presence of the Pcif1 protein in both the cytoplasmic and the nuclear compartments, most probably corresponding to the mutated and wild-type proteins, respectively (Fig. 1E).
Adult Pcif1M6stop mutant flies did not show any obvious morphological defects (Fig. 1F) and displayed normal longevity (Fig. 1G). However, about 20% of eggs issued from crossing Pcif1M6stop mutant siblings did not hatch, most probably due to the lack of egg maturation or fertilization (white eggs) (Supplemental Fig. S1A). We also observed that Pcif1M6stop males, although not the females, had a significantly reduced body weight (Fig. 1H). Further investigation revealed that homozygous Pcif1M6stop mutants showed a significantly reduced fertility that was particularly strong in females (Fig. 1I), although examination of dissected fly ovaries did not reveal any gross morphological differences (Fig. 1F). Hemizygous females carrying the Pcif1M6stop allele over a deficiency covering the Pcif1 locus similarly displayed a strongly decreased fertility (Supplemental Fig. 1B).
Interestingly, Pcif1F9ΔNLS mutant flies displayed similar or even stronger phenotypes compared to Pcif1M6stop mutants. Both Pcif1F9ΔNLS homozygous males and females displayed a reduced body weight and a significantly decreased fertility, which was, as observed for Pcif1M6stop, particularly strong in females compared to a moderate effect in males (Fig. 1H,I; Supplemental Fig. 1B). In contrast to Pcif1M6stop, the Pcif1F9ΔNLS mutants had a significantly reduced longevity (Fig. 1G). Enhanced physiological defects observed in Pcif1F9ΔNLS flies indicate that the cytoplasmic protein cannot compensate for the lack of Pcif1 nuclear protein. Moreover, the Pcif1 protein may have a deleterious effect when present in the cytoplasm in the absence of the nuclear protein.
Taken together, these results show that Pcif1 mutants are viable but display physiological defects such as reduced body weight and decreased fertility. Female fertility decrease was notably observed in the two independent alleles outcrossed for 10 generations with the control line, as well as in trans-heterozygous mutants over a deficiency. Thus, this phenotype can be attributed to the lack of nuclear Pcif1 rather than to a genetic background effect.
Loss of fly Pcif1 up-regulates transcripts involved in mitochondrial ATP synthesis
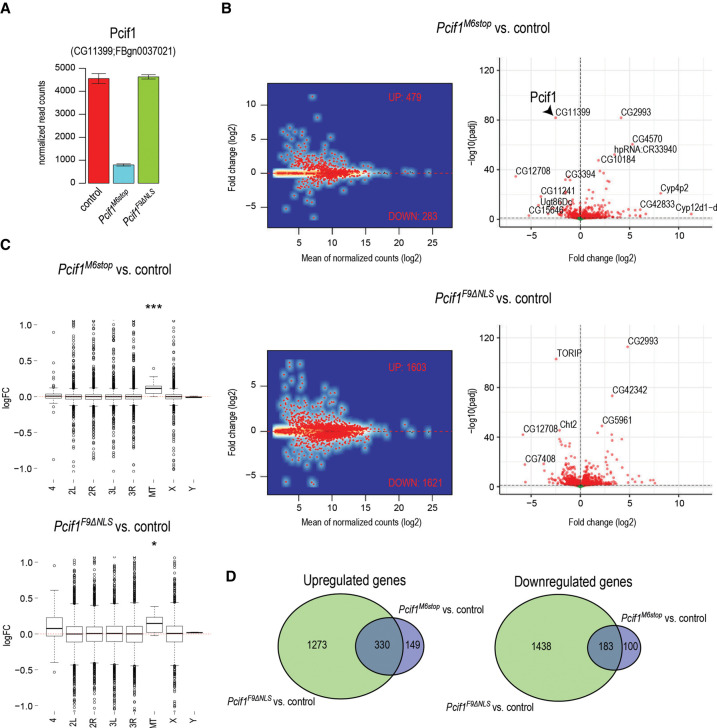
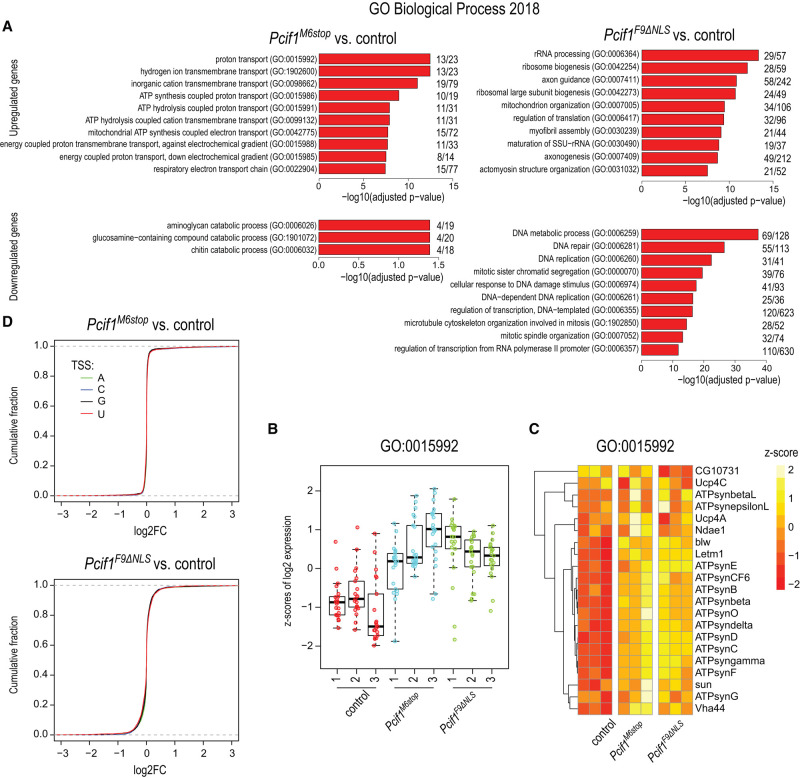
To understand the female fertility defect, we sequenced the mutant ovary transcriptome. We prepared total ovary RNA-seq libraries from 5-d-old homozygous mutant Pcif1M6stop, homozygous mutant Pcif1F9ΔNLS, and control female flies. Consistent with complete absence of the protein, there was a strong reduction in Pcif1 transcript levels in the null mutant where the premature stop codon may induce degradation of the transcript by nonsense mediated decay (NMD) (Baker and Parker 2004), while the levels were unchanged in the Pcif1F9ΔNLS mutant (Fig. 2A). The levels of several hundred transcripts were altered, with ∼500 transcripts being up-regulated and ∼250 transcripts down-regulated in the loss-of-function Pcif1M6stop mutant (Fig. 2B; Supplemental Table S1). The altered genes are distributed across different chromosomes, with a slight overall up-regulation of transcripts from the mitochondrial genome (Fig. 2C). Interestingly, gene ontology analysis showed that nuclear encoded genes involved in proton transport and mitochondrial ATP synthesis were up-regulated in the Pcif1M6stop mutant (Fig. 3A; Supplemental Table S1).
FIGURE 2.
Transcriptomic analyses in Pcif1 mutants. (A) Barplot showing reduced transcript levels of Pcif1 in Pcif1M6stop while the transcript levels in Pcif1F9ΔNLS are not affected. Data shown as mean ± standard deviation from three biological replicas. (B) MA plots and volcano plots showing gene expression changes in Pcif1M6stop and in Pcif1F9ΔNLS. Significantly dysregulated genes (Padj ≤ 0.1) are shown in red. (C) Boxplots showing gene expression changes from individual chromosomes. The view is limited to only changes below twofold to better see the median differences (P-value: [*] P < 0.05, [***] P < 0.001; wilcox.text of symmetric distribution). (D) Venn diagrams showing overlap in sets of up-regulated and down-regulated genes in Pcif1M6stop and Pcif1F9ΔNLS.
FIGURE 3.
Pcif1 regulates gene expression of proton transport genes. (A) Gene ontology analysis for differentially expressed genes. Only the top 10 significantly enriched terms are shown. (B) Boxplot showing the z-scores of expression across individual samples for proton transport genes (GO:0015992). (C) Heatmap showing the z-scores of expression for proton transport genes (GO:0015992). (D) Plots showing the cumulative distributions of gene expression changes in the Pcif1M6stop and in Pcif1F9ΔNLS when compared to the control. Separate curves are plotted based on the transcription start site nucleotide (TSS).
Next, we used the homozygous Pcif1F9ΔNLS mutant ovary transcriptome to examine if the observed transcript changes in the null mutant could be attributed to potential nuclear functions of Pcif1. The RNA sequence analysis from Pcif1F9ΔNLS mutant ovaries revealed a much larger disruption of around 3000 transcripts, with an equal number being up- and down-regulated (Fig. 2B; Supplemental Table S1). We attribute this larger disruption in the transcriptome to potential deleterious effects of the mislocalized Pcif1ΔNLS protein in the cytoplasm (see above and Fig. 1E). Significantly, a large part of the altered ovarian transcriptome in the Pcif1M6stop mutant overlapped with that seen in the Pcif1F9ΔNLS mutant (Fig. 2D). Among this shared set of altered genes are those involved in proton transport and mitochondrial ATP synthesis, and these were uniformly up-regulated in both the null and the Pcif1F9ΔNLS mutant fly ovaries (Fig. 3B,C; Supplemental Table S1).
One of the transcripts up-regulated in Pcif1M6stop and Pcif1F9ΔNLS mutant ovaries encodes the α subunit of the mitochondrial F1F0 ATP synthase of complex V (ATP5A synonym bellwether [blw] in Drosophila). Western blot analyses of protein extracts isolated from ovaries revealed that the ATP5A protein was however not significantly up-regulated (Supplemental Fig. S2A). No obvious defects were detected on the mitochondrial network of the Pcif1M6stop null mutant by electron microscopy, although we cannot exclude subtle morphological or functional differences at the level of this analysis. In the case of the Pcif1F9ΔNLS mutant, we observed some enlarged mitochondria that may however result from the ectopic localization of the protein in the cytoplasm rather than the absence of the nuclear Pcif1 protein. For this reason, a putative effect would not reflect a physiological role of Pcif1and was not further investigated (Supplemental Fig. S2B).
Mammalian PCIF1 targets m7G-capped RNA substrates with an adenosine as the transcription start site (TSS) nucleotide for cotranscriptional catalysis of the m6Am modification, and such modified transcripts are preferentially stabilized in cell culture and mouse testes (Boulias et al. 2019; Pandey et al. 2020). However, we did not find any influence of the TSS nucleotide in regulating transcript levels in the mutant fly ovaries (Fig. 3D). In conclusion, our results show that although catalytically inactive, fly Pcif1 has a functional role in regulating gene expression, and this depends on its nuclear localization. We propose that this collective regulation of a few hundred transcripts (either direct or indirect) may contribute to overall fly fertility.
Pcif1 binds to euchromatic regions along polytene chromosomes
Drosophila Pcif1 lacks the m6Am RNA methylation activity found in its mammalian counterpart, but as observed for the mammalian protein, Drosophila Pcif1 is nuclear and has the ability to interact in vitro with the Ser5-phosphorylated carboxy-terminal domain (CTD) of RNA polymerase II (Pandey et al. 2020). Our experiments above point to a role for Pcif1 in gene regulation, but the mechanism of action is unclear. To obtain further evidence for this nuclear role, we performed Pcif1 immunostaining on larval polytene chromosomes. These experiments showed that Pcif1 localized to polytene chromosome interbands, which are known to contain other proteins characteristic of open euchromatin and to be transcriptionally active regions (Fig. 4A). We propose that Pcif1, in association with RNA Pol II, contributes to gene expression regulation at transcriptionally active sites.
FIGURE 4.
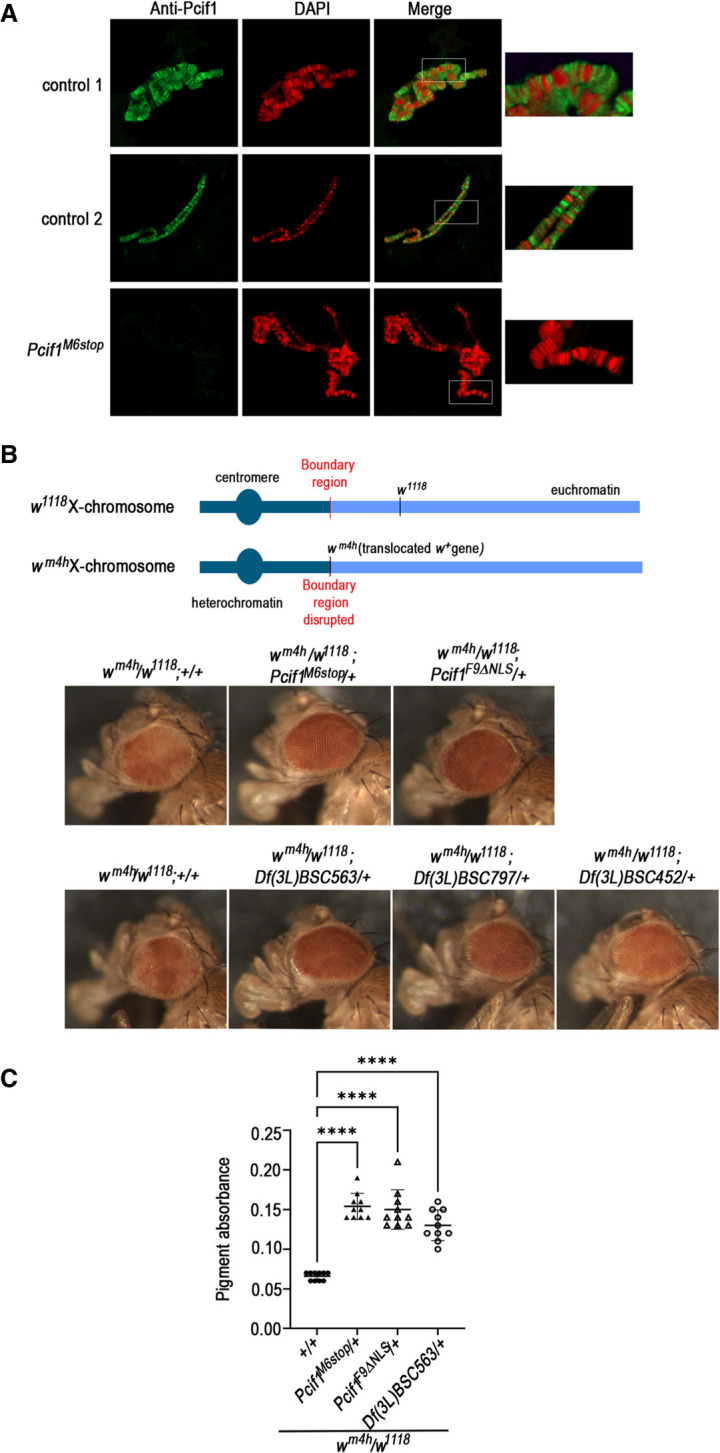
Pcif1 binds to euchromatic regions of polytene chromosomes and behaves as a suppressor of position-effect variegation. (A) Larval polytene chromosome immunostaining was performed for Pcif1BG02557 controls (two samples) and Pcif1M6stop mutants as indicated. (B) PEV assay. Schematic illustration of the position-effect variegation affecting the white gene in the wm4h allele. Representative eye phenotype out of 20 to 50 females of the following genotypes: wm4h/w1118; Pcif1M6stop/+ or wm4h/w1118; Pcif1F9ΔNLS/+ compared to control wm4h/w1118; +/+ flies (first row) and wm4h/w1118; D(3L)/+ female flies carrying a chromosomal deletion encompassing the Pcif1 gene [Df(3L)BSC563, Df(3L)BSC797, or Df(3L)BSC452 as indicated on figure] compared to wm4h/w1118; +/+ flies (second row). (C) Quantification of pigment absorbance was performed for five heads per genotype at a wavelength of 480 nm. Ten replicates were performed. One representative experiment out of two is shown. One-way ANOVA test was performed using GraphPad Prism9. P-value: (****) P < 0.0001.
Loss of Pcif1 suppresses position-effect variegation
The detection of Pcif1 at transcriptionally active euchromatic sites prompted us to use the sensitive position effect variegation (PEV) assay to investigate further whether loss of Pcif1 has functional effects on gene regulation. In this experiment, a variegated eye phenotype is induced by the wm4h allele, where the wild-type white gene (w+) that specifies red eye color is translocated from its normal location in an euchromatin region on the X chromosome to a pericentric heterochromatin zone (Reuter and Wolff 1981; Wang et al. 2014). This induces a clonally inherited epigenetic silencing of the w+ gene that results in a variegated eye due to the different levels of expression of the gene in individual photoreceptors that constitute the compound eye in flies. This translocation renders White protein expression and subsequent eye coloration very sensitive to subtle changes in transcription of the wm4h allele.
As expected, we observed that hemizygous wm4h/w1118 female flies displayed a variegated eye phenotype instead of the uniform red eye observed in w+/w+ females (Fig. 4B). When placed in the heterozygous Pcif1M6Stop/+ background, we observed a significant suppression of PEV, resulting in a uniformly dark red eye color (Fig. 4B,C). We confirmed that this is specifically due to loss of nuclear action of Pcif1, as an identical observation was made in the heterozygous Pcif1F9ΔNLS/+ background (Fig. 4B,C). As an additional support, we showed that a similar suppression of PEV was induced in a different genetic background where flies carried a large genomic deletion covering the Pcif1 locus at the hemizygous state (wm4h/+;Df(3L)BSC563/+) (Fig. 4B,C). Moreover, two other independent genomic deletions (Df(3L)BSC797/+ and Df(3L)BSC7452/+) encompassing the Pcif1 gene locus also displayed similar suppression of wm4h silencing, although with a lower penetrance in the case of Df(3L)BSC452/+ (Fig. 4B).
These results show that, in addition to being present at transcriptionally active euchromatic sites, Pcif1 participates directly or indirectly in the epigenetic phenomenon of PEV.
DISCUSSION
Our genetic analyses revealed that fly Pcif1 is not essential for life and normal development, while a strong and significant reduction in female fertility was observed in the two mutant alleles generated in this study. RNA-seq analysis showed that absence of fly Pcif1 resulted in the alteration of hundreds of transcripts. A set of up-regulated transcripts in Pcif1 mutants ovaries corresponded to nuclear genes encoding mitochondrial proteins involved in the respiratory chain. Functional respiratory chain components are required in the late germarium for the proper replication of mitochondrial DNA (mtDNA) and normal mitochondrial biogenesis (Wang et al. 2019). We detected neither significant changes in the expression level of one of these mitochondrial proteins (ATP5A) by western analysis nor morphological defects in the mitochondrial network of Pcif1M6stop; however, in Pcif1F9ΔNLS some mitochondria appeared slightly enlarged, which might be a consequence of Pcif1 mislocalization while not reflecting the physiological function of Pcif1. Nevertheless, because the observed transcript up-regulation was only around 20%, this difference might be difficult to detect at the protein level by western analysis, and we may still speculate that subtle alteration of mitochondrial function may prevent proper egg maturation or development (Hill et al. 2014; Wang et al. 2019).
Analysis of another fly mutant where the nuclear localization signal (NLS) in Pcif1 is specifically deleted showed an even larger number of transcript changes. We propose that this is a compounded effect of both direct loss of Pcif1-mediated nuclear functions and a potential deleterious effect of an RNA-binding protein that is stranded in the cytoplasm. In fact, compared to the Pcif1 null and control flies which have similar life spans, the Pcif1F9ΔNLS flies have a strikingly reduced longevity. This is likely due to the potential toxicity caused by the cytosolic mislocalization of the protein.
This leads to the most important question of how the catalytic dead Pcif1 protein might directly regulate gene expression in the nucleus. Hypophosphorylated RNA Pol II is recruited to gene promoters, which at initiation of transcription becomes phosphorylated on Ser5 within the consensus heptapeptide YSPTSPS repeats in the CTD of the largest subunit of the RNA Pol II (Buratowski 2009). This modification helps to recruit RNA capping enzymes that use WW domains to interact with the phosphorylated CTD (Haline-Vaz et al. 2008; Ghosh et al. 2011; Galloway and Cowling 2019). Later during elongation, phosphorylation at Ser2 helps to recruit 3′ end processing factors (Licatalosi et al. 2002; Meinhart and Cramer 2004) and histone methyltransferases (Hampsey and Reinberg 2003), and this is accompanied by dephosphorylation at Ser5. Thus, the dynamic phosphorylation of the RNA Pol II CTD can impact gene expression at multiple steps. Human PCIF1 (Akichika et al. 2019; Boulias et al. 2019; Sendinc et al. 2019; Pandey et al. 2020) and fly Pcif1 (Pandey et al. 2020) both interact with Ser5-phosphorylated CTD in vitro. Consistently, we observed that Pcif1 localized along the polytene chromosomes at the level of transcriptionally active sites. Human PCIF1 overexpression is shown to regulate transactivation of gene reporters and this is dependent on its ability to interact with the CTD (Hirose et al. 2008). Interestingly, the interaction of the recombinant WW domain of human PCIF1—that lacks any catalytic activity—inhibits the dephosphorylation of Ser5 in the RNA Pol II CTD in vitro (Hirose et al. 2008), suggesting a potential mechanism by which the fly protein may influence gene expression by similarly interfering with RNA Pol II phosphorylation status. Moreover, our study demonstrated that loss of Pcif1 or mislocalization of Pcif1 to the cytoplasm influenced the epigenetic phenomenon of position effect variegation (PEV). We therefore propose that the nuclear effects of fly Pcif1 are dependent on its WW domain and its ability to interact with Ser5-phosphorylated CTD of RNA Pol II at transcriptionally active sites. In fact, fly Pcif1 was found in immunoprecipitated nuclear complexes consisting of the RNA Pol II subunits and the cap-binding complex (CBP) that binds nascent RNA (Kachaev et al. 2019). Evidently, such a gene regulatory role would be cotranscriptional. Since we did not find any enrichment of genes with a TSS adenosine in these altered gene sets, other mechanisms might specify such targets.
The multifunctionality of RNA modification writer enzymes is a topic that is gaining increasing attention. The focus so far has been on the catalytic activity and the impact these modifications have on gene regulation (Roignant and Soller 2017). Nevertheless, several studies point to noncatalytic roles for some of the major RNA methyltransferases active in human cells. The nuclear m6A writer METTL3 is shown to have a cytosolic role in enhancing translation by promoting formation of the mRNA closed loop structure (Choe et al. 2018). Another nuclear mammalian m6A writer METTL16 methylates nuclear U6 snRNA (Pendleton et al. 2018), but METTL16 is also shown to have a cytosolic function (Nance et al. 2020) as a translation factor (Su et al. 2022). While mammalian PCIF1 is known as an m6Am writer (Akichika et al. 2019), in this study we use the naturally catalytic-dead fly Pcif1 to describe its noncatalytic role in nuclear gene regulation to ensure female fertility and as a modulator of epigenetic processes.
MATERIALS AND METHODS
Generation of Pcif1 mutant flies
The Drosophila melanogaster Pcif1 ortholog (Pcif1, CG11399) was targeted with two gRNAs designed to delete the putative nuclear localization signal (NLS) at the amino terminus of the protein (Fig. 1A). The sgRNAs were identified with the “CRISPR Optimal Target Finder” website (http://tools.flycrispr.molbio.wisc.edu/targetFinder/). They are located within the CDS at the following positions:
chr3L:20783975 (AGCATCATCACCTGAAGCGG, sgRNA1)
chr3L:20783692 (CCTTGCGTTCCATCAGCCAG, sgRNA2)
The two sgRNAs were cloned separately into the vector pU6-BbsI-chiRNA using oligonucleotides RRoligo1161 + RRoligo1162 for sgRNA1 and RRoligo1163 + RRoligo1164 for sgRNA2 (Supplemental Table S1). The positive clones were PCR identified and sequenced to confirm the presence of correct sgRNA sequences.
The resulting plasmids were injected into ∼200 embryos of a nos-Cas9 transgenic strain (genotype: yw;attP40[nos-Cas9]/CyO, Bloomington Drosophila Stock Center #54594) by BestGene.
The adult founder males (G0) were crossed with virgin w/w; MKRS/TM6 females. Ten G1 males emerging from each G0 cross were then individually crossed with five virgin females’ w/w; MKRS/TM6 for 3 d at 25°C, and each male was collected for PCR genotyping using primers CG11399_PCR2_F and CG11399_PCR2_R that flank the targeted region (Supplemental Table S1). This gave PCR products indicative of alleles with either deletion (∼300 bp) or the wild-type allele (582 bp). The deleted PCR products were further characterized by Sanger sequencing which leads to the identification of two independent deletions of the targeted sequence (alleles Pcif1F9ΔNLS, Pcif1M6stop) (Fig. 1B).
In order to remove putative mutations induced by the CRISPR–Cas9 mutagenesis from the genome of Pcif1 mutants, the Pcif1 alleles were outcrossed using the Dmel\P{GT1}CG11399BG02557 (Pcif1BG02557, here referred to as the control) fly line. This fly line carries a P element containing a miniwhite [mw+] gene inserted in the first intron of the Pcif1 gene (Fig. 1C) in a w1118 context, and showed no phenotypic defects. It was used to backcross the Pcif1 mutant alleles for five (longevity assay) to 10 generations (weight monitoring, fertility analysis and PEV assay). Therefore, except if otherwise indicated (PEV assay with wmh4, see below), the phenotypic analysis of Pcif1F9ΔNLS, Pcif1M6stop alleles was performed in the w1118 background of the P{GT1}CG11399BG02557 line.
Other fly lines used in this study
Three deficiency fly lines carrying three different chromosomal deletions encompassing Pcif1 locus on the third chromosome were used in this study to observe the variegation eye phenotype. These lines were provided by the Bloomington Drosophila Stock Center (BDSC):
25721 w[1118]; Df(3L)BSC563/TM6C, cu[1] Sb[1] (Df(3L)BSC563);
27369 w[1118]; Df(3L)BSC797/TM6C, Sb[1] cu[1] (Df(3L)BSC797);
24956 w[1118]; Df(3L)BSC452/TM6C, Sb[1] cu[1] (Df(3L)BSC452).
The wm4h fly line was provided by Dr. C. Grimaud (Montpellier).
Hatching rate analysis
Fifty adult females were mated with males for 24 h at 25°C on media allowing egg collection (335 mL H2O, 15 g Agar, 165 mL grape juice and 8 g sucrose). One-hundred eggs issued from each cross were transferred on fresh media and incubated at 25°C for 24 h. The number of unhatched eggs was counted. Three independent replicates were performed for each cross.
Fertility analysis
Fifty males or virgin females were collected and crossed individually with three wild-type w1118 flies of the opposite sex at 25°C. After a mating period of 2 d, they were transferred to a new vial for 5 d and discarded. The emerging flies were counted and the statistical analysis was performed using the one-way ANOVA test in GraphPad Prism9.
Longevity analysis
A total of 100 to 150 males of the same age were collected and raised at 25°C per group of 10 flies per vial. Flies were flipped into fresh vials every 2–3 d for the duration of the experiment. The number of dead flies was counted every 2 d. The statistical analysis was performed using the Log-rank (Mantel-Cox) test in GraphPad Prism 9.
Weight analysis
The larval density was here controlled by transferring 50 L1 larvae in a new vial. The resulting males and females were weighted by a group of 10 flies 2–3 d after emergence on a Mettler Toledo Classic precision balance. Ten replicates were performed for each sex and genotype. The statistical analysis was performed using the one-way ANOVA test in GraphPad Prism9.
Immunofluorescence analysis
Five to ten ovary pairs were dissected from control, Pcif1F9ΔNLS, Pcif1M6stop, or Pcif1F9ΔNLS/+ females in PBS. They were immediately fixed in 4% PFA for 30 min at room temperature (RT). Fixed tissues were then washed (3 × 5 min) with 1× PBS, permealized for 5 min with PBT (1xPBS containing 0.1% Triton X-100) at RT and blocked in blocking buffer (1× PBS, 0.1% Triton X-100, 5% horse serum) for 1 h at RT. They were incubated with rabbit anti-Pcif1 (1:100) (Pandey et al. 2020) and mouse anti-fibrillarin (1:100; AbCam ab4566) antibodies in blocking buffer overnight at 4°C. They were then washed (3 × 15 min) in PBT at RT and incubated for 4 h with anti-rabbit conjugated to Alexa 488 (1:500; Invitrogen, cat. no. A11029), anti-mouse conjugated to Cyanine 3 (1:500; Jackson ImmunoResearch, cat. no. 111165144) antibodies, and Hoechst (1:1000; Thermo Fisher, cat. no. 33342) in blocking buffer at RT. Ovaries or fat bodies were then washed in PBT (3 × 10 min) at RT and stored in PBS before mounting in Dako (cat. no. S3022). Images were captured with The Nikon A1RSi confocal microscope (CEA, Grenoble).
Western blot
For analyzing Pcif1 expression, 10 heads from 5-d-old males were dissected for western blot analysis. To check the expression of the ATP5A mitochondrial marker, 10 pairs of ovaries from female adult flies were dissected. Samples were lysed in 100 µL of 1× Laemmli buffer (Bio-Rad, cat. no. 1610747) containing 1× β-mercaptoethanol (Sigma, cat. no. 07917KE). The lysates were boiled at 95°C for 5 min and centrifuged at 12,000 rpm at RT for 5 min. An amount of 30 µL of each sample was loaded on 7.5% Mini-PROTEAN TGX (Bio-Rad, cat. no. 456-8025). Gel electrophoresis was performed at 70 V for 200 min. After separation, the proteins were transferred on an Immobilon-P 0.45 µm nitrocellulose membrane (Merck Millipore, cat. no. IPHV00010) overnight at 5 V at RT using the Trans-Blot SD Semi-Dry Transfer Cell system (Bio-Rad, cat. no. 1703940). After the transfer, the membrane was washed in Tris-buffered saline (TBS, 20 mM Tris, 150 mM NaCl, pH 7.6) and blocked for 1 h at room temperature with 5% fat free milk in TBS with 0.05% Tween20 (TTBS) (Sigma, cat. no. P7949). The blocked membranes were incubated with rabbit anti-Pcif1 (1:1000) (Pandey et al. 2020) for 3 h at room temperature in 5% milk with TTBS, or with anti-mouse ATP5A (1:1000, AbCam ab14748) overnight at 4°C. Then, membranes were washed three times for 10 min with TTBS and incubated with HRP-conjugated secondary antibody at either 1:10,000 dilution with anti-rabbit IgG HRP-linked or at 1:2000 dilution with anti-mouse IgG HRP-linked for 1 h at RT in 5% milk in TTBS. The membranes were washed three times for 10 min with TTBS and incubated with SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher, cat. no. 34095) for 5 min at RT or with Immobilon Forte Western HRP Substrate (Merck Millipore, cat. no.WBLUF0500) for 15 min at RT. The signal was detected using Molecular Imager ChemiDoc XRS+ (Bio-Rad, cat. no. 252559) with ImageLab software (v 5.1).
Transcriptome analyses
Ovaries were dissected from 5-d-old homozygous Pcif1M6stop, homozygous Pcif1F9ΔNLS, and control female flies. Three pairs of ovaries were collected per line for RNA extraction with TRIzol reagent. Ribozero-depleted deep sequencing libraries were prepared and sequenced as previously described (Pandey et al. 2020). Specifically, the NEBNext Ultra kit (NEB: E7760L kit) was used to prepare the paired-end libraries which were sequenced in high-mode on Next-seq 500 at GeneCore, EMBL.
Reads were sorted into individual libraries based on the barcodes and mapped to the fly transcriptome (Ensembl release 95) using salmon v1.3.0 (salmon quant with options -l A –validateMappings –gcBias) (Patro et al. 2017). Further analysis was performed using R version 3.6.3 and Bioconductor (Huber et al. 2015). The DESeq function of DESeq2_1.25.10 bioconductor package (Love et al. 2014) was used to obtain log2 fold changes of gene expression between control and mutant samples and the adjusted P-values. An adjusted P-value of 0.1 was used as a threshold for statistical significance. In MA plots the genes with significantly different expression between the mutant and control samples were highlighted in red (Fig. 2B). The sets of genes up-regulated and down-regulated in the mutant ovaries were compared using Venn diagrams (VennDiagram 1.6.20) (Fig. 2D). The Volcano plots were plotted using the EnhancedVolcano function from the EnhancedVolcano 1.3.5 package (https://github.com/kevinblighe/EnhancedVolcano). To see whether whole chromosome expression might be affected, we produced boxplots of log2 fold expression changes for genes lying on individual chromosomes with the y-axes limited to <−1,1> to better see whether the median fold change deviates from zero (Fig. 2C). The wilcox.test was used to test the symmetric distribution of log2 fold changes for each chromosome. To see whether the transcript starting nucleotide affects the overall gene expression changes, we filtered only genes that produce transcripts with a specific starting nucleotide and divided them into the groups based on that nucleotide. Using stats:::ecdf, we then computed the empirical cumulative distribution function on log2 fold changes between the mutant and control samples and plotted the distribution (Fig. 3D). The group of genes found to be significantly up- or down-regulated in the mutant were searched for enriched gene ontology terms in the biological process ontology using ENRICHR (Chen et al. 2013; Kuleshov et al. 2016), and the top 10 enriched categories were shown (Fig. 3A). The proton transport genes (associated with the term GO: 0015992) were found to be enriched among up-regulated genes in the mutant samples. To visualize their expression across all samples, we calculated the z-scores of log2 expression (reads per million) for each of those genes and plotted the boxplots for individual samples (Fig. 3B). Similarly, the heatmap of z-scores was plotted using pheatmap 1.0.12 (Fig. 3C).
Electron microscopy imaging of fly ovaries
For transmission electron microscopy (TEM) imaging, dissected ovaries from 5-d-old fertilized females were fixed for 30 min at room temperature by addition in the culture medium of a 2× fixative solution (4% paraformaldehyde, 0.4% glutaraldehyde, 0.2 M PHEM). They were then fixed for another 30 min at room temperature in 2% paraformaldehyde, 0.2% glutaraldehyde in 0.1 M PHEM (30 mM PIPES, 12.5 mM HEPES, 5 mM MgCl2, 1 mM EGTA, pH 7). Ovaries were rinsed three times in 0.1 M PHEM and post-fixed in 1% osmium tetroxide (OsO4), 1.5% potassium ferrocyanide in 0.1 M PHEM buffer for 1 h at room temperature. After three washes in 0.1 M PHEM, they were post-stained using 0.5% uranyl acetate in 30% ethanol for 30 min at room temperature, in the dark. Ovaries were then dehydrated in a graded ethanol series (50%, 70%, 90%, 5 min each, then 100% for 5 min, three times), and embedded in Epon resin. After polymerization during 48 h at 65°C, the blocs were cut on a Leica UC7 ultramicrotome using a Diatome 35° diamond knife. Ultrathin sections (70 nm) were collected on Formvar carbon coated copper grids and post-stained with 2% uranyl acetate and 2% lead citrate. Images were recorded on a Tecnai G2 Spirit BioTwin (FEI) transmission electron microscope (TEM), operating at 120 kV, using an ORIUS SC1000 CCD camera (Gatan).
Larval chromosome staining from dissected salivary glands
Chromosome squashes were performed with the acid-free technique as described in Johansen et al. (2009). Pcif1BG02557or Pcif1M6stop third-instar larvae were rinsed in PBS and dissected in a formaldehyde dissection buffer (0.15 M PIPES, 3 mM MgSO3, 1.5 mM EGTA, 1.5% NP40, 2% formaldehyde). Then, the salivary glands were incubated 5 min in glycerol 50% and squashed on frosted microscope slides. After squashing, the slides were incubated for 30 min in 0.1% Triton X-100 in PBS (PBST) and blocked for 1 h in PBST-BSA 5%. The slides were incubated in a humid chamber with the rabbit anti-Pcif1 (1:100) overnight. The slides were washed three times in PBST before being incubated with anti-rabbit conjugated to Alexa 488 in a humid chamber for 2 h at RT. Two additional washes in PBS were performed and the samples stained with DAPI. The slides were immediately observed with a microscope confocal LSM800.
PEV observation and pigment quantification
Five virgin wm4h/wm4h; +/+ females were crossed with three males of the following genotype: w1118; Pcif1M6stop, w1118; Pcif1F9ΔNLS, w1118; Df(3L)BSC797/TM3, w1118; Df(3L)BSC563/TM6 or w1118; Df(3L)BSC452/TM6. In the G1 progeny, variegated eyes were checked in trans-heterozygous females. Pictures of 3-d-old adults were taken using a Leica MZFIII microscope (Leica Microsystem CHS).
For pigmentation quantification, five heads from 6-d-old females of the genotypes of interest were dissected and snap-frozen in liquid nitrogen. Heads were lysed in 500 mL of AEA lysis buffer (30% ethanol, 0.2% HCl) and homogenized with an electric pestler, and the mix was stored at 25°C for 48 h in the dark. Each sample was then centrifuged at 10,000 rpm for 5 min at room temperature, and the supernatant was transferred in a new tube and centrifuged again at 10,000 rpm for 5 min at RT. An amount of 150 µL of the supernatant was loaded on a 96-well plate, and the absorbance of the extracted pigments was measured at 480 nm. Ten replicates were analyzed for each genotype. The statistical analysis was performed using the one-way ANOVA test in GraphPad Prism 9.
DATA DEPOSITION
Deep sequencing data generated in this study are deposited with the Gene Expression Omnibus (GEO) under accession number GEO: GSE218059.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. A. Boivin, Dr. W.P. Galej, and Dr. A. Verdel for scientific discussions. We thank Dr. C. Moriscot for electron microscopy analysis. We thank the iGE3 Genomics Platform at the University of Geneva and the Genomics Core at EMBL Heidelberg. This work was supported by the GRAL LabEX (ANR-10-LABX-49-01) financed within the University Grenoble Alpes graduate school (Ecoles Universitaires de Recherche) CBH-EUR-GS (ANR-17-EURE-0003) to M.O.F. and by the “Région Auvergne-Rhône-Alpes” (PAI 2021) to E.T. G.F. is supported by the IDEX PhD Fellowship from the University of Grenoble, and by funding from the Swiss National Science Foundation (ERC Transfer Grant CRETP3_166923) to R.S.P. E.B. is supported by grants from the Agence Nationale pour la Recherche (ANR-20-CE12-0005 CHApiTRE and ANR-21-CE12-0022 BiopiC) and the French government Emergence-IDEX-ISITE initiative 16-IDEX-0001 (CAP20-25). This work used the EM facilities of the Grenoble Instruct–ERIC Center (ISBG; UAR 3518 CNRS-CEA-UGA-EMBL) within the Grenoble Partnership for Structural Biology (PSB), supported by FRISBI (ANR-10-INBS-0005-02) and LabEx GRAL. The IBS Electron Microscope facility is supported by the Auvergne Rhône-Alpes Region, the Fonds Feder, the Fondation pour la Recherche Médicale, and GIS-IBiSA. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study.
Author contributions: G.F. performed most of the experiments for this study; G.F. and E.T. generated fly mutants and performed fly crosses; E.D. and R.R.P. prepared sequencing libraries; D.H. performed computational analyses; N.G. and E.B. conducted immunostaining on polytene chromosomes; E.T., M-O.F., and R.S.P. designed and supervised the study. Manuscript preparation and writing were performed by G.F., E.T., R.S.P., and M-O.F. with input from everyone.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079192.122.
REFERENCES
- Akichika S, Hirano S, Shichino Y, Suzuki T, Nishimasu H, Ishitani R, Sugita A, Hirose Y, Iwasaki S, Nureki O, et al. 2019. Cap-specific terminal N6-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science 363: eaav0080. 10.1126/science.aav0080 [DOI] [PubMed] [Google Scholar]
- Baker KE, Parker R. 2004. Nonsense-mediated mRNA decay: terminating erroneous gene expression. Curr Opin Cell Biol 16: 293–299. 10.1016/j.ceb.2004.03.003 [DOI] [PubMed] [Google Scholar]
- Belanger F, Stepinski J, Darzynkiewicz E, Pelletier J. 2010. Characterization of hMTr1, a human Cap1 2′-O-ribose methyltransferase. J Biol Chem 285: 33037–33044. 10.1074/jbc.M110.155283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulias K, Toczydlowska-Socha D, Hawley BR, Liberman N, Takashima K, Zaccara S, Guez T, Vasseur JJ, Debart F, Aravind L, et al. 2019. Identification of the m6Am methyltransferase PCIF1 reveals the location and functions of m6Am in the transcriptome. Mol Cell 75: 631–643.e638. 10.1016/j.molcel.2019.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. 2009. Progression through the RNA polymerase II CTD cycle. Mol Cell 36: 541–546. 10.1016/j.molcel.2009.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma'ayan A. 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14: 128. 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J, Du P, Kim W, Tang S, Sliz P, et al. 2018. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature 561: 556–560. 10.1038/s41586-018-0538-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Sakuraba K, Komuro A, Kato S, Harada F, Hirose Y. 2003. PCIF1, a novel human WW domain-containing protein, interacts with the phosphorylated RNA polymerase II. Biochem Biophys Res Commun 301: 378–385. 10.1016/S0006-291X(02)03015-2 [DOI] [PubMed] [Google Scholar]
- Galloway A, Cowling VH. 2019. mRNA cap regulation in mammalian cell function and fate. Biochim Biophys Acta 1862: 270–279. 10.1016/j.bbagrm.2018.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Shuman S, Lima CD. 2011. Structural insights to how mammalian capping enzyme reads the CTD code. Mol cell 43: 299–310. 10.1016/j.molcel.2011.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haline-Vaz T, Silva TC, Zanchin NI. 2008. The human interferon-regulated ISG95 protein interacts with RNA polymerase II and shows methyltransferase activity. Biochem Biophys Res Commun 372: 719–724. 10.1016/j.bbrc.2008.05.137 [DOI] [PubMed] [Google Scholar]
- Hampsey M, Reinberg D. 2003. Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation. Cell 113: 429–432. 10.1016/S0092-8674(03)00360-X [DOI] [PubMed] [Google Scholar]
- Haussmann IU, Wu Y, Nallasivan MP, Archer N, Bodi Z, Hebenstreit D, Waddell S, Fray R, Soller M. 2022. CMTr cap-adjacent 2′-O-ribose mRNA methyltransferases are required for reward learning and mRNA localization to synapses. Nat Commun 13: 1209. 10.1038/s41467-022-28549-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JH, Chen Z, Xu H. 2014. Selective propagation of functional mitochondrial DNA during oogenesis restricts the transmission of a deleterious mitochondrial variant. Nat Genet 46: 389–392. 10.1038/ng.2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, Iwamoto Y, Sakuraba K, Yunokuchi I, Harada F, Ohkuma Y. 2008. Human phosphorylated CTD-interacting protein, PCIF1, negatively modulates gene expression by RNA polymerase II. Biochem Biophys Res Commun 369: 449–455. 10.1016/j.bbrc.2008.02.042 [DOI] [PubMed] [Google Scholar]
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, et al. 2015. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 12: 115–121. 10.1038/nmeth.3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inesta-Vaquera F, Cowling VH. 2017. Regulation and function of CMTR1-dependent mRNA cap methylation. Wiley Interdiscip Rev RNA 8: e1450. 10.1002/wrna.1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen KM, Cai W, Deng H, Bao X, Zhang W, Girton J, Johansen J. 2009. Polytene chromosome squash methods for studying transcription and epigenetic chromatin modification in Drosophila using antibodies. Methods 483: 87–97. 10.1016/j.ymeth.2009.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachaev ZM, Lebedeva LA, Shaposhnikov AV, Moresco JJ, Yates JR III, Schedl P, Shidlovskii YV. 2019. Paip2 cooperates with Cbp80 at an active promoter and participates in RNA Polymerase II phosphorylation in Drosophila. FEBS Lett 593: 1102–1112. 10.1002/1873-3468.13391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith JM, Ensinger MJ, Moss B. 1978. HeLa cell RNA (2'-O-methyladenosine-N6-)-methyltransferase specific for the capped 5′-end of messenger RNA. J Biol Chem 253: 5033–5039. 10.1016/S0021-9258(17)34652-5 [DOI] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44: W90–W97. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL. 2002. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol Cell 9: 1101–1111. 10.1016/S1097-2765(02)00518-X [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhart A, Cramer P. 2004. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature 430: 223–226. 10.1038/nature02679 [DOI] [PubMed] [Google Scholar]
- Moyer SA, Banerjee AK. 1976. In vivo methylation of vesicular stomatitis virus and its host-cell messenger RNA species. Virology 70: 339–351. 10.1016/0042-6822(76)90276-2 [DOI] [PubMed] [Google Scholar]
- Nance DJ, Satterwhite ER, Bhaskar B, Misra S, Carraway KR, Mansfield KD. 2020. Characterization of METTL16 as a cytoplasmic RNA binding protein. PLoS One 15: e0227647. 10.1371/journal.pone.0227647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey RR, Delfino E, Homolka D, Roithova A, Chen K-M, Li L, Franco G, Vågbø CB, Taillebourg E, Fauvarque M-O. 2020. The mammalian cap-specific m6Am RNA methyltransferase PCIF1 regulates transcript levels in mouse tissues. Cell Rep 32: 108038. 10.1016/j.celrep.2020.108038 [DOI] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14: 417–419. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton KE, Park S-K, Hunter OV, Bresson SM, Conrad NK. 2018. Balance between MAT2A intron detention and splicing is determined cotranscriptionally. RNA 24: 778–786. 10.1261/rna.064899.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G, Wolff I. 1981. Isolation of dominant suppressor mutations for position-effect variegation in Drosophila melanogaster. Mol Gen Genet 182: 516–519. 10.1007/BF00293947 [DOI] [PubMed] [Google Scholar]
- Roignant JY, Soller M. 2017. m6A in mRNA: an ancient mechanism for fine-tuning gene expression. Trends Genet 33: 380–390. 10.1016/j.tig.2017.04.003 [DOI] [PubMed] [Google Scholar]
- Sendinc E, Valle-Garcia D, Dhall A, Chen H, Henriques T, Navarrete-Perea J, Sheng W, Gygi SP, Adelman K, Shi Y. 2019. PCIF1 catalyzes m6Am mRNA methylation to regulate gene expression. Mol Cell 75: 620–630.e629. 10.1016/j.molcel.2019.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatkin AJ. 1976. Capping of eucaryotic mRNAs. Cell 9: 645–653. 10.1016/0092-8674(76)90128-8 [DOI] [PubMed] [Google Scholar]
- Su R, Dong L, Li Y, Gao M, He PC, Liu W, Wei J, Zhao Z, Gao L, Han L. 2022. METTL16 exerts an m6A-independent function to facilitate translation and tumorigenesis. Nat Cell Biol 24: 205–216. 10.1038/s41556-021-00835-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Lawry ST, Cohen AL, Jia S. 2014. Chromosome boundary elements and regulation of heterochromatin spreading. Cell Mol Life Sci 71: 4841–4852. 10.1007/s00018-014-1725-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZH, Liu Y, Chaitankar V, Pirooznia M, Xu H. 2019. Electron transport chain biogenesis activated by a JNK-insulin-Myc relay primes mitochondrial inheritance in Drosophila. Elife 15: e49309. 10.7554/eLife.49309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C, Gershowitz A, Moss B. 1975. N6, O2′-dimethyladenosine a novel methylated ribonucleoside next to the 5′ terminal of animal cell and virus mRNAs. Nature 257: 251–253. 10.1038/257251a0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.