Abstract
The Malayan pit viper (Calloselasma rhodostoma) is a hemotoxic snake widely found in Southeast Asia and is responsible for the majority of poisoning cases in this region, including Thailand. However, a comprehensive knowledge of the venom protein profile and classification, as well as novel venom proteins, of this viper is still limited. Recently, the detailed composition of several snake venoms has been discovered through the use of transcriptome analysis. Therefore, the aim of this study was to employ a next-generation sequencing platform and bioinformatics analysis to undertake venom-gland de novo transcriptomics of Malayan pit vipers. Furthermore, 21,272 functional coding genes were identified from 36,577 transcripts, of which 314 transcripts were identified as toxin proteins, accounting for 61.41% of total FPKM, which can be categorized into 22 toxin gene families. The most abundant are snake venom metalloproteinase kistomin (P0CB14) and zinc metalloproteinase/disintegrin (P30403), which account for 60.47% of total toxin FPKM and belong to the SVMP toxin family, followed by snake venom serine protease 1 (O13059) and Snaclec rhodocetin subunit beta (P81398), which account for 6.84% and 5.50% of total toxin FPKM and belong to the snake venom serine protease (SVSP) and Snaclec toxin family, respectively. Amino acid sequences of the aforementioned toxins were compared with those identified in other important medical hemotoxic snakes from Southeast Asia, including the Siamese Russell's viper (Daboia siamensis) and green pit viper (Trimeresurus albolabris), in order to analyze their protein homology. The results demonstrated that ranges of 58%–62%, 31%–60%, and 48%–59% identity was observed among the SVMP, Snaclec, and SVSP toxin families, respectively. Understanding the venom protein profile and classification is essential in interpreting clinical symptoms during human envenomation and developing potential therapeutic applications. Moreover, the variability of toxin families and amino acid sequences among related hemotoxic snakes found in this study suggests the use and development of universal antivenom for the treatment of envenomating patients is still challenging.
Keywords: Calloselasma rhodostoma, Venomous snakes, Hemotoxic venom, Venom-gland, Trancriptomics
1. Introduction
Snake-bite envenoming is a disease classified by the World Health Organization (WHO) as a neglected tropical disease that causes morbidity, disability, and mortality in many parts of the world [1]. It has been estimated that 1.8 to 2.7 million cases of snake-bite envenomation result in 81,000 to 138,000 deaths out of 5.4 million snake-bites per year [2]. However, snake-bite envenomation victims are more burdened in sub-Saharan Africa and South and Southeast Asia [3]. In Southeast Asia, an estimated 242,648 snake-bite victims have been counted annually, of which 15,903 died, and 954 were amputated [4].
At least 196 species and subspecies of snakes, of which 59 species and subspecies are considered venomous snakes in Thailand [5]. The National Health Security Office (NHSO) reported 7.9 snake-bite cases per 100,000 people in 2017 [6]. There are seven medically important venomous snakes that can be classified according to the pharmacological effects of their venom, including i) hemotoxic snakes: Malayan pit viper (Calloselasma rhodostoma), Siamese Russell's viper (Daboia siamensis), white-lipped green pit viper (Trimeresurus albolabris) and ii) neurotoxic snakes: king cobra (Ophiophagus hannah), monocled cobra (Naja kaouthia), banded krait (Bungarus fasciatus), Malayan krait (B. candidus) [7]. Among these species, C. rhodostoma was responsible for most cases (38%) of poisoning in Thailand [7].
C. rhodostoma is endemic to Southeast Asia from Thailand to northern Malaysia and on the island of Java [8] and is listed as a category one medically crucial venomous snake of Southeast Asia [7]. The most effective treatment for envenomation is the intravenous administration of antivenom. However, antivenom should be given only to patients in whom its benefits are considered likely to exceed its risks, such as anaphylactic reactions, pyogenic reactions and serum sickness types of reaction. Therefore, antivenom treatment is indicated if and when a patient with proven or suspected snake-bite develops one or more of the following signs, including hemostatic abnormalities, cardiovascular abnormalities, acute kidney injury, hemoglobinuria/myoglobinuria, and rapid extension of swelling. In Thailand, two types of antivenoms are produced by the Thai Red Cross Society, including monovalent antivenoms and polyvalent antivenoms, covered all seven medically important venomous snakes of Thailand.
Clinical manifestations following poisoning with C. rhodostoma are systemic coagulopathy leading to petechiae, epistaxis, hematuria, and hemoptysis, and consumption coagulopathy, which can lead to death due to intracranial hemorrhage [6,9]. It also causes local effects such as local pain, swelling, ecchymosis, bleeding per wound, bleb, tissue necrosis, hematoma, necrotizing fasciitis, and compartment syndrome [10]. Many studies reported the toxins that play key roles for systemic hemorrhagic and coagulopathic effects of the venom are metalloproteinases [11], platelet-disrupting Snaclecs [12], and the thrombin-like enzyme ancrod [13], while phospholipase A2 [14], l-amino acid oxidase and other hemorrhagins contribute to the local tissue damages in C. rhodostoma venom [15].
The very high complexity and diversity of snake venoms result in a wide range of clinical manifestations. Therefore, advanced analytical tools need to be used to investigate and unravel the complexities of these venoms. Transcriptomics and proteomics studies are powerful tools and high throughput technologies to study genes and proteins expression profile of the organism. Several transcriptomics and proteomics analysis have been implemented to study the toxin profile from venom-glands of the hemotoxic snakes in Thailand, including D. siamensis [[16], [17], [18], [19], [20]] and T. albolabris [21,22]. While C. rhodostoma, several proteomics studies have been performed [15,[23], [24], [25], [26]], the protein identification has been relied on databases of other snakes resulting in unable to find particular C. rhodostoma toxins as well as novel venom proteins. Since lacking the C. rhodostoma venom-gland transcriptomic database, this study hypothesized that, the transcriptomics database from venom-gland of C. rhodostoma can be generated using transcriptomics approach. Therefore, the aim of this study is to determine the RNA-Seq transcriptome from the venom-glands of C. rhodostoma by de novo sequencing. The sequenced transcripts were functionally annotated and identified. The toxin gene expression profile from the venom-gland of C. rhodostoma was then compared to other hemotoxic snakes in Thailand. From these pieces of knowledge, the database created in this study will serve as the first step toward a comprehensive understanding of venom profile and classification. This knowledge can then be applied by other omics- and functional researches focusing on the venom of C. rhodostoma and closely related species. Furthermore, this information can be utilized to improve the understanding of the pathological effects of envenomating patients, which can be further applied to improve antivenom production in the future.
2. Results and discussion
2.1. Venom-gland de novo sequencing and assembly
The C. rhodostoma venom-glands transcriptome was sequenced, and a total of 65,944,213 and 64,895,515 raw reads from CR1 and CR2, respectively, were obtained. After data filtering, CR1 and CR2 yielded 64,040,919 and 63,115,504 unpaired clean reads, respectively. Additional file 1 shows the sequencing data output statistics, error rate distribution, and base content of individual samples. Furthermore, using the short reads assembling the program, Trinity, the clean reads (longer than 300 nucleotides) passed the Illumina quality filter for the de novo assembly. Trinity produced 78,051 transcripts (mean length = 1516 nucleotides) with the N50 (shortest contig length needed to cover 50% of the transcriptome) equal to 2892. Transcripts were then further clustered and streamlined to form 46,409 UniGenes (mean length = 1245 nucleotides) with the N50 value equal to 2389. In addition, after filtering low-frequency transcripts (Fragments Per Kilobase of exon model per Million; FPKM <1), the assembled was reduced to 36,577 transcripts, as shown by the transcript length statistic of clean reads (Additional file 2).
2.2. Annotation and gene ontology (GO) term analysis
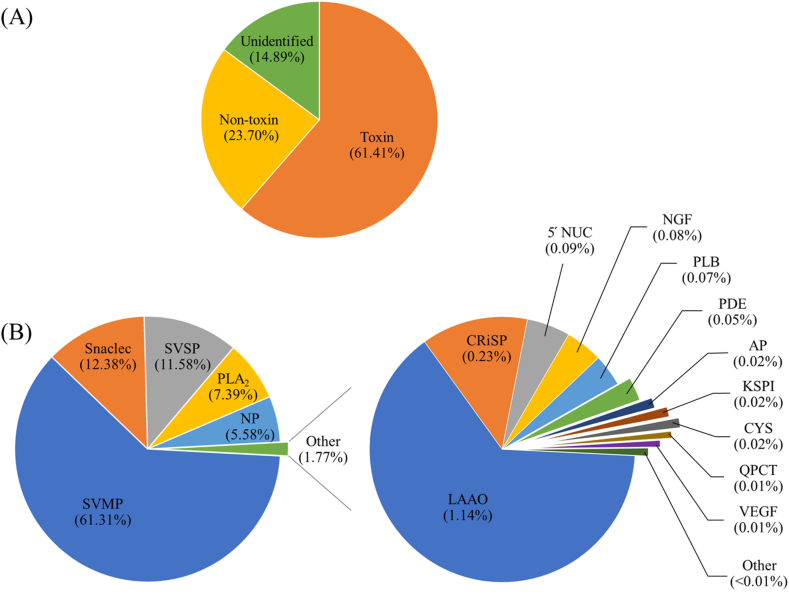
A BLASTx search was performed between the filtered UniGenes and sequences in the NCBI non-redundant (nr) protein, Swiss-Prot, and Pfam databases, yielding 19,395 (53.03%), 16,026 (43.81%), and 14,096 (38.54%) annotated UniGenes, respectively (Additional file 3). These genes were aligned with the genes from Protobothrops mucrosquamatus (Viper snake; 37.97%), and the elapid snakes such as Pseudonaja textilis (Eastern brown snake; 10.50%), Notechis scutatus (Tiger snake; 9.03%), and Ophiophagus Hannah (King Cobra; 8.68%) (Additional file 4). Furthermore, among the 21,272 functional protein encoding genes discovered, 314 transcripts (61.41% of the total FPKM) were identified as toxin proteins. However, 15,305 transcripts (14.89% of the total FPKM) could not be identified from these databases (Fig. 1A).
Fig. 1.
De novo transcriptome of C. rhodostoma venom glands. (A) The pie chart displaying an overview of transcript-cleaned reads with FPKM ≥1 identified by the NCBI non-redundant (nr) protein, Swiss-Prot, and Pfam databases. (B) Pie charts representing the expression abundance (%) of toxin transcripts by gene families. SVMP, snake venom metalloproteinase, demonstrates the most abundance, followed by Snaclec, snake venom C-type lectin/lectin-like protein; SVSP, snake venom serine protease; PLA2, phospholipase A2; and NP, natriuretic peptide, respectively (B, left). The other transcripts from the toxin families were minor (1.77%), and consist of LAAO, L-amino acid oxidase; CRiSP, cysteine-rich secretory protein; NUC, 5′ nucleotidase; NGF, nerve growth factor; PLB; phospholipase B, PDE, phosphodiesterase; AP, aminopeptidase; KSPI, Kunitz-type serine protease inhibitor; CYS, cystatin; QPCT; glutaminyl-peptide cyclotransferase, VEGF, vascular endothelial growth factor. While HYA, hyaluronidase; PLA2I, phospholipase A2 inhibitor; DPP IV, dipeptidylpeptidase IV; CVF, Cobra venom factor; WAP, waprin; and AChE, acetylcholinesterase, were expressed at levels lower than 0.01% of FPKM from identified toxins (B, right).
The transcripts encoding toxin proteins were clustered into 22 different toxin families based on sequence similarity. The three most abundant toxin families identified in the venom gland include snake venom metalloproteinase (SVMP), which accounted for 61.31% of the total toxin FPKM (out of 74 transcripts), followed by snake venom C-type lectin/lectin-like protein (Snaclec), with 12.38% (out of 51 transcripts), and snake venom serine protease (SVSP), with 11.58% (out of 24 transcripts) of the total toxin FPKM. Other toxin families identified in this study include phospholipase A2 (PLA2), natriuretic peptide (NP), L-amino acid oxidase (LAAO), cysteine-rich secretory protein (CRiSP), 5′ nucleotidase (5′ NUC), nerve growth factor (NGF), phospholipase B (PLB), phosphodiesterase (PDE), aminopeptidase (AP), Kunitz-type serine protease inhibitor (KSPI), cystatin, and glutaminyl-peptide cyclotransferase (QPCT), which accounted for 14.70% (out of 136 transcripts) of the total toxin FPKM. Moreover, 29 transcripts in 7 toxin gene families, including vascular endothelial growth factor (VEGF), hyaluronidase (HYA), phospholipase A2 inhibitor (PLA2I), dipeptidylpeptidase IV (DPP IV), Cobra venom factor (CVF), waprin (WAP), and acetylcholinesterase (AChE), were detected in extremely small numbers (less than 0.01% of the total toxin FPKM) (Fig. 2B). However, in the present study, other snake toxins such as three-finger toxin, arginine ester hydrolase, ohanin/vespryns, snake venom growth factor, insulin, trypsinogen, and flavin monoamine oxidase could not be detected (Additional file 5).
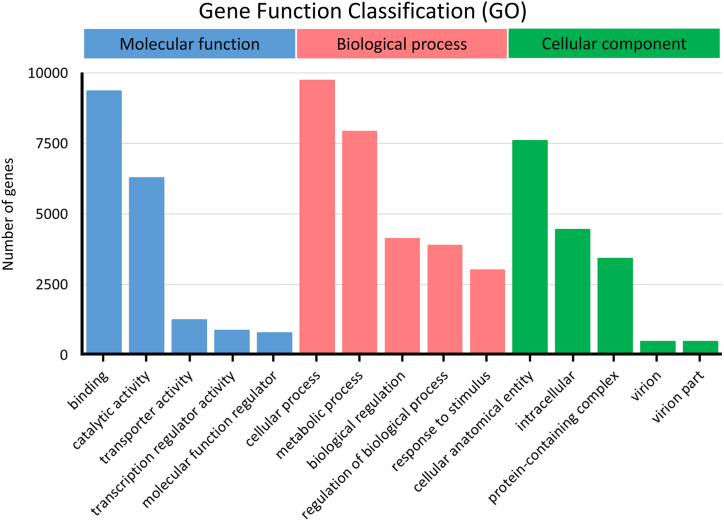
Fig. 2.
Top 5 gene ontology (GO) term classification of the venom-gland transcriptome of C. rhodostoma performed by BLAST2GO. Gene ontology functions are shown on the X-axis. The number of genes is shown on the Y-axis. The full list of GO term classifications of all transcripts is provided in Additional file 6.
The most abundant toxin family found in C. rhodostoma, SVMPs, has been reported to be a diverse group of multi-domain proteins with several biological activities, including the ability to induce hemorrhage, the proteolytic degradation of fibrinogen and fibrin, the induction of apoptosis, and the inhibition of platelet aggregation [27]. This finding was consistent with the clinical presentation of C. rhodostoma envenomation patients. Furthermore, the patient had local effects (93.4%) such as swelling, pain, bleeding per wound, ecchymosis, bleb, necrosis, hematoma, necrotizing fasciitis, and compartment syndrome. The systemic effect (88.0%) included hemorrhage and abnormal hemostasis (prolonged venous clotting time, unclotted 20-min whole blood clotting time, platelets less than 50,000/μL and prolonged INR greater than 1.2), which recovered fully after antivenin administration [10].
These results displayed a similar pattern when compared to the venom protein expression profile of C. rhodostama isolated from Malaysia, Indonesia, Thailand, and Vietnam. The relative abundance of venom protein composition showed SVMP (35.66%–46.30%), SVSP (13.64%–21.28%), and Snaclec (10.22%–20.70%) dominate the venom protein families, followed by LAAO (6.98%–11.08%) and PLA2 (4.40%–13.81%) [15]. A similar profile was observed in T. albolabris, a hemotoxic snake of Thailand. It was found that SVMP (21.9%), Snaclec (18.8%), and SVSP (12.5%) are the toxin families with the greatest abundance, followed by PLA2 (9.4%) and 5′ NUC (6.3%) [21]. In D. siamensis, the transcriptomic analysis of the venom gland showed SVSP (21.9%), 5′ NUC (21.9%), and SVMP (20.5%) are the most observed transcript gene families, followed by LAAO (16.1%) and PLA2 (13.4%) [17]. Moreover, proteomics analysis of D. siamensis venom isolated from Thailand [17,18], Indonesia [18], Guangzi and Taiwan [19,20] showed various patterns of toxin composition. However, the most abundant toxin families include SVMP, SVSP, Snaclec, KSPI, and PLA2.
The protein functional group classifications of the transcript were predicted using a GO. Here 16,062 (34.61%) of the 46,409 (100%) assembled transcripts were successfully allocated into at least one of three GO terms, including 42 GO assignments. The first group, “Molecular function,” contained 13,249 transcripts, which were followed by 11,696 and 8395 transcripts in “Biological process” and “Cellular component,” respectively. Fig. 2 shows that the predominant terms listed under “Molecular function” were binding (58.36%), catalytic activity (39.12%), transporter activity (7.82%), transcription regulator activity (5.45%), and molecular function regulator (4.89%); the predominant terms listed for “Biological process” were cellular process (60.72%), metabolic process (49.38%), biological regulation (25.77%), regulation of biological process (24.27%), and response to stimulus (18.81%), while those for “Cellular component” were cellular anatomical entity (47.41%), intracellular (27.71%), protein-containing complex (21.30%), virion (3.04%), and virion part (3.04%) (Fig. 2). The detailed information about the assembled contig annotation is shown in Additional file 6.
2.3. Toxin transcripts analysis
The three major toxin families, including SVMP, Snaclec, and SVSP, accounted for 85.27% of the total toxin FPKM and were obtained for further analyses. In the SVMP family, 69 of the 74 transcripts were identified by the Swiss-Prot protein database. Furthermore, after subtracting the non-snake proteins, 17 Swiss-Prot protein IDs remained, accounting for 61.29% of the total toxin FPKM. The snake venom metalloproteinase kistomin (P0CB14) and zinc metalloproteinase/disintegrin (P30403) have been identified with 100% identity to C. rhodostoma. These two toxins accounted for the almost toxins in this family (60.47% of the total toxin FPKM). However, in the Snaclec family (the second most identified toxin with 12.38% of the total toxin FPKM), 20 snake proteins were identified from 51 transcripts. There were six proteins matched to the C. rhodostoma protein with 98.31%–100% identity. Among these, Snaclec rhodocetin subunit beta (P81398) is the most abundant toxin identified, accounting for 5.50% of total toxin FPKM. Also, in the SVSP family, 14 snake proteins were identified from 24 transcripts, covering 11.57% of the total toxin FPKM. Only two proteins matched the C. rhodostoma protein with 100% identity, including the thrombin-like enzyme ancrod-2 (P47797) and the thrombin-like enzyme ancrod-2 (P26324), which accounted for 3.93% and 0.54% of total toxin FPKM, respectively. However, the most abundant toxin in this family matched SVSP 1 (O13059) from Trimeresurus gramineus, accounting for 6.84% of the total toxin FPKM (Table 1).
Table 1.
Top 3 toxin gene families expressed in the venom gland of C. rhodostoma.
| Toxin abundance (%) | Swiss-Prot ID | Length (amino acid) | Annotated Description | Species | Identity (%) | E-value |
|---|---|---|---|---|---|---|
| snake venom metalloproteinase (SVMP) (61.29% of total toxin FPKM) | ||||||
| 33.16 | P0CB14 | 417 | Snake venom metalloproteinase kistomin | Calloselasma rhodostoma | 100.00 | 6.40E-141 |
| 27.31 | P30403 | 478 | Zinc metalloproteinase/disintegrin | Calloselasma rhodostoma | 100.00 | 1.30E-108 |
| 0.42 | Q8AWI5 | 610 | Zinc metalloproteinase-disintegrin-like halysase | Gloydius halys | 82.47 | 3.10E-251 |
| 0.18 | Q90ZI3 | 612 | Zinc metalloproteinase-disintegrin-like HV1 | Protobothrops flavoviridis | 90.72 | 8.60E-45 |
| 0.14 | Q7SZD9 | 478 | Zinc metalloproteinase/disintegrin ussurin | Gloydius ussuriensis | 76.86 | 1.90E-159 |
| 0.04 | P0DM87 | 484 | Zinc metalloproteinase-disintegrin stejnitin | Trimeresurus stejnegeri | 76.49 | 2.10E-113 |
| 0.03 | Q1PS45 | 608 | Zinc metalloproteinase-disintegrin-like agkihagin | Deinagkistrodon acutus | 89.32 | 1.10E-117 |
| <0.01 | Q2EI26 | 413 | Snake venom metalloproteinase AaPA | Deinagkistrodon acutus | 77.50 | 1.70E-09 |
| <0.01 | F8S108 | 610 | Zinc metalloproteinase-disintegrin-like 4a | Crotalus adamanteus | 96.00 | 1.40E-07 |
| <0.01 | C5H5D4 | 414 | Zinc metalloproteinase-disintegrin-like batroxstatin-3 (Fragment) | Bothrops atrox | 80.95 | 2.80E-25 |
| <0.01 | C5H5D5 | 421 | Zinc metalloproteinase-disintegrin-like lachestatin-1 | Lachesis muta rhombeata | 88.89 | 5.40E-07 |
| <0.01 | J3S829 | 612 | Zinc metalloproteinase-disintegrin-like 2a | Crotalus adamanteus | 79.26 | 1.30E-68 |
| <0.01 | Q6T271 | 325 | Zinc metalloproteinase/disintegrin (Fragment) | Bitis gabonica | 85.19 | 1.50E-06 |
| <0.01 | Q2LD49 | 621 | Zinc metalloproteinase-disintegrin-like TSV-DM | Trimeresurus stejnegeri | 72.50 | 5.60E-09 |
| <0.01 | Q8AWX7 | 488 | Zinc metalloproteinase-disintegrin agkistin | Gloydius halys | 90.32 | 2.10E-11 |
| <0.01 | Q9W6M5 | 610 | Zinc metalloproteinase-disintegrin-like acurhagin | Deinagkistrodon acutus | 78.13 | 1.20E-06 |
| <0.01 | A8QL59 | 621 | Zinc metalloproteinase-disintegrin-like NaMP | Naja atra | 70.18 | 2.90E-18 |
| snake venom C-type lectin/lectin-like protein (Snaclec) (12.38% of total toxin FPKM) | ||||||
| 5.50 | P81398 | 129 | Snaclec rhodocetin subunit beta | Calloselasma rhodostoma | 100.00 | 1.60E-72 |
| 3.36 | Q9I841 | 136 | Snaclec rhodocytin subunit alpha | Calloselasma rhodostoma | 100.00 | 1.70E-16 |
| 1.07 | P81397 | 133 | Snaclec rhodocetin subunit alpha | Calloselasma rhodostoma | 98.31 | 4.70E-65 |
| 1.07 | D2YW39 | 135 | Snaclec rhodocetin subunit gamma | Calloselasma rhodostoma | 97.04 | 5.20E-78 |
| 0.39 | D2YW40 | 124 | Snaclec rhodocetin subunit delta | Calloselasma rhodostoma | 98.36 | 6.40E-72 |
| 0.35 | Q8AYA5 | 158 | Snaclec agglucetin subunit alpha-2 | Deinagkistrodon acutus | 73.38 | 1.20E-57 |
| 0.21 | P0DM39 | 148 | Snaclec alboaggregin-D subunit beta | Trimeresurus albolabris | 60.61 | 4.40E-46 |
| 0.21 | Q9PSN0 | 135 | C-type lectin PAL | Bitis arietans | 83.46 | 4.40E-70 |
| 0.17 | A7X3Z7 | 163 | C-type lectin lectoxin-Lio2 | Erythrolamprus poecilogyrus | 45.07 | 5.30E-34 |
| 0.05 | Q9I840 | 146 | Snaclec rhodocytin subunit beta | Calloselasma rhodostoma | 100.00 | 4.30E-75 |
| 0.01 | Q6TPH0 | 158 | Snaclec mucrocetin subunit alpha | Protobothrops mucrosquamatus | 62.04 | 9.30E-36 |
| <0.01 | A7X3Z0 | 158 | C-type lectin lectoxin-Thr1 | Thrasops jacksonii | 39.66 | 4.70E-30 |
| <0.01 | D2YVJ8 | 158 | C-type lectin mannose-binding isoform | Tropidechis carinatus | 65.71 | 3.80E-08 |
| <0.01 | Q6QX33 | 158 | C-type lectin BiL | Bothrops insularis | 60.98 | 9.60E-07 |
| <0.01 | J3SBP0 | 157 | C-type lectin 9a | Crotalus adamanteus | 75.00 | 5.70E-14 |
| <0.01 | Q9PRY7 | 135 | C-type lectin BjL | Bothrops jararaca | 100.00 | 1.00E-09 |
| <0.01 | Q696W1 | 158 | Snaclec coagulation factor X-activating enzyme light chain 2 | Macrovipera lebetina | 67.65 | 5.00E-07 |
| <0.01 | C6JUN9 | 158 | C-type lectin | Micrurus corallinus | 42.95 | 3.00E-30 |
| <0.01 | B4XSY9 | 156 | Snaclec A14 | Macrovipera lebetina | 61.22 | 5.90E-11 |
| <0.01 | P81112 | 134 | Snaclec alboaggregin-A subunit alpha | Trimeresurus albolabris | 79.31 | 8.10E-07 |
| snake venom serine protease (SVSP) (11.57% of total toxin FPKM) | ||||||
| 6.84 | O13059 | 258 | Snake venom serine protease 1 | Trimeresurus gramineus | 92.31 | 2.30E-19 |
| 3.93 | P47797 | 258 | Thrombin-like enzyme ancrod-2 | Calloselasma rhodostoma | 100.00 | 1.10E-19 |
| 0.54 | P26324 | 234 | Thrombin-like enzyme ancrod | Calloselasma rhodostoma | 100.00 | 1.40E-57 |
| 0.09 | P05620 | 260 | Thrombin-like enzyme flavoxobin | Protobothrops flavoviridis | 85.37 | 1.70E-12 |
| 0.05 | Q8UVX1 | 260 | Snake venom serine protease gussurobin | Gloydius ussuriensis | 70.18 | 9.90E-43 |
| 0.03 | Q9I8X2 | 260 | Thrombin-like enzyme acutobin | Deinagkistrodon acutus | 52.10 | 2.20E-31 |
| 0.02 | Q8AY78 | 258 | Snake venom serine protease 5 | Trimeresurus stejnegeri | 79.59 | 4.90E-18 |
| 0.02 | O73800 | 260 | Snake venom serine protease salmobin | Gloydius halys | 64.86 | 1.80E-53 |
| 0.02 | Q2QA04 | 262 | Snake venom serine protease | Crotalus durissus durissus | 69.54 | 4.30E-53 |
| 0.01 | Q8AY81 | 260 | Thrombin-like enzyme stejnobin | Trimeresurus stejnegeri | 72.34 | 1.40E-12 |
| 0.01 | Q71QI3 | 257 | Snake venom serine protease CL5 | Trimeresurus stejnegeri | 77.27 | 2.40E-23 |
| 0.01 | E0Y420 | 258 | Serine protease VLSP-3 | Macrovipera lebetina | 88.14 | 2.70E-25 |
| <0.01 | Q802F0 | 258 | Snake venom serine protease PTLE1 | Gloydius halys | 86.21 | 1.90E-06 |
| <0.01 | A7LAC6 | 260 | Thrombin-like enzyme 1 | Trimeresurus albolabris | 62.50 | 5.00E-10 |
Furthermore, similar results were observed in the proteomics analysis of C. rhodostoma venom. The most abundant toxin belongs to the SVMP toxin family, including snake venom metalloproteinase kistomin (P0CB14) and zinc metalloproteinase/disintegrin (P30403), which accounted for 20.42% and 19.78%, respectively. These were followed by toxins in Snaclec family including Snaclec rhodocetin subunit alpha (P81397), Snaclec rhodocetin subunit beta (P81398), Snaclec rhodocetin subunit gamma (D2YW39), Snaclec rhodocetin subunit delta (D2YW40), Snaclec rhodocytin subunit beta (Q9I840), Snaclec rhodocytin subunit alpha (Q9I841), and the toxins in SVSP family including thrombin-like enzyme ancrod-2 (P47797), thrombin-like enzyme ancrod (P26324) and thrombin-like enzyme contortrixobin (P82981) [15,26].
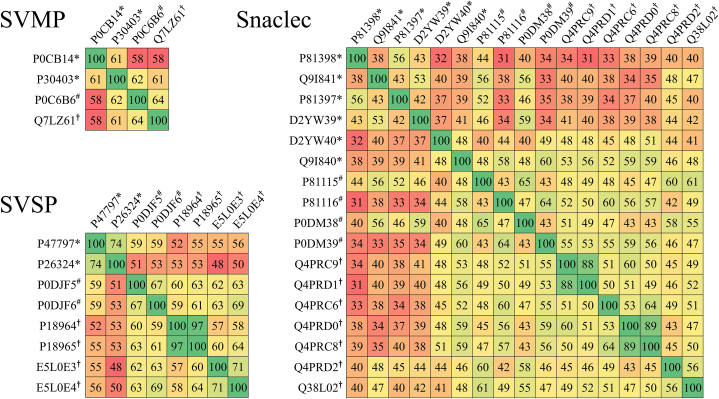
The identities of the toxins in each major toxin family from three medically important hemotoxic snakes in Thailand were determined using a protein identity matrix. Percent identity was calculated by Clustal Omega software using the amino acid sequences retrieved from the Swiss-Prot protein database. In the SVMP family, 61% identity was observed by comparing the snake venom metalloproteinase kistomin (P0CB14) and zinc metalloproteinase/disintegrin (P30403) of C. rhodostoma. Furthermore, 58%–62% identity was observed when comparing these two toxins to major SVMP toxins identified in T. albolabris and D. siamensis. In the Snaclec family, 6 toxins from C. rhodostoma showed 32%–56% identity, and 31%–60% identity was observed after comparing these toxins to Snaclec toxins from T. albolabris and D. siamensis. In the SVSP family, thrombin-like enzyme ancrod-2 (P47797), 74% identity was observed between thrombin-like enzyme ancrod (P26324) and thrombin-like enzyme contortrixobin (P82981) of C. rhodostoma, and 48%–59% identity was observed for the SVSP toxins found in T. albolabris and D. siamensis [15,[18], [19], [20], [21], [22],26] (Fig. 3). It could be suggested that the use of universal antivenom compounds is possible and needs further consideration based on the results of the protein identification of major toxin families among the hemotoxic snakes found in Thailand.
Fig. 3.
Protein identity matrix of the top 3 toxin families including snake venom metalloproteinase (SVMP), snake venom serine protease (SVSP) and snake venom C-type lectin/lectin-like protein (Snaclec). The transcripts detected in each family were annotated, and amino acid sequences were retrieved from the Swiss-Prot protein database. The protein identity (%) of each pair of proteins was calculated by multiple sequence alignment using Clustal Omega software. The source of protein sequences in the Swiss-Prot protein database originated from C. rhodostoma (*), T. albolabris (#) and D. siamensis (†). The Swiss-Prot ID, protein names, and snake species used in this analysis are provided in the Additional file 7.
Presently, the intravenous administration of antivenom is the mainstay treatment for envenomated patients. Antivenom production (purified polyclonal antibodies) is dependent on the hyperimmunization of animals (typically horses or sheep) with whole snake venoms, which bind either toxin or non-toxin substances and act by neutralizing and/or clearing venom toxins from circulation. Therefore, the potency of antivenoms against specific toxins or batch-to-batch variations of antivenom production need to be carefully monitored. According to SVMPs, which play a key role in the overall pathology and pathophysiology of envenoming in viperid venoms. The novel broad-spectrum molecules were developed to inhibit the activity of SVMP toxins, including peptide/peptidomimetic compounds [28], proteins [29,30], or chemical substances such as flavonoids [31,32], CP471474 [33], Marimastat, Prinomastat, Dimercaprol, and DMPS [34]. Among these, only a broad-spectrum matrix metalloproteinase inhibitor (CP471474) mixed with a phospholipase A2 inhibitor (Varespladib) partially inhibited the lethal activity of Bothrops asper and Crotalus durissus cumanensis venoms [33]. Therefore, broad-spectrum compounds for the treatment of envenomated patients among hemotoxic snakes found in Thailand need further investigation.
2.4. SVMP sequence alignment and phylogenetic tree analysis
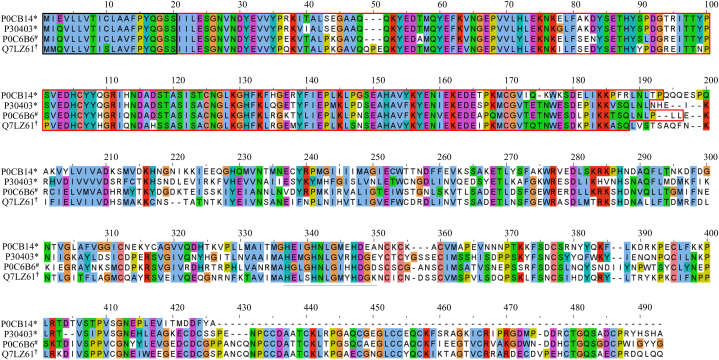
The SVMP toxin family is the most abundant toxin transcript found in this study, which includes snake venom metalloproteinase kistomin (P0CB14) and zinc metalloproteinase/disintegrin (P30403). As mentioned above, the SVMP toxin family also contributes to the major toxin family in T. albolabris (P0C6B6) and D. siamensis (Q7LZ61) [[17], [18], [19], [20], [21]]. In this regard, a comparison of amino acid sequences encoding these toxins by multiple alignment was conducted to determine protein conserved regions, which may be useful for the development of universal antivenom in the future. The result showed the highest amino acid sequence conservation at the N-terminal region, which includes the signal peptide (black box) and propeptide domain (red box). Like other secreted proteins, SVMPs contain a signal peptide, and the propeptide domain is responsible for driving the nascent SVMP precursor protein to the endoplasmic reticulum, where the propeptide domain is removed to obtain the mature form in snake venom and secrete it [35]. Since it is removed from molecules during protein maturation, this conserved region might not have contributed to the neutralization of the universal antivenom of these proteins. The low amino acid sequence conservation was observed in the rest of the sequence (Fig. 4). According to this finding, higher toxin variation was observed among the SVMP toxin from three important medical hemotoxic venoms from Thailand. Therefore, the use of universal antivenom to neutralize the SVMP toxins from those snakes is still challenging and requires further in-depth research.
Fig. 4.
Multiple sequence alignment of partial snake venom metalloproteinase (SVMP) proteins including snake venom metalloproteinase kistomin (P0CB14) and zinc metalloproteinase/disintegrin (P30403) from C. rhodostoma (*), zinc metalloproteinase homolog-disintegrin albolatin (P0C6B6) from T. albolabris (#), and coagulation factor X-activating enzyme heavy chain (Q7LZ61) from D. siamensis (†) using Clustal Omega and Jalview software. The signal peptide (black box) and propeptide (red box). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
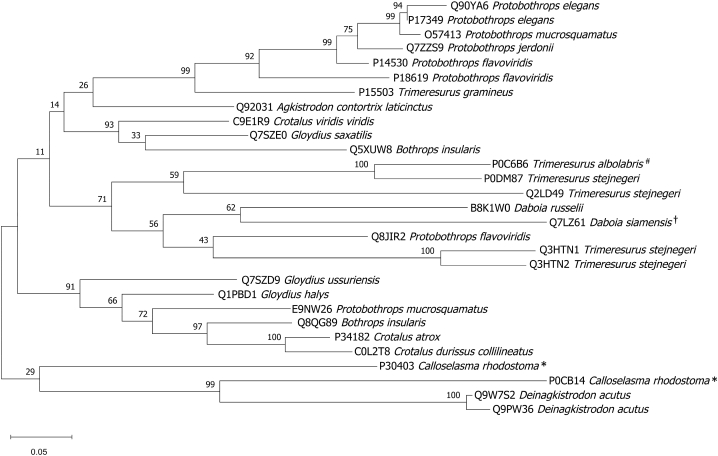
Furthermore, the phylogenetic tree among P0CB14 and P30403 was performed in order to determine the relationship of these toxins against SVMP toxins from the Swiss-Prot protein database. The most hit homologous sequences from BLAST searches were retrieved together with the major SVMP toxins from T. albolabris (P0C6B6) and D. siamensis (Q7LZ61). The result showed that, the major SVMP toxins from C. rhodostoma (P0CB14 and P30403) shared a common ancestor with SVMP toxins from Deinagkistrodon acutus (Q9W7S2 and Q9PW36). While SVMP toxins from T. albolabris (P0C6B6) and D. siamensis (Q7LZ61) have the same common origin as SVMP toxins from Daboia russelii, Protobothrops spp., Agkistrodon contortrix laticinctus, Crotalus viridis viridis, Gloydius saxatilis, and Bothrops insularis (Fig. 5).
Fig. 5.
Phylogenetic analysis of snake venom metalloproteinase kistomin (P0CB14) and zinc metalloproteinase/disintegrin (P30403) from C. rhodostoma (*) against the most hit homologous sequences from BLAST using MEGA11 software. The major snake venom metalloproteinase (SVMP) proteins from T. albolabris (#) and D. siamensis (†) were also included in the phylogenetic tree.
3. Conclusion
This study is the first venom-glands transcriptome analysis of C. rhodostoma, classified as a category 1 medically important venomous snake of Southeast Asia. The result showed that the SVMP toxin family was the most abundant, followed by the Snaclec and SVSP toxin families, respectively, identified from the venom gland of C. rhodostoma. The results also showed the variation of toxin families, the variation of toxin among the SVMP toxin family, and the diversity in amino acid sequences in each toxin of the hemotoxic snake found in Thailand. Therefore, the development of compounds, including universal antibodies and/or inhibitors, for the treatment of envenomating patients is still challenging. Finally, the genome project of C. rhodostoma and other venomous snakes is crucial to comprehend, not only for the biology of snakes but also to understand the expression, regulation, and interaction of venomous toxin genes, which can be further applied for improving antivenom production in the future.
4. Materials and methods
4.1. Ethics and biosafety statement
All experiments in the animal study were approved by the Queen Saovabha Memorial Institute Animal Care and Use Committee with protocol number QSMI-ACUC-09-2020 and the Faculty of Tropical Medicine-Animal Care and Use Committee (FTM-ACUC) with protocol number 015-2021. All methods performed in this study were also approved by the Institutional Biosafety Committee, Faculty of Tropical Medicine, Mahidol University, with submission number FTM-IBC-21-09.
4.2. Snakes and venom-gland dissection
One male and one female of C. rhodostoma were used in this study. The snakes were captured from the Surat Thani province (the Southern part of Thailand) and transported to Queen Saovabha Memorial Institute (QSMI), Thai Red Cross Society, Thailand in May 2021. The male snake (CR1) had a body weight of 85 g with 53 cm in length (head to tail). The female snake (CR2) had a body weight of 105 g with 70.3 cm in length. The venom glands were dissected 3 days after stimulation by milking. The snake venom was kept at −20 °C for further study. The venom gland dissection was gently performed on a sterile surface under general anesthesia by intramuscular injection with 10 mg/kg of ketamine. Approximately 500 mg of venom glands were extracted from each snake. The venom glands were washed three times with sterile normal saline solution and were immediately frozen using dry ice. Then, the extracted venom glands were proceeded for RNA extraction on the same day.
4.3. RNA extraction, cDNA construction and transcriptome sequencing
The right venom gland of each snake was thawed on ice. The tissue was homogenized by grinding in liquid nitrogen using mortar and pestle together with 1 mL of TRIzol™ Reagent (Thermo Fisher Scientific, CA, USA). Then, total RNA was extracted following the manufacturer’s instructions. In brief, DNA, proteins, and polysaccharides were removed, and total RNA was precipitated and washed with ethanol. Finally, RNA pellet was resuspended in 50 μL of RNase-free water. Total RNA quality and concentration were assessed using an Agilent 2100 Bioanalyzer (Agilent technologies, CA, USA). RNA integrity number (RIN) > 7 was used to prepare the library.
The library was constructed using the NEBNext® Ultra Directional RNA Library Prep Kit for Illumina® (New England Biolabs, MA, USA) according to the manufacturer’s protocol. In brief, poly(A) mRNA isolation was performed using the NEBNext Oligo d(T)25 magnetic beads. Enriched poly(A) mRNA isolated from the total venom-gland was fragmented into short fragments, which further acted as templates for cDNA synthesis. First Strand Synthesis Reaction Buffer and NEBNext Random Primers. First-strand cDNA was then synthesized from mRNA using ProtoScript II Reverse Transcriptase, and second strand cDNA was synthesized using Second Strand Synthesis Enzyme Mix. Double-stranded cDNA was purified using AMPure XP beads Mag PCR Clean-up (Beckman Coulter, CA, USA) and treated with End Prep Enzyme Mix to repair both ends and add a dA-tail in a single reaction, followed by T-A ligation to add adaptors to both ends. The adaptor ligated DNA fragments were purified using AMPure XP beads Mag PCR Clean-up (Beckman Coulter, CA, USA). Each sample was then amplified by PCR for 12 cycles using P5 and P7 primers; both primers carry sequences that can anneal to the flow cell to enable bridge PCR, while the P7 primer carries a six-base index that enables multiplexing. The PCR products were cleaned using AMPure XP beads Mag PCR Clean-up (Beckman Coulter, CA, USA), validated using an Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA), and quantified using a Qubit 2.0 Fluorometer (Invitrogen, CA, USA). Next, libraries with different indices were multiplexed and loaded on an Illumina Novaseq 6000 platform according to the manufacturer’s instructions (Illumina, CA, USA). Sequencing was performed using a 2 × 150-bp paired-end.
4.4. Data filtering
The generated mRNA data were transformed by base calling into sequence data, called the raw reads, and stored in a FASTQ format. Raw reads were filtered as part of the quality control process to generate clean reads in the pre-analysis stage [36]. This involved the removal of adapter contamination, uncertain nucleotides constituting (>10%), low quality reads with >50% of low-quality bases (base quality <5) before the downstream process of transcriptome assembly.
4.5. De novo transcriptome assembly
The de novo transcriptome assembly was performed using the Trinity short-read assembly program (version 2.6.6). This program has been shown to recover good transcripts after passing quality filters while also recovering relatively long transcripts with the highest specificity [37]. The trinity program including with three independent software modules, including Inchworm, Chrysalis, and Butterfly [38], to generate long scaffolds from individual reads. The process was based on the de Bruijn graphs construction algorithm, starting with the alignment of k-mers (k = 31), and reads with a certain length of overlap were merged into linear contigs. Reads were mapped back to the generated contigs, then contigs from the same transcript, and the distances between them were determined. The contigs were then divided into clusters, each of which contained a complete set of de Bruijn graphs (representing transcriptional complexity at a particular gene or locus). The graphs were processed independently to obtain full-length transcripts for alternatively spliced isoforms and to separate transcripts corresponding to paralogous genes. The percentage of clean reads (Q20), a reference value for quality control assessment, was determined as a benchmark for successful de novo assembly of the transcriptome. Finally, the transcript sequences obtained from this process were designated as UniGenes.
4.6. Clustering and functional annotation
UniGenes from the transcriptome assembly were further processed for sequence splicing and redundancy removal with Corset (version 4.6) to acquire non-redundant (NR) transcripts [39]. Corset works by clustering contigs based on shared reads and separates contigs when different expression patterns between samples are observed. Corset also uses the read information to filter out contigs with a low number of mapped reads (<10 reads). To remove the redundancy, the longest sequence in each cluster was chosen. Proteins with the highest ranks in the BLASTx results were referred to determine the coding region sequences of UniGenes, followed by a translation into amino acid sequences (using standard codon table). The distributions of the length of contigs, scaffolds, and UniGenes were calculated and the N50 length was set at N50 > 500 for assembling success.
4.7. Transcript abundance quantification
To determine transcript abundances for the identified genes, clean reads were aligned to UniGenes using Bowtie2 (version 2.4.5). Transcript abundances were then calculated using RSEM (version 1.2.28). Fragments per kilobase of exon model per million mapped reads (FPKM) were used to determine transcript abundance for the identified gene [40]. FPKM is the summation of normalized read counts based on gene length and the total number of mapped reads. Data were obtained using RSEM tool in conjunction with Trinity based on a following equation (1).
| (1) |
where B is the number of fragments/reads which are aligned to gene A; N is the total number of fragments/reads that are aligned to all genes; C is the base number in the coding sequence of gene A.
4.8. Transcripts categorization
The UniGenes were categorized into 3 groups which are toxin, non-toxin and unidentified. The toxin transcripts were recruited by manually searching with toxin-related keywords against the annotated transcripts. The non-toxin and unidentified groups contain the cellular proteins or housekeeping genes or the transcripts that could not be identified. The abundance of gene expression was determined by dividing the summation of all FPKM of each group by the total number of FPKM.
4.9. Protein identity matrix and multiple sequence alignment
According to transcript abundance quantification, metalloproteinase (SVMP toxin family), serine protease (SVSP toxin family) and Snaclec rhodocetin (Snaclec toxin family) are the most abundant transcripts encoding venom proteins, respectively. The protein identity in each toxin family mentioned above was compared in order to determine the percentage homology and conserved regions. In summary, the amino acid sequences of representatives from each toxin family were retrieved from the Swiss-Prot protein database (17 December 2022) (https://www.uniprot.org/). The SVMP toxin family was P0CB14 (C. rhodostoma), P30403 (C. rhodostoma), P0C6B6 (T. albolabris), and Q7LZ61 (D. siamensis). The Snaclec toxin family were retrieved from C. rhodostoma (P81398, Q9I841, P81397, D2YW39, D2YW40 and Q9I840), T. albolabris (P81115, P81116, P0DM38 and P0DM39) and D. siamensis (Q4PRC9, Q4PRD1, Q4PRC6, Q4PRD0, Q4PRC8, Q4PRD2 and Q38L02). SVSP toxin family were retrieved from C. rhodostoma (P47797 and P26324), T. albolabris (P0DJF5 and P0DJF6), and D. siamensis (P18964, P18965, E5L0E3, and E5L0E4). The protein identity of each pair of proteins was calculated by multiple sequence alignment using Clustal omega software. Jalview software (version 2.11.2.0) was used to determine the constructed multiple sequence alignment.
4.10. Phylogenetic analysis
The top 10 most hit protein sequences after BLAST search in the Swiss-Prot protein database were retrieved (17 December 2022) (https://www.uniprot.org/). The tree was constructed with Molecular Evolutionary Genetics Analysis (Mega) version 11.
4.11. Statistical analysis
Due to transcriptomic annotation, the longest UniGenes sequence was matched using BLASTx to the protein database in the NCBI non-redundant (NR) with a cut-off value (E < value 10−5) for clustering and functional annotation. Afterwards, the gene ontology classification was performed. The NCBI non-redundant (cut-off value E < 10−2), Pfam (cut-off value E < 10−5), and Swiss-Prot database (cut-off value E < 10−5) databases were searched for UniGenes with an FPKM value of at least one. The confidence limits of the created phylogenetic tree were then calculated for the phylogenetic analysis using the maximum likelihood with Jones-Taylor-Thornton (JTT) substitution model and the Bootstrap test (1000 repetitions).
Authors contribution statement
Poom Adisakwattana, Onrapak Reamtong and Charin Thawornkuno: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper. Lawan Chanhome: Performed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper. Narongsak Chaiyabutr: Contributed reagents, materials, analysis tools or data; Wrote the paper. Orawan Phuphisut: Analyzed and interpreted the data; Wrote the paper.
Funding
This research project is supported by Mahidol University, Thailand.
Data availability statement
The data presented in this study are available in this article or Supplementary Materials.o
Declaration of competing interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Footnotes
Key Contribution: This study pioneeringly focuses on the analysis of transcriptomics profiling and classification of venom proteins from venom-glands of C. rhodostoma. The toxin gene expression profile of C. rhodostoma was then compared to transcriptomics and/or proteomics study of D. siamensis and T. albolabris, a category 1 medically important hemotoxic snake found in Southeast Asia.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e15476.
RNA-Seq reads have been deposited in the NCBI-Sequence Read Archive (SRA) database under accession numbers PRJNA949833.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
The sequencing data output statistic, error rate distribution and base content.
Transcript length statistic of clean reads.
BLASTx search against NCBI non-redundant (nr) protein, Swiss-Prot and Pfam databases.
Species distribution (pptx 94 kb).
Toxin family classification (xlsx 8935 kb).
GO annotation (xlsx 1129 kb).
Swiss-Prot ID for protein identity matrix (xlsx 19 kb).
References
- 1.World Health Organization . WHO Geneva; Switzerland: 2019. Rationale for the Strategy. (In the Snakebite Envenoming: A Strategy for Prevention and Control). [Google Scholar]
- 2.Gutierrez J.M., Calvete J.J., Habib A.G., Harrison R.A., Williams D.J., Warrell D.A. Snakebite envenoming. Nat. Rev. Dis. Prim. 2017;3 doi: 10.1038/nrdp.2017.79. [DOI] [PubMed] [Google Scholar]
- 3.Kasturiratne A., Wickremasinghe A.R., de Silva N., Gunawardena N.K., Pathmeswaran A., Premaratna R., et al. The global burden of snakebite: a literature analysis and modelling based on regional estimates of envenoming and deaths. PLoS Med. 2008;5(11):e218. doi: 10.1371/journal.pmed.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patikorn C., Blessmann J., New M.T., Tiglao P.J.G., Vasaruchapong T., Maharani T., et al. Estimating economic and disease burden of snakebite in ASEAN countries using a decision analytic model. PLoS Neglected Trop. Dis. 2022;16(9) doi: 10.1371/journal.pntd.0010775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chanhome L., Cox M.J., Vasaruchapong T., Chaiyabutra N., Sitprija V. Characterization of venomous snakes of Thailand. Asian Biomed. 2011;5:311–328. [Google Scholar]
- 6.World Health Organization . second ed. WHO, Regional Office for South-East Asia; Delhi, India: 2016. Venomous Snakes of the South-East Asia Region, Their Venoms and Pathophysiology of Human Envenoming: in the Guidelines for the Management of Snake-Bites. [Google Scholar]
- 7.World Health Organization . WHO, Regional Office for South-East Asia; Delhi, India: 2016. Epidemiology of Snakebites in South-East Asia Region Countries: in the Guidelines for the Management of Snake-Bites. [Google Scholar]
- 8.McDiarmid R.W., Campbell J.A., Toure T. vol. 1. Herpetologists' League; Washington, DC, USA: 1999. (Snake Species of the World: A Taxonomic and Geographic Reference). [Google Scholar]
- 9.Wongtongkam N., Wilde H., Sitthi-Amorn C., Ratanabanangkoon K. A study of 225 Malayan pit viper bites in Thailand. Mil. Med. 2005;170(4):342–348. doi: 10.7205/milmed.170.4.342. [DOI] [PubMed] [Google Scholar]
- 10.Tangtrongchitr T., Thumtecho S., Janprasert J., Sanprasert K., Tongpoo A., Tanpudsa Y., et al. Malayan pit viper envenomation and treatment in Thailand. Therapeut. Clin. Risk Manag. 2021;17:1257–1266. doi: 10.2147/TCRM.S337199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan N. The biochemistry of venoms of some venomous snakes of Malaysia-a review. Trop. Biomed. 1991;8:91–103. [Google Scholar]
- 12.Shin Y., Morita T. Rhodocytin, a functional novel platelet agonist belonging to the heterodimeric C-type lectin family, induces platelet aggregation independently of glycoprotein Ib. Biochem. Biophys. Res. Commun. 1998;245(3):741–745. doi: 10.1006/bbrc.1998.8516. [DOI] [PubMed] [Google Scholar]
- 13.Maduwage K., Isbister G.K. Current treatment for venom-induced consumption coagulopathy resulting from snakebite. PLoS Neglected Trop. Dis. 2014;8(10) doi: 10.1371/journal.pntd.0003220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonfim V.L., Ponce-Soto L.A., de Souza M.D., Souza G.H., Baldasso P.A., Eberlin M.N., et al. Structural and functional characterization of myotoxin, Cr-IV 1, a phospholipase A2 D49 from the venom of the snake Calloselasma rhodostoma. Biologicals. 2008;36(3):168–176. doi: 10.1016/j.biologicals.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 15.Tang E.L.H., Tan N.H., Fung S.Y., Tan C.H. Comparative proteomes, immunoreactivities and neutralization of procoagulant activities of Calloselasma rhodostoma (Malayan pit viper) venoms from four regions in Southeast Asia. Toxicon. 2019;169:91–102. doi: 10.1016/j.toxicon.2019.08.004. [DOI] [PubMed] [Google Scholar]
- 16.Yee K.T., Tongsima S., Vasieva O., Ngamphiw C., Wilantho A., Wilkinson M.C., et al. Analysis of snake venom metalloproteinases from Myanmar Russell's viper transcriptome. Toxicon. 2018;146:31–41. doi: 10.1016/j.toxicon.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 17.Saethang T., Somparn P., Payungporn S., Sriswasdi S., Yee K.T., Hodge K., et al. Identification of Daboia siamensis venome using integrated multi-omics data. Sci. Rep. 2022;12(1) doi: 10.1038/s41598-022-17300-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lingam T.M.C., Tan K.Y., Tan C.H. Proteomics and antivenom immunoprofiling of Russell's viper (Daboia siamensis) venoms from Thailand and Indonesia. J. Venom. Anim. Toxins Incl. Trop. Dis. 2020;26 doi: 10.1590/1678-9199-JVATITD-2019-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanz L., Quesada-Bernat S., Chen P.Y., Lee C.D., Chiang J.R., Calvete J.J. Translational venomics: third-generation antivenomics of anti-siamese Russell's viper, Daboia siamensis, antivenom manufactured in taiwan CDC's vaccine center. Trav. Med. Infect. Dis. 2018;3(2):66. doi: 10.3390/tropicalmed3020066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan K.Y., Tan N.H., Tan C.H. Venom proteomics and antivenom neutralization for the Chinese eastern Russell's viper, Daboia siamensis from Guangxi and Taiwan. Sci. Rep. 2018;8(1):8545. doi: 10.1038/s41598-018-25955-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anita S., Sadjuri A.R., Rahmah L., Nugroho H.A., Trilaksono M.W., et al. Venom composition of Trimeresurus albolabris, T. insularis, T. puniceus and T. purpureomaculatus from Indonesia. J. Venom. Anim. Toxins Incl. Trop. Dis. 2022;28 doi: 10.1590/1678-9199-JVATITD-2021-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liew J.L., Tan N.H., Tan C.H. Proteomics and preclinical antivenom neutralization of the mangrove pit viper (Trimeresurus purpureomaculatus, Malaysia) and white-lipped pit viper (Trimeresurus albolabris, Thailand) venoms. Acta Trop. 2020;209 doi: 10.1016/j.actatropica.2020.105528. [DOI] [PubMed] [Google Scholar]
- 23.Vejayan J., Khoon T.L., Ibrahim H. Comparative analysis of the venom proteome of four important Malaysian snake species. J. Venom. Anim. Toxins Incl. Trop. Dis. 2014;20(1):6. doi: 10.1186/1678-9199-20-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kunalan S., Othman I., Hassan S.S., Hodgson W.C. Proteomic characterization of two medically important Malaysian snake venoms, Calloselasma rhodostoma (Malayan pit viper) and Ophiophagus hannah (King Cobra) Toxins. 2018;10(11):434. doi: 10.3390/toxins10110434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ali S.A., Baumann K., Jackson T.N., Wood K., Mason S., Undheim E.A., et al. Proteomic comparison of Hypnale hypnale (hump-nosed pit-viper) and Calloselasma rhodostoma (Malayan pit-viper) venoms. J. Proteonomics. 2013;91:338–343. doi: 10.1016/j.jprot.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 26.Tang E.L., Tan C.H., Fung S.Y., Tan N.H. Venomics of Calloselasma rhodostoma, the Malayan pit viper: a complex toxin arsenal unraveled. J. Proteonomics. 2016;148:44–56. doi: 10.1016/j.jprot.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 27.Luchini L.S.G., Pidde G., Squaiella-Baptistao C.C., Tambourgi D.V. Corrigendum: complement system inhibition modulates the pro-inflammatory effects of a snake venom metalloproteinase. Front. Immunol. 2019;10:1539. doi: 10.3389/fimmu.2019.01137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yee K.T., Pitts M., Tongyoo P., Rojnuckarin P., Wilkinson M.C. Snake venom metalloproteinases and their peptide inhibitors from Myanmar Russell's viper venom. Toxins. 2016;9(1):15. doi: 10.3390/toxins9010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamoto M., Escalante T., Gutiérrez J.M., Shea K.J. A biomimetic of endogenous tissue inhibitors of metalloproteinases: inhibition mechanism and contribution of composition, polymer size, and shape to the inhibitory effect. Nano Lett. 2021;21(13):5663–5670. doi: 10.1021/acs.nanolett.1c01357. [DOI] [PubMed] [Google Scholar]
- 30.Ukken F.P., Dowell N.L., Hajra M., Carroll S.B. A novel broad spectrum venom metalloproteinase autoinhibitor in the rattlesnake Crotalus atrox evolved via a shift in paralog function. Proc. Natl. Acad. Sci. U.S.A. 2022;119(51) doi: 10.1073/pnas.2214880119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Preciado L.M., Comer J., Nunez V., Rey-Suarez P., Pereanez J.A. Inhibition of a snake venom metalloproteinase by the flavonoid myricetin. Molecules. 2018;23(10):2662. doi: 10.3390/molecules23102662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sachetto A.T.A., Miyamoto J.G., Tashima A.K., de Souza A.O., Santoro M.L. The bioflavonoids Rutin and Rutin succinate neutralize the toxins of B. jararaca venom and inhibit its lethality. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.828269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quiroz S., Castaneda I.C., Granados J., Patino A.C., Preciado L.M., Pereanez J.A. Inhibitory effects of varespladib, CP471474, and their potential synergistic activity on Bothrops asper and Crotalus durissus cumanensis venoms. Molecules. 2022;27(23):8588. doi: 10.3390/molecules27238588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menzies S.K., Clare R.H., Xie C., Westhorpe A., Hall S.R., Edge R.J., et al. In vitro and in vivo preclinical venom inhibition assays identify metalloproteinase inhibiting drugs as potential future treatments for snakebite envenoming by Dispholidus typus. Toxicon X. 2022;14 doi: 10.1016/j.toxcx.2022.100118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moura-da-Silva A.M., Almeida M.T., Portes-Junior J.A., Nicolau C.A., Gomes-Neto F., Valente R.H. Processing of snake venom metalloproteinases: generation of toxin diversity and enzyme inactivation. Toxins. 2016;8(6):183. doi: 10.3390/toxins8060183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conesa A., Madrigal P., Tarazona S., Gomez-Cabrero D., Cervera A., McPherson A., et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17:13. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holding M.L., Margres M.J., Mason A.J., Parkinson C.L., Rokyta D.R. Evaluating the performance of de novo assembly methods for venom-gland transcriptomics. Toxins. 2018;10(6):249. doi: 10.3390/toxins10060249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davidson N.M., Oshlack A. Corset: enabling differential gene expression analysis for de novo assembled transcriptomes. Genome Biol. 2014;15(7):410. doi: 10.1186/s13059-014-0410-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The sequencing data output statistic, error rate distribution and base content.
Transcript length statistic of clean reads.
BLASTx search against NCBI non-redundant (nr) protein, Swiss-Prot and Pfam databases.
Species distribution (pptx 94 kb).
Toxin family classification (xlsx 8935 kb).
GO annotation (xlsx 1129 kb).
Swiss-Prot ID for protein identity matrix (xlsx 19 kb).
Data Availability Statement
The data presented in this study are available in this article or Supplementary Materials.o