Abstract
The actin cytoskeleton is a ubiquitous participant in cellular functions that maintain viability, but how it controls programmed cell death is not well understood. Here we show that in response to DNA damage, human cells form a juxtanuclear F-actin–rich territory that coordinates the organized progression of apoptosome assembly to caspase activation. This cytoskeletal compartment is created by the actin nucleation factors JMY, WHAMM, and the Arp2/3 complex, and it excludes proteins that inhibit JMY and WHAMM activity. Within the territory, mitochondria undergo outer membrane permeabilization and JMY localization overlaps with punctate structures containing the core apoptosome components cytochrome c and Apaf-1. The F-actin–rich area also encompasses initiator caspase-9 and clusters of a cleaved form of executioner caspase-3 but restricts accessibility of the caspase inhibitor XIAP. The clustering and potency of caspase-3 activation are positively regulated by the amount of actin polymerized by JMY and WHAMM. These results indicate that JMY-mediated actin reorganization functions in apoptotic signaling by coupling the biogenesis of apoptosomes to the localized processing of caspases.
INTRODUCTION
Actin, one of the most ubiquitous proteins in eukaryotic cells, is polymerized into filaments that can generate forces, create scaffolds, and act as tracks for motor proteins, making the actin cytoskeleton a dynamic participant in virtually all cellular functions. The formation, organization, and turnover of filamentous (F-) actin are regulated by numerous actin-binding proteins (Pollard, 2016). To create branched F-actin networks within cells, the Arp2/3 complex nucleates actin filaments by collaborating with nucleation-promoting factors from the Wiskott–Aldrich Syndrome Protein (WASP) family (Campellone and Welch, 2010; Rotty et al., 2013). The mammalian WASP family is composed of nine members that stimulate Arp2/3-mediated actin assembly at distinct cellular localizations in response to different signals (Kramer et al., 2022; Campellone et al., 2023). Functions for the actin cytoskeleton during cell motility, adhesion, and division have been extensively characterized (Swaney and Li, 2016; Svitkina, 2018; Pollard and O’Shaughnessy, 2019), and newer activities for nucleation factors in membrane trafficking and chromatin dynamics are emerging (Caridi et al., 2019; Chakrabarti et al., 2021), but their roles in programmed cell death remain understudied.
The normal progression of many organismal processes, such as embryogenesis, tissue turnover, and tumor suppression, relies on a tightly regulated form of cell death called apoptosis (Kerr et al., 1972; Galluzzi et al., 2018). Apoptosis can be driven by intrinsic mitochondria-mediated and extrinsic receptor-mediated death mechanisms that converge on a terminal execution program (von Karstedt et al., 2017; Singh et al., 2019). Intrinsic pathways are initiated in response to intracellular damage and are characterized by mitochondrial outer membrane permeabilization, resulting in the export of apoptogenic proteins, including cytochrome c (cyto c), from the mitochondria to the cytosol (Tait and Green, 2013; Bock and Tait, 2020). Once cytosolic, cyto c can interact with the apoptotic protease-activating factor-1 (Apaf-1) to instigate the assembly of macromolecular signaling platforms called apoptosomes (Riedl and Salvesen, 2007). Apoptosomes mediate the multimerization and proteolytic processing of initiator and executioner caspases (Bratton and Salvesen, 2010; Yuan and Akey, 2013; Dorstyn et al., 2018), the latter of which eventually cleave multiple proteins in the cytosol and nucleus to drive a cell suicide program (Green and Llambi, 2015; Julien and Wells, 2017). While the structural and biophysical properties of apoptosomes have come into focus in vitro (Rodriquez and Lazebnik, 1999; Zou et al., 1999; Yin et al., 2006; Yuan et al., 2011, 2013; Hu et al., 2014; Zhou et al., 2015; Cheng et al., 2016; Wu et al., 2016; Li et al., 2017), their biological characteristics within cells remain less clear.
Cytoskeletal studies of apoptotic cell death initially linked the late-stage changes in cell adherence and morphology to actin filament disassembly, rearrangement, and even cleavage by caspases (Gourlay and Ayscough, 2005; Desouza et al., 2012). Actin is also recruited to mitochondria earlier in apoptosis at around the time of mitochondrial permeabilization (Tang et al., 2006; Wang et al., 2008; Rehklau et al., 2012), and the actin turnover machinery affects intrinsic apoptosis at multiple steps. The mitochondrial localization of cofilin, which depolymerizes actin filaments, influences the release of cyto c (Chua et al., 2003; Zhu et al., 2006; Klamt et al., 2009). Moreover, the F-actin–severing protein gelsolin may regulate mitochondrial membrane integrity, be cleaved by caspases, stimulate actin depolymerization, and enable chromatin fragmentation (Kothakota et al., 1997; Kusano et al., 2000; Ahn et al., 2003; Chhabra et al., 2005).
Roles for actin assembly proteins as active participants in DNA damage–induced apoptosis have also been uncovered. One of the WASP family members, JMY, was discovered as a cofactor that affects the function of the key tumor suppressor protein and transcription factor p53 (Shikama et al., 1999). Transient overexpression and depletion experiments suggest that JMY can promote cell death by enhancing p53-mediated transcription of BAX, a proapoptotic gene encoding a mitochondrial permeabilization factor (Shikama et al., 1999; Coutts et al., 2009). However, the activity of JMY in transcriptional regulation via p53 may not be the primary apoptotic driver following acute DNA damage, as the genetic deletion of JMY does not impact nuclear p53 modification or p53-driven transcription of genes that commonly lead to a proliferation arrest or mitochondrial permeabilization (King et al., 2021).
JMY shuttles between the nucleus and cytosol (Coutts et al., 2009; Zuchero et al., 2012), and while endogenous JMY is primarily nuclear in multiple cell lines (Coutts et al., 2007; Zuchero et al., 2009), tagged JMY is often cytoplasmic (Coutts et al., 2009; Firat-Karalar et al., 2011; Zuchero et al., 2012; Schlüter et al., 2014; Hu and Mullins, 2019). Under normal cellular growth conditions, the quantities of JMY and p53 are modulated by Mdm2, which can promote their proteasomal degradation (Coutts et al., 2007). Upon treatment with apoptosis-inducing stressors, JMY and p53 are stabilized and their abundance increases (Demonacos et al., 2001; Coutts et al., 2007, 2009). Although JMY may have proapoptotic functions in the nucleus, its best characterized activities are as an actin nucleation factor in the cytosol (Zuchero et al., 2009). In fact, following DNA damage, JMY, its closest homologue WHAMM, and the Arp2/3 complex are necessary for rapid proapoptotic responses in the cytoplasm (King et al., 2021; Haarer et al., 2023). Cells lacking JMY or WHAMM exhibit kinetic delays or deficiencies in the mitochondrial export of cyto c, activation of caspases, and the terminal proteolytic cleavage events that dismantle the cell (King et al., 2021). In further support of the importance of actin assembly in intrinsic death pathways, apoptotic cells undergo cytoplasmic rearrangements of F-actin, JMY, cyto c, and executioner caspases (King et al., 2021).
Collectively, these studies indicate that the actin cytoskeleton is an important player in the transition from mitochondrial permeabilization to executioner caspase activation. However, the underlying mechanisms by which it impacts progression through this crucial period of intrinsic apoptotic signaling are unclear. In the current study, we characterized the intracellular organization and function of multiple elements of the actin regulatory and apoptotic machinery and discovered that JMY-mediated actin polymerization creates a cytosolic F-actin–rich territory that compartmentalizes the apoptosome biogenesis process and promotes an efficient localized activation of caspases.
RESULTS
Reorganization of JMY within a cytosolic F-actin–rich territory is a more prominent step of the apoptotic response than its import into the nucleus
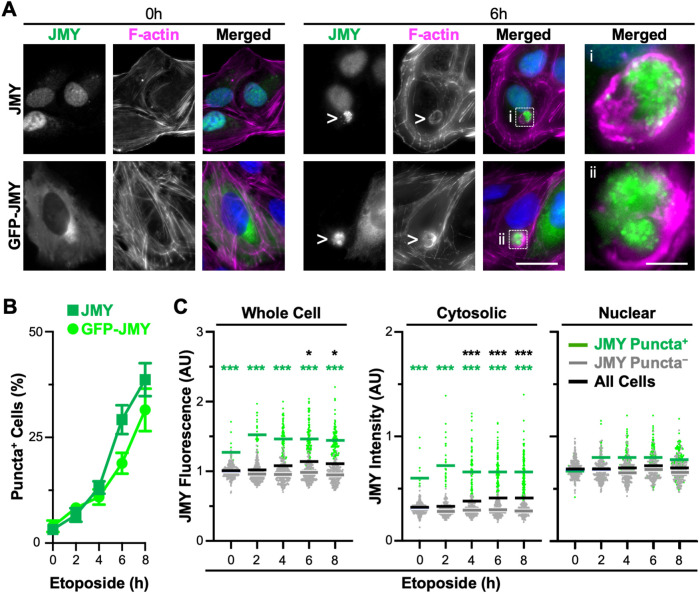
In cells responding to DNA damage, JMY, cyto c, and caspase-3 form clusters of cytosolic puncta that associate with actin filaments (King et al., 2021). To evaluate the timing of JMY puncta formation, we treated U2OS osteosarcoma cells with etoposide, a topoisomerase II inhibitor that induces double-strand DNA breaks. We then visualized the localization of endogenous JMY, or stably encoded GFP-tagged JMY (Supplemental Figure S1), at regular intervals up to 8 h. As seen previously, both JMY and GFP-JMY formed intense juxtanuclear clusters of puncta following etoposide exposure (Figure 1A). Compared to steady state conditions where only ∼3% of cells harbored such structures, the proportion of JMY puncta-positive cells increased to 13% by 4 h of etoposide treatment and reached ∼40% by 8 h (Figure 1B). Each positive cell had a single zone of clustered puncta, and staining with phalloidin demonstrated that F-actin comprised the areas in-between and/or surrounding the JMY or GFP-JMY puncta to create a round overall structure with a typical diameter of 8–16 μm (Figure 1A). Similar JMY and actin rearrangements were also observed in cells treated with the alkylating agent mitomycin C, suggesting that such structures are a shared feature in cells subjected to distinct DNA-damaging agents (Supplemental Figure S2). We refer to these cytoskeleton-associated clusters of JMY puncta as F-actin–rich territories.
FIGURE 1:
JMY forms cytosolic puncta within juxtanuclear F-actin–rich territories during DNA damage–induced apoptosis. (A) U2OS cells were treated with 10 μM etoposide for 0–8 h before being fixed and stained with a JMY antibody to visualize endogenous JMY (green), phalloidin to visualize F-actin (magenta), and DAPI to detect DNA (blue). U2OS cells encoding GFP-JMY (green) were also stained with phalloidin and DAPI. Arrowheads and magnifications (i, ii) highlight examples of cytosolic JMY puncta. Scale bars: 25, 5 μm. (B) The percentage of cells with JMY or GFP-JMY puncta was calculated. Each point represents the mean ± SD from three experiments (n = 2084–3044 cells per point). (C) Whole cell, cytosolic, and nuclear JMY fluorescence values were measured in ImageJ. Each point represents one cell (n = 310–348 per timepoint), where green points are JMY puncta–positive cells and gray points are JMY puncta–negative cells. Green, gray, and black bars are the mean values for the positive, negative, and total populations, each from three experiments. Green significance stars refer to comparisons of the puncta-positive to puncta-negative population. Black stars refer to comparisons of the total population at each timepoint to the 0 h timepoint. AU = arbitrary units. *p < 0.05; ***p < 0.001 (ANOVA, Tukey post-hoc tests).
Because JMY protein levels rise in cells exposed to apoptosis-inducing agents (Demonacos et al., 2001; Coutts et al., 2007, 2009), we hypothesized that such increases could be attributed to an accumulation of the cytoplasmic species of JMY. Consistent with previous observations, immunoblotting of U2OS cell lysates confirmed that JMY was more abundant in etoposide-treated samples (Supplemental Figure S3). To monitor the amount of nuclear versus cytoplasmic JMY in individual cells microscopically, we fixed and stained them to visualize endogenous JMY, F-actin, and DNA. We then quantified whole cell JMY fluorescence as well as JMY intensity in the nucleus and cytosol. Similar to previous findings (Demonacos et al., 2001; Coutts et al., 2007), whole cell JMY fluorescence increased significantly by 6 h of etoposide treatment (Figure 1C and Supplemental Figure S3, black bars and stars). A comparison of JMY intensities in the cytosol versus nucleus revealed a 3:7 ratio of cytosolic:nuclear JMY under basal conditions that was altered following etoposide exposure (Figure 1C and Supplemental Figure S3). After 4–8 h, nuclear JMY intensity remained unchanged, whereas cytosolic JMY intensity increased significantly (Figure 1C and Supplemental Figure S3, black bars and stars). Together, these results show that endogenous JMY exhibits a primarily nuclear localization, but upon introduction of an apoptosis-inducing stressor, JMY expression increases specifically in the cytosol while remaining high in the nucleus.
To test whether the formation of juxtanuclear JMY puncta was responsible for the increased cytosolic JMY intensity, we compared the levels of JMY in cells that contained clusters of JMY puncta to those cells that did not. When evaluating cell populations categorized as JMY puncta positive versus puncta negative, the puncta-positive cells had significantly higher whole cell JMY fluorescence relative to the puncta-negative cells at all timepoints (Figure 1C, left panel, green stars). When assessing the cytosol only, the puncta-positive cells always showed >2-fold higher cytosolic JMY intensity compared with puncta-negative cells (Figure 1C, middle panel). This was in contrast to the nucleus, where puncta-positive and puncta-negative populations displayed statistically indistinguishable levels of nuclear JMY intensity regardless of the duration of etoposide treatment (Figure 1C and Supplemental Figure S3). This difference is further emphasized by comparing the ratio of cytosolic:nuclear JMY intensities in the two populations, as the puncta-negative cells maintained a 3:7 ratio of cytosolic to nuclear JMY intensity with or without etoposide, whereas the puncta-positive cells displayed a nearly 1:1 ratio of cytosolic to nuclear JMY intensity after etoposide exposure (Figure 1C, green bars). Thus, in response to DNA damage, the overall increase in JMY abundance is due to the formation of a juxtanuclear cluster of JMY puncta.
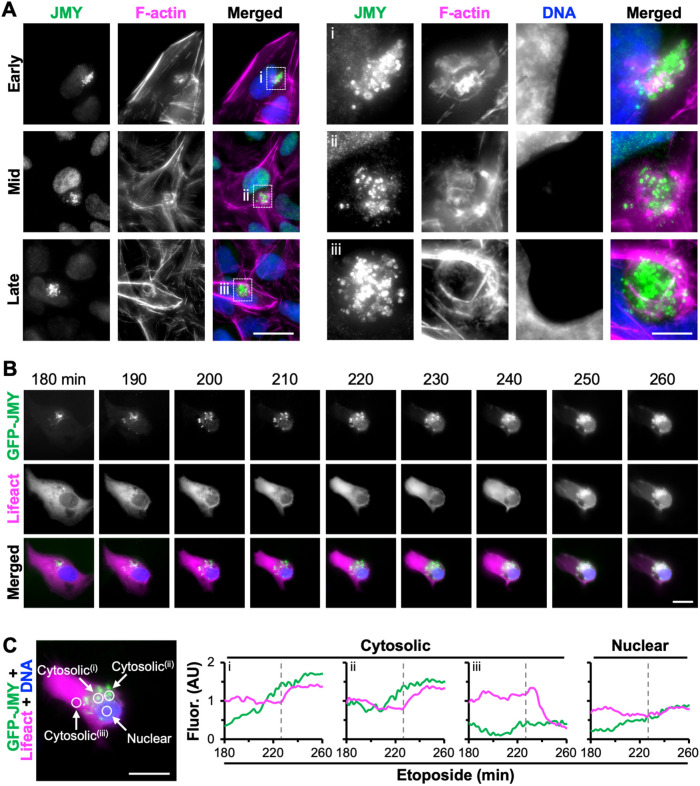
Given that the best understood molecular behavior of JMY is to promote actin assembly, we next investigated the organization of F-actin relative to the JMY puncta during the course of etoposide exposure. Based on imaging of fixed samples, some cells had actin filaments interspersed and overlapping with the juxtanuclear JMY puncta (Figure 2A, top row). In other cells, F-actin staining was more intense at the periphery of the clustered JMY puncta than in-between the puncta, and the nucleus was slightly deformed (Figure 2A, middle row). The etoposide-treated population also included some cells with a prominent ring of actin filaments surrounding the clustered JMY puncta, and such cells had a more exaggerated denting of the adjacent nucleus (Figure 2A, bottom row). These examples appear to reflect a morphological maturation of the juxtanuclear region in which JMY accumulation is accompanied by actin polymerization and the reorganization of filaments into a distinct round territory.
FIGURE 2:
JMY forms cytosolic puncta before the assembly of juxtanuclear F-actin–rich territories. (A) U2OS cells were treated with 10 μM etoposide before being fixed and stained with a JMY antibody (green), phalloidin to visualize F-actin (magenta), and DAPI to detect DNA (blue). Images represent early-, mid-, and late-stage F-actin–rich territories. Magnifications (i–iii) highlight examples of JMY-associated territories. Scale bars: 25, 5 μm. (B) U2OS cells encoding GFP-JMY (green) and transiently expressing Lifeact-mCherry (magenta) were treated with etoposide for 3 h before incubation with Hoescht to visualize DNA (blue). Selected frames (taken from Supplemental Video S1) show the formation of punctate GFP-JMY structures and an F-actin–rich territory. Scale bar: 20 μm. (C) Circles (5 μm diameter) were drawn within a cytosolic region containing punctate GFP-JMY (i and ii), a cytosolic region without GFP-JMY puncta (iii), or the nucleus, and the pixel intensity profiles for GFP-JMY and Lifeact-mCherry were measured. Dashed gray lines show the time when F-actin territory formation begins.
To visualize F-actin–rich territory biogenesis live, we utilized U2OS cells stably encoding GFP-JMY and transiently expressing the F-actin probe Lifeact-mCherry. The cells were treated with etoposide for 3 h and then imaged for an additional 1.5 h. Selected frames taken from Supplemental Video S1 show that the number and intensity of punctate GFP-JMY structures increased in the juxtanuclear region over time (Figure 2B). During the same period, the cell began to change shape, but F-actin was not concentrated in the juxtanuclear area until GFP-JMY levels reached an apparent threshold (Figure 2B). Fluorescence intensity profiles for two cytosolic areas within the portion of the cell containing GFP-JMY puncta showed GFP-JMY intensity increasing steadily and plateauing between 230 and 260 min, while Lifeact-mCherry intensity remained constant initially and then increased beginning at 230 min before achieving its maximum values between 240 and 260 min (Figure 2C, i and ii). In contrast, a cytosolic region lacking GFP-JMY puncta showed a decrease in Lifeact-mCherry intensity after 230 min (Figure 2C, iii), and a nuclear region did not display any dramatic changes in JMY or F-actin intensity over time (Figure 2C, nuclear). These results indicate that increased incorporation of JMY into a cluster of cytosolic puncta precedes the formation of a juxtanuclear F-actin–rich territory.
Movie S1.
Time-lapse movie showing the formation of punctate GFP-JMY structures and an F-actin-rich territory. Live U2OS cells expressing GFP-JMY (green) and Lifeact-mCherry (magenta) and treated with hoechst (blue) were imaged at 2min intervals 180-260min after exposure to 10μM etoposide. Playback rate = 10 frames per second. Scale bar: 20μm. Still frames are shown in Figure 2.
Apoptotic F-actin–rich territories contain the JMY/WHAMM subfamily of Arp2/3-activating, nucleation-promoting factors
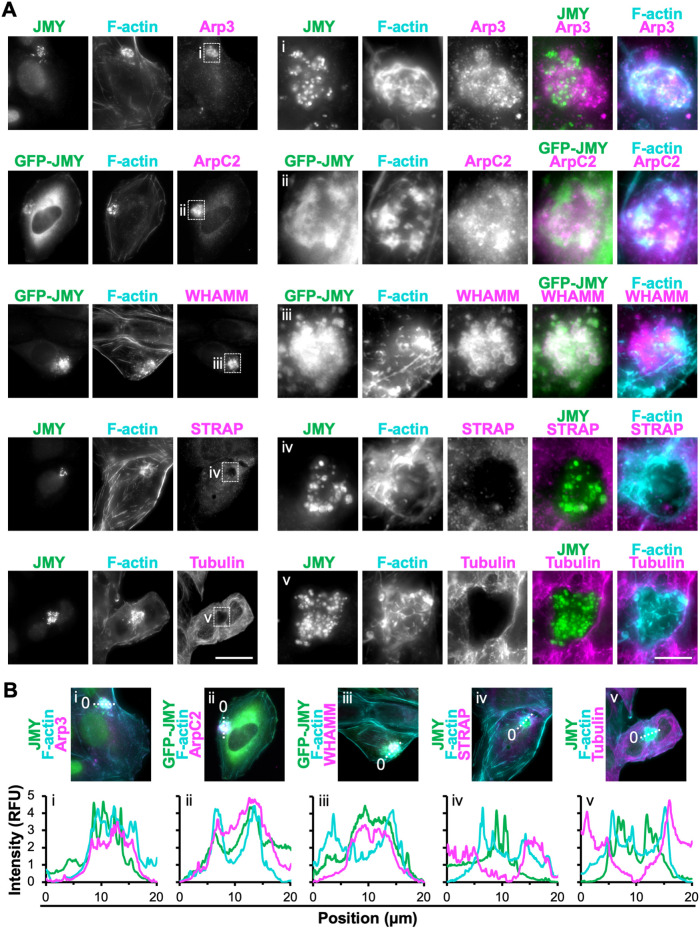
JMY, its closest homologue, WHAMM, and their downstream actin nucleator, the Arp2/3 complex, are all required for efficient intrinsic apoptosis (King et al., 2021). Therefore, we next studied the localization of these and other actin nucleation and branching factors in relation to the F-actin–rich territories. To do this, we treated U2OS cells with etoposide and visualized JMY or GFP-JMY along with F-actin, the Arp2/3 complex, and the branching regulator cortactin. Two subunits representative of the Arp2/3 complex, Arp3 and ArpC2, as well as cortactin, localized to the JMY-containing territories (Figure 3A and Supplemental Figure S4). Fluorescence intensity plot profiles of Arp2/3 subunit localizations showed higher intensity throughout the JMY and F-actin–rich regions compared with adjacent cytosolic areas (Figure 3B), and magnifications indicated that the Arp2/3 complex often overlapped with intense F-actin structures interspersed throughout the territories (Figure 3, A and B). In contrast, fluorescence profiles of cortactin, which binds F-actin and the Arp2/3 complex to modulate branchpoint stability (Schnoor et al., 2018), typically showed higher cortactin intensity at the peripheral actin filaments surrounding interior punctate JMY structures (Supplemental Figure S4). Thus, the apoptotic F-actin–rich regions contain the canonical components of the branched actin assembly machinery, with the Arp2/3 complex localizing throughout the territory and cortactin found mainly at the territory perimeter.
FIGURE 3:
Subset of actin nucleation and branching factors present within apoptotic F-actin–rich territories. (A) U2OS cells (rows 1, 4, and 5) or U2OS cells encoding GFP-JMY (rows 2 and 3; green) were treated with etoposide for 6 h, fixed, and stained with antibodies to JMY (rows 1, 4, and 5; green), antibodies to Arp3, ArpC2, WHAMM, STRAP, or tubulin (magenta), and with phalloidin (F-actin; cyan). Magnifications (i–iii) depict clusters of JMY and other nucleation factors in F-actin territories. Magnifications (iv and v) depict territories that exclude STRAP and tubulin. Scale bars: 25, 5 μm. (B) Lines (20 μm) were drawn through the images in A to measure the pixel intensity profiles. RFU = relative fluorescence units. See Supplemental Figure S3 for nucleation factors that were not enriched within F-actin territories.
Genetic inactivation of each WASP-family Arp2/3 activator previously revealed that only cells lacking JMY or WHAMM have substantial deficiencies in their intrinsic apoptosis responses (King et al., 2021). To characterize which, if any, nucleation-promoting factors other than JMY associate with the apoptotic F-actin territories in U2OS cells, we assessed the localization of WHAMM and representatives of other subgroups within the WASP family. Endogenous and tagged WHAMM, but not N-WASP, WAVE2, or WASH, exhibited strong localization to the JMY- and F-actin–rich regions (Figure 3A and Supplemental Figure S4). Magnifications highlighted the occasional colocalization of JMY, WHAMM, and F-actin (Figure 3A and Supplemental Figure S4), and fluorescence intensity plot profiles showed increased WHAMM intensity in close proximity to JMY within F-actin territories (Figure 3B and Supplemental Figure S4). These results demonstrate that the cytosolic F-actin–rich territories that form during the intrinsic apoptotic response contain a specific subset of nucleation factors, namely JMY, the Arp2/3 complex, and WHAMM.
The different localizations and functions of WASP-family members are controlled via interactions with regulatory complexes, small G-proteins, phospholipids, and microtubules, but little is known about how upstream factors regulate the localization or actin assembly activity of JMY or WHAMM under apoptotic conditions. In certain settings, the stress-response protein STRAP may inhibit JMY-mediated nucleation and interrupt its cytosolic functions while facilitating its role as a coactivator of transcription in the nucleus (Demonacos et al., 2001; Hu and Mullins, 2019). We examined the localization of STRAP relative to JMY and found that, in stark contrast to the enrichment of JMY and the Arp2/3 complex in the apoptotic F-actin territories, STRAP was missing from these regions (Figure 3, A and B). For WHAMM, binding to microtubules inhibits its nucleation-promoting activity (Campellone et al., 2008; Shen et al., 2012). Microtubules were not found within the JMY-containing F-actin territories either (Figure 3A), as magnifications and fluorescence intensity plot profiles showed a clear absence of tubulin staining in such regions (Figure 3, A and B). Hence, the apoptotic F-actin–rich territories not only consist of specific components of the branched actin nucleation machinery, but also exclude inhibitors of both JMY- and WHAMM-mediated actin polymerization.
Mitochondrial permeabilization and apoptosome biogenesis take place within juxtanuclear F-actin–rich territories
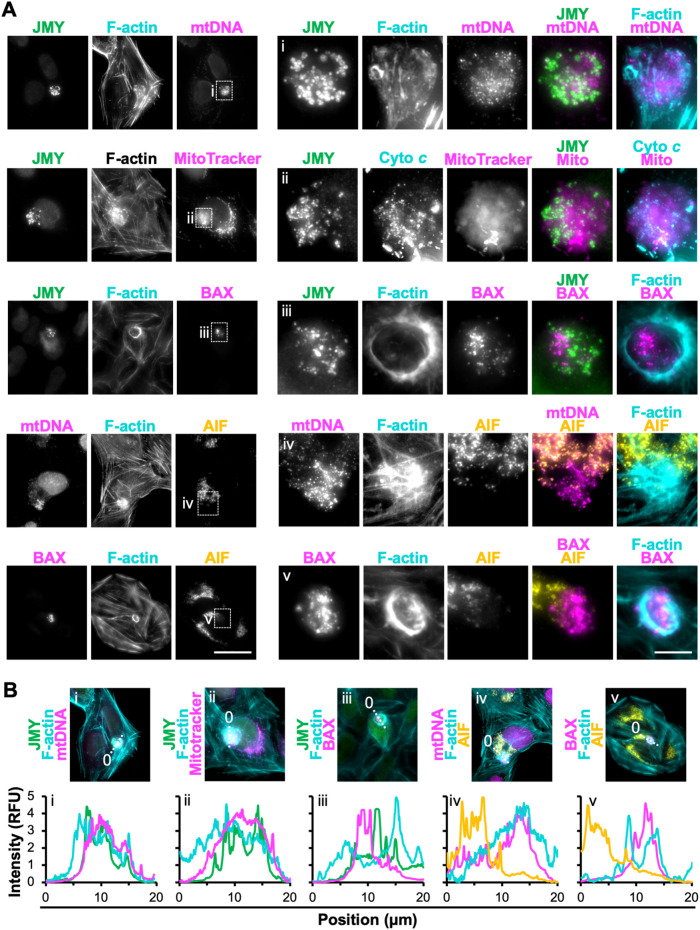
Intrinsic pathways of apoptosis are characterized by the export of apoptogenic proteins such as cyto c from mitochondria to the cytosol (Tait and Green, 2013; Bock and Tait, 2020). Previous work showed that punctate JMY structures can overlap with mitochondria-independent cyto c and that cytosolic cyto c puncta are not detectable in JMY knockout cells (King et al., 2021), so we next evaluated the localization of multiple mitochondrial components in cells undergoing apoptosis. First, to visualize all mitochondria, we stained etoposide-treated cells with an antibody that recognizes mitochondrial DNA (mtDNA). Mitochondria were present throughout the cell, although their DNA was more strongly stained within the F-actin–rich territory than outside of it (Figure 4, A and B). Second, we treated cells with MitoTracker Red, a fluorescent probe that accumulates in mitochondria in a membrane potential–dependent manner. MitoTracker fluorescence appeared in typical chain-like patterns outside of the territories but as diffuse clouds within the territories (Figure 4A). Circular cyto c puncta were found in the region of diffuse MitoTracker staining (Figure 4A), similar to previous observations (King et al., 2021). These results are consistent with the idea that mitochondria may be specifically depolarized and permeabilized within the F-actin–rich territories.
FIGURE 4:
Permeabilized mitochondria located within F-actin–rich territories. (A) U2OS cells were treated with etoposide for 6 h, fixed, and stained with antibodies to JMY (green), mitochondrial DNA (mtDNA; magenta), BAX (magenta), and AIF (yellow) and with phalloidin (F-actin; cyan). In row 2, MitoTracker Red (magenta) was added before fixation. Scale bars: 25, 5 μm. (B) Lines (20 μm) were drawn through the images in A to measure the pixel intensity profiles. RFU = relative fluorescence units.
As a more direct test of whether mitochondrial permeabilization takes place in this juxtanuclear area, we stained cells for BAX, a key apoptotic regulator that resides in the cytosol in healthy cells but accumulates on the mitochondrial outer membrane in apoptotic cells, where it forms pores to promote cyto c export (Wolter et al., 1997; Popgeorgiev et al., 2018). Strikingly, BAX staining was concentrated within the F-actin–rich territories (Figure 4, A and B), suggesting that permeabilized mitochondria were enriched in this cytoskeletal region. For an independent readout of permeabilized mitochondria, we stained cells for apoptosis-inducing factor (AIF), a protein that, like cyto c, resides in the intermembrane space and upon proapoptotic signaling is released from the mitochondria (Candé et al., 2002). Similar to cyto c, AIF was visible in mitochondria outside of F-actin–rich territories (Figure 4A). However, within the territories, cyto c formed mitochondria-independent puncta, whereas AIF was generally absent, even when mtDNA and BAX were there (Figure 4, A and B). Thus, F-actin–rich territories appear to encompass sites of mitochondrial permeabilization, and among multiple released apoptogenic proteins, only cyto c forms prominent puncta.
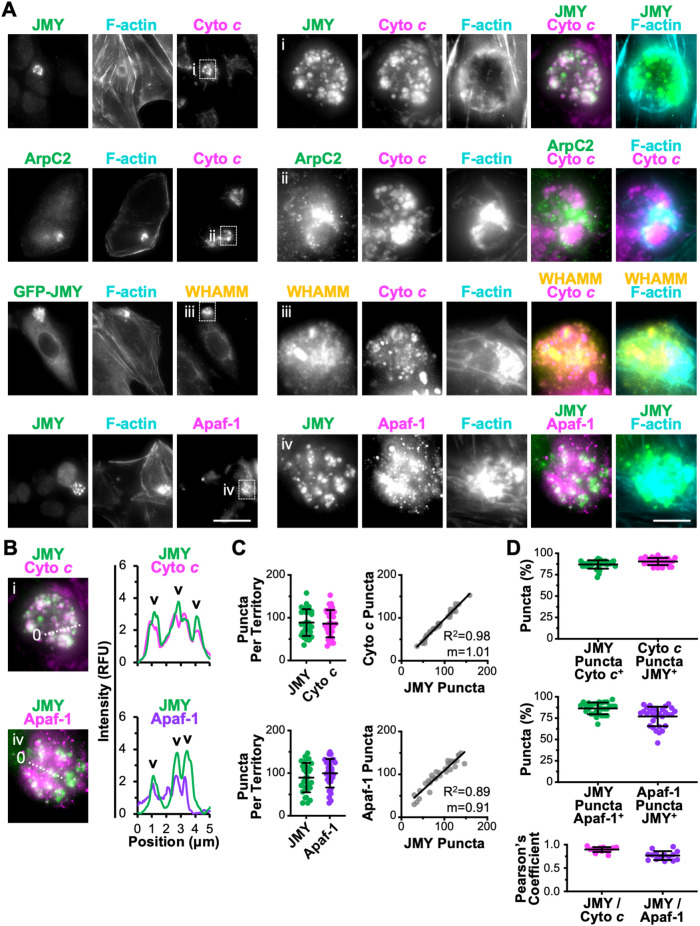
Because exported cyto c had this distinct juxtanuclear localization pattern, we next examined the relationship of cyto c to the cytoskeletal components in the territories. Consistent with earlier results (King et al., 2021), many cytosolic cyto c puncta colocalized with bright JMY puncta that were clustered within the F-actin–rich regions (Figure 5A). Such cyto c puncta also formed in close proximity to Arp2/3 complex and WHAMM structures, but with less overlap (Figure 5A).
FIGURE 5:
Apoptosome components cyto c and Apaf-1 clustered within F-actin–rich territories. (A) U2OS cells (rows 1, 2, and 4) or U2OS cells encoding GFP-JMY (row 3; green) were treated with etoposide for 6 h, fixed, and stained with antibodies to JMY (rows 1, 2, and 4; green), cyto c or Apaf-1 (magenta), and/or WHAMM (yellow) and with phalloidin (F-actin; cyan). Magnifications (i–iv) depict JMY, WHAMM, cyto c, and Apaf-1 within F-actin territories. Scale bars: 25, 5 μm. (B) Lines (5 μm) were drawn through the magnified images in i and iv to measure the pixel intensity profiles. Arrowheads highlight examples of cytosolic cyto c or Apaf-1 puncta that overlap with JMY puncta. (C) For quantification of A, the JMY and cyto c or Apaf-1 puncta were counted per territory. Each point represents one cell (n = 30). The numbers of cyto c or Apaf-1 puncta per territory were plotted against the number of JMY puncta per territory. The slopes (m) in the linear trendline equations for cyto c (Y = 1.01X – 3.61) and Apaf-1 (Y = 0.91X + 18.02) were significantly nonzero (p < 0.001). (D) Quantifications show the percentage of JMY puncta per territory that overlapped with cyto c or Apaf-1 puncta (cyto c or Apaf-1 positive) and the percentage of cyto c or Apaf-1 puncta per territory that overlapped with JMY puncta (JMY-positive). The Pearson’s correlation coefficient for JMY with cyto c or Apaf-1 was calculated for puncta within territories. Each point represents the coefficient within a single territory (n = 15). Pearson’s coefficient for JMY/cyto c (0.90 ± 0.05) was significantly different from that for JMY/mtDNA (0.05 ± 0.04), and Pearson’s coefficient for JMY/Apaf-1 (0.77 ± 0.09) was significantly different from that for JMY/mtDNA (0.06 ± 0.04), p < 0.001 (t tests). See Supplemental Figure S4 for other factors that were not enriched in F-actin territories.
After cyto c becomes cytosolic, its primary proapoptotic function is to interact with the scaffolding protein Apaf-1 and trigger Apaf-1 oligomerization into heptameric complexes called apoptosomes, which serve as platforms for the multimerization and proteolytic processing of caspases (Bratton and Salvesen, 2010; Yuan and Akey, 2013; Dorstyn et al., 2018). To explore whether Apaf-1–containing apoptosomes might form within the F-actin territories, we stained cells with Apaf-1 antibodies. Apaf-1 puncta were indeed enriched within the territories and sometimes overlapped with JMY structures (Figure 5A). Fluorescence intensity plot profiles showed colocalization between JMY and cyto c puncta as well as between JMY and several larger Apaf-1 puncta (Figure 5B). Therefore, the cytosolic JMY-containing F-actin–rich areas contain both of the core apoptosome proteins.
To further investigate whether JMY influences apoptosome formation within the territories, we characterized the local abundance of apoptosome components relative to JMY. Quantification of the number of individual puncta gave median values of 89 JMY puncta and 86 cyto c puncta per F-actin–rich territory (Figure 5C). The numbers of puncta varied from territory to territory, but within individual territories the numbers of cyto c and JMY puncta showed a positive correlation (Figure 5C). When assessing their colocalization by considering both total and partial overlap, 87% of JMY puncta were cyto c positive and 90% of cyto c puncta were JMY positive (Figure 5D). Moreover, the Pearson’s correlation coefficient for JMY and cyto c pixels within F-actin–rich territories was 0.90 (Figure 5D). Assessments of JMY and Apaf-1 structures within F-actin territories revealed similar phenotypes, including median values of 89 JMY puncta and 100 Apaf-1 puncta per territory and a positive correlation between the numbers of JMY and Apaf-1 puncta (Figure 5C). Eighty-seven percent of JMY puncta showed overlap with Apaf-1, while 77% of Apaf-1 puncta showed overlap with JMY, and the Pearson’s correlation coefficient for JMY and Apaf-1 pixels within F-actin–rich territories was 0.77 (Figure 5D), indicating that many JMY and Apaf-1 structures also localize together, although not to the same extent as JMY and cyto c. Collectively, these results suggest that individual F-actin–rich territories harbor relatively stoichiometric amounts of JMY, cyto c, and Apaf-1 structures.
For gauging the specificity with which proapoptotic factors accumulate in the territories, we also examined the localization of other cellular components by visualizing p53 or the Golgi in relation to JMY and F-actin. Similar to AIF, and consistent with a primarily nuclear function during apoptosis, p53 was not enriched within cytosolic JMY-associated F-actin territories (Supplemental Figure S5). WHAMM and JMY promote vesicle trafficking to and from the Golgi (Campellone et al., 2008; Schlüter et al., 2014), and several Golgi proteins can regulate apoptosis (He et al., 2020), but a Golgi marker, GM130, was not found within the F-actin regions either (Supplemental Figure S5). These findings support the idea that F-actin–rich territories function as compartments for grouping specific subsets of proapoptotic factors in close proximity to one another.
Apoptotic F-actin–rich territories concentrate active executioner caspases
If cyto c and Apaf-1 come together in F-actin–rich territories to efficiently form apoptosomes, then the territories should also impact subsequent steps of the intrinsic apoptotic signaling cascade. Apoptosomes activate initiator caspases, leading to the activation of executioner caspases, which cleave multiple proteins in the cytosol and nucleus to drive apoptotic cell death (Dorstyn et al., 2018; Bock and Tait, 2020). While structural and biochemical studies have shown apoptosomes to be important for starting caspase cleavage cascades through interactions between Apaf-1 and initiator procaspase-9 (Li et al., 2017; Su et al., 2017; Wang et al., 2017), the spatiotemporal mechanisms underlying caspase recruitment to, and activation by, apoptosomes in cells are not well understood. Because we previously demonstrated that JMY knockout fibroblasts have defects in the cleavage of initiator caspase-9 and executioner caspase-3 (King et al., 2021), we next evaluated the localization of caspase-9 and caspase-3 in relation to apoptotic F-actin–rich territories. Staining of etoposide-treated cells with antibodies to total caspase-9 (TCasp-9) and total caspase-3 (TCasp-3), which recognize both their inactive full-length and active cleaved forms, showed that both caspases were present in the JMY-containing F-actin regions (Supplemental Figure S6).
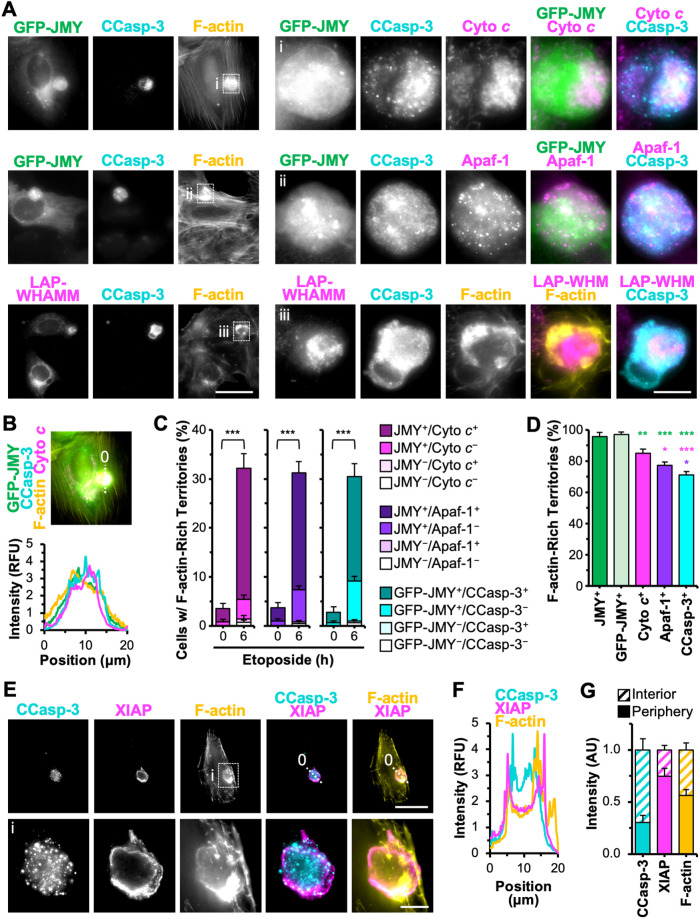
Cleaved caspase-3 (CCasp-3) can associate with JMY and F-actin in apoptotic cells (King et al., 2021), so to next determine whether the apoptosome-containing territories harbored active caspases, we stained U2OS cells with antibodies that specifically recognize active CCasp-3. CCasp-3 fluorescence was usually present entirely within F-actin-rich territories (Figure 6, A and B, and Supplemental Figure S6). CCasp-3 staining sometimes overlapped with JMY and WHAMM structures and was also superimposed with clusters of cyto c and punctate Apaf-1 (Figure 6A), indicating that JMY, WHAMM, apoptosomes, initiator caspases, and active executioner caspases altogether localize within F-actin–rich territories.
FIGURE 6:
Active executioner caspase-3 concentrated within apoptosome-containing, F-actin–rich territories. (A) U2OS cells expressing GFP-JMY (green) or transfected with a plasmid encoding LAP-WHAMM (magenta) were treated with etoposide for 6 h, fixed, and stained with an antibody that recognizes CCasp-3 at Asp175 (cyan), with antibodies against cyto c or Apaf-1 (magenta), and with phalloidin (F-actin; yellow). Magnifications (i–iii) depict GFP-JMY, cyto c, Apaf-1, CCasp-3, and LAP-WHAMM within F-actin territories. Scale bars: 25, 5 μm. (B) A 20 μm line was drawn through an image from A to measure the pixel intensity profiles for GFP-JMY, CCasp-3, F-actin, and cyto c. (C, D) U2OS cells or U2OS cells expressing GFP-JMY were untreated or treated with etoposide, fixed, and stained with antibodies against JMY and cyto c, JMY and Apaf-1, or CCasp-3, all in conjunction with phalloidin. The percentage of cells harboring F-actin–rich territories containing JMY, GFP-JMY, cyto c, or Apaf-1 puncta or with CCasp-3 clusters was quantified. In C, the fractions of F-actin–rich territories that were doubly positive, singly positive, or doubly negative are shown. Each bar represents the mean ± SD from three experiments (n = 504–638 cells per bar). ***p < 0.001 (t tests). In D, each bar represents the mean ± SD from three experiments (n = 182–195 territories per bar). Green, magenta, and purple significance stars refer to comparisons to the JMY+ bar, cyto c+ bar, and Apaf-1+ bar, respectively. (E) U2OS cells were treated with etoposide, fixed, and stained with antibodies to CCasp-3 (cyan) and XIAP (magenta) and with phalloidin (F-actin; yellow). Magnifications (i) depict F-actin territories surrounded by XIAP. Scale bars: 25, 5 μm. (F) A 20 μm line was drawn through the image in E to measure the pixel intensity profiles for CCasp-3, XIAP, and F-actin. (G) CCasp-3, XIAP, and F-actin fluorescence intensities were measured in the whole territory as well as the interior and peripheral portions (n = 15 territories). *p < 0.05; **p < 0.01; ***p < 0.001 (ANOVA, Tukey post-hoc tests).
Given that the canonical intrinsic pathway of apoptosis is considered a stepwise process starting with the mitochondrial release of cyto c, which triggers the assembly of the Apaf-1 apoptosome, allowing for the activation of initiator and then executioner caspases, we next wanted to better understand where JMY and F-actin fit in this pathway. To begin to do this, we measured the frequency with which F-actin–rich territories harbored JMY puncta, apoptosome components, and active caspases after 6 h of etoposide treatment, when F-actin–rich territories were present in >30% of cells (Figures 1B and 6C). The majority of F-actin territories contained a pairwise combination of JMY with cyto c, Apaf-1, or CCasp-3 staining, as ∼25% of cells included F-actin–rich regions that were positive for both JMY and the apoptosome components, while ∼20% were positive for both GFP-JMY and CCasp-3 (Figure 6C). In contrast, only 5–10% of cells had F-actin territories containing JMY puncta without apoptosome components or active caspase-3, and territories that lacked JMY, cyto c, Apaf-1, and CCasp-3 staining were even rarer (Figure 6C). When quantifying the proportion of F-actin territories that stained for JMY, apoptosome proteins, or active caspases, >96% of F-actin territories were JMY puncta positive, 85% were cyto c positive, 77% were Apaf-1 positive, and 71% were CCasp-3 positive (Figure 6D). The frequencies with which F-actin–rich territories include JMY puncta, apoptosomes, and executioner caspases are consistent with the conclusion that a stepwise pathway from mitochondrial permeabilization to apoptosome biogenesis to caspase activation all takes place within this discrete domain of the cytosol.
Progression of the canonical intrinsic apoptotic pathway can be controlled by negative regulators as well, and members of the inhibitor of apoptosis protein (IAP) family prevent apoptosis by blocking caspase activation or activity (Hrdinka and Yabal, 2019). One such caspase inhibitor is XIAP, which binds to caspase-9, -3, and -7, thereby blocking their proteolytic activities (Deveraux and Reed, 1999; Scott et al., 2005; Eckelman et al., 2006). We next monitored the spatial relationship of XIAP to JMY, CCasp-3, and F-actin. XIAP was recruited to JMY-containing, F-actin–rich territories, but in general XIAP staining did not overlap with the JMY puncta (Supplemental Figure S6). Instead, it was found around the perimeters of the territories (Figure 6E and Supplemental Figure S6). Fluorescence intensity plot profiles confirmed XIAP intensity peaking at the peripheral boundaries of the territory rather than in association with the interior JMY and CCasp-3 structures (Figure 6F and Supplemental Figure S6). Measurements of JMY, CCasp-3, XIAP, and F-actin levels at the inner versus outer portions of an assortment of territories revealed that JMY and CCasp-3 quantities were, on average, three times higher in internal regions, whereas XIAP was three times higher at the perimeters (Figure 6G and Supplemental Figure S6). The locations of F-actin enrichment varied from territory to territory (Figure 2A) and averaged out to be similar in the inner and outer zones (Figure 6G and Supplemental Figure S6). These results indicate that the apoptotic F-actin–rich territories not only compartmentalize apoptosomes, initiator caspases, and executioner caspases but also feature a specialized peripheral positioning of the caspase inhibitor XIAP.
Localized JMY-mediated Arp2/3 activation and actin polymerization are crucial for driving executioner caspase activation
Evidence from the intermediate filament system suggests that caspase inclusions may form due to the cleavage of cytoskeletal factors and their collapse around the caspases (MacFarlane et al., 2000; Lee et al., 2002; Dinsdale et al., 2004), while our emerging model is that the formation of caspase-containing cytoskeletal territories is driven by nucleation factor–mediated actin assembly. To further distinguish between these possibilities, we tested whether caspase activity was required for F-actin–rich territory formation by performing etoposide-induced apoptosis experiments in the absence or presence of the broad caspase inhibitor Z-VAD-fmk or the executioner-specific inhibitor Z-DEVD-fmk. The inhibitors did not reduce territory formation; they increased it (Supplemental Figure S7). Z-VAD-fmk, which is capable of inhibiting initiator caspase-9, prevented executioner CCasp-3 accumulation within the territories (Supplemental Figure S7). These findings oppose the idea that caspase activity is necessary for F-actin–rich territory biogenesis.
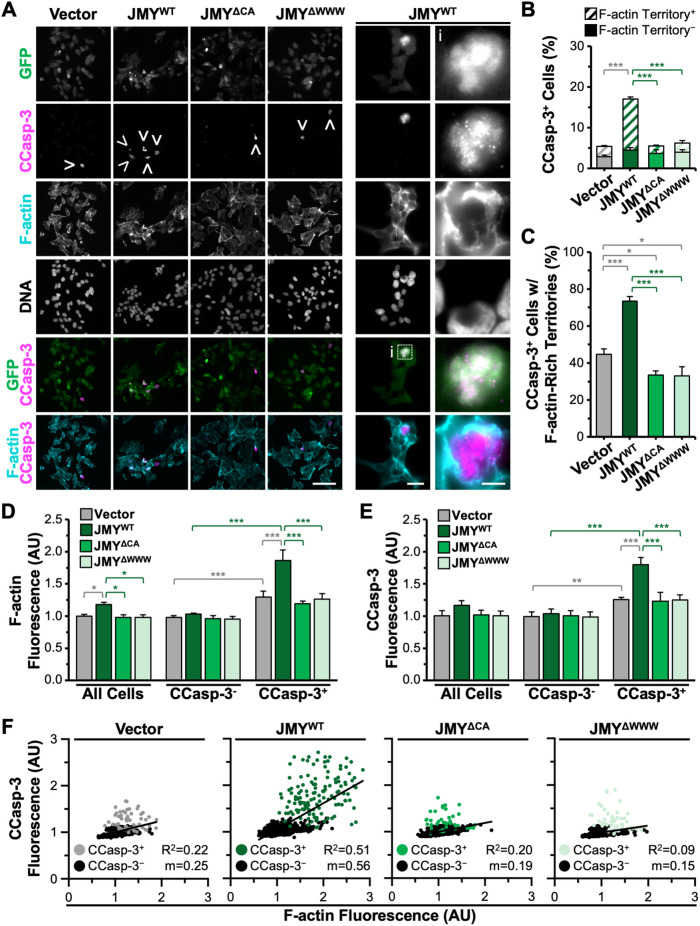
Recent work showed that JMY requires its actin- and Arp2/3-binding domains in order to enable efficient caspase-3 cleavage (King et al., 2021), but the degree to which actin assembly specifically within territories impacts caspase-3 processing is unknown. JMY activates the Arp2/3 complex using a C-terminal domain containing three actin-binding WASP-homology-2 (W) motifs and an Arp2/3-binding connector-acidic (CA) segment and can also use its WWW region to nucleate actin directly (Zuchero et al., 2009). JMY knockout (JMYKO) fibroblasts are deficient in caspase-3 activation (King et al., 2021), so we utilized several GFP-tagged JMY derivatives for performing rescue experiments: wild-type JMY (JMYWT); a JMY mutant missing all three W motifs (JMYΔWWW) that can neither nucleate actin directly nor cooperate with the Arp2/3 complex; and a JMY mutant lacking the CA segment (JMYΔCA) that cannot activate Arp2/3 but retains its actin nucleating WWW region. We introduced the GFP-JMY plasmids, or a vector control, into the JMYKO cells, exposed them to etoposide, and stained them to visualize active CCasp-3 and F-actin (Figure 7A). Similar to previous observations (King et al., 2021), just ∼5% of vector- or JMY mutant–transfected JMYKO cells stained positive for active CCasp-3 (Figure 7, A and B), while >15% of cells transfected with the full-length JMY construct were CCasp-3 positive (Figure 7, A and B), indicating that only wild-type JMY could improve executioner caspase activation in JMYKO cells.
FIGURE 7:
Localized activation of caspase-3 is controlled by JMY-dependent actin polymerization. (A) JMYKO cells encoding GFP (vector) or GFP-tagged JMY constructs (JMYWT, JMYΔCA, JMYΔWWW; green) were treated with etoposide for 5 h, fixed, and stained with an antibody to visualize CCasp-3 (magenta), phalloidin (F-actin; cyan), and DAPI to detect DNA. Arrowheads highlight examples of CCasp-3 staining, and magnifications (i) depict GFP-JMYWT and CCasp-3 surrounded by F-actin. Scale bars: 100, 25, 5 μm. (B) The percentage of cells with CCasp-3 staining was quantified and is displayed as the fraction of CCasp-3–positive cells that were (Territory+) or were not (Territory-) associated with F-actin. Each bar represents the mean ± SD from three experiments (n = 3447–6083 cells analyzed per construct). Gray significance stars refer to comparisons to the vector bar, and green significance stars are comparisons to the JMYWT bar. (C) The percentage of cells with CCasp-3 staining associated with F-actin was quantified from B. Each bar is the mean ± SD from three experiments (n = 1038 cells for WT and 190–230 for vector and mutant samples). (D, E) Whole cell fluorescence values for F-actin and CCasp-3 were measured, and the mean value for the vector sample was set to 1. Each bar is the mean ± SD from three experiments (n = 592–816 cells per sample). (F) Whole cell fluorescence values for F-actin and CCasp-3 were plotted against one another. Each point represents one cell (n = 592–816 cells per sample), where gray/green points are CCasp-3–positive cells and black points are CCasp-3–negative cells. The slopes (m) in the linear trendline regression equations for the total cell populations (vector: Y = 0.25X + 0.75; JMYWT: Y = 0.56X + 0.49; JMYΔCA: Y = 0.19X + 0.81; JMYΔWWW: Y = 0.15X + 0.84) were significantly nonzero (p < 0.001). *p < 0.05; **p < 0.01; ***p < 0.001 (ANOVA, Tukey post-hoc tests).
To explore the precise relationship between caspase-3 activation and F-actin territory formation, we quantified the proportion of CCasp-3–positive cells that had active caspase staining localized within the F-actin–rich regions. Out of the infrequent CCasp-3–positive cells observed in the vector-transfected JMYKO sample, less than half were associated with F-actin–rich territories (Figure 7, B and C). Compared to CCasp-3–positive cells that had strong active caspase staining clustered within F-actin–rich regions, those that were independent of F-actin displayed weaker CCasp-3 staining that was more diffuse throughout the cell. In contrast, for samples transfected with the wild-type JMY construct, 75% of the active CCasp-3–positive cells had their staining encompassed within F-actin–rich territories (Figure 7, B and C). For both JMY mutants, the frequency of active caspase staining that associated with F-actin–rich territories slightly decreased relative to that of vector-transfected cells (Figure 7C). These results show that the actin polymerization capacity of JMY is required for localized caspase activation specifically within F-actin–rich territories.
We next tested whether the amount of F-actin or CCasp-3 differed in the absence or presence of the distinct JMY derivatives by quantifying the F-actin and CCasp-3 fluorescence intensities for individual cells. Mean F-actin fluorescence values were higher in the cells transfected with wild-type JMY compared with the vector- or mutant-transfected JMYKO cells (Figure 7D). Moreover, when total cell populations were split into CCasp-3–positive or –negative groups, the CCasp-3–positive cells showed greater F-actin fluorescence compared with the negative cells, and the JMYWT reexpressing cells contained significantly more F-actin than the vector-transfected JMYKO cells (Figure 7D). The increased whole cell F-actin content observed in the JMYWT sample can therefore be attributed to the apoptotic territories.
Within the CCasp-3–positive population, JMYWT reexpressing cells displayed significantly greater CCasp-3 fluorescence compared with the vector-expressing cells (Figure 7E). In contrast, the JMYΔWWW and JMYΔCA samples behaved identically to vector-transfected cells in assays measuring both F-actin and CCasp-3 levels (Figure 7, D and E). To directly relate caspase-3 activation to F-actin abundance, we plotted CCasp-3 and F-actin fluorescence against one another on a cell-by-cell basis. Such comparisons demonstrated that cells transfected with wild-type JMY exhibited a stronger positive correlation between the intensity of CCasp-3 and F-actin fluorescence than did vector- or mutant-transfected cells (Figure 7F). These results collectively show that a more frequent and more efficient activation of caspase-3 occurs when it is confined within F-actin–rich territories and that the amount of actin polymerized via JMY has a direct positive relationship with the potency of caspase-3 activation.
JMY and WHAMM differentially contribute to F-actin–rich territory assembly and clustering-mediated caspase activation
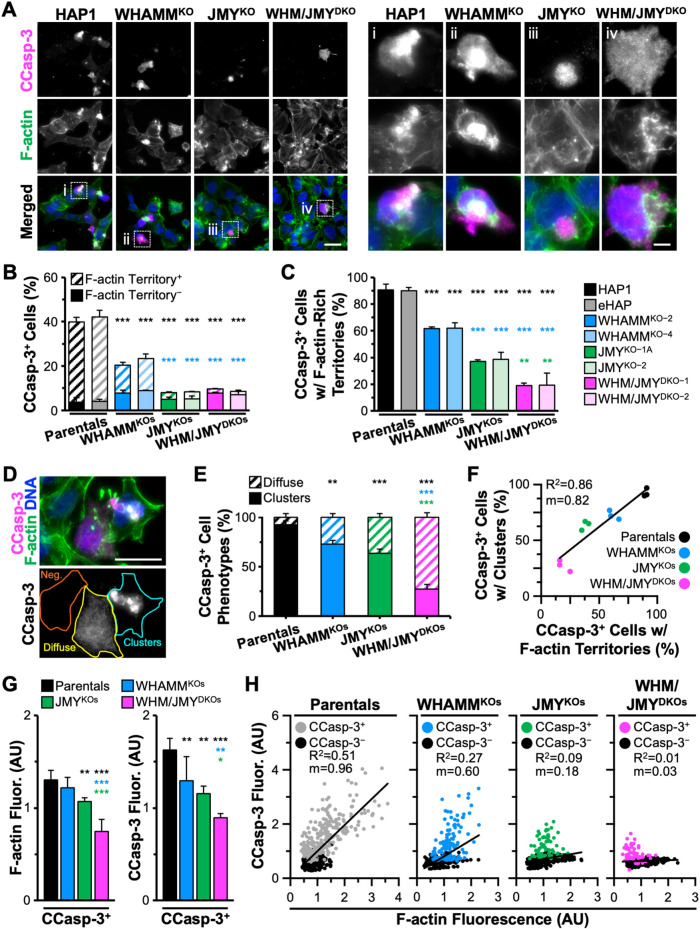
Although JMY is crucial for assembling apoptotic F-actin territories where caspase-3 is activated, some JMY-deficient cells still contain actin-associated caspase-3 staining (Figure 7B). To clarify the molecular basis for forming the cytoskeletal territories, we assessed the relative contributions of JMY and WHAMM. We employed two independent JMY KO fibroblast cell lines, two independent WHAMM KO lines, and two WHAMM/JMY double-knockout (DKO) lines, exposed them to etoposide, and stained them to visualize active CCasp-3 and F-actin. Similar to previous observations (King et al., 2021), ∼40% of parental (HAP1, eHAP) cells stained positive for active CCasp-3, whereas only ∼20% of WHAMMKO and <10% of the JMYKO or WHAMM/JMYDKO cells were positive for CCasp-3 staining (Figure 8, A and B), reaffirming that wild-type cells can execute an efficient cell death program whereas JMYKO or WHAMMKO cells have significant deficiencies in executioner caspase activation.
FIGURE 8:
Amount of F-actin encompassing the caspase activation clusters enhanced by WHAMM. (A) Parental (HAP1, eHAP), WHAMMKO (WHAMMKO-2, WHAMMKO-4), JMYKO (JMYKO-1A, JMYKO-2), and WHAMM/JMYDKO (WHM/JMYDKO-1, WHM/JMYDKO-2) cells were treated with 5 μM etoposide for 6 h before fixing and staining with an antibody to visualize CCasp-3 (magenta), phalloidin (F-actin; green), and DAPI (blue). Magnifications (i–iv) show examples of CCasp-3 staining. Scale bars: 25, 5 μm. (B) The percentage of cells with CCasp-3 staining was quantified and is displayed as the fraction of CCasp-3–positive cells that were (Territory+) or were not (Territory-) associated with F-actin–rich territories. Each bar represents the mean ± SD from three experiments (n = 246–438 cells analyzed per bar). Black significance stars refer to comparisons to the parental bars, and blue significance stars are comparisons to the WHAMMKO bars. (C) The percentage of CCasp-3–positive cells with F-actin–rich territories is displayed. Each bar represents the mean ± SD from three experiments (n = 104–106 cells for parental bars; 66–73 cells for WHAMMKO bars; and 31–36 cells for JMYKO or WHM/JMYDKO bars). Black significance stars are comparisons to the parental bars, blue significance stars are to the WHAMMKO, and green significance stars are to the JMYKO. (D) A representative image of JMYKO cells highlights cells that are CCasp-3 negative (orange), CCasp-3 positive but diffuse (yellow), or CCasp-3 positive in clusters (cyan). Scale bar: 25 μm. (E) The percentage of CCasp-3–positive cells with clustered or diffuse phenotypes was quantified. Each bar represents the mean ± SD from three experiments (n = 63–98 cells per bar). (F) The percentage of CCasp-3–positive cells with a clustered phenotype was plotted against the percentage of CCasp-3–positive cells with F-actin territories. Each point represents the mean from an individual experiment (n = 18–39 cells per point; 63–98 cells per sample). The slope (m) in the linear trendline equation (Y = 0.82X + 20.97) was significantly nonzero (p < 0.001). (G) Whole cell fluorescence values for F-actin and CCasp-3 were measured in individual cells and normalized to the parental sample. Each bar is the mean ± SD from three experiments (n = 607–866 cells per sample). Total and CCasp-3–negative cell data appear in Supplemental Figure S8. (H) Whole cell fluorescence values for CCasp-3 and F-actin were plotted against one another. Each point represents one cell (n = 607–866 cells per plot), where gray, blue, green, or pink points are CCasp-3 positive, and black points are CCasp-3 negative. The slopes (m) in the linear trendline regression equations for Parentals (Y = 0.96X + 0.04), WHAMMKOs (Y = 0.60X + 0.20), and JMYKOs (Y = 0.18X + 0.53) were significantly nonzero (p < 0.001), while the slope for the WHAMM/JMYDKO sample (Y = 0.03X + 0.61) was not (p = 0.070). *p < 0.05; **p < 0.01; ***p < 0.001 (ANOVA, Tukey post-hoc tests in B–D, G; Fisher’s exact test in E).
To determine the extent to which each cell line activated caspase-3 within F-actin–rich territories, we quantified the percentage of CCasp-3–positive cells that were positive for F-actin. In >90% of CCasp-3–positive parental cells, caspase staining was found in F-actin–rich territories (Figure 8, B and C). In contrast, ∼60% of WHAMMKO, <40% of JMYKO, and <20% of WHAMM/JMYDKO cells had CCasp-3 staining in conjunction with actin filaments (Figure 8C). Two staining phenotypes for CCasp-3–positive cells were apparent—clustered or diffuse (Figure 8D)—and classifying cells in these two categories even without phalloidin staining was predictive of the genotype of the cells. Independent assessments of the fraction of CCasp-3–positive cells with each phenotype demonstrated that >90% of CCasp-3–positive wild-type cells displayed a clustered staining pattern, leaving <10% with a diffuse localization (Figure 8E). The proportion of diffuse CCasp-3–positive cells increased to >25% of WHAMMKO, >35% of JMYKO, and >70% of WHAMM/JMYDKO CCasp-3–positive cells (Figure 8E). When reincorporating the F-actin staining data and plotting all genotypes, the percentage of CCasp-3–positive cells showing a clustered phenotype positively correlated with the percentage of CCasp-3–positive cells containing F-actin territories (Figure 8F). Thus, while a WHAMM mutation results in partial deficits in the clustering and frequency of caspase-3 activation and a JMY mutation causes severe impairments, a WHAMM/JMY double mutation causes even more extreme defects.
Finally, to determine how deleting WHAMM and/or JMY influences the amounts of active caspase-3 and actin filaments during apoptosis, we quantified the fluorescence levels of CCasp-3 and F-actin. Cells classified as CCasp-3 positive were not all equivalent, as average CCasp-3 and F-actin intensities each displayed stepwise decreases when WHAMM, JMY, or both factors were deleted (Figure 8G and Supplemental Figure S8). Single cell analyses of wild-type samples further showed a positive correlation between CCasp-3 levels and F-actin abundance (Figure 8H). In contrast, the WHAMMKO and JMYKO cells showed weaker correlations between CCasp-3 and F-actin fluorescence, with a more severe phenotype in the JMYKO cells (Figure 8H). The WHAMM/JMYDKOs did not show any correlation between CCasp-3 and F-actin fluorescence per cell (Figure 8H). These results point to a prominent role for JMY and a secondary role for WHAMM in promoting apoptotic F-actin–rich territory assembly and driving compartmentalized caspase activation.
DISCUSSION
Cytoskeletal studies of apoptotic cell death have often focused on the changes in actin filament disassembly that drive terminal morphological phenotypes. However, roles for actin assembly proteins as active participants earlier in apoptosis have recently been uncovered, as the WASP family members JMY and WHAMM both contribute to a DNA damage–induced apoptosis pathway after p53-mediated cell cycle arrest (King et al., 2021). Our work now shows that JMY- and WHAMM-derived F-actin–rich territories promote programmed cell death by creating a discrete juxtanuclear environment for apoptosome biogenesis and caspase activation (Figure 9). These findings reveal a new regulatory mechanism governing the spatiotemporal progression of intrinsic apoptosis in cells and create a framework for understanding how dynamic cytoskeletal responses can organize specific signaling pathways.
FIGURE 9:
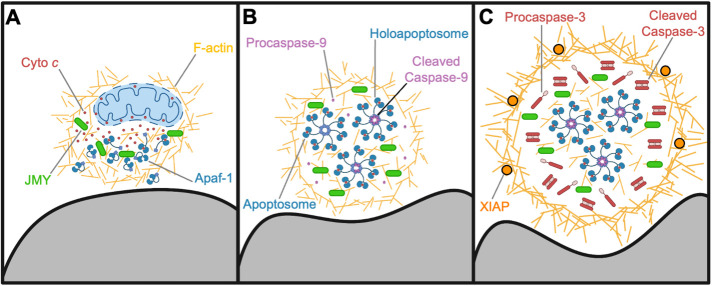
Model for apoptotic F-actin–rich territory assembly in coordinating apoptosome biogenesis and caspase activation. (A) Mitochondrial outer membrane permeabilization and release of cyto c (red circles) into the cytosol result in recruitment of JMY (green) and Arp2/3 complex–mediated assembly of actin filaments (gold) in a juxtanuclear region. WHAMM enhances actin polymerization and branching, and the early F-actin–rich territory sequesters cyto c and enables interactions with Apaf-1 (blue monomers). (B) F-actin remodeling and territory maturation create a microenvironment conducive to the biogenesis and concentration of apoptosomes (blue/red heptamers). Initiator procaspase-9 (light purple) incorporates into holoapoptosomes (heptamers with purple hubs), which are retained within the territory. (C) The F-actin–rich territory increases in density to create a transient subcellular compartment for optimizing holoapoptosome-mediated processing of executioner procaspase-3 (red monomers) into active CCasp-3 (red dimers). F-actin functions to cluster the caspase activation process in the internal portion of the territory, while dense peripheral F-actin networks and associated proteins restrict accessibility of the caspase inhibitor XIAP (orange circles). Eventually, active caspase-3 achieves high enough quantities to bypass XIAP, escape the territory, and trigger the rapid proteolysis of substrates throughout the rest of the cell. Note that the stoichiometry and size of organelles and macromolecules shown in the figure (e.g., mitochondria, F-actin, apoptosomes, proteins) are not drawn to scale.
JMY has the ability to shuttle between the nucleus and cytosol (Zuchero et al., 2012), and we found that endogenous JMY is primarily nuclear at steady state. In contrast, after etoposide-mediated genotoxic damage, the cytosolic abundance of JMY increases significantly—a phenotype that is due to the formation of a cluster of JMY puncta. The accumulation of juxtanuclear JMY puncta is then followed by the polymerization and reorganization of F-actin into a unique cytoskeletal territory. WHAMM and the Arp2/3 complex also localize throughout the JMY-containing, F-actin territories, revealing that these structures contain the specific members of the actin assembly machinery that were previously found to be important for intrinsic apoptosis (King et al., 2021). STRAP, an inhibitor of JMY-mediated actin assembly (Hu and Mullins, 2019), and microtubules, which can repress WHAMM-mediated Arp2/3 activation (Shen et al., 2012), are absent from the F-actin–rich regions. The cis-Golgi is also excluded, whereas the F-actin–binding protein cortactin localizes to the peripheral actin filaments, and other proteins like p53 and WAVE2 are present indiscriminately inside and outside the territories. These data are consistent with the idea that JMY and WHAMM stimulate Arp2/3-mediated branched actin assembly to construct a proapoptotic cytoskeletal compartment for sorting distinct subsets of subcellular components.
Mitochondrial permeabilization, a critical point in the cellular commitment to apoptosis, is also influenced by JMY and WHAMM. Mitochondrial outer membrane remodeling is driven by BCL-2 family proteins, which enable the export of cyto c to the cytosol. Some evidence indicates that this process initially occurs in a subset of mitochondria before propagating throughout the cell (Bhola et al., 2009; Rehm et al., 2009; Bock and Tait, 2020). In etoposide-treated cells, JMY puncta were previously seen to partially colocalize with mitochondria-independent cytosolic cyto c, and JMY was found to be required for the formation of the cyto c puncta themselves (King et al., 2021). These observations, when combined with our current findings that BAX-positive mitochondria along with the JMY and cyto c structures are confined to a single juxtanuclear F-actin–rich territory, imply that the initial site of mitochondrial permeabilization and the location of territory assembly are one and the same (Figure 9A).
Apoptosomes, macromolecular scaffolds composed of cyto c and Apaf-1, are central players in intrinsic apoptosis and another focal point of regulation (Bratton and Salvesen, 2010; Yuan and Akey, 2013; Dorstyn et al., 2018). Biochemical and structural studies have defined the composition, configuration, and basic requirements for apoptosome biogenesis in vitro, but the mechanisms controlling its assembly, organization, and activity are less well understood in cells. Cyto c and Apaf-1 are both enriched in F-actin territories, and JMY puncta also overlap with Apaf-1 puncta, albeit to a lesser extent than with cyto c. These data lead to the conclusions that apoptotic F-actin–rich territories compartmentalize the process of apoptosome biogenesis and that efficient apoptosome formation in cells is dependent on JMY.
Apaf-1 monomers exist in a closed conformation, and cyto c binding triggers structural changes that allow the assembly of apoptosomes composed of seven Apaf-1 and cyto c subunits arranged in a wheel-like structure with spokes that radiate from a central hub (Yuan et al., 2010, 2013; Zhou et al., 2015; Cheng et al., 2016; Sahebazzamani et al., 2021). Apart from this structural information, the cellular mechanisms that dictate cyto c localization in the cytosol as well as its interactions with Apaf-1 are obscure. Our data address this subject, as JMY and F-actin appear to mediate the early sequestration of mitochondria-released cyto c before its incorporation into apoptosomes. How Apaf-1 is recruited to territories is unknown, but the initial presence of cyto c within these nascent compartments likely facilitates Apaf-1 retention and integration into mature apoptosomes. Because most apoptotic F-actin–rich territories contain similar numbers of cyto c and Apaf-1 puncta, we believe that the majority of cyto c and Apaf-1 structures visualized by immunofluorescence microscopy comprise groups of mature apoptosomes (Figure 9B).
Apoptosomes are important for the initiation of the caspase cleavage cascade by serving as platforms that interact with procaspase-9 monomers, resulting in their multimerization and activation (Li et al., 2017). Some evidence suggests that, after activation, caspase-9 remains bound to the apoptosome, creating holoapoptosomes, where it is able to process executioner caspases (Rodriquez and Lazebnik, 1999; Yin et al., 2006; Malladi et al., 2009; Yuan et al., 2011; Hu et al., 2014). However, the intracellular mechanisms underlying caspase recruitment to and activation by apoptosomes are not well characterized. Clusters of initiator caspase-9 and executioner caspase-3 are found in close proximity to collections of punctate JMY and apoptosomal proteins within F-actin–rich territories, leading us to conclude that these cytoskeletal structures are particularly conducive to apoptosome-mediated initiation of the caspase cleavage cascade.
The intense localization of cleaved executioner caspase-3 within the territories further shows that these apoptosome-enriched areas are maintained through the stage of the intrinsic pathway in which the terminal caspases are activated. Aligning with a compartmentalization role for the territory, the caspase inhibitor XIAP is restricted from accessing the internal portion, where the majority of JMY, cyto c, Apaf-1, and active caspase-3 are located. Thus, the F-actin–rich territories serve to concentrate both apoptosomes and caspases while shielding the caspases from a prominent inhibitor. As a consequence, the cytoskeletal rearrangements create a microenvironment that ensures a rapid and efficient transition from the biogenesis of apoptosomes to the induction of the caspase cascade, while simultaneously preventing cleavage events from taking place throughout the rest of the cell (Figure 9C). We speculate that XIAP has affinity for proteins located at the periphery of the mature round territory. We further hypothesize that the temporary physical isolation provided to the active caspases in the center of the territory permits them to reach quantities that can eventually overwhelm the peripheral inhibitors, escape the territory, and execute a sudden or synchronized demolition of the cell.
JMY requires its actin- and Arp2/3-binding domains to enable caspase-3 cleavage (King et al., 2021), and our current work demonstrates not only that JMY-mediated assembly of F-actin territories results in a more frequent triggering of executioner caspase activation on a per-cell basis but that denser actin networks in the territories drive more potent processing of caspase-3 to its cleaved active form. The use of single- and double-knockout cells indicates that JMY and WHAMM contribute to the amount of F-actin–comprising territories. Based on the severity of mutant cell phenotypes, JMY is pivotal for creating these cytoskeletal structures, whereas WHAMM may increase the density of actin filaments within them. In addition to producing greater levels of cleaved executioner caspases, F-actin–rich territories cause a distinct clustering of caspases, as subdued or absent F-actin territories in JMY- and WHAMM-deficient cells correspond to low and/or diffuse cleaved caspase signals. JMY and WHAMM therefore both function in organizing and clustering multiple proapoptotic components within a single cytoskeletal compartment.
The proper spatiotemporal regulation of intracellular processes is achieved by compartmentalization in membrane-bound organelles or less-characterized membraneless organelles (Gabaldón and Pittis, 2015; Zhao and Zhang, 2020). Such subcellular partitioning enhances the rate and specificity of biochemical reactions by selectively increasing the proximity of particular proteins to positive regulators while insulating these factors from negative regulators and competing reactions. The rapid creation of compartments de novo is well illustrated by the formation of viral assembly factories in infected cells (Miller and Krijnse-Locker, 2008; Schmid et al., 2014). Moreover, the biogenesis of membraneless organelles is often driven by multivalent interactions (Li et al., 2012; Gomes and Shorter, 2019), and the cytoskeleton plays important roles in their establishment, remodeling, and disassembly (Kaizuka et al., 2007; Banani et al., 2017; Koppers et al., 2020). Cytoskeletal filaments can also assemble into structures that segregate organelles and intracellular cargo, for example, the actin cages that surround damaged mitochondria (Moore et al., 2016; Kruppa et al., 2018) or the actin cocoons that encapsulate bacteria-containing vacuoles (Kühn et al., 2020).
Consistent with the above themes, key members of the actin polymerization machinery and multiple proapoptotic proteins are selectively sorted into F-actin–rich territories that protect the actin nucleation factors from negative regulators and the caspases from competitive inhibitors. Determining how the apoptosomal and cytoskeletal components physically engage one another will shed light on the dynamic events that take place early in apoptosis, while investigations into the mechanisms by which caspases disseminate from the F-actin–rich territories will clarify how these proteases gain access to their diverse targets later in apoptosis. Thus, further characterizations of the biogenesis, remodeling, dissolution, and broader prevalence of F-actin–rich territories could provide new avenues for understanding and modulating the cellular responses to proapoptotic stressors.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Biological materials
Research conducted with biological materials was approved by the UConn Institutional Biosafety Committee. This study did not include human subjects or live animals.
Cell culture
Cell lines are listed in Supplemental Table S1, plasmids in Supplemental Table S2, and key reagents in Supplemental Table S3. U2OS cells (UC Berkeley cell culture facility) were cultured in DMEM (Invitrogen) supplemented with Glutamax, 10% fetal bovine serum (FBS), and antibiotic–antimycotic. HAP1 cell derivatives (Horizon Genomics) were described previously (King et al., 2021) and were cultured in Iscove’s Modified Dulbecco’s Medium (IMDM; Invitrogen), 10% FBS, and penicillin–streptomycin. All cell lines were grown at 37°C in 5% CO2, and assays were performed using cells that had been in active culture for 2–10 trypsinized passages. Cells were treated with 5–20 μM etoposide (Sigma Aldrich) diluted from an initial stock of 50 μM dissolved in media or with 60 μM mitomycin c (Fisher) diluted from an initial stock of 1.5 mM. Combination treatments used 10 μM etoposide with 50 μM z-VAD-fmk or z-DEVD-fmk (Selleckchem). Equivalent volumes of media without drugs were used as controls.
Transgene expression
Plasmids encoding GFP-tagged mouse JMY (King et al., 2021), LAP (localization and affinity purification)-tagged human WHAMM (Campellone et al., 2008), and mCherry-tagged Lifeact (Ohkawa and Welch, 2018) were described previously. For stable expression of GFP-JMY derivatives, U2OS or JMYKO-1A cells cultured in 12-well plates were transfected with 2–5 μg of linearized plasmid encoding GFP or GFP-JMY derivatives using Lipofectamine LTX (Invitrogen). Twenty-four hours later, the cells were transferred to media containing 0.6–1.5 mg/ml G418 for 12–16 d. Surviving colonies were collected, expanded in 24-well plates, six-well plates, and ultimately T-25 flasks before cryopreservation. U2OS cells were further enriched for GFP+ cells using a FACSAria flow cytometer (BD Biosciences). Upon reanimation, G418 concentrations were reduced to 350 μg/ml in six-well plates. The cell populations were subjected to experimental manipulations within five passages in media containing 350 μg/ml G418. For transient expression of Lifeact-mCherry or LAP-WHAMM, U2OS cells were transfected in six-well plates with 500 ng of DNA 24 h before reseeding onto 12 mm glass coverslips in 24-well plates or into 35 mm glass-bottom microwell dishes.
Immunoblotting
To prepare whole cell extracts, cells were washed with phosphate-buffered saline (PBS), collected, and pelleted via centrifugation. Cell pellets were resuspended in lysis buffer (20 mM HEPES, pH 7.4, 100 mM NaCl, 1% Triton X-100, 1 mM Na3VO4, 1 mM NaF, plus 1 mM phenylmethylsulfonyl fluoride and 10 μg/ml each aprotinin, leupeptin, pepstatin, and chymostatin), diluted in SDS–PAGE sample buffer, boiled, centrifuged, and subjected to SDS–PAGE before transfer to nitrocellulose membranes (GE Healthcare). Membranes were blocked in PBS + 5% milk (PBS-M) before being probed with primary antibodies (Supplemental Table S3) diluted in PBS-M overnight at 4°C plus an additional 2–3 h at room temperature. Membranes were rinsed twice with PBS and washed thrice with PBS + 0.5% Tween-20 (PBS-T). Membranes were then probed with secondary antibodies conjugated to IRDye-800, IRDye-680, or horseradish peroxidase (Supplemental Table S3) and diluted in PBS-M. Membranes were again rinsed with PBS and washed with PBS-T. Blots were imaged using a LI-COR Odyssey Fc imaging system. Band intensities were determined using the Analysis tool in LI-COR Image Studio software, and quantities of proteins of interest were normalized to tubulin, actin, and/or GAPDH loading controls.
Immunostaining
For immunofluorescence microscopy, approximately 1.5–2.5 × 105 cells were seeded onto 12 mm glass coverslips in 24-well plates and allowed to grow for 24 h. After control or etoposide treatments, cells were washed with PBS, fixed in 2.5 or 3.7% paraformaldehyde (PFA) in PBS for 30 min, washed, permeabilized with 0.1% Triton X-100 in PBS, washed, and incubated in blocking buffer (1% FBS + 1% bovine serum albumin [BSA] + 0.02% NaN3 in PBS) for a minimum of 15 min. Cells were probed with primary antibodies (Supplemental Table S3) diluted in blocking buffer for 45 min. Cells were washed and treated with AlexaFluor-conjugated secondary antibodies, 4′,6-diamidino-2-phenylindole (DAPI), and/or AlexaFluor-conjugated phalloidin (Supplemental Table S3) for 45 min, followed by washes and mounting in Prolong Gold antifade reagent (Invitrogen). MitoTracker Red (Invitrogen) was added for 30 min before fixation. For live imaging, approximately 106 cells were seeded into 35 mm glass-bottom dishes (MatTek) 24 h before etoposide treatments.
Fluorescence microscopy
All fixed and live images were captured using a Nikon Eclipse Ti inverted microscope with Plan Apo 100×/1.45, Plan Apo 60×/1.40, or Plan Fluor 20×/0.5 numerical aperture objectives using an Andor Clara-E camera and a computer running NIS Elements software. Fixed cells were viewed in multiple focal planes, and Z-series were captured at 0.2–0.4 μm steps. Images presented in the figures represent one- or two-slice projections. Live cell imaging was performed in a 37°C chamber (Okolab) using the 100× objective, and images were captured at 2 min intervals. Image processing and analyses used ImageJ/FIJI software (Schindelin et al., 2012).
Image processing and quantification
The ImageJ Cell Counter plug-in was used to quantify the percentage of cells with JMY puncta, CCasp-3 positivity, or F-actin–rich territories by manually counting the total cells in the DAPI channel and the cells that were positive for cytosolic punctate JMY staining in the JMY or GFP-JMY channel, the cells that were positive for CCasp-3 staining, or the cells that contained F-actin–rich territories. Similar quantifications were performed to determine the percentage of F-actin–rich territories that associated with JMY, GFP-JMY, cyto c, Apaf-1, or CCasp-3 staining. The Cell Counter plug-in was also used to quantify the percentage of CCasp-3–positive cells that possessed F-actin–rich territories or exhibited clustered versus diffuse staining. Similar methods were used to perform JMY puncta overlap assays by identifying F-actin–rich territories in the phalloidin channel and manually counting the JMY puncta that did and did not overlap with cyto c or Apaf-1 puncta. The JACoP plug-in was used to quantify Pearson’s coefficient values for JMY and cyto c or Apaf-1 puncta within F-actin–rich territories (Bolte and Cordelières, 2006).
For analyses of total cellular JMY, CCasp-3, or F-actin levels, the Selection tool was used in the phalloidin channel to select cells, the Measure tool was used in the JMY, CCasp-3, or phalloidin channel to measure the integrated density and area per cell, and then the integrated density was normalized to the cell area to generate the mean whole cell fluorescence. For analyses of nuclear JMY levels, the Threshold, Watershed, and Analyze tools were used in the DAPI channel to separate individual nuclei, the ROI Manager Measure tool was used in the JMY channel to measure the integrated density and area per nucleus, and then the integrated density was normalized to the nuclear area to generate the mean nuclear fluorescence. For analyses of cytoplasmic JMY levels, the nuclear integrated density or area was subtracted from the whole cell integrated density or area for individual cells, and then the cytosolic integrated density was normalized to the cytosolic area to generate the mean cytosolic fluorescence. The mean cytosolic fluorescence and mean nuclear fluorescence values were further normalized by dividing by their sum and multiplying by the mean whole cell fluorescence to derive the JMY intensities for each compartment on a per-cell basis. For analysis of GFP-JMY or Lifeact-mCherry levels over time, the Selection tool was used in the mCherry and DAPI channel to select cytosolic or nuclear areas and the Measure tool was used in the GFP or mCherry channel to measure the integrated density and area per selection. Then the integrated density was normalized to the selection area to generate the mean fluorescence for each timepoint, and the average mean fluorescence for each channel was set to one.
Pixel intensity plots were generated using the Plot Profile tool. Lines were drawn through F-actin–, CCasp-3–, or JMY-rich regions after identification in the phalloidin, CCasp-3, or JMY channels, and the Plot Profile tool was used to measure the distance and integrated densities for each channel. The mean integrated density for each channel was set to one. For analyses of JMY, CCasp-3, XIAP, and F-actin levels within territories, the Selection tool was used in the phalloidin channel to set the territory area, and the Measure tool was used in the JMY, CCasp-3, XIAP, and phalloidin channels to measure the integrated density per selection. Analyses of interior JMY, CCasp-3, XIAP, or F-actin levels utilized the Enlarge Selection tool to shrink the Selected area by 2 μm, and the Measure tool was used in the JMY, CCasp-3, XIAP, and phalloidin channels to measure the integrated density. In measurements of JMY, CCasp-3, XIAP, or F-actin levels at the territory periphery, the interior territory integrated density was subtracted from the whole territory integrated density for individual cells. The mean whole territory integrated density for each channel was set to one.
Reproducibility and statistics
All conclusions were based on observations made from at least three separate experiments, and quantifications were based on data from three representative experiments except where noted in the figure legends. The sample size used for statistical tests was the number of times an experiment was performed except where noted in the legends. Statistical analyses were performed using GraphPad Prism software. Statistics on data sets with three or more conditions were performed using analyses of variance (ANOVAs) followed by Tukey’s post-hoc test. P values for data sets comparing two conditions were determined using unpaired t tests. P values for data sets with ± scoring used Fisher’s exact test. P values <0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank Wu He at the UConn Flow Cytometry Facility for cell sorting and L.T. Bear for support with experimental design. We also thank Katrina Velle, Nathan Leclair, and Campellone Lab members for their comments on this paper. K.G.C. was supported by National Institutes of Health Grants R01-GM107441 and K02-AG050774. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations used:
- Apaf-1
apoptotic protease-activating factor-1
- CA
connector-acidic
- CCasp-3
cleaved caspase-3
- cyto c
cytochrome c
- DKO
double knockout
- F-actin
filamentous-actin
- IAP
inhibitor of apoptosis protein
- KO
knockout
- mtDNA
mitochondrial DNA
- TCasp-3
total caspase-3
- TCasp-9
total caspase-9
- W
WASP-homology-2
- WASP
Wiskott-Aldrich Syndrome Protein
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E22-04-0119) on March 15, 2023.
REFERENCES
- Ahn JS, Jang IS, Kim DI, Cho KA, Park YH, Kim K, Kwak CS, Chul Park S (2003). Aging-associated increase of gelsolin for apoptosis resistance. Biochem Biophys Res Commun 312, 1335–1341. [DOI] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, Rosen MK (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18, 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhola PD, Mattheyses AL, Simon SM (2009). Spatial and temporal dynamics of mitochondrial membrane permeability waves during apoptosis. Biophys J 97, 2222–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock FJ, Tait SWG (2020). Mitochondria as multifaceted regulators of cell death. Nat Rev Mol Cell Biol 21, 85–100. [DOI] [PubMed] [Google Scholar]
- Bolte S, Cordelières FP (2006). A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224, 213–232. [DOI] [PubMed] [Google Scholar]
- Bratton SB, Salvesen GS (2010). Regulation of the Apaf-1-caspase-9 apoptosome. J Cell Sci 123, 3209–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Lebek NM, King VL (2023). Branching out in different directions: emerging cellular functions for the Arp2/3 complex and WASP-family actin nucleation factors. Eur J Cell Biol 102, 151301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Webb NJ, Znameroski EA, Welch MD (2008). WHAMM is an Arp2/3 complex activator that binds microtubules and functions in ER to Golgi transport. Cell 134, 148–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Welch MD (2010). A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol 11, 237–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candé C, Cohen I, Daugas E, Ravagnan L, Larochette N, Zamzami N, Kroemer G (2002). Apoptosis-inducing factor (AIF): a novel caspase-independent death effector released from mitochondria. Biochimie 84, 215–222. [DOI] [PubMed] [Google Scholar]
- Caridi CP, Plessner M, Grosse R, Chiolo I (2019). Nuclear actin filaments in DNA repair dynamics. Nat Cell Biol 21, 1068–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti R, Lee M, Higgs HN (2021). Multiple roles for actin in secretory and endocytic pathways. Curr Biol 31, R603–R618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng TC, Hong C, Akey IV, Yuan S, Akey CW (2016). A near atomic structure of the active human apoptosome. eLife 5, e17755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra D, Nosworthy NJ, dos Remedios CG (2005). The N-terminal fragment of gelsolin inhibits the interaction of DNase I with isolated actin, but not with the cofilin-actin complex. Proteomics 5, 3131–3136. [DOI] [PubMed] [Google Scholar]
- Chua BT, Volbracht C, Tan KO, Li R, Yu VC, Li P (2003). Mitochondrial translocation of cofilin is an early step in apoptosis induction. Nat Cell Biol 5, 1083–1089. [DOI] [PubMed] [Google Scholar]
- Coutts AS, Boulahbel H, Graham A, La Thangue NB (2007). Mdm2 targets the p53 transcription cofactor JMY for degradation. EMBO Rep 8, 84–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutts AS, Weston L, La Thangue NB (2009). A transcription co-factor integrates cell adhesion and motility with the p53 response. Proc Natl Acad Sci USA 106, 19872–19877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demonacos C, Krstic-Demonacos M, La Thangue NB (2001). A TPR motif cofactor contributes to p300 activity in the p53 response. Mol Cell 8, 71–84. [DOI] [PubMed] [Google Scholar]
- Desouza M, Gunning PW, Stehn JR (2012). The actin cytoskeleton as a sensor and mediator of apoptosis. Bioarchitecture 2, 75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveraux QL, Reed JC (1999). IAP family proteins—suppressors of apoptosis. Genes Dev 13, 239–252. [DOI] [PubMed] [Google Scholar]
- Dinsdale D, Lee JC, Dewson G, Cohen GM, Peter ME (2004). Intermediate filaments control the intracellular distribution of caspases during apoptosis. Am J Pathol 164, 395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorstyn L, Akey CW, Kumar S (2018). New insights into apoptosome structure and function. Cell Death Differ 25, 1194–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckelman BP, Salvesen GS, Scott FL (2006). Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep 7, 988–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firat-Karalar EN, Hsiue PP, Welch MD (2011). The actin nucleation factor JMY is a negative regulator of neuritogenesis. Mol Biol Cell 22, 4563–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldón T, Pittis AA (2015). Origin and evolution of metabolic sub-cellular compartmentalization in eukaryotes. Biochimie 119, 262–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, Alnemri ES, Altucci L, Amelio I, Andrews DW, et al. (2018). Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ 25, 486–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes E, Shorter J (2019). The molecular language of membraneless organelles. J Biol Chem 294, 7115–7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourlay CW, Ayscough KR (2005). The actin cytoskeleton: a key regulator of apoptosis and ageing? Nat Rev Mol Cell Biol 6, 583–589. [DOI] [PubMed] [Google Scholar]
- Green DR, Llambi F (2015). Cell death signaling. Cold Spring Harb Perspect Biol 7, a006080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarer EL, Theodore CJ, Guo S, Frier RB, Campellone KG (2023). Genomic instability caused by Arp2/3 complex inactivation results in micronucleus biogenesis and, cellular senescence. PLoS Genet 19, e1010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Liu H, Deng S, Chen X, Li D, Jiang X, Zeng W, Lu W (2020). The Golgi apparatus may be a potential therapeutic target for apoptosis-related neurological diseases. Front Cell Dev Biol 8, 830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrdinka M, Yabal M (2019). Inhibitor of apoptosis proteins in human health and disease. Genes Immun 20, 641–650. [DOI] [PubMed] [Google Scholar]
- Hu Q, Wu D, Chen W, Yan Z, Yan C, He T, Liang Q, Shi Y (2014). Molecular determinants of caspase-9 activation by the Apaf-1 apoptosome. Proc Natl Acad Sci USA 111, 16254–16261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Mullins RD (2019). LC3 and STRAP regulate actin filament assembly by JMY during autophagosome formation. J Cell Biol 218, 251–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien O, Wells JA (2017). Caspases and their substrates. Cell Death Differ 24, 1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaizuka Y, Douglass AD, Varma R, Dustin ML, Vale RD (2007). Mechanisms for segregating T cell receptor and adhesion molecules during immunological synapse formation in Jurkat T cells. Proc Natl Acad Sci USA 104, 20296–20301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr JF, Wyllie AH, Currie AR (1972). Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer 26, 239–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King VL, Leclair NK, Coulter AM, Campellone KG (2021). The actin nucleation factors JMY and WHAMM enable a rapid Arp2/3 complex-mediated intrinsic pathway of apoptosis. PLoS Genet 17, e1009512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klamt F, Zdanov S, Levine RL, Pariser A, Zhang Y, Zhang B, Yu L-R, Veenstra TD, Shacter E (2009). Oxidant-induced apoptosis is mediated by oxidation of the actin-regulatory protein cofilin. Nat Cell Biol 11, 1241–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppers M, Özkan N, Farías GG (2020). Complex interactions between membrane-bound organelles, biomolecular condensates and the cytoskeleton. Front Cell Dev Biol 8, 618733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothakota S, Azuma T, Reinhard C, Klippel A, Tang J, Chu K, McGarry TJ, Kirschner MW, Koths K, Kwiatkowski DJ, et al. (1997). Caspase-3-generated fragment of gelsolin: effector of morphological change in apoptosis. Science 278, 294–298. [DOI] [PubMed] [Google Scholar]
- Kramer DA, Piper HK, Chen B (2022). WASP family proteins: molecular mechanisms and implications in human disease. Eur J Cell Biol 101, 151244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruppa AJ, Kishi-Itakura C, Masters TA, Rorbach JE, Grice GL, Kendrick-Jones J, Nathan JA, Minczuk M, Buss F (2018). Myosin VI-dependent actin cages encapsulate Parkin-positive damaged mitochondria. Dev Cell 44, 484–499.e486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn S, Bergqvist J, Gil M, Valenzuela C, Barrio L, Lebreton S, Zurzolo C, Enninga J (2020). Actin assembly around the Shigella-containing vacuole promotes successful infection. Cell Rep 31, 107638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano H, Shimizu S, Koya RC, Fujita H, Kamada S, Kuzumaki N, Tsujimoto Y (2000). Human gelsolin prevents apoptosis by inhibiting apoptotic mitochondrial changes via closing VDAC. Oncogene 19, 4807–4814. [DOI] [PubMed] [Google Scholar]
- Lee JC, Schickling O, Stegh AH, Oshima RG, Dinsdale D, Cohen GM, Peter ME (2002). DEDD regulates degradation of intermediate filaments during apoptosis. J Cell Biol 158, 1051–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. (2012). Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhou M, Hu Q, Bai XC, Huang W, Scheres SH, Shi Y (2017). Mechanistic insights into caspase-9 activation by the structure of the apoptosome holoenzyme. Proc Natl Acad Sci USA 114, 1542–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFarlane M, Merrison W, Dinsdale D, Cohen GM (2000). Active caspases and cleaved cytokeratins are sequestered into cytoplasmic inclusions in TRAIL-induced apoptosis. J Cell Biol 148, 1239–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malladi S, Challa-Malladi M, Fearnhead HO, Bratton SB (2009). The Apaf-1*procaspase-9 apoptosome complex functions as a proteolytic-based molecular timer. EMBO J 28, 1916–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S, Krijnse-Locker J (2008). Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol 6, 363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AS, Wong YC, Simpson CL, Holzbaur ELF (2016). Dynamic actin cycling through mitochondrial subpopulations locally regulates the fission–fusion balance within mitochondrial networks. Nat Commun 7, 12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkawa T, Welch MD (2018). Baculovirus actin-based motility drives nuclear envelope disruption and nuclear egress. Curr Biol 28, 2153–2159.e2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD (2016). Actin and actin-binding proteins. Cold Spring Harb Perspect Biol 8, a018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, O’Shaughnessy B (2019). Molecular mechanism of cytokinesis. Annu Rev Biochem 88, 661–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popgeorgiev N, Jabbour L, Gillet G (2018). Subcellular localization and dynamics of the Bcl-2 family of proteins. Front Cell Dev Biol 6, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehklau K, Gurniak CB, Conrad M, Friauf E, Ott M, Rust MB (2012). ADF/cofilin proteins translocate to mitochondria during apoptosis but are not generally required for cell death signaling. Cell Death Differ 19, 958–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm M, Huber HJ, Hellwig CT, Anguissola S, Dussmann H, Prehn JH (2009). Dynamics of outer mitochondrial membrane permeabilization during apoptosis. Cell Death Differ 16, 613–623. [DOI] [PubMed] [Google Scholar]
- Riedl SJ, Salvesen GS (2007). The apoptosome: signalling platform of cell death. Nat Rev Mol Cell Biol 8, 405–413. [DOI] [PubMed] [Google Scholar]
- Rodriquez J, Lazebnik Y (1999). Caspase-9 and APAF-1 form an active holoenzyme. Genes Dev 13, 3179–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotty JD, Wu C, Bear JE (2013). New insights into the regulation and cellular functions of the ARP2/3 complex. Nat Rev Mol Cell Biol 14, 7–12. [DOI] [PubMed] [Google Scholar]
- Sahebazzamani F, Hosseinkhani S, Eriksson LA, Fearnhead HO (2021). Apoptosome formation through disruption of the K192-D616 salt bridge in the Apaf-1 closed form. ACS Omega 6, 22551–22558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlüter K, Waschbüsch D, Anft M, Hügging D, Kind S, Hänisch J, Lakisic G, Gautreau A, Barnekow A, Stradal TEB (2014). JMY is involved in anterograde vesicle trafficking from the trans-Golgi network. Eur J Cell Biol 93, 194–204. [DOI] [PubMed] [Google Scholar]
- Schmid M, Speiseder T, Dobner T, Gonzalez RA (2014). DNA virus replication compartments. J Virol 88, 1404–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnoor M, Stradal TE, Rottner K (2018). Cortactin: cell functions of a multifaceted actin-binding protein. Trends Cell Biol 28, 79–98. [DOI] [PubMed] [Google Scholar]
- Scott FL, Denault JB, Riedl SJ, Shin H, Renatus M, Salvesen GS (2005). XIAP inhibits caspase-3 and -7 using two binding sites: evolutionarily conserved mechanism of IAPs. EMBO J 24, 645–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q-T, Hsiue PP, Sindelar CV, Welch MD, Campellone KG, Wang H-W (2012). Structural insights into WHAMM-mediated cytoskeletal coordination during membrane remodeling. J Cell Biol 199, 111–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikama N, Lee CW, France S, Delavaine L, Lyon J, Krstic-Demonacos M, La Thangue NB (1999). A novel cofactor for p300 that regulates the p53 response. Mol Cell 4, 365–376. [DOI] [PubMed] [Google Scholar]
- Singh R, Letai A, Sarosiek K (2019). Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat Rev Mol Cell Biol 20, 175–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su TW, Yang CY, Kao WP, Kuo BJ, Lin SM, Lin JY, Lo YC, Lin SC (2017). Structural insights into DD-fold assembly and caspase-9 activation by the Apaf-1 apoptosome. Structure 25, 407–420. [DOI] [PubMed] [Google Scholar]
- Svitkina T (2018). The actin cytoskeleton and actin-based motility. Cold Spring Harb Perspect Biol 10, a018267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney KF, Li R (2016). Function and regulation of the Arp2/3 complex during cell migration in diverse environments. Curr Opin Cell Biol 42, 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait SWG, Green DR (2013). Mitochondrial regulation of cell death. Cold Spring Harb Perspect Biol 5, a008706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang HL, Le AH, Lung HL (2006). The increase in mitochondrial association with actin precedes Bax translocation in apoptosis. Biochem J 396, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Karstedt S, Montinaro A, Walczak H (2017). Exploring the TRAILs less travelled: TRAIL in cancer biology and therapy. Nat Rev Cancer 17, 352–366. [DOI] [PubMed] [Google Scholar]
- Wang C, Zhou GL, Vedantam S, Li P, Field J (2008). Mitochondrial shuttling of CAP1 promotes actin- and cofilin-dependent apoptosis. J Cell Sci 121, 2913–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Qiao Q, Wu H (2017). Understanding CARD tricks in apoptosomes. Structure 25, 575–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter KG, Hsu YT, Smith CL, Nechushtan A, Xi XG, Youle RJ (1997). Movement of Bax from the cytosol to mitochondria during apoptosis. J Cell Biol 139, 1281–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CC, Lee S, Malladi S, Chen MD, Mastrandrea NJ, Zhang Z, Bratton SB (2016). The Apaf-1 apoptosome induces formation of caspase-9 homo- and heterodimers with distinct activities. Nat Commun 7, 13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Park HH, Chung JY, Lin SC, Lo YC, da Graca LS, Jiang X, Wu H (2006). Caspase-9 holoenzyme is a specific and optimal procaspase-3 processing machine. Mol Cell 22, 259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Akey CW (2013). Apoptosome structure, assembly, and procaspase activation. Structure 21, 501–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Topf M, Reubold TF, Eschenburg S, Akey CW (2013). Changes in Apaf-1 conformation that drive apoptosome assembly. Biochemistry 52, 2319–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Yu X, Asara JM, Heuser JE, Ludtke SJ, Akey CW (2011). The holo-apoptosome: activation of procaspase-9 and interactions with caspase-3. Structure 19, 1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Yu X, Topf M, Ludtke SJ, Wang X, Akey CW (2010). Structure of an apoptosome-procaspase-9 CARD complex. Structure 18, 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YG, Zhang H (2020). Phase separation in membrane biology: the interplay between membrane-bound organelles and membraneless condensates. Dev Cell 55, 30–44. [DOI] [PubMed] [Google Scholar]
- Zhou M, Li Y, Hu Q, Bai XC, Huang W, Yan C, Scheres SH, Shi Y (2015). Atomic structure of the apoptosome: mechanism of cytochrome c- and dATP-mediated activation of Apaf-1. Genes Dev 29, 2349–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Fukada K, Zhu H, Kyprianou N (2006). Prohibitin and cofilin are intracellular effectors of transforming growth factor beta signaling in human prostate cancer cells. Cancer Res 66, 8640–8647. [DOI] [PubMed] [Google Scholar]
- Zou H, Li Y, Liu X, Wang X (1999). An APAF-1.cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J Biol Chem 274, 11549–11556. [DOI] [PubMed] [Google Scholar]
- Zuchero JB, Belin B, Mullins RD (2012). Actin binding to WH2 domains regulates nuclear import of the multifunctional actin regulator JMY. Mol Biol Cell 23, 853–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchero JB, Coutts AS, Quinlan ME, Thangue NBL, Mullins RD (2009). p53-cofactor JMY is a multifunctional actin nucleation factor. Nat Cell Biol 11, 451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.