Abstract
Plasmodium falciparum, the most virulent agent of human malaria, spread from Africa to all continents following the out-of-Africa human migrations. During the transatlantic slave trade between the 16th and 19th centuries, it was introduced twice independently to the Americas where it adapted to new environmental conditions (new human populations and mosquito species). Here, we analyzed the genome-wide polymorphisms of 2,635 isolates across the current P. falciparum distribution range in Africa, Asia, Oceania, and the Americas to investigate its genetic structure, invasion history, and selective pressures associated with its adaptation to the American environment. We confirmed that American populations originated from Africa with at least two independent introductions that led to two genetically distinct clusters, one in the North (Haiti and Colombia) and one in the South (French Guiana and Brazil), and an admixed Peruvian group. Genome scans revealed recent and more ancient signals of positive selection in the American populations. Particularly, we detected positive selection signals in genes involved in interactions with hosts (human and mosquito) cells and in genes involved in resistance to malaria drugs in both clusters. Analyses suggested that for five genes, adaptive introgression between clusters or selection on standing variation was at the origin of this repeated evolution. This study provides new genetic evidence on P. falciparum colonization history and on its local adaptation in the Americas.
Keywords: population genomics, adaptive evolution, host–pathogen interactions, Americas, Plasmodium falciparum
Introduction
Despite the remarkable advances in medical research and treatments during the 20th century, infectious diseases remain among the leading causes of death in low- and middle-income countries (Geneva: World Health Organization 2022). One of the reasons is the frequent emergence of new infectious diseases and the re-emergence of old ones (Zumla and Hui 2019; Sabin et al. 2020). Emerging infectious diseases are diseases that have recently increased in incidence, in geographic or host range, are newly recognized or are caused by new pathogens. One key aspect of their emergence is the pathogen adaptability to new environmental conditions (e.g., ecosystems, hosts, treatments, and vectors). Explaining how pathogens can adapt to new environments is therefore a prerequisite to better understand the emergence of infectious diseases.
For many pathogen species, the experimental investigation of genetic adaptation to new environments is difficult due to the challenges associated with culturing species in laboratory conditions, especially species with complex life cycles or with long generation times. An alternative strategy is to analyze well-documented past emergence events that provide a natural “experiment” to identify the genetic basis of genetic adaptation associated with the colonization of novel environments. During the evolution of human populations, new infectious diseases have recurrently emerged. Therefore, there are now several examples of well-documented past emergence events that might serve as models to analyze how pathogens adapted to new environmental conditions (Wolfe et al. 2007; Choi and Thines 2015). With human migrations, some parasites have been introduced in new regions and are now established (Bryant et al. 2007; Small et al. 2019; Platt et al. 2022). Plasmodium falciparum colonization of the Americas is one of such examples.
Plasmodium falciparum is a unicellular eukaryote parasite that causes the most severe form of human malaria, leading to the death of approximately half a million people every year, mainly among <5-year-old children (World Health Organization 2020). Therefore, it is a major public health issue. During its life cycle, P. falciparum successively infects two hosts: a vector of the genus Anopheles (mosquito) and Homo sapiens sapiens.
During its evolutionary history, P. falciparum emerged and colonized new geographical areas several times (Tanabe et al. 2010). It was introduced to the Americas from Africa during the transatlantic slave trade that lasted from the 16th to the 19th centuries (Anderson et al. 2000; Yalcindag et al. 2012; Rodrigues et al. 2018). During its colonization of the New World, P. falciparum encountered a new human host environment (Amerindian, European, African, and admixed populations) and new vector species (e.g. Anopheles darlingi, one of the main vector species in South America [Zimmerman 1992; Laporta et al. 2015], and Anopheles albimanus, the main vector in Central America [Zimmerman 1992; Frederick et al. 2016]). Compared with Anopheles gambiae sensu lato species complex and Anopheles funestus (the main African malaria vector species (Gillies and Coetzee 1987)), A. darlingi belong to a different subgenera that might have diverged between 80 and 100 million years ago (Moreno et al. 2010; Neafsey et al. 2015; Martinez-Villegas et al. 2019). Moreover, in the last few decades, P. falciparum went through another dramatic change in its environment: the use of antimalarial drugs (Wongsrichanalai et al. 2002; Mita et al. 2009) to prevent or cure malarial infections. Some of these drugs are still massively used and exert very strong selective pressures on the parasite. Different populations around the world have developed resistance to these drugs, and some new P. falciparum genotypes have spread (Mita et al. 2009; Plowe 2009).
The environmental changes during or after the colonization of the Americas by P. falciparum likely resulted in powerful selective pressures that forced the parasite to adapt to these new local conditions and to evolve toward different phenotypes and genotypes. Previous studies on few candidate genes identified some genes that may have played a role in the successful P. falciparum colonization of the Americas, for instance, the P47 (Molina-Cruz and Barillas-Mury 2014; Canepa et al. 2016; Tagliamonte et al. 2020) and P48/45 genes (van Dijk et al. 2001), two genes involved in the avoidance of the mosquito immune system. Moreover, some studies showed that, in the New World, some P. falciparum genes that encode proteins implicated in red blood cell invasion, such as the erythrocyte binding antigen (eba) surface ligand are either under stronger selection (Yalcindag et al. 2014) or differentially expressed (Lopez-Perez et al. 2012) compared with the African populations. However, no study has performed a genome-wide analysis of P. falciparum response to these new environmental conditions. Yet, due to the large amount of genomic data available, P. falciparum offers a unique opportunity to study its genomic adaptation in the context of the colonization of a novel environment. Moreover, the two independent P. falciparum introductions in the New World from Africa during the transatlantic slave trade (Yalcindag et al. 2012) may be regarded as two potentially independent replicates of the same “natural experiment.” It has been suggested that the first introduction occurred in the south of the continent (through Brazil) and the second, more recent, in the north (through Colombia). These two independent introductions likely coincided with the two main slave trade routes from Africa to the Americas: one organized by the Portuguese empire to bring slaves to the current Brazil (600 years ago) and the other organized by the Spanish empire to Colombia (300 years ago) (Yalcindag et al. 2012). These two independent introductions can give information on whether evolution followed the same path (repeated or parallel evolution) and whether the same genes responded to the novel environmental conditions. As the populations that resulted from these two introductions have historically exchanged migrants (Yalcindag et al. 2012), it is also possible to determine the importance of migration in the adaptive evolution of the introduced populations.
In the present study, we compiled four whole genome polymorphism data sets of P. falciparum from the Americas, the source area (Africa), and other world areas (Asia and Oceania). Using these data, we studied the population genetic structure and colonization history of the American P. falciparum populations. We then scanned the genomes to identify genes and genomic regions that may have responded to the selection imposed by the colonization of novel environments in the New World. As selection may have occurred at different time points in the history of the introduced populations, we used complementary methods to identify recent and also more ancient signals of selection. Then, for genes that showed signals of selection in distinct American populations, we determined whether alleles were independently selected within each location or whether they resulted from a process of adaptive migration between populations.
Results
Compilation of Whole Genome Polymorphism Data
We obtained whole genome polymorphism data for P. falciparum from publicly available data sets. Specifically, we retrieved the single-nucleotide polymorphism (SNP) data (VCF files) from the MalariaGen Project (Pearson et al. 2019) that includes 7,113 isolates from 29 countries, particularly Africa (3,877 isolates), Asia (2,942 isolates), Oceania (231 isolates), and South America (39 isolates) and also laboratory samples (16 isolates) and samples collected from travelers (8 isolates). As this data set only included two countries from the Americas (Colombia and Peru), we also used population data from Brazil (23 isolates) (Moser et al. 2020), French Guiana (36 isolates) (Pelleau et al. 2015), and Haiti (21 isolates) (Tagliamonte et al. 2020). The total data set before filtering included 7,193 infections from 32 countries in the Americas, Africa, Asia, and Oceania.
For the MalariaGen data set, we quality filtered the SNP data and samples following the recommendation by Pearson et al. (2019). We kept only high-quality biallelic SNPs from the core genome (i.e., regions present in all individuals, as defined by Pearson et al. (2019)) and 5,970 samples that corresponded to the analysis set as defined by Pearson et al. (2019). We excluded the only sample from Mozambique, from a traveler, to avoid bias due to a population made of only one individual. For all populations, we removed samples with >20% missing data (11 samples) and with multi-strain infections (i.e., multiple P. falciparum genotypes), reported as FWS >0.95 (supplementary fig. S1, Supplementary Material online). In addition, for all pairs of strains displaying excessive relatedness (pairwise-IBD > 0.5), we excluded one strain (supplementary fig. S1, Supplementary Material online). The final sample included 2,635 individuals (fig. 1A, supplementary table S1 and fig. S2, Supplementary Material online).
Fig. 1.
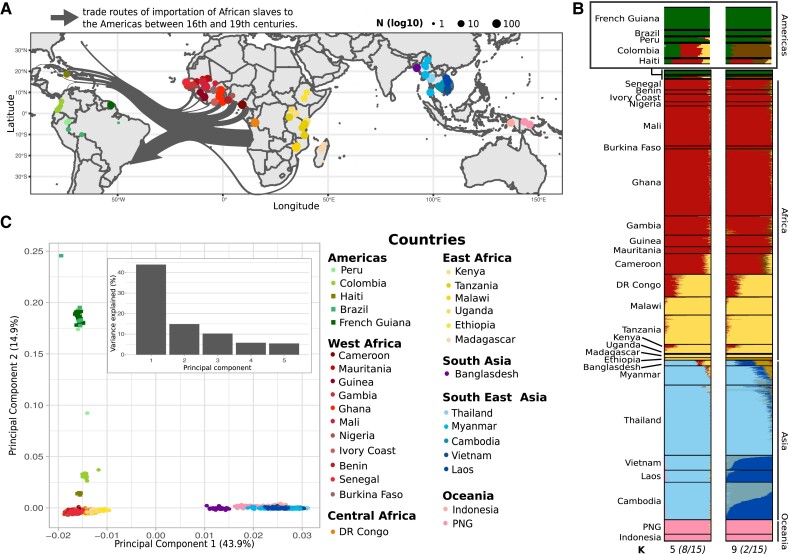
Geographical origin and genetic structure of Plasmodium falciparum isolates. (A) Geographical origin of the 2,635 P. falciparum isolates per region: South America (n = 48), West Africa (n = 1,046), Central Africa (n = 138), East Africa (n = 348), South Asia (n = 36), South East Asia (n = 885), and Oceania (n = 112). The circle size is proportional to the sampling effort per site (log10N). Arrows indicate the slave trade waves, and their width is proportional to the number of slaves transported (source: www.slavevoyages.org, accessed on October 25, 2021). (B) ADMIXTURE results for K = 5 and K = 9 clusters (indicated at the bottom, with the clustering convergence rate for 15 replicated runs). DR Congo, Democratic Republic of Congo. PNG, Papua New Guinea. (C) Graphical representation of the PCA results. Circles correspond to P. falciparum genomes obtained from the MalariaGen Project (Pearson et al. 2019); squares represent the newly added data for this study (Haiti, Brazil, and French Guiana). The histogram represents the percentage of the genetic variance explained by the five first PCs.
Then, from this data set, we removed SNPs with >10% of missing data. We set a minimum allele frequency (MAF) filter at 0.01%, to delete polymorphisms associated with sequencing errors, and a minimum coverage depth at 15×. In the end, our data set included 78,036 SNPs for 2,635 samples, with a mean SNP density of 3.69 per kilobase. The filtering steps are described in supplementary figure S2, Supplementary Material online. Following Pearson et al. (2019), we defined eight geographic regions for the analyses: the Americas (SAM), West Africa (WAF), Central Africa (CAF), East Africa (EAF), South Asia (SAS), West Southeast Asia (WSEA), East Southeast Asia (ESEA), and Oceania (OCE).
Two Distinct Admixed Genetic Clusters of P. falciparum in the Americas
To obtain insights into the genetic relationships among isolates from all over the world, we first investigated P. falciparum population structure using two complementary approaches: (1) a genetic ancestry analysis with the ADMIXTURE v1.3.0 software and (2) principal component analysis (PCA). For these analyses, we used a data set of 41,921 SNPs from 2,434 individuals after linkage disequilibrium (LD) pruning, MAF filtering, and missing data filtering (see Materials and Methods for more details).
The genetic ancestry analysis with ADMIXTURE identified nine distinct genetic pools as the optimal number of clusters (K) of P. falciparum populations worldwide (supplementary fig. S3, Supplementary Material online). With K = 2, Asian populations formed one cluster and African and American populations the other (supplementary fig. S4, Supplementary Material online). From K = 5 onward, the American populations became distinct from the others (fig. 1B). With K = 9, isolates from Brazil and French Guiana formed a distinct genetic cluster (dark green in fig. 1B), whereas isolates from Peru, Colombia, and Haiti displayed evidence of admixed ancestry, especially with African populations (brown in fig. 1B). We obtained similar results by PCA (fig. 1C). The first principal component (PC), which represented 43.9% of the variance explained, separated the Asian populations from the African and American populations. The second PC (14.9% of variance explained) split African isolates from American isolates and also showed the intracontinental structuring of American isolates. Indeed, Brazil and French Guiana isolates were more distant from African isolates than other American populations, and we could not identify any finer subdivision. Conversely, isolates from Colombia and Haiti were quite distinct and very close to Africa. Peruvian isolates were in the middle between the cluster formed by Haitian and Colombian isolates (SAM North cluster) and the cluster formed by Brazilian and French Guianan isolates (SAM South cluster).
To further investigate the genetic relationships between the American populations and populations from the rest of the world as well as their colonization history, we estimated population trees and networks using two approaches implemented in TreeMix (Pickrell and Pritchard 2012)) and ADMIXTOOLS2 (Maier et al. 2023), respectively. These two methods use the shared and private genetic ancestry components to infer the population branching while taking into account also historical migration and admixture events between populations. For both analyses, we used genome data of Plasmodium praefalciparum, the closest known relative of P. falciparum, as an outgroup to root the tree/network, and a LD-pruned data set composed of 20,943 SNPs for 2,638 individuals.
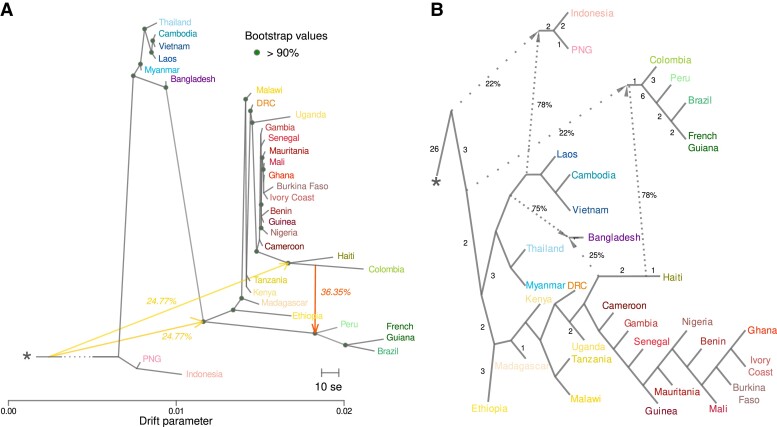
The TreeMix analysis identified three migration edges as optimal for our data set (supplementary fig. S5AandB, Supplementary Material online). The resulting TreeMix consensus tree (fig. 2A) confirmed the global genetic structuration given by the PCA and ADMIXTURE analyses, particularly the close relationship between the African and American populations. Many African populations were very closely related, with very short branches. In other words, they were all part of the same genetic cluster that did not drift much in recent times. The tree showed that the two American clusters (SAM North and SAM South) connected to different positions in the tree. SAM North populations (Haiti–Colombia) were more closely related to populations of West Africa, whereas SAM South populations (Brazil–French Guiana) and Peru were branched deeper in the tree as a sister group to the African cluster. Moreover, the American populations were characterized by a strong drift effect, marked by longer branches. This reflects founder effects. Our results for the American populations also indicated 25% of interbreeding with an older or unsampled population (on the P. praefalciparum branch) and more recent mixing (up to 36.35%) between the Colombian branch and the cluster composed of Peruvian, Brazilian, and French Guianan populations (fig. 2A).
Fig. 2.
Relationships and admixture proportions between P. falciparum populations. (A) TreeMix tree of P. falciparum populations with three migration edges (arrows), rooted with P. praefalciparum, indicated with the asterisk. Filled circles represent nodes supported by bootstrap values >90%. The scale bar shows ten times the mean standard error (se) of the entries in the sample covariance matrix. Arrows show the migration edges between tree branches. The migration weight is indicated as a percentage on the migration edge. (B) Admixture graph of P. falciparum populations, with three admixture events, rooted with P. praefalciparum. Admixture events (and the estimated percentages) are shown with dashed arrows that connect the admixed population with the two source population branches. The number on each branch represents the branch length that indicates the amount of drift accumulated along that branch (in f-statistic units, multiplied by 1,000 and rounded to the nearest integer). Branches without a number are branches with a drift score = 0. PNG, Papua New Guinea; DRC, Democratic Republic of Congo. The asterisk represents the P. praefalciparum outgroup.
The ADMIXTOOLS2 analysis also identified an optimal number of admixture events equivalent to the one found with TreeMix (n = 3) (supplementary fig. S5C, Supplementary Material online). This network of P. falciparum populations confirmed the results of the population structure analyses (fig. 2B). Haiti and the common ancestor of Colombia, Peru, and SAM South cluster shared 78% of ancestry, although 22% came from an older or unsampled population.
Altogether, these results suggest that the American populations are subdivided into two distinct genetic clusters: a southern cluster that includes Brazil and French Guiana (SAM South) and a northern cluster that includes Colombia and Haiti (SAM North). The TreeMix and ADMIXTOOLS2 analyses also suggested that all southern populations (SAM South cluster and Peru) are the result of an ancestral large admixture event between the northern and southern clusters and also with unsampled populations.
Founder Effect Associated with P. falciparum Colonization of the Americas
To assess the demographic history of the American populations, and particularly whether they went through bottleneck events during the colonization process, we calculated Tajima's D values (Tajima 1989) and analyzed the effective population size (Ne) changes through time using Stairway Plot 2 (Liu and Fu 2020). For these analyses, we only kept the American populations grouped by cluster (excluding the highly admixed Peruvian population), three African populations (Democratic Republic of Congo, Tanzania, and Senegal), and one Asian population (Myanmar) for comparison. The polarized data set included 31,892 SNPs for 500 samples, from which we could generate an unfolded site frequency spectrum (SFS).
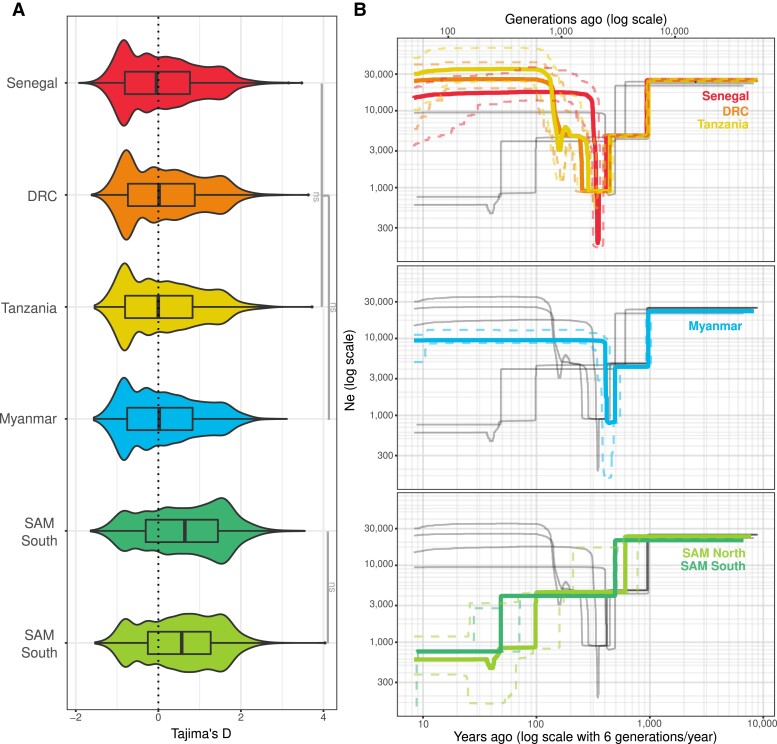
Tajima’s D values of the American populations were positive and not different between the SAM genetic clusters (Wilcox test, Bonferroni adjusted P value: 1.32e−01). Conversely, the African and Asian populations displayed Tajima’s D values close to zero but with mostly negative values (fig. 3A). This relative lack of rare alleles or excess of shared variants in the American populations, characterized by positive Tajima's D values, suggests a historical demographic contraction. Analysis of the population size changes, based on the SFS (fig. 3B), indicated that all American populations showed similar patterns, characterized by a decline or a bottleneck in the population that started between 3,000 and 3,600 generations ago. By considering six generations per year for P. falciparum (Otto et al. 2018), this corresponded approximately to 600 and 500 years ago (i.e., the time of slaves’ arrival in the New World). More recently, SAM North (Haiti–Colombia) underwent a decline (600 generations ago, ∼100 years ago), followed by a bottleneck (240 generations ago, ∼40 years ago). SAM South (Brazil–French Guiana) populations declined 300 generations ago (∼50 years ago). In comparison, the African and Asian populations did not show any decline in the same period (fig. 3B).
Fig. 3.
Demographic history of P. falciparum in South America, compared with African and Asian populations. (A) Comparison of the distributions of Tajima's D values of the two American genetic clusters, three African populations (Senegal, Tanzania, and DRC), and one Asian country (Myanmar). The gray bars with ns indicate that there is no significant differences between the distributions with a Wilcox test (B) Variation in effective population (Ne) size with time back to the most recent common ancestor inferred from Stairway Plot 2 for the two American clusters, three African populations (Senegal, Tanzania, and DRC), and one Asian country (Myanmar). The solid lines correspond to the median, and the dashed lines represent the 95% confidence interval. Gray lines represents the other populations. Note that the axes are in the log10 scale. DRC, Democratic Republic of Congo. SAM North is the cluster with Haiti and Colombia, and SAM South is the cluster composed of Brazil and French Guiana.
Genomic Evidence of Recent and Ancient Local Adaptations in the American Populations
We first used haplotype-based tests (XP-EHH and Rsb) to detect evidences of recent positive selection in the P. falciparum genomes in the Americas compared with Africa. XP-EHH and Rsb allow detecting signals of selection that are specific to the introduced populations compared with the source (here, the African populations). As these tests are based on haplotype length and LD, they tend to detect recent or ongoing positive selection events because signals of more ancient selective events would have been broken by recombination over time (Voight et al. 2006; Sabeti et al. 2007; Tang et al. 2007). We performed all tests for the SAM North and SAM South clusters independently and used Senegal as the reference population from the native African zone. The data set for these analyses included 78,036 SNPs for 98 samples.
In the SAM North cluster (Colombia–Haiti), the XP-EHH and Rsb tools detected respectively 20 and 19 genes with significant SNPs in their coding sequence (CDS) or untranslated region (UTR) (supplementary fig. S6, Supplementary Material online, and fig. 4), of which 17 were in common. In the SAM South cluster (Brazil–French Guiana), 16 genes displayed significant SNPs in the CDS or UTR regions with XP-EHH (supplementary fig. S6, Supplementary Material online) and 16 with Rsb (fig. 4); 14 genes were in common. All candidate genes identified as potentially under positive selection in the American clusters are listed in supplementary tables S2–S4, Supplementary Material online.
Fig. 4.
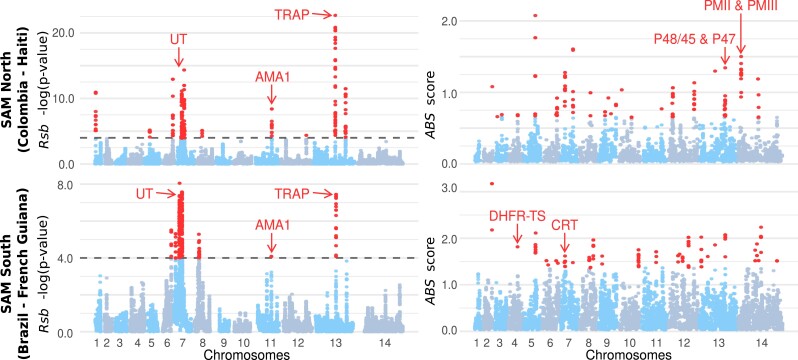
Evidence of selective sweeps in the two American genetic clusters. Manhattan plots showing the Rsb and ABS selection scans for the SAM North (Colombia–Haiti) and SAM South (Brazil–French Guiana) clusters. For the Rsb scores, the dotted lines represent the threshold significance value -log(P value) = 4. The points in red are SNPs marking a selective sweep in the SAM cluster (negative Rsb values). For the ABS scores, all values in red represent the top 1% of values, evidence of past positive selection events. UT, ubiquitin-protein transferase; DHFR-TS, dihydrofolate reductase-thymidylate synthase; CRT, chloroquine resistance transporter; AMA1, apical membrane antigen-1; TRAP, thrombospondin-related adhesive protein; PMII, plasmepsin II; and PMIII, plasmepsin III.
These selection tests provided primary evidence of recent selection signals. Indeed, older signals, dating back to the first generations of colonization history (between 400 and 10,000 generations ago according to Yalcindag et al. (2012) and Anderson et al. (2000)) will not be visible with Rsb and XP-EHH. These signals of selection may have been hidden by recombination events occurring at each generation, breaking up LD tracks along the genome. Therefore, to detect more ancient positive selection signals, we used the ancestral branch statistic (ABS), an FST-like statistic in which two closely related populations from the same American cluster were compared with two outgroup populations (one from Senegal, Africa, and one from Myanmar, Asia) in a quartet population system (Cheng et al. 2017). This approach allowed detecting signals of selection in the ancestral branch that linked the American populations to the African/Asian populations. The outgroup (Senegal and Myanmar) populations were the populations with the least amount of missing data in the two continents. We used the data set already exploited for the XP-EHH and Rsb analyses, to which we added 103 isolates from Myanmar.
We found significant signals of positive selection for 109 and 188 genes with CDS or UTR regions included in outlier windows that putatively underwent positive selection in SAM North (supplementary tables S2 and S4, Supplementary Material online) and in SAM South (supplementary tables S3 and S4, Supplementary Material online), respectively. We identified several genes implicated in interactions with the hosts (Anopheles spp. and/or humans) and with drug resistance (fig. 4). None of these genes overlapped with the results obtained with the haplotype-based tests (XP-EHH and Rsb).
Cluster-Specific Selection, Parallel Evolution, or Adaptive Migration between the Northern and Southern American Clusters
The two independent waves of introduction in the Americas offered the opportunity to determine whether evolution proceeded similarly in the two clusters to adapt to the new environments. To this aim, we looked for different categories of genes or genomic regions that showed (1) a signal of selection in one but not in the other cluster (cluster-specific selection) and (2) evidence of positive selection in both clusters.
Most loci (n = 80/132 in SAM North and n = 154/206 in SAM South) showed evidence of selection in one cluster, but not in the other. For instance, the ABS values showed that some genes involved in the parasite immune evasion from the mosquito immune system (i.e., P48/45 and P47) were under selection exclusively in the SAM North cluster. Moreover, in SAM North populations, genes involved in resistance to dihydroartemisinin-piperaquine combination treatments (plasmepsin II and III, PMII, and PMIII) displayed evidence of positive selection events, independently of the SAM South cluster. Lastly, the chloroquine resistance transporter (CRT) gene, which is implicated in P. falciparum resistance to chloroquine, was under positive selection only in the SAM South cluster.
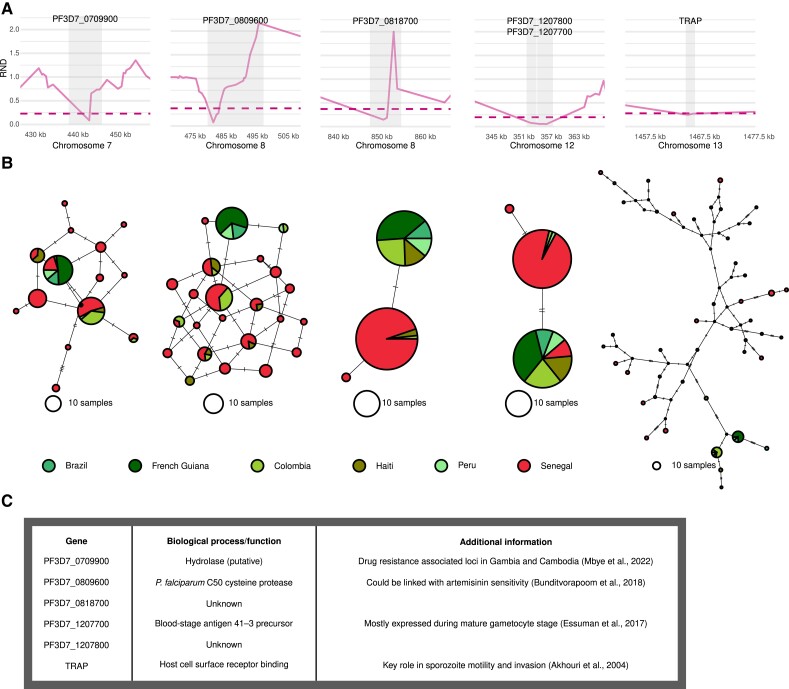
Other genes (n = 52) showed evidence of positive selection in both clusters: 37 genes with ABS and 14 genes with the other haplotype-based tests (supplementary table S4, Supplementary Material online). Some of these genes encode proteins implicated in the parasite–host interactions, for instance, apical membrane antigen 1 (AMA1) and thrombospondin-related adhesive protein (TRAP), and others in drug resistance (e.g., ubiquitin-protein transferase, UT). The finding that the same genes were under selection in both clusters could be explained by different evolutionary mechanisms: (1) parallel evolution, if the two genes independently responded to similar selective pressures in the two clusters; (2) adaptive migration, if the advantageous allele was first positively selected in one cluster and then migrated to the other cluster where it was also positively selected locally; (3) selection on standing variation, if the same allele, from the ancestral population, was selected in both clusters; and (4) selection in the ancestral population, before the cluster divergence (for the genome regions that resulted from introgression or admixture between clusters). To disentangle these different possibilities, for each gene, we used the relative node depth (RND) to measure the haplotype similarity around the selected loci (Feder et al. 2005). This statistic takes into account local diversity variations along the genome. A low RND value compared with the rest of the genome suggests adaptive migration, whereas no difference would indicate parallel evolution, selection on standing variation, or ancestral selection. Most regions fell into the second category. Only five genomic regions had low RND values, suggesting adaptive migration between clusters (fig. 5A). The signal was particularly strong for TRAP, a gene involved in the parasite interaction with mosquitoes and humans. Indeed, we found only few, closely related haplotypes in the two American clusters, compared with the high haplotypic diversity in the African populations (fig. 5B).
Fig. 5.
Candidate genes for convergent evolution or introgression between American clusters. (A) RND values between SAM North (Haiti–Colombia) and SAM South (Brazil–French Guiana) for five regions. The region in gray represents the gene (gene name on top). The dotted line marks the 5% threshold of the lowest values on the chromosome. TRAP, thrombospondin-related adhesive protein. (B) Median-joining haplotype networks of the regions with the lowest RND values between American clusters. TRAP, thrombospondin-related adhesive protein. (C) Table of biological processes and functions for each candidate genes.
Discussion
Plasmodium falciparum was introduced into the Americas from Africa during the transatlantic slave trade from the 16th to the 19th centuries (Anderson et al. 2000; Yalcindag et al. 2012; Rodrigues et al. 2018). This history of colonization of a new continent offers the opportunity to analyze how this parasite genetically adapted to new environmental conditions (new human host populations with distinct characteristics from the source populations as well as new mosquito host species and new abiotic conditions). The different waves of introduction that likely occurred during this colonization history can be potentially considered as replicates of the same “natural experiment” to explore the repeatability of adaptive evolution.
Plasmodium falciparum Population Structure and Demographic History in the Americas
Genomic information on P. falciparum in the Americas allowed us to (re-)explore its population structure and colonization history on this continent. First, we confirmed that the American populations of P. falciparum originated from Africa (Anderson et al. 2000; Yalcindag et al. 2012; Rodrigues et al. 2018). When we modeled two genetic clusters (K = 2) in the ADMIXTURE genetic ancestry analysis (supplementary fig. S3, Supplementary Material online), we observed a distinction mainly between Asian and African/American populations. American populations split from African populations only with K values >5. This conclusion was confirmed by the PCA (fig. 1B). The population branching obtained with TreeMix and ADMIXTOOLS2 (fig. 2) again indicated that American populations were more closely related to African populations or to P. praefalciparum, an African gorilla parasite. Unlike Rodrigues et al. (2018), we did not observe any evidence of introgression from the Asian strains into the American populations. This difference could be explained by the fact that Rodrigues et al. (2018) used mitochondrial markers, whereas we used nuclear SNPs. Indeed, mitochondrial genomes are haploid and clonally transmitted without any recombination among lineages (Galtier et al. 2009; Preston et al. 2014). Therefore, mitochondrial genomes form a single locus potentially subject to selective processes and with very limited resolution and representativeness of the population genetic structure and demographic histories. As these mitochondrial markers have been used to infer ancient colonization or migration events that are undetectable in the nuclear genome (Diez Benavente et al. 2020), this Asian introgression may reflect ancient gene flows or incomplete lineage sorting.
Our results also confirmed previous observations that American P. falciparum populations are subdivided into at least two distinct genetic clusters: one in the North (Colombia and Haiti) and one in the South (Brazil and French Guiana). The Peruvian population was admixed between these clusters. The PCA results (fig. 1B) indicated that the SAM South cluster was more differentiated from the native African populations than the SAM North cluster. Similarly, the ADMIXTURE results showed that the Brazil and French Guiana populations formed a cluster without any evidence of admixture with African populations, even at low K values (fig. 1C). The number of slaves as well as the timing of their arrival was very similar between the Caribbean–Spanish Mainland and Brazil–French Guiana during the transatlantic slave trade (supplementary fig. S7, Supplementary Material online). Also, we did not find any evidence of more recent migration between Africa and Haiti/Colombia than with Brazil/French Guiana during our recent human history. One explanation for the lowest proximity of SAM South to Africa could be that this cluster comes, at least in part, from an unsampled population, genetically differentiated from our sampled populations in Africa. Indeed, the networks obtained by TreeMix and ADMIXTOOLS2 reveal that SAM South branches at the base of the African populations and that admixture (of about 20%) occurred with a nonsampled population close to P. praefalciparum, an African parasite of gorillas (fig. 2). We can therefore deduce that the population of origin would be an unsampled African population (ancestral and extinct or from another region of Africa). Although African populations are poorly differentiated nowadays (see fig. 1B and C and also Anderson et al. 2000; Yalcindag et al. 2012), it would be relevant to expand the sampling geographic coverage in this continent, especially toward Angola which was the largest supplier of slaves to the New World (Klein 1978), in order to get a better picture of the origin of the American populations.
Using 12 microsatellites markers and 384 SNPs, Yalcindag et al. (2012) also found a genetic demarcation between Colombian populations and Brazilian/French Guianan populations and populations with interbreeding profiles in Peru and Venezuela. Scenario-testing using Approximate Bayesian computation suggested that this structuring was the result of several independent introductions of P. falciparum in South America (Yalcindag et al. 2012). In agreement, our TreeMix and ADMIXTOOLS2 analyses indicated at least two independent introductions into the Americas. However, unlike Yalcindag et al. (2012), our analyses suggested that either both Haitian and Colombian populations were introduced independently of the Peruvian, Brazilian, and French Guianan populations or only the Haitian population (fig. 2). Both analyses also suggested introgression from the SAM North cluster (36.35% from Colombia with TreeMix and 76% from Haiti with ADMIXTOOLS2) to the south American common ancestor (fig. 2). So, this suggests that the two clusters exchanged migrants only after their introductions, thus creating in-between admixed populations, as Peru (fig. 1B; see also Yalcindag et al. 2012) and Venezuela (Yalcindag et al. 2012). Larger and more uniform sampling in Latin America would allow us to better understand admixture and gene flows between these clusters as well as the history of the admixed populations.
Concerning the demographic history of the American populations, our analyses (Tajima's D and Stairway Plot 2) suggested that they went through several declines and bottlenecks during or after the colonization of the Americas (fig. 3B). Their consequence is also visible in the longer branch lengths of the TreeMix tree for the American populations compared with populations of the other continents. These results suggest higher genetic drift in American populations compared with the other populations because of founder effects or bottlenecks during the colonization of the new continent and/or intense selection pressure due to new environmental conditions. Indeed, some decline events found with Stairway Plot 2 occurred at times that may correspond to the introduction dates of P. falciparum in the Americas, as proposed by Yalcindag et al. (2012). This pattern is also visible in other parasites that arrived to the Americas during the transatlantic trade, as Leishmania chagasi (Leblois et al. 2011) or Schistosoma mansoni (Platt et al. 2022). Conversely, other events were too recent to have been caused by a founding effect following the parasite introduction into the New World (fig. 3B). Specifically, the SAM South (Brazil–French Guiana) cluster experienced an effective population size (Ne) decrease long after its likely introduction into South America. This recent demographic decline (∼50 years ago) might be explained by the selection imposed by antimalarial drugs. Furthermore, it is possible that the small Ne observed over a recent time in the Americas is the result of successive bottlenecks due to the epidemic transmission that takes place in the region. With cyclic variations of populations size, the estimated Ne should then reflect only the harmonic mean of population sizes: small but stable (Ellegren and Galtier 2016).
Evidence of Adaptation in the Americas
During the colonization of the Americas, P. falciparum faced new environmental conditions that could have exerted strong selection pressures. These adaptation processes have left an imprint in P. falciparum genome.
Adaptation to New Hosts (Mosquitoes and Humans)
Two genes of the 6-cysteine family (P47 and P48/45), expressed during the stages when P. falciparum is present in mosquitoes, showed extreme ABS values (supplementary tables S2, Supplementary Material online), indicating positive selection early in the colonization history of the Americas. In P. falciparum, P47 allows escaping the vector immune system (Molina-Cruz and Barillas-Mury 2014; Canepa et al. 2016), whereas the protein encoded by P48/45 plays an essential role in reproduction, which takes place in the mosquito (van Dijk et al. 2001). This divergent selection for P47 and P48/45 was previously described in worldwide studies on the polymorphism of these genes (P47, Anthony et al. 2007; and P48/45, Conway et al. 2001). Here, P47 and P48/45 showed evidence of positive selection only in the SAM North cluster (Colombia–Haiti), as already reported by Tagliamonte et al. (2020).
For some of the genes under selection, the origin of the selective pressure was less evident because they are expressed at different stages of the parasite life cycle, both in humans and mosquitoes. For instance, TRAP allows crossing the cell barriers in both hosts (Akhouri et al. 2004), and AMA1 is important for the invasion of erythrocytes (Triglia et al. 2000) and hepatocytes (Yang et al. 2017). An adaptation of AMA1 to mosquitoes cannot be entirely ruled out because it was recently shown that this protein is involved in the invasion of the mosquito salivary glands (Fernandes et al. 2022).
Other genes (n = 127), such as RF1, HAD2, and C3AP3, also may have played a role in P. falciparum adaptation to the mosquito and/or to human host, but their function is unknown. Functional analyses may help to better understand P. falciparum adaptive processes in the Americas.
Adaptation to Antimalarial Treatments
Another major selection pressure exerted on P. falciparum population in the Americas is related to the use of antimalarial drugs by human populations to prevent infection. Plasmodium falciparum genomes in the Americas include some evidence of selection concerning known resistance genes (Wongsrichanalai et al. 2002; Mita et al. 2009). We found a common signal in the SAM North and South clusters for UT, a gene potentially involved in resistance to quinine (Sanchez et al. 2014). However, the two American clusters did not present similar profiles for other drugs response genes (supplementary tables S2 and S3, Supplementary Material online). In SAM North (Haiti–Colombia), we identified selection signals for PMII and PMIII, two genes involved in resistance to the dihydroartemisinin-piperaquine combination (Mukherjee et al. 2018) that currently have been detected only in Asia (Amato et al. 2017; Witkowski et al. 2017). In the SAM South cluster (Brazil–French Guiana), as expected, we found selection signals for the dihydrofolate reductase (DHFR-TS) and CRT genes. DHFR-TS is a gene implicated in the resistance to the sulfadoxine-pyrimethamine (SP) combination (Happi et al. 2005). This treatment was introduced in Venezuela in the 1950s (Gabaldon and Guerrero 1959), and in the 1970s, it was used in various American countries as an alternative to chloroquine, although pyrimethamine-resistant strains had been already detected in some regions (Maberti 1960; Walker and Lopez-Antunano 1968). DHFR-mutant parasites, resistant to SP, indigenously evolved in South America (Mita et al. 2009). The first cases of resistance were reported in Venezuela in 1977 (Godoy et al. 1977) and then in Colombia in 1981 (Espinal et al. 1985). From each of these countries, a distinct resistant lineage spread to South America. Currently, the Colombian lineage is also found in Peru, whereas the Venezuelan lineage has spread mainly to Brazil and to Bolivia (Mita et al. 2009). On the other hand, CRT confers resistance to chloroquine and resistant strains appeared in Colombia and Venezuela in 1960 (Moore and Lanier 1961), independently of other world regions (Mita et al. 2009; Plowe 2009). Then, resistance spread throughout the American continent between the 1960s and the 1980s, with two distinct main genotypes (Mita et al. 2009; Plowe 2009). We did not find any evidence of selection signals for CRT in the SAM North cluster. This result highlights one of the main limitations of this study. Because of limited sample size per country in the Americas (less than 20 individuals per population), we had to group together populations which were closely related but also quite distinct (e.g., Colombia and Haiti in SAM North). Thus, for selection signals that are specific to one population but not the other, we were unable to detect them. This is likely the case here with CRT and DHFR in SAM North. Indeed, although selection on these genes is expected to have occurred in the Colombian population where resistances against chloroquine and SP have been observed (Mita et al. 2009; Plowe 2009), it is not the case for Haiti where chloroquine and SP resistance has never been detected (Carter et al. 2012; Neuberger et al. 2012; Vincent et al. 2018; Rogier et al. 2020), although this remains controversial for chloroquine resistance (Londono et al. 2009; Gharbi et al. 2012). Thus, by mixing the two populations, we artificially reduced the signal of selection and were likely not able to detect it with the tools used. It is therefore important for future studies to increase the sample size within each population to be able to use these tools on each of them independently and detect population-specific selection signals. Furthermore, French Guianan populations have recently evolved a compensatory mutation on the CRT gene that makes it sensitive to chloroquine. The frequency of this mutation has increased from 2.7% in 2002 to 58% in 2012 (Pelleau et al. 2015). Such rapid allele frequency increase could suggest a recent adaptation signal that could have been detected with XP-EHH and/or Rsb. Since we used the whole data set from Pelleau et al. (2015), without taking into account the generation gap (from 1994 to 2013) and the resulting difference in haplotype frequency, we could not detect a signal with these analyses. Again, the absence of detection could also be explained by the fact that, for these analyses, we combined the American populations by cluster due to the small sample sizes.
Surprisingly, the majority of significant signals for drug resistance genes were found with ABS (a test based on FST) that is designed to detect ancient signals of selection and not with XP-EHH or Rsb (that are supposed to detect recent or ongoing hard sweep signals). The explanation for that is likely associated to the construction of the statistics. XP-EHH and Rsb compare the size of extended haplotype homozygosity (EHH) for each SNP between populations (Voight et al. 2006; Sabeti et al. 2007; Tang et al. 2007). As the African and American populations experienced the same drug pressure at about the same time (Wongsrichanalai et al. 2002; Mita et al. 2009; Plowe 2009), they both have long EHH. Therefore, selective sweeps cannot be detected with XP-EHH or Rsb. By construction, ABS detects ancient signals of selection in regions where the ancestral branch is longer than the rest of the genome. But if there is recent selection in one of the terminal populations and then migration toward the other population and selection, then this may create the false picture of an ancestral selection (see supplementary fig. S8, Supplementary Material online). So with ABS, we would observe an outlier value, although this is not a signal of ancient selection, but a recent one with adaptive migration. Here, this is likely what happened with some of the drug resistance alleles. Indeed, often drug resistance haplotypes were different between the Old World (outgroup populations) and the New World (target populations), and they were rapidly fixed and spread intracontinentally (Mita et al. 2009; Plowe 2009). This may explain why traces of selection on these genes can be detected with ABS, although they are not ancient selection events.
Cluster-Specific Selection, Parallel Evolution, and Adaptive Migration
Although most genes (n = 234) underwent cluster-specific selection, some (n = 51) experienced selection in both clusters. It is thought that repeated and parallel evolution is infrequent in populations (Bailey et al. 2015), particularly because there are too many different phenotype combinations and even more genotype combinations that can generate higher fitness in a new environment. Consequently, the probability of parallel evolution for a particular phenotype is considered very low because it is very unlikely that the same combinations of de novo mutations might occur twice by chance (Bailey et al. 2015). However, it is more and more acknowledged that much adaptation, especially in the context of a rapid environmental change (e.g., during introduction to a new area), proceeds from the sorting of ancestral standing genetic variation and does not rely completely on de novo mutations (Thompson et al. 2019). In the P. falciparum populations in the Americas, this might have occurred for most genes showing evidence of selection in the New World, at least for the genomic regions that came from two distinct introduction waves in the North and the South (between 20% and 60% of the genome for the SAM South cluster, depending on the analysis). For the other genomic regions that originated from admixture or gene flow from the SAM North cluster, selection might have taken place on haplotypes that were already separated by genetic drift from Africa.
Convergent gene evolution can occur in independently introduced populations also through adaptive migration (i.e., by introducing the identical adaptive alleles from one population into others where they are selected) (Zhang et al. 2021). In this scenario, gene flow plays an important role in moving the same allelic variants that share a single mutational origin among populations (Zhang et al. 2021). In our data set, such signal of adaptive migration between clusters was observed for five genes, including TRAP that plays a key role in sporozoite motility and invasion (fig. 5C). Although haplotypic diversity is very important in Africa, only few dominant haplotypes remain in South America, thus concomitantly confirming the strong positive selection that occurred on this gene in the New World and also suggesting that only few related haplotypes spread throughout South America through migration. It is not known how this variant was selected in this new environment. On the other hand, in Africa and Asia, TRAP is more under balancing selection whereby it maintains high levels of genetic and haplotypic diversity (Naung et al. 2022).
Studies using model-based statistical approaches (see for ex. Lee and Coop 2017, 2019) will be needed to investigate the different modes of convergent adaption in the American populations.
Conclusions
We explored the genomic polymorphism of P. falciparum populations in the Americas and different regions of the world. By analyzing P. falciparum nuclear genome, we could describe its population structure and refine the history of its colonization of the Americas. We confirmed the existence of at least two independent waves of introduction from Africa: one in the North and the other in the South. Unlike previous studies, we found that populations in the SAM South cluster (Brazil–French Guiana) are the results of an ancestral admixture from the first and second waves of migration. By exploring the genomes of American populations of P. falciparum, we also detected many genes that are evolving under positive selection in these populations. Among them, some had already been described as selected in this continent, whereas others are completely new. Most genes showed only signals of selection in one cluster, suggesting that selective pressures vary among locations or that selection has not taken the same path to adapt to similar environments. However, some genes were under selection in both clusters, indicating that adaptive evolution was repeatable. For few of these genes, we found evidence that this adaptive repeated evolution occurred through adaptive migration between clusters or selection on standing ancestral variations. Thus, the history of colonization and adaptation of P. falciparum in the Americas remains to be further explored, with a larger sampling from other regions. A larger data set will enable to increase the power for selection detection, particularly to detect soft sweeps with statistics such as H12 (Garud et al. 2015) and more recent machine-learning tools such as diploSH/IC (Kern and Schrider 2018).
Materials and Methods
Data Mapping, SNP Calling, and Compilation
The sequencing data for the isolates added to the MalariaGen data set and the samples from the outgroup were retrieved as FASTQ files (supplementary table S1, Supplementary Material online). Then, sequencing reads were trimmed to remove adapters and preprocessed to eliminate low-quality reads (−quality-cutoff = 30) using the cutadapt program (Martin 2011). Reads shorter than 50 bp and containing “N” were discarded (−minimum-length = 50 –max-n = 0). Sequenced reads were aligned to the Pf3D7 v3 reference genome of P. falciparum (Gardner et al. 2002) using bwa-mem (Li and Durbin 2009). A first filter was applied to exclude isolates with a mean genome coverage depth lower than 5×. The Genome Analysis Toolkit (GATK, version 3.8.0, McKenna et al. (2010)) was used to call SNPs in each isolate following the GATK best practices. Duplicate reads were marked using the MarkDuplicates tool from the Picard tools 2.5.0 (broadinstitute.github.io/picard/) with default options. Local realignment around indels was performed using the IndelRealigner tool from GATK. Variants were called using the HaplotypeCaller module in GATK and reads mapped with a “reads minimum mapping quality” of 30 (-mmq 30) and minimum base quality of >20 (−min_base_quality_score 20). During SNP calling, the genotypic information was kept for all sites (variants and invariant sites, option ERC) to retain the information carried by the SNPs fixed for the reference allele. Therefore, the VCF files obtained with the MalariaGen Project data set could be merged without losing information for the sites fixed in American populations and with the same nucleotide as the reference genome. VCF files were merged with BCFtools v1.10.2 (Li 2011; Danecek et al. 2021). All these steps are summarized in supplementary figure S2, Supplementary Material online.
SNP Data Filtration
The variant filtration steps were performed using VCFtools v 0.1.16 (Danecek et al. 2011) and BCFtools v1.10.2 (Li 2011; Danecek et al. 2021). The within-host infection complexity was assessed by calculating the FWS values (Amegashie et al. 2020) with vcfdo (github.com/IDEELResearch/vcfdo; last accessed July 2022). An FWS threshold of >0.95 was used as a proxy for monoclonal infection (supplementary fig. S1, Supplementary Material online).
Highly related samples and clones could have generated spurious signals of population structure, biased estimators of population genetic variation, and violated the assumptions of the model-based population genetic approaches used in this study (e.g., ADMIXTURE, TreeMix, and ADMIXTOOLS2) (Wang 2018). Therefore, the relatedness between haploid genotype pairs was measured by estimating the pairwise fraction of the genome identical by descent (IBD) between strains within populations using the hmmIBD program, with the default parameters for recombination and genotyping error rates, and using the allele frequencies estimated by the program (Schaffner et al. 2018). Isolate pairs that shared >50% of IBD were considered highly related (supplementary fig. S1, Supplementary Material online). In each family of related samples, only the strain with the lowest amount of missing data was retained. All the data filtration steps are summarized in supplementary figure S2, Supplementary Material online.
Population Structure, Admixture, and Relationships between Populations
As PCA and ADMIXTURE analyses require a data set with unlinked variants, SNPs were LD-pruned with PLINK v.2 (Chang et al. 2015). All SNPs with a correlation coefficient >0.1 (parameters: –indep-pairwise 50 10 0.1) were removed using a window size of 50 SNPs, and a step of 10. PCA was carried out with PLINK v.2 (Chang et al. 2015) and the following parameters: –geno –maf 0.001 –mind. The MAF was set at 0.1% to remove doubletons. Then, the maximum likelihood clustering method implemented in the ADMIXTURE v 1.3.0 software (Alexander et al. 2009) was used with different cluster (K) numbers, from 2 to 15, with 15 replicates for each K to check consistency among replicates. The optimal K value was estimated using the cross-validation index, and convergence was checked with pong (Behr et al. 2016).
TreeMix and ADMIXTOOLS2 were used to estimate the most likely population tree or network topology and reticulations among them, based on variance–covariance in allele frequency. When adding the outgroup with three P. praefalciparum samples from Otto et al. (2018), only biallelic SNPs with no missing data in at least one P. praefalciparum sample were kept. As TreeMix and ADMIXTOOLS2 require unlinked SNPs, the data set was LD-pruned as done for the PCA and ADMIXTURE analyses. For TreeMix, the number of migration events (m) that best fitted the data was calculated by running TreeMix 15 times for each m value, with m ranging from 0 to 15. The optimal m value (m = 3) was estimated using the OptM R package (Fitak 2021). Then, a consensus tree with bootstrap node support was obtained by running TreeMix 100 times and postprocessing using the BITE R package (Milanesi et al. 2017). To find the best network topology with ADMIXTOOLS2, the function find_graphs was used for 0 to 12 admixture events with 100 replicates each and at most 300 generations for each. For the five best network topologies (i.e., those displaying the likelihood score closest to zero), the goodness of fit was computed with the R package admixture-graph (Leppälä et al. 2017). This approach allows comparing the observed f4 statistics among the different alternatives and identifying the graph(s) that best fit the data. Two graphs with the best goodness of fit and the least f4 statistics outliers were selected (supplementary fig. S9, Supplementary Material online). Figure 2B shows only the graph with the lowest number of f4 statistics outliers.
Demographic History of the American Populations
Tajima's D (Tajima 1989) was measured for both American clusters, three African populations (Senegal, Democratic Republic of Congo, and Tanzania), and one Asian population (Myanmar). These populations were chosen because they had the smallest amount of missing data for each region of interest (West Africa, Central Africa, East Africa, and Asia, respectively). Tajima's D values were estimated using VCFtools v 0.1.16 (Danecek et al. 2011), with a window of 5 kb. The sample size was standardized (i.e., 20 randomly chosen isolates for each population) to obtain values that could be compared. Moreover, the variation in effective population size over time was estimated using Stairway Plot v2.1.1 (Liu and Fu 2020) and the same populations and clusters used to calculate Tajima’s D, but without any sample size standardization. Three P. praefalciparum genomes from the study by Otto et al. (2018) were used to polarize the ancestral versus derived states of SNPs and create an unfolded SFS. Only biallelic SNPs without missing data in at least one P. praefalciparum sample were considered for this analysis. The SFS was generated with easySFS (github.com/isaacovercast/easySFS). For these analyses, a mutation rate of 4.055 × 10−9 (Otto et al. 2018) was assumed and we set an observed number of sites equal to the number of P. praefalciparum sites that are in the core genome of this P. falciparum data set (10,007,378 sites) (Miles et al. 2015).
Detection of Positive Selection
Given the low sample size in each American locality (from n = 5 to n = 18), selection scan analyses were performed at the scale of the genetic clusters defined by ADMIXTURE and PCA, as described by Hupalo et al. (2016). Indeed, the power of XP-EHH and Rsb analyses is influenced by the sample size, and at least 20 haplotypes are recommended by Pickrell et al. (2009). Thus, each American cluster (SAM North and SAM South) was compared with the West African population that had the smallest amount of missing data: Senegal. For ABS, the Senegal population was kept as an outgroup and Myanmar, the Asian population with the smallest amount of missing data, was added. The XP-EHH and Rsb scores were calculated using the R package rehh (Gautier et al. 2017). Following Klassmann and Gautier (2022), to keep the maximum SNP number without compromising the test statistical power, data were not polarized with P. praefalciparum. Thus, the allele present in the reference genome was considered the ancestral allele. The significance threshold was set at -log(P value) = 4, as recommended by Gautier et al. (2017), and only SNPs with negative standardized values (i.e., indicating positive selection for the American populations rather than African population) were considered.
The ABS values were calculated with CalcABS (Cheng et al. 2017) in sliding windows of the genome (a window of 20 kb with a step of 1 kb). All windows with <20 SNPs were removed to avoid extreme values caused by a low SNP number in some windows. The 1% most extreme values were considered as evidence that the genomic regions displayed signs of an ancient selective sweep in our South American populations. Among these extreme values, peaks (i.e., ≥3 consecutive windows with outliers values) were observed in some regions. Due to the large window size and to avoid high artefactual values caused by a very extreme region, only the regions with the maximum values for each peak were kept. In the absence of peaks (less than three consecutive windows with outliers values), windows with the most extreme 1% values were kept.
Once selection signals were detected, the identified genes were annotated using the general feature format (GFF) file available from genedb (genedb.org, January 2021 version) and the intersect function of BEDtools v 2.26.0 (Quinlan and Hall 2010). Additional information (e.g., gene name, function, and biological process) was retrieved from PlasmoDB (plasmodb.org, accessed in February 2022).
Detection of Introgression
RND values (Feder et al. 2005) were calculated between SAM South and SAM North clusters, with the Senegalese population as outgroup, on sliding windows of 5 kb with a step size of 1 kb and at least 20 SNPs per window. To confirm the information given by the RNDs, the haplotype network for the three populations with extremely low RNDs was visualized with popart v1.7 (Leigh and Bryant 2015) and the median-joining method (Bandelt et al. 1999), with ε equals zero.
Supplementary Material
Acknowledgments
We would like to thank the i-trop bioinformatics platform at IRD Montpellier, the South Green platform, and the French Bioinformatics Institute (IFB) for providing access to high-performance computing cluster (HPC) resources. We also thank the Center for Information Technology of the University of Groningen for their support and for providing access to the Peregrine high-performance computing cluster. This work was supported by the Agence Nationale de la Recherche MICETRAL (ANR-19-CE35-0010) and Agence Nationale de la Recherche GENAD (ANR-20-CE35-0003).
Contributor Information
Margaux J M Lefebvre, MiVEGEC, Univ. Montpellier, CNRS, IRD, Montpellier, France.
Josquin Daron, MiVEGEC, Univ. Montpellier, CNRS, IRD, Montpellier, France.
Eric Legrand, Malaria Biology and Vaccine Unit, Institut Pasteur, Paris, France.
Michael C Fontaine, MiVEGEC, Univ. Montpellier, CNRS, IRD, Montpellier, France; Groningen Institute for Evolutionary Life Sciences (GELIFES), University of Groningen, Groningen, The Netherlands.
Virginie Rougeron, REHABS, International Research Laboratory, CNRS-NMU-UCBL, George Campus, Nelson Mandela University, George, South Africa.
Franck Prugnolle, REHABS, International Research Laboratory, CNRS-NMU-UCBL, George Campus, Nelson Mandela University, George, South Africa.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
Conception: F.P., V.R., E.L., M.F. and M.L.; funding acquisition: V.R. and F.P.; method development and data analysis: M.L., J.D., M.F., and F.P.; interpretation of the results: M.L., J.D., M.F., F.P., and V.R.; drafting of the manuscript: M.L. and F.P.; and reviewing and editing of the manuscript: M.L., E.L., M.F., F.P., and V.R.
Data Availability
The majority of data is from the P. falciparum Community Project conducted by MalariaGen (Pearson et al. 2019) and can be downloaded from the Wellcome Trust Sanger Institute public ftp site (ftp://ngs.sanger.ac.uk/production/malaria/pfcommunityproject/CatalogueOfVariations_v4.0/). For the other samples from Brazil, French Guiana, and Haiti, raw sequencing reads are available from the NCBI Sequencing Read Archive under the BioProject accession numbers PRJNA312679, PRJNA242163, and PRJNA603776. Plasmodium praefalciparum samples are accessible from the European Nucleotide Archive under sample accessions SAMEA2464702, SAMEA2073285, and SAMEA2493921. The scripts are available in this github repository: https://github.com/MargauxLefebvre/P.falciparum_americas.
References
- Akhouri RR, Bhattacharyya A, Pattnaik P, Malhotra P, Sharma A. 2004. Structural and functional dissection of the adhesive domains of Plasmodium falciparum thrombospondin-related anonymous protein (TRAP). Biochem J. 379:815–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19:1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato R, Lim P, Miotto O, Amaratunga C, Dek D, Pearson RD, Almagro-Garcia J, Neal AT, Sreng S, Suon S, et al. 2017. Genetic markers associated with dihydroartemisinin–piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype–phenotype association study. Lancet Infect Dis. 17:164–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amegashie EA, Amenga-Etego L, Adobor C, Ogoti P, Mbogo K, Amambua-Ngwa A, Ghansah A. 2020. Population genetic analysis of the Plasmodium falciparum circumsporozoite protein in two distinct ecological regions in Ghana. Malar J. 19:437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson TJC, Haubold B, Williams JT, Estrada-Franco JG, Richardson L, Mollinedo R, Bockarie M, Mokili J, Mharakurwa S, French N, et al. 2000. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol. 17:1467–1482. [DOI] [PubMed] [Google Scholar]
- Anthony TG, Polley SD, Vogler AP, Conway DJ. 2007. Evidence of non-neutral polymorphism in Plasmodium falciparum gamete surface protein genes Pfs47 and Pfs48/45. Mol Biochem Parasitol. 156:117–123. [DOI] [PubMed] [Google Scholar]
- Bailey SF, Rodrigue N, Kassen R. 2015. The effect of selection environment on the probability of parallel evolution. Mol Biol Evol. 32:1436–1448. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 16:37–48. [DOI] [PubMed] [Google Scholar]
- Behr AA, Liu KZ, Liu-Fang G, Nakka P, Ramachandran S. 2016. Pong: fast analysis and visualization of latent clusters in population genetic data. Bioinformatics 32:2817–2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant JE, Holmes EC, Barrett ADT. 2007. Out of Africa: a molecular perspective on the introduction of yellow fever virus into the Americas. PLOS Pathog. 3:e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunditvorapoom D, Kochakarn T, Kotanan N, Modchang C, Kümpornsin K, Loesbanluechai D, Krasae T, Cui L, Chotivanich K, White NJ, et al. 2018. Fitness loss under amino acid starvation in artemisinin-resistant Plasmodium falciparum isolates from Cambodia. Sci Rep. 8:12622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canepa GE, Molina-Cruz A, Barillas-Mury C. 2016. Molecular analysis of Pfs47-mediated Plasmodium evasion of mosquito immunity. PLoS One 11:e0168279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter TE, Warner M, Mulligan CJ, Existe A, Victor YS, Memnon G, Boncy J, Oscar R, Fukuda MM, Okech BA. 2012. Evaluation of dihydrofolate reductase and dihydropteroate synthetase genotypes that confer resistance to sulphadoxine-pyrimethamine in Plasmodium falciparum in Haiti. Malar J. 11:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 4:s13742-015-0047–0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Xu C, DeGiorgio M. 2017. Fast and robust detection of ancestral selective sweeps. Mol Ecol. 26:6871–6891. [DOI] [PubMed] [Google Scholar]
- Choi Y-J, Thines M. 2015. Host jumps and radiation, not co-divergence drives diversification of obligate pathogens. A case study in downy mildews and Asteraceae. PLoS One 10:e0133655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway DJ, Machado RLD, Singh B, Dessert P, Mikes ZS, Povoa MM, Oduola AMJ, Roper C. 2001. Extreme geographical fixation of variation in the Plasmodium falciparum gamete surface protein gene Pfs48/45 compared with microsatellite loci. Mol Biochem Parasitol. 115:145–156. [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM. 2021. Twelve years of SAMtools and BCFtools. Gigascience 10:giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez Benavente E, Campos M, Phelan J, Nolder D, Dombrowski JG, Marinho CRF, Sriprawat K, Taylor AR, Watson J, Roper C, et al. 2020. A molecular barcode to inform the geographical origin and transmission dynamics of Plasmodium vivax malaria. PLoS Genet. 16:e1008576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H, Galtier N. 2016. Determinants of genetic diversity. Nat Rev Genet. 17:422–433. [DOI] [PubMed] [Google Scholar]
- Espinal C, Cortes G, Guerra P, Arias A. 1985. Sensitivity of Plasmodium falciparum to antimalarial drugs in Colombia. Am J Trop Med Hyg. 34:675–680. [DOI] [PubMed] [Google Scholar]
- Essuman E, Grabias B, Verma N, Chorazeczewski JK, Tripathi AK, Mlambo G, Addison EA, Amoah AGB, Quakyi I, Oakley MS, et al. 2017. A novel gametocyte biomarker for superior molecular detection of the Plasmodium falciparum infectious reservoirs. J Infect Dis. 216:1264–1272. [DOI] [PubMed] [Google Scholar]
- Feder JL, Xie X, Rull J, Velez S, Forbes A, Leung B, Dambroski H, Filchak KE, Aluja M. 2005. Mayr, Dobzhansky, and Bush and the complexities of sympatric speciation in Rhagoletis. Proc Natl Acad Sci USA. 102:6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes P, Loubens M, Le Borgne R, Marinach C, Ardin B, Briquet S, Vincensini L, Hamada S, Hoareau-Coudert B, Verbavatz J-M, et al. 2022. The AMA1-RON complex drives Plasmodium sporozoite invasion in the mosquito and mammalian hosts. PLoS Pathog. 18:e1010643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitak RR. 2021. Optm: estimating the optimal number of migration edges on population trees using Treemix. Biol Methods Protoc. 6:bpab017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick J, Saint Jean Y, Lemoine JF, Dotson EM, Mace KE, Chang M, Slutsker L, Le Menach A, Beier JC, Eisele TP, et al. 2016. Malaria vector research and control in Haiti: a systematic review. Malar J. 15:376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldon A, Guerrero L. 1959. An attempt to eradicate malaria by the weekly administration of pyrimethamine in areas of out-of-doors transmission in Venezuela. Am J Trop Med Hyg. 8:433–439. [DOI] [PubMed] [Google Scholar]
- Galtier N, Nabholz B, Glémin S, Hurst GDD. 2009. Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol Ecol. 18:4541–4550. [DOI] [PubMed] [Google Scholar]
- Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, et al. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419:498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garud NR, Messer PW, Buzbas EO, Petrov DA. 2015. Recent selective sweeps in North American Drosophila melanogaster show signatures of soft sweeps. PLoS Genet. 11:e1005004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier M, Klassmann A, Vitalis R. 2017. rehh 2.0: a reimplementation of the R package rehh to detect positive selection from haplotype structure. Mol Ecol Resour. 17:78–90. [DOI] [PubMed] [Google Scholar]
- Geneva: World Health Organization . 2022. World health statistics 2022: monitoring health for the SDGs, sustainable development goals. World Health Organization; Available from:https://www.who.int/data/gho/publications/world-health-statistics.
- Gharbi M, Pillai DR, Lau R, Hubert V, Khairnar K, Existe A, Kendjo E, Dahlström S, Guérin PJ, Le Bras J.. 2012. Chloroquine-resistant malaria in travelers returning from Haiti after 2010 earthquake. Emerg Infect Dis. 18:1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies MT, Coetzee M. 1987. A supplement to the Anophelinae of Africa south of the Sahara (Afrotropical Region). Publ South Afr Inst Med Res. 55:2–2. [Google Scholar]
- Godoy GA, Volcan G, Guevara R, Medrano C, Castro J, Texeira A. 1977. Venezuelan strains of Plasmodium falciparum resistant to sulfa and pyrimethamine as demonstrated by in vitro test. Rev Latinoam Microbiol. 19:229–231. [PubMed] [Google Scholar]
- Happi CT, Gbotosho GO, Folarin OA, Akinboye DO, Yusuf BO, Ebong OO, Sowunmi A, Kyle DE, Milhous W, Wirth DF, et al. 2005. Polymorphisms in Plasmodium falciparum dhfr and dhps genes and age related in vivo sulfadoxine–pyrimethamine resistance in malaria-infected patients from Nigeria. Malar Res Afr Multilater Initiat Malar. 95:183–193. [DOI] [PubMed] [Google Scholar]
- Hupalo DN, Luo Z, Melnikov A, Sutton PL, Rogov P, Escalante A, Vallejo AF, Herrera S, Arévalo-Herrera M, Fan Q, et al. 2016. Population genomics studies identify signatures of global dispersal and drug resistance in Plasmodium vivax. Nat Genet. 48:953–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern AD, Schrider DR. 2018. diploS/HIC: An Updated Approach to Classifying Selective Sweeps. G3-Genes Genom Genet 8(6):1959–1970. 10.1534/g3.118.200262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassmann A, Gautier M. 2022. Detecting selection using extended haplotype homozygosity (EHH)-based statistics in unphased or unpolarized data. PLoS One 17:e0262024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein HS. 1978. The middle passage. Princeton University Press. Available from:http://www.jstor.org/stable/j.ctt1mf6xwn.
- Laporta GZ, Linton Y-M, Wilkerson RC, Bergo ES, Nagaki SS, Sant’Ana DC, Sallum MAM. 2015. Malaria vectors in South America: current and future scenarios. Parasit Vectors 8:426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblois R, Kuhls K, François O, Schönian G, Wirth T. 2011. Guns, germs and dogs: on the origin of Leishmania chagasi. Infect Genet Evol. 11:1091–1095. [DOI] [PubMed] [Google Scholar]
- Lee KM, Coop G. 2017. Distinguishing among modes of convergent adaptation using population genomic data. Genetics 207:1591–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KM, Coop G. 2019. Population genomics perspectives on convergent adaptation. Philos Trans R Soc B Biol Sci. 374:20180236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh JW, Bryant D. 2015. popart: full-feature software for haplotype network construction. Methods Ecol Evol. 6:1110–1116. [Google Scholar]
- Leppälä K, Nielsen SV, Mailund T. 2017. Admixturegraph: an R package for admixture graph manipulation and fitting. Bioinformatics 33:1738–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27:2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Fu Y-X. 2020. Stairway Plot 2: demographic history inference with folded SNP frequency spectra. Genome Biol. 21:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londono BL, Eisele TP, Keating J, Bennett A, Chattopadhyay C, Heyliger G, Mack B, Rawson I, Vely J-F, Désinor O. 2009. Chloroquine-resistant haplotype Plasmodium falciparum parasites, Haiti. Emerg Infect Dis. 15:735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Perez M, Villasis E, Machado RLD, Póvoa MM, Vinetz JM, Blair S, Gamboa D, Lustigman S. 2012. Plasmodium falciparum field isolates from South America use an atypical red blood cell invasion pathway associated with invasion ligand polymorphisms. PLoS One 7:e47913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maberti S. 1960. Development of resistance to pyrimethamine. report of 15 cases studied at Trujillo, Venezuela. Arch Venez Med Trop Parasitol Med. 3:239–259. [PubMed] [Google Scholar]
- Maier R, Flegontov P, Flegontova O, Isildak U, Changmai P, Reich D. 2023. On the limits of fitting complex models of population history to f-statistics. eLife 12:e85492. 10.7554/eLife.85492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17:10–12. [Google Scholar]
- Martinez-Villegas L, Assis-Geraldo J, Koerich LB, Collier TC, Lee Y, Main BJ, Rodrigues NB, Orfano AS, Pires ACAM, Campolina TB, et al. 2019. Characterization of the complete mitogenome of Anopheles aquasalis, and phylogenetic divergences among Anopheles from diverse geographic zones. PLoS One 14:e0219523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbye H, Mane K, Diop MF, Demba MA, Bojang F, Mohammed NI, Jeffries D, Quashie NB, D’Alessandro U, Amambua-Ngwa A. 2022. Plasmodium falciparum merozoite invasion ligands, linked antimalarial resistance loci and ex vivo responses to antimalarials in The Gambia. J Antimicrob Chemother. 77:2946–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milanesi M, Capomaccio S, Vajana E, Bomba L, Garcia JF, Ajmone-Marsan P, Colli L. 2017. BITE: an R package for biodiversity analyses. bioRxiv [Preprint] 2017:181610. doi: 10.1101/181610. [DOI] [PubMed]
- Miles A, Iqbal Z, Vauterin P, Pearson R, Campino S, Theron M, Gould K, Mead D, Drury E, O’Brien J, et al. 2015. Genome variation and meiotic recombination in Plasmodium falciparum: insights from deep sequencing of genetic crosses. bioRxiv [Preprint] 2015:024182. doi: 10.1101/024182. [DOI] [PMC free article] [PubMed]
- Mita T, Tanabe K, Kita K. 2009. Spread and evolution of Plasmodium falciparum drug resistance. Parasitol Int. 58:201–209. [DOI] [PubMed] [Google Scholar]
- Molina-Cruz A, Barillas-Mury C. 2014. The remarkable journey of adaptation of the Plasmodium falciparum malaria parasite to New World anopheline mosquitoes. Mem Inst Oswaldo Cruz 109:662–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DV, Lanier JE. 1961. Observations on two Plasmodium falciparum infections with an abnormal response to chloroquine. Am J Trop Med Hyg. 10:5–9. [DOI] [PubMed] [Google Scholar]
- Moreno M, Marinotti O, Krzywinski J, Tadei WP, James AA, Achee NL, Conn JE. 2010. Complete mtDNA genomes of Anopheles darlingi and an approach to anopheline divergence time. Malar J. 9:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser KA, Drábek EF, Dwivedi A, Stucke EM, Crabtree J, Dara A, Shah Z, Adams M, Li T, Rodrigues PT, et al. 2020. Strains used in whole organism Plasmodium falciparum vaccine trials differ in genome structure, sequence, and immunogenic potential. Genome Med. 12:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Gagnon D, Wirth DF, Richard D. 2018. Inactivation of Plasmepsins 2 and 3 sensitizes Plasmodium falciparum to the antimalarial drug piperaquine. Antimicrob Agents Chemother. 62:e02309-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naung MT, Martin E, Munro J, Mehra S, Guy AJ, Laman M, Harrison GLA, Tavul L, Hetzel M, Kwiatkowski D, et al. 2022. Global diversity and balancing selection of 23 leading Plasmodium falciparum candidate vaccine antigens. PLoS Comput Biol. 18:e1009801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neafsey DE, Waterhouse Robert M, Abai Mohammad R, Aganezov Sergey S, Alekseyev Max A, Allen James E, James A, Bruno A, Peter A, Gleb A, et al. 2015. Highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science 347:1258522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuberger A, Zhong K, Kain KC, Schwartz E. 2012. Lack of evidence for chloroquine-resistant Plasmodium falciparum malaria, Leogane, Haiti. Emerg Infect Dis. 18:1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto TD, Gilabert A, Crellen T, Böhme U, Arnathau C, Sanders M, Oyola SO, Okouga AP, Boundenga L, Willaume E, et al. 2018. Genomes of all known members of a Plasmodium subgenus reveal paths to virulent human malaria. Nat Microbiol. 3:687–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson RD, Amato R, Kwiatkowski DP, MalariaGEN Plasmodium falciparum Community Project . 2019. An open dataset of Plasmodium falciparum genome variation in 7,000 worldwide samples. bioRxiv [Preprint] 2019:824730. doi: 10.1101/824730. [DOI]
- Pelleau S, Moss EL, Dhingra SK, Volney B, Casteras J, Gabryszewski SJ, Volkman SK, Wirth DF, Legrand E, Fidock DA, et al. 2015. Adaptive evolution of malaria parasites in French Guiana: reversal of chloroquine resistance by acquisition of a mutation in pfcrt. Proc Natl Acad Sci USA 112:11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Coop G, Novembre J, Kudaravalli S, Li JZ, Absher D, Srinivasan BS, Barsh GS, Myers RM, Feldman MW. 2009. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 19:826–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell J, Pritchard J. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet 8(11):e1002967. 10.1371/journal.pgen.1002967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt RN, Le Clec’h W, Chevalier FD, McDew-White M, LoVerde PT, Ramiro de Assis R, Oliveira G, Kinung’hi S, Djirmay AG, Steinauer ML. 2022. Genomic analysis of a parasite invasion: colonization of the Americas by the blood fluke Schistosoma mansoni. Mol Ecol. 31:2242–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowe CV. 2009. The evolution of drug-resistant malaria. Trans R Soc Trop Med Hyg. 103:S11–S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston MD, Campino S, Assefa SA, Echeverry DF, Ocholla H, Amambua-Ngwa A, Stewart LB, Conway DJ, Borrmann S, Michon P, et al. 2014. A barcode of organellar genome polymorphisms identifies the geographic origin of Plasmodium falciparum strains. Nat Commun. 5:4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues PT, Valdivia HO, de Oliveira TC, Alves JMP, Duarte AMRC, Cerutti-Junior C, Buery JC, Brito CFA, de Souza JC, Hirano ZMB, et al. 2018. Human migration and the spread of malaria parasites to the New World. Sci Rep. 8:1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogier E, Herman C, Huber CS, Hamre KE, Pierre B, Mace KE, Présumé J, Mondélus G, Romilus I, Elismé T. 2020. Nationwide monitoring for Plasmodium falciparum drug-resistance alleles to chloroquine, sulfadoxine, and pyrimethamine, Haiti, 2016–2017. Emerg Infect Dis. 26:902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R, et al. 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 449:913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabin NS, Calliope AS, Simpson SV, Arima H, Ito H, Nishimura T, Yamamoto T. 2020. Implications of human activities for (re)emerging infectious diseases, including COVID-19. J Physiol Anthropol. 39:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez CP, Liu C-H, Mayer S, Nurhasanah A, Cyrklaff M, Mu J, Ferdig MT, Stein WD, Lanzer M. 2014. A HECT ubiquitin-protein ligase as a novel candidate gene for altered quinine and quinidine responses in Plasmodium falciparum. PLoS Genet. 10:e1004382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner SF, Taylor AR, Wong W, Wirth DF, Neafsey DE. 2018. hmmIBD: software to infer pairwise identity by descent between haploid genotypes. Malar J. 17:196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small ST, Labbé F, Coulibaly YI, Nutman TB, King CL, Serre D, Zimmerman PA. 2019. Human migration and the spread of the nematode parasite Wuchereria bancrofti. Mol Biol Evol. 36:1931–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliamonte MS, Yowell CA, Elbadry MA, Boncy J, Raccurt CP, Okech BA, Goss EM, Salemi M, Dame JB. 2020. Genetic markers of adaptation of Plasmodium falciparum to transmission by American vectors identified in the genomes of parasites from Haiti and South America. mSphere 5:e00937-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe K, Mita T, Jombart T, Eriksson A, Horibe S, Palacpac N, Ranford-Cartwright L, Sawai H, Sakihama N, Ohmae H, et al. 2010. Plasmodium falciparum accompanied the human expansion out of Africa. Curr Biol. 20:1283–1289. [DOI] [PubMed] [Google Scholar]
- Tang K, Thornton KR, Stoneking M. 2007. A new approach for using genome scans to detect recent positive selection in the human genome. PLoS Biol. 5:e171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson KA, Osmond MM, Schluter D. 2019. Parallel genetic evolution and speciation from standing variation. Evol Lett. 3:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triglia T, Healer J, Caruana SR, Hodder AN, Anders RF, Crabb BS, Cowman AF. 2000. Apical membrane antigen 1 plays a central role in erythrocyte invasion by Plasmodium species. Mol Microbiol. 38:706–718. [DOI] [PubMed] [Google Scholar]
- van Dijk MR, Janse CJ, Thompson J, Waters AP, Braks JAM, Dodemont HJ, Stunnenberg HG, van Gemert G-J, Sauerwein RW, Eling W. 2001. A central role for P48/45 in malaria parasite male gamete fertility. Cell 104:153–164. [DOI] [PubMed] [Google Scholar]
- Vincent JP, Komaki-Yasuda K, Existe AV, Boncy J, Kano S. 2018. No Plasmodium falciparum chloroquine resistance transporter and artemisinin resistance mutations, Haiti. Emerg Infect Dis. 24:2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. 2006. A map of recent positive selection in the human genome. PLoS Biol. 4:e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AJ, Lopez-Antunano FJ. 1968. Response to drugs of South American strains of Plasmodium falciparum. Trans R Soc Trop Med Hyg. 62:654–667. [DOI] [PubMed] [Google Scholar]
- Wang J. 2018. Effects of sampling close relatives on some elementary population genetics analyses. Mol Ecol Resour. 18:41–54. [DOI] [PubMed] [Google Scholar]
- Witkowski B, Duru V, Khim N, Ross LS, Saintpierre B, Beghain J, Chy S, Kim S, Ke S, Kloeung N, et al. 2017. A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: a phenotype–genotype association study. Lancet Infect Dis. 17:174–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe ND, Dunavan CP, Diamond J. 2007. Origins of major human infectious diseases. Nature 447:279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongsrichanalai C, Pickard AL, Wernsdorfer WH, Meshnick SR. 2002. Epidemiology of drug-resistant malaria. Lancet Infect Dis. 2:209–218. [DOI] [PubMed] [Google Scholar]
- World Health Organization . 2020. World malaria report 2020 global messaging. Available from:https://www.who.int/publications/m/item/world-malaria-report-2020-global-messaging.
- Yalcindag E, Elguero E, Arnathau C, Durand P, Akiana J, Anderson TJ, Aubouy A, Balloux F, Besnard P, Bogreau H, et al. 2012. Multiple independent introductions of Plasmodium falciparum in South America. Proc Natl Acad Sci USA 109:511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcindag E, Rougeron V, Elguero E, Arnathau C, Durand P, Brisse S, Diancourt L, Aubouy A, Becquart P, D’Alessandro U, et al. 2014. Patterns of selection on Plasmodium falciparum erythrocyte-binding antigens after the colonization of the New World. Mol Ecol. 23:1979–1993. [DOI] [PubMed] [Google Scholar]
- Yang ASP, Lopaticki S, O’Neill MT, Erickson SM, Douglas DN, Kneteman NM, Boddey JA. 2017. AMA1 and MAEBL are important for Plasmodium falciparum sporozoite infection of the liver. Cell Microbiol. 19:e12745. [DOI] [PubMed] [Google Scholar]
- Zhang X, Rayner JG, Blaxter M, Bailey NW. 2021. Rapid parallel adaptation despite gene flow in silent crickets. Nat Commun. 12:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman RH. 1992. Ecology of malaria vectors in the Americas and future direction. Mem Inst Oswaldo Cruz 87:371–383. [DOI] [PubMed] [Google Scholar]
- Zumla A, Hui DSC. 2019. Emerging and reemerging infectious diseases: global overview. Infect Dis Clin. 33:13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The majority of data is from the P. falciparum Community Project conducted by MalariaGen (Pearson et al. 2019) and can be downloaded from the Wellcome Trust Sanger Institute public ftp site (ftp://ngs.sanger.ac.uk/production/malaria/pfcommunityproject/CatalogueOfVariations_v4.0/). For the other samples from Brazil, French Guiana, and Haiti, raw sequencing reads are available from the NCBI Sequencing Read Archive under the BioProject accession numbers PRJNA312679, PRJNA242163, and PRJNA603776. Plasmodium praefalciparum samples are accessible from the European Nucleotide Archive under sample accessions SAMEA2464702, SAMEA2073285, and SAMEA2493921. The scripts are available in this github repository: https://github.com/MargauxLefebvre/P.falciparum_americas.