Abstract
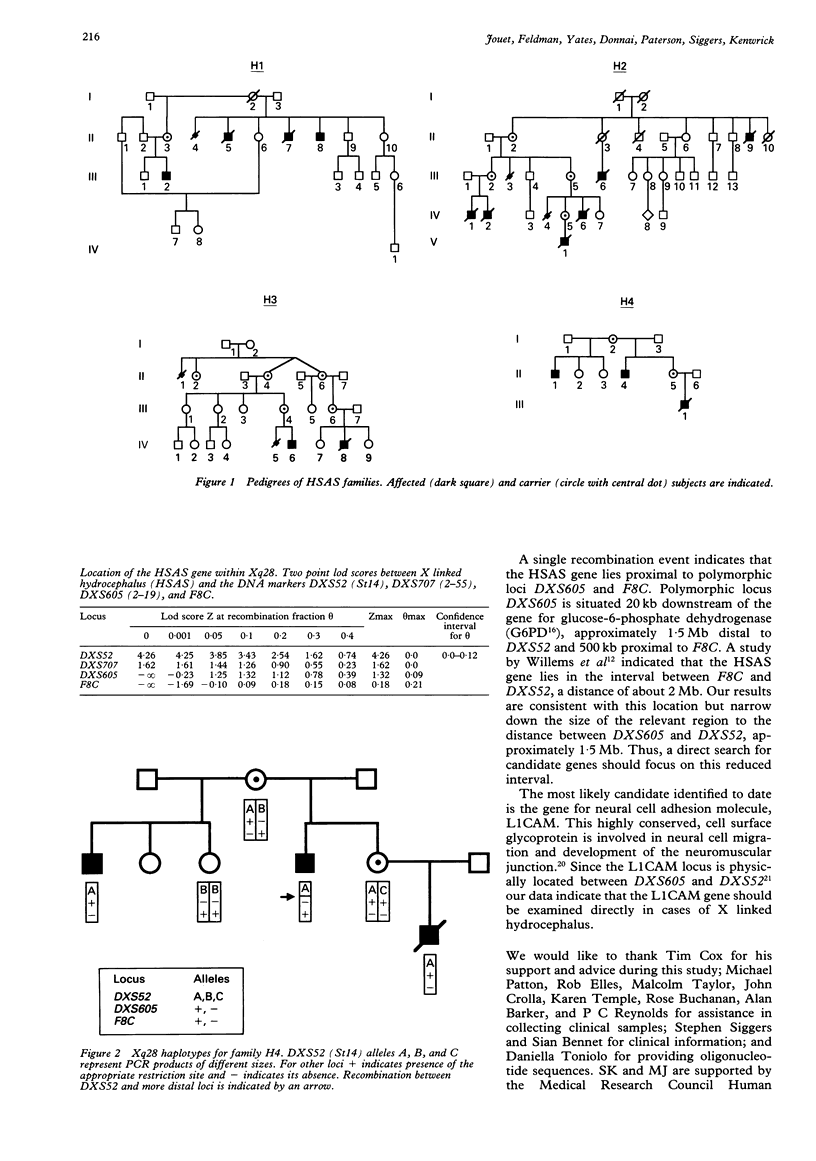
The most common inherited form of hydrocephalus, X linked hydrocephalus (HSAS), is characterised by mental retardation, adducted thumbs, and spastic paraplegia. Genetic analysis has mapped the locus for HSAS to subchromosomal band Xq28 within a region of approximately 2 megabases of DNA. In order to refine the location of the disease gene we have conducted genetic linkage analysis with Xq28 marker loci in four additional HSAS families. A lod score of 4.26 with polymorphic marker DXS52 (St14) confirms the linkage of HSAS to Xq28. Identification of a recombination event between the HSAS gene and Xq28 loci F8C and DXS605 (2-19) reduces the size of the interval likely to contain the disease locus to about 1.5 megabases, the distance between DXS605 and DXS52. The locus for neural cell adhesion molecule, L1CAM, maps within this interval and therefore represents a candidate gene for HSAS.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Hlavin M. L., Lemmon V. Molecular structure and functional testing of human L1CAM: an interspecies comparison. Genomics. 1991 Oct;11(2):416–423. doi: 10.1016/0888-7543(91)90150-d. [DOI] [PubMed] [Google Scholar]
- Keats B., Ott J., Conneally M. Report of the committee on linkage and gene order. Cytogenet Cell Genet. 1989;51(1-4):459–502. doi: 10.1159/000132805. [DOI] [PubMed] [Google Scholar]
- Kenwrick S., Ionasescu V., Ionasescu G., Searby C., King A., Dubowitz M., Davies K. E. Linkage studies of X-linked recessive spastic paraplegia using DNA probes. Hum Genet. 1986 Jul;73(3):264–266. doi: 10.1007/BF00401241. [DOI] [PubMed] [Google Scholar]
- Kogan S. C., Doherty M., Gitschier J. An improved method for prenatal diagnosis of genetic diseases by analysis of amplified DNA sequences. Application to hemophilia A. N Engl J Med. 1987 Oct 15;317(16):985–990. doi: 10.1056/NEJM198710153171603. [DOI] [PubMed] [Google Scholar]
- Landrieu P., Ninane J., Ferrière G., Lyon G. Aqueductal stenosis in X-linked hydrocephalus: a secondary phenomenon? Dev Med Child Neurol. 1979 Oct;21(5):637–642. doi: 10.1111/j.1469-8749.1979.tb01678.x. [DOI] [PubMed] [Google Scholar]
- Lathrop G. M., Lalouel J. M., Julier C., Ott J. Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3443–3446. doi: 10.1073/pnas.81.11.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestrini E., Rivella S., Tribioli C., Rocchi M., Camerino G., Santachiara-Benerecetti S., Parolini O., Notarangelo L. D., Toniolo D. Identification of novel RFLPs in the vicinity of CpG islands in Xq28: application to the analysis of the pattern of X chromosome inactivation. Am J Hum Genet. 1992 Jan;50(1):156–163. [PMC free article] [PubMed] [Google Scholar]
- Ott J. Estimation of the recombination fraction in human pedigrees: efficient computation of the likelihood for human linkage studies. Am J Hum Genet. 1974 Sep;26(5):588–597. [PMC free article] [PubMed] [Google Scholar]
- Poustka A., Dietrich A., Langenstein G., Toniolo D., Warren S. T., Lehrach H. Physical map of human Xq27-qter: localizing the region of the fragile X mutation. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8302–8306. doi: 10.1073/pnas.88.19.8302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards B., Heilig R., Oberlé I., Storjohann L., Horn G. T. Rapid PCR analysis of the St14 (DXS52) VNTR. Nucleic Acids Res. 1991 Apr 25;19(8):1944–1944. doi: 10.1093/nar/19.8.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrander-Stumpel C., Legius E., Fryns J. P., Cassiman J. J. MASA syndrome: new clinical features and linkage analysis using DNA probes. J Med Genet. 1990 Nov;27(11):688–692. doi: 10.1136/jmg.27.11.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serville F., Lyonnet S., Pelet A., Reynaud M., Louail C., Munnich A., Le Merrer M. X-linked hydrocephalus: clinical heterogeneity at a single gene locus. Eur J Pediatr. 1992 Jul;151(7):515–518. doi: 10.1007/BF01957757. [DOI] [PubMed] [Google Scholar]
- Willems P. J., Brouwer O. F., Dijkstra I., Wilmink J. X-linked hydrocephalus. Am J Med Genet. 1987 Aug;27(4):921–928. doi: 10.1002/ajmg.1320270419. [DOI] [PubMed] [Google Scholar]
- Willems P. J., Dijkstra I., Van der Auwera B. J., Vits L., Coucke P., Raeymaekers P., Van Broeckhoven C., Consalez G. G., Freeman S. B., Warren S. T. Assignment of X-linked hydrocephalus to Xq28 by linkage analysis. Genomics. 1990 Oct;8(2):367–370. doi: 10.1016/0888-7543(90)90294-5. [DOI] [PubMed] [Google Scholar]
- Willems P. J., Vits L., Raeymaekers P., Beuten J., Coucke P., Holden J. J., Van Broeckhoven C., Warren S. T., Sagi M., Robinson D. Further localization of X-linked hydrocephalus in the chromosomal region Xq28. Am J Hum Genet. 1992 Aug;51(2):307–315. [PMC free article] [PubMed] [Google Scholar]
- Winter R. M., Davies K. E., Bell M. V., Huson S. M., Patterson M. N. MASA syndrome: further clinical delineation and chromosomal localisation. Hum Genet. 1989 Jul;82(4):367–370. doi: 10.1007/BF00273999. [DOI] [PubMed] [Google Scholar]


