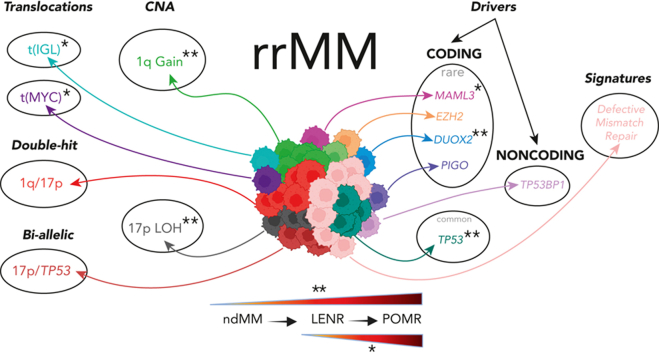
Key Points
-
•
Using the largest WGS data set, we show biallelic and double-hit events plus novel rare drivers are enriched in relapsed/refractory myeloma.
-
•
EZH2, PIGO, and DUOX2 are novel, nearly mutually exclusive candidate driver genes in relapse/refractory myeloma.
Visual Abstract
Abstract
Large-scale analyses of genomic data from patients with newly diagnosed multiple myeloma (ndMM) have been undertaken, however, large-scale analysis of relapsed/refractory MM (rrMM) has not been performed. We hypothesize that somatic variants chronicle the therapeutic exposures and clonal structure of myeloma from ndMM to rrMM stages. We generated whole-genome sequencing (WGS) data from 418 tumors (386 patients) derived from 6 rrMM clinical trials and compared them with WGS from 198 unrelated patients with ndMM in a population-based case-control fashion. We identified significantly enriched events at the rrMM stage, including drivers (DUOX2, EZH2, TP53), biallelic inactivation (TP53), noncoding mutations in bona fide drivers (TP53BP1, BLM), copy number aberrations (CNAs; 1qGain, 17pLOH), and double-hit events (Amp1q-ISS3, 1qGain-17p loss-of-heterozygosity). Mutational signature analysis identified a subclonal defective mismatch repair signature enriched in rrMM and highly active in high mutation burden tumors, a likely feature of therapy-associated expanding subclones. Further analysis focused on the association of genomic aberrations enriched at different stages of resistance to immunomodulatory agent (IMiD)–based therapy. This analysis revealed that TP53, DUOX2, 1qGain, and 17p loss-of-heterozygosity increased in prevalence from ndMM to lenalidomide resistant (LENR) to pomalidomide resistant (POMR) stages, whereas enrichment of MAML3 along with immunoglobulin lambda (IGL) and MYC translocations distinguished POM from the LEN subgroup. Genomic drivers associated with rrMM are those that confer clonal selective advantage under therapeutic pressure. Their role in therapy evasion should be further evaluated in longitudinal patient samples, to confirm these associations with the evolution of clinical resistance and to identify molecular subsets of rrMM for the development of targeted therapies.
Introduction
Despite tremendous advance in therapeutics, multiple myeloma (MM) remains largely incurable, characterized by periods of remission and relapse, with most patients eventually dying from the disease. Currently, combinations of immunomodulatory agents (IMiDs; eg, lenalidomide and pomalidomide), proteasome inhibitors, steroids, anti-CD38 antibodies, and more recently B-cell maturation antigen (BCMA)-targeted therapies are standards of care in newly diagnosed MM (ndMM) and/or relapsed/refractory MM (rrMM) settings. The complex array of combinatorial therapeutic options creates selection pressure that dictates the evolution of genomic resistance in patients through the acquisition of novel and/or expansion of existing genetic drivers. Owing to its higher resolution in accurately detecting genomic events,1 whole-genome sequencing (WGS) analysis of a large cohort of rrMM genomes may uncover evidence regarding the relationship between therapeutic exposure and the selective pressure driving the evolution of therapy-exposed and resistant subclones.
Previously, our ndMM analysis identified an array of genomic aberrations as well as oncogenic dependencies indicating that the genomic landscape in ndMM is characterized by primary events followed by acquisition of additional genetic events.2 The WGS landscape of rrMM has only previously been described in small data sets with limited investigation into the effects of specific therapeutic pressures on tumor genetics.3, 4, 5 We recently demonstrated a highly significant enrichment of genomic CRBN aberrations, and loss of the 2q37 region, which houses COP9 signalosome members following acquisition of lenalidomide or pomalidomide resistance (designated as LENR and POMR, respectively).6,7 Gene expression signatures have also been shown to transition to those that indicate higher risk as disease progresses.8
Here, we present a genome-wide unsupervised analysis of somatic mutations, copy number aberrations (CNAs), and structural variants (SVs) along with mutational signatures in a set of 418 rrMM WGS samples from 386 patients. Drug-specific resistance-associated tumor genomic variants have previously been described (eg, cereblon aberration associated with IMiD resistance,7 proteasome subunit PSMB5 aberrations associated with protease inhibitor resistance,9 and BCMA locus loss after BCMA-directed therapy10), as well as immune cell abnormalities associated with resistance to immunotherapies. However, deciphering how drug-specific genomic evolution enhance the acquisition or enrichment of “high-risk” genetic features during sequential therapeutic exposures has not been explored systematically. By utilizing the latest stage-available samples from rrMM patients, we compared them with ndMM WGS samples from 198 unrelated patients11 to enable an in-depth population-level case-control analysis of how the MM genomes evolve in response to therapeutic exposures of lenalidomide- and pomalidomide-based regimens (Figure 1). We identified mutually exclusive rare driver genes (EZH2, DUOX2, and PIGO), enrichment of existing high-risk features such as 1q Gain and TP53/17p events (suggesting a transition to high-risk biology in rrMM) and enrichment of a novel subclonal DNA mismatch repair signature in high mutation burden tumors.
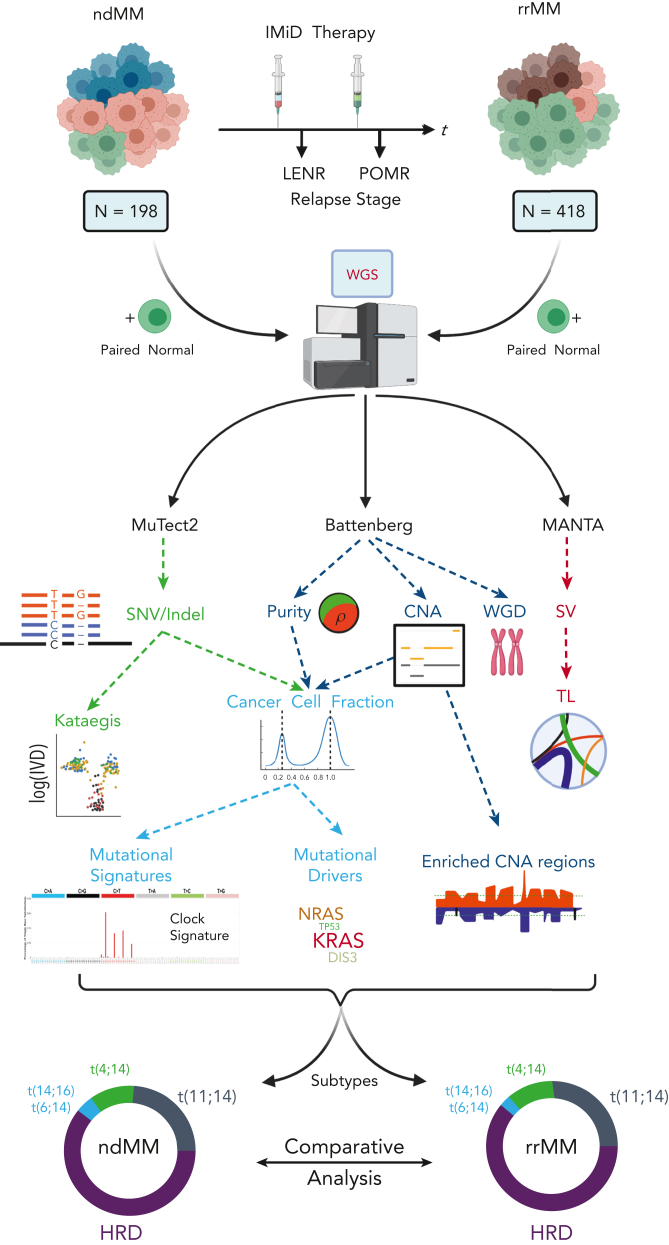
Figure 1.
Overview of collection, processing, and analysis of the WGS data along with the comparative analysis strategy in detecting genomic events associated with the relapse/refractory stage of MM. Myeloma tumors not belonging to HRD and IGH translocation subtypes are not represented in the circular proportion plots. HRD, hyperdiploid; TL, translocation; WGD, whole-genome duplication.
Methods
Patient characteristics
This study is based on an analysis of a set of unrelated ndMM and rrMM cases with WGS data (supplemental Methods and Table 1 for details; available on the Blood website). For our analysis, patients with rrMM were classified based on refractory status LENR or POMR. Because most patients were from pomalidomide trials, which required either exposure or refractoriness to lenalidomide, the LENR cohort (n = 269/386 patients, 70%) is larger than the POMR cohort (n = 55/386 patients, 14%) (Table 1, supplemental Table 1). The ndMM data set comprised WGS data from patients in IFM2009 (n = 198).11
Table 1.
Demographics of the patients with ndMM or rrMM
| ndMM | rrMM |
|||
|---|---|---|---|---|
| Total | LENR | POMR | ||
| Number of patients (number of samples) | 198 (198) | 386 (418) | 269 (273) | 55 (83) |
| Mean study entry age, y (95% CI) | 57.2 (56.3-58.2) | 66.8 (66-67.5) | 66.9 (66-67.8) | 63.5 (61.4-65.6) |
| Mean time since diagnosis, y (95% CI) | - | 5.9 (5.6-6.2) | 5.8 (5.5-6.1) | 6.8 (6.1-7.5) |
| Sex | ||||
| Male/total patients with data (%) | 93/149 (62.4) | 213/386 (55.2) | 147/269 (54.6) | 31/55 (56.4) |
| Female/total patients with data (%) | 56/149 (37.6) | 173/386 (44.8) | 122/269 (45.4) | 24/55 (43.6) |
| ISS | ||||
| I/total patients with data (%) | 55/156 (35.3) | 97/315 (30.8) | 77/226 (34.1) | 9/46 (19.6) |
| II/total patients with data (%) | 66/156 (42.3) | 113/315 (35.9) | 82/226 (36.3) | 24/46 (52.2) |
| III/total patients with data (%) | 35/156 (22.4) | 105/315 (33.3) | 67/226 (29.6) | 13/46 (28.3) |
| Number of patients that received stem cell transplants (%) | 87/187 (46.5) | 232/352 (65.9) | 165/253 (65.2) | 43/55 (78.2) |
| BORT | ||||
| Exposed/total patients with data (%) | - | 320/320 (100) | 239/239 (100) | 54/54 (100) |
| Refractory/total patients with data (%) | - | 231/310 (74.5) | 195/236 (82.6) | 33/47 (70.2) |
| DAR | ||||
| Exposed/total patients with data (%) | - | 38/38 (12.2) | 14/14 (100) | 23/23 (100) |
| Refractory/total patients with data (%) | - | 21/38 (55.3) | 4/14 (28.6) | 17/23 (73.9) |
| Translocation | ||||
| t(4;14) (%) | 24 (12.1) | 45 (11.7) | 37 (13.8) | 5 (9.1) |
| t(8;14) (%) | 12 (6.1) | 25 (6.5) | 18 (6.7) | 3 (5.5) |
| t(11;14) (%) | 41 (20.7) | 80 (20.7) | 52 (19.3) | 14 (25.5) |
| t(14;16) (%) | 5 (2.5) | 6 (1.6) | 5 (1.9) | 1 (1.8) |
| t(14;20) (%) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| t(6;14) (%) | 5 (2.5) | 6 (1.6) | 2 (0.7) | 2 (3.6) |
| t(8;22) (%) | 11 (5.5) | 12 (3.1) | 9 (3.3) | 2 (3.6) |
Patients are classified by disease setting (ndMM vs rrMM) and among patients with rrMM, cohorts are designated by refractory status to lenalidomide or pomalidomide. Translocation prevalence is derived from genomics data. BORT, bortezomib; DAR, daratumumab; ISS, International Staging System.
WGS and genomic analyses
CD138+ plasma cells and matched germline controls (peripheral blood mononuclear cells) were stored in RLT buffer (Qiagen) and DNA was extracted using the Qiagen AllPrep DNA Mini Kit. WGS was performed on tumor/normal pairs at an average of 60×/30× depth. All WGS raw data processing and analyses are described in detail in the supplemental Methods. Briefly, FASTQ files were aligned to the human genome assembly hg38 using BWA-mem. Duplicate reads were removed using Picard MarkDuplicates and base recalibration of alignments was performed using Base Quality Score Recalibration according to Genome Analysis Toolkit Best Practices. Somatic variants were called using MuTect2 and annotated by ANNOVAR.12 Genome-wide copy number aberration analysis was undertaken in the entire data set by using Battenberg.13 In addition to calling clonal and subclonal allele-specific CNAs, it was also used to estimate purity and average ploidy of each tumor. To estimate clonality, we calculated the cancer cell fraction (CCF) of each variant by adjusting the variant allele frequency (VAF) for copy number status at the given locus, multiplicity of the variant, and tumor purity.14 To assign somatic mutations as clonal or subclonal, the observed VAF of each mutation was modeled using a binomial distribution to obtain its 95% confidence interval (CI) and CCF computed for the 95% boundaries. Variants with an upper CCF boundary above and below 1 were considered to be clonal and subclonal, respectively. To identify candidate drivers of the relapse/refractory stage of MM, we used cDriver.15 Assignment of identified drivers into tumor suppressor gene (TSG) and oncogene (ONC) categories was undertaken using the 20/20 principle.16 We analyzed all drivers for gene essentiality as reported in DepMap.17 De novo extraction of single-base substitution (SBS) signatures was independently implemented using SigProfilerExtractor18 and signeR,19 and assigned to COSMIC signatures18 by computing cosine similarities. To fit the mutational spectrum of each tumor with the identified COSMIC signatures, we used mmsig.20 This was implemented to not only minimize mutational signature bleeding across samples21 but to also accurately estimate the relative contribution of each signature. SVs from whole-genome data were detected using MANTA.22 All statistical analyses were implemented in R.23
Results
The demographics and patient characteristic of total, LENR, and POMR cohorts are described in Table 1. Most LENR/POMR patients were also refractory to bortezomib (83% in LENR; 70% in POMR) and nearly all patients had received dexamethasone; therefore, whereas they are classified by their refractory status to LEN or POM in this analysis, these patients were also exposed or refractory to proteasome inhibitors, and/or a corticosteroid. In addition, 66% of the rrMM population received a stem cell transplant, which exposed them to high-dose melphalan. Only a small number of patients exposed (n = 38) or refractory (n = 21) to daratumumab were present. This data set was generated from trials of IMiDs, thus data from patients that were non-IMiD treated are not represented.
We classified the rrMM and ndMM genomes into MM subgroups based on canonical immunoglobulin heavy-chain translocations and hyperdiploidy (HRD, ≥3 copy number gain of odd-number chromosomes). To address the aim of this study and given that no significant difference in the distribution of MM subtypes was observed between ndMM and rrMM, we focused on subgrouping patients based on IMiD resistance (LENR and POMR, Figure 1). Because of sample size limitations, for a number of differential analyses we first compared the rrMM and ndMM patient data sets, and then sought differential enrichment in LENR and POMR subgroups.
Identification of enriched and novel mutational drivers in rrMM tumors
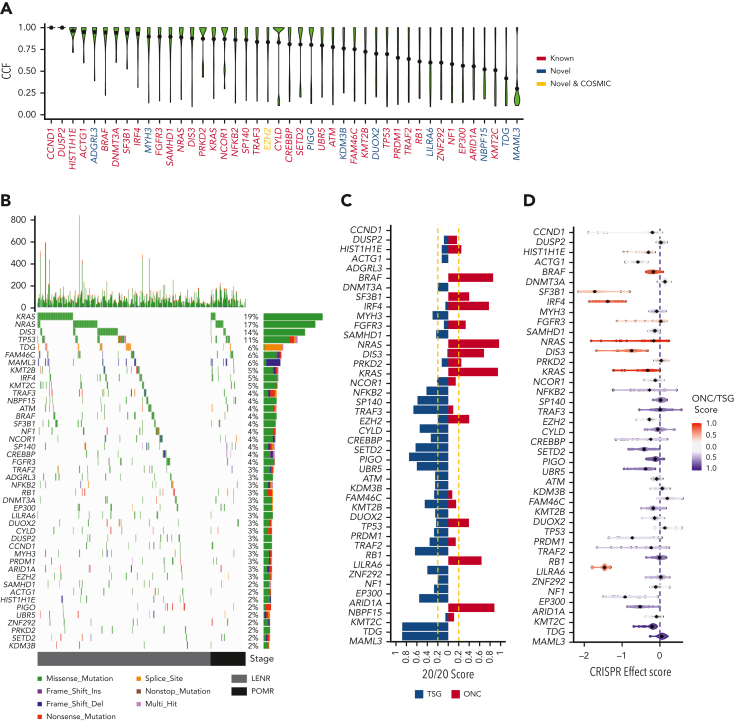
We initially sought ndMM-specific drivers (Walker et al2; N = 1273) in the rrMM data set and detected 34 (out of 63) of these drivers (recurrence ≥2%). Mutational driver analysis on the rrMM data set also detected an additional 10 previously unreported promising candidate driver genes (recurrence ≥2%; false discovery rate [FDR] < 0.05), including a COSMIC cancer gene (https://cancer.sanger.ac.uk/census), EZH2. Analysis of clonality of drivers based on their CCF14,24 distribution showed that novel driver genes were generally subclonal (Figure 2A). Analysis of the distribution of nonsilent mutations showed distinct variant types in TDG and MAML3 where the predicted functional consequence of most mutations was splicing defect and frameshift, respectively (Figure 2B, supplemental Table 2). All 44 drivers were examined to see whether they could be classified as TSG or ONC based on their mutation spectrum (Figure 2C). Among the novel genes, MAML3, TDG, and PIGO showed the strongest TSG signals, whereas NBPF15 showed a strong ONC signal similar to BRAF and NRAS/KRAS. We also analyzed all drivers for gene essentiality using DepMap.17 Novel drivers categorized as TSG such as DUOX2, PIGO, MAML3, and TDG showed overlapping distributions to well-known TSGs such as RB1, SETD2, and TRAF3. In addition, we saw a strong CRISPR effect score signal (median = −1.46) for LILRA6, a driver categorized as an ONC, similar to well-known ONCs such as IRF4 and SF3B1 (Figure 2D). Overall, when comparing ONC and TSG genes, we saw a highly significant elevated CRISPR effect score in ONC genes (mean of −0.63 vs mean of −0.18; P = 3.3 × 10−17).
Figure 2.
The mutational driver landscape of rrMM. (A) CCF distribution of driver genes identified in this cohort ordered by mean CCF (black dot). Gene labels are colored according to their status: known, previously reported in ndMM; novel COSMIC, newly identified but present in the COSMIC cancer gene set; and novel, not previously reported in either ndMM or COSMIC. (B) Mutational landscape of drivers identified in the rrMM cohort. The top panel represents the frequency of nonsilent mutations in each tumor across the genome. The main panel is a waterfall plot of mutations identified in each driver gene across all tumors. The right panel shows the frequency of each driver in the rrMM cohort in percentages and the type of mutations that constitute the mutation burden of each driver gene (length of the bar reflecting the frequency). All 3 panels are colored according to the functional consequence of mutations as predicted by ANNOVAR. Multi-hit: tumors with more than one mutation with different functional consequence. Tumors are ordered by their relapse stage. LENR, lenalidomide resistant; POMR, pomalidomide resistant. (C) Categorization of identified driver genes into ONC or TSG based on the 20/20 rule.16 The ONC and TSG scores are shown with red and blue color, respectively, and the vertical yellow dashed lines represent the cut-off thresholds for the 20/20 rule. Genes are ordered by mean CCF as in panel A. (D) Gene essentiality of driver genes based on the CRISPR effect score in MM cell lines where more negative values represent essential genes. Each dot represents a cell line, and the diamond represents the median value per gene. Each gene is colored based on their mean ONC/TSG score. Scores between −0.2 and 0.2 are considered inconclusive (gray). Genes are ordered by mean CCF as in panel A.
Interestingly, a total of 46 LENR/POMR tumors (14.2%; Figure 2B) were negative for all mutational drivers. No significant difference was observed between LENR and POMR for the prevalence of these tumors (P = .2). These tumors were further assessed for translocations and CNA. In total, 40 tumors (87%) had at least 1 such major event in their genome (supplemental Figure 1), however, the rest were devoid of any known genomic drivers.
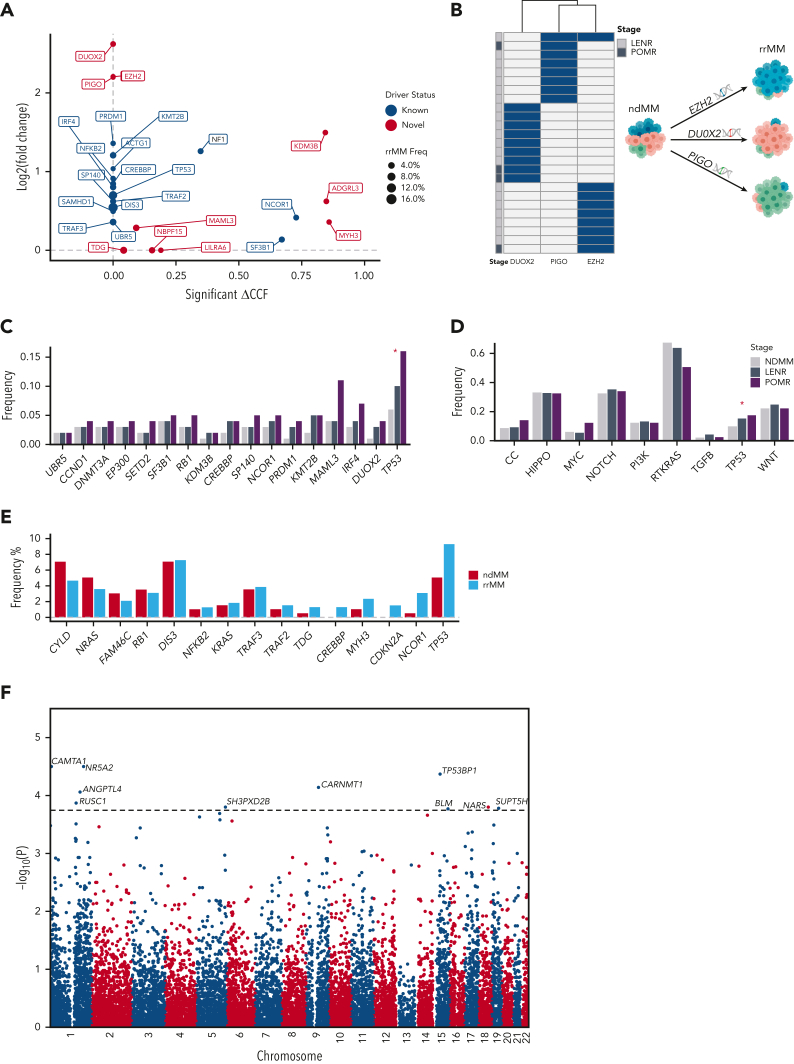
To identify which drivers are unique or enriched in rrMM and thus relevant to therapy-driven evolution, we compared their recurrence and clonality between the rrMM and ndMM data sets. Interestingly, although novel candidate drivers (except PIGO) were not rrMM-specific, when compared with ndMM, the most extreme changes in CCF and recurrence were observed in these drivers (Figure 3A). Specifically, DUOX2, EZH2, and PIGO had the highest enrichment (fold-change > 4.5). Interestingly, these 3 drivers were virtually mutually exclusive, suggesting alternate trajectories of therapeutic resistance (Figure 3B). To ensure robustness of these findings, we included the MMRF CoMMpass25 whole-exome sequencing data set (N = 1001), making a combined ndMM data set (N = 1001 + 198 = 1199). We showed that the frequency of almost all drivers is overestimated in the WGS ndMM data set (supplemental Figure 2A) and, in turn, most enrichment fold-changes are higher when the combined ndMM is used. Interestingly, in addition to the top-enriched novel drivers remaining highly enriched (log2FC ≥ 2; PIGO, DUOX2, and EZH2), MAML3 also became more enriched in rrMM (log2FC = 2.9, supplemental Figure 2B).
Figure 3.
Differential enrichment of mutational drivers in rrMM and its IMiD therapy–defined subgroups. (A) Enrichment analysis of all mutational drivers based on coding mutations. Each driver was compared between rrMM and ndMM cohorts for a significant enrichment in CCF (x-axis) and increase in the prevalence quantified by log2 value of the fold-change (y-axis) in rrMM. Drivers with no statistically significant increase in CCF were assigned a ΔCCF value of 0 (situated on the y-axis). Drivers with no enrichment for either CCF or prevalence are not shown. Genes previously detected in ndMM as drivers2 are shown in blue and genes detected only in the rrMM cohort are shown in red. The size of each point reflects the prevalence of each driver in the rrMM cohort. (B) Mutual exclusivity of the 3 novel drivers, which are virtually unique to patients with rrMM (with no difference in prevalence between LENR and POMR), suggesting alternative evolutionary trajectories to therapeutic resistance. (C) Therapy stage subgroup analysis showed significant enrichment of TP53 from newly diagnosed to lenalidomide resistant to pomalidomide resistant stages based on the proportion trend test (P < .05; red asterisk). Driver genes are ordered based on significance of the proportion trend test with most significant trends on the right-hand side. (D) Per-pathway analysis shows depletion and enrichment of mutations in canonical cancer pathways from newly diagnosed to lenalidomide resistant to pomalidomide resistant stages. The RTKRAS and TP53 pathway showed significant negative and positive cline based on the proportion trend test. (D) Frequency barplot of biallelic drivers in rrMM and ndMM. Biallelic is defined as LOH (ie, copy loss) along with a nonsilent mutation or homozygous deletion. Driver genes are ordered left to right based on the frequency difference between rrMM and ndMM in ascending order. (E) Manhattan plot for a genome-wide scan of mutational drivers based on noncoding somatic variant clustering. The genome was divided into nonoverlapping bins of 100 kb and the rate of somatic mutation in each was compared between the rrMM and ndMM cohorts based on a Fisher exact test, and the –log10 of the corresponding P value was plotted. The dotted horizontal line represents the genome-wide significance threshold (FDR < 0.05). Ten bins showed significant enrichment of noncoding somatic variants in rrMM. Each significant signal is annotated with the name of the most likely gene targeted in each bin. Asterisk denotes statistically significant enrichment (P < .05). LOH, loss of heterozygosity.
We then sought to identify driver genes enriched across IMiD therapy stages (ie, ndMM to LENR to POMR). A positive trend in prevalence was observed for 16 driver genes (Figure 3C), among which TSGs were enriched (5.5-fold vs oncogenes). TP53 showed a statistically significant increase (proportion trend test, P = .016). Although the significance of DUOX2 and IRF4 was borderline owing to lower recurrence (P < .1), both were enriched at the POMR stage when compared with ndMM (7.3- and 2.9-fold increase, respectively). Interestingly, although there was no difference between ndMM and LENR in MAML3 frequency, POMR showed a 2.75-fold higher recurrence of this driver gene (Figure 3C).
To gain more insight into the biology of rrMM and to increase statistical power, we also analyzed the prevalence of nonsilent mutations in 10 canonical cancer pathways26 across IMiD therapy stages. Of 3 pathways related to cell cycle, MYC and TP53 showed an overall positive trend (Figure 3D), however, only TP53 showed a statistically significant increase (proportion trend test, P = .049). Overall, rrMM genomes showed acquisition of novel TSG drivers such as EZH2, PIGO, and DUOX2 or enrichment of high-risk recurrent drivers such as TP5327 and DIS3.28 Furthermore, we undertook network and pathway enrichment analysis based on the 44 drivers to investigate possible mechanisms of disease progression potentially driven by the novel candidate drivers. First, we generated a functional interaction network among all driver genes using STRING version 11.5.29 Of the 10 promising candidate drivers, TDG, MAML3, and EZH2 had 2, 3, and 5 interactions, respectively, with known MM drivers (supplemental Figure 3A), suggesting potential functional association. All 3 also clustered within the same subnetwork with known driver genes such as RB1, CCND1, and CREBBP (supplemental Figure 3B). Next, we used Enrichr30 to identify enriched pathways unique to promising candidate drivers. Among the enriched pathways (supplemental Table 3), base excision repair (TDG), notch signaling pathway (MAML3), and B-cell receptor signaling pathway (LILRA6) were notable.
TP53 and other biallelic events increase in rrMM tumors
At the individual tumor level, we sought to assess the extent of biallelic events given their association with relapse in MM.31 We observed biallelic inactivation of 15 driver genes (Figure 3E), of which one (CDKN2A) was solely inactivated by homozygous deletion (HD) events (supplemental Figure 4). We showed that biallelic CDKN2A and CREBBP are unique to rrMM. This is consistent with that reported previously for ndMM where both were observed at only 0.1% and 0.2%, respectively (N = 1074).2 TP53 was not only the most frequent biallelic event in rrMM (9.3%) but it was also enriched compared with ndMM (1.8 fold). Among the novel drivers, notably, TDG showed enriched biallelic inactivation (2.6 fold) at 1.3% prevalence in rrMM. Given its high TSG signal (Figure 2B) and recent evidence supporting its tumor suppressive role,32 this observation further corroborates that TDG acts as a TSG in MM.
Noncoding mutational hotspot analysis identifies 10 additional bona fide drivers in rrMM
Leveraging the WGS breadth of our rrMM and ndMM data sets, we sought to identify novel rrMM drivers by detecting differentially enriched somatic noncoding mutational hotspots flanking protein-coding genes (refer to Ansari-Pour et al33). For this analysis, we used a construct similar to that of a genome-wide association study and looked for regions with a higher somatic mutation recurrence in the rrMM data set. A total of 10 genome-wide signals (FDR < 0.05) were identified (Figure 3F). Of note, TP53BP1, which is a haploinsufficient TSG,34 has been previously shown to be involved in double-strand DNA break repair in MM35 and its loss has also been reported to induce chemoresistance in colorectal cancer.36 Given that these mutations are noncoding, we hypothesize that, in the context of DNA damaging drugs, acquiring such variants may lead to the dysregulation of genes such as TP53BP1, which may, in turn, confer resistance to tumors that harbor them.
Translocation landscape of rrMM tumors
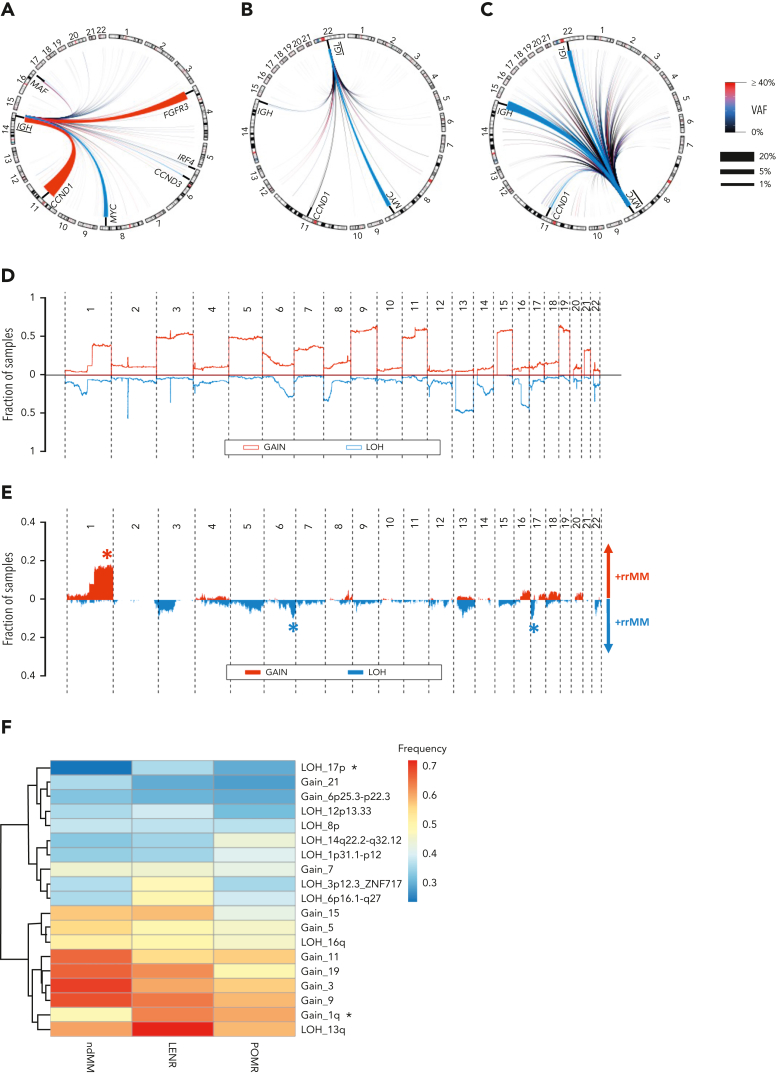
SV analysis was undertaken to specifically detect translocations with a focus on those that involved the IGH, IGL, and MYC loci in the rrMM data set. IGH events (canonical or otherwise) were found in 42% of tumors (Table 1), similar to the ndMM data set (44%) and to that previously reported (41%),37 with the most common IGH events being t(11;14), t(4;14), and t(8;14), whereas t(6;14) and t(14;16) were present in a few cases (Figure 4A). As expected, the first 2 were found to be early events in MM (ie, clonal), however t(8;14) was found to be at subclonal levels across the data set (Figure 4A). IGL- and MYC-centric translocations were present in 11% and 29.5% of tumors, respectively, and were mainly subclonal (Figure 4B-C), consistent with these events being secondary genomic aberrations.38,39 The prevalence of these secondary translocations was then examined across clinical settings. MYC translocations showed an increase in prevalence from LENR to POMR (27% to 44%, odds ration [OR], 2.1; P = .023) but no difference was observed in VAF distribution after adjusting for purity (P = .157). A significant increase in prevalence was also observed for IGL events from LENR to POMR (10% to 22%, OR, 2.5; P = .021) but with no change in clonality (P = .68). This increase in IGL prevalence is consistent with a recent study showing that no significant survival benefit was provided by IMiD-containing regimens for patients with IGL translocations.37
Figure 4.
The translocation (TL) and CNA landscape of rrMM. (A) Circos plot of all IGH-translocated events. Common canonical pairs are annotated. Chromosomes involved in canonical translocations (Chr 4,6,8,11,14,16) are zoomed-in proportionally for better visualization of such events. (B) Circos plot of all IGL-translocated events. Common canonical events are annotated. Chromosomes involved in canonical translocations (Chr 8,11,14, 22) are zoomed-in proportionally for better visualization of such events. (C) Circos plot of all MYC-translocated events. Common canonical events are annotated. Chromosomes involved in canonical translocations (Chr 8,11,14, 22) are zoomed-in proportionally for better visualization of such events. For panels A to C, the color and thickness of the lines connecting the 2 TL breakpoint pairs denote the mean VAF and the prevalence of the TL in this cohort, respectively. Accordingly, thick red lines represent early (ie, high VAF) and frequent TL events in rrMM. (D) Genome-wide landscape of CNA in the form of gain and LOH in the ndMM cohort. The y-axis represents the fraction of samples harboring a particular event at any given chromosomal location. LOH is shown in the opposite direction to the gain events for better visualization. (E) Genome-wide differential landscape of CNA in the form of gain and LOH in the rrMM compared with the ndMM. The y-axis represents the enriched fraction of samples in rrMM harboring a CNA. LOH is shown in the opposite direction to the gain events and depleted events in rrMM are not shown for better visualization. Asterisks denote statistically significant enrichment of common CNA events. (F) Heatmap and dendrogram of enriched CNA at ndMM, LENR, and POMR stages. The rows are the enriched CNA events and columns are the therapeutic stages. Asterisks denote significant positive cline from ndMM to LENR to POMR based on the proportion trend test (FDR < 0.05).
Multiple CNA events are enriched at later therapy stages
We compared the rrMM landscape of CNA events against those in ndMM (Figure 4D) and identified the differentially enriched CNAs. We observed significant overrepresentation of 1q Gain, 6q LOH, and 17p LOH (FDR < 0.01; Figure 4E). To trace CNA enrichment across therapy stages, significantly enriched copy number regions (supplemental Methods) were identified in the rrMM data set across the genome and their recurrence frequency was compared (Figure 4F). Notably, we observed a significant positive cline of 1q Gain and 17p LOH recurrence from ndMM to LENR to POMR (proportion trend test, P < .05). We also observed a positive cline of recurrence of LOH events at 1p and 14q but they did not reach statistical significance. The higher prevalence of these events with later therapy stage is consistent with these events being associated with disease progression and poor prognosis.2,40,41 Although the rrMM patient population is extremely complex with concurrent layers of genomic, immune, clinical, and treatment heterogeneity, we nonetheless examined the prognostic effect of 1q Gain and 17p LOH events per trial. We only observed a significant prognostic effect in the MM-010 trial for 1q Gain based on progression-free survival (P = .031; supplemental Figure 5).
Somatic interaction analysis identifies significant enrichment of Amp1q-ISS3 and 1qGain-17pLOH in rrMM
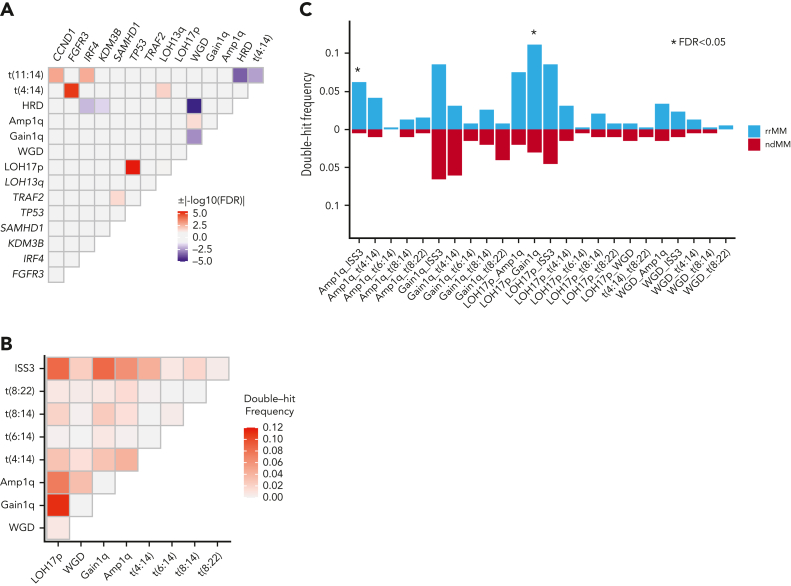
The associations among all mutational drivers, enriched CNAs, WGD, and recurrent translocations (TLs) were analyzed together to identify novel patterns of somatic interaction in the form of significant cooccurrence and mutual exclusivity in the rrMM data set. Genomic events with at least 1 significant pairwise association (FDR < 0.1) were identified (Figure 5A). The most significant interactions were the cooccurrence of TP53-LOH17p and mutual exclusivity of HRD-WGD, of which the latter has not been previously reported. When compared with ndMM (supplemental Figure 6), we observed a significant association between IRF4 and t(11:14) only in the rrMM data set (OR,10.4; P = 8 × 10−6).
Figure 5.
Somatic interactions of genomic aberrations in the rrMM data set. (A) Only genomic events with at least 1 significant pairwise association (FDR < 0.1) are shown. (B) Frequency of double-hit events of high-risk features observed in the rrMM data set. (C) Enrichment analysis of double-hit events in rrMM. Each event was compared between rrMM and ndMM cohorts using the Fisher exact test and those significant after multiple-testing correction (FDR < 0.05) are shown with an asterisk.
We also sought to identify interactions in the form of double-hit events (ie, 2 independent events occurring at 2 different loci in the same genome at the patient level. Double-hit events were identified in 171 (44.3%) patients with rrMM (supplemental Figure 7). The rate of double-hit events was markedly lower in the ndMM cohort (30.8%), thus showing a significant enrichment in rrMM (OR, 1.8; P = 1.7 × 10−3). We calculated the frequency of each double-hit event in rrMM (Figure 5B) and ndMM (supplemental Figure 8). We observed 2 double-hit events enriched significantly in rrMM (FDR < 0.05; Figure 5C), namely Amp1q-ISS3 (12.4 fold) and 1qGain-17pLOH (3.7 fold), of which the latter was the most frequent double-hit event in rrMM (11%; Figure 5B). Interestingly, Amp1q-ISS3, which shows the highest enrichment, has been previously shown to be a high-risk subgroup in ndMM.42 In addition, 1qGain-17pLOH is the cooccurrence of the 2 most frequent enriched CNA in rrMM (Figure 4E). Across clinical settings, the overall rate of double-hit events was not significantly different between POMR and LENR (P = .17), and the same was true for each individual event (supplemental Figure 9).
rrMM tumors show subclonal enrichment of defective mismatch repair signature
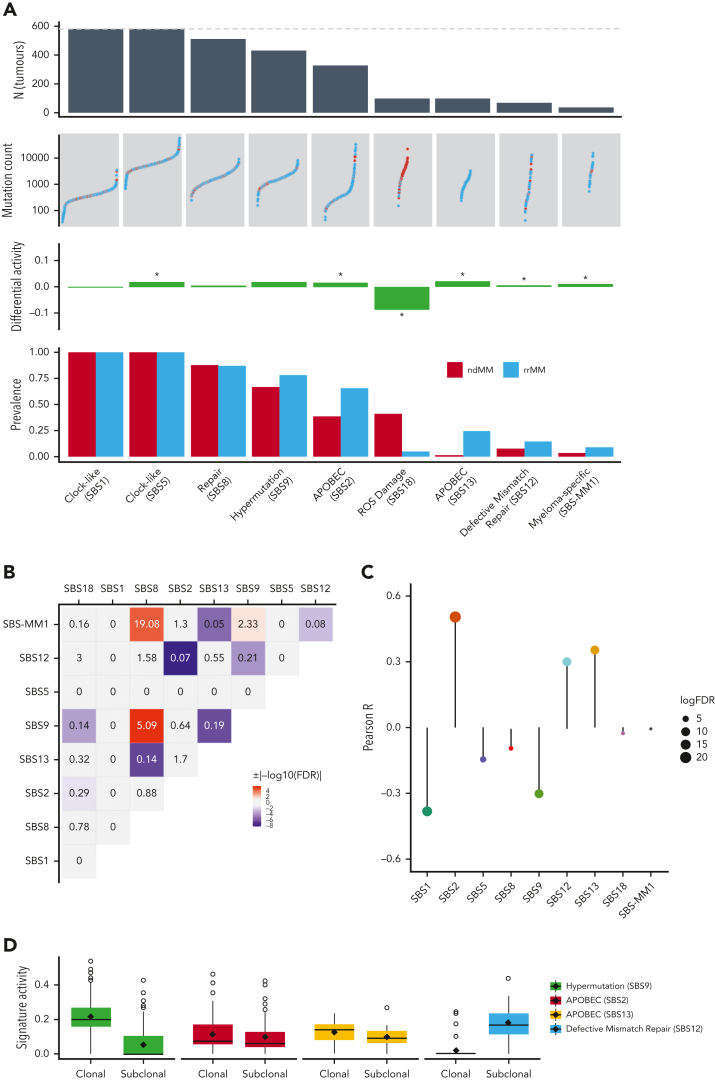
Finally, we analyzed the mutational signatures to detect an etiological basis for exogenous and endogenous factors associated with relapse/refractory status. De novo extraction of mutational signatures detected 8 SBS COSMIC signatures18 (supplemental Figure 10A). All signatures, except SBS12, had been previously observed in ndMM11,43 and rrMM43 tumors. When fitting the identified COSMIC signatures to each tumor, we also included SBS-MM15,24,43 to see whether its inclusion better reconstructs the observed mutational spectrum. We observed an increase in both prevalence and activity of SBS2, SBS13, SBS12, and SBS-MM1 (Figure 6A), all of which, except SBS-MM1, are either associated with APOBEC (SBS2/SBS13) or defective mismatch repair (DMR; SBS12). SBS-MM1 is a relatively flat signature that has been associated with melphalan treatment and generally with rrMM.43 Our analysis suggests that SBS-MM1 is likely to be associated with rrMM owing to higher prevalence and activity. However, we should note that the highly significant co-occurrence of SBS-MM1 with SBS8 (Figure 6B) is elusive and suggests that fine-tuned functional experiments are required to establish the underlying etiology of SBS-MM1 and how it may relate or differ from SBS8. Analysis of signature activity against mutation burden showed strong positive correlations with APOBEC and DMR signatures (Figure 6C). The activity of mutational signatures was also analyzed at clonal and subclonal stages of evolution. Of note, SBS12 (DMR) showed the strongest increase in activity among all signatures at the subclonal level (Figure 6D, supplemental Figure 10B). We therefore examined whether mutations in DMR-related genes led to the increase in DMR signature activity. We curated a list of 6 well-known mismatch repair genes (MSH2, MLH1, PMS2, MSH3, MSH6, and MLH3) and identified tumors with nonsilent mutations in them. We observed a slight enrichment (OR, 1.7) of DMR-related genes in tumors with increased DMR activity but owing to the small sample size of mutant tumors (N = 13), this was not statistically significant (P = .42). We further explored the DMR signature to see whether more genomic aberrations are present in tumors with increased DMR activity. We computed 2 genome-wide metrics, namely (a) proportion of genome altered (PGA) and (b) mutation burden. We observed no significant increase in PGA (mean of 35.1% in increased DMR vs mean of 32.7% in other tumors, P = .48). However, interestingly, a highly significant increase in mutation burden was observed in increased DMR tumors (mean of 35 286 vs mean of 17 998, 2-fold, P = 7.8 × 10−8), which was driven by a 2.2-fold increase in single nucleotide variants (mean of 31 884 vs mean of 14 409, P = 4.8 × 10-9). This suggests that DMR may, in part, be an endogenous factor associated with expanding subclones under therapeutic selection pressure.
Figure 6.
Differential analysis of genome-wide mutational signatures. (A) From top to bottom: number of tumors with SBS signatures across the entire data set (dotted line represents combined ndMM and rrMM sample size) with signatures sorted left to right by descending count, number of mutations per sample (color representing groups) in respective signatures, differential activity of signatures between rrMM and ndMM with values in the positive direction showing enrichment in the rrMM cohort and vice versa, proportion of tumors carrying each signature in each cohort. (B) Heatmap of pairwise associations among SBS signatures as assessed by Fisher exact test. ORs are given for each pair and shades of red and blue colors represents statistically significant cooccurrence (OR > 1) and mutual exclusivity (0 < OR < 1) patterns, respectively, after multiple-testing correction (FDR < 0.05). Gray color indicates nonsignificant association regardless of OR value. (C) Lollipop plot of correlation coefficient values between the activity of each SBS signature and mutation burden in rrMM tumors. The length of the bars represents the Pearson R value and the size of the circles represents the log10 of the FDR-adjusted P value of the correlation (larger circles indicate higher significance). Colors of the signatures match those in supplemental Figure 10. (D) Shift in mutational signature activity between clonal and subclonal mutations for signatures displaying strong positive correlation with mutation burden in the rrMM cohort and SBS9 (hypermutation signature) for comparison. SBS12 shows a strong signal of subclonal increase, whereas SBS9 shows decrease across most tumors.
Discussion
Recent studies characterizing the genomic features of rrMM have either analyzed small numbers of samples3,5,44 or lacked the breadth of WGS.45 In combination with deep WGS ndMM data,11 we performed an unsupervised analysis of the largest deep WGS rrMM data set to date (total N = 616), thus providing, to the best of our knowledge, the most comprehensive analysis of the rrMM genomic landscape to date. We identified novel rrMM driver genes through coding and noncoding variation in the genome. Interestingly, all 10 coding-variant drivers showed either a significant clonal expansion or strong enrichment in recurrence in rrMM compared with ndMM. This observation suggests that these driver genes are associated with therapeutic resistance, either already present before therapeutic exposure but providing clonal advantage upon exposure to therapy or arising de novo and enriching at the rrMM stage at the time of acquisition of resistance. Functional analysis of these promising candidate drivers is nonetheless required to corroborate these findings and provide insight into the exact role of these novel drivers in therapeutic resistance. There is a possibility that we may have not detected these driver genes at the ndMM stage owing to their very low CCF (ie, tiny subclones). However, this is unlikely, given that the average sequencing coverage of the ndMM cohort was higher than that in the rrMM cohort (94× vs 83×; Kolmogorov-Smirnov test, P = 7 × 10−14). In addition to the enrichment of TP53 (and TP53-17pLOH biallelic events, along with its pathway in general) and IRF4, we also observed a positive cline in the prevalence of a novel highly enriched driver (DUOX2) from ndMM to LENR to POMR, which is associated with multiple cancers and involved in the regulation of the AKT pathway.46 Furthermore, by detecting enrichment of somatic variants in noncoding hotspots,33 a total of 10 previously unreported drivers in MM such as TP53BP1 and BLM were identified, which need to be examined at the transcriptomic level to assess whether they dysregulate these bona fide drivers.
When comparing CNA enrichment patterns, 1q Gain and 17p LOH were not only enriched in the rrMM data set, their occurrence as double-hits was also enriched. In addition, they had a significant increasing trend from LENR to POMR. We also observed an increase, albeit statistically nonsignificant, in the prevalence of LOH events at 1p and 14q with rrMM therapy stages. The nonsignificance of the increase may be because of the relatively small sample size of the POMR cohort (N = 55) in this study. Although we found 3 genomic features that distinguished POMR from LENR tumors (MAML3 driver gene and IGL/tMYC translocations), a larger POMR cohort would be beneficial to examining more robustly whether LEN and POM select subclones with distinct genomic drivers or POM generally selects the same subclones already expanding in LENR tumors. For instance, we identified 2 driver genes (NF1 and ADGRL3), which increased in prevalence from ndMM (0.015 and 0.02) to LENR (0.048 and 0.037) but were completely absent from POMR. It is not possible to determine whether their absence is because of favorable response to POM or simply not observed owing to the small size of this subgroup. To evaluate the latter possibility further, we calculated the expected upper limit frequency for an unobserved event47 in POMR (q = 0.053). Because this frequency is above that observed in LENR, the absence of those 2 drivers may be owing to low statistical power.
Mutational signature analysis identified a novel enriched subclonal signature at rrMM (SBS12) with its etiology likely to be DMR. DMR tumors accumulate high levels of mutations (consistent with our findings; Figure 6C), a feature linked to acquisition of drug resistance,48 including in myeloma.49 Alternative therapies such as those based on synthetic lethal approaches50,51 that could exploit DMR in mismatch repair–deficient rrMM tumors may be fruitful.
Our analysis provides insight into the nature of the evolution of myeloma genome under therapeutic pressure. Although in rrMM, therapeutic clonal selection might be expected to reduce the intratumoral heterogeneity, we instead find that ongoing clonal evolution provides genomes with highly diverse enrichment of LOH events, mutational signatures, genetic aberrations, and subclonal complexity. It is therefore likely that ongoing acquisition of novel features is occurring alongside enrichment of those already present. The increase in high-risk changes associated with shortened survival (such as the double-hit events described) alongside direct selection of therapeutic resistance markers are thus unsurprising. However, this reconstruction of the rrMM genome, which blends acquisition of the functional high-risk myeloma state with drug-specific resistance features (such as CRBN loss) pose significant challenges in designing new therapies. Specifically, there must be an even greater imperative to consider clonal evolution in pinpointing which events are connected to disease relapse (ie, present in the most recent common ancestor of the expanding subclone[s]). A limitation of our study was the unrelatedness of the ndMM and rrMM cohorts. We6 and others5,52,53 have shown that sequential sampling is the ideal approach for analyzing clonal dynamics. Detection of subclones that predict relapse might enable investigation of change of therapy and thus their appearance or expansion must be carefully considered as we plan the trials and therapeutic approaches of the future.
Conflicts-of-interest disclosure: Bristol Myers Squibb (BMS) Corporation has employment and equity ownership; funding for data generation, processing, and storage was provided by BMS Corporation; F.T. and A.T. were BMS employees at the time this analysis was conducted; B.W. receives research funding from BMS; and N.A.-P. is a BMS consultant. The remaining authors declare no competing financial interests.
Acknowledgments
The authors thank Marissa Hirst, Hillary Mosso, and Dan Rozelle from Rancho Biosciences for data curation, cleaning, and summary of patient demographics for this analysis. The authors also thank Garth McGrath from Bristol Myers Squibb for significant assistance identifying samples that made this analysis possible and for assistance with data curation. B.W. is partly funded by a grant from the Leukemia & Lymphoma Society.
Authorship
Contribution: E.F. and A.T. conceived and designed the project, and acquired funding; E.F., F.T., and M.O.E., performed project administration; E.F., M.O.E., N.S., S.G., K.M., N.A.-P., F.T. H.A.-L., M.S., N.M., G.M., B.W., and A.T performed oversight and management of resources (data generation, collection, transfer, infrastructure, data processing); N.A.-P. performed computational and statistical analyses; N.A.-P., S.G., M.O.E., N.S., F.T., E.F., B.W., H.A.-L., G.M., N.M., M.S., and A.T. performed analyses and interpretation; N.A.-P., M.S., M.O.E., F.T., N.S., S.G., E.F., and A.T. performed and structured data visualization; A.T. provided supervision and scientific direction; N.A.-P., S.G., M.S., M.O.E., E.F., and A.T. wrote the manuscript; and all authors critically reviewed the manuscript and figures.
Footnotes
Whole-genome sequencing data (FASTQ, BAM, and VCF files) pertaining to the rrMM cohort are deposited on the Amazon Web Services (AWS) S3 cloud storage at s3://celgene-rnd-riku-prismm/.
Data for academic/noncommercial purposes are available on request from the author, Erin Flynt (erin.flynt@bms.com).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Supplementary Material
References
- 1.Jafari M, Guan Y, Wedge DC, Ansari-Pour N. Re-evaluating experimental validation in the Big Data Era: a conceptual argument. Genome Biol. 2021;22(1) doi: 10.1186/s13059-021-02292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walker BA, Mavrommatis K, Wardell CP, et al. Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood. 2018;132(6):587–597. doi: 10.1182/blood-2018-03-840132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoang PH, Cornish AJ, Sherborne AL, et al. An enhanced genetic model of relapsed IGH-translocated multiple myeloma evolutionary dynamics. Blood Cancer J. 2020;10(10):101. doi: 10.1038/s41408-020-00367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoang PH, Dobbins SE, Cornish AJ, et al. Whole-genome sequencing of multiple myeloma reveals oncogenic pathways are targeted somatically through multiple mechanisms. Leukemia. 2018;32(11):2459–2470. doi: 10.1038/s41375-018-0103-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maura F, Bolli N, Angelopoulos N, et al. Genomic landscape and chronological reconstruction of driver events in multiple myeloma. Nat Commun. 2019;10(1):3835. doi: 10.1038/s41467-019-11680-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gooding S, Ansari-Pour N, Kazeroun MH, et al. Loss of COP9-signalosome genes at 2q37 is associated with IMiD agent resistance in multiple myeloma. Blood. 2022;140(16):1816–1821. doi: 10.1182/blood.2022015909. [DOI] [PubMed] [Google Scholar]
- 7.Gooding S, Ansari-Pour N, Towfic F, et al. Multiple cereblon genetic changes are associated with acquired resistance to lenalidomide or pomalidomide in multiple myeloma. Blood. 2021;137(2):232–237. doi: 10.1182/blood.2020007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ortiz-Estevez M, Towfic F, Flynt E, et al. Integrative multi-omics identifies high risk multiple myeloma subgroup associated with significant DNA loss and dysregulated DNA repair and cell cycle pathways. BMC Med Genomics. 2021;14(1):295. doi: 10.1186/s12920-021-01140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrio S, Stuhmer T, Da-Via M, et al. Spectrum and functional validation of PSMB5 mutations in multiple myeloma. Leukemia. 2019;33(2):447–456. doi: 10.1038/s41375-018-0216-8. [DOI] [PubMed] [Google Scholar]
- 10.Samur MK, Fulciniti M, Aktas Samur A, et al. Biallelic loss of BCMA as a resistance mechanism to CAR T cell therapy in a patient with multiple myeloma. Nat Commun. 2021;12(1):868. doi: 10.1038/s41467-021-21177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samur MK, Aktas Samur A, Fulciniti M, et al. Genome-wide somatic alterations in multiple myeloma reveal a superior outcome group. J Clin Oncol. 2020;38(27):3107–3118. doi: 10.1200/JCO.20.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nik-Zainal S, Van Loo P, Wedge DC, et al. The life history of 21 breast cancers. Cell. 2012;149(5):994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rabbie R, Ansari-Pour N, Cast O, et al. Multi-site clonality analysis uncovers pervasive heterogeneity across melanoma metastases. Nat Commun. 2020;11(1):4306. doi: 10.1038/s41467-020-18060-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zapata L, Susak H, Drechsel O, Friedlander MR, Estivill X, Ossowski S. Signatures of positive selection reveal a universal role of chromatin modifiers as cancer driver genes. Sci Rep. 2017;7(1):13124. doi: 10.1038/s41598-017-12888-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr., Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsherniak A, Vazquez F, Montgomery PG, et al. Defining a cancer dependency map. Cell. 2017;170(3):564–576.e16. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexandrov LB, Kim J, Haradhvala NJ, et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578(7793):94–101. doi: 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosales RA, Drummond RD, Valieris R, Dias-Neto E, da Silva IT. signeR: an empirical Bayesian approach to mutational signature discovery. Bioinformatics. 2017;33(1):8–16. doi: 10.1093/bioinformatics/btw572. [DOI] [PubMed] [Google Scholar]
- 20.Rustad EH, Nadeu F, Angelopoulos N, et al. mmsig: a fitting approach to accurately identify somatic mutational signatures in hematological malignancies. Commun Biol. 2021;4(1):424. doi: 10.1038/s42003-021-01938-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maura F, Degasperi A, Nadeu F, et al. A practical guide for mutational signature analysis in hematological malignancies. Nat Commun. 2019;10(1):2969. doi: 10.1038/s41467-019-11037-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Schulz-Trieglaff O, Shaw R, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2015;32(8):1220–1222. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]
- 23.Team RC . 2013. R: a language and environment for statistical computing. [Google Scholar]
- 24.Bolli N, Avet-Loiseau H, Wedge DC, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014;5:2997. doi: 10.1038/ncomms3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keats JJ, Speyer G, Christofferson A, et al. Molecular predictors of outcome and drug response in multiple myeloma: an interim analysis of the Mmrf CoMMpass study. Blood. 2016;128(22):194. [Google Scholar]
- 26.Sanchez-Vega F, Mina M, Armenia J, et al. Oncogenic signaling pathways in The Cancer Genome Atlas. Cell. 2018;173(2):321–337.e310. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martello M, Poletti A, Borsi E, et al. Clonal and subclonal TP53 molecular impairment is associated with prognosis and progression in multiple myeloma. Blood Cancer J. 2022;12(1):1–7. doi: 10.1038/s41408-022-00610-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyle EM, Ashby C, Tytarenko RG, et al. BRAF and DIS3 mutations associate with adverse outcome in a long-term follow-up of patients with multiple myeloma. Clin Cancer Res. 2020;26(10):2422–2432. doi: 10.1158/1078-0432.CCR-19-1507. [DOI] [PubMed] [Google Scholar]
- 29.Szklarczyk D, Gable AL, Nastou KC, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie Z, Bailey A, Kuleshov MV, et al. Gene set knowledge discovery with Enrichr. Curr Protoc. 2021;1(3):e90. doi: 10.1002/cpz1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weinhold N, Ashby C, Rasche L, et al. Clonal selection and double-hit events involving tumor suppressor genes underlie relapse in myeloma. Blood. 2016;128(13):1735–1744. doi: 10.1182/blood-2016-06-723007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hassan HM, Isovic M, Underhill MT, Torchia J. TDG is a novel tumor suppressor of liver malignancies. Mol Cell Oncol. 2020;7(4):1768819. doi: 10.1080/23723556.2020.1768819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ansari-Pour N, Zheng Y, Yoshimatsu TF, et al. Whole-genome analysis of Nigerian patients with breast cancer reveals ethnic-driven somatic evolution and distinct genomic subtypes. Nat Commun. 2021;12(1):6946. doi: 10.1038/s41467-021-27079-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Squatrito M, Vanoli F, Schultz N, Jasin M, Holland EC. 53BP1 is a haploinsufficient tumor suppressor and protects cells from radiation response in glioma. Cancer Res. 2012;72(20):5250–5260. doi: 10.1158/0008-5472.CAN-12-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cottini F, Hideshima T, Suzuki R, et al. Synthetic lethal approaches exploiting DNA damage in aggressive myeloma. Cancer Discov. 2015;5(9):972–987. doi: 10.1158/2159-8290.CD-14-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao J, Huang A, Zheng X, et al. 53BP1 loss induces chemoresistance of colorectal cancer cells to 5-fluorouracil by inhibiting the ATM-CHK2-P53 pathway. J Cancer Res Clin Oncol. 2017;143(3):419–431. doi: 10.1007/s00432-016-2302-5. [DOI] [PubMed] [Google Scholar]
- 37.Barwick BG, Neri P, Bahlis NJ, et al. Multiple myeloma immunoglobulin lambda translocations portend poor prognosis. Nat Commun. 2019;10(1):1911. doi: 10.1038/s41467-019-09555-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bergsagel PL, Kuehl WM. Chromosome translocations in multiple myeloma. Oncogene. 2001;20(40):5611–5622. doi: 10.1038/sj.onc.1204641. [DOI] [PubMed] [Google Scholar]
- 39.Misund K, Keane N, Stein CK, et al. MYC dysregulation in the progression of multiple myeloma. Leukemia. 2020;34(1):322–326. doi: 10.1038/s41375-019-0543-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boyd KD, Ross FM, Walker BA, et al. Mapping of chromosome 1p deletions in myeloma identifies FAM46C at 1p12 and CDKN2C at 1p32.3 as being genes in regions associated with adverse survival. Clin Cancer Res. 2011;17(24):7776–7784. doi: 10.1158/1078-0432.CCR-11-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu KL, Beverloo B, Lokhorst HM, et al. Abnormalities of chromosome 1p/q are highly associated with chromosome 13/13q deletions and are an adverse prognostic factor for the outcome of high-dose chemotherapy in patients with multiple myeloma. Br J Haematol. 2007;136(4):615–623. doi: 10.1111/j.1365-2141.2006.06481.x. [DOI] [PubMed] [Google Scholar]
- 42.Walker BA, Mavrommatis K, Wardell CP, et al. A high-risk, double-hit, group of newly diagnosed myeloma identified by genomic analysis. Leukemia. 2019;33(1):159–170. doi: 10.1038/s41375-018-0196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rustad EH, Yellapantula V, Leongamornlert D, et al. Timing the initiation of multiple myeloma. Nat Commun. 2020;11(1):1917. doi: 10.1038/s41467-020-15740-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giesen N, Paramasivam N, Toprak UH, et al. Comprehensive genomic analysis of refractory multiple myeloma reveals a complex mutational landscape associated with drug resistance and novel therapeutic vulnerabilities. Haematologica. 2022;107(8):1891–1901. doi: 10.3324/haematol.2021.279360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones JR, Weinhold N, Ashby C, et al. Clonal evolution in myeloma: the impact of maintenance lenalidomide and depth of response on the genetics and sub-clonal structure of relapsed disease in uniformly treated newly diagnosed patients. Haematologica. 2019;104(7):1440–1450. doi: 10.3324/haematol.2018.202200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang X, Han J, Feng L, et al. DUOX2 promotes the progression of colorectal cancer cells by regulating the AKT pathway and interacting with RPL3. Carcinogenesis. 2021;42(1):105–117. doi: 10.1093/carcin/bgaa056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ingram C.J.E., Ekong R, Ansari-Pour N, Bradman N, Swallow DM. Group-based pharmacogenetic prediction: is it feasible and do current NHS England ethnic classifications provide appropriate data? Pharmacogenomics J. 2021;21(1):47–59. doi: 10.1038/s41397-020-0175-0. [DOI] [PubMed] [Google Scholar]
- 48.Germano G, Amirouchene-Angelozzi N, Rospo G, Bardelli A. The clinical impact of the genomic landscape of mismatch repair-deficient cancers. Cancer Discov. 2018;8(12):1518–1528. doi: 10.1158/2159-8290.CD-18-0150. [DOI] [PubMed] [Google Scholar]
- 49.Miyashita K, Fujii K, Suehiro Y, et al. Heterochronous occurrence of microsatellite instability in multiple myeloma - an implication for a role of defective DNA mismatch repair in myelomagenesis. Leuk Lymphoma. 2018;59(10):2454–2459. doi: 10.1080/10428194.2018.1427862. [DOI] [PubMed] [Google Scholar]
- 50.Begum R, Martin SA. Targeting mismatch repair defects: a novel strategy for personalized cancer treatment. DNA Repair. 2016;38:135–139. doi: 10.1016/j.dnarep.2015.11.026. [DOI] [PubMed] [Google Scholar]
- 51.Martin SA, Lord CJ, Ashworth A. Therapeutic targeting of the DNA mismatch repair pathway. Clin Cancer Res. 2010;16(21):5107–5113. doi: 10.1158/1078-0432.CCR-10-0821. [DOI] [PubMed] [Google Scholar]
- 52.Misund K, Hofste Op Bruinink D, Coward E, et al. Clonal evolution after treatment pressure in multiple myeloma: heterogenous genomic aberrations and transcriptomic convergence. Leukemia. 2022;36(7):1887–1897. doi: 10.1038/s41375-022-01597-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vo JN, Wu YM, Mishler J, et al. The genetic heterogeneity and drug resistance mechanisms of relapsed refractory multiple myeloma. Nat Commun. 2022;13(1):3750. doi: 10.1038/s41467-022-31430-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.