Abstract
Plasmacytoid dendritic cells (pDCs) have been shown to play an important role during immune responses, ranging from initial viral control through the production of type I interferons (IFNs) to antigen presentation. However, recent studies uncovered unexpected heterogeneity among pDCs. Notably, we identified a new subset, referred to as pDC-like cells, that resembles pDCs but also shares cDC features. Here we show that this subset is a circulating progenitor distinct from common DC progenitors (CDP), with prominent cDC2 precursor potential. Our findings from hCD2 and CD300c lineage tracing mouse models suggest that a significant fraction of cDC2s originate from pDC-like cells, which can therefore be referred to as pre-DC2. This precursor subset responds to homeostatic cytokines, such as macrophage colony stimulating factor (M-CSF), by expanding and differentiating into cDC2 that efficiently prime Th17 cells. Development of pre-DC2 into CX3CR1+ ESAM− cDC2b but not CX3CR1− ESAM+ cDC2a requires the transcription factor KLF4. Finally, we show that under homeostatic conditions this developmental pathway regulates the immune threshold at barrier sites, by controlling the pool of Th17 cells within skin-draining lymph nodes.
One-sentence Summary:
pDC-like cells are circulating precursors for cDC2 that control differentiation of homeostatic CD4+ Th17 cells.
Introduction
Dendritic cells (DCs) are dedicated immune cell subsets which shape the appropriate immune response during infections, while maintaining tissue integrity under steady state conditions1-4. DCs can be subdivided into plasmacytoid DCs (pDCs) and conventional DCs (cDCs), two functionally specialized DC branches each devoted to respond to specific classes of pathogens1, 2, 5-9. pDCs are a unique population capable of producing large amounts of type I and Type III IFNs in response to viral infections 6, 10-12. In humans, three stable subpopulations of pDCs were described based on the expression of PD-L1 and CD80 after activation with influenza virus 13. cDCs are professional antigen presenting cells (APCs) that initiate the adaptive immune response by capturing, processing, and presenting foreign particles to naïve T cells in the draining lymph nodes (LNs)1, 14. Upon activation, cDCs also secrete a variety of inflammatory cytokines that will shape the milieu and promote the activation and recruitment of innate immune cells. Following antigen capture and processing, cDCs will migrate to the draining LNs and prime T cell responses. The type of cytokines that are produced and become prevalent during an immune response will depend on the pathogen or inflammatory insult, as well as the cDC subset that is predominantly or selectively activated, as hypothesized by the “division of labor” hypothesis7, 15. cDCs are subdivided into cDC1 and cDC2. cDC1 are characterized by the expression of the chemokine receptor XCR1 and the transcription factor (TF) IRF816. This subset is specialized in IL-12 production and cross-presentation of antigens that promotes ILC1, CD4+ Th1 and cytotoxic CD8+ T cell (CTL) immunity17-24. cDC1s are therefore essential to promote anti-tumor, anti-viral immunity, as well as initiate immune responses against intracellular pathogens. cDC2s are profoundly heterogeneous comprising multiple subsets all characterized by the expression of the TF IRF4 and mostly responsible for promoting CD4+ T helper responses25-31. cDC2 specifically activate Tregs, Th2, and Th17 T cells, and were shown to depend on PU.1, KLF4, STAT6, RelB, Notch2 and IRF4 amongst others32. The lack of either TF leads to selective subset deficiencies, however since the remaining cells may also be impaired in their function, dissecting the individual contribution of each subset to immune responses in the absence of specific targeting remains an open challenge. Recent single cell sequencing studies in mice and humans revealed the broad heterogeneity within cDC2 under steady state as well as under inflammatory conditions 33-41 further segregating the cDC2 compartment into cDC2a, cDC2b, DC3, inflammatory DCs (iDCs) or myeloid regulatory DCs (mregDCs), these latter characterized by the acquisition of new markers and/or transcriptional profiles following exposure to specific cytokines 13, 38-40. Collectively the cDC2 field has become increasingly more complex to dissect and assigning a specific function to each newly characterized subset has been a main goal with the scope of improving our capacity to harness their properties in clinical settings. 35, 37-40. Among the newly identified subsets we and others recently described a small fraction of cells within the pDC pool that is characterized by cDC features, including the expression of the cDC-specific TF Zbtb46. We referred to this subset as to pDC-like cells 33, 34, 42-44.
Within human peripheral blood mononuclear cells (PBMCs), about 2-3% of pDCs showed transcriptionally mixed cDC and pDC features, and were referred to as Axl+Siglec6+ DC (AS DC) or pre-DCs 35, 37, suggesting parallelism between mouse and human studies. However, the nature of these human subsets and pDC-like cells, their functional and developmental properties, as well as their interrelationship with previously described DC subsets remained to be addressed32. Here we show that pDC-like cells are present at different frequencies in all tissues and appear as a subset of circulating cDC progenitors, with prominent in vivo and in vitro cDC2 developmental potential. We therefore relabeled them as pre-DC2. We show that differentiation of pre-DC2 into CX3CR1+ESAM− cDC2b strictly depends on the TF KLF4. Further, they respond to homeostatic cytokines, in particular M-CSF, and their differentiation into cDC2 controls the pool of Th17 under steady state conditions.
Results
pDC-like cells are highly abundant in the heart and at epithelial sites
We have recently identified a new DC subset with phenotypic pDC features but functional cDC properties that we referred to as pDC-like cells43. Heterogeneity within the pDC compartment was also reported by several groups using specific markers or through single cell sequencing experiments 33-35, 37, 42, however the interrelationships among all previously identified subsets has been complicated because of the different markers or conditions used, with some researchers referring to mature or immature stages. We have therefore performed a multidimensional flow cytometry analysis that included all previously established gating strategies for subsets considered to be progenitors (fig. S1A-C) as well as subsets considered mature (fig. S1D-I). For progenitor subsets we included, besides mature pDC and pDC-like cells, Lin−B220−FLT3+Ly6C−CD115+CD127− CDPs (CD115+ CDPs)45-47, Lin−B220−FLT3+Ly6C−CD115−CD127−CD115− CDPs (CD115− CDPs)48, Lin−Zbtb46-GFP+-cKitintFLT3+ pre-cDC149, and the pre-DC subsets identified by Schlitzer et al. as Lin−B220−FLT3+CD11c+Ly6C+SiglecH+ pre-cDCs, Lin−B220−FLT3+CD11c+Ly6C−SiglecH− pre-cDC1, Lin−B220−FLT3+CD11c+Ly6C+SiglecH− pre-cDC2, Lin−B220−FLT3+CD11c+Ly6C−SiglecH+ common DC/pDC precursors50, as well as our previously identified committed Lin− B220−FLT3+Ly6C−CD115−IL-7R+SiglecH+Ly6D+ pre-pDC43 (fig. S1B). As shown in the UMAP projection, there is a strong overlap between IL7R-expressing pre-pDCs43 and the Ly6C−SiglecH+ common DC/pDC precursors 50, suggesting that the cDC precursor potential contained within this subset is likely originating from CD115-expressing myeloid precursors and are therefore also included within the CDP gate (fig. S1C). However, pDC-like cells represent a small fraction that is contained only within the Ly6C+SiglecH+ pre-cDCs gate 50. CD226 as well as Zbtb46-GFP were both described as specific markers for pre-cDC1, however while pDC-like cells were Zbtb46-GFP positive they did not express CD226 (fig. S1A and S1D). Similarly, the expression of CD2 that in human samples segregates pDCs into two subsets 51 is expressed at low levels within both pDC and pDC-like cells (fig. S2E). CCR9 that among other receptors was used to identified CCR9− immature pDCs from CCR9+ mature pDCs42, 52 53, is not expressed on pDC-like cells and remains heterogeneous on conventional pDCs (fig. S1E-F). The only marker that separated pDC from pDC-like cells that had been used before is CX3CR1 which however labels all bone marrow (BM) but around 70% of splenic and peripheral pDC-like cells (fig. S1F-H). This marker was recently used to identify transitional DCs (tDCs) 33. As shown in the UMAP overlay, pDC-like cells largely overlap with the CD11clow tDCs but not with CD11chi tDCs that further segregate in at least two subsets (fig. S1G-I). Collectively, each gating strategy results in a partial overlap in subsets, suggesting that identification of new markers, or the development of reporter lines would be essential to improve resolution.
Therefore, to better relate this subset to conventional pDCs and cDCs and potentially identify new specific markers, we performed a high dimensional screening for surface proteins on pDCs and cDCs by flow cytometry and applied the “Infinity Flow” algorithm that uses machine learning to determine the co-expression of different markers across clusters36, 54. CD19−CD3−CD11c+ splenocytes isolated from Zbtb46-GFP expressing mice were screened for the expression of 255 markers. Nine markers were selected as backbone staining to allow specific gating on pDCs, pDC-like cells, and cDCs. Non-linear dimensionality reduction via Uniform manifold approximation and projection (UMAP) on a two-dimensional (2D) space of the backbone markers revealed subclusters for all known DC subsets (Fig. 1A and fig. S2A). pDCs and pDC-like cells were defined by the expression of BST2 and SIGLEC-H, however segregated into two subsets based on Zbtb46-GFP; cDC1 and cDC2 could be identified by the expression of XCR1 and SIRP-α, respectively (Fig. 1A). Within CD19−CD3−CD11c+ splenocytes we also had NK1.1-expressing NK cells as well as Ter119+ erythroid lineage cell precursors (fig. S2A), which were not considered for further analysis.
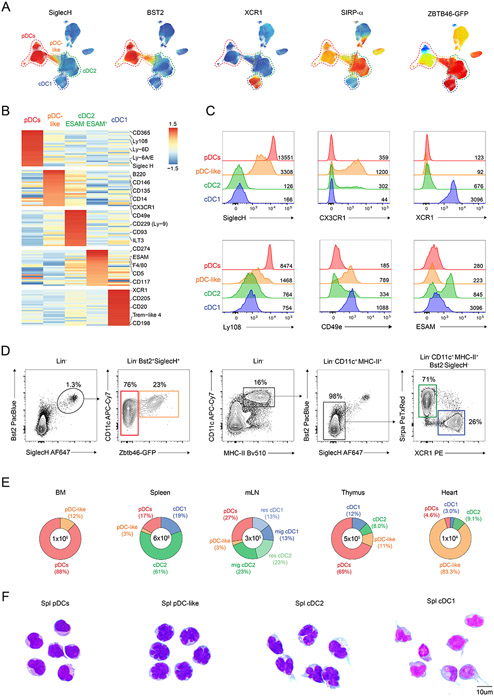
Figure 1. pDC-like cells are highly abundant in the heart and at epithelial sites.
(A-B) Splenocytes from three Zbtb46GFP/WT mice were enriched for CD11c and stained with 9 backbone markers followed by the individual 255 PE-conjugated markers provided by the LEGENDscreen™ kit as explained in the methods. (A) A non-linear dimensionality reduction was performed on the backbone markers and projected on a UMAP space. Shown is the relative expression for the indicated marker across the UMAP. (B) Shown is an expression heatmap obtained for the top 25 differentially expressed markers on the clustered DC subsets obtained as explained in the methods. (C) Shown are single-color histograms for the indicated markers expressed by pDCs (red), pDC-like cells (orange), cDC2 (green) and cDC1 (blue). (D) Gating strategy for splenic pDCs (red), pDC-like cells (orange), cDC2 (green) and cDC1 (blue) (n=6 mice). (E) Shown are the total cell number and percent distribution of all DC subsets across the indicated lymphoid and non-lymphoid tissues (n=3 mice, two independent experiments). (F) May–Grünwald staining was performed on cytospins of sort-purified splenic pDCs, pDC-like, cDC2 and cDC1 (n = 3 representative images taken from 2 independent experiments: scale bars, 10 μm).
Importantly, pDC-like cells defined as Zbtb46-GFP+ expressed low- to mid-levels of the pDC markers SIGLECH and BST2 and clustered between conventional pDCs and cDC2 within the UMAP space, further supporting the intermediate features of this subset (Fig. 1A and fig. S2C). Taking advantage of all the 255 surface markers, we clustered all Lin−NK1.1−Ter119− splenocytes using the Self-Organizing Maps algorithm on flow cytometry data FlowSOM32 and identified 5 major DC subsets: cDC1, conventional pDCs, pDC-like cells, ESAM+ and ESAM− cDC2 (Fig. 1B and fig. S2B). This clustering analysis on one hand highlighted the previously identified heterogeneity within the cDC2 compartment and on the other hand showed the unique identity of the pDC-like compartment that segregated apart from conventional pDCs and cDCs. The top 25 differentially expressed markers across these 5 clusters show selective expression for only few markers and receptors, which were only differentially expressed and not exclusive as shown in the heatmap (Fig. 1B). Some of the cluster-defining markers were further validated by flow cytometry using the mean fluorescent intensity (MFI) across subsets (Fig. 1C). Conventional pDCs expressed the well-defined markers LY6D, SIGLEC-H, CD45R/B220 as well as the newly identified markers LY108 and SCA1 (Fig. 1B-C and fig. S2D-E). pDC-like cells mostly expressed SIGLECH, CD146, FLT3 (CD135), CD2 and CD14 amongst others (Fig. 1B-C and fig. S2D-E). Besides those, there were also several markers that showed high levels on ESAM− cDC2 and were higher on pDC-like cells compared to pDCs such as the fractalkine receptor CX3CR1 and the integrin α5 CD49e (Fig. 1B-C). ESAM+ cDC2 that were previously shown to depend on Notch2, expressed CD5, CD117 and CD274, while ESAM− cDC2 were characterized by high expression of CD229, ILT3 and CX3CR1 (Fig. 1B). The cDC1 cluster showed as expected high expression of XCR1, CD205 and TREM-like 4. Additionally, we identified the chemokine receptor CCR8 (CD198) that promotes CCL1-mediated migration (Fig. 1B and fig. S2D).
To determine the tissue distribution of cDCs and pDCs, we applied the best gating strategy, as shown in Fig. 1D and fig. S2C including CD45 for non-lymphoid tissues. cDCs were mostly absent from the BM, where absolute numbers and relative abundance of pDCs and pDC-like cells were the highest (Fig. 1E). Interestingly, pDC-like cells accounted for over 80% of all DCs in the heart. Unexpectedly, pDCs outnumbered other subsets in intraepithelial lymphoid tissues (IEL), liver and thymus (Fig. 1E and fig. S2F). We next compared the morphology of pDC-like cells to all DC subsets using the May-Grünwald Giemsa staining. pDC-like cells exhibited a round-shaped morphology reminiscent of conventional pDCs, but appeared to have a polylobated nucleus, as observed for cDC2s (Fig. 1F). Dendrites were evident only on cDC1 and cDC2 but not on pDCs or pDC-like cells (Fig. 1F).
Collectively, pDC-like cells appear as an independent subset which has a unique tissue distribution, differential surface marker expression, and unique morphology when compared to cDCs and pDCs.
pDC-like cells belong to the DC lineage
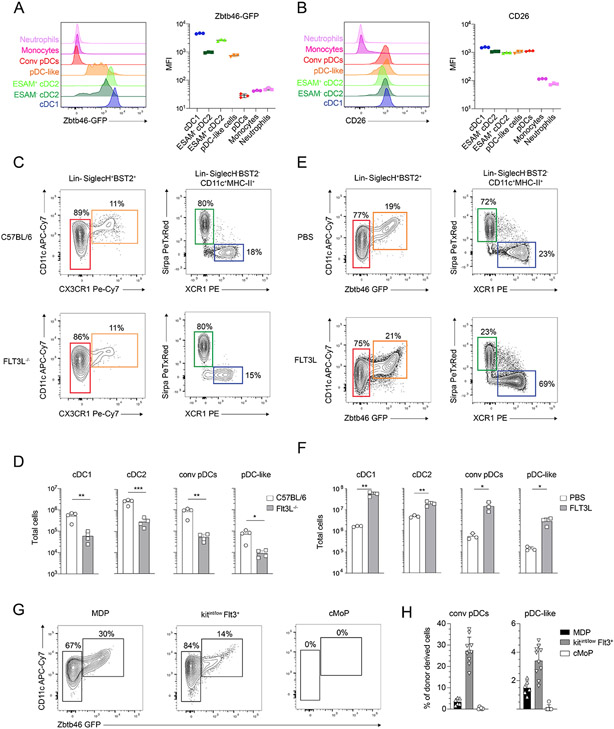
We and others have recently shown that pDCs are mostly derived from a lymphoid progenitor that expresses IL-7R, LY6D and SIGLECH43, 55, 56, while cDCs are considered to differentiate from common dendritic cell precursors (CDPs) that express CSF1R45, 47, 49, 50, 57, 58. Given the common phenotypical and morphological features of pDC-like cells that were shared across conventional pDCs, cDCs and monocytes, we wanted to determine their lineage. cDCs and pDCs are defined by their developmental dependency on FLT3-FLT3L, and so far, the best cDC-defining features are the expression of the TF ZBTB46 and of the dipeptidyl peptidase-4 (CD26)15, 26, 59. pDC-like cells, that are defined by the expression of Zbtb46-GFP, also expressed CD26, which does not label CD11b+Ly6Chi monocytes nor CD11b+Ly6Cint granulocytes (Figure 2A-B). To determine the developmental requirement of pDC-like cells on FLT3-FLT3L, we analyzed Flt3l−/− mice and injected FLT3L into Zbtb46-GFP mice (Fig. 2C-F). However, since Zbtb46-GFP could not be used in Flt3l−/− mice, we optimized an alternative gating strategy based on the results obtained above by replacing the expression of GFP with CX3CR1. Approximately 70% of SIGLECH+BST2+ZBTB46+ pDC-like cells co-express CX3CR1, allowing for the identification of most pDC-like cells across all strains (fig. S3A). All DC subsets were strongly reduced in Flt3l−/−mice, including pDC-like cells, validating their affiliation to the DC lineage (Fig. 2C and 2D). Additionally, exogenous administration of FLT3L in Zbtb46-GFP mice resulted in about 20-fold expansion of pDC-like cells, similarly as cDC1 (20-fold), cDC2 (4-fold) and conventional pDCs (20-fold) (Fig. 2E and 2F).
Figure 2. pDC-like cells are bona fide dendritic cells.
(A-B) Shown are single-color histograms and the mean fluorescent intensity (MFI) for Zbtb46-GFP (A) or CD26 (B) expression across the indicated leukocytes. (C-D) C57BL/6 and Flt3L−/− mice were analyzed for splenic cDC subsets, pDC and pDC-like cells. Cells were gated as shown in (C). Total cells gated as in (C) are shown in (D). (E-F) Zbtb46GFP/WT mice were injected with PBS or FLT3L as explained in methods. Shown is the frequency (E) and number (F) of splenic cDC subsets, pDC and pDC-like cells gated as shown in (E) (n=3-4 mice, two or three independent experiments, each dot represents a sample, and thin lines represent the mean ± s.d.). (G-H) MDPs (black bars), cKITint/lowFLT3+DC precursors (gray bars) and cMoPs (white bars), gated as defined in methods isolated from Zbtb46-GFP mice were adoptively transferred into sub-lethally irradiated congenic mice. Mice were analyzed for donor derived cells four days after injection. Shown are two color plots (G) or percent donor derived (H) pDC (CD11clowGFP−) and pDC-like cells (CD11c+GFP+), pre-gated as 7-AAD−Lin−SiglecH+Bst2+. (n=6-8 mice, two or three independent experiments, each dot represents a sample, and thin lines represent the mean ± s.d.). Statistical analysis was done with unpaired two-tailed t-test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
FLT3L is an instructive cytokine for the development of DCs as it is sufficient to induce their differentiation from BM cells in vitro. We therefore cultured BM cells isolated from Zbtb46-GFP mice and determined the output of pDCs, cDCs and both DC subsets, which showed slightly different developmental kinetics (fig. S3B-C). pDCs reached the highest frequency in FLT3L cultures by day 6, accounting for approximately half of the total CD11c+ cells (fig. S3C). From day 6 onwards, pDC frequency declined, consistent with their progressive differentiation from declining progenitors, and their short survival capacity in vitro. pDC-like cells however, showed similar in vitro kinetics as cDCs, where the output remained stable from day 6 to day 9, hinting to a shared developmental pathway or similar survival properties (fig. S3C).
To dissect the ontogeny and lineage relationships of pDC-like cells, we transferred Monocyte and DC progenitors (MDPs), common Monocyte progenitors (cMoP) 57, 60-63 and FLT3+cKitint/low DC progenitors into sub-lethally irradiated recipient mice and analyzed their progeny at day 4 using congenic markers. MDPs, which represent the most immature subset among the three sorted populations, could generate cDCs, pDCs, pDC-like cells as well as monocytes. cMoPs showed no cDC, pDC or pDC-like potential, while being the most efficient progenitors for monocytes, as reported 61 (Fig. 2G-H and fig S3D). DC progenitors gave rise to all DC subsets including pDC-like cells, while exhibiting limited monocyte potential (Fig. 2G-H and fig S3E).
Collectively, both in vitro as well as in vivo transfer experiments showed that pDC-like cells develop from a FLT3-expressing DC precursor, are dependent on FLT3-FLT3L signaling, and share similar in vitro developmental kinetics with cDCs but not pDCs, corroborating their close relationship to the DC lineage.
pDC-like cells are transcriptionally similar to pDCs
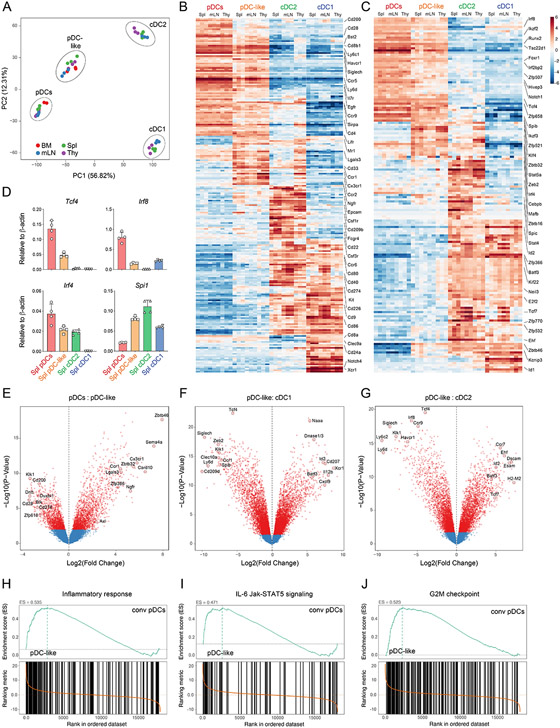
To compare the transcriptional signature of pDC-like cells with the other DC subsets, and gain insights into their functional properties, we decided to perform bulk RNA sequencing on pDC, pDC-like cells and cDCs isolated from BM, spleen, thymus and mesenteric lymph nodes (mLNs) isolated from Zbtb46-GFP reporter mice. Principal component analysis (PCA) showed individual segregation of each subset, independent of their tissue origin (Fig. 3A), suggesting that subset identity was defining over tissue specificity. pDC-like cells clustered closer to conventional pDCs than to cDC1 or cDC2, suggesting higher similarity to pDCs. We subsequently mapped the most differentially expressed genes (Log2FC > 2.0) across the 4 subsets to identify the ones which were uniquely expressed (Fig. 3B-C, fig. S4A and Data file S1). We narrowed this analysis for surface markers (Fig. 3B) and TFs (Fig. 3C). The highest expressed surface markers detected on pDC-like cells were common to conventional pDCs, and were Bst2, SiglecH, Sirpa and Lifr, (Fig. 3B), validating their phenotypic proximity obtained by our flowSOM analysis (Fig. 1B). Interestingly, we also identified Ccr1, Ccr2, Cd33, Lgals3, and Ngfr, that showed high expression on thymic cDC2s. Collectively, besides a few surface markers including CX3CR1, most surface receptors expressed by pDC-like cells were either in common with conventional pDCs or cDCs (Fig. 3B and fig. S4B).
Figure 3. pDC-like cells are transcriptionally similar to pDCs.
(A-J) Bulk RNA Sequencing was performed on sort purified pDCs, pDC-like cells, cDC2 and cDC1 isolated from the BM (red), Spleen (green), Thymus (purple) and mLN (blue). RNA was isolate on three consecutive days using each time cells obtained from a single mouse (A) Principal Component Analysis (PCA) on the DC subsets calculated as explained in methods. (B-C) Shown are heatmaps with relative expression values, centered and normalized on the mean for the most differentially expressed cell-surface genes (B) or transcription factors (C) (log2FC >2.0) on the indicated subsets and tissues. (D) qRT-PCR on the indicated genes for the indicated subsets (n=4 mice, two independent experiments, shown are mean values +/−s.d.). (E-G) Volcano plots showing pair-wise comparisons as Log2FC expression on transcripts for pDCs versus pDC-like cells (E), pDC-like cells versus cDC1 (F) and pDC-like cells vs cDC2 pooled across all tissues (G). (H-J) Gene set enrichment analysis, performed on the Molecular Signature Database (MSigDb v5.2) showing the enrichment score (ES) for splenic pDC-like cells versus splenic pDCs, as depicted.
Similarly, we identified 45 pDC-like differentially expressed TFs, such as Klf4, Spi1 and Zbtb32, however while their expression was higher on pDC-like cells, these TFs also characterized cDC2 (Fig. 3C and fig. S4C). pDCs as well as pDC-like cells expressed the TFs Irf8, Runx2, SpiB as well as the pDC master regulator Tcf4 (Fig. 3C-D and S4C). Collectively, most of the differentially expressed genes (Data file S1) were not unique to one subset, likely reflecting shared ontogeny or developmental convergence (Fig. 3B-C, fig. S4A and Data file S1).
Given the high transcriptional similarities between subsets, we focused on pairwise comparison between pDC-like cells and other subsets applying stringent criteria (log2 FC > 2.0) (Fig. 3E-G and fig. S4B-C). Axl, Lgals3, Csf1r, and Cd80 as well as the transcription factors Zbtb32, Zbtb46, Myc, Klf4, and Spi1 showed higher expression in pDC-like cells compared to pDCs (Data file S2), while conventional pDCs expressed Cd200, Ly6d and Ccr9 and the DNA binding proteins or TFs Oasl1, Dntt, Duxbl1 as well as Gcm2 (Fig. 3E, fig. S4B-C and Data file S2). Pairwise comparison between pDC-like cells and cDC1 or cDC2 showed higher expression of pDC-related genes such as Tcf4, Irf8, Zeb2, Siglech, Ccr9 and Klk1 on pDC-like cells, and higher expression of Naaa, Id2, Xcr1 and Batf3 or Ccr7, Ehf, Esam and Dscam, on cDC1 or cDC2, respectively (Fig 3F-G). The expression of thymic cDC2-related markers on pDC-like cells could potentially reflect some functional or developmental features of this subset, however more studies are required to dissect this aspect (Fig. 3B-C).
Gene set enrichment analysis (GSEA) on all cells of each subset, performed independently of their tissue of origin, showed an enrichment in genes of the set “Inflammatory responses” and “IL-6/Jak-STAT5 pathways” suggesting a more inflammatory profile for pDC-like cells as compared to conventional pDCs (Fig. 3H-I and fig. S4D). pDC-like cells were also highly enriched in gene sets related to cell cycle and proliferation (Fig. 3J and fig. S4D).
Collectively, these data suggest that pDC-like cells are a unique dendritic cell subset, characterized by myeloid hallmark genes which share the expression of a large number of TFs and surface markers with conventional pDCs.
pDC-like cells respond to homeostatic cytokines and differentiate into cDC2
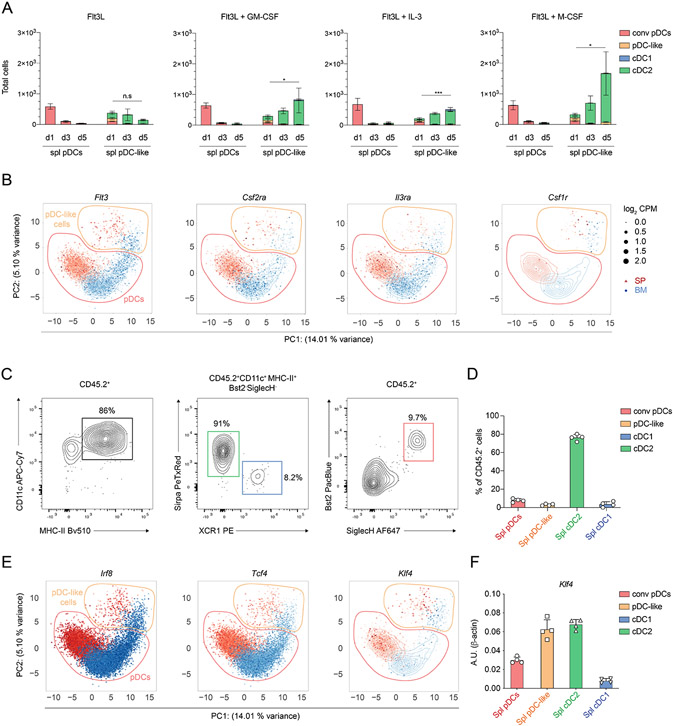
Based on previous findings and the expression of proliferative markers as well as cell cycle associated genes, we hypothesized that pDC-like cells could represent an immature or a precursor population, as reported for human pre-DCs 35. We therefore assessed their developmental potential in vitro and compared it to conventional pDCs. Sort-purified splenic pDCs or pDC-like cells were cultured in the presence of FLT3L alone or in combination with other cytokines, such as GM-CSF, IL-3, M-CSF. Splenic as well as BM pDCs did not survive beyond day 1 (spleen) and 3 (BM) in culture consistent with previous reports (Fig. 4A and fig. S5A). pDC-like cells, however, expanded progressively over 5 days, with BM-isolated cells showing overall a larger output in numbers compared to splenic pDC-like cells (fig. S5A). To our surprise, pDC-like cells generated almost exclusively cDC2 under all conditions tested, suggesting that they may represent a specific precursor for this subset (Fig. 4A and fig. S5A). These results are in line with the hypothesis that Axl-expressing pre-pDCs are precursors of human CD1c+ cDC2 35. The proliferative potential of pDC-like cells was linked to their capacity to respond to homeostatic and inflammatory cytokines, and mirrored their transcriptional profile with the expression of Flt3, Csf2ra, Csf1r and Il3ra cytokine receptors (Fig. 4B and S5C). Splenic pDC-like cells output was significantly higher when cells were cultured with FLT3L and GM-CSF, IL-3 or M-CSF (Fig. 4A and fig. S5B).
Figure 4. pDC-like cells respond to homeostatic cytokines and differentiate into cDC2.
(A) Sort purified splenic pDCs and pDC-like cells isolated from C57BL/6 mice were cultured for four days in the presence of FLT3L alone (left panel) or FLT3L in combination with GM-CSF, IL-3 or M-CSF as indicated. cDC1 (blue), cDC2 (green), pDCs (red) and pDC-like cells (orange) were determined at the indicated timepoints. (B) Shown is the UMAP projection depicting the expression levels for the indicated genes obtained from scRNA seq data previously generated on sort purified BM and splenic Bst2+ SiglecH+ cells43. BM (blue) and splenic (red) pDC and pDC-like cells were determined based on clustering analysis and are shown by the dotted contour lines. The size of each dot corresponds to the relative expression of a given gene for each cell. The contour lines indicate the density of the BM (blue) and splenic (red) cells in the PCA space calculated as previously described 43. (C and D) Sort purified pDC-like cells isolated from Zbtb46-GFP mice were transferred into sub-lethally irradiated congenic mice. Donor cells were analyzed 4 days after transfer based on the expression of DC markers. The relative contribution was determined (D) for donor derived cells gated as in (C). (E) scRNA-seq expression of the indicated TFs as in (B). (F) qRT-PCR for the expression of Klf4 on sort purified splenic pDCs (red), pDC-like (orange), cDC2 (green), and cDC1 (blue) (n=4 mice, two independent experiments, shown are mean values +/−s.d.).
To define whether the observed developmental potential is extendable to in vivo settings, we transferred BM or splenic pDCs or pDC-like cells into sublethally irradiated congenic recipient mice and analyzed their progeny. When pDCs were transferred, 80% of the recovered cells were pDCs (fig. S5D-E), suggesting that their identity was defined and that they represented a developmental end stage with limited to no proliferative potential. However, when pDC-like cells were transferred over 80% of the progeny identified as cDC2s (Fig. 4C-D). We could still detect a small fraction of CD45.2 cells that were included within the cDC1 (5-8%) or pDC (5-10%) gates respectively (Fig. 4C-D). These data suggested that most pDC-like cells will develop into cDC2, but may maintain the capacity to generate cDC1 and pDCs upon transfer.
Collectively, pDC-like cells are precursors that are highly responsive to homeostatic cytokines and mostly differentiate into CD11c+MHC-IIhiSIRP-α+ cDC2, despite having a transcriptional profile that aligned to pDCs.
Selective KLF4 deficiency profoundly alters pDC-like signature
KLF4, that was shown to be expressed on cDC244, was one of the most DEG between pDC-like cells and conventional pDCs, which also express most pDC related TFs (fig. S4C). We therefore evaluated the expression levels at single cell resolution for Klf4, Irf8 and Tcf4 that were previously linked to cDC and pDC development 1, 43, 44, 64. Klf4 was mostly restricted to pDC-like cells, while Irf8 and Tcf4 were equally expressed by pDCs and pDC-like cells independently whether cells were isolated from BM or spleen (Fig. 4E). This result was confirmed by qRT-PCR (Fig. 4F).
Given the heterogeneity of cDC2, and to ensure an unbiased approach, we performed single cell RNA sequencing (scRNA-seq) on enriched and sort purified CD11c+ cells, that would include all subsets isolated from spleen, mLNs and skin-draining lymph nodes (sLN) of WT and Klf4 conditional KO (Klf4-cKO) mice.
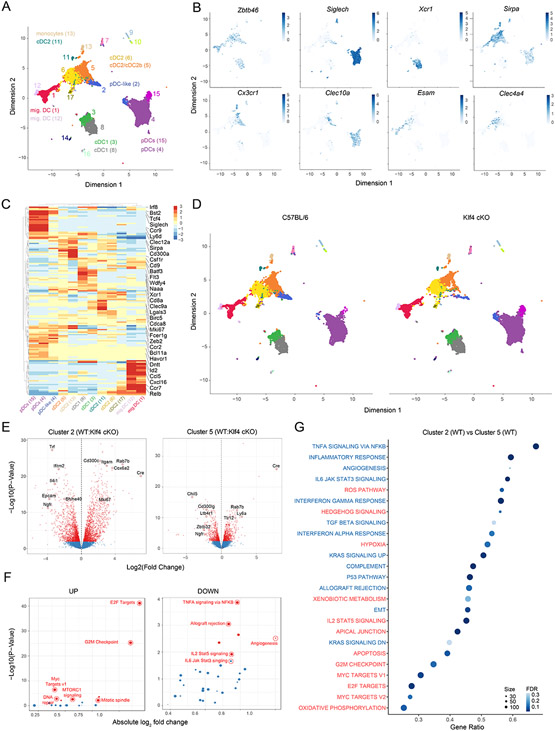
We captured approximately 33,000 cells that resulted in 17 clusters using a tree-cut/hierarchical method and projected all cells on a UMAP space (Fig. 5A). Clusters 7, 14 and 16 could be excluded from further analysis based on computational duplet scoring (fig. S5F). All other clusters were analyzed for transcriptional similarity to the ImmGen database 65. Clusters 9 and 10 identified with CD11c+ ILC and NK cells and were therefore not further considered. All other 12 clusters were either pDCs or myeloid subset based on similarity scores to the ImmGen hallmark gene signature (fig. S5G). We could assign each cluster to a specific DC subset by using the generated similarity heatmap and combining it with Zbtb46, SiglecH, Bst2, Xcr1 and Sirpa expression. Cluster 3 and 8 identified as Xcr1+ cDC1; clusters 1, 6, 11, 12, and 17 as the heterogeneous group of cDC2; cluster 13 was mostly similar to monocytes and did not express the transcript for Dpp4 (CD26) (fig. S6A); clusters 4, and 15 could be assigned to pDCs; finally, cluster 2 identified with pDC-like cells based on the co-expression of Zbtb46, SiglecH and Bst2 (Fig. 5B and fig. S5G). Cluster 5 aligned with several ImmGen-based cDC2 and monocyte subsets, suggesting heterogeneity or a rather complex transcriptional profile for these cells, as it was shown for human DC3 that share features and markers across both populations 66. Also pDC-like cells defined as cluster 2, broadly overlapped with multiple ImmGen subsets, specifically pDCs, cDC2, monocytes and CDPs (Fig. 5B and fig. S5G). Using the algorithm FindMarkers, we defined cluster specific transcripts and performed a hierarchical distribution for the most DEGs (LogFC >1.5) (Fig. 5C). Using the expression of Ccr7 we could assign clusters 1 and 12 to the migratory DC2 compartment (fig. S6A). The expression of Cx3cr1, Clec10a, and Ccr2 aligned cluster 5 with human DC366, or murine iDC 38, or cDC2b39. Esam and Clec4a4 identified Cluster 6 as the Notch2-dependent ESAMhi cDC2 subset67, 68 (Fig. 5B).
Figure 5. Selective Klf4 deficiency profoundly alters pDC-like signature.
(A-G) scRNA-seq on CD11c+ enriched and sort-purified DCs isolated from spleens, sLN and mLN derived from C57BL/6 (WT) or Klf4fl/flCD11cCre+ (Klf4-cKO) mice performed and analyzed as explained in methods. (A) Shown are all cells colored based on cluster distribution and projected on a UMAP 2D-space. (B) Shown are UMAP plots depicting the expression for the indicated genes obtained for WT cells. (C) Shown is a relative expression heatmap of the top 50 DEGs across all clusters. (D) Shown are UMAP plots depicting cells obtained from WT or Klf4-cKO mice as indicated. (E) Volcano plots showing pair-wise comparisons between WT and Klf4-cKO cells from cluster 2 (left) or cluster 5 (right). (F) GSEA performed on the Molecular Signature Database (MSigDb v5.2) comparing the enrichment score (ES) obtained for WT and Klf4-cKO cells from cluster 2. Pathways were plotted based on up (left) or down-regulation in Klf4-cKO versus WT cells (right). (G) GSEA dot plot analysis showing the most enriched pathways between cluster 2 and 5 for cells obtained from WT mice. Pathways are colored based on their down (blue) or up-regulation (red) in cluster 2 (FDR= false discovery rate).
We subsequently projected WT and Klf4-cKO cells separately in the UMAP space including all tissues grouped together (Fig. 5D) or individually (fig. S6B). Across all clusters, pDC-like cells (cluster 2) were heavily reduced with a 70% to 30% contribution of WT versus Klf4-cKO cells (fig. S6D). Further, pDC-like cells showed a peculiar distribution on the UMAP space, which split the subset into WT cells projecting close to cluster 5 cDC2, and Klf4-cKO cells in proximity of cluster 4 pDCs. Therefore, and not surprisingly, pDC-like cells had the highest number of differentially expressed genes between WT and Klf4-cKO cells (fig. S6C and Data file S3), suggesting that KLF4 might be essential in regulating the transcriptional profile and function of pDC-like cells.
Klf4-cKO mice were previously shown to lack CD11b−CD24− double negative (DN) cDC2 within skin-draining LNs and to have reduced cDC2 in spleen 44. The lack of these subset was suggested to be the reason for the impaired Th2 immunity 44, 69. We could computationally identify the missing DN cDC2 as cluster 11, which was the only absent cluster in sLN of Klf4-cKO mice (fig. S6B). Moreover, this subset best aligned with the ImmGen profile for DN cDC (fig. S5G). However, despite being transcriptionally altered in the absence of KLF4, pDC-like cells, were still present despite reduced, suggesting a more subtle defect (fig. S6B and D).
To perform pairwise comparisons across subsets, we computationally re-bulked all clusters across genotypes. Among the most DEGs between WT and Klf4-cKO pDC-like cells, we identified Cd300c, Il4i1, and Rab7b which were suggested to regulate migration of DCs 70 (Fig. 5E and Data file S3). Further, similar to the bulk RNA data, GSEA highlighted an enrichment for genes related to inflammatory responses in WT cells, while Klf4-cKO pDC-like cells showed high expression of genes associated to cell cycle (Fig. 5F and fig. S6E).
Given the evidence that pDC-like cells may represent a precursor population, we reasoned that pDC-like cells could differentiate into cluster 5 cDC2, which was also reduced in Klf4 KO mice, suggesting a possible differentiation block in the absence of KLF4. While pDC-like cells from WT animals were capable to mature to a transcriptional profile that better aligned with cDC2 and therefore projected in the UMAP space close to cDC2, Klf4-cKO cells projected next to pDCs, reflecting the more immature transcriptional profile (Fig. 5D). PCA projection of the clusters/subsets showed a gap between pDC and cDC2 for the cells belonging to Klf4 KO mice (fig. S6G), that could potentially reflect a developmental block. WT pDC-like cells projected near WT cluster 5, which based on transcripts similarity to ImmGen subsets overlaps with cDC2 and monocytes and based on surface markers resembles the previously described cDC2b subset 39. Therefore, assuming that pDC-like cells would differentiate into cDC2b (cluster 5), we directly compared the transcriptional profile of WT pDC-like cells and WT cDC2b (Data file S3) in order to identify genes necessary for this developmental progression. WT pDC-like cells were highly enriched in proliferation-related transcripts compared to cDC2b cells (Fig. 5G). Further, KLF4 was among the highest expressed TF on cluster 2 and 5 as shown in fig. S6F, making this TF a likely candidate gene, necessary for this developmental progression.
Collectively, scRNA-seq analysis confirmed the identify of pDC-like cells as a unique subset within pDC and cDCs and highlighted the well-known subset heterogeneity across DCs. Further it showed the profound impact that the lack of a TF, in this case KLF4, had on some of the clusters as well as tissues. Collectively, the broad expression across multiple clusters suggested that more cells were impaired in Klf4-cKO mice besides the previously identified DN cDCs 44, including pDC-like cells and cDC2b.
pDC-like cells require KLF4 for cDC2 development
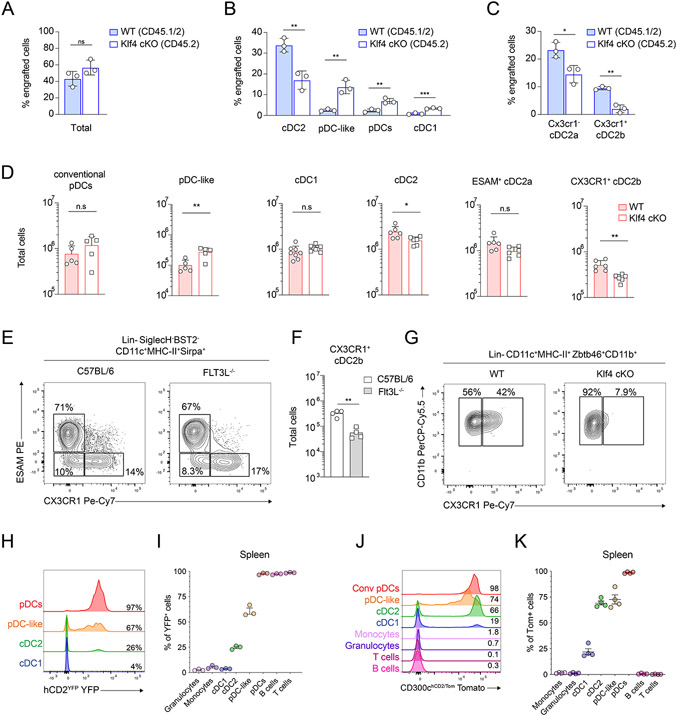
Absence of KLF4 translated transcriptionally into the accumulation of pDC-like cells next to conventional pDCs within the UMAP space (Fig. 5D) and resulted in a clear gap between cluster 2 and cluster 5 in the PCA projection, supporting the hypothesis of a developmental block (fig. S6G). Further, based on the ImmGen similarity heatmap (Fig. S5G), pDC-like cells had some transcriptional similarity with CDPs and GSEA highlighted enrichment for genes related to cell cycle and proliferation (Fig. 3J). Therefore, hypothesizing that absence of KLF4 would result in a developmental block that would prevent pDC-like cells to give rise to cDC2b, we compared WT and Klf4-cKO pDC and pDC-like cells for their potential to generate progeny. Sort purified pDC-like cells isolated from BM and spleen of WT (CD45.1/.2) and Klf4-cKO mice (CD45.2) were transferred in competitive settings into sublethally irradiated congenic CD45.1 recipient mice. pDC which do not survive in vitro, when co-transferred remained as expected pDCs and there was no difference in the proportions of WT or Klf4-cKO derived pDC, suggesting that this TF had no evident impact on pDCs (fig. S7B and S7D). pDC-like cells isolated from KLF4-deficient or WT mice were equally efficient in generating progeny (Fig. 6A). However, donor-derived cDC2 were mostly of WT origin, suggesting an impaired ability of Klf4-cKO pDC-like cells to develop into cDC2 (Fig. 6B, fig. S7A and S7C). Importantly, when cDC2 progeny was subdivided into ESAM+ cDC2a and CX3CR1-expressing cDC2b, development of cDC2b from KLF4-deficient pDC-like cells was almost completely abrogated, suggesting that KLF4 is essential for the generation of CX3CR1-expressing cDC2b (Fig. 6C, fig. S7A and 7C).
Figure 6. pDC-like cells require KLF4 for cDC2 development.
(A-C) pDC-like cells isolated from Cd45.1/.2Zbtb46GFP/WT (WT) and Cd45.2Zbtb46GFP/WT Klf4-cKO were co-transferred into sub-lethally irradiated recipient Cd45.1 recipient mice. Shown are percentages of total donor-derived cells (A) or percentage donor-derived cDC2, pDC-like, pDCs and cDC1 cells (B), or CX3CR1− cDC2a and CX3CR1+ cDC2b cells (C). (n=3 mice, three independent experiments, each dot represents a sample, and thin lines represent the mean ± s.d.). (D) Shown are total cells present in Klf4fl/flCD11ccre− (WT) or Klf4fl/flCD11cCre+ (Klf4-cKO) mice. Each subset is gated as shown in Fig 1D and fig. S2A. (n=6 mice, three independent experiments, each dot represents a sample, and thin lines represent the mean ± s.d.). (E) Shown is the expression of CX3CR1 and ESAM on splenic cDC2 pre-gated as 7-AAD−Lin−SiglecH−Bst2−CD11c+MHC-II+ obtained from C57BL/6 or Flt3L−/− mice. (F) Total cell number of splenic cDC2b obtained from C57BL/6 and Flt3L−/− mice. (G) Sort purified splenic pDC-like cells obtained from Zbtb46GFP/WTKlf4fl/flCre− or Zbtb46GFP/WTKlf4-cKO mice were cultured for four days in presence of FLT3L. Shown is the expression of CX3CR1 on cDC2 pre-gated as 7-AAD−Lin−SiglecH−Bst2−CD11c+MHC-II+GFP+CD11b+. (H-I) The expression of YFP as single-color histograms (H) or cumulative bar graph (I) is shown for the indicated subsets on splenocytes isolated from hCD2YFP mice. (J-K) Splenocytes isolated from CD300chCD2/Tom mice were analyzed for the expression of Tomato. Shown are single-color histograms (J) and cumulative bar graphs (K) of Tomato positive cells across the indicated immune cell subsets gated as shown in Fig. 1D or as CD19+B220+B cells, CD3+ T cells, Ly6ChiCD11b+ monocytes and Ly6CintCD11b+ granulocytes. Statistical analysis was done with unpaired two-tailed t-test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
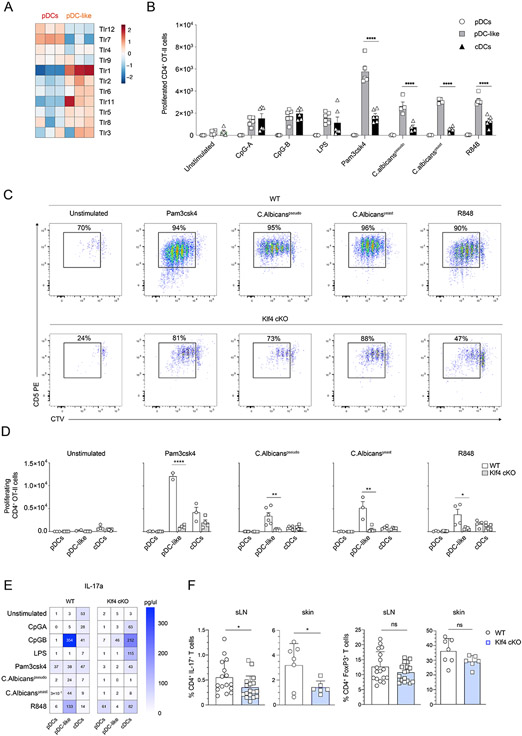
Figure 7. KLF4-dependent cDC2 regulate homeostatic Th17 and Tregs in the skin.
(A) Relative expression heat map on pDCs and pDC-like cells sorted and analyzed as in Fig.3A for the indicated TLRs. (B) OT-II T cells were co-cultured in the presence of activate cDCs, pDCs or pDC-like cells isolated from Zbtb46GFP/WT (WT) mice as indicated. Shown are total cell numbers of proliferating OT-II T cells gated as CD4+TCRβ+CD5+CTVint/neg at day 4. (n=4-6 mice, two independent experiments, each dot represents a sample, and thin lines represent the mean ± s.d.). (C-D) OT-II T cells were co-cultured in the presence of activated cDCs, pDCs or pDC-like cells isolated from Zbtb46GFP/WT (WT), or Klf4fl/flCD11cCre+Zbtb46GFP/WT (Klf4-cKO) as indicated. (C) Shown are representative flow cytometry plots showing the expression of CD5 and CTV on OT-II T cells. (D) Shown are total cell numbers of proliferating OT-II T cells gated as CD4+TCRβ+CD5+CTVint/neg. (E) CD4+ T cells obtained from DBA/2 mice were co-cultured in the presence of activated cDCs, pDCs or pDC-like cells isolated from Zbtb46GFP/WT (WT) or Klf4fl/flCD11cCre+Zbtb46GFP/WT (Klf4-cKO) mice, as indicated. The concentration of IL-17a was measured in the supernatant using the BioLegend T Helper cytometric bead array panel according to the manufacturer’s instructions (n=3 mice, three independent experiments). (F) Ex-vivo isolated cells from the skin or the sLNs obtained from Klf4fl/flCD11cCre−Zbtb46GFP/WT (WT) or Klf4fl/flCD11cCre+Zbtb46GFP/WT (Klf4-cKO), as indicated, were stimulated with PMA, ionomycin and brefeldin A and analyzed for the percent CD4+IL-17+ T cells (left) or CD4+Foxp3+Tregs (right). Statistical analysis was done with unpaired two-tailed t-test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Surprisingly, under homeostatic conditions Klf4-cKO mice showed higher numbers of pDC-like cells and equal numbers of pDCs as well as cDC2a, but consistent with the competitive transfer data, lower abundancy of CX3CR+ cDC2b in the spleen (Fig. 6D). Considering that cDC2b express several monocyte markers including CCR2 (Fig. 5C and fig. S6A), that pDC-like cells expanded in the presence of M-CSF (Fig. 4A), and that the expression of CX3CR1 and CCR2 was historically associated to monocytes and monocyte-derived DCs, we wanted to ensure their DC-lineage identity as a DC subset. Indeed, cDC2b were significantly reduced in Flt3l−/− mice, similar to all DC subsets (Fig. 6E-F and fig. S7E). FLT3L injection expanded all cDC subsets including CX3CR1+ cDC2b, confirming their dependency on FLT3-FLT3L signaling and thereby their DC lineage (fig. S7F-G). And when cultured with FLT3L alone or in combination with M-CSF, only Klf4-cKO but not WT pDC-like cells were impaired in their ability to differentiate into CX3CR1+ cDC2b but not cDC2a (Fig. 6G and fig. S7H-J).
Collectively, pDC-like cells are a cDC2 circulating precursor population, different than CDPs, that can generate ESAM+ cDC2a and CX3CR1+ cDC2b. Further, the development of cDC2b but not cDC2a from pDC-like cells is strictly dependent on KLF4.
hCD2Cre and Cd300Cre specifically label pDC-like cells and cDC2
To gain insights into the ontogeny of pDC-like derived cDC2, we looked for genes that would result in the labeling or tracing of pDC-like cells and allow us to track their progeny in vivo also under homeostatic conditions. We identified Cd300c as highly expressed within pDCs and pDC-like cells at single-cell resolution (fig. S7K) as well as in bulk RNA-seq datasets. We therefore generated a new mouse model by inserting the IRES-iCre cassette linked to the self-cleaving P2A peptide, followed by the extracellular domain of the human CD2 coding sequence (hCD2), in the 3’untranslated region of Cd300c, hereafter referred to as CD300iCre-hCD2 (fig. S7L). Staining with the anti-human CD2 antibody allowed us to trace the expression of Cd300c, while induction of the iCre cassette would allow to trace the progeny following recombination of loxP sites when crossed to a Rosa26Tomato line. As shown by hCD2 staining, all pDCs and 70% of pDC-like cells expressed the construct (fig. S7M).
In parallel, we explored the possibility that the transgenic hCD2-cre line 71 (hereafter referred to as hCD2Cre), in which cre-recombinase is driven by the human CD2 promoter, may potentially trace pDCs and pDC-like cells given that hCD2 was highly expressed in human pre-DCs35, 56. In this mouse model the expression of Cre cannot be followed as there is no reporter co-expression. However, to look at the progeny of pDC-like cells we crossed the hCD2Cre line to Rosa26YFP mice, generating hCD2YFP, and the Cd300ciCre-hCD2 line to Rosa26Tomato mice, generating the Cd300chCD2/Tomato mice. Direct comparison of the two lines showed YFP labeling on all lymphocytes, pDCs and on about 60-70% pDC-like cells in hCD2YFP mice (Fig. 6H-I). In Cd300chCD2/Tomato mice all pDCs, 75% pDC-like cells as well as cDC2s, and about 25% of cDC1 expressed Tomato (Fig. 6J-6K). These data corroborated the hypothesis that pDC-like cells are precursors to at least a fraction of cDC2. Moreover, the absence of labeling on CDPs in hCD2YFP mice suggested that the contribution of pDC-like cell to the cDC2 pool was at least 25%. However, since labeling within the pDC-like cells is not complete in either strain, stringent conclusions on the ontogeny of any DC subsets are not possible.
pDC-like derived cDC2 regulate homeostatic CD4+ Th17 in the skin-draining LNs
To understand the functional properties of pDC-like cells and their cDC2 progeny we determined their pathogen recognition receptor expression profiles and performed cognate T cell proliferation assays after activation using different types of stimulations. TLR profiles were highly different between pDCs and pDC-like cells with high relative expression of TLR1 in the latter, suggesting that pDCs were more prone to respond to viral products, while pDC-like cells to bacterial and fungal pathogens (Fig. 7A). We then compared T cell stimulatory potential for pDCs, cDCs and pDC-like cells activated with CpG, LPS, Pam3CSK4, Candida albicans, or resiquimod (R848). pDC-like cells were unexpectedly better at inducing T cell proliferation than cDCs not only after TLR1/2 stimulation using Pam3CSK4 but also following TLR7/8 agonist R848 and C. albicans that triggers TLR1/2/4 and 6 (Fig. 7B). TLR9 agonist CpG induced an equal OT-II T cell response by pDC-like cells and cDCs (Fig. 7B). Cross-presentation potential was as expected prominent in cDC1 but limited in other subsets if cells were stimulated appropriately in particular using TLR7 ligands (fig. S8A). The capacity of pDC-like cells to induce higher T cell proliferation as compared to cDCs was likely linked to their proliferative capacity, which would result in higher numbers of cDC2 within the culture following activation. To address this possibility, we looked at the endpoint of T cell proliferation for Zbtb46-GFP cells that identify all cDCs that remained live. As shown in fig. S7B, the number of GFP-expressing DCs was much higher in most conditions where pDC-like cells were used when compared to cDCs or pDCs.
Since pDC-like cells depend on KLF4 for their development into CX3CR1+ cDC2b, we asked whether the observed functional properties could be abrogated by the lack of this transcription factor, given that pDC-like cells would not be able to differentiate into cDC2b when activated. KLF4 deficiency almost completely abrogated the capacity of pDC-like cells to fully mature into CX3CR1+ cDC2b and thereby induce proliferation of T cells in all conditions (Fig. 7C-7D). cDC2 were previously shown to express and depend on the TF IRF4 25, 28. Surprisingly, despite the expression of IRF4 by pDC-like cells, their T cell stimulatory capacity was not affected in the absence of this TF (fig. S8C).
Stimulation with C. albicans, Pam3CSK4 and R848 will trigger different types of T cell polarization, such as Th17 or Th1 72. We therefore analyzed the cytokine production by pDC, cDC or pDC-like cell activated T cells in all conditions. T cells activated by pDC-like cells showed higher production of IL-17A, IL-17F, IL-22, IL-6 and IL-10 (Fig. 7E and fig. S8D) when compared to cDC-primed T cells, which more efficiently polarized towards IL-4, IL-9 and IFN- γ-producing T cells. T cells stimulated by either pDC-like cells or cDCs produced equal amounts of IL-2, IL-5, IL-21 and IL-13, while pDCs induced TNF-α production by T cells when activated via TLR7 and TLR9 ligands.
Lack of KLF4 and therefore inability to differentiate/mature into cDC2b abrogated T cell secretion of IL-17, IL-6, and IL-22, while reducing their capacity to produce IL-10 (Fig. 7E and fig. S8D). Surprisingly, cDCs from KLF4-deficient mice showed increased Th17 polarization when compared to WT cDC (Fig. 7E). It can be hypothesized that in the absence of pDC-like derived cDC2b the pool of cDC is unbalanced and may favor Th17 priming, potentially contributing to the defective Th2 response previously observed in Klf4-cKO mice 44.
The capacity of pDC-like cells to differentiate into cDCs, to respond to a variety of stimuli, activate T cells, and to proliferate in response to homeostatic cytokines such as FLT3L and M-CSF, prompted us to determine if this subset was relevant in controlling the homeostatic triggering that occurs in response to microbiota at epithelial sites, such as skin. We detected a significant reduction of Th17 polarized T cells at steady state within skin-draining LNs (Fig. 7F), but no differences in γδ+ IL-17 T cells (fig. S8E). Importantly in the context of strong inflammation, such as in imiquimod-induced contact hypersensitivity, ear thickness as well as T cell activation was not impaired (fig. S8F).
Collectively we believe that Th17 priming may occur by pDC-like-derived as well as CDP-derived cDC2b. Furthermore, these results suggest that KLF4-dependent cDC2b likely control homeostatic Th17 induction, but that under strong inflammatory settings CDP-derived cDCs can compensate for the lack of this subset.
Discussion
In this study, we phenotypically, developmentally, and functionally characterized the recently identified subset referred to as pDC-like cells. In human PBMCs, a corresponding population was independently described by two groups as Axl+Siglec6+ DCs (AS-DC)37 and as IL-3R+CD2+CX3CR1+CD33+ pre-DCs35. Several other studies identified a small subset of cells expressing cDC and pDC markers in mice33, 34, 42, 43, 73, however, the developmental and functional properties of these cells remained to be addressed. We show that pDC-like cells are a circulating precursor subset that will undergo proliferation and mostly differentiate to cDC2 (Fig. 4D and 6A-K). Based on multiple lines of evidence, pDC-like cells can be ascribed to the DC lineage, despite the expression of several monocyte markers, such as CX3CR1, CCR2 and CD14. The progeny of pDC-like cells comprises mostly cDC2 which can be further subdivided by the expression of ESAM and CX3CR1 into 3 subsets: ESAM−CX3CR1+; ESAM−CX3CR1− and ESAM+CX3CR1−. In human studies, the expression of CX3CR1 was mostly associated with monocyte-derived as well as iDCs that shared monocyte and DC features and were referred to as DC3 35, 37, 66. In mice, the nomenclature and subset characterization became controversial and without a broad consensus. What is well recognized is that the cDC2 compartment segregates into Notch2-dependent ESAM+ cDC2 and Notch2-independent ESAM− cDC2. Within the Notch2-independent cDC2 subset, we and others identified the CD11b−CD24− double negative (DN) subset in the skin-draining LNs69 and showed that their development is KLF4-dependent 44. In a recent report, Rudensky’s group characterized the transcriptional profile of DCs by single cell transcriptomics. They identified multiple cDC2 clusters: the Tbx21+ cDC2a, which were shown to be part of the ESAM+ cDC2, and the Rorc-expressing cDC2b subset that is included within ESAM− cDC2 39. During inflammatory conditions the subset discrimination becomes even more complex 38. However, Tbx21+ cDC2a were suggested to be anti-inflammatory while Rorc+ cDC2b pro-inflammatory, based on reduced up-regulation of CD86 and MHC-II upon LPS stimulation39. These Rorc+ cDC2b cells seem to correspond to our cluster 5 given the expression of Clec10a, Clec12a, Ccr2 and Cx3cr1 (Fig. 5b and fig. S5a) 39. Cluster 5 shares features with inflammatory DCs (iDCs) and DC3 38, 66, 74, such as the expression of monocyte markers. Nevertheless, independently of their nomenclature, CX3CR1+ cDC2b could be generated from pDC-like cells and this developmental pathway was abrogated in the absence of KLF4. But lack of this TF did not alter their differentiation capacity to cDC2a. Nevertheless, the contribution to the mature DC pool by CDP and pDC-like cells remains an open question. To discriminate CDP from pDC-like derived cDC2, we generated a new Cd300ciCre-hCD2 lineage tracer/reporter mouse model. However, since pDC-like cells were not all labelled it remains impossible to determine if unlabeled DC derive from CDPs or unlabeled pDC-like cells. Nevertheless, the fact that pDC-like cells and cDC2 are equally labelled suggests that a fraction of cDC2 is indeed pDC-like cell derived. It remains possible, though unanswered, that pDC-like cells originate from CDP. Given that hCD2Cre lineage tracer labels all pDCs, most pDC-like cells and about 25% of cDC2 but no cDC1, we can hypothesize that at least all labeled cDC2 are pDC-like derived, since CD115+ CDPs are unlabeled in these mice. It is nevertheless important to mention that cDC2 could also be derived from common lymphoid progenitors (CLPs) 75 that are partially labeled in hCD2YFP mice76. Further studies will be necessary to systematically address the ontogeny of each DC subset and clearly dissect the branching at the progenitor level.
The development or differentiation of pDC-like cells into cDCs occurs in response to the homeostatic cytokines such as FLT3L and M-CSF, suggesting that the pool of generated cells is constantly replenished from circulating pDC-like precursors. It is intriguing that the highest abundance of pDC-like cells that also express CX3CR1, across all tissues analyzed, is within the heart and the whole brain including meninges, where homing requires this chemokine. It is possible to envision that in the absence of inflammation pDC-like cells and their progeny maintain an anti-inflammatory environment to preserve tissue integrity.
Using Klf4-cKO mice that have impaired cDC2 development from pDC-like cells, we were able to partially dissect their functional properties and show that polarization of T cells stimulated by pDC-like-derived cDCs was towards IL-17A- and IL-10-producing T cells. KLF4 deficiency completely abrogated the capacity of pDC-like-derived DCs to activate T cells and to produce polarizing cytokines, likely because cDC2b differentiation was impaired. However, since cDC2a could still differentiate in the absence of KLF4, it is possible to speculate that the signals driving T cell priming are the result of complex interactions from multiple DC subsets competing for antigen and growth factors, further complicating the functional map of DC subsets. It also remains questionable if pDC-like cells are bona fide circulating precursors or if they can be defined as an immature DC subset that will upregulate functional markers upon stimulation, similar to circulating monocytes which acquire different functional properties depending on the type of activation. We would rather suggest that these cells are precursors, based on their increased capacity to proliferate and survive in culture when compared to pDCs, as well as their localization within the BM. Nevertheless, describing them as an immature DC subset or DC progenitors remains mostly a semantic question, until more stringent lineage tracers are developed.
Under homeostatic conditions, the impairment of cDC2 development from pDC-like cells resulted in reduced IL-17A-producing CD4+ T cells in vivo (Fig. 7F). KLF4 was previously described as required for the development of skin CD11b−CD24− DN cDC2, and essential to promote Th2 immunity44, 69, 77. More recently the group of Ronchese showed that Th2 driven immunity by DN cDC2 required IL-13 signaling for the induction of KLF4 via STAT6. While we hypothesized that migratory DN cDC2 correspond to cluster 11 by single cell transcriptomic analysis, we observed selective expression of Il13ra1 by cluster 5 or cDC2b, which identified with pDC-like-derived cells. It is possible to envision that maturation of pDC-like cells to an IL-13 responsive cDC2 subset may require KLF4 (figure S8F). However, an alternative explanation is that the lack of KLF4 within the cDC compartment could shift the balance between Th17 and Th2 priming. We show that under homeostatic conditions lack of KLF4 impairs Th17 formation. However, upon inflammation Th17 priming by KLF4-deficient cDCs is increased as shown by the in vitro data (Fig. 7E). Therefore, it is possible that in the absence of KLF4 the increased propensity of cDCs for Th17 polarization may prevent Th2 immunity. Future studies will be required to specifically address this aspect.
Collectively, we here show that pDC-like cells are circulating progenitors, which differentiate into cDC2 under homeostatic conditions. This developmental pathway is strictly dependent on the expression of the TF KLF4, highlighting the heterogeneity of the DC compartment as well as its transcriptional complexity. Furthermore, pDC-like-derived cDC2 prime Th17 and IL-22-producing T cells. In the absence of KLF4 the pool of Th17 cells is reduced at epithelial sites, such as the skin, suggesting that this DC subset is responsible for the homeostatic activation of Th17 cells.
Materials and Methods
Mice
All animals were bred and maintained in a specific pathogen-free animal facility according to institutional guidelines (Veterinäramt BS, License number 2786_26606 and NIH license number 19-896 and animal study proposal LHIM3E). Mice of the following genotypes as well as their intercrosses were bred in house: C57BL/6J, Klf4fl/flCD11cCre, Irf4fl/flCD11cCre, Zbtb46GFP/WT, hCD2creRosa26YFP. Klf4fl/fl mice were obtained from MMRC (MMRRC line 29877) and backcrossed to Itgax-cre and Zbtb46GFP/WT. Sperm of hCD2cre mice was obtained from R. Treisman Lab and used for mouse rederivation. For BM chimera experiments, the B6.SJL mice were purchased from Jackson Laboratories and maintained in house. Unless otherwise indicated, experiments used sex and age matched littermates between 6 to 14 weeks of age. CD300cicre-hCD2 were a generous gift of Prof. M. Colonna. CD300ciCre- hCD2 were generated at the University School of medicine in St. Louis. Briefly, IRES-iCre-P2A-hCD2 construct was inserted after the endogenous CD300c STOP codon using CRISPR-Cas9 technology. Correct integration of the reporter gene was confirmed by conventional and long-range PCR. The founders were bred to C57BL/6, and breeders were maintained as CD300ciCre-hCD2 heterozygous mice.
Immune cell harvest
Dendritic cells were harvested as follows: BM resident conventional pDCs and pDC-like cells were collected from femurs, tibia and pelvic bones by mechanical fragmentation of bones and single cell filtration. DCs from the spleen, LNs, thymus, lungs, liver, brain, Peyer’s patches (PP), and kidney were obtained by 30 min enzymatic digestion with Collagenase B (0.25 mg/ml) and DNase I (0.1 mg/ml) at 37°C. Red blood cell lysis of BM, spleen, lung, thymus, liver and kidney cell suspensions was performed using ACK lysis buffer. Mononuclear cells from brain were further enriched by a Percoll® gradient (70/30%). Intra epithelial lymphocytes (IEL) were obtained by mechanical separation. Lamina propria (LP) cells were obtained by 60 min enzymatic digestion with 0.25 mg/ml Collagenase 8 (Sigma) and 0.1 mg/ml DNase I (Sigma) at 37°C. All samples were filtered through a 70 μm cell strainer and used for further analysis. Single-cell suspension from the skin-draining lymph nodes or the ear pinnae were isolated as previously described 78. Conventional pDCs (Lin−Bst2+SiglecH+Zbtb46−), pDC-like cells (Lin−Bst2+SiglecH+Zbtb46+), cDC1 (Lin−Bst2−SiglecH−CD11c+MHC-II+SIRP-α−XCR1+) and cDC2 (Lin−Bst2−SiglecH−CD11c+MHC-II+SIRP-α−XCR1−) were sorted as indicated in Figure 1D (Lineage markers used are CD3, CD19). For single cell RNA-seq, CD11c+ cells from the spleen, sLN and mLN were enriched and sort purified as follows: Lin− CD11c+. For cell sorting, a BD FACSAria II with a custom built-in violet laser was used. Cells were sorted into PBS supplemented with 0.5% BSA and 2.5 mM EDTA. Cell purities of at least 95% were confirmed by post-sort analysis.
Infinity Flow
ACK lysed splenocytes from three Zbtb46GFP/WT mice were isolated and enriched for CD19− CD3− cells. Cells were stained for 30 min at 4°C with the following 9 backbone antibodies and then washed: 7-AAD, SiglecH-AF647 (clone 551), CD11c-APC/Cy7 (clone N418), Bst2-PacBlue (clone 927), MHC-II-BV510 (clone M5/114.15.2), CD11b-BV785 (clone M1/70), XCR1-BV650 (clone ZET), CD19-Bio (clone 6D5), CD3-Bio (clone 17A2). The backbone labeled cells were then stained with 255 different PE-conjugated antibodies using the LegendScreen Mouse PE kit (BioLegend) following the manufacturer’s instructions. We used the Infinity Flow pipeline to combine overlapping flow cytometry panels and simultaneous analyze the co-expression patterns of hundreds of surface-expressed proteins across millions of individual cells as described in Becht et al. 54. Clustering was performed using the Flowsom clustering method, with the parameters FlowSOM metacluster k = 7.
May-Grünwald Giemsa staining
Cytospins of 5x103 sort-purified cells were stained with May-Grünwald Giemsa staining kits, according to the manufacturer’s instructions (Sigma Aldrich). Slides were air-dried, sealed with Eukit quick hardening mounting medium, (Sigma Aldrich) and images were taken using a Leica DMI 4000 microscope.
In vivo transfer
0.5x105-3x105 sort-purified CD45.2 conventional pDCs or pDC-like cells were injected i.v. into sublethally irradiated CD45.1 mice. Reconstitution capacity of progenitors was assessed 4 days after injection by FACS. For competitive transplantation experiments, 0.5x105-2x105 sort-purified CD45.1/.2Klf4WT/WTCD11cCre−Zbtb46GFP/WT or CD45.2/.2 Klf4fl/flCD11cCreZbtb46GFP/WT progenitors were co-injected intravenously at a 1:1 ratio into sublethal irradiated CD45.1 mice.
Cell culture
1x103-5x103 sort-purified DCs were cultured in IMDM (Gibco) supplemented with 10% fetal calf serum (MPbio). To induce pDC or cDC development, cells were cultured in the presence of 100 ng/ml recombinant hFLT3L. Recombinant mIL-7 (100 ng/ml), M-CSF (100 ng/ml), GM-CSF (100 ng/ml), mLIF (100 ng/ml) or IL-3 (50 ng/ml) was supplemented as indicated.
Allogeneic T cell stimulation:
4x103 sort-purified DC subsets were co-cultured with 2x104 CD4+ T cells derived from LNs of DBA/2 mice. The CD4 T cells were enriched by using anti-biotin Miltenyi beads. In short: total LN derived lymphocytes were stained for Lineage positive cells (MHC-II, B220, Ter119, CD11b, CD11c, Gr-1, NK1.1 and CD8), enriched with anti-biotin Miltenyi beads and further sort purified by gating on Lineage negative cells. The sort purified CD4+ T cells were labeled with CellTrace™ Violet (ThermoFisher Scientific) according to the manufacturer’s instructions. Supernatant was collected and proliferation rates were measured by flow after 4 days in culture. Supernatant was analyzed using the LEGENDplex murine Th Panel bead-based immunoassay (BioLegend).
OT-II proliferation assay:
2x103 sort-purified conventional pDCs, pDC-like cells and splenic cDCs of the indicated mouse strains were co-cultured with 1x104 labeled OT-II CD4+ T-cells in the presence of OVA-protein (1ug/ml) and stimulated with: LPS (500 ng/ml), CpG-A (6 μg/ml), CpG-B (6 μg/ml), Pam3CSK4 (300 ng/ml), heat inactivated C. albicans yeast or C. albicans pseudohyphae (both at 10x103 cfu/ml) and R848 (500 ng/ml). OT-II T cells were labeled with CellTrace™ Violet (ThermoFisher Scientific) according to the manufacturer’s instructions. Proliferation rates were measured after 4 days in culture. As controls, labeled and unlabeled OT-II T cells were cultured on anti-CD3 (1 μg/ml) and anti-CD28 (0.5 μg/ml) pre-coated wells.
Imiquimod treatment
Mice were treated daily for 5 consecutive days with 10 mg of 5% imiquimod (IMQ) cream (Aldara cream; Valeant Pharmaceuticals) topically applied on each ear. Ear-skin thickness was measured using a digital caliper (Mitutoyo) the day of the first IMQ application, then daily during IMQ application and the day after the last application of IMQ. For each mouse, a daily ear-skin-thickness value was calculated by averaging the thickness of both ears. The change in ear-skin thickness over time is reported as the change in ear-skin thickness (mean ± SEM) relative to baseline at day 0 (first day of IMQ application).
Antibodies and flow cytometry
Cells were stained as previously described10 using the antibodies listed in Table S1. Cells were analyzed on a BD LSR Fortessa and data were analyzed with FlowJo X software (TreeStar).
Quantitative PCR (qPCR)
RNA of sort-purified progenitors was extracted, and cDNA generated as previously described44. KAPA SYBR Fast universal qPCR kit (Kapa Biosystems) was used, and samples were run on Applied Biosystems StepOnePlus qPCR machine. The primers used are listed in Table S2.
Statistical analysis
Analysis of all data was done with an unpaired, two-tailed Student’s t-test or one-way ANOVA with Tukey post-test with a 95% confidence interval (Prism; GraphPad Software, Inc.). P values less than 0.05 were considered significant. *0.01 < P < 0.05; **0.001 < P < 0.01; ***P < 0.001; ****P < 0.0001.
Supplementary Material
Data file S1: DEGs across pDCs, pDC-like cells, cDC1 and cDC2.
Data file S2: Genes differentially expressed between pDC-like cells vs pDCs/cDC1/cDC2 depicted in the Volcano plots.
Data file S3: DEGs between WT and Klf4-cKO derived clusters.
Data file S4: Raw data file
Acknowledgments
We thank Marco Colonna for generously sharing the newly generated CD300cicre-hCD2 mice, R. Treisman for donating the sperm of hCD2cre mice, Florent Ginhoux and Etienne K. Becht for sharing the Infinity Flow pipeline, The Genome Engineering and iPSC Center (GEiC), Mike White, and the transgenic core facility at the Washington University in St. Louis, L. Davidson and the NIDCR Animal facility, A. Brülhart and the W1060 University of Basel animal facility, M. Abshari and the combined technical core at NIDCR, G. Capoferri, and C. Engdahl, M. Burgunder, S. Sikanjic and Joanne Shi for technical help, and C. Beisel and the Genomics Facility Basel (D-BSSE ETH Zürich). Calculations were performed at sciCORE (http://scicore.unibas.ch/).
Funding.
This work was supported in part by the SNF grants PP00P3_179056 and 310030_185193. PFR is currently sponsored by the SNF grants P2BSP3_191754 and P500PB_211033. This research was also supported by the Intramural Research Program of the NIH, NIDCR (ZIADE000752-02). NB, YB and MSL are supported by the Intramural Research Program of the NIH, NIAID. Finally, we would like to thank G. Trinchieri for insightful discussions and intellectual input into the study.
Footnotes
Competing interests. The authors declare no competing interests.
Data and materials availability. All RNA sequencing data are deposited at the Gene Expression Omnibus database as a Super Series GSE223842. Bulk RNA-seq data are available under accession number GSE223100. CITE-seq dataset are available under accession number GSE223841. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes
- 1.Cabeza-Cabrerizo M, Cardoso A, Minutti CM, Pereira da Costa M & Reis e Sousa C Dendritic Cells Revisited. Annu Rev Immunol 39, 131–166 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Eisenbarth SC Dendritic cell subsets in T cell programming: location dictates function. Nat Rev Immunol 19, 89–103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shortman K & Liu YJ Mouse and human dendritic cell subtypes. Nat.Rev.Immunol 2, 151–161 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Banchereau J et al. Immunobiology of dendritic cells [Review]. Annual Review of Immunology 18, 767–811 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Guilliams M et al. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat.Rev.Immunol 14, 571–578 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swiecki M & Colonna M The multifaceted biology of plasmacytoid dendritic cells. Nat Rev Immunol 15, 471–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy TL et al. Transcriptional Control of Dendritic Cell Development. Annu Rev Immunol 34, 93–119 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson DA 3rd, Dutertre CA, Ginhoux F & Murphy KM Genetic models of human and mouse dendritic cell development and function. Nat Rev Immunol 21, 101–115 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tussiwand R & Gautier EL Transcriptional Regulation of Mononuclear Phagocyte Development. Front Immunol 6, 533 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodrigues PF & Tussiwand R Novel concepts in plasmacytoid dendritic cell (pDC) development and differentiation. Mol Immunol 126, 25–30 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Reizis B Plasmacytoid Dendritic Cells: Development, Regulation, and Function. Immunity 50, 37–50 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allman D et al. Ikaros is required for plasmacytoid dendritic cell differentiation. Blood 108, 4025–4034 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alculumbre SG et al. Diversification of human plasmacytoid predendritic cells in response to a single stimulus. Nat Immunol 19, 63–75 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Roquilly A, Mintern JD & Villadangos JA Spatiotemporal Adaptations of Macrophage and Dendritic Cell Development and Function. Annu Rev Immunol (2022). [DOI] [PubMed] [Google Scholar]
- 15.Satpathy AT et al. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. Journal of Experimental Medicine 209, 1135–1152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanno Y, Levi BZ, Tamura T & Ozato K Immune cell-specific amplification of interferon signaling by the IRF-4/8-PU.1 complex. J.Interferon Cytokine Res 25, 770–779 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Hildner K et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science 322, 1097–1100 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferris ST et al. cDC1 prime and are licensed by CD4(+) T cells to induce anti-tumour immunity. Nature 584, 624–629 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tussiwand R et al. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature 490, 502–507 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashok D et al. Cross-presenting dendritic cells are required for control of Leishmania major infection. European Journal of Immunology 44, 1422–1432 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Everts B et al. Migratory CD103+ dendritic cells suppress helminth-driven type 2 immunity through constitutive expression of IL-12. J Exp Med 213, 35–51 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bottcher JP et al. NK Cells Stimulate Recruitment of cDC1 into the Tumor Microenvironment Promoting Cancer Immune Control. Cell 172, 1022–1037 e1014 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Theisen DJ et al. WDFY4 is required for cross-presentation in response to viral and tumor antigens. Science 362, 694–699 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kretzer NM et al. RAB43 facilitates cross-presentation of cell-associated antigens by CD8alpha+ dendritic cells. J Exp Med 213, 2871–2883 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Persson EK et al. IRF4 Transcription-Factor-Dependent CD103(+)CD11b(+) Dendritic Cells Drive Mucosal T Helper 17 Cell Differentiation. Immunity 38, 958–969 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Guilliams M et al. Unsupervised High-Dimensional Analysis Aligns Dendritic Cells across Tissues and Species. Immunity 45, 669–684 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shin JY, Wang CY, Lin CC & Chu CL A recently described type 2 conventional dendritic cell (cDC2) subset mediates inflammation. Cell Mol Immunol 17, 1215–1217 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlitzer A et al. IRF4 Transcription Factor-Dependent CD11b(+) Dendritic Cells in Human and Mouse Control Mucosal IL-17 Cytokine Responses. Immunity 38, 970–983 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao Y et al. Control of T helper 2 responses by transcription factor IRF4-dependent dendritic cells. Immunity 39, 722–732 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bajana S, Roach K, Turner S, Paul J & Kovats S IRF4 promotes cutaneous dendritic cell migration to lymph nodes during homeostasis and inflammation. Journal of Immunology 189, 3368–3377 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams JW et al. Transcription factor IRF4 drives dendritic cells to promote Th2 differentiation. Nat.Commun 4, 2990 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nutt SL & Chopin M Transcriptional Networks Driving Dendritic Cell Differentiation and Function. Immunity 52, 942–956 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Leylek R et al. Integrated Cross-Species Analysis Identifies a Conserved Transitional Dendritic Cell Population. Cell Rep 29, 3736–3750 e3738 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alcantara-Hernandez M et al. High-Dimensional Phenotypic Mapping of Human Dendritic Cells Reveals Interindividual Variation and Tissue Specialization. Immunity 47, 1037–1050 e1036 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.See P et al. Mapping the human DC lineage through the integration of high-dimensional techniques. Science 356 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dutertre CA et al. Single-Cell Analysis of Human Mononuclear Phagocytes Reveals Subset-Defining Markers and Identifies Circulating Inflammatory Dendritic Cells. Immunity 51, 573–589 e578 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Villani AC et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bosteels C et al. Inflammatory Type 2 cDCs Acquire Features of cDC1s and Macrophages to Orchestrate Immunity to Respiratory Virus Infection. Immunity 52, 1039–1056 e1039 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown CC et al. Transcriptional Basis of Mouse and Human Dendritic Cell Heterogeneity. Cell 179, 846–863 e824 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maier B et al. A conserved dendritic-cell regulatory program limits antitumour immunity. Nature 580, 257–262 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alculumbre S et al. Plasmacytoid pre-dendritic cells (pDC): from molecular pathways to function and disease association. Semin Cell Dev Biol 86, 24–35 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Schlitzer A et al. Identification of CCR9− murine plasmacytoid DC precursors with plasticity to differentiate into conventional DCs. Blood 117, 6562–6570 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Rodrigues PF et al. Distinct progenitor lineages contribute to the heterogeneity of plasmacytoid dendritic cells. Nat Immunol 19, 711–722 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tussiwand R et al. Klf4 expression in conventional dendritic cells is required for T helper 2 cell responses. Immunity 42, 916–928 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Onai N et al. Identification of clonogenic common Flt3(+) M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nature Immunology 8, 1207–1216 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Liu K et al. Origin of dendritic cells in peripheral lymphoid organs of mice. Nat Immunol 8, 578–583 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Naik SH et al. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat Immunol 8, 1217–1226 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Onai N et al. A clonogenic progenitor with prominent plasmacytoid dendritic cell developmental potential. Immunity 38, 943–957 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Grajales-Reyes GE et al. Batf3 maintains autoactivation of Irf8 for commitment of a CD8alpha(+) conventional DC clonogenic progenitor. Nat Immunol 16, 708–717 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schlitzer A et al. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat Immunol 16, 718–728 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Matsui T et al. CD2 distinguishes two subsets of human plasmacytoid dendritic cells with distinct phenotype and functions. J Immunol 182, 6815–6823 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schlitzer A et al. Tissue-specific differentiation of a circulating CCR9− pDC-like common dendritic cell precursor. Blood 119, 6063–6071 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Lutz K et al. Ly6D(+)Siglec-H(+) precursors contribute to conventional dendritic cells via a Zbtb46(+)Ly6D(+) intermediary stage. Nat Commun 13, 3456 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Becht E et al. High-throughput single-cell quantification of hundreds of proteins using conventional flow cytometry and machine learning. Sci Adv 7, eabg0505 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herman JS, Sagar & Grun D FateID infers cell fate bias in multipotent progenitors from single-cell RNA-seq data. Nat Methods 15, 379–386 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Dress RJ et al. Plasmacytoid dendritic cells develop from Ly6D(+) lymphoid progenitors distinct from the myeloid lineage. Nat Immunol 20, 852–864 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Liu K et al. In vivo analysis of dendritic cell development and homeostasis. Science 324, 392–397 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bagadia P et al. An Nfil3-Zeb2-Id2 pathway imposes Irf8 enhancer switching during cDC1 development. Nat Immunol 20, 1174–1185 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meredith MM et al. Zinc finger transcription factor zDC is a negative regulator required to prevent activation of classical dendritic cells in the steady state. Journal of Experimental Medicine 209, 1583–1593 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yanez A et al. Granulocyte-Monocyte Progenitors and Monocyte-Dendritic Cell Progenitors Independently Produce Functionally Distinct Monocytes. Immunity 47, 890–902 e894 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hettinger J et al. Origin of monocytes and macrophages in a committed progenitor. Nat.Immunol 14, 821–830 (2013). [DOI] [PubMed] [Google Scholar]
- 62.Naik SH et al. Intrasplenic steady-state dendritic cell precursors that are distinct from monocytes. Nat Immunol 7, 663–671 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Fogg DK et al. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science 311, 83–87 (2006). [DOI] [PubMed] [Google Scholar]
- 64.Cisse B et al. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development 1. Cell 135, 37–48 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoshida H et al. The cis-Regulatory Atlas of the Mouse Immune System. Cell 176, 897–912 e820 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bourdely P et al. Transcriptional and Functional Analysis of CD1c(+) Human Dendritic Cells Identifies a CD163(+) Subset Priming CD8(+)CD103(+) T Cells. Immunity 53, 335–352 e338 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Satpathy AT et al. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat.Immunol 14, 937–948 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lewis KL et al. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity 35, 780–791 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Connor LM, Tang SC, Camberis M, Le Gros G & Ronchese F Helminth-Conditioned Dendritic Cells Prime CD4+ T Cells to IL-4 Production In Vivo. Journal of Immunology (2014). [DOI] [PubMed] [Google Scholar]
- 70.Vestre K et al. Rab7b regulates dendritic cell migration by linking lysosomes to the actomyosin cytoskeleton. J Cell Sci 134 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhumabekov T, Corbella P, Tolaini M & Kioussis D Improved version of a human CD2 minigene based vector for T cell-specific expression in transgenic mice. J Immunol Methods 185, 133–140 (1995). [DOI] [PubMed] [Google Scholar]
- 72.Sallusto F & Lanzavecchia A Heterogeneity of CD4+ memory T cells: functional modules for tailored immunity. Eur J Immunol 39, 2076–2082 (2009). [DOI] [PubMed] [Google Scholar]
- 73.Bar-On L et al. CX3CR1+ CD8alpha+ dendritic cells are a steady-state population related to plasmacytoid dendritic cells. Proc.Natl.Acad.Sci.U.S A 107, 14745–14750 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cabeza-Cabrerizo M et al. Recruitment of dendritic cell progenitors to foci of influenza A virus infection sustains immunity. Sci Immunol 6, eabi9331 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sathe P, Vremec D, Wu L, Corcoran L & Shortman K Convergent differentiation: myeloid and lymphoid pathways to murine plasmacytoid dendritic cells. Blood 121, 11–19 (2013). [DOI] [PubMed] [Google Scholar]
- 76.Siegemund S, Shepherd J, Xiao C & Sauer K hCD2-iCre and Vav-iCre mediated gene recombination patterns in murine hematopoietic cells. PLoS One 10, e0124661 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mayer JU et al. Homeostatic IL-13 in healthy skin directs dendritic cell differentiation to promote TH2 and inhibit TH17 cell polarization. Nat Immunol 22, 1538–1550 (2021). [DOI] [PubMed] [Google Scholar]
- 78.Naik S et al. Compartmentalized control of skin immunity by resident commensals. Science 337, 1115–1119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gaidatzis D, Lerch A, Hahne F & Stadler MB QuasR: quantification and annotation of short reads in R. Bioinformatics 31, 1130–1132 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lun AT, Bach K & Marioni JC Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol 17, 75 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ritchie ME et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Germain PL, Lun A, Garcia Meixide C, Macnair W & Robinson MD Doublet identification in single-cell sequencing data using scDblFinder. F1000Res 10, 979 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data file S1: DEGs across pDCs, pDC-like cells, cDC1 and cDC2.
Data file S2: Genes differentially expressed between pDC-like cells vs pDCs/cDC1/cDC2 depicted in the Volcano plots.
Data file S3: DEGs between WT and Klf4-cKO derived clusters.
Data file S4: Raw data file