Abstract
Human-ribosomal s6 kinase 1 (h-RSK1) is an effector kinase of the Ras/MAPK signaling pathway, which is involved in the regulation of the cell cycle, proliferation, and survival. RSKs comprise two functionally distinct kinase domains at the N-terminal (NTKD) and C-terminal (CTKD) separated by a linker region. The mutations in RSK1 may have the potential to provide an extra benefit to the cancer cell to proliferate, migrate, and survive. The present study focuses on evaluating the structural basis for the missense mutations identified at the C-terminal kinase domain of human-RSK1. A total of 139 mutations reported on RSK1 were retrieved from cBioPortal, where 62 were located at the CTKD region. Furthermore, 10 missense mutations Arg434Pro, Thr701Met, Ala704Thr, Arg725Trp, Arg726Gln, His533Asn, Pro613Leu, Ser720Cys, Arg725Gln, and Ser732Phe were predicted to be deleterious using in silico tools. To our observation, these mutations are located in the evolutionarily conserved region of RSK1 and shown to alter the inter- and intramolecular interactions and also the conformational stability of RSK1-CTKD. The molecular dynamics (MD) simulation study further revealed that the five mutations Arg434Pro, Thr701Met, Ala704Thr, Arg725Trp, and Arg726Gln showed maximum structural alterations in RSK1-CTKD. Thus, based on the in silico and MD simulation analysis, it can be concluded that the reported mutations may serve as potential candidates for further functional studies.
1. Introduction
The Ras/mitogen-activated protein kinase (MAPK) pathway regulates fundamental cellular processes like cell cycle control and progression, apoptosis, cell proliferation, and differentiation.1−3 The ribosomal S6 kinases (RSKs) act downstream to the Ras/MAPK signaling pathway to generate the ultimate functional response. In humans, four RSK isoforms, termed RSK1, RSK2, RSK3, and RSK4, and two structural homologs, mitogen and stress-activated kinase (MSK1 and MSK2), have been reported.4 All the RSK isoforms share 70–80% sequence identity and are composed of two distinct nonidentical functional kinase domains at the N-terminal and C-terminal separated by a linker of ∼100 amino acids.4,5 The C-terminal kinase domain (CTKD) belongs to the calcium/calmodulin-dependent protein kinase (CaMK) family, and the N-terminal kinase domain belongs to the AGC kinase family, which interacts with different substrates upon activation.5−7 Extracellular signal-regulated kinase 1/2 (ERK1/2) is the activator of RSKs and interacts with the CTKD on binding at the ERK docking site (D-domain). The C-terminal tail (CTT) adjacent to the CTKD is reported to play a regulatory role in RSK activity as well as ERK-RSK interactions.8,9 The crystal structure of the CTKD was observed to have a small N-lobe rich in β-sheets and α-helices, and a large C-lobe rich in α-helices.7 RSK activation cascade involves a series of phosphorylation events. Activation starts after the binding of ERK1/2, which phosphorylates Thr 573 in the activation loop of the CTKD, followed by autophosphorylation of Ser 380 located in the linker region. This further leads to the recruitment of phosphoinositidine-dependent kinase 1 (PDK1) on the NTKD, which phosphorylates Ser 221 of the activation loop at the NTKD, thereby leading to the complete activation of RSK1.4,5,9 RSKs are ubiquitously expressed in human tissues and are reported to be functionally redundant. Intriguingly, in cancer conditions, RSK1 and RSK2 lead to tumor growth and promotion, while RSK3 and RSK4 act as tumor suppressors.10 However, the role of different isoforms of RSKs in cancer development and progression is still not well-defined. Therefore, it is of utmost important to understand the functional aspects of all the RSK isoforms for better therapeutic interventions.11,12 Recently, the aberrant activity of RSK1 was reported in pancreatic cancer,7 nodular melanoma,13 breast cancer,14 prostate cancer,15 and high-grade glioma,16 and it acts as a key modulator of lung metastasis.17 Nevertheless, structural and functional consequences of cancer-associated missense mutations reported in human-RSK1 have not been evaluated. Therefore, understanding the pathogenicity and structural dynamics of reported missense mutations of RSK1 could provide better insights for functional studies and targeted inhibitor design.
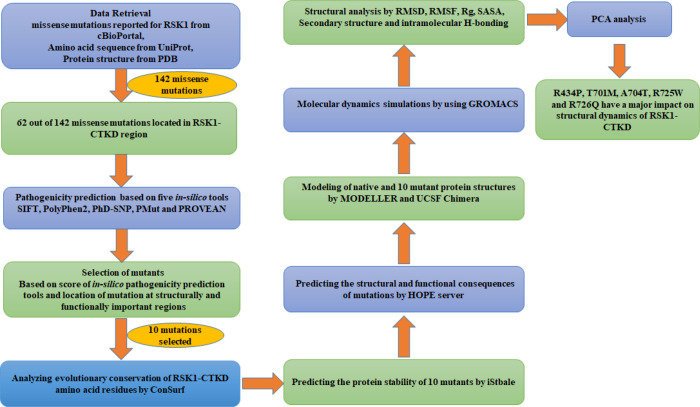
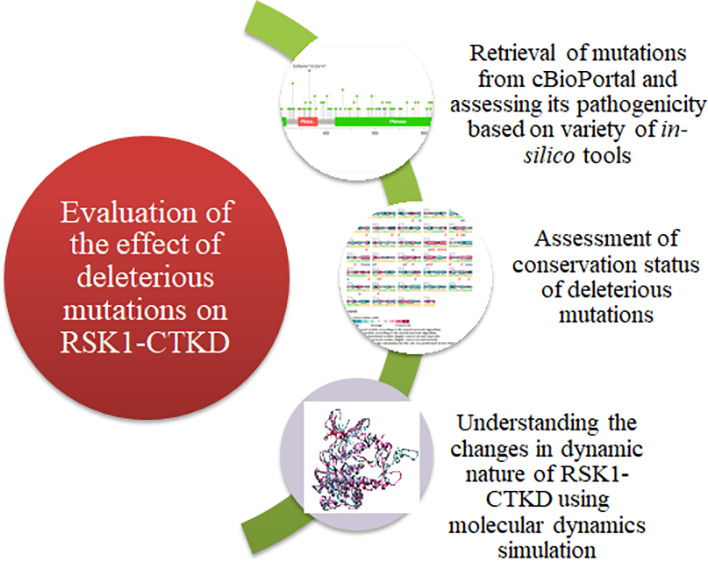
Recent advancement in in silico pathogenicity prediction tools has eased the process of screening large mutational data to prioritize and validate mutations using structural and functional aspects.18−20 Moreover, molecular dynamics simulation is a computational approach to explore the structural changes in protein structure brought about by mutation. In addition, these investigations contribute to the understanding of how mutations affect protein–protein interactions, folding pattern, and ligand binding for targeted drug design.21−23 Therefore, after screening the pathogenicity using different in silico tools, molecular dynamics (MD) simulation was performed to gain more insights into the effect of mutations on protein structure and dynamics.24−29 The CTKD plays a crucial role in RSK1 activation. Therefore, the present study aimed to understand the effect of cancer-associated missense mutations identified in h-RSK1-CTKD. The objectives of the current study are to use different in silico tools for screening the missense mutations identified in RSK1-CTKD and further explore molecular dynamics simulations to comprehend how putative pathogenic mutations affect the structural dynamics of proteins. Furthermore, based on available methodologies and in silico tools, the workflow was designed to screen the mutations located in h-RSK1-CTKD (Figure 1). The study reported here will provide the basis to understand the functional consequences of mutations identified in h-RSK1-CTKD.
Figure 1.
Workflow for mutation assessment.
2. Methods
2.1. Data Retrieval
Mutation data reported for h-RSK1 were retrieved from cBioPortal (https://www.cbioportal.org/), which is an open-access platform and the repository for all the mutations identified from a larger cohort of patient populations. It provides access to the molecular profile of tumor samples as well as clinical attributes from an extensive cancer study.30 The protein sequence of RSK1 was obtained from the UniProt database (UniProt ID Q15418)31 for in silico analyses. The three-dimensional structures of RSK1-CTKD and RSK1-ERK2 complexes were downloaded from the Protein Data Bank with PDB IDs 3RNY(7) and 4NIF(32) (https://www.rcsb.org/).33
2.2. In Silico Screening of Mutations Identified in h-RSK1
Different in silico tools such as SIFT (Sorting Intolerant From Tolerant) (https://sift.jcvi.org/www/SIFT_seq_submit2.html), PolyPhen2 (http://genetics.bwh.harvard.edu/pph2/), PhD-SNP (Predictor of human Deleterious Single Nucleotide Polymorphism) (https://snps.biofold.org/phd-snp/phd-snp.html), PMut (http://mmb.irbbarcelona.org/PMut/analyses/new/), and PROVEAN (http://provean.jcvi.org/seq_submit.php) were used to screen pathogenicity of mutations. SIFT predicts whether the substitution is deleterious or tolerated based on the location of substitution by the sequence alignment. If the alteration is at a highly conserved region, then it will be categorized as deleterious with a score less than 0.05 and as tolerated with a score more than 0.01.34 PolyPhen2 (Polymorphism Phenotyping v2) utilizes an integrative approach of structural trait and sequence alignment to generate an evolutionary conservation profile to sort mutations as benign, probably damaging, and possibly damaging.35 PhD-SNP generates a comparative conservation score of multiple sequence alignment, which further identifies the effect of an SNP as a disease or neutral.36 PMut is based on a neural network and predicts the pathogenicity of single-point mutation. The prediction score ranges from 0 to 1, and the cutoff value is set to be 0.5. The neutral substitution value ranges from 0 to 0.5, whereas the pathogenic is from 0.5 to 1.37 Based on the alignment score for a given protein query, PROVEAN predicts an effect of substitution on protein function. The PROVEAN cutoff scores were set to −2.5 to discriminate between deleterious and neutral substitutions.38
2.3. Analysis of Evolutionary Conserved Amino Acid Sequences of h-RSK1
The amino acid sequence of h-RSK1-CTKD was analyzed with ConSurf (https://consurf.tau.ac.il/), which predicts the evolutionary conservation status of amino acid residues by determining the phylogenetic relations. Along with the conservation status, ConSurf also provides the buried or exposed status of amino acid residues in the given sequences.39
2.4. Determining the Effect of Mutation on Protein Stability
The iStable (http://predictor.nchu.edu.tw/iStable/) tool was used to predict the effect of selected mutations on the structural stability of h-RSK1-CTKD. iStable is a support vector machine-based meta predictor, which predicts the effect of mutation on the stability of a protein structure. It provides a combined outcome from I-Mutant and MUpro and predicts the change in protein stability. The amino acid sequence for RSK1 and default iStable parameters were used to evaluate the stability of native protein conformation caused by mutations.40
2.5. Prediction of the Structural and Functional Impact of Mutation on RSK1-CTKD
Have Y(Our) Protein Explained (HOPE) (https://www3.cmbi.umcn.nl/hope/) is a fully automated program to predict the effect of mutation on protein structure and function. It predicts the disease-related phenotype as well as explains any sort of alterations in protein structure due to mutation.41 The amino acid sequence for RSK1 and mutations were used for HOPE prediction.
2.6. Modeling of the RSK1-CTKD Structure
A few regions in the RSK1-CTKD in PDB ID 3RNY structure were missing. Therefore, the complete RSK1-CTKD structure including activation loops (577–585) and phosphate-binding loops (P-loop, 425–431) along with Lys 438, Ala 439, and Thr 440 was reconstructed using MODELLER 10.0.42 The missing region of CTT (Ser 709 to Leu 735) was adopted from the AlphaFold model (AF-Q15418-F1)43 and used as a template to construct the complete structure of RSK1-CTKD using the homology modeling by MODELLER. All mutants were generated from the complete structure using the UCSF Chimera command line.44
2.7. Molecular Dynamics Simulations
All molecular dynamics simulations (MDs) were conducted for a time scale of 200 ns using the GROMACS 2019.5 package with the AMBER 99SB ILDN all-atom force field at 310 K.45 MD simulations were run on a DELL PowerEdge R740 Rack server machine comprising 40 physical processor cores within two 2nd generation Intel Xeon Gold scalable processors accelerated by an NVIDIA Tesla V100 Tensor Core graphic processor unit (GPU). Protein structures of each system were placed at the center of a cubic box in such a way that the distance between the protein surface and each side of the box was at least 5 Å. An explicit solvent TIP3P water model was used to hydrate all the systems within the box. The systems were neutralized by adding counterions, and energy minimizations were performed using the steepest descent minimizer for 50,000 steps. The simulations were performed under periodic boundary conditions with 50,000 steps of NVT (constant number of particles, volume, and temperature) followed by 50,000 steps of NPT (constant number of particles, pressure, and temperature) ensemble. During these position restraint equilibration runs, V-rescale and Berendsen’s coupling algorithms were used to keep the temperature (310 K) and pressure (1 bar) constant, respectively. Finally, 200 ns production MD runs were performed, allowing all molecules to move in all directions according to a classical Newtonian leap-frog MD integrator. The pressure of all the systems was maintained at 1 bar by isotropic pressure coupling in x, y, and z components to a Parrinello–Rahman barostat with a time constant τ = 2.0 ps and a compressibility of 4.5 × 10–5 bar–1 in all three dimensions. The time steps for the simulations were 2 fs, and the coordinates were stored every 10 ps. The analyses were performed using the GROMACS 2019.5 package.46−48
2.8. MD Trajectory Analysis and Visualization
Comparative examination of structural alterations between the RSK1-CTKD wild-type (WT) and mutants was performed by analyzing the MD trajectories. The root-mean-square deviation (RMSD), radius of gyration (Rg), root-mean-square fluctuations (RMSF), solvent-accessible surface area (SASA), secondary structure, and intramolecular hydrogen bonds were analyzed using gmx rms, gmx gyrate, gmx rmsf, gmx sasa, gmx do_dssp, and gmx hbond of GROMACS. The plots were generated using Grace software.
2.9. Principal Component Analysis
To identify the configuration space of anharmonic motion with only a few degrees of freedom, principal component analysis (PCA) or essential dynamics (ED) was used. The PCA extracts dominant modes in the overall molecular motion from MD trajectories. The motion of structures was identified by the most vital eigenvector projection in Cartesian trajectory coordinates. Cartesian principal components were calculated from a 3N × 3N covariance matrix of the three-dimensional coordinates of proteins’ backbone (N being the atom of the backbone) using the gmx covar tool of GROMACS. The eigenvectors and their corresponding eigenvalues were analyzed by the gmx anaeig tool to obtain representative eigenvectors that exhibited essential motion of biological relevance. The initial six eigenvectors and their corresponding eigenvalues were extracted from each trajectory, out of which the first two eigenvectors, commonly regarded as principal components (PCs) 1 and 2, were analyzed.
3. Results and Discussion
Overall, 221 mutations were reported for h-RSK1 in cBioPortal out of which 183 were missense mutations. However, some mutations were also reported more than once as they were associated with different cancer studies. Subsequently, sorting missense mutations resulted in the identification of 139 unique mutations. Finally, 62 out of 139 mutations were located in the CTKD region and chosen for in silico and MD analysis (Table S1).
3.1. Screening of Missense Mutations for Pathogenicity
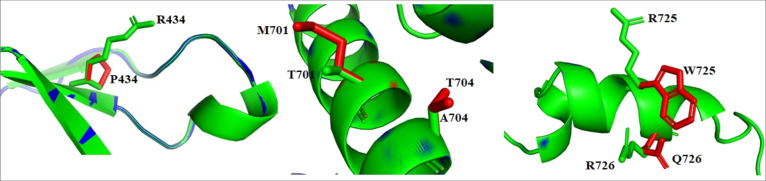
Five different in silico tools, namely, SIFT, PhD-SNP, PMut, PROVEAN, and PolyPhen2, were used to screen 62 missense variants located in RSK1-CTKD for pathogenicity. A total of 21 mutations were categorized as deleterious by at least four prediction tools. The overall kinase domain was found to be dynamically active and regulated by different structural and functional regions present within the domain.49,50 The P-loop region, catalytic loop, activation loop, and αL helix play a crucial role in kinase activity, and hence, the mutations associated with these regions may affect the functions of a kinase domain.7,49 h-RSK1-CTT (696–735) acts as a phospho-switch to regulate the interaction with ERK1/2 and subsequent activity of RSK1-CTKD.9 Therefore, out of 21 predicted pathogenic mutations, 10 mutations were observed to be associated with functionally important regions of the kinase domain (Table 1). Arg434 is located near the P-loop region, which plays an important role in ATP binding to the kinase domain. His533 in the catalytic loop plays an essential role in the kinase activity, whereas Pro613 may be involved in stabilizing the catalytic loop.7,49 Furthermore, Thr701Met, Ala704Thr, Ser720Cys, Arg725Trp, Arg725Gln, Arg726Gln, and Ser732Phe mutations are part of regulatory CTT and located at the binding interface of ERK-RSK. Though Ser732Phe was categorized as deleterious by only two in silico prediction tools, it has been reported that Ser732 undergoes autophosphorylation, playing a key role in the regulation of RSK1 interactions with ERK1/2.9 Side chain conformations of few mutants and the corresponding wild-type residue are shown in comparison in Figure 2.
Table 1. Prioritized Mutations and Pathogenicity Predicted by Five In Silico Tools.
| protein change | phD-SNP | PMut | PROVEAN | SIFT | PolyPhen2 |
|---|---|---|---|---|---|
| R434P | disease | disease | deleterious | deleterious | probably damaging |
| H533N | disease | disease | deleterious | deleterious | benign |
| P613L | disease | disease | deleterious | deleterious | probably damaging |
| T701M | disease | disease | deleterious | deleterious | probably damaging |
| A704T | disease | disease | deleterious | deleterious | probably damaging |
| S720C | neutral | disease | deleterious | deleterious | probably damaging |
| R725W | disease | disease | deleterious | deleterious | probably damaging |
| R725Q | disease | disease | deleterious | deleterious | possibly damaging |
| R726Q | disease | disease | deleterious | deleterious | benign |
| S732F | neutral | neutral | deleterious | deleterious | benign |
Figure 2.
Comparative representation of alteration in side chain conformation of WT (green) and mutant (red) residues represented in a stick model.
3.2. Mutations Located in the Evolutionarily Conserved Region Affect the Structural Stability
ConSurf was used to determine the evolutionary conservancy of amino acid residues of RSK1-CTKD. Selected 10 mutations were analyzed for the conservation status of their corresponding wild-type amino acid residue. His533, Ala704, and Thr701 were predicted to be highly conserved and buried. Residues like Pro613, Ser720, Arg725, Arg726, and Ser732 were predicted to be highly conserved and exposed, whereas only Arg434 was averagely conserved and exposed (Figure S1). The iStable tool determined the effect of mutations on the structural stability of h-RSK1-CTKD. The RSK1 Pro613Leu, Ser720Cys, and Ser732Phe mutants were reported to increase the structural stability, while the rest of the 7 mutations were predicted to decrease the stability (Table 2).
Table 2. Effect of Mutation on Protein Stability Predicted by iStable.
| protein change | iStable | iStable conf |
|---|---|---|
| R434P | decrease | 0.8 |
| H533N | decrease | 0.7 |
| P613L | increased | 0.5 |
| T701M | decrease | 0.5 |
| A704T | decrease | 0.7 |
| S720C | increased | 0.5 |
| R725W | decrease | 0.8 |
| R725Q | decrease | 0.8 |
| R726Q | decrease | 0.8 |
| S732F | increased | 0.5 |
3.3. Mutations Lead to the Loss of the h-RSK1-CTKD Interaction Network
The HOPE server revealed that selected mutations were located in functionally important regions and protein–protein interacting interfaces. Changes in hydrophobicity, charge, size of an amino acid residue, and disruption of hydrogen bonds were predicted for all 10 mutations. The positively charged wild-type residue was mutated into a neutral residue, and the loss of salt bridge interactions between Glu422 and Glu443 was predicted in Arg434Pro. His533Asn may create space and disrupt the interactions in the protein core as Asn is smaller than His. Alteration in Pro613 to Leu can create a structural distortion because of the larger size of a mutated residue than wild-type. Thr701Met, Ala704Thr, Ser720Cys, and Ser732Phe mutated to more hydrophobic residues resulting in the loss of hydrogen bonds and hydrophobic interactions, thereby affecting the structural integrity. Like Arg434Pro, mutations Arg725Trp, Arg725Gln, and Arg726Gln show alteration of positively charged wild-type residues into neutral mutant residues leading to the loss of external and internal interactions, thereby affecting the protein function and conformation (Table S2). Overall, the HOPE server predicted that the mutations alter the interaction network and structural and functional properties of an RSK1-CTKD.
3.4. Mutations Differentiate into Two Groups Based on Molecular Dynamics Simulations
To understand the structural and functional behavior of the mutations, molecular dynamics simulation was performed. Analyses of RMSD, Rg, RMSF, SASA, secondary structure, intramolecular hydrogen bonds, and PCA were done for wild-type and mutant proteins. All mutations were clustered into two groups based on the outcome of MD analyses. Mutations Arg434Pro, Thr701Met, Ala704Thr, Arg725Trp, and Arg726Gln showing major structural deviation from the wild-type were represented in one group. Meanwhile, the other group consisting of His533Asn, Pro613Leu, Ser720Cys, Arg725Gln, and Ser732Phe showed less structural deviation than the wild-type.
3.4.1. Mutation Alters the Stability of the h-RSK1-CTKD Structure
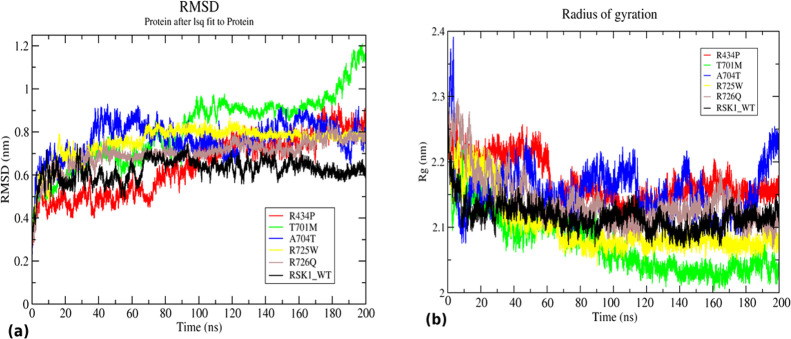
The RMSD value was calculated to understand the conformational stability of wild-type and mutants during the simulations. RMSD values of RSK1 Arg439Pro, Thr701Met, Ala704Thr, Arg725Trp, and Arg726Gln mutants and wild-type were compared (Figure 3a). The wild-type structure was observed to have slightly unstable RMSD values till 70 ns before stabilization. After 70 ns, the RMSD value was maintained at 0.6 nm throughout the simulation. Arg434Pro and Thr701Met showed very unstable RMSD throughout the simulation. The RMSD values of Arg434Pro and Thr701Met were gradually increased from 0.4 to 0.7 and 1 nm, respectively. Similarly, the RMSD value for the mutant Ala704Thr was observed to be 0.8 nm indicating unstable behavior during the simulation. Mutants Arg725Trp and Arg726Gln exhibited more stable but slightly higher RMSD than the wild-type maintaining a value at 0.7 nm. The time-averaged RMSD values were determined and ranked in increasing order as Thr701Met (0.81 nm) > Ala704Thr (0.76 nm) > Arg725Trp (0.75 nm) > Arg726Trp (0.69 nm) > Arg434Pro (0.64 nm) > WT (0.62 nm) (Table 3). Meanwhile, mutants His533Asn, Pro613Leu, Ser720Cys, Arg725Gln, and Ser732Phe exhibited reasonably stable RMSD, all of which were converged around 0.6–0.7 nm at the end of 200 ns (Figure S2a).
Figure 3.
Structural stability and compactness analysis of RSK1-CTKD WT (black) and mutants R434P (red), T701M (green), A704T (blue), R725W (yellow), and R726Q (brown). (a) RMSD of WT and mutants showing alteration in structural conformation and (b) Rg of WT and mutants indicating an alteration in compactness of a structure.
Table 3. Time-Averaged Structural Properties Calculated for Wild-Type, Arg434Pro, Thr701Met, Ala704Thr, Arg725Trp, and Arg726Glna.
| analyses | WT | R434P | T701M | A704T | R725W | R726Q |
|---|---|---|---|---|---|---|
| RMSD (nm) | 0.62 | 0.64 | 0.81 | 0.76 | 0.75 | 0.69 |
| RMSF (nm) | 0.22 | 0.25 | 0.27 | 0.26 | 0.22 | 0.21 |
| Rg (nm) | 2.11 | 2.16 | 2.07 | 2.15 | 2.10 | 2.13 |
| SASA (nm2) | 101.46 | 176.87 | 177.19 | 175.43 | 169.24 | 172.36 |
RMSD: root-mean-square deviation, RMSF: root-mean-square fluctuation, Rg: radius of gyration, and SASA: solvent-accessible surface area.
RSK1 wild-type was observed to have a lower average RMSD value as compared to Arg439Pro, Thr701Met, Ala704Thr, Arg725Trp, and Arg726Gln mutants. Based on the RMSD analysis, it can be concluded that mutations alter the native structure of a protein and its stability. Since stability and native conformation are essential for the protein activity and functions, deviation in any such property due to mutation may have functional consequences.
3.4.2. Mutations Affect the Structural Compactness of h-RSK1-CTKD
The radius of gyration depicts the overall protein dimension, thereby acting as an important parameter to evaluate the compactness of a protein. To study the effect of mutation on the structural integrity of h-RSK1-CTKD, the Rg value was determined for wild-type and mutants. The mutations Arg34Pro, Thr701Met, Ala704Thr, Arg725Trp, and Arg726Gln cause major deviation in the structural compactness of the wild-type protein (Figure 3b). The wild-type structure was observed to be stable and compact as the Rg value is stably maintained around 2.1 nm. A stable Rg value was observed for Arg434Pro till 60 ns and then a gradual drop in the Rg value from 2.2 to 2.15 nm and a further drop to 2.1 nm up to 100 ns. Thereafter, Rg again increased to 2.1 nm and was maintained till the end of the simulation. Thr701Met implied considerable folding and compactness as the Rg value dropped gradually from 2.25 to 2.03 nm. A mutant structure, Ala704Thr, exhibited Rg in an interesting manner, which indicates the elevation of the Rg value up to 2.4 nm at the beginning of the simulation that immediately dropped down to 2.1 nm within 10 ns. From 20 ns onward, the Rg value of Ala704Thr showed several ups and down with short time intervals within the range of 2.1–2.2 nm. It was reflected that the structure was rapidly folded, unfolded, and refolded at different intervals of time. From 180 ns onward, the Rg value of Ala704Thr was elevated beyond 2.2 nm with the trend of a further increment of Rg at the end of the simulation. Like Thr701Met, the mutant Arg725Trp also achieved a compact structure with a stable 2.07 nm Rg value after 80 ns till 200 ns. In the case of the mutant Arg726Gln, Rg dropped initially from 2.3 to 2.15 nm up to 40 ns, and then, it was observed to be unstable and fluctuating within 2.1–2.2 nm during the simulation time. Overall, the average Rg values were calculated and arranged in increasing order as follows: Arg434Pro (2.16) > Ala704Thr (2.15) > Arg726Gln (2.13) > WT (2.11) > Arg725Trp (2.10) > Thr701Met (2.07) (Table 3). Except for Pro613Leu and Ser732Phe, mutants His533Asn, Ser720Cys, and Arg725Gln showed resemblance to the wild-type (Figure S2b). The result indicated that mutants Arg434Pro, Ala704Thr, and Arg726Gln achieved less compact structures as their Rg value was higher than the wild-type. The Rg analysis revealed that the mutations affect the structural compactness of mutant proteins.
3.4.3. Mutation Perturbs the Structural Flexibility of h-RSK1-CTKD
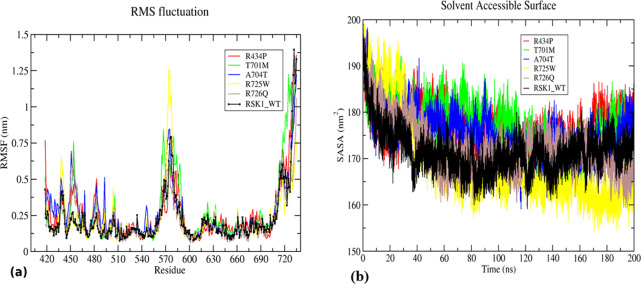
RMSF analysis was carried out to understand the alteration in flexibility of an amino acid residue in RSK1-CTKD and mutants. The RMSF value for each residue number was calculated for all mutants and wild-type. The loop region provides flexibility to a protein structure. The activation loop (570–580 amino acids) and the terminal region were observed to give a prominent peak throughout the simulation in WT as well as mutants. Along with the activation loop, several smaller peaks appeared at residue positions 431–436, 450–460, 480, and 500. The highest RMSF value of approximately 1.25 nm was observed in only the Arg725Trp mutant at the activation loop compared to other mutants and wild-type. Mutants Thr701Met, Arg726Gln, Ala704Thr, and Arg434Pro were observed to be more flexible in the residue range 450–460 with RMSF values of 0.76, 0.74, 0.72, and 0.47 nm, respectively (Figure 4a). According to the average fluctuation score, the values were arranged as follows: Thr701Met (0.27 nm) > Ala704Thr (0.26 nm) > Arg434Pro (0.25 nm) > Arg725Trp (0.22 nm) = WT (0.22 nm) > Arg726Gln (0.21 nm) (Table 3). Meanwhile, slight variation in flexibility was observed in His533Asn, Ser720Cys, Arg725Gln, Pro613Leu, and Ser732Phe (Figure S3a). Increasing and decreasing trends in flexibility were observed for the entire residue range irrespective of the position of a mutation. Thus, RMSF analysis indicated that the mutation alters the overall flexibility of a protein structure.
Figure 4.
Structural flexibility and solvent-accessible surface area (SASA) analysis of RSK1-CTKD WT (black) and mutants R434P (red), T701M (green), A704T (blue), R725W (yellow), and R726Q (brown). (a) RMSF of WT and mutants showing changes in the flexibility of a protein and (b) SASA of WT and mutants indicating an alteration in an area accessible to the solvent.
3.4.4. Mutation Alters the Solvent-Accessible Surface Area (SASA)
To predict the effect of mutation on protein structural integrity, SASA analysis was performed. The wild-type structure showed a drop in the SASA value from 195 to 162 nm2 during the first 40 ns of simulation, and then, it was steadily fluctuating between 165 and 180 nm2 (Figure 4b). Similarly, Arg434Pro mutant SASA values were decreased, but from 20 ns onward, it was increased up to 190 nm2, and then, they were seen fluctuating between 175 and 185 nm2 throughout the simulation. The SASA values for mutants Thr701Met and Ala704Thr were raised up to 180 nm2 till 140 ns and later achieved the same SASA value as that of the wild-type. The mutant Arg725Trp was shown to drop the SASA value considerably from 195 and 190 nm2 and achieved the lowest value of approximately 160 nm2 at the end of the simulation; meanwhile, the mutant Arg726Gln showed a slight fluctuation in the SASA value ranging from 160 to 180 nm2 after 40 ns (Figure 4b). It has been indicated that the average value of SASA for wild-type (101.46 nm2) is less than those of the mutants Arg434Pro (176.87 nm2), Thr701Met (177.19 nm2), Ala704Thr (175.43 nm2), Arg725Trp (169.24 nm2), and Arg726Gln (172.36 nm2) indicating the changes in the structural integrity of a native protein (Table 3). Mutants Pro613Leu and Ser732Phe and mutants His533Asn, Ser720Cys, and Arg725Gln showed similar SASA to that of wild-type (Figure S3b). The SASA values of mutants Arg434Pro, Thr701Met, Ala704Thr, Arg725Trp, and Arg726Gln were higher than that of the wild-type. Hence, it could be concluded that these mutations may achieve more expanded structures than the wild-type.
3.4.5. Mutation Introduces Changes in the Secondary Structure of h-RSK1-CTKD
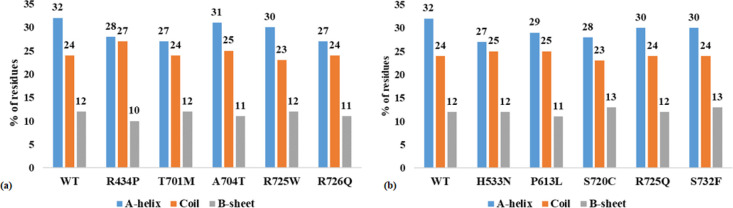
The secondary structure analysis was performed to explore the general alterations in the secondary structure introduced by mutations In the current analysis, the % of residue involved in the formation of the α-helix, β-sheet, and coil were considered and plotted for wild-type and mutants (Figure 5a,b). Slight changes in the % of residues in the α-helix and β-sheet were observed in Ala704Thr and Arg725Trp, while notable changes in the α-helix forming residues were observed in six mutants Arg434Pro, Thr701Met, Arg726Gln, His533Asn, Pro613Leu, and Ser720Cys. No major alterations in α-helix and β-sheet forming residues were observed in Arg725Gln and Ser732phe. However, all mutations had the least effect on coil forming residues. Secondary structures play an important role in protein folding and structural integrity. Aberration in the secondary structure will lead to the loss of native conformation and function of a protein. The analysis revealed that Arg434Pro, Thr701Met, Arg726Gln, His533Asn, Pro613Leu, and Ser720Cys disrupt the α-helix formation, which may cause drift in the native structure.
Figure 5.
Secondary structure analysis. Percentage of residues involved in the formation of α-helix (blue), β-sheet (brown), and coil (orange) in WT and mutants (a) R434P, T701M, A704T, R725W, and R726Q and (b) mutants H533N, P613L, S720C, R725Q, and S732F showing slight alterations in the secondary structure introduced due to the mutations.
3.4.6. Mutation Alters the Intramolecular Hydrogen Bonding
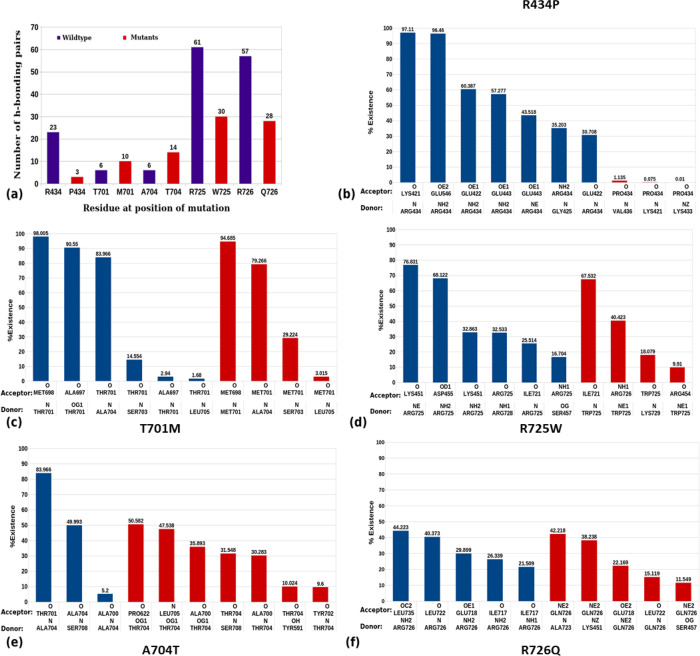
Mutation leads to the disruption of several intramolecular interactions established by wild-type. It can also form new interactions with other residues, which are generally not observed in wild-type conditions. The total number of H-bonds was plotted for mutant residues in comparison with the wild-type residue. A considerable decrease in hydrogen bonding pairs was observed in the case of all arginine mutants such as Arg434Pro, Arg725Trp, and Arg726Gln, while a slight increase in hydrogen bonding pairs was noticed in Thr701Met and Ala704Thr (Figure 6a). A drastic loss in % of H-bond existence was observed in Arg434Pro mutation as the interactions made by the Arg434 residue were not formed by Pro434 (Figure 6b). The majority of interactions formed by the Thr701 residue were maintained in Met701 alteration along with the loss of interactions with the Ala697 residue (Figure 6c). Alteration of the Ala704 residue into Thr was observed to form several new hydrogen bonds with Pro622, Leu705, Tyr591, and Tyr702 while maintaining interactions with Ala700 and Ser708 (Figure 6e). The interaction of Arg725 with Ile721 was consistent even in the Trp725 alteration. The increased existence of Arg725-Ile721 interactions from 25 to 67% in Trp725-Ile721 was noticed, whereas all-new interactions were formed by the altered Trp725 (Figure 6d). Mutation of the Arg residue at 726 into Gln forms new interactions with Ala 723, Lys 451, and Ser 457. It was also observed that the interaction of Leu722 and Glu 718 was not disturbed due to the mutation (Figure 6f). The mutant His533Asn showed increased H-bonding pairs, while a decrease in Pro613Leu, Ser720Cys, Arg725Gln, and Ser732Phe was observed (Figure S4). Hydrogen bonds are important for the stability of a protein as well as direct protein folding. The loss of hydrogen bonds may cause a loss of protein stability and conformation. In the current analysis, it was found that all other mutants except Thr701Met have a reduced number of hydrogen bonding pairs, which may lead to a gain in overall structural flexibility.
Figure 6.
Number of intramolecular hydrogen bonding pairs and their % existence throughout the simulation of WT and mutated residues: (a) number of H-bonding pairs formed by WT (blue) and mutated residues (red) and (b–f) highly scored H-bond existence formed by the WT residue and mutated residue in (b) R434P, (c) T701M, (d) R725W, (e) A704T, and (f) R726Q.
3.5. Mutations Affect the Dynamic Behavior of RSK1-CTKD
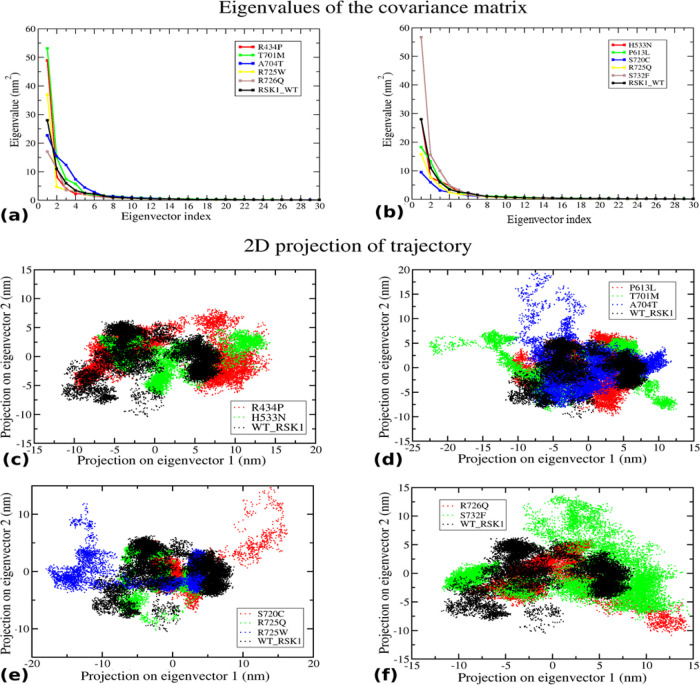
Principal component analysis (PCA) was performed to understand the alteration in the dynamic behavior of a protein. PCs are eigenvectors having larger eigenvalues that represent a significant role in the overall motion of the structure. The eigenvalues were plotted against the corresponding eigenvector index for the first 30 modes of motion. Significantly, dominant motions with higher eigenvalues were observed for the first five eigenvectors, whereas the remaining eigenvectors exhibited remarkably low eigenvalues in the overall system (Figure 7a,b). Here, the first two PCs (PC1 and PC2) were selected to analyze their projection of trajectories in the phase space during simulation. Mutants Arg434Pro and His533Asn, Thr701Met and Ala704Thr, Arg725Trp and Ser720Cys, and Arg726Gln and Ser732Phe were observed to be unstable and flexible as they occupied more space than the wild-type; meanwhile, low space was observed to be occupied by Pro613Leu and Arg725Gln indicating the reduced stability of the mutant protein (Figure 7a–f). Therefore, the PCA analysis suggested that the mutants perturb the structural stability and flexibility of a protein.
Figure 7.
Principal component analysis: (a,b) eigenvalue for the first 30 modes of motion of WT and mutants and (c–f) comparative eigenvector projection profile of WT and mutants. In all four plots, mutants were observed to cover an area less than or more than the WT.
Overall, the results implied that mutants Arg434P, Thr701Met, Ala704Thr, Arg725Trp, and Arg726Gln considerably alter the structural flexibility, integrity, and stability of h-RSK1-CTKD. It is well-known that RSK1 acts as a response element of the Ras/MAPK pathway that regulates diverse cellular processes such as growth, proliferation, differentiation, and survival. The five mutations Arg434P, Thr701Met, Ala704Thr, Arg725Trp, and Arg726Gln prioritized in the current study may affect the interaction of RSK1 with ERK1/2, which could subsequently alter the activation and functions of RSK1. These mutations may alter the paradigm of RSK1 activation and its kinetic potential, which can lead to subsequent functional consequences. Molecular dynamics simulation was used in the current study to analyze the structural dynamics of a mutated as well as wild-type protein, which is of great importance for a follow-up functional study.
4. Conclusions
In this study, combined in silico screening approaches were employed to identify pathogenic mutations in h-RSK1 retrieved from cBioPortal. Based on the pathogenic score predicted by computational tools and the location of substitution, 10 missense mutations Arg434Pro, Thr701Met, Ala704Thr, Arg725Trp, Arg726Gln, His533Asn, Pro613Leu, Ser720Cys, Arg725Gln, and Ser732Phe could be potentially deleterious. Furthermore, the molecular dynamics simulation analysis was used to explore the effect of these deleterious missense mutations on the conformation of the h-RSK1-CTKD structure. The RSMD, RMSF, and Rg analysis revealed that mutations alter the stability, flexibility, and compactness of the native protein. Based on SASA and secondary structure analysis, it was observed that the mutations affect the solvent-accessible surface as well as slight alteration in the secondary structure. Superimposed structures extracted at various time points during the simulations depict that Arg434Pro, Thr701Met, Ala704Thr, Arg725Trp, and Arg726Gln show dynamic conformational changes in the P-loop, activation loop, catalytic loop, and CTT. Alterations at such structural and functional regions may cause functional impairment in the kinase activity of RSK1-CTKD (Figure S5). Moreover, calculation of hydrogen bonding pairs suggested that the mutations resulted in the loss of intramolecular hydrogen bonding, thereby affecting the structural integrity of the protein. These findings were further supported by PCA analysis, which also revealed that the mutations had an effect on the stability and flexibility of the native protein. It was observed that Arg434Pro, Thr701Met, Ala704Thr, Arg725Trp, and Arg726Gln mutations perturb the structural integrity, stability, and flexibility of the native protein to a great extent. Overall, the study provided significant insights into the structural consequences of reported RSK1 mutations in cancer.
Acknowledgments
The authors would like to thank the Bioinformatics Centre (BIC) at ACTREC and BIF College of Veterinary Science, AAU, Guwahati for providing the necessary computational facility.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c00722.
Evolutionary conservancy of RSK1-CTKD generated by ConSurf; structural stability and compactness analysis of RSK1-CTKD; structural flexibility and solvent-accessible surface area analysis; several intramolecular hydrogen bonding pairs and their % existence throughout the simulation; snapshots of WT and mutants; functional consequences of missense mutations; effect of 10 mutations over RSK1-CTKD (PDF)
The study has been supported by DBT (BT/PR40181/BTIS/137/15/2021) to A.K.V.
The authors declare no competing financial interest.
Supplementary Material
References
- Dhillon A. S.; Hagan S.; Rath O.; Kolch W. MAP kinase signalling pathways in cancer. Oncogene 2007, 26, 3279–3290. 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- Zhang W.; Liu H. T. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002, 12, 9–18. 10.1038/sj.cr.7290105. [DOI] [PubMed] [Google Scholar]
- Chang F.; Steelman L. S.; Shelton J. G.; Lee J. T.; Navolanic P. M.; Blalock W. L.; Franklin R.; McCubrey J. A. Regulation of cell cycle progression and apoptosis by the Ras/Raf/MEK/ERK pathway (Review). Int. J. Oncol. 2003, 22, 469–480. [PubMed] [Google Scholar]
- Romeo Y.; Zhang X.; Roux P. P. Regulation and function of the RSK family of protein kinases. Biochem. J. 2012, 441, 553–569. 10.1042/BJ20110289. [DOI] [PubMed] [Google Scholar]
- Anjum R.; Blenis J. The RSK family of kinases: emerging roles in cellular signalling. Nat. Rev. Mol. Cell Biol. 2008, 9, 747–758. 10.1038/nrm2509. [DOI] [PubMed] [Google Scholar]
- Ikuta M.; Kornienko M.; Byrne N.; Reid J. C.; Mizuarai S.; Kotani H.; Munshi S. K. Crystal structures of the N-terminal kinase domain of human RSK1 bound to three different ligands: Implications for the design of RSK1 specific inhibitors. Protein Sci. 2007, 16, 2626–2635. 10.1110/ps.073123707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D.; Fu T. M.; Nan J.; Liu C.; Li L. F.; Su X. D. Structural basis for the autoinhibition of the C-terminal kinase domain of human RSK1. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2012, 68, 680–685. 10.1107/S0907444912007457. [DOI] [PubMed] [Google Scholar]
- Smith J. A.; Poteet-Smith C. E.; Malarkey K.; Sturgill T. W. Identification of an extracellular signal-regulated kinase (ERK) docking site in ribosomal S6 kinase, a sequence critical for activation by ERK in vivo. J. Biol. Chem. 1999, 274, 2893–2898. 10.1074/jbc.274.5.2893. [DOI] [PubMed] [Google Scholar]
- Gógl G.; Biri-Kovács B.; Póti A. L.; Vadászi H.; Szeder B.; Bodor A.; Schlosser G.; Ács A.; Turiák L.; Buday L.; Alexa A.; Nyitray L.; Reményi A. Dynamic control of RSK complexes by phosphoswitch-based regulation. FEBS J. 2018, 285, 46–71. 10.1111/febs.14311. [DOI] [PubMed] [Google Scholar]
- Casalvieri K. A.; Matheson C. J.; Backos D. S.; Reigan P. Selective Targeting of RSK Isoforms in Cancer. Trends Cancer 2017, 3, 302–312. 10.1016/j.trecan.2017.03.004. [DOI] [PubMed] [Google Scholar]
- Sulzmaier F. J.; Ramos J. W. RSK isoforms in cancer cell invasion and metastasis. Cancer Res. 2013, 73, 6099–6105. 10.1158/0008-5472.CAN-13-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houles T.; Roux P. P. Defining the role of the RSK isoforms in cancer. Semin. Cancer Biol. 2018, 48, 53–61. 10.1016/j.semcancer.2017.04.016. [DOI] [PubMed] [Google Scholar]
- Salhi A.; Farhadian J. A.; Giles K. M.; Vega-Saenz de Miera E.; Silva I. P.; Bourque C.; Yeh K.; Chhangawala S.; Wang J.; Ye F.; Zhang D. Y.; Hernando-Monge E.; Houvras Y.; Osman I. RSK1 activation promotes invasion in nodular melanoma. Am. J. Pathol. 2015, 185, 704–716. 10.1016/j.ajpath.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H.; Martin T. A.; Davies E. L.; Ruge F.; Yu H.; Zhang Y.; Teng X. U.; Jiang W. G. The Clinical Implications of RSK1-3 in Human Breast Cancer. Anticancer Res. 2016, 36, 1267–1274. [PubMed] [Google Scholar]
- Yu G.; Lee Y. C.; Cheng C. J.; Wu C. F.; Song J. H.; Gallick G. E.; Yu-Lee L. Y.; Kuang J.; Lin S. H. RSK promotes prostate cancer progression in bone through ING3, CKAP2, and PTK6-mediated cell survival. Mol. Cancer Res. 2015, 13, 348–357. 10.1158/1541-7786.MCR-14-0384-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GN M. H.; Da Silva F. F.; De Bellis B.; Lupinacci F. C. S.; Bellato H. M.; Cruz J. R.; Segundo C. N. C.; Faquini I. V.; Torres L. C.; Sanematsu P. I.; Begnami M. D.; Martins V. R.; Roffe M. Aberrant expression of RSK1 characterizes high-grade gliomas with immune infiltration. Mol. Oncol. 2020, 14, 159–179. 10.1002/1878-0261.12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara R.; Mauri F. A.; Taylor H.; Derua R.; Shia A.; Gray C.; Nicols A.; Shiner R. J.; Schofield E.; Bates P. A.; Waelkens E.; Dallman M.; Lamb J.; Zicha D.; Downward J.; Seckl M. J.; Pardo O. E. An siRNA screen identifies RSK1 as a key modulator of lung cancer metastasis. Oncogene 2011, 30, 3513–3521. 10.1038/onc.2011.61. [DOI] [PubMed] [Google Scholar]
- Li M.; Goncearenco A.; Panchenko A. R. Annotating Mutational Effects on Proteins and Protein Interactions: Designing Novel and Revisiting Existing Protocols. Methods Mol. Biol. 2017, 1550, 235–260. 10.1007/978-1-4939-6747-6_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte A.; Ribeiro D.; Moreira L.; Amaral O. In Silico Analysis of Missense Mutations as a First Step in Functional Studies: Examples from Two Sphingolipidoses. Int. J. Mol. Sci. 2018, 19, 3409. 10.3390/ijms19113409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N.; Khan M. A.; Capasso G.; Zacchia M. Computational and Structural Analysis to Assess the Pathogenicity of Bardet-Biedl Syndrome Related Missense Variants Identified in Bardet-Biedl Syndrome 10 Gene (BBS10). ACS Omega 2022, 7, 37654–37662. 10.1021/acsomega.2c04522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj V. K.; Purohit R. Targeting the protein-protein interface pocket of Aurora-A-TPX2 complex: rational drug design and validation. J. Biomol. Struct. Dyn. 2021, 39, 3882–3891. 10.1080/07391102.2020.1772109. [DOI] [PubMed] [Google Scholar]
- Kumar S.; Bhardwaj V. K.; Singh R.; Purohit R. Structure restoration and aggregate inhibition of V30M mutant transthyretin protein by potential quinoline molecules. Int. J. Biol. Macromol. 2023, 231, 123318. 10.1016/j.ijbiomac.2023.123318. [DOI] [PubMed] [Google Scholar]
- Singh R.; Purohit R. Computational analysis of protein-ligand interaction by targeting a cell cycle restrainer. Comput. Methods Programs Biomed. 2023, 231, 107367. 10.1016/j.cmpb.2023.107367. [DOI] [PubMed] [Google Scholar]
- Navapour L.; Mogharrab N. In silico screening and analysis of nonsynonymous SNPs in human CYP1A2 to assess possible associations with pathogenicity and cancer susceptibility. Sci. Rep. 2021, 11, 4977. 10.1038/s41598-021-83696-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.; Mehmood A.; Wang H.; Xu Q.; Xiong Y.; Wei D. Q. Computational Screening and Analysis of Lung Cancer Related Non-Synonymous Single Nucleotide Polymorphisms on the Human Kirsten Rat Sarcoma Gene. Molecules 2019, 24, 1951. 10.3390/molecules24101951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M. S.; Roy A. S.; Islam M. S. In silico analysis predicting effects of deleterious SNPs of human RASSF5 gene on its structure and functions. Sci. Rep. 2020, 10, 14542. 10.1038/s41598-020-71457-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. A.; Siddiqui M. Q.; Kuligina E.; Varma A. K. Evaluation of conformational transitions of h-BRCA2 functional domain and unclassified variant Arg2502Cys using multimodal approach. Int. J. Biol. Macromol. 2022, 209, 716–724. 10.1016/j.ijbiomac.2022.04.049. [DOI] [PubMed] [Google Scholar]
- Khan M. A.; Varma A. K. In silico and structure-based assessment to classify VUS identified in the alpha-helical domain of BRCA2. J. Biomol. Struct. Dyn. 2022, 1–11. 10.1080/07391102.2022.2148127. [DOI] [PubMed] [Google Scholar]
- Das L.; Shekhar S.; Chandrani P.; Varma A. K. In silico structural analysis of secretory clusterin to assess pathogenicity of mutations identified in the evolutionarily conserved regions. J. Biomol. Struct. Dyn. 2023, 41, 469–478. 10.1080/07391102.2021.2007791. [DOI] [PubMed] [Google Scholar]
- Cerami E.; Gao J.; Dogrusoz U.; Gross B. E.; Sumer S. O.; Aksoy B. A.; Jacobsen A.; Byrne C. J.; Heuer M. L.; Larsson E.; Antipin Y.; Reva B.; Goldberg A. P.; Sander C.; Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery 2012, 2, 401–404. 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R.; Bairoch A.; Wu C. H.; Barker W. C.; Boeckmann B.; Ferro S.; Gasteiger E.; Huang H.; Lopez R.; Magrane M.; Martin M. J.; Natale D. A.; O’Donovan C.; Redaschi N.; Yeh L. S. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004, 32, 115D–1119D. 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa A.; Gogl G.; Glatz G.; Garai A.; Zeke A.; Varga J.; Dudas E.; Jeszenoi N.; Bodor A.; Hetenyi C.; Remenyi A. Structural assembly of the signaling competent ERK2-RSK1 heterodimeric protein kinase complex. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 2711–2716. 10.1073/pnas.1417571112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H. M.; Westbrook J.; Feng Z.; Gilliland G.; Bhat T. N.; Weissig H.; Shindyalov I. N.; Bourne P. E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P. C.; Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei I.; Jordan D. M.; Sunyaev S. R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013, 7, Unit7.20. 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E.; Fariselli P. PhD-SNPg: a webserver and lightweight tool for scoring single nucleotide variants. Nucleic Acids Res. 2017, 45, W247–W252. 10.1093/nar/gkx369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Ferrando V.; Gazzo A.; de la Cruz X.; Orozco M.; Gelpi J. L. PMut: a web-based tool for the annotation of pathological variants on proteins, 2017 update. Nucleic Acids Res. 2017, 45, W222–W228. 10.1093/nar/gkx313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.; Chan A. P. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015, 31, 2745–2747. 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H.; Abadi S.; Martz E.; Chay O.; Mayrose I.; Pupko T.; Ben-Tal N. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016, 44, W344–W350. 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-W.; Lin J.; Chu Y.-W. iStable: off-the-shelf predictor integration for predicting protein stability changes. BMC Bioinf. 2013, 14, S5. 10.1186/1471-2105-14-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venselaar H.; Te Beek T. A.; Kuipers R. K.; Hekkelman M. L.; Vriend G. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinf. 2010, 11, 548. 10.1186/1471-2105-11-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B.; Sali A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinf. 2016, 54, Unit-5.6. 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J.; Evans R.; Pritzel A.; Green T.; Figurnov M.; Ronneberger O.; Tunyasuvunakool K.; Bates R.; Zidek A.; Potapenko A.; Bridgland A.; Meyer C.; Kohl S. A. A.; Ballard A. J.; Cowie A.; Romera-Paredes B.; Nikolov S.; Jain R.; Adler J.; Back T.; Petersen S.; Reiman D.; Clancy E.; Zielinski M.; Steinegger M.; Pacholska M.; Berghammer T.; Bodenstein S.; Silver D.; Vinyals O.; Senior A. W.; Kavukcuoglu K.; Kohli P.; Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E. F.; Goddard T. D.; Huang C. C.; Couch G. S.; Greenblatt D. M.; Meng E. C.; Ferrin T. E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Lindorff-Larsen K.; Piana S.; Palmo K.; Maragakis P.; Klepeis J. L.; Dror R. O.; Shaw D. E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 2010, 78, 1950–1958. 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Spoel D.; Lindahl E.; Hess B.; Groenhof G.; Mark A. E.; Berendsen H. J. C. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Hess B.; Kutzner C.; van der Spoel D.; Lindahl E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 435–447. 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Pronk S.; Pall S.; Schulz R.; Larsson P.; Bjelkmar P.; Apostolov R.; Shirts M. R.; Smith J. C.; Kasson P. M.; van der Spoel D.; Hess B.; Lindahl E. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit A.; Yi L.; Gowthaman R.; Torkamani A.; Schork N. J.; Verkhivker G. M. Sequence and structure signatures of cancer mutation hotspots in protein kinases. PLoS One 2009, 4, e7485 10.1371/journal.pone.0007485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClendon C. L.; Kornev A. P.; Gilson M. K.; Taylor S. S. Dynamic architecture of a protein kinase. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, E4623–E4631. 10.1073/pnas.1418402111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.