Abstract
Heparin-like sulfated polysaccharide, acharan sulfate, was purified from the mucus of an African giant snail with unique sulfated glycosaminoglycans (GAGs). This study reported on finding novel and safe heparin resources from Achatina fulica for further use as well as easy isolation and purification of the active fraction from the initial raw material. Its structure was characterised by a strong-anion exchange combined with high-performance liquid chromatography (HPLC) and nuclear magnetic resonance (NMR) spectroscopy. The results indicated that the potential acharan sulfate fraction is a glycosaminoglycan composed of several repeating disaccharide units, namely, of , and hence, presents heterogeneity regarding negative net charge density. Furthermore, the heparinase digests inhibit the binding of SARS-CoV-2 spike protein to the ACE2 receptor. In summary, the acharan sulfate presented in this work has shown its great potential for application in the preparation of sulfated polysaccharides as an alternative to heparin with important biological activity.
Keywords: Heparin, Sulfated glycosaminoglycans, Achatina fulica, Acharan sulfate, Snail mucus, Strong-anion exchange chromatography, Antiviral screening
Graphical abstract
1. Introduction
Heparin is a highly sulfated form of heparan sulfate (HS) glycosaminoglycans (GAGs) that are found in various tissues throughout the body [1,2]. It is primarily produced by mast cells, which are found to mainly originate from the intestinal mucosa of animals, particularly the porcine (pig) small intestine and bovine (cow) lung, but can also be derived from other animals such as the horse, sheep and donkey [3]. As a medication, it is used as an anti-coagulant [4,5]. It is worth noting that porcine heparin is the most common source of heparin for commercial use [6]. Although the process is considered safe and effective, it has been suggested to exhibit potential risk among groups with different religious beliefs regarding lethal diseases and pathogenic agents. Therefore, the novel and safe sources of heparin should be able to be farmed in house-holds and organic environments. Furthermore, it should also be manageable for agricultural harvesting subject to further uses.
While pursuing long-term studies on heparin, acharan sulfate have been constructed in large amounts from the whole body of the giant African snail with a unique structure [[7], [8], [9], [10]]. They are the major glycoconjugate that has a repeating disaccharide unit, , where X refers to sulfation site. This structure is related but instead represents a new type of linked GAGs significantly different from heparin [10]. The search for new sources for application in medicine and cosmetics from different parts of the giant African snail is a trend in biotechnology. One of the sources of such active compounds is the snail mucus, which does not show any the risk of contamination with microorganisms such as viruses and bacteria, suggesting that it might be a good precursor for heparin polysaccharide preparation having important biological activities.
The highly sulfated domains in heparan sulfate are ubiquitous in the glycocalyx of epithelial cells in the nasal passage and act as a co-receptor in binding with SARS-CoV-2 spike protein at its receptor binding domain (RBD). In the current work, we also focus to examine the structural characterisation of acharan sulfate, which might be involved in important biological roles for the ability of this sulfated GAGs to inhibit the protein binding efficacy. Thus, we propose a new information on the property of this interesting GAGs molecule, acharan sulfate, as a heparin-like substances in place of heparin for the potential anti-SARS-CoV-2 activity.
2. Results and discussion
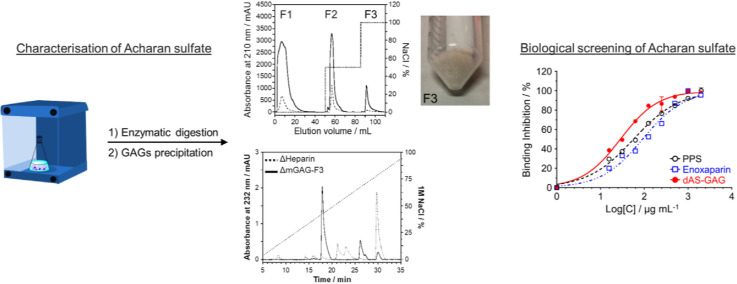
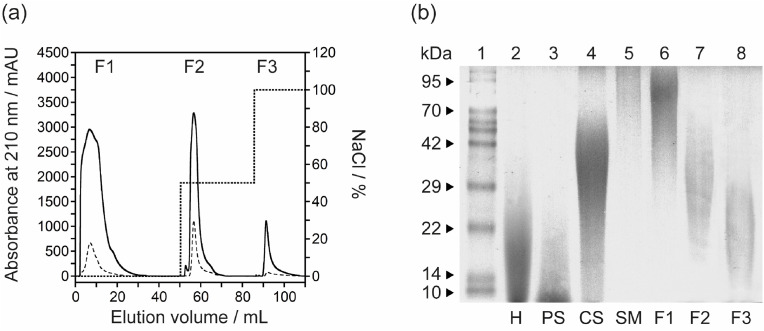
Here, we prepared the sulfated polysaccharides derived from the mucus on the surface of a live giant African snail (Achatina fulica). The thus-obtained a pale-white powders consisting of the sulfated mucus-GAGs (mGAG) polysaccharides were performed. The amount of protein and sulfated GAGs (uronic acid) contents in the mGAG was measured to be around ∼8 and 83% as determined by using a standard curve prepared with bovine serum albumin and purified chondroitin sulfate, respectively. As a result, the mGAG was composed primarily of glycosaminoglycans [7]. The yield of mGAG polysaccharide was approximately ∼20% by dry weight of the mucus. After fractionation and recovery of mGAG on a DEAE-Sepharose anion-exchange chromatography (Fig. 1 a), mGAG and their fractions was analysed based on charge and molecular weight using polyacrylamide gel electrophoresis (PAGE). Three fractions were obtained, collected and named the (1) non-interacting fraction (F1, 62.6% w/w yield), (2) eluted fraction at 0.5 M NaCl (F2, 30.1%w/w yield), and (3) eluted fraction at 1.0 M NaCl (F3, 7.3%w/w yield). According to the low content of protein but high sulfated-GAGs content, F3 fraction (mGAG-F3) was further desalted and freeze-dried to give a white-coloured powder. Although the GAGs component of the mGAG was isolated in the only three metachromatic elutions when compared with previous literature [[10], [11], [12]], all eluted peaks were observed at highest salt concentration (1.0 M NaCl) and differences in the proportion of eluted peaks were obtained [7]. As seen in Fig. 1b, Lanes 1–4 show the molecular weight protein markers, heparin, pentosan polysulfate, and chondroitin sulfate standard, respectively. For mGAG, Lanes 5–8 represent the GAGs of snail mucus (SM), the non-interacting fraction (F1), and the papain-digested snail GAGs from DEAE-Sepharose at 0.5 M NaCl (F2) and 1.0 M NaCl (F3), respectively. The average molecular weight of the mGAG-F3 fraction (Lane 8) was considerably larger than that of an oligosaccharide standard, heparin and pentosan polysulfate, having an average molecular weight of ∼22,000 Da [10]. The mGAG-F2 band (Lane 7), with an electrophoretic migration similar to that of the chondroitin sulfate, was observed in the mucus which is a bit different from the study of Vieira et al. [12]. As acharan sulfate was shown to be the only GAGs component expressed by the Achatina fulica [10], we can assign this band as acharan sulfate. According to the increasing molecular mass of mGAG based on increasing salt concentration in the anion-exchange chromatography, the obtained results by electrophoresis revealed marked difference patterns of the polysaccharide extracted from the mucus compared to the whole body as reported by previous report [12]. We found each eluted band with decreasing molecular mass from SM (Lane 5) to mGAG-F3 (Lane 8), indicating that acharan sulfate is composed of heterogeneous and polydisperse GAGs chains. Nevertheless, both mGAG-F1 and mGAG-F2 will not be used to analyse in further study due to the presence of a contaminating brown pigment that may cause by non-covalent complexes such as proteins. Furthermore, in view of the scarce amount of purified material, we did not further characterize these two GAGs fractions by NMR analysis. Thus, the electrophoretic mobility of the various sulfated polysaccharides is the first indication of the distinctive structures, suggesting the structure of the mGAG correspond to those of chondroitin sulfate standard in this system with molecular mass range of 20–40 kDa [7,9,10,12].
Fig. 1.
Establishment, purification and characterisation of the acharan sulfate from the mucus of A. fulica. (a) The purification of mGAG (solid line) from snail mucus by DEAE-Sepharose anion-exchange chromatography. The column was eluted with a stepwise salt gradient of 0.5 M–1.0 M NaCl (dotted line) in 50 mM sodium acetate buffer (pH 5.5). The fractions were monitored at 210 nm and the flow rate was set at 2.5 mL/min. Three eluent fractions were collected; F1, non-interacting fraction; F2, eluted fraction at 0.5 M NaCl; and F3, eluted fraction at 1.0 M NaCl. The contamination of protein sample can be detected by measuring tis optical absorbance at 280 nm (dashed line). (b) PAGE analysis of mGAG-derived polysaccharides which visualized with alcian blue. Lane 1: Molecular weight protein markers, Lane 2: heparin (H), Lane 3: pentosan polysulfate (PS), Lane 4: chondroitin sulfate (CS), Lane 5: dried mGAG of snail mucus (SM), Lane 6: non-interacting fraction (F1), Lane 7: the papain-digested mGAG from DEAE-Sepharose at 0.5 M NaCl (F2), and Lane 8: the papain-digested mGAG from DEAE-Sepharose at 1.0 M NaCl (F3), respectively. The samples are loaded at the top of the gel (cathode) and migrate towards the anode at the bottom of the gel.
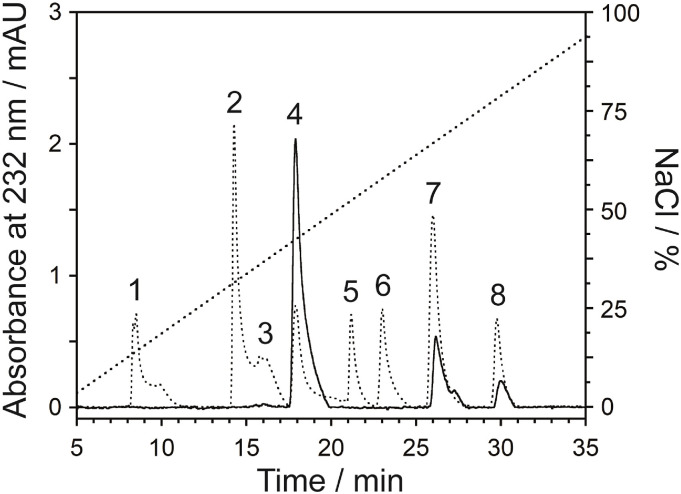
After complete enzymatic digestion (Fig. S1) to basic disaccharide-building blocks using a cocktail of heparinase enzymes, the enzymatic depolymerization of mGAG-F3 was characterised by the strong-anion exchange (SAX)-HPLC column (Fig. 2 and Table 1 ) [13,14]. In order to verify whether the conditions used to depolymerize the mGAG-F3 molecule would alter the disaccharide composition, a control experiment using low molecular weight heparin was carried out to show that it was stable under this condition (Fig. S2). In the disaccharides, ΔUA that denotes a Δ(4,5) unsaturated hexuronic acid, and N-acetylated glucosamine (GlcNAc) were observed as expected [14], containing mostly ΔUA(2S)-GlcNAc (72.92%), ΔUA(2S)-GlcNAc(6S) (14.94%), and ΔUA(2S)-GlcNSO3(6S) (12.14%). Each disaccharide in mGAG-F3 fraction was at a ratio of; 6 (ΔUA(2S)-GlcNAc): 1 (ΔUA(2S)-GlcNAc(6S)): 1 (ΔUA(2S)-GlcNSO3(6S)). This sulfated polysaccharide has a high level of negative charge according to the presence of N- and O-sulfo and carboxyl groups in both glucosamine and iduronic acid units [15]. The mGAG-F3 polysaccharides had a different disaccharide composition by data from literature, which shows that α-l-Iduronic acid (IdoA) residues from the three types of disaccharide units composed of 2-O-sulfated IdoA linked to either non-sulfated or 6-O-sulfated N-acetyl/N-sulfated GlcNAc [9,10,14,16]. Variation in the composition could be expected between different lots of raw materials, and potentially between different enzyme sources as well as the methods used to analyse the digests [17]. In our results, it was notable that minimal changes were observed in the composition, even with significantly different amounts of depolymerization.
Fig. 2.
SAX-HPLC chromatogram of mGAG-F3 (solid line) disaccharides by treatment with a mixing of heparinase enzymes, eluting with a linear gradient of 0.1–1.0 M NaCl (dotted line) at a flow rate of 1.0 mL/min. The standard disaccharide (dashed line) structures are indicated by number labels; 1: ΔUA-GlcNAc, 2: ΔUA-GlcNSO3, 3: ΔUA-GlcNAc(6S), 4: ΔUA(2S)-GlcNAc, 5: ΔUA-GlcNSO3(6S), 6: ΔUA(2S)-GlcNSO3, 7: ΔUA(2S)-GlcNAc(6S), 8: ΔUA(2S)-GlcNSO3(6S) (Fig. S3).
Table 1.
Composition analysis of the disaccharide repeating units in the AS-GAG polysaccharide by SAX-HPLC analysis.
| Peak No. | Disaccharide structures | Composition/% | Ratio |
|---|---|---|---|
| 4 | ΔUA(2S)-GlcNAc | 72.92 | 6.00 |
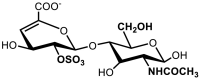 | |||
| 7 | ΔUA(2S)-GlcNAc(6S) | 14.94 | 1.23 |
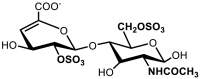 | |||
| 8 | ΔUA(2S)-GlcNSO3(6S) | 12.14 | 1.00 |
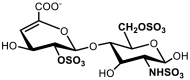 |
Abbreviations: ΔUA, Δ(4,5) unsaturated hexuronic acid; GlcNAc, N-acetylated glucosamine; GlcNSO3, N-sulfated glucosamine; 2S and 6S are 2-O- and 6-O-sulfate groups, respectively.
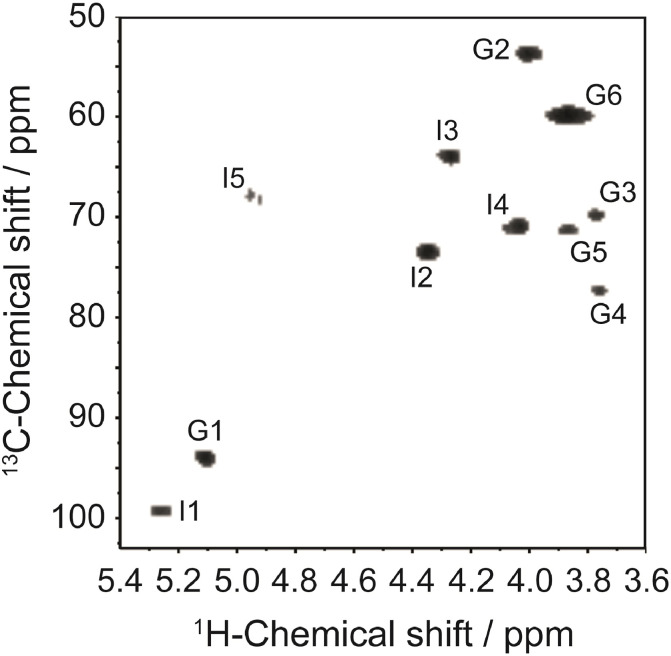
The 1H and 13C one-dimensional NMR spectra (Fig. S4) and the interpretations of 1H/13C HSQC spectra (Fig. 3 ) of the mGAG-F3 confirm the identification of this GAGs fraction as the previously described acharan sulfate [10,12,14]. The assignment was achieved by comparing with the previously literatures (Table 2 and Table S1). Combining these NMR data, the mGAG-F3 structure was confidently proposed, which composed of IdoA (labeled as I) and GlcNAc (labeled as G) units. The intense 1H peak with the most upfield resonance at δ2.061 indicated the substitution of the acetyl methyl groups in the amino sugar, suggesting that the GlcNAc residues in this GAGs fraction are essentially N-acetylated [18]. The presence of two peaks at δ5.237 and δ5.097 also confirmed the anomeric proton resonance corresponding to the H-1 of the IdoA and GlcNAc units, respectively. Because of the downfield resonances of their anomeric proton signals, the proton chemical shifts are in agreement with a 2-O-sulfated α-l-iduronic acid residue as the major disaccharide unit. Compared with H-1 glucosamine of heparan sulfate (at δ5.4) [19], the observation of these upfield shifts may cause on the anomeric proton of GlcNAc appeared to be attributable to the unusual sequence () [10]. The downfield 1H signal of H-5 of IdoA at δ4.932 confirmed the C5 epimerization. These features are somewhat typical for GAGs-like molecules.
Fig. 3.
1H–13C HSQC spectra of the AS-GAG from the mucus of A. fulica. The assignment was based on comparison with the previously described acharan sulfate. The peaks are labeled as I or G for Iduronic acid (IdoA) and N-acetyl glucosamine (GlcNAc) units, respectively. The number after these letters indicate the positions of 1H and 13C.
Table 2.
Proton chemical shifts for acharan sulfate from snail mucus (AS-GAG). Protons labeled as I and G refer to Iduronic acid (IdoA) and N-acetyl glucosamine (GlcNAc) units, respectively.
| Protona | Chemical Shifts/ppm |
|||||
|---|---|---|---|---|---|---|
| Acharan sulfate (This work) | Acharan sulfate [10] | Acharan sulfate [12] | Acharan sulfate [14] | IdoA(2S)-GlcNS(6S) [28] | IdoA-GlcNAc [29] | |
| I-1 | 5.237 | 5.19 | 5.20 | 5.17 | 5.22 | 4.89 |
| I-2 | 4.339b | 4.34b | 4.34b | 4.33b | 4.35b | 3.65 |
| I-3 | 4.270 | 4.28 | 4.19 | 4.27 | 4.20 | 3.83 |
| I-4 | 4.020 | 4.03 | 4.05 | 4.01 | 4.10 | 4.04 |
| I-5 | 4.932 | 4.93 | 5.08 | 4.90 | 4.81 | 4.68 |
| G-1 | 5.097 | 5.11 | 5.07 | 5.10 | 5.39 | 5.11 |
| G-2 | 4.015 | 4.02 | 3.97 | 4.01 | 3.29 | 4.00 |
| G-3 | 3.751 | 3.74 | 3.70 | 3.72 | 3.67 | 3.73 |
| G-4 | 3.758 | 3.74 | 3.70 | 3.73 | 3.77 | 3.73 |
| G-5 | 3.863 | 3.87 | 3.71 | 3.85 | 4.03 | 4.00 |
| G-6 | 3.864, 3.948 | 3.87 | 3.82 | 3.85, 4.00 | 4.27b | 3.85 |
| Acetyl | 2.061 | 2.08 | 2.04 | 2.06 | – | 2.04 |
Although the mGAG-F3 fraction, characterised as acharan sulfate (AS-GAG), shown weak anticlotting activities (data not shown) [14]. The increasing research papers have revealed that non-anticoagulant heparin species possess potential applications for treating inflammation and act as co-receptors for binding to the host cell receptor [20,21]. Recent studies have shown that SARS-CoV-2 spike protein can bind sulfated GAGs depending on the degree of sulfation [22], facilitating the conformational change in spike subunit necessary for binding to the angiotensin-converting enzyme-2 (ACE2) receptor [21]. The binding of spike protein on the surface of cells involves cooperation between ACE2 and heparan sulfate receptors, in which the interaction apparently occurs in a co-dependent manner. The expression of specific oligosaccharide epitopes by cells makes it possible to recruit specific binding. The exact mechanism by which the low molecular weight heparin (LMWH) binds to the virus is not yet fully understood, but it is thought to involve the binding of the specific GAGs motifs to the receptor binding domain of SARS-CoV-2 spike protein that is separate from the site involved in ACE2 binding [21,23,24]. Thus, in the experiments performed, we firstly investigated the effect of AS-GAG as a heparin-like substances for the potential anti-SARS-CoV-2 activity.
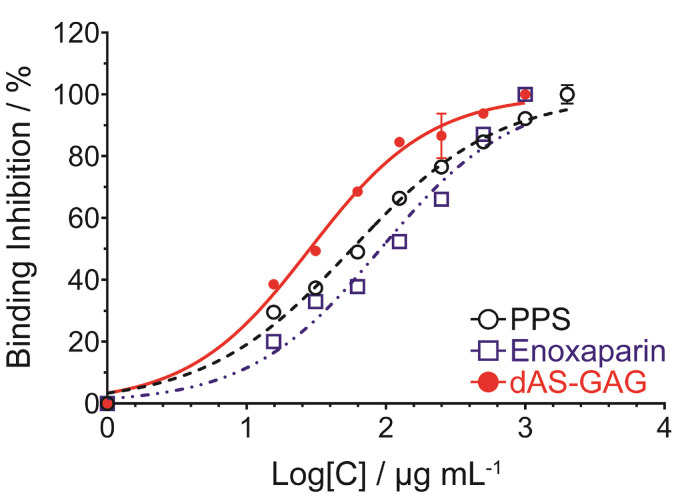
To evaluate the potency and efficacy of the polysulfated GAGs on binding, we first prepared the AS-GAG oligosaccharide by partial digestion with a mix of heparinase enzymes, after which the final digest AS-GAG (dAS-GAG) was resolved into its constituent oligosaccharide size fractions by gel filtration chromatography (Fig. S5). Compared with an average molecular weight of LMWH (Enoxaparin, Clexane®), the unfractionated oligosaccharides of dAS-GAG (tetrasaccharide through octasaccharide) were pooled together, desalted and then freeze-dried. These individual oligosaccharide species of dAS-GAG were very similar to the oligosaccharide distribution of LMWH-Enoxaparin, which were used to investigate the inhibition of SARS spike variant-ACE2 binding by competitive immunoassay assay. Based on the potential activity with a mean of 50% inhibitory concentration (IC50) value, the concentration-dependent inhibition at an inhibitor concentration ranging from 4000–30 μg mL−1 was tested for the dAS-GAG oligosaccharides in comparison with other sulfated polysaccharides, i.e. Enoxaparin and pentosan polysulfate (PPS) (Fig. 4 ). Detailed IC50 results are summarized in Table S2. The unfractionated dAS-GAG gave a low IC50 value of 28.4 μg mL−1 compared with PPS (IC50 = 57.1 μg mL−1), and Enoxaparin (IC50 = 91.5 μg mL−1). Although the IC50 curve fits for this figure have substantial uncertainty due to a lack of sufficient data at a concentration below 30 μg mL−1, the trend is clear. All of these compounds interfered with SARS-CoV-2 spike-ACE2 interaction in a dose-dependent manner, which is comparable to the measurement of the binding kinetics and affinity of SARS-CoV-2 S-RBD interaction with heparin using a Surface Plasmon Resonance [24,25]. The weaker inhibitory activity of LMWH was observed to give less potent inhibition than unfractionated heparin (UFH) polysaccharide [22], supporting the notion that the patterning of the sulfated domains in the chains may play a role in binding. These remains considerable potential for non-systematic use of the sulfated polysaccharides as anti-viral treatment. According to the previous literatures [[25], [26], [27]], even very large doses of UFH and Enoxaparin administered via inhalation has very poor serum bioavailability. However, longer term toxicology and pharmacokinetic studies of intranasal of these heparin-like substances are currently underway. Based on a purpose of SARS-CoV-2 attachment and entry, we can imagine that the mechanism of the potent anti-SARS-CoV-2 activity from acharan sulfate involves the ability of this GAGs interaction to the surface of the viral spike protein. The spike protein is responsible for binding to the ACE2 receptor on the surface of host cells, which allows the virus to enter and infect the cells. It can be noted that the acharan sulfate can compete with the ACE2 receptor for binding to the spike protein, effectively blocking the virus from entering the host cell. This competition occurs because the spike protein has a binding site for heparan sulfate that is located adjacent to the ACE2 binding domain [21,25]. It can be noted that heparin-like acharan sulfate may have potential as a therapeutic agent for COVID-19.
Fig. 4.
Normalized binding inhibition (%) for concentration-dependent inhibition of the S-RBD and ACE2 interaction by; pentosan polysulfate (PPS), Enoxaparin, and dAS-GAG oligosaccharide. A competition of S-ACE2 binding assay was performed by the RayBio® COVID-19 assay kit which uses a 96-well plate coated with recombinantly-expressed human ACE2 protein. The biological analysis was treated by ANOVA, p < 0.05 was selected prior to experiments to reflect statistical significance.
3. Conclusion
Heparin is derived from mammalian sources and might be contaminated with animal proteins and apathogenic agents. Here, we prepared the sulfated GAGs from the mucus of Achatina fulica, characterised its structure, and investigated its anti-viral effects by analysing its binding inhibitory activity. The results were able to show that acharan sulfate from the snail mucus presents heterogeneity regarding negative net charge density and/or molecular mass. The key enzymatic depolymerization process was performed using a previously described procedure with some modification, which could easily isolate and purify the active fraction from the starting raw material. Furthermore, we identified the presence of a GAG disaccharide component, not described before, that arises in minor amounts, showing that this heparin-related GAG presents N-sulfate groups at the glucosamine unit [10]. The purified AS-GAG is mainly composed of , with a proportion of 6:1:1. Since hexasaccharides are less inhibitory against the target protein binding, there is a clear need for the octasaccharide size in optimal binding. As we know, acharan sulfate is widely distributed in the whole body of Achatina fulica, indicating that this GAGs from the snail mucus may be extremely important to the physiology of this gastropod [12]. In addition, competition Surface Plasmon Resonance studies using acharan sulfate oligosaccharides reviewed that efficient binding of pseudotype particles requires both chain length-dependent manner and sulfation probability (data not shown). Thus, we propose a model for the acharan sulfate oligosaccharide mediated cell binding and entry. The study was carried out to investigate the inhibitory effect of AS-GAG oligosaccharide on the binding of the spike protein to the ACE2 receptor. Importantly, it should be noted that this study was conducted in laboratory settings and that the results are not conclusive as a treatment for COVID-19.
4. Experimental
Treatment and purification of large snail mucus polysaccharides–50 g of snail mucus collected from the surface of live snails was freeze-dried. The thus-obtained powder was defatted by 3-times of acetone extraction for 24 h, and then was dried in heating oven at 37 °C. The dried powder was dissolved in 3-vol of sodium acetate buffer (pH 5) containing 100 mg pronase (7.0 U/mg solid), 5 mM EDTA, and 5 mM cysteine. The mixture was incubated at 60 °C for 24 h with sharking at 400 rpm in an incubator. After boiling for 10 min, the cooled mixture was centrifuged at 7000 g for 30 min at 10 °C, and then the polysaccharide in the clear supernatant was precipitated with 4-vol of 5% sodium acetate in cold ethanol. After standing at 4 °C for another 24 h, the pellet formed was collected by centrifugation at 7000 g for 30 min at 4 °C. The resulting residues were reconstituted with water and dialyzed against distilled water (with three changes of distilled water in 24 h). The solution was collected and lyophilized to obtain a pale-white powder consisting of the sulfated mucus-GAGs polysaccharides, it was designated mGAG, that was used for the purification process in next further study.
The crude mGAG polysaccharide was fractionated by an anion-exchange chromatography on a DEAE cross-linked beaded Sepharose column (Hitrap DEAE FF 5 mL, 160 × 250 mm, Cytiva) with elution by a stepwise concentration of 0.5 M–1.0 M NaCl in 50 mM sodium acetate buffer (pH 5.5). The fractions were monitored at 210 nm and the flow rate was set at 2.5 mL/min. Each fraction was desalted using a Hitrap cross-linked dextran column (14.5 × 50 mm, Cytiva), and lyophilized to give a sulfated GAGs polysaccharide as called by F1 to F3 fractions.
The homogeneity of the high mGAG content were determined using liquid chromatography on Superdex 30 Increased 10/300 GL column (Cytiva) by elution with 50 mM phosphate buffer with 0.15 M NaCl (pH 7.0) at 1.5 CV at flow rate 0.5 mL/min operated by ÄKTA Avant 25 (Cytiva). The standards curve for Low molecular weight protein was obtained by applied standard mixtures (1 mg/mL blue dextran, 1 mg/mL aprotinin, 2 mg/mL ribonuclease A (28-4038-41), LMW gel filtration calibration kits (Cytiva), 1 mg/mL cobalamine (Sigma-Aldrich), 0.5 mg/mL phenol red, and 1 mg/mL glycine (Vivantis)). A calibration curve was plotted by the logarithm of molecular weights versus retention time in each standard position. The molecular weight of mGAG fraction was estimated by compared with the calibration curve.
Gel electrophoresis– Polyacrylamide gel electrophoresis (PAGE) was performed for the analysis of the GAGs containing in the digested snail GAGs sample after treatment with protease. Briefly, a polyacrylamide gel (10 × 10 cm, 1.5-mm-thick of 15% acrylamide) was prepared and 20 μg of each oligosaccharide was subjected to electrophoresis at 90 V for 90 min 15 μg of heparin oligosaccharide standard was loaded on the same gel. Tris-Borate-EDTA buffer (pH 8.3) was used as running electrophoresis buffer. Oligosaccharides were visualized by 0.5% (w/v) Alcian Blue in 2% acetic acid for overnight. Destaining was achieved with several washing with by distilled water.
Disaccharide analysis using SAX-HPLC– Heparin and snail GAG oligosaccharides were depolymerized by 0.1 U of the mixing of heparin lyase I, II, and III, in 0.3 mL of 50 mM Tris-HCl buffer (pH 7.2) and 10 mM CaCl2 for 24 h at 37 °C, after which the reaction samples were heated in a boiling water for 3 min. The digested products were injected on an analytical SAX-HPLC column to monitor the reaction. A linear gradient of 0.1–1.0 M NaCl (pH 3.5) was performed over a 50 min period at a flow rate of 1.0 mL/min and the detection was set at 232 nm.
NMR analysis. – 40 mg of the lyophilized samples were dissolved in 99% deuterium oxide (D2O) as a solvent for NMR spectroscopy using a Bruker NEO™ 500 MHz NMR spectrometer (Bruker, Avance III™ HD, Karlsruhe, Germany) with a 5 mm multinuclear inverse probe at 296 K. The 1H and 13C spectra were observed at 500.18 and 125.78 MHz, respectively. The interpretation of 1H/13C heteronuclear single quantum coherence (HSQC) spectra was recorded using state-time proportion phase incrementation for quadrature detection in the indirect dimension. All chemical shifts were relative to the deuterium signal of the solvent.
Spike-ACE2 binding assay– A competition of Spike-ACE2 binding assay was performed by the RayBio® COVID-19 assay kit which uses a 96-well plate coated with recombinantly-expressed human ACE2 protein. The mixture of the testing reagent is then added to the wells in the presence of the recombinant Fc tagged SARS-CoV-2 spike RBD protein and incubated for 3 h at room temperature with shaking. Unbound RBD is removed with washing. HRP-conjugated anti-mouse IgG is applied to the wells and incubated for 1 h at room temperature with shaking. The reagent of 3,3′,5,5′-tetramethylbenzidine (TMB) substrate was added to the wells and incubated for 15 min at room temperature in the dark with shaking. The HRP-conjugated IgG binds to Fc tagged RBD protein and reacts with the TMB solution, producing a blue color that is proportional to the amount of bound RBD. Then, the HRP-TMB reaction is halted with the addition of 2.0 M sulfuric acid for stop-reaction, resulting in a blue-to-yellow color change. The intensity of the yellow color is measured at 450 nm. The biological analysis was treated by ANOVA, p < 0.05 was selected prior to experiments to reflect statistical significance. Data are the means ± SD of 3 experiments performed in duplicate.
Author contributions
K.K. and P.K. contributed toward the conceptualization of methodology and visualization; K.K. and T.C. performed the investigation, validation and formal analysis; K.K. wrote the original manuscript draft; K.K. and P.K. supervised the study and project administration; P.K. was responsible for funding acquisition. All authors contributed to editing and reviewing the manuscript and agreed to the published version of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
We would like to thank ADEN International Co.,Ltd., Thailand (ACHA Snail Project, Mr. Kitpong Puttarathuvanun, CEO (kitpong1@windowslive.com)) as patent's owner and providing the sources of snail mucus (Achatina fulica), and Bene pharmaChem, Germany for providing the pentosan polysulfate (PPS) for this research project. We are particularly grateful to Thailand Excellence Center for Tissue Engineering and Stem Cells (P.K.), Department of Biochemistry, Faculty of Medicine, Chiang Mai University, Chiang Mai, Thailand for helpful discussions concerning analysis and providing access to their experimental resources.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.carres.2023.108832.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
The authors do not have permission to share data.
References
- 1.Bishop J.R., Schuksz M., Esko J.D. Nature. 2007;446:1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- 2.Muñoz E.M., Linhardt R.J. Arterioscler. Thromb. Vasc. Biol. 2004;24:1549–1557. doi: 10.1161/01.ATV.0000137189.22999.3f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charles A.F., Scott D.A. J. Biol. Chem. 1933;102:431–435. [Google Scholar]
- 4.McLean J. American Physiological Society. 1916;41:250–257. [Google Scholar]
- 5.Lindahl U., Thunberg L., Bäckström G., Riesenfeld J., Nordling K., Björk I. J. Biol. Chem. 1984;259:12368–12376. [PubMed] [Google Scholar]
- 6.Zhang F., Yang B., Ly M., Solakyildirim K., Xiao Z., Wang Z., Beaudet J.M., Torelli A.Y., Dordick J.S., Linhardt R.J. Anal. Bioanal. Chem. 2011;401:2793. doi: 10.1007/s00216-011-5367-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeong J., Toida T., Muneta Y., Kosiishi I., Imanari T., Linhardt R.J., Choi H.S., Wu S.J., Kim Y.S. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2001;130:513–519. doi: 10.1016/s1096-4959(01)00468-7. [DOI] [PubMed] [Google Scholar]
- 8.Carnachan S.M., Bell T.J., Sims I.M., Smith R.A.A., Nurcombe V., Cool S.M., Hinkley S.F.R. Carbohydrate Polym. 2016;152:592–597. doi: 10.1016/j.carbpol.2016.07.024. [DOI] [PubMed] [Google Scholar]
- 9.Chi L., Munoz E.M., Choi H.S., Ha Y.W., Kim Y.S., Toida T., Linhardt R.J. Carbohydr. Res. 2006;341:864–869. doi: 10.1016/j.carres.2006.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim Y.S., Jo Y.Y., Chang I.M., Toida T., Park Y., Linhardt R.J. J. Biol. Chem. 1996;271:11750–11755. doi: 10.1074/jbc.271.20.11750. [DOI] [PubMed] [Google Scholar]
- 11.Park Y., Zhang Z., Laremore T.N., Li B., Sim J.-S., Im A.R., Ahn M.Y., Kim Y.S., Linhardt R.J. Glycoconj. J. 2008;25:863–877. doi: 10.1007/s10719-008-9149-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vieira T.C., Costa-Filho A., Salgado N.C., Allodi S., Valente A.P., Nasciutti L.E., Silva L.C. Eur. J. Biochem. 2004;271:845–854. doi: 10.1111/j.1432-1033.2004.03989.x. [DOI] [PubMed] [Google Scholar]
- 13.Spelta F., Liverani L., Peluso A., Marinozzi M., Urso E., Guerrini M., Naggi A. Original Research. 2019;6 doi: 10.3389/fmed.2019.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J., Zhou L., He Z., Gao N., Shang F., Xu J., Li Z., Yang Z., Wu M., Zhao J. Carbohydrate Polym. 2018;181:433–441. doi: 10.1016/j.carbpol.2017.10.091. [DOI] [PubMed] [Google Scholar]
- 15.Linhardt R.J., Toida T. Acc. Chem. Res. 2004;37:431–438. doi: 10.1021/ar030138x. [DOI] [PubMed] [Google Scholar]
- 16.Linhardt R.J. J. Med. Chem. 2003;46:2551–2564. doi: 10.1021/jm030176m. [DOI] [PubMed] [Google Scholar]
- 17.Karamanos N.K., Vanky P., Tzanakakis G.N., Tsegenidis T., Hjerpe A. J. Chromatogr. A. 1997;765:169–179. doi: 10.1016/s0021-9673(96)00930-2. [DOI] [PubMed] [Google Scholar]
- 18.Pomin V.H. Anal. Chem. 2014;86:65–94. doi: 10.1021/ac401791h. [DOI] [PubMed] [Google Scholar]
- 19.Griffin C.C., Linhardt R.J., Van Gorp C.L., Toida T., Hileman R.E., Schubert R.L., 2nd, Brown S.E. Carbohydr. Res. 1995;276:183–197. doi: 10.1016/0008-6215(95)00166-q. [DOI] [PubMed] [Google Scholar]
- 20.Kim S.H., Kearns F.L., Rosenfeld M.A., Casalino L., Papanikolas M.J., Simmerling C., Amaro R.E., Freeman R. ACS Cent. Sci. 2022;8:22–42. doi: 10.1021/acscentsci.1c01080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., Narayanan A., Majowicz S.A., Kwong E.M., McVicar R.N., Thacker B.E., Glass C.A., Yang Z., Torres J.L., Golden G.J., Bartels P.L., Porell R.N., Garretson A.F., Laubach L., Feldman J., Yin X., Pu Y., Hauser B.M., Caradonna T.M., Kellman B.P., Martino C., Gordts P.L.S.M., Chanda S.K., Schmidt A.G., Godula K., Leibel S.L., Jose J., Corbett K.D., Ward A.B., Carlin A.F., Esko J.D. Cell. 2020;183:1043–1057.e1015. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Partridge L.J., Urwin L., Nicklin M.J.H., James D.C., Green L.R., Monk P.N. Cells. 2021;10 doi: 10.3390/cells10061419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cuker A., Tseng E.K., Nieuwlaat R., Angchaisuksiri P., Blair C., Dane K., Davila J., DeSancho M.T., Diuguid D., Griffin D.O., Kahn S.R., Klok F.A., Lee A.I., Neumann I., Pai A., Pai M., Righini M., Sanfilippo K.M., Siegal D., Skara M., Touri K., Akl E.A., Bou Akl I., Boulos M., Brignardello-Petersen R., Charide R., Chan M., Dearness K., Darzi A.J., Kolb P., Colunga-Lozano L.E., Mansour R., Morgano G.P., Morsi R.Z., Noori A., Piggott T., Qiu Y., Roldan Y., Schünemann F., Stevens A., Solo K., Ventresca M., Wiercioch W., Mustafa R.A., Schünemann H.J. Blood Advances. 2021;5:872–888. doi: 10.1182/bloodadvances.2020003763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gelbach A.L., Zhang F., Kwon S.-J., Bates J.T., Farmer A.P., Dordick J.S., Wang C., Linhardt R.J. Original Research. 2022;9 doi: 10.3389/fmolb.2022.912887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tandon R., Sharp J.S., Zhang F., Pomin V.H., Ashpole N.M., Mitra D., McCandless M.G., Jin W., Liu H., Sharma P., Linhardt R.J. J. Virol. 2021;95 doi: 10.1128/JVI.01987-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 27.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaseja M., Rej R.N., Sauriol F., Perlin A.S. Can. J. Chem. 1989;67:1449–1456. [Google Scholar]
- 29.Mulloy B., Forster M.J., Jones C., Drake A.F., Johnson E.A., Davies D.B. Carbohydr. Res. 1994;255:1–26. doi: 10.1016/s0008-6215(00)90968-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors do not have permission to share data.