Abstract
Genetic studies of Campylobacter jejuni have been limited due to the lack of a transposon mutagenesis method. Here, we describe a novel technique for random transposon mutagenesis using a mariner-based transposon into C. jejuni strain 480. Insertions were random, as demonstrated by Southern blot analysis and insertional junction sequencing. We have demonstrated, for the first time, random in vivo transposon mutagenesis of C. jejuni.
Campylobacter jejuni has been identified as the leading cause of acute bacterial diarrhea in the United States, and yet the mechanisms by which this bacterium causes disease in humans are not well understood (11). This paucity of information is partially due to a lack of genetic tools and the relatively recent understanding of Campylobacter spp. as important human pathogens. In particular, the lack of an in vivo transposon mutagenesis method for the efficient generation of random mutants of Campylobacter spp. has restricted molecular genetic studies. To date, the generation of C. jejuni mutant libraries has been limited to transposon shuttle mutagenesis (4) and homologous insertional mutagenesis (2, 16). Unsuccessful attempts have been made to introduce both Gram-positive- and Gram-negative-organism-based transposons into Campylobacter spp. (5). For example, nonrandom site-specific insertional mutants were generated by a TnphoA transposon in C. jejuni (13). Recently, a highly permissive mariner-based transposon known as Himar1 has been utilized for efficient in vivo random transposon mutagenesis in Escherichia coli and mycobacteria (10). We therefore designed and tested an in vivo mariner-based transposon mutagenesis system for the production of random insertional mutants of C. jejuni.
Bacterial strains, media, and growth conditions.
C. jejuni 480 is a highly electrocompetent strain isolated during an outbreak of campylobacteriosis (provided by B. A. M. van der Zeijst) (3). Strain 480 was grown routinely on Mueller-Hinton (MH) agar supplemented with 5% sheep's blood, vancomycin (10 μg/ml), polymyxin B (2.6 U/ml), and trimethoprim (5 μg/ml) at 42°C under microaerobic conditions (5% O2, 10% CO2, 85% N2 gas).
Construction of C. jejuni mini-transposon.
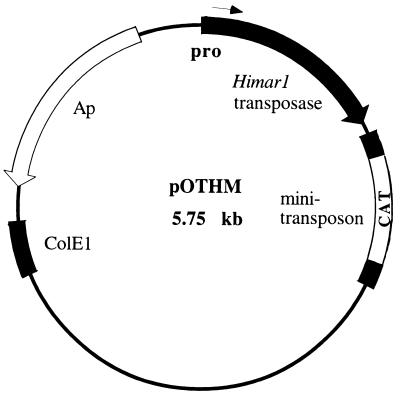
We constructed an in vivo mini-transposon system for C. jejuni using the mariner family of mini-transposons previously applied to other bacteria (1, 10). Primers (5′-CCAACGCGTGGGCTGCAGGGGAGATCTTCTAGATGCTCGGCGGTGTTCCTTTCCAAG-3′ and 5′-CCAACGCGTTGCGCCCTTTAGTTCCTAAAGGGT-3′) were used to amplify the E. coli/C. jejuni-compatible chloramphenicol resistance (Cmr) cassette of pRY111 (provided by P. Guerry) (15). This product was cut with MluI and subcloned in MluI-digested plasmid pEMCAT (1; A. Camelli, unpublished results), replacing the resident Cmr gene within the mini-transposon to create pEMCjCAT. To allow Himar1 transposase expression in C. jejuni, a C. jejuni-specific promoter was used to replace the original Himar1 promoter. A C. jejuni promoter, GenBank accession no. AJ002027, was used for this purpose. This promoter was found to be active in C. jejuni and inactive in E. coli, as indicated by β-galactosidase activity (14), and was chosen due to concerns that an overactive transposase might be toxic to E. coli (10). The C. jejuni promoter was constructed using partially complementary primers (P1, 5′-CCATCTAGAAAGCTTACTTATGTTAAATTTAATTTATCTTATTTTTGCTATATTAACGCCATAAA-3′; P2, 5′-CCAGCATGCCCCCATATGAGCCTTTCTTAAATGTT AATTTTATGGCGTTAATATAGCAAA-3′). P1 and P2 were annealed and extended with Taq and Pfu (Stratagene) DNA polymerase (10:1), and the product was cut with NdeI. A promoterless Himar1 transposase was amplified from a plasmid containing the C9 hyperactive mutant of the Himar1 transposase (6) by using primers (H1, 5′-CCAAAGCTTCCCATATGGGAAAAAAAGGAATTTCGTG-3′; H2, 5′-CCAGCATGCTTATTATTCAACATAGTTCCCTTC-3′). This product was cut with NdeI and ligated to the NdeI-digested promoter product. Primers P1 and H2 were then used to amplify the C. jejuni-compatible promoter-Himar1 fusion by PCR, and the product was cloned in pCR2.1 (Invitrogen) to generate pCRPH. The C. jejuni-compatible promoter-transposase construct and mini-transposon sequences were moved into pUC19 in separate ligation steps following double digestions of pCRPH and pEMCjCAT with HindIII-SphI and SphI-BamHI, respectively. This resulted in the construction of pOTHM (Fig. 1), which is incapable of replicating in C. jejuni and thus serves as a suicide delivery vector.
FIG. 1.
Structure of pOTHM mini-transposon mariner-based delivery system. Himar1 transposase, mariner-based transposase under C. jejuni-specific promoter; mini-transposon, mini-transposon of short inverted repeats flanking antibiotic resistance marker; pro, C. jejuni promoter; CAT, chloramphenicol acetyltransferase antibiotic marker functional in E. coli and C. jejuni; ColE1, pUC19-derived origin of replication; Ap, E. coli ampicillin resistance gene.
Mini-transposon delivery into C. jejuni.
pOTHM was introduced by electroporation into the C. jejuni strain 480 as described by Wassenaar et al. (13). This strain was chosen because of its ability to readily accept exogenous DNA. One microgram of pOTHM was used to electroporate approximately 50 μl of a solution at 5 × 1011 bacteria/ml at 1.25 V, 600 Ω, and 25 μF in a 0.1-cm cuvette. Cells were allowed to recover on Columbia blood agar plates (8) for 4 to 5 h at 37°C under microaerobic conditions. The bacteria were harvested from the plate surface and resuspended in 0.5 ml of MH broth. Transformants were selected at 37°C for 72 h on MH agar supplemented with 5% sheep's blood and 5 μg of chloramphenicol per ml. This procedure typically resulted in 37 ± 4 (mean ± standard deviation) chloramphenicol-resistant colonies per electroporation (n = 5). Transformants were picked and characterized as described below.
Analysis of pOTHM insertions.
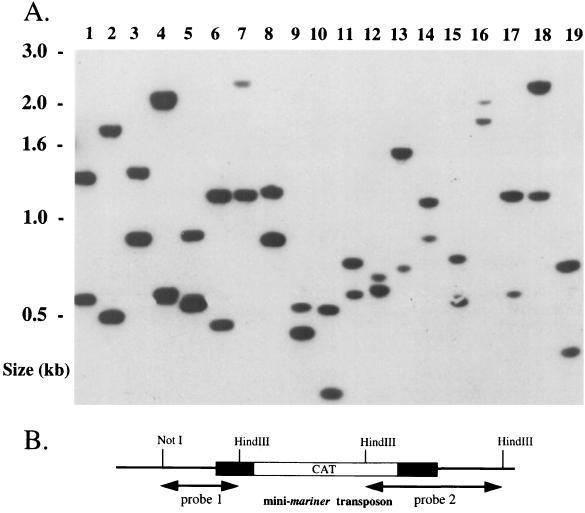
Southern blot analysis was performed on 19 randomly picked transformants obtained from a single electroporation experiment. Genomic DNA was isolated using the G-nome DNA kit (Bio101), digested with HindIII, electrophoretically separated on a 1.8% agarose gel, and transferred to a Hybond N+ membrane (Amersham). The membrane was then probed with DNA containing the 5′ and 3′ portions of the mini-transposon (Fig. 2B) by using the ECL RPN 3000 detection system (Amersham). Two fragments of various sizes would be expected for each transformant with these probes, provided that a single random transposition event had occurred. C. jejuni 480 wild-type chromosomal DNA was used as a negative control and failed to hybridize with the probes as expected (data not shown). As demonstrated in Fig. 2A, each of the 19 transformants had two uniquely sized bands, suggesting that a single random transposon insertion had occurred in each transformant.
FIG. 2.
Southern blot analysis of Campylobacter mini-transposon insertion strains. (A) Lanes 1 to 19, randomly chosen transformants isolated from one transformation experiment. (B) Plasmid pOTHM was cut with HindIII and NotI, and fragments were separated by agarose gel electrophoresis. DNA fragments containing the 5′ and 3′ mini-transposon junctional sequences, probes 1 and 2, respectively, were then isolated and used together as a probe. We believe differences in band intensities relate to variations in DNA transfer.
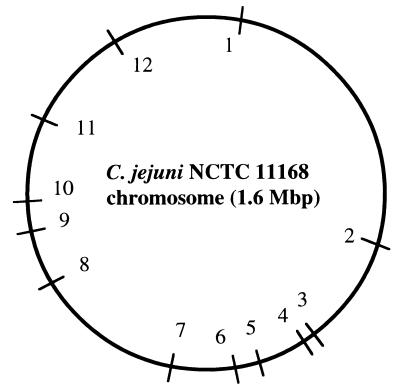
To further confirm the randomness of insertion, the mini-transposon chromosomal junctions were sequenced from 12 mutants obtained from two independent transposition experiments and the sites of insertion were mapped on the complete chromosomal sequence of C. jejuni NCTC 11168 (C. jejuni NCTC 11168 Sequencing Group at the Sanger Centre [http://www.sanger.ac.uk/Projects/C_jejuni/]) (Fig. 3). Inverse PCR of the 12 transformants was performed using HindIII-cut circularized transformant genomic DNA. Primer 1 (5′-CTTCCCAAACGTAAATATCGGCAGTAG-3′) and primer 2 (5′TATCGCTCTTGAAGGGAACTATGTTG3′) extended outward from within the Cmr cassette and were used to determine the insertion site within the C. jejuni genome. Primer walking-directed sequencing of inverse PCR products (using primer 1) was performed with the ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction Kit with Amplitaq DNA Polymerase, FS (Perkin-Elmer). Cycling of oligonucleotides was performed in the thermal cycler (GeneAmp 9600; Perkin-Elmer) by following the instructions in Perkin-Elmer protocol P/N 402078. Reaction products were run on a 373 DNA Sequencer, Stretch (Applied Biosystems). The sequence data obtained from each of the 12 junctions demonstrated a high degree of identity to different segments of the C. jejuni NCTC 11168 chromosome. Furthermore, each of the 12 derived sequences is of either known Campylobacter genes or genes encoding putative proteins homologous to other bacterial species and ORFs of unknown function (Table 1). Analysis of the 30-bp sequence flanking each of the 12 insertion sites revealed no consensus sequence other than the invariant TA dinucleotide which the Himar1 mariner transposon recognizes (7, 10), which might indicate insertion site preference. Based on these results, we conclude that the mini-transposon inserts with a high degree of randomness throughout the C. jejuni genome.
FIG. 3.
Locations of mini-transposon insertion sites within the C. jejuni genome. The nucleotide sequences flanking the insertion sites of 12 transformants were determined; their relative positions within the C. jejuni NCTC 11168 chromosome (9) are shown. Transformants 1 to 12 represent individual C. jejuni 480 mutants selected randomly from two independent transposon mutagenesis experiments. The corresponding C. jejuni genes disrupted by the insertion are as follows: 1, Cj0035c; 2, Cj0558c; 3, Cj0719c; 4, Cj0737; 5, Cj0823; 6, Cj0863c; 7, Cj0967; 8, Cj1205c; 9, Cj1283; 10, Cj1338c; 11, Cj1457c; 12, Cj1624c.
TABLE 1.
Mini-transposon insertion into putative C. jejuni NCTC 11168 ORFs
| Transformant | Homologous ORFa (GenBank accession no.) | E valuea | Organismb |
|---|---|---|---|
| 1 | Unknown | NA | NA |
| 2 | proA (AAA23030) | 0.0 | C. jejuni |
| 3 | orfX (CAA73267) | 8.0E−21 | Lactococcus lactis |
| 4 | hxuA (U08349) | 1.0E−10 | Haemophilus influenzae |
| 5 | Unknown | NA | NA |
| 6 | xprB (AAA62787) | 1.0E−18 | E. coli |
| 7 | Unknown | NA | NA |
| 8 | radA (AAC33293) | 6.0E−85 | Listeria monocytogenes |
| 9 | ktrB (BAA32063) | 1.0E−49 | Vibrio alginolyticus |
| 10 | flaB (AAA23027) | 0.0 | E. coli |
| 11 | Unknown | NA | NA |
| 12 | sdaB (L07763) | 3.0E−97 | E. coli |
DNA sequences of mini-transposon junction sites were used to determine C. jejuni open reading frames (ORFs) by using the Sanger Center C. jejuni NCTC 11168 BLAST program. Homologous ORFs and E values were determined using the GenBank database BLAST program.
Organism in which the homologous ORF was found. NA, not applicable.
Significance.
We have been able to demonstrate, for the first time, random in vivo transposon mutagenesis of C. jejuni. Prior attempts to mutagenize C. jejuni by in vivo transposition have been hindered by a combination of incompatibilities with promoter usage resulting in little or no expression of the transposase (12), the absence of species-specific cofactors necessary for transposition, and a lack of natural transposons (7). For these reasons, we chose to test the ability of the highly permissive Himar1 transposase to mediate random insertional mutagenesis in C. jejuni. As described above, this novel method of transposon mutagenesis in C. jejuni has resulted in the generation of highly random insertional mutations. The efficiency of the procedure is high enough that an insertion library of high complexity can be readily generated. This will greatly aid genetic analysis of this intestinal pathogen by screening for putative virulence determinants that have been inactivated by the insertional mutagenesis process.
Acknowledgments
Research support for this study includes the following grants from the National Institutes of Health (NIH), Bethesda, Md.: HL-55660, AI-16242, and AI-39067 (D.W.K.A.) and AI-40262 and P 30 DK-34928 (A.C.) for the Center of Gastroenterology Research on Absorptive and Secretory Processes.
We thank P. Guerry for her generous gift of pRY111 and B. A. M. van der Zeijst for kindly providing C. jejuni strain 480. We thank Xioping Zhang and Elizabeth A. Joyce for helpful discussions and Abraham L. Sonenshein for the reading of the manuscript. We also thank Anne V. Kane and Ka Ly for their technical assistance.
REFERENCES
- 1.Akerley B J, Rubin E J, Camilli A, Lampe D J, Robertson H M, Mekalanos J J. Systematic identification of essential genes by in vitro mariner mutagenesis. Proc Natl Acad Sci USA. 1998;95:8927–8932. doi: 10.1073/pnas.95.15.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bleumink-Pluym N M, Verschoor F, Gaastra W, van der Zeijst B A, Fry B N. A novel approach for the construction of a Campylobacter mutant library. Microbiology. 1999;145:2145–2151. doi: 10.1099/13500872-145-8-2145. [DOI] [PubMed] [Google Scholar]
- 3.King V, Wassenaar T, van der Zeijst B A M, Newell D G. Variations in Campylobacter jejuni flagellin, and flagellin genes, during in vivo and in vitro passage. Microb Ecol Health Dis. 1991;4:135–140. [Google Scholar]
- 4.Labigne A, Courcoux P, Tompkins L. Cloning of Campylobacter jejuni genes required for leucine biosynthesis, and construction of leu-negative mutant of C. jejuni by shuttle transposon mutagenesis. Res Microbiol. 1992;143:15–26. doi: 10.1016/0923-2508(92)90030-r. [DOI] [PubMed] [Google Scholar]
- 5.Labigne-Roussel A, Courcoux P, Tompkins L. Gene disruption and replacement as a feasible approach for mutagenesis of Campylobacter jejuni. J Bacteriol. 1988;170:1704–1708. doi: 10.1128/jb.170.4.1704-1708.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lampe D J, Akerley B J, Rubin E J, Mekalanos J J, Robertson H M. Hyperactive transposase mutants of the Himar1 mariner transposon. Proc Natl Acad Sci USA. 1999;96:11428–11433. doi: 10.1073/pnas.96.20.11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lampe D J, Churchill M E, Robertson H M. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 1996;15:5470–5479. [PMC free article] [PubMed] [Google Scholar]
- 8.Nuijten P J, Bleumink-Pluym N M, Gaastra W, van der Zeijst B A. Flagellin expression in Campylobacter jejuni is regulated at the transcriptional level. Infect Immun. 1989;57:1084–1088. doi: 10.1128/iai.57.4.1084-1088.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parkhill J, Wren B W, Mungall K, Ketley J M, Churcher C, Basham D, Chillingworth T, Davies R M, Feltwell T, Holroyd S, Jagels K, Karlyshev A V, Moule S, Pallen M J, Penn C W, Quail M A, Rajandream M A, Rutherford K M, van Vliet A H, Whitehead S, Barrell B G. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 10.Rubin E J, Akerley B J, Novik V N, Lampe D J, Husson R N, Mekalanos J J. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc Natl Acad Sci USA. 1999;96:1645–1650. doi: 10.1073/pnas.96.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tauxe R V. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations. In: Nachamkin I, Blaser M J, Tompkins L S, editors. Campylobacter jejuni: current status and future trends. Washington, D.C.: American Society for Microbiology; 1992. pp. 9–19. [Google Scholar]
- 12.Tompkins L S. Genetic and molecular approach to Campylobacter pathogenesis. In: Nachamkin I, Blaser M J, Tompkins L S, editors. Campylobacter jejuni: current status and future trends. Washington, D.C.: American Society for Microbiology; 1992. pp. 241–254. [Google Scholar]
- 13.Wassenaar T M, Fry B N, van der Zeijst B A. Genetic manipulation of Campylobacter: evaluation of natural transformation and electro-transformation. Gene. 1993;132:131–135. doi: 10.1016/0378-1119(93)90525-8. [DOI] [PubMed] [Google Scholar]
- 14.Wosten M M, Boeve M, Koot M G, van Nuene A C, van der Zeijst B A. Identification of Campylobacter jejuni promoter sequences. J Bacteriol. 1998;180:594–599. doi: 10.1128/jb.180.3.594-599.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao R, Alm R A, Trust T J, Guerry P. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene. 1993;130:127–130. doi: 10.1016/0378-1119(93)90355-7. [DOI] [PubMed] [Google Scholar]
- 16.Yao R, Burr D H, Doig P, Trust T J, Niu H, Guerry P. Isolation of motile and non-motile insertional mutants of Campylobacter jejuni: the role of motility in adherence and invasion of eukaryotic cells. Mol Microbiol. 1994;14:883–893. doi: 10.1111/j.1365-2958.1994.tb01324.x. [DOI] [PubMed] [Google Scholar]