Abstract
Graphical modeling of multivariate functional data is becoming increasingly important in a wide variety of applications. The changes of graph structure can often be attributed to external variables, such as the diagnosis status or time, the latter of which gives rise to the problem of dynamic graphical modeling. Most existing methods focus on estimating the graph by aggregating samples, but largely ignore the subject-level heterogeneity due to the external variables. In this article, we introduce a conditional graphical model for multivariate random functions, where we treat the external variables as conditioning set, and allow the graph structure to vary with the external variables. Our method is built on two new linear operators, the conditional precision operator and the conditional partial correlation operator, which extend the precision matrix and the partial correlation matrix to both the conditional and functional settings. We show that their nonzero elements can be used to characterize the conditional graphs, and develop the corresponding estimators. We establish the uniform convergence of the proposed estimators and the consistency of the estimated graph, while allowing the graph size to grow with the sample size, and accommodating both completely and partially observed data. We demonstrate the efficacy of the method through both simulations and a study of brain functional connectivity network.
Keywords: Brain connectivity analysis, Functional magnetic resonance imaging, Graphical model, Karhunen–Loève expansion, Linear operator, Reproducing kernel Hilbert space
1. Introduction
Functional graphical modeling is gaining increasing attention in the recent years, where the central goal is to investigate the interdependence among multivariate random functions. Applications include time course gene expression data in genomics (Wei and Li 2008), multivariate time series data in finance (Tsay and Pourahmadi 2017), electrocorticography, and functional magnetic resonance data in neuroimaging (Zhang et al. 2015), among many others.
Our motivation is brain functional connectivity analysis based on functional magnetic resonance imaging (fMRI). Functional MRI measures brain neural activities via blood oxygenlevel-dependent signals. It depicts brain functional connectivity network, which is shown to alter under different disorders or during different brain developmental stages. Such alterations contain crucial insights of both disorder pathology and development of the brain (Fox and Greicius 2010). The fMRI data are often summarized in the form of a location by time matrix for each individual subject. The rows correspond to a set of brain regions, and the columns correspond to time points that are usually 500 msec to 2 sec apart and span a few minutes in total. From the fMRI scans, a graph is constructed, where nodes represent brain regions, and links represent interactions and dependencies among the regions (Fornito, Zalesky, and Breakspear 2013). Numerous statistical methods have been developed to estimate functional connectivity network. Most of these methods treat the fMRI data as multivariate random variables with repeated observations, where each region is represented by a random variable and the time-course data are taken as repeated measures of that variable (e.g., Bullmore and Sporns 2009; Wang et al. 2016b). There are recent proposals to model the fMRI data as multivariate functions, where the time-course data of each region are taken as a function (e.g., Li and Solea 2018). Given the continuous nature and the short-time interval between the adjacent sampling points of fMRI, we treat the data as multivariate functions, and formulate the connectivity network estimation as a functional graphical modeling problem in this article.
Nearly, all existing graph estimation methods tackle the problem by aggregating samples, sometimes according to the diagnostic groups. However, there is considerable subject-level heterogeneity, which may contain crucial information for our understanding of the network, but has been largely untapped or ignored by existing methods (Fornito, Zalesky, and Breakspear 2013). Heterogeneity could arise due to a subject’s phenotype profile; for example, in our study in Section 7, the brain connectivity network may vary with an individual’s intelligence score. It could also arise due to a time variable, for example, the subject’s age, and the connectivity network may vary with age, which leads to dynamic graphical modeling. In this article, we introduce a conditional functional graphical model for a set of random functions, by modeling the external variables such as the phenotype or age as the conditioning set. Our proposal thus extends two lines of existing and relevant research: from conditional and dynamic graphical modeling of random variables to that of random functions, and from unconditional functional graphical modeling to conditional functional graphical modeling.
The first line of relevant research is the class of graphical models of random variables. Within this class, there is a rich literature on unconditional graphical models (Yuan and Lin 2007; Friedman et al. 2008; Peng et al. 2009, among others). There are extensions to joint estimation of multiple graphs, which arise from a small number of groups, typically two or three, according to an external variable such as the diagnostic status (Danaher, Wang, and Witten 2011; Chun, Zhang, and Zhao 2015; Lee and Liu 2015; Zhu and Li 2018). There have also been some recent proposals of dynamic graphical models (Kolar et al. 2010; Xu and Hero 2014; Zhang and Cao 2017). However, they only considered a discrete-time setting, in which the network is estimated only at a small and discrete number of time points. The most relevant work to ours is Qiu et al. (2016), who also targeted estimation of the individual graph according to an external variable, for example, age. Although both Qiu et al. (2016) and our method are designed for graph estimation at the individual level, the two solutions differ in many ways. Particularly, Qiu et al. (2016) assumed that the repeated observations of each individual come from the same Gaussian distribution, whose dependency was required to follow a certain stationary time series structure. By contrast, we treat the repeated measurements as realizations from a random function, and do not impose any structural or relational assumption on the entire function. More importantly, compared to the random variable setting in Qiu et al. (2016), our functional setting involves an utterly different and new set of modeling techniques and theoretical tools.
The second line of relevant research is the class of unconditional graphical models of random functions, which appeared only recently. Qiao, Guo, and James (2019) introduced a functional Gaussian graphical model, by assuming that the random functions follow a multivariate Gaussian distribution. Li and Solea (2018) relaxed the Gaussian assumption, developed a precision operator that generalizes the concept of precision matrix to the functional setting, and used it to estimate the unconditional functional graph. However, their precision operator is nonrandom, and their graph dimension is fixed. By contrast, we introduce a conditional precision operator (CPO), which is a function of the conditioning variable and is thus random, and we allow the graph dimension to diverge with the sample size. These differences bring extra challenges in analyzing the operator-based statistics. Moreover, because the relation between the CPO and the conditioning variable can be nonlinear, its estimation requires the construction of a reproducing kernel Hilbert space (RKHS) of the conditioning variable, which leads to a more complex asymptotic analysis than that of Li and Solea (2018). We also derive a number of concentration bounds and uniform convergence for our proposed estimators, while such results are not available in Li and Solea (2018).
To address the problem of conditional graphical modeling of random functions, we introduce two new linear operators: the CPO, and the conditional partial correlation operator (CPCO), which extend the precision matrix and the partial correlation matrix from the random variable setting to both the conditional and functional settings. We show that, when the conditional distribution is Gaussian, the conditional graph can be thoroughly captured by the nonzero elements of CPO or CPCO. This property echoes the classic result where a static graph can be inferred by the precision matrix or the partial correlation matrix under the Gaussian assumption. We note that, some early works, such as Lee, Li, and Zhao (2016a,b), also estimated the parameters of interest through linear operators. However, we studied utterly different problems: Lee, Li, and Zhao (2016b) targeted variable selection in classical regressions, Lee, Li, and Zhao (2016a) targeted unconditional graph estimation for random variables, while we target conditional graph estimation for random functions. Both the methodology and theory involved are thus substantially different.
Our proposal makes useful contributions at multiple fronts. On the method side, it offers a new class of statistical models to study conditional graph estimation for multivariate functional data, a problem that remains largely unaddressed. We investigate the parallels between the random variable-based and random function-based graphs, and between the unconditional and conditional graphs. On the theory side, our work develops new tools for operator-based functional data analysis. We establish the conditional graph estimation consistency, along with a set of concentration inequalities and error bounds, for our proposed method. To our knowledge, very little work has investigated function-on-function dependency at such a level of complexity that involves estimating the linear operators under a conditional framework. The tools we develop are general, and can be applied to other settings in high-dimensional functional data analysis. On the computation side, under a properly defined coordinate system, the proposed operators are functions of the sample covariance operator of dimension n × n, with n being the sample size. It is relatively easy to calculate, and the accompanying estimation algorithm can be scaled up to large graphs.
The rest of the article is organized as follows. We begin with a formal definition of conditional functional graphical model in Section 2. We introduce a series of linear operators in Section 3, develop their estimators in Section 4, and study their asymptotic properties in Section 5. We report the simulations in Section 6, and an analysis of an fMRI dataset in Section 7. We relegate all proofs and some additional results to the supplementary appendix.
2. Model
In this section, we formally define the conditional functional graphical model. Let (ΩX, ) be a measurable space. Suppose is a Hilbert space of -valued functions on an interval , for i = 1, …, p, and ΩX is the Cartesian product . Suppose X = (X1, …, Xp)⊤ is a p-dimensional random element on ΩX. Let G = (V, E) be an undirected graph, where V = {1, …, p} represents the set of vertices corresponding to the p random functions, and E = {(i, j) ∈ V × V, i = j} represents the set of undirected edges. A common approach to modeling an undirected graph is to associate the separation on the graph with the conditional independence; in other words, nodes i and j are separated in G if and only if Xi and Xj are independent given the rest of X, that is,
| (1) |
where X−(i,j) represents X with its ith and jth components removed, and ⫫ represents statistical independence. Based on Equation (1), Qiao, Guo, and James (2019) proposed a functional graphical model, which assumed that X follows a multivariate Gaussian distribution.
Next, we introduce a conditional functional graphical model that allows the graph links to vary with the external variables. We focus on the univariate external variable case, but our method can be generalized to multivariate external variables, or external functions. Let Y be a random element defined on ΩY, and be the Borel σ-field generated by the open sets in ΩY. Let and be the distribution of Y and conditional distribution of X | Y, respectively. We next give our formal definition.
Definition 1.
Suppose a random graph Ey for each y ∈ ΩY is defined via the mapping ΩY → 2V×V, y → Ey, where 2V×V is the power set of V × V. We say X follows a conditional functional graphical model with respect to Ey if and only if, for y ∈ ΩY,
| (2) |
We note that Li, Chun, and Zhao (2012) introduced the notion of conditional graphical model. However, our notion of conditional functional graphical model is considerably different, in that our model extends theirs not only from the setting of random variables to random functions, but also from the setting of static graphs to random graphs. Specifically, letting X = (X1, …, Xp)⊤ and Y = (Y1, …, Yq)⊤ denote two random vectors, Li, Chun, and Zhao (2012) considered the model,
for all . In this model, E0 ⊆ 2V×V is a fixed graph, and does not change with the value of Y. In comparison, our model in Equation (2) allows X to be a p-variate random function, and the graph Ey to vary with Y.
3. Linear Operators
In this section, we first introduce a series of linear operators, based on which we then formally define the CPO and the CPCO. Finally, we study the relation between these two operators and the conditional functional graph.
We adopt the following notation throughout this article. For two generic Hilbert spaces Ω, Ω′, let and denote the class of all bounded and Hilbert–Schmidt operators from Ω to Ω′, respectively. We abbreviate and as and whenever they are appropriate. Moreover, let ∥·∥ and ∥·∥HS be the operator norm in and Hilbert-Schmidt norm in . Moreover, let ker(A), range(A), denote the null space, the range, and the closure of the range of an operator A, respectively.
3.1. Conditional Covariance and Correlation Operators
We first define three covariance operators, , , and . We then define the conditional covariance operator , and the conditional correlation operator , which is the building block of CPO and CPCO.
Let be a positive-definite kernel, be its corresponding RKHS, and L2(PY) be the collection of all square-integrable functions of Y under PY. The next assumption ensures the square-integrability of Xi under PX|Y, and that is a subset of L2(PY).
Assumption 1.
There exist M0 > 0 and MY > 0 such that , for i = 1, …, p, and κy(Y, Y) ≤ MY.
We comment that, we choose RKHS as the modeling space, so that the relation of X on Y can be very flexible, and the kernel matrix of Y is of dimension n × n, an attractive feature when the dimension of Y is large compared with n. In addition, a good number of asymptotic tools for RKHS operators have been developed (see, Bach 2009; Lee, Li, and Zhao 2016a). That being said, our theoretical development can also be easily extended to the spaces beyond RKHS. In fact, our population development only requires that is a proper subset of L2(PY), which can be ensured by the square-integrable condition, for an M > 0 and every .
Let ⊗ denote the tensor product; then κy(·, Y) ⊗ κy(·, Y), Xi ⊗ Xj and κy(·, Y) ⊗ Xi ⊗ Xj are random elements in , , and , respectively. Their expectations uniquely define the covariance operators,
| (3) |
for all , , and h, h1, . The next proposition justifies the existence of , , and .
Proposition 1.
If Assumption 1 holds, then there exist linear operators , , and satisfying the relations in Equation (3).
We next introduce a regression operator, through the relation, , where † is the Moore–Penrose inverse. We first need an assumption on the ranges of and , and is sufficiently rich in L2(PY).
Assumption 2.
For every (i, j) ∈ V × V, . Moreover, is dense in L2(PY).
By Assumption 2, for any , there exists a unique such that . Therefore, the inverse is defined as , h ↦ h′, which implies that is well-defined. The range condition that is satisfied generally. For instance, it holds when the rank of is finite, which is reasonable, because in practice can often be approximated by the spanning space of a few leading eigenfunctions of .
The next proposition shows that maps every to E(〈f, Xi〉 〈g, Xj〉 | y), that is, the conditional expectation, and thus this operator can be viewed as a regression operator.
Proposition 2.
If Assumptions 1 and 2 hold, then for all y ∈ ΩY and , we have .
Now we are ready to define the conditional covariance operator, whose existence is justified by Proposition 2 and Riesz representation theorem.
Definition 2.
For each y ∈ ΩY, the bilinear form, , , uniquely defines an operator , via for all . We call the conditional covariance operator.
Note that the mapping , defines a random operator. If the conditional expectations E(〈f, Xi〉| y) = 0, then induces the conditional covariance cov(〈f, Xi〉, 〈g, Xj〉 | y). When Xi and Xj are random vectors, Fukumizu, Bach, and Jordan (2009) introduced the homoscedastic conditional covariance operator, which induces E[cov(〈f, Xi〉, 〈g, Xj〉 | Y)]. Our conditional covariance operator is different from that of Fukumizu, Bach, and Jordan (2009), as it extends the classical conditional covariance to the functional setting, and it deals directly with cov(〈f, Xi〉, 〈g, Xj〉 | y), instead of its expectation. We write the joint operator as the block matrix of operators whose (i, j)th element is ,1 ≤ i, j ≤ p, or more explicitly, , for any f = (f1, …, fp)⊤ ∈ ΩX.
Given the conditional covariance operator , we next define the conditional correlation operator, , for each y ∈ ΩY via,
| (4) |
Its existence and uniqueness are ensured by Baker (1973). Similar to the construction of , we write the joint operator as the block matrix of operators whose (i, j)th element is , 1 ≤ i, j ≤ p. Let denote the block diagonal matrix , for i ∈ V. We then have .
Next, we impose the distributional assumption on X | y.
Assumption 3.
Suppose X | y follows the conditional functional graphical model as defined via (2), and the conditional distribution of X = (X1, …, Xp)⊤ given Y = y follows a centered Gaussian distribution.
The zero-mean condition is imposed to simplify both methodological and theoretical development, and can be relaxed with some modifications. Moreover, we require the conditional distribution X | y to follow a Gaussian distribution. It is possible to relax this Gaussian assumption, by extending the notion of functional additive conditional independence (Li and Solea 2018), or the copula graphical model (Liu et al. 2012). However, we feel that the Gaussian case itself is worthy of a full investigation, and we leave the non-Gaussian extension as future research.
Assumption 3, together with Definition 2, imply that, for f = (f1, …, fp)⊤ ∈ ΩX, , where .
3.2. Conditional Precision and Partial CorrelationOperators
We next formally define the operators CPO and CPCO, then establish their relations with the conditional functional graph. We introduce two assumptions, which ensure that is Hilbert-Schmidt and is invertible. We present some intuitions here, but relegate the detailed technical discussion to Section S.3 in the appendix.
Assumption 4.
For each y ∈ ΩY and i ∈ V, let denote the collection of eigenvalue and eigenfunction pairs of . Let . There exists c1 > 0 such that,
| (5) |
Assumption 5.
For each y ∈ ΩY, .
Assumption 4 characterizes the level of smoothness for the underlying distributions of the random functions. Assumption 5 is to prevent the existence of a constant function consisting of linear combination of nonconstant functions. It can be viewed as the generalization of the nonexistence of collinearity in linear models, or the empty concavity space in generalized liner models (Hastie and Tibshirani 1990). Assumption 4 ensures that is Hilbert-Schmidt, and thus compact. Mean-while, Assumptions 4 and 5 together ensure that is lower-bounded by a strictly positive constant. This implies that is invertible, and that is bounded, which justifies the following definition.
Definition 3.
Define the CPO as the inverse of the joint conditional correlation operator, , for any y ∈ ΩY.
The operator generalizes the precision matrix to the functional and conditional settings. We should clarify that, unlike the standard definition where the precision matrix is defined as the inverse of the covariance matrix (Cai, Liu, and Luo 2011), our CPO is defined as the inverse of the conditional correlation operator. This is to avoid taking inversion on the covariance operator, which is usually not invertible because of its compactness. Next, we develop an operator that generalizes the partial correlation matrix to the functional and conditional setting. Similar to the definition for , we define for any subsets A, B ⊆ V. Therefore, for any subsets A ⊆ V\{i, j}, We define an intermediate operator, through the relation, , for any (i, j) ∈ V × V. We then have the following result if A= −(i, j).
Proposition 3.
There uniquely exists which satisfies that , and .
Its proof is similar to that of (Lee, Li, and Zhao 2016a, theo. 1) and is thus omitted. It justifies the definition of the following operator.
Definition 4.
We call the operator in Proposition 3 the CPCO between Xi and Xj given X−(i,j) and Y.
3.3. Relation With Conditional Functional Graph
We first show that the conditional covariance operator can be constructed by the functions of conditional covariances between the Karhunen-Loève coefficients and the associated eigenfunctions. This simple form provides a convenient way to estimate the conditional covariance operator later. Let denote the collection of eigenvalue and eigenvector pairs of , with . Then Xi can almost surely be represented as , where , for all . This expression is known as the Karhunen-Loève (K-L) expansion (Bosq 2000).
Proposition 4.
Suppose the same conditions in Proposition 2 hold. Then we have
where (, ) and (, ) are from the Karhunen-Loève expansion.
We next show that CPO and CPCO fully characterize the conditional functional independence, and are thus crucial for our estimation of conditional functional graph.
Theorem 1.
If Assumptions 1–5 hold, then we have, for any y ∈ ΩY,
where denotes the (i, j)th element of .
Under the Gaussian distribution, the equivalence between the conditional independence and the zero element of a nonrandom precision matrix is well known in the classical random variables setting. By contrast, Theorem 1 extends to the setting of random functions and also allows the precision operator to vary with Y.
Theorem 2.
If Assumptions 1–3 hold, then we have, for any y ∈ ΩY,
Theorems 1 and 2 suggest that one can estimate the conditional functional graph Ey in Equation (2) through the proposed operators, CPO or CPCO. In the following, we primarily focus on the graph estimation based on CPO, and investigate the corresponding asymptotics. The results based on CPCO can be derived in a parallel fashion, which we only briefly discuss in Section S.4 in the Appendix.
4. Estimation
In this section, we first derive the sample estimate of CPO and the conditional graph at the operator level. We then construct empirical bases and develop coordinate representations for the functions observed at a finite set of time points. Using these coordinate representations, we are able to compute our estimated linear operators. Last, we provide a step-by-step summary of our proposed estimation procedure.
4.1. Operator-Level Estimation
We first derive the sample Karhunen-Loève expansion. We and then sequentially develop the estimators of , , , and finally Ey.
Let (Y1, …, Yn) denote iid samples of Y, and (X1, …, Xn) denote iid samples of X, with , for k = 1, …, n. Let En denote the sample mean operator; that is, for a sample (ω1, …, ωn) from Ω, . We estimate the covariance operators, , , and , by
for any (i, j) ∈ V × V. For i ∈ V, let denote the collection of eigenvalue and eigenfunction pairs of . Then, we have , where is the ath coefficient from the K-L expansion of for the kth subject. Furthermore, we use the leading d terms to approximate ; in other words, we have, , for all k = 1, …, n, and i ∈ V.
By its definition, we estimate the regression operator by
| (6) |
where ϵY > 0 is a prespecified ridge parameter, and it imposes a level of smoothness on the regression structure. Next, by Propositions 2 and 4, given y, d, i, j, we estimate the conditional covariance operator by
| (7) |
where and are from the sample Karhunen-Loève expansion. Let denote the block matrix of operators . Similarly, we can define , for any A, B ⊆ V.
Therefore, by Equation (4), we estimate the conditional correlation operator by
| (8) |
for i ≠ j, i, j ∈ V, and , for i ∈ V, which is the identity mapping from to , and ϵ1 is a ridge regularization parameter imposed on the inverses of and . Let denote the block matrix of operators . The next result shows that the norm of is bounded by one, which resembles the property of the correlation in the classical setting.
Proposition 5.
If κY(y1, y2) ≥ 0 for any y1, y2 ∈ ΩY, then , for any (i, j) ∈ V × V.
Finally, we estimate the conditional precision matrix operator by
| (9) |
where ϵ2 > 0 is another ridge parameter. Using , for each y ∈ ΩY, we estimate the graph Ey by
| (10) |
where ρCPO > 0 is a thresholding parameter. That is, we can obtain an estimator of the conditional graph by hard thresholding. For notational simplicity, hereafter we abbreviate , , , , , , and , by , , , , , , and , respectively.
4.2. Empirical Bases and Coordinate Representation
We next introduce a set of empirical bases and the corresponding coordinate representations. We adopt the following notation. Let Ω be a Hilbert space of functions of T, spanned by a set of bases . For any ω ∈ Ω, let denote its coordinate with respect to . Then ω can be represented as . Let denote the Gram kernel matrix, which implies that, for any (ω1, ω2) ∈ Ω, the inner product . Throughout the article, we use the symbol ⌊·⌋ exclusively for a chosen coordinate system. Let Ω and Ω′ be two Hilbert spaces spanned by and , respectively. Let A : Ω → Ω′ be a linear operator. Then the coordinate representation of A with respect to and is . For a third Hilbert space Ω″ with base , and another linear operator A′ : Ω′ → Ω″, it is easy to see that . For simplicity, we use ⌊A⌋ instead of when there is no confusion.
Note that the random function can only be observed at a finite set of points. To enable computation, we need to approximate the random functions using the partially observed data. We adopt the construction used in Li and Solea (2018), which assumes the sample path of Xk lies on an RKHS of the time variable T with a finite basis. Specifically, suppose is observed on a finite set of time points , k = 1, … n. Let which pools together all the unique time points across all subjects, and N is the length of . Letting be a positive-definite kernel, then can be constructed through , for i ∈ V. Let be the N × N Gram matrix of κT, and its eigen-decomposition, , where is associated with the m leading eigenvalues, and associated with the last N − m eigenvalues. Here we require m ≤ N. Therefore, we can construct an orthonormal basis of ΩN via
| (11) |
where . Then can be represented as . Note that for the kth individual, the function is observed at nk time points, which implies that , where is the m × nk matrix . Therefore, for a given ridge parameter ϵT, the coordinate can be estimated by
| (12) |
We next derive the coordinate representations of , , , which then lead to the coordinates of , , and . Recall is the kernel used to build the RKHS of Y. Let be the corresponding n × n Gram matrix of κY, and .
Proposition 6.
For (f, g) ∈ ΩN × ΩN, , , and .
Moreover, for each i ∈ V, let denote the collection of eigenvalue and eigenvector pairs of ; that is . Then for each i ∈ V, k = 1, …, n, the sample Karhunen-Loève expansion of is of the form,
| (13) |
Proposition 7.
Let , for each i ∈ V. For y ∈ ΩY, the coordinate representation of with respect to and is
| (14) |
where c(y) = diag[ℓ(y)] with , and . Moreover, if is the collection , then,
| (15) |
where , and Id is the d × d identity matrix.
Finally, we compute the squared Hilbert–Schmidt norm of as
for (i, j) ∈ V × V with i ≠ j, and ∥ · ∥F is the Frobenius norm.
4.3. Algorithm
We now summarize our conditional functional graph estimation algorithm based on CPO. The algorithm based on CPCO is similar and is thus omitted. Let λi(A) be the ith largest eigenvalues of an a × a matrix A, for i = 1, …, a.
Choose the kernel function κT. Some commonly used kernel functions include the Brownian motion function κT(s, t) = min(s, t), or the radial basis function (RBF) κT(s, t) = exp[−γT(s − t)2], . For RBF, the bandwidth γT is determined by .
Compute the N × N Gram matrix KT of κT, and let be its eigen-decomposition associated with its m leading eigenvalues. Then use (11) to construct the reduced basis , and the matrix . We suggest choosing m by , in the sense that the cumulative percentage of total variation of KT explained exceeds 99%.
Calculate the coordinate , using (12) with a given ϵT. We choose ϵT = N−1/5λ1(KT) (see, for example Lee, Li, and Zhao 2016a).
Perform eigen-decomposition of , for i ∈ V, and obtain the ath eigenvector , and the ath K-L coefficient using (13), a = 1, …, d. Stack to form , a = 1, …, d, i = 1, …, p. We choose d according to .
Choose the kernel function κY, and compute the corresponding n × n Gram matrix KY. Compute the coordinate using (14), with the ridge parameter ϵY = n−1/5λ1(KY).
Compute the representation of using (15), for a given y ∈ ΩY and the ridge parameters , and .
- Estimate the conditional functional graph for a given y ∈ ΩY by , with a given threshold ρCPO. We determine ρCPO by minimizing the following generalized cross-validation (GCV) criterion over a set of grid points,
where is the sample estimate of , and its coordinate representation is derived in Section S.4 of the appendix, and DFi(ρ) = d2 card [Ni(ρ)], with card(·) being the cardinality and the neighborhood of the ith node in .
Our graph estimation algorithm involves multiple tuning parameters, and their choices are given in the above algorithm. We further study their effects in Section S.6 of the appendix. In general, we have found that the estimated graph is not overly sensitive to the tuning parameters as long as they are within a reasonable range.
We also remark that, the above algorithm assumes the zero-mean condition. In practice, if this does not hold, we can easily modify the algorithm. Specifically, from the coordinates of the estimated conditional covariance operator, we can estimate by , where 1n is the n-dimensional vector with all ones. By redefining the conditional covariance operator as , we can estimate the coordinates of by , where c′ (y) = diag[ℓ(y)] − ℓ(y) ℓ(y)⊤. We then replace in Step 5 of the algorithm, and all subsequent procedures, by .
5. Asymptotic Theory
We begin with some useful concentration inequalities and uniform convergence for several relevant operators. We next establish the uniform convergence of the CPO, and the consistency of the estimated graph. For simplicity, we first assume that the trajectory of the random functions X(t) = [X1(t), …, Xp(t)]⊤ is fully observed for all t ∈ T. We then discuss the setting when Xi is only partially observed in Section 5.3. We also remark that, all our theoretical results allow the dimension of the graph to diverge with the sample size.
5.1. Concentration Inequalities and Uniform Convergence
We first derive the concentration bound and uniform convergence rate for the sample estimators , , and . For two positive sequences {an} and {bn}, write an ≺ bn if an = o(bn), an ⪯ bn if an = O(bn), and an ≍ bn if an ⪯ bn and bn ⪯ an; moreover, let an ∨ bn = bn if an ⪯ bn. Similarly, if cn is a third positive sequence, then we let an ∨ bn ∨ cn = (an ∨ bn) ∨ cn.
Theorem 3.
If Assumptions 1 and 3 hold, then there exist positive constants C1 to C6, such that, (i) ; (ii) ; (iii) , for any t ≥ 0 and any (i, j) ∈ V × V. Moreover, if log p/n → 0, then (iv) ; (v) ; (vi) , as n →∞.
For any (i, j) ∈ V × V and y ∈ Ω, let be the truncated version of . Next, we establish the uniform convergence rate for . We define the exponent of a compact and self-adjoint operator as , for any β > 0, where is the collection of eigenvalue and eigenfunction pairs of A. We need another assumption.
Assumption 6.
For any (i, j) ∈ V × V, there exist β ∈ (0, 1), c2 > 0, and such that with .
This assumption regulates the complexity of (Xi, Xj)⊤ given Y. To see this, note that, as β increases, the regression relation from (Xi, Xj)⊤ on Y are more concentrated on those eigenfunctions corresponding to larger eigenvalues of . Also, we define as the minimal isolation distance among all d + 1 leading eigenvalues of , for all i ∈ V. Note that we allow d to grow with n.
Theorem 4.
If Assumptions 1–3 and 6 hold, ϵY ≺ 1, and , then .
Next, we establish the uniform convergence of the estimated conditional correlation operator . We need an assumption on the tail behavior of random functions.
Assumption 7.
There exists γy > 0 such that , for any y ∈ ΩY.
Assumption 7 is on the decaying rate of the tail eigenvalues of , which, in a sense, characterizes the smoothness of the distribution of Xi given Y.
Theorem 5.
If Assumptions 1–4, 6, and 7 hold, ϵY, ϵ1 ≺ 1, and , then, for any y ∈ ΩY,
where .
To better understand Theorem 5, suppose , , , and , for a1, …, a4 > 0. Then we have
where π = 2(a1 + a2 + a4) + 6a3 < 1. This implies that the graph dimension p can diverge with the sample size n at an exponential rate. In comparison, in the classical random variable setting, the uniform convergence rate of the sample covariance is (log p/n)1/2 (Bickel and Levina 2008). Theorem 5 thus can be viewed as an extension to both the functional and conditional settings where the parameter of interest is a random RKHS operator.
5.2. Uniform Convergence of CPO and Graph Consistency
We next derive the convergence of the estimated CPO, . We need an assumption to regulate the relation between Xi and Xj when conditioning on X−(i,j) and Y.
Assumption 8.
There exists c3 > 0 such that, , where , for any y ∈ ΩY
The following proposition provides a condition under which Assumption 8 is satisfied. Its proof is similar to Proposition S2 and is omitted.
Proposition 8.
Suppose Assumptions 1, 3–5 hold. Then, for any y ∈ ΩY, there exists c3 > 0 such that, , if there exists c4 > 0 satisfying that
| (16) |
where , and are the pairs of eigenvalue and eigenfunction of , and , for any i ∈ V and y ∈ ΩY.
Both conditions (5) and (16) introduce certain levels of smoothness on the conditional distribution of X given Y. Nevertheless, they target different subjects: Equation (5) is about the relation between Xi and Xj given Y, which is required for the consistency of , whereas Equation (16) is about the relation between Xi and Xj given X−(i,j) and Y, which is required for the consistency of .
Theorem 6.
If Assumptions 1–8 hold, ϵY, ϵ1 ≺ 1, and , then for any y ∈ ΩY,
Moreover, if , and , then,
We make a few remarks. First, Li and Solea (2018) established the consistency of their precision operator, as well as the unconditional graph estimation consistency. Note that their operator is nonrandom, and their results were derived with the graph size p fixed. By contrast, Theorem 6 establishes the uniform convergence of the operator , which is random, and the graph estimation consistency is obtained with a diverging p. For instance, if ϵ2 O(n−π′/2) with π′ > 0, then Theorem 6 says that the uniform convergence rate of the estimated CPO depends on p(log p/n1−π−π′)1/2. This implies that we allow p to grow at the polynomial rate of n.
Second, a careful inspection of our proof reveals that, the convergence rate of the empirical CPO depends on the difference , whose order, by Theorem 5, can be no faster than p(log p/n1−π′)1/2. We note that, under the classical random variable setting, the convergence rate of the hard thresholding sample covariance matrix in terms of Frobenius norm is [pc0(p) log p/n]1/2, where the term c0(p) imposes a sparsity structure on the covariance matrix and satisfies that , for i = 1, …, p, with 1(·) being the indicator function (Bickel and Levina 2008, theo. 2). We feel our rate of p(log p/n1−π)1/2 is reasonable and is comparable to the classical result. We also note that there is a difference between p and p1/2 in our rate and the rate of (Bickel and Levina 2008, theo. 2). This difference is mainly due to different sparsity settings imposed by our method and by Bickel and Levina (2008). Note that we do not impose any sparsity structure on the conditional correlation operator . This means we need to estimate all the off-diagonal elements of , whose cardinality grows in the order of p2. In comparison, Bickel and Levina (2008) imposed a sparsity structure on the covariance matrix and the cardinality of nonzero covariances grows only in the order of pc0(p). Moreover, we are dealing with a more complicated setting of random functions and random operators. On the other hand, we show in Section S.5 in the appendix that, if we introduce additional sparsity and regularization, we can further improve the rates in Lemma S8 in the appendix and Theorem 6, so that p can grow at an exponential rate of n. Actually, we have developed a theoretical platform that is not only limited to the present setting. The concept of using vanishing CPO to identify the conditional independence between random functions in a conditional graph can have divarication beyond the scenarios studied in this article; for example, when there are additional sparsity or bendable structures.
Finally, in our theory development, we have imposed a series of technical conditions, which are commonly imposed in the literature, and are usually easy to satisfy.
5.3. Consistency for Partially Observed Random Functions
We next derive the consistency under the scenario when the random functions are only partially observed. Partially observed functions are collected via a dense or a non-dense measurement schedule; see Wang, Chiou, and Muller (2016a) for more discussion on measurement schedule. To avoid digressing from the main context, in this article, we do not go after any specific measurement schedule or regularity setting on the partially observed random function. For partially observed functions X(t), suppose is the estimate of X(t) using the empirical bases developed in Section 4.2. We then estimate the series of the operators and the graph by
where , is the eigenfunction of , and is the block matrix of operators with (i, j)th element being .
Theorems 5 and 6 show that the convergence of the estimated CCO and CPO depends on the uniform convergence of the sample covariance operators in Theorem 3. In particular, it relies on the convergence rates of and . When the random functions are completely observed, both rates, by Theorem 3, are equal to (log p/n)1/2. When the random functions are only partially observed, we let the convergence rates of and to be slower than (log p/n)1/2. More specifically, as specified in Equation (17), we introduce a quantity a that reflects how dense the time points are observed in the random functions. The denser the time points are, the closer a is to zero. Correspondingly, the next theorem extends Theorem 6 to partially observed functions. Its proof is similar to that of Theorem 6, and is thus omitted.
Theorem 7.
If Assumptions 1–8 hold, ϵY, ϵ1 ≺ 1, and there exists a ∈ [0, 1) such that , with , and
| (17) |
Then, , and , for any y ∈ ΩY, as n →∞.
The first condition of Equation (17) is satisfied if the tail of behaves as a sub-Exponential distribution. That is, when there exist c′, c″ > 0 and a ∈ [0, 1), such that , for any t ≥ 0. A similar condition also holds for the second condition of Equation (17). Recall in Theorem 3, we have shown that, for completely observed functions, and behave as a sub-Gaussian distribution for a small t. As such, Equation (17) is more appropriate for partially observed functions.
6. Simulations
Next, we study the finite-sample performance of our method through simulations. Specifically, we consider three graph structures of E: a hub, a tree, and a random graph, as shown in Table 1. We consider p = 10 nodes with the sample size n = 100 first, and consider larger graphs later. Given the graph structure E and the conditional variable y, we generate Xj(t) and its parent nodes based on the following model:
where X(t) | y =[X1(t) | y, …, Xp(t) | y]⊤ is constructed sequentially via the given graph, Pj is the set of the parent nodes of j, and cj is the scale parameter as specified in Table 1. We generate the error function, εi(t) as , where ξ1, …, ξJ are iid standard normal variables, t1, …, tJ are equally spaced points between [0, 1] with J = 50, and κT is a RBF or a Brownian motion covariance function. We then generate the conditioning variable, Y1, …, Yn, as iid Uniform(0, 1). We generate each , k = 1, …, n, i = 1, …, p, with nk = 50 time points. In this model, there are two types of edge patterns: when νij = 1, the strength of edges grows with y, and when νij = 0, the strength of edges decays with y. The tuning parameters are determined by the rules suggested in Section 4.3. In Section S.6 of the appendix, we discuss in detail the effect of the tuning parameters, and show that our method is relatively robust to a range of tuning parameters.
Table 1.
Simulation setup: the edges under three graph structures.
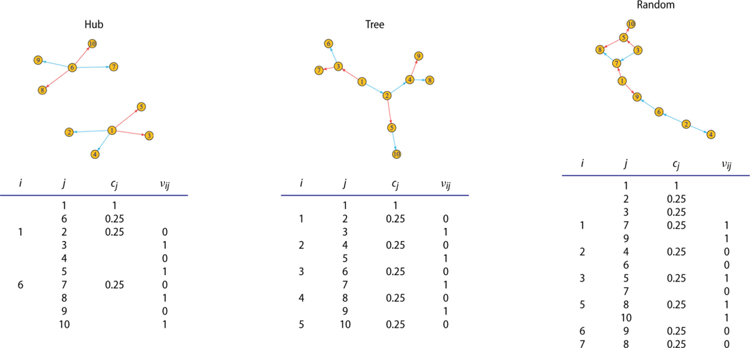
|
We compare our method with three alternative solutions, all of which are variants of graphical Lasso (Friedman et al. 2008, gLASSO). The first solution, which we refer as “Average,” is similar to Kolar et al. (2010). It first estimates the conditional covariance matrix by , where , j = 1, …, 50, k = 1, …, n. It then estimates the conditional precision matrix by applying gLASSO to . The second solution, which we refer as “Majority,” is similar to a procedure in Qiao, Guo, and James (2019). It first estimates the covariance matrix at each time point by . It then estimates the conditional precision matrix at each time point, by applying gLASSO to . It selects those edges that are detected by the majority of the estimates among all the estimated graphs. The third solution, referred as “Unconditional,” is similar to the naive procedure in Qiu et al. (2016), which, without using the information of Y, estimates the covariance matrix by . It then estimates the conditional precision matrix by applying gLASSO to . For the penalty parameter in gLASSO, we adopt the empirical rule suggested by Rothman et al. (2008), and set it to (log p/n)1/2. We use the RBF kernel for both κT and κY in all simulations.
We evaluate the accuracy of the estimated graph using the area under ROC curve (AUC). We first compute the false positive (FP) and true positive (TP) as,
for a given ρ and y ∈ ΩY, and E0 is the true graph. We then compute the pairs of (TPy,ρ, FPy,ρ) over a set of values of ρ, which we set to be the sorted norms of empirical CPO.
Figure 1 reports the AUC for the estimated graph with respect to the external variable Y, under three graph structures, and by the four methods. It is seen that the methods “Majority” and “Unconditional” perform consistently the worst, while “Average” and our proposed CPO methods perform similarly in this example.
Figure 1.
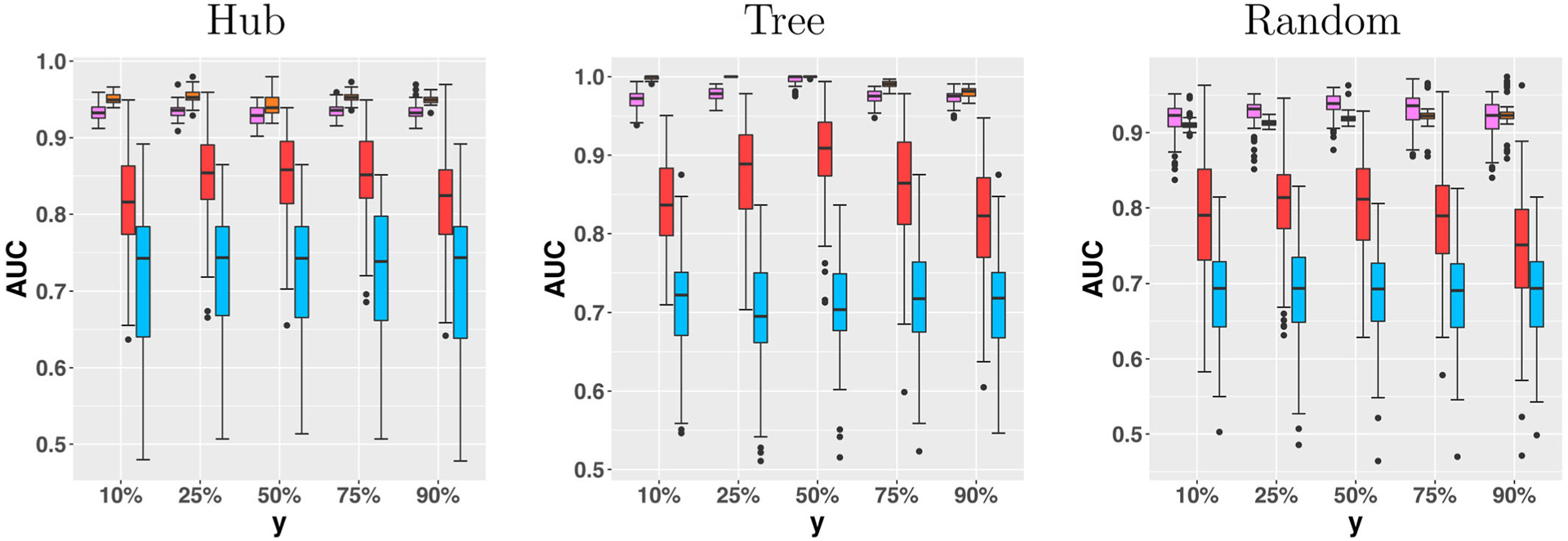
Area under the ROC curve for the estimated graph with respect to the external variable Y, under three graph structures, and by the four methods. From left to right: CPO, Average, Unconditional, Majority. The graph size p = 10.
We next extend our simulations to larger graphs, where we set the graph size p = {30, 50, 100}, with the sample size n = 30 and nk = 30 time points, k = 1, …, n. We then generate the graph in a similar way as before. Specifically, for the hub structure, we generate p/5 independent hubs, and within each hub, two edges have their strength growing with y, and two edges decaying with y. For the tree structure, we expand the tree until the graph reaches the designated size, and in each subsequent layer of the tree, one edge has growing strength and the other has decaying strength with y. For the random structure, we select the edges randomly following a Bernoulli distribution with probability 1/(p−1), and also randomly select half of the selected edges to have growing strength and the other half to have decaying strength. Due to the poor performance of “Majority” and “Unconditional” in the previous simulation setting, we only compare our CPO method with “Average” in the large-graph setting. Figure 2 reports the AUC. It is seen that our CPO method clearly outperforms the “Average” method in some settings, and is comparable in other settings. In particular, for a large graph with p = 100, the improvements of CPO over “Average” in both the hub and random structures are substantial.
Figure 2.
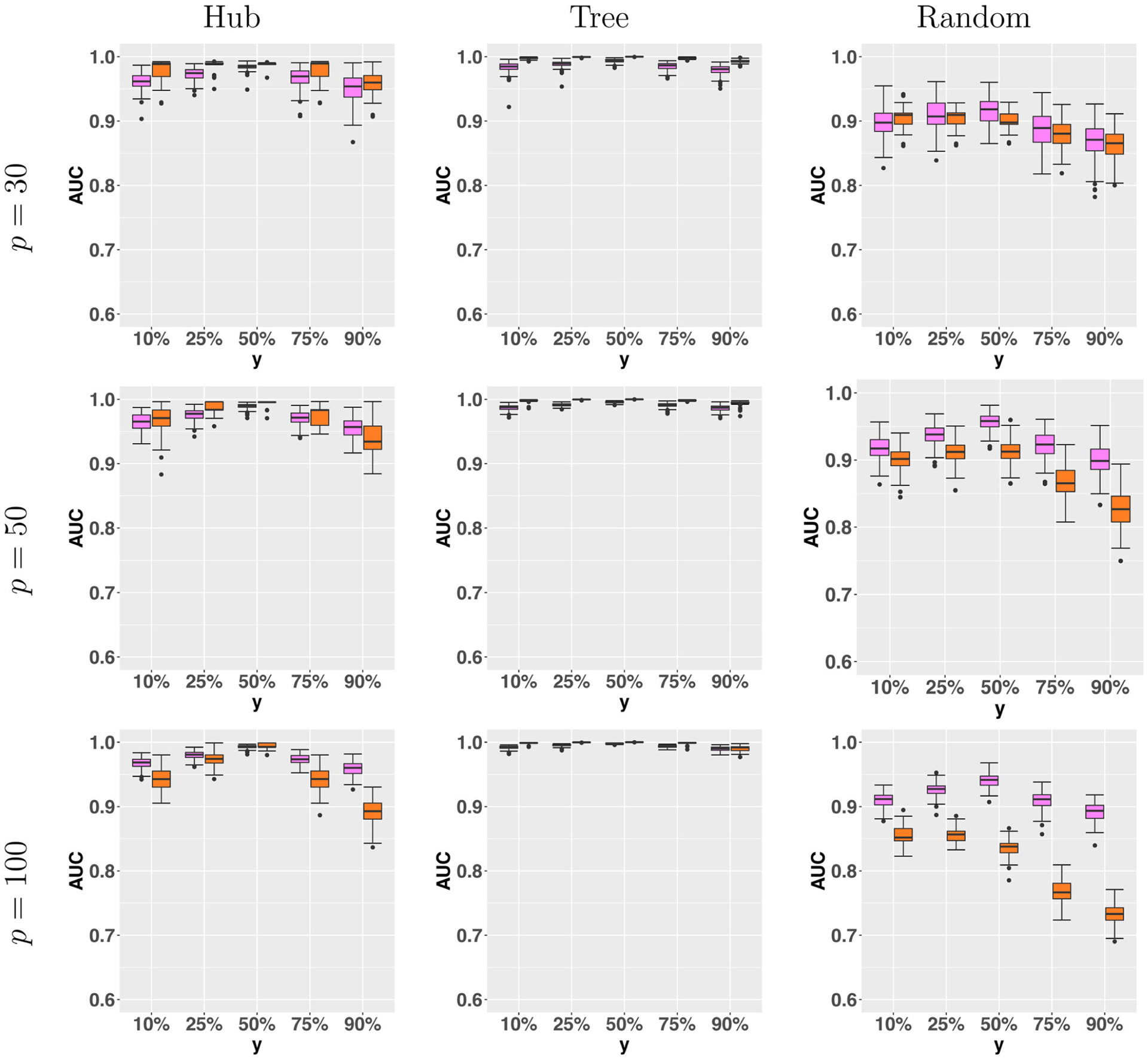
Area under the ROC curve for the estimated graph with respect to the external variable Y, under three graph structures, and by the two methods. From left to right: CPO, Average. The graph size p = {30, 50, 100}.
7. Application
In this section, we illustrate our conditional functional graph estimation method with a brain functional connectivity analysis example. We analyze a dataset from the Human Connectome Project (HCP), which consists of resting-state fMRI scans of 549 individuals. Each fMRI scan has been processed and summarized as a spatial temporal matrix, with rows corresponding to 268 brain regions-of-interest, and columns corresponding to 1200 time points (Greene et al. 2018). Additionally, each subject is collected with a score of the Penn Progressive Matrices, which is a nonverbal group test typically used in educational settings and is generally taken as a surrogate of fluid intelligence. It is of scientific interest to unravel the connectivity patterns of the brain regions conditioning on the intelligence measure.
We apply our conditional functional graphical modeling approach for the whole brain of 268 × 268 connectivity network. Applying the hard thresholding approach would yield a sparse estimate of the conditional graph at each value of Y = y. To identify edges that vary along with the conditional variable Y, we further employ a permutation test approach. Specifically, we permute the observations of Y five hundred times. For each permuted sample, we compute the Hilbert-Schmidt (H-S) norm of the sample CPO for each edge. We then compute the variance of the H-S norms based on 500 permutations. We treat those edges whose corresponding variances above 95% percentile as significant, since in this context a nonzero variance implies that the CPO varies with the conditional variable. In addition, those 268 brain regions have been partitioned into eight functional modules: medial frontal, frontoparietal, default mode, motor, visual, limbic, basal ganglia, and cerebellum (Finn et al. 2015). We count the number of significant edges from the permutation test within each functional module, then use the Fisher’s exact test to determine if a module is significantly enriched.
We have found that the medial frontal module is significantly enriched with numerous significant edges that vary along with the intelligence score. Figure 3 shows the identified significant edges. This finding agrees with the literature, as this module has been reported to contribute to high-level cognitive functions such as emotional responses (Smith et al. 2018) and learning (Zanto and Gazzaley 2013). Moreover, this module was found to have more impact on fluid intelligence compared to other modules (Finn et al. 2015). We have also observed more cross-lobe and inter-hemisphere interactions, which suggests the importance of these edges in cognitive functions. This again complies with the existing literature in neuroscience that the altered interhemispheric interactions of prefrontal lobe is closely related to higher level cognitive traits such as the Internet gaming disorder (Wang et al. 2015). Figure 4 reports the changes of the identified significant edges for the medial frontal module at five different values of the intelligence score. We see from the plot that both increasing and decreasing patterns exist for the strength of the significantly varying edges.
Figure 3.
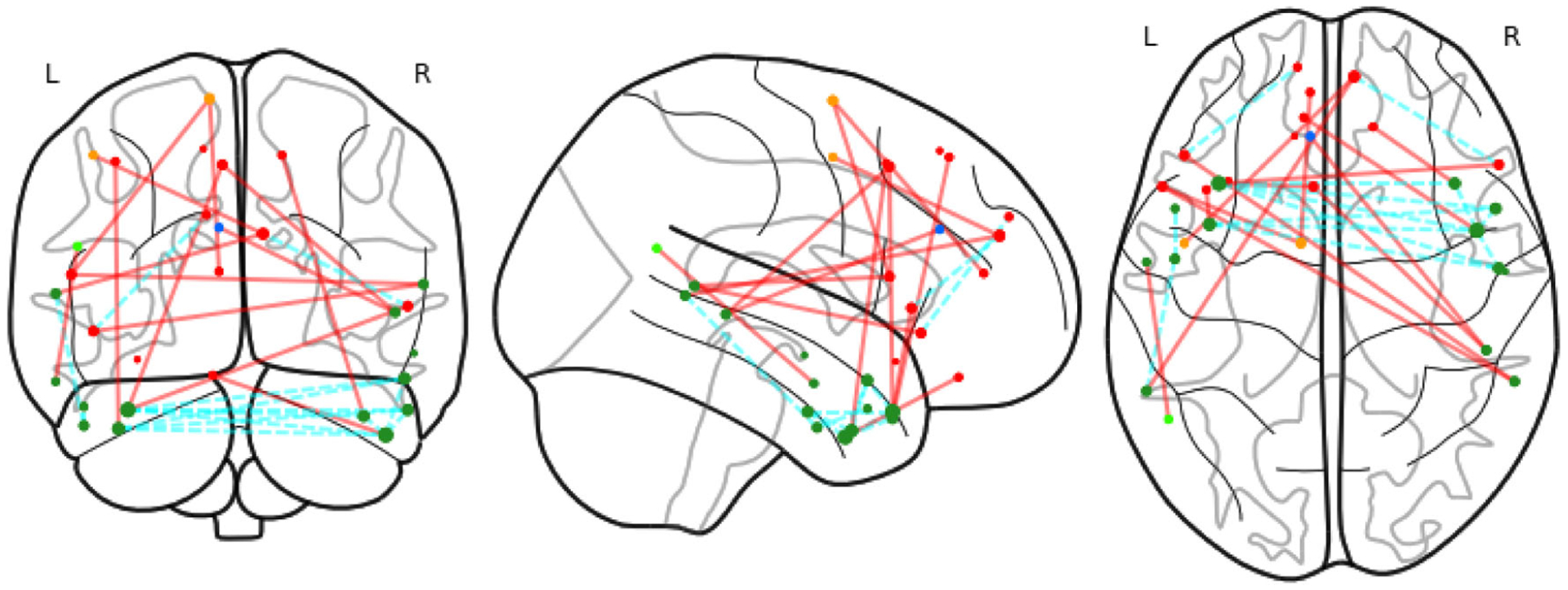
Medial frontal network, with significant edges. Different colors of nodes indicate the ROIs in different lobes: prefrontal, motor, parietal, temporal, limbic and cerebellum. Red lines indicate inter-lobe edges and cyan ones for inner-lobe edges.
Figure 4.
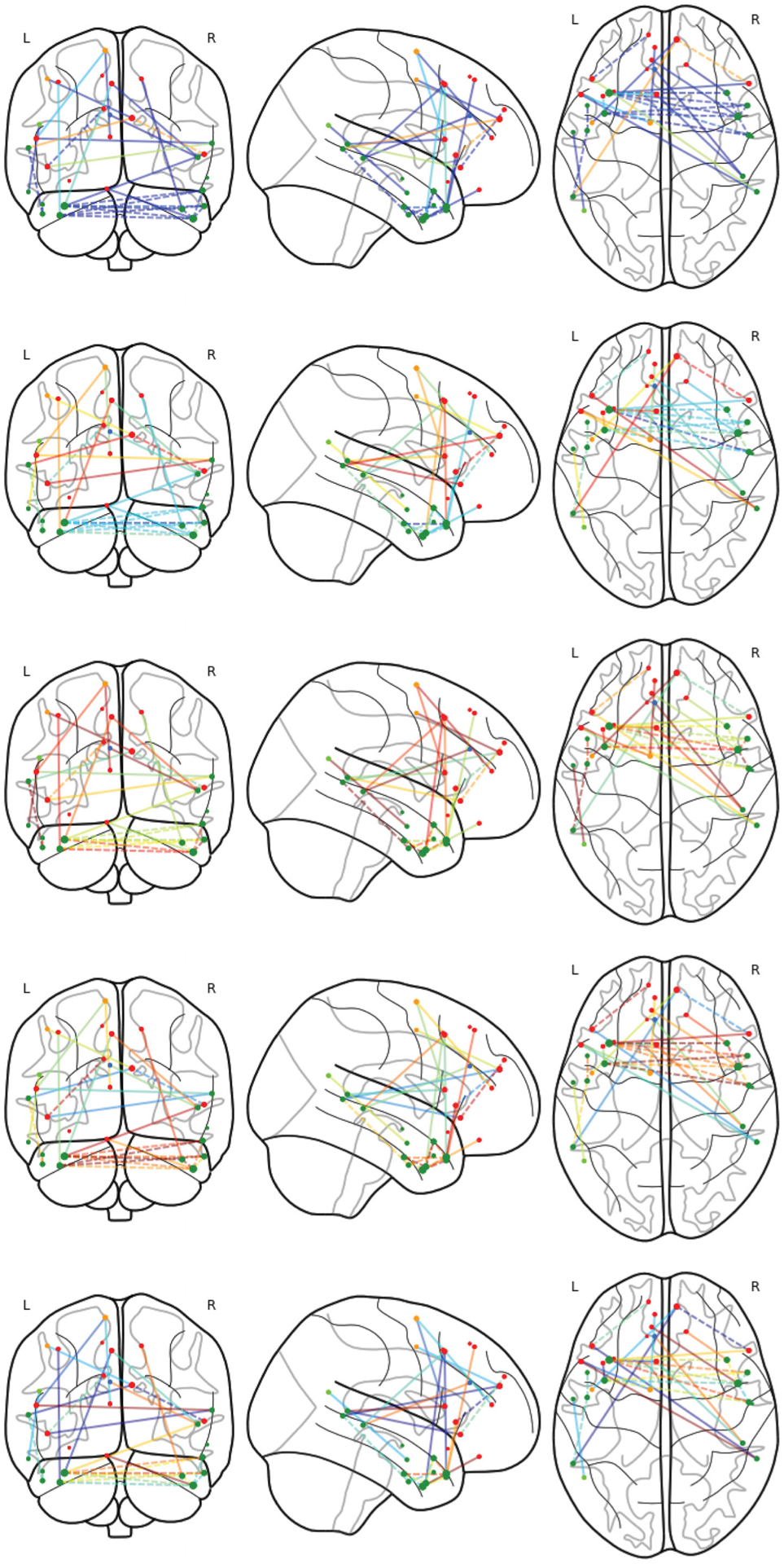
Medial frontal network changes, with respect to the intelligence score at 7, 11, 15, 19, 23. Blue color represents the small H-S norm value of CPO, green the medium norm value, and red the high H-S norm value.
As an independent validation, we replicate the analysis using another resting-state fMRI data of 828 individuals in HCP. We report the changes of the identified significant edges for the medial frontal module from the new dataset in Figure S6 of the appendix. We again find that the medial frontal module is enriched. Hearne, Mattingley, and Cocchi (2016) identified positive correlations between functional connectivity and intelligence in general. Song et al. (2008) reported that most of the brain regions whose connectivity patterns are negatively correlated with the intelligence are around medial frontal gyrus, or part of motor regions, which are part of medial frontal module. Combined with our findings, it suggests that the medial frontal module may play a unique role in intelligence compared to other brain modules.
Supplementary Material
Acknowledgments
We are grateful to two referees and an associate editor for their constructive comments and helpful suggestions.
Funding
Lexin Li’s research was partially supported by NIH (grants nos. R01AG061303, R01AG062542, and R01AG034570). Hongyu Zhao’s research was partially supported by NSF (grant nos. DMS1713120 and DMS1902903), and NIH (grant no. R01 GM134005).
Footnotes
Supplementary Materials
The Supplementary Appendix contains discussions of some assumptions, all the proofs of the theoretical results, and some results from additional asymptotic and numerical analyses.
References
- Bach F (2009), “High-Dimensional Non-Linear Variable Selection Through Hierarchical Kernel Learning,” arXiv no. 0909.0844. [Google Scholar]
- Baker CR (1973), “Joint Measures and Cross-Covariance Operators,” Transactions of the American Mathematical Society, 186, 273–289. [Google Scholar]
- Bickel PJ, and Levina E (2008), “Covariance Regularization by Thresholding,” The Annals of Statistics, 36, 2577–2604. [Google Scholar]
- Bosq D (2000), Linear Processes in Function Spaces, New York: Springer. [Google Scholar]
- Bullmore E, and Sporns O (2009), “Complex Brain Networks: Graph Theoretical Analysis of Structural and Functional Systems,” Nature Reviews. Neuroscience, 10, 186–198. [DOI] [PubMed] [Google Scholar]
- Cai TT, Liu W, and Luo X (2011), “A Constrained ℓ1 Minimization Approach to Sparse Precision Matrix Estimation,” Journal of the American Statistical Association, 106(494):594–607. [Google Scholar]
- Chun H, Zhang X, and Zhao H (2015), “Gene Regulation Network Inference With Joint Sparse Gaussian Graphical Models,” Journal of Computational and Graphical Statistics, 24, 954–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danaher P, Wang P, and Witten DM (2011), “The joint Graphical Lasso for Inverse Covariance Estimation Across Multiple Classes,” arXiv no. 1111.0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn ES, Shen X, Scheinost D, Rosenberg MD, Huang J, Chun MM, Papademetris X, and Constable RT (2015), “Functional Connectome Fingerprinting: Identifying Individuals Using Patterns of Brain Connectivity,” Nature Neuroscience, 18, 1664–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornito A, Zalesky A, and Breakspear M (2013), “Graph Analysis of the Human Connectome: Promise, Progress, and Pitfalls,” NeuroImage, 80, 426–444. [DOI] [PubMed] [Google Scholar]
- Fox MD, and Greicius M (2010), “Clinical Applications of Resting State Functional Connectivity,” Frontiers in Systems Neuroscience, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JH, Hastie TJ, and Tibshirani RJ (2008), “Sparse Inverse Covariance Estimation With the Graphical Lasso,” Biostatistics, 9, 432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumizu K, Bach FR, and Jordan MI (2009), “Kernel Dimension Reduction in Regression,” The Annals of Statistics, 37, 1871–1905. [Google Scholar]
- Greene AS, Gao S, Scheinost D, and Constable RT (2018), “Task-induced Brain State Manipulation Improves Prediction of Individual Traits,” A Nature Communications, 9, 2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie TJ and Tibshirani RJ (1990), Generalized Additive Models, New York: Chapman & Hall. [Google Scholar]
- Hearne LJ, Mattingley JB, and Cocchi L (2016), “Functional Brain Networks Related to Individual Differences in Human Intelligence at Rest,” Scientific Reports, 6, 32328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolar M, Song L, Ahmed A, and Xing EP (2010), “Estimating Time-Varying Networks,” The Annals of Applied Statistics, 4, 94–123. [Google Scholar]
- Lee K-Y, Li B, and Zhao H (2016a), “On an Additive Partial Correlation Operator and Nonparametric Estimation of Graphical Models,” Biometrika, 103, 513–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ______ (2016b), “Variable Selection Via Additive Conditional Independence,” Journal of the Royal Statistical Society, Series B, 78, 1037–1055. [Google Scholar]
- Lee W, and Liu Y (2015), “Joint Estimation of Multiple Precision Matrices With Common Structures,” The Journal of Machine Learning Research, 16, 1035–1062. [PMC free article] [PubMed] [Google Scholar]
- Li B, Chun H, and Zhao H (2012), “Sparse Estimation of Conditional Graphical Models With Application to Gene Networks,” Journal of the American Statistical Association, 107, 152–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, and Solea E (2018), “A Nonparametric Graphical Model for Functional Data With Application to Brain Networks Based on fMRI,” Journal of the American Statistical Association, 113, 1637–1655. [Google Scholar]
- Liu H, Han F, Yuan M, Lafferty J, and Wasserman L (2012), “ High-Dimensional Semiparametric Gaussian Copula Graphical Models,” The Annals of Statistics, 40, 2293–2326. [Google Scholar]
- Peng J, Wang P, Zhou N, and Zhu J (2009), “Partial Correlation Estimation by Joint Sparse Regression Models,” Journal of the American Statistical Association, 104, 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao X, Guo S, and James GM (2019), “Functional Graphical Models,” Journal of the American Statistical Association, 114, 211–222. [Google Scholar]
- Qiu H, Han F, Liu H, and Caffo B (2016), “Joint Estimation of Multiple Graphical Models from High Dimensional Time Series,” Journal of the Royal Statistical Society, Series B, 78, 487–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman AJ, Bickel PJ, Levina E, and Zhu J (2008), “Sparse Permutation Invariant Covariance Estimation,” Electronic Journal of Statistics, 2, 494–515. [Google Scholar]
- Smith R, Lane RD, Alkozei A, Bao J, Smith C, Sanova A, Nettles M, and Killgore WDS (2018), “The Role of Medial Prefrontal Cortex in the Working Memory Maintenance of One’s Own Emotional Responses,” Science Reports, 8, 3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M, Zhou Y, Li J, Liu Y, Tian L, Yu C, and Jiang T (2008), “Brain Spontaneous Functional Connectivity and Intelligence,” Neuroimage, 41, 1168–1176. [DOI] [PubMed] [Google Scholar]
- Tsay RS, and Pourahmadi M (2017), “Modelling Structured Correlation Matrices,” Biometrika, 104, 237–242. [Google Scholar]
- Wang J-L, Chiou J-M, and Muller H-G (2016a), “Functional Data Analysis,” Annual Review of Statistics and Its Application, 3, 257–295. [Google Scholar]
- Wang Y, Kang J, Kemmer PB, and Guo Y (2016b), “An Efficient and Reliable Statistical Method for Estimating Functional Connectivity in Large Scale Brain Networks Using Partial Correlation,” Frontiers in Neuroscience, 10, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yin Y, Sun YW, Zhou Y, Chen X, Ding WN, Wang W, Li W, Xu JR, and Du YS (2015), “Decreased Prefrontal Lobe Interhemispheric Functional Connectivity in Adolescents With Internet Gaming Disorder: A Primary Study Using Resting-State fMRI,” PLoS ONE, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z and Li H (2008), “A Hidden Spatial-Temporal Markov Random Field Model for Network-Based Analysis of Time Course Gene Expression Data,” Annals of Applied Statistics, 2, 408–429. [Google Scholar]
- Xu KS, and Hero AO (2014), “Dynamic Stochastic Blockmodels for Time-Evolving Social Networks,” IEEE Journal of Selected Topics in Signal Processing, 8, 552–562. [Google Scholar]
- Yuan M, and Lin Y (2007), “Model Selection and Estimation in the Gaussian Graphical Model,” Biometrika, 94, 19–35. [Google Scholar]
- Zanto TP, and Gazzaley A (2013), “Fronto-Parietal Network: Flexible Hub of Cognitive Control,” Trends in Cognitive Sciences, 17, 602–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, and Cao J (2017), “Finding Common Modules in a Time-Varying Network With Application to the Drosophila Melanogaster Gene Regulation Network,” Journal of the American Statistical Association, 112, 994–1008. [Google Scholar]
- Zhang T, Wu J, Li F, Caffo B, and Boatman-Reich D (2015), “A Dynamic Directional Model for Effective Brain Connectivity Using Electrocorticographic (ECoG) Time Series,” Journal of the American Statistical Association, 110, 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, and Li L (2018), “Multiple Matrix Gaussian Graphs Estimation,” Journal of the Royal Statistical Society, Series B, 80, 927–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


