Abstract
Background and Objectives:
Concomitant carriage of blaNDM-1 and plasmid mediated quinolone resistance determinants (PMQRs) by multi drug resistant (MDR) Klebsiella pneumoniae (K. pneumoniae) has increased globally, often related to their presence on transmissible plasmids. In this study, we hypothesized the presence of blaNDM-1 and PMQRs on a single conjugative plasmid that circulates among K. pneumoniae strains isolated from Assiut University Hospital.
Materials and Methods:
Twenty-two clinical MDR K. pneumoniae strains harboring both blaNDM-1 and PMQRs were genotyped using pulsed field gel electrophoresis. Horizontal transfer of blaNDM-1 and PMQRs was evaluated by conjugation and trans-conjugants were screened for the presence of both genes and integron by PCR. Trans-conjugant’s plasmid DNA bands were purified using agarose gel electrophoresis and different DNA bands were screened for blaNDM-1 and PMQRs. Plasmids carrying blaNDM-1 and PMQRs were typed by PCR based replicon typing.
Results:
All MDR K. pneumoniae contained class 1 integron and belonged to 15 pulsotypes. BlaNDM-1 and PMQRs were co-transferred in each conjugation process. Multiple replicons (5–9 types) were detected in each trans-conjugant; with IncFIIK and IncFIB-KQ replicons being common among all trans-conjugants. Both blaNDM-1 and PMQRs were detected on a pKpQIL-like multi-replicon plasmid that was present in all K. pneumoniae strains.
Conclusion:
In view of these results, the presence of blaNDM-1 and PMQRs on pKpQIL-like plasmid that existed in multiple unrelated K. pneumoniae isolates is highly suggestive of the circulation of pKpQIL-like MDR plasmids in our hospitals. Moreover, carriage of integrons by the-circulating MDR plasmids increases the risk of dissemination of antimicrobial resistance among pathogens.
Keywords: Klebsiella pneumoniae, Plasmids, New Delhi metalo beta-lactamase, Drug resistance, Polymerase chain reaction
INTRODUCTION
Klebsiella pneumoniae is a major cause of opportunistic nosocomial infections including; urinary tract infections, pneumonia, blood stream infections, wound infections and sepsis. It usually colonizes the gastrointestinal tract, nasopharynx and skin (1). Emergence of multidrug resistance (MDR) [defined as a lack of susceptibility to three or more antimicrobial categories (2)] among K. pneumoniae strains is considered an increasingly threatening problem globally. Infections caused by such organism are associated with major therapeutic problems resulting in increased morbidity and mortality (3).
The main mechanism associated with acquisition of MDR by bacteria is the dissemination of resistance genes by mobile genetic elements (insertion sequences, transposons and gene cassettes/integrons) carried by resistance plasmids. These resistance plasmids are the major players in the spread of MDR as they often carry multiple resistance elements and capable of transferring them simultaneously between different bacterial strains (4). Integrons are gene-capturing platforms that have an important role in the dissemination of antimicrobial resistance genes by plasmids (5). They are composed of two major components; the first is the integron-integrase gene (intI) and its promoter (PintI), an integration site named attachment site of the integron (attI), and a constitutive promoter (Pc) for integrated gene cassettes. The second component is a cluster of gene cassettes carrying antimicrobial resistance genes (6).
Carbapenems and/or fluoroquinolones (FQs) are often the best options for managing infections due to MDR K. pneumoniae (7). Unluckily, co-emergence of carbapenems and fluoroquinolone resistance has caused major difficulty in treating such infections (8). Plasmids play a main role in spreading of both carbapenem and FQs resistance. Plasmid-mediated quinolone resistance (PMQR) is conferred by qnr genes (qnrA, qnrB, qnrS, qnrC, qnrD) and aac(6′)-Ib-cr (9). Likewise, many plasmid mediated carbapenemases (IMP, VIM, SIM, SPM, GIM, KPC, SME) have been detected in K. pneumoniae, with New Delhi metallo-β-lactamase-1 (NDM-1) has been accepted as the most widely disseminating carbapenemase worldwide (10).
NDM-1-encoding plasmids often co-carry other resistance determinants including PMQRs (9). Previous studies reported that blaNDM-1 carrying plasmids belongs to diverse incompatibility (Inc) groups, including IncN, IncC, IncFIB, IncFIB (K), IncR, IncFII, IncFIA, IncFII (K), IncHI1 and Col4401 (11). Recently, a pKpQIL-like plasmid was reported as a carrier of blaNDM-1 (12).
PKpQIL, a bla KPC-3-encoding plasmid, was firstly isolated in 2006 from K. pneumoniae strain sequence type 258 (ST 258) (13). PKpQIL is a multi-replicon, self-transmissible plasmid with size of 113,637-bp, belongs to the IncFII group and carries two replicons IncFIIK and IncFIB-KQ (14). Sequencing of pKpQIL suggested that it was formed as a result of recombination between a pKPN4-like plasmid and a pNYC-like plasmid thus it carries 2 replicons. New variants of pKpQIL have emerged worldwide including; pKpQIL-IT isolated in Italy but mediated resistance to kanamycin that was nonexistent on the initial pKpQIL plasmid (15) and pKpQIL-UK that has few nucleotide substitutions from the original pKpQIL (16).
In Egypt, high prevalence of PMQRs and blaNDM-1 has been identified among MDR K. pneumoniae strains (17, 18). Nevertheless, the existence and genetic characterization of circulating plasmids co-carrying PMQRs and blaNDM-1 among K. pneumoniae is still not yet investigated. As far as we know, this is the first study to declare the role of pKpQIL-like plasmid in dissemination of PMQRs and blaNDM-1 among K. pneumoniae isolated from Assiut University Hospital, Egypt.
MATERIALS AND METHODS
The Ethics Committee of the Faculty of Medicine, Assiut University approved this study according to the latest revision of the Declaration of Helsinki, and informed consent was obtained from the participants (IBR no: 17200282).
Bacterial isolates
The current work is a retrospective study investigating 22 MDR K. pneumoniae strains collected from the Infection Control Laboratory at Assiut University Hospital from the period of January 2019 to January 2020. K. pneumoniae strains were retrieved from patients with hospital-acquired infections at different ICUs: chest (n= 10), paediatric (n= 8) and neurology (n= 4). Infections were considered as hospital-acquired when the symptoms and signs appeared 48 hrs. or more after hospitalization. All K. pneumoniae strains were chosen based on dual acquisition of blaNDM-1 and PMQR determinants (qnrB, qnrS1 and/or aac(6′)-Ib-cr).
Resistance phenotype determination
Resistance phenotype was determined to all isolates using Kirby–Bauer disk diffusion test (19) with antibiotic discs (Oxoid, UK) for different classes of antibiotics including; penicillin derivatives [amoxicillin (AML10 μg), amoxicillin/clavulanic acid (AMC 20/10 μg), and piperacillins (PI 100 μg)], cephalosporins [cefazolin (CZ 30 μg), cefpodoxime (CPD 30 μg), cefoperazone (CPZ 75 μg) and ceftriaxone (CTR 30 μg)], aminoglycosides [gentamicin (GE 10 μg) and amikacin (AK 30 μg)], tetracycline (TE 30 μg), chloramphenicol (C 30 μg) and trimethoprim-sulfonamide (SXT 1.25 /23.75 μg). The minimum inhibitory concentrations (MICs) of imipenem and ciprofloxacin were determined by E-tests (BioMérieux, Solna, Sweden). Results were interpreted based on the Clinical and Laboratory Standard Institute guidelines (CLSI) 2019 (20).
Genotyping of K. pneumoniae isolates by PFGE
Clonal relatedness of K. pneumoniae isolates was determined by pulsed field gel electrophoresis (PFGE) following the PulseNet protocol of the Centres for Disease Control (CDC) 2017 (21). Briefly, total genomic DNA from each strain was digested with XbaI (New England Biolabs, Beverly, MA, USA) for 16 h at 37°C. Fragments separation was performed using 1% certified Mega base agarose in a CHEF-DR III system (Bio-Rad). Size was estimated by Lambda Ladder PFGE Marker (New England Biolabs, Beverly, MA, USA). The subsequent Conditions were selected: Initial switch time: 6.7 Sec., Final switch time: 35.3 Sec., Voltage: 6 V/cm, included angle: 120° and Run time: 22 hours. The PFGE patterns were analyzed by a transient BioNumerics software evaluation license (Version 7.6, Applied Maths, Belgium), and agreement to publish was received. The dendrogram was created using the un-weighted pair group method with arithmetic mean (UPGMA) clustering method with Dice’s Similarity coefficient. A similarity coefficient of 80% or more was used as the threshold of a cluster.
Conjugation experiment
Conjugal transfer of plasmids carrying blaNDM-1 and PMQR genes from K. pneumoniae strains was implemented according to the method modified by Miller (22) using E. coli J53 (recA−, Azr) as the recipient. Trans-conjugants were selected on Luria–Bertani (LB) agar plates to which sodium azide (200 μg/mL) and nalidixic acid (20 μg/mL) were added. Trans-conjugants were Replica-plated on 2 LB agar plates; one with meropenem (0.5 μg/mL) and the other containing nalidixic acid (20 μg/mL). Colonies that showed growth on both replica LB agar plates were selected.
Screening for PMQRs, blaNDM-1 and integrons among trans-conjugants
Plasmids from trans-conjugants were extracted using PureLinkTM Quick Plasmid Miniprep Kit (Thermo Fisher Scientific, Massachusetts, USA) following the manufacturer’s instructions. The existence of PMQRs and blaNDM-1 in trans-conjugants was determined by PCR. To assess the capability of plasmids to capture new antibiotic resistance gene cassettes, the three classes of integrons (Int1, Int2 and Int3) were screened among trans-conjugants by conventional PCR. PCR amplification was done in a thermal cycler (BioRadT100, USA) using GoTaq® Flexi DNA Polymerase Kit (Promega, USA) in a 50-μl volume containing 100 pmol of each primer, 1×PCR buffer, 0.2 mM dNTPs, 1.5 mM MgCl2, 1.25 U of Taq polymerase and 100 ng of DNA template. The amplification programs were conducted as follows: an initial denaturation step at 95°C for 5 min, followed by 30 cycles of DNA denaturation at 95°C for 30 sec, primer annealing for 30 sec, and primer extension at 72°C, then a final extension step was done at 72°C for 5 min. Annealing temperatures, time required for primer extension and sequences of oligonucleotide primers used for PCR amplification are listed in Table (1).
Plasmid typing from trans-conjugants
PCR based replicon typing (PBRT) was used for detection of plasmid replicons in trans-conjugants, using PBRT 2.0 kit (DIATHEVA, Italy). This new kit affords a set of eight specific PCR tests adjusted to do eight multiplex PCRs for the amplification of 30 replicons: HI1, HI2, I1 alpha, I1 gamma, I2, X1, X2, X3, X4, L, M, N, N2, FIA, FIB, FII, FIIS, FIIK, FIB-KN, FIB-KQ, W, P1, A/C, T, K, U, R, B/O, HIB-M and FIB-M, that are representatives of the main plasmid incompatibility groups among Enterobacteriaceae. Moreover, FIB-KN recognizes the pKPN3-like plasmids; FIB-KQ recognizes the pKpQIL-like plasmid which are common in K. pneumoniae (29). The kit included positive control plasmid’s DNA for each mix (supplementary Table 1). The eight multiplex PCRs were performed in a 50 μL volume containing the reaction mix, 1.25 U of Taq polymerase and 100 ng of DNA template. The amplification program was conducted as follows: an initial denaturation step at 95°C for 10 min, followed by 30 cycles of DNA denaturation at 95°C for 60 sec, primer annealing at 60°C for 30 sec, and primer extension at 72°C for 60 sec, then a final extension step was done at 72°C for 5 min. The resulting PCR products were analyzed in a 2% agarose gel with ethidium bromide staining and visualized under ultraviolet light.
Table 1.
Annealing temperatures, time required for primer extension and sequences of oligonucleotide primers used in PCR amplification of blaNDM-1, PMQRs and integrons
| Gene | Sequence (5′-3′) | Size (bp) | Annealing temperature | Time for primer extension | Reference |
|---|---|---|---|---|---|
| bla NDM1 | F: GGTTTGGCGATCTGGTTTTC R: CGGAATGGCTCATCACGATC |
621 bp | 55°C | 30 Sec. | (23) |
| qnrB | F: GATCGTGAAAGCCAGAAAGG R: ACGATGCCTGGTAGTTGTCC |
469 bp | 54°C | 30 Sec. | (24) |
| qnrS | F: CAATCATACATATCGGCACC R: TCAGGATAAACAACAATACCC |
641 bp | 56°C | 30 Sec. | (25) |
| aac(6′)-Ib | F: TTGCGATGCTCTATGAGTGGCTA R: CTCGAATGCCTGGCGTGTTT |
482 bp | 58°C | 30 Sec. | (26) |
| Int1 | F: CTG CGT TCG GTC AAG GTT CT R: GGA ATG GCC GAG CAG ATC CT |
882 bp | 58°C | 50 Sec. | (27) |
| Int2 | F: CACGGATATGCGACAAAAAGGT R: GTAGCAAACGAGTGACGAAATG |
788 bp | 54°C | 40 Sec. | (28) |
| Int3 | F: GCCTCCGGCAGCGACTTTCAG R: ACGGATCTGCCAAACCTGACT |
979 bp | 56°C | 60 Sec. | (28) |
Bp = base pair
Replicon typing of plasmids carrying blaNDM-1 and PMQRs
Gel electrophoresis of each trans-conjugant’s plasmid DNA-using a 1Kb DNA ladder (New England BioLabs, USA) - was employed thus plasmids of different sizes separate into distinct bands. In an attempt to identify which plasmid/s carry blaNDM-1 and PMQRs, each plasmid DNA band was purified from the gel using PureLinkTM Quick gel extraction Kit (Thermo Fisher Scientific, Massachusetts, USA) according to the manufacturer’s instructions. Each band was screened for the presence of blaNDM1 and PMQR genes by PCR. Determination of replicon type/s of the blaNDM-1, PMQRs containing plasmid was performed by PBRT using the previously mentioned kit.
RESULTS
Resistance phenotypes
All 22 K. pneumoniae isolates were found to be MDR. Moreover, seventeen (77.3%) K. pneumoniae isolates were imipenem resistant, while 12 (54.5%) isolates were ciprofloxacin resistant detected by E-test®. Ciprofloxacin and imipenem-MICs as well as resistance phenotype of K. pneumoniae isolates are shown in Table (2).
Table 2.
Antimicrobial susceptibility of K. pneumoniae isolates
| Code | MIC IMP | MIC CIP | Resistance phenotype* |
|---|---|---|---|
| KP 1 | >32 (R) | >32 (R) | MDR1,2,3,6,7 |
| KP 2 | >32 (R) | >32 (R) | MDR1,2,3,4,5, 6,7,8 |
| KP 3 | 4 (R) | 0.75 (I) | MDR1,2,3,4,5,7 |
| KP 4 | 4 (R) | 0.75 (I) | MDR1,2,3,4,5,7 |
| KP 5 | 8 (R) | 0.125 (S) | MDR1,2,3,5,7,8 |
| KP 6 | 12 (R) | >32 (R) | MDR1,2,3,5,6,7 |
| KP 7 | 16 (R) | >32 (R) | MDR1,2,3,5,6,7 |
| KP 8 | 1 (S) | >32 (R) | MDR1,2,4,5,6,7,8 |
| KP 9 | 0.75 (S) | >32 (R) | MDR1,2,4,5,6,7,8 |
| KP 10 | 4 (R) | 0.75 (I) | MDR1,2,3,7,8 |
| KP 11 | 4 (R) | >32 (R) | MDR1,2,3,4,5, 6,7,8 |
| KP 12 | 4 (R) | >32 (R) | MDR1,2,3, 6,7,8 |
| KP 13 | >32 (R) | >32 (R) | MDR1,2,3,4, 6,7,8 |
| KP 14 | 0.125(S) | 4 (R) | MDR1,2,4,5,6,7,8 |
| KP 15 | 4 (R) | .064 (S) | MDR1,2,3,7,8 |
| KP 16 | >32 (R) | >32 (R) | MDR1,2,3,6,7,8 |
| KP 17 | 12 (R) | >32 (R) | MDR1,2,3,6,7,8 |
| KP 18 | 3 (I) | 0.75 (I) | MDR1,2,7,8 |
| KP 19 | 4 (R) | 0.38 (I) | MDR1,2,3,7,8 |
| KP 20 | 1.5 (I) | 0.125 (S) | MDR1,2,5,7,8 |
| KP 21 | 4 (R) | .047 (S) | MDR1,2,3,7,8 |
| KP 22 | 4 (R) | .064 (S) | MDR1,2,3,7,8 |
Key: 1=Penicillins; 2=Cephalosporins; 3= Carbapenems; 4=Tetracyclines; 5=Chloramphenicol; 6=Fluoroquinolone; 7= Trimethoprim sulfonadmide; 8 = Aminoglycosides.
Genotyping of K. pneumoniae strains by PFGE
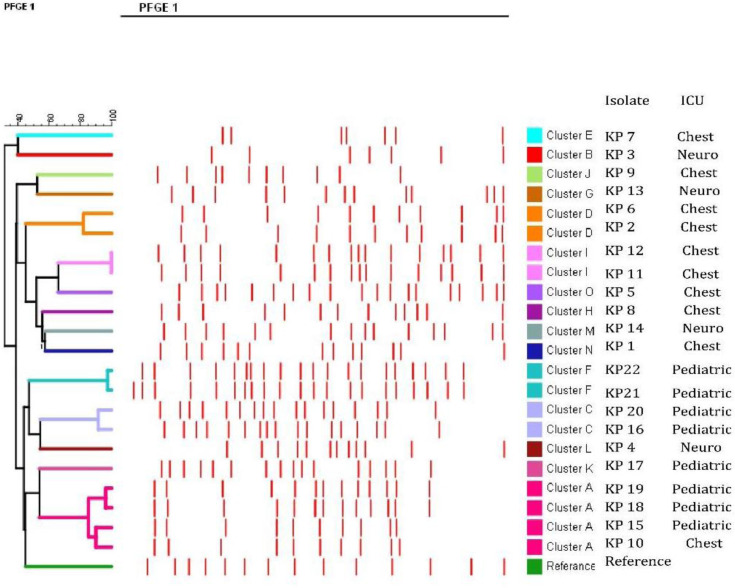
PFGE analyses of the 22 K. pneumoniae isolates identified 15 different clusters (A-O) (Supplementary Fig. 1) and a dendrogram was constructed using BioNumerics software (Version 7.6, Applied Maths, Belgium) (a transient evaluation license) with isolates sharing ≥ 80% of the bands were assigned to the same cluster (Fig. 1). Five clusters comprised more than one K. pneumoniae strain; cluster A (No.= 4; KP10, KP15, KP18 and KP19), KP10 was isolated from chest ICU while the others were isolated from paediatric ICU, cluster C (No.= 2; KP16, KP20) both were isolated from paediatric ICU, cluster D (No.= 2; KP2, KP6) both were isolated from chest ICU, cluster F (No= 2; KP21, KP22) both were isolated from paediatric ICU and cluster I (No.= 2; KP11, KP12) both were isolated from chest ICU. The remaining isolates (No.= 10) were clonally unrelated and were categorized into different clusters.
Fig. 1.
Dendrogram comparing PFGE profiles of K. pneumoniae isolates co-harboring blaNDM1 and PMQRs
Screening for blaNDM-1, PMQRs and integrons among trans-conjugants
Both blaNDM-1 and PMQRs were co-transferred in each conjugation event from all K. pneumoniae isolates. All the 22 trans-conjugants were found to be (Int1) positive, while (Int2 and Int3) were negative among all trans-conjugants.
PBRT of trans-conjugant’s plasmid DNA
PBRT revealed the presence of multiple plasmid replicons (5–9 types) in each trans-conjugant as shown in Table 3. IncFIIK and IncFIB-KQ were the most common replicon types identified among all trans-conjugants, indicating that each trans-conjugant harbored a plasmid with pKpQIL-like backbone. Moreover, 10 K. pneumoniae–belonging to 8 pulsotypes- were isolated from chest ICU between January 2019 and May 2019, and all contained IncI1gamma and IncFII replicons as well. Similarly, four K. pneumoniae- comprised different pulsotypes-isolated from neurology ICU between January 2019 and January 2020 also contained IncI1gamma and IncFII replicons.
Table 3.
Resistance genes, replicon types, source and PFGE type of K. pneumoniae isolates.
| Code | NDM | PMQRs | Int1 | Replicon types | ICU | Pulsotype |
|---|---|---|---|---|---|---|
| KP 1 | + | qnr B, qnrS | + | IncFIIK, IncFIB-KQ, IncFIB, IncFII, IncI1gamma | Chest | N |
| KP 2 | + | qnr B, qnrS | + | IncFIIK, IncFIB-KQ, IncHI1, IncM, IncFIB-M, IncFII, IncI1gamma | Chest | D |
| KP 3 | + | qnr B, qnrS | + | IncFIIK, IncFIB-KQ, IncHI1, IncL, IncFIB-M, IncFII, IncI1gamma, IncFIA | Neuro | B |
| KP 4 | + | qnr B, qnrS | + | IncFIIK, IncFIB-KQ, IncHI1, IncFIB, IncFIB-M, IncFII, IncI1gamma | Neuro | L |
| KP 5 | + | qnr B, qnrS, aac(6′)-Ib-cr | + | IncFIIK, IncFIB-KQ, IncHI1, IncM, IncL, IncFIB-M, IncFII, IncI1gamma | Chest | O |
| KP 6 | + | qnr B, qnrS, aac(6′)-Ib-cr | + | IncFIIK, IncFIB-KQ, IncHI1, IncFIB-M, IncFII, IncI1gamma | Chest | D |
| KP 7 | + | qnr B | + | IncFIIK, IncFIB-KQ, IncHI1, IncM, IncFIA, IncL, IncFIB-M, IncFII, IncI1gamma | Chest | E |
| KP 8 | + | qnr B, qnrS | + | IncFIIK, IncFIB-KQ, IncHI1, IncM, IncFIA, IncFII, IncI1gamma | Chest | H |
| KP 9 | + | qnr B, qnrS | + | IncFIIK, IncFIB-KQ, IncHI1, IncM, IncFII, IncI1gamma | Chest | J |
| KP 10 | + | qnr B, qnrS | + | IncFIIK, IncFIB-KQ, IncHI1, IncM, IncFII, IncI1gamma | Chest | A |
| KP 11 | + | qnr B, qnrS, aac(6′)-Ib-cr | + | IncFIIK, IncFIB-KQ, IncHI1, IncM, IncFIA, IncFII, IncI1gamma | Chest | I |
| KP 12 | + | qnr B, qnrS | + | IncFIIK, IncFIB-KQ, IncM, IncFIA, IncFII, IncI1gamma | Chest | I |
| KP 13 | + | qnr B, qnrS | + | IncFIIK, IncFIB-KQ, IncHI1, IncM, IncFIB, IncFII, IncI1gamma | Neuro | G |
| KP 14 | + | qnr B, qnrS | + | IncFIIK, IncFIB-KQ IncM, IncFIA, IncFII, IncI1gamma | Neuro | M |
| KP 15 | + | qnr B, qnrS | + | IncFIIK, IncFIB-KQ, IncHI1, IncFIB, IncFIB-M, IncFII, IncI1gamma | Pediatric | A |
| KP 16 | + | qnr B, qnrS, aac(6′)-Ib-cr | + | IncFIIK, IncFIB-KQ, IncFIA, IncFIB, IncFII | Pediatric | C |
| KP 17 | + | qnr B, qnrS | + | IncFIIK, IncFIB-KQ, IncHI1, IncFIA, IncFIB, IncFII, IncL IncFIIK, IncFIB-KQ, IncHI1, IncFIA, IncFII, IncI1gamma, IncFIB-M IncFIIK, IncFIB-KQ, IncHI1, IncM, IncFIA, IncFIB, IncI1gamma, IncFIB-M IncFIIK, IncFIB-KQ, IncHI1, IncFIB, IncFIB-M, IncFII | Pediatric | K |
| KP 18 | + | qnr B, qnrS | + | Pediatric | A | |
| KP 19 | + | qnr B, qnrS, aac(6′)-Ib-cr | + | Pediatric | A | |
| KP 20 | + | qnr B, qnrS | + | Pediatric | C | |
| KP 21 | + | qnr B, qnrS | + | IncFIIK, IncFIB-KQ, IncHI1, IncM, IncFIB, IncFIB-M, IncFII | Pediatric | F |
| KP 22 | + | qnr B | + | IncFIIK, IncFIB-KQ, IncHI1, IncM, IncL, IncI1gamma | Pediatric | F |
Replicon typing of plasmids carrying blaNDM-1 and PMQRs
Gel electrophoresis revealed that each trans-conjugant harbored multiple plasmids appeared as discrete DNA bands on the gel. Screening the various plasmid DNA bands for blaNDM-1 and PMQRs, revealed that- in all trans-conjugants- a single plasmid DNA band was found to be positive for both blaNDM-1 and PMQRs. PBRT of the plasmid’s DNA co-harboring blaNDM-1 and PMQRs revealed that in all trans-conjugants, each plasmid carried 2 replicons (IncFIIK and IncFIB-KQ) signifying that blaNDM-1 and PMQRs might be carried by a multi-replicon pKpQIL-like plasmid.
DISCUSSION
Global emergence and dissemination of MDR K. pneumoniae pose a serious public health burden worldwide (30). Various genes entangled in multiple drug resistances are plasmid mediated; including PMQRs and blaNDM-1 genes (4). Horizontal transfer of plasmids coding multiple resistance determinants is associated with dissemination of MDR among bacterial pathogens (31). In this study, we hypothesized the existence of blaNDM-1 and PMQRs on a single transmissible plasmid that circulates among MDR K. pneumoniae isolated from Assiut University Hospital, Egypt. To test this hypothesis, we performed plasmid typing of 22 clinical K. pneumoniae isolated from 3 different ICUs; chest ICU, paediatric ICU, and neurology ICU; all contained one or more of the PMQRs as well as blaNDM-1.
Twenty-two MDR K. pneumoniae were included in this study. These MDR K. pneumoniae possessed at least resistance to four drug classes, with 2 K. pneumoniae found to be resistant to the all tested drug classes. These results suggest that ICUs in Assiut University Hospitals serve as a source for MDR K. pneumoniae probably due to increased consumption of antibiotics (32). Previous studies investigating K. pneumoniae isolated from Assiut University Hospitals reported multidrug resistance among K. pneumoniae strains, however, the resistance rates are higher in this study (18, 33). The predominance of MDR bacteria in the community is of great concern, as it is associated with increased length of hospital stay, deaths, healthcare charges, and antimicrobial use (34).
In the present study, the resistance rate against imipenem in blaNDM positive K. pneumoniae isolates, as detected by E-test, was 77.3%. However, Huang et al. (2018) reported higher imipenem resistance rates (100%) among blaNDM positive K. pneumoniae isolates (35). On the other hand, the resistance rate against ciprofloxacin in PMQRs positive K. pneumoniae isolates, as detected by E-test, was 54.5%. However, Xue et al. (2017) reported that only 8% of PMQRs positive K. pneumoniae isolates were ciprofloxacin resistant (36). The variation in the resistance determinants rates could be attributed to the differences in the geographic localities.
Plasmids circulate between different bacterial species and are capable of acquiring resistance genes through small mobile elements such as integrons and transposons. Integrons carrying resistance genes, when jump on a plasmid, bring about acquisition and dissemination of MDR (37). In this study, Int1 was identified in all K. pneumoniae isolates, which is in consistent with previous studies reporting Int1 as the most prevalent type of integrons associated with blaNDM-1 and PMQRs in K. pneumoniae (17, 38). The high frequency of integrons in plasmids extracted from K. pneumoniae isolated from our hospital increases the danger of acquisition of more resistance determinants by those plasmids and thus spread of resistance to more antibiotics.
PFGE typing of the 22 K. pneumoniae isolates displayed that these strains comprised 15 pulsotypes; hence they were polyclonal in origin. Nevertheless, a number of K. pneumoniae sharing the same pulsotype were recovered from different patients admitted to the same ICU at different times, denoting that some K. pneumoniae strains are endemic and circulate between patients admitted to such ICU. Surprisingly, one isolate (KP10) recovered from chest ICU and 3 isolates recovered from paediatric ICU (KP15, KP18 and KP19) were found to belong to the same cluster. The presence of pathogens of the same pulsotypes in different hospital wards emphasizes the circulation of K. pneumoniae in the hospital environment (39).
Transmissible plasmids are capable of causing major changes in bacterial populations by simultaneously disseminating resistances to multiple anti-bacterial agents (40). Both PMQRs and blaNDM-1 were reported to be present on transmissible plasmids (18, 41). In the present study, blaNDM-1 and PMQRs were successfully transferred at a rate of 100% to the recipient (E. coli J53) through conjugation. The high rate of transfer of blaNDM-1 and PMQRs contributes to the fast spread of those genes among clonally unrelated bacterial strains (42).
PBRT was used for typing of plasmids isolated from trans-conjugants; PBRT is a method for plasmid typing which helps understanding their distribution and relationships (43). Multiple plasmid replicons (5–9 types) were detected in every trans-conjugant denoting the existence of several transmissible plasmids in each K. pneumoniae pathogen. The most common detected replicon type was IncFIIK, which was found in all trans-conjugants and hence implying its existence in all of the 22 K. pneumoniae isolates as well. This finding is in accordance with previously published data reporting IncFIIK as being the most prevalent replicon type in Klebsiella species (44–46). Moreover, all trans-conjugants were also positive for IncFIB-KQ replicon signifying that each of the 22 K. pneumoniae pathogens contained a plasmid with pKpQIL-like backbone (29). Sharing pKpQIL-like plasmids (and other plasmid replicons) between clonally unrelated K. pneumoniae strains isolated from different ICUs throughout the whole duration of the study is highly suggestive for the circulation and the endemicity of such plasmid in the hospital environment.
In order to determine whether blaNDM-1 and PMQRs are co-carried by a single plasmid, we purified plasmid DNA bands- after gel electrophoresis- from all trans-conjugants and screened them for the presence of such resistance determinants. For all trans-conjugants, both blaNDM-1 and PMQRs were detected on a single plasmid DNA band. PBRT of such plasmid revealed that it is multi-replicon plasmid carrying 2 replicons; IncFIIK and IncFIB-KQ. These findings imply that blaNDM-1 and PMQRs are highly suggested to be co-carried by a multi-replicon pKpQIL-like plasmid that was shared among the 22 K. pneumoniae strains. Similarly, a recent study reported the discovery of pKpQIL-like plasmid co-harboring blaNDM1 and PMQR (qnrS1) and carried by 98% of the K. pneumoniae isolates (12).
In agreement with our results, a study implemented in USA revealed that (35.6%) of K. pneumoniae isolates were harboring pKpQIL-like plasmids (47). Furthermore, several studies established the major role of pKpQIL-like plasmid in the dissemination of carbapenem resistance among K. pneumoniae strains. Interestingly, in 2010, a study reported that loss of pKpQIL plasmid from K. pneumoniae isolates resulted in complete loss of carbapenem resistance demonstrating its fundamental role in disseminating carbapenem resistance (48). Doumith et al. stated that pKpQIL-like plasmids participated by a main role in the propagation of KPC enzymes in the UK (16). Similarly, Reyes et al. reported the presence of the bla KPC-2 gene on the Tn4401a transposon carried by pKpQIL-like plasmid (49).
CONCLUSION
Findings of this study suggest that MDR pK-pQIL-like plasmid is among the major players in the spread of blaNDM-1 and PMQRs among K. pneumoniae strains in Assiut University Hospital. The presence of pKpQIL-like plasmid together with other plasmid replicons in many clonally unrelated K. pneumoniae isolates is strongly suggestive of the presence of circulating MDR plasmids in the hospital environment. As far as we know this is the first report of pK-pQIL-like plasmid co-carrying PMQRs and blaNDM-1 in K. pneumoniae isolated from Egypt. Plasmid sequencing is recommended for better understanding the genetic aspects of circulating plasmids and the relatedness of plasmids discovered in this study with the worldwide plasmid epidemiology.
Determining the Sequence Type of K. pneumoniae using multi locus sequence typing (MLST) would further identify the clonal linage and the relatedness of the isolates. Also, plasmid sequencing would have supported the results of this study. Unfortunately, due to the restricted funding resources we were not able to perform either of these tests.
ACKNOWLEDGEMENTS
This study was funded by the Faculty of Medicine Grant Office, Assiut University, grant number 2018-12-24-002.
REFERENCES
- 1.Gorrie CL, Mirčeta M, Wick RR, Judd LM, Lam MMC, Gomi R, et al. Genomic dissection of Klebsiella pneumoniae infections in hospital patients reveals insights into an opportunistic pathogen. Nat Commun 2022; 13: 3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hawkey PM, Warren RE, Livermore DM, McNulty CAM, Enoch DA, Otter JA, et al. Treatment of infections caused by multidrug-resistant Gram-negative bacteria: report of the British Society for Antimicrobial Chemotherapy/Healthcare Infection Society/British Infection Association Joint Working Party. J Antimicrob Chemother 2018; 73(suppl_3): iii2–iii78. [DOI] [PubMed] [Google Scholar]
- 3.Effah CY, Sun T, Liu S, Wu Y. Klebsiella pneumoniae: an increasing threat to public health. Ann Clin Microbiol Antimicrob 2020; 19: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Partridge SR, Kwong SM, Firth N, Jensen SO. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev 2018; 31(4): e00088–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fonseca ÉL, Vicente AC. Integron functionality, genome innovation: an update on the subtle and smart strategy of integrase and gene cassette expression regulation. Microorganisms 2022; 10: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cury J, Jove T, Touchon M, Neron B, Rocha EP. Identification and analysis of integrons and cassette arrays in bacterial genomes. Nucleic Acids Res 2016; 44: 4539–4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreira RL, da Silva B, Rezende GS, Nakamura-Silva R, Pitondo-Silva A, Campanini EB, et al. High prevalence of multidrug-resistant Klebsiella pneumoniae harboring several virulence and β-lactamase encoding genes in a Brazilian intensive care unit. Front Microbiol 2019; 9: 3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhan Q, Xu Y, Wang B, Yu J, Shen X, Liu L, et al. Distribution of fluoroquinolone resistance determinants in Carbapenem-resistant Klebsiella pneumoniae clinical isolates associated with bloodstream infections in China. BMC Microbiol 2021; 21: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitra S, Mukherjee S, Naha S, Chattopadhyay P, Dutta S, Basu S. Evaluation of co-transfer of plasmid-mediated fluoroquinolone resistance genes and blaNDM gene in Enterobacteriaceae causing neonatal septicaemia. Antimicrob Resist Infect Control 2019; 8: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datta S, Mitra S, Chattopadhyay P, Som T, Mukherjee S, Basu S. Spread and exchange of blaNDM-1 in hospitalized neonates: role of mobilizable genetic elements. Eur J Clin Microbiol Infect Dis 2017; 36: 255–265. [DOI] [PubMed] [Google Scholar]
- 11.Kk S, Ekedahl E, Hoang NTB, Sewunet T, Berglund B, Lundberg L, et al. High diversity of blaNDM-1-encoding plasmids in Klebsiella pneumoniae isolated from neonates in a Vietnamese hospital. Int J Antimicrob Agents 2022; 59: 106496. [DOI] [PubMed] [Google Scholar]
- 12.Di Pilato V, Henrici De Angelis L, Aiezza N, Baccani I, Niccolai C, Parisio EM, et al. Resistome and virulome accretion in an NDM-1-producing ST147 sub lineage of Klebsiella pneumoniae associated with an outbreak in Tuscany, Italy: a genotypic and phenotypic characterization. Lancet Microbe 2022; 3(3): e224–e234. [DOI] [PubMed] [Google Scholar]
- 13.Leavitt A, Navon-Venezia S, Chmelnitsky I, Schwaber MJ, Carmeli Y. Emergence of KPC-2 and KPC-3 in carbapenem-resistant Klebsiella pneumoniae strains in an Israeli hospital. Antimicrob Agents Chemother 2007; 51: 3026–3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leavitt A, Chmelnitsky I, Carmeli Y, Navon-Venezia S. Complete nucleotide sequence of KPC-3-encoding plasmid pKpQIL in the epidemic Klebsiella pneumoniae sequence Type 258. Antimicrob Agents Chemother 2010; 54: 4493–4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García-Fernández A, Villa L, Carta C, Venditti C, Giordano A, Venditti M, et al. Klebsiella pneumoniae ST258 producing KPC-3 identified in italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob Agents Chemother 2012; 56: 2143–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doumith M, Findlay J, Hirani H, Hopkins KL, Livermore DM, Dodgson A, et al. Major role of pKpQIL-like plasmids in the early dissemination of KPC-type carbapenemases in the UK. J Antimicrob Chemother 2017; 72: 2241–2248. [DOI] [PubMed] [Google Scholar]
- 17.Khalil MAF, Elgaml A, El-Mowafy M. Emergence of multidrug-resistant New Delhi metallo-β-lactamase-1-producing Klebsiella pneumoniae in Egypt. Microb Drug Resist 2017; 23: 480–487. [DOI] [PubMed] [Google Scholar]
- 18.Abdel-Rahim MH, El-Badawy O, Hadiya S, Daef EA, Suh S-J, Boothe DM, et al. Patterns of fluoroquinolone resistance in enterobacteriaceae isolated from the Assiut University Hospitals, Egypt: A comparative study. Microb Drug Resist 2019; 25: 509–519. [DOI] [PubMed] [Google Scholar]
- 19.Yao H, Liu J, Jiang X, Chen F, Lu X, Zhang J. Analysis of the clinical effect of combined drug susceptibility to guide medication for carbapenem-resistant Klebsiella pneumoniae patients based on the Kirby-Bauer disk diffusion method. Infect Drug Resist 2021; 14: 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.CLSI (2019). Performance standards for antimicrobial susceptibility testing, 29th informational supplement (M100-S29). Clinical and Laboratory Standards Institute, Wayne, PA, USA. [Google Scholar]
- 21.Centers for Disease Control and Prevention (CDC) (2017). Standard operating procedure for PulseNet PFGE of Escherichia coli O157: H7, Escherichia coli non-O157 (STEC), Salmonella serotypes, Shigella sonnei and Shigella flexneri. http://www.cdc.gov/pulsenet/PDF/ecoli-shigella-salmonella-pfge-protocol-508c.pdf
- 22.Toledano-Tableros JE, Gayosso-Vázquez C, Jarillo-Quijada MD, Fernández-Vázquez JL, Morfin-Otero R, Rodríguez-Noriega E, et al. Dissemination of blaNDM-1 Gene among several Klebsiella pneumoniae sequence types in Mexico associated with horizontal transfer mediated by IncF-like plasmids. Front Microbiol 2021; 12: 611274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poirel L, Revathi G, Bernabeu S, Nordmann P. Detection of NDM-1-producing Klebsiella pneumoniae in Kenya. Antimicrob Agents Chemother 2011; 55: 934–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gay K, Robicsek A, Strahilevitz J, Park CH, Jacoby G, Barrett TJ, et al. Plasmid-mediated quinolone resistance in non-Typhi serotypes of Salmonella enterica. Clin Infect Dis 2006; 43: 297–304. [DOI] [PubMed] [Google Scholar]
- 25.Wu J-J, Ko W-C, Tsai S-H, Yan J-J. Prevalence of plasmid-mediated quinolone resistance determinants QnrA, QnrB, and QnrS among clinical isolates of Enterobacter cloacae in a Taiwanese hospital. Antimicrob Agents Chemother 2007; 51: 1223–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park CH, Robicsek A, Jacoby GA, Sahm D, Hooper DC. Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin modifying enzyme. Antimicrob Agents Chemother 2006; 50: 3953–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cocchi S, Grasselli E, Gutacker M, Benagli C, Convert M, Piffaretti J. Distribution and characterization of integrons in Escherichia coli strains of animal and human origin. FEMS Immunol Med Microbiol 2007; 50: 126–132. [DOI] [PubMed] [Google Scholar]
- 28.Kaushik M, Khare N, Kumar S, Gulati P. High prevalence of antibiotic resistance and integrons in E. coli isolated from an urban river water, India. Microb Drug Resist 2019; 25: 359–370. [DOI] [PubMed] [Google Scholar]
- 29.Magi G, Tontaerlli F, Caucci S, Sante LD, Brenciani A, Morroni G, et al. High prevalence of carbapenem-resistant Klebsiella pneumoniae ST307 recovered from fecal samples in an Italian hospital. Future Microbiol 2021; 16: 703–711. [DOI] [PubMed] [Google Scholar]
- 30.Martin RM, Bachman MA. Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front Cell Infect Microbiol 2018; 8: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugawara Y, Akeda Y, Hagiya H, Sakamoto N, Takeuchi D, Shanmugakani RK, et al. Spreading patterns of NDM-producing Enterobacteriaceae in clinical and environmental settings in Yangon, Myanmar. Antimicrob Agents Chemother 2019; 63(3): e01924–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reygaert WC. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol 2018; 4: 482–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohamed ER, Aly SA, Halby HM, Ahmed SH, Zakaria AM, El-Asheer OM. Epidemiological typing of multidrug-resistant Klebsiella pneumoniae, which causes paediatric ventilator-associated pneumonia in Egypt. J Med Microbiol 2017; 66: 628–634. [DOI] [PubMed] [Google Scholar]
- 34.Bassetti M, Righi E, Carnelutti A, Graziano E, Russo A. Multidrug-resistant Klebsiella pneumoniae: challenges for treatment, prevention and infection control. Expert Rev Anti Infect Ther 2018; 16: 749–761. [DOI] [PubMed] [Google Scholar]
- 35.Huang X, Cheng X, Sun P, Tang C, Ni F, Liu G. Characteristics of NDM-1-producing Klebsiella pneumoniae ST234 and ST1412 isolates spread in a neonatal unit. BMC Microbiol 2018; 18: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xue G, Li J, Feng Y, Xu W, Li S, Yan C, et al. High prevalence of plasmid-mediated quinolone resistance determinants in Escherichia coli and Klebsiella pneumoniae isolates from pediatric patients in China. Microb Drug Resist 2017; 23: 107–114. [DOI] [PubMed] [Google Scholar]
- 37.Tang M, Kong X, Hao J, Liu J. Epidemiological characteristics and formation mechanisms of multidrug-resistant hypervirulent Klebsiella pneumoniae. Front Microbiol 2020; 11: 581543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moghadam MT, Shariati A, Mirkalantari S, Karmostaji A. The complex genetic region conferring transferable antibiotic resistance in multidrug-resistant and extremely drug-resistant Klebsiella pneumoniae clinical isolates. New Microbes New Infect 2020; 36: 100693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Facciolà A, Pellicanò GF, Visalli G, Paolucci IA, Venanzi Rullo E, Ceccarelli M, et al. The role of the hospital environment in the healthcare-associated infections: a general review of the literature. Eur Rev Med Pharmacol Sci 2019; 23: 1266–1278. [DOI] [PubMed] [Google Scholar]
- 40.Vrancianu CO, Popa LI, Bleotu C, Chifiriuc MC. Targeting plasmids to limit acquisition and transmission of antimicrobial resistance. Front Microbiol 2020; 11: 761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bi R, Kong Z, Qian H, Jiang F, Kang H, Gu B, et al. High prevalence of blaNDM variants among carbapenem-resistant Escherichia coli in Northern Jiangsu province, China. Front Microbiol 2018; 9: 2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toledano-Tableros JE, Gayosso-Vázquez C, Jarillo-Quijada MD, Fernández-Vázquez JL, Morfin-Otero R, Rodríguez-Noriega E, et al. Dissemination of blaNDM-1 Gene among several Klebsiella pneumoniae sequence types in Mexico associated with horizontal transfer mediated by IncF-Like plasmids. Front Microbiol 2021; 12: 611274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang C-Y, Lin H-J, Chang L-L, Ma L, Siu LK, Tung Y-C, et al. Characterization of extended-spectrum β-lactamase-carrying plasmids in clinical isolates of Klebsiella pneumoniae from Taiwan. Microb Drug Resist 2017; 23: 98–106. [DOI] [PubMed] [Google Scholar]
- 44.Al-Marzooq F, Mohd Yusof MY, Tay ST. Molecular analysis of antibiotic resistance determinants and plasmids in Malaysian isolates of multidrug resistant Klebsiella pneumoniae. PLoS One 2015; 10(7): e0133654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zaman TU, Alrodayyan M, Albladi M, Aldrees M, Siddique MI, Aljohani S, et al. Clonal diversity and genetic profiling of antibiotic resistance among multidrug/carbapenem-resistant Klebsiella pneumoniae isolates from a tertiary care hospital in Saudi Arabia. BMC Infect Dis 2018; 18: 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mohamed ER, Ali MY, Waly NGFM, Halby HM, El-Baky RMA. The Inc FII Plasmid and its Contribution in the Transmission of blaNDM1 and blaKPC2 in Klebsiella pneumoniae in Egypt. Antibiotics (Basel) 2019; 8: 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen L, Chavda KD, Melano RG, Jacobs MR, Koll B, Hong T, et al. Comparative genomic analysis of KPC-encoding pKpQIL-like plasmids and their distribution in New Jersey and New York Hospitals. Antimicrob Agents Chemother 2014; 58: 2871–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leavitt A, Chmelnitsky I, Ofek I, Carmeli Y, Navon-Venezia S. Plasmid pKpQIL encoding KPC-3 and TEM-1 confers carbapenem resistance in an extremely drug-resistant epidemic Klebsiella pneumoniae strain. J Antimicrob Chemother 2010; 65: 243–248. [DOI] [PubMed] [Google Scholar]
- 49.Reyes J, Cárdenas P, Tamayo R, Villavicencio F, Aguilar A, Melano RG, et al. Characterization of blaKPC-2-Harboring Klebsiella pneumoniae isolates and mobile genetic elements from outbreaks in a Hospital in Ecuador. Microb Drug Resist 2021; 27: 752–759. [DOI] [PubMed] [Google Scholar]